Project
PRJNA375882: Comprehensive Mouse Transcriptomic BodyMap
Navigation
Downloads
Results for Nkx6-2
Z-value: 0.98
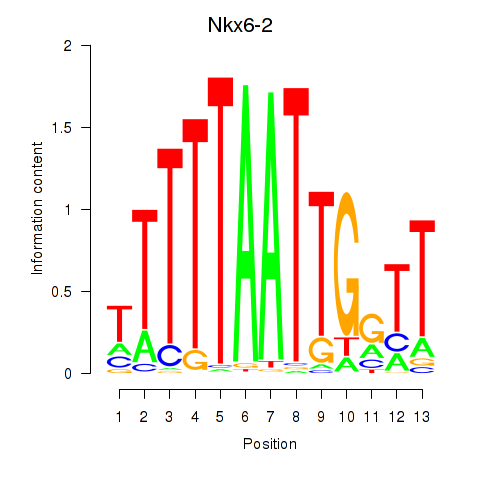
Transcription factors associated with Nkx6-2
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Nkx6-2
|
ENSMUSG00000041309.18 | Nkx6-2 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Nkx6-2 | mm39_v1_chr7_-_139162706_139162724 | 0.23 | 5.3e-02 | Click! |
Activity profile of Nkx6-2 motif
Sorted Z-values of Nkx6-2 motif
Network of associatons between targets according to the STRING database.
First level regulatory network of Nkx6-2
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr8_-_62576140 | 11.36 |
ENSMUST00000034052.14
ENSMUST00000034054.9 |
Anxa10
|
annexin A10 |
| chr3_+_138019040 | 7.85 |
ENSMUST00000013455.13
ENSMUST00000106247.2 |
Adh6a
|
alcohol dehydrogenase 6A (class V) |
| chr3_+_106020545 | 7.46 |
ENSMUST00000079132.12
ENSMUST00000139086.2 |
Chia1
|
chitinase, acidic 1 |
| chr3_+_69129745 | 6.99 |
ENSMUST00000183126.2
|
Arl14
|
ADP-ribosylation factor-like 14 |
| chr10_+_115653152 | 6.76 |
ENSMUST00000080630.11
ENSMUST00000179196.3 ENSMUST00000035563.15 |
Tspan8
|
tetraspanin 8 |
| chr8_-_65582206 | 5.55 |
ENSMUST00000098713.5
|
Smim31
|
small integral membrane protein 31 |
| chr19_+_58717319 | 5.54 |
ENSMUST00000048644.6
ENSMUST00000236445.2 |
Pnliprp1
|
pancreatic lipase related protein 1 |
| chr16_+_96268806 | 4.92 |
ENSMUST00000061739.9
|
Pcp4
|
Purkinje cell protein 4 |
| chr5_+_66833434 | 4.73 |
ENSMUST00000031131.11
|
Uchl1
|
ubiquitin carboxy-terminal hydrolase L1 |
| chr8_-_65146079 | 4.67 |
ENSMUST00000048967.9
|
Cpe
|
carboxypeptidase E |
| chr5_+_92957231 | 3.86 |
ENSMUST00000113054.9
|
Shroom3
|
shroom family member 3 |
| chr8_+_21515561 | 3.74 |
ENSMUST00000076754.3
|
Defa21
|
defensin, alpha, 21 |
| chr5_-_87682972 | 3.60 |
ENSMUST00000120150.2
|
Sult1b1
|
sulfotransferase family 1B, member 1 |
| chr5_+_92831150 | 3.60 |
ENSMUST00000113055.9
|
Shroom3
|
shroom family member 3 |
| chr14_+_80237691 | 3.57 |
ENSMUST00000228749.2
ENSMUST00000088735.4 |
Olfm4
|
olfactomedin 4 |
| chr8_+_21652293 | 3.56 |
ENSMUST00000098897.2
|
Defa22
|
defensin, alpha, 22 |
| chr2_-_103114105 | 3.55 |
ENSMUST00000111174.8
|
Ehf
|
ets homologous factor |
| chr5_+_92831468 | 3.52 |
ENSMUST00000168878.8
|
Shroom3
|
shroom family member 3 |
| chr8_+_22055402 | 3.46 |
ENSMUST00000084040.3
|
Defa37
|
defensin, alpha, 37 |
| chr9_-_123507847 | 3.36 |
ENSMUST00000170591.2
ENSMUST00000171647.9 |
Slc6a20a
|
solute carrier family 6 (neurotransmitter transporter), member 20A |
| chr8_+_21917427 | 3.35 |
ENSMUST00000095424.6
|
Defa36
|
defensin, alpha, 36 |
| chrX_+_138464065 | 3.23 |
ENSMUST00000113027.8
|
Rnf128
|
ring finger protein 128 |
| chr8_+_21787455 | 3.16 |
ENSMUST00000098892.5
|
Defa5
|
defensin, alpha, 5 |
| chr13_+_31740117 | 3.07 |
ENSMUST00000042118.11
|
Foxq1
|
forkhead box Q1 |
| chr8_+_22155813 | 2.99 |
ENSMUST00000075268.5
|
Defa34
|
defensin, alpha, 34 |
| chr8_+_94537910 | 2.92 |
ENSMUST00000138659.9
|
Gnao1
|
guanine nucleotide binding protein, alpha O |
| chr2_+_24043159 | 2.83 |
ENSMUST00000028363.2
|
Il1f8
|
interleukin 1 family, member 8 |
| chr9_-_21223631 | 2.48 |
ENSMUST00000115433.11
|
Ap1m2
|
adaptor protein complex AP-1, mu 2 subunit |
| chr16_-_21980200 | 2.40 |
ENSMUST00000115379.2
|
Igf2bp2
|
insulin-like growth factor 2 mRNA binding protein 2 |
| chr2_+_125876883 | 2.31 |
ENSMUST00000110442.2
|
Fgf7
|
fibroblast growth factor 7 |
| chr2_+_83642910 | 2.22 |
ENSMUST00000051454.4
|
Fam171b
|
family with sequence similarity 171, member B |
| chr5_+_75312939 | 2.14 |
ENSMUST00000202681.4
ENSMUST00000000476.15 |
Pdgfra
|
platelet derived growth factor receptor, alpha polypeptide |
| chr2_+_22512195 | 2.02 |
ENSMUST00000028123.4
|
Gad2
|
glutamic acid decarboxylase 2 |
| chr17_-_43003135 | 1.93 |
ENSMUST00000170723.8
ENSMUST00000164524.2 ENSMUST00000024711.11 ENSMUST00000167993.8 |
Adgrf4
|
adhesion G protein-coupled receptor F4 |
| chr18_+_34973605 | 1.90 |
ENSMUST00000043484.8
|
Reep2
|
receptor accessory protein 2 |
| chr1_+_66360865 | 1.90 |
ENSMUST00000114013.8
|
Map2
|
microtubule-associated protein 2 |
| chr1_+_177557380 | 1.80 |
ENSMUST00000016106.6
|
1700016C15Rik
|
RIKEN cDNA 1700016C15 gene |
| chr18_-_43032359 | 1.78 |
ENSMUST00000117687.8
|
Ppp2r2b
|
protein phosphatase 2, regulatory subunit B, beta |
| chr5_-_66211842 | 1.76 |
ENSMUST00000200852.4
|
Rbm47
|
RNA binding motif protein 47 |
| chr4_-_154721288 | 1.76 |
ENSMUST00000030902.13
ENSMUST00000105637.8 ENSMUST00000070313.14 ENSMUST00000105636.8 ENSMUST00000105638.9 ENSMUST00000097759.9 ENSMUST00000124771.2 |
Prdm16
|
PR domain containing 16 |
| chr2_+_125876566 | 1.62 |
ENSMUST00000064794.14
|
Fgf7
|
fibroblast growth factor 7 |
| chr1_+_170136372 | 1.57 |
ENSMUST00000056991.6
|
Spata46
|
spermatogenesis associated 46 |
| chr5_-_51711237 | 1.55 |
ENSMUST00000132734.8
|
Ppargc1a
|
peroxisome proliferative activated receptor, gamma, coactivator 1 alpha |
| chr5_-_131336914 | 1.54 |
ENSMUST00000160609.2
|
Galnt17
|
polypeptide N-acetylgalactosaminyltransferase 17 |
| chr3_+_96508400 | 1.53 |
ENSMUST00000062058.5
|
Lix1l
|
Lix1-like |
| chr7_-_44711771 | 1.45 |
ENSMUST00000210101.2
ENSMUST00000209219.2 |
Prrg2
|
proline-rich Gla (G-carboxyglutamic acid) polypeptide 2 |
| chr11_+_100902572 | 1.41 |
ENSMUST00000092663.4
|
Atp6v0a1
|
ATPase, H+ transporting, lysosomal V0 subunit A1 |
| chr7_+_43339842 | 1.40 |
ENSMUST00000056329.7
|
Klk14
|
kallikrein related-peptidase 14 |
| chr2_-_32976378 | 1.40 |
ENSMUST00000049618.9
|
Garnl3
|
GTPase activating RANGAP domain-like 3 |
| chr5_-_51711204 | 1.39 |
ENSMUST00000196968.5
|
Ppargc1a
|
peroxisome proliferative activated receptor, gamma, coactivator 1 alpha |
| chr1_-_149836974 | 1.38 |
ENSMUST00000190507.2
ENSMUST00000070200.15 |
Pla2g4a
|
phospholipase A2, group IVA (cytosolic, calcium-dependent) |
| chr18_-_79152504 | 1.37 |
ENSMUST00000025430.11
|
Setbp1
|
SET binding protein 1 |
| chr2_-_84481058 | 1.37 |
ENSMUST00000111670.9
ENSMUST00000111697.9 ENSMUST00000111696.8 ENSMUST00000111678.8 ENSMUST00000111690.8 ENSMUST00000111695.8 ENSMUST00000111677.8 ENSMUST00000111698.8 ENSMUST00000099941.9 ENSMUST00000111676.8 ENSMUST00000111694.8 ENSMUST00000111675.8 ENSMUST00000111689.8 ENSMUST00000111687.8 ENSMUST00000111692.8 ENSMUST00000111685.8 ENSMUST00000111686.8 ENSMUST00000111688.8 ENSMUST00000111693.8 ENSMUST00000111684.8 |
Ctnnd1
|
catenin (cadherin associated protein), delta 1 |
| chr1_+_34044940 | 1.36 |
ENSMUST00000187486.7
ENSMUST00000182697.8 |
Dst
|
dystonin |
| chr16_+_45355654 | 1.35 |
ENSMUST00000159945.8
|
Slc9c1
|
solute carrier family 9, subfamily C (Na+-transporting carboxylic acid decarboxylase), member 1 |
| chr9_-_35469818 | 1.34 |
ENSMUST00000034612.7
|
Ddx25
|
DEAD box helicase 25 |
| chr1_+_43484895 | 1.30 |
ENSMUST00000086421.9
|
Nck2
|
non-catalytic region of tyrosine kinase adaptor protein 2 |
| chr12_-_80807454 | 1.27 |
ENSMUST00000073251.8
|
Ccdc177
|
coiled-coil domain containing 177 |
| chr18_+_37568647 | 1.26 |
ENSMUST00000055495.6
|
Pcdhb12
|
protocadherin beta 12 |
| chr3_+_55369149 | 1.24 |
ENSMUST00000199585.5
ENSMUST00000070418.9 |
Dclk1
|
doublecortin-like kinase 1 |
| chr11_-_101062111 | 1.24 |
ENSMUST00000164474.8
ENSMUST00000043397.14 |
Plekhh3
|
pleckstrin homology domain containing, family H (with MyTH4 domain) member 3 |
| chr6_+_40619913 | 1.18 |
ENSMUST00000238599.2
|
Mgam
|
maltase-glucoamylase |
| chr2_+_157209506 | 1.17 |
ENSMUST00000081202.6
|
Manbal
|
mannosidase, beta A, lysosomal-like |
| chr3_+_85946145 | 1.17 |
ENSMUST00000238331.2
|
Sh3d19
|
SH3 domain protein D19 |
| chr3_+_103482591 | 1.15 |
ENSMUST00000090697.11
ENSMUST00000239027.2 |
Syt6
|
synaptotagmin VI |
| chr6_-_102441628 | 1.12 |
ENSMUST00000032159.7
|
Cntn3
|
contactin 3 |
| chr15_+_18819033 | 1.12 |
ENSMUST00000166873.9
|
Cdh10
|
cadherin 10 |
| chr15_-_13173736 | 1.11 |
ENSMUST00000036439.6
|
Cdh6
|
cadherin 6 |
| chr2_-_32977182 | 1.09 |
ENSMUST00000102810.10
|
Garnl3
|
GTPase activating RANGAP domain-like 3 |
| chr5_-_87739442 | 1.07 |
ENSMUST00000031201.9
|
Sult1e1
|
sulfotransferase family 1E, member 1 |
| chr18_-_43032514 | 1.03 |
ENSMUST00000236238.2
|
Ppp2r2b
|
protein phosphatase 2, regulatory subunit B, beta |
| chr12_+_117807607 | 0.98 |
ENSMUST00000176735.8
ENSMUST00000177339.2 |
Cdca7l
|
cell division cycle associated 7 like |
| chr7_+_4925781 | 0.98 |
ENSMUST00000207527.2
ENSMUST00000207687.2 ENSMUST00000208754.2 |
Nat14
|
N-acetyltransferase 14 |
| chr11_+_104983022 | 0.97 |
ENSMUST00000021029.6
|
Efcab3
|
EF-hand calcium binding domain 3 |
| chr4_-_106588122 | 0.93 |
ENSMUST00000148281.2
|
Mroh7
|
maestro heat-like repeat family member 7 |
| chr4_-_117039809 | 0.93 |
ENSMUST00000065896.9
|
Kif2c
|
kinesin family member 2C |
| chr6_-_57367651 | 0.92 |
ENSMUST00000164732.2
|
Vmn1r18
|
vomeronasal 1 receptor 18 |
| chr2_-_84481020 | 0.90 |
ENSMUST00000067232.10
|
Ctnnd1
|
catenin (cadherin associated protein), delta 1 |
| chr1_+_66361252 | 0.89 |
ENSMUST00000123647.8
|
Map2
|
microtubule-associated protein 2 |
| chr8_-_62355690 | 0.87 |
ENSMUST00000121785.9
ENSMUST00000034057.14 |
Palld
|
palladin, cytoskeletal associated protein |
| chr16_-_44153288 | 0.85 |
ENSMUST00000136381.8
|
Sidt1
|
SID1 transmembrane family, member 1 |
| chr6_-_66656668 | 0.84 |
ENSMUST00000071414.2
|
Vmn1r35
|
vomeronasal 1 receptor 35 |
| chr4_-_83203388 | 0.84 |
ENSMUST00000150522.8
|
Ttc39b
|
tetratricopeptide repeat domain 39B |
| chr10_+_18283405 | 0.83 |
ENSMUST00000037341.14
|
Nhsl1
|
NHS-like 1 |
| chr2_-_84481101 | 0.81 |
ENSMUST00000111691.2
|
Ctnnd1
|
catenin (cadherin associated protein), delta 1 |
| chr5_+_148239975 | 0.81 |
ENSMUST00000152105.8
ENSMUST00000085554.5 |
Mtus2
|
microtubule associated tumor suppressor candidate 2 |
| chr6_-_124746468 | 0.80 |
ENSMUST00000204896.3
|
Eno2
|
enolase 2, gamma neuronal |
| chr2_+_85715984 | 0.77 |
ENSMUST00000213441.3
|
Olfr1023
|
olfactory receptor 1023 |
| chr6_-_13839914 | 0.76 |
ENSMUST00000060442.14
|
Gpr85
|
G protein-coupled receptor 85 |
| chr14_+_27598021 | 0.76 |
ENSMUST00000211684.2
ENSMUST00000210924.2 |
Erc2
|
ELKS/RAB6-interacting/CAST family member 2 |
| chr4_+_145595364 | 0.76 |
ENSMUST00000123460.2
|
Zfp986
|
zinc finger protein 986 |
| chr4_+_3940747 | 0.72 |
ENSMUST00000119403.2
|
Chchd7
|
coiled-coil-helix-coiled-coil-helix domain containing 7 |
| chr2_-_84480804 | 0.71 |
ENSMUST00000066177.10
|
Ctnnd1
|
catenin (cadherin associated protein), delta 1 |
| chr4_-_138641225 | 0.70 |
ENSMUST00000097830.4
|
Otud3
|
OTU domain containing 3 |
| chr4_-_97472844 | 0.67 |
ENSMUST00000107067.8
ENSMUST00000107068.9 |
E130114P18Rik
|
RIKEN cDNA E130114P18 gene |
| chr4_+_147106307 | 0.67 |
ENSMUST00000075775.6
|
Rex2
|
reduced expression 2 |
| chr15_-_79718423 | 0.67 |
ENSMUST00000109623.8
ENSMUST00000109625.8 ENSMUST00000023060.13 ENSMUST00000089299.6 |
Cbx6
Npcd
|
chromobox 6 neuronal pentraxin chromo domain |
| chr11_+_98754434 | 0.66 |
ENSMUST00000142414.8
ENSMUST00000037480.9 |
Wipf2
|
WAS/WASL interacting protein family, member 2 |
| chr19_+_32597379 | 0.66 |
ENSMUST00000236290.2
ENSMUST00000025833.7 |
Papss2
|
3'-phosphoadenosine 5'-phosphosulfate synthase 2 |
| chr11_-_73215442 | 0.66 |
ENSMUST00000021119.9
|
Aspa
|
aspartoacylase |
| chr4_-_91264670 | 0.66 |
ENSMUST00000107109.9
ENSMUST00000107111.9 ENSMUST00000107120.8 |
Elavl2
|
ELAV like RNA binding protein 1 |
| chr10_+_26698556 | 0.64 |
ENSMUST00000135866.2
|
Arhgap18
|
Rho GTPase activating protein 18 |
| chr7_-_142215027 | 0.64 |
ENSMUST00000105936.8
|
Igf2
|
insulin-like growth factor 2 |
| chr17_-_48739874 | 0.64 |
ENSMUST00000046549.5
|
Apobec2
|
apolipoprotein B mRNA editing enzyme, catalytic polypeptide 2 |
| chr2_-_164493683 | 0.62 |
ENSMUST00000099095.4
|
Wfdc9
|
WAP four-disulfide core domain 9 |
| chr10_-_102866076 | 0.62 |
ENSMUST00000218282.2
ENSMUST00000170026.2 |
Alx1
Gm17028
|
ALX homeobox 1 predicted gene 17028 |
| chr16_-_44153498 | 0.61 |
ENSMUST00000047446.13
|
Sidt1
|
SID1 transmembrane family, member 1 |
| chr4_-_146993984 | 0.60 |
ENSMUST00000238583.2
ENSMUST00000049821.4 |
Gm21411
|
predicted gene, 21411 |
| chr4_+_146586445 | 0.60 |
ENSMUST00000105735.9
|
Zfp981
|
zinc finger protein 981 |
| chr3_-_96359622 | 0.59 |
ENSMUST00000093126.11
ENSMUST00000098841.4 |
BC107364
|
cDNA sequence BC107364 |
| chr11_+_62770275 | 0.57 |
ENSMUST00000014321.5
|
Tvp23b
|
trans-golgi network vesicle protein 23B |
| chr4_+_147390131 | 0.57 |
ENSMUST00000148762.4
|
Zfp988
|
zinc finger protein 988 |
| chr4_+_145241454 | 0.55 |
ENSMUST00000105741.2
|
Zfp990
|
zinc finger protein 990 |
| chr4_+_90107057 | 0.52 |
ENSMUST00000107129.2
|
Zfp352
|
zinc finger protein 352 |
| chr19_+_13208692 | 0.52 |
ENSMUST00000207246.4
|
Olfr1463
|
olfactory receptor 1463 |
| chr1_+_63215976 | 0.51 |
ENSMUST00000129339.8
|
Eef1b2
|
eukaryotic translation elongation factor 1 beta 2 |
| chr19_-_33728759 | 0.51 |
ENSMUST00000147153.4
|
Lipo2
|
lipase, member O2 |
| chr19_-_14575395 | 0.51 |
ENSMUST00000052011.15
ENSMUST00000167776.3 |
Tle4
|
transducin-like enhancer of split 4 |
| chr5_-_86780277 | 0.51 |
ENSMUST00000116553.9
|
Tmprss11f
|
transmembrane protease, serine 11f |
| chr7_-_85985625 | 0.50 |
ENSMUST00000069279.5
|
Olfr307
|
olfactory receptor 307 |
| chr4_+_146033882 | 0.50 |
ENSMUST00000105730.2
ENSMUST00000091878.6 |
Zfp987
|
zinc finger protein 987 |
| chr4_+_146093394 | 0.50 |
ENSMUST00000168483.9
|
Zfp600
|
zinc finger protein 600 |
| chr1_-_63215812 | 0.49 |
ENSMUST00000185847.2
ENSMUST00000185732.7 ENSMUST00000188370.7 ENSMUST00000168099.9 |
Ndufs1
|
NADH:ubiquinone oxidoreductase core subunit S1 |
| chr15_+_39255185 | 0.48 |
ENSMUST00000228839.2
|
Rims2
|
regulating synaptic membrane exocytosis 2 |
| chr4_-_147787010 | 0.46 |
ENSMUST00000117638.2
|
Zfp534
|
zinc finger protein 534 |
| chr8_-_49008305 | 0.46 |
ENSMUST00000110346.9
ENSMUST00000211976.2 |
Tenm3
|
teneurin transmembrane protein 3 |
| chr2_+_3115250 | 0.46 |
ENSMUST00000072955.12
|
Fam171a1
|
family with sequence similarity 171, member A1 |
| chr4_+_145237329 | 0.44 |
ENSMUST00000105742.8
ENSMUST00000136309.8 |
Zfp990
|
zinc finger protein 990 |
| chrX_+_151922936 | 0.43 |
ENSMUST00000039720.11
ENSMUST00000144175.3 |
Rragb
|
Ras-related GTP binding B |
| chr4_+_147445744 | 0.42 |
ENSMUST00000133078.8
ENSMUST00000154154.2 |
Zfp978
|
zinc finger protein 978 |
| chr10_+_79746690 | 0.42 |
ENSMUST00000181321.2
|
Gm26602
|
predicted gene, 26602 |
| chr6_+_132739094 | 0.40 |
ENSMUST00000069268.3
|
Tas2r102
|
taste receptor, type 2, member 102 |
| chr4_+_140428777 | 0.40 |
ENSMUST00000138808.8
ENSMUST00000038893.6 |
Rcc2
|
regulator of chromosome condensation 2 |
| chr5_+_110987839 | 0.39 |
ENSMUST00000200172.2
ENSMUST00000066160.3 |
Chek2
|
checkpoint kinase 2 |
| chr1_+_63216281 | 0.39 |
ENSMUST00000188524.2
|
Eef1b2
|
eukaryotic translation elongation factor 1 beta 2 |
| chr17_-_30107544 | 0.38 |
ENSMUST00000171691.9
|
Mdga1
|
MAM domain containing glycosylphosphatidylinositol anchor 1 |
| chr7_+_3648264 | 0.37 |
ENSMUST00000206287.2
ENSMUST00000038913.16 |
Cnot3
|
CCR4-NOT transcription complex, subunit 3 |
| chr6_-_119940694 | 0.36 |
ENSMUST00000161512.3
|
Wnk1
|
WNK lysine deficient protein kinase 1 |
| chr11_-_101857831 | 0.36 |
ENSMUST00000001534.7
|
Sost
|
sclerostin |
| chr3_-_75072319 | 0.35 |
ENSMUST00000124618.2
|
Zbbx
|
zinc finger, B-box domain containing |
| chr11_-_4391082 | 0.33 |
ENSMUST00000109949.8
ENSMUST00000130174.2 |
Hormad2
|
HORMA domain containing 2 |
| chr1_-_63215952 | 0.33 |
ENSMUST00000185412.7
ENSMUST00000027111.15 ENSMUST00000189664.2 |
Ndufs1
|
NADH:ubiquinone oxidoreductase core subunit S1 |
| chr3_+_103482547 | 0.32 |
ENSMUST00000121834.8
|
Syt6
|
synaptotagmin VI |
| chr7_-_102805563 | 0.32 |
ENSMUST00000218483.2
|
Olfr589
|
olfactory receptor 589 |
| chr2_-_89675315 | 0.31 |
ENSMUST00000099762.2
|
Olfr48
|
olfactory receptor 48 |
| chr16_+_48877762 | 0.31 |
ENSMUST00000168680.2
|
Myh15
|
myosin, heavy chain 15 |
| chr5_-_110987604 | 0.30 |
ENSMUST00000056937.12
|
Hscb
|
HscB iron-sulfur cluster co-chaperone |
| chr15_+_34453432 | 0.29 |
ENSMUST00000060894.9
|
Erich5
|
glutamate rich 5 |
| chr8_-_22396428 | 0.29 |
ENSMUST00000051965.5
|
Defb11
|
defensin beta 11 |
| chrX_-_111315519 | 0.29 |
ENSMUST00000124335.8
|
Satl1
|
spermidine/spermine N1-acetyl transferase-like 1 |
| chr11_+_52251687 | 0.29 |
ENSMUST00000102758.8
|
Vdac1
|
voltage-dependent anion channel 1 |
| chr2_+_163535925 | 0.28 |
ENSMUST00000109400.3
|
Pkig
|
protein kinase inhibitor, gamma |
| chr18_-_84104507 | 0.28 |
ENSMUST00000060303.10
|
Tshz1
|
teashirt zinc finger family member 1 |
| chr14_+_65504067 | 0.27 |
ENSMUST00000224629.2
|
Fbxo16
|
F-box protein 16 |
| chr18_+_57275854 | 0.27 |
ENSMUST00000139892.2
|
Megf10
|
multiple EGF-like-domains 10 |
| chr8_+_41128099 | 0.27 |
ENSMUST00000051614.5
|
Adam24
|
a disintegrin and metallopeptidase domain 24 (testase 1) |
| chrX_+_109857866 | 0.27 |
ENSMUST00000078229.5
|
Pou3f4
|
POU domain, class 3, transcription factor 4 |
| chr7_-_10009278 | 0.26 |
ENSMUST00000060374.5
|
Vmn1r66
|
vomeronasal 1 receptor 66 |
| chr3_+_68479578 | 0.26 |
ENSMUST00000170788.9
|
Schip1
|
schwannomin interacting protein 1 |
| chr7_+_107679062 | 0.25 |
ENSMUST00000213601.2
|
Olfr481
|
olfactory receptor 481 |
| chr2_+_85838122 | 0.24 |
ENSMUST00000062166.2
|
Olfr1032
|
olfactory receptor 1032 |
| chr2_+_87609827 | 0.24 |
ENSMUST00000105210.3
|
Olfr152
|
olfactory receptor 152 |
| chr4_-_43823866 | 0.24 |
ENSMUST00000215406.2
ENSMUST00000079234.6 ENSMUST00000214843.2 |
Olfr156
|
olfactory receptor 156 |
| chr18_+_4993795 | 0.24 |
ENSMUST00000153016.8
|
Svil
|
supervillin |
| chr3_+_5815863 | 0.24 |
ENSMUST00000192045.2
|
Gm8797
|
predicted pseudogene 8797 |
| chr7_-_19449319 | 0.24 |
ENSMUST00000032555.10
ENSMUST00000093552.12 |
Tomm40
|
translocase of outer mitochondrial membrane 40 |
| chr5_-_110987441 | 0.23 |
ENSMUST00000145318.2
|
Hscb
|
HscB iron-sulfur cluster co-chaperone |
| chr18_+_37554471 | 0.23 |
ENSMUST00000053073.6
|
Pcdhb11
|
protocadherin beta 11 |
| chr16_-_59166089 | 0.22 |
ENSMUST00000084791.4
|
Olfr206
|
olfactory receptor 206 |
| chr2_+_85600147 | 0.22 |
ENSMUST00000065626.3
|
Olfr1013
|
olfactory receptor 1013 |
| chr15_-_84804239 | 0.22 |
ENSMUST00000189185.2
|
Gm29666
|
predicted gene 29666 |
| chr11_-_73382303 | 0.21 |
ENSMUST00000119863.2
ENSMUST00000215358.2 ENSMUST00000214623.2 |
Olfr381
|
olfactory receptor 381 |
| chr2_-_69619864 | 0.20 |
ENSMUST00000094942.4
|
Ccdc173
|
coiled-coil domain containing 173 |
| chr2_-_87467879 | 0.20 |
ENSMUST00000216082.2
|
Olfr1132
|
olfactory receptor 1132 |
| chr15_+_98335351 | 0.19 |
ENSMUST00000075851.4
|
Olfr282
|
olfactory receptor 282 |
| chr2_+_109522781 | 0.19 |
ENSMUST00000111050.10
|
Bdnf
|
brain derived neurotrophic factor |
| chr6_+_125026865 | 0.19 |
ENSMUST00000112413.8
|
Acrbp
|
proacrosin binding protein |
| chr9_-_113537277 | 0.18 |
ENSMUST00000111861.4
ENSMUST00000035086.13 |
Pdcd6ip
|
programmed cell death 6 interacting protein |
| chr13_+_41302272 | 0.18 |
ENSMUST00000141292.9
|
Sycp2l
|
synaptonemal complex protein 2-like |
| chr1_+_88062508 | 0.18 |
ENSMUST00000113134.8
ENSMUST00000140092.8 |
Ugt1a6a
|
UDP glucuronosyltransferase 1 family, polypeptide A6A |
| chrX_-_36485911 | 0.17 |
ENSMUST00000115210.2
|
Rhox1
|
reproductive homeobox 1 |
| chr4_+_145397238 | 0.17 |
ENSMUST00000105738.9
|
Zfp980
|
zinc finger protein 980 |
| chr2_-_89671899 | 0.16 |
ENSMUST00000213833.2
|
Olfr1256
|
olfactory receptor 1256 |
| chr14_-_32110312 | 0.15 |
ENSMUST00000100723.4
|
1700024G13Rik
|
RIKEN cDNA 1700024G13 gene |
| chr4_-_147726953 | 0.14 |
ENSMUST00000133006.2
ENSMUST00000037565.14 ENSMUST00000105720.8 |
Zfp979
|
zinc finger protein 979 |
| chr6_-_81942906 | 0.14 |
ENSMUST00000032124.9
|
Mrpl19
|
mitochondrial ribosomal protein L19 |
| chr7_-_5325456 | 0.14 |
ENSMUST00000207520.2
|
Nlrp2
|
NLR family, pyrin domain containing 2 |
| chr5_+_140404997 | 0.14 |
ENSMUST00000100507.8
|
Eif3b
|
eukaryotic translation initiation factor 3, subunit B |
| chr2_-_37537224 | 0.13 |
ENSMUST00000028279.10
|
Strbp
|
spermatid perinuclear RNA binding protein |
| chr18_-_84104574 | 0.13 |
ENSMUST00000175783.3
|
Tshz1
|
teashirt zinc finger family member 1 |
| chr4_+_147056433 | 0.13 |
ENSMUST00000146688.3
|
Zfp989
|
zinc finger protein 989 |
| chr16_+_44215136 | 0.12 |
ENSMUST00000099742.9
|
Cfap44
|
cilia and flagella associated protein 44 |
| chr7_-_102998876 | 0.12 |
ENSMUST00000215042.2
|
Olfr600
|
olfactory receptor 600 |
| chr6_-_66614736 | 0.10 |
ENSMUST00000074381.6
|
Vmn1r34
|
vomeronasal 1 receptor 34 |
| chr11_-_4390745 | 0.09 |
ENSMUST00000109948.8
|
Hormad2
|
HORMA domain containing 2 |
| chr9_+_91250864 | 0.09 |
ENSMUST00000173054.8
|
Zic4
|
zinc finger protein of the cerebellum 4 |
| chr7_-_142215595 | 0.09 |
ENSMUST00000145896.3
|
Igf2
|
insulin-like growth factor 2 |
| chr19_-_13827773 | 0.09 |
ENSMUST00000215350.2
|
Olfr1501
|
olfactory receptor 1501 |
| chr11_-_115824290 | 0.09 |
ENSMUST00000021097.10
|
Recql5
|
RecQ protein-like 5 |
| chr5_+_20112704 | 0.09 |
ENSMUST00000115267.7
|
Magi2
|
membrane associated guanylate kinase, WW and PDZ domain containing 2 |
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.6 | 4.7 | GO:0030070 | insulin processing(GO:0030070) |
| 1.3 | 3.9 | GO:0061033 | secretion by lung epithelial cell involved in lung growth(GO:0061033) |
| 1.0 | 2.9 | GO:1904635 | glomerular visceral epithelial cell apoptotic process(GO:1903210) regulation of glomerular visceral epithelial cell apoptotic process(GO:1904633) positive regulation of glomerular visceral epithelial cell apoptotic process(GO:1904635) positive regulation of progesterone biosynthetic process(GO:2000184) |
| 0.8 | 3.4 | GO:0035524 | proline transmembrane transport(GO:0035524) |
| 0.8 | 7.5 | GO:0006032 | chitin metabolic process(GO:0006030) chitin catabolic process(GO:0006032) |
| 0.8 | 4.9 | GO:0007412 | axon target recognition(GO:0007412) |
| 0.8 | 4.7 | GO:0006068 | ethanol catabolic process(GO:0006068) |
| 0.6 | 3.8 | GO:0060690 | epithelial cell differentiation involved in salivary gland development(GO:0060690) |
| 0.5 | 2.1 | GO:0072277 | metanephric glomerulus morphogenesis(GO:0072275) metanephric glomerulus vasculature morphogenesis(GO:0072276) metanephric glomerular capillary formation(GO:0072277) |
| 0.4 | 2.0 | GO:0006538 | glutamate catabolic process(GO:0006538) |
| 0.3 | 1.9 | GO:0050916 | sensory perception of sweet taste(GO:0050916) |
| 0.3 | 4.9 | GO:0033603 | positive regulation of dopamine secretion(GO:0033603) |
| 0.3 | 1.3 | GO:0036493 | positive regulation of translation in response to endoplasmic reticulum stress(GO:0036493) negative regulation of PERK-mediated unfolded protein response(GO:1903898) |
| 0.2 | 1.5 | GO:0060478 | acrosomal vesicle exocytosis(GO:0060478) |
| 0.2 | 0.7 | GO:1990167 | protein K27-linked deubiquitination(GO:1990167) |
| 0.2 | 1.4 | GO:2001280 | positive regulation of prostaglandin biosynthetic process(GO:0031394) positive regulation of unsaturated fatty acid biosynthetic process(GO:2001280) |
| 0.2 | 2.4 | GO:0016554 | cytidine to uridine editing(GO:0016554) |
| 0.2 | 0.7 | GO:0034036 | purine ribonucleoside bisphosphate biosynthetic process(GO:0034036) 3'-phosphoadenosine 5'-phosphosulfate biosynthetic process(GO:0050428) |
| 0.2 | 2.8 | GO:0043653 | mitochondrial fragmentation involved in apoptotic process(GO:0043653) |
| 0.2 | 0.9 | GO:0030951 | establishment or maintenance of microtubule cytoskeleton polarity(GO:0030951) |
| 0.1 | 1.5 | GO:0033227 | dsRNA transport(GO:0033227) |
| 0.1 | 0.4 | GO:0072356 | chromosome passenger complex localization to kinetochore(GO:0072356) |
| 0.1 | 1.8 | GO:0090336 | positive regulation of brown fat cell differentiation(GO:0090336) |
| 0.1 | 1.3 | GO:0098719 | sodium ion import across plasma membrane(GO:0098719) sodium ion import into cell(GO:1990118) |
| 0.1 | 0.7 | GO:0006083 | acetate metabolic process(GO:0006083) |
| 0.1 | 1.4 | GO:0007320 | insemination(GO:0007320) |
| 0.1 | 11.0 | GO:0002066 | columnar/cuboidal epithelial cell development(GO:0002066) |
| 0.1 | 6.8 | GO:1900047 | negative regulation of blood coagulation(GO:0030195) negative regulation of hemostasis(GO:1900047) |
| 0.1 | 1.4 | GO:0070072 | vacuolar proton-transporting V-type ATPase complex assembly(GO:0070072) |
| 0.1 | 1.6 | GO:0007342 | fusion of sperm to egg plasma membrane(GO:0007342) |
| 0.1 | 0.8 | GO:0097475 | motor neuron migration(GO:0097475) |
| 0.1 | 0.4 | GO:0023016 | signal transduction by trans-phosphorylation(GO:0023016) |
| 0.1 | 0.4 | GO:0060023 | soft palate development(GO:0060023) |
| 0.1 | 0.5 | GO:0097428 | protein maturation by iron-sulfur cluster transfer(GO:0097428) |
| 0.1 | 1.4 | GO:0008090 | retrograde axonal transport(GO:0008090) |
| 0.1 | 0.7 | GO:2000467 | positive regulation of glycogen (starch) synthase activity(GO:2000467) |
| 0.1 | 0.4 | GO:2000054 | negative regulation of Wnt signaling pathway involved in dorsal/ventral axis specification(GO:2000054) |
| 0.1 | 2.6 | GO:0031069 | hair follicle morphogenesis(GO:0031069) |
| 0.1 | 0.3 | GO:0032918 | polyamine acetylation(GO:0032917) spermidine acetylation(GO:0032918) |
| 0.1 | 0.3 | GO:0060468 | prevention of polyspermy(GO:0060468) |
| 0.1 | 2.8 | GO:0007026 | negative regulation of microtubule depolymerization(GO:0007026) |
| 0.1 | 3.6 | GO:1900026 | positive regulation of substrate adhesion-dependent cell spreading(GO:1900026) |
| 0.0 | 1.2 | GO:0051044 | positive regulation of membrane protein ectodomain proteolysis(GO:0051044) |
| 0.0 | 0.4 | GO:0051964 | negative regulation of synapse assembly(GO:0051964) |
| 0.0 | 0.2 | GO:0000915 | assembly of actomyosin apparatus involved in cytokinesis(GO:0000912) actomyosin contractile ring assembly(GO:0000915) actomyosin contractile ring organization(GO:0044837) |
| 0.0 | 2.9 | GO:0007212 | dopamine receptor signaling pathway(GO:0007212) |
| 0.0 | 0.9 | GO:0071803 | positive regulation of podosome assembly(GO:0071803) |
| 0.0 | 0.4 | GO:1990253 | cellular response to leucine starvation(GO:1990253) |
| 0.0 | 1.5 | GO:0097352 | autophagosome maturation(GO:0097352) |
| 0.0 | 0.5 | GO:0098828 | spontaneous neurotransmitter secretion(GO:0061669) positive regulation of inhibitory postsynaptic potential(GO:0097151) spontaneous synaptic transmission(GO:0098814) modulation of inhibitory postsynaptic potential(GO:0098828) |
| 0.0 | 0.2 | GO:0022601 | menstrual cycle phase(GO:0022601) |
| 0.0 | 0.3 | GO:0043652 | engulfment of apoptotic cell(GO:0043652) recognition of apoptotic cell(GO:0043654) |
| 0.0 | 0.4 | GO:0051177 | meiotic sister chromatid cohesion(GO:0051177) |
| 0.0 | 2.8 | GO:0032755 | positive regulation of interleukin-6 production(GO:0032755) |
| 0.0 | 1.3 | GO:0010501 | RNA secondary structure unwinding(GO:0010501) |
| 0.0 | 1.2 | GO:0021952 | central nervous system projection neuron axonogenesis(GO:0021952) |
| 0.0 | 0.5 | GO:0097264 | self proteolysis(GO:0097264) |
| 0.0 | 0.1 | GO:1990414 | replication-born double-strand break repair via sister chromatid exchange(GO:1990414) |
| 0.0 | 0.1 | GO:0075525 | viral translational termination-reinitiation(GO:0075525) |
| 0.0 | 7.6 | GO:0042742 | defense response to bacterium(GO:0042742) |
| 0.0 | 0.3 | GO:2001054 | negative regulation of mesenchymal cell apoptotic process(GO:2001054) |
| 0.0 | 2.3 | GO:0007156 | homophilic cell adhesion via plasma membrane adhesion molecules(GO:0007156) |
| 0.0 | 0.3 | GO:0006851 | mitochondrial calcium ion transport(GO:0006851) |
| 0.0 | 0.2 | GO:0052696 | flavonoid glucuronidation(GO:0052696) xenobiotic glucuronidation(GO:0052697) |
| 0.0 | 0.9 | GO:0006414 | translational elongation(GO:0006414) |
| 0.0 | 2.0 | GO:0019236 | response to pheromone(GO:0019236) |
| 0.0 | 0.1 | GO:0071043 | CUT catabolic process(GO:0071034) CUT metabolic process(GO:0071043) |
| 0.0 | 0.3 | GO:2000480 | negative regulation of cAMP-dependent protein kinase activity(GO:2000480) |
| 0.0 | 0.2 | GO:0030150 | protein import into mitochondrial matrix(GO:0030150) |
| 0.0 | 0.6 | GO:0001755 | neural crest cell migration(GO:0001755) |
| 0.0 | 0.3 | GO:0008210 | estrogen metabolic process(GO:0008210) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.0 | 2.9 | GO:1990843 | subsarcolemmal mitochondrion(GO:1990843) interfibrillar mitochondrion(GO:1990844) |
| 0.4 | 4.9 | GO:0005883 | neurofilament(GO:0005883) |
| 0.3 | 2.8 | GO:0097442 | CA3 pyramidal cell dendrite(GO:0097442) |
| 0.3 | 3.8 | GO:0005915 | zonula adherens(GO:0005915) |
| 0.2 | 4.7 | GO:0031045 | dense core granule(GO:0031045) |
| 0.2 | 1.4 | GO:0000220 | vacuolar proton-transporting V-type ATPase, V0 domain(GO:0000220) |
| 0.2 | 1.4 | GO:0031673 | H zone(GO:0031673) |
| 0.1 | 3.6 | GO:0042581 | specific granule(GO:0042581) |
| 0.1 | 0.9 | GO:0005853 | eukaryotic translation elongation factor 1 complex(GO:0005853) |
| 0.1 | 0.4 | GO:1990131 | Gtr1-Gtr2 GTPase complex(GO:1990131) |
| 0.1 | 1.3 | GO:0033391 | chromatoid body(GO:0033391) |
| 0.1 | 0.8 | GO:0000015 | phosphopyruvate hydratase complex(GO:0000015) |
| 0.1 | 2.5 | GO:0060077 | inhibitory synapse(GO:0060077) |
| 0.1 | 1.8 | GO:0000159 | protein phosphatase type 2A complex(GO:0000159) |
| 0.1 | 1.5 | GO:0097038 | perinuclear endoplasmic reticulum(GO:0097038) |
| 0.0 | 9.9 | GO:0043296 | apical junction complex(GO:0043296) |
| 0.0 | 1.4 | GO:0042588 | zymogen granule(GO:0042588) |
| 0.0 | 0.4 | GO:0030015 | CCR4-NOT core complex(GO:0030015) |
| 0.0 | 1.8 | GO:0016235 | aggresome(GO:0016235) |
| 0.0 | 0.9 | GO:0035371 | microtubule plus-end(GO:0035371) |
| 0.0 | 2.7 | GO:0005881 | cytoplasmic microtubule(GO:0005881) |
| 0.0 | 2.0 | GO:0005834 | heterotrimeric G-protein complex(GO:0005834) |
| 0.0 | 0.4 | GO:1990023 | mitotic spindle midzone(GO:1990023) |
| 0.0 | 0.9 | GO:0030131 | clathrin adaptor complex(GO:0030131) |
| 0.0 | 0.5 | GO:0046930 | pore complex(GO:0046930) |
| 0.0 | 5.7 | GO:0044306 | neuron projection terminus(GO:0044306) |
| 0.0 | 0.9 | GO:0002102 | podosome(GO:0002102) axonal growth cone(GO:0044295) |
| 0.0 | 3.2 | GO:0005770 | late endosome(GO:0005770) |
| 0.0 | 0.1 | GO:0071541 | eukaryotic translation initiation factor 3 complex, eIF3m(GO:0071541) |
| 0.0 | 0.2 | GO:0000940 | condensed chromosome outer kinetochore(GO:0000940) |
| 0.0 | 0.3 | GO:0001891 | phagocytic cup(GO:0001891) |
| 0.0 | 0.6 | GO:0030173 | integral component of Golgi membrane(GO:0030173) |
| 0.0 | 0.0 | GO:1990716 | axonemal central apparatus(GO:1990716) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.2 | 4.7 | GO:0031694 | alpha-2A adrenergic receptor binding(GO:0031694) |
| 0.8 | 7.5 | GO:0004568 | chitinase activity(GO:0004568) |
| 0.7 | 3.6 | GO:0004062 | aryl sulfotransferase activity(GO:0004062) |
| 0.7 | 2.1 | GO:0005017 | platelet-derived growth factor-activated receptor activity(GO:0005017) |
| 0.6 | 3.9 | GO:0005111 | type 2 fibroblast growth factor receptor binding(GO:0005111) |
| 0.4 | 4.7 | GO:0004185 | serine-type carboxypeptidase activity(GO:0004185) |
| 0.4 | 2.9 | GO:0031821 | G-protein coupled serotonin receptor binding(GO:0031821) |
| 0.3 | 3.4 | GO:0015193 | L-proline transmembrane transporter activity(GO:0015193) |
| 0.3 | 2.8 | GO:0005519 | cytoskeletal regulatory protein binding(GO:0005519) |
| 0.3 | 2.0 | GO:0004351 | glutamate decarboxylase activity(GO:0004351) |
| 0.3 | 5.5 | GO:0004806 | triglyceride lipase activity(GO:0004806) |
| 0.2 | 1.5 | GO:0051033 | nucleic acid transmembrane transporter activity(GO:0051032) RNA transmembrane transporter activity(GO:0051033) |
| 0.2 | 0.7 | GO:0019807 | aspartoacylase activity(GO:0019807) |
| 0.2 | 1.2 | GO:0032450 | maltose alpha-glucosidase activity(GO:0032450) |
| 0.2 | 14.2 | GO:0005544 | calcium-dependent phospholipid binding(GO:0005544) |
| 0.2 | 0.7 | GO:0004781 | adenylylsulfate kinase activity(GO:0004020) sulfate adenylyltransferase activity(GO:0004779) sulfate adenylyltransferase (ATP) activity(GO:0004781) |
| 0.2 | 1.1 | GO:0050294 | steroid sulfotransferase activity(GO:0050294) |
| 0.1 | 1.3 | GO:0015386 | potassium:proton antiporter activity(GO:0015386) |
| 0.1 | 2.8 | GO:0005149 | interleukin-1 receptor binding(GO:0005149) |
| 0.1 | 2.9 | GO:0042975 | peroxisome proliferator activated receptor binding(GO:0042975) |
| 0.1 | 2.4 | GO:0048027 | mRNA 5'-UTR binding(GO:0048027) |
| 0.1 | 0.3 | GO:0019809 | spermidine binding(GO:0019809) |
| 0.1 | 0.6 | GO:0004126 | cytidine deaminase activity(GO:0004126) |
| 0.1 | 0.2 | GO:0005169 | neurotrophin TRKB receptor binding(GO:0005169) |
| 0.1 | 0.2 | GO:0031871 | proteinase activated receptor binding(GO:0031871) |
| 0.1 | 0.8 | GO:0004634 | phosphopyruvate hydratase activity(GO:0004634) |
| 0.1 | 0.5 | GO:0015288 | porin activity(GO:0015288) |
| 0.0 | 1.1 | GO:0004653 | polypeptide N-acetylgalactosaminyltransferase activity(GO:0004653) |
| 0.0 | 0.1 | GO:0032090 | Pyrin domain binding(GO:0032090) |
| 0.0 | 1.4 | GO:0046961 | proton-transporting ATPase activity, rotational mechanism(GO:0046961) |
| 0.0 | 0.9 | GO:0051010 | microtubule plus-end binding(GO:0051010) |
| 0.0 | 0.9 | GO:0003746 | translation elongation factor activity(GO:0003746) |
| 0.0 | 10.8 | GO:0051015 | actin filament binding(GO:0051015) |
| 0.0 | 0.3 | GO:0001849 | complement component C1q binding(GO:0001849) |
| 0.0 | 2.8 | GO:0004722 | protein serine/threonine phosphatase activity(GO:0004722) |
| 0.0 | 5.2 | GO:0005178 | integrin binding(GO:0005178) |
| 0.0 | 1.2 | GO:0070064 | proline-rich region binding(GO:0070064) |
| 0.0 | 0.4 | GO:0019869 | chloride channel inhibitor activity(GO:0019869) |
| 0.0 | 0.8 | GO:0051537 | 2 iron, 2 sulfur cluster binding(GO:0051537) |
| 0.0 | 1.3 | GO:0005070 | SH3/SH2 adaptor activity(GO:0005070) |
| 0.0 | 0.4 | GO:0033038 | bitter taste receptor activity(GO:0033038) |
| 0.0 | 0.7 | GO:0005159 | insulin-like growth factor receptor binding(GO:0005159) |
| 0.0 | 1.3 | GO:0003724 | RNA helicase activity(GO:0003724) ATP-dependent RNA helicase activity(GO:0004004) |
| 0.0 | 2.0 | GO:0005550 | pheromone binding(GO:0005550) |
| 0.0 | 3.9 | GO:0005516 | calmodulin binding(GO:0005516) |
| 0.0 | 3.4 | GO:0004252 | serine-type endopeptidase activity(GO:0004252) |
| 0.0 | 5.8 | GO:0005525 | GTP binding(GO:0005525) |
| 0.0 | 0.5 | GO:0001671 | ATPase activator activity(GO:0001671) |
| 0.0 | 0.3 | GO:0004862 | cAMP-dependent protein kinase inhibitor activity(GO:0004862) |
| 0.0 | 0.1 | GO:0009378 | four-way junction helicase activity(GO:0009378) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 2.9 | PID S1P S1P4 PATHWAY | S1P4 pathway |
| 0.1 | 8.5 | PID ARF6 TRAFFICKING PATHWAY | Arf6 trafficking events |
| 0.1 | 4.7 | PID ALPHA SYNUCLEIN PATHWAY | Alpha-synuclein signaling |
| 0.1 | 2.9 | PID HDAC CLASSIII PATHWAY | Signaling events mediated by HDAC Class III |
| 0.1 | 2.1 | PID PDGFRA PATHWAY | PDGFR-alpha signaling pathway |
| 0.1 | 2.8 | PID ATR PATHWAY | ATR signaling pathway |
| 0.0 | 3.7 | PID NFAT TFPATHWAY | Calcineurin-regulated NFAT-dependent transcription in lymphocytes |
| 0.0 | 1.3 | PID EPHRINB REV PATHWAY | Ephrin B reverse signaling |
| 0.0 | 7.4 | NABA ECM AFFILIATED | Genes encoding proteins affiliated structurally or functionally to extracellular matrix proteins |
| 0.0 | 2.8 | PID LKB1 PATHWAY | LKB1 signaling events |
| 0.0 | 1.4 | PID ENDOTHELIN PATHWAY | Endothelins |
| 0.0 | 0.9 | PID AURORA B PATHWAY | Aurora B signaling |
| 0.0 | 3.9 | NABA SECRETED FACTORS | Genes encoding secreted soluble factors |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 5.3 | REACTOME CYTOSOLIC SULFONATION OF SMALL MOLECULES | Genes involved in Cytosolic sulfonation of small molecules |
| 0.3 | 1.2 | REACTOME DIGESTION OF DIETARY CARBOHYDRATE | Genes involved in Digestion of dietary carbohydrate |
| 0.2 | 3.2 | REACTOME DEFENSINS | Genes involved in Defensins |
| 0.2 | 4.7 | REACTOME INSULIN SYNTHESIS AND PROCESSING | Genes involved in Insulin Synthesis and Processing |
| 0.2 | 6.0 | REACTOME ADHERENS JUNCTIONS INTERACTIONS | Genes involved in Adherens junctions interactions |
| 0.2 | 2.5 | REACTOME NEF MEDIATED DOWNREGULATION OF MHC CLASS I COMPLEX CELL SURFACE EXPRESSION | Genes involved in Nef mediated downregulation of MHC class I complex cell surface expression |
| 0.1 | 3.9 | REACTOME ACTIVATED POINT MUTANTS OF FGFR2 | Genes involved in Activated point mutants of FGFR2 |
| 0.1 | 2.0 | REACTOME GABA SYNTHESIS RELEASE REUPTAKE AND DEGRADATION | Genes involved in GABA synthesis, release, reuptake and degradation |
| 0.1 | 2.9 | REACTOME CIRCADIAN REPRESSION OF EXPRESSION BY REV ERBA | Genes involved in Circadian Repression of Expression by REV-ERBA |
| 0.1 | 1.4 | REACTOME ACYL CHAIN REMODELLING OF PS | Genes involved in Acyl chain remodelling of PS |
| 0.1 | 1.3 | REACTOME ACTIVATION OF RAC | Genes involved in Activation of Rac |
| 0.1 | 2.9 | REACTOME INHIBITION OF INSULIN SECRETION BY ADRENALINE NORADRENALINE | Genes involved in Inhibition of Insulin Secretion by Adrenaline/Noradrenaline |
| 0.0 | 0.7 | REACTOME REGULATION OF INSULIN LIKE GROWTH FACTOR IGF ACTIVITY BY INSULIN LIKE GROWTH FACTOR BINDING PROTEINS IGFBPS | Genes involved in Regulation of Insulin-like Growth Factor (IGF) Activity by Insulin-like Growth Factor Binding Proteins (IGFBPs) |
| 0.0 | 0.9 | REACTOME KINESINS | Genes involved in Kinesins |
| 0.0 | 2.4 | REACTOME DIABETES PATHWAYS | Genes involved in Diabetes pathways |
| 0.0 | 1.1 | REACTOME MITOCHONDRIAL PROTEIN IMPORT | Genes involved in Mitochondrial Protein Import |
| 0.0 | 0.8 | REACTOME GLYCOLYSIS | Genes involved in Glycolysis |
| 0.0 | 0.4 | REACTOME DEADENYLATION OF MRNA | Genes involved in Deadenylation of mRNA |
| 0.0 | 0.8 | REACTOME RESPIRATORY ELECTRON TRANSPORT | Genes involved in Respiratory electron transport |