Project
PRJNA375882: Comprehensive Mouse Transcriptomic BodyMap
Navigation
Downloads
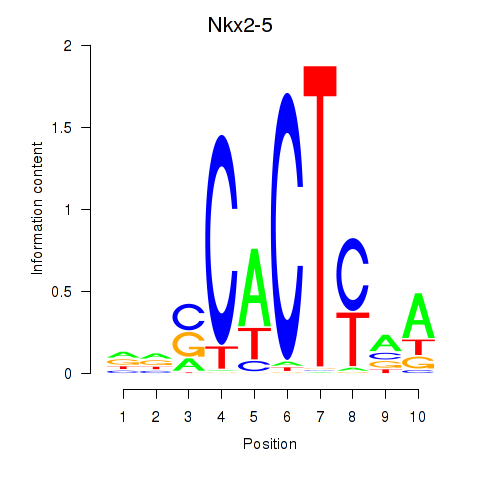
Results for Nkx2-5
Z-value: 0.63
Transcription factors associated with Nkx2-5
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Nkx2-5
|
ENSMUSG00000015579.6 | Nkx2-5 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Nkx2-5 | mm39_v1_chr17_-_27060539_27060565 | 0.13 | 2.8e-01 | Click! |
Activity profile of Nkx2-5 motif
Sorted Z-values of Nkx2-5 motif
Network of associatons between targets according to the STRING database.
First level regulatory network of Nkx2-5
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr14_+_40853734 | 6.73 |
ENSMUST00000022314.10
ENSMUST00000170719.2 |
Sftpa1
|
surfactant associated protein A1 |
| chr6_+_30639217 | 5.29 |
ENSMUST00000031806.10
|
Cpa1
|
carboxypeptidase A1, pancreatic |
| chr6_+_41279199 | 4.93 |
ENSMUST00000031913.5
|
Try4
|
trypsin 4 |
| chr6_+_72281587 | 4.53 |
ENSMUST00000183018.8
ENSMUST00000182014.10 |
Sftpb
|
surfactant associated protein B |
| chr4_-_137137088 | 2.99 |
ENSMUST00000024200.7
|
Cela3a
|
chymotrypsin-like elastase family, member 3A |
| chr6_+_30541581 | 2.93 |
ENSMUST00000096066.5
|
Cpa2
|
carboxypeptidase A2, pancreatic |
| chr12_-_115611981 | 2.35 |
ENSMUST00000103540.3
ENSMUST00000199266.2 |
Ighv8-12
|
immunoglobulin heavy variable V8-12 |
| chr12_-_114117264 | 2.31 |
ENSMUST00000103461.5
|
Ighv7-3
|
immunoglobulin heavy variable 7-3 |
| chr12_-_113958518 | 2.26 |
ENSMUST00000103467.2
|
Ighv14-2
|
immunoglobulin heavy variable 14-2 |
| chr12_-_114286421 | 2.13 |
ENSMUST00000103483.3
|
Ighv3-8
|
immunoglobulin heavy variable V3-8 |
| chr15_-_82108531 | 1.81 |
ENSMUST00000109535.3
ENSMUST00000089161.10 |
Tnfrsf13c
|
tumor necrosis factor receptor superfamily, member 13c |
| chr1_-_170755136 | 1.69 |
ENSMUST00000046322.14
ENSMUST00000159171.2 |
Fcrla
|
Fc receptor-like A |
| chr1_-_170755109 | 1.61 |
ENSMUST00000162136.2
ENSMUST00000162887.2 |
Fcrla
|
Fc receptor-like A |
| chr6_-_136899167 | 1.52 |
ENSMUST00000032343.7
|
Erp27
|
endoplasmic reticulum protein 27 |
| chr4_+_119494901 | 1.49 |
ENSMUST00000024015.3
|
Guca2a
|
guanylate cyclase activator 2a (guanylin) |
| chr12_-_115299134 | 1.45 |
ENSMUST00000195359.6
ENSMUST00000103530.3 |
Ighv1-59
|
immunoglobulin heavy variable V1-59 |
| chr6_+_127554931 | 1.29 |
ENSMUST00000071563.5
|
Cracr2a
|
calcium release activated channel regulator 2A |
| chr14_+_51366306 | 1.14 |
ENSMUST00000226210.2
|
Rnase6
|
ribonuclease, RNase A family, 6 |
| chr14_+_51366512 | 1.13 |
ENSMUST00000095923.4
|
Rnase6
|
ribonuclease, RNase A family, 6 |
| chr9_+_123635529 | 1.11 |
ENSMUST00000049810.9
|
Cxcr6
|
chemokine (C-X-C motif) receptor 6 |
| chr9_+_123635556 | 1.05 |
ENSMUST00000216072.2
|
Cxcr6
|
chemokine (C-X-C motif) receptor 6 |
| chr3_+_3699205 | 0.98 |
ENSMUST00000108394.3
|
Hnf4g
|
hepatocyte nuclear factor 4, gamma |
| chr6_-_41535322 | 0.87 |
ENSMUST00000193003.2
|
Trbv31
|
T cell receptor beta, variable 31 |
| chr5_+_28370687 | 0.76 |
ENSMUST00000036177.9
|
En2
|
engrailed 2 |
| chr11_+_32483290 | 0.69 |
ENSMUST00000102821.4
|
Stk10
|
serine/threonine kinase 10 |
| chr3_-_69767208 | 0.68 |
ENSMUST00000171529.4
ENSMUST00000051239.13 |
Sptssb
|
serine palmitoyltransferase, small subunit B |
| chrX_-_140508177 | 0.64 |
ENSMUST00000067841.8
|
Irs4
|
insulin receptor substrate 4 |
| chr14_+_53743184 | 0.62 |
ENSMUST00000103583.5
|
Trav10
|
T cell receptor alpha variable 10 |
| chr12_-_114186874 | 0.61 |
ENSMUST00000103477.4
ENSMUST00000192499.3 |
Ighv7-4
|
immunoglobulin heavy variable 7-4 |
| chr11_+_87457479 | 0.60 |
ENSMUST00000239011.2
|
Septin4
|
septin 4 |
| chr1_+_172168764 | 0.53 |
ENSMUST00000056136.4
|
Kcnj10
|
potassium inwardly-rectifying channel, subfamily J, member 10 |
| chr14_+_53698556 | 0.48 |
ENSMUST00000181728.3
|
Trav7-4
|
T cell receptor alpha variable 7-4 |
| chr5_+_16758777 | 0.48 |
ENSMUST00000030683.8
|
Hgf
|
hepatocyte growth factor |
| chr15_-_5273659 | 0.46 |
ENSMUST00000047379.15
|
Ptger4
|
prostaglandin E receptor 4 (subtype EP4) |
| chr11_+_64326141 | 0.46 |
ENSMUST00000058652.6
|
Hs3st3a1
|
heparan sulfate (glucosamine) 3-O-sulfotransferase 3A1 |
| chr15_-_5273645 | 0.45 |
ENSMUST00000120563.2
|
Ptger4
|
prostaglandin E receptor 4 (subtype EP4) |
| chr17_+_27152276 | 0.43 |
ENSMUST00000237412.2
|
Phf1
|
PHD finger protein 1 |
| chr19_+_53131187 | 0.41 |
ENSMUST00000050096.15
ENSMUST00000237832.2 |
Add3
|
adducin 3 (gamma) |
| chr11_+_67061908 | 0.41 |
ENSMUST00000018641.8
|
Myh2
|
myosin, heavy polypeptide 2, skeletal muscle, adult |
| chr15_+_66542598 | 0.40 |
ENSMUST00000065916.14
|
Tg
|
thyroglobulin |
| chr4_-_45532470 | 0.40 |
ENSMUST00000147448.2
|
Shb
|
src homology 2 domain-containing transforming protein B |
| chr14_-_8798841 | 0.40 |
ENSMUST00000061045.3
|
Sntn
|
sentan, cilia apical structure protein |
| chr9_-_53447908 | 0.39 |
ENSMUST00000150244.2
|
Atm
|
ataxia telangiectasia mutated |
| chr11_-_78441584 | 0.39 |
ENSMUST00000103242.5
|
Tmem97
|
transmembrane protein 97 |
| chr10_-_116732813 | 0.39 |
ENSMUST00000048229.9
|
Myrfl
|
myelin regulatory factor-like |
| chr11_+_67061837 | 0.39 |
ENSMUST00000170159.8
|
Myh2
|
myosin, heavy polypeptide 2, skeletal muscle, adult |
| chr12_+_103281188 | 0.38 |
ENSMUST00000189885.2
ENSMUST00000179363.2 |
Fam181a
|
family with sequence similarity 181, member A |
| chr7_-_97794679 | 0.38 |
ENSMUST00000098281.4
|
Omp
|
olfactory marker protein |
| chr3_-_49711765 | 0.37 |
ENSMUST00000035931.13
|
Pcdh18
|
protocadherin 18 |
| chr6_-_23650205 | 0.36 |
ENSMUST00000115354.2
|
Rnf133
|
ring finger protein 133 |
| chr16_+_43993599 | 0.36 |
ENSMUST00000119746.8
ENSMUST00000088356.10 ENSMUST00000169582.3 |
Usf3
|
upstream transcription factor family member 3 |
| chr6_-_23650297 | 0.35 |
ENSMUST00000063548.4
|
Rnf133
|
ring finger protein 133 |
| chr5_+_16758897 | 0.33 |
ENSMUST00000196645.2
|
Hgf
|
hepatocyte growth factor |
| chr6_-_41535292 | 0.33 |
ENSMUST00000103300.3
|
Trbv31
|
T cell receptor beta, variable 31 |
| chr8_+_114369838 | 0.32 |
ENSMUST00000095173.3
ENSMUST00000034219.12 ENSMUST00000212269.2 |
Syce1l
|
synaptonemal complex central element protein 1 like |
| chr14_+_80237691 | 0.31 |
ENSMUST00000228749.2
ENSMUST00000088735.4 |
Olfm4
|
olfactomedin 4 |
| chr8_-_33374282 | 0.31 |
ENSMUST00000209107.4
ENSMUST00000209022.3 |
Nrg1
|
neuregulin 1 |
| chr14_-_78326435 | 0.30 |
ENSMUST00000118785.3
ENSMUST00000066437.5 |
Fam216b
|
family with sequence similarity 216, member B |
| chr5_-_124465875 | 0.28 |
ENSMUST00000184951.8
|
Mphosph9
|
M-phase phosphoprotein 9 |
| chr13_-_64645606 | 0.28 |
ENSMUST00000180282.2
|
Fam240b
|
family with sequence similarity 240 member B |
| chr10_+_60235628 | 0.27 |
ENSMUST00000218974.2
|
Gm17455
|
predicted gene, 17455 |
| chr12_-_114909863 | 0.26 |
ENSMUST00000103517.2
ENSMUST00000195417.2 |
Ighv1-43
|
immunoglobulin heavy variable V1-43 |
| chr9_+_53212871 | 0.26 |
ENSMUST00000051014.2
|
Exph5
|
exophilin 5 |
| chr1_-_163552693 | 0.26 |
ENSMUST00000159679.8
|
Mettl11b
|
methyltransferase like 11B |
| chr3_-_153430741 | 0.26 |
ENSMUST00000064460.7
ENSMUST00000200397.5 |
St6galnac3
|
ST6 (alpha-N-acetyl-neuraminyl-2,3-beta-galactosyl-1,3)-N-acetylgalactosaminide alpha-2,6-sialyltransferase 3 |
| chr8_+_125519751 | 0.26 |
ENSMUST00000041106.9
ENSMUST00000167588.9 |
Trim67
|
tripartite motif-containing 67 |
| chr10_-_60667161 | 0.25 |
ENSMUST00000218637.2
|
Unc5b
|
unc-5 netrin receptor B |
| chr4_-_87724533 | 0.24 |
ENSMUST00000126353.8
ENSMUST00000149357.8 |
Mllt3
|
myeloid/lymphoid or mixed-lineage leukemia; translocated to, 3 |
| chr15_+_101071948 | 0.23 |
ENSMUST00000000544.12
|
Acvr1b
|
activin A receptor, type 1B |
| chr11_+_101877876 | 0.23 |
ENSMUST00000010985.8
|
Cfap97d1
|
CFAP97 domain containing 1 |
| chr7_+_28869770 | 0.22 |
ENSMUST00000033886.8
ENSMUST00000209019.2 ENSMUST00000208330.2 |
Ggn
|
gametogenetin |
| chr11_-_96868483 | 0.22 |
ENSMUST00000107624.8
|
Sp2
|
Sp2 transcription factor |
| chr5_-_124465946 | 0.21 |
ENSMUST00000031344.13
|
Mphosph9
|
M-phase phosphoprotein 9 |
| chr7_-_83304698 | 0.21 |
ENSMUST00000145610.8
|
Il16
|
interleukin 16 |
| chr14_-_60488894 | 0.21 |
ENSMUST00000225111.2
|
Gm49336
|
predicted gene, 49336 |
| chr7_-_126496534 | 0.21 |
ENSMUST00000120007.8
|
Tmem219
|
transmembrane protein 219 |
| chr4_-_19122526 | 0.20 |
ENSMUST00000137780.2
|
Cnbd1
|
cyclic nucleotide binding domain containing 1 |
| chr14_+_53328803 | 0.20 |
ENSMUST00000103609.2
|
Trav7n-4
|
T cell receptor alpha variable 7N-4 |
| chr13_+_111391544 | 0.18 |
ENSMUST00000054716.4
|
Actbl2
|
actin, beta-like 2 |
| chr4_-_87724512 | 0.18 |
ENSMUST00000148059.2
|
Mllt3
|
myeloid/lymphoid or mixed-lineage leukemia; translocated to, 3 |
| chr5_-_82271183 | 0.17 |
ENSMUST00000186079.2
ENSMUST00000185607.2 |
1700031L13Rik
|
RIKEN cDNA 1700031L13 gene |
| chr10_+_36383008 | 0.17 |
ENSMUST00000168572.8
|
Hs3st5
|
heparan sulfate (glucosamine) 3-O-sulfotransferase 5 |
| chr14_+_53007210 | 0.17 |
ENSMUST00000178768.4
|
Trav7d-4
|
T cell receptor alpha variable 7D-4 |
| chrX_+_12937714 | 0.17 |
ENSMUST00000169594.9
ENSMUST00000089302.11 |
Usp9x
|
ubiquitin specific peptidase 9, X chromosome |
| chr15_+_76235503 | 0.16 |
ENSMUST00000023212.15
ENSMUST00000160172.8 |
Maf1
|
MAF1 homolog, negative regulator of RNA polymerase III |
| chr7_+_28869629 | 0.16 |
ENSMUST00000098609.4
|
Ggn
|
gametogenetin |
| chr18_+_49965689 | 0.16 |
ENSMUST00000180611.8
|
Dmxl1
|
Dmx-like 1 |
| chr16_-_89368059 | 0.15 |
ENSMUST00000171542.2
|
Krtap11-1
|
keratin associated protein 11-1 |
| chr1_-_133589020 | 0.15 |
ENSMUST00000193504.6
ENSMUST00000195067.2 ENSMUST00000191896.6 ENSMUST00000194668.6 ENSMUST00000195424.6 ENSMUST00000179598.4 ENSMUST00000027736.13 |
Zc3h11a
Gm38394
Zc3h11a
|
zinc finger CCCH type containing 11A predicted gene, 38394 zinc finger CCCH type containing 11A |
| chrX_+_41149264 | 0.15 |
ENSMUST00000224454.2
|
Xiap
|
X-linked inhibitor of apoptosis |
| chr9_-_119852624 | 0.14 |
ENSMUST00000111635.4
|
Xirp1
|
xin actin-binding repeat containing 1 |
| chr14_+_53073315 | 0.14 |
ENSMUST00000197128.2
|
Trav7d-5
|
T cell receptor alpha variable 7D-5 |
| chr6_+_42263644 | 0.14 |
ENSMUST00000163936.8
|
Clcn1
|
chloride channel, voltage-sensitive 1 |
| chr11_+_87457544 | 0.14 |
ENSMUST00000060360.7
|
Septin4
|
septin 4 |
| chr16_+_33504908 | 0.14 |
ENSMUST00000126532.2
|
Heg1
|
heart development protein with EGF-like domains 1 |
| chr14_-_60488922 | 0.13 |
ENSMUST00000225311.2
ENSMUST00000041905.8 |
Gm49336
|
predicted gene, 49336 |
| chr6_-_52222776 | 0.13 |
ENSMUST00000048026.10
|
Hoxa11
|
homeobox A11 |
| chr11_+_23234644 | 0.13 |
ENSMUST00000150750.3
|
Xpo1
|
exportin 1 |
| chr6_-_90013823 | 0.13 |
ENSMUST00000073415.2
|
Vmn1r48
|
vomeronasal 1 receptor 48 |
| chr11_+_68858942 | 0.12 |
ENSMUST00000102606.10
ENSMUST00000018884.6 |
Slc25a35
|
solute carrier family 25, member 35 |
| chrX_-_91164377 | 0.12 |
ENSMUST00000088137.3
|
Mageb18
|
MAGE family member B18 |
| chr11_+_49334680 | 0.12 |
ENSMUST00000055584.2
|
Olfr1388
|
olfactory receptor 1388 |
| chr11_+_49321254 | 0.11 |
ENSMUST00000203369.2
|
Olfr1389
|
olfactory receptor 1389 |
| chrX_-_154123743 | 0.11 |
ENSMUST00000130349.3
|
Prdx4
|
peroxiredoxin 4 |
| chr2_-_120439764 | 0.10 |
ENSMUST00000102496.8
|
Lrrc57
|
leucine rich repeat containing 57 |
| chr10_+_60236532 | 0.10 |
ENSMUST00000218360.2
|
Gm17455
|
predicted gene, 17455 |
| chr6_-_5193816 | 0.10 |
ENSMUST00000002663.12
|
Pon1
|
paraoxonase 1 |
| chr5_-_71815218 | 0.10 |
ENSMUST00000198138.2
|
Gabra4
|
gamma-aminobutyric acid (GABA) A receptor, subunit alpha 4 |
| chrX_-_7765096 | 0.09 |
ENSMUST00000154552.2
|
Pqbp1
|
polyglutamine binding protein 1 |
| chr2_-_73316809 | 0.09 |
ENSMUST00000141264.8
|
Wipf1
|
WAS/WASL interacting protein family, member 1 |
| chr6_-_115830249 | 0.09 |
ENSMUST00000122816.3
|
Mbd4
|
methyl-CpG binding domain protein 4 |
| chr6_-_28421678 | 0.09 |
ENSMUST00000090511.4
|
Gcc1
|
golgi coiled coil 1 |
| chr8_-_46747629 | 0.09 |
ENSMUST00000058636.9
|
Helt
|
helt bHLH transcription factor |
| chr7_-_134100659 | 0.08 |
ENSMUST00000238294.2
|
D7Ertd443e
|
DNA segment, Chr 7, ERATO Doi 443, expressed |
| chr15_-_10470575 | 0.08 |
ENSMUST00000136591.8
|
Dnajc21
|
DnaJ heat shock protein family (Hsp40) member C21 |
| chr9_-_50657800 | 0.07 |
ENSMUST00000239417.2
ENSMUST00000034564.4 |
2310030G06Rik
|
RIKEN cDNA 2310030G06 gene |
| chr11_+_115865535 | 0.07 |
ENSMUST00000021107.14
ENSMUST00000169928.8 ENSMUST00000106461.8 |
Itgb4
|
integrin beta 4 |
| chr16_-_56748424 | 0.07 |
ENSMUST00000210579.2
|
Lnp1
|
leukemia NUP98 fusion partner 1 |
| chr15_-_76419131 | 0.07 |
ENSMUST00000230604.2
|
Tmem249
|
transmembrane protein 249 |
| chr2_+_89642395 | 0.06 |
ENSMUST00000214508.2
|
Olfr1255
|
olfactory receptor 1255 |
| chr11_+_73125118 | 0.06 |
ENSMUST00000102526.9
|
Trpv1
|
transient receptor potential cation channel, subfamily V, member 1 |
| chr17_-_56916771 | 0.05 |
ENSMUST00000052832.6
|
Micos13
|
mitochondrial contact site and cristae organizing system subunit 13 |
| chr10_+_79824418 | 0.05 |
ENSMUST00000004784.11
ENSMUST00000105374.2 |
Cnn2
|
calponin 2 |
| chr10_+_22520910 | 0.05 |
ENSMUST00000042261.5
|
Slc2a12
|
solute carrier family 2 (facilitated glucose transporter), member 12 |
| chr3_-_129597679 | 0.05 |
ENSMUST00000185462.7
ENSMUST00000179187.2 |
Lrit3
|
leucine-rich repeat, immunoglobulin-like and transmembrane domains 3 |
| chr5_-_8417982 | 0.04 |
ENSMUST00000088761.11
ENSMUST00000115386.8 ENSMUST00000050166.14 ENSMUST00000046838.14 ENSMUST00000115388.9 ENSMUST00000088744.12 ENSMUST00000115385.2 |
Adam22
|
a disintegrin and metallopeptidase domain 22 |
| chrX_-_74460168 | 0.04 |
ENSMUST00000033543.14
ENSMUST00000149863.3 ENSMUST00000114081.2 |
Cmc4
Mtcp1
|
C-x(9)-C motif containing 4 mature T cell proliferation 1 |
| chr12_-_115531211 | 0.04 |
ENSMUST00000103536.4
|
Ighv8-11
|
immunoglobulin heavy variable V8-11 |
| chr6_+_136530970 | 0.03 |
ENSMUST00000189535.2
|
Atf7ip
|
activating transcription factor 7 interacting protein |
| chr7_-_4909515 | 0.03 |
ENSMUST00000210663.2
|
Gm36210
|
predicted gene, 36210 |
| chr9_+_7347369 | 0.03 |
ENSMUST00000005950.12
ENSMUST00000065079.6 |
Mmp12
|
matrix metallopeptidase 12 |
| chr16_+_44764028 | 0.03 |
ENSMUST00000114613.9
ENSMUST00000114612.8 ENSMUST00000077178.13 ENSMUST00000048479.14 ENSMUST00000114611.10 |
Cd200r3
|
CD200 receptor 3 |
| chr6_+_92846338 | 0.03 |
ENSMUST00000113434.2
|
Gm15737
|
predicted gene 15737 |
| chr11_-_99907030 | 0.03 |
ENSMUST00000018399.3
|
Krt33a
|
keratin 33A |
| chrX_-_164110372 | 0.02 |
ENSMUST00000058787.9
|
Glra2
|
glycine receptor, alpha 2 subunit |
| chr2_-_111251294 | 0.02 |
ENSMUST00000099617.2
|
Olfr1286
|
olfactory receptor 1286 |
| chr3_-_129834788 | 0.01 |
ENSMUST00000168644.3
|
Sec24b
|
Sec24 related gene family, member B (S. cerevisiae) |
| chr16_+_44764099 | 0.00 |
ENSMUST00000114622.10
ENSMUST00000166731.2 |
Cd200r3
|
CD200 receptor 3 |
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.6 | 6.7 | GO:0008228 | opsonization(GO:0008228) |
| 0.3 | 0.9 | GO:1904464 | negative regulation of integrin activation(GO:0033624) regulation of matrix metallopeptidase secretion(GO:1904464) matrix metallopeptidase secretion(GO:1990773) positive regulation of ovarian follicle development(GO:2000386) regulation of antral ovarian follicle growth(GO:2000387) positive regulation of antral ovarian follicle growth(GO:2000388) negative regulation of eosinophil migration(GO:2000417) |
| 0.3 | 1.8 | GO:0031296 | B cell costimulation(GO:0031296) |
| 0.2 | 1.3 | GO:0032237 | activation of store-operated calcium channel activity(GO:0032237) |
| 0.1 | 0.5 | GO:0051935 | amino acid neurotransmitter reuptake(GO:0051933) glutamate reuptake(GO:0051935) |
| 0.1 | 0.4 | GO:0072434 | signal transduction involved in G2 DNA damage checkpoint(GO:0072425) signal transduction involved in mitotic G2 DNA damage checkpoint(GO:0072434) |
| 0.1 | 0.4 | GO:0061086 | negative regulation of histone H3-K27 methylation(GO:0061086) |
| 0.1 | 0.2 | GO:0021698 | cerebellar cortex structural organization(GO:0021698) |
| 0.1 | 0.7 | GO:1904220 | regulation of serine C-palmitoyltransferase activity(GO:1904220) |
| 0.1 | 0.8 | GO:0060665 | regulation of branching involved in salivary gland morphogenesis by mesenchymal-epithelial signaling(GO:0060665) |
| 0.1 | 8.8 | GO:0006910 | phagocytosis, recognition(GO:0006910) |
| 0.1 | 4.5 | GO:0007585 | respiratory gaseous exchange(GO:0007585) |
| 0.0 | 0.2 | GO:1901165 | positive regulation of trophoblast cell migration(GO:1901165) |
| 0.0 | 0.8 | GO:0001778 | plasma membrane repair(GO:0001778) |
| 0.0 | 0.8 | GO:1990403 | embryonic brain development(GO:1990403) |
| 0.0 | 0.3 | GO:0071985 | multivesicular body sorting pathway(GO:0071985) |
| 0.0 | 0.1 | GO:1902528 | regulation of protein linear polyubiquitination(GO:1902528) positive regulation of protein linear polyubiquitination(GO:1902530) |
| 0.0 | 0.4 | GO:0015705 | iodide transport(GO:0015705) |
| 0.0 | 0.4 | GO:1900194 | negative regulation of oocyte maturation(GO:1900194) |
| 0.0 | 0.1 | GO:0003017 | lymph circulation(GO:0003017) |
| 0.0 | 0.1 | GO:1902617 | response to fluoride(GO:1902617) |
| 0.0 | 0.3 | GO:0021840 | directional guidance of interneurons involved in migration from the subpallium to the cortex(GO:0021840) chemorepulsion involved in interneuron migration from the subpallium to the cortex(GO:0021842) |
| 0.0 | 0.2 | GO:0015015 | heparan sulfate proteoglycan biosynthetic process, enzymatic modification(GO:0015015) |
| 0.0 | 2.3 | GO:0090501 | RNA phosphodiester bond hydrolysis(GO:0090501) |
| 0.0 | 0.4 | GO:0007379 | segment specification(GO:0007379) |
| 0.0 | 0.2 | GO:0033564 | anterior/posterior axon guidance(GO:0033564) |
| 0.0 | 0.1 | GO:0022417 | protein maturation by protein folding(GO:0022417) |
| 0.0 | 3.7 | GO:0007586 | digestion(GO:0007586) |
| 0.0 | 0.3 | GO:0051324 | meiotic prophase I(GO:0007128) prophase(GO:0051324) |
| 0.0 | 0.2 | GO:0016480 | negative regulation of transcription from RNA polymerase III promoter(GO:0016480) |
| 0.0 | 2.3 | GO:0008203 | cholesterol metabolic process(GO:0008203) |
| 0.0 | 0.1 | GO:0000056 | ribosomal small subunit export from nucleus(GO:0000056) |
| 0.0 | 0.4 | GO:0006491 | N-glycan processing(GO:0006491) |
| 0.0 | 0.7 | GO:2000401 | regulation of lymphocyte migration(GO:2000401) |
| 0.0 | 0.2 | GO:0070072 | vacuolar proton-transporting V-type ATPase complex assembly(GO:0070072) |
| 0.0 | 0.4 | GO:0016339 | calcium-dependent cell-cell adhesion via plasma membrane cell adhesion molecules(GO:0016339) |
| 0.0 | 0.2 | GO:0050930 | induction of positive chemotaxis(GO:0050930) |
| 0.0 | 0.1 | GO:0060272 | embryonic skeletal joint morphogenesis(GO:0060272) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 4.5 | GO:0097208 | alveolar lamellar body(GO:0097208) |
| 0.1 | 6.7 | GO:0005771 | multivesicular body(GO:0005771) |
| 0.1 | 0.8 | GO:0005826 | actomyosin contractile ring(GO:0005826) |
| 0.1 | 0.2 | GO:0048179 | activin receptor complex(GO:0048179) |
| 0.1 | 0.7 | GO:0031211 | serine C-palmitoyltransferase complex(GO:0017059) endoplasmic reticulum palmitoyltransferase complex(GO:0031211) |
| 0.1 | 8.8 | GO:0042571 | immunoglobulin complex, circulating(GO:0042571) |
| 0.1 | 0.2 | GO:0043291 | RAVE complex(GO:0043291) |
| 0.0 | 0.5 | GO:0097449 | astrocyte projection(GO:0097449) |
| 0.0 | 0.4 | GO:0035098 | ESC/E(Z) complex(GO:0035098) |
| 0.0 | 0.3 | GO:0042581 | specific granule(GO:0042581) |
| 0.0 | 0.1 | GO:0005642 | annulate lamellae(GO:0005642) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 2.2 | GO:0019958 | C-X-C chemokine binding(GO:0019958) |
| 0.3 | 8.2 | GO:0004181 | metallocarboxypeptidase activity(GO:0004181) |
| 0.2 | 1.5 | GO:0010853 | cyclase activator activity(GO:0010853) guanylate cyclase activator activity(GO:0030250) |
| 0.1 | 0.7 | GO:0004758 | serine C-palmitoyltransferase activity(GO:0004758) C-palmitoyltransferase activity(GO:0016454) |
| 0.1 | 0.4 | GO:0004677 | DNA-dependent protein kinase activity(GO:0004677) |
| 0.1 | 0.9 | GO:0004957 | prostaglandin E receptor activity(GO:0004957) |
| 0.1 | 0.6 | GO:0008467 | [heparan sulfate]-glucosamine 3-sulfotransferase 1 activity(GO:0008467) |
| 0.1 | 0.5 | GO:0015272 | ATP-activated inward rectifier potassium channel activity(GO:0015272) |
| 0.1 | 8.8 | GO:0034987 | immunoglobulin receptor binding(GO:0034987) |
| 0.0 | 8.4 | GO:0004252 | serine-type endopeptidase activity(GO:0004252) |
| 0.0 | 0.2 | GO:0016361 | activin receptor activity, type I(GO:0016361) |
| 0.0 | 0.2 | GO:0001032 | RNA polymerase III type 3 promoter DNA binding(GO:0001032) |
| 0.0 | 0.2 | GO:0005042 | netrin receptor activity(GO:0005042) |
| 0.0 | 0.4 | GO:0004571 | mannosyl-oligosaccharide 1,2-alpha-mannosidase activity(GO:0004571) |
| 0.0 | 0.1 | GO:0004063 | aryldialkylphosphatase activity(GO:0004063) |
| 0.0 | 0.3 | GO:0005176 | ErbB-2 class receptor binding(GO:0005176) |
| 0.0 | 0.2 | GO:0001665 | alpha-N-acetylgalactosaminide alpha-2,6-sialyltransferase activity(GO:0001665) |
| 0.0 | 2.3 | GO:0004540 | ribonuclease activity(GO:0004540) |
| 0.0 | 0.2 | GO:0042609 | CD4 receptor binding(GO:0042609) |
| 0.0 | 0.6 | GO:0043548 | phosphatidylinositol 3-kinase binding(GO:0043548) |
| 0.0 | 3.9 | GO:0001664 | G-protein coupled receptor binding(GO:0001664) |
| 0.0 | 0.1 | GO:0005049 | nuclear export signal receptor activity(GO:0005049) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 6.7 | PID TOLL ENDOGENOUS PATHWAY | Endogenous TLR signaling |
| 0.0 | 4.5 | NABA ECM AFFILIATED | Genes encoding proteins affiliated structurally or functionally to extracellular matrix proteins |
| 0.0 | 0.8 | PID INTEGRIN A4B1 PATHWAY | Alpha4 beta1 integrin signaling events |
| 0.0 | 0.5 | PID NFKAPPAB CANONICAL PATHWAY | Canonical NF-kappaB pathway |
| 0.0 | 0.8 | PID UPA UPAR PATHWAY | Urokinase-type plasminogen activator (uPA) and uPAR-mediated signaling |
| 0.0 | 0.3 | PID ERBB NETWORK PATHWAY | ErbB receptor signaling network |
| 0.0 | 2.8 | NABA ECM REGULATORS | Genes encoding enzymes and their regulators involved in the remodeling of the extracellular matrix |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.9 | REACTOME PROSTANOID LIGAND RECEPTORS | Genes involved in Prostanoid ligand receptors |
| 0.0 | 2.2 | REACTOME CHEMOKINE RECEPTORS BIND CHEMOKINES | Genes involved in Chemokine receptors bind chemokines |
| 0.0 | 0.8 | REACTOME IL 7 SIGNALING | Genes involved in Interleukin-7 signaling |
| 0.0 | 0.4 | REACTOME G2 M DNA DAMAGE CHECKPOINT | Genes involved in G2/M DNA damage checkpoint |
| 0.0 | 0.2 | REACTOME ROLE OF DCC IN REGULATING APOPTOSIS | Genes involved in Role of DCC in regulating apoptosis |
| 0.0 | 0.6 | REACTOME HS GAG BIOSYNTHESIS | Genes involved in HS-GAG biosynthesis |
| 0.0 | 0.3 | REACTOME DOWNREGULATION OF ERBB2 ERBB3 SIGNALING | Genes involved in Downregulation of ERBB2:ERBB3 signaling |