Project
PRJNA375882: Comprehensive Mouse Transcriptomic BodyMap
Navigation
Downloads
Results for Nr0b1
Z-value: 0.56
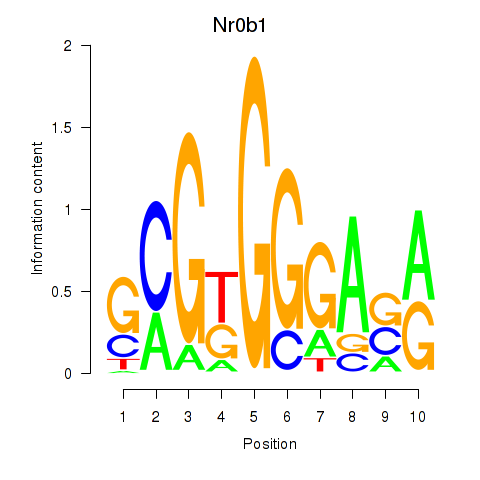
Transcription factors associated with Nr0b1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Nr0b1
|
ENSMUSG00000025056.5 | Nr0b1 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Nr0b1 | mm39_v1_chrX_+_85235370_85235388 | 0.13 | 2.7e-01 | Click! |
Activity profile of Nr0b1 motif
Sorted Z-values of Nr0b1 motif
Network of associatons between targets according to the STRING database.
First level regulatory network of Nr0b1
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr1_+_88015524 | 4.44 |
ENSMUST00000113139.2
|
Ugt1a8
|
UDP glucuronosyltransferase 1 family, polypeptide A8 |
| chr9_-_119812042 | 2.48 |
ENSMUST00000214058.2
|
Csrnp1
|
cysteine-serine-rich nuclear protein 1 |
| chr5_-_107873883 | 2.48 |
ENSMUST00000159263.3
|
Gfi1
|
growth factor independent 1 transcription repressor |
| chr9_-_110571645 | 2.47 |
ENSMUST00000006005.12
|
Pth1r
|
parathyroid hormone 1 receptor |
| chr2_+_27567213 | 2.05 |
ENSMUST00000077257.12
|
Rxra
|
retinoid X receptor alpha |
| chr2_+_4722956 | 2.05 |
ENSMUST00000056914.7
|
Bend7
|
BEN domain containing 7 |
| chr11_-_69260203 | 1.86 |
ENSMUST00000092971.13
ENSMUST00000108661.8 |
Chd3
|
chromodomain helicase DNA binding protein 3 |
| chr5_+_3393893 | 1.72 |
ENSMUST00000165117.8
ENSMUST00000197385.2 |
Cdk6
|
cyclin-dependent kinase 6 |
| chr11_-_115503704 | 1.66 |
ENSMUST00000106506.8
|
Mif4gd
|
MIF4G domain containing |
| chr18_+_35963353 | 1.63 |
ENSMUST00000235169.2
|
Cxxc5
|
CXXC finger 5 |
| chr6_-_72212547 | 1.59 |
ENSMUST00000042646.8
|
Atoh8
|
atonal bHLH transcription factor 8 |
| chr4_+_148215339 | 1.54 |
ENSMUST00000084129.9
|
Mad2l2
|
MAD2 mitotic arrest deficient-like 2 |
| chr8_-_71834543 | 1.53 |
ENSMUST00000002466.9
|
Nr2f6
|
nuclear receptor subfamily 2, group F, member 6 |
| chr11_-_52165682 | 1.51 |
ENSMUST00000238914.2
|
Tcf7
|
transcription factor 7, T cell specific |
| chr12_-_104831335 | 1.47 |
ENSMUST00000109936.3
|
Clmn
|
calmin |
| chr2_-_144174066 | 1.45 |
ENSMUST00000037423.4
|
Ovol2
|
ovo like zinc finger 2 |
| chr10_-_127504416 | 1.45 |
ENSMUST00000129252.2
|
Nab2
|
Ngfi-A binding protein 2 |
| chr3_+_86131970 | 1.42 |
ENSMUST00000192145.6
ENSMUST00000194759.6 ENSMUST00000107635.7 |
Lrba
|
LPS-responsive beige-like anchor |
| chr2_-_157121440 | 1.40 |
ENSMUST00000143663.2
|
Mroh8
|
maestro heat-like repeat family member 8 |
| chr13_+_44884200 | 1.30 |
ENSMUST00000173704.8
ENSMUST00000044608.14 ENSMUST00000173367.8 |
Jarid2
|
jumonji, AT rich interactive domain 2 |
| chr11_-_88608958 | 1.29 |
ENSMUST00000107908.2
|
Msi2
|
musashi RNA-binding protein 2 |
| chr4_-_123644091 | 1.26 |
ENSMUST00000102636.4
|
Akirin1
|
akirin 1 |
| chr6_+_120643323 | 1.25 |
ENSMUST00000112686.8
|
Cecr2
|
CECR2, histone acetyl-lysine reader |
| chr15_-_85918378 | 1.24 |
ENSMUST00000016172.10
|
Celsr1
|
cadherin, EGF LAG seven-pass G-type receptor 1 |
| chr17_-_23964807 | 1.24 |
ENSMUST00000046525.10
|
Kremen2
|
kringle containing transmembrane protein 2 |
| chr5_+_31070739 | 1.24 |
ENSMUST00000031055.8
|
Emilin1
|
elastin microfibril interfacer 1 |
| chr11_-_115503316 | 1.18 |
ENSMUST00000106507.9
|
Mif4gd
|
MIF4G domain containing |
| chr11_-_88609048 | 1.13 |
ENSMUST00000107909.8
|
Msi2
|
musashi RNA-binding protein 2 |
| chr19_-_24533183 | 1.11 |
ENSMUST00000112673.9
ENSMUST00000025800.15 |
Pip5k1b
|
phosphatidylinositol-4-phosphate 5-kinase, type 1 beta |
| chr10_-_128425519 | 1.10 |
ENSMUST00000082059.7
|
Erbb3
|
erb-b2 receptor tyrosine kinase 3 |
| chr2_-_144173615 | 1.10 |
ENSMUST00000103171.10
|
Ovol2
|
ovo like zinc finger 2 |
| chr19_+_37423198 | 1.05 |
ENSMUST00000025944.9
|
Hhex
|
hematopoietically expressed homeobox |
| chr13_+_94495457 | 1.01 |
ENSMUST00000022196.5
|
Ap3b1
|
adaptor-related protein complex 3, beta 1 subunit |
| chr2_+_153684901 | 0.95 |
ENSMUST00000175856.3
|
Efcab8
|
EF-hand calcium binding domain 8 |
| chr11_+_107438751 | 0.93 |
ENSMUST00000100305.8
ENSMUST00000075012.8 ENSMUST00000106746.8 |
Helz
|
helicase with zinc finger domain |
| chr10_+_36383008 | 0.93 |
ENSMUST00000168572.8
|
Hs3st5
|
heparan sulfate (glucosamine) 3-O-sulfotransferase 5 |
| chr11_-_69871320 | 0.90 |
ENSMUST00000143175.2
|
Elp5
|
elongator acetyltransferase complex subunit 5 |
| chr4_-_129590609 | 0.90 |
ENSMUST00000102588.10
|
Tmem39b
|
transmembrane protein 39b |
| chr6_+_134012916 | 0.87 |
ENSMUST00000164648.2
|
Etv6
|
ets variant 6 |
| chrX_-_37653396 | 0.85 |
ENSMUST00000016681.15
|
Cul4b
|
cullin 4B |
| chr2_+_157120946 | 0.84 |
ENSMUST00000116380.9
ENSMUST00000029171.6 |
Rpn2
|
ribophorin II |
| chrX_+_13147209 | 0.83 |
ENSMUST00000000804.7
|
Ddx3x
|
DEAD box helicase 3, X-linked |
| chr10_-_81186137 | 0.81 |
ENSMUST00000167481.8
|
Hmg20b
|
high mobility group 20B |
| chr10_-_81186025 | 0.74 |
ENSMUST00000122993.8
|
Hmg20b
|
high mobility group 20B |
| chr7_+_79675727 | 0.73 |
ENSMUST00000049680.10
|
Zfp710
|
zinc finger protein 710 |
| chr10_-_81186222 | 0.71 |
ENSMUST00000020454.11
ENSMUST00000105324.9 ENSMUST00000154609.3 ENSMUST00000105323.8 |
Hmg20b
|
high mobility group 20B |
| chr8_-_106434565 | 0.69 |
ENSMUST00000013299.11
|
Enkd1
|
enkurin domain containing 1 |
| chr11_+_85723377 | 0.67 |
ENSMUST00000000095.7
|
Tbx2
|
T-box 2 |
| chr18_+_7869066 | 0.67 |
ENSMUST00000171486.8
ENSMUST00000170932.8 ENSMUST00000167020.8 |
Wac
|
WW domain containing adaptor with coiled-coil |
| chr6_+_85164420 | 0.67 |
ENSMUST00000045942.9
|
Emx1
|
empty spiracles homeobox 1 |
| chr17_+_23898223 | 0.65 |
ENSMUST00000024699.4
ENSMUST00000232719.2 |
Cldn6
|
claudin 6 |
| chr2_-_131194754 | 0.52 |
ENSMUST00000059372.11
|
Rnf24
|
ring finger protein 24 |
| chr11_+_105072619 | 0.52 |
ENSMUST00000092537.10
ENSMUST00000015107.13 ENSMUST00000145048.8 |
Tlk2
|
tousled-like kinase 2 (Arabidopsis) |
| chr7_-_133826817 | 0.42 |
ENSMUST00000067680.11
|
Adam12
|
a disintegrin and metallopeptidase domain 12 (meltrin alpha) |
| chr11_+_3152874 | 0.42 |
ENSMUST00000179770.8
ENSMUST00000110048.8 |
Eif4enif1
|
eukaryotic translation initiation factor 4E nuclear import factor 1 |
| chr11_-_62348115 | 0.41 |
ENSMUST00000069456.11
ENSMUST00000018645.13 |
Ncor1
|
nuclear receptor co-repressor 1 |
| chr8_+_106434901 | 0.39 |
ENSMUST00000013302.7
ENSMUST00000211852.2 |
4933405L10Rik
|
RIKEN cDNA 4933405L10 gene |
| chr5_-_137530214 | 0.38 |
ENSMUST00000140139.2
|
Gnb2
|
guanine nucleotide binding protein (G protein), beta 2 |
| chr2_+_129040677 | 0.37 |
ENSMUST00000028880.10
|
Slc20a1
|
solute carrier family 20, member 1 |
| chr7_-_30560989 | 0.35 |
ENSMUST00000052700.6
|
Ffar1
|
free fatty acid receptor 1 |
| chr17_+_88837540 | 0.34 |
ENSMUST00000038551.8
|
Ppp1r21
|
protein phosphatase 1, regulatory subunit 21 |
| chr5_-_108134869 | 0.31 |
ENSMUST00000145239.2
ENSMUST00000031198.11 |
Dipk1a
|
divergent protein kinase domain 1A |
| chrX_-_47123719 | 0.24 |
ENSMUST00000039026.8
|
Apln
|
apelin |
| chr6_+_83771985 | 0.21 |
ENSMUST00000113851.8
|
Nagk
|
N-acetylglucosamine kinase |
| chr18_+_7868823 | 0.17 |
ENSMUST00000171042.8
ENSMUST00000166378.8 ENSMUST00000074919.11 |
Wac
|
WW domain containing adaptor with coiled-coil |
| chr4_-_148215135 | 0.16 |
ENSMUST00000030862.5
|
Draxin
|
dorsal inhibitory axon guidance protein |
| chr11_-_62348599 | 0.14 |
ENSMUST00000127471.9
|
Ncor1
|
nuclear receptor co-repressor 1 |
| chr2_-_52314837 | 0.10 |
ENSMUST00000036541.8
|
Arl5a
|
ADP-ribosylation factor-like 5A |
| chr8_-_85807308 | 0.10 |
ENSMUST00000093357.12
|
Wdr83
|
WD repeat domain containing 83 |
| chr5_-_31350449 | 0.09 |
ENSMUST00000166769.8
|
Eif2b4
|
eukaryotic translation initiation factor 2B, subunit 4 delta |
| chr16_+_56942050 | 0.07 |
ENSMUST00000166897.3
|
Tomm70a
|
translocase of outer mitochondrial membrane 70A |
| chr15_-_73295074 | 0.06 |
ENSMUST00000226893.2
|
Ptk2
|
PTK2 protein tyrosine kinase 2 |
| chr16_+_8647959 | 0.06 |
ENSMUST00000023150.7
|
1810013L24Rik
|
RIKEN cDNA 1810013L24 gene |
| chr8_-_85807281 | 0.02 |
ENSMUST00000152785.8
|
Wdr83
|
WD repeat domain containing 83 |
| chr5_-_31350352 | 0.02 |
ENSMUST00000202758.4
ENSMUST00000114603.8 |
Eif2b4
|
eukaryotic translation initiation factor 2B, subunit 4 delta |
| chr19_-_46028060 | 0.01 |
ENSMUST00000056931.14
|
Ldb1
|
LIM domain binding 1 |
| chr9_-_108888779 | 0.01 |
ENSMUST00000061973.5
|
Trex1
|
three prime repair exonuclease 1 |
| chr11_+_19874354 | 0.00 |
ENSMUST00000093299.13
|
Spred2
|
sprouty-related EVH1 domain containing 2 |
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.6 | 2.5 | GO:0070103 | regulation of interleukin-6-mediated signaling pathway(GO:0070103) |
| 0.6 | 1.7 | GO:0036275 | response to 5-fluorouracil(GO:0036275) |
| 0.4 | 1.2 | GO:0060448 | dichotomous subdivision of terminal units involved in lung branching(GO:0060448) |
| 0.4 | 2.1 | GO:0045994 | regulation of translational initiation by iron(GO:0006447) positive regulation of translational initiation by iron(GO:0045994) |
| 0.4 | 4.4 | GO:0052697 | flavonoid glucuronidation(GO:0052696) xenobiotic glucuronidation(GO:0052697) |
| 0.4 | 1.1 | GO:0061010 | gall bladder development(GO:0061010) |
| 0.3 | 1.0 | GO:0048007 | antigen processing and presentation of lipid antigen via MHC class Ib(GO:0048003) antigen processing and presentation, exogenous lipid antigen via MHC class Ib(GO:0048007) |
| 0.3 | 1.5 | GO:2000048 | negative regulation of cell-cell adhesion mediated by cadherin(GO:2000048) |
| 0.3 | 2.6 | GO:0060214 | endocardium formation(GO:0060214) |
| 0.2 | 0.8 | GO:0071894 | histone H2B conserved C-terminal lysine ubiquitination(GO:0071894) |
| 0.2 | 1.4 | GO:0016480 | negative regulation of transcription from RNA polymerase III promoter(GO:0016480) |
| 0.2 | 0.9 | GO:0015015 | heparan sulfate proteoglycan biosynthetic process, enzymatic modification(GO:0015015) |
| 0.2 | 1.5 | GO:0002568 | somatic diversification of T cell receptor genes(GO:0002568) somatic recombination of T cell receptor gene segments(GO:0002681) T cell receptor V(D)J recombination(GO:0033153) |
| 0.1 | 0.9 | GO:0007296 | vitellogenesis(GO:0007296) |
| 0.1 | 0.5 | GO:0072362 | regulation of glycolytic process by negative regulation of transcription from RNA polymerase II promoter(GO:0072362) |
| 0.1 | 0.8 | GO:0042256 | mature ribosome assembly(GO:0042256) |
| 0.1 | 2.5 | GO:0060732 | positive regulation of inositol phosphate biosynthetic process(GO:0060732) |
| 0.1 | 0.7 | GO:0060596 | mammary placode formation(GO:0060596) |
| 0.1 | 2.3 | GO:0033234 | negative regulation of protein sumoylation(GO:0033234) |
| 0.1 | 1.3 | GO:0001842 | neural fold formation(GO:0001842) |
| 0.1 | 1.3 | GO:0051574 | positive regulation of histone H3-K9 methylation(GO:0051574) |
| 0.1 | 0.2 | GO:1904020 | regulation of G-protein coupled receptor internalization(GO:1904020) |
| 0.1 | 0.9 | GO:0070914 | UV-damage excision repair(GO:0070914) |
| 0.1 | 1.5 | GO:0050961 | detection of temperature stimulus involved in sensory perception(GO:0050961) detection of temperature stimulus involved in sensory perception of pain(GO:0050965) |
| 0.1 | 1.6 | GO:0045603 | positive regulation of endothelial cell differentiation(GO:0045603) |
| 0.0 | 0.7 | GO:0060019 | radial glial cell differentiation(GO:0060019) |
| 0.0 | 1.1 | GO:0070886 | positive regulation of calcineurin-NFAT signaling cascade(GO:0070886) |
| 0.0 | 0.5 | GO:0001672 | regulation of chromatin assembly or disassembly(GO:0001672) |
| 0.0 | 2.5 | GO:0060325 | face morphogenesis(GO:0060325) |
| 0.0 | 0.2 | GO:0019262 | N-acetylneuraminate catabolic process(GO:0019262) |
| 0.0 | 1.2 | GO:0070207 | protein homotrimerization(GO:0070207) |
| 0.0 | 1.9 | GO:0006333 | chromatin assembly or disassembly(GO:0006333) |
| 0.0 | 0.4 | GO:0006817 | phosphate ion transport(GO:0006817) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.6 | 1.7 | GO:0097132 | cyclin D2-CDK6 complex(GO:0097132) |
| 0.6 | 2.3 | GO:0002111 | BRCA2-BRAF35 complex(GO:0002111) |
| 0.4 | 1.3 | GO:0090537 | CERF complex(GO:0090537) |
| 0.4 | 1.5 | GO:0016035 | zeta DNA polymerase complex(GO:0016035) |
| 0.1 | 0.9 | GO:0033588 | Elongator holoenzyme complex(GO:0033588) |
| 0.1 | 0.9 | GO:0031465 | Cul4B-RING E3 ubiquitin ligase complex(GO:0031465) |
| 0.1 | 0.8 | GO:0008250 | oligosaccharyltransferase complex(GO:0008250) |
| 0.1 | 1.9 | GO:0090545 | NuRD complex(GO:0016581) CHD-type complex(GO:0090545) |
| 0.1 | 1.1 | GO:0031254 | uropod(GO:0001931) cell trailing edge(GO:0031254) |
| 0.1 | 1.0 | GO:0030123 | AP-3 adaptor complex(GO:0030123) |
| 0.1 | 1.3 | GO:0035098 | ESC/E(Z) complex(GO:0035098) |
| 0.0 | 0.8 | GO:0005852 | eukaryotic translation initiation factor 3 complex(GO:0005852) |
| 0.0 | 0.5 | GO:0016580 | Sin3 complex(GO:0016580) |
| 0.0 | 1.5 | GO:0005719 | nuclear euchromatin(GO:0005719) |
| 0.0 | 2.4 | GO:0005844 | polysome(GO:0005844) |
| 0.0 | 1.2 | GO:0008305 | integrin complex(GO:0008305) |
| 0.0 | 2.5 | GO:0017053 | transcriptional repressor complex(GO:0017053) |
| 0.0 | 0.7 | GO:0016327 | apicolateral plasma membrane(GO:0016327) |
| 0.0 | 2.5 | GO:0031526 | brush border membrane(GO:0031526) |
| 0.0 | 0.1 | GO:0005851 | eukaryotic translation initiation factor 2B complex(GO:0005851) |
| 0.0 | 1.1 | GO:0016328 | lateral plasma membrane(GO:0016328) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 1.7 | GO:0098770 | FBXO family protein binding(GO:0098770) |
| 0.4 | 1.2 | GO:0098640 | integrin binding involved in cell-matrix adhesion(GO:0098640) |
| 0.4 | 2.5 | GO:0004991 | parathyroid hormone receptor activity(GO:0004991) |
| 0.3 | 2.1 | GO:0070644 | vitamin D response element binding(GO:0070644) |
| 0.2 | 0.9 | GO:0008467 | [heparan sulfate]-glucosamine 3-sulfotransferase 1 activity(GO:0008467) |
| 0.1 | 1.1 | GO:0016308 | 1-phosphatidylinositol-4-phosphate 5-kinase activity(GO:0016308) |
| 0.1 | 1.1 | GO:0004716 | receptor signaling protein tyrosine kinase activity(GO:0004716) |
| 0.1 | 1.9 | GO:0008190 | eukaryotic initiation factor 4E binding(GO:0008190) |
| 0.1 | 4.4 | GO:0015020 | glucuronosyltransferase activity(GO:0015020) |
| 0.1 | 2.3 | GO:0000400 | four-way junction DNA binding(GO:0000400) |
| 0.1 | 0.8 | GO:0004576 | oligosaccharyl transferase activity(GO:0004576) dolichyl-diphosphooligosaccharide-protein glycotransferase activity(GO:0004579) |
| 0.1 | 2.4 | GO:0008266 | poly(U) RNA binding(GO:0008266) |
| 0.1 | 0.4 | GO:0015319 | sodium:inorganic phosphate symporter activity(GO:0015319) |
| 0.1 | 0.2 | GO:0045127 | N-acetylglucosamine kinase activity(GO:0045127) |
| 0.1 | 0.5 | GO:0035033 | histone deacetylase regulator activity(GO:0035033) |
| 0.0 | 1.1 | GO:0008432 | JUN kinase binding(GO:0008432) |
| 0.0 | 1.3 | GO:0032452 | histone demethylase activity(GO:0032452) |
| 0.0 | 1.0 | GO:0030742 | GTP-dependent protein binding(GO:0030742) |
| 0.0 | 0.8 | GO:0000993 | RNA polymerase II core binding(GO:0000993) |
| 0.0 | 0.7 | GO:0001618 | virus receptor activity(GO:0001618) |
| 0.0 | 1.6 | GO:0070888 | E-box binding(GO:0070888) |
| 0.0 | 6.2 | GO:0001227 | transcriptional repressor activity, RNA polymerase II transcription regulatory region sequence-specific binding(GO:0001227) |
| 0.0 | 1.4 | GO:0051018 | protein kinase A binding(GO:0051018) |
| 0.0 | 0.3 | GO:0045125 | bioactive lipid receptor activity(GO:0045125) |
| 0.0 | 0.9 | GO:0003684 | damaged DNA binding(GO:0003684) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 3.2 | PID A6B1 A6B4 INTEGRIN PATHWAY | a6b1 and a6b4 Integrin signaling |
| 0.0 | 1.2 | PID WNT SIGNALING PATHWAY | Wnt signaling network |
| 0.0 | 1.1 | PID WNT CANONICAL PATHWAY | Canonical Wnt signaling pathway |
| 0.0 | 1.7 | PID IL2 STAT5 PATHWAY | IL2 signaling events mediated by STAT5 |
| 0.0 | 2.3 | PID MYC REPRESS PATHWAY | Validated targets of C-MYC transcriptional repression |
| 0.0 | 2.4 | PID HDAC CLASSI PATHWAY | Signaling events mediated by HDAC Class I |
| 0.0 | 1.5 | WNT SIGNALING | Genes related to Wnt-mediated signal transduction |
| 0.0 | 0.4 | PID INTEGRIN A9B1 PATHWAY | Alpha9 beta1 integrin signaling events |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 4.4 | REACTOME GLUCURONIDATION | Genes involved in Glucuronidation |
| 0.1 | 2.6 | REACTOME CIRCADIAN REPRESSION OF EXPRESSION BY REV ERBA | Genes involved in Circadian Repression of Expression by REV-ERBA |
| 0.0 | 1.1 | REACTOME DOWNREGULATION OF ERBB2 ERBB3 SIGNALING | Genes involved in Downregulation of ERBB2:ERBB3 signaling |
| 0.0 | 1.7 | REACTOME G1 PHASE | Genes involved in G1 Phase |
| 0.0 | 0.9 | REACTOME HS GAG BIOSYNTHESIS | Genes involved in HS-GAG biosynthesis |
| 0.0 | 0.7 | REACTOME TIGHT JUNCTION INTERACTIONS | Genes involved in Tight junction interactions |
| 0.0 | 0.3 | REACTOME SYNTHESIS SECRETION AND INACTIVATION OF GIP | Genes involved in Synthesis, Secretion, and Inactivation of Glucose-dependent Insulinotropic Polypeptide (GIP) |
| 0.0 | 2.1 | REACTOME CLASS B 2 SECRETIN FAMILY RECEPTORS | Genes involved in Class B/2 (Secretin family receptors) |
| 0.0 | 2.3 | REACTOME FACTORS INVOLVED IN MEGAKARYOCYTE DEVELOPMENT AND PLATELET PRODUCTION | Genes involved in Factors involved in megakaryocyte development and platelet production |
| 0.0 | 1.0 | REACTOME GOLGI ASSOCIATED VESICLE BIOGENESIS | Genes involved in Golgi Associated Vesicle Biogenesis |
| 0.0 | 1.5 | REACTOME DNA REPAIR | Genes involved in DNA Repair |