Project
PRJNA375882: Comprehensive Mouse Transcriptomic BodyMap
Navigation
Downloads
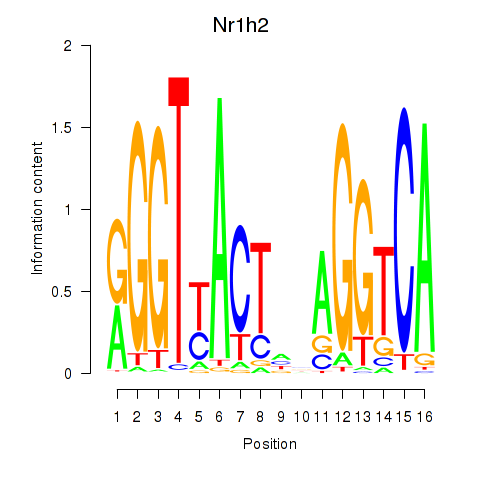
Results for Nr1h2
Z-value: 1.00
Transcription factors associated with Nr1h2
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Nr1h2
|
ENSMUSG00000060601.14 | Nr1h2 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Nr1h2 | mm39_v1_chr7_-_44203319_44203392 | -0.59 | 3.6e-08 | Click! |
Activity profile of Nr1h2 motif
Sorted Z-values of Nr1h2 motif
Network of associatons between targets according to the STRING database.
First level regulatory network of Nr1h2
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr3_+_146302832 | 9.20 |
ENSMUST00000029837.14
ENSMUST00000147409.2 ENSMUST00000121133.2 |
Uox
|
urate oxidase |
| chr17_+_25023263 | 7.79 |
ENSMUST00000234372.2
ENSMUST00000024972.7 |
Meiob
|
meiosis specific with OB domains |
| chr17_-_84154173 | 7.34 |
ENSMUST00000000687.9
|
Haao
|
3-hydroxyanthranilate 3,4-dioxygenase |
| chr1_-_139487951 | 7.04 |
ENSMUST00000023965.8
|
Cfhr1
|
complement factor H-related 1 |
| chr17_-_84154196 | 6.86 |
ENSMUST00000234214.2
|
Haao
|
3-hydroxyanthranilate 3,4-dioxygenase |
| chr18_-_9619460 | 6.36 |
ENSMUST00000234003.2
ENSMUST00000062769.7 |
Cetn1
|
centrin 1 |
| chr4_-_63072367 | 6.34 |
ENSMUST00000030041.5
|
Ambp
|
alpha 1 microglobulin/bikunin precursor |
| chr7_-_97066937 | 6.32 |
ENSMUST00000043077.8
|
Thrsp
|
thyroid hormone responsive |
| chr2_+_102488985 | 5.56 |
ENSMUST00000080210.10
|
Slc1a2
|
solute carrier family 1 (glial high affinity glutamate transporter), member 2 |
| chr3_-_146302343 | 5.38 |
ENSMUST00000029836.9
|
Dnase2b
|
deoxyribonuclease II beta |
| chr3_-_95148909 | 5.23 |
ENSMUST00000090815.6
ENSMUST00000107197.2 |
Gm128
|
predicted gene 128 |
| chr7_-_12732067 | 5.08 |
ENSMUST00000032539.14
ENSMUST00000120903.8 |
Slc27a5
|
solute carrier family 27 (fatty acid transporter), member 5 |
| chr15_+_88484484 | 4.98 |
ENSMUST00000066949.9
|
Zdhhc25
|
zinc finger, DHHC domain containing 25 |
| chr7_-_103778992 | 4.77 |
ENSMUST00000053743.6
|
Ubqln5
|
ubiquilin 5 |
| chr2_+_155224105 | 4.71 |
ENSMUST00000134218.2
|
Trp53inp2
|
transformation related protein 53 inducible nuclear protein 2 |
| chr7_-_12731594 | 4.63 |
ENSMUST00000133977.3
|
Slc27a5
|
solute carrier family 27 (fatty acid transporter), member 5 |
| chr7_-_140367737 | 4.32 |
ENSMUST00000211616.2
ENSMUST00000026553.6 |
Syce1
|
synaptonemal complex central element protein 1 |
| chr12_+_112073261 | 4.11 |
ENSMUST00000223412.2
|
Aspg
|
asparaginase |
| chr4_+_120389415 | 4.05 |
ENSMUST00000062990.4
|
Slfnl1
|
schlafen like 1 |
| chr6_+_54016543 | 4.04 |
ENSMUST00000046856.14
|
Chn2
|
chimerin 2 |
| chr6_+_90439596 | 4.04 |
ENSMUST00000203039.3
|
Klf15
|
Kruppel-like factor 15 |
| chr6_+_90439544 | 4.03 |
ENSMUST00000032174.12
|
Klf15
|
Kruppel-like factor 15 |
| chr16_+_17712061 | 3.97 |
ENSMUST00000046937.4
|
Tssk1
|
testis-specific serine kinase 1 |
| chr5_-_146107531 | 3.94 |
ENSMUST00000174320.2
|
Gm6309
|
predicted gene 6309 |
| chr9_+_110673565 | 3.80 |
ENSMUST00000176403.8
|
Prss46
|
protease, serine 46 |
| chr5_+_144979796 | 3.78 |
ENSMUST00000031624.5
|
1700018F24Rik
|
RIKEN cDNA 1700018F24 gene |
| chr7_+_43600038 | 3.77 |
ENSMUST00000072204.5
|
Klk1b8
|
kallikrein 1-related peptidase b8 |
| chr5_-_144969564 | 3.74 |
ENSMUST00000071421.6
|
Gm4871
|
predicted gene 4871 |
| chr10_-_68377672 | 3.65 |
ENSMUST00000020103.9
|
Cabcoco1
|
ciliary associated calcium binding coiled-coil 1 |
| chr4_-_152122891 | 3.61 |
ENSMUST00000030792.2
|
Tas1r1
|
taste receptor, type 1, member 1 |
| chr2_+_154390808 | 3.45 |
ENSMUST00000045116.11
ENSMUST00000109709.4 |
1700003F12Rik
|
RIKEN cDNA 1700003F12 gene |
| chr5_-_146122114 | 3.45 |
ENSMUST00000073721.7
|
1700001J03Rik
|
RIKEN cDNA 1700001J03 gene |
| chr15_-_79212400 | 3.43 |
ENSMUST00000173163.8
ENSMUST00000047816.15 ENSMUST00000172403.9 ENSMUST00000173632.8 |
Pla2g6
|
phospholipase A2, group VI |
| chr14_-_101437750 | 3.33 |
ENSMUST00000187304.2
|
Prr30
|
proline rich 30 |
| chr2_+_155223728 | 3.31 |
ENSMUST00000043237.14
ENSMUST00000174685.8 |
Trp53inp2
|
transformation related protein 53 inducible nuclear protein 2 |
| chr7_-_44828962 | 3.30 |
ENSMUST00000211004.2
ENSMUST00000179443.3 |
Gfy
|
golgi-associated olfactory signaling regulator |
| chr7_+_114367971 | 3.20 |
ENSMUST00000117543.3
ENSMUST00000151464.2 |
Insc
|
INSC spindle orientation adaptor protein |
| chr4_-_123217391 | 3.13 |
ENSMUST00000102640.2
|
Oxct2a
|
3-oxoacid CoA transferase 2A |
| chr5_-_21850539 | 3.13 |
ENSMUST00000115234.2
|
Fbxl13
|
F-box and leucine-rich repeat protein 13 |
| chr15_-_79212323 | 3.03 |
ENSMUST00000166977.9
|
Pla2g6
|
phospholipase A2, group VI |
| chr7_-_42097503 | 3.00 |
ENSMUST00000032648.5
|
4933421I07Rik
|
RIKEN cDNA 4933421I07 gene |
| chrX_-_66965036 | 2.98 |
ENSMUST00000026325.2
|
4933436I01Rik
|
RIKEN cDNA 4933436I01 gene |
| chr15_-_89294434 | 2.88 |
ENSMUST00000109314.9
|
Syce3
|
synaptonemal complex central element protein 3 |
| chrX_-_134985958 | 2.87 |
ENSMUST00000138878.2
ENSMUST00000080929.13 |
Nxf3
|
nuclear RNA export factor 3 |
| chr3_-_137892434 | 2.86 |
ENSMUST00000012186.9
ENSMUST00000199293.2 |
4930579F01Rik
|
RIKEN cDNA 4930579F01 gene |
| chr11_+_84070593 | 2.86 |
ENSMUST00000137500.9
ENSMUST00000130012.9 |
Acaca
|
acetyl-Coenzyme A carboxylase alpha |
| chr17_-_56607286 | 2.80 |
ENSMUST00000097303.3
|
Arrdc5
|
arrestin domain containing 5 |
| chr5_-_21850579 | 2.78 |
ENSMUST00000051358.11
|
Fbxl13
|
F-box and leucine-rich repeat protein 13 |
| chr5_+_146450933 | 2.74 |
ENSMUST00000200228.5
ENSMUST00000036715.16 ENSMUST00000077133.7 |
Gm3402
|
predicted gene 3402 |
| chr8_-_106863423 | 2.73 |
ENSMUST00000146940.2
|
Esrp2
|
epithelial splicing regulatory protein 2 |
| chr5_+_146428173 | 2.72 |
ENSMUST00000110611.8
ENSMUST00000198912.2 |
Gm6370
|
predicted gene 6370 |
| chr5_-_146097347 | 2.68 |
ENSMUST00000199463.2
|
Gm5565
|
predicted gene 5565 |
| chr14_+_40794817 | 2.57 |
ENSMUST00000189865.7
|
Dydc1
|
DPY30 domain containing 1 |
| chr8_-_106863521 | 2.56 |
ENSMUST00000115979.9
|
Esrp2
|
epithelial splicing regulatory protein 2 |
| chr11_+_58854616 | 2.54 |
ENSMUST00000075141.7
|
Trim17
|
tripartite motif-containing 17 |
| chr2_-_152438356 | 2.52 |
ENSMUST00000109834.3
|
Defb45
|
defensin beta 45 |
| chr5_+_146439209 | 2.49 |
ENSMUST00000110598.3
|
4930449I24Rik
|
RIKEN cDNA 4930449I24 gene |
| chr17_-_56607250 | 2.48 |
ENSMUST00000233911.2
|
Arrdc5
|
arrestin domain containing 5 |
| chr11_+_84070678 | 2.48 |
ENSMUST00000136463.9
|
Acaca
|
acetyl-Coenzyme A carboxylase alpha |
| chr9_-_122673080 | 2.47 |
ENSMUST00000203176.3
ENSMUST00000203656.3 ENSMUST00000204619.2 |
Gm35549
|
predicted gene, 35549 |
| chr14_+_51790247 | 2.44 |
ENSMUST00000096170.5
|
Gm5622
|
predicted gene 5622 |
| chr2_-_65068917 | 2.42 |
ENSMUST00000090896.10
ENSMUST00000155082.2 |
Cobll1
|
Cobl-like 1 |
| chr6_+_124639990 | 2.39 |
ENSMUST00000004381.14
|
Lpcat3
|
lysophosphatidylcholine acyltransferase 3 |
| chr6_-_119521243 | 2.33 |
ENSMUST00000119369.2
ENSMUST00000178696.8 |
Wnt5b
|
wingless-type MMTV integration site family, member 5B |
| chr19_-_5452521 | 2.31 |
ENSMUST00000235569.2
|
Tsga10ip
|
testis specific 10 interacting protein |
| chr11_+_54988866 | 2.31 |
ENSMUST00000000608.8
|
Gm2a
|
GM2 ganglioside activator protein |
| chr5_+_146462611 | 2.28 |
ENSMUST00000110596.2
|
Gm3404
|
predicted gene 3404 |
| chr1_+_190769010 | 2.24 |
ENSMUST00000077889.8
|
Spata45
|
spermatogenesis associated 45 |
| chr7_+_49408847 | 2.23 |
ENSMUST00000085272.7
ENSMUST00000207895.2 |
Htatip2
|
HIV-1 Tat interactive protein 2 |
| chr4_+_119280002 | 2.18 |
ENSMUST00000094819.5
|
Zmynd12
|
zinc finger, MYND domain containing 12 |
| chr2_+_158148413 | 2.17 |
ENSMUST00000109491.8
ENSMUST00000016168.9 |
Lbp
|
lipopolysaccharide binding protein |
| chr2_-_65068960 | 2.15 |
ENSMUST00000112429.9
ENSMUST00000102726.8 ENSMUST00000112430.8 |
Cobll1
|
Cobl-like 1 |
| chr9_-_63306497 | 2.14 |
ENSMUST00000168665.3
|
2300009A05Rik
|
RIKEN cDNA 2300009A05 gene |
| chr5_+_107479023 | 2.12 |
ENSMUST00000031215.15
ENSMUST00000112677.10 |
Brdt
|
bromodomain, testis-specific |
| chr5_-_138562933 | 2.05 |
ENSMUST00000031501.2
|
1700123K08Rik
|
RIKEN cDNA 1700123K08 gene |
| chrX_-_73689241 | 1.90 |
ENSMUST00000114119.2
|
Pwwp4c
|
PWWP domain containing 4C |
| chr5_+_146418775 | 1.84 |
ENSMUST00000179032.3
|
Gm6408
|
predicted gene 6408 |
| chr7_-_45375205 | 1.82 |
ENSMUST00000094424.7
|
Spaca4
|
sperm acrosome associated 4 |
| chr4_-_115875055 | 1.79 |
ENSMUST00000049095.6
|
Faah
|
fatty acid amide hydrolase |
| chr14_+_47069667 | 1.78 |
ENSMUST00000140114.3
ENSMUST00000133989.8 |
Cgrrf1
|
cell growth regulator with ring finger domain 1 |
| chr15_+_82439273 | 1.77 |
ENSMUST00000229103.2
ENSMUST00000068861.8 ENSMUST00000229904.2 |
Cyp2d12
|
cytochrome P450, family 2, subfamily d, polypeptide 12 |
| chr1_+_57416752 | 1.75 |
ENSMUST00000042734.3
|
1700066M21Rik
|
RIKEN cDNA 1700066M21 gene |
| chr11_-_94867153 | 1.72 |
ENSMUST00000103162.8
ENSMUST00000166320.8 |
Sgca
|
sarcoglycan, alpha (dystrophin-associated glycoprotein) |
| chr8_-_71085097 | 1.70 |
ENSMUST00000110103.2
|
Gdf15
|
growth differentiation factor 15 |
| chr17_-_65847731 | 1.69 |
ENSMUST00000233117.2
ENSMUST00000062161.7 |
Tmem232
|
transmembrane protein 232 |
| chr17_-_65847777 | 1.67 |
ENSMUST00000086722.10
|
Tmem232
|
transmembrane protein 232 |
| chr14_-_20133246 | 1.64 |
ENSMUST00000059666.6
|
Saysd1
|
SAYSVFN motif domain containing 1 |
| chr13_+_23879775 | 1.51 |
ENSMUST00000041052.5
|
H1f6
|
H1.6 linker histone, cluster member |
| chr11_-_99494134 | 1.48 |
ENSMUST00000072306.4
|
Gm11938
|
predicted gene 11938 |
| chr16_+_13758494 | 1.43 |
ENSMUST00000141971.8
ENSMUST00000124947.8 ENSMUST00000023360.14 ENSMUST00000143697.8 |
Mpv17l
|
Mpv17 transgene, kidney disease mutant-like |
| chr5_+_117495337 | 1.41 |
ENSMUST00000031309.16
|
Wsb2
|
WD repeat and SOCS box-containing 2 |
| chr8_+_104837939 | 1.38 |
ENSMUST00000209911.2
|
Cdh5
|
cadherin 5 |
| chr7_+_28937746 | 1.37 |
ENSMUST00000108238.8
ENSMUST00000032809.10 |
Yif1b
|
Yip1 interacting factor homolog B (S. cerevisiae) |
| chr17_-_36207965 | 1.37 |
ENSMUST00000150056.2
ENSMUST00000156817.2 ENSMUST00000146451.8 ENSMUST00000148482.8 |
2310061I04Rik
|
RIKEN cDNA 2310061I04 gene |
| chr1_+_135980639 | 1.36 |
ENSMUST00000112064.8
|
Cacna1s
|
calcium channel, voltage-dependent, L type, alpha 1S subunit |
| chr15_+_74586682 | 1.36 |
ENSMUST00000023265.5
|
Psca
|
prostate stem cell antigen |
| chr1_+_66214431 | 1.36 |
ENSMUST00000156636.9
|
Map2
|
microtubule-associated protein 2 |
| chr7_-_4633186 | 1.35 |
ENSMUST00000205360.2
ENSMUST00000206610.2 |
Tmem86b
|
transmembrane protein 86B |
| chr3_-_51316347 | 1.33 |
ENSMUST00000193279.2
ENSMUST00000038108.12 |
Ndufc1
|
NADH:ubiquinone oxidoreductase subunit C1 |
| chr13_+_102830104 | 1.31 |
ENSMUST00000172138.2
|
Cd180
|
CD180 antigen |
| chr14_-_72840373 | 1.29 |
ENSMUST00000162825.8
|
Fndc3a
|
fibronectin type III domain containing 3A |
| chr7_+_28937859 | 1.27 |
ENSMUST00000108237.2
|
Yif1b
|
Yip1 interacting factor homolog B (S. cerevisiae) |
| chr3_+_59637203 | 1.26 |
ENSMUST00000168156.3
|
Aadacl2fm2
|
AADACL2 family member 2 |
| chr16_-_37205302 | 1.23 |
ENSMUST00000114781.8
ENSMUST00000114780.8 |
Stxbp5l
|
syntaxin binding protein 5-like |
| chr16_-_37205277 | 1.20 |
ENSMUST00000114787.8
ENSMUST00000114782.8 ENSMUST00000114775.8 |
Stxbp5l
|
syntaxin binding protein 5-like |
| chr2_-_32977182 | 1.19 |
ENSMUST00000102810.10
|
Garnl3
|
GTPase activating RANGAP domain-like 3 |
| chr14_+_15495955 | 1.18 |
ENSMUST00000166618.2
|
Gm6356
|
predicted gene 6356 |
| chr11_-_60101235 | 1.16 |
ENSMUST00000144942.2
|
Srebf1
|
sterol regulatory element binding transcription factor 1 |
| chr15_+_76238632 | 1.16 |
ENSMUST00000208833.3
|
Gm35339
|
predicted gene, 35339 |
| chr1_+_135980488 | 1.12 |
ENSMUST00000160641.8
|
Cacna1s
|
calcium channel, voltage-dependent, L type, alpha 1S subunit |
| chr2_+_153760311 | 1.11 |
ENSMUST00000109760.2
|
Bpifb3
|
BPI fold containing family B, member 3 |
| chr2_+_30156733 | 1.11 |
ENSMUST00000113645.8
ENSMUST00000133877.8 ENSMUST00000139719.8 ENSMUST00000113643.8 ENSMUST00000150695.8 |
Phyhd1
|
phytanoyl-CoA dioxygenase domain containing 1 |
| chr7_+_27879650 | 1.11 |
ENSMUST00000172467.8
|
Dyrk1b
|
dual-specificity tyrosine-(Y)-phosphorylation regulated kinase 1b |
| chr1_+_135980508 | 1.11 |
ENSMUST00000112068.10
|
Cacna1s
|
calcium channel, voltage-dependent, L type, alpha 1S subunit |
| chr5_+_123153072 | 1.10 |
ENSMUST00000051016.5
ENSMUST00000121652.8 |
Orai1
|
ORAI calcium release-activated calcium modulator 1 |
| chr15_+_78761360 | 1.03 |
ENSMUST00000041587.8
|
Gga1
|
golgi associated, gamma adaptin ear containing, ARF binding protein 1 |
| chr16_+_16688692 | 1.02 |
ENSMUST00000232547.2
|
Top3b
|
topoisomerase (DNA) III beta |
| chr12_-_110945376 | 1.01 |
ENSMUST00000142012.2
|
Ankrd9
|
ankyrin repeat domain 9 |
| chrX_-_52610946 | 1.01 |
ENSMUST00000123034.3
|
4933416I08Rik
|
RIKEN cDNA 4933416I08 gene |
| chr4_-_137512682 | 0.98 |
ENSMUST00000133473.2
|
Alpl
|
alkaline phosphatase, liver/bone/kidney |
| chr10_+_129375031 | 0.98 |
ENSMUST00000215436.2
|
Olfr792
|
olfactory receptor 792 |
| chr2_-_160714749 | 0.98 |
ENSMUST00000176141.8
|
Zhx3
|
zinc fingers and homeoboxes 3 |
| chr13_-_100688949 | 0.96 |
ENSMUST00000159515.2
ENSMUST00000160859.8 ENSMUST00000069756.11 |
Ocln
|
occludin |
| chr12_-_21336285 | 0.88 |
ENSMUST00000076260.12
|
Itgb1bp1
|
integrin beta 1 binding protein 1 |
| chr12_-_21336098 | 0.85 |
ENSMUST00000173729.8
|
Itgb1bp1
|
integrin beta 1 binding protein 1 |
| chr6_+_54406588 | 0.84 |
ENSMUST00000132855.8
ENSMUST00000126637.8 |
Wipf3
|
WAS/WASL interacting protein family, member 3 |
| chr4_-_53159885 | 0.83 |
ENSMUST00000030010.4
|
Abca1
|
ATP-binding cassette, sub-family A (ABC1), member 1 |
| chrX_-_153709111 | 0.83 |
ENSMUST00000076986.4
|
Magea6
|
MAGE family member A6 |
| chr7_+_28937898 | 0.82 |
ENSMUST00000138128.3
ENSMUST00000142519.3 |
Yif1b
|
Yip1 interacting factor homolog B (S. cerevisiae) |
| chr6_-_90201420 | 0.82 |
ENSMUST00000076086.3
|
Vmn1r53
|
vomeronasal 1 receptor 53 |
| chr9_-_35111172 | 0.81 |
ENSMUST00000176021.8
ENSMUST00000176531.8 ENSMUST00000176685.8 ENSMUST00000177129.8 |
Tirap
|
toll-interleukin 1 receptor (TIR) domain-containing adaptor protein |
| chr16_-_17711950 | 0.79 |
ENSMUST00000155943.9
|
Dgcr2
|
DiGeorge syndrome critical region gene 2 |
| chr12_-_110945415 | 0.77 |
ENSMUST00000135131.2
ENSMUST00000043459.13 ENSMUST00000128353.8 |
Ankrd9
|
ankyrin repeat domain 9 |
| chr2_+_30156523 | 0.77 |
ENSMUST00000091132.13
|
Phyhd1
|
phytanoyl-CoA dioxygenase domain containing 1 |
| chr6_-_42524521 | 0.77 |
ENSMUST00000217978.2
|
Olfr455
|
olfactory receptor 455 |
| chr11_-_101010640 | 0.74 |
ENSMUST00000107295.10
|
Retreg3
|
reticulophagy regulator family member 3 |
| chr7_-_46902575 | 0.74 |
ENSMUST00000179005.3
|
Mrgpra9
|
MAS-related GPR, member A9 |
| chr12_-_21336176 | 0.71 |
ENSMUST00000172834.2
ENSMUST00000232072.2 |
Itgb1bp1
|
integrin beta 1 binding protein 1 |
| chr13_+_112454981 | 0.71 |
ENSMUST00000223871.2
|
Ankrd55
|
ankyrin repeat domain 55 |
| chr7_-_46902594 | 0.68 |
ENSMUST00000098436.4
|
Mrgpra9
|
MAS-related GPR, member A9 |
| chr11_+_101010764 | 0.66 |
ENSMUST00000043680.9
|
Tubg1
|
tubulin, gamma 1 |
| chr2_-_86940289 | 0.66 |
ENSMUST00000215828.3
|
Olfr259
|
olfactory receptor 259 |
| chr7_+_37885660 | 0.65 |
ENSMUST00000179503.5
|
1600014C10Rik
|
RIKEN cDNA 1600014C10 gene |
| chr17_+_38082190 | 0.64 |
ENSMUST00000217119.2
|
Olfr122
|
olfactory receptor 122 |
| chr17_+_49735386 | 0.62 |
ENSMUST00000165390.9
ENSMUST00000024797.16 |
Mocs1
|
molybdenum cofactor synthesis 1 |
| chr5_+_130248547 | 0.62 |
ENSMUST00000202305.4
ENSMUST00000065329.13 ENSMUST00000200802.4 |
Tmem248
|
transmembrane protein 248 |
| chr1_+_31215482 | 0.62 |
ENSMUST00000062560.14
|
Lgsn
|
lengsin, lens protein with glutamine synthetase domain |
| chr13_+_111391544 | 0.61 |
ENSMUST00000054716.4
|
Actbl2
|
actin, beta-like 2 |
| chr3_-_90421557 | 0.61 |
ENSMUST00000107340.2
ENSMUST00000060738.9 |
S100a1
|
S100 calcium binding protein A1 |
| chr10_+_61253751 | 0.59 |
ENSMUST00000049339.7
|
Nodal
|
nodal |
| chr7_-_27146024 | 0.59 |
ENSMUST00000011895.14
|
Sptbn4
|
spectrin beta, non-erythrocytic 4 |
| chr15_+_85716503 | 0.58 |
ENSMUST00000146088.8
|
Ttc38
|
tetratricopeptide repeat domain 38 |
| chr8_+_55024446 | 0.56 |
ENSMUST00000239166.2
ENSMUST00000239106.2 ENSMUST00000239152.2 |
Asb5
|
ankyrin repeat and SOCs box-containing 5 |
| chr17_+_49735413 | 0.54 |
ENSMUST00000173033.8
|
Mocs1
|
molybdenum cofactor synthesis 1 |
| chr1_+_131526977 | 0.54 |
ENSMUST00000027690.7
|
Avpr1b
|
arginine vasopressin receptor 1B |
| chr13_-_12476313 | 0.51 |
ENSMUST00000143693.8
ENSMUST00000144283.2 ENSMUST00000099820.10 ENSMUST00000135166.8 |
Lgals8
|
lectin, galactose binding, soluble 8 |
| chr7_-_70009669 | 0.51 |
ENSMUST00000208081.2
|
Nr2f2
|
nuclear receptor subfamily 2, group F, member 2 |
| chr6_-_69969961 | 0.51 |
ENSMUST00000200160.5
ENSMUST00000103373.3 |
Igkv18-36
|
immunoglobulin kappa chain variable 18-36 |
| chr5_+_9163244 | 0.50 |
ENSMUST00000198935.2
|
Tmem243
|
transmembrane protein 243, mitochondrial |
| chr7_+_44984723 | 0.50 |
ENSMUST00000211327.2
|
Hrc
|
histidine rich calcium binding protein |
| chr1_+_6285082 | 0.49 |
ENSMUST00000160062.8
|
Rb1cc1
|
RB1-inducible coiled-coil 1 |
| chr2_+_62494622 | 0.47 |
ENSMUST00000028257.3
|
Gca
|
grancalcin |
| chr5_-_136273691 | 0.47 |
ENSMUST00000196447.2
ENSMUST00000196397.5 ENSMUST00000005188.14 |
Sh2b2
|
SH2B adaptor protein 2 |
| chr7_-_85065128 | 0.46 |
ENSMUST00000171213.3
ENSMUST00000233336.2 |
Vmn2r69
|
vomeronasal 2, receptor 69 |
| chr7_+_141988714 | 0.46 |
ENSMUST00000118276.8
ENSMUST00000105976.8 ENSMUST00000097939.9 |
Syt8
|
synaptotagmin VIII |
| chr2_+_154065657 | 0.43 |
ENSMUST00000045959.8
|
Bpifb5
|
BPI fold containing family B, member 5 |
| chr3_-_116217579 | 0.41 |
ENSMUST00000106491.7
ENSMUST00000090464.7 |
Cdc14a
|
CDC14 cell division cycle 14A |
| chrX_-_48877080 | 0.40 |
ENSMUST00000114893.8
|
Igsf1
|
immunoglobulin superfamily, member 1 |
| chr4_-_119279551 | 0.40 |
ENSMUST00000106316.2
ENSMUST00000030385.13 |
Ppcs
|
phosphopantothenoylcysteine synthetase |
| chr2_+_110551976 | 0.40 |
ENSMUST00000090332.5
|
Muc15
|
mucin 15 |
| chr1_+_6284823 | 0.39 |
ENSMUST00000027040.13
|
Rb1cc1
|
RB1-inducible coiled-coil 1 |
| chr1_+_78635591 | 0.36 |
ENSMUST00000134566.8
ENSMUST00000142704.8 ENSMUST00000053760.12 |
Acsl3
Utp14b
|
acyl-CoA synthetase long-chain family member 3 UTP14B small subunit processome component |
| chr1_-_14374842 | 0.35 |
ENSMUST00000188857.7
ENSMUST00000185453.7 |
Eya1
|
EYA transcriptional coactivator and phosphatase 1 |
| chr9_+_106337849 | 0.35 |
ENSMUST00000189099.2
|
Pcbp4
|
poly(rC) binding protein 4 |
| chr16_-_59092995 | 0.35 |
ENSMUST00000216834.2
|
Olfr201
|
olfactory receptor 201 |
| chr5_+_115604321 | 0.33 |
ENSMUST00000145785.8
ENSMUST00000031495.11 ENSMUST00000112071.8 ENSMUST00000125568.2 |
Pla2g1b
|
phospholipase A2, group IB, pancreas |
| chr6_-_24956296 | 0.32 |
ENSMUST00000127247.4
|
Tmem229a
|
transmembrane protein 229A |
| chr8_+_14145848 | 0.30 |
ENSMUST00000152652.8
ENSMUST00000133298.8 |
Dlgap2
|
DLG associated protein 2 |
| chr1_+_78635542 | 0.29 |
ENSMUST00000035779.15
|
Acsl3
|
acyl-CoA synthetase long-chain family member 3 |
| chr1_-_14374794 | 0.26 |
ENSMUST00000190337.7
|
Eya1
|
EYA transcriptional coactivator and phosphatase 1 |
| chr17_-_24388621 | 0.25 |
ENSMUST00000148541.8
ENSMUST00000098862.9 |
Atp6v0c
|
ATPase, H+ transporting, lysosomal V0 subunit C |
| chr17_-_26099832 | 0.24 |
ENSMUST00000176696.3
|
Wfikkn1
|
WAP, FS, Ig, KU, and NTR-containing protein 1 |
| chr9_+_77848556 | 0.24 |
ENSMUST00000134072.2
|
Elovl5
|
ELOVL family member 5, elongation of long chain fatty acids (yeast) |
| chr7_+_139913930 | 0.24 |
ENSMUST00000214858.2
|
Olfr527
|
olfactory receptor 527 |
| chr11_-_120618052 | 0.23 |
ENSMUST00000106148.10
ENSMUST00000026144.5 |
Dcxr
|
dicarbonyl L-xylulose reductase |
| chr5_+_135178509 | 0.20 |
ENSMUST00000153183.8
|
Tbl2
|
transducin (beta)-like 2 |
| chr5_-_124465875 | 0.20 |
ENSMUST00000184951.8
|
Mphosph9
|
M-phase phosphoprotein 9 |
| chr15_+_38662158 | 0.19 |
ENSMUST00000022904.8
ENSMUST00000228820.2 |
Atp6v1c1
|
ATPase, H+ transporting, lysosomal V1 subunit C1 |
| chr16_-_59040130 | 0.17 |
ENSMUST00000214186.2
|
Olfr199
|
olfactory receptor 199 |
| chr3_+_92325386 | 0.14 |
ENSMUST00000029533.3
|
Sprr2j-ps
|
small proline-rich protein 2J, pseudogene |
| chr6_+_85564506 | 0.13 |
ENSMUST00000072018.6
|
Alms1
|
ALMS1, centrosome and basal body associated |
| chr15_-_98215234 | 0.12 |
ENSMUST00000216901.2
|
Olfr285
|
olfactory receptor 285 |
| chr11_-_99474480 | 0.12 |
ENSMUST00000100479.3
|
Krtap1-4
|
keratin associated protein 1-4 |
| chr4_+_43641262 | 0.11 |
ENSMUST00000123351.8
ENSMUST00000128549.3 |
Npr2
|
natriuretic peptide receptor 2 |
| chr6_+_85564566 | 0.11 |
ENSMUST00000213058.2
|
Alms1
|
ALMS1, centrosome and basal body associated |
| chr6_+_116185077 | 0.11 |
ENSMUST00000204051.2
|
Washc2
|
WASH complex subunit 2` |
| chr7_-_118304930 | 0.10 |
ENSMUST00000207323.2
ENSMUST00000038791.15 |
Gde1
|
glycerophosphodiester phosphodiesterase 1 |
| chr11_-_101010715 | 0.09 |
ENSMUST00000017946.6
|
Retreg3
|
reticulophagy regulator family member 3 |
| chr9_-_119652926 | 0.08 |
ENSMUST00000215718.2
|
Scn11a
|
sodium channel, voltage-gated, type XI, alpha |
| chr5_-_124465946 | 0.08 |
ENSMUST00000031344.13
|
Mphosph9
|
M-phase phosphoprotein 9 |
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.4 | 9.7 | GO:0046951 | ketone body biosynthetic process(GO:0046951) |
| 1.6 | 14.2 | GO:0046874 | quinolinate metabolic process(GO:0046874) |
| 1.4 | 4.1 | GO:0006530 | asparagine catabolic process(GO:0006530) |
| 1.1 | 6.5 | GO:2000304 | positive regulation of sphingolipid biosynthetic process(GO:0090154) positive regulation of ceramide biosynthetic process(GO:2000304) |
| 1.1 | 5.3 | GO:0044268 | multicellular organismal protein metabolic process(GO:0044268) |
| 1.0 | 9.2 | GO:0045919 | positive regulation of cytolysis(GO:0045919) |
| 0.9 | 7.8 | GO:0007144 | female meiosis I(GO:0007144) |
| 0.8 | 5.6 | GO:0070777 | D-aspartate transport(GO:0070777) D-aspartate import(GO:0070779) |
| 0.7 | 2.2 | GO:0046022 | regulation of transcription from RNA polymerase II promoter, mitotic(GO:0046021) positive regulation of transcription from RNA polymerase II promoter during mitosis(GO:0046022) |
| 0.6 | 3.1 | GO:0046952 | ketone body catabolic process(GO:0046952) |
| 0.6 | 3.6 | GO:0050917 | sensory perception of umami taste(GO:0050917) |
| 0.4 | 6.3 | GO:0018298 | protein-chromophore linkage(GO:0018298) |
| 0.4 | 9.2 | GO:0046415 | urate metabolic process(GO:0046415) |
| 0.4 | 3.6 | GO:0002074 | extraocular skeletal muscle development(GO:0002074) |
| 0.3 | 2.4 | GO:0006933 | negative regulation of cell adhesion involved in substrate-bound cell migration(GO:0006933) |
| 0.3 | 0.6 | GO:0060460 | left lung morphogenesis(GO:0060460) |
| 0.3 | 0.9 | GO:0061723 | glycophagy(GO:0061723) |
| 0.3 | 2.3 | GO:0009313 | ganglioside catabolic process(GO:0006689) oligosaccharide catabolic process(GO:0009313) |
| 0.3 | 4.3 | GO:0007128 | meiotic prophase I(GO:0007128) prophase(GO:0051324) |
| 0.3 | 0.8 | GO:0032618 | interleukin-15 production(GO:0032618) |
| 0.3 | 8.1 | GO:0072112 | renal filtration cell differentiation(GO:0061318) glomerular visceral epithelial cell differentiation(GO:0072112) glomerular epithelial cell differentiation(GO:0072311) |
| 0.2 | 5.4 | GO:0006309 | apoptotic DNA fragmentation(GO:0006309) |
| 0.2 | 1.4 | GO:0010730 | negative regulation of hydrogen peroxide biosynthetic process(GO:0010730) |
| 0.2 | 0.6 | GO:0006542 | glutamine biosynthetic process(GO:0006542) |
| 0.2 | 2.6 | GO:0006348 | chromatin silencing at telomere(GO:0006348) |
| 0.2 | 7.5 | GO:0010866 | regulation of triglyceride biosynthetic process(GO:0010866) |
| 0.2 | 0.6 | GO:0042998 | positive regulation of Golgi to plasma membrane protein transport(GO:0042998) positive regulation of establishment of protein localization to plasma membrane(GO:0090004) regulation of phosphatidylcholine biosynthetic process(GO:2001245) |
| 0.2 | 1.4 | GO:1901552 | positive regulation of endothelial cell development(GO:1901552) positive regulation of establishment of endothelial barrier(GO:1903142) |
| 0.1 | 1.2 | GO:0006777 | Mo-molybdopterin cofactor biosynthetic process(GO:0006777) Mo-molybdopterin cofactor metabolic process(GO:0019720) |
| 0.1 | 0.8 | GO:0032489 | regulation of Cdc42 protein signal transduction(GO:0032489) response to high density lipoprotein particle(GO:0055099) |
| 0.1 | 2.9 | GO:0016973 | poly(A)+ mRNA export from nucleus(GO:0016973) |
| 0.1 | 0.5 | GO:0060849 | regulation of transcription involved in lymphatic endothelial cell fate commitment(GO:0060849) |
| 0.1 | 2.1 | GO:0007141 | male meiosis I(GO:0007141) |
| 0.1 | 2.9 | GO:0007130 | synaptonemal complex assembly(GO:0007130) |
| 0.1 | 3.3 | GO:0097499 | protein localization to nonmotile primary cilium(GO:0097499) |
| 0.1 | 5.3 | GO:0060445 | branching involved in salivary gland morphogenesis(GO:0060445) |
| 0.1 | 0.3 | GO:0044240 | multicellular organism lipid catabolic process(GO:0044240) |
| 0.1 | 1.8 | GO:0016998 | cell wall macromolecule catabolic process(GO:0016998) |
| 0.1 | 1.2 | GO:0046485 | ether lipid metabolic process(GO:0046485) |
| 0.1 | 2.4 | GO:0051639 | actin filament network formation(GO:0051639) |
| 0.1 | 0.5 | GO:0001992 | regulation of systemic arterial blood pressure by vasopressin(GO:0001992) |
| 0.1 | 0.2 | GO:0042732 | D-xylose metabolic process(GO:0042732) |
| 0.1 | 0.5 | GO:0002317 | plasma cell differentiation(GO:0002317) |
| 0.1 | 0.3 | GO:1902163 | negative regulation of DNA damage response, signal transduction by p53 class mediator resulting in transcription of p21 class mediator(GO:1902163) |
| 0.1 | 8.0 | GO:0000045 | autophagosome assembly(GO:0000045) |
| 0.1 | 0.5 | GO:0006265 | DNA topological change(GO:0006265) |
| 0.1 | 0.6 | GO:0045162 | clustering of voltage-gated sodium channels(GO:0045162) |
| 0.1 | 2.6 | GO:0000132 | establishment of mitotic spindle orientation(GO:0000132) |
| 0.1 | 1.0 | GO:0070673 | response to interleukin-18(GO:0070673) |
| 0.1 | 1.7 | GO:1901741 | positive regulation of myoblast fusion(GO:1901741) |
| 0.0 | 0.5 | GO:0001922 | B-1 B cell homeostasis(GO:0001922) |
| 0.0 | 1.3 | GO:0060009 | Sertoli cell development(GO:0060009) |
| 0.0 | 0.2 | GO:1902081 | regulation of calcium ion import into sarcoplasmic reticulum(GO:1902080) negative regulation of calcium ion import into sarcoplasmic reticulum(GO:1902081) |
| 0.0 | 0.6 | GO:1901387 | positive regulation of voltage-gated calcium channel activity(GO:1901387) |
| 0.0 | 1.1 | GO:0002115 | store-operated calcium entry(GO:0002115) |
| 0.0 | 0.4 | GO:0000022 | mitotic spindle elongation(GO:0000022) mitotic spindle midzone assembly(GO:0051256) |
| 0.0 | 0.7 | GO:0000212 | meiotic spindle organization(GO:0000212) |
| 0.0 | 2.4 | GO:0046676 | negative regulation of insulin secretion(GO:0046676) |
| 0.0 | 3.5 | GO:0006888 | ER to Golgi vesicle-mediated transport(GO:0006888) |
| 0.0 | 2.5 | GO:0051865 | protein autoubiquitination(GO:0051865) |
| 0.0 | 1.3 | GO:0010257 | NADH dehydrogenase complex assembly(GO:0010257) mitochondrial respiratory chain complex I assembly(GO:0032981) mitochondrial respiratory chain complex I biogenesis(GO:0097031) |
| 0.0 | 2.3 | GO:0045600 | positive regulation of fat cell differentiation(GO:0045600) |
| 0.0 | 0.4 | GO:0015937 | coenzyme A biosynthetic process(GO:0015937) |
| 0.0 | 4.2 | GO:0007286 | spermatid development(GO:0007286) |
| 0.0 | 1.0 | GO:0036075 | endochondral ossification(GO:0001958) replacement ossification(GO:0036075) |
| 0.0 | 0.2 | GO:0034626 | fatty acid elongation, saturated fatty acid(GO:0019367) fatty acid elongation, unsaturated fatty acid(GO:0019368) fatty acid elongation, monounsaturated fatty acid(GO:0034625) fatty acid elongation, polyunsaturated fatty acid(GO:0034626) |
| 0.0 | 0.6 | GO:0051560 | mitochondrial calcium ion homeostasis(GO:0051560) |
| 0.0 | 0.3 | GO:0007042 | lysosomal lumen acidification(GO:0007042) |
| 0.0 | 1.0 | GO:1901998 | toxin transport(GO:1901998) |
| 0.0 | 1.4 | GO:0099601 | regulation of neurotransmitter receptor activity(GO:0099601) |
| 0.0 | 0.1 | GO:0002322 | B cell proliferation involved in immune response(GO:0002322) |
| 0.0 | 0.9 | GO:0034605 | cellular response to heat(GO:0034605) |
| 0.0 | 0.1 | GO:0007168 | receptor guanylyl cyclase signaling pathway(GO:0007168) |
| 0.0 | 1.0 | GO:0045669 | positive regulation of osteoblast differentiation(GO:0045669) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.0 | 2.9 | GO:0042272 | nuclear RNA export factor complex(GO:0042272) |
| 0.6 | 7.2 | GO:0000801 | central element(GO:0000801) |
| 0.4 | 1.2 | GO:0019008 | molybdopterin synthase complex(GO:0019008) |
| 0.2 | 1.7 | GO:0016012 | sarcoglycan complex(GO:0016012) |
| 0.2 | 1.4 | GO:0097442 | CA3 pyramidal cell dendrite(GO:0097442) |
| 0.1 | 6.8 | GO:0030173 | integral component of Golgi membrane(GO:0030173) |
| 0.1 | 2.6 | GO:0048188 | Set1C/COMPASS complex(GO:0048188) |
| 0.1 | 6.4 | GO:0032391 | photoreceptor connecting cilium(GO:0032391) |
| 0.1 | 9.7 | GO:0009925 | basal plasma membrane(GO:0009925) |
| 0.1 | 0.6 | GO:0008091 | spectrin(GO:0008091) |
| 0.1 | 5.6 | GO:0030673 | axolemma(GO:0030673) |
| 0.1 | 0.9 | GO:1990316 | ATG1/ULK1 kinase complex(GO:1990316) |
| 0.1 | 0.7 | GO:0005827 | polar microtubule(GO:0005827) |
| 0.1 | 7.8 | GO:0005776 | autophagosome(GO:0005776) |
| 0.1 | 0.2 | GO:0097447 | dendritic tree(GO:0097447) |
| 0.1 | 3.6 | GO:0005891 | voltage-gated calcium channel complex(GO:0005891) |
| 0.0 | 12.1 | GO:0005777 | peroxisome(GO:0005777) microbody(GO:0042579) |
| 0.0 | 13.4 | GO:0072562 | blood microparticle(GO:0072562) |
| 0.0 | 1.0 | GO:0065010 | extracellular membrane-bounded organelle(GO:0065010) |
| 0.0 | 0.2 | GO:0000221 | vacuolar proton-transporting V-type ATPase, V1 domain(GO:0000221) |
| 0.0 | 0.8 | GO:0034364 | high-density lipoprotein particle(GO:0034364) |
| 0.0 | 0.3 | GO:0000220 | vacuolar proton-transporting V-type ATPase, V0 domain(GO:0000220) |
| 0.0 | 1.0 | GO:0016327 | apicolateral plasma membrane(GO:0016327) |
| 0.0 | 1.3 | GO:0045271 | mitochondrial respiratory chain complex I(GO:0005747) NADH dehydrogenase complex(GO:0030964) respiratory chain complex I(GO:0045271) |
| 0.0 | 1.0 | GO:0030131 | clathrin adaptor complex(GO:0030131) |
| 0.0 | 4.5 | GO:0001650 | fibrillar center(GO:0001650) |
| 0.0 | 12.2 | GO:0031966 | mitochondrial membrane(GO:0031966) |
| 0.0 | 2.9 | GO:0001669 | acrosomal vesicle(GO:0001669) |
| 0.0 | 3.2 | GO:0031225 | anchored component of membrane(GO:0031225) |
| 0.0 | 0.6 | GO:0031430 | M band(GO:0031430) |
| 0.0 | 0.1 | GO:0044299 | C-fiber(GO:0044299) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.6 | 6.3 | GO:0019862 | IgA binding(GO:0019862) |
| 1.4 | 4.1 | GO:0004067 | asparaginase activity(GO:0004067) |
| 1.1 | 5.4 | GO:0004531 | deoxyribonuclease II activity(GO:0004531) |
| 1.1 | 5.3 | GO:0003989 | acetyl-CoA carboxylase activity(GO:0003989) |
| 0.9 | 5.6 | GO:0015501 | glutamate:sodium symporter activity(GO:0015501) |
| 0.8 | 0.8 | GO:0035663 | Toll-like receptor 2 binding(GO:0035663) |
| 0.8 | 3.1 | GO:0008260 | 3-oxoacid CoA-transferase activity(GO:0008260) |
| 0.8 | 7.8 | GO:0008310 | single-stranded DNA 3'-5' exodeoxyribonuclease activity(GO:0008310) |
| 0.7 | 9.2 | GO:0016661 | oxidoreductase activity, acting on other nitrogenous compounds as donors(GO:0016661) |
| 0.7 | 6.4 | GO:0032795 | heterotrimeric G-protein binding(GO:0032795) |
| 0.6 | 9.7 | GO:0031957 | very long-chain fatty acid-CoA ligase activity(GO:0031957) |
| 0.6 | 6.5 | GO:0047499 | calcium-independent phospholipase A2 activity(GO:0047499) |
| 0.6 | 13.2 | GO:0019825 | oxygen binding(GO:0019825) |
| 0.5 | 2.3 | GO:0004563 | beta-N-acetylhexosaminidase activity(GO:0004563) |
| 0.3 | 2.4 | GO:0047184 | 1-acylglycerophosphocholine O-acyltransferase activity(GO:0047184) |
| 0.3 | 2.2 | GO:0071723 | lipopeptide binding(GO:0071723) |
| 0.3 | 0.8 | GO:0034188 | apolipoprotein receptor activity(GO:0030226) apolipoprotein A-I receptor activity(GO:0034188) phosphatidylserine-translocating ATPase activity(GO:0090556) |
| 0.2 | 0.6 | GO:0004356 | glutamate-ammonia ligase activity(GO:0004356) ammonia ligase activity(GO:0016211) |
| 0.2 | 0.6 | GO:0070698 | type I activin receptor binding(GO:0070698) |
| 0.2 | 1.2 | GO:0032810 | sterol response element binding(GO:0032810) |
| 0.2 | 1.3 | GO:0016803 | ether hydrolase activity(GO:0016803) |
| 0.2 | 1.4 | GO:0005519 | cytoskeletal regulatory protein binding(GO:0005519) |
| 0.2 | 5.0 | GO:0019707 | protein-cysteine S-palmitoyltransferase activity(GO:0019706) protein-cysteine S-acyltransferase activity(GO:0019707) |
| 0.1 | 1.0 | GO:0004035 | alkaline phosphatase activity(GO:0004035) |
| 0.1 | 3.6 | GO:0008331 | high voltage-gated calcium channel activity(GO:0008331) |
| 0.1 | 3.6 | GO:0008527 | taste receptor activity(GO:0008527) |
| 0.1 | 1.8 | GO:0047372 | acylglycerol lipase activity(GO:0047372) |
| 0.1 | 1.0 | GO:0030306 | ADP-ribosylation factor binding(GO:0030306) |
| 0.1 | 2.6 | GO:0042800 | histone methyltransferase activity (H3-K4 specific)(GO:0042800) |
| 0.1 | 1.8 | GO:0003796 | lysozyme activity(GO:0003796) |
| 0.1 | 2.4 | GO:0005092 | GDP-dissociation inhibitor activity(GO:0005092) |
| 0.1 | 4.6 | GO:0003785 | actin monomer binding(GO:0003785) |
| 0.1 | 0.5 | GO:0003917 | DNA topoisomerase type I activity(GO:0003917) |
| 0.1 | 0.5 | GO:0008269 | JAK pathway signal transduction adaptor activity(GO:0008269) |
| 0.1 | 1.1 | GO:0015279 | store-operated calcium channel activity(GO:0015279) |
| 0.1 | 2.1 | GO:0070577 | lysine-acetylated histone binding(GO:0070577) |
| 0.1 | 0.5 | GO:0005000 | vasopressin receptor activity(GO:0005000) |
| 0.0 | 0.6 | GO:0004467 | long-chain fatty acid-CoA ligase activity(GO:0004467) |
| 0.0 | 2.3 | GO:0005109 | frizzled binding(GO:0005109) |
| 0.0 | 0.4 | GO:0034711 | inhibin binding(GO:0034711) |
| 0.0 | 1.4 | GO:0033130 | acetylcholine receptor binding(GO:0033130) |
| 0.0 | 0.3 | GO:0047498 | calcium-dependent phospholipase A2 activity(GO:0047498) |
| 0.0 | 5.4 | GO:0043130 | ubiquitin binding(GO:0043130) |
| 0.0 | 2.2 | GO:0016620 | oxidoreductase activity, acting on the aldehyde or oxo group of donors, NAD or NADP as acceptor(GO:0016620) |
| 0.0 | 7.6 | GO:0004252 | serine-type endopeptidase activity(GO:0004252) |
| 0.0 | 1.7 | GO:0005160 | transforming growth factor beta receptor binding(GO:0005160) |
| 0.0 | 0.6 | GO:0044548 | S100 protein binding(GO:0044548) |
| 0.0 | 1.8 | GO:0008395 | steroid hydroxylase activity(GO:0008395) |
| 0.0 | 2.9 | GO:0019905 | syntaxin binding(GO:0019905) |
| 0.0 | 1.2 | GO:0051539 | 4 iron, 4 sulfur cluster binding(GO:0051539) |
| 0.0 | 0.5 | GO:0001972 | retinoic acid binding(GO:0001972) |
| 0.0 | 5.4 | GO:0030674 | protein binding, bridging(GO:0030674) |
| 0.0 | 8.6 | GO:0003729 | mRNA binding(GO:0003729) poly(A) RNA binding(GO:0044822) |
| 0.0 | 0.4 | GO:0008553 | hydrogen-exporting ATPase activity, phosphorylative mechanism(GO:0008553) |
| 0.0 | 0.2 | GO:0009922 | fatty acid elongase activity(GO:0009922) 3-oxo-arachidoyl-CoA synthase activity(GO:0102336) 3-oxo-cerotoyl-CoA synthase activity(GO:0102337) 3-oxo-lignoceronyl-CoA synthase activity(GO:0102338) |
| 0.0 | 0.1 | GO:0016941 | natriuretic peptide receptor activity(GO:0016941) |
| 0.0 | 0.6 | GO:0030506 | ankyrin binding(GO:0030506) |
| 0.0 | 1.6 | GO:0051213 | dioxygenase activity(GO:0051213) |
| 0.0 | 0.4 | GO:0016881 | acid-amino acid ligase activity(GO:0016881) |
| 0.0 | 3.6 | GO:0000287 | magnesium ion binding(GO:0000287) |
| 0.0 | 0.2 | GO:0008191 | metalloendopeptidase inhibitor activity(GO:0008191) |
| 0.0 | 0.1 | GO:0008889 | glycerophosphodiester phosphodiesterase activity(GO:0008889) |
| 0.0 | 1.4 | GO:0008013 | beta-catenin binding(GO:0008013) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 4.0 | PID RAC1 REG PATHWAY | Regulation of RAC1 activity |
| 0.0 | 1.4 | PID S1P S1P2 PATHWAY | S1P2 pathway |
| 0.0 | 1.2 | SA CASPASE CASCADE | Apoptosis is mediated by caspases, cysteine proteases arranged in a proteolytic cascade. |
| 0.0 | 0.8 | PID TOLL ENDOGENOUS PATHWAY | Endogenous TLR signaling |
| 0.0 | 6.3 | NABA ECM REGULATORS | Genes encoding enzymes and their regulators involved in the remodeling of the extracellular matrix |
| 0.0 | 1.5 | PID TAP63 PATHWAY | Validated transcriptional targets of TAp63 isoforms |
| 0.0 | 1.4 | PID LKB1 PATHWAY | LKB1 signaling events |
| 0.0 | 2.3 | WNT SIGNALING | Genes related to Wnt-mediated signal transduction |
| 0.0 | 1.1 | PID CD8 TCR PATHWAY | TCR signaling in naïve CD8+ T cells |
| 0.0 | 1.0 | PID TGFBR PATHWAY | TGF-beta receptor signaling |
| 0.0 | 0.6 | PID IL6 7 PATHWAY | IL6-mediated signaling events |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.0 | 14.2 | REACTOME TRYPTOPHAN CATABOLISM | Genes involved in Tryptophan catabolism |
| 0.9 | 9.7 | REACTOME RECYCLING OF BILE ACIDS AND SALTS | Genes involved in Recycling of bile acids and salts |
| 0.3 | 9.2 | REACTOME ACYL CHAIN REMODELLING OF PE | Genes involved in Acyl chain remodelling of PE |
| 0.2 | 3.6 | REACTOME CLASS C 3 METABOTROPIC GLUTAMATE PHEROMONE RECEPTORS | Genes involved in Class C/3 (Metabotropic glutamate/pheromone receptors) |
| 0.1 | 6.1 | REACTOME FATTY ACYL COA BIOSYNTHESIS | Genes involved in Fatty Acyl-CoA Biosynthesis |
| 0.1 | 1.1 | REACTOME ELEVATION OF CYTOSOLIC CA2 LEVELS | Genes involved in Elevation of cytosolic Ca2+ levels |
| 0.1 | 5.6 | REACTOME AMINO ACID AND OLIGOPEPTIDE SLC TRANSPORTERS | Genes involved in Amino acid and oligopeptide SLC transporters |
| 0.1 | 3.6 | REACTOME NCAM1 INTERACTIONS | Genes involved in NCAM1 interactions |
| 0.1 | 2.3 | REACTOME GLYCOSPHINGOLIPID METABOLISM | Genes involved in Glycosphingolipid metabolism |
| 0.0 | 0.8 | REACTOME HDL MEDIATED LIPID TRANSPORT | Genes involved in HDL-mediated lipid transport |
| 0.0 | 0.7 | REACTOME RECRUITMENT OF NUMA TO MITOTIC CENTROSOMES | Genes involved in Recruitment of NuMA to mitotic centrosomes |
| 0.0 | 1.4 | REACTOME ADHERENS JUNCTIONS INTERACTIONS | Genes involved in Adherens junctions interactions |
| 0.0 | 0.6 | REACTOME APOPTOTIC CLEAVAGE OF CELL ADHESION PROTEINS | Genes involved in Apoptotic cleavage of cell adhesion proteins |
| 0.0 | 1.2 | REACTOME RORA ACTIVATES CIRCADIAN EXPRESSION | Genes involved in RORA Activates Circadian Expression |
| 0.0 | 0.5 | REACTOME TRANSCRIPTIONAL REGULATION OF WHITE ADIPOCYTE DIFFERENTIATION | Genes involved in Transcriptional Regulation of White Adipocyte Differentiation |
| 0.0 | 0.4 | REACTOME VITAMIN B5 PANTOTHENATE METABOLISM | Genes involved in Vitamin B5 (pantothenate) metabolism |
| 0.0 | 0.6 | REACTOME SIGNALING BY NODAL | Genes involved in Signaling by NODAL |
| 0.0 | 0.4 | REACTOME CONVERSION FROM APC C CDC20 TO APC C CDH1 IN LATE ANAPHASE | Genes involved in Conversion from APC/C:Cdc20 to APC/C:Cdh1 in late anaphase |
| 0.0 | 1.3 | REACTOME RESPIRATORY ELECTRON TRANSPORT | Genes involved in Respiratory electron transport |
| 0.0 | 1.2 | REACTOME METABOLISM OF VITAMINS AND COFACTORS | Genes involved in Metabolism of vitamins and cofactors |
| 0.0 | 0.6 | REACTOME NCAM SIGNALING FOR NEURITE OUT GROWTH | Genes involved in NCAM signaling for neurite out-growth |
| 0.0 | 0.4 | REACTOME INSULIN RECEPTOR RECYCLING | Genes involved in Insulin receptor recycling |
| 0.0 | 0.5 | REACTOME REGULATION OF KIT SIGNALING | Genes involved in Regulation of KIT signaling |