Project
PRJNA375882: Comprehensive Mouse Transcriptomic BodyMap
Navigation
Downloads
Results for Nr1h4
Z-value: 2.31
Transcription factors associated with Nr1h4
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Nr1h4
|
ENSMUSG00000047638.16 | Nr1h4 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Nr1h4 | mm39_v1_chr10_-_89369432_89369452 | 0.62 | 8.3e-09 | Click! |
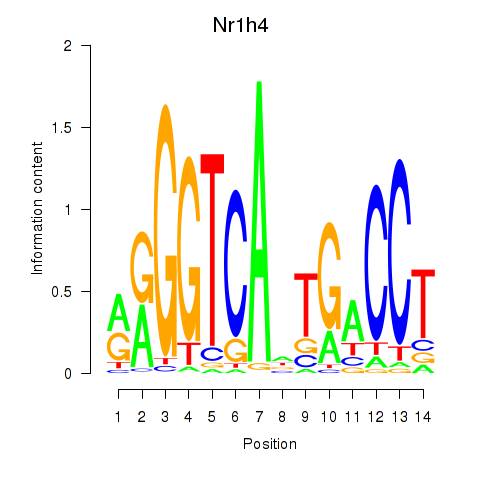
Activity profile of Nr1h4 motif
Sorted Z-values of Nr1h4 motif
Network of associatons between targets according to the STRING database.
First level regulatory network of Nr1h4
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr2_-_69172944 | 64.10 |
ENSMUST00000102709.8
ENSMUST00000102710.10 ENSMUST00000180142.2 |
Abcb11
|
ATP-binding cassette, sub-family B (MDR/TAP), member 11 |
| chr7_-_105249308 | 43.78 |
ENSMUST00000210531.2
ENSMUST00000033185.10 |
Hpx
|
hemopexin |
| chr1_+_67162176 | 35.59 |
ENSMUST00000027144.8
|
Cps1
|
carbamoyl-phosphate synthetase 1 |
| chr14_-_30645711 | 35.10 |
ENSMUST00000006697.17
|
Itih3
|
inter-alpha trypsin inhibitor, heavy chain 3 |
| chr6_+_141575226 | 32.37 |
ENSMUST00000042812.9
|
Slco1b2
|
solute carrier organic anion transporter family, member 1b2 |
| chr14_-_30645503 | 31.97 |
ENSMUST00000227995.2
|
Itih3
|
inter-alpha trypsin inhibitor, heavy chain 3 |
| chr8_-_93806593 | 29.80 |
ENSMUST00000109582.3
|
Ces1b
|
carboxylesterase 1B |
| chr1_+_171052623 | 29.07 |
ENSMUST00000111321.8
ENSMUST00000005824.12 ENSMUST00000111320.8 ENSMUST00000111319.2 |
Apoa2
|
apolipoprotein A-II |
| chr15_-_82678490 | 26.77 |
ENSMUST00000006094.6
|
Cyp2d26
|
cytochrome P450, family 2, subfamily d, polypeptide 26 |
| chr3_+_137983250 | 22.82 |
ENSMUST00000004232.10
|
Adh1
|
alcohol dehydrogenase 1 (class I) |
| chr17_-_84154173 | 22.64 |
ENSMUST00000000687.9
|
Haao
|
3-hydroxyanthranilate 3,4-dioxygenase |
| chr19_-_38113056 | 21.44 |
ENSMUST00000236283.2
|
Rbp4
|
retinol binding protein 4, plasma |
| chr9_+_46179899 | 20.34 |
ENSMUST00000121598.8
|
Apoa5
|
apolipoprotein A-V |
| chr16_-_22847808 | 19.03 |
ENSMUST00000115349.9
|
Kng2
|
kininogen 2 |
| chr16_-_22847760 | 18.45 |
ENSMUST00000039338.13
|
Kng2
|
kininogen 2 |
| chr11_-_116089866 | 18.22 |
ENSMUST00000066587.12
|
Acox1
|
acyl-Coenzyme A oxidase 1, palmitoyl |
| chr11_-_116089595 | 18.14 |
ENSMUST00000072948.11
|
Acox1
|
acyl-Coenzyme A oxidase 1, palmitoyl |
| chr8_+_13110921 | 17.95 |
ENSMUST00000211363.2
ENSMUST00000033822.4 |
Proz
|
protein Z, vitamin K-dependent plasma glycoprotein |
| chr16_-_22847829 | 17.81 |
ENSMUST00000100046.9
|
Kng2
|
kininogen 2 |
| chr15_-_82648376 | 17.13 |
ENSMUST00000055721.6
|
Cyp2d40
|
cytochrome P450, family 2, subfamily d, polypeptide 40 |
| chr17_-_84154196 | 16.60 |
ENSMUST00000234214.2
|
Haao
|
3-hydroxyanthranilate 3,4-dioxygenase |
| chr12_-_103597663 | 15.57 |
ENSMUST00000121625.2
ENSMUST00000044231.12 |
Serpina10
|
serine (or cysteine) peptidase inhibitor, clade A (alpha-1 antiproteinase, antitrypsin), member 10 |
| chr17_-_32639936 | 15.31 |
ENSMUST00000170392.9
ENSMUST00000237165.2 ENSMUST00000235892.2 ENSMUST00000114455.3 |
Pglyrp2
|
peptidoglycan recognition protein 2 |
| chr4_+_133280680 | 15.19 |
ENSMUST00000042706.3
|
Nr0b2
|
nuclear receptor subfamily 0, group B, member 2 |
| chr10_-_80934708 | 15.03 |
ENSMUST00000117422.2
|
Creb3l3
|
cAMP responsive element binding protein 3-like 3 |
| chr1_+_163979384 | 14.69 |
ENSMUST00000086040.6
|
F5
|
coagulation factor V |
| chr9_+_107957621 | 14.48 |
ENSMUST00000035211.14
|
Mst1
|
macrophage stimulating 1 (hepatocyte growth factor-like) |
| chr9_+_107957640 | 14.25 |
ENSMUST00000162886.2
|
Mst1
|
macrophage stimulating 1 (hepatocyte growth factor-like) |
| chr16_-_22848153 | 14.11 |
ENSMUST00000232459.2
|
Kng2
|
kininogen 2 |
| chr17_-_35081456 | 13.76 |
ENSMUST00000025229.11
ENSMUST00000176203.9 ENSMUST00000128767.8 |
Cfb
|
complement factor B |
| chr13_+_91889626 | 12.95 |
ENSMUST00000022120.5
|
Acot12
|
acyl-CoA thioesterase 12 |
| chr10_+_23770586 | 12.71 |
ENSMUST00000041416.8
|
Vnn1
|
vanin 1 |
| chr5_+_137568982 | 12.64 |
ENSMUST00000196471.5
ENSMUST00000198783.5 |
Tfr2
|
transferrin receptor 2 |
| chr17_-_35081129 | 12.48 |
ENSMUST00000154526.8
|
Cfb
|
complement factor B |
| chr16_-_20549294 | 12.13 |
ENSMUST00000231826.2
ENSMUST00000076422.13 ENSMUST00000232217.2 |
Thpo
|
thrombopoietin |
| chr7_-_30755007 | 12.01 |
ENSMUST00000206474.2
ENSMUST00000205807.2 ENSMUST00000039909.13 ENSMUST00000206305.2 ENSMUST00000205439.2 |
Fxyd1
|
FXYD domain-containing ion transport regulator 1 |
| chr19_+_44980565 | 11.85 |
ENSMUST00000179305.2
|
Sema4g
|
sema domain, immunoglobulin domain (Ig), transmembrane domain (TM) and short cytoplasmic domain, (semaphorin) 4G |
| chr1_-_162687254 | 11.55 |
ENSMUST00000131058.8
|
Fmo1
|
flavin containing monooxygenase 1 |
| chr11_+_72326337 | 11.51 |
ENSMUST00000076443.10
|
Ggt6
|
gamma-glutamyltransferase 6 |
| chr9_+_108539296 | 11.19 |
ENSMUST00000035222.6
|
Slc25a20
|
solute carrier family 25 (mitochondrial carnitine/acylcarnitine translocase), member 20 |
| chr1_-_162687369 | 10.51 |
ENSMUST00000193078.6
|
Fmo1
|
flavin containing monooxygenase 1 |
| chr17_-_57535003 | 10.18 |
ENSMUST00000177046.2
ENSMUST00000024988.15 |
C3
|
complement component 3 |
| chrX_-_51254129 | 9.94 |
ENSMUST00000033450.3
|
Gpc4
|
glypican 4 |
| chr11_+_72326391 | 9.48 |
ENSMUST00000100903.3
|
Ggt6
|
gamma-glutamyltransferase 6 |
| chr7_-_30754193 | 9.34 |
ENSMUST00000205778.2
|
Fxyd1
|
FXYD domain-containing ion transport regulator 1 |
| chr3_+_154302311 | 9.34 |
ENSMUST00000192462.6
ENSMUST00000029850.15 |
Cryz
|
crystallin, zeta |
| chr1_-_162641495 | 9.16 |
ENSMUST00000144916.8
ENSMUST00000140274.2 |
Fmo4
|
flavin containing monooxygenase 4 |
| chr7_-_30754223 | 8.91 |
ENSMUST00000206012.2
ENSMUST00000108110.5 |
Fxyd1
|
FXYD domain-containing ion transport regulator 1 |
| chr7_-_30754240 | 8.71 |
ENSMUST00000206860.2
ENSMUST00000071697.11 |
Fxyd1
|
FXYD domain-containing ion transport regulator 1 |
| chr4_-_141345549 | 8.60 |
ENSMUST00000053263.9
|
Tmem82
|
transmembrane protein 82 |
| chr11_+_72326358 | 8.57 |
ENSMUST00000108499.2
|
Ggt6
|
gamma-glutamyltransferase 6 |
| chr11_+_70410445 | 8.21 |
ENSMUST00000179000.2
|
Gltpd2
|
glycolipid transfer protein domain containing 2 |
| chr13_+_41013230 | 8.19 |
ENSMUST00000110191.10
|
Gcnt2
|
glucosaminyl (N-acetyl) transferase 2, I-branching enzyme |
| chr11_+_101442961 | 8.04 |
ENSMUST00000103099.8
|
Nbr1
|
NBR1, autophagy cargo receptor |
| chr11_-_120715351 | 8.03 |
ENSMUST00000055655.9
|
Fasn
|
fatty acid synthase |
| chr12_-_16660960 | 7.69 |
ENSMUST00000239165.2
ENSMUST00000111067.10 |
Lpin1
|
lipin 1 |
| chr4_-_15149755 | 7.69 |
ENSMUST00000108273.2
|
Necab1
|
N-terminal EF-hand calcium binding protein 1 |
| chr10_+_116137277 | 7.44 |
ENSMUST00000092167.7
|
Ptprb
|
protein tyrosine phosphatase, receptor type, B |
| chr13_+_25127127 | 7.27 |
ENSMUST00000021773.13
|
Gpld1
|
glycosylphosphatidylinositol specific phospholipase D1 |
| chr9_-_20791012 | 7.04 |
ENSMUST00000043726.8
|
Angptl6
|
angiopoietin-like 6 |
| chr18_+_61058716 | 6.82 |
ENSMUST00000115297.8
|
Camk2a
|
calcium/calmodulin-dependent protein kinase II alpha |
| chr15_-_31367872 | 6.64 |
ENSMUST00000123325.9
|
Ankrd33b
|
ankyrin repeat domain 33B |
| chr11_+_66915969 | 6.63 |
ENSMUST00000079077.12
ENSMUST00000061786.6 |
Tmem220
|
transmembrane protein 220 |
| chr17_-_87573294 | 6.42 |
ENSMUST00000145895.8
ENSMUST00000129616.8 ENSMUST00000155904.2 ENSMUST00000151155.8 ENSMUST00000144236.9 ENSMUST00000024963.11 |
Mcfd2
|
multiple coagulation factor deficiency 2 |
| chr18_+_61058684 | 6.21 |
ENSMUST00000102888.10
ENSMUST00000025519.11 |
Camk2a
|
calcium/calmodulin-dependent protein kinase II alpha |
| chr15_-_31367668 | 5.97 |
ENSMUST00000110410.10
ENSMUST00000076942.5 |
Ankrd33b
|
ankyrin repeat domain 33B |
| chr11_+_97576619 | 5.94 |
ENSMUST00000107584.8
ENSMUST00000107585.9 |
Cisd3
|
CDGSH iron sulfur domain 3 |
| chr6_+_72391283 | 5.68 |
ENSMUST00000065906.9
|
Ggcx
|
gamma-glutamyl carboxylase |
| chr17_-_74354844 | 5.49 |
ENSMUST00000043458.9
|
Srd5a2
|
steroid 5 alpha-reductase 2 |
| chr7_-_99345016 | 5.39 |
ENSMUST00000107086.9
|
Slco2b1
|
solute carrier organic anion transporter family, member 2b1 |
| chr17_-_79328157 | 5.35 |
ENSMUST00000168887.8
ENSMUST00000119284.8 |
Prkd3
|
protein kinase D3 |
| chr6_-_24528012 | 5.24 |
ENSMUST00000023851.9
|
Ndufa5
|
NADH:ubiquinone oxidoreductase subunit A5 |
| chr11_+_97576724 | 5.09 |
ENSMUST00000107583.3
|
Cisd3
|
CDGSH iron sulfur domain 3 |
| chr13_+_41040657 | 5.07 |
ENSMUST00000069958.15
|
Gcnt2
|
glucosaminyl (N-acetyl) transferase 2, I-branching enzyme |
| chr19_+_38995463 | 5.06 |
ENSMUST00000025966.5
|
Cyp2c55
|
cytochrome P450, family 2, subfamily c, polypeptide 55 |
| chr15_+_102010632 | 5.05 |
ENSMUST00000229592.2
|
Tns2
|
tensin 2 |
| chrX_-_137985960 | 4.71 |
ENSMUST00000033626.15
ENSMUST00000060824.4 |
Serpina7
|
serine (or cysteine) peptidase inhibitor, clade A (alpha-1 antiproteinase, antitrypsin), member 7 |
| chr5_-_147662798 | 4.48 |
ENSMUST00000110529.6
ENSMUST00000031653.12 |
Flt1
|
FMS-like tyrosine kinase 1 |
| chr4_+_117706390 | 4.35 |
ENSMUST00000132043.9
ENSMUST00000169990.8 |
Slc6a9
|
solute carrier family 6 (neurotransmitter transporter, glycine), member 9 |
| chr9_-_21900724 | 4.33 |
ENSMUST00000045726.8
|
Rgl3
|
ral guanine nucleotide dissociation stimulator-like 3 |
| chr11_+_101443014 | 4.14 |
ENSMUST00000147239.8
|
Nbr1
|
NBR1, autophagy cargo receptor |
| chr2_-_174305856 | 4.00 |
ENSMUST00000016396.8
|
Atp5e
|
ATP synthase, H+ transporting, mitochondrial F1 complex, epsilon subunit |
| chr11_+_101443674 | 3.98 |
ENSMUST00000107213.8
ENSMUST00000107208.8 ENSMUST00000107212.8 ENSMUST00000127421.8 |
Nbr1
|
NBR1, autophagy cargo receptor |
| chr15_-_90934059 | 3.97 |
ENSMUST00000109288.9
ENSMUST00000100304.11 |
Kif21a
|
kinesin family member 21A |
| chr16_+_7011580 | 3.94 |
ENSMUST00000231194.2
|
Rbfox1
|
RNA binding protein, fox-1 homolog (C. elegans) 1 |
| chr4_+_117706559 | 3.88 |
ENSMUST00000163288.2
|
Slc6a9
|
solute carrier family 6 (neurotransmitter transporter, glycine), member 9 |
| chr15_-_90934021 | 3.80 |
ENSMUST00000109287.4
ENSMUST00000067205.16 ENSMUST00000088614.13 |
Kif21a
|
kinesin family member 21A |
| chr7_-_126944578 | 3.71 |
ENSMUST00000060783.7
|
Zfp768
|
zinc finger protein 768 |
| chr3_+_121761471 | 3.70 |
ENSMUST00000196479.5
ENSMUST00000197155.5 |
Arhgap29
|
Rho GTPase activating protein 29 |
| chr12_+_76593799 | 3.50 |
ENSMUST00000218380.2
ENSMUST00000219751.2 |
Plekhg3
|
pleckstrin homology domain containing, family G (with RhoGef domain) member 3 |
| chr11_+_75404356 | 3.45 |
ENSMUST00000042808.13
ENSMUST00000118243.2 |
Scarf1
|
scavenger receptor class F, member 1 |
| chr7_-_44320244 | 3.33 |
ENSMUST00000048102.15
|
Myh14
|
myosin, heavy polypeptide 14 |
| chr2_-_130480014 | 3.24 |
ENSMUST00000089561.10
ENSMUST00000110260.8 |
Lzts3
|
leucine zipper, putative tumor suppressor family member 3 |
| chr4_-_137509493 | 3.23 |
ENSMUST00000153588.8
|
Alpl
|
alkaline phosphatase, liver/bone/kidney |
| chr10_-_78134026 | 3.17 |
ENSMUST00000105389.8
|
Agpat3
|
1-acylglycerol-3-phosphate O-acyltransferase 3 |
| chr17_+_3532455 | 3.16 |
ENSMUST00000227604.2
|
Tiam2
|
T cell lymphoma invasion and metastasis 2 |
| chr2_+_175971970 | 3.10 |
ENSMUST00000135430.2
|
2210418O10Rik
|
RIKEN cDNA 2210418O10 gene |
| chr3_-_107424637 | 3.07 |
ENSMUST00000166892.2
|
Slc6a17
|
solute carrier family 6 (neurotransmitter transporter), member 17 |
| chr7_-_103142086 | 3.06 |
ENSMUST00000055787.8
|
Olfr609
|
olfactory receptor 609 |
| chr10_+_43777777 | 3.03 |
ENSMUST00000054418.12
|
Rtn4ip1
|
reticulon 4 interacting protein 1 |
| chr9_+_44018551 | 3.00 |
ENSMUST00000114821.9
ENSMUST00000114818.9 |
C1qtnf5
|
C1q and tumor necrosis factor related protein 5 |
| chr9_-_106353792 | 2.86 |
ENSMUST00000214682.2
ENSMUST00000112479.9 |
Parp3
|
poly (ADP-ribose) polymerase family, member 3 |
| chrX_-_137985979 | 2.84 |
ENSMUST00000152457.2
|
Serpina7
|
serine (or cysteine) peptidase inhibitor, clade A (alpha-1 antiproteinase, antitrypsin), member 7 |
| chr1_+_153528689 | 2.80 |
ENSMUST00000041776.12
|
Rgs8
|
regulator of G-protein signaling 8 |
| chr7_+_128213084 | 2.78 |
ENSMUST00000043138.13
|
Inpp5f
|
inositol polyphosphate-5-phosphatase F |
| chr17_+_24888704 | 2.72 |
ENSMUST00000047179.7
|
Zfp598
|
zinc finger protein 598 |
| chr18_-_67582191 | 2.68 |
ENSMUST00000025408.10
|
Afg3l2
|
AFG3-like AAA ATPase 2 |
| chr9_+_44018520 | 2.59 |
ENSMUST00000114816.8
|
C1qtnf5
|
C1q and tumor necrosis factor related protein 5 |
| chr8_+_94905710 | 2.56 |
ENSMUST00000034215.8
ENSMUST00000212291.2 ENSMUST00000211807.2 |
Mt1
|
metallothionein 1 |
| chr10_+_4561974 | 2.54 |
ENSMUST00000105590.8
ENSMUST00000067086.14 |
Esr1
|
estrogen receptor 1 (alpha) |
| chr10_-_89568106 | 2.50 |
ENSMUST00000020109.5
|
Actr6
|
ARP6 actin-related protein 6 |
| chr1_-_171074652 | 2.47 |
ENSMUST00000013737.13
ENSMUST00000111318.8 |
Ndufs2
|
NADH:ubiquinone oxidoreductase core subunit S2 |
| chr4_-_129015493 | 2.45 |
ENSMUST00000135763.2
ENSMUST00000149763.3 ENSMUST00000164649.8 |
Hpca
|
hippocalcin |
| chr9_-_106533279 | 2.45 |
ENSMUST00000023959.13
ENSMUST00000201681.2 |
Grm2
|
glutamate receptor, metabotropic 2 |
| chr17_+_24888641 | 2.38 |
ENSMUST00000234956.2
|
Zfp598
|
zinc finger protein 598 |
| chr6_-_41752111 | 2.36 |
ENSMUST00000214976.3
|
Olfr459
|
olfactory receptor 459 |
| chr19_+_4644365 | 2.33 |
ENSMUST00000113825.4
|
Pcx
|
pyruvate carboxylase |
| chr3_-_120965327 | 2.31 |
ENSMUST00000170781.2
ENSMUST00000039761.12 ENSMUST00000106467.8 ENSMUST00000106466.10 ENSMUST00000164925.9 |
Rwdd3
|
RWD domain containing 3 |
| chr2_-_85917726 | 2.25 |
ENSMUST00000216886.2
ENSMUST00000213333.2 ENSMUST00000216020.2 |
Olfr1037
|
olfactory receptor 1037 |
| chr11_+_95304903 | 2.25 |
ENSMUST00000107724.9
ENSMUST00000150884.8 ENSMUST00000107722.8 ENSMUST00000127713.2 |
Spop
|
speckle-type BTB/POZ protein |
| chr6_-_41751648 | 2.24 |
ENSMUST00000214752.2
|
Olfr459
|
olfactory receptor 459 |
| chr4_-_129015682 | 2.22 |
ENSMUST00000139450.8
ENSMUST00000125931.9 ENSMUST00000116444.10 |
Hpca
|
hippocalcin |
| chr1_-_63215952 | 2.21 |
ENSMUST00000185412.7
ENSMUST00000027111.15 ENSMUST00000189664.2 |
Ndufs1
|
NADH:ubiquinone oxidoreductase core subunit S1 |
| chr6_-_29380423 | 2.20 |
ENSMUST00000147483.3
|
Opn1sw
|
opsin 1 (cone pigments), short-wave-sensitive (color blindness, tritan) |
| chr1_-_63215812 | 2.14 |
ENSMUST00000185847.2
ENSMUST00000185732.7 ENSMUST00000188370.7 ENSMUST00000168099.9 |
Ndufs1
|
NADH:ubiquinone oxidoreductase core subunit S1 |
| chr5_-_129916283 | 2.14 |
ENSMUST00000094280.4
|
Chchd2
|
coiled-coil-helix-coiled-coil-helix domain containing 2 |
| chr7_+_44091822 | 2.13 |
ENSMUST00000058667.15
|
Lrrc4b
|
leucine rich repeat containing 4B |
| chr2_+_177404691 | 2.13 |
ENSMUST00000119838.9
|
Gm14322
|
predicted gene 14322 |
| chr15_-_77811935 | 2.08 |
ENSMUST00000174529.2
ENSMUST00000173631.8 |
Txn2
|
thioredoxin 2 |
| chr17_+_37716368 | 2.07 |
ENSMUST00000077008.4
|
Olfr107
|
olfactory receptor 107 |
| chr3_-_108133914 | 2.05 |
ENSMUST00000141387.4
|
Sypl2
|
synaptophysin-like 2 |
| chr7_-_4973960 | 2.00 |
ENSMUST00000144863.8
|
Sbk3
|
SH3 domain binding kinase family, member 3 |
| chr9_-_22043083 | 1.88 |
ENSMUST00000069330.14
ENSMUST00000217643.2 |
Acp5
|
acid phosphatase 5, tartrate resistant |
| chr8_+_93400916 | 1.87 |
ENSMUST00000034185.13
|
Irx6
|
Iroquois homeobox 6 |
| chr4_+_53440389 | 1.85 |
ENSMUST00000107646.9
ENSMUST00000102911.10 |
Slc44a1
|
solute carrier family 44, member 1 |
| chr11_-_102588536 | 1.85 |
ENSMUST00000164506.3
ENSMUST00000092569.13 |
Ccdc43
|
coiled-coil domain containing 43 |
| chr9_+_44018583 | 1.83 |
ENSMUST00000152956.8
ENSMUST00000114815.3 ENSMUST00000206295.2 ENSMUST00000206769.2 ENSMUST00000205500.2 |
C1qtnf5
|
C1q and tumor necrosis factor related protein 5 |
| chr19_+_4644425 | 1.83 |
ENSMUST00000238089.2
|
Pcx
|
pyruvate carboxylase |
| chr7_-_47631843 | 1.82 |
ENSMUST00000087092.4
|
Mrgpra4
|
MAS-related GPR, member A4 |
| chr11_-_115704447 | 1.78 |
ENSMUST00000041684.11
ENSMUST00000156812.2 |
Caskin2
|
CASK-interacting protein 2 |
| chr1_+_75213114 | 1.77 |
ENSMUST00000188290.7
|
Dnajb2
|
DnaJ heat shock protein family (Hsp40) member B2 |
| chr4_-_129015027 | 1.77 |
ENSMUST00000030572.10
|
Hpca
|
hippocalcin |
| chr7_-_142215027 | 1.73 |
ENSMUST00000105936.8
|
Igf2
|
insulin-like growth factor 2 |
| chr7_-_4974167 | 1.70 |
ENSMUST00000133272.2
|
Sbk3
|
SH3 domain binding kinase family, member 3 |
| chr1_+_75213258 | 1.68 |
ENSMUST00000185654.3
|
Dnajb2
|
DnaJ heat shock protein family (Hsp40) member B2 |
| chr8_+_85807369 | 1.68 |
ENSMUST00000079764.14
|
Wdr83os
|
WD repeat domain 83 opposite strand |
| chr16_+_20354225 | 1.53 |
ENSMUST00000090023.13
ENSMUST00000007216.9 ENSMUST00000232001.2 |
Ap2m1
|
adaptor-related protein complex 2, mu 1 subunit |
| chr14_-_52704547 | 1.50 |
ENSMUST00000205811.2
|
Olfr1508
|
olfactory receptor 1508 |
| chr15_-_98465516 | 1.49 |
ENSMUST00000012104.7
|
Ccnt1
|
cyclin T1 |
| chr5_-_37494179 | 1.48 |
ENSMUST00000114148.2
|
Evc
|
EvC ciliary complex subunit 1 |
| chr7_-_4517559 | 1.48 |
ENSMUST00000163538.8
|
Tnnt1
|
troponin T1, skeletal, slow |
| chr7_-_46783432 | 1.47 |
ENSMUST00000102626.10
|
Ptpn5
|
protein tyrosine phosphatase, non-receptor type 5 |
| chr5_-_37494213 | 1.47 |
ENSMUST00000031005.11
|
Evc
|
EvC ciliary complex subunit 1 |
| chr9_+_56902172 | 1.41 |
ENSMUST00000034832.8
|
Ptpn9
|
protein tyrosine phosphatase, non-receptor type 9 |
| chr3_+_99147677 | 1.36 |
ENSMUST00000151606.8
|
Tbx15
|
T-box 15 |
| chrM_+_8603 | 1.32 |
ENSMUST00000082409.1
|
mt-Co3
|
mitochondrially encoded cytochrome c oxidase III |
| chrX_-_105528972 | 1.27 |
ENSMUST00000140707.2
|
Fndc3c1
|
fibronectin type III domain containing 3C1 |
| chr16_+_18247666 | 1.26 |
ENSMUST00000144233.3
|
Txnrd2
|
thioredoxin reductase 2 |
| chr18_-_35064906 | 1.25 |
ENSMUST00000025218.8
|
Etf1
|
eukaryotic translation termination factor 1 |
| chr11_-_120520954 | 1.20 |
ENSMUST00000106180.2
|
Mafg
|
v-maf musculoaponeurotic fibrosarcoma oncogene family, protein G (avian) |
| chr17_+_25352353 | 1.19 |
ENSMUST00000162862.3
ENSMUST00000040729.9 |
Clcn7
|
chloride channel, voltage-sensitive 7 |
| chr1_-_120197979 | 1.18 |
ENSMUST00000112639.8
|
Steap3
|
STEAP family member 3 |
| chr6_+_24528143 | 1.16 |
ENSMUST00000031696.10
|
Asb15
|
ankyrin repeat and SOCS box-containing 15 |
| chr18_+_36662276 | 1.16 |
ENSMUST00000237595.2
|
Slc4a9
|
solute carrier family 4, sodium bicarbonate cotransporter, member 9 |
| chr4_+_149188585 | 1.15 |
ENSMUST00000103216.10
ENSMUST00000030816.4 |
Dffa
|
DNA fragmentation factor, alpha subunit |
| chr9_-_22042930 | 1.13 |
ENSMUST00000213815.2
|
Acp5
|
acid phosphatase 5, tartrate resistant |
| chr16_+_32250043 | 1.11 |
ENSMUST00000115140.2
ENSMUST00000104893.10 |
Pcyt1a
|
phosphate cytidylyltransferase 1, choline, alpha isoform |
| chr5_+_147894121 | 1.09 |
ENSMUST00000085558.11
ENSMUST00000129092.2 |
Mtus2
|
microtubule associated tumor suppressor candidate 2 |
| chrX_+_20714782 | 1.08 |
ENSMUST00000001155.11
ENSMUST00000122312.8 ENSMUST00000120356.8 ENSMUST00000122850.2 |
Araf
|
Araf proto-oncogene, serine/threonine kinase |
| chrX_-_18327610 | 1.08 |
ENSMUST00000044188.5
|
Dipk2b
|
divergent protein kinase domain 2B |
| chr17_+_24939072 | 1.07 |
ENSMUST00000054289.13
|
Rps2
|
ribosomal protein S2 |
| chr4_+_143076327 | 1.05 |
ENSMUST00000052458.3
|
Lrrc38
|
leucine rich repeat containing 38 |
| chr13_-_40882417 | 1.01 |
ENSMUST00000225180.2
|
Tfap2a
|
transcription factor AP-2, alpha |
| chr6_-_29380467 | 0.99 |
ENSMUST00000080428.13
|
Opn1sw
|
opsin 1 (cone pigments), short-wave-sensitive (color blindness, tritan) |
| chr2_+_128907854 | 0.98 |
ENSMUST00000035812.14
|
Ttl
|
tubulin tyrosine ligase |
| chr16_-_94171556 | 0.97 |
ENSMUST00000113906.9
|
Pigp
|
phosphatidylinositol glycan anchor biosynthesis, class P |
| chr2_-_90748331 | 0.96 |
ENSMUST00000111461.13
|
Ptpmt1
|
protein tyrosine phosphatase, mitochondrial 1 |
| chr6_-_28449250 | 0.96 |
ENSMUST00000164519.9
ENSMUST00000171089.9 ENSMUST00000031718.14 |
Pax4
|
paired box 4 |
| chr11_+_100225233 | 0.93 |
ENSMUST00000017309.2
|
Gast
|
gastrin |
| chr2_+_181139016 | 0.91 |
ENSMUST00000108799.10
|
Tpd52l2
|
tumor protein D52-like 2 |
| chr7_-_4517608 | 0.89 |
ENSMUST00000166959.8
|
Tnnt1
|
troponin T1, skeletal, slow |
| chr1_+_152830720 | 0.83 |
ENSMUST00000043313.15
ENSMUST00000186621.2 |
Nmnat2
|
nicotinamide nucleotide adenylyltransferase 2 |
| chr12_-_87346479 | 0.82 |
ENSMUST00000125733.8
|
Ism2
|
isthmin 2 |
| chr10_-_89522112 | 0.81 |
ENSMUST00000092227.12
ENSMUST00000174252.8 |
Scyl2
|
SCY1-like 2 (S. cerevisiae) |
| chr7_+_11608557 | 0.81 |
ENSMUST00000227611.2
ENSMUST00000226622.2 ENSMUST00000228646.2 ENSMUST00000226855.2 ENSMUST00000228268.2 ENSMUST00000228463.2 |
Vmn1r75
|
vomeronasal 1 receptor 75 |
| chrX_-_166638057 | 0.81 |
ENSMUST00000238211.2
|
Frmpd4
|
FERM and PDZ domain containing 4 |
| chr2_+_181138958 | 0.80 |
ENSMUST00000149163.8
ENSMUST00000000844.15 ENSMUST00000184849.8 ENSMUST00000108800.8 ENSMUST00000069712.9 |
Tpd52l2
|
tumor protein D52-like 2 |
| chrX_-_138683102 | 0.80 |
ENSMUST00000101217.4
|
Ripply1
|
ripply transcriptional repressor 1 |
| chr7_+_140226365 | 0.79 |
ENSMUST00000084456.6
ENSMUST00000211057.2 ENSMUST00000211399.2 |
Olfr53
|
olfactory receptor 53 |
| chr15_+_101371353 | 0.73 |
ENSMUST00000088049.5
|
Krt86
|
keratin 86 |
| chr9_+_50664207 | 0.73 |
ENSMUST00000034562.9
|
Cryab
|
crystallin, alpha B |
| chr13_+_38041910 | 0.72 |
ENSMUST00000138110.8
|
Rreb1
|
ras responsive element binding protein 1 |
| chr2_-_176894622 | 0.72 |
ENSMUST00000133301.8
|
Gm14410
|
predicted gene 14410 |
| chr7_-_45362867 | 0.66 |
ENSMUST00000211340.2
|
Sphk2
|
sphingosine kinase 2 |
| chr9_+_50664288 | 0.64 |
ENSMUST00000214962.2
ENSMUST00000216755.2 |
Cryab
|
crystallin, alpha B |
| chr3_+_87457406 | 0.61 |
ENSMUST00000238894.2
|
Etv3l
|
ets variant 3-like |
| chr13_-_18167890 | 0.61 |
ENSMUST00000099735.6
|
Yae1d1
|
Yae1 domain containing 1 |
| chr2_+_175664894 | 0.57 |
ENSMUST00000109012.4
|
Gm6710
|
predicted gene 6710 |
| chr2_-_175044854 | 0.55 |
ENSMUST00000165892.8
|
Gm14391
|
predicted gene 14391 |
| chr16_+_32249713 | 0.54 |
ENSMUST00000115137.8
ENSMUST00000079791.11 |
Pcyt1a
|
phosphate cytidylyltransferase 1, choline, alpha isoform |
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 12.4 | 49.4 | GO:0010901 | regulation of very-low-density lipoprotein particle remodeling(GO:0010901) |
| 9.2 | 64.1 | GO:0046618 | drug export(GO:0046618) |
| 6.3 | 43.8 | GO:0015886 | positive regulation of humoral immune response mediated by circulating immunoglobulin(GO:0002925) heme transport(GO:0015886) positive regulation of response to interferon-gamma(GO:0060332) positive regulation of interferon-gamma-mediated signaling pathway(GO:0060335) |
| 6.1 | 36.4 | GO:0033540 | fatty acid beta-oxidation using acyl-CoA oxidase(GO:0033540) |
| 5.7 | 22.8 | GO:0006069 | ethanol oxidation(GO:0006069) |
| 5.1 | 35.6 | GO:0019856 | 'de novo' pyrimidine nucleobase biosynthetic process(GO:0006207) pyrimidine nucleobase biosynthetic process(GO:0019856) |
| 4.4 | 39.2 | GO:0046874 | quinolinate metabolic process(GO:0046874) |
| 4.3 | 21.4 | GO:0048807 | female genitalia morphogenesis(GO:0048807) |
| 3.5 | 39.0 | GO:1903278 | positive regulation of sodium ion export(GO:1903275) positive regulation of sodium ion export from cell(GO:1903278) positive regulation of inorganic anion transmembrane transport(GO:1903797) |
| 3.2 | 28.7 | GO:0060763 | mammary duct terminal end bud growth(GO:0060763) |
| 3.0 | 12.1 | GO:0038163 | thrombopoietin-mediated signaling pathway(GO:0038163) |
| 2.7 | 8.2 | GO:0061536 | glycine secretion(GO:0061536) glycine secretion, neurotransmission(GO:0061537) |
| 2.6 | 15.3 | GO:0032827 | negative regulation of natural killer cell differentiation(GO:0032824) negative regulation of natural killer cell differentiation involved in immune response(GO:0032827) |
| 2.5 | 10.2 | GO:0001970 | positive regulation of activation of membrane attack complex(GO:0001970) |
| 2.5 | 7.6 | GO:0033189 | response to vitamin A(GO:0033189) |
| 2.1 | 6.4 | GO:0030827 | negative regulation of cGMP metabolic process(GO:0030824) negative regulation of cGMP biosynthetic process(GO:0030827) negative regulation of guanylate cyclase activity(GO:0031283) |
| 2.1 | 29.6 | GO:0006751 | glutathione catabolic process(GO:0006751) |
| 2.0 | 32.4 | GO:0006857 | oligopeptide transport(GO:0006857) |
| 1.9 | 13.3 | GO:0036438 | maintenance of lens transparency(GO:0036438) |
| 1.8 | 7.3 | GO:0010982 | GPI anchor release(GO:0006507) regulation of high-density lipoprotein particle clearance(GO:0010982) |
| 1.7 | 26.2 | GO:0006957 | complement activation, alternative pathway(GO:0006957) |
| 1.4 | 67.1 | GO:0030212 | hyaluronan metabolic process(GO:0030212) |
| 1.4 | 5.7 | GO:0017187 | peptidyl-glutamic acid carboxylation(GO:0017187) |
| 1.3 | 87.4 | GO:1900047 | negative regulation of blood coagulation(GO:0030195) negative regulation of hemostasis(GO:1900047) |
| 1.3 | 9.2 | GO:0042737 | drug catabolic process(GO:0042737) |
| 1.2 | 22.1 | GO:0070995 | NADPH oxidation(GO:0070995) |
| 1.1 | 12.6 | GO:0097460 | ferrous iron import into cell(GO:0097460) ferrous iron import across plasma membrane(GO:0098707) |
| 1.1 | 5.4 | GO:0071718 | sodium-independent icosanoid transport(GO:0071718) |
| 1.0 | 11.2 | GO:0015879 | carnitine transport(GO:0015879) |
| 1.0 | 8.0 | GO:1901503 | ether lipid biosynthetic process(GO:0008611) glycerol ether biosynthetic process(GO:0046504) ether biosynthetic process(GO:1901503) |
| 1.0 | 15.0 | GO:1990440 | positive regulation of transcription from RNA polymerase II promoter in response to endoplasmic reticulum stress(GO:1990440) |
| 0.9 | 13.0 | GO:0098970 | receptor diffusion trapping(GO:0098953) postsynaptic neurotransmitter receptor diffusion trapping(GO:0098970) neurotransmitter receptor diffusion trapping(GO:0099628) |
| 0.8 | 4.2 | GO:0019072 | viral genome packaging(GO:0019072) viral RNA genome packaging(GO:0019074) |
| 0.8 | 2.3 | GO:1900039 | positive regulation of cellular response to hypoxia(GO:1900039) regulation of hypoxia-inducible factor-1alpha signaling pathway(GO:1902071) |
| 0.8 | 5.4 | GO:0089700 | protein kinase D signaling(GO:0089700) |
| 0.7 | 5.1 | GO:0072344 | rescue of stalled ribosome(GO:0072344) |
| 0.7 | 12.7 | GO:2000400 | positive regulation of T cell differentiation in thymus(GO:0033089) positive regulation of thymocyte aggregation(GO:2000400) |
| 0.7 | 49.0 | GO:0019369 | arachidonic acid metabolic process(GO:0019369) |
| 0.6 | 7.7 | GO:0006642 | triglyceride mobilization(GO:0006642) |
| 0.6 | 5.5 | GO:0061370 | testosterone biosynthetic process(GO:0061370) |
| 0.6 | 4.5 | GO:0048597 | post-embryonic camera-type eye morphogenesis(GO:0048597) |
| 0.5 | 2.5 | GO:0060523 | prostate epithelial cord elongation(GO:0060523) |
| 0.5 | 3.5 | GO:1903644 | regulation of chaperone-mediated protein folding(GO:1903644) |
| 0.5 | 1.5 | GO:2001025 | positive regulation of response to drug(GO:2001025) |
| 0.5 | 2.7 | GO:0031444 | slow-twitch skeletal muscle fiber contraction(GO:0031444) |
| 0.4 | 9.3 | GO:0042178 | xenobiotic catabolic process(GO:0042178) |
| 0.4 | 2.4 | GO:1990166 | protein localization to site of double-strand break(GO:1990166) |
| 0.4 | 2.8 | GO:0031161 | phosphatidylinositol catabolic process(GO:0031161) |
| 0.4 | 3.1 | GO:0015803 | branched-chain amino acid transport(GO:0015803) leucine transport(GO:0015820) |
| 0.4 | 3.0 | GO:0032929 | negative regulation of superoxide anion generation(GO:0032929) |
| 0.4 | 2.2 | GO:2000676 | positive regulation of type B pancreatic cell apoptotic process(GO:2000676) |
| 0.3 | 12.9 | GO:0006084 | acetyl-CoA metabolic process(GO:0006084) |
| 0.3 | 2.6 | GO:1990169 | detoxification of copper ion(GO:0010273) stress response to copper ion(GO:1990169) |
| 0.3 | 1.5 | GO:0016185 | synaptic vesicle budding from presynaptic endocytic zone membrane(GO:0016185) |
| 0.2 | 2.7 | GO:0036444 | calcium ion transmembrane import into mitochondrion(GO:0036444) |
| 0.2 | 1.9 | GO:0015871 | choline transport(GO:0015871) |
| 0.2 | 1.2 | GO:0032074 | negative regulation of nuclease activity(GO:0032074) |
| 0.2 | 3.4 | GO:0048680 | positive regulation of axon regeneration(GO:0048680) |
| 0.2 | 3.2 | GO:0018298 | protein-chromophore linkage(GO:0018298) |
| 0.2 | 16.2 | GO:0045668 | negative regulation of osteoblast differentiation(GO:0045668) |
| 0.2 | 11.8 | GO:0050919 | negative chemotaxis(GO:0050919) |
| 0.2 | 1.0 | GO:0003409 | optic cup structural organization(GO:0003409) |
| 0.2 | 2.8 | GO:0007213 | G-protein coupled acetylcholine receptor signaling pathway(GO:0007213) |
| 0.2 | 4.6 | GO:0014850 | response to muscle activity(GO:0014850) |
| 0.2 | 2.1 | GO:0006662 | glycerol ether metabolic process(GO:0006662) ether metabolic process(GO:0018904) |
| 0.2 | 0.7 | GO:0006669 | sphinganine-1-phosphate biosynthetic process(GO:0006669) |
| 0.2 | 0.8 | GO:0051835 | positive regulation of synapse structural plasticity(GO:0051835) |
| 0.2 | 1.9 | GO:0060040 | retinal bipolar neuron differentiation(GO:0060040) |
| 0.1 | 15.2 | GO:0032024 | positive regulation of insulin secretion(GO:0032024) |
| 0.1 | 1.7 | GO:2000467 | positive regulation of glycogen (starch) synthase activity(GO:2000467) |
| 0.1 | 2.1 | GO:1900037 | regulation of cellular response to hypoxia(GO:1900037) |
| 0.1 | 0.8 | GO:0034628 | nicotinamide nucleotide biosynthetic process from aspartate(GO:0019355) 'de novo' NAD biosynthetic process(GO:0034627) 'de novo' NAD biosynthetic process from aspartate(GO:0034628) |
| 0.1 | 12.6 | GO:0022904 | respiratory electron transport chain(GO:0022904) |
| 0.1 | 1.5 | GO:1900364 | negative regulation of mRNA 3'-end processing(GO:0031441) negative regulation of mRNA polyadenylation(GO:1900364) positive regulation of phosphorylation of RNA polymerase II C-terminal domain(GO:1901409) |
| 0.1 | 4.0 | GO:0015986 | energy coupled proton transport, down electrochemical gradient(GO:0015985) ATP synthesis coupled proton transport(GO:0015986) |
| 0.1 | 1.4 | GO:0007021 | tubulin complex assembly(GO:0007021) |
| 0.1 | 1.0 | GO:0032049 | cardiolipin biosynthetic process(GO:0032049) |
| 0.1 | 0.7 | GO:1903691 | positive regulation of wound healing, spreading of epidermal cells(GO:1903691) |
| 0.1 | 0.8 | GO:2000286 | receptor internalization involved in canonical Wnt signaling pathway(GO:2000286) |
| 0.1 | 3.9 | GO:2001014 | regulation of skeletal muscle cell differentiation(GO:2001014) |
| 0.1 | 3.3 | GO:0071625 | vocalization behavior(GO:0071625) |
| 0.1 | 19.0 | GO:0007596 | blood coagulation(GO:0007596) |
| 0.1 | 3.2 | GO:0001958 | endochondral ossification(GO:0001958) replacement ossification(GO:0036075) |
| 0.1 | 2.9 | GO:0003416 | endochondral bone growth(GO:0003416) |
| 0.1 | 1.3 | GO:0006415 | translational termination(GO:0006415) |
| 0.1 | 1.7 | GO:0006656 | phosphatidylcholine biosynthetic process(GO:0006656) |
| 0.1 | 0.5 | GO:1902412 | regulation of mitotic cytokinesis(GO:1902412) |
| 0.1 | 0.4 | GO:0010968 | regulation of microtubule nucleation(GO:0010968) |
| 0.1 | 1.2 | GO:0015677 | copper ion import(GO:0015677) |
| 0.1 | 1.1 | GO:0001731 | formation of translation preinitiation complex(GO:0001731) |
| 0.1 | 1.0 | GO:1903818 | positive regulation of voltage-gated potassium channel activity(GO:1903818) |
| 0.1 | 4.8 | GO:0070206 | protein trimerization(GO:0070206) |
| 0.0 | 0.8 | GO:0032525 | somite rostral/caudal axis specification(GO:0032525) |
| 0.0 | 2.4 | GO:0014047 | glutamate secretion(GO:0014047) |
| 0.0 | 1.3 | GO:0000305 | response to oxygen radical(GO:0000305) |
| 0.0 | 0.2 | GO:0046010 | positive regulation of circadian sleep/wake cycle, non-REM sleep(GO:0046010) |
| 0.0 | 2.1 | GO:0046426 | negative regulation of JAK-STAT cascade(GO:0046426) negative regulation of STAT cascade(GO:1904893) |
| 0.0 | 1.4 | GO:0048701 | embryonic cranial skeleton morphogenesis(GO:0048701) |
| 0.0 | 0.2 | GO:1900740 | positive regulation of thymocyte apoptotic process(GO:0070245) regulation of protein insertion into mitochondrial membrane involved in apoptotic signaling pathway(GO:1900739) positive regulation of protein insertion into mitochondrial membrane involved in apoptotic signaling pathway(GO:1900740) |
| 0.0 | 1.2 | GO:0009268 | response to pH(GO:0009268) |
| 0.0 | 2.4 | GO:0030641 | regulation of cellular pH(GO:0030641) |
| 0.0 | 0.1 | GO:0022601 | menstrual cycle phase(GO:0022601) |
| 0.0 | 0.5 | GO:0006491 | N-glycan processing(GO:0006491) |
| 0.0 | 7.6 | GO:0001525 | angiogenesis(GO:0001525) |
| 0.0 | 0.4 | GO:0006903 | vesicle targeting(GO:0006903) |
| 0.0 | 0.3 | GO:0001833 | inner cell mass cell proliferation(GO:0001833) |
| 0.0 | 2.5 | GO:0006338 | chromatin remodeling(GO:0006338) |
| 0.0 | 1.0 | GO:0031016 | pancreas development(GO:0031016) |
| 0.0 | 1.1 | GO:0033138 | positive regulation of peptidyl-serine phosphorylation(GO:0033138) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 3.6 | 64.1 | GO:0046581 | intercellular canaliculus(GO:0046581) |
| 3.3 | 13.0 | GO:0099573 | glutamatergic postsynaptic density(GO:0099573) |
| 2.1 | 49.4 | GO:0042627 | chylomicron(GO:0042627) |
| 1.0 | 12.6 | GO:1990712 | HFE-transferrin receptor complex(GO:1990712) |
| 0.9 | 9.7 | GO:0042587 | glycogen granule(GO:0042587) |
| 0.7 | 2.7 | GO:0005745 | m-AAA complex(GO:0005745) |
| 0.6 | 35.6 | GO:0009295 | nucleoid(GO:0009295) mitochondrial nucleoid(GO:0042645) |
| 0.5 | 4.0 | GO:0000275 | mitochondrial proton-transporting ATP synthase complex, catalytic core F(1)(GO:0000275) proton-transporting ATP synthase complex, catalytic core F(1)(GO:0045261) |
| 0.5 | 133.6 | GO:0072562 | blood microparticle(GO:0072562) |
| 0.5 | 14.7 | GO:0031091 | platelet alpha granule(GO:0031091) |
| 0.5 | 3.3 | GO:0097513 | myosin II filament(GO:0097513) |
| 0.4 | 34.6 | GO:0034707 | chloride channel complex(GO:0034707) |
| 0.4 | 6.4 | GO:0044327 | dendritic spine head(GO:0044327) |
| 0.4 | 7.3 | GO:0034364 | high-density lipoprotein particle(GO:0034364) |
| 0.4 | 36.4 | GO:0005778 | peroxisomal membrane(GO:0005778) microbody membrane(GO:0031903) |
| 0.4 | 2.5 | GO:0097550 | transcriptional preinitiation complex(GO:0097550) |
| 0.3 | 16.2 | GO:0000407 | pre-autophagosomal structure(GO:0000407) |
| 0.3 | 1.4 | GO:0070722 | Tle3-Aes complex(GO:0070722) |
| 0.3 | 5.5 | GO:0070852 | cell body fiber(GO:0070852) |
| 0.2 | 12.1 | GO:0005747 | mitochondrial respiratory chain complex I(GO:0005747) NADH dehydrogenase complex(GO:0030964) respiratory chain complex I(GO:0045271) |
| 0.2 | 1.5 | GO:0008024 | cyclin/CDK positive transcription elongation factor complex(GO:0008024) |
| 0.2 | 3.2 | GO:0065010 | extracellular membrane-bounded organelle(GO:0065010) |
| 0.1 | 2.7 | GO:0005861 | troponin complex(GO:0005861) |
| 0.1 | 3.7 | GO:0005665 | DNA-directed RNA polymerase II, core complex(GO:0005665) |
| 0.1 | 7.4 | GO:0016328 | lateral plasma membrane(GO:0016328) |
| 0.1 | 0.4 | GO:0097441 | basilar dendrite(GO:0097441) |
| 0.1 | 1.5 | GO:1990635 | proximal dendrite(GO:1990635) |
| 0.1 | 1.4 | GO:0097512 | cardiac myofibril(GO:0097512) |
| 0.1 | 2.1 | GO:0044300 | cerebellar mossy fiber(GO:0044300) |
| 0.1 | 0.5 | GO:0017177 | glucosidase II complex(GO:0017177) |
| 0.1 | 4.3 | GO:0045334 | clathrin-coated endocytic vesicle(GO:0045334) |
| 0.1 | 34.1 | GO:0016323 | basolateral plasma membrane(GO:0016323) |
| 0.1 | 20.5 | GO:0031225 | anchored component of membrane(GO:0031225) |
| 0.1 | 7.8 | GO:0005871 | kinesin complex(GO:0005871) |
| 0.1 | 60.6 | GO:0031966 | mitochondrial membrane(GO:0031966) |
| 0.1 | 2.1 | GO:0030285 | integral component of synaptic vesicle membrane(GO:0030285) intrinsic component of synaptic vesicle membrane(GO:0098563) |
| 0.1 | 2.8 | GO:0032809 | neuronal cell body membrane(GO:0032809) |
| 0.1 | 0.5 | GO:0000235 | astral microtubule(GO:0000235) |
| 0.1 | 0.4 | GO:0005964 | phosphorylase kinase complex(GO:0005964) |
| 0.1 | 1.0 | GO:0000506 | glycosylphosphatidylinositol-N-acetylglucosaminyltransferase (GPI-GnT) complex(GO:0000506) |
| 0.1 | 2.4 | GO:0035861 | site of double-strand break(GO:0035861) |
| 0.0 | 2.1 | GO:0005758 | mitochondrial intermembrane space(GO:0005758) |
| 0.0 | 91.0 | GO:0005615 | extracellular space(GO:0005615) |
| 0.0 | 1.2 | GO:0005771 | multivesicular body(GO:0005771) |
| 0.0 | 16.7 | GO:0098793 | presynapse(GO:0098793) |
| 0.0 | 3.5 | GO:0016234 | inclusion body(GO:0016234) |
| 0.0 | 7.4 | GO:0043235 | receptor complex(GO:0043235) |
| 0.0 | 78.7 | GO:0070062 | extracellular exosome(GO:0070062) |
| 0.0 | 2.2 | GO:0031463 | Cul3-RING ubiquitin ligase complex(GO:0031463) |
| 0.0 | 3.8 | GO:0005802 | trans-Golgi network(GO:0005802) |
| 0.0 | 1.1 | GO:0005881 | cytoplasmic microtubule(GO:0005881) |
| 0.0 | 0.3 | GO:0071006 | U2-type catalytic step 1 spliceosome(GO:0071006) |
| 0.0 | 3.2 | GO:0045211 | postsynaptic membrane(GO:0045211) |
| 0.0 | 0.2 | GO:0005740 | mitochondrial envelope(GO:0005740) |
| 0.0 | 20.3 | GO:0005783 | endoplasmic reticulum(GO:0005783) |
| 0.0 | 15.6 | GO:0005739 | mitochondrion(GO:0005739) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 11.9 | 35.6 | GO:0004088 | carbamoyl-phosphate synthase (ammonia) activity(GO:0004087) carbamoyl-phosphate synthase (glutamine-hydrolyzing) activity(GO:0004088) |
| 7.6 | 22.8 | GO:0004024 | alcohol dehydrogenase activity, zinc-dependent(GO:0004024) |
| 7.3 | 43.8 | GO:0015232 | heme transporter activity(GO:0015232) |
| 7.3 | 29.1 | GO:0070653 | high-density lipoprotein particle receptor binding(GO:0070653) |
| 7.1 | 21.4 | GO:0034632 | retinol transporter activity(GO:0034632) |
| 5.2 | 36.4 | GO:0016401 | palmitoyl-CoA oxidase activity(GO:0016401) |
| 4.4 | 101.9 | GO:0015125 | bile acid transmembrane transporter activity(GO:0015125) |
| 3.4 | 20.3 | GO:0060228 | phosphatidylcholine-sterol O-acyltransferase activator activity(GO:0060228) lipoprotein lipase activator activity(GO:0060230) |
| 3.3 | 13.3 | GO:0008109 | N-acetyllactosaminide beta-1,6-N-acetylglucosaminyltransferase activity(GO:0008109) |
| 2.7 | 8.0 | GO:0016296 | [acyl-carrier-protein] S-malonyltransferase activity(GO:0004314) oleoyl-[acyl-carrier-protein] hydrolase activity(GO:0004320) myristoyl-[acyl-carrier-protein] hydrolase activity(GO:0016295) palmitoyl-[acyl-carrier-protein] hydrolase activity(GO:0016296) acyl-[acyl-carrier-protein] hydrolase activity(GO:0016297) S-acetyltransferase activity(GO:0016418) S-malonyltransferase activity(GO:0016419) malonyltransferase activity(GO:0016420) phosphopantetheine binding(GO:0031177) |
| 2.6 | 31.2 | GO:0004499 | N,N-dimethylaniline monooxygenase activity(GO:0004499) |
| 2.6 | 12.9 | GO:0003986 | acetyl-CoA hydrolase activity(GO:0003986) |
| 2.6 | 15.3 | GO:0016019 | N-acetylmuramoyl-L-alanine amidase activity(GO:0008745) peptidoglycan receptor activity(GO:0016019) |
| 2.5 | 12.6 | GO:0004998 | transferrin receptor activity(GO:0004998) |
| 2.5 | 29.6 | GO:0036374 | glutathione hydrolase activity(GO:0036374) |
| 2.3 | 9.3 | GO:0003960 | NADPH:quinone reductase activity(GO:0003960) |
| 1.9 | 11.2 | GO:0015199 | amino-acid betaine transmembrane transporter activity(GO:0015199) carnitine transmembrane transporter activity(GO:0015226) |
| 1.6 | 12.7 | GO:0034235 | GPI anchor binding(GO:0034235) |
| 1.5 | 39.2 | GO:0019825 | oxygen binding(GO:0019825) |
| 1.4 | 5.5 | GO:0047751 | 3-oxo-5-alpha-steroid 4-dehydrogenase activity(GO:0003865) cholestenone 5-alpha-reductase activity(GO:0047751) |
| 1.3 | 5.1 | GO:0071614 | linoleic acid epoxygenase activity(GO:0071614) |
| 1.0 | 4.2 | GO:0004736 | pyruvate carboxylase activity(GO:0004736) |
| 0.9 | 26.8 | GO:0070330 | aromatase activity(GO:0070330) |
| 0.8 | 2.5 | GO:0038052 | type 1 metabotropic glutamate receptor binding(GO:0031798) RNA polymerase II transcription factor activity, estrogen-activated sequence-specific DNA binding(GO:0038052) |
| 0.8 | 7.3 | GO:0004630 | phospholipase D activity(GO:0004630) |
| 0.8 | 22.9 | GO:0042975 | peroxisome proliferator activated receptor binding(GO:0042975) |
| 0.8 | 15.0 | GO:0035497 | cAMP response element binding(GO:0035497) |
| 0.7 | 4.3 | GO:0008321 | Ral guanyl-nucleotide exchange factor activity(GO:0008321) |
| 0.6 | 4.5 | GO:0005021 | vascular endothelial growth factor-activated receptor activity(GO:0005021) |
| 0.6 | 8.2 | GO:0015187 | glycine transmembrane transporter activity(GO:0015187) |
| 0.6 | 90.2 | GO:0004867 | serine-type endopeptidase inhibitor activity(GO:0004867) |
| 0.6 | 67.7 | GO:0004869 | cysteine-type endopeptidase inhibitor activity(GO:0004869) |
| 0.6 | 2.8 | GO:0052834 | inositol monophosphate 1-phosphatase activity(GO:0008934) inositol monophosphate 3-phosphatase activity(GO:0052832) inositol monophosphate 4-phosphatase activity(GO:0052833) inositol monophosphate phosphatase activity(GO:0052834) |
| 0.5 | 8.8 | GO:0009931 | calcium-dependent protein serine/threonine kinase activity(GO:0009931) |
| 0.5 | 2.1 | GO:0008113 | peptide-methionine (S)-S-oxide reductase activity(GO:0008113) |
| 0.5 | 39.0 | GO:0017080 | sodium channel regulator activity(GO:0017080) |
| 0.5 | 3.2 | GO:0004035 | alkaline phosphatase activity(GO:0004035) |
| 0.4 | 10.2 | GO:0004866 | endopeptidase inhibitor activity(GO:0004866) |
| 0.4 | 15.4 | GO:0051537 | 2 iron, 2 sulfur cluster binding(GO:0051537) |
| 0.4 | 1.7 | GO:0004105 | choline-phosphate cytidylyltransferase activity(GO:0004105) |
| 0.4 | 17.1 | GO:0008395 | steroid hydroxylase activity(GO:0008395) |
| 0.3 | 72.9 | GO:0004252 | serine-type endopeptidase activity(GO:0004252) |
| 0.3 | 9.9 | GO:0043395 | heparan sulfate proteoglycan binding(GO:0043395) |
| 0.3 | 7.7 | GO:0050136 | NADH dehydrogenase (ubiquinone) activity(GO:0008137) NADH dehydrogenase (quinone) activity(GO:0050136) |
| 0.3 | 3.2 | GO:0008020 | G-protein coupled photoreceptor activity(GO:0008020) |
| 0.3 | 2.7 | GO:0031014 | troponin T binding(GO:0031014) |
| 0.3 | 1.9 | GO:0015220 | choline transmembrane transporter activity(GO:0015220) |
| 0.2 | 2.4 | GO:0001640 | adenylate cyclase inhibiting G-protein coupled glutamate receptor activity(GO:0001640) |
| 0.2 | 5.4 | GO:0004697 | protein kinase C activity(GO:0004697) |
| 0.2 | 2.4 | GO:0003910 | DNA ligase activity(GO:0003909) DNA ligase (ATP) activity(GO:0003910) |
| 0.2 | 13.8 | GO:0051019 | mitogen-activated protein kinase binding(GO:0051019) |
| 0.2 | 4.0 | GO:0046933 | proton-transporting ATP synthase activity, rotational mechanism(GO:0046933) |
| 0.2 | 17.2 | GO:0005507 | copper ion binding(GO:0005507) |
| 0.2 | 3.0 | GO:0003993 | acid phosphatase activity(GO:0003993) |
| 0.2 | 3.4 | GO:0030169 | low-density lipoprotein particle binding(GO:0030169) |
| 0.2 | 1.3 | GO:0003747 | translation release factor activity(GO:0003747) translation termination factor activity(GO:0008079) |
| 0.1 | 1.2 | GO:0000293 | ferric-chelate reductase activity(GO:0000293) cupric reductase activity(GO:0008823) ferric-chelate reductase (NADPH) activity(GO:0052851) |
| 0.1 | 3.5 | GO:0070628 | proteasome binding(GO:0070628) |
| 0.1 | 3.2 | GO:0003841 | 1-acylglycerol-3-phosphate O-acyltransferase activity(GO:0003841) |
| 0.1 | 0.8 | GO:0000309 | nicotinamide-nucleotide adenylyltransferase activity(GO:0000309) nicotinate-nucleotide adenylyltransferase activity(GO:0004515) |
| 0.1 | 26.4 | GO:0052689 | carboxylic ester hydrolase activity(GO:0052689) |
| 0.1 | 3.3 | GO:0030898 | actin-dependent ATPase activity(GO:0030898) |
| 0.1 | 1.5 | GO:0097322 | 7SK snRNA binding(GO:0097322) |
| 0.1 | 5.7 | GO:0016831 | carboxy-lyase activity(GO:0016831) |
| 0.1 | 2.2 | GO:0005328 | neurotransmitter:sodium symporter activity(GO:0005328) |
| 0.1 | 1.0 | GO:0004439 | phosphatidylinositol-4,5-bisphosphate 5-phosphatase activity(GO:0004439) |
| 0.1 | 0.7 | GO:0017050 | sphinganine kinase activity(GO:0008481) D-erythro-sphingosine kinase activity(GO:0017050) |
| 0.1 | 1.3 | GO:0004791 | thioredoxin-disulfide reductase activity(GO:0004791) |
| 0.1 | 1.0 | GO:0017176 | phosphatidylinositol N-acetylglucosaminyltransferase activity(GO:0017176) |
| 0.1 | 5.1 | GO:0043022 | ribosome binding(GO:0043022) |
| 0.1 | 0.4 | GO:0004689 | phosphorylase kinase activity(GO:0004689) |
| 0.1 | 1.0 | GO:0001206 | transcriptional repressor activity, RNA polymerase II distal enhancer sequence-specific binding(GO:0001206) |
| 0.1 | 1.2 | GO:0005247 | voltage-gated chloride channel activity(GO:0005247) |
| 0.1 | 1.2 | GO:0008510 | sodium:bicarbonate symporter activity(GO:0008510) |
| 0.1 | 2.4 | GO:0001106 | RNA polymerase II transcription corepressor activity(GO:0001106) |
| 0.0 | 7.2 | GO:0005179 | hormone activity(GO:0005179) |
| 0.0 | 5.6 | GO:0003777 | microtubule motor activity(GO:0003777) |
| 0.0 | 2.8 | GO:0001965 | G-protein alpha-subunit binding(GO:0001965) |
| 0.0 | 1.1 | GO:0031434 | mitogen-activated protein kinase kinase binding(GO:0031434) |
| 0.0 | 6.9 | GO:0030165 | PDZ domain binding(GO:0030165) |
| 0.0 | 1.5 | GO:0050750 | low-density lipoprotein particle receptor binding(GO:0050750) |
| 0.0 | 1.8 | GO:0008188 | neuropeptide receptor activity(GO:0008188) |
| 0.0 | 1.0 | GO:0016881 | acid-amino acid ligase activity(GO:0016881) |
| 0.0 | 1.2 | GO:0044183 | protein binding involved in protein folding(GO:0044183) |
| 0.0 | 1.1 | GO:0017134 | fibroblast growth factor binding(GO:0017134) |
| 0.0 | 2.7 | GO:0004222 | metalloendopeptidase activity(GO:0004222) |
| 0.0 | 0.6 | GO:0016676 | cytochrome-c oxidase activity(GO:0004129) heme-copper terminal oxidase activity(GO:0015002) oxidoreductase activity, acting on a heme group of donors, oxygen as acceptor(GO:0016676) |
| 0.0 | 5.3 | GO:0004721 | phosphoprotein phosphatase activity(GO:0004721) |
| 0.0 | 0.3 | GO:0004634 | phosphopyruvate hydratase activity(GO:0004634) |
| 0.0 | 2.1 | GO:0004860 | protein kinase inhibitor activity(GO:0004860) |
| 0.0 | 1.4 | GO:0051082 | unfolded protein binding(GO:0051082) |
| 0.0 | 0.5 | GO:0016702 | oxidoreductase activity, acting on single donors with incorporation of molecular oxygen, incorporation of two atoms of oxygen(GO:0016702) |
| 0.0 | 0.3 | GO:0033038 | bitter taste receptor activity(GO:0033038) |
| 0.0 | 0.0 | GO:0003976 | UDP-N-acetylglucosamine-lysosomal-enzyme N-acetylglucosaminephosphotransferase activity(GO:0003976) |
| 0.0 | 2.2 | GO:0001085 | RNA polymerase II transcription factor binding(GO:0001085) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.3 | 25.4 | PID ERB GENOMIC PATHWAY | Validated nuclear estrogen receptor beta network |
| 1.1 | 39.3 | ST WNT CA2 CYCLIC GMP PATHWAY | Wnt/Ca2+/cyclic GMP signaling. |
| 0.4 | 4.5 | PID VEGF VEGFR PATHWAY | VEGF and VEGFR signaling network |
| 0.4 | 28.7 | PID AMB2 NEUTROPHILS PATHWAY | amb2 Integrin signaling |
| 0.3 | 70.0 | NABA ECM AFFILIATED | Genes encoding proteins affiliated structurally or functionally to extracellular matrix proteins |
| 0.3 | 90.2 | NABA ECM REGULATORS | Genes encoding enzymes and their regulators involved in the remodeling of the extracellular matrix |
| 0.2 | 0.9 | ST PAC1 RECEPTOR PATHWAY | PAC1 Receptor Pathway |
| 0.2 | 7.3 | PID UPA UPAR PATHWAY | Urokinase-type plasminogen activator (uPA) and uPAR-mediated signaling |
| 0.1 | 8.0 | PID DELTA NP63 PATHWAY | Validated transcriptional targets of deltaNp63 isoforms |
| 0.0 | 2.5 | PID FOXM1 PATHWAY | FOXM1 transcription factor network |
| 0.0 | 1.2 | SA CASPASE CASCADE | Apoptosis is mediated by caspases, cysteine proteases arranged in a proteolytic cascade. |
| 0.0 | 12.4 | NABA MATRISOME ASSOCIATED | Ensemble of genes encoding ECM-associated proteins including ECM-affilaited proteins, ECM regulators and secreted factors |
| 0.0 | 2.2 | PID HEDGEHOG GLI PATHWAY | Hedgehog signaling events mediated by Gli proteins |
| 0.0 | 0.7 | PID S1P META PATHWAY | Sphingosine 1-phosphate (S1P) pathway |
| 0.0 | 1.1 | ST ERK1 ERK2 MAPK PATHWAY | ERK1/ERK2 MAPK Pathway |
| 0.0 | 1.2 | PID AP1 PATHWAY | AP-1 transcription factor network |
| 0.0 | 1.0 | PID CASPASE PATHWAY | Caspase cascade in apoptosis |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 5.4 | 59.3 | REACTOME RECYCLING OF BILE ACIDS AND SALTS | Genes involved in Recycling of bile acids and salts |
| 2.8 | 39.2 | REACTOME TRYPTOPHAN CATABOLISM | Genes involved in Tryptophan catabolism |
| 2.6 | 36.4 | REACTOME REGULATION OF COMPLEMENT CASCADE | Genes involved in Regulation of Complement cascade |
| 1.8 | 23.6 | REACTOME GAMMA CARBOXYLATION TRANSPORT AND AMINO TERMINAL CLEAVAGE OF PROTEINS | Genes involved in Gamma-carboxylation, transport, and amino-terminal cleavage of proteins |
| 1.7 | 49.4 | REACTOME CHYLOMICRON MEDIATED LIPID TRANSPORT | Genes involved in Chylomicron-mediated lipid transport |
| 1.5 | 36.4 | REACTOME ALPHA LINOLENIC ACID ALA METABOLISM | Genes involved in alpha-linolenic acid (ALA) metabolism |
| 1.2 | 14.7 | REACTOME COMMON PATHWAY | Genes involved in Common Pathway |
| 0.5 | 9.9 | REACTOME HS GAG DEGRADATION | Genes involved in HS-GAG degradation |
| 0.5 | 8.0 | REACTOME VITAMIN B5 PANTOTHENATE METABOLISM | Genes involved in Vitamin B5 (pantothenate) metabolism |
| 0.5 | 11.2 | REACTOME ACTIVATED AMPK STIMULATES FATTY ACID OXIDATION IN MUSCLE | Genes involved in Activated AMPK stimulates fatty-acid oxidation in muscle |
| 0.4 | 4.5 | REACTOME VEGF LIGAND RECEPTOR INTERACTIONS | Genes involved in VEGF ligand-receptor interactions |
| 0.4 | 5.4 | REACTOME TRANSPORT OF ORGANIC ANIONS | Genes involved in Transport of organic anions |
| 0.4 | 13.0 | REACTOME CREB PHOSPHORYLATION THROUGH THE ACTIVATION OF CAMKII | Genes involved in CREB phosphorylation through the activation of CaMKII |
| 0.4 | 7.7 | REACTOME SYNTHESIS OF PE | Genes involved in Synthesis of PE |
| 0.4 | 5.5 | REACTOME ANDROGEN BIOSYNTHESIS | Genes involved in Androgen biosynthesis |
| 0.4 | 22.1 | REACTOME PHASE1 FUNCTIONALIZATION OF COMPOUNDS | Genes involved in Phase 1 - Functionalization of compounds |
| 0.3 | 7.4 | REACTOME NA CL DEPENDENT NEUROTRANSMITTER TRANSPORTERS | Genes involved in Na+/Cl- dependent neurotransmitter transporters |
| 0.3 | 3.2 | REACTOME OPSINS | Genes involved in Opsins |
| 0.2 | 3.5 | REACTOME SYNTHESIS OF PC | Genes involved in Synthesis of PC |
| 0.2 | 4.0 | REACTOME FORMATION OF ATP BY CHEMIOSMOTIC COUPLING | Genes involved in Formation of ATP by chemiosmotic coupling |
| 0.2 | 17.7 | REACTOME NUCLEAR RECEPTOR TRANSCRIPTION PATHWAY | Genes involved in Nuclear Receptor transcription pathway |
| 0.2 | 12.1 | REACTOME RESPIRATORY ELECTRON TRANSPORT | Genes involved in Respiratory electron transport |
| 0.1 | 6.4 | REACTOME TRANSPORT TO THE GOLGI AND SUBSEQUENT MODIFICATION | Genes involved in Transport to the Golgi and subsequent modification |
| 0.1 | 2.4 | REACTOME CLASS C 3 METABOTROPIC GLUTAMATE PHEROMONE RECEPTORS | Genes involved in Class C/3 (Metabotropic glutamate/pheromone receptors) |
| 0.1 | 27.8 | REACTOME METABOLISM OF AMINO ACIDS AND DERIVATIVES | Genes involved in Metabolism of amino acids and derivatives |
| 0.1 | 3.3 | REACTOME SEMA4D INDUCED CELL MIGRATION AND GROWTH CONE COLLAPSE | Genes involved in Sema4D induced cell migration and growth-cone collapse |
| 0.1 | 1.2 | REACTOME APOPTOSIS INDUCED DNA FRAGMENTATION | Genes involved in Apoptosis induced DNA fragmentation |
| 0.1 | 1.7 | REACTOME REGULATION OF INSULIN LIKE GROWTH FACTOR IGF ACTIVITY BY INSULIN LIKE GROWTH FACTOR BINDING PROTEINS IGFBPS | Genes involved in Regulation of Insulin-like Growth Factor (IGF) Activity by Insulin-like Growth Factor Binding Proteins (IGFBPs) |
| 0.1 | 5.1 | REACTOME PLATELET AGGREGATION PLUG FORMATION | Genes involved in Platelet Aggregation (Plug Formation) |
| 0.1 | 1.5 | REACTOME RETROGRADE NEUROTROPHIN SIGNALLING | Genes involved in Retrograde neurotrophin signalling |
| 0.1 | 3.2 | REACTOME SYNTHESIS OF PA | Genes involved in Synthesis of PA |
| 0.1 | 3.8 | REACTOME METABOLISM OF VITAMINS AND COFACTORS | Genes involved in Metabolism of vitamins and cofactors |
| 0.0 | 1.5 | REACTOME ELONGATION ARREST AND RECOVERY | Genes involved in Elongation arrest and recovery |
| 0.0 | 2.7 | REACTOME STRIATED MUSCLE CONTRACTION | Genes involved in Striated Muscle Contraction |
| 0.0 | 0.5 | REACTOME CALNEXIN CALRETICULIN CYCLE | Genes involved in Calnexin/calreticulin cycle |
| 0.0 | 1.2 | REACTOME TRANSFERRIN ENDOCYTOSIS AND RECYCLING | Genes involved in Transferrin endocytosis and recycling |
| 0.0 | 0.4 | REACTOME GLYCOGEN BREAKDOWN GLYCOGENOLYSIS | Genes involved in Glycogen breakdown (glycogenolysis) |
| 0.0 | 1.1 | REACTOME FORMATION OF THE TERNARY COMPLEX AND SUBSEQUENTLY THE 43S COMPLEX | Genes involved in Formation of the ternary complex, and subsequently, the 43S complex |
| 0.0 | 0.7 | REACTOME SPHINGOLIPID DE NOVO BIOSYNTHESIS | Genes involved in Sphingolipid de novo biosynthesis |
| 0.0 | 1.3 | REACTOME NONSENSE MEDIATED DECAY ENHANCED BY THE EXON JUNCTION COMPLEX | Genes involved in Nonsense Mediated Decay Enhanced by the Exon Junction Complex |
| 0.0 | 0.2 | REACTOME GLUCAGON TYPE LIGAND RECEPTORS | Genes involved in Glucagon-type ligand receptors |
| 0.0 | 2.8 | REACTOME G ALPHA I SIGNALLING EVENTS | Genes involved in G alpha (i) signalling events |
| 0.0 | 0.9 | REACTOME PEPTIDE LIGAND BINDING RECEPTORS | Genes involved in Peptide ligand-binding receptors |