Project
PRJNA375882: Comprehensive Mouse Transcriptomic BodyMap
Navigation
Downloads
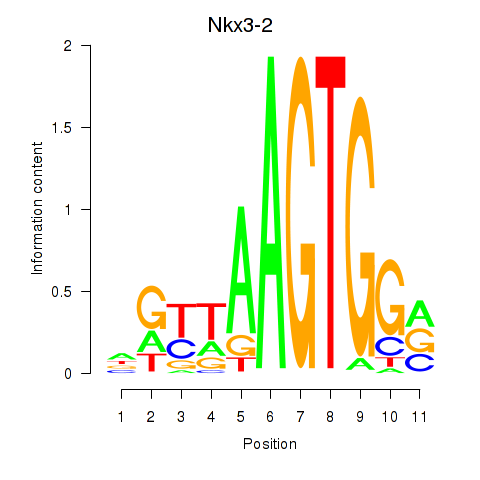
Results for Nkx3-2
Z-value: 0.92
Transcription factors associated with Nkx3-2
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Nkx3-2
|
ENSMUSG00000049691.9 | Nkx3-2 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Nkx3-2 | mm39_v1_chr5_-_41921834_41921844 | 0.03 | 8.2e-01 | Click! |
Activity profile of Nkx3-2 motif
Sorted Z-values of Nkx3-2 motif
Network of associatons between targets according to the STRING database.
First level regulatory network of Nkx3-2
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr11_-_120622770 | 25.25 |
ENSMUST00000154565.2
ENSMUST00000026148.9 |
Cbr2
|
carbonyl reductase 2 |
| chr15_+_75468473 | 8.34 |
ENSMUST00000189944.7
ENSMUST00000023243.11 |
Gpihbp1
|
GPI-anchored HDL-binding protein 1 |
| chr15_+_75468494 | 6.24 |
ENSMUST00000189874.2
|
Gpihbp1
|
GPI-anchored HDL-binding protein 1 |
| chr9_+_122980006 | 4.70 |
ENSMUST00000026890.6
|
Clec3b
|
C-type lectin domain family 3, member b |
| chr11_+_43365103 | 4.43 |
ENSMUST00000173002.8
ENSMUST00000057679.10 |
C1qtnf2
|
C1q and tumor necrosis factor related protein 2 |
| chr6_-_133830613 | 3.80 |
ENSMUST00000203168.2
ENSMUST00000048032.5 |
Kap
|
kidney androgen regulated protein |
| chr9_+_48073296 | 3.58 |
ENSMUST00000216998.2
ENSMUST00000215780.2 |
Nxpe4
|
neurexophilin and PC-esterase domain family, member 4 |
| chr1_-_162726234 | 3.55 |
ENSMUST00000111510.8
ENSMUST00000045902.13 |
Fmo2
|
flavin containing monooxygenase 2 |
| chr11_-_69686756 | 3.39 |
ENSMUST00000045971.9
|
Chrnb1
|
cholinergic receptor, nicotinic, beta polypeptide 1 (muscle) |
| chr11_-_101877832 | 3.34 |
ENSMUST00000107173.9
ENSMUST00000107172.8 |
Dusp3
|
dual specificity phosphatase 3 (vaccinia virus phosphatase VH1-related) |
| chr8_+_46338498 | 3.33 |
ENSMUST00000034053.7
|
Pdlim3
|
PDZ and LIM domain 3 |
| chr8_+_46338557 | 3.32 |
ENSMUST00000210422.2
|
Pdlim3
|
PDZ and LIM domain 3 |
| chr4_-_141325517 | 3.28 |
ENSMUST00000131317.8
ENSMUST00000006381.11 ENSMUST00000129602.8 |
Fblim1
|
filamin binding LIM protein 1 |
| chr4_+_148085179 | 3.26 |
ENSMUST00000103230.5
|
Nppa
|
natriuretic peptide type A |
| chr8_+_104828253 | 3.19 |
ENSMUST00000034339.10
|
Cdh5
|
cadherin 5 |
| chr15_+_34306812 | 3.19 |
ENSMUST00000226766.2
ENSMUST00000163455.9 ENSMUST00000022947.7 ENSMUST00000228570.2 ENSMUST00000227759.2 |
Matn2
|
matrilin 2 |
| chrX_-_96420756 | 3.17 |
ENSMUST00000113832.2
ENSMUST00000037353.10 |
Eda2r
|
ectodysplasin A2 receptor |
| chr10_+_116111441 | 3.07 |
ENSMUST00000218553.2
|
Ptprb
|
protein tyrosine phosphatase, receptor type, B |
| chr7_-_67409432 | 2.99 |
ENSMUST00000208815.2
ENSMUST00000074233.12 ENSMUST00000208231.2 ENSMUST00000051389.10 |
Synm
|
synemin, intermediate filament protein |
| chr10_+_127350820 | 2.92 |
ENSMUST00000035735.11
|
Ndufa4l2
|
Ndufa4, mitochondrial complex associated like 2 |
| chr9_-_44646487 | 2.87 |
ENSMUST00000034611.15
|
Phldb1
|
pleckstrin homology like domain, family B, member 1 |
| chr3_-_81981946 | 2.79 |
ENSMUST00000029635.14
ENSMUST00000193597.2 |
Gucy1b1
|
guanylate cyclase 1, soluble, beta 1 |
| chr17_-_82045800 | 2.77 |
ENSMUST00000235015.2
ENSMUST00000163123.3 |
Slc8a1
|
solute carrier family 8 (sodium/calcium exchanger), member 1 |
| chr17_+_44114894 | 2.75 |
ENSMUST00000044895.13
|
Rcan2
|
regulator of calcineurin 2 |
| chr9_-_58220469 | 2.72 |
ENSMUST00000061799.10
|
Loxl1
|
lysyl oxidase-like 1 |
| chr7_-_48493388 | 2.52 |
ENSMUST00000167786.4
|
Csrp3
|
cysteine and glycine-rich protein 3 |
| chr5_+_149335214 | 2.39 |
ENSMUST00000093110.12
|
Medag
|
mesenteric estrogen dependent adipogenesis |
| chr8_-_86107593 | 2.37 |
ENSMUST00000122452.8
|
Mylk3
|
myosin light chain kinase 3 |
| chrX_-_151820545 | 2.33 |
ENSMUST00000051484.5
|
Mageh1
|
MAGE family member H1 |
| chr17_-_43854530 | 2.30 |
ENSMUST00000178772.3
|
Ankrd66
|
ankyrin repeat domain 66 |
| chr3_-_57483330 | 2.26 |
ENSMUST00000120977.2
|
Wwtr1
|
WW domain containing transcription regulator 1 |
| chr3_-_57483175 | 2.20 |
ENSMUST00000029380.14
|
Wwtr1
|
WW domain containing transcription regulator 1 |
| chr15_+_97990431 | 2.19 |
ENSMUST00000229280.2
ENSMUST00000163507.8 ENSMUST00000230445.2 |
Pfkm
|
phosphofructokinase, muscle |
| chr4_-_151129020 | 2.19 |
ENSMUST00000103204.11
|
Per3
|
period circadian clock 3 |
| chrX_-_158921370 | 2.14 |
ENSMUST00000033662.9
|
Pdha1
|
pyruvate dehydrogenase E1 alpha 1 |
| chr3_+_89344006 | 2.12 |
ENSMUST00000038942.10
ENSMUST00000130858.8 ENSMUST00000146630.8 ENSMUST00000145753.2 |
Pbxip1
|
pre B cell leukemia transcription factor interacting protein 1 |
| chr3_-_37473715 | 2.12 |
ENSMUST00000138949.2
ENSMUST00000149449.8 ENSMUST00000108118.9 ENSMUST00000108117.3 ENSMUST00000099130.9 ENSMUST00000052645.13 |
Nudt6
|
nudix (nucleoside diphosphate linked moiety X)-type motif 6 |
| chr6_+_21985902 | 2.08 |
ENSMUST00000115383.9
|
Cped1
|
cadherin-like and PC-esterase domain containing 1 |
| chr16_+_57173456 | 2.07 |
ENSMUST00000159816.8
|
Filip1l
|
filamin A interacting protein 1-like |
| chr2_+_69500444 | 2.06 |
ENSMUST00000100050.4
|
Klhl41
|
kelch-like 41 |
| chr7_+_127503251 | 1.98 |
ENSMUST00000071056.14
|
Bckdk
|
branched chain ketoacid dehydrogenase kinase |
| chr8_-_76133212 | 1.97 |
ENSMUST00000212864.2
|
Gm10358
|
predicted gene 10358 |
| chr4_-_58206596 | 1.95 |
ENSMUST00000042850.9
|
Svep1
|
sushi, von Willebrand factor type A, EGF and pentraxin domain containing 1 |
| chr15_-_5093222 | 1.94 |
ENSMUST00000110689.5
|
C7
|
complement component 7 |
| chr8_-_86112938 | 1.93 |
ENSMUST00000137290.2
|
Mylk3
|
myosin light chain kinase 3 |
| chr1_+_135764092 | 1.91 |
ENSMUST00000188028.7
ENSMUST00000178204.8 ENSMUST00000190451.7 ENSMUST00000189732.7 ENSMUST00000189355.7 |
Tnnt2
|
troponin T2, cardiac |
| chr3_-_30194559 | 1.89 |
ENSMUST00000108271.10
|
Mecom
|
MDS1 and EVI1 complex locus |
| chr15_-_98193995 | 1.88 |
ENSMUST00000023722.12
ENSMUST00000169721.3 |
Zfp641
|
zinc finger protein 641 |
| chr10_-_13350106 | 1.87 |
ENSMUST00000105545.12
|
Phactr2
|
phosphatase and actin regulator 2 |
| chr18_-_36648850 | 1.85 |
ENSMUST00000025363.7
|
Hbegf
|
heparin-binding EGF-like growth factor |
| chr6_+_113449237 | 1.85 |
ENSMUST00000204447.3
|
Il17rc
|
interleukin 17 receptor C |
| chr7_+_113365356 | 1.79 |
ENSMUST00000084696.6
|
Spon1
|
spondin 1, (f-spondin) extracellular matrix protein |
| chr8_+_27575611 | 1.78 |
ENSMUST00000178514.8
ENSMUST00000033876.14 |
Adgra2
|
adhesion G protein-coupled receptor A2 |
| chr14_-_79539063 | 1.78 |
ENSMUST00000022595.8
|
Rgcc
|
regulator of cell cycle |
| chr4_-_148021159 | 1.78 |
ENSMUST00000105712.2
|
Plod1
|
procollagen-lysine, 2-oxoglutarate 5-dioxygenase 1 |
| chr6_+_97187650 | 1.77 |
ENSMUST00000044681.7
|
Arl6ip5
|
ADP-ribosylation factor-like 6 interacting protein 5 |
| chr10_+_79824418 | 1.75 |
ENSMUST00000004784.11
ENSMUST00000105374.2 |
Cnn2
|
calponin 2 |
| chr9_-_103980200 | 1.74 |
ENSMUST00000188000.2
|
Ackr4
|
atypical chemokine receptor 4 |
| chr4_+_149670889 | 1.70 |
ENSMUST00000105691.8
|
Clstn1
|
calsyntenin 1 |
| chr14_+_52131067 | 1.70 |
ENSMUST00000047726.12
|
Slc39a2
|
solute carrier family 39 (zinc transporter), member 2 |
| chr15_+_11064887 | 1.68 |
ENSMUST00000061318.9
|
Adamts12
|
a disintegrin-like and metallopeptidase (reprolysin type) with thrombospondin type 1 motif, 12 |
| chr16_+_21023505 | 1.62 |
ENSMUST00000006112.7
|
Ephb3
|
Eph receptor B3 |
| chr5_-_8417982 | 1.61 |
ENSMUST00000088761.11
ENSMUST00000115386.8 ENSMUST00000050166.14 ENSMUST00000046838.14 ENSMUST00000115388.9 ENSMUST00000088744.12 ENSMUST00000115385.2 |
Adam22
|
a disintegrin and metallopeptidase domain 22 |
| chr1_+_179788675 | 1.59 |
ENSMUST00000076687.12
ENSMUST00000097450.10 ENSMUST00000212756.2 |
Cdc42bpa
|
CDC42 binding protein kinase alpha |
| chr8_+_95498822 | 1.58 |
ENSMUST00000211956.2
ENSMUST00000211947.2 |
Cx3cl1
|
chemokine (C-X3-C motif) ligand 1 |
| chr18_+_23885390 | 1.58 |
ENSMUST00000170802.8
ENSMUST00000155708.8 ENSMUST00000118826.9 |
Mapre2
|
microtubule-associated protein, RP/EB family, member 2 |
| chr16_-_38533597 | 1.56 |
ENSMUST00000023487.5
|
Arhgap31
|
Rho GTPase activating protein 31 |
| chr9_-_43027809 | 1.53 |
ENSMUST00000216126.2
ENSMUST00000213544.2 ENSMUST00000061833.6 |
Tlcd5
|
TLC domain containing 5 |
| chr5_-_31311496 | 1.51 |
ENSMUST00000201353.4
ENSMUST00000200864.4 ENSMUST00000200833.4 ENSMUST00000201491.2 ENSMUST00000200744.4 ENSMUST00000202241.4 ENSMUST00000154241.8 |
Mpv17
|
MpV17 mitochondrial inner membrane protein |
| chr8_-_123404811 | 1.50 |
ENSMUST00000006525.14
ENSMUST00000064674.13 |
Cbfa2t3
|
CBFA2/RUNX1 translocation partner 3 |
| chr11_+_110956980 | 1.49 |
ENSMUST00000042970.3
|
Kcnj2
|
potassium inwardly-rectifying channel, subfamily J, member 2 |
| chr12_-_103660946 | 1.49 |
ENSMUST00000118101.2
|
Serpina1f
|
serine (or cysteine) peptidase inhibitor, clade A, member 1F |
| chr2_-_35351259 | 1.48 |
ENSMUST00000113001.9
ENSMUST00000113002.9 |
Ggta1
|
glycoprotein galactosyltransferase alpha 1, 3 |
| chr7_-_79492091 | 1.47 |
ENSMUST00000049004.8
|
Anpep
|
alanyl (membrane) aminopeptidase |
| chr15_-_102533494 | 1.44 |
ENSMUST00000184077.8
ENSMUST00000184906.8 ENSMUST00000169033.8 |
Atf7
|
activating transcription factor 7 |
| chr8_-_41494890 | 1.43 |
ENSMUST00000051379.14
|
Mtus1
|
mitochondrial tumor suppressor 1 |
| chr11_+_118324652 | 1.43 |
ENSMUST00000106286.3
|
C1qtnf1
|
C1q and tumor necrosis factor related protein 1 |
| chr11_+_35011953 | 1.41 |
ENSMUST00000069837.4
|
Slit3
|
slit guidance ligand 3 |
| chr5_+_30360246 | 1.41 |
ENSMUST00000026841.15
ENSMUST00000123980.8 ENSMUST00000114783.6 ENSMUST00000114786.8 |
Hadhb
|
hydroxyacyl-CoA dehydrogenase trifunctional multienzyme complex subunit beta |
| chr11_+_87471867 | 1.39 |
ENSMUST00000018544.12
ENSMUST00000063156.11 ENSMUST00000107960.8 |
Septin4
|
septin 4 |
| chr2_-_35351408 | 1.39 |
ENSMUST00000102794.8
|
Ggta1
|
glycoprotein galactosyltransferase alpha 1, 3 |
| chr4_-_119031050 | 1.35 |
ENSMUST00000052715.10
ENSMUST00000179290.8 ENSMUST00000154226.2 |
Zfp691
|
zinc finger protein 691 |
| chr8_-_123278054 | 1.35 |
ENSMUST00000156333.9
ENSMUST00000067252.14 |
Piezo1
|
piezo-type mechanosensitive ion channel component 1 |
| chr16_+_4704166 | 1.34 |
ENSMUST00000230990.2
|
Mgrn1
|
mahogunin, ring finger 1 |
| chr8_+_75940572 | 1.33 |
ENSMUST00000139848.8
|
Rasd2
|
RASD family, member 2 |
| chr1_+_43769750 | 1.31 |
ENSMUST00000027217.9
|
Ecrg4
|
ECRG4 augurin precursor |
| chr10_-_12702674 | 1.26 |
ENSMUST00000219130.2
|
Utrn
|
utrophin |
| chr5_+_136112765 | 1.25 |
ENSMUST00000042135.14
|
Rasa4
|
RAS p21 protein activator 4 |
| chr2_+_120459593 | 1.24 |
ENSMUST00000180041.9
|
Stard9
|
START domain containing 9 |
| chr7_-_109559593 | 1.22 |
ENSMUST00000106722.2
|
Dennd5a
|
DENN/MADD domain containing 5A |
| chr7_-_109559671 | 1.19 |
ENSMUST00000080437.13
|
Dennd5a
|
DENN/MADD domain containing 5A |
| chr10_-_12745109 | 1.18 |
ENSMUST00000218635.2
|
Utrn
|
utrophin |
| chr11_+_51654511 | 1.16 |
ENSMUST00000020653.6
|
Sar1b
|
secretion associated Ras related GTPase 1B |
| chr3_+_14545751 | 1.16 |
ENSMUST00000037321.8
ENSMUST00000120484.8 ENSMUST00000120801.2 |
Slc7a12
|
solute carrier family 7 (cationic amino acid transporter, y+ system), member 12 |
| chr18_+_50186349 | 1.15 |
ENSMUST00000148159.3
|
Tnfaip8
|
tumor necrosis factor, alpha-induced protein 8 |
| chr2_-_164246526 | 1.14 |
ENSMUST00000109359.8
ENSMUST00000109358.8 ENSMUST00000103103.10 |
Matn4
|
matrilin 4 |
| chr4_-_130253703 | 1.13 |
ENSMUST00000134159.3
|
Zcchc17
|
zinc finger, CCHC domain containing 17 |
| chr9_+_106325828 | 1.12 |
ENSMUST00000217496.2
|
Abhd14b
|
abhydrolase domain containing 14b |
| chr9_-_35111172 | 1.11 |
ENSMUST00000176021.8
ENSMUST00000176531.8 ENSMUST00000176685.8 ENSMUST00000177129.8 |
Tirap
|
toll-interleukin 1 receptor (TIR) domain-containing adaptor protein |
| chr4_-_148021217 | 1.11 |
ENSMUST00000019199.14
|
Plod1
|
procollagen-lysine, 2-oxoglutarate 5-dioxygenase 1 |
| chr19_-_4062656 | 1.10 |
ENSMUST00000134479.8
ENSMUST00000128787.8 ENSMUST00000237862.2 ENSMUST00000236203.2 ENSMUST00000133474.8 |
Ndufv1
|
NADH:ubiquinone oxidoreductase core subunit V1 |
| chr7_+_26821266 | 1.10 |
ENSMUST00000206552.2
|
Cyp2f2
|
cytochrome P450, family 2, subfamily f, polypeptide 2 |
| chr4_-_40239700 | 1.08 |
ENSMUST00000142055.2
|
Ddx58
|
DEAD/H box helicase 58 |
| chr5_+_136112817 | 1.08 |
ENSMUST00000100570.10
|
Rasa4
|
RAS p21 protein activator 4 |
| chr12_-_103660916 | 1.07 |
ENSMUST00000117053.8
|
Serpina1f
|
serine (or cysteine) peptidase inhibitor, clade A, member 1F |
| chrX_+_149829131 | 1.05 |
ENSMUST00000112685.8
|
Fgd1
|
FYVE, RhoGEF and PH domain containing 1 |
| chr1_+_17797257 | 1.04 |
ENSMUST00000159958.8
ENSMUST00000160305.8 ENSMUST00000095075.5 |
Crispld1
|
cysteine-rich secretory protein LCCL domain containing 1 |
| chr3_+_123061094 | 1.03 |
ENSMUST00000047923.12
ENSMUST00000200333.2 |
Sec24d
|
Sec24 related gene family, member D (S. cerevisiae) |
| chr6_-_92511836 | 1.03 |
ENSMUST00000113446.8
|
Prickle2
|
prickle planar cell polarity protein 2 |
| chr14_+_54598283 | 1.02 |
ENSMUST00000000985.7
|
Oxa1l
|
oxidase assembly 1-like |
| chr19_-_4062738 | 1.02 |
ENSMUST00000136921.2
ENSMUST00000042497.14 |
Ndufv1
|
NADH:ubiquinone oxidoreductase core subunit V1 |
| chr9_+_69902697 | 1.01 |
ENSMUST00000165389.8
|
Bnip2
|
BCL2/adenovirus E1B interacting protein 2 |
| chr6_+_83011154 | 1.01 |
ENSMUST00000000707.9
ENSMUST00000101257.4 |
Loxl3
|
lysyl oxidase-like 3 |
| chr2_+_20742115 | 1.00 |
ENSMUST00000114606.8
ENSMUST00000114608.3 |
Etl4
|
enhancer trap locus 4 |
| chr17_+_36134450 | 1.00 |
ENSMUST00000172846.2
|
Flot1
|
flotillin 1 |
| chr9_+_57847387 | 0.99 |
ENSMUST00000043059.9
|
Sema7a
|
sema domain, immunoglobulin domain (Ig), and GPI membrane anchor, (semaphorin) 7A |
| chr16_+_57173632 | 0.98 |
ENSMUST00000099667.3
|
Filip1l
|
filamin A interacting protein 1-like |
| chr17_+_37313502 | 0.95 |
ENSMUST00000173921.8
ENSMUST00000172580.8 |
Zfp57
|
zinc finger protein 57 |
| chr15_-_102533357 | 0.94 |
ENSMUST00000183765.2
|
Atf7
|
activating transcription factor 7 |
| chr1_-_168259839 | 0.93 |
ENSMUST00000188912.7
|
Pbx1
|
pre B cell leukemia homeobox 1 |
| chr13_-_35147771 | 0.91 |
ENSMUST00000164155.2
ENSMUST00000021853.12 |
Eci3
|
enoyl-Coenzyme A delta isomerase 3 |
| chr16_+_4704103 | 0.91 |
ENSMUST00000023159.10
ENSMUST00000070658.16 ENSMUST00000229038.2 |
Mgrn1
|
mahogunin, ring finger 1 |
| chr13_-_104953631 | 0.91 |
ENSMUST00000154165.2
|
Cwc27
|
CWC27 spliceosome-associated protein |
| chr8_+_105066924 | 0.91 |
ENSMUST00000212081.2
|
Cmtm3
|
CKLF-like MARVEL transmembrane domain containing 3 |
| chrX_-_135104589 | 0.89 |
ENSMUST00000066819.11
|
Tceal5
|
transcription elongation factor A (SII)-like 5 |
| chr7_-_30855295 | 0.89 |
ENSMUST00000186723.3
|
Gramd1a
|
GRAM domain containing 1A |
| chr6_-_72357398 | 0.88 |
ENSMUST00000101285.10
ENSMUST00000074231.6 |
Vamp5
|
vesicle-associated membrane protein 5 |
| chr7_-_110581652 | 0.87 |
ENSMUST00000005751.13
|
Irag1
|
inositol 1,4,5-triphosphate receptor associated 1 |
| chr8_+_75941130 | 0.86 |
ENSMUST00000132133.2
|
Rasd2
|
RASD family, member 2 |
| chr6_-_21851827 | 0.86 |
ENSMUST00000202353.2
ENSMUST00000134635.2 ENSMUST00000123116.8 ENSMUST00000120965.8 ENSMUST00000143531.2 |
Tspan12
|
tetraspanin 12 |
| chr2_+_122146153 | 0.84 |
ENSMUST00000099461.4
|
Duox1
|
dual oxidase 1 |
| chr4_+_63477018 | 0.84 |
ENSMUST00000077709.11
|
Tmem268
|
transmembrane protein 268 |
| chr11_+_49141410 | 0.83 |
ENSMUST00000129588.2
|
Mgat1
|
mannoside acetylglucosaminyltransferase 1 |
| chr6_+_83112793 | 0.82 |
ENSMUST00000065512.11
|
Rtkn
|
rhotekin |
| chr7_+_43228999 | 0.82 |
ENSMUST00000058104.8
ENSMUST00000205769.2 |
Zfp719
|
zinc finger protein 719 |
| chr5_-_30360113 | 0.82 |
ENSMUST00000156859.3
|
Hadha
|
hydroxyacyl-CoA dehydrogenase trifunctional multienzyme complex subunit alpha |
| chr9_-_100916946 | 0.82 |
ENSMUST00000035116.12
|
Pccb
|
propionyl Coenzyme A carboxylase, beta polypeptide |
| chr1_-_75110511 | 0.81 |
ENSMUST00000027405.6
|
Slc23a3
|
solute carrier family 23 (nucleobase transporters), member 3 |
| chr15_+_54434576 | 0.79 |
ENSMUST00000025356.4
|
Mal2
|
mal, T cell differentiation protein 2 |
| chr2_+_128540822 | 0.78 |
ENSMUST00000014505.5
|
Mertk
|
MER proto-oncogene tyrosine kinase |
| chr14_-_54648057 | 0.77 |
ENSMUST00000200545.2
ENSMUST00000227967.2 |
Slc7a7
|
solute carrier family 7 (cationic amino acid transporter, y+ system), member 7 |
| chr8_-_94825556 | 0.77 |
ENSMUST00000034206.6
|
Bbs2
|
Bardet-Biedl syndrome 2 (human) |
| chr9_+_40180726 | 0.77 |
ENSMUST00000171835.9
|
Scn3b
|
sodium channel, voltage-gated, type III, beta |
| chr11_-_94440025 | 0.77 |
ENSMUST00000040487.4
|
Rsad1
|
radical S-adenosyl methionine domain containing 1 |
| chr2_+_125876883 | 0.76 |
ENSMUST00000110442.2
|
Fgf7
|
fibroblast growth factor 7 |
| chr5_-_113163339 | 0.75 |
ENSMUST00000197776.2
ENSMUST00000065167.9 |
Grk3
|
G protein-coupled receptor kinase 3 |
| chr1_-_168259710 | 0.74 |
ENSMUST00000072863.6
|
Pbx1
|
pre B cell leukemia homeobox 1 |
| chr19_-_24178000 | 0.74 |
ENSMUST00000233658.3
|
Tjp2
|
tight junction protein 2 |
| chr9_+_106325860 | 0.73 |
ENSMUST00000185527.3
|
Abhd14b
|
abhydrolase domain containing 14b |
| chr11_-_72380730 | 0.72 |
ENSMUST00000045303.10
|
Spns2
|
spinster homolog 2 |
| chr18_+_37580692 | 0.71 |
ENSMUST00000052387.5
|
Pcdhb14
|
protocadherin beta 14 |
| chr12_+_112940086 | 0.70 |
ENSMUST00000165079.8
ENSMUST00000221500.2 ENSMUST00000221104.2 ENSMUST00000002880.7 ENSMUST00000222209.2 |
Btbd6
|
BTB (POZ) domain containing 6 |
| chrX_-_99669507 | 0.69 |
ENSMUST00000059099.7
|
Pdzd11
|
PDZ domain containing 11 |
| chr9_+_122863791 | 0.68 |
ENSMUST00000026893.6
ENSMUST00000215247.2 |
Tgm4
|
transglutaminase 4 (prostate) |
| chr12_+_51737775 | 0.67 |
ENSMUST00000218820.2
ENSMUST00000021338.10 |
Ap4s1
|
adaptor-related protein complex AP-4, sigma 1 |
| chr11_+_69891398 | 0.67 |
ENSMUST00000019362.15
ENSMUST00000190940.2 |
Dvl2
|
dishevelled segment polarity protein 2 |
| chr19_-_60862964 | 0.66 |
ENSMUST00000025961.7
|
Prdx3
|
peroxiredoxin 3 |
| chr14_-_121742659 | 0.66 |
ENSMUST00000088386.8
|
Slc15a1
|
solute carrier family 15 (oligopeptide transporter), member 1 |
| chr14_-_119336809 | 0.65 |
ENSMUST00000156203.8
|
Uggt2
|
UDP-glucose glycoprotein glucosyltransferase 2 |
| chr9_-_56151334 | 0.65 |
ENSMUST00000188142.7
|
Peak1
|
pseudopodium-enriched atypical kinase 1 |
| chr1_+_63769772 | 0.64 |
ENSMUST00000027103.7
|
Fastkd2
|
FAST kinase domains 2 |
| chr2_-_36026785 | 0.64 |
ENSMUST00000028251.10
|
Rbm18
|
RNA binding motif protein 18 |
| chr18_+_74401322 | 0.63 |
ENSMUST00000224047.2
|
Mbd1
|
methyl-CpG binding domain protein 1 |
| chr17_+_15616464 | 0.62 |
ENSMUST00000055352.8
|
Fam120b
|
family with sequence similarity 120, member B |
| chr5_+_32616187 | 0.62 |
ENSMUST00000015100.15
|
Ppp1cb
|
protein phosphatase 1 catalytic subunit beta |
| chr2_+_32253016 | 0.61 |
ENSMUST00000132028.8
ENSMUST00000136079.8 |
Ciz1
|
CDKN1A interacting zinc finger protein 1 |
| chr7_+_86175967 | 0.61 |
ENSMUST00000071112.4
|
Olfr297
|
olfactory receptor 297 |
| chr8_-_69541852 | 0.60 |
ENSMUST00000037478.13
ENSMUST00000148856.2 |
Slc18a1
|
solute carrier family 18 (vesicular monoamine), member 1 |
| chr11_+_97594220 | 0.60 |
ENSMUST00000103147.5
|
Psmb3
|
proteasome (prosome, macropain) subunit, beta type 3 |
| chr1_+_39940189 | 0.59 |
ENSMUST00000191761.6
ENSMUST00000193682.6 ENSMUST00000195860.6 ENSMUST00000195259.6 ENSMUST00000195636.6 ENSMUST00000192509.6 |
Map4k4
|
mitogen-activated protein kinase kinase kinase kinase 4 |
| chr14_-_54647647 | 0.59 |
ENSMUST00000228488.2
ENSMUST00000195970.5 |
Slc7a7
|
solute carrier family 7 (cationic amino acid transporter, y+ system), member 7 |
| chr7_-_19093383 | 0.59 |
ENSMUST00000047036.10
|
Cd3eap
|
CD3E antigen, epsilon polypeptide associated protein |
| chr19_+_45035942 | 0.59 |
ENSMUST00000237222.2
ENSMUST00000111954.11 |
Sfxn3
|
sideroflexin 3 |
| chr4_+_156110621 | 0.58 |
ENSMUST00000103173.10
ENSMUST00000040274.13 ENSMUST00000122001.3 |
Tnfrsf18
|
tumor necrosis factor receptor superfamily, member 18 |
| chr14_-_20844074 | 0.58 |
ENSMUST00000080440.14
ENSMUST00000100837.11 ENSMUST00000071816.7 |
Camk2g
|
calcium/calmodulin-dependent protein kinase II gamma |
| chr16_+_10298011 | 0.58 |
ENSMUST00000230450.2
|
Ciita
|
class II transactivator |
| chr2_+_160722562 | 0.57 |
ENSMUST00000109456.9
|
Lpin3
|
lipin 3 |
| chrX_+_138585728 | 0.57 |
ENSMUST00000096313.4
|
Tbc1d8b
|
TBC1 domain family, member 8B |
| chr17_+_12803019 | 0.54 |
ENSMUST00000046959.9
ENSMUST00000233066.2 |
Slc22a2
|
solute carrier family 22 (organic cation transporter), member 2 |
| chr10_-_53255959 | 0.54 |
ENSMUST00000220443.2
|
Cep85l
|
centrosomal protein 85-like |
| chr2_-_119493237 | 0.54 |
ENSMUST00000028768.2
ENSMUST00000110801.8 ENSMUST00000110802.8 |
Ndufaf1
|
NADH:ubiquinone oxidoreductase complex assembly factor 1 |
| chr10_-_128755127 | 0.53 |
ENSMUST00000149961.2
ENSMUST00000026406.14 |
Rdh5
|
retinol dehydrogenase 5 |
| chr16_-_32306683 | 0.53 |
ENSMUST00000042042.9
|
Slc51a
|
solute carrier family 51, alpha subunit |
| chr3_-_108306267 | 0.52 |
ENSMUST00000147251.2
|
Celsr2
|
cadherin, EGF LAG seven-pass G-type receptor 2 |
| chr5_-_86475595 | 0.51 |
ENSMUST00000122377.2
|
Tmprss11d
|
transmembrane protease, serine 11d |
| chr4_-_123611974 | 0.51 |
ENSMUST00000137312.2
ENSMUST00000106206.8 |
Ndufs5
|
NADH:ubiquinone oxidoreductase core subunit S5 |
| chr13_+_43938251 | 0.51 |
ENSMUST00000015540.4
|
Cd83
|
CD83 antigen |
| chr7_+_43000838 | 0.49 |
ENSMUST00000206299.2
ENSMUST00000121494.2 |
Siglecf
|
sialic acid binding Ig-like lectin F |
| chr4_-_40239778 | 0.49 |
ENSMUST00000037907.13
|
Ddx58
|
DEAD/H box helicase 58 |
| chr1_-_20688196 | 0.48 |
ENSMUST00000088448.12
|
Pkhd1
|
polycystic kidney and hepatic disease 1 |
| chr19_-_4927910 | 0.48 |
ENSMUST00000006626.5
|
Actn3
|
actinin alpha 3 |
| chr4_-_143176874 | 0.47 |
ENSMUST00000073641.4
ENSMUST00000061277.11 |
Oog4
|
oogenesin 4 |
| chr2_-_160701523 | 0.47 |
ENSMUST00000103112.8
|
Zhx3
|
zinc fingers and homeoboxes 3 |
| chr19_+_45991907 | 0.46 |
ENSMUST00000099393.4
|
Hps6
|
HPS6, biogenesis of lysosomal organelles complex 2 subunit 3 |
| chr19_+_24853039 | 0.45 |
ENSMUST00000073080.7
|
Gm10053
|
predicted gene 10053 |
| chr2_+_11710101 | 0.45 |
ENSMUST00000138349.8
ENSMUST00000135341.8 ENSMUST00000128156.9 |
Il15ra
|
interleukin 15 receptor, alpha chain |
| chr11_+_59433508 | 0.45 |
ENSMUST00000101148.9
|
Nlrp3
|
NLR family, pyrin domain containing 3 |
| chr17_-_34847013 | 0.44 |
ENSMUST00000166040.9
|
Ppt2
|
palmitoyl-protein thioesterase 2 |
| chr10_+_78410180 | 0.44 |
ENSMUST00000218061.2
ENSMUST00000218787.2 ENSMUST00000105384.5 ENSMUST00000218875.2 |
Ilvbl
|
ilvB (bacterial acetolactate synthase)-like |
| chr3_+_99203818 | 0.43 |
ENSMUST00000150756.3
|
Tbx15
|
T-box 15 |
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 4.9 | 14.6 | GO:0090320 | regulation of chylomicron remnant clearance(GO:0090320) |
| 4.2 | 25.2 | GO:0006116 | NADH oxidation(GO:0006116) |
| 1.5 | 1.5 | GO:0090076 | relaxation of skeletal muscle(GO:0090076) |
| 1.1 | 3.2 | GO:0031104 | dendrite regeneration(GO:0031104) |
| 0.8 | 2.5 | GO:1903920 | positive regulation of actin filament severing(GO:1903920) |
| 0.8 | 3.3 | GO:1902304 | positive regulation of potassium ion export(GO:1902304) |
| 0.7 | 2.9 | GO:0033575 | protein glycosylation at cell surface(GO:0033575) protein galactosylation at cell surface(GO:0033580) protein galactosylation(GO:0042125) |
| 0.7 | 4.3 | GO:0002528 | regulation of vascular permeability involved in acute inflammatory response(GO:0002528) |
| 0.7 | 3.5 | GO:0031443 | fast-twitch skeletal muscle fiber contraction(GO:0031443) |
| 0.6 | 1.9 | GO:0051542 | elastin biosynthetic process(GO:0051542) |
| 0.6 | 3.6 | GO:0072592 | oxygen metabolic process(GO:0072592) |
| 0.6 | 1.8 | GO:0003331 | regulation of extracellular matrix constituent secretion(GO:0003330) positive regulation of extracellular matrix constituent secretion(GO:0003331) |
| 0.6 | 2.9 | GO:0046946 | hydroxylysine metabolic process(GO:0046946) hydroxylysine biosynthetic process(GO:0046947) |
| 0.5 | 4.7 | GO:0010756 | positive regulation of plasminogen activation(GO:0010756) |
| 0.5 | 1.9 | GO:0032972 | regulation of muscle filament sliding speed(GO:0032972) |
| 0.4 | 4.5 | GO:0072307 | metanephric nephron tubule epithelial cell differentiation(GO:0072257) regulation of metanephric nephron tubule epithelial cell differentiation(GO:0072307) |
| 0.4 | 2.2 | GO:0061622 | glycolytic process through glucose-1-phosphate(GO:0061622) |
| 0.4 | 1.7 | GO:0030167 | proteoglycan catabolic process(GO:0030167) regulation of hepatocyte growth factor receptor signaling pathway(GO:1902202) |
| 0.4 | 1.1 | GO:0032618 | interleukin-15 production(GO:0032618) |
| 0.4 | 1.1 | GO:0018931 | naphthalene metabolic process(GO:0018931) naphthalene-containing compound metabolic process(GO:0090420) |
| 0.4 | 1.8 | GO:0002036 | regulation of L-glutamate transport(GO:0002036) |
| 0.4 | 3.2 | GO:1901552 | positive regulation of endothelial cell development(GO:1901552) positive regulation of establishment of endothelial barrier(GO:1903142) |
| 0.4 | 1.4 | GO:0070100 | negative regulation of chemokine-mediated signaling pathway(GO:0070100) |
| 0.3 | 2.4 | GO:0007527 | adult somatic muscle development(GO:0007527) |
| 0.3 | 1.6 | GO:0050902 | leukocyte adhesive activation(GO:0050902) |
| 0.3 | 1.6 | GO:0009597 | detection of virus(GO:0009597) |
| 0.3 | 2.8 | GO:0071313 | cellular response to caffeine(GO:0071313) positive regulation of the force of heart contraction(GO:0098735) |
| 0.3 | 1.3 | GO:0033634 | positive regulation of cell-cell adhesion mediated by integrin(GO:0033634) |
| 0.3 | 1.0 | GO:0033615 | mitochondrial proton-transporting ATP synthase complex assembly(GO:0033615) |
| 0.3 | 0.8 | GO:0061033 | secretion by lung epithelial cell involved in lung growth(GO:0061033) |
| 0.3 | 1.0 | GO:0006049 | UDP-N-acetylglucosamine catabolic process(GO:0006049) nucleotide-sugar catabolic process(GO:0009227) |
| 0.2 | 3.4 | GO:0035095 | behavioral response to nicotine(GO:0035095) |
| 0.2 | 2.1 | GO:0061732 | mitochondrial acetyl-CoA biosynthetic process from pyruvate(GO:0061732) |
| 0.2 | 1.8 | GO:1902548 | negative regulation of cellular response to vascular endothelial growth factor stimulus(GO:1902548) |
| 0.2 | 0.8 | GO:0042335 | cuticle development(GO:0042335) |
| 0.2 | 1.0 | GO:0001830 | trophectodermal cell fate commitment(GO:0001830) |
| 0.2 | 0.8 | GO:0038108 | negative regulation of appetite by leptin-mediated signaling pathway(GO:0038108) |
| 0.2 | 2.9 | GO:1904261 | regulation of basement membrane assembly involved in embryonic body morphogenesis(GO:1904259) positive regulation of basement membrane assembly involved in embryonic body morphogenesis(GO:1904261) basement membrane assembly involved in embryonic body morphogenesis(GO:2001197) |
| 0.2 | 5.9 | GO:0070208 | protein heterotrimerization(GO:0070208) |
| 0.2 | 1.5 | GO:0048069 | eye pigmentation(GO:0048069) |
| 0.2 | 1.4 | GO:0000821 | regulation of arginine metabolic process(GO:0000821) |
| 0.2 | 0.7 | GO:0045297 | mating plug formation(GO:0042628) single-organism reproductive behavior(GO:0044704) post-mating behavior(GO:0045297) |
| 0.2 | 1.0 | GO:0060355 | positive regulation of cell adhesion molecule production(GO:0060355) |
| 0.2 | 0.7 | GO:0014022 | neural plate elongation(GO:0014022) convergent extension involved in neural plate elongation(GO:0022007) |
| 0.2 | 2.3 | GO:0043951 | negative regulation of cAMP-mediated signaling(GO:0043951) |
| 0.2 | 0.8 | GO:0097350 | neutrophil clearance(GO:0097350) |
| 0.1 | 3.3 | GO:0000188 | inactivation of MAPK activity(GO:0000188) |
| 0.1 | 1.5 | GO:1903715 | regulation of aerobic respiration(GO:1903715) |
| 0.1 | 0.6 | GO:1902775 | mitochondrial large ribosomal subunit assembly(GO:1902775) |
| 0.1 | 2.1 | GO:0031272 | regulation of pseudopodium assembly(GO:0031272) |
| 0.1 | 1.7 | GO:0018158 | protein oxidation(GO:0018158) |
| 0.1 | 1.9 | GO:0090336 | positive regulation of brown fat cell differentiation(GO:0090336) |
| 0.1 | 0.3 | GO:0046005 | positive regulation of circadian sleep/wake cycle, REM sleep(GO:0046005) |
| 0.1 | 0.3 | GO:2001176 | mediator complex assembly(GO:0036034) regulation of mediator complex assembly(GO:2001176) positive regulation of mediator complex assembly(GO:2001178) |
| 0.1 | 3.0 | GO:0007614 | short-term memory(GO:0007614) |
| 0.1 | 0.2 | GO:1900625 | regulation of monocyte aggregation(GO:1900623) positive regulation of monocyte aggregation(GO:1900625) |
| 0.1 | 0.7 | GO:2001205 | negative regulation of osteoclast development(GO:2001205) |
| 0.1 | 0.3 | GO:0019085 | early viral transcription(GO:0019085) |
| 0.1 | 1.5 | GO:0032836 | glomerular basement membrane development(GO:0032836) |
| 0.1 | 0.6 | GO:0051581 | negative regulation of neurotransmitter uptake(GO:0051581) negative regulation of serotonin uptake(GO:0051612) |
| 0.1 | 1.6 | GO:0033623 | regulation of integrin activation(GO:0033623) |
| 0.1 | 1.9 | GO:0006883 | cellular sodium ion homeostasis(GO:0006883) |
| 0.1 | 0.4 | GO:0071348 | cellular response to interleukin-11(GO:0071348) |
| 0.1 | 2.0 | GO:0009083 | branched-chain amino acid catabolic process(GO:0009083) |
| 0.1 | 0.7 | GO:0042938 | dipeptide transport(GO:0042938) |
| 0.1 | 0.8 | GO:1903208 | neuron death in response to hydrogen peroxide(GO:0036476) regulation of hydrogen peroxide-induced neuron death(GO:1903207) negative regulation of hydrogen peroxide-induced neuron death(GO:1903208) |
| 0.1 | 0.3 | GO:2000978 | auditory receptor cell fate determination(GO:0042668) negative regulation of forebrain neuron differentiation(GO:2000978) |
| 0.1 | 0.5 | GO:0033326 | cerebrospinal fluid secretion(GO:0033326) |
| 0.1 | 1.5 | GO:0035814 | negative regulation of renal sodium excretion(GO:0035814) |
| 0.1 | 0.3 | GO:0019388 | galactose catabolic process(GO:0019388) |
| 0.1 | 0.4 | GO:0043490 | malate-aspartate shuttle(GO:0043490) |
| 0.1 | 0.9 | GO:0050861 | positive regulation of B cell receptor signaling pathway(GO:0050861) |
| 0.1 | 1.0 | GO:0060907 | positive regulation of macrophage cytokine production(GO:0060907) |
| 0.1 | 1.8 | GO:1900017 | positive regulation of cytokine production involved in inflammatory response(GO:1900017) |
| 0.1 | 0.5 | GO:1903361 | protein localization to basolateral plasma membrane(GO:1903361) |
| 0.1 | 0.8 | GO:0001766 | membrane raft polarization(GO:0001766) membrane raft distribution(GO:0031580) |
| 0.1 | 0.4 | GO:0050713 | negative regulation of interleukin-1 beta secretion(GO:0050713) |
| 0.1 | 2.2 | GO:0033235 | positive regulation of protein sumoylation(GO:0033235) |
| 0.1 | 2.3 | GO:0034260 | negative regulation of GTPase activity(GO:0034260) |
| 0.1 | 1.8 | GO:0050765 | negative regulation of phagocytosis(GO:0050765) |
| 0.1 | 0.8 | GO:0060373 | regulation of ventricular cardiac muscle cell membrane depolarization(GO:0060373) |
| 0.1 | 0.5 | GO:0032713 | negative regulation of interleukin-4 production(GO:0032713) |
| 0.1 | 2.2 | GO:0045187 | regulation of circadian sleep/wake cycle, sleep(GO:0045187) |
| 0.1 | 0.6 | GO:0005981 | regulation of glycogen catabolic process(GO:0005981) |
| 0.1 | 2.0 | GO:0007097 | nuclear migration(GO:0007097) establishment of nucleus localization(GO:0040023) |
| 0.1 | 0.6 | GO:0045343 | MHC class I biosynthetic process(GO:0045341) regulation of MHC class I biosynthetic process(GO:0045343) positive regulation of MHC class I biosynthetic process(GO:0045345) |
| 0.1 | 1.6 | GO:0007413 | axonal fasciculation(GO:0007413) retinal ganglion cell axon guidance(GO:0031290) |
| 0.1 | 1.0 | GO:0090110 | cargo loading into COPII-coated vesicle(GO:0090110) |
| 0.0 | 0.1 | GO:0072695 | negative regulation of DNA recombination at telomere(GO:0048239) regulation of DNA recombination at telomere(GO:0072695) |
| 0.0 | 1.7 | GO:0010971 | positive regulation of G2/M transition of mitotic cell cycle(GO:0010971) |
| 0.0 | 1.4 | GO:0010758 | regulation of macrophage chemotaxis(GO:0010758) |
| 0.0 | 0.2 | GO:0017185 | peptidyl-lysine hydroxylation(GO:0017185) |
| 0.0 | 1.9 | GO:0014044 | Schwann cell development(GO:0014044) |
| 0.0 | 0.5 | GO:0051608 | histamine transport(GO:0051608) |
| 0.0 | 1.2 | GO:0000413 | protein peptidyl-prolyl isomerization(GO:0000413) |
| 0.0 | 0.6 | GO:0071712 | ER-associated misfolded protein catabolic process(GO:0071712) |
| 0.0 | 0.9 | GO:0043045 | DNA methylation involved in embryo development(GO:0043045) changes to DNA methylation involved in embryo development(GO:1901538) |
| 0.0 | 0.3 | GO:0097428 | protein maturation by iron-sulfur cluster transfer(GO:0097428) |
| 0.0 | 2.1 | GO:0042775 | mitochondrial ATP synthesis coupled electron transport(GO:0042775) |
| 0.0 | 1.7 | GO:0006829 | zinc II ion transport(GO:0006829) |
| 0.0 | 0.5 | GO:0051574 | positive regulation of histone H3-K9 methylation(GO:0051574) |
| 0.0 | 0.6 | GO:0061179 | negative regulation of insulin secretion involved in cellular response to glucose stimulus(GO:0061179) |
| 0.0 | 0.4 | GO:0044387 | negative regulation of protein kinase activity by regulation of protein phosphorylation(GO:0044387) |
| 0.0 | 0.6 | GO:0048712 | negative regulation of astrocyte differentiation(GO:0048712) |
| 0.0 | 0.4 | GO:0050957 | equilibrioception(GO:0050957) |
| 0.0 | 0.1 | GO:0014886 | transition between slow and fast fiber(GO:0014886) |
| 0.0 | 0.9 | GO:0010842 | retina layer formation(GO:0010842) |
| 0.0 | 0.6 | GO:1901897 | regulation of relaxation of cardiac muscle(GO:1901897) |
| 0.0 | 3.2 | GO:0072332 | intrinsic apoptotic signaling pathway by p53 class mediator(GO:0072332) |
| 0.0 | 0.7 | GO:0032825 | positive regulation of natural killer cell differentiation(GO:0032825) |
| 0.0 | 0.3 | GO:0030007 | cellular potassium ion homeostasis(GO:0030007) |
| 0.0 | 7.5 | GO:0007519 | skeletal muscle tissue development(GO:0007519) |
| 0.0 | 0.3 | GO:0034773 | histone H4-K20 trimethylation(GO:0034773) |
| 0.0 | 0.6 | GO:0035357 | peroxisome proliferator activated receptor signaling pathway(GO:0035357) |
| 0.0 | 0.3 | GO:0097264 | self proteolysis(GO:0097264) |
| 0.0 | 0.5 | GO:0015721 | bile acid and bile salt transport(GO:0015721) |
| 0.0 | 1.3 | GO:0045600 | positive regulation of fat cell differentiation(GO:0045600) |
| 0.0 | 0.6 | GO:0009303 | rRNA transcription(GO:0009303) |
| 0.0 | 1.0 | GO:0060325 | face morphogenesis(GO:0060325) |
| 0.0 | 1.9 | GO:0042147 | retrograde transport, endosome to Golgi(GO:0042147) |
| 0.0 | 0.8 | GO:0006779 | porphyrin-containing compound biosynthetic process(GO:0006779) |
| 0.0 | 1.1 | GO:0048678 | response to axon injury(GO:0048678) |
| 0.0 | 1.1 | GO:0032611 | interleukin-1 beta production(GO:0032611) |
| 0.0 | 0.4 | GO:0071425 | hematopoietic stem cell proliferation(GO:0071425) |
| 0.0 | 0.2 | GO:0018298 | protein-chromophore linkage(GO:0018298) |
| 0.0 | 1.7 | GO:0051965 | positive regulation of synapse assembly(GO:0051965) |
| 0.0 | 0.0 | GO:0097089 | methyl-branched fatty acid metabolic process(GO:0097089) |
| 0.0 | 0.2 | GO:0018026 | peptidyl-lysine monomethylation(GO:0018026) |
| 0.0 | 0.5 | GO:0010257 | NADH dehydrogenase complex assembly(GO:0010257) mitochondrial respiratory chain complex I assembly(GO:0032981) mitochondrial respiratory chain complex I biogenesis(GO:0097031) |
| 0.0 | 0.1 | GO:2000675 | negative regulation of type B pancreatic cell apoptotic process(GO:2000675) |
| 0.0 | 1.0 | GO:0051057 | positive regulation of small GTPase mediated signal transduction(GO:0051057) |
| 0.0 | 0.1 | GO:0070072 | vacuolar proton-transporting V-type ATPase complex assembly(GO:0070072) |
| 0.0 | 1.1 | GO:0046847 | filopodium assembly(GO:0046847) |
| 0.0 | 0.2 | GO:0046007 | negative regulation of activated T cell proliferation(GO:0046007) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.6 | 4.7 | GO:0001652 | granular component(GO:0001652) |
| 0.4 | 2.2 | GO:0005945 | 6-phosphofructokinase complex(GO:0005945) |
| 0.4 | 14.6 | GO:0034364 | high-density lipoprotein particle(GO:0034364) |
| 0.3 | 2.0 | GO:0005947 | mitochondrial alpha-ketoglutarate dehydrogenase complex(GO:0005947) |
| 0.3 | 3.0 | GO:0060053 | neurofilament cytoskeleton(GO:0060053) |
| 0.3 | 3.3 | GO:0042629 | mast cell granule(GO:0042629) |
| 0.3 | 1.9 | GO:1990584 | cardiac Troponin complex(GO:1990584) |
| 0.3 | 2.1 | GO:0045254 | pyruvate dehydrogenase complex(GO:0045254) |
| 0.2 | 1.9 | GO:0005579 | membrane attack complex(GO:0005579) |
| 0.2 | 3.4 | GO:0005892 | acetylcholine-gated channel complex(GO:0005892) |
| 0.2 | 2.9 | GO:0005751 | mitochondrial respiratory chain complex IV(GO:0005751) |
| 0.2 | 2.9 | GO:0045180 | basal cortex(GO:0045180) |
| 0.1 | 2.6 | GO:0031235 | intrinsic component of the cytoplasmic side of the plasma membrane(GO:0031235) |
| 0.1 | 0.5 | GO:0031084 | BLOC-2 complex(GO:0031084) |
| 0.1 | 0.4 | GO:0070722 | Tle3-Aes complex(GO:0070722) |
| 0.1 | 1.5 | GO:0031983 | vesicle lumen(GO:0031983) |
| 0.1 | 2.1 | GO:0031143 | pseudopodium(GO:0031143) |
| 0.1 | 2.4 | GO:0070938 | contractile ring(GO:0070938) |
| 0.1 | 0.6 | GO:0072357 | PTW/PP1 phosphatase complex(GO:0072357) |
| 0.1 | 0.9 | GO:0031254 | uropod(GO:0001931) cell trailing edge(GO:0031254) |
| 0.1 | 0.4 | GO:0072559 | NLRP3 inflammasome complex(GO:0072559) |
| 0.1 | 0.8 | GO:0034464 | BBSome(GO:0034464) |
| 0.1 | 3.5 | GO:0001772 | immunological synapse(GO:0001772) |
| 0.1 | 1.0 | GO:0030127 | COPII vesicle coat(GO:0030127) |
| 0.1 | 13.0 | GO:0030018 | Z disc(GO:0030018) |
| 0.1 | 5.9 | GO:0005581 | collagen trimer(GO:0005581) |
| 0.1 | 2.6 | GO:0005747 | mitochondrial respiratory chain complex I(GO:0005747) NADH dehydrogenase complex(GO:0030964) respiratory chain complex I(GO:0045271) |
| 0.1 | 0.7 | GO:0008385 | IkappaB kinase complex(GO:0008385) |
| 0.1 | 2.1 | GO:0030904 | retromer complex(GO:0030904) |
| 0.0 | 2.9 | GO:0009295 | nucleoid(GO:0009295) mitochondrial nucleoid(GO:0042645) |
| 0.0 | 0.1 | GO:0043291 | RAVE complex(GO:0043291) |
| 0.0 | 6.5 | GO:0042641 | actomyosin(GO:0042641) |
| 0.0 | 1.0 | GO:0031616 | spindle pole centrosome(GO:0031616) |
| 0.0 | 5.7 | GO:0005604 | basement membrane(GO:0005604) |
| 0.0 | 1.6 | GO:0035371 | microtubule plus-end(GO:0035371) |
| 0.0 | 0.6 | GO:0005736 | DNA-directed RNA polymerase I complex(GO:0005736) |
| 0.0 | 1.0 | GO:0016327 | apicolateral plasma membrane(GO:0016327) |
| 0.0 | 2.4 | GO:0014704 | intercalated disc(GO:0014704) |
| 0.0 | 0.2 | GO:0061689 | tricellular tight junction(GO:0061689) |
| 0.0 | 5.5 | GO:0070160 | bicellular tight junction(GO:0005923) occluding junction(GO:0070160) |
| 0.0 | 1.0 | GO:0042588 | zymogen granule(GO:0042588) |
| 0.0 | 0.1 | GO:0030870 | Mre11 complex(GO:0030870) |
| 0.0 | 0.6 | GO:0005839 | proteasome core complex(GO:0005839) |
| 0.0 | 1.7 | GO:0031985 | Golgi cisterna(GO:0031985) |
| 0.0 | 0.5 | GO:0035098 | ESC/E(Z) complex(GO:0035098) |
| 0.0 | 0.2 | GO:0008250 | oligosaccharyltransferase complex(GO:0008250) |
| 0.0 | 0.2 | GO:0071439 | clathrin complex(GO:0071439) |
| 0.0 | 2.9 | GO:0034399 | nuclear periphery(GO:0034399) |
| 0.0 | 0.9 | GO:0005720 | nuclear heterochromatin(GO:0005720) |
| 0.0 | 0.2 | GO:0030126 | COPI vesicle coat(GO:0030126) |
| 0.0 | 1.1 | GO:0022625 | cytosolic large ribosomal subunit(GO:0022625) |
| 0.0 | 3.5 | GO:0005578 | proteinaceous extracellular matrix(GO:0005578) |
| 0.0 | 1.0 | GO:0032592 | integral component of mitochondrial membrane(GO:0032592) |
| 0.0 | 2.6 | GO:0030027 | lamellipodium(GO:0030027) |
| 0.0 | 23.7 | GO:0005739 | mitochondrion(GO:0005739) |
| 0.0 | 0.1 | GO:0032797 | SMN complex(GO:0032797) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 3.6 | 25.2 | GO:0004090 | carbonyl reductase (NADPH) activity(GO:0004090) |
| 2.9 | 14.6 | GO:0035478 | chylomicron binding(GO:0035478) |
| 0.9 | 2.8 | GO:0099580 | ion antiporter activity involved in regulation of postsynaptic membrane potential(GO:0099580) |
| 0.7 | 2.2 | GO:0016509 | long-chain-enoyl-CoA hydratase activity(GO:0016508) long-chain-3-hydroxyacyl-CoA dehydrogenase activity(GO:0016509) |
| 0.7 | 2.1 | GO:0004739 | pyruvate dehydrogenase (acetyl-transferring) activity(GO:0004739) |
| 0.6 | 2.9 | GO:0008475 | procollagen-lysine 5-dioxygenase activity(GO:0008475) |
| 0.5 | 4.3 | GO:0004687 | myosin light chain kinase activity(GO:0004687) |
| 0.4 | 2.2 | GO:0003872 | 6-phosphofructokinase activity(GO:0003872) |
| 0.4 | 2.9 | GO:0047276 | N-acetyllactosaminide 3-alpha-galactosyltransferase activity(GO:0047276) |
| 0.4 | 2.0 | GO:0004365 | glyceraldehyde-3-phosphate dehydrogenase (NAD+) (phosphorylating) activity(GO:0004365) glyceraldehyde-3-phosphate dehydrogenase (NAD(P)+) (phosphorylating) activity(GO:0043891) |
| 0.4 | 2.5 | GO:0061629 | RNA polymerase II sequence-specific DNA binding transcription factor binding(GO:0061629) |
| 0.3 | 1.9 | GO:0031013 | troponin I binding(GO:0031013) |
| 0.3 | 2.8 | GO:0008597 | calcium-dependent protein serine/threonine phosphatase regulator activity(GO:0008597) |
| 0.3 | 3.6 | GO:0004499 | N,N-dimethylaniline monooxygenase activity(GO:0004499) |
| 0.3 | 1.1 | GO:0035663 | Toll-like receptor 2 binding(GO:0035663) |
| 0.3 | 0.8 | GO:0004658 | propionyl-CoA carboxylase activity(GO:0004658) |
| 0.3 | 1.0 | GO:0003827 | alpha-1,3-mannosylglycoprotein 2-beta-N-acetylglucosaminyltransferase activity(GO:0003827) |
| 0.2 | 1.4 | GO:0048495 | Roundabout binding(GO:0048495) |
| 0.2 | 1.6 | GO:0005005 | transmembrane-ephrin receptor activity(GO:0005005) |
| 0.2 | 0.7 | GO:0042936 | dipeptide transporter activity(GO:0042936) |
| 0.2 | 1.8 | GO:0030368 | interleukin-17 receptor activity(GO:0030368) |
| 0.2 | 0.5 | GO:0005333 | acetylcholine transmembrane transporter activity(GO:0005277) norepinephrine transmembrane transporter activity(GO:0005333) acetate ester transmembrane transporter activity(GO:1901375) |
| 0.2 | 3.4 | GO:0042166 | acetylcholine binding(GO:0042166) |
| 0.2 | 3.3 | GO:0033549 | MAP kinase phosphatase activity(GO:0033549) |
| 0.2 | 1.0 | GO:0004720 | protein-lysine 6-oxidase activity(GO:0004720) |
| 0.2 | 5.5 | GO:0017166 | vinculin binding(GO:0017166) |
| 0.2 | 0.8 | GO:0004165 | dodecenoyl-CoA delta-isomerase activity(GO:0004165) |
| 0.2 | 2.7 | GO:0016641 | oxidoreductase activity, acting on the CH-NH2 group of donors, oxygen as acceptor(GO:0016641) |
| 0.2 | 0.8 | GO:0047696 | beta-adrenergic receptor kinase activity(GO:0047696) |
| 0.2 | 0.6 | GO:0015222 | serotonin transmembrane transporter activity(GO:0015222) |
| 0.1 | 0.8 | GO:0005095 | GTPase inhibitor activity(GO:0005095) |
| 0.1 | 2.2 | GO:0031681 | G-protein beta-subunit binding(GO:0031681) |
| 0.1 | 0.7 | GO:0046624 | sphingolipid transporter activity(GO:0046624) |
| 0.1 | 3.3 | GO:0031005 | filamin binding(GO:0031005) |
| 0.1 | 0.8 | GO:0005111 | type 2 fibroblast growth factor receptor binding(GO:0005111) |
| 0.1 | 3.3 | GO:0051428 | peptide hormone receptor binding(GO:0051428) |
| 0.1 | 6.0 | GO:0008307 | structural constituent of muscle(GO:0008307) |
| 0.1 | 2.1 | GO:0010181 | FMN binding(GO:0010181) |
| 0.1 | 2.9 | GO:0004129 | cytochrome-c oxidase activity(GO:0004129) heme-copper terminal oxidase activity(GO:0015002) oxidoreductase activity, acting on a heme group of donors(GO:0016675) oxidoreductase activity, acting on a heme group of donors, oxygen as acceptor(GO:0016676) |
| 0.1 | 1.2 | GO:0008381 | mechanically-gated ion channel activity(GO:0008381) mechanically gated channel activity(GO:0022833) |
| 0.1 | 1.5 | GO:0086008 | voltage-gated potassium channel activity involved in cardiac muscle cell action potential repolarization(GO:0086008) |
| 0.1 | 0.8 | GO:0086006 | voltage-gated sodium channel activity involved in cardiac muscle cell action potential(GO:0086006) |
| 0.1 | 0.3 | GO:0071820 | N-box binding(GO:0071820) |
| 0.1 | 0.6 | GO:0010385 | double-stranded methylated DNA binding(GO:0010385) |
| 0.1 | 0.6 | GO:0004111 | creatine kinase activity(GO:0004111) |
| 0.1 | 0.4 | GO:0015183 | L-aspartate transmembrane transporter activity(GO:0015183) |
| 0.1 | 0.2 | GO:0016206 | catechol O-methyltransferase activity(GO:0016206) |
| 0.1 | 0.3 | GO:0036313 | phosphatidylinositol 3-kinase catalytic subunit binding(GO:0036313) |
| 0.1 | 0.4 | GO:0033192 | calmodulin-dependent protein phosphatase activity(GO:0033192) |
| 0.1 | 1.4 | GO:0015174 | basic amino acid transmembrane transporter activity(GO:0015174) |
| 0.1 | 0.7 | GO:0008474 | palmitoyl-(protein) hydrolase activity(GO:0008474) palmitoyl hydrolase activity(GO:0098599) |
| 0.1 | 0.7 | GO:0003810 | protein-glutamine gamma-glutamyltransferase activity(GO:0003810) |
| 0.1 | 0.6 | GO:0035251 | UDP-glucosyltransferase activity(GO:0035251) |
| 0.1 | 1.5 | GO:0070006 | metalloaminopeptidase activity(GO:0070006) |
| 0.1 | 0.2 | GO:0050501 | hyaluronan synthase activity(GO:0050501) |
| 0.1 | 1.0 | GO:0038191 | neuropilin binding(GO:0038191) |
| 0.1 | 0.8 | GO:0016634 | oxidoreductase activity, acting on the CH-CH group of donors, oxygen as acceptor(GO:0016634) |
| 0.1 | 1.7 | GO:0019956 | G-protein coupled chemoattractant receptor activity(GO:0001637) chemokine receptor activity(GO:0004950) chemokine binding(GO:0019956) |
| 0.0 | 1.8 | GO:0070412 | R-SMAD binding(GO:0070412) |
| 0.0 | 0.7 | GO:0008379 | thioredoxin peroxidase activity(GO:0008379) |
| 0.0 | 0.3 | GO:0004614 | phosphoglucomutase activity(GO:0004614) |
| 0.0 | 1.7 | GO:0005385 | zinc ion transmembrane transporter activity(GO:0005385) |
| 0.0 | 0.2 | GO:0004060 | arylamine N-acetyltransferase activity(GO:0004060) |
| 0.0 | 2.5 | GO:0051019 | mitogen-activated protein kinase binding(GO:0051019) |
| 0.0 | 0.1 | GO:0008967 | phosphoglycolate phosphatase activity(GO:0008967) |
| 0.0 | 0.3 | GO:0042799 | histone methyltransferase activity (H4-K20 specific)(GO:0042799) |
| 0.0 | 0.4 | GO:0030976 | thiamine pyrophosphate binding(GO:0030976) |
| 0.0 | 0.9 | GO:0042813 | Wnt-activated receptor activity(GO:0042813) |
| 0.0 | 1.1 | GO:0008392 | arachidonic acid epoxygenase activity(GO:0008392) |
| 0.0 | 1.1 | GO:0048020 | CCR chemokine receptor binding(GO:0048020) |
| 0.0 | 0.4 | GO:0070492 | oligosaccharide binding(GO:0070492) |
| 0.0 | 3.2 | GO:0004222 | metalloendopeptidase activity(GO:0004222) |
| 0.0 | 1.4 | GO:0005154 | epidermal growth factor receptor binding(GO:0005154) |
| 0.0 | 1.7 | GO:0001540 | beta-amyloid binding(GO:0001540) |
| 0.0 | 7.9 | GO:0003714 | transcription corepressor activity(GO:0003714) |
| 0.0 | 0.2 | GO:0046974 | histone methyltransferase activity (H3-K9 specific)(GO:0046974) |
| 0.0 | 1.2 | GO:0003755 | peptidyl-prolyl cis-trans isomerase activity(GO:0003755) |
| 0.0 | 0.6 | GO:0070003 | threonine-type endopeptidase activity(GO:0004298) threonine-type peptidase activity(GO:0070003) |
| 0.0 | 0.6 | GO:0008195 | phosphatidate phosphatase activity(GO:0008195) |
| 0.0 | 0.5 | GO:0004745 | retinol dehydrogenase activity(GO:0004745) |
| 0.0 | 1.1 | GO:0043027 | cysteine-type endopeptidase inhibitor activity involved in apoptotic process(GO:0043027) |
| 0.0 | 7.0 | GO:0030246 | carbohydrate binding(GO:0030246) |
| 0.0 | 0.1 | GO:0010859 | calcium-dependent cysteine-type endopeptidase inhibitor activity(GO:0010859) |
| 0.0 | 0.2 | GO:0008020 | G-protein coupled photoreceptor activity(GO:0008020) |
| 0.0 | 0.2 | GO:0004579 | oligosaccharyl transferase activity(GO:0004576) dolichyl-diphosphooligosaccharide-protein glycotransferase activity(GO:0004579) |
| 0.0 | 1.1 | GO:0017112 | Rab guanyl-nucleotide exchange factor activity(GO:0017112) |
| 0.0 | 0.5 | GO:0050136 | NADH dehydrogenase (ubiquinone) activity(GO:0008137) NADH dehydrogenase (quinone) activity(GO:0050136) |
| 0.0 | 0.7 | GO:0001103 | RNA polymerase II repressing transcription factor binding(GO:0001103) |
| 0.0 | 0.6 | GO:0005035 | tumor necrosis factor-activated receptor activity(GO:0005031) death receptor activity(GO:0005035) |
| 0.0 | 1.6 | GO:0005518 | collagen binding(GO:0005518) |
| 0.0 | 0.7 | GO:0005109 | frizzled binding(GO:0005109) |
| 0.0 | 0.8 | GO:0004714 | transmembrane receptor protein tyrosine kinase activity(GO:0004714) |
| 0.0 | 0.6 | GO:0004683 | calmodulin-dependent protein kinase activity(GO:0004683) |
| 0.0 | 1.0 | GO:0043022 | ribosome binding(GO:0043022) |
| 0.0 | 0.3 | GO:0046965 | retinoid X receptor binding(GO:0046965) |
| 0.0 | 0.2 | GO:0008603 | cAMP-dependent protein kinase regulator activity(GO:0008603) |
| 0.0 | 0.5 | GO:0030742 | GTP-dependent protein binding(GO:0030742) |
| 0.0 | 0.6 | GO:0005484 | SNAP receptor activity(GO:0005484) |
| 0.0 | 2.6 | GO:0019903 | protein phosphatase binding(GO:0019903) |
| 0.0 | 0.5 | GO:0032452 | histone demethylase activity(GO:0032452) |
| 0.0 | 0.3 | GO:0004198 | calcium-dependent cysteine-type endopeptidase activity(GO:0004198) |
| 0.0 | 0.1 | GO:0098505 | G-rich strand telomeric DNA binding(GO:0098505) |
| 0.0 | 0.6 | GO:0004715 | non-membrane spanning protein tyrosine kinase activity(GO:0004715) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 3.2 | PID S1P S1P2 PATHWAY | S1P2 pathway |
| 0.1 | 2.8 | PID TCR CALCIUM PATHWAY | Calcium signaling in the CD4+ TCR pathway |
| 0.0 | 10.2 | NABA ECM AFFILIATED | Genes encoding proteins affiliated structurally or functionally to extracellular matrix proteins |
| 0.0 | 1.3 | PID ERBB NETWORK PATHWAY | ErbB receptor signaling network |
| 0.0 | 2.3 | PID RAS PATHWAY | Regulation of Ras family activation |
| 0.0 | 2.3 | PID EPHB FWD PATHWAY | EPHB forward signaling |
| 0.0 | 8.5 | NABA ECM GLYCOPROTEINS | Genes encoding structural ECM glycoproteins |
| 0.0 | 9.4 | NABA ECM REGULATORS | Genes encoding enzymes and their regulators involved in the remodeling of the extracellular matrix |
| 0.0 | 1.1 | PID TOLL ENDOGENOUS PATHWAY | Endogenous TLR signaling |
| 0.0 | 2.4 | PID RB 1PATHWAY | Regulation of retinoblastoma protein |
| 0.0 | 2.2 | PID AP1 PATHWAY | AP-1 transcription factor network |
| 0.0 | 2.1 | PID MYC REPRESS PATHWAY | Validated targets of C-MYC transcriptional repression |
| 0.0 | 2.9 | PID MYC ACTIV PATHWAY | Validated targets of C-MYC transcriptional activation |
| 0.0 | 0.7 | PID BETA CATENIN DEG PATHWAY | Degradation of beta catenin |
| 0.0 | 0.6 | ST WNT CA2 CYCLIC GMP PATHWAY | Wnt/Ca2+/cyclic GMP signaling. |
| 0.0 | 1.1 | PID CDC42 REG PATHWAY | Regulation of CDC42 activity |
| 0.0 | 1.7 | PID P75 NTR PATHWAY | p75(NTR)-mediated signaling |
| 0.0 | 2.0 | PID CDC42 PATHWAY | CDC42 signaling events |
| 0.0 | 1.0 | SIG BCR SIGNALING PATHWAY | Members of the BCR signaling pathway |
| 0.0 | 1.7 | PID CMYB PATHWAY | C-MYB transcription factor network |
| 0.0 | 0.5 | PID CONE PATHWAY | Visual signal transduction: Cones |
| 0.0 | 1.2 | PID REG GR PATHWAY | Glucocorticoid receptor regulatory network |
| 0.0 | 2.1 | PID P53 DOWNSTREAM PATHWAY | Direct p53 effectors |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 3.3 | REACTOME ERKS ARE INACTIVATED | Genes involved in ERKs are inactivated |
| 0.2 | 7.7 | REACTOME YAP1 AND WWTR1 TAZ STIMULATED GENE EXPRESSION | Genes involved in YAP1- and WWTR1 (TAZ)-stimulated gene expression |
| 0.2 | 3.0 | REACTOME MITOCHONDRIAL FATTY ACID BETA OXIDATION | Genes involved in Mitochondrial Fatty Acid Beta-Oxidation |
| 0.2 | 2.1 | REACTOME REGULATION OF PYRUVATE DEHYDROGENASE PDH COMPLEX | Genes involved in Regulation of pyruvate dehydrogenase (PDH) complex |
| 0.1 | 2.8 | REACTOME PLATELET CALCIUM HOMEOSTASIS | Genes involved in Platelet calcium homeostasis |
| 0.1 | 3.3 | REACTOME CELL EXTRACELLULAR MATRIX INTERACTIONS | Genes involved in Cell-extracellular matrix interactions |
| 0.1 | 2.2 | REACTOME ANTIGEN PRESENTATION FOLDING ASSEMBLY AND PEPTIDE LOADING OF CLASS I MHC | Genes involved in Antigen Presentation: Folding, assembly and peptide loading of class I MHC |
| 0.1 | 1.6 | REACTOME NFKB ACTIVATION THROUGH FADD RIP1 PATHWAY MEDIATED BY CASPASE 8 AND10 | Genes involved in NF-kB activation through FADD/RIP-1 pathway mediated by caspase-8 and -10 |
| 0.1 | 3.2 | REACTOME ADHERENS JUNCTIONS INTERACTIONS | Genes involved in Adherens junctions interactions |
| 0.1 | 1.7 | REACTOME ZINC TRANSPORTERS | Genes involved in Zinc transporters |
| 0.1 | 1.9 | REACTOME COMPLEMENT CASCADE | Genes involved in Complement cascade |
| 0.1 | 1.9 | REACTOME SHC1 EVENTS IN ERBB4 SIGNALING | Genes involved in SHC1 events in ERBB4 signaling |
| 0.0 | 0.6 | REACTOME CALNEXIN CALRETICULIN CYCLE | Genes involved in Calnexin/calreticulin cycle |
| 0.0 | 2.2 | REACTOME GLYCOLYSIS | Genes involved in Glycolysis |
| 0.0 | 1.1 | REACTOME NA CL DEPENDENT NEUROTRANSMITTER TRANSPORTERS | Genes involved in Na+/Cl- dependent neurotransmitter transporters |
| 0.0 | 1.4 | REACTOME SIGNALING BY HIPPO | Genes involved in Signaling by Hippo |
| 0.0 | 2.7 | REACTOME PHASE1 FUNCTIONALIZATION OF COMPOUNDS | Genes involved in Phase 1 - Functionalization of compounds |
| 0.0 | 2.9 | REACTOME COLLAGEN FORMATION | Genes involved in Collagen formation |
| 0.0 | 1.0 | REACTOME OTHER SEMAPHORIN INTERACTIONS | Genes involved in Other semaphorin interactions |
| 0.0 | 2.6 | REACTOME RESPIRATORY ELECTRON TRANSPORT | Genes involved in Respiratory electron transport |
| 0.0 | 1.4 | REACTOME BASIGIN INTERACTIONS | Genes involved in Basigin interactions |
| 0.0 | 1.6 | REACTOME CHEMOKINE RECEPTORS BIND CHEMOKINES | Genes involved in Chemokine receptors bind chemokines |
| 0.0 | 0.6 | REACTOME SYNTHESIS OF PE | Genes involved in Synthesis of PE |
| 0.0 | 0.4 | REACTOME THE NLRP3 INFLAMMASOME | Genes involved in The NLRP3 inflammasome |
| 0.0 | 0.6 | REACTOME HORMONE SENSITIVE LIPASE HSL MEDIATED TRIACYLGLYCEROL HYDROLYSIS | Genes involved in Hormone-sensitive lipase (HSL)-mediated triacylglycerol hydrolysis |
| 0.0 | 0.4 | REACTOME GLUCONEOGENESIS | Genes involved in Gluconeogenesis |
| 0.0 | 1.0 | REACTOME TRANSPORT TO THE GOLGI AND SUBSEQUENT MODIFICATION | Genes involved in Transport to the Golgi and subsequent modification |
| 0.0 | 0.1 | REACTOME PACKAGING OF TELOMERE ENDS | Genes involved in Packaging Of Telomere Ends |
| 0.0 | 0.2 | REACTOME OPSINS | Genes involved in Opsins |
| 0.0 | 1.3 | REACTOME STRIATED MUSCLE CONTRACTION | Genes involved in Striated Muscle Contraction |
| 0.0 | 0.2 | REACTOME COPI MEDIATED TRANSPORT | Genes involved in COPI Mediated Transport |
| 0.0 | 1.1 | REACTOME NETRIN1 SIGNALING | Genes involved in Netrin-1 signaling |
| 0.0 | 1.0 | REACTOME MYOGENESIS | Genes involved in Myogenesis |
| 0.0 | 0.8 | REACTOME INTERACTION BETWEEN L1 AND ANKYRINS | Genes involved in Interaction between L1 and Ankyrins |
| 0.0 | 0.2 | REACTOME HYALURONAN METABOLISM | Genes involved in Hyaluronan metabolism |
| 0.0 | 1.1 | REACTOME MYD88 MAL CASCADE INITIATED ON PLASMA MEMBRANE | Genes involved in MyD88:Mal cascade initiated on plasma membrane |
| 0.0 | 0.4 | REACTOME ACTIVATED POINT MUTANTS OF FGFR2 | Genes involved in Activated point mutants of FGFR2 |
| 0.0 | 1.1 | REACTOME NRAGE SIGNALS DEATH THROUGH JNK | Genes involved in NRAGE signals death through JNK |
| 0.0 | 0.3 | REACTOME GLYCOGEN BREAKDOWN GLYCOGENOLYSIS | Genes involved in Glycogen breakdown (glycogenolysis) |
| 0.0 | 0.7 | REACTOME AMINO ACID AND OLIGOPEPTIDE SLC TRANSPORTERS | Genes involved in Amino acid and oligopeptide SLC transporters |