Project
PRJNA375882: Comprehensive Mouse Transcriptomic BodyMap
Navigation
Downloads
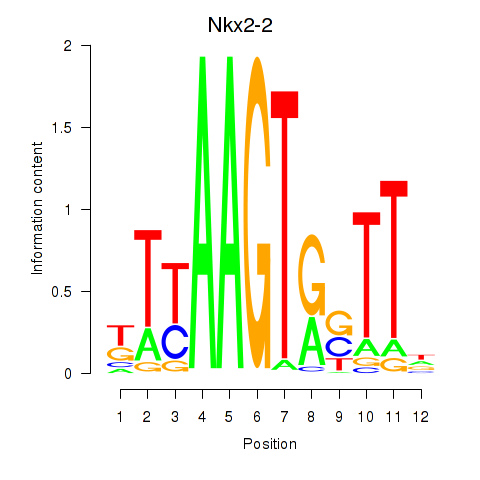
Results for Nkx2-2
Z-value: 0.90
Transcription factors associated with Nkx2-2
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Nkx2-2
|
ENSMUSG00000027434.12 | Nkx2-2 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Nkx2-2 | mm39_v1_chr2_-_147028309_147028331 | -0.75 | 3.9e-14 | Click! |
Activity profile of Nkx2-2 motif
Sorted Z-values of Nkx2-2 motif
Network of associatons between targets according to the STRING database.
First level regulatory network of Nkx2-2
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr2_+_174292471 | 6.89 |
ENSMUST00000016399.6
|
Tubb1
|
tubulin, beta 1 class VI |
| chr1_-_173703424 | 5.40 |
ENSMUST00000186442.7
|
Mndal
|
myeloid nuclear differentiation antigen like |
| chr13_-_113237505 | 5.34 |
ENSMUST00000224282.2
ENSMUST00000023897.7 |
Gzma
|
granzyme A |
| chr6_+_67873135 | 5.18 |
ENSMUST00000103310.3
|
Igkv14-126
|
immunoglobulin kappa variable 14-126 |
| chr9_+_51124983 | 4.42 |
ENSMUST00000034554.9
|
Pou2af1
|
POU domain, class 2, associating factor 1 |
| chr19_-_11243530 | 4.32 |
ENSMUST00000169159.3
|
Ms4a1
|
membrane-spanning 4-domains, subfamily A, member 1 |
| chr6_-_41681273 | 4.10 |
ENSMUST00000031899.14
|
Kel
|
Kell blood group |
| chr17_+_18108102 | 4.08 |
ENSMUST00000054871.12
ENSMUST00000064068.5 |
Fpr3
Fpr2
|
formyl peptide receptor 3 formyl peptide receptor 2 |
| chr1_-_73055043 | 3.94 |
ENSMUST00000027374.7
|
Tnp1
|
transition protein 1 |
| chr10_-_117681864 | 3.94 |
ENSMUST00000064667.9
|
Rap1b
|
RAS related protein 1b |
| chr17_+_18108086 | 3.81 |
ENSMUST00000149944.2
|
Fpr2
|
formyl peptide receptor 2 |
| chr2_-_58050494 | 3.69 |
ENSMUST00000028175.7
|
Cytip
|
cytohesin 1 interacting protein |
| chr6_-_136834725 | 3.61 |
ENSMUST00000032341.3
|
Art4
|
ADP-ribosyltransferase 4 |
| chr6_+_68233361 | 3.50 |
ENSMUST00000103320.3
|
Igkv14-111
|
immunoglobulin kappa variable 14-111 |
| chr6_+_65929546 | 3.38 |
ENSMUST00000043382.9
|
4930544G11Rik
|
RIKEN cDNA 4930544G11 gene |
| chr5_-_88675700 | 3.33 |
ENSMUST00000087033.6
|
Jchain
|
immunoglobulin joining chain |
| chr7_+_44485704 | 3.22 |
ENSMUST00000033015.8
|
Il4i1
|
interleukin 4 induced 1 |
| chrX_+_106192510 | 3.19 |
ENSMUST00000147521.8
ENSMUST00000167673.2 |
P2ry10b
|
purinergic receptor P2Y, G-protein coupled 10B |
| chr10_+_87926932 | 3.17 |
ENSMUST00000048621.8
|
Pmch
|
pro-melanin-concentrating hormone |
| chr2_-_84605732 | 3.00 |
ENSMUST00000023994.10
|
Serping1
|
serine (or cysteine) peptidase inhibitor, clade G, member 1 |
| chr9_+_72345801 | 2.98 |
ENSMUST00000184604.8
ENSMUST00000034746.10 |
Mns1
|
meiosis-specific nuclear structural protein 1 |
| chr16_+_14523696 | 2.96 |
ENSMUST00000023356.8
|
Snai2
|
snail family zinc finger 2 |
| chr11_-_79414542 | 2.95 |
ENSMUST00000179322.2
|
Evi2b
|
ecotropic viral integration site 2b |
| chr15_-_5093222 | 2.85 |
ENSMUST00000110689.5
|
C7
|
complement component 7 |
| chr2_-_84605764 | 2.84 |
ENSMUST00000111641.2
|
Serping1
|
serine (or cysteine) peptidase inhibitor, clade G, member 1 |
| chr5_-_82271183 | 2.80 |
ENSMUST00000186079.2
ENSMUST00000185607.2 |
1700031L13Rik
|
RIKEN cDNA 1700031L13 gene |
| chr10_-_59277570 | 2.77 |
ENSMUST00000009798.5
|
Oit3
|
oncoprotein induced transcript 3 |
| chr1_-_173426640 | 2.73 |
ENSMUST00000150649.9
ENSMUST00000180215.2 ENSMUST00000097462.9 |
Ifi213
|
interferon activated gene 213 |
| chr8_-_79975199 | 2.70 |
ENSMUST00000034109.6
|
1700011L22Rik
|
RIKEN cDNA 1700011L22 gene |
| chr12_-_112824506 | 2.65 |
ENSMUST00000021729.9
|
Gpr132
|
G protein-coupled receptor 132 |
| chr2_-_83771564 | 2.65 |
ENSMUST00000038223.8
ENSMUST00000152829.2 |
Zswim2
|
zinc finger SWIM-type containing 2 |
| chr1_-_85664246 | 2.64 |
ENSMUST00000064788.14
|
A630001G21Rik
|
RIKEN cDNA A630001G21 gene |
| chr8_-_25438784 | 2.61 |
ENSMUST00000119720.8
ENSMUST00000121438.9 |
Adam32
|
a disintegrin and metallopeptidase domain 32 |
| chr6_-_23650205 | 2.43 |
ENSMUST00000115354.2
|
Rnf133
|
ring finger protein 133 |
| chr11_+_96209093 | 2.41 |
ENSMUST00000049241.9
|
Hoxb4
|
homeobox B4 |
| chr2_+_3705824 | 2.39 |
ENSMUST00000115054.9
|
Fam107b
|
family with sequence similarity 107, member B |
| chr7_+_45067747 | 2.33 |
ENSMUST00000238638.2
|
Lhb
|
luteinizing hormone beta |
| chr8_-_3976808 | 2.32 |
ENSMUST00000188386.2
ENSMUST00000084086.9 ENSMUST00000171635.8 |
Cd209b
|
CD209b antigen |
| chr6_-_3494587 | 2.29 |
ENSMUST00000049985.15
|
Hepacam2
|
HEPACAM family member 2 |
| chr1_-_165830160 | 2.26 |
ENSMUST00000111429.11
ENSMUST00000176800.2 ENSMUST00000177358.8 |
Pou2f1
|
POU domain, class 2, transcription factor 1 |
| chr18_+_43897354 | 2.21 |
ENSMUST00000187157.7
ENSMUST00000043803.13 ENSMUST00000189750.2 |
Scgb3a2
|
secretoglobin, family 3A, member 2 |
| chr11_+_105885461 | 2.19 |
ENSMUST00000190995.2
|
Ace3
|
angiotensin I converting enzyme (peptidyl-dipeptidase A) 3 |
| chr6_+_67586695 | 2.18 |
ENSMUST00000103303.3
|
Igkv1-135
|
immunoglobulin kappa variable 1-135 |
| chr14_-_49482846 | 2.13 |
ENSMUST00000227113.2
ENSMUST00000130853.2 ENSMUST00000228936.2 ENSMUST00000022398.15 |
ccdc198
|
coiled-coil domain containing 198 |
| chr3_-_107893676 | 2.13 |
ENSMUST00000066530.7
ENSMUST00000012348.9 |
Gstm2
|
glutathione S-transferase, mu 2 |
| chr1_+_173501215 | 2.10 |
ENSMUST00000085876.12
|
Ifi208
|
interferon activated gene 208 |
| chr2_+_127750978 | 2.04 |
ENSMUST00000110344.2
|
Acoxl
|
acyl-Coenzyme A oxidase-like |
| chr6_-_23650297 | 2.04 |
ENSMUST00000063548.4
|
Rnf133
|
ring finger protein 133 |
| chr4_-_45532470 | 2.03 |
ENSMUST00000147448.2
|
Shb
|
src homology 2 domain-containing transforming protein B |
| chr11_-_79971750 | 2.03 |
ENSMUST00000103233.10
ENSMUST00000061283.15 |
Crlf3
|
cytokine receptor-like factor 3 |
| chr7_-_122969064 | 1.99 |
ENSMUST00000207010.2
ENSMUST00000167309.8 ENSMUST00000205262.2 ENSMUST00000106442.9 ENSMUST00000098060.5 |
Arhgap17
|
Rho GTPase activating protein 17 |
| chr13_-_20008397 | 1.97 |
ENSMUST00000222664.2
ENSMUST00000065335.3 |
Gpr141
|
G protein-coupled receptor 141 |
| chr11_+_48691175 | 1.93 |
ENSMUST00000020640.8
|
Rack1
|
receptor for activated C kinase 1 |
| chr7_+_122969004 | 1.91 |
ENSMUST00000206721.2
|
Lcmt1
|
leucine carboxyl methyltransferase 1 |
| chr3_+_108646974 | 1.90 |
ENSMUST00000133931.9
|
Aknad1
|
AKNA domain containing 1 |
| chr7_+_121333700 | 1.87 |
ENSMUST00000000221.6
|
Scnn1g
|
sodium channel, nonvoltage-gated 1 gamma |
| chr10_+_57521930 | 1.86 |
ENSMUST00000177325.8
|
Pkib
|
protein kinase inhibitor beta, cAMP dependent, testis specific |
| chr12_+_78240999 | 1.84 |
ENSMUST00000211288.3
|
Gm6657
|
predicted gene 6657 |
| chr1_-_165830187 | 1.82 |
ENSMUST00000184643.8
ENSMUST00000160908.8 ENSMUST00000027850.15 ENSMUST00000160260.9 |
Pou2f1
|
POU domain, class 2, transcription factor 1 |
| chr3_-_49711765 | 1.81 |
ENSMUST00000035931.13
|
Pcdh18
|
protocadherin 18 |
| chr14_-_101846459 | 1.80 |
ENSMUST00000161991.8
|
Tbc1d4
|
TBC1 domain family, member 4 |
| chr13_-_19917092 | 1.80 |
ENSMUST00000151029.3
|
Gpr141b
|
G protein-coupled receptor 141B |
| chr16_-_85347305 | 1.78 |
ENSMUST00000175700.8
ENSMUST00000114174.3 |
Cyyr1
|
cysteine and tyrosine-rich protein 1 |
| chr8_+_46387647 | 1.76 |
ENSMUST00000095326.10
|
Ccdc110
|
coiled-coil domain containing 110 |
| chrX_+_106193167 | 1.75 |
ENSMUST00000137107.2
ENSMUST00000067249.3 |
P2ry10b
|
purinergic receptor P2Y, G-protein coupled 10B |
| chr5_+_67125759 | 1.74 |
ENSMUST00000238993.2
ENSMUST00000038188.14 |
Limch1
|
LIM and calponin homology domains 1 |
| chr14_-_56026266 | 1.73 |
ENSMUST00000168716.8
ENSMUST00000178399.3 ENSMUST00000022830.14 |
Ripk3
|
receptor-interacting serine-threonine kinase 3 |
| chr5_+_11392646 | 1.72 |
ENSMUST00000179482.2
|
Speer1
|
spermatogenesis associated glutamate (E)-rich protein 1 |
| chr5_+_11821653 | 1.72 |
ENSMUST00000178158.2
|
Gm8926
|
predicted gene 8926 |
| chr6_-_73446560 | 1.68 |
ENSMUST00000070163.6
|
4931417E11Rik
|
RIKEN cDNA 4931417E11 gene |
| chr9_+_108225026 | 1.65 |
ENSMUST00000035237.12
ENSMUST00000194959.6 |
Usp4
|
ubiquitin specific peptidase 4 (proto-oncogene) |
| chr5_+_149363168 | 1.63 |
ENSMUST00000201683.4
ENSMUST00000202920.4 |
Tex26
|
testis expressed 26 |
| chr18_-_44308126 | 1.63 |
ENSMUST00000066328.5
|
Spinkl
|
serine protease inhibitor, Kazal type-like |
| chr14_+_32713387 | 1.63 |
ENSMUST00000123822.8
ENSMUST00000120951.2 |
Lrrc18
|
leucine rich repeat containing 18 |
| chr18_-_3309858 | 1.62 |
ENSMUST00000144496.8
ENSMUST00000154715.8 |
Crem
|
cAMP responsive element modulator |
| chr13_-_76453191 | 1.62 |
ENSMUST00000208418.2
|
Fam81b
|
family with sequence similarity 81, member B |
| chr14_-_42872221 | 1.58 |
ENSMUST00000100694.5
|
Gm10376
|
predicted gene 10376 |
| chr5_-_110987441 | 1.58 |
ENSMUST00000145318.2
|
Hscb
|
HscB iron-sulfur cluster co-chaperone |
| chr4_+_102843540 | 1.57 |
ENSMUST00000030248.12
ENSMUST00000125417.9 ENSMUST00000169211.3 |
Dynlt5
|
dynein light chain Tctex-type 5 |
| chr5_+_11733203 | 1.55 |
ENSMUST00000178989.2
|
Gm8922
|
predicted gene 8922 |
| chrX_+_106193060 | 1.54 |
ENSMUST00000125676.8
ENSMUST00000180182.2 |
P2ry10b
|
purinergic receptor P2Y, G-protein coupled 10B |
| chr14_+_32713336 | 1.52 |
ENSMUST00000038956.12
|
Lrrc18
|
leucine rich repeat containing 18 |
| chr3_-_130524024 | 1.52 |
ENSMUST00000079085.11
|
Rpl34
|
ribosomal protein L34 |
| chr5_-_34326847 | 1.50 |
ENSMUST00000202042.2
ENSMUST00000060049.8 |
Haus3
|
HAUS augmin-like complex, subunit 3 |
| chr5_+_11178835 | 1.50 |
ENSMUST00000178863.2
|
Gm8879
|
predicted gene 8879 |
| chr5_+_67125902 | 1.50 |
ENSMUST00000127184.8
|
Limch1
|
LIM and calponin homology domains 1 |
| chr5_+_11467593 | 1.50 |
ENSMUST00000179679.2
|
Gm8897
|
predicted gene 8897 |
| chr8_+_46984016 | 1.48 |
ENSMUST00000152423.2
|
Acsl1
|
acyl-CoA synthetase long-chain family member 1 |
| chr2_-_140231618 | 1.47 |
ENSMUST00000122367.8
ENSMUST00000120133.2 |
Sel1l2
|
sel-1 suppressor of lin-12-like 2 (C. elegans) |
| chr13_-_67080968 | 1.44 |
ENSMUST00000172597.8
ENSMUST00000173773.2 |
Mterf3
|
mitochondrial transcription termination factor 3 |
| chr17_+_7592045 | 1.44 |
ENSMUST00000095726.11
ENSMUST00000128533.8 ENSMUST00000129709.8 ENSMUST00000147803.8 ENSMUST00000140192.8 ENSMUST00000138222.8 ENSMUST00000144861.2 |
Tcp10a
|
t-complex protein 10a |
| chr3_-_49711706 | 1.44 |
ENSMUST00000191794.2
|
Pcdh18
|
protocadherin 18 |
| chr6_+_68402550 | 1.44 |
ENSMUST00000103323.3
|
Igkv16-104
|
immunoglobulin kappa variable 16-104 |
| chr11_+_23234644 | 1.43 |
ENSMUST00000150750.3
|
Xpo1
|
exportin 1 |
| chr14_+_24540745 | 1.42 |
ENSMUST00000112384.10
|
Rps24
|
ribosomal protein S24 |
| chr10_-_13053733 | 1.42 |
ENSMUST00000019954.6
|
Zc2hc1b
|
zinc finger, C2HC-type containing 1B |
| chrX_+_110154017 | 1.41 |
ENSMUST00000210720.3
|
Cylc1
|
cylicin, basic protein of sperm head cytoskeleton 1 |
| chr19_+_6097083 | 1.41 |
ENSMUST00000134667.8
|
Syvn1
|
synovial apoptosis inhibitor 1, synoviolin |
| chr14_-_59632830 | 1.41 |
ENSMUST00000166912.3
|
Phf11c
|
PHD finger protein 11C |
| chr11_+_75521800 | 1.40 |
ENSMUST00000006286.9
|
Inpp5k
|
inositol polyphosphate 5-phosphatase K |
| chr3_-_105940130 | 1.40 |
ENSMUST00000200146.2
|
Chil5
|
chitinase-like 5 |
| chr6_-_120334382 | 1.39 |
ENSMUST00000032283.12
|
Ccdc77
|
coiled-coil domain containing 77 |
| chr5_-_35897331 | 1.37 |
ENSMUST00000201511.2
|
Sh3tc1
|
SH3 domain and tetratricopeptide repeats 1 |
| chr10_+_79500387 | 1.37 |
ENSMUST00000020554.8
|
Madcam1
|
mucosal vascular addressin cell adhesion molecule 1 |
| chr5_+_11234174 | 1.37 |
ENSMUST00000168407.3
|
Gm5861
|
predicted gene 5861 |
| chr19_+_6097111 | 1.35 |
ENSMUST00000025723.9
|
Syvn1
|
synovial apoptosis inhibitor 1, synoviolin |
| chr6_-_70769135 | 1.34 |
ENSMUST00000066134.6
|
Rpia
|
ribose 5-phosphate isomerase A |
| chr5_+_11553780 | 1.34 |
ENSMUST00000179375.2
|
Gm8906
|
predicted gene 8906 |
| chr18_+_20443795 | 1.33 |
ENSMUST00000077146.4
|
Dsg1a
|
desmoglein 1 alpha |
| chr12_+_78243846 | 1.32 |
ENSMUST00000188791.2
|
Gm6657
|
predicted gene 6657 |
| chr17_+_43671314 | 1.32 |
ENSMUST00000226087.2
|
Adgrf5
|
adhesion G protein-coupled receptor F5 |
| chr9_-_107512566 | 1.30 |
ENSMUST00000055704.12
|
Gnai2
|
guanine nucleotide binding protein (G protein), alpha inhibiting 2 |
| chr5_+_149363114 | 1.30 |
ENSMUST00000031667.8
|
Tex26
|
testis expressed 26 |
| chr4_+_115420876 | 1.30 |
ENSMUST00000126645.8
ENSMUST00000030480.4 |
Cyp4a31
|
cytochrome P450, family 4, subfamily a, polypeptide 31 |
| chr12_+_33003882 | 1.29 |
ENSMUST00000076698.13
|
Sypl
|
synaptophysin-like protein |
| chr6_+_137731526 | 1.28 |
ENSMUST00000203216.3
ENSMUST00000087675.9 ENSMUST00000203693.3 |
Dera
|
deoxyribose-phosphate aldolase (putative) |
| chr6_+_21985902 | 1.27 |
ENSMUST00000115383.9
|
Cped1
|
cadherin-like and PC-esterase domain containing 1 |
| chr16_+_32315076 | 1.27 |
ENSMUST00000064192.8
|
Zdhhc19
|
zinc finger, DHHC domain containing 19 |
| chr11_+_68393845 | 1.24 |
ENSMUST00000102613.8
ENSMUST00000060441.7 |
Pik3r6
|
phosphoinositide-3-kinase regulatory subunit 5 |
| chr11_-_115078147 | 1.24 |
ENSMUST00000103038.8
ENSMUST00000103039.2 ENSMUST00000103040.11 |
Nat9
|
N-acetyltransferase 9 (GCN5-related, putative) |
| chr14_+_54000594 | 1.23 |
ENSMUST00000103589.6
|
Trav14-3
|
T cell receptor alpha variable 14-3 |
| chr15_-_10470575 | 1.23 |
ENSMUST00000136591.8
|
Dnajc21
|
DnaJ heat shock protein family (Hsp40) member C21 |
| chr16_+_23338960 | 1.22 |
ENSMUST00000211460.2
ENSMUST00000210658.2 ENSMUST00000209198.2 ENSMUST00000210371.2 ENSMUST00000211499.2 ENSMUST00000210795.2 ENSMUST00000209422.2 |
Gm45338
Rtp4
|
predicted gene 45338 receptor transporter protein 4 |
| chr13_+_47347301 | 1.22 |
ENSMUST00000110111.4
|
Rnf144b
|
ring finger protein 144B |
| chr6_+_122967309 | 1.21 |
ENSMUST00000079379.3
|
Clec4a4
|
C-type lectin domain family 4, member a4 |
| chr13_-_103901010 | 1.20 |
ENSMUST00000210489.2
|
Srek1
|
splicing regulatory glutamine/lysine-rich protein 1 |
| chrX_+_67864699 | 1.20 |
ENSMUST00000096420.3
|
Gm14698
|
predicted gene 14698 |
| chr6_+_67816777 | 1.19 |
ENSMUST00000200578.5
ENSMUST00000103308.3 |
Igkv9-129
|
immunoglobulin kappa variable 9-129 |
| chr3_-_75177378 | 1.19 |
ENSMUST00000039047.5
|
Serpini2
|
serine (or cysteine) peptidase inhibitor, clade I, member 2 |
| chr1_-_161807205 | 1.18 |
ENSMUST00000162676.2
|
4930558K02Rik
|
RIKEN cDNA 4930558K02 gene |
| chr6_+_5390386 | 1.18 |
ENSMUST00000183358.2
|
Asb4
|
ankyrin repeat and SOCS box-containing 4 |
| chr6_-_120334400 | 1.18 |
ENSMUST00000112703.8
|
Ccdc77
|
coiled-coil domain containing 77 |
| chr5_+_11307124 | 1.18 |
ENSMUST00000177727.2
|
Gm8890
|
predicted gene 8890 |
| chr16_+_32315090 | 1.17 |
ENSMUST00000231510.2
|
Zdhhc19
|
zinc finger, DHHC domain containing 19 |
| chr3_-_6685492 | 1.17 |
ENSMUST00000091364.4
|
1700008P02Rik
|
RIKEN cDNA 1700008P02 gene |
| chrX_+_106836189 | 1.17 |
ENSMUST00000101292.9
|
Tent5d
|
terminal nucleotidyltransferase 5D |
| chr14_+_53994813 | 1.17 |
ENSMUST00000180380.3
|
Trav13-4-dv7
|
T cell receptor alpha variable 13-4-DV7 |
| chr6_+_68026941 | 1.16 |
ENSMUST00000103316.2
|
Igkv9-120
|
immunoglobulin kappa chain variable 9-120 |
| chr15_-_102259158 | 1.16 |
ENSMUST00000231061.2
ENSMUST00000041208.9 |
Aaas
|
achalasia, adrenocortical insufficiency, alacrimia |
| chr1_-_69723316 | 1.15 |
ENSMUST00000190855.7
ENSMUST00000188110.7 ENSMUST00000191262.7 |
Ikzf2
|
IKAROS family zinc finger 2 |
| chr4_-_108075119 | 1.13 |
ENSMUST00000223127.2
ENSMUST00000043793.7 ENSMUST00000106690.9 |
Zyg11a
|
zyg-11 family member A, cell cycle regulator |
| chr6_+_137731599 | 1.12 |
ENSMUST00000204356.2
|
Dera
|
deoxyribose-phosphate aldolase (putative) |
| chr2_+_51962831 | 1.12 |
ENSMUST00000112693.10
|
Rif1
|
replication timing regulatory factor 1 |
| chr1_-_155910567 | 1.11 |
ENSMUST00000141878.8
|
Tor1aip1
|
torsin A interacting protein 1 |
| chr11_+_78192355 | 1.10 |
ENSMUST00000045026.4
|
Spag5
|
sperm associated antigen 5 |
| chr9_-_106768601 | 1.09 |
ENSMUST00000069036.14
|
Manf
|
mesencephalic astrocyte-derived neurotrophic factor |
| chr17_+_79934096 | 1.08 |
ENSMUST00000224618.2
|
Rmdn2
|
regulator of microtubule dynamics 2 |
| chr15_+_66542598 | 1.08 |
ENSMUST00000065916.14
|
Tg
|
thyroglobulin |
| chr17_+_79919267 | 1.08 |
ENSMUST00000223924.2
|
Rmdn2
|
regulator of microtubule dynamics 2 |
| chr2_-_152857239 | 1.06 |
ENSMUST00000028972.9
|
Pdrg1
|
p53 and DNA damage regulated 1 |
| chr13_+_23940964 | 1.05 |
ENSMUST00000102965.4
|
H4c2
|
H4 clustered histone 2 |
| chr5_-_110987604 | 1.05 |
ENSMUST00000056937.12
|
Hscb
|
HscB iron-sulfur cluster co-chaperone |
| chr11_-_45845992 | 1.05 |
ENSMUST00000109254.2
|
Thg1l
|
tRNA-histidine guanylyltransferase 1-like (S. cerevisiae) |
| chr19_-_46950948 | 1.04 |
ENSMUST00000236924.2
|
Nt5c2
|
5'-nucleotidase, cytosolic II |
| chr3_-_104771716 | 1.03 |
ENSMUST00000195912.2
ENSMUST00000094028.10 |
Capza1
|
capping protein (actin filament) muscle Z-line, alpha 1 |
| chrX_+_61511597 | 1.02 |
ENSMUST00000033537.2
|
4931400O07Rik
|
RIKEN cDNA 4931400O07 gene |
| chr6_-_50631418 | 1.02 |
ENSMUST00000031853.8
|
Npvf
|
neuropeptide VF precursor |
| chr12_+_105996961 | 1.02 |
ENSMUST00000220629.2
|
Vrk1
|
vaccinia related kinase 1 |
| chr5_-_83502966 | 1.01 |
ENSMUST00000053543.11
|
Tecrl
|
trans-2,3-enoyl-CoA reductase-like |
| chr15_-_74869684 | 1.01 |
ENSMUST00000190188.2
ENSMUST00000189068.7 ENSMUST00000186526.7 ENSMUST00000187171.2 ENSMUST00000187994.7 |
Ly6a
|
lymphocyte antigen 6 complex, locus A |
| chr9_+_39853511 | 1.00 |
ENSMUST00000062833.6
|
Olfr974
|
olfactory receptor 974 |
| chr15_+_6552270 | 1.00 |
ENSMUST00000226412.2
|
Fyb
|
FYN binding protein |
| chr7_+_19079058 | 0.99 |
ENSMUST00000160369.8
|
Ercc1
|
excision repair cross-complementing rodent repair deficiency, complementation group 1 |
| chr5_+_110988095 | 0.98 |
ENSMUST00000198373.2
|
Chek2
|
checkpoint kinase 2 |
| chrX_+_100317629 | 0.98 |
ENSMUST00000117706.8
|
Med12
|
mediator complex subunit 12 |
| chr6_+_21986445 | 0.97 |
ENSMUST00000115382.8
|
Cped1
|
cadherin-like and PC-esterase domain containing 1 |
| chr14_+_24540777 | 0.96 |
ENSMUST00000169826.3
ENSMUST00000225023.2 ENSMUST00000223999.2 |
Rps24
|
ribosomal protein S24 |
| chr3_-_98496123 | 0.95 |
ENSMUST00000178221.4
|
Gm10681
|
predicted gene 10681 |
| chr3_+_137849189 | 0.95 |
ENSMUST00000040321.13
|
Trmt10a
|
tRNA methyltransferase 10A |
| chr14_-_73286504 | 0.94 |
ENSMUST00000044664.12
|
Cysltr2
|
cysteinyl leukotriene receptor 2 |
| chr5_+_110987839 | 0.94 |
ENSMUST00000200172.2
ENSMUST00000066160.3 |
Chek2
|
checkpoint kinase 2 |
| chr1_+_40478787 | 0.93 |
ENSMUST00000097772.10
|
Il1rl1
|
interleukin 1 receptor-like 1 |
| chr7_-_43906802 | 0.92 |
ENSMUST00000107945.8
ENSMUST00000118216.8 |
Acp4
|
acid phosphatase 4 |
| chr14_+_53666165 | 0.92 |
ENSMUST00000180549.3
|
Trav6-3
|
T cell receptor alpha variable 6-3 |
| chr14_-_73286535 | 0.90 |
ENSMUST00000169168.3
|
Cysltr2
|
cysteinyl leukotriene receptor 2 |
| chr18_-_3280999 | 0.89 |
ENSMUST00000049942.13
|
Crem
|
cAMP responsive element modulator |
| chr1_-_66984178 | 0.88 |
ENSMUST00000027151.12
|
Myl1
|
myosin, light polypeptide 1 |
| chr6_-_67931692 | 0.87 |
ENSMUST00000103313.3
|
Igkv9-123
|
immunoglobulin kappa variable 9-123 |
| chr6_+_68495964 | 0.87 |
ENSMUST00000199510.5
ENSMUST00000103325.3 |
Igkv14-100
|
immunoglobulin kappa chain variable 14-100 |
| chr2_-_140012447 | 0.87 |
ENSMUST00000046030.8
|
Esf1
|
ESF1 nucleolar pre-rRNA processing protein homolog |
| chr11_-_115078653 | 0.87 |
ENSMUST00000103041.8
|
Nat9
|
N-acetyltransferase 9 (GCN5-related, putative) |
| chrX_+_115358631 | 0.87 |
ENSMUST00000101269.2
|
Cpxcr1
|
CPX chromosome region, candidate 1 |
| chr14_+_52962756 | 0.87 |
ENSMUST00000181483.3
|
Trav6d-3
|
T cell receptor alpha variable 6D-3 |
| chr1_-_155910546 | 0.86 |
ENSMUST00000169241.8
|
Tor1aip1
|
torsin A interacting protein 1 |
| chr17_-_3746536 | 0.85 |
ENSMUST00000115800.2
|
Nox3
|
NADPH oxidase 3 |
| chr9_+_35470752 | 0.85 |
ENSMUST00000034615.10
ENSMUST00000121246.2 |
Pus3
|
pseudouridine synthase 3 |
| chr17_-_34846323 | 0.84 |
ENSMUST00000168709.3
ENSMUST00000064953.15 ENSMUST00000170345.8 ENSMUST00000171121.9 ENSMUST00000168391.9 ENSMUST00000169067.9 |
Gm20460
Ppt2
|
predicted gene 20460 palmitoyl-protein thioesterase 2 |
| chr3_-_98471301 | 0.84 |
ENSMUST00000058728.10
|
Gm10681
|
predicted gene 10681 |
| chrX_+_125947591 | 0.84 |
ENSMUST00000096332.2
|
Gm382
|
predicted gene 382 |
| chr6_-_120334302 | 0.84 |
ENSMUST00000163827.8
|
Ccdc77
|
coiled-coil domain containing 77 |
| chr12_-_83534482 | 0.84 |
ENSMUST00000177959.8
ENSMUST00000178756.8 |
Dpf3
|
D4, zinc and double PHD fingers, family 3 |
| chr6_-_142268667 | 0.83 |
ENSMUST00000128446.2
|
Slco1a5
|
solute carrier organic anion transporter family, member 1a5 |
| chr18_-_3281752 | 0.82 |
ENSMUST00000140332.8
ENSMUST00000147138.8 |
Crem
|
cAMP responsive element modulator |
| chr9_-_119151428 | 0.82 |
ENSMUST00000040853.11
|
Oxsr1
|
oxidative-stress responsive 1 |
| chr3_-_57202301 | 0.82 |
ENSMUST00000171384.8
|
Tm4sf1
|
transmembrane 4 superfamily member 1 |
| chr2_-_73284262 | 0.81 |
ENSMUST00000102679.8
|
Wipf1
|
WAS/WASL interacting protein family, member 1 |
| chr13_-_38220899 | 0.81 |
ENSMUST00000110233.8
ENSMUST00000074969.11 ENSMUST00000131066.2 |
Cage1
|
cancer antigen 1 |
| chrX_+_143239577 | 0.80 |
ENSMUST00000155206.8
|
Trpc5os
|
transient receptor potential cation channel, subfamily C, member 5, opposite strand |
| chr11_+_67061908 | 0.79 |
ENSMUST00000018641.8
|
Myh2
|
myosin, heavy polypeptide 2, skeletal muscle, adult |
| chr11_-_69791756 | 0.79 |
ENSMUST00000018714.13
ENSMUST00000128046.2 |
2810408A11Rik
|
RIKEN cDNA 2810408A11 gene |
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.9 | 5.8 | GO:0001869 | regulation of complement activation, lectin pathway(GO:0001868) negative regulation of complement activation, lectin pathway(GO:0001869) |
| 1.2 | 8.5 | GO:0002430 | complement receptor mediated signaling pathway(GO:0002430) |
| 1.1 | 3.2 | GO:0046005 | positive regulation of circadian sleep/wake cycle, REM sleep(GO:0046005) |
| 1.0 | 3.0 | GO:0070563 | negative regulation of vitamin D receptor signaling pathway(GO:0070563) |
| 0.8 | 3.9 | GO:0007290 | spermatid nucleus elongation(GO:0007290) |
| 0.7 | 4.1 | GO:0031133 | regulation of axon diameter(GO:0031133) |
| 0.6 | 4.1 | GO:0071698 | olfactory placode formation(GO:0030910) olfactory placode development(GO:0071698) olfactory placode morphogenesis(GO:0071699) |
| 0.5 | 5.3 | GO:0032074 | negative regulation of nuclease activity(GO:0032074) |
| 0.5 | 1.9 | GO:0090004 | positive regulation of Golgi to plasma membrane protein transport(GO:0042998) positive regulation of establishment of protein localization to plasma membrane(GO:0090004) |
| 0.5 | 1.9 | GO:0051325 | interphase(GO:0051325) mitotic interphase(GO:0051329) |
| 0.4 | 3.9 | GO:2000301 | negative regulation of synaptic vesicle exocytosis(GO:2000301) |
| 0.4 | 1.7 | GO:2000449 | regulation of CD8-positive, alpha-beta T cell extravasation(GO:2000449) |
| 0.4 | 1.6 | GO:1902490 | regulation of sperm capacitation(GO:1902490) |
| 0.4 | 2.4 | GO:0046121 | deoxyribonucleoside catabolic process(GO:0046121) |
| 0.4 | 1.9 | GO:0006481 | C-terminal protein methylation(GO:0006481) |
| 0.4 | 1.1 | GO:2001034 | positive regulation of double-strand break repair via nonhomologous end joining(GO:2001034) |
| 0.4 | 1.1 | GO:0002014 | vasoconstriction of artery involved in ischemic response to lowering of systemic arterial blood pressure(GO:0002014) |
| 0.4 | 2.1 | GO:0070458 | detoxification of nitrogen compound(GO:0051410) cellular detoxification of nitrogen compound(GO:0070458) |
| 0.4 | 1.4 | GO:0045869 | negative regulation of single stranded viral RNA replication via double stranded DNA intermediate(GO:0045869) |
| 0.3 | 1.3 | GO:0006014 | D-ribose metabolic process(GO:0006014) |
| 0.3 | 3.3 | GO:0060267 | positive regulation of respiratory burst(GO:0060267) |
| 0.3 | 2.3 | GO:0001878 | response to yeast(GO:0001878) |
| 0.3 | 1.0 | GO:0006295 | pyrimidine dimer repair by nucleotide-excision repair(GO:0000720) nucleotide-excision repair, DNA incision, 3'-to lesion(GO:0006295) nucleotide-excision repair, DNA incision, 5'-to lesion(GO:0006296) positive regulation of t-circle formation(GO:1904431) |
| 0.3 | 2.6 | GO:0097428 | protein maturation by iron-sulfur cluster transfer(GO:0097428) |
| 0.3 | 2.4 | GO:0048539 | bone marrow development(GO:0048539) |
| 0.3 | 0.9 | GO:0009629 | response to gravity(GO:0009629) |
| 0.3 | 0.8 | GO:2000686 | regulation of rubidium ion transmembrane transporter activity(GO:2000686) |
| 0.3 | 1.0 | GO:0072355 | histone H3-T3 phosphorylation(GO:0072355) |
| 0.2 | 0.7 | GO:0060197 | epidermal cell division(GO:0010481) regulation of epidermal cell division(GO:0010482) cloacal septation(GO:0060197) |
| 0.2 | 0.7 | GO:0051542 | elastin biosynthetic process(GO:0051542) regulation of cytochrome-c oxidase activity(GO:1904959) |
| 0.2 | 2.0 | GO:0071763 | nuclear membrane organization(GO:0071763) |
| 0.2 | 0.5 | GO:0046949 | fatty-acyl-CoA biosynthetic process(GO:0046949) malonyl-CoA metabolic process(GO:2001293) |
| 0.2 | 1.8 | GO:0031339 | negative regulation of vesicle fusion(GO:0031339) |
| 0.2 | 1.1 | GO:0090235 | regulation of metaphase plate congression(GO:0090235) |
| 0.2 | 1.4 | GO:0006032 | chitin metabolic process(GO:0006030) chitin catabolic process(GO:0006032) |
| 0.1 | 0.6 | GO:0090472 | dibasic protein processing(GO:0090472) |
| 0.1 | 1.6 | GO:1900194 | negative regulation of oocyte maturation(GO:1900194) |
| 0.1 | 1.4 | GO:0000056 | ribosomal small subunit export from nucleus(GO:0000056) |
| 0.1 | 0.4 | GO:0071726 | response to diacyl bacterial lipopeptide(GO:0071724) cellular response to diacyl bacterial lipopeptide(GO:0071726) |
| 0.1 | 3.0 | GO:0070986 | left/right axis specification(GO:0070986) |
| 0.1 | 0.8 | GO:1990481 | mRNA pseudouridine synthesis(GO:1990481) |
| 0.1 | 1.0 | GO:0090245 | axis elongation involved in somitogenesis(GO:0090245) |
| 0.1 | 0.4 | GO:0010845 | positive regulation of reciprocal meiotic recombination(GO:0010845) |
| 0.1 | 3.5 | GO:1902236 | negative regulation of endoplasmic reticulum stress-induced intrinsic apoptotic signaling pathway(GO:1902236) |
| 0.1 | 0.4 | GO:1905168 | positive regulation of double-strand break repair via homologous recombination(GO:1905168) |
| 0.1 | 1.6 | GO:0000244 | spliceosomal tri-snRNP complex assembly(GO:0000244) |
| 0.1 | 1.2 | GO:2001214 | positive regulation of vasculogenesis(GO:2001214) |
| 0.1 | 1.5 | GO:0044539 | long-chain fatty acid import(GO:0044539) |
| 0.1 | 1.3 | GO:0071073 | positive regulation of phospholipid biosynthetic process(GO:0071073) |
| 0.1 | 2.6 | GO:0033539 | fatty acid beta-oxidation using acyl-CoA dehydrogenase(GO:0033539) |
| 0.1 | 0.3 | GO:0006788 | heme oxidation(GO:0006788) |
| 0.1 | 1.0 | GO:0032277 | negative regulation of gonadotropin secretion(GO:0032277) |
| 0.1 | 2.4 | GO:0006883 | cellular sodium ion homeostasis(GO:0006883) |
| 0.1 | 1.1 | GO:0015705 | iodide transport(GO:0015705) |
| 0.1 | 1.9 | GO:2000480 | negative regulation of cAMP-dependent protein kinase activity(GO:2000480) |
| 0.1 | 0.9 | GO:0070535 | histone H2A K63-linked ubiquitination(GO:0070535) |
| 0.1 | 0.6 | GO:0030263 | apoptotic chromosome condensation(GO:0030263) |
| 0.1 | 0.6 | GO:0071630 | nucleus-associated proteasomal ubiquitin-dependent protein catabolic process(GO:0071630) |
| 0.1 | 3.2 | GO:0016339 | calcium-dependent cell-cell adhesion via plasma membrane cell adhesion molecules(GO:0016339) |
| 0.1 | 1.3 | GO:0007213 | G-protein coupled acetylcholine receptor signaling pathway(GO:0007213) |
| 0.1 | 3.2 | GO:0009072 | aromatic amino acid family metabolic process(GO:0009072) |
| 0.1 | 1.0 | GO:0019471 | 4-hydroxyproline metabolic process(GO:0019471) |
| 0.1 | 2.3 | GO:0030728 | ovulation(GO:0030728) |
| 0.1 | 0.7 | GO:2000857 | positive regulation of mineralocorticoid secretion(GO:2000857) positive regulation of aldosterone secretion(GO:2000860) |
| 0.1 | 0.4 | GO:0090070 | positive regulation of ribosome biogenesis(GO:0090070) positive regulation of rRNA processing(GO:2000234) |
| 0.1 | 0.3 | GO:0001732 | formation of cytoplasmic translation initiation complex(GO:0001732) |
| 0.1 | 2.5 | GO:0010960 | magnesium ion homeostasis(GO:0010960) |
| 0.1 | 3.3 | GO:0006471 | protein ADP-ribosylation(GO:0006471) |
| 0.1 | 0.4 | GO:0002826 | negative regulation of T-helper 1 type immune response(GO:0002826) |
| 0.1 | 2.7 | GO:0000462 | maturation of SSU-rRNA from tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000462) |
| 0.1 | 1.6 | GO:1904707 | positive regulation of vascular smooth muscle cell proliferation(GO:1904707) |
| 0.1 | 0.6 | GO:0001867 | complement activation, lectin pathway(GO:0001867) |
| 0.1 | 0.3 | GO:0060082 | response to carbon monoxide(GO:0034465) eye blink reflex(GO:0060082) |
| 0.1 | 2.7 | GO:0010972 | negative regulation of G2/M transition of mitotic cell cycle(GO:0010972) |
| 0.1 | 4.4 | GO:0051865 | protein autoubiquitination(GO:0051865) |
| 0.1 | 1.8 | GO:0035313 | wound healing, spreading of epidermal cells(GO:0035313) |
| 0.1 | 1.0 | GO:0046085 | adenosine metabolic process(GO:0046085) |
| 0.0 | 0.7 | GO:0051026 | chiasma assembly(GO:0051026) |
| 0.0 | 5.6 | GO:0051225 | spindle assembly(GO:0051225) |
| 0.0 | 11.1 | GO:0002377 | immunoglobulin production(GO:0002377) |
| 0.0 | 2.0 | GO:0071158 | positive regulation of cell cycle arrest(GO:0071158) |
| 0.0 | 0.8 | GO:0071688 | striated muscle myosin thick filament assembly(GO:0071688) |
| 0.0 | 0.8 | GO:0001778 | plasma membrane repair(GO:0001778) |
| 0.0 | 0.3 | GO:0035735 | intraciliary transport involved in cilium morphogenesis(GO:0035735) |
| 0.0 | 0.7 | GO:0044336 | canonical Wnt signaling pathway involved in negative regulation of apoptotic process(GO:0044336) |
| 0.0 | 3.3 | GO:0006687 | glycosphingolipid metabolic process(GO:0006687) |
| 0.0 | 1.0 | GO:0042761 | very long-chain fatty acid biosynthetic process(GO:0042761) |
| 0.0 | 0.2 | GO:0046813 | receptor-mediated virion attachment to host cell(GO:0046813) |
| 0.0 | 0.7 | GO:0048387 | negative regulation of retinoic acid receptor signaling pathway(GO:0048387) |
| 0.0 | 2.2 | GO:0043666 | regulation of phosphoprotein phosphatase activity(GO:0043666) |
| 0.0 | 0.5 | GO:0006577 | amino-acid betaine metabolic process(GO:0006577) |
| 0.0 | 0.4 | GO:0006228 | UTP biosynthetic process(GO:0006228) |
| 0.0 | 0.5 | GO:0033617 | mitochondrial respiratory chain complex IV assembly(GO:0033617) |
| 0.0 | 0.8 | GO:0046827 | positive regulation of protein export from nucleus(GO:0046827) |
| 0.0 | 0.1 | GO:0070125 | mitochondrial translational elongation(GO:0070125) |
| 0.0 | 0.8 | GO:0015721 | bile acid and bile salt transport(GO:0015721) |
| 0.0 | 0.7 | GO:0045898 | regulation of RNA polymerase II transcriptional preinitiation complex assembly(GO:0045898) |
| 0.0 | 2.3 | GO:0007098 | centrosome cycle(GO:0007098) |
| 0.0 | 1.4 | GO:0042255 | ribosome assembly(GO:0042255) |
| 0.0 | 1.8 | GO:0007218 | neuropeptide signaling pathway(GO:0007218) |
| 0.0 | 1.0 | GO:0043551 | regulation of phosphatidylinositol 3-kinase activity(GO:0043551) |
| 0.0 | 1.1 | GO:0051438 | regulation of ubiquitin-protein transferase activity(GO:0051438) |
| 0.0 | 0.4 | GO:0016540 | protein autoprocessing(GO:0016540) |
| 0.0 | 1.4 | GO:0034113 | heterotypic cell-cell adhesion(GO:0034113) |
| 0.0 | 0.3 | GO:0015747 | urate transport(GO:0015747) |
| 0.0 | 0.4 | GO:0045741 | positive regulation of epidermal growth factor-activated receptor activity(GO:0045741) |
| 0.0 | 0.3 | GO:0018206 | peptidyl-methionine modification(GO:0018206) |
| 0.0 | 0.1 | GO:0061143 | alveolar primary septum development(GO:0061143) |
| 0.0 | 0.6 | GO:0042438 | melanin biosynthetic process(GO:0042438) |
| 0.0 | 3.9 | GO:0000209 | protein polyubiquitination(GO:0000209) |
| 0.0 | 5.4 | GO:0042113 | B cell activation(GO:0042113) |
| 0.0 | 1.2 | GO:0000381 | regulation of alternative mRNA splicing, via spliceosome(GO:0000381) |
| 0.0 | 3.4 | GO:0030308 | negative regulation of cell growth(GO:0030308) |
| 0.0 | 0.7 | GO:0055090 | acylglycerol homeostasis(GO:0055090) triglyceride homeostasis(GO:0070328) |
| 0.0 | 0.5 | GO:0010043 | response to zinc ion(GO:0010043) |
| 0.0 | 0.5 | GO:0061014 | positive regulation of mRNA catabolic process(GO:0061014) |
| 0.0 | 0.3 | GO:0000380 | alternative mRNA splicing, via spliceosome(GO:0000380) |
| 0.0 | 0.7 | GO:0055008 | cardiac muscle tissue morphogenesis(GO:0055008) |
| 0.0 | 0.5 | GO:0007520 | myoblast fusion(GO:0007520) |
| 0.0 | 0.5 | GO:0001580 | detection of chemical stimulus involved in sensory perception of bitter taste(GO:0001580) |
| 0.0 | 0.4 | GO:0007602 | phototransduction(GO:0007602) |
| 0.0 | 1.0 | GO:0051289 | protein homotetramerization(GO:0051289) |
| 0.0 | 2.6 | GO:0031032 | actomyosin structure organization(GO:0031032) |
| 0.0 | 1.8 | GO:0007605 | sensory perception of sound(GO:0007605) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.6 | 1.9 | GO:1990630 | IRE1-RACK1-PP2A complex(GO:1990630) |
| 0.4 | 2.1 | GO:0000839 | Hrd1p ubiquitin ligase ERAD-L complex(GO:0000839) |
| 0.4 | 1.4 | GO:0043159 | acrosomal matrix(GO:0043159) |
| 0.3 | 1.0 | GO:0000110 | nucleotide-excision repair factor 1 complex(GO:0000110) |
| 0.3 | 2.8 | GO:0005579 | membrane attack complex(GO:0005579) |
| 0.3 | 3.9 | GO:0000788 | nuclear nucleosome(GO:0000788) |
| 0.2 | 1.2 | GO:0005944 | phosphatidylinositol 3-kinase complex, class IB(GO:0005944) |
| 0.2 | 1.7 | GO:0097342 | ripoptosome(GO:0097342) |
| 0.2 | 1.2 | GO:0031466 | Cul5-RING ubiquitin ligase complex(GO:0031466) |
| 0.2 | 1.5 | GO:0070652 | HAUS complex(GO:0070652) |
| 0.2 | 2.3 | GO:0000164 | protein phosphatase type 1 complex(GO:0000164) |
| 0.1 | 0.7 | GO:0005713 | recombination nodule(GO:0005713) |
| 0.1 | 1.4 | GO:0005642 | annulate lamellae(GO:0005642) |
| 0.1 | 0.8 | GO:0005826 | actomyosin contractile ring(GO:0005826) |
| 0.1 | 0.3 | GO:0043614 | multi-eIF complex(GO:0043614) |
| 0.1 | 1.1 | GO:0001940 | male pronucleus(GO:0001940) |
| 0.1 | 1.1 | GO:0031462 | Cul2-RING ubiquitin ligase complex(GO:0031462) |
| 0.1 | 0.6 | GO:0005827 | polar microtubule(GO:0005827) |
| 0.1 | 0.9 | GO:0016272 | prefoldin complex(GO:0016272) |
| 0.1 | 0.6 | GO:0001674 | female germ cell nucleus(GO:0001674) |
| 0.1 | 1.0 | GO:0071203 | WASH complex(GO:0071203) |
| 0.1 | 0.6 | GO:0061574 | ASAP complex(GO:0061574) |
| 0.1 | 0.5 | GO:0005744 | mitochondrial inner membrane presequence translocase complex(GO:0005744) |
| 0.1 | 1.9 | GO:0034706 | sodium channel complex(GO:0034706) |
| 0.1 | 0.4 | GO:0046696 | lipopolysaccharide receptor complex(GO:0046696) |
| 0.0 | 3.1 | GO:0022627 | cytosolic small ribosomal subunit(GO:0022627) |
| 0.0 | 1.3 | GO:0030285 | integral component of synaptic vesicle membrane(GO:0030285) intrinsic component of synaptic vesicle membrane(GO:0098563) |
| 0.0 | 1.7 | GO:0016460 | myosin II complex(GO:0016460) |
| 0.0 | 1.5 | GO:0030057 | desmosome(GO:0030057) |
| 0.0 | 1.4 | GO:0034451 | centriolar satellite(GO:0034451) |
| 0.0 | 7.9 | GO:0090575 | RNA polymerase II transcription factor complex(GO:0090575) |
| 0.0 | 1.3 | GO:0030140 | trans-Golgi network transport vesicle(GO:0030140) |
| 0.0 | 0.4 | GO:0030688 | preribosome, small subunit precursor(GO:0030688) |
| 0.0 | 0.5 | GO:0000177 | cytoplasmic exosome (RNase complex)(GO:0000177) |
| 0.0 | 3.8 | GO:0042571 | immunoglobulin complex, circulating(GO:0042571) |
| 0.0 | 3.3 | GO:0005811 | lipid particle(GO:0005811) |
| 0.0 | 0.8 | GO:0071565 | nBAF complex(GO:0071565) |
| 0.0 | 2.0 | GO:0022625 | cytosolic large ribosomal subunit(GO:0022625) |
| 0.0 | 1.7 | GO:0005834 | heterotrimeric G-protein complex(GO:0005834) |
| 0.0 | 1.0 | GO:0016592 | mediator complex(GO:0016592) |
| 0.0 | 3.0 | GO:0005882 | intermediate filament(GO:0005882) |
| 0.0 | 3.4 | GO:0072562 | blood microparticle(GO:0072562) |
| 0.0 | 0.3 | GO:0048787 | presynaptic active zone membrane(GO:0048787) |
| 0.0 | 0.9 | GO:0035861 | site of double-strand break(GO:0035861) |
| 0.0 | 1.3 | GO:0016459 | myosin complex(GO:0016459) |
| 0.0 | 3.3 | GO:0001669 | acrosomal vesicle(GO:0001669) |
| 0.0 | 7.5 | GO:0009897 | external side of plasma membrane(GO:0009897) |
| 0.0 | 1.9 | GO:0016605 | PML body(GO:0016605) |
| 0.0 | 1.5 | GO:0031526 | brush border membrane(GO:0031526) |
| 0.0 | 4.5 | GO:0031965 | nuclear membrane(GO:0031965) |
| 0.0 | 1.5 | GO:0005791 | rough endoplasmic reticulum(GO:0005791) |
| 0.0 | 1.5 | GO:0032587 | ruffle membrane(GO:0032587) |
| 0.0 | 1.6 | GO:0000932 | cytoplasmic mRNA processing body(GO:0000932) |
| 0.0 | 0.2 | GO:0043240 | Fanconi anaemia nuclear complex(GO:0043240) |
| 0.0 | 0.7 | GO:0031941 | filamentous actin(GO:0031941) |
| 0.0 | 2.0 | GO:0005923 | bicellular tight junction(GO:0005923) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.6 | 7.9 | GO:0004982 | N-formyl peptide receptor activity(GO:0004982) |
| 1.1 | 3.2 | GO:0001716 | L-amino-acid oxidase activity(GO:0001716) |
| 0.8 | 3.3 | GO:0019862 | IgA binding(GO:0019862) |
| 0.5 | 1.4 | GO:0052658 | inositol-1,4,5-trisphosphate 5-phosphatase activity(GO:0052658) |
| 0.5 | 1.4 | GO:0098640 | integrin binding involved in cell-matrix adhesion(GO:0098640) |
| 0.4 | 1.6 | GO:1904408 | dihydronicotinamide riboside quinone reductase activity(GO:0001512) melatonin binding(GO:1904408) |
| 0.4 | 1.9 | GO:0003880 | protein C-terminal carboxyl O-methyltransferase activity(GO:0003880) |
| 0.4 | 1.8 | GO:0004974 | leukotriene receptor activity(GO:0004974) |
| 0.4 | 2.2 | GO:0008241 | peptidyl-dipeptidase activity(GO:0008241) |
| 0.4 | 2.5 | GO:0009019 | tRNA (guanine-N1-)-methyltransferase activity(GO:0009019) |
| 0.3 | 1.0 | GO:0008192 | RNA guanylyltransferase activity(GO:0008192) |
| 0.3 | 1.0 | GO:0035175 | histone kinase activity (H3-S10 specific)(GO:0035175) |
| 0.3 | 1.5 | GO:0043758 | acetate-CoA ligase (ADP-forming) activity(GO:0043758) |
| 0.3 | 0.6 | GO:0004875 | complement receptor activity(GO:0004875) |
| 0.3 | 3.6 | GO:0003956 | NAD(P)+-protein-arginine ADP-ribosyltransferase activity(GO:0003956) |
| 0.2 | 1.7 | GO:0004704 | NF-kappaB-inducing kinase activity(GO:0004704) |
| 0.2 | 1.9 | GO:0030292 | protein tyrosine kinase inhibitor activity(GO:0030292) |
| 0.2 | 2.6 | GO:0003997 | acyl-CoA oxidase activity(GO:0003997) |
| 0.2 | 5.8 | GO:0001848 | complement binding(GO:0001848) |
| 0.2 | 3.5 | GO:1904264 | ubiquitin protein ligase activity involved in ERAD pathway(GO:1904264) |
| 0.2 | 1.3 | GO:0018685 | alkane 1-monooxygenase activity(GO:0018685) leukotriene-B4 20-monooxygenase activity(GO:0050051) |
| 0.2 | 1.6 | GO:0031685 | adenosine receptor binding(GO:0031685) |
| 0.2 | 4.3 | GO:0023026 | MHC class II protein complex binding(GO:0023026) |
| 0.2 | 0.7 | GO:0043682 | copper-exporting ATPase activity(GO:0004008) copper-transporting ATPase activity(GO:0043682) |
| 0.2 | 1.0 | GO:1990599 | 3' overhang single-stranded DNA endodeoxyribonuclease activity(GO:1990599) |
| 0.2 | 0.6 | GO:0031208 | POZ domain binding(GO:0031208) |
| 0.2 | 1.9 | GO:0015280 | ligand-gated sodium channel activity(GO:0015280) |
| 0.2 | 1.4 | GO:0004568 | chitinase activity(GO:0004568) |
| 0.1 | 0.7 | GO:0005275 | amine transmembrane transporter activity(GO:0005275) |
| 0.1 | 1.2 | GO:0031849 | olfactory receptor binding(GO:0031849) |
| 0.1 | 0.4 | GO:0002113 | interleukin-33 binding(GO:0002113) |
| 0.1 | 1.2 | GO:0046935 | 1-phosphatidylinositol-3-kinase regulator activity(GO:0046935) |
| 0.1 | 2.4 | GO:0016832 | aldehyde-lyase activity(GO:0016832) |
| 0.1 | 1.4 | GO:0005049 | nuclear export signal receptor activity(GO:0005049) |
| 0.1 | 1.1 | GO:0008474 | palmitoyl-(protein) hydrolase activity(GO:0008474) palmitoyl hydrolase activity(GO:0098599) |
| 0.1 | 0.3 | GO:0051916 | granulocyte colony-stimulating factor binding(GO:0051916) |
| 0.1 | 4.6 | GO:0001671 | ATPase activator activity(GO:0001671) |
| 0.1 | 0.8 | GO:0015349 | thyroid hormone transmembrane transporter activity(GO:0015349) |
| 0.1 | 0.5 | GO:0003989 | acetyl-CoA carboxylase activity(GO:0003989) |
| 0.1 | 0.3 | GO:0004392 | heme oxygenase (decyclizing) activity(GO:0004392) |
| 0.1 | 4.1 | GO:0000983 | transcription factor activity, RNA polymerase II core promoter sequence-specific(GO:0000983) |
| 0.1 | 2.3 | GO:0005537 | mannose binding(GO:0005537) |
| 0.1 | 1.9 | GO:0004862 | cAMP-dependent protein kinase inhibitor activity(GO:0004862) |
| 0.1 | 2.6 | GO:0019706 | protein-cysteine S-palmitoyltransferase activity(GO:0019706) protein-cysteine S-acyltransferase activity(GO:0019707) |
| 0.1 | 0.5 | GO:0070180 | large ribosomal subunit rRNA binding(GO:0070180) |
| 0.1 | 2.1 | GO:0043295 | glutathione binding(GO:0043295) oligopeptide binding(GO:1900750) |
| 0.1 | 5.4 | GO:0001105 | RNA polymerase II transcription coactivator activity(GO:0001105) |
| 0.1 | 3.2 | GO:0071855 | neuropeptide receptor binding(GO:0071855) |
| 0.1 | 0.5 | GO:0008172 | S-methyltransferase activity(GO:0008172) |
| 0.1 | 0.4 | GO:0071723 | lipopeptide binding(GO:0071723) |
| 0.1 | 0.9 | GO:0016175 | superoxide-generating NADPH oxidase activity(GO:0016175) |
| 0.1 | 13.5 | GO:0001078 | transcriptional repressor activity, RNA polymerase II core promoter proximal region sequence-specific binding(GO:0001078) |
| 0.1 | 3.1 | GO:0031369 | translation initiation factor binding(GO:0031369) |
| 0.1 | 0.7 | GO:0030983 | mismatched DNA binding(GO:0030983) |
| 0.1 | 0.9 | GO:0003993 | acid phosphatase activity(GO:0003993) |
| 0.0 | 1.0 | GO:0097493 | structural molecule activity conferring elasticity(GO:0097493) |
| 0.0 | 1.5 | GO:0045295 | gamma-catenin binding(GO:0045295) |
| 0.0 | 1.0 | GO:0008253 | 5'-nucleotidase activity(GO:0008253) |
| 0.0 | 0.8 | GO:0009982 | pseudouridine synthase activity(GO:0009982) |
| 0.0 | 2.3 | GO:0004864 | protein phosphatase inhibitor activity(GO:0004864) |
| 0.0 | 1.3 | GO:0031683 | G-protein beta/gamma-subunit complex binding(GO:0031683) |
| 0.0 | 2.6 | GO:0019003 | GDP binding(GO:0019003) |
| 0.0 | 0.7 | GO:0097371 | MDM2/MDM4 family protein binding(GO:0097371) |
| 0.0 | 0.6 | GO:0048406 | nerve growth factor binding(GO:0048406) |
| 0.0 | 0.3 | GO:0016861 | intramolecular oxidoreductase activity, interconverting aldoses and ketoses(GO:0016861) |
| 0.0 | 0.1 | GO:0070287 | ferritin receptor activity(GO:0070287) |
| 0.0 | 3.7 | GO:0004222 | metalloendopeptidase activity(GO:0004222) |
| 0.0 | 0.3 | GO:0008420 | CTD phosphatase activity(GO:0008420) |
| 0.0 | 3.4 | GO:0005179 | hormone activity(GO:0005179) |
| 0.0 | 1.6 | GO:0005070 | SH3/SH2 adaptor activity(GO:0005070) |
| 0.0 | 0.2 | GO:0001601 | peptide YY receptor activity(GO:0001601) pancreatic polypeptide receptor activity(GO:0001602) |
| 0.0 | 2.6 | GO:0043621 | protein self-association(GO:0043621) |
| 0.0 | 1.2 | GO:0031624 | ubiquitin conjugating enzyme binding(GO:0031624) |
| 0.0 | 0.6 | GO:0005031 | tumor necrosis factor-activated receptor activity(GO:0005031) death receptor activity(GO:0005035) |
| 0.0 | 0.1 | GO:0004525 | ribonuclease III activity(GO:0004525) double-stranded RNA-specific ribonuclease activity(GO:0032296) |
| 0.0 | 0.5 | GO:0042800 | histone methyltransferase activity (H3-K4 specific)(GO:0042800) |
| 0.0 | 1.4 | GO:0019843 | rRNA binding(GO:0019843) |
| 0.0 | 4.2 | GO:0004252 | serine-type endopeptidase activity(GO:0004252) |
| 0.0 | 0.8 | GO:1990841 | promoter-specific chromatin binding(GO:1990841) |
| 0.0 | 0.3 | GO:1901702 | urate transmembrane transporter activity(GO:0015143) salt transmembrane transporter activity(GO:1901702) |
| 0.0 | 0.4 | GO:0051920 | peroxiredoxin activity(GO:0051920) |
| 0.0 | 1.0 | GO:0016627 | oxidoreductase activity, acting on the CH-CH group of donors(GO:0016627) |
| 0.0 | 1.9 | GO:0008080 | N-acetyltransferase activity(GO:0008080) |
| 0.0 | 0.8 | GO:0051539 | 4 iron, 4 sulfur cluster binding(GO:0051539) |
| 0.0 | 0.7 | GO:0051393 | alpha-actinin binding(GO:0051393) |
| 0.0 | 0.6 | GO:0015036 | disulfide oxidoreductase activity(GO:0015036) |
| 0.0 | 2.1 | GO:0004867 | serine-type endopeptidase inhibitor activity(GO:0004867) |
| 0.0 | 0.5 | GO:0005385 | zinc ion transmembrane transporter activity(GO:0005385) |
| 0.0 | 0.6 | GO:0051721 | protein phosphatase 2A binding(GO:0051721) |
| 0.0 | 1.8 | GO:0005200 | structural constituent of cytoskeleton(GO:0005200) |
| 0.0 | 1.1 | GO:0051082 | unfolded protein binding(GO:0051082) |
| 0.0 | 1.0 | GO:0005518 | collagen binding(GO:0005518) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 7.4 | PID TCR CALCIUM PATHWAY | Calcium signaling in the CD4+ TCR pathway |
| 0.1 | 7.9 | PID UPA UPAR PATHWAY | Urokinase-type plasminogen activator (uPA) and uPAR-mediated signaling |
| 0.1 | 3.9 | PID NECTIN PATHWAY | Nectin adhesion pathway |
| 0.1 | 1.4 | PID INTEGRIN5 PATHWAY | Beta5 beta6 beta7 and beta8 integrin cell surface interactions |
| 0.1 | 2.8 | PID ATM PATHWAY | ATM pathway |
| 0.1 | 1.4 | PID RANBP2 PATHWAY | Sumoylation by RanBP2 regulates transcriptional repression |
| 0.1 | 5.4 | PID IL12 2PATHWAY | IL12-mediated signaling events |
| 0.1 | 1.3 | PID S1P S1P4 PATHWAY | S1P4 pathway |
| 0.1 | 1.6 | PID PDGFRA PATHWAY | PDGFR-alpha signaling pathway |
| 0.0 | 3.7 | PID KIT PATHWAY | Signaling events mediated by Stem cell factor receptor (c-Kit) |
| 0.0 | 1.2 | PID EPHA FWDPATHWAY | EPHA forward signaling |
| 0.0 | 0.6 | PID WNT CANONICAL PATHWAY | Canonical Wnt signaling pathway |
| 0.0 | 0.7 | PID EPHRINB REV PATHWAY | Ephrin B reverse signaling |
| 0.0 | 1.6 | PID CDC42 REG PATHWAY | Regulation of CDC42 activity |
| 0.0 | 0.8 | PID AMB2 NEUTROPHILS PATHWAY | amb2 Integrin signaling |
| 0.0 | 1.4 | PID ENDOTHELIN PATHWAY | Endothelins |
| 0.0 | 0.8 | PID FCER1 PATHWAY | Fc-epsilon receptor I signaling in mast cells |
| 0.0 | 4.7 | NABA ECM REGULATORS | Genes encoding enzymes and their regulators involved in the remodeling of the extracellular matrix |
| 0.0 | 0.4 | PID TOLL ENDOGENOUS PATHWAY | Endogenous TLR signaling |
| 0.0 | 1.0 | PID TCR PATHWAY | TCR signaling in naïve CD4+ T cells |
| 0.0 | 0.3 | PID SMAD2 3PATHWAY | Regulation of cytoplasmic and nuclear SMAD2/3 signaling |
| 0.0 | 3.0 | NABA ECM GLYCOPROTEINS | Genes encoding structural ECM glycoproteins |
| 0.0 | 1.0 | PID BETA CATENIN NUC PATHWAY | Regulation of nuclear beta catenin signaling and target gene transcription |
| 0.0 | 0.4 | PID PTP1B PATHWAY | Signaling events mediated by PTP1B |
| 0.0 | 0.3 | PID IL2 PI3K PATHWAY | IL2 signaling events mediated by PI3K |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 4.9 | REACTOME INTRINSIC PATHWAY | Genes involved in Intrinsic Pathway |
| 0.2 | 2.3 | REACTOME GLYCOPROTEIN HORMONES | Genes involved in Glycoprotein hormones |
| 0.2 | 3.9 | REACTOME P130CAS LINKAGE TO MAPK SIGNALING FOR INTEGRINS | Genes involved in p130Cas linkage to MAPK signaling for integrins |
| 0.1 | 1.9 | REACTOME G2 M DNA DAMAGE CHECKPOINT | Genes involved in G2/M DNA damage checkpoint |
| 0.1 | 0.4 | REACTOME OLFACTORY SIGNALING PATHWAY | Genes involved in Olfactory Signaling Pathway |
| 0.1 | 1.8 | REACTOME EICOSANOID LIGAND BINDING RECEPTORS | Genes involved in Eicosanoid ligand-binding receptors |
| 0.1 | 2.3 | REACTOME POST CHAPERONIN TUBULIN FOLDING PATHWAY | Genes involved in Post-chaperonin tubulin folding pathway |
| 0.1 | 1.4 | REACTOME CYCLIN A B1 ASSOCIATED EVENTS DURING G2 M TRANSITION | Genes involved in Cyclin A/B1 associated events during G2/M transition |
| 0.1 | 2.8 | REACTOME RNA POL III TRANSCRIPTION INITIATION FROM TYPE 3 PROMOTER | Genes involved in RNA Polymerase III Transcription Initiation From Type 3 Promoter |
| 0.1 | 0.4 | REACTOME IKK COMPLEX RECRUITMENT MEDIATED BY RIP1 | Genes involved in IKK complex recruitment mediated by RIP1 |
| 0.1 | 2.4 | REACTOME COMPLEMENT CASCADE | Genes involved in Complement cascade |
| 0.1 | 2.0 | REACTOME ACTIVATED AMPK STIMULATES FATTY ACID OXIDATION IN MUSCLE | Genes involved in Activated AMPK stimulates fatty-acid oxidation in muscle |
| 0.1 | 1.0 | REACTOME ADVANCED GLYCOSYLATION ENDPRODUCT RECEPTOR SIGNALING | Genes involved in Advanced glycosylation endproduct receptor signaling |
| 0.1 | 1.7 | REACTOME RIP MEDIATED NFKB ACTIVATION VIA DAI | Genes involved in RIP-mediated NFkB activation via DAI |
| 0.1 | 0.6 | REACTOME GAMMA CARBOXYLATION TRANSPORT AND AMINO TERMINAL CLEAVAGE OF PROTEINS | Genes involved in Gamma-carboxylation, transport, and amino-terminal cleavage of proteins |
| 0.1 | 1.0 | REACTOME ABACAVIR TRANSPORT AND METABOLISM | Genes involved in Abacavir transport and metabolism |
| 0.1 | 1.3 | REACTOME ADENYLATE CYCLASE INHIBITORY PATHWAY | Genes involved in Adenylate cyclase inhibitory pathway |
| 0.1 | 11.1 | REACTOME PEPTIDE LIGAND BINDING RECEPTORS | Genes involved in Peptide ligand-binding receptors |
| 0.1 | 3.1 | REACTOME ACTIVATION OF CHAPERONE GENES BY XBP1S | Genes involved in Activation of Chaperone Genes by XBP1(S) |
| 0.1 | 3.4 | REACTOME FORMATION OF THE TERNARY COMPLEX AND SUBSEQUENTLY THE 43S COMPLEX | Genes involved in Formation of the ternary complex, and subsequently, the 43S complex |
| 0.0 | 1.0 | REACTOME FORMATION OF INCISION COMPLEX IN GG NER | Genes involved in Formation of incision complex in GG-NER |
| 0.0 | 2.6 | REACTOME SYNTHESIS OF PIPS AT THE PLASMA MEMBRANE | Genes involved in Synthesis of PIPs at the plasma membrane |
| 0.0 | 1.0 | REACTOME SIGNAL REGULATORY PROTEIN SIRP FAMILY INTERACTIONS | Genes involved in Signal regulatory protein (SIRP) family interactions |
| 0.0 | 0.9 | REACTOME TERMINATION OF O GLYCAN BIOSYNTHESIS | Genes involved in Termination of O-glycan biosynthesis |
| 0.0 | 0.6 | REACTOME MITOCHONDRIAL FATTY ACID BETA OXIDATION | Genes involved in Mitochondrial Fatty Acid Beta-Oxidation |
| 0.0 | 0.7 | REACTOME NA CL DEPENDENT NEUROTRANSMITTER TRANSPORTERS | Genes involved in Na+/Cl- dependent neurotransmitter transporters |
| 0.0 | 1.2 | REACTOME NEP NS2 INTERACTS WITH THE CELLULAR EXPORT MACHINERY | Genes involved in NEP/NS2 Interacts with the Cellular Export Machinery |
| 0.0 | 1.1 | REACTOME MEIOTIC RECOMBINATION | Genes involved in Meiotic Recombination |
| 0.0 | 2.0 | REACTOME PEPTIDE CHAIN ELONGATION | Genes involved in Peptide chain elongation |
| 0.0 | 0.5 | REACTOME ZINC TRANSPORTERS | Genes involved in Zinc transporters |
| 0.0 | 0.7 | REACTOME DOWNREGULATION OF SMAD2 3 SMAD4 TRANSCRIPTIONAL ACTIVITY | Genes involved in Downregulation of SMAD2/3:SMAD4 transcriptional activity |
| 0.0 | 2.4 | REACTOME G ALPHA Q SIGNALLING EVENTS | Genes involved in G alpha (q) signalling events |
| 0.0 | 0.3 | REACTOME METABOLISM OF PORPHYRINS | Genes involved in Metabolism of porphyrins |
| 0.0 | 0.7 | REACTOME MITOCHONDRIAL PROTEIN IMPORT | Genes involved in Mitochondrial Protein Import |
| 0.0 | 0.2 | REACTOME FANCONI ANEMIA PATHWAY | Genes involved in Fanconi Anemia pathway |
| 0.0 | 0.7 | REACTOME ION TRANSPORT BY P TYPE ATPASES | Genes involved in Ion transport by P-type ATPases |