Project
PRJNA375882: Comprehensive Mouse Transcriptomic BodyMap
Navigation
Downloads
Results for Nkx6-3_Dbx2_Barx2
Z-value: 0.90
Transcription factors associated with Nkx6-3_Dbx2_Barx2
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
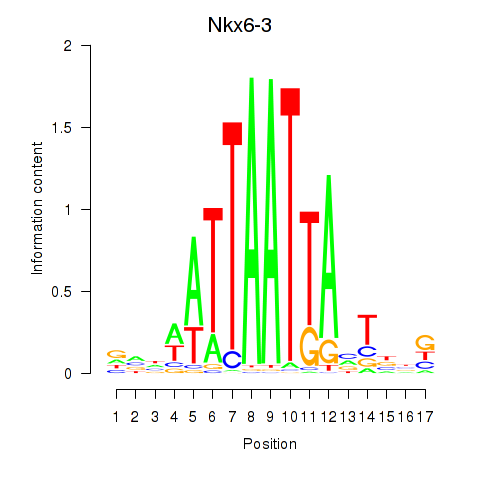
Nkx6-3
|
ENSMUSG00000063672.8 | Nkx6-3 |
|
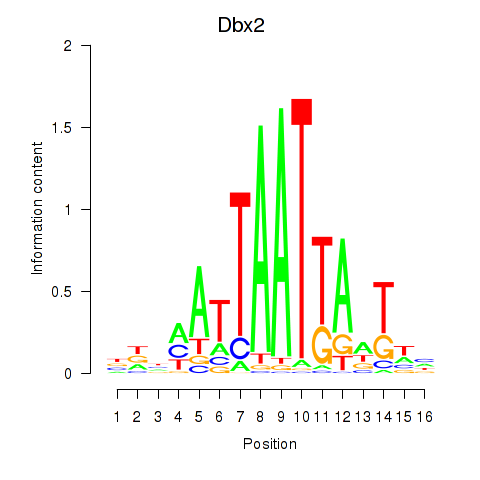
Dbx2
|
ENSMUSG00000045608.8 | Dbx2 |
|
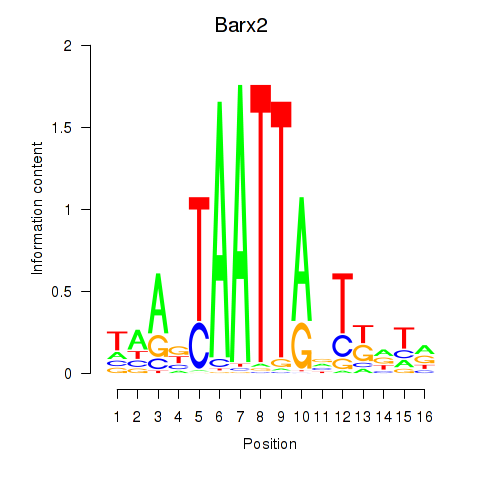
Barx2
|
ENSMUSG00000032033.12 | Barx2 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Barx2 | mm39_v1_chr9_-_31824758_31824779 | -0.50 | 7.2e-06 | Click! |
| Dbx2 | mm39_v1_chr15_-_95553841_95553841 | -0.25 | 3.3e-02 | Click! |
| Nkx6-3 | mm39_v1_chr8_+_23643279_23643292 | -0.07 | 5.7e-01 | Click! |
Activity profile of Nkx6-3_Dbx2_Barx2 motif
Sorted Z-values of Nkx6-3_Dbx2_Barx2 motif
Network of associatons between targets according to the STRING database.
First level regulatory network of Nkx6-3_Dbx2_Barx2
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr13_-_63036096 | 13.38 |
ENSMUST00000092888.11
|
Fbp1
|
fructose bisphosphatase 1 |
| chr19_-_39801188 | 12.40 |
ENSMUST00000162507.2
ENSMUST00000160476.9 ENSMUST00000239028.2 |
Cyp2c40
|
cytochrome P450, family 2, subfamily c, polypeptide 40 |
| chr19_-_39875192 | 12.04 |
ENSMUST00000168838.3
|
Cyp2c69
|
cytochrome P450, family 2, subfamily c, polypeptide 69 |
| chr13_+_24023428 | 9.44 |
ENSMUST00000091698.12
ENSMUST00000110422.3 ENSMUST00000166467.9 |
Slc17a3
|
solute carrier family 17 (sodium phosphate), member 3 |
| chr2_-_34990689 | 8.62 |
ENSMUST00000226631.2
ENSMUST00000045776.5 ENSMUST00000226972.2 |
AI182371
|
expressed sequence AI182371 |
| chr1_+_139429430 | 7.25 |
ENSMUST00000027615.7
|
F13b
|
coagulation factor XIII, beta subunit |
| chr19_-_39637489 | 7.06 |
ENSMUST00000067328.7
|
Cyp2c67
|
cytochrome P450, family 2, subfamily c, polypeptide 67 |
| chr7_+_30193047 | 6.90 |
ENSMUST00000058280.13
ENSMUST00000133318.8 ENSMUST00000142575.8 ENSMUST00000131040.2 |
Prodh2
|
proline dehydrogenase (oxidase) 2 |
| chr13_+_24023386 | 6.87 |
ENSMUST00000039721.14
|
Slc17a3
|
solute carrier family 17 (sodium phosphate), member 3 |
| chr16_+_37400500 | 6.77 |
ENSMUST00000160847.2
|
Hgd
|
homogentisate 1, 2-dioxygenase |
| chr16_+_37400590 | 6.55 |
ENSMUST00000159787.8
|
Hgd
|
homogentisate 1, 2-dioxygenase |
| chr5_+_90708962 | 6.38 |
ENSMUST00000094615.8
ENSMUST00000200765.2 |
Albfm1
|
albumin superfamily member 1 |
| chr19_-_7943365 | 6.16 |
ENSMUST00000182102.8
ENSMUST00000075619.5 |
Slc22a27
|
solute carrier family 22, member 27 |
| chr12_-_25147139 | 6.01 |
ENSMUST00000221761.2
|
Id2
|
inhibitor of DNA binding 2 |
| chr9_-_15212849 | 5.44 |
ENSMUST00000034414.10
|
4931406C07Rik
|
RIKEN cDNA 4931406C07 gene |
| chr11_+_109434519 | 4.89 |
ENSMUST00000106696.2
|
Arsg
|
arylsulfatase G |
| chr2_-_134396268 | 4.24 |
ENSMUST00000028704.3
|
Hao1
|
hydroxyacid oxidase 1, liver |
| chr14_+_26722319 | 3.92 |
ENSMUST00000035433.10
|
Hesx1
|
homeobox gene expressed in ES cells |
| chr7_-_12829100 | 3.80 |
ENSMUST00000209822.3
ENSMUST00000235753.2 |
Vmn1r85
|
vomeronasal 1 receptor 85 |
| chr9_-_15212745 | 3.57 |
ENSMUST00000217042.2
|
4931406C07Rik
|
RIKEN cDNA 4931406C07 gene |
| chr9_+_21634779 | 3.49 |
ENSMUST00000034713.9
|
Ldlr
|
low density lipoprotein receptor |
| chr3_+_59989282 | 3.35 |
ENSMUST00000029326.6
|
Sucnr1
|
succinate receptor 1 |
| chr3_-_67422821 | 3.24 |
ENSMUST00000054825.5
|
Rarres1
|
retinoic acid receptor responder (tazarotene induced) 1 |
| chr17_-_36343573 | 3.17 |
ENSMUST00000102678.5
|
H2-T23
|
histocompatibility 2, T region locus 23 |
| chr8_+_22329942 | 3.12 |
ENSMUST00000006745.4
|
Defb2
|
defensin beta 2 |
| chr19_+_37425180 | 2.92 |
ENSMUST00000128184.3
|
Hhex
|
hematopoietically expressed homeobox |
| chr6_-_129449739 | 2.81 |
ENSMUST00000112076.9
ENSMUST00000184581.3 |
Clec7a
|
C-type lectin domain family 7, member a |
| chr2_+_110427643 | 2.72 |
ENSMUST00000045972.13
ENSMUST00000111026.3 |
Slc5a12
|
solute carrier family 5 (sodium/glucose cotransporter), member 12 |
| chr16_+_22769822 | 2.64 |
ENSMUST00000023590.9
|
Hrg
|
histidine-rich glycoprotein |
| chr19_-_39729431 | 2.49 |
ENSMUST00000099472.4
|
Cyp2c68
|
cytochrome P450, family 2, subfamily c, polypeptide 68 |
| chr1_+_87983099 | 2.24 |
ENSMUST00000138182.8
ENSMUST00000113142.10 |
Ugt1a10
|
UDP glycosyltransferase 1 family, polypeptide A10 |
| chr7_-_44753168 | 2.20 |
ENSMUST00000211085.2
ENSMUST00000210642.2 ENSMUST00000003512.9 |
Fcgrt
|
Fc fragment of IgG receptor and transporter |
| chr16_+_22769844 | 2.12 |
ENSMUST00000232422.2
|
Hrg
|
histidine-rich glycoprotein |
| chr6_+_38086190 | 2.11 |
ENSMUST00000031851.5
|
Tmem213
|
transmembrane protein 213 |
| chr5_+_87148697 | 2.10 |
ENSMUST00000031186.9
|
Ugt2b35
|
UDP glucuronosyltransferase 2 family, polypeptide B35 |
| chr6_+_40619913 | 1.97 |
ENSMUST00000238599.2
|
Mgam
|
maltase-glucoamylase |
| chr13_+_24985640 | 1.81 |
ENSMUST00000019276.12
|
BC005537
|
cDNA sequence BC005537 |
| chr8_-_65489791 | 1.79 |
ENSMUST00000124790.8
|
Apela
|
apelin receptor early endogenous ligand |
| chr16_+_11224481 | 1.75 |
ENSMUST00000122168.8
|
Snx29
|
sorting nexin 29 |
| chr7_+_78922947 | 1.74 |
ENSMUST00000037315.13
|
Abhd2
|
abhydrolase domain containing 2 |
| chr14_+_47535717 | 1.63 |
ENSMUST00000166743.9
|
Mapk1ip1l
|
mitogen-activated protein kinase 1 interacting protein 1-like |
| chr13_-_73826124 | 1.62 |
ENSMUST00000022105.15
ENSMUST00000109680.10 ENSMUST00000221026.2 ENSMUST00000109679.4 |
Slc6a18
|
solute carrier family 6 (neurotransmitter transporter), member 18 |
| chr6_-_38086484 | 1.62 |
ENSMUST00000114908.5
|
Atp6v0a4
|
ATPase, H+ transporting, lysosomal V0 subunit A4 |
| chr1_+_87983189 | 1.60 |
ENSMUST00000173325.2
|
Ugt1a10
|
UDP glycosyltransferase 1 family, polypeptide A10 |
| chr8_-_65489834 | 1.58 |
ENSMUST00000142822.4
|
Apela
|
apelin receptor early endogenous ligand |
| chr18_+_36661198 | 1.55 |
ENSMUST00000237174.2
ENSMUST00000236124.2 ENSMUST00000236779.2 ENSMUST00000235181.2 ENSMUST00000074298.13 ENSMUST00000115694.3 ENSMUST00000236126.2 |
Slc4a9
|
solute carrier family 4, sodium bicarbonate cotransporter, member 9 |
| chr13_-_73826108 | 1.49 |
ENSMUST00000222029.2
ENSMUST00000223074.2 ENSMUST00000220650.2 ENSMUST00000221987.2 |
Slc6a18
|
solute carrier family 6 (neurotransmitter transporter), member 18 |
| chr6_-_115569504 | 1.45 |
ENSMUST00000112957.2
|
Mkrn2os
|
makorin, ring finger protein 2, opposite strand |
| chr13_+_24986003 | 1.42 |
ENSMUST00000155575.2
|
BC005537
|
cDNA sequence BC005537 |
| chr2_+_22959223 | 1.41 |
ENSMUST00000114523.10
|
Acbd5
|
acyl-Coenzyme A binding domain containing 5 |
| chr15_-_101422054 | 1.41 |
ENSMUST00000230067.3
|
Gm49425
|
predicted gene, 49425 |
| chr8_-_22396428 | 1.30 |
ENSMUST00000051965.5
|
Defb11
|
defensin beta 11 |
| chr3_-_88317601 | 1.27 |
ENSMUST00000193338.6
ENSMUST00000056370.13 |
Pmf1
|
polyamine-modulated factor 1 |
| chr17_-_37404764 | 1.27 |
ENSMUST00000087144.5
|
Olfr91
|
olfactory receptor 91 |
| chr6_-_50631418 | 1.26 |
ENSMUST00000031853.8
|
Npvf
|
neuropeptide VF precursor |
| chr4_-_14621669 | 1.26 |
ENSMUST00000143105.2
|
Slc26a7
|
solute carrier family 26, member 7 |
| chr2_+_69050315 | 1.25 |
ENSMUST00000005364.12
ENSMUST00000112317.3 |
G6pc2
|
glucose-6-phosphatase, catalytic, 2 |
| chr11_-_99979052 | 1.25 |
ENSMUST00000107419.2
|
Krt32
|
keratin 32 |
| chr2_+_36120438 | 1.14 |
ENSMUST00000062069.6
|
Ptgs1
|
prostaglandin-endoperoxide synthase 1 |
| chr5_-_65855511 | 1.13 |
ENSMUST00000201948.4
|
Pds5a
|
PDS5 cohesin associated factor A |
| chr11_-_99265721 | 1.11 |
ENSMUST00000006963.3
|
Krt28
|
keratin 28 |
| chr9_+_118892497 | 1.08 |
ENSMUST00000141185.8
ENSMUST00000126251.8 ENSMUST00000136561.2 |
Vill
|
villin-like |
| chr11_+_73489420 | 1.08 |
ENSMUST00000214228.2
|
Olfr384
|
olfactory receptor 384 |
| chr6_-_41752111 | 1.07 |
ENSMUST00000214976.3
|
Olfr459
|
olfactory receptor 459 |
| chr18_+_47245204 | 1.07 |
ENSMUST00000234633.2
|
Hspe1-rs1
|
heat shock protein 1 (chaperonin 10), related sequence 1 |
| chr5_-_116162415 | 1.05 |
ENSMUST00000031486.14
ENSMUST00000111999.8 |
Prkab1
|
protein kinase, AMP-activated, beta 1 non-catalytic subunit |
| chrX_-_142716085 | 1.02 |
ENSMUST00000087313.10
|
Dcx
|
doublecortin |
| chr11_+_94218810 | 1.01 |
ENSMUST00000107818.9
ENSMUST00000051221.13 |
Ankrd40
|
ankyrin repeat domain 40 |
| chr2_-_69542805 | 0.95 |
ENSMUST00000102706.4
ENSMUST00000073152.13 |
Fastkd1
|
FAST kinase domains 1 |
| chr18_+_11766333 | 0.94 |
ENSMUST00000115861.9
|
Rbbp8
|
retinoblastoma binding protein 8, endonuclease |
| chr10_-_129630735 | 0.89 |
ENSMUST00000217283.2
ENSMUST00000214206.2 ENSMUST00000214878.2 |
Olfr810
|
olfactory receptor 810 |
| chr8_-_70664901 | 0.89 |
ENSMUST00000063788.8
ENSMUST00000110127.8 |
Slc25a42
|
solute carrier family 25, member 42 |
| chr1_+_133237516 | 0.88 |
ENSMUST00000094557.7
ENSMUST00000192465.2 ENSMUST00000193888.6 ENSMUST00000194044.6 ENSMUST00000184603.8 |
Golt1a
Gm28040
Gm28040
|
golgi transport 1A predicted gene, 28040 predicted gene, 28040 |
| chr16_-_58620631 | 0.87 |
ENSMUST00000206205.3
|
Olfr173
|
olfactory receptor 173 |
| chrX_+_106299484 | 0.86 |
ENSMUST00000101294.9
ENSMUST00000118820.8 ENSMUST00000120971.8 |
Gpr174
|
G protein-coupled receptor 174 |
| chr5_-_5564730 | 0.85 |
ENSMUST00000115445.8
ENSMUST00000179804.8 ENSMUST00000125110.2 ENSMUST00000115446.8 |
Cldn12
|
claudin 12 |
| chr11_-_73217298 | 0.83 |
ENSMUST00000155630.9
|
Aspa
|
aspartoacylase |
| chr18_+_50184769 | 0.81 |
ENSMUST00000134348.8
ENSMUST00000153873.3 |
Tnfaip8
|
tumor necrosis factor, alpha-induced protein 8 |
| chr4_+_34893772 | 0.81 |
ENSMUST00000029975.10
ENSMUST00000135871.8 ENSMUST00000108130.2 |
Cga
|
glycoprotein hormones, alpha subunit |
| chr2_-_168608949 | 0.79 |
ENSMUST00000075044.10
|
Sall4
|
spalt like transcription factor 4 |
| chr2_+_22959452 | 0.77 |
ENSMUST00000155602.4
|
Acbd5
|
acyl-Coenzyme A binding domain containing 5 |
| chrX_+_100473161 | 0.75 |
ENSMUST00000033673.7
|
Nono
|
non-POU-domain-containing, octamer binding protein |
| chrX_+_138464065 | 0.71 |
ENSMUST00000113027.8
|
Rnf128
|
ring finger protein 128 |
| chr4_-_14621497 | 0.71 |
ENSMUST00000149633.2
|
Slc26a7
|
solute carrier family 26, member 7 |
| chr15_-_101801351 | 0.71 |
ENSMUST00000100179.2
|
Krt76
|
keratin 76 |
| chr6_+_37847721 | 0.70 |
ENSMUST00000031859.14
ENSMUST00000120428.8 |
Trim24
|
tripartite motif-containing 24 |
| chr17_+_37977879 | 0.68 |
ENSMUST00000215811.2
|
Olfr118
|
olfactory receptor 118 |
| chr1_+_150195158 | 0.67 |
ENSMUST00000165062.8
ENSMUST00000191228.7 ENSMUST00000186572.7 ENSMUST00000185698.2 |
Pdc
|
phosducin |
| chrX_+_151909893 | 0.67 |
ENSMUST00000163801.2
|
Foxr2
|
forkhead box R2 |
| chr16_+_22676589 | 0.66 |
ENSMUST00000004574.14
ENSMUST00000178320.2 ENSMUST00000166487.10 |
Dnajb11
|
DnaJ heat shock protein family (Hsp40) member B11 |
| chr5_-_5564873 | 0.66 |
ENSMUST00000060947.14
|
Cldn12
|
claudin 12 |
| chr7_-_133304244 | 0.64 |
ENSMUST00000209636.2
ENSMUST00000153698.3 |
Uros
|
uroporphyrinogen III synthase |
| chr6_-_3399451 | 0.64 |
ENSMUST00000120087.6
|
Samd9l
|
sterile alpha motif domain containing 9-like |
| chr14_+_47536075 | 0.63 |
ENSMUST00000227554.2
|
Mapk1ip1l
|
mitogen-activated protein kinase 1 interacting protein 1-like |
| chr5_-_3697806 | 0.61 |
ENSMUST00000119783.2
ENSMUST00000007559.15 |
Gatad1
|
GATA zinc finger domain containing 1 |
| chr7_-_80055168 | 0.58 |
ENSMUST00000107362.10
ENSMUST00000135306.3 |
Furin
|
furin (paired basic amino acid cleaving enzyme) |
| chr1_+_173983199 | 0.57 |
ENSMUST00000213748.2
|
Olfr420
|
olfactory receptor 420 |
| chr9_+_40092216 | 0.57 |
ENSMUST00000218134.2
ENSMUST00000216720.2 ENSMUST00000214763.2 |
Olfr986
|
olfactory receptor 986 |
| chr10_-_126866682 | 0.56 |
ENSMUST00000040560.11
|
Tsfm
|
Ts translation elongation factor, mitochondrial |
| chr5_+_88523967 | 0.55 |
ENSMUST00000073363.2
|
Amtn
|
amelotin |
| chr11_+_58062467 | 0.52 |
ENSMUST00000020820.2
|
Mrpl22
|
mitochondrial ribosomal protein L22 |
| chr12_-_55061117 | 0.52 |
ENSMUST00000172875.8
|
Baz1a
|
bromodomain adjacent to zinc finger domain 1A |
| chr2_-_85632888 | 0.51 |
ENSMUST00000217410.3
ENSMUST00000216425.3 |
Olfr1016
|
olfactory receptor 1016 |
| chr4_-_14621805 | 0.51 |
ENSMUST00000042221.14
|
Slc26a7
|
solute carrier family 26, member 7 |
| chr14_-_86986541 | 0.51 |
ENSMUST00000226254.2
|
Diaph3
|
diaphanous related formin 3 |
| chr16_-_22676264 | 0.50 |
ENSMUST00000232075.2
ENSMUST00000004576.8 |
Tbccd1
|
TBCC domain containing 1 |
| chr1_-_138103021 | 0.50 |
ENSMUST00000182755.8
ENSMUST00000193650.2 ENSMUST00000182283.8 |
Ptprc
|
protein tyrosine phosphatase, receptor type, C |
| chr2_+_85868891 | 0.50 |
ENSMUST00000218397.2
|
Olfr1033
|
olfactory receptor 1033 |
| chr3_+_94744844 | 0.50 |
ENSMUST00000107270.9
|
Pogz
|
pogo transposable element with ZNF domain |
| chr3_-_62414391 | 0.50 |
ENSMUST00000029336.6
|
Dhx36
|
DEAH (Asp-Glu-Ala-His) box polypeptide 36 |
| chr7_+_18853778 | 0.49 |
ENSMUST00000053109.5
|
Fbxo46
|
F-box protein 46 |
| chr9_+_72714156 | 0.48 |
ENSMUST00000055535.9
|
Prtg
|
protogenin |
| chr17_+_29493049 | 0.48 |
ENSMUST00000149405.4
|
BC004004
|
cDNA sequence BC004004 |
| chr10_-_23968192 | 0.47 |
ENSMUST00000092654.4
|
Taar8b
|
trace amine-associated receptor 8B |
| chr7_-_106279791 | 0.47 |
ENSMUST00000215541.2
|
Olfr693
|
olfactory receptor 693 |
| chr2_+_87686206 | 0.47 |
ENSMUST00000217376.2
|
Olfr1151
|
olfactory receptor 1151 |
| chr12_-_11258973 | 0.46 |
ENSMUST00000049877.3
|
Msgn1
|
mesogenin 1 |
| chr6_-_68968278 | 0.46 |
ENSMUST00000197966.2
|
Igkv4-81
|
immunoglobulin kappa variable 4-81 |
| chr6_-_138020409 | 0.45 |
ENSMUST00000111873.8
ENSMUST00000141280.3 |
Slc15a5
|
solute carrier family 15, member 5 |
| chr5_-_53864595 | 0.45 |
ENSMUST00000200691.4
|
Cckar
|
cholecystokinin A receptor |
| chr2_+_83554868 | 0.44 |
ENSMUST00000111740.9
|
Itgav
|
integrin alpha V |
| chr3_+_5283577 | 0.43 |
ENSMUST00000175866.8
|
Zfhx4
|
zinc finger homeodomain 4 |
| chr17_+_29493113 | 0.43 |
ENSMUST00000234326.2
ENSMUST00000235117.2 |
BC004004
|
cDNA sequence BC004004 |
| chr1_+_180878797 | 0.42 |
ENSMUST00000036819.7
|
9130409I23Rik
|
RIKEN cDNA 9130409I23 gene |
| chr8_-_112458723 | 0.42 |
ENSMUST00000212349.2
|
Bcar1
|
breast cancer anti-estrogen resistance 1 |
| chr17_-_8366536 | 0.41 |
ENSMUST00000231927.2
|
Rnaset2a
|
ribonuclease T2A |
| chr2_-_164080172 | 0.41 |
ENSMUST00000109374.2
|
Svs2
|
seminal vesicle secretory protein 2 |
| chrX_+_159551009 | 0.41 |
ENSMUST00000033650.14
|
Rs1
|
retinoschisis (X-linked, juvenile) 1 (human) |
| chr1_-_138102972 | 0.41 |
ENSMUST00000195533.6
ENSMUST00000183301.8 |
Ptprc
|
protein tyrosine phosphatase, receptor type, C |
| chr2_+_85597442 | 0.40 |
ENSMUST00000216397.3
|
Olfr1013
|
olfactory receptor 1013 |
| chr7_-_106354591 | 0.40 |
ENSMUST00000214306.2
ENSMUST00000216255.2 |
Olfr698
|
olfactory receptor 698 |
| chr2_-_84545504 | 0.40 |
ENSMUST00000035840.6
|
Zdhhc5
|
zinc finger, DHHC domain containing 5 |
| chr3_-_86827640 | 0.39 |
ENSMUST00000195561.6
|
Dclk2
|
doublecortin-like kinase 2 |
| chr11_+_116734104 | 0.38 |
ENSMUST00000106370.10
|
Mettl23
|
methyltransferase like 23 |
| chr17_+_38104635 | 0.38 |
ENSMUST00000214770.3
|
Olfr123
|
olfactory receptor 123 |
| chr16_+_45044678 | 0.38 |
ENSMUST00000102802.10
ENSMUST00000063654.6 |
Btla
|
B and T lymphocyte associated |
| chr11_-_59466995 | 0.38 |
ENSMUST00000215339.2
ENSMUST00000214351.2 |
Olfr222
|
olfactory receptor 222 |
| chr14_-_63221950 | 0.38 |
ENSMUST00000100493.3
|
Defb48
|
defensin beta 48 |
| chr11_-_99412162 | 0.37 |
ENSMUST00000107445.8
|
Krt39
|
keratin 39 |
| chr5_+_124045238 | 0.37 |
ENSMUST00000023869.15
|
Denr
|
density-regulated protein |
| chr7_-_103094646 | 0.37 |
ENSMUST00000215417.2
|
Olfr605
|
olfactory receptor 605 |
| chr2_-_111880531 | 0.37 |
ENSMUST00000213582.2
ENSMUST00000213961.3 ENSMUST00000215531.2 |
Olfr1312
|
olfactory receptor 1312 |
| chr6_-_136918885 | 0.36 |
ENSMUST00000111891.4
|
Arhgdib
|
Rho, GDP dissociation inhibitor (GDI) beta |
| chrX_+_159551171 | 0.36 |
ENSMUST00000112368.3
|
Rs1
|
retinoschisis (X-linked, juvenile) 1 (human) |
| chr2_-_87570322 | 0.35 |
ENSMUST00000214573.2
|
Olfr1138
|
olfactory receptor 1138 |
| chr3_-_14843512 | 0.35 |
ENSMUST00000094365.11
|
Car1
|
carbonic anhydrase 1 |
| chr6_-_136918495 | 0.35 |
ENSMUST00000111892.8
|
Arhgdib
|
Rho, GDP dissociation inhibitor (GDI) beta |
| chr1_-_37955569 | 0.35 |
ENSMUST00000078307.7
|
Lyg2
|
lysozyme G-like 2 |
| chr11_+_94219046 | 0.35 |
ENSMUST00000021227.6
|
Ankrd40
|
ankyrin repeat domain 40 |
| chr12_-_113542610 | 0.34 |
ENSMUST00000195468.6
ENSMUST00000103442.3 |
Ighv5-2
|
immunoglobulin heavy variable 5-2 |
| chr7_-_107857554 | 0.34 |
ENSMUST00000215173.2
|
Olfr488
|
olfactory receptor 488 |
| chr1_+_34478932 | 0.34 |
ENSMUST00000027303.14
|
Imp4
|
IMP4, U3 small nucleolar ribonucleoprotein |
| chrX_+_110154017 | 0.33 |
ENSMUST00000210720.3
|
Cylc1
|
cylicin, basic protein of sperm head cytoskeleton 1 |
| chr6_-_136918671 | 0.33 |
ENSMUST00000032344.12
|
Arhgdib
|
Rho, GDP dissociation inhibitor (GDI) beta |
| chr3_+_94745009 | 0.33 |
ENSMUST00000107266.8
ENSMUST00000042402.12 ENSMUST00000107269.2 |
Pogz
|
pogo transposable element with ZNF domain |
| chr6_+_113419530 | 0.33 |
ENSMUST00000101070.5
ENSMUST00000204254.2 |
Jagn1
|
jagunal homolog 1 |
| chr4_+_145397238 | 0.33 |
ENSMUST00000105738.9
|
Zfp980
|
zinc finger protein 980 |
| chr6_+_129554868 | 0.32 |
ENSMUST00000053708.9
|
Klre1
|
killer cell lectin-like receptor family E member 1 |
| chr7_+_63835154 | 0.32 |
ENSMUST00000177102.8
|
Trpm1
|
transient receptor potential cation channel, subfamily M, member 1 |
| chr10_-_126866658 | 0.32 |
ENSMUST00000120547.2
ENSMUST00000152054.8 |
Tsfm
|
Ts translation elongation factor, mitochondrial |
| chr17_+_38104420 | 0.32 |
ENSMUST00000216051.3
|
Olfr123
|
olfactory receptor 123 |
| chr7_+_63835285 | 0.31 |
ENSMUST00000206263.2
ENSMUST00000206107.2 ENSMUST00000205731.2 ENSMUST00000206706.2 ENSMUST00000205690.2 |
Trpm1
|
transient receptor potential cation channel, subfamily M, member 1 |
| chr13_-_43634695 | 0.31 |
ENSMUST00000144326.4
|
Ranbp9
|
RAN binding protein 9 |
| chr6_-_136918844 | 0.31 |
ENSMUST00000204934.2
|
Arhgdib
|
Rho, GDP dissociation inhibitor (GDI) beta |
| chr12_+_87993774 | 0.31 |
ENSMUST00000178852.2
|
Eif1ad11
|
eukaryotic translation initiation factor 1A domain containing 11 |
| chr3_+_5283606 | 0.30 |
ENSMUST00000026284.13
|
Zfhx4
|
zinc finger homeodomain 4 |
| chr9_+_20148415 | 0.30 |
ENSMUST00000086474.6
|
Olfr872
|
olfactory receptor 872 |
| chr5_-_52347826 | 0.30 |
ENSMUST00000199321.5
ENSMUST00000195922.2 ENSMUST00000031061.12 |
Dhx15
|
DEAH (Asp-Glu-Ala-His) box polypeptide 15 |
| chr2_-_86257093 | 0.29 |
ENSMUST00000217481.2
|
Olfr1062
|
olfactory receptor 1062 |
| chr8_-_85389470 | 0.29 |
ENSMUST00000060427.6
|
Ier2
|
immediate early response 2 |
| chr6_+_30611028 | 0.29 |
ENSMUST00000115138.8
|
Cpa5
|
carboxypeptidase A5 |
| chr9_-_19275301 | 0.28 |
ENSMUST00000214810.2
|
Olfr846
|
olfactory receptor 846 |
| chr4_+_147056433 | 0.28 |
ENSMUST00000146688.3
|
Zfp989
|
zinc finger protein 989 |
| chr16_-_58695131 | 0.28 |
ENSMUST00000217377.2
|
Olfr177
|
olfactory receptor 177 |
| chr10_+_26698556 | 0.28 |
ENSMUST00000135866.2
|
Arhgap18
|
Rho GTPase activating protein 18 |
| chr7_-_107813843 | 0.28 |
ENSMUST00000216489.2
|
Olfr487
|
olfactory receptor 487 |
| chr7_-_103191924 | 0.27 |
ENSMUST00000214269.3
|
Olfr612
|
olfactory receptor 612 |
| chr8_+_121842902 | 0.27 |
ENSMUST00000054691.8
|
Foxc2
|
forkhead box C2 |
| chr2_+_85648823 | 0.26 |
ENSMUST00000214416.2
|
Olfr1018
|
olfactory receptor 1018 |
| chrX_-_8118541 | 0.26 |
ENSMUST00000115594.8
ENSMUST00000115595.8 ENSMUST00000033513.10 |
Ftsj1
|
FtsJ RNA methyltransferase homolog 1 (E. coli) |
| chr4_+_109092610 | 0.26 |
ENSMUST00000106628.8
|
Calr4
|
calreticulin 4 |
| chr2_+_83554741 | 0.26 |
ENSMUST00000028499.11
|
Itgav
|
integrin alpha V |
| chr2_-_86109346 | 0.25 |
ENSMUST00000217294.2
ENSMUST00000217245.2 ENSMUST00000216432.2 |
Olfr1051
|
olfactory receptor 1051 |
| chr3_-_27208010 | 0.25 |
ENSMUST00000108300.8
ENSMUST00000108298.9 |
Ect2
|
ect2 oncogene |
| chr2_+_110551976 | 0.25 |
ENSMUST00000090332.5
|
Muc15
|
mucin 15 |
| chr4_-_133972890 | 0.24 |
ENSMUST00000030644.8
|
Zfp593
|
zinc finger protein 593 |
| chr7_+_102526329 | 0.24 |
ENSMUST00000098216.2
|
Olfr568
|
olfactory receptor 568 |
| chr7_+_103205286 | 0.24 |
ENSMUST00000214345.2
ENSMUST00000215673.2 |
Olfr615
|
olfactory receptor 615 |
| chr8_-_84976330 | 0.24 |
ENSMUST00000019506.9
|
D8Ertd738e
|
DNA segment, Chr 8, ERATO Doi 738, expressed |
| chr16_-_19443851 | 0.23 |
ENSMUST00000079891.4
|
Olfr171
|
olfactory receptor 171 |
| chr6_-_38331482 | 0.23 |
ENSMUST00000031850.10
ENSMUST00000114898.3 |
Zc3hav1
|
zinc finger CCCH type, antiviral 1 |
| chr7_+_84562283 | 0.23 |
ENSMUST00000216367.2
ENSMUST00000214501.3 |
Olfr290
|
olfactory receptor 290 |
| chr4_-_146993984 | 0.23 |
ENSMUST00000238583.2
ENSMUST00000049821.4 |
Gm21411
|
predicted gene, 21411 |
| chr10_+_50468765 | 0.23 |
ENSMUST00000035606.10
|
Ascc3
|
activating signal cointegrator 1 complex subunit 3 |
| chr10_+_129601351 | 0.23 |
ENSMUST00000203236.3
|
Olfr808
|
olfactory receptor 808 |
| chr7_+_20402843 | 0.23 |
ENSMUST00000179079.2
|
Vmn1r107
|
vomeronasal 1 receptor 107 |
| chr7_+_22357443 | 0.23 |
ENSMUST00000177632.2
|
Vmn1r155
|
vomeronasal 1 receptor 155 |
| chr4_+_147390131 | 0.23 |
ENSMUST00000148762.4
|
Zfp988
|
zinc finger protein 988 |
| chr2_-_84481020 | 0.22 |
ENSMUST00000067232.10
|
Ctnnd1
|
catenin (cadherin associated protein), delta 1 |
| chr13_-_59970383 | 0.22 |
ENSMUST00000225987.2
|
Tut7
|
terminal uridylyl transferase 7 |
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 3.3 | 13.4 | GO:0046351 | disaccharide biosynthetic process(GO:0046351) |
| 2.3 | 6.9 | GO:0010133 | proline catabolic process to glutamate(GO:0010133) |
| 2.0 | 6.0 | GO:0001966 | thigmotaxis(GO:0001966) |
| 1.6 | 4.8 | GO:0097037 | heme export(GO:0097037) |
| 1.5 | 22.5 | GO:0015747 | urate transport(GO:0015747) |
| 1.5 | 13.3 | GO:0006572 | tyrosine catabolic process(GO:0006572) |
| 1.1 | 3.2 | GO:0002476 | antigen processing and presentation of endogenous peptide antigen via MHC class Ib(GO:0002476) positive regulation of CD8-positive, alpha-beta T cell proliferation(GO:2000566) |
| 1.0 | 34.0 | GO:0019373 | epoxygenase P450 pathway(GO:0019373) |
| 0.9 | 3.5 | GO:0090118 | regulation of phosphatidylcholine catabolic process(GO:0010899) receptor-mediated endocytosis of low-density lipoprotein particle involved in cholesterol transport(GO:0090118) positive regulation of lysosomal protein catabolic process(GO:1905167) |
| 0.8 | 4.2 | GO:0009441 | glycolate metabolic process(GO:0009441) |
| 0.5 | 2.9 | GO:0061017 | hepatoblast differentiation(GO:0061017) |
| 0.4 | 3.9 | GO:0030916 | otic vesicle formation(GO:0030916) |
| 0.3 | 3.8 | GO:0052697 | flavonoid glucuronidation(GO:0052696) xenobiotic glucuronidation(GO:0052697) |
| 0.3 | 0.9 | GO:2000536 | negative regulation of entry of bacterium into host cell(GO:2000536) |
| 0.3 | 0.9 | GO:1901642 | purine nucleoside transmembrane transport(GO:0015860) nucleoside transmembrane transport(GO:1901642) |
| 0.3 | 2.5 | GO:0019532 | oxalate transport(GO:0019532) |
| 0.2 | 0.9 | GO:2000471 | regulation of hematopoietic stem cell migration(GO:2000471) positive regulation of hematopoietic stem cell migration(GO:2000473) |
| 0.2 | 0.9 | GO:0070125 | mitochondrial translational elongation(GO:0070125) |
| 0.2 | 0.6 | GO:0006780 | uroporphyrinogen III biosynthetic process(GO:0006780) |
| 0.2 | 2.2 | GO:0030242 | pexophagy(GO:0030242) |
| 0.2 | 0.8 | GO:0036367 | adaptation of rhodopsin mediated signaling(GO:0016062) light adaption(GO:0036367) |
| 0.2 | 0.6 | GO:0070175 | positive regulation of enamel mineralization(GO:0070175) |
| 0.2 | 1.1 | GO:0019371 | cyclooxygenase pathway(GO:0019371) maintenance of blood-brain barrier(GO:0035633) |
| 0.2 | 0.8 | GO:1903377 | negative regulation of oxidative stress-induced neuron intrinsic apoptotic signaling pathway(GO:1903377) |
| 0.1 | 1.3 | GO:1901164 | negative regulation of trophoblast cell migration(GO:1901164) |
| 0.1 | 0.6 | GO:0090472 | dibasic protein processing(GO:0090472) |
| 0.1 | 0.4 | GO:1990859 | cellular response to endothelin(GO:1990859) |
| 0.1 | 0.9 | GO:0006083 | acetate metabolic process(GO:0006083) |
| 0.1 | 0.3 | GO:1902256 | apoptotic process involved in outflow tract morphogenesis(GO:0003275) regulation of apoptotic process involved in outflow tract morphogenesis(GO:1902256) |
| 0.1 | 1.3 | GO:0032277 | negative regulation of gonadotropin secretion(GO:0032277) |
| 0.1 | 1.6 | GO:0070072 | vacuolar proton-transporting V-type ATPase complex assembly(GO:0070072) |
| 0.1 | 1.1 | GO:1901727 | positive regulation of histone deacetylase activity(GO:1901727) |
| 0.1 | 0.9 | GO:0010792 | DNA double-strand break processing involved in repair via single-strand annealing(GO:0010792) |
| 0.1 | 0.5 | GO:0051684 | maintenance of Golgi location(GO:0051684) |
| 0.1 | 0.5 | GO:0090669 | telomerase RNA stabilization(GO:0090669) |
| 0.1 | 0.7 | GO:0046116 | queuosine biosynthetic process(GO:0008616) queuosine metabolic process(GO:0046116) |
| 0.1 | 0.8 | GO:0046884 | follicle-stimulating hormone secretion(GO:0046884) |
| 0.1 | 0.7 | GO:0070562 | regulation of vitamin D receptor signaling pathway(GO:0070562) |
| 0.1 | 1.0 | GO:0035878 | nail development(GO:0035878) |
| 0.1 | 1.7 | GO:0032570 | response to progesterone(GO:0032570) |
| 0.1 | 0.3 | GO:0002856 | stimulatory C-type lectin receptor signaling pathway(GO:0002223) negative regulation of natural killer cell mediated immune response to tumor cell(GO:0002856) negative regulation of natural killer cell mediated cytotoxicity directed against tumor cell target(GO:0002859) |
| 0.1 | 0.4 | GO:0002023 | reduction of food intake in response to dietary excess(GO:0002023) positive regulation of somatostatin secretion(GO:0090274) |
| 0.1 | 0.6 | GO:0035726 | common myeloid progenitor cell proliferation(GO:0035726) |
| 0.1 | 0.3 | GO:0031120 | snRNA pseudouridine synthesis(GO:0031120) |
| 0.1 | 0.4 | GO:0002188 | translation reinitiation(GO:0002188) |
| 0.1 | 0.3 | GO:0060690 | epithelial cell differentiation involved in salivary gland development(GO:0060690) |
| 0.1 | 1.8 | GO:0007064 | mitotic sister chromatid cohesion(GO:0007064) |
| 0.0 | 1.3 | GO:0051156 | glucose 6-phosphate metabolic process(GO:0051156) |
| 0.0 | 0.3 | GO:0009253 | peptidoglycan metabolic process(GO:0000270) peptidoglycan catabolic process(GO:0009253) |
| 0.0 | 0.8 | GO:0048672 | positive regulation of collateral sprouting(GO:0048672) |
| 0.0 | 3.4 | GO:0060395 | SMAD protein signal transduction(GO:0060395) |
| 0.0 | 0.8 | GO:0001833 | inner cell mass cell proliferation(GO:0001833) |
| 0.0 | 0.7 | GO:0048733 | sebaceous gland development(GO:0048733) |
| 0.0 | 0.2 | GO:1903026 | negative regulation of RNA polymerase II regulatory region sequence-specific DNA binding(GO:1903026) |
| 0.0 | 0.2 | GO:0006307 | DNA dealkylation involved in DNA repair(GO:0006307) |
| 0.0 | 0.4 | GO:0046642 | negative regulation of alpha-beta T cell proliferation(GO:0046642) |
| 0.0 | 0.7 | GO:0016556 | mRNA modification(GO:0016556) |
| 0.0 | 6.3 | GO:0007596 | blood coagulation(GO:0007596) |
| 0.0 | 0.5 | GO:0007379 | segment specification(GO:0007379) |
| 0.0 | 0.3 | GO:0030223 | neutrophil differentiation(GO:0030223) |
| 0.0 | 0.1 | GO:1990416 | cellular response to brain-derived neurotrophic factor stimulus(GO:1990416) |
| 0.0 | 0.2 | GO:0071360 | cellular response to exogenous dsRNA(GO:0071360) |
| 0.0 | 0.7 | GO:0042036 | negative regulation of cytokine biosynthetic process(GO:0042036) |
| 0.0 | 1.6 | GO:0051453 | regulation of intracellular pH(GO:0051453) |
| 0.0 | 0.4 | GO:0018345 | protein palmitoylation(GO:0018345) |
| 0.0 | 0.2 | GO:0051988 | regulation of attachment of spindle microtubules to kinetochore(GO:0051988) |
| 0.0 | 0.9 | GO:0043029 | T cell homeostasis(GO:0043029) |
| 0.0 | 0.4 | GO:0048240 | sperm capacitation(GO:0048240) |
| 0.0 | 4.7 | GO:0042742 | defense response to bacterium(GO:0042742) |
| 0.0 | 0.0 | GO:0052695 | cellular glucuronidation(GO:0052695) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.6 | 4.8 | GO:0061474 | phagolysosome membrane(GO:0061474) |
| 1.2 | 3.5 | GO:1990666 | PCSK9-LDLR complex(GO:1990666) |
| 0.3 | 1.3 | GO:0000444 | MIS12/MIND type complex(GO:0000444) |
| 0.2 | 3.2 | GO:0042612 | MHC class I protein complex(GO:0042612) |
| 0.2 | 0.9 | GO:0034686 | integrin alphav-beta3 complex(GO:0034683) integrin alphav-beta8 complex(GO:0034686) |
| 0.2 | 1.6 | GO:0000220 | vacuolar proton-transporting V-type ATPase, V0 domain(GO:0000220) |
| 0.1 | 1.7 | GO:0097524 | sperm plasma membrane(GO:0097524) |
| 0.1 | 0.5 | GO:0008623 | CHRAC(GO:0008623) |
| 0.1 | 0.6 | GO:0035841 | new growing cell tip(GO:0035841) |
| 0.1 | 0.4 | GO:0070992 | translation initiation complex(GO:0070992) |
| 0.1 | 0.3 | GO:0034457 | Mpp10 complex(GO:0034457) |
| 0.1 | 0.7 | GO:0005726 | perichromatin fibrils(GO:0005726) |
| 0.1 | 0.3 | GO:0043159 | acrosomal matrix(GO:0043159) |
| 0.1 | 0.8 | GO:0042382 | paraspeckles(GO:0042382) |
| 0.1 | 3.1 | GO:0031526 | brush border membrane(GO:0031526) |
| 0.1 | 0.2 | GO:0005594 | collagen type IX trimer(GO:0005594) |
| 0.0 | 0.3 | GO:0071008 | U2-type post-mRNA release spliceosomal complex(GO:0071008) |
| 0.0 | 0.7 | GO:0034663 | endoplasmic reticulum chaperone complex(GO:0034663) |
| 0.0 | 0.2 | GO:0097149 | centralspindlin complex(GO:0097149) |
| 0.0 | 0.3 | GO:0031429 | box H/ACA snoRNP complex(GO:0031429) box H/ACA RNP complex(GO:0072588) |
| 0.0 | 0.6 | GO:0012510 | trans-Golgi network transport vesicle membrane(GO:0012510) |
| 0.0 | 1.0 | GO:0031588 | nucleotide-activated protein kinase complex(GO:0031588) |
| 0.0 | 15.4 | GO:0016324 | apical plasma membrane(GO:0016324) |
| 0.0 | 0.6 | GO:0005605 | basal lamina(GO:0005605) |
| 0.0 | 1.5 | GO:0016328 | lateral plasma membrane(GO:0016328) |
| 0.0 | 4.7 | GO:0042579 | peroxisome(GO:0005777) microbody(GO:0042579) |
| 0.0 | 0.5 | GO:0031616 | spindle pole centrosome(GO:0031616) |
| 0.0 | 0.7 | GO:0045095 | keratin filament(GO:0045095) |
| 0.0 | 5.3 | GO:0000323 | lytic vacuole(GO:0000323) lysosome(GO:0005764) |
| 0.0 | 1.6 | GO:0032993 | protein-DNA complex(GO:0032993) |
| 0.0 | 8.7 | GO:0000785 | chromatin(GO:0000785) |
| 0.0 | 45.4 | GO:0070062 | extracellular exosome(GO:0070062) |
| 0.0 | 0.7 | GO:0005770 | late endosome(GO:0005770) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.6 | 16.4 | GO:0015562 | efflux transmembrane transporter activity(GO:0015562) |
| 1.1 | 4.2 | GO:0016899 | oxidoreductase activity, acting on the CH-OH group of donors, oxygen as acceptor(GO:0016899) |
| 0.9 | 34.0 | GO:0008392 | arachidonic acid epoxygenase activity(GO:0008392) |
| 0.9 | 14.6 | GO:0050308 | sugar-phosphatase activity(GO:0050308) |
| 0.4 | 3.5 | GO:0030229 | very-low-density lipoprotein particle receptor activity(GO:0030229) |
| 0.4 | 6.2 | GO:1901702 | urate transmembrane transporter activity(GO:0015143) salt transmembrane transporter activity(GO:1901702) |
| 0.4 | 13.3 | GO:0016702 | oxidoreductase activity, acting on single donors with incorporation of molecular oxygen, incorporation of two atoms of oxygen(GO:0016702) |
| 0.3 | 2.3 | GO:0019770 | IgG receptor activity(GO:0019770) |
| 0.3 | 2.0 | GO:0032450 | maltose alpha-glucosidase activity(GO:0032450) |
| 0.3 | 0.9 | GO:0019807 | aspartoacylase activity(GO:0019807) |
| 0.3 | 2.7 | GO:0015129 | lactate transmembrane transporter activity(GO:0015129) |
| 0.3 | 4.9 | GO:0008484 | sulfuric ester hydrolase activity(GO:0008484) |
| 0.2 | 3.2 | GO:0030881 | beta-2-microglobulin binding(GO:0030881) |
| 0.2 | 2.9 | GO:0008190 | eukaryotic initiation factor 4E binding(GO:0008190) |
| 0.2 | 0.9 | GO:0051185 | coenzyme transporter activity(GO:0051185) |
| 0.2 | 2.5 | GO:0019531 | oxalate transmembrane transporter activity(GO:0019531) |
| 0.2 | 6.9 | GO:0016645 | oxidoreductase activity, acting on the CH-NH group of donors(GO:0016645) |
| 0.1 | 1.3 | GO:0005094 | Rho GDP-dissociation inhibitor activity(GO:0005094) |
| 0.1 | 6.0 | GO:0015020 | glucuronosyltransferase activity(GO:0015020) |
| 0.1 | 1.7 | GO:0047372 | acylglycerol lipase activity(GO:0047372) |
| 0.1 | 0.6 | GO:0004852 | uroporphyrinogen-III synthase activity(GO:0004852) |
| 0.1 | 0.4 | GO:0042284 | sphingolipid delta-4 desaturase activity(GO:0042284) |
| 0.1 | 0.7 | GO:0034056 | estrogen response element binding(GO:0034056) |
| 0.1 | 0.9 | GO:0019960 | C-X3-C chemokine binding(GO:0019960) |
| 0.1 | 3.1 | GO:0005328 | neurotransmitter:sodium symporter activity(GO:0005328) |
| 0.1 | 0.3 | GO:0030519 | snoRNP binding(GO:0030519) |
| 0.1 | 1.6 | GO:0008553 | hydrogen-exporting ATPase activity, phosphorylative mechanism(GO:0008553) |
| 0.1 | 1.3 | GO:0043522 | leucine zipper domain binding(GO:0043522) |
| 0.1 | 0.7 | GO:0008479 | queuine tRNA-ribosyltransferase activity(GO:0008479) |
| 0.1 | 1.5 | GO:0008510 | sodium:bicarbonate symporter activity(GO:0008510) |
| 0.1 | 0.8 | GO:0008429 | phosphatidylethanolamine binding(GO:0008429) |
| 0.1 | 0.9 | GO:0000014 | single-stranded DNA endodeoxyribonuclease activity(GO:0000014) |
| 0.1 | 2.2 | GO:0000062 | fatty-acyl-CoA binding(GO:0000062) |
| 0.0 | 0.5 | GO:0051880 | G-quadruplex DNA binding(GO:0051880) |
| 0.0 | 0.5 | GO:0001594 | trace-amine receptor activity(GO:0001594) |
| 0.0 | 5.6 | GO:0004869 | cysteine-type endopeptidase inhibitor activity(GO:0004869) |
| 0.0 | 1.0 | GO:0004679 | AMP-activated protein kinase activity(GO:0004679) |
| 0.0 | 0.3 | GO:0034513 | box H/ACA snoRNA binding(GO:0034513) |
| 0.0 | 0.4 | GO:0004064 | arylesterase activity(GO:0004064) |
| 0.0 | 0.9 | GO:0045125 | bioactive lipid receptor activity(GO:0045125) |
| 0.0 | 0.9 | GO:0003746 | translation elongation factor activity(GO:0003746) |
| 0.0 | 0.6 | GO:0048406 | nerve growth factor binding(GO:0048406) |
| 0.0 | 0.9 | GO:0043395 | heparan sulfate proteoglycan binding(GO:0043395) |
| 0.0 | 0.1 | GO:0019864 | IgG binding(GO:0019864) |
| 0.0 | 0.2 | GO:0043140 | ATP-dependent 3'-5' DNA helicase activity(GO:0043140) |
| 0.0 | 0.2 | GO:0043023 | ribosomal large subunit binding(GO:0043023) |
| 0.0 | 0.3 | GO:0016427 | tRNA (cytosine) methyltransferase activity(GO:0016427) |
| 0.0 | 3.5 | GO:0005179 | hormone activity(GO:0005179) |
| 0.0 | 0.3 | GO:0003796 | lysozyme activity(GO:0003796) |
| 0.0 | 3.7 | GO:0047485 | protein N-terminus binding(GO:0047485) |
| 0.0 | 1.1 | GO:0004601 | peroxidase activity(GO:0004601) |
| 0.0 | 0.2 | GO:0017151 | DEAD/H-box RNA helicase binding(GO:0017151) |
| 0.0 | 0.4 | GO:0004181 | metallocarboxypeptidase activity(GO:0004181) |
| 0.0 | 0.4 | GO:0019706 | protein-cysteine S-palmitoyltransferase activity(GO:0019706) protein-cysteine S-acyltransferase activity(GO:0019707) |
| 0.0 | 2.7 | GO:0044325 | ion channel binding(GO:0044325) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 6.6 | PID HIF1 TFPATHWAY | HIF-1-alpha transcription factor network |
| 0.0 | 10.6 | NABA ECM REGULATORS | Genes encoding enzymes and their regulators involved in the remodeling of the extracellular matrix |
| 0.0 | 0.9 | PID INTEGRIN5 PATHWAY | Beta5 beta6 beta7 and beta8 integrin cell surface interactions |
| 0.0 | 0.8 | PID CIRCADIAN PATHWAY | Circadian rhythm pathway |
| 0.0 | 0.9 | PID ATM PATHWAY | ATM pathway |
| 0.0 | 0.4 | PID INTEGRIN A9B1 PATHWAY | Alpha9 beta1 integrin signaling events |
| 0.0 | 0.8 | PID LIS1 PATHWAY | Lissencephaly gene (LIS1) in neuronal migration and development |
| 0.0 | 0.9 | SIG PIP3 SIGNALING IN B LYMPHOCYTES | Genes related to PIP3 signaling in B lymphocytes |
| 0.0 | 1.6 | PID RHOA REG PATHWAY | Regulation of RhoA activity |
| 0.0 | 1.0 | PID LKB1 PATHWAY | LKB1 signaling events |
| 0.0 | 0.3 | PID ECADHERIN KERATINOCYTE PATHWAY | E-cadherin signaling in keratinocytes |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 4.9 | REACTOME THE ACTIVATION OF ARYLSULFATASES | Genes involved in The activation of arylsulfatases |
| 0.5 | 2.0 | REACTOME DIGESTION OF DIETARY CARBOHYDRATE | Genes involved in Digestion of dietary carbohydrate |
| 0.4 | 7.3 | REACTOME COMMON PATHWAY | Genes involved in Common Pathway |
| 0.2 | 13.4 | REACTOME GLUCONEOGENESIS | Genes involved in Gluconeogenesis |
| 0.1 | 3.1 | REACTOME NA CL DEPENDENT NEUROTRANSMITTER TRANSPORTERS | Genes involved in Na+/Cl- dependent neurotransmitter transporters |
| 0.1 | 3.5 | REACTOME CHYLOMICRON MEDIATED LIPID TRANSPORT | Genes involved in Chylomicron-mediated lipid transport |
| 0.1 | 1.0 | REACTOME REGULATION OF RHEB GTPASE ACTIVITY BY AMPK | Genes involved in Regulation of Rheb GTPase activity by AMPK |
| 0.1 | 0.8 | REACTOME GLYCOPROTEIN HORMONES | Genes involved in Glycoprotein hormones |
| 0.1 | 17.0 | REACTOME METABOLISM OF AMINO ACIDS AND DERIVATIVES | Genes involved in Metabolism of amino acids and derivatives |
| 0.1 | 0.6 | REACTOME GAMMA CARBOXYLATION TRANSPORT AND AMINO TERMINAL CLEAVAGE OF PROTEINS | Genes involved in Gamma-carboxylation, transport, and amino-terminal cleavage of proteins |
| 0.1 | 0.9 | REACTOME PHOSPHORYLATION OF CD3 AND TCR ZETA CHAINS | Genes involved in Phosphorylation of CD3 and TCR zeta chains |
| 0.0 | 1.6 | REACTOME INSULIN RECEPTOR RECYCLING | Genes involved in Insulin receptor recycling |
| 0.0 | 0.9 | REACTOME PECAM1 INTERACTIONS | Genes involved in PECAM1 interactions |
| 0.0 | 0.4 | REACTOME P130CAS LINKAGE TO MAPK SIGNALING FOR INTEGRINS | Genes involved in p130Cas linkage to MAPK signaling for integrins |
| 0.0 | 4.4 | REACTOME RESPONSE TO ELEVATED PLATELET CYTOSOLIC CA2 | Genes involved in Response to elevated platelet cytosolic Ca2+ |
| 0.0 | 1.5 | REACTOME TIGHT JUNCTION INTERACTIONS | Genes involved in Tight junction interactions |
| 0.0 | 0.6 | REACTOME METABOLISM OF PORPHYRINS | Genes involved in Metabolism of porphyrins |
| 0.0 | 0.9 | REACTOME MEIOTIC RECOMBINATION | Genes involved in Meiotic Recombination |
| 0.0 | 1.3 | REACTOME GLUCOSE TRANSPORT | Genes involved in Glucose transport |
| 0.0 | 0.7 | REACTOME SIGNALING BY FGFR1 FUSION MUTANTS | Genes involved in Signaling by FGFR1 fusion mutants |
| 0.0 | 1.4 | REACTOME MITOTIC PROMETAPHASE | Genes involved in Mitotic Prometaphase |