Project
PRJNA375882: Comprehensive Mouse Transcriptomic BodyMap
Navigation
Downloads
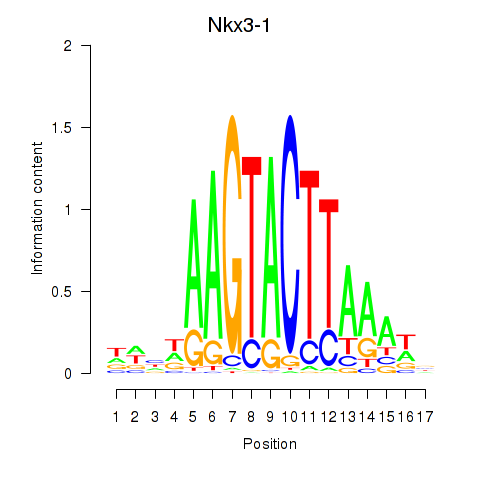
Results for Nkx3-1
Z-value: 1.27
Transcription factors associated with Nkx3-1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Nkx3-1
|
ENSMUSG00000022061.9 | Nkx3-1 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Nkx3-1 | mm39_v1_chr14_+_69428087_69428140 | -0.05 | 6.8e-01 | Click! |
Activity profile of Nkx3-1 motif
Sorted Z-values of Nkx3-1 motif
Network of associatons between targets according to the STRING database.
First level regulatory network of Nkx3-1
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr19_+_58748132 | 10.49 |
ENSMUST00000026081.5
|
Pnliprp2
|
pancreatic lipase-related protein 2 |
| chr3_-_113325938 | 7.34 |
ENSMUST00000132353.2
|
Amy2a1
|
amylase 2a1 |
| chr4_-_137157824 | 7.34 |
ENSMUST00000102522.5
|
Cela3b
|
chymotrypsin-like elastase family, member 3B |
| chr6_-_41012435 | 6.46 |
ENSMUST00000031931.6
|
2210010C04Rik
|
RIKEN cDNA 2210010C04 gene |
| chr3_-_113198765 | 6.16 |
ENSMUST00000179568.3
|
Amy2a4
|
amylase 2a4 |
| chr3_-_113231368 | 5.77 |
ENSMUST00000179314.3
|
Amy2a3
|
amylase 2a3 |
| chr3_-_113263974 | 5.68 |
ENSMUST00000098667.5
|
Amy2a2
|
amylase 2a2 |
| chr6_+_78402956 | 5.32 |
ENSMUST00000079926.6
|
Reg1
|
regenerating islet-derived 1 |
| chr3_-_113166153 | 5.04 |
ENSMUST00000098673.5
|
Amy2a5
|
amylase 2a5 |
| chr8_+_114860375 | 4.57 |
ENSMUST00000147605.8
ENSMUST00000134593.2 |
Nudt7
|
nudix (nucleoside diphosphate linked moiety X)-type motif 7 |
| chr8_+_46984016 | 4.55 |
ENSMUST00000152423.2
|
Acsl1
|
acyl-CoA synthetase long-chain family member 1 |
| chr8_+_114860297 | 4.46 |
ENSMUST00000073521.12
ENSMUST00000066514.13 |
Nudt7
|
nudix (nucleoside diphosphate linked moiety X)-type motif 7 |
| chr3_-_92659644 | 4.21 |
ENSMUST00000029527.6
|
Lce1g
|
late cornified envelope 1G |
| chr8_+_114860342 | 4.17 |
ENSMUST00000109109.8
|
Nudt7
|
nudix (nucleoside diphosphate linked moiety X)-type motif 7 |
| chr3_+_146276147 | 4.08 |
ENSMUST00000199489.5
|
Uox
|
urate oxidase |
| chr1_+_139429430 | 3.97 |
ENSMUST00000027615.7
|
F13b
|
coagulation factor XIII, beta subunit |
| chr10_-_59277570 | 3.89 |
ENSMUST00000009798.5
|
Oit3
|
oncoprotein induced transcript 3 |
| chr4_-_108075119 | 3.77 |
ENSMUST00000223127.2
ENSMUST00000043793.7 ENSMUST00000106690.9 |
Zyg11a
|
zyg-11 family member A, cell cycle regulator |
| chr11_-_58504307 | 3.72 |
ENSMUST00000048801.8
|
Lypd8l
|
LY6/PLAUR domain containing 8 like |
| chr14_-_21798678 | 3.69 |
ENSMUST00000075040.9
|
Dusp13
|
dual specificity phosphatase 13 |
| chr15_-_96918203 | 3.69 |
ENSMUST00000166223.2
|
Slc38a4
|
solute carrier family 38, member 4 |
| chr11_-_21320452 | 3.56 |
ENSMUST00000102875.11
|
Ugp2
|
UDP-glucose pyrophosphorylase 2 |
| chr4_-_137137088 | 3.48 |
ENSMUST00000024200.7
|
Cela3a
|
chymotrypsin-like elastase family, member 3A |
| chr12_+_8062331 | 3.43 |
ENSMUST00000171239.2
|
Apob
|
apolipoprotein B |
| chr1_-_190897012 | 3.43 |
ENSMUST00000171798.2
|
Fam71a
|
family with sequence similarity 71, member A |
| chr6_-_83504471 | 3.25 |
ENSMUST00000141904.8
|
Actg2
|
actin, gamma 2, smooth muscle, enteric |
| chr2_-_84605764 | 3.24 |
ENSMUST00000111641.2
|
Serping1
|
serine (or cysteine) peptidase inhibitor, clade G, member 1 |
| chr2_-_84605732 | 3.17 |
ENSMUST00000023994.10
|
Serping1
|
serine (or cysteine) peptidase inhibitor, clade G, member 1 |
| chr6_-_83504756 | 3.15 |
ENSMUST00000152029.2
|
Actg2
|
actin, gamma 2, smooth muscle, enteric |
| chr5_-_145521533 | 3.10 |
ENSMUST00000075837.8
|
Cyp3a41b
|
cytochrome P450, family 3, subfamily a, polypeptide 41B |
| chr14_-_21798694 | 3.04 |
ENSMUST00000183943.2
|
Dusp13
|
dual specificity phosphatase 13 |
| chr11_-_121410152 | 2.85 |
ENSMUST00000092298.6
|
Zfp750
|
zinc finger protein 750 |
| chr5_-_145816774 | 2.76 |
ENSMUST00000035918.8
|
Cyp3a11
|
cytochrome P450, family 3, subfamily a, polypeptide 11 |
| chr4_-_115353326 | 2.72 |
ENSMUST00000030487.3
|
Cyp4a14
|
cytochrome P450, family 4, subfamily a, polypeptide 14 |
| chrX_+_149330371 | 2.71 |
ENSMUST00000066337.13
ENSMUST00000112715.2 |
Alas2
|
aminolevulinic acid synthase 2, erythroid |
| chrX_+_163052367 | 2.65 |
ENSMUST00000145412.8
ENSMUST00000033749.9 |
Pir
|
pirin |
| chr3_-_92686198 | 2.64 |
ENSMUST00000090866.2
|
Lce1i
|
late cornified envelope 1I |
| chr5_-_145742672 | 2.61 |
ENSMUST00000067479.6
|
Cyp3a44
|
cytochrome P450, family 3, subfamily a, polypeptide 44 |
| chr5_-_21629661 | 2.59 |
ENSMUST00000115245.8
ENSMUST00000030552.7 |
Ccdc146
|
coiled-coil domain containing 146 |
| chr6_+_54016543 | 2.54 |
ENSMUST00000046856.14
|
Chn2
|
chimerin 2 |
| chr13_-_41981893 | 2.51 |
ENSMUST00000137905.2
|
Adtrp
|
androgen dependent TFPI regulating protein |
| chr3_+_57332735 | 2.47 |
ENSMUST00000029377.8
|
Tm4sf4
|
transmembrane 4 superfamily member 4 |
| chr5_-_145406533 | 2.46 |
ENSMUST00000031633.5
|
Cyp3a16
|
cytochrome P450, family 3, subfamily a, polypeptide 16 |
| chr1_+_157334347 | 2.39 |
ENSMUST00000027881.15
|
Sec16b
|
SEC16 homolog B (S. cerevisiae) |
| chr3_+_93184854 | 2.38 |
ENSMUST00000180308.3
|
Flg
|
filaggrin |
| chr5_-_145656934 | 2.37 |
ENSMUST00000094111.6
|
Cyp3a41a
|
cytochrome P450, family 3, subfamily a, polypeptide 41A |
| chr1_+_157334298 | 2.36 |
ENSMUST00000086130.9
|
Sec16b
|
SEC16 homolog B (S. cerevisiae) |
| chr13_-_41981812 | 2.27 |
ENSMUST00000223337.2
ENSMUST00000221691.2 |
Adtrp
|
androgen dependent TFPI regulating protein |
| chr19_-_10217968 | 2.27 |
ENSMUST00000189897.2
ENSMUST00000186056.7 ENSMUST00000088013.12 |
Myrf
|
myelin regulatory factor |
| chr14_+_40853734 | 2.26 |
ENSMUST00000022314.10
ENSMUST00000170719.2 |
Sftpa1
|
surfactant associated protein A1 |
| chr9_+_78099229 | 2.18 |
ENSMUST00000034903.7
|
Gsta4
|
glutathione S-transferase, alpha 4 |
| chr7_-_133203838 | 2.15 |
ENSMUST00000033275.4
|
Tex36
|
testis expressed 36 |
| chr2_+_79538124 | 2.12 |
ENSMUST00000090760.9
ENSMUST00000040863.11 ENSMUST00000111780.3 |
Ppp1r1c
|
protein phosphatase 1, regulatory inhibitor subunit 1C |
| chr18_+_43897354 | 2.11 |
ENSMUST00000187157.7
ENSMUST00000043803.13 ENSMUST00000189750.2 |
Scgb3a2
|
secretoglobin, family 3A, member 2 |
| chr5_+_73071607 | 2.09 |
ENSMUST00000144843.8
|
Slain2
|
SLAIN motif family, member 2 |
| chr13_-_42000958 | 1.96 |
ENSMUST00000072012.10
|
Adtrp
|
androgen dependent TFPI regulating protein |
| chr18_-_12995681 | 1.93 |
ENSMUST00000121808.8
ENSMUST00000118313.8 |
Osbpl1a
|
oxysterol binding protein-like 1A |
| chr12_+_101370932 | 1.89 |
ENSMUST00000055156.5
|
Catsperb
|
cation channel sperm associated auxiliary subunit beta |
| chr1_-_107206091 | 1.78 |
ENSMUST00000166100.2
ENSMUST00000027565.5 |
Serpinb3b
Serpinb3c
|
serine (or cysteine) peptidase inhibitor, clade B (ovalbumin), member 3B serine (or cysteine) peptidase inhibitor, clade B, member 3C |
| chr9_-_103099262 | 1.75 |
ENSMUST00000170904.2
|
Trf
|
transferrin |
| chr14_+_64331130 | 1.74 |
ENSMUST00000224112.2
ENSMUST00000165710.2 ENSMUST00000170709.2 |
Prss51
|
protease, serine 51 |
| chr2_-_52225763 | 1.74 |
ENSMUST00000238288.2
ENSMUST00000238749.2 |
Neb
|
nebulin |
| chr6_-_88604404 | 1.71 |
ENSMUST00000120933.5
|
Kbtbd12
|
kelch repeat and BTB (POZ) domain containing 12 |
| chr3_-_54962899 | 1.71 |
ENSMUST00000199144.5
|
Ccna1
|
cyclin A1 |
| chr3_-_54962922 | 1.69 |
ENSMUST00000197238.5
|
Ccna1
|
cyclin A1 |
| chr15_+_102058936 | 1.67 |
ENSMUST00000023806.14
|
Soat2
|
sterol O-acyltransferase 2 |
| chr19_-_8382424 | 1.61 |
ENSMUST00000064507.12
ENSMUST00000120540.2 ENSMUST00000096269.11 |
Slc22a30
|
solute carrier family 22, member 30 |
| chr12_+_17316546 | 1.61 |
ENSMUST00000057288.7
ENSMUST00000239402.2 |
Pdia6
|
protein disulfide isomerase associated 6 |
| chrY_+_12273932 | 1.58 |
ENSMUST00000187922.2
|
Gm29554
|
predicted gene 29554 |
| chr18_+_57666852 | 1.58 |
ENSMUST00000079738.10
ENSMUST00000135806.8 ENSMUST00000127130.9 |
Ccdc192
|
coiled-coil domain containing 192 |
| chr7_-_103799746 | 1.55 |
ENSMUST00000059121.5
|
Ubqlnl
|
ubiquilin-like |
| chr8_-_41494890 | 1.54 |
ENSMUST00000051379.14
|
Mtus1
|
mitochondrial tumor suppressor 1 |
| chr11_-_61269131 | 1.53 |
ENSMUST00000148671.2
|
Slc47a1
|
solute carrier family 47, member 1 |
| chr17_-_57289121 | 1.52 |
ENSMUST00000056113.5
|
Acer1
|
alkaline ceramidase 1 |
| chr9_+_103917821 | 1.51 |
ENSMUST00000216593.2
ENSMUST00000147249.3 |
Nphp3
Gm28305
|
nephronophthisis 3 (adolescent) predicted gene 28305 |
| chr5_-_24732200 | 1.51 |
ENSMUST00000088311.6
|
Gbx1
|
gastrulation brain homeobox 1 |
| chrY_+_57195716 | 1.50 |
ENSMUST00000189109.7
ENSMUST00000191355.7 ENSMUST00000190292.2 |
Sly
|
Sycp3 like Y-linked |
| chr2_+_22959223 | 1.50 |
ENSMUST00000114523.10
|
Acbd5
|
acyl-Coenzyme A binding domain containing 5 |
| chr5_-_134776101 | 1.48 |
ENSMUST00000015138.13
|
Eln
|
elastin |
| chr10_+_97442727 | 1.47 |
ENSMUST00000105286.4
|
Kera
|
keratocan |
| chr1_+_170983081 | 1.47 |
ENSMUST00000111334.2
|
Mpz
|
myelin protein zero |
| chr9_-_40873629 | 1.45 |
ENSMUST00000160120.2
|
Jhy
|
junctional cadherin complex regulator |
| chrY_+_65386218 | 1.42 |
ENSMUST00000190282.7
ENSMUST00000185550.7 |
Gm20736
|
predicted gene, 20736 |
| chrX_+_14077387 | 1.39 |
ENSMUST00000105137.4
|
H2al1n
|
H2A histone family member L1N |
| chrX_-_117390434 | 1.39 |
ENSMUST00000073857.6
|
Tgif2lx1
|
TGFB-induced factor homeobox 2-like, X-linked 1 |
| chr1_+_106908709 | 1.38 |
ENSMUST00000027564.8
|
Serpinb13
|
serine (or cysteine) peptidase inhibitor, clade B (ovalbumin), member 13 |
| chr4_+_134124691 | 1.38 |
ENSMUST00000105870.8
|
Pafah2
|
platelet-activating factor acetylhydrolase 2 |
| chrY_-_1459771 | 1.35 |
ENSMUST00000091188.7
|
Usp9y
|
ubiquitin specific peptidase 9, Y chromosome |
| chr9_+_106325860 | 1.33 |
ENSMUST00000185527.3
|
Abhd14b
|
abhydrolase domain containing 14b |
| chr10_-_128047658 | 1.32 |
ENSMUST00000061995.10
|
Spryd4
|
SPRY domain containing 4 |
| chrY_-_68306157 | 1.31 |
ENSMUST00000189084.7
ENSMUST00000189422.7 |
Gm20937
|
predicted gene, 20937 |
| chr5_-_86893645 | 1.30 |
ENSMUST00000161306.2
|
Tmprss11e
|
transmembrane protease, serine 11e |
| chr17_+_29487881 | 1.30 |
ENSMUST00000234845.2
ENSMUST00000235038.2 ENSMUST00000235050.2 ENSMUST00000120346.9 ENSMUST00000234377.2 ENSMUST00000235074.2 ENSMUST00000235040.2 ENSMUST00000234256.2 ENSMUST00000234459.2 |
BC004004
|
cDNA sequence BC004004 |
| chrX_+_156481906 | 1.26 |
ENSMUST00000136141.2
ENSMUST00000190091.7 |
Smpx
|
small muscle protein, X-linked |
| chr3_+_62327089 | 1.26 |
ENSMUST00000161057.2
|
Arhgef26
|
Rho guanine nucleotide exchange factor (GEF) 26 |
| chr1_-_130380153 | 1.24 |
ENSMUST00000140400.2
|
Cd55
|
CD55 molecule, decay accelerating factor for complement |
| chrY_+_14400710 | 1.23 |
ENSMUST00000192683.2
|
Gm30737
|
predicted gene, 30737 |
| chrY_+_81469276 | 1.23 |
ENSMUST00000187135.2
|
Gm20911
|
predicted gene, 20911 |
| chrY_+_19113007 | 1.23 |
ENSMUST00000194086.2
|
Gm35134
|
predicted gene, 35134 |
| chrY_+_71480718 | 1.21 |
ENSMUST00000188958.2
|
Gm20869
|
predicted gene, 20869 |
| chrY_+_87128072 | 1.21 |
ENSMUST00000189543.7
|
Gm21477
|
predicted gene, 21477 |
| chrY_-_35087178 | 1.21 |
ENSMUST00000186996.7
|
Gm20855
|
predicted gene, 20855 |
| chrY_+_88064584 | 1.21 |
ENSMUST00000187146.2
|
Gm28102
|
predicted gene 28102 |
| chrY_+_7722941 | 1.21 |
ENSMUST00000189007.2
|
Gm28576
|
predicted gene 28576 |
| chrY_+_78224262 | 1.20 |
ENSMUST00000185575.2
|
Gm29564
|
predicted gene 29564 |
| chrY_+_80145048 | 1.20 |
ENSMUST00000187433.7
|
Gm21760
|
predicted gene, 21760 |
| chrY_+_61655274 | 1.20 |
ENSMUST00000189455.2
|
Gm21497
|
predicted gene, 21497 |
| chrY_-_40272248 | 1.20 |
ENSMUST00000191443.7
|
Gm21865
|
predicted gene, 21865 |
| chrY_+_48861106 | 1.20 |
ENSMUST00000185924.2
|
Gm28553
|
predicted gene 28553 |
| chrY_+_7253787 | 1.20 |
ENSMUST00000186194.2
|
Gm28998
|
predicted gene 28998 |
| chrY_+_76772869 | 1.20 |
ENSMUST00000189238.2
|
Gm21173
|
predicted gene, 21173 |
| chrY_-_68843997 | 1.20 |
ENSMUST00000191123.2
|
Gm20817
|
predicted gene, 20817 |
| chrY_-_35876871 | 1.20 |
ENSMUST00000188585.7
|
Gm20896
|
predicted gene, 20896 |
| chrY_+_80938264 | 1.20 |
ENSMUST00000185340.2
|
Gm28897
|
predicted gene 28897 |
| chrY_+_83042216 | 1.20 |
ENSMUST00000187165.2
|
Gm28827
|
predicted gene 28827 |
| chrX_+_133088238 | 1.19 |
ENSMUST00000064476.5
|
Arl13a
|
ADP-ribosylation factor-like 13A |
| chrY_+_74481800 | 1.19 |
ENSMUST00000189924.2
|
Gm29110
|
predicted gene 29110 |
| chrY_-_22046833 | 1.19 |
ENSMUST00000192521.2
|
Gm31571
|
predicted gene, 31571 |
| chrY_+_86073041 | 1.18 |
ENSMUST00000188754.2
|
Gm20820
|
predicted gene, 20820 |
| chrY_+_87562235 | 1.18 |
ENSMUST00000186493.2
|
Gm20906
|
predicted gene, 20906 |
| chrX_+_117336912 | 1.18 |
ENSMUST00000072518.8
|
Tgif2lx2
|
TGFB-induced factor homeobox 2-like, X-linked 2 |
| chr10_-_129751466 | 1.18 |
ENSMUST00000213438.2
|
Olfr816
|
olfactory receptor 816 |
| chrY_+_53840528 | 1.18 |
ENSMUST00000186578.2
|
Gm20929
|
predicted gene, 20929 |
| chrY_+_82236496 | 1.18 |
ENSMUST00000185636.2
|
Gm21317
|
predicted gene, 21317 |
| chrY_-_22856736 | 1.18 |
ENSMUST00000192892.2
|
Gm21366
|
predicted gene, 21366 |
| chrY_+_84757954 | 1.18 |
ENSMUST00000186110.2
|
Gm21409
|
predicted gene, 21409 |
| chrY_-_36690635 | 1.18 |
ENSMUST00000185565.2
|
Gm20835
|
predicted gene, 20835 |
| chrY_-_37214948 | 1.18 |
ENSMUST00000190782.2
|
Gm20905
|
predicted gene, 20905 |
| chrY_+_52776619 | 1.18 |
ENSMUST00000191553.7
|
Gm21258
|
predicted gene, 21258 |
| chrY_+_75726591 | 1.18 |
ENSMUST00000188672.2
|
Gm20850
|
predicted gene, 20850 |
| chrY_-_20993657 | 1.17 |
ENSMUST00000191675.2
|
Gm29866
|
predicted gene, 29866 |
| chrY_+_49567735 | 1.17 |
ENSMUST00000189354.7
|
Gm21209
|
predicted gene, 21209 |
| chrY_+_62199394 | 1.17 |
ENSMUST00000186938.7
|
Gm21518
|
predicted gene, 21518 |
| chrY_+_63236955 | 1.17 |
ENSMUST00000187768.2
|
Gm21627
|
predicted gene, 21627 |
| chrY_+_79330834 | 1.17 |
ENSMUST00000185338.7
|
Gm20916
|
predicted gene, 20916 |
| chrY_+_71703312 | 1.17 |
ENSMUST00000189315.2
|
Gm20870
|
predicted gene, 20870 |
| chrY_+_50604830 | 1.17 |
ENSMUST00000185245.2
|
Gm20883
|
predicted gene, 20883 |
| chrY_+_51121799 | 1.17 |
ENSMUST00000185327.7
|
Gm21117
|
predicted gene, 21117 |
| chrY_+_55728335 | 1.17 |
ENSMUST00000185713.7
|
Gm21858
|
predicted gene, 21858 |
| chrY_-_72258387 | 1.17 |
ENSMUST00000191305.7
|
Gm20843
|
predicted gene, 20843 |
| chrY_+_73680584 | 1.16 |
ENSMUST00000189346.2
|
Gm20903
|
predicted gene, 20903 |
| chrY_+_57714225 | 1.16 |
ENSMUST00000190573.2
|
Gm28961
|
predicted gene 28961 |
| chrY_+_77704077 | 1.16 |
ENSMUST00000190551.7
|
Gm21650
|
predicted gene, 21650 |
| chrY_+_84108548 | 1.16 |
ENSMUST00000185776.7
|
Gm21095
|
predicted gene, 21095 |
| chrY_+_51848699 | 1.15 |
ENSMUST00000187477.2
|
Gm20920
|
predicted gene, 20920 |
| chrY_+_64481794 | 1.15 |
ENSMUST00000186004.2
|
Gm20908
|
predicted gene, 20908 |
| chrY_+_55210299 | 1.14 |
ENSMUST00000187293.7
|
Gm20931
|
predicted gene, 20931 |
| chrY_+_75207125 | 1.14 |
ENSMUST00000190173.2
|
Gm20814
|
predicted gene, 20814 |
| chrY_+_70453151 | 1.13 |
ENSMUST00000186890.7
|
Gm20888
|
predicted gene, 20888 |
| chrY_+_8257907 | 1.12 |
ENSMUST00000186739.2
|
Gm20824
|
predicted gene, 20824 |
| chrX_+_61511597 | 1.12 |
ENSMUST00000033537.2
|
4931400O07Rik
|
RIKEN cDNA 4931400O07 gene |
| chr9_+_40785277 | 1.12 |
ENSMUST00000067375.5
|
Bsx
|
brain specific homeobox |
| chr16_-_37681508 | 1.10 |
ENSMUST00000205931.2
|
Gm36028
|
predicted gene, 36028 |
| chrY_+_89064075 | 1.10 |
ENSMUST00000186443.2
|
Gm21294
|
predicted gene, 21294 |
| chrY_-_28226857 | 1.09 |
ENSMUST00000188864.2
|
Gm20894
|
predicted gene, 20894 |
| chr5_-_72716942 | 1.09 |
ENSMUST00000074948.5
ENSMUST00000087216.12 |
Nfxl1
|
nuclear transcription factor, X-box binding-like 1 |
| chrX_+_110154017 | 1.09 |
ENSMUST00000210720.3
|
Cylc1
|
cylicin, basic protein of sperm head cytoskeleton 1 |
| chrY_-_26732144 | 1.09 |
ENSMUST00000189518.2
|
Gm20890
|
predicted gene, 20890 |
| chrY_-_27477265 | 1.09 |
ENSMUST00000188744.2
|
Gm21488
|
predicted gene, 21488 |
| chrY_+_6583546 | 1.07 |
ENSMUST00000190026.2
|
Gm28919
|
predicted gene 28919 |
| chrY_-_42537595 | 1.06 |
ENSMUST00000190453.2
|
Gm29276
|
predicted gene 29276 |
| chr10_+_69055215 | 1.06 |
ENSMUST00000172261.3
|
Rhobtb1
|
Rho-related BTB domain containing 1 |
| chrY_+_13494026 | 1.05 |
ENSMUST00000193306.2
|
Gm20773
|
predicted gene, 20773 |
| chrY_-_38457583 | 1.05 |
ENSMUST00000185240.2
|
Gm20897
|
predicted gene, 20897 |
| chrY_+_17874741 | 1.03 |
ENSMUST00000188928.3
ENSMUST00000177639.7 |
Gm20772
Gm20831
|
predicted gene, 20772 predicted gene, 20831 |
| chrY_+_13252391 | 1.03 |
ENSMUST00000192749.2
ENSMUST00000178599.7 |
Gm21440
Gm21454
|
predicted gene, 21440 predicted gene, 21454 |
| chr18_+_12732951 | 1.02 |
ENSMUST00000234255.2
ENSMUST00000169401.8 |
Ttc39c
|
tetratricopeptide repeat domain 39C |
| chr18_+_44237577 | 1.01 |
ENSMUST00000239465.2
|
Spink12
|
serine peptidase inhibitor, Kazal type 12 |
| chr7_+_12568647 | 1.00 |
ENSMUST00000004614.15
|
Zfp110
|
zinc finger protein 110 |
| chr3_-_95595157 | 0.99 |
ENSMUST00000015994.4
ENSMUST00000148854.2 ENSMUST00000117782.8 |
Adamtsl4
|
ADAMTS-like 4 |
| chrY_+_13042891 | 0.98 |
ENSMUST00000194687.2
|
Gm20807
|
predicted gene, 20807 |
| chrY_-_10536042 | 0.97 |
ENSMUST00000187245.2
|
Gm20737
|
predicted gene, 20737 |
| chrY_+_13376362 | 0.95 |
ENSMUST00000192204.2
|
Gm21454
|
predicted gene, 21454 |
| chrY_+_18089116 | 0.95 |
ENSMUST00000186035.2
|
Ssty1
|
spermiogenesis specific transcript on the Y 1 |
| chrY_+_40760259 | 0.95 |
ENSMUST00000190909.2
|
Gm20795
|
predicted gene, 20795 |
| chrX_+_156482116 | 0.95 |
ENSMUST00000112521.8
|
Smpx
|
small muscle protein, X-linked |
| chr1_-_173569301 | 0.94 |
ENSMUST00000042610.15
ENSMUST00000127730.2 |
Ifi207
|
interferon activated gene 207 |
| chr3_+_14928561 | 0.93 |
ENSMUST00000029076.6
|
Car3
|
carbonic anhydrase 3 |
| chrX_-_4194587 | 0.93 |
ENSMUST00000179325.2
|
Btbd35f20
|
BTB domain containing 35, family member 20 |
| chr2_+_22959452 | 0.92 |
ENSMUST00000155602.4
|
Acbd5
|
acyl-Coenzyme A binding domain containing 5 |
| chrY_+_17965160 | 0.92 |
ENSMUST00000194411.2
|
Gm20831
|
predicted gene, 20831 |
| chrY_+_13161968 | 0.92 |
ENSMUST00000195626.2
|
Gm21425
|
predicted gene, 21425 |
| chr9_+_39933648 | 0.90 |
ENSMUST00000059859.5
|
Olfr981
|
olfactory receptor 981 |
| chr2_-_151510453 | 0.89 |
ENSMUST00000180195.8
ENSMUST00000096439.4 |
Rad21l
|
RAD21-like (S. pombe) |
| chr6_-_40521911 | 0.85 |
ENSMUST00000089490.3
|
Olfr461
|
olfactory receptor 461 |
| chr9_-_20887967 | 0.85 |
ENSMUST00000214218.2
|
S1pr2
|
sphingosine-1-phosphate receptor 2 |
| chr3_-_146391230 | 0.84 |
ENSMUST00000197989.2
ENSMUST00000125965.8 |
Samd13
|
sterile alpha motif domain containing 13 |
| chr2_+_22958179 | 0.83 |
ENSMUST00000227663.2
ENSMUST00000028121.15 ENSMUST00000227809.2 ENSMUST00000144088.2 |
Acbd5
|
acyl-Coenzyme A binding domain containing 5 |
| chr18_-_84854841 | 0.81 |
ENSMUST00000236689.2
|
Gm17266
|
predicted gene, 17266 |
| chr2_+_37029334 | 0.80 |
ENSMUST00000214905.2
ENSMUST00000217298.2 ENSMUST00000104995.3 |
Olfr364-ps1
|
olfactory receptor 364, pseudogene 1 |
| chrX_+_163221035 | 0.80 |
ENSMUST00000033755.6
|
Asb11
|
ankyrin repeat and SOCS box-containing 11 |
| chr14_+_51599213 | 0.77 |
ENSMUST00000162998.2
|
Gm7247
|
predicted gene 7247 |
| chr17_+_35354172 | 0.76 |
ENSMUST00000172571.8
ENSMUST00000173491.8 |
Bag6
|
BCL2-associated athanogene 6 |
| chr17_-_15784582 | 0.75 |
ENSMUST00000147532.8
|
Prdm9
|
PR domain containing 9 |
| chr7_+_103205286 | 0.73 |
ENSMUST00000214345.2
ENSMUST00000215673.2 |
Olfr615
|
olfactory receptor 615 |
| chr3_+_90155479 | 0.73 |
ENSMUST00000015467.9
|
Slc39a1
|
solute carrier family 39 (zinc transporter), member 1 |
| chr4_-_156340713 | 0.73 |
ENSMUST00000219393.2
|
Samd11
|
sterile alpha motif domain containing 11 |
| chrX_-_74918122 | 0.72 |
ENSMUST00000033547.14
|
Pls3
|
plastin 3 (T-isoform) |
| chr7_-_107857554 | 0.71 |
ENSMUST00000215173.2
|
Olfr488
|
olfactory receptor 488 |
| chrX_-_74918709 | 0.71 |
ENSMUST00000114059.10
|
Pls3
|
plastin 3 (T-isoform) |
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 3.3 | 13.2 | GO:0034034 | coenzyme A catabolic process(GO:0015938) nucleoside bisphosphate catabolic process(GO:0033869) ribonucleoside bisphosphate catabolic process(GO:0034031) purine nucleoside bisphosphate catabolic process(GO:0034034) acetyl-CoA catabolic process(GO:0046356) |
| 2.1 | 6.4 | GO:0001868 | regulation of complement activation, lectin pathway(GO:0001868) negative regulation of complement activation, lectin pathway(GO:0001869) |
| 1.2 | 3.6 | GO:0006011 | UDP-glucose metabolic process(GO:0006011) glucose 1-phosphate metabolic process(GO:0019255) |
| 0.6 | 10.5 | GO:0019374 | galactolipid metabolic process(GO:0019374) |
| 0.5 | 1.5 | GO:0097374 | sensory neuron axon guidance(GO:0097374) |
| 0.5 | 4.8 | GO:0048208 | vesicle targeting, rough ER to cis-Golgi(GO:0048207) COPII vesicle coating(GO:0048208) |
| 0.3 | 3.4 | GO:0010886 | positive regulation of cholesterol storage(GO:0010886) |
| 0.3 | 0.7 | GO:0060300 | regulation of cytokine activity(GO:0060300) |
| 0.3 | 5.3 | GO:0061469 | regulation of type B pancreatic cell proliferation(GO:0061469) |
| 0.3 | 1.2 | GO:0030450 | regulation of complement activation, classical pathway(GO:0030450) |
| 0.3 | 1.5 | GO:0010446 | response to alkaline pH(GO:0010446) |
| 0.3 | 3.2 | GO:0030242 | pexophagy(GO:0030242) |
| 0.3 | 4.6 | GO:0044539 | long-chain fatty acid import(GO:0044539) |
| 0.3 | 2.3 | GO:0032286 | central nervous system myelin maintenance(GO:0032286) |
| 0.3 | 0.8 | GO:0010845 | positive regulation of reciprocal meiotic recombination(GO:0010845) |
| 0.2 | 1.4 | GO:0032053 | ciliary basal body organization(GO:0032053) cerebrospinal fluid secretion(GO:0033326) |
| 0.2 | 1.8 | GO:0070447 | positive regulation of oligodendrocyte progenitor proliferation(GO:0070447) |
| 0.2 | 1.5 | GO:0035469 | determination of pancreatic left/right asymmetry(GO:0035469) maintenance of organ identity(GO:0048496) |
| 0.2 | 1.7 | GO:0034379 | very-low-density lipoprotein particle assembly(GO:0034379) |
| 0.2 | 2.3 | GO:0008228 | opsonization(GO:0008228) |
| 0.2 | 1.9 | GO:0015747 | urate transport(GO:0015747) |
| 0.2 | 3.7 | GO:0046415 | urate metabolic process(GO:0046415) |
| 0.2 | 2.7 | GO:0042541 | hemoglobin biosynthetic process(GO:0042541) |
| 0.2 | 1.4 | GO:1902172 | keratinocyte apoptotic process(GO:0097283) regulation of keratinocyte apoptotic process(GO:1902172) |
| 0.2 | 1.2 | GO:0036506 | maintenance of unfolded protein(GO:0036506) maintenance of unfolded protein involved in ERAD pathway(GO:1904378) |
| 0.1 | 1.0 | GO:1990009 | retinal cell apoptotic process(GO:1990009) |
| 0.1 | 0.3 | GO:0021828 | cerebral cortex tangential migration using cell-axon interactions(GO:0021824) gonadotrophin-releasing hormone neuronal migration to the hypothalamus(GO:0021828) hypothalamic tangential migration using cell-axon interactions(GO:0021856) neural crest cell migration involved in sympathetic nervous system development(GO:1903045) facioacoustic ganglion development(GO:1903375) |
| 0.1 | 1.3 | GO:1901727 | positive regulation of histone deacetylase activity(GO:1901727) |
| 0.1 | 0.8 | GO:0003376 | sphingosine-1-phosphate signaling pathway(GO:0003376) |
| 0.1 | 0.6 | GO:0046878 | positive regulation of saliva secretion(GO:0046878) |
| 0.1 | 0.4 | GO:0010510 | regulation of acetyl-CoA biosynthetic process from pyruvate(GO:0010510) |
| 0.1 | 0.9 | GO:0044821 | meiotic telomere tethering at nuclear periphery(GO:0044821) meiotic attachment of telomere to nuclear envelope(GO:0070197) chromosome attachment to the nuclear envelope(GO:0097240) |
| 0.1 | 1.5 | GO:0061303 | cornea development in camera-type eye(GO:0061303) |
| 0.1 | 6.9 | GO:0018149 | peptide cross-linking(GO:0018149) |
| 0.1 | 1.5 | GO:0045217 | cell-cell junction maintenance(GO:0045217) |
| 0.1 | 38.3 | GO:0051321 | meiotic cell cycle(GO:0051321) |
| 0.1 | 2.4 | GO:0061436 | establishment of skin barrier(GO:0061436) |
| 0.1 | 0.7 | GO:0034316 | negative regulation of Arp2/3 complex-mediated actin nucleation(GO:0034316) |
| 0.1 | 0.4 | GO:0036016 | response to interleukin-3(GO:0036015) cellular response to interleukin-3(GO:0036016) positive regulation of adrenergic receptor signaling pathway(GO:0071879) |
| 0.1 | 2.1 | GO:0007020 | microtubule nucleation(GO:0007020) |
| 0.1 | 0.7 | GO:2000857 | positive regulation of mineralocorticoid secretion(GO:2000857) positive regulation of aldosterone secretion(GO:2000860) |
| 0.1 | 0.6 | GO:0072307 | metanephric nephron tubule epithelial cell differentiation(GO:0072257) regulation of metanephric nephron tubule epithelial cell differentiation(GO:0072307) |
| 0.1 | 3.7 | GO:0003333 | amino acid transmembrane transport(GO:0003333) |
| 0.1 | 0.5 | GO:0006499 | N-terminal protein myristoylation(GO:0006499) |
| 0.1 | 10.8 | GO:0008203 | cholesterol metabolic process(GO:0008203) |
| 0.1 | 0.6 | GO:0010994 | regulation of ubiquitin homeostasis(GO:0010993) free ubiquitin chain polymerization(GO:0010994) |
| 0.1 | 3.8 | GO:0051438 | regulation of ubiquitin-protein transferase activity(GO:0051438) |
| 0.1 | 1.4 | GO:0051639 | actin filament network formation(GO:0051639) |
| 0.1 | 1.9 | GO:0048240 | sperm capacitation(GO:0048240) |
| 0.1 | 1.3 | GO:0001886 | endothelial cell morphogenesis(GO:0001886) |
| 0.0 | 0.6 | GO:0035845 | photoreceptor cell outer segment organization(GO:0035845) |
| 0.0 | 1.5 | GO:0007530 | sex determination(GO:0007530) |
| 0.0 | 0.3 | GO:0035553 | oxidative single-stranded RNA demethylation(GO:0035553) |
| 0.0 | 1.1 | GO:0060056 | mammary gland involution(GO:0060056) |
| 0.0 | 0.6 | GO:0017121 | phospholipid scrambling(GO:0017121) |
| 0.0 | 0.4 | GO:0002826 | negative regulation of T-helper 1 type immune response(GO:0002826) |
| 0.0 | 1.5 | GO:0010758 | regulation of macrophage chemotaxis(GO:0010758) |
| 0.0 | 0.5 | GO:0070131 | positive regulation of mitochondrial translation(GO:0070131) |
| 0.0 | 0.1 | GO:0006429 | leucyl-tRNA aminoacylation(GO:0006429) |
| 0.0 | 1.8 | GO:1900087 | positive regulation of G1/S transition of mitotic cell cycle(GO:1900087) |
| 0.0 | 0.7 | GO:0071577 | zinc II ion transmembrane transport(GO:0071577) |
| 0.0 | 1.1 | GO:0050911 | detection of chemical stimulus involved in sensory perception of smell(GO:0050911) |
| 0.0 | 0.3 | GO:0032049 | cardiolipin biosynthetic process(GO:0032049) |
| 0.0 | 0.3 | GO:0010606 | positive regulation of cytoplasmic mRNA processing body assembly(GO:0010606) |
| 0.0 | 0.2 | GO:0042427 | serotonin biosynthetic process(GO:0042427) primary amino compound biosynthetic process(GO:1901162) |
| 0.0 | 0.4 | GO:0046520 | sphingoid biosynthetic process(GO:0046520) |
| 0.0 | 0.6 | GO:1990126 | retrograde transport, endosome to plasma membrane(GO:1990126) |
| 0.0 | 0.6 | GO:0030502 | negative regulation of bone mineralization(GO:0030502) |
| 0.0 | 0.7 | GO:0007202 | activation of phospholipase C activity(GO:0007202) |
| 0.0 | 0.2 | GO:0070995 | NADPH oxidation(GO:0070995) |
| 0.0 | 1.6 | GO:0045454 | cell redox homeostasis(GO:0045454) |
| 0.0 | 0.3 | GO:0033617 | mitochondrial respiratory chain complex IV assembly(GO:0033617) |
| 0.0 | 0.1 | GO:0007468 | regulation of rhodopsin gene expression(GO:0007468) |
| 0.0 | 0.9 | GO:0006730 | one-carbon metabolic process(GO:0006730) |
| 0.0 | 0.3 | GO:0000338 | protein deneddylation(GO:0000338) |
| 0.0 | 0.7 | GO:0030224 | monocyte differentiation(GO:0030224) |
| 0.0 | 1.3 | GO:0045214 | sarcomere organization(GO:0045214) |
| 0.0 | 0.3 | GO:2000480 | negative regulation of cAMP-dependent protein kinase activity(GO:2000480) |
| 0.0 | 3.6 | GO:0050907 | detection of chemical stimulus involved in sensory perception(GO:0050907) |
| 0.0 | 0.1 | GO:0015705 | iodide transport(GO:0015705) |
| 0.0 | 1.5 | GO:0043149 | contractile actin filament bundle assembly(GO:0030038) stress fiber assembly(GO:0043149) |
| 0.0 | 0.4 | GO:0016339 | calcium-dependent cell-cell adhesion via plasma membrane cell adhesion molecules(GO:0016339) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.1 | 3.4 | GO:0034359 | mature chylomicron(GO:0034359) |
| 0.8 | 3.4 | GO:0097123 | cyclin A1-CDK2 complex(GO:0097123) |
| 0.5 | 10.5 | GO:0042589 | zymogen granule membrane(GO:0042589) |
| 0.4 | 1.5 | GO:0097543 | ciliary inversin compartment(GO:0097543) |
| 0.3 | 0.7 | GO:0098842 | postsynaptic early endosome(GO:0098842) |
| 0.3 | 5.3 | GO:0042588 | zymogen granule(GO:0042588) |
| 0.3 | 45.1 | GO:0000800 | lateral element(GO:0000800) |
| 0.3 | 1.5 | GO:0071953 | elastic fiber(GO:0071953) |
| 0.3 | 2.2 | GO:0005927 | muscle tendon junction(GO:0005927) |
| 0.3 | 1.1 | GO:0043159 | acrosomal matrix(GO:0043159) |
| 0.2 | 3.8 | GO:0031462 | Cul2-RING ubiquitin ligase complex(GO:0031462) |
| 0.2 | 9.2 | GO:0001533 | cornified envelope(GO:0001533) |
| 0.2 | 1.8 | GO:0097433 | dense body(GO:0097433) |
| 0.1 | 1.2 | GO:0071818 | BAT3 complex(GO:0071818) ER membrane insertion complex(GO:0072379) |
| 0.1 | 1.9 | GO:0036128 | CatSper complex(GO:0036128) |
| 0.1 | 0.4 | GO:0005967 | mitochondrial pyruvate dehydrogenase complex(GO:0005967) |
| 0.1 | 0.6 | GO:0030906 | retromer, cargo-selective complex(GO:0030906) |
| 0.1 | 1.6 | GO:0034663 | endoplasmic reticulum chaperone complex(GO:0034663) |
| 0.1 | 25.4 | GO:0005777 | peroxisome(GO:0005777) microbody(GO:0042579) |
| 0.1 | 0.6 | GO:0097542 | ciliary tip(GO:0097542) |
| 0.0 | 2.1 | GO:0035371 | microtubule plus-end(GO:0035371) |
| 0.0 | 2.3 | GO:0005771 | multivesicular body(GO:0005771) |
| 0.0 | 0.4 | GO:0017059 | serine C-palmitoyltransferase complex(GO:0017059) endoplasmic reticulum palmitoyltransferase complex(GO:0031211) |
| 0.0 | 11.0 | GO:0072562 | blood microparticle(GO:0072562) |
| 0.0 | 1.0 | GO:0005614 | interstitial matrix(GO:0005614) |
| 0.0 | 0.1 | GO:0033165 | interphotoreceptor matrix(GO:0033165) |
| 0.0 | 13.0 | GO:0005789 | endoplasmic reticulum membrane(GO:0005789) |
| 0.0 | 1.5 | GO:0005791 | rough endoplasmic reticulum(GO:0005791) |
| 0.0 | 0.9 | GO:0005865 | striated muscle thin filament(GO:0005865) |
| 0.0 | 1.4 | GO:0032420 | stereocilium(GO:0032420) |
| 0.0 | 2.2 | GO:0005814 | centriole(GO:0005814) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 3.2 | 22.7 | GO:0004556 | alpha-amylase activity(GO:0004556) |
| 2.6 | 13.2 | GO:0003986 | acetyl-CoA hydrolase activity(GO:0003986) |
| 1.7 | 13.3 | GO:0050649 | testosterone 6-beta-hydroxylase activity(GO:0050649) |
| 0.9 | 4.6 | GO:0043758 | acetate-CoA ligase (ADP-forming) activity(GO:0043758) |
| 0.9 | 2.7 | GO:0003870 | 5-aminolevulinate synthase activity(GO:0003870) N-succinyltransferase activity(GO:0016749) |
| 0.9 | 2.6 | GO:0008127 | quercetin 2,3-dioxygenase activity(GO:0008127) |
| 0.7 | 10.5 | GO:0047372 | acylglycerol lipase activity(GO:0047372) |
| 0.5 | 3.6 | GO:0070569 | uridylyltransferase activity(GO:0070569) |
| 0.4 | 2.4 | GO:0030280 | structural constituent of epidermis(GO:0030280) |
| 0.4 | 2.7 | GO:0018685 | alkane 1-monooxygenase activity(GO:0018685) |
| 0.3 | 4.1 | GO:0016661 | oxidoreductase activity, acting on other nitrogenous compounds as donors(GO:0016661) |
| 0.3 | 2.1 | GO:0004865 | protein serine/threonine phosphatase inhibitor activity(GO:0004865) |
| 0.3 | 3.4 | GO:0035473 | lipase binding(GO:0035473) |
| 0.3 | 1.8 | GO:0072510 | ferric iron transmembrane transporter activity(GO:0015091) trivalent inorganic cation transmembrane transporter activity(GO:0072510) |
| 0.2 | 6.4 | GO:0001848 | complement binding(GO:0001848) |
| 0.2 | 0.7 | GO:0001607 | neuromedin U receptor activity(GO:0001607) |
| 0.2 | 1.5 | GO:0017040 | ceramidase activity(GO:0017040) |
| 0.1 | 0.4 | GO:0002113 | interleukin-33 binding(GO:0002113) |
| 0.1 | 4.9 | GO:0000062 | fatty-acyl-CoA binding(GO:0000062) |
| 0.1 | 1.0 | GO:0005166 | neurotrophin p75 receptor binding(GO:0005166) |
| 0.1 | 1.5 | GO:0005451 | monovalent cation:proton antiporter activity(GO:0005451) |
| 0.1 | 2.0 | GO:1901702 | urate transmembrane transporter activity(GO:0015143) salt transmembrane transporter activity(GO:1901702) |
| 0.1 | 6.7 | GO:0008138 | protein tyrosine/serine/threonine phosphatase activity(GO:0008138) |
| 0.1 | 0.4 | GO:0004740 | pyruvate dehydrogenase (acetyl-transferring) kinase activity(GO:0004740) |
| 0.1 | 1.4 | GO:0003847 | 1-alkyl-2-acetylglycerophosphocholine esterase activity(GO:0003847) |
| 0.1 | 1.6 | GO:0015037 | peptide disulfide oxidoreductase activity(GO:0015037) |
| 0.1 | 20.9 | GO:0004252 | serine-type endopeptidase activity(GO:0004252) |
| 0.1 | 0.4 | GO:0004758 | serine C-palmitoyltransferase activity(GO:0004758) C-palmitoyltransferase activity(GO:0016454) |
| 0.1 | 0.9 | GO:0016151 | nickel cation binding(GO:0016151) |
| 0.1 | 0.2 | GO:0004510 | tryptophan 5-monooxygenase activity(GO:0004510) |
| 0.1 | 0.7 | GO:0036122 | BMP binding(GO:0036122) |
| 0.1 | 0.6 | GO:1990430 | extracellular matrix protein binding(GO:1990430) |
| 0.1 | 1.2 | GO:0015643 | toxic substance binding(GO:0015643) |
| 0.1 | 2.8 | GO:1990841 | promoter-specific chromatin binding(GO:1990841) |
| 0.0 | 0.7 | GO:0071933 | Arp2/3 complex binding(GO:0071933) |
| 0.0 | 0.3 | GO:0035515 | oxidative RNA demethylase activity(GO:0035515) |
| 0.0 | 2.2 | GO:0004364 | glutathione transferase activity(GO:0004364) |
| 0.0 | 0.1 | GO:0004819 | glutamine-tRNA ligase activity(GO:0004819) leucine-tRNA ligase activity(GO:0004823) |
| 0.0 | 0.8 | GO:0042800 | histone methyltransferase activity (H3-K4 specific)(GO:0042800) |
| 0.0 | 5.3 | GO:0008083 | growth factor activity(GO:0008083) |
| 0.0 | 1.9 | GO:0015485 | cholesterol binding(GO:0015485) |
| 0.0 | 3.7 | GO:0015171 | amino acid transmembrane transporter activity(GO:0015171) |
| 0.0 | 0.6 | GO:0017128 | phospholipid scramblase activity(GO:0017128) |
| 0.0 | 1.5 | GO:0050840 | extracellular matrix binding(GO:0050840) |
| 0.0 | 0.3 | GO:0038191 | neuropilin binding(GO:0038191) |
| 0.0 | 0.5 | GO:0042923 | neuropeptide binding(GO:0042923) |
| 0.0 | 0.4 | GO:0050321 | tau-protein kinase activity(GO:0050321) |
| 0.0 | 0.7 | GO:0005385 | zinc ion transmembrane transporter activity(GO:0005385) |
| 0.0 | 0.4 | GO:0005527 | macrolide binding(GO:0005527) FK506 binding(GO:0005528) |
| 0.0 | 0.5 | GO:0004993 | G-protein coupled serotonin receptor activity(GO:0004993) |
| 0.0 | 0.2 | GO:0004499 | N,N-dimethylaniline monooxygenase activity(GO:0004499) |
| 0.0 | 0.3 | GO:0070016 | armadillo repeat domain binding(GO:0070016) |
| 0.0 | 0.1 | GO:0004035 | alkaline phosphatase activity(GO:0004035) |
| 0.0 | 0.3 | GO:0004653 | polypeptide N-acetylgalactosaminyltransferase activity(GO:0004653) |
| 0.0 | 0.2 | GO:0003993 | acid phosphatase activity(GO:0003993) |
| 0.0 | 0.3 | GO:0004862 | cAMP-dependent protein kinase inhibitor activity(GO:0004862) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 3.4 | SA REG CASCADE OF CYCLIN EXPR | Expression of cyclins regulates progression through the cell cycle by activating cyclin-dependent kinases. |
| 0.1 | 5.7 | PID HNF3A PATHWAY | FOXA1 transcription factor network |
| 0.1 | 5.9 | SIG REGULATION OF THE ACTIN CYTOSKELETON BY RHO GTPASES | Genes related to regulation of the actin cytoskeleton |
| 0.1 | 20.4 | NABA ECM REGULATORS | Genes encoding enzymes and their regulators involved in the remodeling of the extracellular matrix |
| 0.0 | 6.5 | NABA ECM GLYCOPROTEINS | Genes encoding structural ECM glycoproteins |
| 0.0 | 2.5 | PID RAC1 REG PATHWAY | Regulation of RAC1 activity |
| 0.0 | 0.8 | PID S1P S1P2 PATHWAY | S1P2 pathway |
| 0.0 | 1.6 | ST PHOSPHOINOSITIDE 3 KINASE PATHWAY | PI3K Pathway |
| 0.0 | 1.1 | NABA PROTEOGLYCANS | Genes encoding proteoglycans |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 3.6 | REACTOME GLUCURONIDATION | Genes involved in Glucuronidation |
| 0.3 | 4.0 | REACTOME COMMON PATHWAY | Genes involved in Common Pathway |
| 0.3 | 6.4 | REACTOME INTRINSIC PATHWAY | Genes involved in Intrinsic Pathway |
| 0.2 | 3.4 | REACTOME CYCLIN A B1 ASSOCIATED EVENTS DURING G2 M TRANSITION | Genes involved in Cyclin A/B1 associated events during G2/M transition |
| 0.2 | 4.6 | REACTOME ACTIVATED AMPK STIMULATES FATTY ACID OXIDATION IN MUSCLE | Genes involved in Activated AMPK stimulates fatty-acid oxidation in muscle |
| 0.1 | 9.6 | REACTOME LIPID DIGESTION MOBILIZATION AND TRANSPORT | Genes involved in Lipid digestion, mobilization, and transport |
| 0.1 | 6.4 | REACTOME SMOOTH MUSCLE CONTRACTION | Genes involved in Smooth Muscle Contraction |
| 0.1 | 2.7 | REACTOME METABOLISM OF PORPHYRINS | Genes involved in Metabolism of porphyrins |
| 0.1 | 1.5 | REACTOME KERATAN SULFATE DEGRADATION | Genes involved in Keratan sulfate degradation |
| 0.1 | 1.2 | REACTOME REGULATION OF COMPLEMENT CASCADE | Genes involved in Regulation of Complement cascade |
| 0.1 | 1.5 | REACTOME BILE SALT AND ORGANIC ANION SLC TRANSPORTERS | Genes involved in Bile salt and organic anion SLC transporters |
| 0.1 | 3.7 | REACTOME AMINO ACID TRANSPORT ACROSS THE PLASMA MEMBRANE | Genes involved in Amino acid transport across the plasma membrane |
| 0.0 | 1.5 | REACTOME SPHINGOLIPID DE NOVO BIOSYNTHESIS | Genes involved in Sphingolipid de novo biosynthesis |
| 0.0 | 0.4 | REACTOME REGULATION OF PYRUVATE DEHYDROGENASE PDH COMPLEX | Genes involved in Regulation of pyruvate dehydrogenase (PDH) complex |
| 0.0 | 0.7 | REACTOME ZINC TRANSPORTERS | Genes involved in Zinc transporters |
| 0.0 | 0.7 | REACTOME SIGNALING BY BMP | Genes involved in Signaling by BMP |
| 0.0 | 0.9 | REACTOME STRIATED MUSCLE CONTRACTION | Genes involved in Striated Muscle Contraction |
| 0.0 | 0.8 | REACTOME MEIOTIC RECOMBINATION | Genes involved in Meiotic Recombination |
| 0.0 | 1.1 | REACTOME ACTIVATION OF CHAPERONE GENES BY XBP1S | Genes involved in Activation of Chaperone Genes by XBP1(S) |
| 0.0 | 0.3 | REACTOME SEMA3A PLEXIN REPULSION SIGNALING BY INHIBITING INTEGRIN ADHESION | Genes involved in SEMA3A-Plexin repulsion signaling by inhibiting Integrin adhesion |
| 0.0 | 0.9 | REACTOME GLUTATHIONE CONJUGATION | Genes involved in Glutathione conjugation |
| 0.0 | 0.6 | REACTOME SIGNALING BY HIPPO | Genes involved in Signaling by Hippo |
| 0.0 | 0.3 | REACTOME SYNTHESIS OF SUBSTRATES IN N GLYCAN BIOSYTHESIS | Genes involved in Synthesis of substrates in N-glycan biosythesis |
| 0.0 | 0.3 | REACTOME ACYL CHAIN REMODELLING OF PG | Genes involved in Acyl chain remodelling of PG |