Project
PRJNA375882: Comprehensive Mouse Transcriptomic BodyMap
Navigation
Downloads
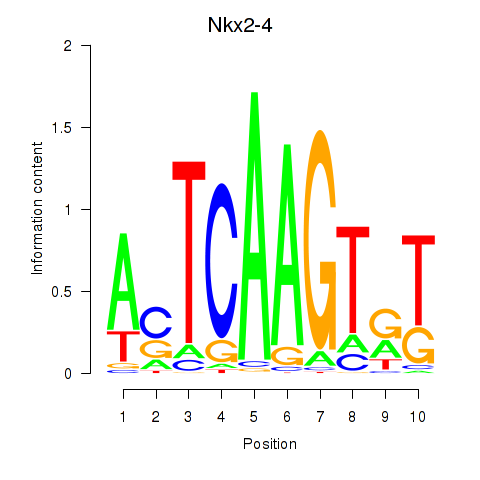
Results for Nkx2-4
Z-value: 0.93
Transcription factors associated with Nkx2-4
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Nkx2-4
|
ENSMUSG00000054160.3 | Nkx2-4 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Nkx2-4 | mm39_v1_chr2_-_146927365_146927406 | 0.25 | 3.1e-02 | Click! |
Activity profile of Nkx2-4 motif
Sorted Z-values of Nkx2-4 motif
Network of associatons between targets according to the STRING database.
First level regulatory network of Nkx2-4
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr6_+_41107047 | 6.82 |
ENSMUST00000103271.2
|
Trbv13-3
|
T cell receptor beta, variable 13-3 |
| chr6_+_41098273 | 4.55 |
ENSMUST00000103270.4
|
Trbv13-2
|
T cell receptor beta, variable 13-2 |
| chrX_+_106132840 | 3.74 |
ENSMUST00000118666.8
ENSMUST00000053375.4 |
P2ry10
|
purinergic receptor P2Y, G-protein coupled 10 |
| chr15_-_100567377 | 3.58 |
ENSMUST00000182814.8
ENSMUST00000238935.2 ENSMUST00000182068.8 ENSMUST00000182574.2 ENSMUST00000182775.8 |
Bin2
|
bridging integrator 2 |
| chr15_-_100567412 | 3.45 |
ENSMUST00000183211.8
|
Bin2
|
bridging integrator 2 |
| chr4_-_129467430 | 3.28 |
ENSMUST00000102596.8
|
Lck
|
lymphocyte protein tyrosine kinase |
| chr15_-_78449172 | 3.23 |
ENSMUST00000230952.2
|
Rac2
|
Rac family small GTPase 2 |
| chr9_-_14815228 | 3.03 |
ENSMUST00000034409.14
ENSMUST00000117620.8 |
Izumo1r
|
IZUMO1 receptor, JUNO |
| chr2_+_154390808 | 2.79 |
ENSMUST00000045116.11
ENSMUST00000109709.4 |
1700003F12Rik
|
RIKEN cDNA 1700003F12 gene |
| chr2_+_174485327 | 2.72 |
ENSMUST00000059452.6
|
Zfp831
|
zinc finger protein 831 |
| chr1_+_59669482 | 2.71 |
ENSMUST00000190490.2
|
Gm973
|
predicted gene 973 |
| chr17_-_57501170 | 2.66 |
ENSMUST00000005976.8
|
Tnfsf14
|
tumor necrosis factor (ligand) superfamily, member 14 |
| chr8_-_110305672 | 2.59 |
ENSMUST00000074898.8
|
Hp
|
haptoglobin |
| chr5_+_35156389 | 2.35 |
ENSMUST00000114281.8
ENSMUST00000114280.8 |
Rgs12
|
regulator of G-protein signaling 12 |
| chr1_+_100025486 | 2.35 |
ENSMUST00000188735.2
|
Cntnap5b
|
contactin associated protein-like 5B |
| chr2_+_61634797 | 2.28 |
ENSMUST00000048934.15
|
Tbr1
|
T-box brain transcription factor 1 |
| chr6_+_29398919 | 2.18 |
ENSMUST00000181464.8
ENSMUST00000180829.8 |
Ccdc136
|
coiled-coil domain containing 136 |
| chrX_-_35755483 | 2.14 |
ENSMUST00000169499.2
|
Gm14569
|
predicted gene 14569 |
| chr13_+_83723255 | 2.07 |
ENSMUST00000199167.5
ENSMUST00000195904.5 |
Mef2c
|
myocyte enhancer factor 2C |
| chr7_+_54485336 | 2.06 |
ENSMUST00000082373.8
|
Luzp2
|
leucine zipper protein 2 |
| chr1_-_175319842 | 2.00 |
ENSMUST00000195324.6
ENSMUST00000192227.6 ENSMUST00000194555.6 |
Rgs7
|
regulator of G protein signaling 7 |
| chr1_-_28819331 | 1.98 |
ENSMUST00000059937.5
|
Gm597
|
predicted gene 597 |
| chr15_+_6552270 | 1.95 |
ENSMUST00000226412.2
|
Fyb
|
FYN binding protein |
| chr2_-_84865831 | 1.90 |
ENSMUST00000028465.14
|
P2rx3
|
purinergic receptor P2X, ligand-gated ion channel, 3 |
| chr10_+_87926932 | 1.90 |
ENSMUST00000048621.8
|
Pmch
|
pro-melanin-concentrating hormone |
| chr13_+_83723743 | 1.89 |
ENSMUST00000198217.5
ENSMUST00000199210.5 |
Mef2c
|
myocyte enhancer factor 2C |
| chr17_+_70828649 | 1.81 |
ENSMUST00000233283.2
|
Dlgap1
|
DLG associated protein 1 |
| chr17_+_70829050 | 1.80 |
ENSMUST00000133717.9
ENSMUST00000148486.8 |
Dlgap1
|
DLG associated protein 1 |
| chr7_-_80554807 | 1.80 |
ENSMUST00000119428.2
ENSMUST00000026817.11 |
Nmb
|
neuromedin B |
| chr5_+_35156454 | 1.77 |
ENSMUST00000114283.8
|
Rgs12
|
regulator of G-protein signaling 12 |
| chr14_+_53853772 | 1.75 |
ENSMUST00000180972.3
|
Trav12-2
|
T cell receptor alpha variable 12-2 |
| chr3_-_92496293 | 1.74 |
ENSMUST00000098888.7
|
Smcp
|
sperm mitochondria-associated cysteine-rich protein |
| chr19_-_39637489 | 1.69 |
ENSMUST00000067328.7
|
Cyp2c67
|
cytochrome P450, family 2, subfamily c, polypeptide 67 |
| chr18_+_37085673 | 1.66 |
ENSMUST00000192512.6
ENSMUST00000192295.2 ENSMUST00000115661.5 |
Pcdha4
Gm42416
|
protocadherin alpha 4 predicted gene, 42416 |
| chr11_+_110858842 | 1.66 |
ENSMUST00000180023.8
ENSMUST00000106636.8 |
Kcnj16
|
potassium inwardly-rectifying channel, subfamily J, member 16 |
| chr1_-_88133472 | 1.62 |
ENSMUST00000119972.4
|
Dnajb3
|
DnaJ heat shock protein family (Hsp40) member B3 |
| chr3_-_102843406 | 1.56 |
ENSMUST00000199930.2
ENSMUST00000029448.11 ENSMUST00000196988.5 |
Sycp1
|
synaptonemal complex protein 1 |
| chr18_+_44096300 | 1.55 |
ENSMUST00000069245.8
|
Spink5
|
serine peptidase inhibitor, Kazal type 5 |
| chr14_+_53878403 | 1.53 |
ENSMUST00000184874.2
|
Trav14-2
|
T cell receptor alpha variable 14-2 |
| chr13_+_47347301 | 1.52 |
ENSMUST00000110111.4
|
Rnf144b
|
ring finger protein 144B |
| chr17_+_70829144 | 1.47 |
ENSMUST00000140728.8
|
Dlgap1
|
DLG associated protein 1 |
| chr3_-_92926364 | 1.46 |
ENSMUST00000193944.2
ENSMUST00000029520.9 |
Lce1m
|
late cornified envelope 1M |
| chr6_-_122587005 | 1.45 |
ENSMUST00000032211.5
|
Gdf3
|
growth differentiation factor 3 |
| chr5_-_23821523 | 1.45 |
ENSMUST00000088392.9
|
Srpk2
|
serine/arginine-rich protein specific kinase 2 |
| chr1_+_85454323 | 1.44 |
ENSMUST00000239236.2
|
Gm7592
|
predicted gene 7592 |
| chr3_+_153549846 | 1.43 |
ENSMUST00000044089.4
|
Asb17
|
ankyrin repeat and SOCS box-containing 17 |
| chr6_-_97037366 | 1.41 |
ENSMUST00000089295.6
|
Tafa4
|
TAFA chemokine like family member 4 |
| chr13_+_35843816 | 1.37 |
ENSMUST00000075220.14
|
Cdyl
|
chromodomain protein, Y chromosome-like |
| chr10_+_4561974 | 1.35 |
ENSMUST00000105590.8
ENSMUST00000067086.14 |
Esr1
|
estrogen receptor 1 (alpha) |
| chr8_-_79539838 | 1.34 |
ENSMUST00000146824.2
|
Lsm6
|
LSM6 homolog, U6 small nuclear RNA and mRNA degradation associated |
| chr5_+_14564932 | 1.32 |
ENSMUST00000182407.8
ENSMUST00000030691.17 |
Pclo
|
piccolo (presynaptic cytomatrix protein) |
| chr6_-_98319684 | 1.32 |
ENSMUST00000164491.3
|
Mdfic2
|
MyoD family inhibitor domain containing 2 |
| chr6_+_142359099 | 1.28 |
ENSMUST00000126521.9
ENSMUST00000211094.2 |
Spx
|
spexin hormone |
| chr17_+_69071546 | 1.26 |
ENSMUST00000233625.2
|
L3mbtl4
|
L3MBTL4 histone methyl-lysine binding protein |
| chrX_+_102674181 | 1.24 |
ENSMUST00000033692.9
|
Zcchc13
|
zinc finger, CCHC domain containing 13 |
| chr1_+_175526152 | 1.21 |
ENSMUST00000094288.10
ENSMUST00000171939.8 |
Wdr64
|
WD repeat domain 64 |
| chr6_-_29164981 | 1.21 |
ENSMUST00000007993.16
|
Rbm28
|
RNA binding motif protein 28 |
| chr8_+_46081213 | 1.18 |
ENSMUST00000130850.8
|
Sorbs2
|
sorbin and SH3 domain containing 2 |
| chr13_+_83672965 | 1.16 |
ENSMUST00000199432.5
ENSMUST00000198069.5 ENSMUST00000197681.5 ENSMUST00000197722.5 ENSMUST00000197938.5 |
Mef2c
|
myocyte enhancer factor 2C |
| chr3_-_108108113 | 1.15 |
ENSMUST00000106655.2
ENSMUST00000065664.7 |
Cyb561d1
|
cytochrome b-561 domain containing 1 |
| chr15_-_76419131 | 1.15 |
ENSMUST00000230604.2
|
Tmem249
|
transmembrane protein 249 |
| chr5_-_122188165 | 1.12 |
ENSMUST00000154139.8
|
Cux2
|
cut-like homeobox 2 |
| chr2_+_29951859 | 1.10 |
ENSMUST00000102866.10
|
Set
|
SET nuclear oncogene |
| chrX_-_142716200 | 1.07 |
ENSMUST00000112851.8
ENSMUST00000112856.3 ENSMUST00000033642.10 |
Dcx
|
doublecortin |
| chr2_-_34645241 | 1.05 |
ENSMUST00000102800.9
|
Gapvd1
|
GTPase activating protein and VPS9 domains 1 |
| chr3_-_10366229 | 1.02 |
ENSMUST00000119761.2
ENSMUST00000029043.13 |
Fabp12
|
fatty acid binding protein 12 |
| chr6_-_28133324 | 1.02 |
ENSMUST00000131897.2
|
Grm8
|
glutamate receptor, metabotropic 8 |
| chr8_-_3904309 | 1.00 |
ENSMUST00000033888.5
|
Cd209e
|
CD209e antigen |
| chr19_+_5497575 | 1.00 |
ENSMUST00000025850.7
ENSMUST00000236774.2 |
Fosl1
|
fos-like antigen 1 |
| chr6_+_30568366 | 0.98 |
ENSMUST00000049251.6
|
Cpa4
|
carboxypeptidase A4 |
| chr10_+_128067964 | 0.96 |
ENSMUST00000125289.8
ENSMUST00000105242.8 |
Timeless
|
timeless circadian clock 1 |
| chr11_+_101623776 | 0.95 |
ENSMUST00000039152.14
|
Dhx8
|
DEAH (Asp-Glu-Ala-His) box polypeptide 8 |
| chr2_-_27397668 | 0.94 |
ENSMUST00000154316.8
|
Brd3
|
bromodomain containing 3 |
| chr18_-_3281752 | 0.94 |
ENSMUST00000140332.8
ENSMUST00000147138.8 |
Crem
|
cAMP responsive element modulator |
| chr15_+_99568208 | 0.94 |
ENSMUST00000023758.9
|
Asic1
|
acid-sensing (proton-gated) ion channel 1 |
| chr6_+_96090127 | 0.89 |
ENSMUST00000122120.8
|
Tafa1
|
TAFA chemokine like family member 1 |
| chr15_+_21111428 | 0.89 |
ENSMUST00000075132.8
|
Cdh12
|
cadherin 12 |
| chr12_-_81468705 | 0.88 |
ENSMUST00000085319.4
|
Adam4
|
a disintegrin and metallopeptidase domain 4 |
| chrY_+_17400761 | 0.88 |
ENSMUST00000179408.3
|
Rbm31y
|
RNA binding motif 31, Y-linked |
| chr2_+_131792774 | 0.87 |
ENSMUST00000110170.8
ENSMUST00000110172.9 ENSMUST00000110171.9 |
Prnd
|
prion like protein doppel |
| chr14_-_32110312 | 0.87 |
ENSMUST00000100723.4
|
1700024G13Rik
|
RIKEN cDNA 1700024G13 gene |
| chr1_+_85177316 | 0.86 |
ENSMUST00000161424.5
ENSMUST00000113402.4 |
Gm7609
|
predicted pseudogene 7609 |
| chr19_-_39875192 | 0.85 |
ENSMUST00000168838.3
|
Cyp2c69
|
cytochrome P450, family 2, subfamily c, polypeptide 69 |
| chr6_+_41331039 | 0.85 |
ENSMUST00000072103.7
|
Try10
|
trypsin 10 |
| chr4_-_136620376 | 0.83 |
ENSMUST00000046332.6
|
C1qc
|
complement component 1, q subcomponent, C chain |
| chr8_+_46080840 | 0.78 |
ENSMUST00000135336.9
|
Sorbs2
|
sorbin and SH3 domain containing 2 |
| chr14_+_53061814 | 0.77 |
ENSMUST00000103648.4
|
Trav11d
|
T cell receptor alpha variable 11D |
| chr11_+_87483723 | 0.74 |
ENSMUST00000119628.8
|
Mtmr4
|
myotubularin related protein 4 |
| chr12_+_105996961 | 0.72 |
ENSMUST00000220629.2
|
Vrk1
|
vaccinia related kinase 1 |
| chr3_+_76500857 | 0.71 |
ENSMUST00000162471.2
|
Fstl5
|
follistatin-like 5 |
| chr5_+_24890810 | 0.71 |
ENSMUST00000068825.8
|
Nub1
|
negative regulator of ubiquitin-like proteins 1 |
| chr14_+_73376192 | 0.68 |
ENSMUST00000171070.8
|
Rcbtb2
|
regulator of chromosome condensation (RCC1) and BTB (POZ) domain containing protein 2 |
| chr10_+_128067934 | 0.67 |
ENSMUST00000055539.11
ENSMUST00000105244.8 ENSMUST00000105243.9 |
Timeless
|
timeless circadian clock 1 |
| chr11_-_101490011 | 0.67 |
ENSMUST00000209862.2
ENSMUST00000154147.2 |
Gm11634
|
predicted gene 11634 |
| chr18_-_3281089 | 0.67 |
ENSMUST00000139537.2
ENSMUST00000124747.8 |
Crem
|
cAMP responsive element modulator |
| chr9_-_39618413 | 0.66 |
ENSMUST00000215192.2
|
Olfr149
|
olfactory receptor 149 |
| chr2_-_85027041 | 0.63 |
ENSMUST00000099930.9
ENSMUST00000111601.2 |
Lrrc55
|
leucine rich repeat containing 55 |
| chr14_+_53081922 | 0.62 |
ENSMUST00000181360.3
ENSMUST00000183652.2 |
Trav12d-1
|
T cell receptor alpha variable 12D-1 |
| chr15_-_98796373 | 0.60 |
ENSMUST00000229775.2
ENSMUST00000023737.6 |
Dhh
|
desert hedgehog |
| chr1_-_63153675 | 0.60 |
ENSMUST00000097718.9
|
Ino80d
|
INO80 complex subunit D |
| chr3_+_88536805 | 0.59 |
ENSMUST00000175745.2
|
Arhgef2
|
rho/rac guanine nucleotide exchange factor (GEF) 2 |
| chr6_+_121320339 | 0.58 |
ENSMUST00000168295.2
|
Slc6a12
|
solute carrier family 6 (neurotransmitter transporter, betaine/GABA), member 12 |
| chr2_-_63014622 | 0.58 |
ENSMUST00000075052.10
ENSMUST00000112454.8 |
Kcnh7
|
potassium voltage-gated channel, subfamily H (eag-related), member 7 |
| chr2_+_153716958 | 0.58 |
ENSMUST00000028983.3
|
Bpifb2
|
BPI fold containing family B, member 2 |
| chr11_+_110858882 | 0.57 |
ENSMUST00000125692.2
|
Kcnj16
|
potassium inwardly-rectifying channel, subfamily J, member 16 |
| chrX_-_142716085 | 0.57 |
ENSMUST00000087313.10
|
Dcx
|
doublecortin |
| chr11_+_101623836 | 0.56 |
ENSMUST00000129741.2
|
Dhx8
|
DEAH (Asp-Glu-Ala-His) box polypeptide 8 |
| chr6_-_28133128 | 0.56 |
ENSMUST00000132755.2
|
Grm8
|
glutamate receptor, metabotropic 8 |
| chr15_-_27788693 | 0.52 |
ENSMUST00000226287.2
|
Trio
|
triple functional domain (PTPRF interacting) |
| chr2_-_35995283 | 0.50 |
ENSMUST00000112960.8
ENSMUST00000112967.12 ENSMUST00000112963.8 |
Lhx6
|
LIM homeobox protein 6 |
| chr14_-_8798841 | 0.50 |
ENSMUST00000061045.3
|
Sntn
|
sentan, cilia apical structure protein |
| chr18_+_34675366 | 0.50 |
ENSMUST00000012426.3
|
Wnt8a
|
wingless-type MMTV integration site family, member 8A |
| chr2_-_112089627 | 0.50 |
ENSMUST00000043970.2
|
Nutm1
|
NUT midline carcinoma, family member 1 |
| chr1_+_85577766 | 0.49 |
ENSMUST00000066427.11
|
Sp100
|
nuclear antigen Sp100 |
| chrX_+_36528991 | 0.49 |
ENSMUST00000115194.5
|
Rhox4a
|
reproductive homeobox 4A |
| chr4_+_62443606 | 0.47 |
ENSMUST00000062145.2
|
4933430I17Rik
|
RIKEN cDNA 4933430I17 gene |
| chr5_-_120887864 | 0.47 |
ENSMUST00000053909.13
ENSMUST00000081491.13 |
Oas2
|
2'-5' oligoadenylate synthetase 2 |
| chr18_-_3281727 | 0.47 |
ENSMUST00000154705.8
ENSMUST00000151084.8 |
Crem
|
cAMP responsive element modulator |
| chr19_-_39801188 | 0.46 |
ENSMUST00000162507.2
ENSMUST00000160476.9 ENSMUST00000239028.2 |
Cyp2c40
|
cytochrome P450, family 2, subfamily c, polypeptide 40 |
| chrX_+_36572501 | 0.46 |
ENSMUST00000179915.4
|
Rhox4a2
|
reproductive homeobox 4A2 |
| chr9_+_108437485 | 0.45 |
ENSMUST00000081111.14
ENSMUST00000193421.2 |
Impdh2
|
inosine monophosphate dehydrogenase 2 |
| chr7_-_34512101 | 0.45 |
ENSMUST00000078686.8
ENSMUST00000205259.2 ENSMUST00000154629.3 |
Chst8
|
carbohydrate sulfotransferase 8 |
| chr9_-_57347366 | 0.45 |
ENSMUST00000214144.2
ENSMUST00000085709.6 ENSMUST00000214624.2 ENSMUST00000215883.2 ENSMUST00000214339.2 ENSMUST00000215299.2 ENSMUST00000214166.2 ENSMUST00000214065.2 |
Ppcdc
|
phosphopantothenoylcysteine decarboxylase |
| chr5_-_137531471 | 0.43 |
ENSMUST00000143495.8
ENSMUST00000111020.8 ENSMUST00000111023.8 ENSMUST00000111038.8 |
Gnb2
Epo
|
guanine nucleotide binding protein (G protein), beta 2 erythropoietin |
| chr11_+_53611611 | 0.43 |
ENSMUST00000048605.3
|
Il5
|
interleukin 5 |
| chr15_+_54274151 | 0.43 |
ENSMUST00000036737.4
|
Colec10
|
collectin sub-family member 10 |
| chr10_-_33662700 | 0.41 |
ENSMUST00000223295.2
|
Sult3a2
|
sulfotransferase family 3A, member 2 |
| chr6_+_123206880 | 0.40 |
ENSMUST00000205129.2
|
Clec4n
|
C-type lectin domain family 4, member n |
| chr12_-_91712783 | 0.40 |
ENSMUST00000166967.2
|
Ston2
|
stonin 2 |
| chr8_+_67312847 | 0.39 |
ENSMUST00000118009.2
|
Naf1
|
nuclear assembly factor 1 ribonucleoprotein |
| chr6_-_91784299 | 0.36 |
ENSMUST00000159684.8
|
Grip2
|
glutamate receptor interacting protein 2 |
| chr6_+_123206802 | 0.36 |
ENSMUST00000112554.9
ENSMUST00000024118.11 ENSMUST00000117130.8 |
Clec4n
|
C-type lectin domain family 4, member n |
| chr2_+_131792931 | 0.36 |
ENSMUST00000110169.2
|
Prnd
|
prion like protein doppel |
| chr14_-_52273600 | 0.36 |
ENSMUST00000214342.2
|
Olfr221
|
olfactory receptor 221 |
| chr6_+_43212512 | 0.33 |
ENSMUST00000078057.4
|
Olfr47
|
olfactory receptor 47 |
| chr14_-_79461316 | 0.32 |
ENSMUST00000040802.5
|
Zfp957
|
zinc finger protein 957 |
| chrX_+_111513971 | 0.31 |
ENSMUST00000071814.13
|
Zfp711
|
zinc finger protein 711 |
| chrX_+_147446594 | 0.30 |
ENSMUST00000101186.10
ENSMUST00000101190.11 |
Gm15127
|
predicted gene 15127 |
| chr7_-_132414823 | 0.29 |
ENSMUST00000106165.8
|
Fam53b
|
family with sequence similarity 53, member B |
| chr17_+_37253916 | 0.28 |
ENSMUST00000173072.2
|
Rnf39
|
ring finger protein 39 |
| chrX_+_55493325 | 0.27 |
ENSMUST00000079663.7
|
Gm2174
|
predicted gene 2174 |
| chrX_+_146963720 | 0.27 |
ENSMUST00000112796.2
|
Gm15107
|
predicted gene 15107 |
| chr6_+_70332836 | 0.26 |
ENSMUST00000103390.3
|
Igkv8-18
|
immunoglobulin kappa variable 8-18 |
| chrX_+_147665572 | 0.26 |
ENSMUST00000163338.2
|
Luzp4
|
leucine zipper protein 4 |
| chrX_-_72158969 | 0.25 |
ENSMUST00000114520.2
|
Xlr5a
|
X-linked lymphocyte-regulated 5A |
| chr18_+_36431732 | 0.25 |
ENSMUST00000210490.3
|
Igip
|
IgA inducing protein |
| chrX_+_148567469 | 0.24 |
ENSMUST00000101180.8
|
Gm15097
|
predicted gene 15097 |
| chrX_+_148377566 | 0.24 |
ENSMUST00000101181.5
|
Gm10439
|
predicted gene 10439 |
| chrX_+_146572724 | 0.24 |
ENSMUST00000178833.2
|
Gm15128
|
predicted gene 15128 |
| chr16_-_63684477 | 0.24 |
ENSMUST00000232654.2
ENSMUST00000064405.8 |
Epha3
|
Eph receptor A3 |
| chrX_+_72194740 | 0.23 |
ENSMUST00000097221.4
|
Xlr5b
|
X-linked lymphocyte-regulated 5B |
| chrX_+_148230613 | 0.23 |
ENSMUST00000178169.2
|
Gm15085
|
predicted gene 15085 |
| chrX_+_147871311 | 0.23 |
ENSMUST00000177606.8
|
Ott
|
ovary testis transcribed |
| chr3_-_110158280 | 0.22 |
ENSMUST00000190378.2
ENSMUST00000106567.2 |
Prmt6
|
protein arginine N-methyltransferase 6 |
| chr11_-_83320281 | 0.22 |
ENSMUST00000052521.9
|
Gas2l2
|
growth arrest-specific 2 like 2 |
| chr8_+_46944000 | 0.20 |
ENSMUST00000110372.9
ENSMUST00000130563.2 |
Acsl1
|
acyl-CoA synthetase long-chain family member 1 |
| chr5_-_143211632 | 0.20 |
ENSMUST00000085733.9
|
Spdye4a
|
speedy/RINGO cell cycle regulator family, member E4A |
| chrX_-_72334121 | 0.19 |
ENSMUST00000088450.4
|
Xlr5c
|
X-linked lymphocyte-regulated 5C |
| chr15_+_81350842 | 0.18 |
ENSMUST00000230062.2
|
Rbx1
|
ring-box 1 |
| chr1_+_85577709 | 0.17 |
ENSMUST00000155094.8
ENSMUST00000054279.15 ENSMUST00000147552.8 ENSMUST00000153574.8 ENSMUST00000150967.8 |
Sp100
|
nuclear antigen Sp100 |
| chr8_-_3798922 | 0.17 |
ENSMUST00000208960.2
ENSMUST00000207979.2 |
Cd209a
|
CD209a antigen |
| chr14_+_26414422 | 0.17 |
ENSMUST00000022433.12
|
Dnah12
|
dynein, axonemal, heavy chain 12 |
| chr3_+_96604415 | 0.17 |
ENSMUST00000107077.4
|
Pias3
|
protein inhibitor of activated STAT 3 |
| chr17_-_3746536 | 0.17 |
ENSMUST00000115800.2
|
Nox3
|
NADPH oxidase 3 |
| chrX_+_146548987 | 0.16 |
ENSMUST00000112814.8
|
Gm15128
|
predicted gene 15128 |
| chrX_+_147880948 | 0.16 |
ENSMUST00000177554.2
|
Ott
|
ovary testis transcribed |
| chr11_+_49213569 | 0.16 |
ENSMUST00000215671.2
|
Olfr1391
|
olfactory receptor 1391 |
| chr19_-_45731312 | 0.16 |
ENSMUST00000026241.12
ENSMUST00000026240.14 ENSMUST00000111928.8 |
Fgf8
|
fibroblast growth factor 8 |
| chr9_-_71803354 | 0.13 |
ENSMUST00000184448.8
|
Tcf12
|
transcription factor 12 |
| chr15_+_68800261 | 0.12 |
ENSMUST00000022954.7
|
Khdrbs3
|
KH domain containing, RNA binding, signal transduction associated 3 |
| chr10_-_4338032 | 0.11 |
ENSMUST00000100078.10
|
Zbtb2
|
zinc finger and BTB domain containing 2 |
| chr5_-_137531463 | 0.11 |
ENSMUST00000170293.8
|
Gnb2
|
guanine nucleotide binding protein (G protein), beta 2 |
| chr3_-_64317478 | 0.10 |
ENSMUST00000170280.3
|
Vmn2r4
|
vomeronasal 2, receptor 4 |
| chrX_+_36739065 | 0.10 |
ENSMUST00000089075.6
|
Rhox4e
|
reproductive homeobox 4E |
| chr6_-_91784405 | 0.09 |
ENSMUST00000162300.8
|
Grip2
|
glutamate receptor interacting protein 2 |
| chr1_-_138547403 | 0.09 |
ENSMUST00000027642.5
ENSMUST00000186017.7 |
Nek7
|
NIMA (never in mitosis gene a)-related expressed kinase 7 |
| chr2_+_88217406 | 0.08 |
ENSMUST00000214040.3
|
Olfr1178
|
olfactory receptor 1178 |
| chr1_+_173986288 | 0.07 |
ENSMUST00000068403.4
|
Olfr420
|
olfactory receptor 420 |
| chr16_-_92622659 | 0.07 |
ENSMUST00000186296.2
|
Runx1
|
runt related transcription factor 1 |
| chr12_+_38830812 | 0.06 |
ENSMUST00000160856.8
|
Etv1
|
ets variant 1 |
| chr12_+_38831093 | 0.05 |
ENSMUST00000161513.9
|
Etv1
|
ets variant 1 |
| chr7_-_81216687 | 0.05 |
ENSMUST00000042318.6
|
Fsd2
|
fibronectin type III and SPRY domain containing 2 |
| chr6_+_132739094 | 0.04 |
ENSMUST00000069268.3
|
Tas2r102
|
taste receptor, type 2, member 102 |
| chr12_+_17740831 | 0.03 |
ENSMUST00000071858.5
|
Hpcal1
|
hippocalcin-like 1 |
| chr11_+_98754434 | 0.03 |
ENSMUST00000142414.8
ENSMUST00000037480.9 |
Wipf2
|
WAS/WASL interacting protein family, member 2 |
| chr5_-_137531413 | 0.02 |
ENSMUST00000168746.8
|
Gnb2
|
guanine nucleotide binding protein (G protein), beta 2 |
| chr11_+_49138278 | 0.01 |
ENSMUST00000109194.2
|
Mgat1
|
mannoside acetylglucosaminyltransferase 1 |
| chr12_+_87921198 | 0.01 |
ENSMUST00000110145.12
ENSMUST00000181843.2 ENSMUST00000180706.8 ENSMUST00000181394.8 ENSMUST00000181326.8 ENSMUST00000181300.2 |
Gm2042
|
predicted gene 2042 |
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.9 | 2.7 | GO:0008588 | release of cytoplasmic sequestered NF-kappaB(GO:0008588) |
| 0.9 | 2.6 | GO:2000295 | regulation of hydrogen peroxide catabolic process(GO:2000295) |
| 0.8 | 4.1 | GO:0038032 | termination of G-protein coupled receptor signaling pathway(GO:0038032) |
| 0.6 | 1.9 | GO:0046005 | positive regulation of circadian sleep/wake cycle, REM sleep(GO:0046005) |
| 0.5 | 1.6 | GO:0051878 | lateral element assembly(GO:0051878) |
| 0.4 | 1.3 | GO:0099526 | presynaptic signal transduction(GO:0098928) presynapse to nucleus signaling pathway(GO:0099526) |
| 0.4 | 5.1 | GO:0003138 | primary heart field specification(GO:0003138) sinoatrial valve development(GO:0003172) sinoatrial valve morphogenesis(GO:0003185) |
| 0.4 | 5.1 | GO:0070842 | aggresome assembly(GO:0070842) |
| 0.4 | 1.9 | GO:1904058 | positive regulation of sensory perception of pain(GO:1904058) |
| 0.4 | 2.3 | GO:0001661 | conditioned taste aversion(GO:0001661) |
| 0.3 | 1.6 | GO:1904976 | response to bleomycin(GO:1904975) cellular response to bleomycin(GO:1904976) |
| 0.3 | 3.2 | GO:0060753 | regulation of mast cell chemotaxis(GO:0060753) |
| 0.3 | 0.6 | GO:0030908 | intein-mediated protein splicing(GO:0016539) protein splicing(GO:0030908) |
| 0.3 | 1.6 | GO:0002786 | regulation of antimicrobial peptide production(GO:0002784) regulation of antibacterial peptide production(GO:0002786) |
| 0.3 | 3.3 | GO:0070474 | positive regulation of uterine smooth muscle contraction(GO:0070474) |
| 0.2 | 1.4 | GO:0035063 | nuclear speck organization(GO:0035063) |
| 0.2 | 1.4 | GO:0060523 | prostate epithelial cord elongation(GO:0060523) |
| 0.2 | 1.7 | GO:0007341 | penetration of zona pellucida(GO:0007341) |
| 0.2 | 7.0 | GO:0097320 | membrane tubulation(GO:0097320) |
| 0.2 | 1.3 | GO:1904306 | positive regulation of gastro-intestinal system smooth muscle contraction(GO:1904306) |
| 0.2 | 1.5 | GO:0048859 | formation of anatomical boundary(GO:0048859) |
| 0.2 | 0.7 | GO:0072355 | histone H3-T3 phosphorylation(GO:0072355) |
| 0.2 | 1.6 | GO:0007196 | adenylate cyclase-inhibiting G-protein coupled glutamate receptor signaling pathway(GO:0007196) |
| 0.2 | 1.0 | GO:0007296 | vitellogenesis(GO:0007296) |
| 0.2 | 3.0 | GO:0007342 | fusion of sperm to egg plasma membrane(GO:0007342) |
| 0.2 | 0.9 | GO:0050915 | sensory perception of sour taste(GO:0050915) |
| 0.1 | 0.4 | GO:0045645 | regulation of eosinophil differentiation(GO:0045643) positive regulation of eosinophil differentiation(GO:0045645) |
| 0.1 | 6.8 | GO:0001562 | response to protozoan(GO:0001562) |
| 0.1 | 0.4 | GO:1902219 | negative regulation of intrinsic apoptotic signaling pathway in response to osmotic stress(GO:1902219) |
| 0.1 | 3.7 | GO:0051482 | positive regulation of cytosolic calcium ion concentration involved in phospholipase C-activating G-protein coupled signaling pathway(GO:0051482) |
| 0.1 | 2.2 | GO:0001675 | acrosome assembly(GO:0001675) |
| 0.1 | 0.5 | GO:0009946 | proximal/distal axis specification(GO:0009946) |
| 0.1 | 0.6 | GO:0009992 | cellular water homeostasis(GO:0009992) |
| 0.1 | 1.6 | GO:0048672 | positive regulation of collateral sprouting(GO:0048672) |
| 0.1 | 0.8 | GO:0045650 | negative regulation of macrophage differentiation(GO:0045650) |
| 0.1 | 0.5 | GO:0021853 | cerebral cortex GABAergic interneuron migration(GO:0021853) interneuron migration(GO:1904936) |
| 0.1 | 0.4 | GO:0000454 | snoRNA guided rRNA pseudouridine synthesis(GO:0000454) telomerase RNA stabilization(GO:0090669) |
| 0.1 | 0.6 | GO:0071802 | negative regulation of podosome assembly(GO:0071802) |
| 0.1 | 0.2 | GO:0034970 | histone H3-R2 methylation(GO:0034970) |
| 0.1 | 0.5 | GO:0006177 | GMP biosynthetic process(GO:0006177) |
| 0.1 | 0.5 | GO:0018377 | protein myristoylation(GO:0018377) |
| 0.1 | 0.2 | GO:0009629 | response to gravity(GO:0009629) |
| 0.0 | 1.2 | GO:0006878 | cellular copper ion homeostasis(GO:0006878) |
| 0.0 | 0.5 | GO:0014824 | artery smooth muscle contraction(GO:0014824) |
| 0.0 | 1.1 | GO:0007614 | short-term memory(GO:0007614) |
| 0.0 | 1.8 | GO:0050482 | arachidonic acid secretion(GO:0050482) arachidonate transport(GO:1903963) |
| 0.0 | 2.2 | GO:0010107 | potassium ion import(GO:0010107) |
| 0.0 | 0.6 | GO:1903818 | positive regulation of voltage-gated potassium channel activity(GO:1903818) |
| 0.0 | 2.0 | GO:0003301 | physiological muscle hypertrophy(GO:0003298) physiological cardiac muscle hypertrophy(GO:0003301) cell growth involved in cardiac muscle cell development(GO:0061049) |
| 0.0 | 2.1 | GO:0006687 | glycosphingolipid metabolic process(GO:0006687) |
| 0.0 | 0.4 | GO:0015937 | coenzyme A biosynthetic process(GO:0015937) |
| 0.0 | 0.8 | GO:0019373 | epoxygenase P450 pathway(GO:0019373) |
| 0.0 | 1.5 | GO:0018149 | peptide cross-linking(GO:0018149) |
| 0.0 | 1.3 | GO:0030490 | maturation of SSU-rRNA(GO:0030490) |
| 0.0 | 2.0 | GO:0045576 | mast cell activation(GO:0045576) |
| 0.0 | 0.8 | GO:0050832 | defense response to fungus(GO:0050832) |
| 0.0 | 0.1 | GO:0071336 | regulation of hair follicle cell proliferation(GO:0071336) positive regulation of progesterone secretion(GO:2000872) |
| 0.0 | 0.4 | GO:0016486 | peptide hormone processing(GO:0016486) |
| 0.0 | 2.1 | GO:0032436 | positive regulation of proteasomal ubiquitin-dependent protein catabolic process(GO:0032436) |
| 0.0 | 2.0 | GO:0008277 | regulation of G-protein coupled receptor protein signaling pathway(GO:0008277) |
| 0.0 | 0.2 | GO:0044539 | long-chain fatty acid import(GO:0044539) |
| 0.0 | 1.1 | GO:0006334 | nucleosome assembly(GO:0006334) |
| 0.0 | 4.5 | GO:0019221 | cytokine-mediated signaling pathway(GO:0019221) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 2.6 | GO:0031838 | haptoglobin-hemoglobin complex(GO:0031838) |
| 0.4 | 1.3 | GO:0044317 | rod spherule(GO:0044317) |
| 0.3 | 1.6 | GO:0000802 | transverse filament(GO:0000802) |
| 0.3 | 1.6 | GO:0097209 | epidermal lamellar body(GO:0097209) |
| 0.2 | 7.0 | GO:0001891 | phagocytic cup(GO:0001891) |
| 0.2 | 1.4 | GO:0097550 | transcriptional preinitiation complex(GO:0097550) |
| 0.1 | 3.3 | GO:0000242 | pericentriolar material(GO:0000242) |
| 0.1 | 1.3 | GO:0005688 | U6 snRNP(GO:0005688) |
| 0.1 | 1.9 | GO:0005639 | integral component of nuclear inner membrane(GO:0005639) intrinsic component of nuclear inner membrane(GO:0031229) |
| 0.1 | 4.1 | GO:0097440 | apical dendrite(GO:0097440) |
| 0.1 | 2.0 | GO:0044292 | dendrite terminus(GO:0044292) |
| 0.1 | 2.2 | GO:0002080 | acrosomal membrane(GO:0002080) |
| 0.1 | 1.3 | GO:0031045 | dense core granule(GO:0031045) |
| 0.0 | 0.2 | GO:0043224 | nuclear SCF ubiquitin ligase complex(GO:0043224) |
| 0.0 | 4.4 | GO:0001669 | acrosomal vesicle(GO:0001669) |
| 0.0 | 1.5 | GO:0001533 | cornified envelope(GO:0001533) |
| 0.0 | 2.6 | GO:0042734 | presynaptic membrane(GO:0042734) |
| 0.0 | 2.0 | GO:0043198 | dendritic shaft(GO:0043198) |
| 0.0 | 4.3 | GO:0031225 | anchored component of membrane(GO:0031225) |
| 0.0 | 3.2 | GO:0005884 | actin filament(GO:0005884) |
| 0.0 | 7.0 | GO:0014069 | postsynaptic density(GO:0014069) |
| 0.0 | 4.4 | GO:0030017 | sarcomere(GO:0030017) |
| 0.0 | 0.4 | GO:0005732 | small nucleolar ribonucleoprotein complex(GO:0005732) |
| 0.0 | 0.2 | GO:0036156 | inner dynein arm(GO:0036156) |
| 0.0 | 0.2 | GO:0043020 | NADPH oxidase complex(GO:0043020) |
| 0.0 | 1.5 | GO:0071013 | catalytic step 2 spliceosome(GO:0071013) |
| 0.0 | 1.3 | GO:0005581 | collagen trimer(GO:0005581) |
| 0.0 | 1.1 | GO:0005811 | lipid particle(GO:0005811) |
| 0.0 | 1.2 | GO:0005681 | spliceosomal complex(GO:0005681) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.7 | 3.3 | GO:0042610 | CD8 receptor binding(GO:0042610) |
| 0.5 | 1.4 | GO:0038052 | type 1 metabotropic glutamate receptor binding(GO:0031798) RNA polymerase II transcription factor activity, estrogen-activated sequence-specific DNA binding(GO:0038052) |
| 0.4 | 2.6 | GO:0030492 | hemoglobin binding(GO:0030492) |
| 0.4 | 1.8 | GO:0031705 | bombesin receptor binding(GO:0031705) |
| 0.2 | 0.7 | GO:0035175 | histone kinase activity (H3-S10 specific)(GO:0035175) |
| 0.2 | 1.6 | GO:0001642 | group III metabotropic glutamate receptor activity(GO:0001642) |
| 0.2 | 1.9 | GO:0035381 | extracellular ATP-gated cation channel activity(GO:0004931) ATP-gated ion channel activity(GO:0035381) |
| 0.2 | 5.1 | GO:0003680 | AT DNA binding(GO:0003680) |
| 0.2 | 3.7 | GO:0001608 | G-protein coupled nucleotide receptor activity(GO:0001608) G-protein coupled purinergic nucleotide receptor activity(GO:0045028) |
| 0.2 | 2.7 | GO:0001206 | transcriptional repressor activity, RNA polymerase II distal enhancer sequence-specific binding(GO:0001206) |
| 0.1 | 0.4 | GO:0005137 | interleukin-5 receptor binding(GO:0005137) |
| 0.1 | 0.4 | GO:0005128 | erythropoietin receptor binding(GO:0005128) |
| 0.1 | 3.0 | GO:0005542 | folic acid binding(GO:0005542) |
| 0.1 | 0.4 | GO:0001537 | N-acetylgalactosamine 4-O-sulfotransferase activity(GO:0001537) |
| 0.1 | 0.6 | GO:0005332 | gamma-aminobutyric acid:sodium symporter activity(GO:0005332) |
| 0.1 | 4.1 | GO:0001965 | G-protein alpha-subunit binding(GO:0001965) |
| 0.1 | 0.9 | GO:0015280 | ligand-gated sodium channel activity(GO:0015280) |
| 0.1 | 1.3 | GO:0005522 | profilin binding(GO:0005522) |
| 0.1 | 0.5 | GO:0003938 | IMP dehydrogenase activity(GO:0003938) |
| 0.1 | 2.8 | GO:0005242 | inward rectifier potassium channel activity(GO:0005242) |
| 0.1 | 2.0 | GO:0031681 | G-protein beta-subunit binding(GO:0031681) |
| 0.1 | 1.6 | GO:0005537 | mannose binding(GO:0005537) |
| 0.1 | 1.9 | GO:0071855 | neuropeptide receptor binding(GO:0071855) |
| 0.1 | 0.6 | GO:0005113 | patched binding(GO:0005113) |
| 0.1 | 1.3 | GO:0005184 | neuropeptide hormone activity(GO:0005184) |
| 0.1 | 2.7 | GO:0005164 | tumor necrosis factor receptor binding(GO:0005164) |
| 0.0 | 1.1 | GO:0032794 | GTPase activating protein binding(GO:0032794) |
| 0.0 | 0.2 | GO:0043758 | acetate-CoA ligase (ADP-forming) activity(GO:0043758) |
| 0.0 | 0.5 | GO:0001730 | 2'-5'-oligoadenylate synthetase activity(GO:0001730) |
| 0.0 | 0.2 | GO:0044020 | protein-arginine omega-N monomethyltransferase activity(GO:0035241) histone methyltransferase activity (H4-R3 specific)(GO:0044020) |
| 0.0 | 1.0 | GO:0004181 | metallocarboxypeptidase activity(GO:0004181) |
| 0.0 | 0.9 | GO:0070577 | lysine-acetylated histone binding(GO:0070577) |
| 0.0 | 1.4 | GO:0071889 | 14-3-3 protein binding(GO:0071889) |
| 0.0 | 1.5 | GO:0031624 | ubiquitin conjugating enzyme binding(GO:0031624) |
| 0.0 | 0.2 | GO:0008093 | cytoskeletal adaptor activity(GO:0008093) |
| 0.0 | 1.5 | GO:0005160 | transforming growth factor beta receptor binding(GO:0005160) |
| 0.0 | 0.8 | GO:0008392 | arachidonic acid epoxygenase activity(GO:0008392) |
| 0.0 | 4.4 | GO:0001046 | core promoter sequence-specific DNA binding(GO:0001046) |
| 0.0 | 0.4 | GO:0070034 | telomerase RNA binding(GO:0070034) |
| 0.0 | 1.5 | GO:0003724 | RNA helicase activity(GO:0003724) ATP-dependent RNA helicase activity(GO:0004004) |
| 0.0 | 1.4 | GO:0004402 | histone acetyltransferase activity(GO:0004402) |
| 0.0 | 0.2 | GO:0019788 | NEDD8 transferase activity(GO:0019788) |
| 0.0 | 0.6 | GO:0030676 | Rac guanyl-nucleotide exchange factor activity(GO:0030676) |
| 0.0 | 1.2 | GO:0005507 | copper ion binding(GO:0005507) |
| 0.0 | 0.2 | GO:0016175 | superoxide-generating NADPH oxidase activity(GO:0016175) |
| 0.0 | 7.0 | GO:0005543 | phospholipid binding(GO:0005543) |
| 0.0 | 3.2 | GO:0019887 | protein kinase regulator activity(GO:0019887) |
| 0.0 | 0.5 | GO:0005109 | frizzled binding(GO:0005109) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 3.2 | PID IL8 CXCR2 PATHWAY | IL8- and CXCR2-mediated signaling events |
| 0.1 | 6.5 | PID P38 ALPHA BETA DOWNSTREAM PATHWAY | Signaling mediated by p38-alpha and p38-beta |
| 0.1 | 3.3 | PID NFKAPPAB ATYPICAL PATHWAY | Atypical NF-kappaB pathway |
| 0.1 | 1.6 | PID CIRCADIAN PATHWAY | Circadian rhythm pathway |
| 0.1 | 3.1 | PID TCR CALCIUM PATHWAY | Calcium signaling in the CD4+ TCR pathway |
| 0.0 | 1.6 | PID LIS1 PATHWAY | Lissencephaly gene (LIS1) in neuronal migration and development |
| 0.0 | 0.6 | PID HEDGEHOG 2PATHWAY | Signaling events mediated by the Hedgehog family |
| 0.0 | 0.4 | PID IL5 PATHWAY | IL5-mediated signaling events |
| 0.0 | 2.0 | PID TCR PATHWAY | TCR signaling in naïve CD4+ T cells |
| 0.0 | 0.4 | PID EPO PATHWAY | EPO signaling pathway |
| 0.0 | 0.6 | PID REELIN PATHWAY | Reelin signaling pathway |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 3.3 | REACTOME TRANSLOCATION OF ZAP 70 TO IMMUNOLOGICAL SYNAPSE | Genes involved in Translocation of ZAP-70 to Immunological synapse |
| 0.2 | 3.7 | REACTOME P2Y RECEPTORS | Genes involved in P2Y receptors |
| 0.1 | 0.8 | REACTOME CREATION OF C4 AND C2 ACTIVATORS | Genes involved in Creation of C4 and C2 activators |
| 0.1 | 4.2 | REACTOME ERK MAPK TARGETS | Genes involved in ERK/MAPK targets |
| 0.1 | 1.6 | REACTOME CLASS C 3 METABOTROPIC GLUTAMATE PHEROMONE RECEPTORS | Genes involved in Class C/3 (Metabotropic glutamate/pheromone receptors) |
| 0.1 | 2.0 | REACTOME GENERATION OF SECOND MESSENGER MOLECULES | Genes involved in Generation of second messenger molecules |
| 0.1 | 1.3 | REACTOME MRNA DECAY BY 5 TO 3 EXORIBONUCLEASE | Genes involved in mRNA Decay by 5' to 3' Exoribonuclease |
| 0.1 | 3.2 | REACTOME GPVI MEDIATED ACTIVATION CASCADE | Genes involved in GPVI-mediated activation cascade |
| 0.1 | 2.8 | REACTOME INHIBITION OF VOLTAGE GATED CA2 CHANNELS VIA GBETA GAMMA SUBUNITS | Genes involved in Inhibition of voltage gated Ca2+ channels via Gbeta/gamma subunits |
| 0.0 | 0.7 | REACTOME SYNTHESIS OF PIPS AT THE LATE ENDOSOME MEMBRANE | Genes involved in Synthesis of PIPs at the late endosome membrane |
| 0.0 | 8.0 | REACTOME G ALPHA I SIGNALLING EVENTS | Genes involved in G alpha (i) signalling events |
| 0.0 | 1.6 | REACTOME MEIOTIC SYNAPSIS | Genes involved in Meiotic Synapsis |
| 0.0 | 0.5 | REACTOME PURINE RIBONUCLEOSIDE MONOPHOSPHATE BIOSYNTHESIS | Genes involved in Purine ribonucleoside monophosphate biosynthesis |
| 0.0 | 0.4 | REACTOME VITAMIN B5 PANTOTHENATE METABOLISM | Genes involved in Vitamin B5 (pantothenate) metabolism |
| 0.0 | 0.6 | REACTOME NA CL DEPENDENT NEUROTRANSMITTER TRANSPORTERS | Genes involved in Na+/Cl- dependent neurotransmitter transporters |
| 0.0 | 0.9 | REACTOME ADHERENS JUNCTIONS INTERACTIONS | Genes involved in Adherens junctions interactions |
| 0.0 | 0.5 | REACTOME DCC MEDIATED ATTRACTIVE SIGNALING | Genes involved in DCC mediated attractive signaling |
| 0.0 | 1.8 | REACTOME PEPTIDE LIGAND BINDING RECEPTORS | Genes involved in Peptide ligand-binding receptors |
| 0.0 | 0.5 | REACTOME TRAFFICKING OF GLUR2 CONTAINING AMPA RECEPTORS | Genes involved in Trafficking of GluR2-containing AMPA receptors |
| 0.0 | 1.3 | REACTOME NUCLEAR SIGNALING BY ERBB4 | Genes involved in Nuclear signaling by ERBB4 |
| 0.0 | 1.1 | REACTOME INTERFERON GAMMA SIGNALING | Genes involved in Interferon gamma signaling |
| 0.0 | 1.6 | REACTOME L1CAM INTERACTIONS | Genes involved in L1CAM interactions |
| 0.0 | 0.4 | REACTOME IL RECEPTOR SHC SIGNALING | Genes involved in Interleukin receptor SHC signaling |