Project
PRJNA375882: Comprehensive Mouse Transcriptomic BodyMap
Navigation
Downloads
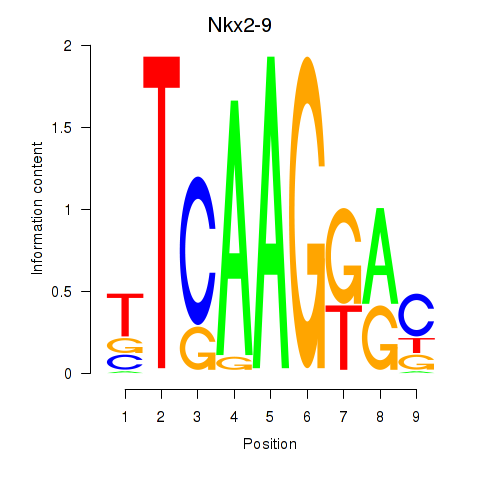
Results for Nkx2-9
Z-value: 1.20
Transcription factors associated with Nkx2-9
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Nkx2-9
|
ENSMUSG00000058669.8 | Nkx2-9 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Nkx2-9 | mm39_v1_chr12_-_56660054_56660113 | -0.32 | 6.2e-03 | Click! |
Activity profile of Nkx2-9 motif
Sorted Z-values of Nkx2-9 motif
Network of associatons between targets according to the STRING database.
First level regulatory network of Nkx2-9
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr6_-_78445846 | 13.06 |
ENSMUST00000032089.3
|
Reg3g
|
regenerating islet-derived 3 gamma |
| chr9_+_98372575 | 11.37 |
ENSMUST00000035029.3
|
Rbp2
|
retinol binding protein 2, cellular |
| chr6_+_135339929 | 8.40 |
ENSMUST00000032330.16
|
Emp1
|
epithelial membrane protein 1 |
| chr16_-_48232770 | 8.29 |
ENSMUST00000212197.2
|
Gm5485
|
predicted gene 5485 |
| chr12_-_114487525 | 7.27 |
ENSMUST00000103495.3
|
Ighv10-3
|
immunoglobulin heavy variable V10-3 |
| chr7_-_144761806 | 7.01 |
ENSMUST00000208788.2
|
Smim38
|
small integral membrane protein 38 |
| chr9_-_99592116 | 6.94 |
ENSMUST00000035048.12
|
Cldn18
|
claudin 18 |
| chr2_+_172864153 | 6.79 |
ENSMUST00000173997.2
|
Rbm38
|
RNA binding motif protein 38 |
| chr5_+_91222470 | 5.88 |
ENSMUST00000031324.6
|
Ereg
|
epiregulin |
| chr2_+_67004178 | 5.51 |
ENSMUST00000239009.2
ENSMUST00000238912.2 |
Xirp2
|
xin actin-binding repeat containing 2 |
| chr12_-_113561594 | 5.43 |
ENSMUST00000103444.3
|
Ighv5-4
|
immunoglobulin heavy variable 5-4 |
| chr16_-_58344548 | 5.29 |
ENSMUST00000114357.10
|
St3gal6
|
ST3 beta-galactoside alpha-2,3-sialyltransferase 6 |
| chr16_-_58344602 | 5.27 |
ENSMUST00000114358.9
|
St3gal6
|
ST3 beta-galactoside alpha-2,3-sialyltransferase 6 |
| chr4_+_135975243 | 5.26 |
ENSMUST00000102533.11
ENSMUST00000143942.2 |
Tcea3
|
transcription elongation factor A (SII), 3 |
| chr13_-_32960379 | 5.22 |
ENSMUST00000230119.2
|
Mylk4
|
myosin light chain kinase family, member 4 |
| chr8_-_13446769 | 5.21 |
ENSMUST00000033826.4
|
Atp4b
|
ATPase, H+/K+ exchanging, beta polypeptide |
| chrX_+_21581135 | 5.15 |
ENSMUST00000033414.8
|
Slc6a14
|
solute carrier family 6 (neurotransmitter transporter), member 14 |
| chr12_-_115410489 | 5.14 |
ENSMUST00000194581.2
|
Ighv1-62-2
|
immunoglobulin heavy variable 1-62-2 |
| chr9_-_99592058 | 5.06 |
ENSMUST00000136429.8
|
Cldn18
|
claudin 18 |
| chr12_-_113802603 | 4.95 |
ENSMUST00000103458.3
ENSMUST00000193652.2 |
Ighv5-16
|
immunoglobulin heavy variable 5-16 |
| chr6_-_3487369 | 4.88 |
ENSMUST00000201607.4
|
Hepacam2
|
HEPACAM family member 2 |
| chr16_-_58344497 | 4.84 |
ENSMUST00000151510.2
ENSMUST00000126978.8 ENSMUST00000123918.8 |
St3gal6
|
ST3 beta-galactoside alpha-2,3-sialyltransferase 6 |
| chr18_-_65527078 | 4.78 |
ENSMUST00000035548.16
|
Alpk2
|
alpha-kinase 2 |
| chr7_-_44318710 | 4.66 |
ENSMUST00000208131.2
|
Myh14
|
myosin, heavy polypeptide 14 |
| chr12_-_113896002 | 4.65 |
ENSMUST00000103463.3
|
Ighv14-1
|
immunoglobulin heavy variable 14-1 |
| chr17_+_34482183 | 4.51 |
ENSMUST00000040828.7
ENSMUST00000237342.2 ENSMUST00000237866.2 |
H2-Ab1
|
histocompatibility 2, class II antigen A, beta 1 |
| chr12_-_113589576 | 4.44 |
ENSMUST00000103446.2
|
Ighv5-6
|
immunoglobulin heavy variable 5-6 |
| chr7_-_110462446 | 4.35 |
ENSMUST00000033050.5
|
Lyve1
|
lymphatic vessel endothelial hyaluronan receptor 1 |
| chr12_-_114502585 | 4.27 |
ENSMUST00000103496.4
|
Ighv1-7
|
immunoglobulin heavy variable V1-7 |
| chr11_+_117740111 | 4.25 |
ENSMUST00000093906.5
|
Birc5
|
baculoviral IAP repeat-containing 5 |
| chr6_+_68414401 | 4.24 |
ENSMUST00000103324.3
|
Igkv15-103
|
immunoglobulin kappa chain variable 15-103 |
| chr4_-_138484889 | 4.15 |
ENSMUST00000030526.7
|
Pla2g2f
|
phospholipase A2, group IIF |
| chr12_-_114443071 | 4.11 |
ENSMUST00000103492.2
|
Ighv10-1
|
immunoglobulin heavy variable 10-1 |
| chr9_+_75682637 | 3.97 |
ENSMUST00000012281.8
|
Bmp5
|
bone morphogenetic protein 5 |
| chr11_+_117740077 | 3.77 |
ENSMUST00000081387.11
|
Birc5
|
baculoviral IAP repeat-containing 5 |
| chr11_-_99383938 | 3.67 |
ENSMUST00000006969.8
|
Krt23
|
keratin 23 |
| chr16_+_32090286 | 3.62 |
ENSMUST00000093183.5
|
Smco1
|
single-pass membrane protein with coiled-coil domains 1 |
| chr6_-_113411489 | 3.54 |
ENSMUST00000133348.2
|
Cidec
|
cell death-inducing DFFA-like effector c |
| chr12_-_113823290 | 3.46 |
ENSMUST00000103459.5
|
Ighv5-17
|
immunoglobulin heavy variable 5-17 |
| chr12_-_114793177 | 3.32 |
ENSMUST00000103511.2
ENSMUST00000195735.2 |
Ighv1-31
|
immunoglobulin heavy variable 1-31 |
| chr5_+_117457126 | 3.20 |
ENSMUST00000111967.8
|
Vsig10
|
V-set and immunoglobulin domain containing 10 |
| chr7_-_19411866 | 2.97 |
ENSMUST00000142352.9
|
Apoc2
|
apolipoprotein C-II |
| chr4_+_129714494 | 2.91 |
ENSMUST00000165853.2
|
Ptp4a2
|
protein tyrosine phosphatase 4a2 |
| chr6_-_78355834 | 2.90 |
ENSMUST00000089667.8
ENSMUST00000167492.4 |
Reg3d
|
regenerating islet-derived 3 delta |
| chr12_-_114579763 | 2.86 |
ENSMUST00000103500.2
|
Ighv1-12
|
immunoglobulin heavy variable V1-12 |
| chr12_-_113700190 | 2.85 |
ENSMUST00000103452.3
ENSMUST00000192264.2 |
Ighv5-9-1
|
immunoglobulin heavy variable 5-9-1 |
| chr13_+_75987987 | 2.50 |
ENSMUST00000022082.8
ENSMUST00000223120.2 ENSMUST00000220523.2 |
Glrx
|
glutaredoxin |
| chr17_+_35284315 | 2.45 |
ENSMUST00000173207.8
|
Ly6g6c
|
lymphocyte antigen 6 complex, locus G6C |
| chr11_-_46280281 | 2.37 |
ENSMUST00000101306.4
|
Itk
|
IL2 inducible T cell kinase |
| chr11_-_46280298 | 2.26 |
ENSMUST00000109237.9
|
Itk
|
IL2 inducible T cell kinase |
| chr16_-_92262969 | 2.14 |
ENSMUST00000232239.2
ENSMUST00000060005.15 |
Rcan1
|
regulator of calcineurin 1 |
| chr11_-_46280336 | 2.13 |
ENSMUST00000020664.13
|
Itk
|
IL2 inducible T cell kinase |
| chr14_-_24054352 | 2.06 |
ENSMUST00000190339.2
|
Kcnma1
|
potassium large conductance calcium-activated channel, subfamily M, alpha member 1 |
| chr3_+_99203818 | 2.03 |
ENSMUST00000150756.3
|
Tbx15
|
T-box 15 |
| chr7_+_45084300 | 1.97 |
ENSMUST00000211150.2
|
Gys1
|
glycogen synthase 1, muscle |
| chr1_+_131671751 | 1.89 |
ENSMUST00000049027.10
|
Slc26a9
|
solute carrier family 26, member 9 |
| chr12_-_113860566 | 1.89 |
ENSMUST00000103474.5
|
Ighv7-1
|
immunoglobulin heavy variable 7-1 |
| chr5_-_29683468 | 1.87 |
ENSMUST00000165512.4
ENSMUST00000001608.8 |
Mnx1
|
motor neuron and pancreas homeobox 1 |
| chr10_-_62258195 | 1.87 |
ENSMUST00000020277.9
|
Hkdc1
|
hexokinase domain containing 1 |
| chr1_+_61017057 | 1.86 |
ENSMUST00000027162.12
ENSMUST00000102827.4 |
Icos
|
inducible T cell co-stimulator |
| chr10_+_79518138 | 1.84 |
ENSMUST00000166603.2
ENSMUST00000219791.2 ENSMUST00000219930.2 ENSMUST00000218964.2 |
Cdc34
|
cell division cycle 34 |
| chrX_-_99638466 | 1.83 |
ENSMUST00000053373.2
|
P2ry4
|
pyrimidinergic receptor P2Y, G-protein coupled, 4 |
| chrX_-_101232978 | 1.79 |
ENSMUST00000033683.8
|
Rps4x
|
ribosomal protein S4, X-linked |
| chrX_-_47763355 | 1.73 |
ENSMUST00000053970.4
|
Gpr119
|
G-protein coupled receptor 119 |
| chr10_+_121477699 | 1.73 |
ENSMUST00000120642.9
ENSMUST00000132744.2 |
D930020B18Rik
|
RIKEN cDNA D930020B18 gene |
| chr12_-_113666198 | 1.64 |
ENSMUST00000103450.4
|
Ighv5-12
|
immunoglobulin heavy variable 5-12 |
| chr11_-_80670815 | 1.64 |
ENSMUST00000041065.14
ENSMUST00000070997.6 |
Myo1d
|
myosin ID |
| chr12_+_24758724 | 1.64 |
ENSMUST00000153058.8
|
Rrm2
|
ribonucleotide reductase M2 |
| chr2_-_76698725 | 1.63 |
ENSMUST00000149616.8
ENSMUST00000152185.8 ENSMUST00000130915.8 ENSMUST00000155365.8 ENSMUST00000128071.8 |
Ttn
|
titin |
| chr1_-_66902429 | 1.61 |
ENSMUST00000027153.6
|
Acadl
|
acyl-Coenzyme A dehydrogenase, long-chain |
| chr3_-_92577621 | 1.60 |
ENSMUST00000029530.6
|
Lce1a2
|
late cornified envelope 1A2 |
| chr7_+_45084257 | 1.53 |
ENSMUST00000003964.17
|
Gys1
|
glycogen synthase 1, muscle |
| chr7_-_103928939 | 1.52 |
ENSMUST00000051795.10
|
Trim5
|
tripartite motif-containing 5 |
| chr3_-_59009231 | 1.51 |
ENSMUST00000085040.5
|
Gpr171
|
G protein-coupled receptor 171 |
| chr12_-_115825934 | 1.50 |
ENSMUST00000198777.2
|
Ighv1-77
|
immunoglobulin heavy variable 1-77 |
| chr1_-_181669891 | 1.50 |
ENSMUST00000193028.2
ENSMUST00000191878.6 ENSMUST00000005003.12 |
Lbr
|
lamin B receptor |
| chr9_-_56068282 | 1.46 |
ENSMUST00000034876.10
|
Tspan3
|
tetraspanin 3 |
| chr10_-_89342493 | 1.46 |
ENSMUST00000058126.15
ENSMUST00000105296.9 |
Nr1h4
|
nuclear receptor subfamily 1, group H, member 4 |
| chr6_-_126826077 | 1.46 |
ENSMUST00000205002.3
ENSMUST00000088194.7 |
Ndufa9
|
NADH:ubiquinone oxidoreductase subunit A9 |
| chr11_+_76792977 | 1.45 |
ENSMUST00000102495.8
|
Tmigd1
|
transmembrane and immunoglobulin domain containing 1 |
| chrX_+_41238410 | 1.37 |
ENSMUST00000127618.8
|
Stag2
|
stromal antigen 2 |
| chr15_+_58805605 | 1.30 |
ENSMUST00000022980.5
|
Ndufb9
|
NADH:ubiquinone oxidoreductase subunit B9 |
| chr10_+_121477493 | 1.26 |
ENSMUST00000142501.8
|
D930020B18Rik
|
RIKEN cDNA D930020B18 gene |
| chr10_+_59057767 | 1.25 |
ENSMUST00000182161.2
|
Sowahc
|
sosondowah ankyrin repeat domain family member C |
| chr4_+_135870808 | 1.25 |
ENSMUST00000008016.3
|
Id3
|
inhibitor of DNA binding 3 |
| chr19_-_40576782 | 1.23 |
ENSMUST00000176939.8
|
Aldh18a1
|
aldehyde dehydrogenase 18 family, member A1 |
| chr2_+_121337195 | 1.22 |
ENSMUST00000028676.12
|
Wdr76
|
WD repeat domain 76 |
| chr11_+_68858942 | 1.21 |
ENSMUST00000102606.10
ENSMUST00000018884.6 |
Slc25a35
|
solute carrier family 25, member 35 |
| chrX_+_41238193 | 1.18 |
ENSMUST00000115073.9
ENSMUST00000115072.8 |
Stag2
|
stromal antigen 2 |
| chr19_+_43741550 | 1.17 |
ENSMUST00000153295.2
|
Cutc
|
cutC copper transporter |
| chr14_+_53007210 | 1.15 |
ENSMUST00000178768.4
|
Trav7d-4
|
T cell receptor alpha variable 7D-4 |
| chr4_+_43401232 | 1.11 |
ENSMUST00000125399.2
|
Rusc2
|
RUN and SH3 domain containing 2 |
| chr9_+_102503815 | 1.10 |
ENSMUST00000038673.14
ENSMUST00000186693.2 |
Anapc13
|
anaphase promoting complex subunit 13 |
| chr9_-_60429300 | 1.09 |
ENSMUST00000140824.2
|
Thsd4
|
thrombospondin, type I, domain containing 4 |
| chr13_+_109397184 | 1.07 |
ENSMUST00000153234.8
|
Pde4d
|
phosphodiesterase 4D, cAMP specific |
| chr7_-_126859790 | 1.03 |
ENSMUST00000035276.5
|
Dctpp1
|
dCTP pyrophosphatase 1 |
| chr4_-_21685781 | 1.03 |
ENSMUST00000076206.11
|
Prdm13
|
PR domain containing 13 |
| chr10_+_44943262 | 1.00 |
ENSMUST00000099858.4
|
Prep
|
prolyl endopeptidase |
| chr7_-_33929667 | 1.00 |
ENSMUST00000206415.2
|
Gpi1
|
glucose-6-phosphate isomerase 1 |
| chr12_+_24758968 | 0.95 |
ENSMUST00000154588.2
|
Rrm2
|
ribonucleotide reductase M2 |
| chr2_+_80145805 | 0.94 |
ENSMUST00000028392.8
|
Dnajc10
|
DnaJ heat shock protein family (Hsp40) member C10 |
| chr11_-_68864666 | 0.93 |
ENSMUST00000038644.5
|
Rangrf
|
RAN guanine nucleotide release factor |
| chr11_+_87590720 | 0.92 |
ENSMUST00000040089.5
|
Rnf43
|
ring finger protein 43 |
| chr6_-_42442130 | 0.88 |
ENSMUST00000216650.2
|
Olfr458
|
olfactory receptor 458 |
| chr1_+_63215976 | 0.86 |
ENSMUST00000129339.8
|
Eef1b2
|
eukaryotic translation elongation factor 1 beta 2 |
| chr11_+_114559350 | 0.85 |
ENSMUST00000106602.10
ENSMUST00000077915.10 ENSMUST00000106599.8 ENSMUST00000082092.5 |
Rpl38
|
ribosomal protein L38 |
| chr7_-_103814019 | 0.84 |
ENSMUST00000154555.2
ENSMUST00000051137.15 |
Olfm5
|
olfactomedin 5 |
| chr11_+_72498029 | 0.83 |
ENSMUST00000021148.13
ENSMUST00000138247.8 |
Ube2g1
|
ubiquitin-conjugating enzyme E2G 1 |
| chr3_+_159545309 | 0.82 |
ENSMUST00000068952.10
ENSMUST00000198878.2 |
Wls
|
wntless WNT ligand secretion mediator |
| chr12_-_56583582 | 0.81 |
ENSMUST00000001536.9
|
Nkx2-1
|
NK2 homeobox 1 |
| chr6_-_37419030 | 0.81 |
ENSMUST00000041093.6
|
Creb3l2
|
cAMP responsive element binding protein 3-like 2 |
| chr11_+_59503792 | 0.78 |
ENSMUST00000055276.6
|
Olfr225
|
olfactory receptor 225 |
| chr9_+_44238089 | 0.78 |
ENSMUST00000054708.5
|
Dpagt1
|
dolichyl-phosphate (UDP-N-acetylglucosamine) acetylglucosaminephosphotransferase 1 (GlcNAc-1-P transferase) |
| chr7_-_33929708 | 0.75 |
ENSMUST00000038027.6
|
Gpi1
|
glucose-6-phosphate isomerase 1 |
| chr1_-_194813631 | 0.75 |
ENSMUST00000194111.6
ENSMUST00000193094.6 |
Cr1l
|
complement component (3b/4b) receptor 1-like |
| chr1_-_63215812 | 0.74 |
ENSMUST00000185847.2
ENSMUST00000185732.7 ENSMUST00000188370.7 ENSMUST00000168099.9 |
Ndufs1
|
NADH:ubiquinone oxidoreductase core subunit S1 |
| chr1_-_63215952 | 0.73 |
ENSMUST00000185412.7
ENSMUST00000027111.15 ENSMUST00000189664.2 |
Ndufs1
|
NADH:ubiquinone oxidoreductase core subunit S1 |
| chr11_-_22932090 | 0.72 |
ENSMUST00000160826.2
ENSMUST00000093270.6 ENSMUST00000071068.9 ENSMUST00000159081.8 |
Commd1b
Commd1
|
COMM domain containing 1B COMM domain containing 1 |
| chr6_-_42453259 | 0.71 |
ENSMUST00000204324.3
ENSMUST00000203396.3 |
Olfr457
|
olfactory receptor 457 |
| chr15_-_98675142 | 0.71 |
ENSMUST00000166022.2
|
Wnt10b
|
wingless-type MMTV integration site family, member 10B |
| chr7_-_101714251 | 0.70 |
ENSMUST00000130074.2
ENSMUST00000131104.3 ENSMUST00000096639.12 |
Rnf121
|
ring finger protein 121 |
| chr14_+_54440591 | 0.70 |
ENSMUST00000103725.2
|
Traj16
|
T cell receptor alpha joining 16 |
| chr16_+_24266829 | 0.68 |
ENSMUST00000078988.10
|
Lpp
|
LIM domain containing preferred translocation partner in lipoma |
| chr11_-_106679312 | 0.68 |
ENSMUST00000021062.12
|
Ddx5
|
DEAD box helicase 5 |
| chr14_+_53209905 | 0.63 |
ENSMUST00000196023.2
|
Trav3d-3
|
T cell receptor alpha variable 3D-3 |
| chr14_+_53903370 | 0.63 |
ENSMUST00000181768.3
|
Trav3-3
|
T cell receptor alpha variable 3-3 |
| chrX_+_84091915 | 0.60 |
ENSMUST00000239019.2
ENSMUST00000113992.3 ENSMUST00000113991.8 |
Dmd
|
dystrophin, muscular dystrophy |
| chr6_-_38331482 | 0.59 |
ENSMUST00000031850.10
ENSMUST00000114898.3 |
Zc3hav1
|
zinc finger CCCH type, antiviral 1 |
| chr8_-_46747629 | 0.58 |
ENSMUST00000058636.9
|
Helt
|
helt bHLH transcription factor |
| chr3_+_137983250 | 0.58 |
ENSMUST00000004232.10
|
Adh1
|
alcohol dehydrogenase 1 (class I) |
| chr6_-_42437951 | 0.57 |
ENSMUST00000090156.2
|
Olfr458
|
olfactory receptor 458 |
| chr4_-_72770859 | 0.57 |
ENSMUST00000179234.2
ENSMUST00000078617.5 |
Aldoart1
|
aldolase 1 A, retrogene 1 |
| chr10_-_34294461 | 0.56 |
ENSMUST00000213269.2
ENSMUST00000099973.4 ENSMUST00000105512.8 ENSMUST00000047885.14 |
Nt5dc1
|
5'-nucleotidase domain containing 1 |
| chr11_-_106679671 | 0.56 |
ENSMUST00000123339.2
|
Ddx5
|
DEAD box helicase 5 |
| chr15_+_79575046 | 0.55 |
ENSMUST00000046463.10
|
Gtpbp1
|
GTP binding protein 1 |
| chr13_-_21476849 | 0.54 |
ENSMUST00000110491.9
|
Gpx5
|
glutathione peroxidase 5 |
| chr7_-_110681402 | 0.54 |
ENSMUST00000159305.2
|
Eif4g2
|
eukaryotic translation initiation factor 4, gamma 2 |
| chr2_+_121337226 | 0.53 |
ENSMUST00000099473.10
ENSMUST00000110602.9 |
Wdr76
|
WD repeat domain 76 |
| chr6_+_42263609 | 0.52 |
ENSMUST00000238845.2
ENSMUST00000031894.13 |
Clcn1
|
chloride channel, voltage-sensitive 1 |
| chr7_-_85971258 | 0.51 |
ENSMUST00000044256.6
|
Olfr308
|
olfactory receptor 308 |
| chr2_+_139993963 | 0.51 |
ENSMUST00000104994.3
|
Gm17374
|
predicted gene, 17374 |
| chr19_+_29675224 | 0.50 |
ENSMUST00000025719.5
|
Mlana
|
melan-A |
| chr19_+_43741513 | 0.48 |
ENSMUST00000112047.10
|
Cutc
|
cutC copper transporter |
| chr7_-_81439173 | 0.42 |
ENSMUST00000026092.9
|
3110040N11Rik
|
RIKEN cDNA 3110040N11 gene |
| chr12_+_55646209 | 0.42 |
ENSMUST00000051857.5
|
Insm2
|
insulinoma-associated 2 |
| chr11_-_106679983 | 0.42 |
ENSMUST00000129585.8
|
Ddx5
|
DEAD box helicase 5 |
| chr19_-_43741363 | 0.41 |
ENSMUST00000045562.6
|
Cox15
|
cytochrome c oxidase assembly protein 15 |
| chr8_-_81607109 | 0.38 |
ENSMUST00000034150.10
|
Gab1
|
growth factor receptor bound protein 2-associated protein 1 |
| chr6_+_42263644 | 0.36 |
ENSMUST00000163936.8
|
Clcn1
|
chloride channel, voltage-sensitive 1 |
| chr10_+_94034817 | 0.36 |
ENSMUST00000020209.16
ENSMUST00000179990.8 |
Ndufa12
|
NADH:ubiquinone oxidoreductase subunit A12 |
| chr19_+_43741431 | 0.36 |
ENSMUST00000026199.14
|
Cutc
|
cutC copper transporter |
| chr7_+_75259778 | 0.36 |
ENSMUST00000207923.2
|
Akap13
|
A kinase (PRKA) anchor protein 13 |
| chr14_+_53088747 | 0.36 |
ENSMUST00000103588.4
|
Trav13d-1
|
T cell receptor alpha variable 13D-1 |
| chr7_-_67022520 | 0.35 |
ENSMUST00000156690.8
ENSMUST00000107476.8 ENSMUST00000076325.12 ENSMUST00000032776.15 ENSMUST00000133074.2 |
Mef2a
|
myocyte enhancer factor 2A |
| chr12_-_114576295 | 0.35 |
ENSMUST00000191801.2
|
Ighv1-11
|
immunoglobulin heavy variable V1-11 |
| chr2_+_20742115 | 0.33 |
ENSMUST00000114606.8
ENSMUST00000114608.3 |
Etl4
|
enhancer trap locus 4 |
| chr3_-_154760978 | 0.31 |
ENSMUST00000064076.6
|
Tnni3k
|
TNNI3 interacting kinase |
| chr7_+_88079465 | 0.29 |
ENSMUST00000107256.4
|
Rab38
|
RAB38, member RAS oncogene family |
| chr10_+_18345706 | 0.29 |
ENSMUST00000162891.8
ENSMUST00000100054.4 |
Nhsl1
|
NHS-like 1 |
| chr13_-_64645606 | 0.28 |
ENSMUST00000180282.2
|
Fam240b
|
family with sequence similarity 240 member B |
| chr7_-_45829353 | 0.28 |
ENSMUST00000033123.8
|
Abcc8
|
ATP-binding cassette, sub-family C (CFTR/MRP), member 8 |
| chr16_+_33201164 | 0.27 |
ENSMUST00000165418.9
|
Zfp148
|
zinc finger protein 148 |
| chr5_+_92135294 | 0.26 |
ENSMUST00000178614.3
|
Odaph
|
odontogenesis associated phosphoprotein |
| chr16_-_92622659 | 0.26 |
ENSMUST00000186296.2
|
Runx1
|
runt related transcription factor 1 |
| chr18_+_37102969 | 0.24 |
ENSMUST00000194544.2
|
Gm37013
|
predicted gene, 37013 |
| chr17_+_24939225 | 0.24 |
ENSMUST00000146867.2
|
Rps2
|
ribosomal protein S2 |
| chrM_+_7779 | 0.24 |
ENSMUST00000082408.1
|
mt-Atp6
|
mitochondrially encoded ATP synthase 6 |
| chr10_+_87926932 | 0.23 |
ENSMUST00000048621.8
|
Pmch
|
pro-melanin-concentrating hormone |
| chr16_+_36514386 | 0.23 |
ENSMUST00000119464.2
|
Ildr1
|
immunoglobulin-like domain containing receptor 1 |
| chr17_-_37548054 | 0.22 |
ENSMUST00000213328.2
ENSMUST00000214848.2 ENSMUST00000214622.2 ENSMUST00000073667.6 ENSMUST00000216225.2 |
Olfr97
|
olfactory receptor 97 |
| chr6_-_40976413 | 0.22 |
ENSMUST00000166306.3
|
Gm2663
|
predicted gene 2663 |
| chr17_+_35878332 | 0.22 |
ENSMUST00000044326.5
|
2300002M23Rik
|
RIKEN cDNA 2300002M23 gene |
| chr12_-_75463555 | 0.21 |
ENSMUST00000051079.4
|
Gphb5
|
glycoprotein hormone beta 5 |
| chr18_-_60981981 | 0.21 |
ENSMUST00000177172.8
ENSMUST00000175934.8 ENSMUST00000176630.8 |
Tcof1
|
treacle ribosome biogenesis factor 1 |
| chr4_-_119047180 | 0.21 |
ENSMUST00000150864.3
ENSMUST00000141227.9 |
Ermap
|
erythroblast membrane-associated protein |
| chr2_-_105832353 | 0.20 |
ENSMUST00000155811.2
|
Dnajc24
|
DnaJ heat shock protein family (Hsp40) member C24 |
| chr17_-_29738764 | 0.19 |
ENSMUST00000168877.2
|
Gm17657
|
predicted gene, 17657 |
| chr16_+_36514334 | 0.18 |
ENSMUST00000023617.13
ENSMUST00000089618.10 |
Ildr1
|
immunoglobulin-like domain containing receptor 1 |
| chr11_+_106680062 | 0.15 |
ENSMUST00000103068.10
ENSMUST00000018516.11 |
Cep95
|
centrosomal protein 95 |
| chr6_+_43143944 | 0.15 |
ENSMUST00000060243.5
|
Olfr437
|
olfactory receptor 437 |
| chr19_-_12209960 | 0.14 |
ENSMUST00000207710.3
|
Olfr1432
|
olfactory receptor 1432 |
| chr3_-_37180093 | 0.13 |
ENSMUST00000029275.6
|
Il2
|
interleukin 2 |
| chrM_+_7758 | 0.11 |
ENSMUST00000082407.1
|
mt-Atp8
|
mitochondrially encoded ATP synthase 8 |
| chr19_+_46045675 | 0.08 |
ENSMUST00000126127.8
ENSMUST00000147640.2 |
Pprc1
|
peroxisome proliferative activated receptor, gamma, coactivator-related 1 |
| chr17_+_28910393 | 0.08 |
ENSMUST00000124886.9
ENSMUST00000114758.9 |
Mapk14
|
mitogen-activated protein kinase 14 |
| chr14_+_53210216 | 0.07 |
ENSMUST00000103599.3
|
Trav3d-3
|
T cell receptor alpha variable 3D-3 |
| chr3_-_64467239 | 0.07 |
ENSMUST00000165012.3
|
Vmn2r6
|
vomeronasal 2, receptor 6 |
| chr4_+_34893772 | 0.05 |
ENSMUST00000029975.10
ENSMUST00000135871.8 ENSMUST00000108130.2 |
Cga
|
glycoprotein hormones, alpha subunit |
| chr5_-_24650164 | 0.05 |
ENSMUST00000115043.8
ENSMUST00000115041.2 ENSMUST00000030800.13 |
Fastk
|
Fas-activated serine/threonine kinase |
| chr7_-_138447933 | 0.05 |
ENSMUST00000118810.2
ENSMUST00000075667.5 ENSMUST00000119664.2 |
Mapk1ip1
|
mitogen-activated protein kinase 1 interacting protein 1 |
| chr19_+_12257218 | 0.04 |
ENSMUST00000207186.4
ENSMUST00000207915.2 |
Olfr1434
|
olfactory receptor 1434 |
| chr2_+_128971620 | 0.04 |
ENSMUST00000035481.5
|
Chchd5
|
coiled-coil-helix-coiled-coil-helix domain containing 5 |
| chr9_-_48822399 | 0.02 |
ENSMUST00000003826.8
|
Htr3a
|
5-hydroxytryptamine (serotonin) receptor 3A |
| chr4_-_119047146 | 0.02 |
ENSMUST00000124626.9
|
Ermap
|
erythroblast membrane-associated protein |
| chr12_-_115567853 | 0.02 |
ENSMUST00000103538.3
ENSMUST00000198646.2 |
Ighv1-67
|
immunoglobulin heavy variable V1-67 |
| chr6_+_33226020 | 0.01 |
ENSMUST00000052266.15
ENSMUST00000090381.11 ENSMUST00000115080.2 |
Exoc4
|
exocyst complex component 4 |
| chr15_-_98816012 | 0.01 |
ENSMUST00000023736.10
|
Lmbr1l
|
limb region 1 like |
| chr6_+_122684448 | 0.01 |
ENSMUST00000112581.8
ENSMUST00000112580.8 ENSMUST00000012540.5 |
Nanog
|
Nanog homeobox |
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.7 | 5.2 | GO:0033577 | protein glycosylation in endoplasmic reticulum(GO:0033577) |
| 1.7 | 12.0 | GO:2001205 | negative regulation of osteoclast development(GO:2001205) |
| 1.6 | 13.1 | GO:0051715 | cytolysis in other organism(GO:0051715) |
| 1.5 | 4.5 | GO:0002343 | peripheral B cell selection(GO:0002343) B cell affinity maturation(GO:0002344) |
| 1.3 | 11.4 | GO:0006776 | vitamin A metabolic process(GO:0006776) |
| 1.2 | 5.9 | GO:0048165 | ovarian cumulus expansion(GO:0001550) fused antrum stage(GO:0048165) |
| 1.0 | 8.0 | GO:0031536 | positive regulation of exit from mitosis(GO:0031536) |
| 1.0 | 4.0 | GO:0021502 | neural fold elevation formation(GO:0021502) |
| 0.7 | 3.0 | GO:0060697 | positive regulation of phospholipid catabolic process(GO:0060697) |
| 0.4 | 2.5 | GO:2000587 | negative regulation of platelet-derived growth factor receptor-beta signaling pathway(GO:2000587) |
| 0.4 | 1.2 | GO:0006592 | ornithine biosynthetic process(GO:0006592) |
| 0.4 | 6.8 | GO:0006977 | DNA damage response, signal transduction by p53 class mediator resulting in cell cycle arrest(GO:0006977) |
| 0.4 | 55.6 | GO:0006910 | phagocytosis, recognition(GO:0006910) |
| 0.4 | 1.5 | GO:0034971 | histone H3-R17 methylation(GO:0034971) bile acid signaling pathway(GO:0038183) |
| 0.3 | 2.1 | GO:0034465 | response to carbon monoxide(GO:0034465) eye blink reflex(GO:0060082) |
| 0.3 | 1.0 | GO:0042262 | DNA protection(GO:0042262) |
| 0.3 | 6.8 | GO:0001865 | NK T cell differentiation(GO:0001865) |
| 0.3 | 1.6 | GO:0043056 | forward locomotion(GO:0043056) |
| 0.3 | 0.9 | GO:0038018 | Wnt receptor catabolic process(GO:0038018) |
| 0.3 | 0.8 | GO:0061357 | positive regulation of Wnt protein secretion(GO:0061357) |
| 0.3 | 8.4 | GO:0032060 | bleb assembly(GO:0032060) |
| 0.3 | 0.8 | GO:0021759 | globus pallidus development(GO:0021759) |
| 0.3 | 1.6 | GO:0042758 | long-chain fatty acid catabolic process(GO:0042758) |
| 0.3 | 2.5 | GO:0032876 | negative regulation of DNA endoreduplication(GO:0032876) |
| 0.2 | 15.4 | GO:0018279 | protein N-linked glycosylation via asparagine(GO:0018279) |
| 0.2 | 0.7 | GO:1902071 | regulation of hypoxia-inducible factor-1alpha signaling pathway(GO:1902071) |
| 0.2 | 4.1 | GO:0006027 | glycosaminoglycan catabolic process(GO:0006027) |
| 0.2 | 1.8 | GO:0061621 | NADH regeneration(GO:0006735) canonical glycolysis(GO:0061621) glucose catabolic process to pyruvate(GO:0061718) |
| 0.2 | 1.8 | GO:0030321 | transepithelial chloride transport(GO:0030321) |
| 0.2 | 0.6 | GO:0021629 | olfactory nerve morphogenesis(GO:0021627) olfactory nerve structural organization(GO:0021629) |
| 0.2 | 0.9 | GO:0034975 | protein folding in endoplasmic reticulum(GO:0034975) |
| 0.1 | 1.6 | GO:1903800 | positive regulation of production of miRNAs involved in gene silencing by miRNA(GO:1903800) |
| 0.1 | 0.6 | GO:0006069 | ethanol oxidation(GO:0006069) |
| 0.1 | 3.5 | GO:0034389 | lipid particle organization(GO:0034389) |
| 0.1 | 0.7 | GO:0051885 | positive regulation of anagen(GO:0051885) |
| 0.1 | 1.8 | GO:0043951 | negative regulation of cAMP-mediated signaling(GO:0043951) |
| 0.1 | 4.7 | GO:0070584 | mitochondrion morphogenesis(GO:0070584) |
| 0.1 | 2.6 | GO:0009263 | deoxyribonucleotide biosynthetic process(GO:0009263) |
| 0.1 | 0.8 | GO:0019348 | dolichol metabolic process(GO:0019348) |
| 0.1 | 0.9 | GO:0098909 | regulation of cardiac muscle cell action potential involved in regulation of contraction(GO:0098909) |
| 0.1 | 4.1 | GO:1903963 | arachidonic acid secretion(GO:0050482) arachidonate transport(GO:1903963) |
| 0.1 | 2.1 | GO:0002517 | T cell tolerance induction(GO:0002517) |
| 0.1 | 1.9 | GO:0008272 | sulfate transport(GO:0008272) |
| 0.1 | 0.3 | GO:1901082 | regulation of relaxation of smooth muscle(GO:1901080) positive regulation of relaxation of smooth muscle(GO:1901082) |
| 0.1 | 1.1 | GO:0048251 | elastic fiber assembly(GO:0048251) |
| 0.1 | 1.9 | GO:0021520 | spinal cord motor neuron cell fate specification(GO:0021520) |
| 0.1 | 2.1 | GO:0031987 | locomotion involved in locomotory behavior(GO:0031987) |
| 0.1 | 1.9 | GO:0046835 | carbohydrate phosphorylation(GO:0046835) |
| 0.1 | 1.1 | GO:1901898 | negative regulation of relaxation of cardiac muscle(GO:1901898) |
| 0.1 | 2.0 | GO:0055070 | copper ion homeostasis(GO:0055070) |
| 0.1 | 1.7 | GO:2000001 | regulation of DNA damage checkpoint(GO:2000001) |
| 0.1 | 0.8 | GO:0048318 | axial mesoderm development(GO:0048318) |
| 0.1 | 0.2 | GO:0046005 | positive regulation of circadian sleep/wake cycle, REM sleep(GO:0046005) |
| 0.1 | 0.7 | GO:2000258 | negative regulation of complement activation(GO:0045916) negative regulation of protein activation cascade(GO:2000258) |
| 0.1 | 0.4 | GO:0055005 | ventricular cardiac myofibril assembly(GO:0055005) |
| 0.1 | 1.3 | GO:0006120 | mitochondrial electron transport, NADH to ubiquinone(GO:0006120) |
| 0.1 | 1.5 | GO:0006744 | ubiquinone biosynthetic process(GO:0006744) |
| 0.1 | 0.9 | GO:0019227 | neuronal action potential propagation(GO:0019227) action potential propagation(GO:0098870) |
| 0.0 | 0.4 | GO:0071883 | activation of MAPK activity by adrenergic receptor signaling pathway(GO:0071883) |
| 0.0 | 3.5 | GO:0009250 | glycogen biosynthetic process(GO:0005978) glucan biosynthetic process(GO:0009250) |
| 0.0 | 0.3 | GO:1903232 | platelet dense granule organization(GO:0060155) phagosome acidification(GO:0090383) melanosome assembly(GO:1903232) |
| 0.0 | 1.2 | GO:0030903 | notochord development(GO:0030903) |
| 0.0 | 1.8 | GO:0042773 | ATP synthesis coupled electron transport(GO:0042773) |
| 0.0 | 0.6 | GO:1900246 | positive regulation of RIG-I signaling pathway(GO:1900246) |
| 0.0 | 0.6 | GO:0097154 | GABAergic neuron differentiation(GO:0097154) |
| 0.0 | 4.0 | GO:0055008 | cardiac muscle tissue morphogenesis(GO:0055008) |
| 0.0 | 0.3 | GO:0097475 | motor neuron migration(GO:0097475) |
| 0.0 | 4.9 | GO:0007098 | centrosome cycle(GO:0007098) |
| 0.0 | 5.2 | GO:0018107 | peptidyl-threonine phosphorylation(GO:0018107) |
| 0.0 | 1.1 | GO:0070979 | protein K11-linked ubiquitination(GO:0070979) |
| 0.0 | 2.0 | GO:0048701 | embryonic cranial skeleton morphogenesis(GO:0048701) |
| 0.0 | 0.1 | GO:2000320 | T cell homeostatic proliferation(GO:0001777) regulation of T cell homeostatic proliferation(GO:0046013) negative regulation of T-helper 17 type immune response(GO:2000317) negative regulation of T-helper 17 cell differentiation(GO:2000320) |
| 0.0 | 5.2 | GO:0009636 | response to toxic substance(GO:0009636) |
| 0.0 | 0.2 | GO:0014029 | neural crest formation(GO:0014029) |
| 0.0 | 0.4 | GO:0008535 | respiratory chain complex IV assembly(GO:0008535) |
| 0.0 | 1.5 | GO:0030968 | endoplasmic reticulum unfolded protein response(GO:0030968) |
| 0.0 | 0.3 | GO:0086069 | bundle of His cell to Purkinje myocyte communication(GO:0086069) |
| 0.0 | 0.2 | GO:0002155 | thyroid hormone mediated signaling pathway(GO:0002154) regulation of thyroid hormone mediated signaling pathway(GO:0002155) |
| 0.0 | 1.5 | GO:0016126 | sterol biosynthetic process(GO:0016126) |
| 0.0 | 0.8 | GO:0070534 | protein K63-linked ubiquitination(GO:0070534) |
| 0.0 | 1.6 | GO:0010923 | negative regulation of phosphatase activity(GO:0010923) |
| 0.0 | 0.1 | GO:0014835 | myoblast differentiation involved in skeletal muscle regeneration(GO:0014835) |
| 0.0 | 0.3 | GO:0035728 | response to hepatocyte growth factor(GO:0035728) |
| 0.0 | 0.2 | GO:0001731 | formation of translation preinitiation complex(GO:0001731) |
| 0.0 | 0.9 | GO:0006414 | translational elongation(GO:0006414) |
| 0.0 | 0.6 | GO:0061014 | positive regulation of mRNA catabolic process(GO:0061014) |
| 0.0 | 0.3 | GO:0015986 | energy coupled proton transport, down electrochemical gradient(GO:0015985) ATP synthesis coupled proton transport(GO:0015986) |
| 0.0 | 0.5 | GO:0006446 | regulation of translational initiation(GO:0006446) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.0 | 8.0 | GO:0031021 | interphase microtubule organizing center(GO:0031021) |
| 0.7 | 4.7 | GO:0097513 | myosin II filament(GO:0097513) |
| 0.5 | 5.2 | GO:0005890 | sodium:potassium-exchanging ATPase complex(GO:0005890) |
| 0.5 | 2.0 | GO:0070722 | Tle3-Aes complex(GO:0070722) |
| 0.5 | 55.6 | GO:0042571 | immunoglobulin complex, circulating(GO:0042571) |
| 0.4 | 2.6 | GO:0005971 | ribonucleoside-diphosphate reductase complex(GO:0005971) |
| 0.4 | 4.5 | GO:0042613 | MHC class II protein complex(GO:0042613) |
| 0.4 | 13.1 | GO:0044215 | other organism(GO:0044215) other organism cell(GO:0044216) other organism part(GO:0044217) |
| 0.3 | 0.8 | GO:0070992 | translation initiation complex(GO:0070992) |
| 0.3 | 3.0 | GO:0034363 | intermediate-density lipoprotein particle(GO:0034363) |
| 0.2 | 1.6 | GO:0071141 | SMAD protein complex(GO:0071141) |
| 0.2 | 1.5 | GO:1990462 | omegasome(GO:1990462) |
| 0.1 | 0.9 | GO:0005853 | eukaryotic translation elongation factor 1 complex(GO:0005853) |
| 0.1 | 2.1 | GO:0048787 | presynaptic active zone membrane(GO:0048787) |
| 0.1 | 4.6 | GO:0045271 | mitochondrial respiratory chain complex I(GO:0005747) NADH dehydrogenase complex(GO:0030964) respiratory chain complex I(GO:0045271) |
| 0.1 | 1.5 | GO:0005639 | integral component of nuclear inner membrane(GO:0005639) nuclear lamina(GO:0005652) intrinsic component of nuclear inner membrane(GO:0031229) |
| 0.1 | 0.9 | GO:0034663 | endoplasmic reticulum chaperone complex(GO:0034663) |
| 0.1 | 1.6 | GO:0005859 | muscle myosin complex(GO:0005859) |
| 0.1 | 12.4 | GO:0070160 | occluding junction(GO:0070160) |
| 0.1 | 4.1 | GO:0031941 | filamentous actin(GO:0031941) |
| 0.0 | 0.7 | GO:0031462 | Cul2-RING ubiquitin ligase complex(GO:0031462) |
| 0.0 | 1.6 | GO:0005790 | smooth endoplasmic reticulum(GO:0005790) |
| 0.0 | 0.3 | GO:0033162 | melanosome membrane(GO:0033162) ER-mitochondrion membrane contact site(GO:0044233) chitosome(GO:0045009) |
| 0.0 | 0.5 | GO:0097524 | sperm plasma membrane(GO:0097524) |
| 0.0 | 0.5 | GO:0016281 | eukaryotic translation initiation factor 4F complex(GO:0016281) |
| 0.0 | 0.7 | GO:0001527 | microfibril(GO:0001527) fibril(GO:0043205) |
| 0.0 | 5.2 | GO:0031526 | brush border membrane(GO:0031526) |
| 0.0 | 1.1 | GO:0005680 | anaphase-promoting complex(GO:0005680) |
| 0.0 | 0.6 | GO:0000177 | cytoplasmic exosome (RNase complex)(GO:0000177) |
| 0.0 | 2.0 | GO:0022627 | cytosolic small ribosomal subunit(GO:0022627) |
| 0.0 | 3.5 | GO:0005811 | lipid particle(GO:0005811) |
| 0.0 | 0.3 | GO:0008282 | ATP-sensitive potassium channel complex(GO:0008282) |
| 0.0 | 1.8 | GO:0060170 | ciliary membrane(GO:0060170) |
| 0.0 | 2.5 | GO:0005758 | mitochondrial intermembrane space(GO:0005758) |
| 0.0 | 1.5 | GO:0005719 | nuclear euchromatin(GO:0005719) |
| 0.0 | 3.5 | GO:0016234 | inclusion body(GO:0016234) |
| 0.0 | 1.6 | GO:0001533 | cornified envelope(GO:0001533) |
| 0.0 | 4.9 | GO:0030496 | midbody(GO:0030496) |
| 0.0 | 3.2 | GO:0005882 | intermediate filament(GO:0005882) |
| 0.0 | 9.3 | GO:0000139 | Golgi membrane(GO:0000139) |
| 0.0 | 0.3 | GO:0045263 | proton-transporting ATP synthase complex, coupling factor F(o)(GO:0045263) |
| 0.0 | 0.6 | GO:0031527 | filopodium membrane(GO:0031527) |
| 0.0 | 0.5 | GO:0042470 | melanosome(GO:0042470) pigment granule(GO:0048770) |
| 0.0 | 1.1 | GO:0005891 | voltage-gated calcium channel complex(GO:0005891) |
| 0.0 | 0.4 | GO:0070069 | cytochrome complex(GO:0070069) |
| 0.0 | 4.9 | GO:0005911 | cell-cell junction(GO:0005911) |
| 0.0 | 2.4 | GO:0031225 | anchored component of membrane(GO:0031225) |
| 0.0 | 3.7 | GO:0016324 | apical plasma membrane(GO:0016324) |
| 0.0 | 0.9 | GO:0014704 | intercalated disc(GO:0014704) |
| 0.0 | 0.9 | GO:0034707 | chloride channel complex(GO:0034707) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.2 | 15.4 | GO:0052798 | beta-galactoside alpha-2,3-sialyltransferase activity(GO:0052798) |
| 1.7 | 5.2 | GO:0008900 | hydrogen:potassium-exchanging ATPase activity(GO:0008900) |
| 1.2 | 3.5 | GO:0004373 | glycogen (starch) synthase activity(GO:0004373) |
| 1.0 | 5.2 | GO:0005275 | amine transmembrane transporter activity(GO:0005275) |
| 1.0 | 13.1 | GO:0042834 | peptidoglycan binding(GO:0042834) |
| 0.8 | 2.5 | GO:0019153 | protein-disulfide reductase (glutathione) activity(GO:0019153) |
| 0.6 | 11.4 | GO:0019841 | retinol binding(GO:0019841) |
| 0.6 | 1.8 | GO:0045030 | UTP-activated nucleotide receptor activity(GO:0045030) |
| 0.6 | 0.6 | GO:0004024 | alcohol dehydrogenase activity, zinc-dependent(GO:0004024) |
| 0.6 | 5.2 | GO:0004687 | myosin light chain kinase activity(GO:0004687) |
| 0.5 | 1.6 | GO:0035500 | MH2 domain binding(GO:0035500) |
| 0.5 | 3.0 | GO:0060230 | lipoprotein lipase activator activity(GO:0060230) |
| 0.4 | 2.6 | GO:0016728 | ribonucleoside-diphosphate reductase activity, thioredoxin disulfide as acceptor(GO:0004748) oxidoreductase activity, acting on CH or CH2 groups, disulfide as acceptor(GO:0016728) ribonucleoside-diphosphate reductase activity(GO:0061731) |
| 0.4 | 55.6 | GO:0034987 | immunoglobulin receptor binding(GO:0034987) |
| 0.4 | 1.2 | GO:0004350 | glutamate 5-kinase activity(GO:0004349) glutamate-5-semialdehyde dehydrogenase activity(GO:0004350) delta1-pyrroline-5-carboxylate synthetase activity(GO:0017084) amino acid kinase activity(GO:0019202) |
| 0.4 | 1.6 | GO:0004466 | long-chain-acyl-CoA dehydrogenase activity(GO:0004466) |
| 0.3 | 4.0 | GO:0070700 | BMP receptor binding(GO:0070700) |
| 0.3 | 1.5 | GO:0038181 | bile acid receptor activity(GO:0038181) |
| 0.3 | 4.5 | GO:1990405 | protein antigen binding(GO:1990405) |
| 0.3 | 2.1 | GO:0060072 | large conductance calcium-activated potassium channel activity(GO:0060072) |
| 0.2 | 6.3 | GO:0030898 | actin-dependent ATPase activity(GO:0030898) |
| 0.2 | 2.1 | GO:0008597 | calcium-dependent protein serine/threonine phosphatase regulator activity(GO:0008597) |
| 0.2 | 1.9 | GO:0019158 | fructokinase activity(GO:0008865) mannokinase activity(GO:0019158) |
| 0.2 | 0.8 | GO:0003976 | UDP-N-acetylglucosamine-lysosomal-enzyme N-acetylglucosaminephosphotransferase activity(GO:0003976) |
| 0.2 | 1.8 | GO:0016861 | intramolecular oxidoreductase activity, interconverting aldoses and ketoses(GO:0016861) |
| 0.2 | 8.9 | GO:0008536 | Ran GTPase binding(GO:0008536) |
| 0.2 | 4.6 | GO:0005540 | hyaluronic acid binding(GO:0005540) |
| 0.1 | 1.5 | GO:0070087 | chromo shadow domain binding(GO:0070087) |
| 0.1 | 1.6 | GO:0031433 | telethonin binding(GO:0031433) |
| 0.1 | 4.1 | GO:0004623 | phospholipase A2 activity(GO:0004623) |
| 0.1 | 1.5 | GO:0008329 | signaling pattern recognition receptor activity(GO:0008329) pattern recognition receptor activity(GO:0038187) |
| 0.1 | 1.9 | GO:0019531 | oxalate transmembrane transporter activity(GO:0019531) |
| 0.1 | 5.9 | GO:0005154 | epidermal growth factor receptor binding(GO:0005154) |
| 0.1 | 3.3 | GO:0003954 | NADH dehydrogenase activity(GO:0003954) |
| 0.1 | 0.4 | GO:0070506 | high-density lipoprotein particle receptor activity(GO:0070506) |
| 0.1 | 0.3 | GO:0036461 | BLOC-2 complex binding(GO:0036461) |
| 0.1 | 6.8 | GO:0004715 | non-membrane spanning protein tyrosine kinase activity(GO:0004715) |
| 0.1 | 1.5 | GO:0001608 | G-protein coupled nucleotide receptor activity(GO:0001608) G-protein coupled purinergic nucleotide receptor activity(GO:0045028) |
| 0.1 | 1.0 | GO:0047429 | nucleoside-triphosphate diphosphatase activity(GO:0047429) |
| 0.1 | 1.7 | GO:0031210 | phosphatidylcholine binding(GO:0031210) |
| 0.1 | 0.2 | GO:0031531 | thyrotropin-releasing hormone receptor binding(GO:0031531) |
| 0.1 | 0.2 | GO:0001042 | RNA polymerase I core binding(GO:0001042) |
| 0.1 | 0.8 | GO:0031852 | mu-type opioid receptor binding(GO:0031852) |
| 0.1 | 0.9 | GO:0016671 | oxidoreductase activity, acting on a sulfur group of donors, disulfide as acceptor(GO:0016671) |
| 0.1 | 3.9 | GO:0051393 | alpha-actinin binding(GO:0051393) |
| 0.1 | 1.0 | GO:0070008 | serine-type exopeptidase activity(GO:0070008) |
| 0.1 | 6.8 | GO:0003730 | mRNA 3'-UTR binding(GO:0003730) |
| 0.1 | 0.3 | GO:0031013 | troponin I binding(GO:0031013) |
| 0.1 | 0.8 | GO:0001161 | intronic transcription regulatory region sequence-specific DNA binding(GO:0001161) |
| 0.1 | 2.7 | GO:0061631 | ubiquitin conjugating enzyme activity(GO:0061631) |
| 0.0 | 0.6 | GO:0017151 | DEAD/H-box RNA helicase binding(GO:0017151) |
| 0.0 | 0.9 | GO:0005247 | voltage-gated chloride channel activity(GO:0005247) |
| 0.0 | 0.8 | GO:0035497 | cAMP response element binding(GO:0035497) |
| 0.0 | 2.0 | GO:0001106 | RNA polymerase II transcription corepressor activity(GO:0001106) |
| 0.0 | 0.9 | GO:0003746 | translation elongation factor activity(GO:0003746) |
| 0.0 | 0.4 | GO:0004691 | cAMP-dependent protein kinase activity(GO:0004691) |
| 0.0 | 1.6 | GO:0005109 | frizzled binding(GO:0005109) |
| 0.0 | 2.7 | GO:0005507 | copper ion binding(GO:0005507) |
| 0.0 | 0.6 | GO:0002162 | dystroglycan binding(GO:0002162) |
| 0.0 | 1.1 | GO:0004115 | 3',5'-cyclic-AMP phosphodiesterase activity(GO:0004115) |
| 0.0 | 0.6 | GO:0008253 | 5'-nucleotidase activity(GO:0008253) |
| 0.0 | 0.1 | GO:0005134 | interleukin-2 receptor binding(GO:0005134) |
| 0.0 | 1.2 | GO:0005504 | fatty acid binding(GO:0005504) |
| 0.0 | 2.9 | GO:0004725 | protein tyrosine phosphatase activity(GO:0004725) |
| 0.0 | 0.5 | GO:0004602 | glutathione peroxidase activity(GO:0004602) |
| 0.0 | 0.4 | GO:0015002 | cytochrome-c oxidase activity(GO:0004129) heme-copper terminal oxidase activity(GO:0015002) oxidoreductase activity, acting on a heme group of donors, oxygen as acceptor(GO:0016676) |
| 0.0 | 2.9 | GO:0003735 | structural constituent of ribosome(GO:0003735) |
| 0.0 | 1.0 | GO:0042054 | histone methyltransferase activity(GO:0042054) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 5.9 | PID ERBB NETWORK PATHWAY | ErbB receptor signaling network |
| 0.2 | 8.0 | PID AURORA A PATHWAY | Aurora A signaling |
| 0.1 | 3.5 | PID INSULIN GLUCOSE PATHWAY | Insulin-mediated glucose transport |
| 0.1 | 11.4 | PID RB 1PATHWAY | Regulation of retinoblastoma protein |
| 0.1 | 2.9 | PID PRL SIGNALING EVENTS PATHWAY | Signaling events mediated by PRL |
| 0.1 | 6.8 | ST T CELL SIGNAL TRANSDUCTION | T Cell Signal Transduction |
| 0.1 | 11.5 | NABA ECM AFFILIATED | Genes encoding proteins affiliated structurally or functionally to extracellular matrix proteins |
| 0.0 | 2.5 | PID PLK1 PATHWAY | PLK1 signaling events |
| 0.0 | 1.8 | SIG REGULATION OF THE ACTIN CYTOSKELETON BY RHO GTPASES | Genes related to regulation of the actin cytoskeleton |
| 0.0 | 2.6 | PID E2F PATHWAY | E2F transcription factor network |
| 0.0 | 2.1 | PID HES HEY PATHWAY | Notch-mediated HES/HEY network |
| 0.0 | 1.5 | PID RXR VDR PATHWAY | RXR and RAR heterodimerization with other nuclear receptor |
| 0.0 | 1.1 | PID HNF3B PATHWAY | FOXA2 and FOXA3 transcription factor networks |
| 0.0 | 0.6 | PID DELTA NP63 PATHWAY | Validated transcriptional targets of deltaNp63 isoforms |
| 0.0 | 1.6 | PID P53 DOWNSTREAM PATHWAY | Direct p53 effectors |
| 0.0 | 0.9 | PID P73PATHWAY | p73 transcription factor network |
| 0.0 | 0.7 | PID ECADHERIN STABILIZATION PATHWAY | Stabilization and expansion of the E-cadherin adherens junction |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 15.4 | REACTOME PRE NOTCH PROCESSING IN GOLGI | Genes involved in Pre-NOTCH Processing in Golgi |
| 0.4 | 4.4 | REACTOME HYALURONAN UPTAKE AND DEGRADATION | Genes involved in Hyaluronan uptake and degradation |
| 0.3 | 12.0 | REACTOME TIGHT JUNCTION INTERACTIONS | Genes involved in Tight junction interactions |
| 0.2 | 4.1 | REACTOME ACYL CHAIN REMODELLING OF PS | Genes involved in Acyl chain remodelling of PS |
| 0.2 | 11.4 | REACTOME METABOLISM OF STEROID HORMONES AND VITAMINS A AND D | Genes involved in Metabolism of steroid hormones and vitamins A and D |
| 0.2 | 5.2 | REACTOME NA CL DEPENDENT NEUROTRANSMITTER TRANSPORTERS | Genes involved in Na+/Cl- dependent neurotransmitter transporters |
| 0.2 | 6.8 | REACTOME GENERATION OF SECOND MESSENGER MOLECULES | Genes involved in Generation of second messenger molecules |
| 0.2 | 3.0 | REACTOME HDL MEDIATED LIPID TRANSPORT | Genes involved in HDL-mediated lipid transport |
| 0.2 | 5.1 | REACTOME SYNTHESIS AND INTERCONVERSION OF NUCLEOTIDE DI AND TRIPHOSPHATES | Genes involved in Synthesis and interconversion of nucleotide di- and triphosphates |
| 0.1 | 5.9 | REACTOME SHC1 EVENTS IN ERBB4 SIGNALING | Genes involved in SHC1 events in ERBB4 signaling |
| 0.1 | 4.7 | REACTOME SEMA4D INDUCED CELL MIGRATION AND GROWTH CONE COLLAPSE | Genes involved in Sema4D induced cell migration and growth-cone collapse |
| 0.1 | 1.8 | REACTOME P2Y RECEPTORS | Genes involved in P2Y receptors |
| 0.1 | 5.2 | REACTOME ION TRANSPORT BY P TYPE ATPASES | Genes involved in Ion transport by P-type ATPases |
| 0.1 | 1.6 | REACTOME MITOCHONDRIAL FATTY ACID BETA OXIDATION | Genes involved in Mitochondrial Fatty Acid Beta-Oxidation |
| 0.1 | 10.6 | REACTOME MITOTIC PROMETAPHASE | Genes involved in Mitotic Prometaphase |
| 0.1 | 1.7 | REACTOME SYNTHESIS SECRETION AND INACTIVATION OF GIP | Genes involved in Synthesis, Secretion, and Inactivation of Glucose-dependent Insulinotropic Polypeptide (GIP) |
| 0.1 | 4.6 | REACTOME RESPIRATORY ELECTRON TRANSPORT | Genes involved in Respiratory electron transport |
| 0.0 | 1.5 | REACTOME CHOLESTEROL BIOSYNTHESIS | Genes involved in Cholesterol biosynthesis |
| 0.0 | 1.2 | REACTOME AMINO ACID SYNTHESIS AND INTERCONVERSION TRANSAMINATION | Genes involved in Amino acid synthesis and interconversion (transamination) |
| 0.0 | 1.1 | REACTOME DARPP 32 EVENTS | Genes involved in DARPP-32 events |
| 0.0 | 2.0 | REACTOME FORMATION OF THE TERNARY COMPLEX AND SUBSEQUENTLY THE 43S COMPLEX | Genes involved in Formation of the ternary complex, and subsequently, the 43S complex |
| 0.0 | 3.0 | REACTOME GLUCOSE METABOLISM | Genes involved in Glucose metabolism |
| 0.0 | 1.9 | REACTOME COSTIMULATION BY THE CD28 FAMILY | Genes involved in Costimulation by the CD28 family |
| 0.0 | 0.8 | REACTOME BIOSYNTHESIS OF THE N GLYCAN PRECURSOR DOLICHOL LIPID LINKED OLIGOSACCHARIDE LLO AND TRANSFER TO A NASCENT PROTEIN | Genes involved in Biosynthesis of the N-glycan precursor (dolichol lipid-linked oligosaccharide, LLO) and transfer to a nascent protein |
| 0.0 | 0.3 | REACTOME SIGNALING BY CONSTITUTIVELY ACTIVE EGFR | Genes involved in Signaling by constitutively active EGFR |
| 0.0 | 1.7 | REACTOME TRANSLATION | Genes involved in Translation |
| 0.0 | 0.4 | REACTOME ERK MAPK TARGETS | Genes involved in ERK/MAPK targets |