Project
PRJNA375882: Comprehensive Mouse Transcriptomic BodyMap
Navigation
Downloads
Results for Nkx2-6
Z-value: 0.45
Transcription factors associated with Nkx2-6
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Nkx2-6
|
ENSMUSG00000044186.11 | Nkx2-6 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Nkx2-6 | mm39_v1_chr14_+_69409251_69409414 | -0.17 | 1.5e-01 | Click! |
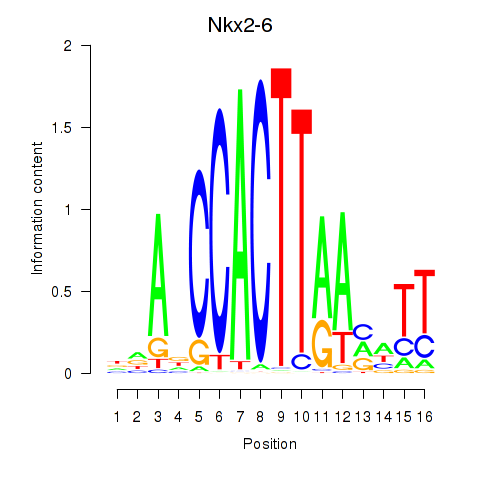
Activity profile of Nkx2-6 motif
Sorted Z-values of Nkx2-6 motif
Network of associatons between targets according to the STRING database.
First level regulatory network of Nkx2-6
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr2_-_119060366 | 3.22 |
ENSMUST00000076084.6
|
Ppp1r14d
|
protein phosphatase 1, regulatory inhibitor subunit 14D |
| chr6_-_41681273 | 3.19 |
ENSMUST00000031899.14
|
Kel
|
Kell blood group |
| chr2_-_119060330 | 2.92 |
ENSMUST00000110820.3
|
Ppp1r14d
|
protein phosphatase 1, regulatory inhibitor subunit 14D |
| chr12_-_11485639 | 2.62 |
ENSMUST00000220506.2
|
Vsnl1
|
visinin-like 1 |
| chr4_+_46039202 | 2.54 |
ENSMUST00000156200.8
|
Tmod1
|
tropomodulin 1 |
| chr12_-_114263874 | 2.20 |
ENSMUST00000103482.2
ENSMUST00000194159.2 |
Ighv9-4
|
immunoglobulin heavy variable 9-4 |
| chr5_-_8417982 | 1.50 |
ENSMUST00000088761.11
ENSMUST00000115386.8 ENSMUST00000050166.14 ENSMUST00000046838.14 ENSMUST00000115388.9 ENSMUST00000088744.12 ENSMUST00000115385.2 |
Adam22
|
a disintegrin and metallopeptidase domain 22 |
| chr15_+_73594965 | 1.41 |
ENSMUST00000165541.8
ENSMUST00000167582.8 |
Ptp4a3
|
protein tyrosine phosphatase 4a3 |
| chr15_-_37734579 | 1.29 |
ENSMUST00000145909.9
ENSMUST00000153775.9 |
Gm49397
Ncald
|
predicted gene, 49397 neurocalcin delta |
| chr15_+_73595012 | 1.26 |
ENSMUST00000230044.2
|
Ptp4a3
|
protein tyrosine phosphatase 4a3 |
| chr4_+_62537750 | 1.01 |
ENSMUST00000084521.11
ENSMUST00000107424.2 |
Rgs3
|
regulator of G-protein signaling 3 |
| chr12_-_114073050 | 0.97 |
ENSMUST00000103472.4
|
Ighv9-2
|
immunoglobulin heavy variable V9-2 |
| chr19_+_55240357 | 0.85 |
ENSMUST00000225551.2
|
Acsl5
|
acyl-CoA synthetase long-chain family member 5 |
| chr11_+_103664976 | 0.84 |
ENSMUST00000000127.6
|
Wnt3
|
wingless-type MMTV integration site family, member 3 |
| chr3_+_87432879 | 0.84 |
ENSMUST00000170036.8
ENSMUST00000117293.2 |
Etv3
|
ets variant 3 |
| chr5_+_110478558 | 0.76 |
ENSMUST00000112481.2
|
Pole
|
polymerase (DNA directed), epsilon |
| chr3_+_62327089 | 0.69 |
ENSMUST00000161057.2
|
Arhgef26
|
Rho guanine nucleotide exchange factor (GEF) 26 |
| chr5_+_145063568 | 0.63 |
ENSMUST00000138922.2
|
Arpc1b
|
actin related protein 2/3 complex, subunit 1B |
| chr19_+_8779903 | 0.62 |
ENSMUST00000172175.3
|
Zbtb3
|
zinc finger and BTB domain containing 3 |
| chr1_+_179788675 | 0.61 |
ENSMUST00000076687.12
ENSMUST00000097450.10 ENSMUST00000212756.2 |
Cdc42bpa
|
CDC42 binding protein kinase alpha |
| chr9_-_10904714 | 0.55 |
ENSMUST00000162484.8
ENSMUST00000160216.8 |
Cntn5
|
contactin 5 |
| chr2_+_85715984 | 0.49 |
ENSMUST00000213441.3
|
Olfr1023
|
olfactory receptor 1023 |
| chr2_+_3115250 | 0.49 |
ENSMUST00000072955.12
|
Fam171a1
|
family with sequence similarity 171, member A1 |
| chr2_-_147028309 | 0.43 |
ENSMUST00000067075.7
|
Nkx2-2
|
NK2 homeobox 2 |
| chr7_-_115630282 | 0.42 |
ENSMUST00000206034.2
ENSMUST00000106612.8 |
Sox6
|
SRY (sex determining region Y)-box 6 |
| chr15_-_78657640 | 0.41 |
ENSMUST00000018313.6
|
Mfng
|
MFNG O-fucosylpeptide 3-beta-N-acetylglucosaminyltransferase |
| chr4_-_91288221 | 0.39 |
ENSMUST00000102799.10
|
Elavl2
|
ELAV like RNA binding protein 1 |
| chr10_+_4297762 | 0.31 |
ENSMUST00000215696.2
|
Akap12
|
A kinase (PRKA) anchor protein (gravin) 12 |
| chr14_-_73286535 | 0.28 |
ENSMUST00000169168.3
|
Cysltr2
|
cysteinyl leukotriene receptor 2 |
| chr14_-_73286504 | 0.13 |
ENSMUST00000044664.12
|
Cysltr2
|
cysteinyl leukotriene receptor 2 |
| chr14_+_54598283 | 0.01 |
ENSMUST00000000985.7
|
Oxa1l
|
oxidase assembly 1-like |
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 3.2 | GO:0031133 | regulation of axon diameter(GO:0031133) |
| 0.4 | 2.5 | GO:0030241 | skeletal muscle myosin thick filament assembly(GO:0030241) |
| 0.3 | 0.8 | GO:1904954 | Spemann organizer formation(GO:0060061) canonical Wnt signaling pathway involved in midbrain dopaminergic neuron differentiation(GO:1904954) |
| 0.3 | 0.8 | GO:0045004 | DNA replication proofreading(GO:0045004) |
| 0.2 | 0.9 | GO:0010747 | positive regulation of plasma membrane long-chain fatty acid transport(GO:0010747) |
| 0.1 | 0.4 | GO:0021530 | spinal cord oligodendrocyte cell differentiation(GO:0021529) spinal cord oligodendrocyte cell fate specification(GO:0021530) |
| 0.1 | 2.7 | GO:0043117 | positive regulation of vascular permeability(GO:0043117) |
| 0.0 | 2.6 | GO:0035774 | positive regulation of insulin secretion involved in cellular response to glucose stimulus(GO:0035774) |
| 0.0 | 0.8 | GO:0097011 | cellular response to granulocyte macrophage colony-stimulating factor stimulus(GO:0097011) |
| 0.0 | 0.4 | GO:0021780 | oligodendrocyte cell fate specification(GO:0021778) oligodendrocyte cell fate commitment(GO:0021779) glial cell fate specification(GO:0021780) |
| 0.0 | 1.5 | GO:0032292 | myelination in peripheral nervous system(GO:0022011) peripheral nervous system axon ensheathment(GO:0032292) |
| 0.0 | 0.4 | GO:0002315 | marginal zone B cell differentiation(GO:0002315) |
| 0.0 | 6.1 | GO:0035304 | regulation of protein dephosphorylation(GO:0035304) |
| 0.0 | 0.7 | GO:0001886 | endothelial cell morphogenesis(GO:0001886) |
| 0.0 | 3.2 | GO:0006910 | phagocytosis, recognition(GO:0006910) |
| 0.0 | 0.3 | GO:0010739 | positive regulation of protein kinase A signaling(GO:0010739) |
| 0.0 | 0.6 | GO:0040023 | nuclear migration(GO:0007097) establishment of nucleus localization(GO:0040023) |
| 0.0 | 0.6 | GO:0034314 | Arp2/3 complex-mediated actin nucleation(GO:0034314) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.6 | GO:0036284 | tubulobulbar complex(GO:0036284) |
| 0.2 | 0.8 | GO:0008622 | epsilon DNA polymerase complex(GO:0008622) |
| 0.1 | 0.8 | GO:0090571 | RNA polymerase II transcription repressor complex(GO:0090571) |
| 0.0 | 2.5 | GO:0005865 | striated muscle thin filament(GO:0005865) |
| 0.0 | 3.2 | GO:0042571 | immunoglobulin complex, circulating(GO:0042571) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.4 | GO:0033829 | O-fucosylpeptide 3-beta-N-acetylglucosaminyltransferase activity(GO:0033829) |
| 0.1 | 6.1 | GO:0004864 | protein phosphatase inhibitor activity(GO:0004864) |
| 0.1 | 2.5 | GO:0005523 | tropomyosin binding(GO:0005523) |
| 0.1 | 0.4 | GO:0004974 | leukotriene receptor activity(GO:0004974) |
| 0.1 | 0.8 | GO:0008310 | single-stranded DNA 3'-5' exodeoxyribonuclease activity(GO:0008310) |
| 0.1 | 0.8 | GO:0017151 | DEAD/H-box RNA helicase binding(GO:0017151) |
| 0.0 | 0.9 | GO:0102391 | decanoate--CoA ligase activity(GO:0102391) |
| 0.0 | 4.7 | GO:0004222 | metalloendopeptidase activity(GO:0004222) |
| 0.0 | 0.8 | GO:0048018 | receptor agonist activity(GO:0048018) |
| 0.0 | 1.3 | GO:0043014 | alpha-tubulin binding(GO:0043014) |
| 0.0 | 3.2 | GO:0034987 | immunoglobulin receptor binding(GO:0034987) |
| 0.0 | 2.7 | GO:0004725 | protein tyrosine phosphatase activity(GO:0004725) |
| 0.0 | 0.3 | GO:0008179 | adenylate cyclase binding(GO:0008179) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 2.7 | PID PRL SIGNALING EVENTS PATHWAY | Signaling events mediated by PRL |
| 0.0 | 0.8 | PID WNT SIGNALING PATHWAY | Wnt signaling network |
| 0.0 | 1.0 | PID EPHRINB REV PATHWAY | Ephrin B reverse signaling |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 1.3 | REACTOME IONOTROPIC ACTIVITY OF KAINATE RECEPTORS | Genes involved in Ionotropic activity of Kainate Receptors |
| 0.0 | 0.8 | REACTOME REPAIR SYNTHESIS FOR GAP FILLING BY DNA POL IN TC NER | Genes involved in Repair synthesis for gap-filling by DNA polymerase in TC-NER |
| 0.0 | 2.5 | REACTOME STRIATED MUSCLE CONTRACTION | Genes involved in Striated Muscle Contraction |
| 0.0 | 0.4 | REACTOME EICOSANOID LIGAND BINDING RECEPTORS | Genes involved in Eicosanoid ligand-binding receptors |
| 0.0 | 0.9 | REACTOME SYNTHESIS OF VERY LONG CHAIN FATTY ACYL COAS | Genes involved in Synthesis of very long-chain fatty acyl-CoAs |
| 0.0 | 0.4 | REACTOME PRE NOTCH PROCESSING IN GOLGI | Genes involved in Pre-NOTCH Processing in Golgi |
| 0.0 | 0.4 | REACTOME REGULATION OF GENE EXPRESSION IN BETA CELLS | Genes involved in Regulation of gene expression in beta cells |