Project
PRJNA375882: Comprehensive Mouse Transcriptomic BodyMap
Navigation
Downloads
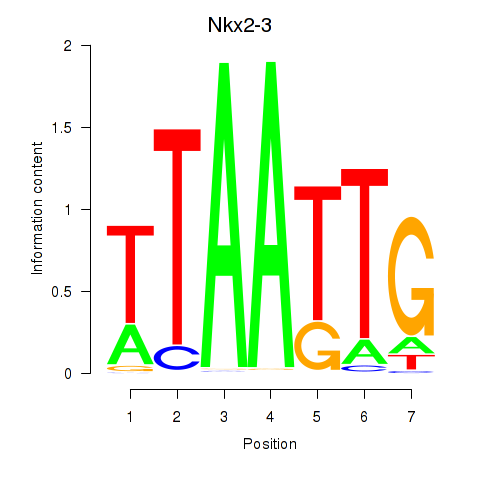
Results for Nkx2-3
Z-value: 1.16
Transcription factors associated with Nkx2-3
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Nkx2-3
|
ENSMUSG00000044220.14 | Nkx2-3 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Nkx2-3 | mm39_v1_chr19_+_43600738_43600764 | -0.44 | 1.2e-04 | Click! |
Activity profile of Nkx2-3 motif
Sorted Z-values of Nkx2-3 motif
Network of associatons between targets according to the STRING database.
First level regulatory network of Nkx2-3
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr3_+_67799510 | 9.14 |
ENSMUST00000063263.5
ENSMUST00000182006.4 |
Iqcj
Iqschfp
|
IQ motif containing J Iqcj and Schip1 fusion protein |
| chr3_+_55369149 | 6.88 |
ENSMUST00000199585.5
ENSMUST00000070418.9 |
Dclk1
|
doublecortin-like kinase 1 |
| chr15_-_96929086 | 6.65 |
ENSMUST00000230086.2
|
Slc38a4
|
solute carrier family 38, member 4 |
| chr2_-_32976378 | 6.44 |
ENSMUST00000049618.9
|
Garnl3
|
GTPase activating RANGAP domain-like 3 |
| chr14_+_27598021 | 5.87 |
ENSMUST00000211684.2
ENSMUST00000210924.2 |
Erc2
|
ELKS/RAB6-interacting/CAST family member 2 |
| chr13_+_42834039 | 5.34 |
ENSMUST00000128646.8
|
Phactr1
|
phosphatase and actin regulator 1 |
| chr11_-_97944239 | 5.20 |
ENSMUST00000017544.9
|
Stac2
|
SH3 and cysteine rich domain 2 |
| chr2_+_22512195 | 5.00 |
ENSMUST00000028123.4
|
Gad2
|
glutamic acid decarboxylase 2 |
| chr16_+_6887689 | 4.95 |
ENSMUST00000229741.2
|
Rbfox1
|
RNA binding protein, fox-1 homolog (C. elegans) 1 |
| chr11_-_119438569 | 4.92 |
ENSMUST00000026670.5
|
Nptx1
|
neuronal pentraxin 1 |
| chr6_-_138398376 | 4.91 |
ENSMUST00000163065.8
|
Lmo3
|
LIM domain only 3 |
| chr1_+_66360865 | 4.85 |
ENSMUST00000114013.8
|
Map2
|
microtubule-associated protein 2 |
| chr12_-_11486544 | 4.78 |
ENSMUST00000072299.7
|
Vsnl1
|
visinin-like 1 |
| chr8_-_65146079 | 4.66 |
ENSMUST00000048967.9
|
Cpe
|
carboxypeptidase E |
| chr11_-_79394904 | 4.64 |
ENSMUST00000164465.3
|
Omg
|
oligodendrocyte myelin glycoprotein |
| chr12_+_38833501 | 4.58 |
ENSMUST00000159334.8
|
Etv1
|
ets variant 1 |
| chr16_-_23339329 | 4.51 |
ENSMUST00000230040.2
ENSMUST00000229619.2 |
Masp1
|
mannan-binding lectin serine peptidase 1 |
| chr13_+_83652352 | 4.49 |
ENSMUST00000198916.5
ENSMUST00000200123.5 ENSMUST00000005722.14 ENSMUST00000163888.8 |
Mef2c
|
myocyte enhancer factor 2C |
| chr2_-_164013033 | 4.45 |
ENSMUST00000045196.4
|
Kcns1
|
K+ voltage-gated channel, subfamily S, 1 |
| chr13_-_78344492 | 4.43 |
ENSMUST00000125176.3
|
Nr2f1
|
nuclear receptor subfamily 2, group F, member 1 |
| chr3_+_55369384 | 4.42 |
ENSMUST00000200352.2
|
Dclk1
|
doublecortin-like kinase 1 |
| chr13_+_43019718 | 4.39 |
ENSMUST00000131942.2
|
Phactr1
|
phosphatase and actin regulator 1 |
| chr11_+_16207705 | 4.25 |
ENSMUST00000109645.9
ENSMUST00000109647.3 |
Vstm2a
|
V-set and transmembrane domain containing 2A |
| chr19_+_38252984 | 4.18 |
ENSMUST00000198518.5
ENSMUST00000199812.5 |
Lgi1
|
leucine-rich repeat LGI family, member 1 |
| chr18_+_37888770 | 4.11 |
ENSMUST00000061279.10
|
Pcdhga11
|
protocadherin gamma subfamily A, 11 |
| chr3_-_126792056 | 3.97 |
ENSMUST00000044443.15
|
Ank2
|
ankyrin 2, brain |
| chr3_+_55369288 | 3.92 |
ENSMUST00000198412.5
ENSMUST00000199169.5 ENSMUST00000199702.5 ENSMUST00000198437.5 |
Dclk1
|
doublecortin-like kinase 1 |
| chr12_+_38833454 | 3.90 |
ENSMUST00000161980.8
ENSMUST00000160701.8 |
Etv1
|
ets variant 1 |
| chr2_-_113844100 | 3.90 |
ENSMUST00000090275.5
|
Gjd2
|
gap junction protein, delta 2 |
| chr12_+_38830081 | 3.89 |
ENSMUST00000095767.11
|
Etv1
|
ets variant 1 |
| chr8_+_55407872 | 3.87 |
ENSMUST00000033915.9
|
Gpm6a
|
glycoprotein m6a |
| chr5_-_87572060 | 3.85 |
ENSMUST00000072818.6
|
Ugt2b38
|
UDP glucuronosyltransferase 2 family, polypeptide B38 |
| chr2_+_135899847 | 3.85 |
ENSMUST00000057503.7
|
Lamp5
|
lysosomal-associated membrane protein family, member 5 |
| chr19_+_40078132 | 3.81 |
ENSMUST00000068094.13
ENSMUST00000080171.3 |
Cyp2c50
|
cytochrome P450, family 2, subfamily c, polypeptide 50 |
| chr13_-_113800172 | 3.81 |
ENSMUST00000054650.5
|
Hspb3
|
heat shock protein 3 |
| chr10_+_29087658 | 3.80 |
ENSMUST00000213489.2
|
9330159F19Rik
|
RIKEN cDNA 9330159F19 gene |
| chr16_+_20511991 | 3.73 |
ENSMUST00000149543.9
ENSMUST00000232207.2 ENSMUST00000118919.9 |
Fam131a
|
family with sequence similarity 131, member A |
| chr17_-_90395771 | 3.54 |
ENSMUST00000197268.5
ENSMUST00000173917.8 |
Nrxn1
|
neurexin I |
| chr19_+_38253105 | 3.51 |
ENSMUST00000196090.2
|
Lgi1
|
leucine-rich repeat LGI family, member 1 |
| chr18_-_31580436 | 3.50 |
ENSMUST00000025110.5
|
Syt4
|
synaptotagmin IV |
| chr1_+_66361252 | 3.50 |
ENSMUST00000123647.8
|
Map2
|
microtubule-associated protein 2 |
| chr19_+_38253077 | 3.50 |
ENSMUST00000198045.5
|
Lgi1
|
leucine-rich repeat LGI family, member 1 |
| chr13_+_93444514 | 3.49 |
ENSMUST00000079086.8
|
Homer1
|
homer scaffolding protein 1 |
| chr10_+_29019645 | 3.48 |
ENSMUST00000092629.4
|
Soga3
|
SOGA family member 3 |
| chr15_+_21111428 | 3.47 |
ENSMUST00000075132.8
|
Cdh12
|
cadherin 12 |
| chr2_-_10135449 | 3.47 |
ENSMUST00000042290.14
|
Itih2
|
inter-alpha trypsin inhibitor, heavy chain 2 |
| chr15_+_92495007 | 3.46 |
ENSMUST00000035399.10
|
Pdzrn4
|
PDZ domain containing RING finger 4 |
| chr10_+_29087602 | 3.43 |
ENSMUST00000092627.6
|
9330159F19Rik
|
RIKEN cDNA 9330159F19 gene |
| chr7_+_119217004 | 3.43 |
ENSMUST00000047929.13
ENSMUST00000135683.3 |
Acsm1
|
acyl-CoA synthetase medium-chain family member 1 |
| chr2_+_67578556 | 3.42 |
ENSMUST00000180887.2
|
B3galt1
|
UDP-Gal:betaGlcNAc beta 1,3-galactosyltransferase, polypeptide 1 |
| chr11_+_29668563 | 3.41 |
ENSMUST00000060992.6
|
Rtn4
|
reticulon 4 |
| chr8_+_24159669 | 3.41 |
ENSMUST00000042352.11
ENSMUST00000207301.2 |
Zmat4
|
zinc finger, matrin type 4 |
| chr12_+_52746158 | 3.38 |
ENSMUST00000095737.5
|
Akap6
|
A kinase (PRKA) anchor protein 6 |
| chr12_+_38830812 | 3.35 |
ENSMUST00000160856.8
|
Etv1
|
ets variant 1 |
| chr2_-_23939401 | 3.28 |
ENSMUST00000051416.12
|
Hnmt
|
histamine N-methyltransferase |
| chr12_+_38830283 | 3.21 |
ENSMUST00000162563.8
ENSMUST00000161164.8 ENSMUST00000160996.8 |
Etv1
|
ets variant 1 |
| chr15_+_91949032 | 3.17 |
ENSMUST00000169825.8
|
Cntn1
|
contactin 1 |
| chr13_+_83652150 | 3.15 |
ENSMUST00000198199.5
|
Mef2c
|
myocyte enhancer factor 2C |
| chr18_-_43820759 | 3.15 |
ENSMUST00000082254.8
|
Jakmip2
|
janus kinase and microtubule interacting protein 2 |
| chr13_+_83652280 | 3.12 |
ENSMUST00000199450.5
|
Mef2c
|
myocyte enhancer factor 2C |
| chr4_-_91264670 | 3.07 |
ENSMUST00000107109.9
ENSMUST00000107111.9 ENSMUST00000107120.8 |
Elavl2
|
ELAV like RNA binding protein 1 |
| chr15_+_100768776 | 3.05 |
ENSMUST00000108909.9
|
Scn8a
|
sodium channel, voltage-gated, type VIII, alpha |
| chr2_-_64806106 | 3.02 |
ENSMUST00000156765.2
|
Grb14
|
growth factor receptor bound protein 14 |
| chr13_-_32960379 | 3.02 |
ENSMUST00000230119.2
|
Mylk4
|
myosin light chain kinase family, member 4 |
| chr5_+_81169049 | 3.01 |
ENSMUST00000117253.8
ENSMUST00000120128.8 |
Adgrl3
|
adhesion G protein-coupled receptor L3 |
| chr15_-_37734579 | 3.00 |
ENSMUST00000145909.9
ENSMUST00000153775.9 |
Gm49397
Ncald
|
predicted gene, 49397 neurocalcin delta |
| chr4_-_41640321 | 3.00 |
ENSMUST00000127306.2
|
Enho
|
energy homeostasis associated |
| chr15_-_43733389 | 2.97 |
ENSMUST00000067469.6
|
Tmem74
|
transmembrane protein 74 |
| chr4_+_127066667 | 2.95 |
ENSMUST00000106094.9
|
Dlgap3
|
DLG associated protein 3 |
| chr8_+_46080840 | 2.93 |
ENSMUST00000135336.9
|
Sorbs2
|
sorbin and SH3 domain containing 2 |
| chr9_+_32027335 | 2.87 |
ENSMUST00000174641.8
|
Arhgap32
|
Rho GTPase activating protein 32 |
| chr12_-_46863726 | 2.85 |
ENSMUST00000219330.2
|
Nova1
|
NOVA alternative splicing regulator 1 |
| chr10_+_112151245 | 2.84 |
ENSMUST00000218445.2
|
Kcnc2
|
potassium voltage gated channel, Shaw-related subfamily, member 2 |
| chr15_-_77813123 | 2.78 |
ENSMUST00000109748.9
ENSMUST00000109747.9 ENSMUST00000100486.6 ENSMUST00000005487.12 |
Txn2
|
thioredoxin 2 |
| chr6_-_138399896 | 2.75 |
ENSMUST00000161450.8
ENSMUST00000163024.8 ENSMUST00000162185.8 |
Lmo3
|
LIM domain only 3 |
| chr4_+_99812912 | 2.74 |
ENSMUST00000102783.5
|
Pgm1
|
phosphoglucomutase 1 |
| chr6_-_138404076 | 2.69 |
ENSMUST00000203435.3
|
Lmo3
|
LIM domain only 3 |
| chr1_+_81055201 | 2.67 |
ENSMUST00000123285.2
|
Nyap2
|
neuronal tyrosine-phophorylated phosphoinositide 3-kinase adaptor 2 |
| chr1_-_14380418 | 2.66 |
ENSMUST00000027066.13
ENSMUST00000168081.9 |
Eya1
|
EYA transcriptional coactivator and phosphatase 1 |
| chr3_+_137770813 | 2.66 |
ENSMUST00000163080.3
|
1110002E22Rik
|
RIKEN cDNA 1110002E22 gene |
| chr12_+_38831093 | 2.66 |
ENSMUST00000161513.9
|
Etv1
|
ets variant 1 |
| chr12_-_72455708 | 2.65 |
ENSMUST00000078505.14
|
Rtn1
|
reticulon 1 |
| chr18_-_39000056 | 2.64 |
ENSMUST00000236630.2
ENSMUST00000237356.2 |
Fgf1
|
fibroblast growth factor 1 |
| chr3_-_26187883 | 2.62 |
ENSMUST00000108308.10
ENSMUST00000075054.10 |
Nlgn1
|
neuroligin 1 |
| chr16_-_34083549 | 2.62 |
ENSMUST00000114949.8
ENSMUST00000114954.8 |
Kalrn
|
kalirin, RhoGEF kinase |
| chr12_+_103524690 | 2.58 |
ENSMUST00000187155.7
|
Ppp4r4
|
protein phosphatase 4, regulatory subunit 4 |
| chr4_-_73869071 | 2.57 |
ENSMUST00000095023.2
ENSMUST00000030101.4 |
2310002L09Rik
|
RIKEN cDNA 2310002L09 gene |
| chr18_+_37138256 | 2.55 |
ENSMUST00000115658.6
|
Pcdha11
|
protocadherin alpha 11 |
| chr4_-_91264727 | 2.54 |
ENSMUST00000107124.10
|
Elavl2
|
ELAV like RNA binding protein 1 |
| chr19_-_4927910 | 2.49 |
ENSMUST00000006626.5
|
Actn3
|
actinin alpha 3 |
| chr19_-_8382424 | 2.47 |
ENSMUST00000064507.12
ENSMUST00000120540.2 ENSMUST00000096269.11 |
Slc22a30
|
solute carrier family 22, member 30 |
| chr18_+_37433852 | 2.47 |
ENSMUST00000051754.2
|
Pcdhb3
|
protocadherin beta 3 |
| chr2_-_63014622 | 2.45 |
ENSMUST00000075052.10
ENSMUST00000112454.8 |
Kcnh7
|
potassium voltage-gated channel, subfamily H (eag-related), member 7 |
| chr13_+_83723255 | 2.45 |
ENSMUST00000199167.5
ENSMUST00000195904.5 |
Mef2c
|
myocyte enhancer factor 2C |
| chr13_+_83672654 | 2.40 |
ENSMUST00000199019.5
|
Mef2c
|
myocyte enhancer factor 2C |
| chr3_+_102981352 | 2.39 |
ENSMUST00000176440.2
|
Ampd1
|
adenosine monophosphate deaminase 1 |
| chr19_+_39980868 | 2.38 |
ENSMUST00000049178.3
|
Cyp2c37
|
cytochrome P450, family 2. subfamily c, polypeptide 37 |
| chr9_-_48747474 | 2.37 |
ENSMUST00000216150.2
|
Zbtb16
|
zinc finger and BTB domain containing 16 |
| chr3_+_68479578 | 2.36 |
ENSMUST00000170788.9
|
Schip1
|
schwannomin interacting protein 1 |
| chr1_-_193052533 | 2.36 |
ENSMUST00000169907.8
|
Camk1g
|
calcium/calmodulin-dependent protein kinase I gamma |
| chrX_+_156481906 | 2.36 |
ENSMUST00000136141.2
ENSMUST00000190091.7 |
Smpx
|
small muscle protein, X-linked |
| chr4_-_110143777 | 2.35 |
ENSMUST00000138972.8
|
Elavl4
|
ELAV like RNA binding protein 4 |
| chr15_-_13173736 | 2.35 |
ENSMUST00000036439.6
|
Cdh6
|
cadherin 6 |
| chr2_-_32314017 | 2.35 |
ENSMUST00000113307.9
|
Slc25a25
|
solute carrier family 25 (mitochondrial carrier, phosphate carrier), member 25 |
| chr19_-_39637489 | 2.34 |
ENSMUST00000067328.7
|
Cyp2c67
|
cytochrome P450, family 2, subfamily c, polypeptide 67 |
| chr3_+_137329791 | 2.34 |
ENSMUST00000165845.2
|
Ddit4l
|
DNA-damage-inducible transcript 4-like |
| chr15_+_100768806 | 2.32 |
ENSMUST00000201549.4
ENSMUST00000108908.6 |
Scn8a
|
sodium channel, voltage-gated, type VIII, alpha |
| chr17_+_3447465 | 2.31 |
ENSMUST00000072156.7
|
Tiam2
|
T cell lymphoma invasion and metastasis 2 |
| chr18_+_69726431 | 2.29 |
ENSMUST00000201091.4
ENSMUST00000201037.4 ENSMUST00000114977.5 |
Tcf4
|
transcription factor 4 |
| chr2_+_22959452 | 2.29 |
ENSMUST00000155602.4
|
Acbd5
|
acyl-Coenzyme A binding domain containing 5 |
| chr16_-_22258469 | 2.28 |
ENSMUST00000079601.13
|
Etv5
|
ets variant 5 |
| chr12_+_37930169 | 2.28 |
ENSMUST00000221176.2
|
Dgkb
|
diacylglycerol kinase, beta |
| chr3_+_62327089 | 2.25 |
ENSMUST00000161057.2
|
Arhgef26
|
Rho guanine nucleotide exchange factor (GEF) 26 |
| chr14_-_4506874 | 2.25 |
ENSMUST00000224934.2
|
Thrb
|
thyroid hormone receptor beta |
| chr1_-_162687254 | 2.25 |
ENSMUST00000131058.8
|
Fmo1
|
flavin containing monooxygenase 1 |
| chr10_+_86599836 | 2.23 |
ENSMUST00000218802.2
|
Gm49358
|
predicted gene, 49358 |
| chr13_+_83723743 | 2.23 |
ENSMUST00000198217.5
ENSMUST00000199210.5 |
Mef2c
|
myocyte enhancer factor 2C |
| chr1_-_14380327 | 2.23 |
ENSMUST00000080664.14
|
Eya1
|
EYA transcriptional coactivator and phosphatase 1 |
| chr3_-_141687987 | 2.22 |
ENSMUST00000029948.15
|
Bmpr1b
|
bone morphogenetic protein receptor, type 1B |
| chr10_-_127024641 | 2.21 |
ENSMUST00000218654.2
|
Arhgef25
|
Rho guanine nucleotide exchange factor (GEF) 25 |
| chr5_+_8943677 | 2.21 |
ENSMUST00000003717.13
|
Abcb4
|
ATP-binding cassette, sub-family B (MDR/TAP), member 4 |
| chr9_-_58108988 | 2.18 |
ENSMUST00000163200.3
ENSMUST00000165276.2 ENSMUST00000214647.2 |
Islr2
|
immunoglobulin superfamily containing leucine-rich repeat 2 |
| chr12_+_29584560 | 2.17 |
ENSMUST00000021009.10
|
Myt1l
|
myelin transcription factor 1-like |
| chr15_+_100768551 | 2.16 |
ENSMUST00000082209.13
|
Scn8a
|
sodium channel, voltage-gated, type VIII, alpha |
| chr16_+_8288627 | 2.15 |
ENSMUST00000046470.16
ENSMUST00000150790.2 ENSMUST00000142899.2 |
Mettl22
|
methyltransferase like 22 |
| chr16_-_23339548 | 2.14 |
ENSMUST00000089883.7
|
Masp1
|
mannan-binding lectin serine peptidase 1 |
| chr1_-_162687369 | 2.12 |
ENSMUST00000193078.6
|
Fmo1
|
flavin containing monooxygenase 1 |
| chr7_-_19432933 | 2.11 |
ENSMUST00000174355.8
ENSMUST00000172983.8 ENSMUST00000174710.2 ENSMUST00000003066.16 ENSMUST00000174064.9 |
Apoe
|
apolipoprotein E |
| chr9_-_99302205 | 2.10 |
ENSMUST00000123771.2
|
Mras
|
muscle and microspikes RAS |
| chr7_+_143838191 | 2.09 |
ENSMUST00000097929.4
|
Shank2
|
SH3 and multiple ankyrin repeat domains 2 |
| chr14_+_75693396 | 2.09 |
ENSMUST00000164848.3
|
Siah3
|
siah E3 ubiquitin protein ligase family member 3 |
| chrX_+_99019176 | 2.09 |
ENSMUST00000113781.8
ENSMUST00000113783.8 ENSMUST00000113779.8 ENSMUST00000113776.8 ENSMUST00000113775.8 ENSMUST00000113780.8 ENSMUST00000113778.8 ENSMUST00000113777.8 ENSMUST00000071453.3 |
Eda
|
ectodysplasin-A |
| chr18_-_44308126 | 2.08 |
ENSMUST00000066328.5
|
Spinkl
|
serine protease inhibitor, Kazal type-like |
| chr12_+_102094977 | 2.08 |
ENSMUST00000159329.8
|
Slc24a4
|
solute carrier family 24 (sodium/potassium/calcium exchanger), member 4 |
| chr3_-_110051253 | 2.07 |
ENSMUST00000133268.9
ENSMUST00000051253.4 |
Ntng1
|
netrin G1 |
| chr2_+_73102269 | 2.07 |
ENSMUST00000090813.6
|
Sp9
|
trans-acting transcription factor 9 |
| chr4_+_48663504 | 2.06 |
ENSMUST00000030033.5
|
Cavin4
|
caveolae associated 4 |
| chr1_-_163552693 | 2.03 |
ENSMUST00000159679.8
|
Mettl11b
|
methyltransferase like 11B |
| chr1_+_179938904 | 2.02 |
ENSMUST00000145181.2
|
Cdc42bpa
|
CDC42 binding protein kinase alpha |
| chr1_-_165955445 | 1.98 |
ENSMUST00000085992.4
ENSMUST00000192369.6 |
Dusp27
|
dual specificity phosphatase 27 (putative) |
| chr17_+_93506435 | 1.98 |
ENSMUST00000234646.2
ENSMUST00000234081.2 |
Adcyap1
|
adenylate cyclase activating polypeptide 1 |
| chr4_-_110149916 | 1.97 |
ENSMUST00000106601.8
|
Elavl4
|
ELAV like RNA binding protein 4 |
| chr1_-_193052568 | 1.96 |
ENSMUST00000016323.11
|
Camk1g
|
calcium/calmodulin-dependent protein kinase I gamma |
| chr7_+_143792455 | 1.96 |
ENSMUST00000239495.2
|
Shank2
|
SH3 and multiple ankyrin repeat domains 2 |
| chr7_-_48497771 | 1.94 |
ENSMUST00000032658.14
|
Csrp3
|
cysteine and glycine-rich protein 3 |
| chr2_+_69727599 | 1.93 |
ENSMUST00000131553.2
|
Ubr3
|
ubiquitin protein ligase E3 component n-recognin 3 |
| chr10_-_8632519 | 1.91 |
ENSMUST00000212869.2
|
Sash1
|
SAM and SH3 domain containing 1 |
| chrX_-_142716200 | 1.91 |
ENSMUST00000112851.8
ENSMUST00000112856.3 ENSMUST00000033642.10 |
Dcx
|
doublecortin |
| chr6_+_141575226 | 1.89 |
ENSMUST00000042812.9
|
Slco1b2
|
solute carrier organic anion transporter family, member 1b2 |
| chr17_-_30107544 | 1.89 |
ENSMUST00000171691.9
|
Mdga1
|
MAM domain containing glycosylphosphatidylinositol anchor 1 |
| chr13_-_36918424 | 1.89 |
ENSMUST00000037623.15
|
Nrn1
|
neuritin 1 |
| chr13_-_76091931 | 1.86 |
ENSMUST00000022078.12
ENSMUST00000109606.3 |
Rhobtb3
|
Rho-related BTB domain containing 3 |
| chr1_-_63215952 | 1.85 |
ENSMUST00000185412.7
ENSMUST00000027111.15 ENSMUST00000189664.2 |
Ndufs1
|
NADH:ubiquinone oxidoreductase core subunit S1 |
| chr10_+_69761784 | 1.84 |
ENSMUST00000181974.8
ENSMUST00000182795.8 ENSMUST00000182437.8 |
Ank3
|
ankyrin 3, epithelial |
| chr1_+_72323526 | 1.83 |
ENSMUST00000027380.12
ENSMUST00000141783.2 |
Tmem169
|
transmembrane protein 169 |
| chrM_+_10167 | 1.82 |
ENSMUST00000082414.1
|
mt-Nd4
|
mitochondrially encoded NADH dehydrogenase 4 |
| chr12_+_58258558 | 1.82 |
ENSMUST00000110671.3
ENSMUST00000044299.3 |
Sstr1
|
somatostatin receptor 1 |
| chr8_+_45960931 | 1.81 |
ENSMUST00000171337.10
ENSMUST00000067107.15 |
Sorbs2
|
sorbin and SH3 domain containing 2 |
| chr6_-_138403732 | 1.81 |
ENSMUST00000162932.2
|
Lmo3
|
LIM domain only 3 |
| chr18_+_37651393 | 1.80 |
ENSMUST00000097609.3
|
Pcdhb22
|
protocadherin beta 22 |
| chr9_+_77543776 | 1.80 |
ENSMUST00000057781.8
|
Klhl31
|
kelch-like 31 |
| chr5_+_89175815 | 1.80 |
ENSMUST00000130041.8
|
Slc4a4
|
solute carrier family 4 (anion exchanger), member 4 |
| chr1_-_63215812 | 1.79 |
ENSMUST00000185847.2
ENSMUST00000185732.7 ENSMUST00000188370.7 ENSMUST00000168099.9 |
Ndufs1
|
NADH:ubiquinone oxidoreductase core subunit S1 |
| chr11_+_67061908 | 1.79 |
ENSMUST00000018641.8
|
Myh2
|
myosin, heavy polypeptide 2, skeletal muscle, adult |
| chrX_+_92698469 | 1.76 |
ENSMUST00000113933.9
|
Pcyt1b
|
phosphate cytidylyltransferase 1, choline, beta isoform |
| chr15_+_102885467 | 1.75 |
ENSMUST00000001706.7
|
Hoxc9
|
homeobox C9 |
| chr10_-_25076008 | 1.73 |
ENSMUST00000100012.3
|
Akap7
|
A kinase (PRKA) anchor protein 7 |
| chr10_+_79650496 | 1.72 |
ENSMUST00000218857.2
ENSMUST00000220365.2 |
Palm
|
paralemmin |
| chr2_+_69727563 | 1.72 |
ENSMUST00000055758.16
ENSMUST00000112251.9 |
Ubr3
|
ubiquitin protein ligase E3 component n-recognin 3 |
| chr9_+_88462944 | 1.71 |
ENSMUST00000164661.4
ENSMUST00000215498.2 ENSMUST00000216686.2 |
Trim43a
|
tripartite motif-containing 43A |
| chr7_-_27252543 | 1.69 |
ENSMUST00000127240.8
ENSMUST00000117095.8 ENSMUST00000117611.8 |
Pld3
|
phospholipase D family, member 3 |
| chr17_+_56312672 | 1.69 |
ENSMUST00000133998.8
|
Mpnd
|
MPN domain containing |
| chr13_-_23806530 | 1.67 |
ENSMUST00000062045.4
|
H1f4
|
H1.4 linker histone, cluster member |
| chr12_-_80807454 | 1.66 |
ENSMUST00000073251.8
|
Ccdc177
|
coiled-coil domain containing 177 |
| chr6_-_101176147 | 1.64 |
ENSMUST00000239140.2
|
Pdzrn3
|
PDZ domain containing RING finger 3 |
| chr13_+_23991010 | 1.64 |
ENSMUST00000006786.11
ENSMUST00000099697.3 |
Slc17a2
|
solute carrier family 17 (sodium phosphate), member 2 |
| chr2_+_3115250 | 1.62 |
ENSMUST00000072955.12
|
Fam171a1
|
family with sequence similarity 171, member A1 |
| chr5_+_89175894 | 1.62 |
ENSMUST00000113216.9
ENSMUST00000134303.2 |
Slc4a4
|
solute carrier family 4 (anion exchanger), member 4 |
| chr6_+_112250719 | 1.61 |
ENSMUST00000032376.6
|
Lmcd1
|
LIM and cysteine-rich domains 1 |
| chr15_+_28203872 | 1.60 |
ENSMUST00000067048.8
|
Dnah5
|
dynein, axonemal, heavy chain 5 |
| chr3_+_68598757 | 1.60 |
ENSMUST00000107816.4
|
Il12a
|
interleukin 12a |
| chr10_-_88440869 | 1.58 |
ENSMUST00000119185.8
ENSMUST00000238199.2 |
Mybpc1
|
myosin binding protein C, slow-type |
| chr13_+_83672389 | 1.58 |
ENSMUST00000200394.5
|
Mef2c
|
myocyte enhancer factor 2C |
| chr7_-_47677345 | 1.57 |
ENSMUST00000094390.3
|
Mrgprx1
|
MAS-related GPR, member X1 |
| chr17_-_78725510 | 1.56 |
ENSMUST00000234029.2
ENSMUST00000234530.2 ENSMUST00000234052.2 ENSMUST00000070039.14 ENSMUST00000112487.3 |
Fez2
|
fasciculation and elongation protein zeta 2 (zygin II) |
| chr9_+_113641615 | 1.55 |
ENSMUST00000111838.10
ENSMUST00000166734.10 ENSMUST00000214522.2 ENSMUST00000163895.3 |
Clasp2
|
CLIP associating protein 2 |
| chr11_+_67061837 | 1.55 |
ENSMUST00000170159.8
|
Myh2
|
myosin, heavy polypeptide 2, skeletal muscle, adult |
| chr10_+_69816153 | 1.54 |
ENSMUST00000182207.8
|
Ank3
|
ankyrin 3, epithelial |
| chr19_+_13208692 | 1.54 |
ENSMUST00000207246.4
|
Olfr1463
|
olfactory receptor 1463 |
| chr5_+_57875309 | 1.52 |
ENSMUST00000191837.6
ENSMUST00000068110.10 |
Pcdh7
|
protocadherin 7 |
| chr13_-_23041731 | 1.51 |
ENSMUST00000228645.2
|
Vmn1r211
|
vomeronasal 1 receptor 211 |
| chr16_-_74208395 | 1.49 |
ENSMUST00000227347.2
|
Robo2
|
roundabout guidance receptor 2 |
| chr11_+_67090878 | 1.49 |
ENSMUST00000124516.8
ENSMUST00000018637.15 ENSMUST00000129018.8 |
Myh1
|
myosin, heavy polypeptide 1, skeletal muscle, adult |
| chr13_+_41013230 | 1.48 |
ENSMUST00000110191.10
|
Gcnt2
|
glucosaminyl (N-acetyl) transferase 2, I-branching enzyme |
| chr3_-_141875070 | 1.48 |
ENSMUST00000106230.2
|
Bmpr1b
|
bone morphogenetic protein receptor, type 1B |
| chr1_+_134637031 | 1.47 |
ENSMUST00000121990.2
|
Syt2
|
synaptotagmin II |
| chr10_-_20600797 | 1.47 |
ENSMUST00000020165.14
|
Pde7b
|
phosphodiesterase 7B |
| chr19_-_44541270 | 1.45 |
ENSMUST00000166808.2
|
Gm20538
|
predicted gene 20538 |
| chr1_+_179788675 | 1.44 |
ENSMUST00000076687.12
ENSMUST00000097450.10 ENSMUST00000212756.2 |
Cdc42bpa
|
CDC42 binding protein kinase alpha |
| chr7_+_27252658 | 1.43 |
ENSMUST00000067386.14
ENSMUST00000191126.7 ENSMUST00000187960.7 |
2310022A10Rik
|
RIKEN cDNA 2310022A10 gene |
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.6 | 19.4 | GO:0003185 | primary heart field specification(GO:0003138) sinoatrial valve development(GO:0003172) sinoatrial valve morphogenesis(GO:0003185) |
| 1.6 | 4.7 | GO:0030070 | insulin processing(GO:0030070) |
| 1.1 | 21.6 | GO:0048935 | peripheral nervous system neuron differentiation(GO:0048934) peripheral nervous system neuron development(GO:0048935) |
| 1.1 | 5.3 | GO:0070346 | positive regulation of fat cell proliferation(GO:0070346) |
| 1.0 | 3.0 | GO:1903537 | meiotic cell cycle process involved in oocyte maturation(GO:1903537) regulation of meiotic cell cycle process involved in oocyte maturation(GO:1903538) |
| 1.0 | 5.0 | GO:0006538 | glutamate catabolic process(GO:0006538) |
| 0.9 | 2.8 | GO:0038060 | nitric oxide-cGMP-mediated signaling pathway(GO:0038060) |
| 0.9 | 2.8 | GO:0050925 | negative regulation of negative chemotaxis(GO:0050925) |
| 0.9 | 3.7 | GO:0071596 | ubiquitin-dependent protein catabolic process via the N-end rule pathway(GO:0071596) |
| 0.9 | 2.6 | GO:0048789 | cytoskeletal matrix organization at active zone(GO:0048789) neurexin clustering involved in presynaptic membrane assembly(GO:0097115) retrograde trans-synaptic signaling by trans-synaptic protein complex(GO:0098942) |
| 0.9 | 5.1 | GO:1902261 | positive regulation of delayed rectifier potassium channel activity(GO:1902261) |
| 0.8 | 2.5 | GO:0014862 | regulation of the force of skeletal muscle contraction(GO:0014728) regulation of skeletal muscle contraction by chemo-mechanical energy conversion(GO:0014862) |
| 0.8 | 2.4 | GO:1903173 | phytol metabolic process(GO:0033306) fatty alcohol metabolic process(GO:1903173) |
| 0.7 | 3.7 | GO:0001550 | ovarian cumulus expansion(GO:0001550) fused antrum stage(GO:0048165) |
| 0.7 | 2.2 | GO:1903412 | response to bile acid(GO:1903412) cellular response to bile acid(GO:1903413) |
| 0.7 | 2.1 | GO:1903000 | regulation of lipid transport across blood brain barrier(GO:1903000) positive regulation of lipid transport across blood brain barrier(GO:1903002) |
| 0.7 | 4.9 | GO:0035865 | response to potassium ion(GO:0035864) cellular response to potassium ion(GO:0035865) |
| 0.7 | 1.3 | GO:0099543 | retrograde trans-synaptic signaling by soluble gas(GO:0098923) trans-synaptic signaling by soluble gas(GO:0099543) |
| 0.7 | 3.3 | GO:0001692 | histamine metabolic process(GO:0001692) |
| 0.6 | 1.9 | GO:1903920 | positive regulation of actin filament severing(GO:1903920) |
| 0.6 | 2.6 | GO:0080163 | regulation of protein serine/threonine phosphatase activity(GO:0080163) |
| 0.6 | 1.8 | GO:1900135 | positive regulation of renin secretion into blood stream(GO:1900135) |
| 0.6 | 3.5 | GO:0090126 | protein complex assembly involved in synapse maturation(GO:0090126) |
| 0.5 | 2.1 | GO:0061153 | trachea submucosa development(GO:0061152) trachea gland development(GO:0061153) |
| 0.5 | 2.1 | GO:1902490 | regulation of sperm capacitation(GO:1902490) |
| 0.5 | 4.6 | GO:0098728 | germ-line stem cell division(GO:0042078) male germ-line stem cell asymmetric division(GO:0048133) germline stem cell asymmetric division(GO:0098728) |
| 0.5 | 6.6 | GO:0001867 | complement activation, lectin pathway(GO:0001867) |
| 0.5 | 5.6 | GO:0072513 | positive regulation of secondary heart field cardioblast proliferation(GO:0072513) |
| 0.5 | 1.9 | GO:1902498 | regulation of protein autoubiquitination(GO:1902498) |
| 0.4 | 4.0 | GO:0036371 | protein localization to M-band(GO:0036309) protein localization to T-tubule(GO:0036371) |
| 0.4 | 2.1 | GO:1903215 | negative regulation of protein targeting to mitochondrion(GO:1903215) |
| 0.4 | 2.0 | GO:0071651 | positive regulation of chemokine (C-C motif) ligand 5 production(GO:0071651) |
| 0.4 | 3.5 | GO:2000301 | negative regulation of synaptic vesicle exocytosis(GO:2000301) |
| 0.4 | 1.2 | GO:2001034 | positive regulation of double-strand break repair via nonhomologous end joining(GO:2001034) |
| 0.4 | 1.1 | GO:0015881 | creatine transport(GO:0015881) |
| 0.4 | 2.7 | GO:0045196 | establishment or maintenance of neuroblast polarity(GO:0045196) establishment of neuroblast polarity(GO:0045200) |
| 0.4 | 3.4 | GO:0019375 | galactosylceramide biosynthetic process(GO:0006682) galactolipid biosynthetic process(GO:0019375) |
| 0.4 | 3.4 | GO:0071787 | endoplasmic reticulum tubular network assembly(GO:0071787) |
| 0.4 | 2.6 | GO:1901509 | regulation of endothelial tube morphogenesis(GO:1901509) |
| 0.4 | 2.2 | GO:0007621 | negative regulation of female receptivity(GO:0007621) |
| 0.4 | 1.1 | GO:0006530 | asparagine catabolic process(GO:0006530) |
| 0.3 | 1.3 | GO:2001226 | negative regulation of chloride transport(GO:2001226) |
| 0.3 | 1.0 | GO:2000536 | negative regulation of entry of bacterium into host cell(GO:2000536) |
| 0.3 | 2.6 | GO:0061366 | behavioral response to chemical pain(GO:0061366) behavioral response to formalin induced pain(GO:0061368) |
| 0.3 | 17.3 | GO:0021952 | central nervous system projection neuron axonogenesis(GO:0021952) |
| 0.3 | 3.8 | GO:0043651 | linoleic acid metabolic process(GO:0043651) |
| 0.3 | 3.0 | GO:0032264 | IMP salvage(GO:0032264) |
| 0.3 | 2.5 | GO:0045919 | positive regulation of cytolysis(GO:0045919) |
| 0.3 | 2.9 | GO:0030242 | pexophagy(GO:0030242) |
| 0.3 | 0.8 | GO:0021759 | globus pallidus development(GO:0021759) |
| 0.3 | 3.4 | GO:1900827 | positive regulation of cell communication by electrical coupling(GO:0010650) positive regulation of membrane depolarization during cardiac muscle cell action potential(GO:1900827) |
| 0.3 | 1.0 | GO:0046951 | ketone body biosynthetic process(GO:0046951) |
| 0.3 | 0.5 | GO:1900377 | negative regulation of melanin biosynthetic process(GO:0048022) negative regulation of secondary metabolite biosynthetic process(GO:1900377) |
| 0.2 | 0.5 | GO:0021762 | substantia nigra development(GO:0021762) |
| 0.2 | 0.9 | GO:0051344 | negative regulation of cyclic-nucleotide phosphodiesterase activity(GO:0051344) |
| 0.2 | 0.7 | GO:2001055 | positive regulation of mesenchymal cell apoptotic process(GO:2001055) |
| 0.2 | 0.7 | GO:0006597 | spermine biosynthetic process(GO:0006597) |
| 0.2 | 4.2 | GO:0070995 | NADPH oxidation(GO:0070995) |
| 0.2 | 0.4 | GO:0060166 | olfactory pit development(GO:0060166) |
| 0.2 | 1.5 | GO:0036438 | maintenance of lens transparency(GO:0036438) |
| 0.2 | 1.0 | GO:0099566 | regulation of postsynaptic cytosolic calcium ion concentration(GO:0099566) |
| 0.2 | 0.6 | GO:2000118 | regulation of sodium-dependent phosphate transport(GO:2000118) |
| 0.2 | 4.0 | GO:1902259 | regulation of delayed rectifier potassium channel activity(GO:1902259) |
| 0.2 | 1.2 | GO:1904936 | cerebral cortex GABAergic interneuron migration(GO:0021853) interneuron migration(GO:1904936) |
| 0.2 | 0.6 | GO:0044878 | mitotic cytokinesis checkpoint(GO:0044878) |
| 0.2 | 1.9 | GO:0051964 | negative regulation of synapse assembly(GO:0051964) |
| 0.2 | 4.6 | GO:0032291 | central nervous system myelination(GO:0022010) axon ensheathment in central nervous system(GO:0032291) |
| 0.2 | 3.3 | GO:0001778 | plasma membrane repair(GO:0001778) |
| 0.2 | 4.4 | GO:0021796 | cerebral cortex regionalization(GO:0021796) |
| 0.2 | 1.3 | GO:0090074 | negative regulation of protein homodimerization activity(GO:0090074) |
| 0.2 | 3.5 | GO:2001256 | regulation of store-operated calcium entry(GO:2001256) |
| 0.2 | 0.9 | GO:0051684 | maintenance of Golgi location(GO:0051684) |
| 0.2 | 8.8 | GO:0007026 | negative regulation of microtubule depolymerization(GO:0007026) |
| 0.2 | 4.5 | GO:0040023 | nuclear migration(GO:0007097) establishment of nucleus localization(GO:0040023) |
| 0.2 | 1.6 | GO:2000510 | positive regulation of dendritic cell chemotaxis(GO:2000510) |
| 0.2 | 2.5 | GO:0070886 | positive regulation of calcineurin-NFAT signaling cascade(GO:0070886) |
| 0.2 | 0.9 | GO:0038032 | termination of G-protein coupled receptor signaling pathway(GO:0038032) |
| 0.2 | 2.5 | GO:0015747 | urate transport(GO:0015747) |
| 0.2 | 2.8 | GO:0071688 | striated muscle myosin thick filament assembly(GO:0071688) |
| 0.2 | 4.7 | GO:0019373 | epoxygenase P450 pathway(GO:0019373) |
| 0.2 | 1.1 | GO:0060287 | determination of pancreatic left/right asymmetry(GO:0035469) epithelial cilium movement involved in determination of left/right asymmetry(GO:0060287) |
| 0.1 | 0.4 | GO:0021530 | spinal cord oligodendrocyte cell differentiation(GO:0021529) spinal cord oligodendrocyte cell fate specification(GO:0021530) |
| 0.1 | 0.9 | GO:0001757 | somite specification(GO:0001757) |
| 0.1 | 0.6 | GO:1904861 | excitatory synapse assembly(GO:1904861) |
| 0.1 | 2.8 | GO:0006662 | glycerol ether metabolic process(GO:0006662) ether metabolic process(GO:0018904) |
| 0.1 | 1.6 | GO:0036158 | outer dynein arm assembly(GO:0036158) |
| 0.1 | 5.8 | GO:1900746 | regulation of vascular endothelial growth factor signaling pathway(GO:1900746) regulation of cellular response to vascular endothelial growth factor stimulus(GO:1902547) |
| 0.1 | 0.9 | GO:0015676 | vanadium ion transport(GO:0015676) lead ion transport(GO:0015692) |
| 0.1 | 1.6 | GO:0033227 | dsRNA transport(GO:0033227) |
| 0.1 | 2.7 | GO:0055003 | cardiac myofibril assembly(GO:0055003) |
| 0.1 | 1.1 | GO:0032020 | ISG15-protein conjugation(GO:0032020) |
| 0.1 | 0.7 | GO:0060160 | negative regulation of dopamine receptor signaling pathway(GO:0060160) |
| 0.1 | 1.8 | GO:0097264 | self proteolysis(GO:0097264) |
| 0.1 | 0.5 | GO:0016999 | antibiotic metabolic process(GO:0016999) |
| 0.1 | 1.4 | GO:0045760 | positive regulation of action potential(GO:0045760) |
| 0.1 | 1.9 | GO:0006857 | oligopeptide transport(GO:0006857) |
| 0.1 | 8.8 | GO:0019228 | neuronal action potential(GO:0019228) |
| 0.1 | 0.5 | GO:1902683 | regulation of receptor localization to synapse(GO:1902683) |
| 0.1 | 0.9 | GO:0046013 | regulation of T cell homeostatic proliferation(GO:0046013) |
| 0.1 | 3.7 | GO:2001014 | regulation of skeletal muscle cell differentiation(GO:2001014) |
| 0.1 | 3.4 | GO:0015701 | bicarbonate transport(GO:0015701) |
| 0.1 | 0.7 | GO:1903336 | endosome localization(GO:0032439) negative regulation of vacuolar transport(GO:1903336) |
| 0.1 | 0.7 | GO:0050916 | protein transport into membrane raft(GO:0032596) sensory perception of sweet taste(GO:0050916) |
| 0.1 | 4.1 | GO:0071625 | vocalization behavior(GO:0071625) |
| 0.1 | 10.5 | GO:0045600 | positive regulation of fat cell differentiation(GO:0045600) |
| 0.1 | 1.6 | GO:1902902 | negative regulation of autophagosome assembly(GO:1902902) |
| 0.1 | 0.9 | GO:0031179 | peptide modification(GO:0031179) |
| 0.1 | 1.1 | GO:1902412 | regulation of mitotic cytokinesis(GO:1902412) |
| 0.1 | 0.8 | GO:0048743 | positive regulation of skeletal muscle fiber development(GO:0048743) |
| 0.1 | 0.9 | GO:1902965 | regulation of protein localization to early endosome(GO:1902965) positive regulation of protein localization to early endosome(GO:1902966) |
| 0.1 | 0.7 | GO:0014004 | microglia differentiation(GO:0014004) microglia development(GO:0014005) |
| 0.1 | 2.4 | GO:0001553 | luteinization(GO:0001553) |
| 0.1 | 2.1 | GO:0031987 | locomotion involved in locomotory behavior(GO:0031987) |
| 0.1 | 4.8 | GO:0035774 | positive regulation of insulin secretion involved in cellular response to glucose stimulus(GO:0035774) |
| 0.1 | 0.8 | GO:2000766 | negative regulation of cytoplasmic translation(GO:2000766) |
| 0.1 | 0.5 | GO:0032918 | polyamine acetylation(GO:0032917) spermidine acetylation(GO:0032918) |
| 0.1 | 1.4 | GO:0018298 | protein-chromophore linkage(GO:0018298) |
| 0.1 | 0.6 | GO:0033540 | fatty acid beta-oxidation using acyl-CoA oxidase(GO:0033540) |
| 0.1 | 1.8 | GO:0006120 | mitochondrial electron transport, NADH to ubiquinone(GO:0006120) |
| 0.1 | 0.5 | GO:0002188 | translation reinitiation(GO:0002188) |
| 0.1 | 1.3 | GO:0048739 | cardiac muscle fiber development(GO:0048739) |
| 0.1 | 2.3 | GO:0014823 | response to activity(GO:0014823) |
| 0.1 | 0.5 | GO:0003199 | endocardial cushion to mesenchymal transition involved in heart valve formation(GO:0003199) |
| 0.1 | 4.2 | GO:0042773 | ATP synthesis coupled electron transport(GO:0042773) |
| 0.1 | 2.2 | GO:0001886 | endothelial cell morphogenesis(GO:0001886) |
| 0.1 | 1.1 | GO:1901898 | negative regulation of relaxation of muscle(GO:1901078) negative regulation of relaxation of cardiac muscle(GO:1901898) |
| 0.1 | 1.6 | GO:0007635 | chemosensory behavior(GO:0007635) |
| 0.1 | 0.5 | GO:0015867 | ATP transport(GO:0015867) |
| 0.1 | 2.6 | GO:0015986 | energy coupled proton transport, down electrochemical gradient(GO:0015985) ATP synthesis coupled proton transport(GO:0015986) |
| 0.1 | 3.2 | GO:0007205 | protein kinase C-activating G-protein coupled receptor signaling pathway(GO:0007205) |
| 0.1 | 0.2 | GO:0090063 | positive regulation of microtubule nucleation(GO:0090063) |
| 0.1 | 1.8 | GO:0071392 | cellular response to estradiol stimulus(GO:0071392) |
| 0.1 | 3.5 | GO:0030212 | hyaluronan metabolic process(GO:0030212) |
| 0.1 | 4.2 | GO:0003333 | amino acid transmembrane transport(GO:0003333) |
| 0.1 | 0.4 | GO:0003310 | pancreatic A cell differentiation(GO:0003310) intestinal epithelial structure maintenance(GO:0060729) |
| 0.1 | 3.9 | GO:0051491 | positive regulation of filopodium assembly(GO:0051491) |
| 0.1 | 11.7 | GO:0050806 | positive regulation of synaptic transmission(GO:0050806) |
| 0.1 | 3.0 | GO:0045747 | positive regulation of Notch signaling pathway(GO:0045747) |
| 0.1 | 9.6 | GO:0030038 | contractile actin filament bundle assembly(GO:0030038) stress fiber assembly(GO:0043149) |
| 0.1 | 0.7 | GO:0006335 | DNA replication-dependent nucleosome assembly(GO:0006335) ribosomal protein import into nucleus(GO:0006610) DNA replication-dependent nucleosome organization(GO:0034723) |
| 0.1 | 1.8 | GO:1903861 | positive regulation of dendrite extension(GO:1903861) |
| 0.1 | 0.2 | GO:0072708 | response to sorbitol(GO:0072708) response to dithiothreitol(GO:0072720) |
| 0.1 | 0.5 | GO:0090520 | sphingolipid mediated signaling pathway(GO:0090520) |
| 0.1 | 1.3 | GO:0035563 | positive regulation of chromatin binding(GO:0035563) |
| 0.1 | 0.3 | GO:0070309 | lens fiber cell morphogenesis(GO:0070309) |
| 0.1 | 7.5 | GO:0019236 | response to pheromone(GO:0019236) |
| 0.1 | 2.1 | GO:0050911 | detection of chemical stimulus involved in sensory perception of smell(GO:0050911) |
| 0.1 | 2.9 | GO:0010765 | positive regulation of sodium ion transport(GO:0010765) |
| 0.1 | 0.4 | GO:0048852 | diencephalon morphogenesis(GO:0048852) |
| 0.1 | 5.5 | GO:1900006 | positive regulation of dendrite development(GO:1900006) |
| 0.0 | 0.7 | GO:0030828 | positive regulation of cGMP biosynthetic process(GO:0030828) |
| 0.0 | 0.2 | GO:0006421 | asparaginyl-tRNA aminoacylation(GO:0006421) |
| 0.0 | 0.8 | GO:0043508 | negative regulation of JUN kinase activity(GO:0043508) |
| 0.0 | 1.3 | GO:0035455 | response to interferon-alpha(GO:0035455) |
| 0.0 | 5.8 | GO:0007416 | synapse assembly(GO:0007416) |
| 0.0 | 2.2 | GO:0045773 | positive regulation of axon extension(GO:0045773) |
| 0.0 | 4.8 | GO:0007156 | homophilic cell adhesion via plasma membrane adhesion molecules(GO:0007156) |
| 0.0 | 0.2 | GO:2000054 | negative regulation of Wnt signaling pathway involved in dorsal/ventral axis specification(GO:2000054) |
| 0.0 | 0.3 | GO:0003406 | retinal pigment epithelium development(GO:0003406) |
| 0.0 | 1.9 | GO:0007528 | neuromuscular junction development(GO:0007528) |
| 0.0 | 5.7 | GO:0018107 | peptidyl-threonine phosphorylation(GO:0018107) |
| 0.0 | 1.7 | GO:0003301 | physiological muscle hypertrophy(GO:0003298) physiological cardiac muscle hypertrophy(GO:0003301) cell growth involved in cardiac muscle cell development(GO:0061049) |
| 0.0 | 0.6 | GO:0045899 | positive regulation of RNA polymerase II transcriptional preinitiation complex assembly(GO:0045899) |
| 0.0 | 0.2 | GO:1903818 | positive regulation of voltage-gated potassium channel activity(GO:1903818) |
| 0.0 | 7.5 | GO:1990830 | response to leukemia inhibitory factor(GO:1990823) cellular response to leukemia inhibitory factor(GO:1990830) |
| 0.0 | 0.8 | GO:0097186 | amelogenesis(GO:0097186) |
| 0.0 | 0.3 | GO:0060836 | lymphatic endothelial cell differentiation(GO:0060836) |
| 0.0 | 0.2 | GO:0060837 | blood vessel endothelial cell differentiation(GO:0060837) |
| 0.0 | 0.5 | GO:0043248 | proteasome assembly(GO:0043248) |
| 0.0 | 0.3 | GO:0006883 | cellular sodium ion homeostasis(GO:0006883) |
| 0.0 | 1.1 | GO:0006346 | methylation-dependent chromatin silencing(GO:0006346) |
| 0.0 | 2.9 | GO:0003073 | regulation of systemic arterial blood pressure(GO:0003073) |
| 0.0 | 0.6 | GO:0051044 | positive regulation of membrane protein ectodomain proteolysis(GO:0051044) |
| 0.0 | 0.1 | GO:0006627 | protein processing involved in protein targeting to mitochondrion(GO:0006627) |
| 0.0 | 0.7 | GO:0010818 | T cell chemotaxis(GO:0010818) |
| 0.0 | 1.0 | GO:0035335 | peptidyl-tyrosine dephosphorylation(GO:0035335) |
| 0.0 | 0.4 | GO:0032331 | negative regulation of chondrocyte differentiation(GO:0032331) |
| 0.0 | 0.4 | GO:0046040 | IMP metabolic process(GO:0046040) |
| 0.0 | 0.1 | GO:1901475 | pyruvate transport(GO:0006848) pyruvate transmembrane transport(GO:1901475) |
| 0.0 | 1.0 | GO:0007588 | excretion(GO:0007588) |
| 0.0 | 0.6 | GO:2000785 | regulation of autophagosome assembly(GO:2000785) |
| 0.0 | 2.3 | GO:0072332 | intrinsic apoptotic signaling pathway by p53 class mediator(GO:0072332) |
| 0.0 | 1.0 | GO:0001541 | ovarian follicle development(GO:0001541) |
| 0.0 | 0.1 | GO:1902224 | ketone body metabolic process(GO:1902224) |
| 0.0 | 0.6 | GO:0030513 | positive regulation of BMP signaling pathway(GO:0030513) |
| 0.0 | 0.6 | GO:0050919 | negative chemotaxis(GO:0050919) |
| 0.0 | 2.2 | GO:0014065 | phosphatidylinositol 3-kinase signaling(GO:0014065) |
| 0.0 | 0.4 | GO:0080111 | DNA demethylation(GO:0080111) |
| 0.0 | 1.2 | GO:0055001 | muscle cell development(GO:0055001) |
| 0.0 | 0.6 | GO:0043392 | negative regulation of DNA binding(GO:0043392) |
| 0.0 | 2.8 | GO:0016236 | macroautophagy(GO:0016236) |
| 0.0 | 0.3 | GO:0006783 | heme biosynthetic process(GO:0006783) |
| 0.0 | 0.5 | GO:0006471 | protein ADP-ribosylation(GO:0006471) |
| 0.0 | 1.2 | GO:0035383 | acyl-CoA metabolic process(GO:0006637) thioester metabolic process(GO:0035383) |
| 0.0 | 1.2 | GO:0031032 | actomyosin structure organization(GO:0031032) |
| 0.0 | 0.1 | GO:0000290 | deadenylation-dependent decapping of nuclear-transcribed mRNA(GO:0000290) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.9 | 8.4 | GO:0097442 | CA3 pyramidal cell dendrite(GO:0097442) |
| 0.8 | 3.4 | GO:0014701 | junctional sarcoplasmic reticulum membrane(GO:0014701) |
| 0.5 | 1.6 | GO:1904511 | cortical microtubule plus-end(GO:1903754) cytoplasmic microtubule plus-end(GO:1904511) |
| 0.5 | 3.8 | GO:0005927 | muscle tendon junction(GO:0005927) |
| 0.4 | 1.3 | GO:0097233 | lamellar body membrane(GO:0097232) alveolar lamellar body membrane(GO:0097233) |
| 0.4 | 2.1 | GO:0034365 | discoidal high-density lipoprotein particle(GO:0034365) |
| 0.4 | 3.3 | GO:0005826 | actomyosin contractile ring(GO:0005826) |
| 0.4 | 8.2 | GO:0031045 | dense core granule(GO:0031045) |
| 0.4 | 3.8 | GO:0033116 | endoplasmic reticulum-Golgi intermediate compartment membrane(GO:0033116) |
| 0.3 | 5.5 | GO:0042788 | polysomal ribosome(GO:0042788) |
| 0.3 | 8.8 | GO:0001518 | voltage-gated sodium channel complex(GO:0001518) |
| 0.3 | 4.1 | GO:0005883 | neurofilament(GO:0005883) |
| 0.3 | 6.6 | GO:0017146 | NMDA selective glutamate receptor complex(GO:0017146) |
| 0.2 | 1.0 | GO:0034683 | integrin alphav-beta3 complex(GO:0034683) integrin alphav-beta8 complex(GO:0034686) |
| 0.2 | 9.1 | GO:0060077 | inhibitory synapse(GO:0060077) |
| 0.2 | 0.9 | GO:0034751 | aryl hydrocarbon receptor complex(GO:0034751) |
| 0.2 | 0.5 | GO:0070992 | translation initiation complex(GO:0070992) |
| 0.2 | 4.3 | GO:0005859 | muscle myosin complex(GO:0005859) |
| 0.2 | 1.6 | GO:0036157 | outer dynein arm(GO:0036157) |
| 0.2 | 3.4 | GO:0014731 | spectrin-associated cytoskeleton(GO:0014731) |
| 0.2 | 2.6 | GO:0045263 | proton-transporting ATP synthase complex, coupling factor F(o)(GO:0045263) |
| 0.2 | 2.7 | GO:0046581 | intercellular canaliculus(GO:0046581) |
| 0.1 | 3.9 | GO:0005922 | connexon complex(GO:0005922) |
| 0.1 | 0.9 | GO:0070826 | paraferritin complex(GO:0070826) |
| 0.1 | 1.2 | GO:0016012 | sarcoglycan complex(GO:0016012) |
| 0.1 | 5.9 | GO:0048786 | presynaptic active zone(GO:0048786) |
| 0.1 | 1.7 | GO:0032591 | dendritic spine membrane(GO:0032591) |
| 0.1 | 6.5 | GO:0030673 | axolemma(GO:0030673) |
| 0.1 | 4.0 | GO:0031430 | M band(GO:0031430) |
| 0.1 | 0.4 | GO:0071007 | U2-type catalytic step 2 spliceosome(GO:0071007) |
| 0.1 | 0.7 | GO:0097443 | sorting endosome(GO:0097443) |
| 0.1 | 0.6 | GO:1990130 | Iml1 complex(GO:1990130) |
| 0.1 | 1.8 | GO:0042582 | primary lysosome(GO:0005766) azurophil granule(GO:0042582) |
| 0.1 | 3.4 | GO:0071782 | endoplasmic reticulum tubular network(GO:0071782) |
| 0.1 | 1.1 | GO:0000235 | astral microtubule(GO:0000235) |
| 0.1 | 0.9 | GO:1990357 | terminal web(GO:1990357) |
| 0.1 | 27.1 | GO:0030017 | sarcomere(GO:0030017) |
| 0.1 | 1.2 | GO:0008540 | proteasome regulatory particle, base subcomplex(GO:0008540) |
| 0.1 | 5.9 | GO:0046658 | anchored component of plasma membrane(GO:0046658) |
| 0.1 | 2.6 | GO:0005790 | smooth endoplasmic reticulum(GO:0005790) |
| 0.1 | 3.9 | GO:0044295 | axonal growth cone(GO:0044295) |
| 0.1 | 3.6 | GO:0030964 | mitochondrial respiratory chain complex I(GO:0005747) NADH dehydrogenase complex(GO:0030964) respiratory chain complex I(GO:0045271) |
| 0.1 | 1.0 | GO:0005869 | dynactin complex(GO:0005869) |
| 0.1 | 1.1 | GO:0031235 | intrinsic component of the cytoplasmic side of the plasma membrane(GO:0031235) |
| 0.1 | 0.4 | GO:0000120 | RNA polymerase I transcription factor complex(GO:0000120) |
| 0.1 | 0.3 | GO:0097057 | TRAF2-GSTP1 complex(GO:0097057) |
| 0.1 | 2.6 | GO:0030315 | T-tubule(GO:0030315) |
| 0.1 | 22.0 | GO:0099572 | postsynaptic density(GO:0014069) postsynaptic specialization(GO:0099572) |
| 0.1 | 1.0 | GO:0043198 | dendritic shaft(GO:0043198) |
| 0.1 | 5.6 | GO:0008076 | voltage-gated potassium channel complex(GO:0008076) |
| 0.1 | 2.4 | GO:0070469 | respiratory chain(GO:0070469) |
| 0.1 | 2.7 | GO:0014704 | intercalated disc(GO:0014704) |
| 0.0 | 0.6 | GO:0000815 | ESCRT III complex(GO:0000815) |
| 0.0 | 7.2 | GO:0031225 | anchored component of membrane(GO:0031225) |
| 0.0 | 1.3 | GO:0071565 | nBAF complex(GO:0071565) |
| 0.0 | 0.1 | GO:0042720 | mitochondrial inner membrane peptidase complex(GO:0042720) |
| 0.0 | 1.9 | GO:0016235 | aggresome(GO:0016235) |
| 0.0 | 0.3 | GO:0005579 | membrane attack complex(GO:0005579) |
| 0.0 | 3.4 | GO:0005778 | peroxisomal membrane(GO:0005778) microbody membrane(GO:0031903) |
| 0.0 | 0.9 | GO:0097440 | apical dendrite(GO:0097440) |
| 0.0 | 1.9 | GO:0031594 | neuromuscular junction(GO:0031594) |
| 0.0 | 1.9 | GO:0008287 | protein serine/threonine phosphatase complex(GO:0008287) phosphatase complex(GO:1903293) |
| 0.0 | 24.8 | GO:0045202 | synapse(GO:0045202) |
| 0.0 | 3.1 | GO:0030426 | growth cone(GO:0030426) |
| 0.0 | 1.2 | GO:0035861 | site of double-strand break(GO:0035861) |
| 0.0 | 0.2 | GO:0001674 | female germ cell nucleus(GO:0001674) |
| 0.0 | 0.7 | GO:0005891 | voltage-gated calcium channel complex(GO:0005891) |
| 0.0 | 14.8 | GO:0043005 | neuron projection(GO:0043005) |
| 0.0 | 1.8 | GO:0031463 | Cul3-RING ubiquitin ligase complex(GO:0031463) |
| 0.0 | 0.1 | GO:0030669 | clathrin-coated endocytic vesicle membrane(GO:0030669) |
| 0.0 | 0.7 | GO:0032391 | photoreceptor connecting cilium(GO:0032391) |
| 0.0 | 1.0 | GO:0009925 | basal plasma membrane(GO:0009925) |
| 0.0 | 0.2 | GO:0016272 | prefoldin complex(GO:0016272) |
| 0.0 | 3.0 | GO:0005802 | trans-Golgi network(GO:0005802) |
| 0.0 | 1.1 | GO:0042641 | actomyosin(GO:0042641) |
| 0.0 | 3.5 | GO:0005759 | mitochondrial matrix(GO:0005759) |
| 0.0 | 0.7 | GO:0005881 | cytoplasmic microtubule(GO:0005881) |
| 0.0 | 0.6 | GO:0001750 | photoreceptor outer segment(GO:0001750) |
| 0.0 | 3.3 | GO:0000151 | ubiquitin ligase complex(GO:0000151) |
| 0.0 | 1.3 | GO:0005777 | peroxisome(GO:0005777) microbody(GO:0042579) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.2 | 3.5 | GO:0030348 | syntaxin-3 binding(GO:0030348) |
| 1.0 | 3.8 | GO:0071614 | linoleic acid epoxygenase activity(GO:0071614) |
| 0.9 | 8.4 | GO:0005519 | cytoskeletal regulatory protein binding(GO:0005519) |
| 0.9 | 3.4 | GO:0047275 | glucosaminylgalactosylglucosylceramide beta-galactosyltransferase activity(GO:0047275) |
| 0.7 | 19.4 | GO:0003680 | AT DNA binding(GO:0003680) |
| 0.7 | 5.0 | GO:0004351 | glutamate decarboxylase activity(GO:0004351) |
| 0.7 | 2.1 | GO:0046911 | metal chelating activity(GO:0046911) |
| 0.7 | 2.8 | GO:0008113 | peptide-methionine (S)-S-oxide reductase activity(GO:0008113) |
| 0.6 | 2.4 | GO:0019166 | trans-2-enoyl-CoA reductase (NADPH) activity(GO:0019166) |
| 0.5 | 2.7 | GO:0004614 | phosphoglucomutase activity(GO:0004614) |
| 0.5 | 7.7 | GO:0050897 | cobalt ion binding(GO:0050897) |
| 0.4 | 1.3 | GO:0046870 | cadmium ion binding(GO:0046870) |
| 0.4 | 1.8 | GO:0004105 | choline-phosphate cytidylyltransferase activity(GO:0004105) |
| 0.4 | 3.4 | GO:0003996 | acyl-CoA ligase activity(GO:0003996) |
| 0.4 | 2.1 | GO:0008273 | calcium, potassium:sodium antiporter activity(GO:0008273) |
| 0.4 | 1.1 | GO:0005308 | creatine transmembrane transporter activity(GO:0005308) |
| 0.4 | 1.5 | GO:0008109 | N-acetyllactosaminide beta-1,6-N-acetylglucosaminyltransferase activity(GO:0008109) |
| 0.4 | 2.2 | GO:0090554 | phosphatidylcholine-translocating ATPase activity(GO:0090554) |
| 0.4 | 2.5 | GO:0001851 | complement component C3b binding(GO:0001851) |
| 0.4 | 5.8 | GO:0001011 | transcription factor activity, sequence-specific DNA binding, RNA polymerase recruiting(GO:0001011) transcription factor activity, TFIIB-class binding(GO:0001087) |
| 0.4 | 1.1 | GO:0004067 | asparaginase activity(GO:0004067) |
| 0.4 | 8.8 | GO:0031402 | sodium ion binding(GO:0031402) |
| 0.3 | 4.2 | GO:0004499 | N,N-dimethylaniline monooxygenase activity(GO:0004499) |
| 0.3 | 3.0 | GO:0004687 | myosin light chain kinase activity(GO:0004687) |
| 0.3 | 1.6 | GO:0005143 | interleukin-12 receptor binding(GO:0005143) |
| 0.3 | 3.7 | GO:0005025 | transforming growth factor beta receptor activity, type I(GO:0005025) |
| 0.3 | 1.8 | GO:0004994 | somatostatin receptor activity(GO:0004994) |
| 0.3 | 0.9 | GO:0036004 | GAF domain binding(GO:0036004) |
| 0.3 | 3.0 | GO:0047623 | AMP deaminase activity(GO:0003876) adenosine-phosphate deaminase activity(GO:0047623) |
| 0.3 | 2.8 | GO:0008046 | axon guidance receptor activity(GO:0008046) |
| 0.3 | 1.9 | GO:0061629 | RNA polymerase II sequence-specific DNA binding transcription factor binding(GO:0061629) |
| 0.3 | 1.1 | GO:0031694 | beta-adrenergic receptor activity(GO:0004939) alpha-2A adrenergic receptor binding(GO:0031694) |
| 0.3 | 3.5 | GO:0031802 | type 5 metabotropic glutamate receptor binding(GO:0031802) |
| 0.3 | 1.6 | GO:0051032 | nucleic acid transmembrane transporter activity(GO:0051032) RNA transmembrane transporter activity(GO:0051033) |
| 0.3 | 0.8 | GO:0008330 | protein tyrosine/threonine phosphatase activity(GO:0008330) |
| 0.3 | 2.8 | GO:0005347 | ATP transmembrane transporter activity(GO:0005347) ADP transmembrane transporter activity(GO:0015217) |
| 0.3 | 3.5 | GO:0097109 | neuroligin family protein binding(GO:0097109) |
| 0.2 | 1.5 | GO:0043533 | inositol 1,3,4,5 tetrakisphosphate binding(GO:0043533) |
| 0.2 | 1.7 | GO:0031750 | D3 dopamine receptor binding(GO:0031750) |
| 0.2 | 6.8 | GO:0005251 | delayed rectifier potassium channel activity(GO:0005251) |
| 0.2 | 3.2 | GO:0044323 | retinoic acid-responsive element binding(GO:0044323) |
| 0.2 | 9.7 | GO:0008157 | protein phosphatase 1 binding(GO:0008157) |
| 0.2 | 0.9 | GO:0016842 | amidine-lyase activity(GO:0016842) |
| 0.2 | 6.5 | GO:0008137 | NADH dehydrogenase (ubiquinone) activity(GO:0008137) NADH dehydrogenase (quinone) activity(GO:0050136) |
| 0.2 | 1.7 | GO:0070290 | N-acylphosphatidylethanolamine-specific phospholipase D activity(GO:0070290) |
| 0.2 | 1.9 | GO:0015198 | oligopeptide transporter activity(GO:0015198) |
| 0.2 | 0.6 | GO:0015321 | sodium-dependent phosphate transmembrane transporter activity(GO:0015321) |
| 0.2 | 1.2 | GO:0004692 | cGMP-dependent protein kinase activity(GO:0004692) |
| 0.2 | 3.2 | GO:0004143 | diacylglycerol kinase activity(GO:0004143) |
| 0.2 | 1.1 | GO:0042296 | ISG15 transferase activity(GO:0042296) |
| 0.2 | 3.4 | GO:0043495 | protein anchor(GO:0043495) |
| 0.2 | 2.6 | GO:0042043 | neurexin family protein binding(GO:0042043) |
| 0.2 | 0.7 | GO:0003998 | acylphosphatase activity(GO:0003998) |
| 0.2 | 2.2 | GO:0004887 | thyroid hormone receptor activity(GO:0004887) |
| 0.2 | 1.1 | GO:0019808 | polyamine binding(GO:0019808) |
| 0.2 | 5.9 | GO:0005452 | inorganic anion exchanger activity(GO:0005452) |
| 0.2 | 2.4 | GO:0001206 | transcriptional repressor activity, RNA polymerase II distal enhancer sequence-specific binding(GO:0001206) |
| 0.2 | 4.7 | GO:0008392 | arachidonic acid epoxygenase activity(GO:0008392) |
| 0.2 | 0.8 | GO:0008390 | testosterone 16-alpha-hydroxylase activity(GO:0008390) |
| 0.1 | 1.6 | GO:0005436 | sodium:phosphate symporter activity(GO:0005436) |
| 0.1 | 1.0 | GO:0031749 | D2 dopamine receptor binding(GO:0031749) |
| 0.1 | 5.5 | GO:0017091 | AU-rich element binding(GO:0017091) |
| 0.1 | 2.8 | GO:0097493 | structural molecule activity conferring elasticity(GO:0097493) |
| 0.1 | 2.6 | GO:0044548 | S100 protein binding(GO:0044548) |
| 0.1 | 2.7 | GO:0008170 | N-methyltransferase activity(GO:0008170) |
| 0.1 | 1.4 | GO:0008020 | G-protein coupled photoreceptor activity(GO:0008020) |
| 0.1 | 0.7 | GO:0048248 | CXCR3 chemokine receptor binding(GO:0048248) |
| 0.1 | 0.3 | GO:0003870 | 5-aminolevulinate synthase activity(GO:0003870) N-succinyltransferase activity(GO:0016749) |
| 0.1 | 1.0 | GO:0019960 | C-X3-C chemokine binding(GO:0019960) |
| 0.1 | 0.3 | GO:0035731 | S-nitrosoglutathione binding(GO:0035730) dinitrosyl-iron complex binding(GO:0035731) |
| 0.1 | 3.6 | GO:0015020 | glucuronosyltransferase activity(GO:0015020) |
| 0.1 | 4.0 | GO:0004115 | 3',5'-cyclic-AMP phosphodiesterase activity(GO:0004115) |
| 0.1 | 1.2 | GO:0015194 | L-serine transmembrane transporter activity(GO:0015194) serine transmembrane transporter activity(GO:0022889) |
| 0.1 | 0.6 | GO:0016401 | palmitoyl-CoA oxidase activity(GO:0016401) |
| 0.1 | 1.6 | GO:0002162 | dystroglycan binding(GO:0002162) |
| 0.1 | 4.3 | GO:0004683 | calmodulin-dependent protein kinase activity(GO:0004683) |
| 0.1 | 0.4 | GO:0004126 | cytidine deaminase activity(GO:0004126) |
| 0.1 | 0.6 | GO:0036402 | proteasome-activating ATPase activity(GO:0036402) |
| 0.1 | 1.3 | GO:0048273 | mitogen-activated protein kinase p38 binding(GO:0048273) |
| 0.1 | 2.9 | GO:0000062 | fatty-acyl-CoA binding(GO:0000062) |
| 0.1 | 4.1 | GO:0035255 | ionotropic glutamate receptor binding(GO:0035255) |
| 0.1 | 2.0 | GO:0051428 | peptide hormone receptor binding(GO:0051428) |
| 0.1 | 2.6 | GO:0005242 | inward rectifier potassium channel activity(GO:0005242) |
| 0.1 | 3.0 | GO:0043014 | alpha-tubulin binding(GO:0043014) |
| 0.1 | 2.6 | GO:0030676 | Rac guanyl-nucleotide exchange factor activity(GO:0030676) |
| 0.1 | 1.0 | GO:0017017 | MAP kinase tyrosine/serine/threonine phosphatase activity(GO:0017017) |
| 0.1 | 0.8 | GO:0000900 | translation repressor activity, nucleic acid binding(GO:0000900) |
| 0.1 | 1.0 | GO:0031957 | very long-chain fatty acid-CoA ligase activity(GO:0031957) |
| 0.1 | 1.7 | GO:0016208 | AMP binding(GO:0016208) |
| 0.1 | 5.8 | GO:0030507 | spectrin binding(GO:0030507) |
| 0.1 | 7.5 | GO:0005550 | pheromone binding(GO:0005550) |
| 0.1 | 6.6 | GO:0048306 | calcium-dependent protein binding(GO:0048306) |
| 0.1 | 0.5 | GO:0017034 | Rap guanyl-nucleotide exchange factor activity(GO:0017034) |
| 0.1 | 0.8 | GO:0008097 | 5S rRNA binding(GO:0008097) |
| 0.1 | 1.2 | GO:0032454 | histone demethylase activity (H3-K9 specific)(GO:0032454) |
| 0.1 | 2.1 | GO:0030742 | GTP-dependent protein binding(GO:0030742) |
| 0.0 | 0.8 | GO:0001161 | intronic transcription regulatory region sequence-specific DNA binding(GO:0001161) intronic transcription regulatory region DNA binding(GO:0044213) |
| 0.0 | 1.0 | GO:0033038 | bitter taste receptor activity(GO:0033038) |
| 0.0 | 0.3 | GO:0005111 | type 2 fibroblast growth factor receptor binding(GO:0005111) |
| 0.0 | 2.0 | GO:0008138 | protein tyrosine/serine/threonine phosphatase activity(GO:0008138) |
| 0.0 | 0.9 | GO:0045499 | chemorepellent activity(GO:0045499) |
| 0.0 | 1.1 | GO:0008327 | methyl-CpG binding(GO:0008327) |
| 0.0 | 3.0 | GO:0001540 | beta-amyloid binding(GO:0001540) |
| 0.0 | 2.1 | GO:0005164 | tumor necrosis factor receptor binding(GO:0005164) |
| 0.0 | 3.0 | GO:0017080 | sodium channel regulator activity(GO:0017080) |
| 0.0 | 0.3 | GO:0034711 | inhibin binding(GO:0034711) |
| 0.0 | 0.5 | GO:0003956 | NAD(P)+-protein-arginine ADP-ribosyltransferase activity(GO:0003956) |
| 0.0 | 1.4 | GO:0031435 | mitogen-activated protein kinase kinase kinase binding(GO:0031435) |
| 0.0 | 3.9 | GO:0004725 | protein tyrosine phosphatase activity(GO:0004725) |
| 0.0 | 4.5 | GO:0005089 | Rho guanyl-nucleotide exchange factor activity(GO:0005089) |
| 0.0 | 4.2 | GO:0015171 | amino acid transmembrane transporter activity(GO:0015171) |
| 0.0 | 3.0 | GO:0030971 | receptor tyrosine kinase binding(GO:0030971) |
| 0.0 | 0.4 | GO:0019869 | chloride channel inhibitor activity(GO:0019869) |
| 0.0 | 1.0 | GO:0008569 | ATP-dependent microtubule motor activity, minus-end-directed(GO:0008569) dynein light chain binding(GO:0045503) |
| 0.0 | 1.3 | GO:0008307 | structural constituent of muscle(GO:0008307) |
| 0.0 | 3.5 | GO:0008276 | protein methyltransferase activity(GO:0008276) |
| 0.0 | 3.7 | GO:0030165 | PDZ domain binding(GO:0030165) |
| 0.0 | 0.2 | GO:0008508 | bile acid:sodium symporter activity(GO:0008508) |
| 0.0 | 3.9 | GO:0005262 | calcium channel activity(GO:0005262) |
| 0.0 | 0.1 | GO:0030371 | translation repressor activity(GO:0030371) |
| 0.0 | 4.4 | GO:0004867 | serine-type endopeptidase inhibitor activity(GO:0004867) |
| 0.0 | 0.4 | GO:0003708 | retinoic acid receptor activity(GO:0003708) |
| 0.0 | 0.4 | GO:0017070 | U6 snRNA binding(GO:0017070) |
| 0.0 | 8.5 | GO:0005549 | odorant binding(GO:0005549) |
| 0.0 | 0.9 | GO:0005057 | receptor signaling protein activity(GO:0005057) |
| 0.0 | 1.0 | GO:0005109 | frizzled binding(GO:0005109) |
| 0.0 | 1.1 | GO:0004722 | protein serine/threonine phosphatase activity(GO:0004722) |
| 0.0 | 1.8 | GO:0019888 | protein phosphatase regulator activity(GO:0019888) |
| 0.0 | 12.1 | GO:0004674 | protein serine/threonine kinase activity(GO:0004674) |
| 0.0 | 0.1 | GO:0016716 | oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, another compound as one donor, and incorporation of one atom of oxygen(GO:0016716) |
| 0.0 | 5.9 | GO:0044822 | mRNA binding(GO:0003729) poly(A) RNA binding(GO:0044822) |
| 0.0 | 0.7 | GO:0008139 | nuclear localization sequence binding(GO:0008139) |
| 0.0 | 2.4 | GO:0005179 | hormone activity(GO:0005179) |
| 0.0 | 0.9 | GO:0003785 | actin monomer binding(GO:0003785) |
| 0.0 | 0.4 | GO:0042923 | neuropeptide binding(GO:0042923) |
| 0.0 | 2.1 | GO:0015078 | hydrogen ion transmembrane transporter activity(GO:0015078) |
| 0.0 | 1.0 | GO:0017022 | myosin binding(GO:0017022) |
| 0.0 | 1.2 | GO:0002039 | p53 binding(GO:0002039) |
| 0.0 | 0.5 | GO:0042056 | chemoattractant activity(GO:0042056) |
| 0.0 | 0.4 | GO:0008253 | 5'-nucleotidase activity(GO:0008253) |
| 0.0 | 1.4 | GO:0005200 | structural constituent of cytoskeleton(GO:0005200) |
| 0.0 | 1.2 | GO:0042626 | ATPase activity, coupled to transmembrane movement of substances(GO:0042626) |
| 0.0 | 0.1 | GO:0050833 | pyruvate transmembrane transporter activity(GO:0050833) |
| 0.0 | 0.4 | GO:0071837 | HMG box domain binding(GO:0071837) |
| 0.0 | 0.9 | GO:0051117 | ATPase binding(GO:0051117) |
| 0.0 | 1.4 | GO:0047485 | protein N-terminus binding(GO:0047485) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 21.1 | PID P38 MK2 PATHWAY | p38 signaling mediated by MAPKAP kinases |
| 0.3 | 19.4 | PID MAPK TRK PATHWAY | Trk receptor signaling mediated by the MAPK pathway |
| 0.1 | 8.4 | PID LKB1 PATHWAY | LKB1 signaling events |
| 0.1 | 8.0 | PID RAC1 REG PATHWAY | Regulation of RAC1 activity |
| 0.1 | 1.0 | SA PTEN PATHWAY | PTEN is a tumor suppressor that dephosphorylates the lipid messenger phosphatidylinositol triphosphate. |
| 0.1 | 1.7 | ST GA12 PATHWAY | G alpha 12 Pathway |
| 0.1 | 1.0 | PID REELIN PATHWAY | Reelin signaling pathway |
| 0.1 | 7.3 | PID P75 NTR PATHWAY | p75(NTR)-mediated signaling |
| 0.1 | 3.3 | PID INTEGRIN A4B1 PATHWAY | Alpha4 beta1 integrin signaling events |
| 0.1 | 2.7 | PID LIS1 PATHWAY | Lissencephaly gene (LIS1) in neuronal migration and development |
| 0.1 | 4.6 | PID ARF6 TRAFFICKING PATHWAY | Arf6 trafficking events |
| 0.1 | 3.0 | PID INSULIN PATHWAY | Insulin Pathway |
| 0.1 | 4.4 | PID CDC42 PATHWAY | CDC42 signaling events |
| 0.1 | 12.4 | NABA ECM GLYCOPROTEINS | Genes encoding structural ECM glycoproteins |
| 0.1 | 1.0 | PID NEPHRIN NEPH1 PATHWAY | Nephrin/Neph1 signaling in the kidney podocyte |
| 0.0 | 1.6 | PID IL27 PATHWAY | IL27-mediated signaling events |
| 0.0 | 2.6 | PID TRKR PATHWAY | Neurotrophic factor-mediated Trk receptor signaling |
| 0.0 | 3.1 | PID FGF PATHWAY | FGF signaling pathway |
| 0.0 | 2.1 | ST INTEGRIN SIGNALING PATHWAY | Integrin Signaling Pathway |
| 0.0 | 0.9 | PID HIF2PATHWAY | HIF-2-alpha transcription factor network |
| 0.0 | 0.9 | ST GA13 PATHWAY | G alpha 13 Pathway |
| 0.0 | 0.6 | PID P38 ALPHA BETA DOWNSTREAM PATHWAY | Signaling mediated by p38-alpha and p38-beta |
| 0.0 | 1.1 | ST MYOCYTE AD PATHWAY | Myocyte Adrenergic Pathway is a specific case of the generalized Adrenergic Pathway. |
| 0.0 | 0.9 | PID ERBB NETWORK PATHWAY | ErbB receptor signaling network |
| 0.0 | 2.7 | PID NOTCH PATHWAY | Notch signaling pathway |
| 0.0 | 8.7 | NABA ECM REGULATORS | Genes encoding enzymes and their regulators involved in the remodeling of the extracellular matrix |
| 0.0 | 2.6 | PID HIF1 TFPATHWAY | HIF-1-alpha transcription factor network |
| 0.0 | 1.7 | PID BMP PATHWAY | BMP receptor signaling |
| 0.0 | 1.4 | PID RXR VDR PATHWAY | RXR and RAR heterodimerization with other nuclear receptor |
| 0.0 | 0.9 | ST WNT BETA CATENIN PATHWAY | Wnt/beta-catenin Pathway |
| 0.0 | 0.8 | PID IL4 2PATHWAY | IL4-mediated signaling events |
| 0.0 | 0.3 | PID ALK1 PATHWAY | ALK1 signaling events |
| 0.0 | 0.8 | PID HNF3B PATHWAY | FOXA2 and FOXA3 transcription factor networks |
| 0.0 | 0.2 | PID NCADHERIN PATHWAY | N-cadherin signaling events |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.8 | 6.6 | REACTOME CREATION OF C4 AND C2 ACTIVATORS | Genes involved in Creation of C4 and C2 activators |
| 0.4 | 21.0 | REACTOME ERK MAPK TARGETS | Genes involved in ERK/MAPK targets |
| 0.2 | 12.2 | REACTOME INTERACTION BETWEEN L1 AND ANKYRINS | Genes involved in Interaction between L1 and Ankyrins |
| 0.2 | 5.0 | REACTOME GABA SYNTHESIS RELEASE REUPTAKE AND DEGRADATION | Genes involved in GABA synthesis, release, reuptake and degradation |
| 0.2 | 4.7 | REACTOME INSULIN SYNTHESIS AND PROCESSING | Genes involved in Insulin Synthesis and Processing |
| 0.2 | 2.5 | REACTOME REGULATION OF COMPLEMENT CASCADE | Genes involved in Regulation of Complement cascade |
| 0.2 | 3.9 | REACTOME GAP JUNCTION ASSEMBLY | Genes involved in Gap junction assembly |
| 0.2 | 9.9 | REACTOME VOLTAGE GATED POTASSIUM CHANNELS | Genes involved in Voltage gated Potassium channels |
| 0.2 | 2.7 | REACTOME GLYCOGEN BREAKDOWN GLYCOGENOLYSIS | Genes involved in Glycogen breakdown (glycogenolysis) |
| 0.1 | 3.0 | REACTOME IONOTROPIC ACTIVITY OF KAINATE RECEPTORS | Genes involved in Ionotropic activity of Kainate Receptors |
| 0.1 | 3.1 | REACTOME FGFR4 LIGAND BINDING AND ACTIVATION | Genes involved in FGFR4 ligand binding and activation |
| 0.1 | 3.8 | REACTOME ADHERENS JUNCTIONS INTERACTIONS | Genes involved in Adherens junctions interactions |
| 0.1 | 3.2 | REACTOME ACTIVATED NOTCH1 TRANSMITS SIGNAL TO THE NUCLEUS | Genes involved in Activated NOTCH1 Transmits Signal to the Nucleus |
| 0.1 | 1.4 | REACTOME OPSINS | Genes involved in Opsins |
| 0.1 | 2.0 | REACTOME BOTULINUM NEUROTOXICITY | Genes involved in Botulinum neurotoxicity |
| 0.1 | 6.1 | REACTOME MYOGENESIS | Genes involved in Myogenesis |
| 0.1 | 4.0 | REACTOME SIGNALING BY BMP | Genes involved in Signaling by BMP |
| 0.1 | 3.0 | REACTOME PURINE SALVAGE | Genes involved in Purine salvage |
| 0.1 | 2.1 | REACTOME HDL MEDIATED LIPID TRANSPORT | Genes involved in HDL-mediated lipid transport |
| 0.1 | 2.6 | REACTOME TIE2 SIGNALING | Genes involved in Tie2 Signaling |
| 0.1 | 12.7 | REACTOME P75 NTR RECEPTOR MEDIATED SIGNALLING | Genes involved in p75 NTR receptor-mediated signalling |
| 0.1 | 3.2 | REACTOME EFFECTS OF PIP2 HYDROLYSIS | Genes involved in Effects of PIP2 hydrolysis |
| 0.1 | 1.3 | REACTOME TETRAHYDROBIOPTERIN BH4 SYNTHESIS RECYCLING SALVAGE AND REGULATION | Genes involved in Tetrahydrobiopterin (BH4) synthesis, recycling, salvage and regulation |
| 0.1 | 1.0 | REACTOME SYNTHESIS OF BILE ACIDS AND BILE SALTS VIA 24 HYDROXYCHOLESTEROL | Genes involved in Synthesis of bile acids and bile salts via 24-hydroxycholesterol |
| 0.1 | 4.2 | REACTOME AMINO ACID TRANSPORT ACROSS THE PLASMA MEMBRANE | Genes involved in Amino acid transport across the plasma membrane |
| 0.1 | 2.8 | REACTOME SIGNALING BY ROBO RECEPTOR | Genes involved in Signaling by Robo receptor |
| 0.1 | 6.2 | REACTOME NUCLEAR RECEPTOR TRANSCRIPTION PATHWAY | Genes involved in Nuclear Receptor transcription pathway |
| 0.1 | 4.2 | REACTOME RESPIRATORY ELECTRON TRANSPORT | Genes involved in Respiratory electron transport |
| 0.1 | 5.1 | REACTOME TRANSPORT OF INORGANIC CATIONS ANIONS AND AMINO ACIDS OLIGOPEPTIDES | Genes involved in Transport of inorganic cations/anions and amino acids/oligopeptides |
| 0.1 | 0.9 | REACTOME DOWNREGULATION OF ERBB2 ERBB3 SIGNALING | Genes involved in Downregulation of ERBB2:ERBB3 signaling |
| 0.1 | 1.8 | REACTOME SYNTHESIS OF PC | Genes involved in Synthesis of PC |
| 0.0 | 3.6 | REACTOME L1CAM INTERACTIONS | Genes involved in L1CAM interactions |
| 0.0 | 1.9 | REACTOME REGULATION OF BETA CELL DEVELOPMENT | Genes involved in Regulation of beta-cell development |
| 0.0 | 2.8 | REACTOME STRIATED MUSCLE CONTRACTION | Genes involved in Striated Muscle Contraction |
| 0.0 | 1.0 | REACTOME RAS ACTIVATION UOPN CA2 INFUX THROUGH NMDA RECEPTOR | Genes involved in Ras activation uopn Ca2+ infux through NMDA receptor |
| 0.0 | 3.4 | REACTOME PHASE II CONJUGATION | Genes involved in Phase II conjugation |
| 0.0 | 0.3 | REACTOME COMPLEMENT CASCADE | Genes involved in Complement cascade |
| 0.0 | 1.2 | REACTOME NITRIC OXIDE STIMULATES GUANYLATE CYCLASE | Genes involved in Nitric oxide stimulates guanylate cyclase |
| 0.0 | 1.0 | REACTOME PEROXISOMAL LIPID METABOLISM | Genes involved in Peroxisomal lipid metabolism |
| 0.0 | 1.1 | REACTOME DARPP 32 EVENTS | Genes involved in DARPP-32 events |
| 0.0 | 0.4 | REACTOME ABACAVIR TRANSPORT AND METABOLISM | Genes involved in Abacavir transport and metabolism |
| 0.0 | 1.2 | REACTOME CDK MEDIATED PHOSPHORYLATION AND REMOVAL OF CDC6 | Genes involved in CDK-mediated phosphorylation and removal of Cdc6 |
| 0.0 | 0.2 | REACTOME THE ACTIVATION OF ARYLSULFATASES | Genes involved in The activation of arylsulfatases |
| 0.0 | 2.0 | REACTOME NGF SIGNALLING VIA TRKA FROM THE PLASMA MEMBRANE | Genes involved in NGF signalling via TRKA from the plasma membrane |
| 0.0 | 0.6 | REACTOME ENDOSOMAL SORTING COMPLEX REQUIRED FOR TRANSPORT ESCRT | Genes involved in Endosomal Sorting Complex Required For Transport (ESCRT) |
| 0.0 | 0.3 | REACTOME METABOLISM OF PORPHYRINS | Genes involved in Metabolism of porphyrins |
| 0.0 | 3.5 | REACTOME ANTIGEN PROCESSING UBIQUITINATION PROTEASOME DEGRADATION | Genes involved in Antigen processing: Ubiquitination & Proteasome degradation |
| 0.0 | 0.5 | REACTOME ABC FAMILY PROTEINS MEDIATED TRANSPORT | Genes involved in ABC-family proteins mediated transport |
| 0.0 | 0.4 | REACTOME RNA POL I TRANSCRIPTION TERMINATION | Genes involved in RNA Polymerase I Transcription Termination |
| 0.0 | 0.6 | REACTOME GOLGI ASSOCIATED VESICLE BIOGENESIS | Genes involved in Golgi Associated Vesicle Biogenesis |
| 0.0 | 0.6 | REACTOME BIOLOGICAL OXIDATIONS | Genes involved in Biological oxidations |
| 0.0 | 0.2 | REACTOME MITOCHONDRIAL TRNA AMINOACYLATION | Genes involved in Mitochondrial tRNA aminoacylation |
| 0.0 | 0.5 | REACTOME CLASS B 2 SECRETIN FAMILY RECEPTORS | Genes involved in Class B/2 (Secretin family receptors) |