Project
PRJNA375882: Comprehensive Mouse Transcriptomic BodyMap
Navigation
Downloads
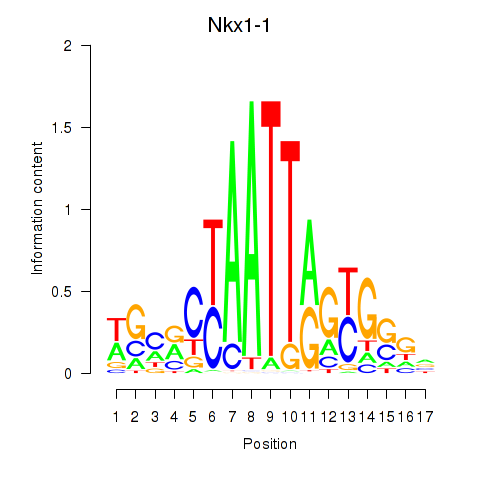
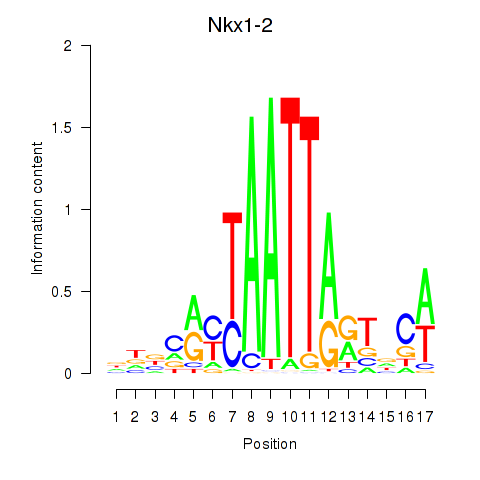
Results for Nkx1-1_Nkx1-2
Z-value: 0.66
Transcription factors associated with Nkx1-1_Nkx1-2
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Nkx1-1
|
ENSMUSG00000029112.6 | Nkx1-1 |
|
Nkx1-2
|
ENSMUSG00000048528.8 | Nkx1-2 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Nkx1-1 | mm39_v1_chr5_-_33591320_33591320 | 0.35 | 2.4e-03 | Click! |
| Nkx1-2 | mm39_v1_chr7_-_132201344_132201366 | 0.02 | 8.8e-01 | Click! |
Activity profile of Nkx1-1_Nkx1-2 motif
Sorted Z-values of Nkx1-1_Nkx1-2 motif
Network of associatons between targets according to the STRING database.
First level regulatory network of Nkx1-1_Nkx1-2
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr16_+_22965286 | 6.63 |
ENSMUST00000023593.6
|
Adipoq
|
adiponectin, C1Q and collagen domain containing |
| chr16_+_22965330 | 5.42 |
ENSMUST00000171309.2
|
Adipoq
|
adiponectin, C1Q and collagen domain containing |
| chr6_-_124756645 | 4.01 |
ENSMUST00000147669.2
ENSMUST00000128697.8 ENSMUST00000032218.10 ENSMUST00000112475.9 |
Lrrc23
|
leucine rich repeat containing 23 |
| chr1_-_171854818 | 3.82 |
ENSMUST00000138714.2
ENSMUST00000027837.13 ENSMUST00000111264.8 |
Vangl2
|
VANGL planar cell polarity 2 |
| chr14_-_70666513 | 3.80 |
ENSMUST00000226426.2
ENSMUST00000048129.6 |
Piwil2
|
piwi-like RNA-mediated gene silencing 2 |
| chr11_+_87000032 | 3.46 |
ENSMUST00000020794.6
|
Ska2
|
spindle and kinetochore associated complex subunit 2 |
| chr18_+_12874390 | 3.00 |
ENSMUST00000121018.8
ENSMUST00000119108.8 ENSMUST00000186263.2 ENSMUST00000191078.7 |
Cabyr
|
calcium-binding tyrosine-(Y)-phosphorylation regulated (fibrousheathin 2) |
| chr19_+_8779903 | 2.97 |
ENSMUST00000172175.3
|
Zbtb3
|
zinc finger and BTB domain containing 3 |
| chr8_-_3744167 | 2.75 |
ENSMUST00000005678.6
|
Fcer2a
|
Fc receptor, IgE, low affinity II, alpha polypeptide |
| chr11_-_86999481 | 2.27 |
ENSMUST00000051395.9
|
Prr11
|
proline rich 11 |
| chr3_-_90297187 | 2.15 |
ENSMUST00000029541.12
|
Slc27a3
|
solute carrier family 27 (fatty acid transporter), member 3 |
| chr3_+_94320548 | 1.99 |
ENSMUST00000166032.8
ENSMUST00000200486.5 ENSMUST00000196386.5 ENSMUST00000045245.10 ENSMUST00000197901.5 ENSMUST00000198041.2 |
Tdrkh
Gm42463
|
tudor and KH domain containing protein predicted gene 42463 |
| chr18_+_12874368 | 1.95 |
ENSMUST00000235000.2
ENSMUST00000115857.9 |
Cabyr
|
calcium-binding tyrosine-(Y)-phosphorylation regulated (fibrousheathin 2) |
| chr14_+_65612788 | 1.93 |
ENSMUST00000224687.2
|
Zfp395
|
zinc finger protein 395 |
| chr11_+_104468107 | 1.83 |
ENSMUST00000106956.10
|
Myl4
|
myosin, light polypeptide 4 |
| chr7_-_24423715 | 1.83 |
ENSMUST00000081657.6
|
Lypd11
|
Ly6/PLAUR domain containing 11 |
| chr11_+_104467791 | 1.78 |
ENSMUST00000106957.8
|
Myl4
|
myosin, light polypeptide 4 |
| chr4_-_140501507 | 1.78 |
ENSMUST00000026381.7
|
Padi4
|
peptidyl arginine deiminase, type IV |
| chr18_-_24736848 | 1.70 |
ENSMUST00000070726.10
|
Slc39a6
|
solute carrier family 39 (metal ion transporter), member 6 |
| chr5_-_82272549 | 1.70 |
ENSMUST00000188072.2
ENSMUST00000185410.2 |
1700031L13Rik
|
RIKEN cDNA 1700031L13 gene |
| chr1_-_144427302 | 1.70 |
ENSMUST00000184189.3
|
Rgs21
|
regulator of G-protein signalling 21 |
| chr6_+_29694181 | 1.70 |
ENSMUST00000046750.14
ENSMUST00000115250.4 |
Tspan33
|
tetraspanin 33 |
| chr16_+_51851588 | 1.65 |
ENSMUST00000114471.3
|
Cblb
|
Casitas B-lineage lymphoma b |
| chr15_+_97990431 | 1.63 |
ENSMUST00000229280.2
ENSMUST00000163507.8 ENSMUST00000230445.2 |
Pfkm
|
phosphofructokinase, muscle |
| chr3_-_15902583 | 1.59 |
ENSMUST00000108354.8
ENSMUST00000108349.2 ENSMUST00000108352.9 ENSMUST00000108350.8 ENSMUST00000050623.11 |
Sirpb1c
|
signal-regulatory protein beta 1C |
| chr5_+_138115165 | 1.53 |
ENSMUST00000062350.15
ENSMUST00000110961.9 ENSMUST00000080732.10 ENSMUST00000110960.9 ENSMUST00000142185.8 ENSMUST00000136425.2 ENSMUST00000110959.2 |
Zscan21
|
zinc finger and SCAN domain containing 21 |
| chr4_-_88595161 | 1.34 |
ENSMUST00000105148.2
|
Ifna16
|
interferon alpha 16 |
| chr18_-_24736521 | 1.31 |
ENSMUST00000154205.2
|
Slc39a6
|
solute carrier family 39 (metal ion transporter), member 6 |
| chr2_+_154390808 | 1.30 |
ENSMUST00000045116.11
ENSMUST00000109709.4 |
1700003F12Rik
|
RIKEN cDNA 1700003F12 gene |
| chr9_-_20871081 | 1.29 |
ENSMUST00000177754.9
|
Dnmt1
|
DNA methyltransferase (cytosine-5) 1 |
| chr11_+_116734104 | 1.26 |
ENSMUST00000106370.10
|
Mettl23
|
methyltransferase like 23 |
| chr19_-_40371016 | 1.26 |
ENSMUST00000225766.3
|
Sorbs1
|
sorbin and SH3 domain containing 1 |
| chr3_-_116217579 | 1.24 |
ENSMUST00000106491.7
ENSMUST00000090464.7 |
Cdc14a
|
CDC14 cell division cycle 14A |
| chr17_+_7437500 | 1.20 |
ENSMUST00000024575.8
|
Rps6ka2
|
ribosomal protein S6 kinase, polypeptide 2 |
| chr6_+_145561483 | 1.20 |
ENSMUST00000087445.7
|
Tuba3b
|
tubulin, alpha 3B |
| chr15_-_79626719 | 1.13 |
ENSMUST00000089311.11
ENSMUST00000046259.14 |
Sun2
|
Sad1 and UNC84 domain containing 2 |
| chr1_-_150341911 | 1.10 |
ENSMUST00000162367.8
ENSMUST00000161611.8 ENSMUST00000161320.8 ENSMUST00000159035.2 |
Prg4
|
proteoglycan 4 (megakaryocyte stimulating factor, articular superficial zone protein) |
| chr7_-_30251680 | 1.10 |
ENSMUST00000215288.2
ENSMUST00000108165.8 ENSMUST00000153594.3 |
Proser3
|
proline and serine rich 3 |
| chr15_-_79626694 | 1.09 |
ENSMUST00000100439.10
|
Sun2
|
Sad1 and UNC84 domain containing 2 |
| chr4_-_14621805 | 1.04 |
ENSMUST00000042221.14
|
Slc26a7
|
solute carrier family 26, member 7 |
| chr6_-_23650297 | 1.03 |
ENSMUST00000063548.4
|
Rnf133
|
ring finger protein 133 |
| chr1_-_131441962 | 1.02 |
ENSMUST00000185445.3
|
Srgap2
|
SLIT-ROBO Rho GTPase activating protein 2 |
| chr2_+_91095597 | 1.01 |
ENSMUST00000028691.7
|
Arfgap2
|
ADP-ribosylation factor GTPase activating protein 2 |
| chr4_-_14621497 | 1.00 |
ENSMUST00000149633.2
|
Slc26a7
|
solute carrier family 26, member 7 |
| chr14_+_46997984 | 0.99 |
ENSMUST00000067426.6
|
Cdkn3
|
cyclin-dependent kinase inhibitor 3 |
| chr12_+_4819277 | 0.96 |
ENSMUST00000020967.11
|
Pfn4
|
profilin family, member 4 |
| chr14_-_70666821 | 0.94 |
ENSMUST00000226229.2
|
Piwil2
|
piwi-like RNA-mediated gene silencing 2 |
| chr12_+_4819339 | 0.93 |
ENSMUST00000178879.3
|
Pfn4
|
profilin family, member 4 |
| chrX_+_159551009 | 0.92 |
ENSMUST00000033650.14
|
Rs1
|
retinoschisis (X-linked, juvenile) 1 (human) |
| chr6_-_23650205 | 0.91 |
ENSMUST00000115354.2
|
Rnf133
|
ring finger protein 133 |
| chr5_-_137101108 | 0.91 |
ENSMUST00000077523.4
ENSMUST00000041388.11 |
Serpine1
|
serine (or cysteine) peptidase inhibitor, clade E, member 1 |
| chr8_-_87307294 | 0.90 |
ENSMUST00000131423.8
ENSMUST00000152438.2 |
Abcc12
|
ATP-binding cassette, sub-family C (CFTR/MRP), member 12 |
| chr4_-_131802606 | 0.90 |
ENSMUST00000146021.8
|
Epb41
|
erythrocyte membrane protein band 4.1 |
| chr9_-_26717686 | 0.90 |
ENSMUST00000162702.8
ENSMUST00000040398.14 ENSMUST00000066560.13 |
Glb1l2
|
galactosidase, beta 1-like 2 |
| chr2_-_45002902 | 0.89 |
ENSMUST00000076836.13
ENSMUST00000176732.8 ENSMUST00000200844.4 |
Zeb2
|
zinc finger E-box binding homeobox 2 |
| chr4_-_43710231 | 0.88 |
ENSMUST00000217544.2
ENSMUST00000107862.3 |
Olfr71
|
olfactory receptor 71 |
| chr12_+_108145802 | 0.83 |
ENSMUST00000221167.2
|
Ccnk
|
cyclin K |
| chr4_-_14621669 | 0.82 |
ENSMUST00000143105.2
|
Slc26a7
|
solute carrier family 26, member 7 |
| chr6_-_128339775 | 0.80 |
ENSMUST00000112152.8
ENSMUST00000057421.15 ENSMUST00000112151.2 |
Rhno1
|
RAD9-HUS1-RAD1 interacting nuclear orphan 1 |
| chr15_-_77840856 | 0.80 |
ENSMUST00000117725.2
ENSMUST00000016696.13 |
Foxred2
|
FAD-dependent oxidoreductase domain containing 2 |
| chr12_+_4819022 | 0.80 |
ENSMUST00000219503.2
|
Pfn4
|
profilin family, member 4 |
| chr12_-_4819216 | 0.77 |
ENSMUST00000053458.7
ENSMUST00000218575.2 |
Fam228b
|
family with sequence similarity 228, member B |
| chr1_+_54289833 | 0.77 |
ENSMUST00000027128.11
|
Ccdc150
|
coiled-coil domain containing 150 |
| chr12_+_4819151 | 0.76 |
ENSMUST00000219438.2
|
Pfn4
|
profilin family, member 4 |
| chr12_-_40087393 | 0.76 |
ENSMUST00000146905.2
|
Arl4a
|
ADP-ribosylation factor-like 4A |
| chr6_-_128339458 | 0.75 |
ENSMUST00000155573.3
|
Rhno1
|
RAD9-HUS1-RAD1 interacting nuclear orphan 1 |
| chr4_-_131802561 | 0.75 |
ENSMUST00000105970.8
ENSMUST00000105975.8 |
Epb41
|
erythrocyte membrane protein band 4.1 |
| chr2_-_160950936 | 0.73 |
ENSMUST00000039782.14
ENSMUST00000134178.8 |
Chd6
|
chromodomain helicase DNA binding protein 6 |
| chr2_+_68490186 | 0.71 |
ENSMUST00000055930.6
|
4932414N04Rik
|
RIKEN cDNA 4932414N04 gene |
| chr15_+_41694317 | 0.70 |
ENSMUST00000166917.3
ENSMUST00000230127.2 ENSMUST00000230131.2 |
Oxr1
|
oxidation resistance 1 |
| chr19_+_29344846 | 0.70 |
ENSMUST00000016640.8
|
Cd274
|
CD274 antigen |
| chr7_+_101545547 | 0.69 |
ENSMUST00000035395.14
ENSMUST00000106973.8 ENSMUST00000144207.9 |
Anapc15
|
anaphase promoting complex C subunit 15 |
| chr7_-_12829100 | 0.69 |
ENSMUST00000209822.3
ENSMUST00000235753.2 |
Vmn1r85
|
vomeronasal 1 receptor 85 |
| chr5_-_122959361 | 0.66 |
ENSMUST00000086216.9
|
Anapc5
|
anaphase-promoting complex subunit 5 |
| chr11_-_99134885 | 0.64 |
ENSMUST00000103132.10
ENSMUST00000038214.7 |
Krt222
|
keratin 222 |
| chr9_+_40092216 | 0.63 |
ENSMUST00000218134.2
ENSMUST00000216720.2 ENSMUST00000214763.2 |
Olfr986
|
olfactory receptor 986 |
| chr7_+_140181182 | 0.62 |
ENSMUST00000214180.2
ENSMUST00000211771.2 |
Olfr46
|
olfactory receptor 46 |
| chr3_+_121746862 | 0.61 |
ENSMUST00000037958.14
ENSMUST00000196904.5 |
Arhgap29
|
Rho GTPase activating protein 29 |
| chr2_-_27365633 | 0.60 |
ENSMUST00000138693.8
ENSMUST00000113941.9 ENSMUST00000077737.13 |
Brd3
|
bromodomain containing 3 |
| chr9_-_44024767 | 0.58 |
ENSMUST00000216511.2
ENSMUST00000056328.6 ENSMUST00000185479.2 |
Rnf26
Gm49380
|
ring finger protein 26 predicted gene, 49380 |
| chr19_-_41921676 | 0.57 |
ENSMUST00000075280.12
ENSMUST00000112123.4 |
Exosc1
|
exosome component 1 |
| chr4_-_112632013 | 0.56 |
ENSMUST00000060327.4
|
Skint10
|
selection and upkeep of intraepithelial T cells 10 |
| chrX_+_56257374 | 0.55 |
ENSMUST00000033466.2
|
Cd40lg
|
CD40 ligand |
| chr9_-_100368841 | 0.55 |
ENSMUST00000098458.4
|
Il20rb
|
interleukin 20 receptor beta |
| chr15_+_16728842 | 0.55 |
ENSMUST00000228307.2
|
Cdh9
|
cadherin 9 |
| chr7_-_110673269 | 0.54 |
ENSMUST00000163014.2
|
Eif4g2
|
eukaryotic translation initiation factor 4, gamma 2 |
| chr5_+_30824121 | 0.54 |
ENSMUST00000144742.6
ENSMUST00000149759.2 ENSMUST00000199320.5 |
Cenpa
|
centromere protein A |
| chr19_+_60744385 | 0.54 |
ENSMUST00000088237.6
|
Nanos1
|
nanos C2HC-type zinc finger 1 |
| chr14_+_46998004 | 0.52 |
ENSMUST00000227149.2
|
Cdkn3
|
cyclin-dependent kinase inhibitor 3 |
| chr17_-_57338468 | 0.52 |
ENSMUST00000007814.10
ENSMUST00000233480.2 |
Khsrp
|
KH-type splicing regulatory protein |
| chr11_+_43572825 | 0.52 |
ENSMUST00000061070.6
ENSMUST00000094294.5 |
Pwwp2a
|
PWWP domain containing 2A |
| chr10_+_97442727 | 0.48 |
ENSMUST00000105286.4
|
Kera
|
keratocan |
| chr2_-_111843053 | 0.46 |
ENSMUST00000213559.3
|
Olfr1310
|
olfactory receptor 1310 |
| chr12_+_84463970 | 0.45 |
ENSMUST00000183146.2
|
Rnf113a2
|
ring finger protein 113A2 |
| chrX_-_73129195 | 0.45 |
ENSMUST00000140399.3
ENSMUST00000123362.8 ENSMUST00000100750.10 ENSMUST00000033770.13 |
Mecp2
|
methyl CpG binding protein 2 |
| chr14_-_59602882 | 0.45 |
ENSMUST00000160425.8
ENSMUST00000095157.11 |
Phf11d
|
PHD finger protein 11D |
| chr9_-_97252011 | 0.45 |
ENSMUST00000035026.5
|
Trim42
|
tripartite motif-containing 42 |
| chr11_-_43638790 | 0.44 |
ENSMUST00000048578.3
ENSMUST00000109278.8 |
Ttc1
|
tetratricopeptide repeat domain 1 |
| chr5_+_143846782 | 0.43 |
ENSMUST00000148011.8
ENSMUST00000110709.7 |
Pms2
|
PMS1 homolog2, mismatch repair system component |
| chr6_-_57512355 | 0.43 |
ENSMUST00000042766.6
|
Ppm1k
|
protein phosphatase 1K (PP2C domain containing) |
| chr11_-_99121822 | 0.42 |
ENSMUST00000103133.4
|
Smarce1
|
SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily e, member 1 |
| chr1_+_174329361 | 0.42 |
ENSMUST00000030039.13
|
Fmn2
|
formin 2 |
| chr2_-_102230602 | 0.41 |
ENSMUST00000152929.2
|
Trim44
|
tripartite motif-containing 44 |
| chr3_+_75982890 | 0.40 |
ENSMUST00000160261.8
|
Fstl5
|
follistatin-like 5 |
| chr2_+_89642395 | 0.39 |
ENSMUST00000214508.2
|
Olfr1255
|
olfactory receptor 1255 |
| chr6_-_8180174 | 0.39 |
ENSMUST00000213284.2
|
Col28a1
|
collagen, type XXVIII, alpha 1 |
| chr11_-_97040858 | 0.38 |
ENSMUST00000118375.8
|
Tbkbp1
|
TBK1 binding protein 1 |
| chr13_-_103042554 | 0.36 |
ENSMUST00000171791.8
|
Mast4
|
microtubule associated serine/threonine kinase family member 4 |
| chrX_-_8391193 | 0.36 |
ENSMUST00000115573.2
|
Gm14459
|
predicted gene 14459 |
| chr15_+_75865604 | 0.35 |
ENSMUST00000089669.6
|
Mapk15
|
mitogen-activated protein kinase 15 |
| chr14_+_65504067 | 0.35 |
ENSMUST00000224629.2
|
Fbxo16
|
F-box protein 16 |
| chr6_-_57340712 | 0.34 |
ENSMUST00000228156.2
ENSMUST00000227186.2 ENSMUST00000228294.2 ENSMUST00000228342.2 |
Vmn1r17
|
vomeronasal 1 receptor 17 |
| chr7_+_103628383 | 0.34 |
ENSMUST00000098185.2
|
Olfr635
|
olfactory receptor 635 |
| chr5_-_84565218 | 0.34 |
ENSMUST00000113401.4
|
Epha5
|
Eph receptor A5 |
| chr16_-_29363671 | 0.33 |
ENSMUST00000039090.9
|
Atp13a4
|
ATPase type 13A4 |
| chr6_-_128339654 | 0.32 |
ENSMUST00000203719.2
|
Rhno1
|
RAD9-HUS1-RAD1 interacting nuclear orphan 1 |
| chr4_+_136038263 | 0.30 |
ENSMUST00000105850.8
ENSMUST00000148843.10 |
Hnrnpr
|
heterogeneous nuclear ribonucleoprotein R |
| chr1_+_82817794 | 0.29 |
ENSMUST00000186043.2
|
Agfg1
|
ArfGAP with FG repeats 1 |
| chr1_-_37996838 | 0.29 |
ENSMUST00000027254.10
ENSMUST00000114894.2 |
Lyg1
|
lysozyme G-like 1 |
| chr13_-_103042294 | 0.28 |
ENSMUST00000167462.8
|
Mast4
|
microtubule associated serine/threonine kinase family member 4 |
| chr6_+_57405915 | 0.28 |
ENSMUST00000227909.2
|
Vmn1r20
|
vomeronasal 1 receptor 20 |
| chr10_+_129539079 | 0.27 |
ENSMUST00000213331.2
|
Olfr804
|
olfactory receptor 804 |
| chr18_+_24737009 | 0.26 |
ENSMUST00000234266.2
ENSMUST00000025120.8 |
Elp2
|
elongator acetyltransferase complex subunit 2 |
| chr14_-_52341426 | 0.26 |
ENSMUST00000227536.2
ENSMUST00000227195.2 ENSMUST00000228815.2 ENSMUST00000228198.2 ENSMUST00000227458.2 ENSMUST00000228232.2 ENSMUST00000227242.2 ENSMUST00000228748.2 |
Hnrnpc
|
heterogeneous nuclear ribonucleoprotein C |
| chr14_-_52277310 | 0.26 |
ENSMUST00000216907.2
ENSMUST00000214071.2 ENSMUST00000216188.2 |
Olfr221
|
olfactory receptor 221 |
| chrX_+_159551171 | 0.24 |
ENSMUST00000112368.3
|
Rs1
|
retinoschisis (X-linked, juvenile) 1 (human) |
| chr19_-_29344694 | 0.24 |
ENSMUST00000138051.2
|
Plgrkt
|
plasminogen receptor, C-terminal lysine transmembrane protein |
| chr8_+_34089597 | 0.23 |
ENSMUST00000009774.11
|
Ppp2cb
|
protein phosphatase 2 (formerly 2A), catalytic subunit, beta isoform |
| chr2_+_87725306 | 0.23 |
ENSMUST00000217436.2
|
Olfr1153
|
olfactory receptor 1153 |
| chr5_+_134961205 | 0.22 |
ENSMUST00000047196.14
ENSMUST00000111221.9 ENSMUST00000111219.8 ENSMUST00000068617.12 |
Mettl27
|
methyltransferase like 27 |
| chr7_+_101546059 | 0.22 |
ENSMUST00000143835.8
|
Anapc15
|
anaphase promoting complex C subunit 15 |
| chr4_+_136038301 | 0.22 |
ENSMUST00000084219.12
|
Hnrnpr
|
heterogeneous nuclear ribonucleoprotein R |
| chr17_-_37974666 | 0.21 |
ENSMUST00000215414.2
ENSMUST00000213638.3 |
Olfr117
|
olfactory receptor 117 |
| chr14_-_44987797 | 0.21 |
ENSMUST00000226900.2
|
Gm8267
|
predicted gene 8267 |
| chr17_+_46807637 | 0.21 |
ENSMUST00000046497.8
|
Dnph1
|
2'-deoxynucleoside 5'-phosphate N-hydrolase 1 |
| chr14_-_52341472 | 0.20 |
ENSMUST00000111610.12
ENSMUST00000164655.2 |
Hnrnpc
|
heterogeneous nuclear ribonucleoprotein C |
| chr4_-_112291169 | 0.20 |
ENSMUST00000058605.3
|
Skint9
|
selection and upkeep of intraepithelial T cells 9 |
| chr2_-_168607166 | 0.19 |
ENSMUST00000137536.2
|
Sall4
|
spalt like transcription factor 4 |
| chr2_-_132089667 | 0.19 |
ENSMUST00000110163.8
ENSMUST00000180286.2 ENSMUST00000028816.9 |
Tmem230
|
transmembrane protein 230 |
| chr16_+_51851917 | 0.18 |
ENSMUST00000227062.2
|
Cblb
|
Casitas B-lineage lymphoma b |
| chr4_+_136038243 | 0.17 |
ENSMUST00000131671.8
|
Hnrnpr
|
heterogeneous nuclear ribonucleoprotein R |
| chr10_+_129493563 | 0.17 |
ENSMUST00000217094.2
|
Olfr800
|
olfactory receptor 800 |
| chr4_-_132191181 | 0.17 |
ENSMUST00000102567.4
|
Med18
|
mediator complex subunit 18 |
| chr2_+_85804239 | 0.16 |
ENSMUST00000217244.2
|
Olfr1029
|
olfactory receptor 1029 |
| chr14_+_13803477 | 0.16 |
ENSMUST00000102996.4
|
Cfap20dc
|
CFAP20 domain containing |
| chr2_+_85809620 | 0.16 |
ENSMUST00000056849.3
|
Olfr1030
|
olfactory receptor 1030 |
| chr17_-_37404764 | 0.15 |
ENSMUST00000087144.5
|
Olfr91
|
olfactory receptor 91 |
| chr2_-_89855921 | 0.14 |
ENSMUST00000216616.3
|
Olfr1264
|
olfactory receptor 1264 |
| chr7_+_29683373 | 0.13 |
ENSMUST00000148442.8
|
Zfp568
|
zinc finger protein 568 |
| chr7_-_11414074 | 0.13 |
ENSMUST00000227010.2
ENSMUST00000209638.3 |
Vmn1r72
|
vomeronasal 1 receptor 72 |
| chr17_+_21446349 | 0.13 |
ENSMUST00000235895.2
|
Vmn1r234
|
vomeronasal 1 receptor 234 |
| chr1_+_190660689 | 0.13 |
ENSMUST00000066632.14
ENSMUST00000110899.7 |
Angel2
|
angel homolog 2 |
| chr10_-_129000620 | 0.13 |
ENSMUST00000214271.2
|
Olfr771
|
olfactory receptor 771 |
| chr7_-_26895141 | 0.13 |
ENSMUST00000163311.9
ENSMUST00000126211.2 |
Snrpa
|
small nuclear ribonucleoprotein polypeptide A |
| chr11_-_100653754 | 0.12 |
ENSMUST00000107360.3
ENSMUST00000055083.4 |
Hcrt
|
hypocretin |
| chr11_-_22236795 | 0.12 |
ENSMUST00000180360.8
ENSMUST00000109563.9 |
Ehbp1
|
EH domain binding protein 1 |
| chr1_+_83022653 | 0.12 |
ENSMUST00000222567.2
|
Gm47969
|
predicted gene, 47969 |
| chr8_-_22396428 | 0.11 |
ENSMUST00000051965.5
|
Defb11
|
defensin beta 11 |
| chr2_-_111100733 | 0.10 |
ENSMUST00000099619.6
|
Olfr1277
|
olfactory receptor 1277 |
| chr7_+_103652466 | 0.10 |
ENSMUST00000209757.3
|
Olfr638
|
olfactory receptor 638 |
| chr1_+_88234454 | 0.09 |
ENSMUST00000040210.14
|
Trpm8
|
transient receptor potential cation channel, subfamily M, member 8 |
| chr6_+_37847721 | 0.09 |
ENSMUST00000031859.14
ENSMUST00000120428.8 |
Trim24
|
tripartite motif-containing 24 |
| chr7_-_44578834 | 0.09 |
ENSMUST00000107857.11
ENSMUST00000167930.8 ENSMUST00000085399.13 ENSMUST00000166972.9 |
Ap2a1
|
adaptor-related protein complex 2, alpha 1 subunit |
| chr1_-_173161069 | 0.07 |
ENSMUST00000038227.6
|
Ackr1
|
atypical chemokine receptor 1 (Duffy blood group) |
| chr16_+_13176238 | 0.07 |
ENSMUST00000149359.2
|
Mrtfb
|
myocardin related transcription factor B |
| chr15_+_81548090 | 0.06 |
ENSMUST00000023029.15
ENSMUST00000174229.8 ENSMUST00000172748.8 |
L3mbtl2
|
L3MBTL2 polycomb repressive complex 1 subunit |
| chr2_-_89972637 | 0.06 |
ENSMUST00000215659.2
ENSMUST00000215765.2 ENSMUST00000213293.2 ENSMUST00000214973.2 ENSMUST00000215153.3 |
Olfr32
|
olfactory receptor 32 |
| chr18_+_4993795 | 0.06 |
ENSMUST00000153016.8
|
Svil
|
supervillin |
| chr16_+_51851948 | 0.05 |
ENSMUST00000226593.2
|
Cblb
|
Casitas B-lineage lymphoma b |
| chr1_+_172383499 | 0.04 |
ENSMUST00000061835.10
|
Vsig8
|
V-set and immunoglobulin domain containing 8 |
| chr9_-_85631361 | 0.02 |
ENSMUST00000039213.15
|
Ibtk
|
inhibitor of Bruton agammaglobulinemia tyrosine kinase |
| chr19_-_12313274 | 0.02 |
ENSMUST00000208398.3
|
Olfr1438-ps1
|
olfactory receptor 1438, pseudogene 1 |
| chr10_-_129965752 | 0.02 |
ENSMUST00000215217.2
ENSMUST00000214192.2 |
Olfr824
|
olfactory receptor 824 |
| chr2_-_157188568 | 0.01 |
ENSMUST00000029172.2
|
Ghrh
|
growth hormone releasing hormone |
| chr8_+_114362419 | 0.01 |
ENSMUST00000035777.10
|
Mon1b
|
MON1 homolog B, secretory traffciking associated |
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 4.0 | 12.1 | GO:0072312 | metanephric glomerular visceral epithelial cell differentiation(GO:0072248) metanephric glomerular visceral epithelial cell development(GO:0072249) metanephric glomerular epithelial cell differentiation(GO:0072312) metanephric glomerular epithelial cell development(GO:0072313) |
| 1.3 | 3.8 | GO:0060448 | dichotomous subdivision of terminal units involved in lung branching(GO:0060448) |
| 1.2 | 4.7 | GO:0000239 | pachytene(GO:0000239) |
| 0.4 | 2.2 | GO:0031022 | nuclear migration along microfilament(GO:0031022) |
| 0.4 | 2.8 | GO:0002925 | positive regulation of humoral immune response mediated by circulating immunoglobulin(GO:0002925) |
| 0.4 | 1.8 | GO:0018101 | protein citrullination(GO:0018101) histone citrullination(GO:0036414) |
| 0.3 | 1.6 | GO:0061622 | glycolytic process through glucose-1-phosphate(GO:0061622) |
| 0.3 | 1.3 | GO:0090116 | C-5 methylation of cytosine(GO:0090116) positive regulation of methylation-dependent chromatin silencing(GO:0090309) |
| 0.3 | 2.9 | GO:0019532 | oxalate transport(GO:0019532) |
| 0.3 | 0.9 | GO:2000097 | regulation of smooth muscle cell-matrix adhesion(GO:2000097) |
| 0.3 | 1.2 | GO:0016062 | adaptation of rhodopsin mediated signaling(GO:0016062) light adaption(GO:0036367) |
| 0.3 | 1.0 | GO:0021815 | modulation of microtubule cytoskeleton involved in cerebral cortex radial glia guided migration(GO:0021815) |
| 0.2 | 3.5 | GO:0042989 | sequestering of actin monomers(GO:0042989) |
| 0.2 | 1.9 | GO:0002669 | positive regulation of T cell anergy(GO:0002669) positive regulation of lymphocyte anergy(GO:0002913) |
| 0.2 | 0.6 | GO:2001200 | positive regulation of dendritic cell differentiation(GO:2001200) |
| 0.2 | 0.6 | GO:0001807 | regulation of type IV hypersensitivity(GO:0001807) |
| 0.2 | 0.7 | GO:0036091 | positive regulation of transcription from RNA polymerase II promoter in response to oxidative stress(GO:0036091) |
| 0.2 | 0.5 | GO:0098749 | cerebellar neuron development(GO:0098749) |
| 0.2 | 3.0 | GO:0071578 | zinc II ion transmembrane import(GO:0071578) |
| 0.2 | 0.5 | GO:0090367 | negative regulation of mRNA modification(GO:0090367) |
| 0.1 | 0.4 | GO:2000820 | negative regulation of transcription from RNA polymerase II promoter involved in smooth muscle cell differentiation(GO:2000820) |
| 0.1 | 0.9 | GO:1902748 | positive regulation of lens fiber cell differentiation(GO:1902748) |
| 0.1 | 1.2 | GO:0051256 | mitotic spindle midzone assembly(GO:0051256) |
| 0.1 | 1.1 | GO:0045409 | negative regulation of interleukin-6 biosynthetic process(GO:0045409) |
| 0.1 | 0.4 | GO:0070649 | formin-nucleated actin cable assembly(GO:0070649) |
| 0.1 | 0.8 | GO:0044828 | negative regulation by host of viral genome replication(GO:0044828) |
| 0.1 | 2.0 | GO:0034587 | piRNA metabolic process(GO:0034587) |
| 0.1 | 1.2 | GO:0002035 | brain renin-angiotensin system(GO:0002035) |
| 0.1 | 1.6 | GO:1904778 | regulation of protein localization to cell cortex(GO:1904776) positive regulation of protein localization to cell cortex(GO:1904778) |
| 0.1 | 0.7 | GO:2001181 | positive regulation of interleukin-10 secretion(GO:2001181) |
| 0.1 | 3.6 | GO:0002026 | regulation of the force of heart contraction(GO:0002026) |
| 0.1 | 0.4 | GO:0048298 | positive regulation of isotype switching to IgA isotypes(GO:0048298) |
| 0.0 | 0.9 | GO:0090266 | regulation of mitotic cell cycle spindle assembly checkpoint(GO:0090266) regulation of mitotic spindle checkpoint(GO:1903504) |
| 0.0 | 1.3 | GO:0033141 | positive regulation of peptidyl-serine phosphorylation of STAT protein(GO:0033141) |
| 0.0 | 0.1 | GO:0022007 | neural plate elongation(GO:0014022) convergent extension involved in neural plate elongation(GO:0022007) |
| 0.0 | 0.1 | GO:1900126 | negative regulation of hyaluronan biosynthetic process(GO:1900126) |
| 0.0 | 0.7 | GO:1902083 | negative regulation of peptidyl-cysteine S-nitrosylation(GO:1902083) |
| 0.0 | 0.5 | GO:2000628 | regulation of miRNA metabolic process(GO:2000628) |
| 0.0 | 1.5 | GO:0048240 | sperm capacitation(GO:0048240) |
| 0.0 | 0.4 | GO:0043144 | snoRNA processing(GO:0043144) |
| 0.0 | 0.3 | GO:0009253 | peptidoglycan metabolic process(GO:0000270) peptidoglycan catabolic process(GO:0009253) |
| 0.0 | 0.5 | GO:0061303 | cornea development in camera-type eye(GO:0061303) |
| 0.0 | 0.5 | GO:0051382 | kinetochore assembly(GO:0051382) |
| 0.0 | 1.2 | GO:0070979 | protein K11-linked ubiquitination(GO:0070979) |
| 0.0 | 1.3 | GO:0045725 | positive regulation of glycogen biosynthetic process(GO:0045725) |
| 0.0 | 0.2 | GO:0010756 | positive regulation of plasminogen activation(GO:0010756) |
| 0.0 | 1.9 | GO:0051865 | protein autoubiquitination(GO:0051865) |
| 0.0 | 3.5 | GO:0031110 | regulation of microtubule polymerization or depolymerization(GO:0031110) |
| 0.0 | 0.3 | GO:1904322 | response to forskolin(GO:1904321) cellular response to forskolin(GO:1904322) |
| 0.0 | 0.4 | GO:0006337 | nucleosome disassembly(GO:0006337) |
| 0.0 | 0.7 | GO:0061157 | mRNA destabilization(GO:0061157) |
| 0.0 | 0.2 | GO:0009162 | deoxyribonucleoside monophosphate metabolic process(GO:0009162) |
| 0.0 | 1.9 | GO:0071479 | cellular response to ionizing radiation(GO:0071479) |
| 0.0 | 2.2 | GO:0015908 | fatty acid transport(GO:0015908) |
| 0.0 | 0.2 | GO:0006369 | termination of RNA polymerase II transcription(GO:0006369) |
| 0.0 | 0.1 | GO:0051970 | negative regulation of transmission of nerve impulse(GO:0051970) |
| 0.0 | 1.5 | GO:0048477 | oogenesis(GO:0048477) |
| 0.0 | 0.1 | GO:0050955 | thermoception(GO:0050955) |
| 0.0 | 0.1 | GO:0070562 | regulation of vitamin D receptor signaling pathway(GO:0070562) |
| 0.0 | 0.8 | GO:0050873 | brown fat cell differentiation(GO:0050873) |
| 0.0 | 2.1 | GO:0007050 | cell cycle arrest(GO:0007050) |
| 0.0 | 0.2 | GO:0001675 | acrosome assembly(GO:0001675) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.3 | 3.8 | GO:0060187 | cell pole(GO:0060187) |
| 0.9 | 4.7 | GO:0010370 | perinucleolar chromocenter(GO:0010370) |
| 0.3 | 2.0 | GO:0071547 | piP-body(GO:0071547) |
| 0.3 | 1.6 | GO:0005945 | 6-phosphofructokinase complex(GO:0005945) |
| 0.3 | 0.8 | GO:0002945 | cyclin K-CDK12 complex(GO:0002944) cyclin K-CDK13 complex(GO:0002945) |
| 0.2 | 3.5 | GO:0000940 | condensed chromosome outer kinetochore(GO:0000940) |
| 0.2 | 3.0 | GO:0031258 | lamellipodium membrane(GO:0031258) |
| 0.2 | 1.3 | GO:0005899 | insulin receptor complex(GO:0005899) |
| 0.2 | 2.2 | GO:0034992 | microtubule organizing center attachment site(GO:0034992) LINC complex(GO:0034993) |
| 0.2 | 4.9 | GO:0035686 | sperm fibrous sheath(GO:0035686) |
| 0.2 | 1.7 | GO:0097197 | tetraspanin-enriched microdomain(GO:0097197) |
| 0.2 | 0.9 | GO:0042583 | chromaffin granule(GO:0042583) |
| 0.1 | 0.5 | GO:0000939 | condensed chromosome inner kinetochore(GO:0000939) |
| 0.1 | 12.4 | GO:0005581 | collagen trimer(GO:0005581) |
| 0.1 | 1.0 | GO:0044327 | dendritic spine head(GO:0044327) |
| 0.1 | 0.4 | GO:0032389 | MutLalpha complex(GO:0032389) |
| 0.0 | 1.6 | GO:0005680 | anaphase-promoting complex(GO:0005680) |
| 0.0 | 0.6 | GO:0000176 | nuclear exosome (RNase complex)(GO:0000176) |
| 0.0 | 3.6 | GO:0031672 | A band(GO:0031672) |
| 0.0 | 0.5 | GO:0016281 | eukaryotic translation initiation factor 4F complex(GO:0016281) |
| 0.0 | 0.3 | GO:0033588 | Elongator holoenzyme complex(GO:0033588) |
| 0.0 | 1.3 | GO:0005721 | pericentric heterochromatin(GO:0005721) |
| 0.0 | 0.7 | GO:0005665 | DNA-directed RNA polymerase II, core complex(GO:0005665) |
| 0.0 | 1.6 | GO:0099738 | cell cortex region(GO:0099738) |
| 0.0 | 0.2 | GO:0070847 | core mediator complex(GO:0070847) |
| 0.0 | 0.4 | GO:0071564 | npBAF complex(GO:0071564) |
| 0.0 | 0.5 | GO:0005697 | telomerase holoenzyme complex(GO:0005697) |
| 0.0 | 0.1 | GO:0005726 | perichromatin fibrils(GO:0005726) |
| 0.0 | 0.4 | GO:0000930 | gamma-tubulin complex(GO:0000930) |
| 0.0 | 0.8 | GO:0005788 | endoplasmic reticulum lumen(GO:0005788) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.1 | 12.1 | GO:0033691 | sialic acid binding(GO:0033691) |
| 0.9 | 4.7 | GO:0034584 | piRNA binding(GO:0034584) |
| 0.4 | 1.8 | GO:0004668 | protein-arginine deiminase activity(GO:0004668) |
| 0.3 | 2.8 | GO:0019863 | IgE binding(GO:0019863) |
| 0.3 | 1.6 | GO:0003872 | 6-phosphofructokinase activity(GO:0003872) |
| 0.3 | 1.2 | GO:0004711 | ribosomal protein S6 kinase activity(GO:0004711) |
| 0.2 | 2.2 | GO:0004321 | fatty-acyl-CoA synthase activity(GO:0004321) |
| 0.2 | 0.6 | GO:0005174 | CD40 receptor binding(GO:0005174) |
| 0.2 | 0.6 | GO:0042015 | interleukin-20 binding(GO:0042015) |
| 0.1 | 7.1 | GO:0003785 | actin monomer binding(GO:0003785) |
| 0.1 | 2.2 | GO:0043495 | protein anchor(GO:0043495) |
| 0.1 | 1.3 | GO:0003886 | DNA (cytosine-5-)-methyltransferase activity(GO:0003886) |
| 0.1 | 0.4 | GO:0032138 | single base insertion or deletion binding(GO:0032138) |
| 0.1 | 1.2 | GO:0008429 | phosphatidylethanolamine binding(GO:0008429) oligosaccharide binding(GO:0070492) |
| 0.1 | 0.9 | GO:0004565 | beta-galactosidase activity(GO:0004565) |
| 0.1 | 0.4 | GO:0045322 | unmethylated CpG binding(GO:0045322) |
| 0.1 | 3.0 | GO:0005385 | zinc ion transmembrane transporter activity(GO:0005385) |
| 0.1 | 1.3 | GO:0005132 | type I interferon receptor binding(GO:0005132) |
| 0.0 | 1.6 | GO:0005545 | 1-phosphatidylinositol binding(GO:0005545) |
| 0.0 | 2.7 | GO:0008138 | protein tyrosine/serine/threonine phosphatase activity(GO:0008138) |
| 0.0 | 0.5 | GO:1990247 | N6-methyladenosine-containing RNA binding(GO:1990247) |
| 0.0 | 1.1 | GO:0001871 | pattern binding(GO:0001871) polysaccharide binding(GO:0030247) |
| 0.0 | 0.3 | GO:0005004 | GPI-linked ephrin receptor activity(GO:0005004) |
| 0.0 | 0.8 | GO:0008353 | RNA polymerase II carboxy-terminal domain kinase activity(GO:0008353) |
| 0.0 | 0.5 | GO:0035925 | mRNA 3'-UTR AU-rich region binding(GO:0035925) |
| 0.0 | 1.3 | GO:0005070 | SH3/SH2 adaptor activity(GO:0005070) |
| 0.0 | 0.1 | GO:0035614 | snRNA stem-loop binding(GO:0035614) |
| 0.0 | 1.0 | GO:0031492 | nucleosomal DNA binding(GO:0031492) |
| 0.0 | 0.7 | GO:0070577 | lysine-acetylated histone binding(GO:0070577) |
| 0.0 | 1.9 | GO:0001784 | phosphotyrosine binding(GO:0001784) |
| 0.0 | 0.9 | GO:0070412 | R-SMAD binding(GO:0070412) |
| 0.0 | 0.4 | GO:0004707 | MAP kinase activity(GO:0004707) |
| 0.0 | 0.3 | GO:0003796 | lysozyme activity(GO:0003796) |
| 0.0 | 0.3 | GO:0005388 | calcium-transporting ATPase activity(GO:0005388) |
| 0.0 | 1.9 | GO:0001158 | enhancer sequence-specific DNA binding(GO:0001158) |
| 0.0 | 1.0 | GO:0048365 | Rac GTPase binding(GO:0048365) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 12.1 | NABA ECM GLYCOPROTEINS | Genes encoding structural ECM glycoproteins |
| 0.0 | 2.4 | PID CD40 PATHWAY | CD40/CD40L signaling |
| 0.0 | 1.6 | NABA PROTEOGLYCANS | Genes encoding proteoglycans |
| 0.0 | 1.3 | SIG INSULIN RECEPTOR PATHWAY IN CARDIAC MYOCYTES | Genes related to the insulin receptor pathway |
| 0.0 | 1.8 | ST PHOSPHOINOSITIDE 3 KINASE PATHWAY | PI3K Pathway |
| 0.0 | 1.2 | PID SYNDECAN 2 PATHWAY | Syndecan-2-mediated signaling events |
| 0.0 | 0.4 | PID ERB GENOMIC PATHWAY | Validated nuclear estrogen receptor beta network |
| 0.0 | 0.3 | ST INTERFERON GAMMA PATHWAY | Interferon gamma pathway. |
| 0.0 | 1.3 | PID CD8 TCR DOWNSTREAM PATHWAY | Downstream signaling in naïve CD8+ T cells |
| 0.0 | 0.9 | PID UPA UPAR PATHWAY | Urokinase-type plasminogen activator (uPA) and uPAR-mediated signaling |
| 0.0 | 0.5 | PID FOXM1 PATHWAY | FOXM1 transcription factor network |
| 0.0 | 1.3 | PID RB 1PATHWAY | Regulation of retinoblastoma protein |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 3.0 | REACTOME ZINC TRANSPORTERS | Genes involved in Zinc transporters |
| 0.1 | 12.2 | REACTOME TRANSCRIPTIONAL REGULATION OF WHITE ADIPOCYTE DIFFERENTIATION | Genes involved in Transcriptional Regulation of White Adipocyte Differentiation |
| 0.1 | 1.9 | REACTOME CONVERSION FROM APC C CDC20 TO APC C CDH1 IN LATE ANAPHASE | Genes involved in Conversion from APC/C:Cdc20 to APC/C:Cdh1 in late anaphase |
| 0.1 | 3.6 | REACTOME STRIATED MUSCLE CONTRACTION | Genes involved in Striated Muscle Contraction |
| 0.1 | 1.3 | REACTOME REGULATION OF IFNA SIGNALING | Genes involved in Regulation of IFNA signaling |
| 0.1 | 0.6 | REACTOME MRNA DECAY BY 3 TO 5 EXORIBONUCLEASE | Genes involved in mRNA Decay by 3' to 5' Exoribonuclease |
| 0.0 | 0.7 | REACTOME PD1 SIGNALING | Genes involved in PD-1 signaling |
| 0.0 | 2.2 | REACTOME MEIOTIC SYNAPSIS | Genes involved in Meiotic Synapsis |
| 0.0 | 4.2 | REACTOME MITOTIC PROMETAPHASE | Genes involved in Mitotic Prometaphase |
| 0.0 | 1.6 | REACTOME GLYCOLYSIS | Genes involved in Glycolysis |
| 0.0 | 0.5 | REACTOME DESTABILIZATION OF MRNA BY KSRP | Genes involved in Destabilization of mRNA by KSRP |
| 0.0 | 0.5 | REACTOME KERATAN SULFATE DEGRADATION | Genes involved in Keratan sulfate degradation |
| 0.0 | 1.9 | REACTOME ANTIGEN ACTIVATES B CELL RECEPTOR LEADING TO GENERATION OF SECOND MESSENGERS | Genes involved in Antigen Activates B Cell Receptor Leading to Generation of Second Messengers |
| 0.0 | 1.3 | REACTOME MUSCLE CONTRACTION | Genes involved in Muscle contraction |
| 0.0 | 1.2 | REACTOME ERK MAPK TARGETS | Genes involved in ERK/MAPK targets |
| 0.0 | 1.0 | REACTOME SIGNALING BY ROBO RECEPTOR | Genes involved in Signaling by Robo receptor |
| 0.0 | 0.9 | REACTOME SMAD2 SMAD3 SMAD4 HETEROTRIMER REGULATES TRANSCRIPTION | Genes involved in SMAD2/SMAD3:SMAD4 heterotrimer regulates transcription |
| 0.0 | 0.5 | REACTOME ADHERENS JUNCTIONS INTERACTIONS | Genes involved in Adherens junctions interactions |
| 0.0 | 0.6 | REACTOME IMMUNOREGULATORY INTERACTIONS BETWEEN A LYMPHOID AND A NON LYMPHOID CELL | Genes involved in Immunoregulatory interactions between a Lymphoid and a non-Lymphoid cell |