Project
PRJNA375882: Comprehensive Mouse Transcriptomic BodyMap
Navigation
Downloads
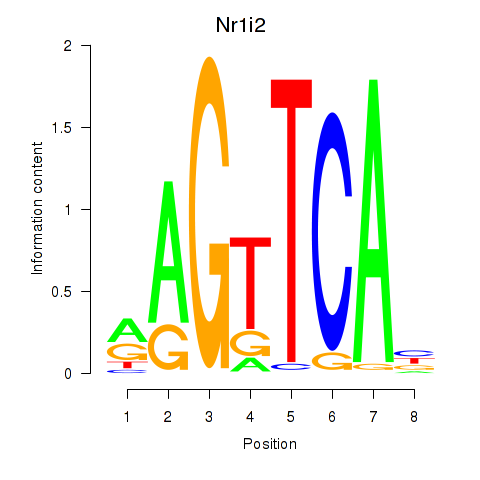
Results for Nr1i2
Z-value: 1.02
Transcription factors associated with Nr1i2
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Nr1i2
|
ENSMUSG00000022809.5 | Nr1i2 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Nr1i2 | mm39_v1_chr16_-_38115172_38115200 | -0.26 | 2.5e-02 | Click! |
Activity profile of Nr1i2 motif
Sorted Z-values of Nr1i2 motif
Network of associatons between targets according to the STRING database.
First level regulatory network of Nr1i2
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr2_-_25359752 | 5.78 |
ENSMUST00000114259.3
ENSMUST00000015234.13 |
Ptgds
|
prostaglandin D2 synthase (brain) |
| chr2_-_25360043 | 5.56 |
ENSMUST00000114251.8
|
Ptgds
|
prostaglandin D2 synthase (brain) |
| chr3_-_92528480 | 4.51 |
ENSMUST00000170676.3
|
Lce6a
|
late cornified envelope 6A |
| chr2_+_135899847 | 3.71 |
ENSMUST00000057503.7
|
Lamp5
|
lysosomal-associated membrane protein family, member 5 |
| chr10_+_126914755 | 3.70 |
ENSMUST00000039259.7
ENSMUST00000217941.2 |
Agap2
|
ArfGAP with GTPase domain, ankyrin repeat and PH domain 2 |
| chr15_-_44978223 | 3.67 |
ENSMUST00000022967.7
|
Kcnv1
|
potassium channel, subfamily V, member 1 |
| chr9_-_53882530 | 3.61 |
ENSMUST00000048409.14
|
Elmod1
|
ELMO/CED-12 domain containing 1 |
| chr1_+_17215581 | 3.58 |
ENSMUST00000026879.8
|
Gdap1
|
ganglioside-induced differentiation-associated-protein 1 |
| chr17_-_37334240 | 3.49 |
ENSMUST00000102665.11
|
Mog
|
myelin oligodendrocyte glycoprotein |
| chr8_-_58249608 | 3.46 |
ENSMUST00000204067.3
|
Galntl6
|
UDP-N-acetyl-alpha-D-galactosamine:polypeptide N-acetylgalactosaminyltransferase-like 6 |
| chr14_-_30637344 | 3.41 |
ENSMUST00000226547.2
|
Itih3
|
inter-alpha trypsin inhibitor, heavy chain 3 |
| chr16_+_41353360 | 3.30 |
ENSMUST00000099761.10
|
Lsamp
|
limbic system-associated membrane protein |
| chr3_-_92594516 | 3.20 |
ENSMUST00000029524.4
|
Lce1d
|
late cornified envelope 1D |
| chr9_+_119978773 | 3.20 |
ENSMUST00000068698.15
ENSMUST00000215512.2 ENSMUST00000111627.3 ENSMUST00000093773.8 |
Mobp
|
myelin-associated oligodendrocytic basic protein |
| chr1_+_72863641 | 3.18 |
ENSMUST00000047328.11
|
Igfbp2
|
insulin-like growth factor binding protein 2 |
| chr16_+_20513658 | 3.17 |
ENSMUST00000056518.13
|
Fam131a
|
family with sequence similarity 131, member A |
| chr3_-_91990439 | 3.14 |
ENSMUST00000058150.8
|
Lor
|
loricrin |
| chr17_+_44263890 | 3.12 |
ENSMUST00000177857.9
ENSMUST00000044792.6 |
Rcan2
|
regulator of calcineurin 2 |
| chr4_+_110254858 | 2.98 |
ENSMUST00000106589.9
ENSMUST00000106587.9 ENSMUST00000106591.8 ENSMUST00000106592.8 |
Agbl4
|
ATP/GTP binding protein-like 4 |
| chr1_-_22386016 | 2.97 |
ENSMUST00000164877.8
|
Rims1
|
regulating synaptic membrane exocytosis 1 |
| chr7_+_43460718 | 2.95 |
ENSMUST00000032955.7
|
Klk7
|
kallikrein related-peptidase 7 (chymotryptic, stratum corneum) |
| chr7_+_3352159 | 2.91 |
ENSMUST00000172109.4
|
Prkcg
|
protein kinase C, gamma |
| chr16_+_17149235 | 2.90 |
ENSMUST00000023450.15
ENSMUST00000231884.2 |
Serpind1
|
serine (or cysteine) peptidase inhibitor, clade D, member 1 |
| chr7_+_3352019 | 2.87 |
ENSMUST00000100301.11
|
Prkcg
|
protein kinase C, gamma |
| chr7_+_24048613 | 2.86 |
ENSMUST00000032683.6
|
Lypd5
|
Ly6/Plaur domain containing 5 |
| chr6_-_60805873 | 2.85 |
ENSMUST00000114268.5
|
Snca
|
synuclein, alpha |
| chr2_+_129854256 | 2.83 |
ENSMUST00000110299.3
|
Tgm3
|
transglutaminase 3, E polypeptide |
| chr18_-_61147272 | 2.80 |
ENSMUST00000025520.10
|
Slc6a7
|
solute carrier family 6 (neurotransmitter transporter, L-proline), member 7 |
| chr12_+_103354915 | 2.78 |
ENSMUST00000101094.9
ENSMUST00000021620.13 |
Otub2
|
OTU domain, ubiquitin aldehyde binding 2 |
| chr7_+_25005510 | 2.76 |
ENSMUST00000119703.8
ENSMUST00000205639.3 ENSMUST00000108409.2 |
Tmem145
|
transmembrane protein 145 |
| chr3_-_92758591 | 2.75 |
ENSMUST00000054426.5
|
Lce1l
|
late cornified envelope 1L |
| chr7_+_43361930 | 2.74 |
ENSMUST00000066834.8
|
Klk13
|
kallikrein related-peptidase 13 |
| chr3_-_92734546 | 2.72 |
ENSMUST00000072363.5
|
Kprp
|
keratinocyte expressed, proline-rich |
| chr17_-_37334091 | 2.68 |
ENSMUST00000167275.3
|
Mog
|
myelin oligodendrocyte glycoprotein |
| chr3_-_127019496 | 2.63 |
ENSMUST00000182064.9
ENSMUST00000182452.8 |
Ank2
|
ankyrin 2, brain |
| chr11_+_67477347 | 2.61 |
ENSMUST00000108682.9
|
Gas7
|
growth arrest specific 7 |
| chr5_+_14075281 | 2.58 |
ENSMUST00000073957.8
|
Sema3e
|
sema domain, immunoglobulin domain (Ig), short basic domain, secreted, (semaphorin) 3E |
| chr2_+_121125918 | 2.52 |
ENSMUST00000110639.8
|
Map1a
|
microtubule-associated protein 1 A |
| chrX_+_10351360 | 2.47 |
ENSMUST00000076354.13
ENSMUST00000115526.2 |
Tspan7
|
tetraspanin 7 |
| chr4_+_110254907 | 2.45 |
ENSMUST00000097920.9
ENSMUST00000080744.13 |
Agbl4
|
ATP/GTP binding protein-like 4 |
| chr16_+_91066602 | 2.43 |
ENSMUST00000056882.7
|
Olig1
|
oligodendrocyte transcription factor 1 |
| chr2_+_25132941 | 2.42 |
ENSMUST00000114355.2
ENSMUST00000060818.2 |
Rnf208
|
ring finger protein 208 |
| chr11_+_115225557 | 2.41 |
ENSMUST00000106543.8
ENSMUST00000019006.5 |
Otop3
|
otopetrin 3 |
| chr10_-_5144699 | 2.40 |
ENSMUST00000215467.2
|
Syne1
|
spectrin repeat containing, nuclear envelope 1 |
| chr2_+_71811526 | 2.37 |
ENSMUST00000090826.12
ENSMUST00000102698.10 |
Rapgef4
|
Rap guanine nucleotide exchange factor (GEF) 4 |
| chr4_-_62005498 | 2.37 |
ENSMUST00000107488.4
ENSMUST00000107472.8 ENSMUST00000084531.11 |
Mup3
|
major urinary protein 3 |
| chr15_-_74624811 | 2.37 |
ENSMUST00000189128.2
ENSMUST00000023259.15 |
Lynx1
|
Ly6/neurotoxin 1 |
| chr8_-_70573465 | 2.33 |
ENSMUST00000002412.9
|
Ncan
|
neurocan |
| chr5_+_37242714 | 2.32 |
ENSMUST00000121010.9
ENSMUST00000174629.2 ENSMUST00000232332.3 |
Jakmip1
Gm1043
|
janus kinase and microtubule interacting protein 1 predicted gene 1043 |
| chr1_-_84673903 | 2.32 |
ENSMUST00000049126.13
|
Dner
|
delta/notch-like EGF repeat containing |
| chr7_-_126062272 | 2.30 |
ENSMUST00000032974.13
|
Atp2a1
|
ATPase, Ca++ transporting, cardiac muscle, fast twitch 1 |
| chr8_+_55024446 | 2.27 |
ENSMUST00000239166.2
ENSMUST00000239106.2 ENSMUST00000239152.2 |
Asb5
|
ankyrin repeat and SOCs box-containing 5 |
| chr7_-_81143631 | 2.27 |
ENSMUST00000082090.15
|
Ap3b2
|
adaptor-related protein complex 3, beta 2 subunit |
| chr17_+_55752370 | 2.27 |
ENSMUST00000133899.2
ENSMUST00000086878.10 |
St6gal2
|
beta galactoside alpha 2,6 sialyltransferase 2 |
| chr9_-_77159111 | 2.26 |
ENSMUST00000184322.8
ENSMUST00000184316.2 |
Mlip
|
muscular LMNA-interacting protein |
| chr2_+_32518402 | 2.25 |
ENSMUST00000156578.8
|
Ak1
|
adenylate kinase 1 |
| chr11_-_3672188 | 2.22 |
ENSMUST00000102950.10
ENSMUST00000101632.4 |
Osbp2
|
oxysterol binding protein 2 |
| chr9_-_103097022 | 2.21 |
ENSMUST00000168142.8
|
Trf
|
transferrin |
| chr13_-_91053440 | 2.17 |
ENSMUST00000109541.4
|
Atp6ap1l
|
ATPase, H+ transporting, lysosomal accessory protein 1-like |
| chr8_-_100143029 | 2.15 |
ENSMUST00000155527.8
ENSMUST00000142129.8 ENSMUST00000093249.11 ENSMUST00000142475.3 ENSMUST00000128860.8 |
Cdh8
|
cadherin 8 |
| chr5_-_118382926 | 2.13 |
ENSMUST00000117177.8
ENSMUST00000133372.2 ENSMUST00000154786.8 ENSMUST00000121369.8 |
Rnft2
|
ring finger protein, transmembrane 2 |
| chr11_+_42310557 | 2.13 |
ENSMUST00000007797.10
|
Gabrb2
|
gamma-aminobutyric acid (GABA) A receptor, subunit beta 2 |
| chr17_+_55752485 | 2.11 |
ENSMUST00000025000.4
|
St6gal2
|
beta galactoside alpha 2,6 sialyltransferase 2 |
| chr4_-_41774097 | 2.09 |
ENSMUST00000108036.8
ENSMUST00000108037.9 ENSMUST00000108032.3 ENSMUST00000173865.9 ENSMUST00000155240.2 |
Ccl27a
|
chemokine (C-C motif) ligand 27A |
| chr5_-_8417982 | 2.08 |
ENSMUST00000088761.11
ENSMUST00000115386.8 ENSMUST00000050166.14 ENSMUST00000046838.14 ENSMUST00000115388.9 ENSMUST00000088744.12 ENSMUST00000115385.2 |
Adam22
|
a disintegrin and metallopeptidase domain 22 |
| chr2_+_164802766 | 2.05 |
ENSMUST00000202223.4
|
Slc12a5
|
solute carrier family 12, member 5 |
| chr2_+_164802729 | 2.05 |
ENSMUST00000202623.4
|
Slc12a5
|
solute carrier family 12, member 5 |
| chr19_-_46315543 | 2.05 |
ENSMUST00000223917.2
ENSMUST00000224447.2 ENSMUST00000041391.5 ENSMUST00000096029.12 |
Psd
|
pleckstrin and Sec7 domain containing |
| chr5_-_87074380 | 2.04 |
ENSMUST00000031183.3
|
Ugt2b1
|
UDP glucuronosyltransferase 2 family, polypeptide B1 |
| chr11_+_67477501 | 2.04 |
ENSMUST00000108680.2
|
Gas7
|
growth arrest specific 7 |
| chrX_+_139243012 | 2.03 |
ENSMUST00000208130.2
|
Frmpd3
|
FERM and PDZ domain containing 3 |
| chr3_+_92874366 | 2.03 |
ENSMUST00000098886.5
|
Lce3e
|
late cornified envelope 3E |
| chr5_+_57875309 | 2.00 |
ENSMUST00000191837.6
ENSMUST00000068110.10 |
Pcdh7
|
protocadherin 7 |
| chr17_-_90763300 | 1.99 |
ENSMUST00000159778.8
ENSMUST00000174337.8 ENSMUST00000172466.8 |
Nrxn1
|
neurexin I |
| chr5_+_13448833 | 1.98 |
ENSMUST00000137798.10
|
Sema3a
|
sema domain, immunoglobulin domain (Ig), short basic domain, secreted, (semaphorin) 3A |
| chrX_+_145745496 | 1.98 |
ENSMUST00000036303.9
ENSMUST00000096299.9 ENSMUST00000156697.8 |
Htr2c
|
5-hydroxytryptamine (serotonin) receptor 2C |
| chr4_-_116982804 | 1.97 |
ENSMUST00000183310.2
|
Btbd19
|
BTB (POZ) domain containing 19 |
| chr1_+_170472092 | 1.93 |
ENSMUST00000046792.9
|
Olfml2b
|
olfactomedin-like 2B |
| chr4_+_156300325 | 1.92 |
ENSMUST00000105572.3
|
Perm1
|
PPARGC1 and ESRR induced regulator, muscle 1 |
| chr4_-_4138817 | 1.91 |
ENSMUST00000133567.2
|
Penk
|
preproenkephalin |
| chr1_-_162305573 | 1.91 |
ENSMUST00000086074.12
ENSMUST00000070330.14 |
Dnm3
|
dynamin 3 |
| chr11_-_119907884 | 1.90 |
ENSMUST00000132575.8
|
Aatk
|
apoptosis-associated tyrosine kinase |
| chr6_-_37276885 | 1.89 |
ENSMUST00000101532.10
|
Dgki
|
diacylglycerol kinase, iota |
| chr17_+_44264130 | 1.88 |
ENSMUST00000229240.2
|
Rcan2
|
regulator of calcineurin 2 |
| chr5_+_36361360 | 1.87 |
ENSMUST00000052224.6
|
Psapl1
|
prosaposin-like 1 |
| chr1_+_135710803 | 1.87 |
ENSMUST00000132795.8
|
Tnni1
|
troponin I, skeletal, slow 1 |
| chr9_+_110595224 | 1.86 |
ENSMUST00000136695.3
|
Myl3
|
myosin, light polypeptide 3 |
| chr11_+_69945157 | 1.86 |
ENSMUST00000108585.9
ENSMUST00000018699.13 |
Asgr1
|
asialoglycoprotein receptor 1 |
| chr18_+_37610858 | 1.85 |
ENSMUST00000051442.7
|
Pcdhb16
|
protocadherin beta 16 |
| chr4_+_42438970 | 1.84 |
ENSMUST00000238328.2
|
Gm21586
|
predicted gene, 21586 |
| chr10_+_128030315 | 1.83 |
ENSMUST00000044776.13
|
Gls2
|
glutaminase 2 (liver, mitochondrial) |
| chrX_+_134894573 | 1.82 |
ENSMUST00000058119.9
|
Arxes2
|
adipocyte-related X-chromosome expressed sequence 2 |
| chr11_-_99313078 | 1.81 |
ENSMUST00000017741.4
|
Krt12
|
keratin 12 |
| chr3_-_80710097 | 1.80 |
ENSMUST00000075316.10
ENSMUST00000107745.8 |
Gria2
|
glutamate receptor, ionotropic, AMPA2 (alpha 2) |
| chr9_-_121324744 | 1.79 |
ENSMUST00000035120.6
|
Cck
|
cholecystokinin |
| chr2_+_148237258 | 1.78 |
ENSMUST00000109962.4
|
Sstr4
|
somatostatin receptor 4 |
| chr3_+_153679073 | 1.78 |
ENSMUST00000089948.6
|
Slc44a5
|
solute carrier family 44, member 5 |
| chr4_-_144447974 | 1.77 |
ENSMUST00000036876.8
|
AAdacl4fm3
|
AADACL4 family member 3 |
| chr13_-_93281065 | 1.77 |
ENSMUST00000062122.4
|
Cmya5
|
cardiomyopathy associated 5 |
| chr8_-_124621483 | 1.76 |
ENSMUST00000034453.6
ENSMUST00000212584.2 |
Acta1
|
actin, alpha 1, skeletal muscle |
| chr6_-_5496261 | 1.76 |
ENSMUST00000203347.3
ENSMUST00000019721.7 |
Pdk4
|
pyruvate dehydrogenase kinase, isoenzyme 4 |
| chr1_+_134121170 | 1.76 |
ENSMUST00000038445.13
ENSMUST00000191577.2 |
Mybph
|
myosin binding protein H |
| chr13_+_19132375 | 1.75 |
ENSMUST00000239207.2
ENSMUST00000003345.10 ENSMUST00000200466.5 |
Amph
|
amphiphysin |
| chr4_-_61259997 | 1.75 |
ENSMUST00000071005.9
ENSMUST00000075206.12 |
Mup14
|
major urinary protein 14 |
| chr6_-_136150076 | 1.75 |
ENSMUST00000053880.13
|
Grin2b
|
glutamate receptor, ionotropic, NMDA2B (epsilon 2) |
| chr11_+_104467791 | 1.74 |
ENSMUST00000106957.8
|
Myl4
|
myosin, light polypeptide 4 |
| chr7_-_105249308 | 1.74 |
ENSMUST00000210531.2
ENSMUST00000033185.10 |
Hpx
|
hemopexin |
| chr3_+_59914164 | 1.73 |
ENSMUST00000169794.2
|
Aadacl2
|
arylacetamide deacetylase like 2 |
| chr2_+_49956441 | 1.72 |
ENSMUST00000112712.10
ENSMUST00000128451.8 ENSMUST00000053208.14 |
Lypd6
|
LY6/PLAUR domain containing 6 |
| chr2_+_177760768 | 1.72 |
ENSMUST00000108917.8
|
Phactr3
|
phosphatase and actin regulator 3 |
| chr10_-_5500636 | 1.71 |
ENSMUST00000215295.2
ENSMUST00000041639.7 |
Syne1
|
spectrin repeat containing, nuclear envelope 1 |
| chr9_-_77159152 | 1.70 |
ENSMUST00000183955.8
|
Mlip
|
muscular LMNA-interacting protein |
| chr9_-_21874802 | 1.68 |
ENSMUST00000006397.7
|
Epor
|
erythropoietin receptor |
| chr7_+_27222678 | 1.68 |
ENSMUST00000108353.9
|
Hipk4
|
homeodomain interacting protein kinase 4 |
| chr11_+_7013422 | 1.68 |
ENSMUST00000020706.5
|
Adcy1
|
adenylate cyclase 1 |
| chr2_-_73605387 | 1.68 |
ENSMUST00000166199.9
|
Chn1
|
chimerin 1 |
| chr5_-_86616849 | 1.67 |
ENSMUST00000101073.3
|
Tmprss11a
|
transmembrane protease, serine 11a |
| chr12_+_74044435 | 1.67 |
ENSMUST00000221220.2
|
Syt16
|
synaptotagmin XVI |
| chr4_-_152402915 | 1.65 |
ENSMUST00000170820.3
ENSMUST00000076183.12 |
Rnf207
|
ring finger protein 207 |
| chr14_-_70867588 | 1.65 |
ENSMUST00000228009.2
|
Dmtn
|
dematin actin binding protein |
| chr4_+_41941572 | 1.63 |
ENSMUST00000108028.9
ENSMUST00000153997.8 |
Gm20878
|
predicted gene, 20878 |
| chr4_-_60777462 | 1.62 |
ENSMUST00000211875.2
|
Mup22
|
major urinary protein 22 |
| chr4_+_124779592 | 1.62 |
ENSMUST00000149146.2
|
Epha10
|
Eph receptor A10 |
| chrX_-_166638057 | 1.61 |
ENSMUST00000238211.2
|
Frmpd4
|
FERM and PDZ domain containing 4 |
| chr12_+_69954506 | 1.60 |
ENSMUST00000223456.2
|
Atl1
|
atlastin GTPase 1 |
| chr1_-_130589321 | 1.57 |
ENSMUST00000137276.3
|
C4bp
|
complement component 4 binding protein |
| chr7_+_43473943 | 1.56 |
ENSMUST00000107966.10
ENSMUST00000177514.8 |
Klk6
|
kallikrein related-peptidase 6 |
| chr6_-_55658242 | 1.56 |
ENSMUST00000044767.10
|
Neurod6
|
neurogenic differentiation 6 |
| chr2_+_96148418 | 1.56 |
ENSMUST00000135431.8
ENSMUST00000162807.9 |
Lrrc4c
|
leucine rich repeat containing 4C |
| chr17_-_56861383 | 1.56 |
ENSMUST00000209718.2
|
Tincr
|
TINCR ubiquitin domain containing |
| chr8_-_59154041 | 1.55 |
ENSMUST00000188531.7
|
Galntl6
|
UDP-N-acetyl-alpha-D-galactosamine:polypeptide N-acetylgalactosaminyltransferase-like 6 |
| chr2_+_109522781 | 1.55 |
ENSMUST00000111050.10
|
Bdnf
|
brain derived neurotrophic factor |
| chr16_-_89940652 | 1.54 |
ENSMUST00000114124.9
|
Tiam1
|
T cell lymphoma invasion and metastasis 1 |
| chr13_+_93444514 | 1.54 |
ENSMUST00000079086.8
|
Homer1
|
homer scaffolding protein 1 |
| chr12_-_41536430 | 1.54 |
ENSMUST00000043884.6
|
Lrrn3
|
leucine rich repeat protein 3, neuronal |
| chr2_+_61634797 | 1.53 |
ENSMUST00000048934.15
|
Tbr1
|
T-box brain transcription factor 1 |
| chr2_+_122146153 | 1.51 |
ENSMUST00000099461.4
|
Duox1
|
dual oxidase 1 |
| chr18_+_20643325 | 1.51 |
ENSMUST00000070892.8
ENSMUST00000234945.2 |
Dsg3
|
desmoglein 3 |
| chr4_-_60457902 | 1.51 |
ENSMUST00000084548.11
ENSMUST00000103012.10 ENSMUST00000107499.4 |
Mup1
|
major urinary protein 1 |
| chr10_-_63926044 | 1.51 |
ENSMUST00000105439.2
|
Lrrtm3
|
leucine rich repeat transmembrane neuronal 3 |
| chr17_+_3532455 | 1.50 |
ENSMUST00000227604.2
|
Tiam2
|
T cell lymphoma invasion and metastasis 2 |
| chr1_-_79836344 | 1.49 |
ENSMUST00000027467.11
|
Serpine2
|
serine (or cysteine) peptidase inhibitor, clade E, member 2 |
| chr1_-_130589349 | 1.48 |
ENSMUST00000027657.14
|
C4bp
|
complement component 4 binding protein |
| chr4_-_60697274 | 1.48 |
ENSMUST00000117932.2
|
Mup12
|
major urinary protein 12 |
| chr12_-_4891435 | 1.47 |
ENSMUST00000219880.2
ENSMUST00000020964.7 |
Fkbp1b
|
FK506 binding protein 1b |
| chr2_-_53975501 | 1.47 |
ENSMUST00000100089.3
|
Rprm
|
reprimo, TP53 dependent G2 arrest mediator candidate |
| chr15_-_63932176 | 1.46 |
ENSMUST00000226675.2
ENSMUST00000228226.2 ENSMUST00000227024.2 |
Cyrib
|
CYFIP related Rac1 interactor B |
| chrX_-_133062677 | 1.46 |
ENSMUST00000033611.5
|
Xkrx
|
X-linked Kx blood group related, X-linked |
| chr4_-_135300934 | 1.45 |
ENSMUST00000105855.2
|
Grhl3
|
grainyhead like transcription factor 3 |
| chr3_-_127202635 | 1.44 |
ENSMUST00000182959.8
|
Ank2
|
ankyrin 2, brain |
| chr14_-_65662609 | 1.43 |
ENSMUST00000224594.2
|
Pnoc
|
prepronociceptin |
| chr6_+_96092230 | 1.43 |
ENSMUST00000075080.6
|
Tafa1
|
TAFA chemokine like family member 1 |
| chr6_+_115111860 | 1.43 |
ENSMUST00000169345.4
|
Syn2
|
synapsin II |
| chr15_+_30457772 | 1.43 |
ENSMUST00000228282.2
|
Ctnnd2
|
catenin (cadherin associated protein), delta 2 |
| chr17_-_56428968 | 1.43 |
ENSMUST00000041357.9
|
Lrg1
|
leucine-rich alpha-2-glycoprotein 1 |
| chr18_-_66993567 | 1.42 |
ENSMUST00000057942.4
|
Mc4r
|
melanocortin 4 receptor |
| chr18_+_42186713 | 1.42 |
ENSMUST00000072008.11
|
Sh3rf2
|
SH3 domain containing ring finger 2 |
| chr3_-_97731826 | 1.42 |
ENSMUST00000200232.3
|
Pde4dip
|
phosphodiesterase 4D interacting protein (myomegalin) |
| chr1_+_182591425 | 1.41 |
ENSMUST00000155229.7
ENSMUST00000153348.8 |
Susd4
|
sushi domain containing 4 |
| chr6_+_49799690 | 1.41 |
ENSMUST00000031843.7
|
Npy
|
neuropeptide Y |
| chr4_+_42293921 | 1.40 |
ENSMUST00000238203.2
|
Gm21953
|
predicted gene, 21953 |
| chr13_-_78345932 | 1.40 |
ENSMUST00000127137.3
|
Nr2f1
|
nuclear receptor subfamily 2, group F, member 1 |
| chr8_-_86107593 | 1.40 |
ENSMUST00000122452.8
|
Mylk3
|
myosin light chain kinase 3 |
| chr9_-_54568950 | 1.40 |
ENSMUST00000128624.2
|
Acsbg1
|
acyl-CoA synthetase bubblegum family member 1 |
| chr9_+_87026337 | 1.39 |
ENSMUST00000113149.8
ENSMUST00000049457.14 ENSMUST00000179313.3 |
Mrap2
|
melanocortin 2 receptor accessory protein 2 |
| chr9_+_109865810 | 1.39 |
ENSMUST00000163190.8
|
Map4
|
microtubule-associated protein 4 |
| chr3_-_26207456 | 1.39 |
ENSMUST00000191835.2
|
Nlgn1
|
neuroligin 1 |
| chr4_-_60222580 | 1.39 |
ENSMUST00000095058.5
ENSMUST00000163931.8 |
Mup8
|
major urinary protein 8 |
| chrX_-_101952278 | 1.37 |
ENSMUST00000119624.8
ENSMUST00000033686.2 |
Dmrtc1a
|
DMRT-like family C1a |
| chr4_-_134099840 | 1.37 |
ENSMUST00000030643.3
|
Extl1
|
exostosin-like glycosyltransferase 1 |
| chr8_-_85500998 | 1.35 |
ENSMUST00000109762.8
|
Nfix
|
nuclear factor I/X |
| chr6_+_115111872 | 1.34 |
ENSMUST00000009538.12
ENSMUST00000203450.2 |
Syn2
|
synapsin II |
| chr17_-_36220924 | 1.34 |
ENSMUST00000141662.8
ENSMUST00000056034.13 ENSMUST00000077494.13 ENSMUST00000149277.8 ENSMUST00000061052.12 |
Atat1
|
alpha tubulin acetyltransferase 1 |
| chr3_+_96432479 | 1.34 |
ENSMUST00000049208.11
|
Hjv
|
hemojuvelin BMP co-receptor |
| chr18_+_42186757 | 1.34 |
ENSMUST00000074679.4
|
Sh3rf2
|
SH3 domain containing ring finger 2 |
| chr6_+_141575226 | 1.33 |
ENSMUST00000042812.9
|
Slco1b2
|
solute carrier organic anion transporter family, member 1b2 |
| chr3_+_129974531 | 1.31 |
ENSMUST00000080335.11
ENSMUST00000106353.2 |
Col25a1
|
collagen, type XXV, alpha 1 |
| chr6_+_21215472 | 1.31 |
ENSMUST00000081542.6
|
Kcnd2
|
potassium voltage-gated channel, Shal-related family, member 2 |
| chr2_+_177783713 | 1.29 |
ENSMUST00000103066.10
|
Phactr3
|
phosphatase and actin regulator 3 |
| chr4_-_61259801 | 1.29 |
ENSMUST00000125461.8
|
Mup14
|
major urinary protein 14 |
| chr11_-_69088635 | 1.29 |
ENSMUST00000094078.4
ENSMUST00000021262.10 |
Alox8
|
arachidonate 8-lipoxygenase |
| chr5_+_121358254 | 1.28 |
ENSMUST00000042614.13
|
Hectd4
|
HECT domain E3 ubiquitin protein ligase 4 |
| chr17_+_14087827 | 1.28 |
ENSMUST00000239324.2
|
Afdn
|
afadin, adherens junction formation factor |
| chr1_+_75526225 | 1.27 |
ENSMUST00000154101.8
|
Slc4a3
|
solute carrier family 4 (anion exchanger), member 3 |
| chr4_+_13743424 | 1.27 |
ENSMUST00000006761.10
|
Runx1t1
|
RUNX1 translocation partner 1 |
| chr16_+_84571011 | 1.27 |
ENSMUST00000114195.8
|
Jam2
|
junction adhesion molecule 2 |
| chr3_+_129326004 | 1.26 |
ENSMUST00000199910.5
ENSMUST00000197070.5 ENSMUST00000071402.7 |
Elovl6
|
ELOVL family member 6, elongation of long chain fatty acids (yeast) |
| chr10_-_8394546 | 1.26 |
ENSMUST00000061601.9
|
Ust
|
uronyl-2-sulfotransferase |
| chr11_+_4936824 | 1.26 |
ENSMUST00000109897.8
ENSMUST00000009234.16 |
Ap1b1
|
adaptor protein complex AP-1, beta 1 subunit |
| chr7_+_43473917 | 1.26 |
ENSMUST00000107968.10
|
Klk6
|
kallikrein related-peptidase 6 |
| chr19_-_21449916 | 1.25 |
ENSMUST00000087600.10
|
Gda
|
guanine deaminase |
| chr1_-_170886924 | 1.25 |
ENSMUST00000164044.8
ENSMUST00000169017.8 |
Fcgr3
|
Fc receptor, IgG, low affinity III |
| chr10_+_90412827 | 1.24 |
ENSMUST00000182550.8
ENSMUST00000099364.12 |
Anks1b
|
ankyrin repeat and sterile alpha motif domain containing 1B |
| chr12_+_37292029 | 1.24 |
ENSMUST00000160390.2
|
Agmo
|
alkylglycerol monooxygenase |
| chr18_+_65715460 | 1.24 |
ENSMUST00000169679.8
ENSMUST00000183326.2 |
Zfp532
|
zinc finger protein 532 |
| chr15_-_98118858 | 1.24 |
ENSMUST00000142443.8
ENSMUST00000170618.8 |
Gm44579
Olfr287
|
predicted gene 44579 olfactory receptor 287 |
| chr4_-_63072367 | 1.23 |
ENSMUST00000030041.5
|
Ambp
|
alpha 1 microglobulin/bikunin precursor |
| chr7_-_142220553 | 1.23 |
ENSMUST00000105935.8
|
Igf2
|
insulin-like growth factor 2 |
| chr4_+_127881786 | 1.22 |
ENSMUST00000184063.3
|
Csmd2
|
CUB and Sushi multiple domains 2 |
| chr7_+_143792455 | 1.22 |
ENSMUST00000239495.2
|
Shank2
|
SH3 and multiple ankyrin repeat domains 2 |
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.9 | 11.3 | GO:2000255 | negative regulation of male germ cell proliferation(GO:2000255) |
| 1.4 | 4.1 | GO:0040040 | thermosensory behavior(GO:0040040) |
| 1.1 | 5.4 | GO:0035609 | C-terminal protein deglutamylation(GO:0035609) |
| 1.0 | 5.8 | GO:0032423 | regulation of mismatch repair(GO:0032423) |
| 0.9 | 2.8 | GO:2000295 | regulation of hydrogen peroxide catabolic process(GO:2000295) |
| 0.9 | 2.6 | GO:1903045 | neural crest cell migration involved in sympathetic nervous system development(GO:1903045) |
| 0.8 | 2.3 | GO:0071929 | alpha-tubulin acetylation(GO:0071929) |
| 0.8 | 2.3 | GO:0090076 | maintenance of mitochondrion location(GO:0051659) relaxation of skeletal muscle(GO:0090076) |
| 0.7 | 2.2 | GO:0061107 | seminal vesicle development(GO:0061107) |
| 0.7 | 2.8 | GO:0035524 | proline transmembrane transport(GO:0035524) |
| 0.7 | 2.0 | GO:0018879 | biphenyl metabolic process(GO:0018879) |
| 0.6 | 1.9 | GO:0098884 | postsynaptic neurotransmitter receptor internalization(GO:0098884) |
| 0.6 | 3.0 | GO:0010808 | positive regulation of synaptic vesicle priming(GO:0010808) |
| 0.6 | 1.8 | GO:0033082 | regulation of extrathymic T cell differentiation(GO:0033082) |
| 0.6 | 3.0 | GO:0002803 | positive regulation of antimicrobial peptide production(GO:0002225) positive regulation of antibacterial peptide production(GO:0002803) |
| 0.6 | 4.1 | GO:0090292 | nuclear matrix organization(GO:0043578) nuclear matrix anchoring at nuclear membrane(GO:0090292) |
| 0.6 | 1.7 | GO:0045963 | negative regulation of catecholamine metabolic process(GO:0045914) negative regulation of dopamine metabolic process(GO:0045963) |
| 0.6 | 1.7 | GO:1903116 | positive regulation of actin filament-based movement(GO:1903116) |
| 0.5 | 4.9 | GO:0036371 | protein localization to M-band(GO:0036309) protein localization to T-tubule(GO:0036371) |
| 0.5 | 0.5 | GO:0031448 | regulation of fast-twitch skeletal muscle fiber contraction(GO:0031446) positive regulation of fast-twitch skeletal muscle fiber contraction(GO:0031448) |
| 0.5 | 4.2 | GO:0070447 | positive regulation of oligodendrocyte progenitor proliferation(GO:0070447) |
| 0.5 | 1.5 | GO:0061193 | taste bud development(GO:0061193) |
| 0.5 | 1.5 | GO:1904266 | regulation of Schwann cell chemotaxis(GO:1904266) positive regulation of Schwann cell chemotaxis(GO:1904268) Schwann cell chemotaxis(GO:1990751) |
| 0.5 | 2.4 | GO:1904457 | positive regulation of neuronal action potential(GO:1904457) |
| 0.5 | 1.4 | GO:2000821 | regulation of grooming behavior(GO:2000821) |
| 0.5 | 1.4 | GO:0048789 | cytoskeletal matrix organization at active zone(GO:0048789) neurexin clustering involved in presynaptic membrane assembly(GO:0097115) retrograde trans-synaptic signaling by trans-synaptic protein complex(GO:0098942) |
| 0.4 | 1.8 | GO:0010510 | regulation of acetyl-CoA biosynthetic process from pyruvate(GO:0010510) regulation of acyl-CoA biosynthetic process(GO:0050812) |
| 0.4 | 1.2 | GO:0001788 | antibody-dependent cellular cytotoxicity(GO:0001788) |
| 0.4 | 2.5 | GO:0035585 | calcium-mediated signaling using extracellular calcium source(GO:0035585) |
| 0.4 | 1.5 | GO:0042335 | cuticle development(GO:0042335) |
| 0.4 | 2.3 | GO:0006172 | ADP biosynthetic process(GO:0006172) |
| 0.4 | 0.4 | GO:0002760 | positive regulation of antimicrobial humoral response(GO:0002760) |
| 0.4 | 2.5 | GO:0021764 | amygdala development(GO:0021764) |
| 0.4 | 1.1 | GO:0002030 | inhibitory G-protein coupled receptor phosphorylation(GO:0002030) activation of meiosis involved in egg activation(GO:0060466) |
| 0.4 | 2.1 | GO:0006543 | glutamine catabolic process(GO:0006543) |
| 0.4 | 1.4 | GO:0030450 | regulation of complement activation, classical pathway(GO:0030450) |
| 0.4 | 1.8 | GO:0099566 | regulation of postsynaptic cytosolic calcium ion concentration(GO:0099566) |
| 0.3 | 2.1 | GO:0071677 | positive regulation of mononuclear cell migration(GO:0071677) |
| 0.3 | 2.0 | GO:0090126 | protein complex assembly involved in synapse maturation(GO:0090126) |
| 0.3 | 1.0 | GO:0006597 | spermine biosynthetic process(GO:0006597) |
| 0.3 | 1.9 | GO:0001980 | regulation of systemic arterial blood pressure by ischemic conditions(GO:0001980) |
| 0.3 | 1.3 | GO:0046098 | guanine metabolic process(GO:0046098) |
| 0.3 | 0.9 | GO:0090420 | naphthalene metabolic process(GO:0018931) naphthalene-containing compound metabolic process(GO:0090420) |
| 0.3 | 0.6 | GO:1905006 | negative regulation of epithelial to mesenchymal transition involved in endocardial cushion formation(GO:1905006) |
| 0.3 | 1.8 | GO:0070885 | negative regulation of calcineurin-NFAT signaling cascade(GO:0070885) |
| 0.3 | 1.7 | GO:0002925 | positive regulation of humoral immune response mediated by circulating immunoglobulin(GO:0002925) |
| 0.3 | 0.9 | GO:0015891 | iron chelate transport(GO:0015688) siderophore transport(GO:0015891) |
| 0.3 | 1.3 | GO:0060385 | axonogenesis involved in innervation(GO:0060385) |
| 0.3 | 0.8 | GO:1902871 | cerebral cortex GABAergic interneuron fate commitment(GO:0021893) positive regulation of neural retina development(GO:0061075) positive regulation of retina development in camera-type eye(GO:1902868) positive regulation of amacrine cell differentiation(GO:1902871) |
| 0.3 | 1.0 | GO:1902268 | negative regulation of polyamine transmembrane transport(GO:1902268) |
| 0.3 | 1.3 | GO:0051122 | hepoxilin metabolic process(GO:0051121) hepoxilin biosynthetic process(GO:0051122) |
| 0.3 | 1.5 | GO:0071918 | urea transmembrane transport(GO:0071918) |
| 0.3 | 3.8 | GO:0035871 | protein K11-linked deubiquitination(GO:0035871) |
| 0.2 | 4.4 | GO:0051823 | regulation of synapse structural plasticity(GO:0051823) |
| 0.2 | 1.2 | GO:0042494 | detection of bacterial lipoprotein(GO:0042494) |
| 0.2 | 0.7 | GO:0006500 | N-terminal protein palmitoylation(GO:0006500) |
| 0.2 | 1.4 | GO:0002528 | regulation of vascular permeability involved in acute inflammatory response(GO:0002528) |
| 0.2 | 8.1 | GO:0031424 | keratinization(GO:0031424) |
| 0.2 | 5.2 | GO:0007614 | short-term memory(GO:0007614) |
| 0.2 | 1.3 | GO:0034334 | adherens junction maintenance(GO:0034334) |
| 0.2 | 0.8 | GO:0032901 | positive regulation of neurotrophin production(GO:0032901) |
| 0.2 | 0.8 | GO:0036343 | psychomotor behavior(GO:0036343) |
| 0.2 | 2.6 | GO:0046485 | ether lipid metabolic process(GO:0046485) |
| 0.2 | 1.4 | GO:0051012 | microtubule sliding(GO:0051012) |
| 0.2 | 3.7 | GO:0033601 | positive regulation of mammary gland epithelial cell proliferation(GO:0033601) |
| 0.2 | 5.2 | GO:0035641 | locomotory exploration behavior(GO:0035641) |
| 0.2 | 1.9 | GO:0046959 | habituation(GO:0046959) |
| 0.2 | 3.3 | GO:0006054 | N-acetylneuraminate metabolic process(GO:0006054) |
| 0.2 | 1.9 | GO:0033227 | dsRNA transport(GO:0033227) |
| 0.2 | 0.5 | GO:0002148 | hypochlorous acid metabolic process(GO:0002148) hypochlorous acid biosynthetic process(GO:0002149) |
| 0.2 | 0.8 | GO:1903054 | negative regulation of extracellular matrix organization(GO:1903054) |
| 0.2 | 1.5 | GO:0051775 | response to redox state(GO:0051775) |
| 0.2 | 1.8 | GO:0030240 | skeletal muscle thin filament assembly(GO:0030240) |
| 0.2 | 1.8 | GO:0071688 | striated muscle myosin thick filament assembly(GO:0071688) |
| 0.2 | 1.9 | GO:0034626 | fatty acid elongation, saturated fatty acid(GO:0019367) fatty acid elongation, unsaturated fatty acid(GO:0019368) fatty acid elongation, monounsaturated fatty acid(GO:0034625) fatty acid elongation, polyunsaturated fatty acid(GO:0034626) |
| 0.1 | 1.9 | GO:0032482 | Rab protein signal transduction(GO:0032482) |
| 0.1 | 0.6 | GO:0014873 | response to muscle activity involved in regulation of muscle adaptation(GO:0014873) |
| 0.1 | 0.4 | GO:1990705 | intrahepatic bile duct development(GO:0035622) cholangiocyte proliferation(GO:1990705) |
| 0.1 | 0.7 | GO:0061141 | lung ciliated cell differentiation(GO:0061141) |
| 0.1 | 0.9 | GO:0097105 | presynaptic membrane assembly(GO:0097105) |
| 0.1 | 0.6 | GO:0090403 | oxidative stress-induced premature senescence(GO:0090403) |
| 0.1 | 1.4 | GO:2000580 | regulation of ATP-dependent microtubule motor activity, plus-end-directed(GO:2000580) positive regulation of ATP-dependent microtubule motor activity, plus-end-directed(GO:2000582) |
| 0.1 | 1.1 | GO:0021778 | oligodendrocyte cell fate specification(GO:0021778) oligodendrocyte cell fate commitment(GO:0021779) glial cell fate specification(GO:0021780) |
| 0.1 | 1.5 | GO:0030050 | vesicle transport along actin filament(GO:0030050) |
| 0.1 | 1.6 | GO:0021684 | cerebellar granular layer formation(GO:0021684) cerebellar granule cell differentiation(GO:0021707) |
| 0.1 | 1.8 | GO:0061303 | cornea development in camera-type eye(GO:0061303) |
| 0.1 | 1.0 | GO:0008050 | female courtship behavior(GO:0008050) |
| 0.1 | 3.9 | GO:0043567 | regulation of insulin-like growth factor receptor signaling pathway(GO:0043567) |
| 0.1 | 2.2 | GO:1902004 | positive regulation of beta-amyloid formation(GO:1902004) |
| 0.1 | 0.5 | GO:1901526 | negative regulation of mitochondrial membrane permeability(GO:0035795) positive regulation of macromitophagy(GO:1901526) positive regulation of mitophagy in response to mitochondrial depolarization(GO:1904925) |
| 0.1 | 0.8 | GO:0045650 | negative regulation of macrophage differentiation(GO:0045650) |
| 0.1 | 2.9 | GO:0060384 | innervation(GO:0060384) |
| 0.1 | 1.2 | GO:1900383 | regulation of synaptic plasticity by receptor localization to synapse(GO:1900383) |
| 0.1 | 1.7 | GO:0061032 | visceral serous pericardium development(GO:0061032) |
| 0.1 | 3.2 | GO:0032098 | regulation of appetite(GO:0032098) |
| 0.1 | 0.3 | GO:0098583 | thorax and anterior abdomen determination(GO:0007356) mastication(GO:0071626) learned vocalization behavior(GO:0098583) |
| 0.1 | 0.4 | GO:0015904 | tetracycline transport(GO:0015904) |
| 0.1 | 3.6 | GO:0008053 | mitochondrial fusion(GO:0008053) |
| 0.1 | 0.5 | GO:0034371 | chylomicron remodeling(GO:0034371) |
| 0.1 | 3.9 | GO:0032011 | ARF protein signal transduction(GO:0032011) regulation of ARF protein signal transduction(GO:0032012) |
| 0.1 | 0.8 | GO:0034651 | cortisol biosynthetic process(GO:0034651) |
| 0.1 | 3.5 | GO:0095500 | acetylcholine receptor signaling pathway(GO:0095500) signal transduction involved in cellular response to ammonium ion(GO:1903831) response to acetylcholine(GO:1905144) cellular response to acetylcholine(GO:1905145) |
| 0.1 | 5.9 | GO:0018149 | peptide cross-linking(GO:0018149) |
| 0.1 | 1.4 | GO:0061436 | establishment of skin barrier(GO:0061436) |
| 0.1 | 0.6 | GO:0016557 | peroxisome membrane biogenesis(GO:0016557) |
| 0.1 | 1.2 | GO:2000467 | positive regulation of glycogen (starch) synthase activity(GO:2000467) |
| 0.1 | 1.9 | GO:0014850 | response to muscle activity(GO:0014850) |
| 0.1 | 2.8 | GO:0016339 | calcium-dependent cell-cell adhesion via plasma membrane cell adhesion molecules(GO:0016339) |
| 0.1 | 1.5 | GO:0045721 | negative regulation of gluconeogenesis(GO:0045721) |
| 0.1 | 0.6 | GO:0098909 | regulation of cardiac muscle cell action potential involved in regulation of contraction(GO:0098909) |
| 0.1 | 1.0 | GO:1990573 | potassium ion import across plasma membrane(GO:1990573) |
| 0.1 | 0.6 | GO:1903566 | positive regulation of protein localization to cilium(GO:1903566) |
| 0.1 | 1.8 | GO:0046855 | inositol phosphate dephosphorylation(GO:0046855) |
| 0.1 | 0.5 | GO:0046125 | pyrimidine deoxyribonucleoside metabolic process(GO:0046125) |
| 0.1 | 0.6 | GO:2000348 | regulation of CD40 signaling pathway(GO:2000348) |
| 0.1 | 0.8 | GO:0051683 | establishment of Golgi localization(GO:0051683) |
| 0.1 | 0.4 | GO:0002071 | glandular epithelial cell maturation(GO:0002071) |
| 0.1 | 0.4 | GO:0072365 | regulation of cellular ketone metabolic process by negative regulation of transcription from RNA polymerase II promoter(GO:0072365) |
| 0.1 | 0.3 | GO:0046881 | positive regulation of follicle-stimulating hormone secretion(GO:0046881) |
| 0.1 | 0.3 | GO:0000350 | generation of catalytic spliceosome for second transesterification step(GO:0000350) |
| 0.1 | 2.3 | GO:0099514 | anterograde synaptic vesicle transport(GO:0048490) synaptic vesicle cytoskeletal transport(GO:0099514) synaptic vesicle transport along microtubule(GO:0099517) |
| 0.1 | 3.7 | GO:0010614 | negative regulation of cardiac muscle hypertrophy(GO:0010614) |
| 0.1 | 0.6 | GO:0071787 | endoplasmic reticulum tubular network assembly(GO:0071787) |
| 0.1 | 1.9 | GO:0014883 | transition between fast and slow fiber(GO:0014883) |
| 0.1 | 0.3 | GO:1902498 | regulation of protein autoubiquitination(GO:1902498) |
| 0.1 | 0.4 | GO:0097498 | endothelial tube lumen extension(GO:0097498) |
| 0.1 | 0.9 | GO:0007021 | tubulin complex assembly(GO:0007021) |
| 0.1 | 2.8 | GO:0045745 | positive regulation of G-protein coupled receptor protein signaling pathway(GO:0045745) |
| 0.1 | 3.6 | GO:0002026 | regulation of the force of heart contraction(GO:0002026) |
| 0.1 | 0.6 | GO:0075071 | autophagy of host cells involved in interaction with symbiont(GO:0075044) autophagy involved in symbiotic interaction(GO:0075071) |
| 0.1 | 0.7 | GO:0002441 | histamine production involved in inflammatory response(GO:0002349) histamine secretion involved in inflammatory response(GO:0002441) histamine secretion by mast cell(GO:0002553) |
| 0.1 | 0.2 | GO:0061763 | multivesicular body-lysosome fusion(GO:0061763) |
| 0.1 | 1.5 | GO:2001256 | regulation of store-operated calcium entry(GO:2001256) |
| 0.1 | 1.7 | GO:0007190 | activation of adenylate cyclase activity(GO:0007190) |
| 0.1 | 1.2 | GO:0045161 | neuronal ion channel clustering(GO:0045161) |
| 0.1 | 1.8 | GO:0042118 | endothelial cell activation(GO:0042118) |
| 0.1 | 3.2 | GO:0001953 | negative regulation of cell-matrix adhesion(GO:0001953) |
| 0.1 | 0.5 | GO:0090521 | glomerular visceral epithelial cell migration(GO:0090521) |
| 0.1 | 1.8 | GO:0035235 | ionotropic glutamate receptor signaling pathway(GO:0035235) |
| 0.1 | 0.1 | GO:0002339 | B cell selection(GO:0002339) peripheral B cell selection(GO:0002343) B cell affinity maturation(GO:0002344) |
| 0.1 | 0.9 | GO:0014807 | regulation of somitogenesis(GO:0014807) |
| 0.1 | 0.6 | GO:0035589 | G-protein coupled purinergic nucleotide receptor signaling pathway(GO:0035589) |
| 0.1 | 1.2 | GO:0071625 | vocalization behavior(GO:0071625) |
| 0.1 | 0.8 | GO:0045835 | negative regulation of meiotic nuclear division(GO:0045835) |
| 0.1 | 0.4 | GO:0043652 | engulfment of apoptotic cell(GO:0043652) |
| 0.1 | 1.1 | GO:0060742 | epithelial cell differentiation involved in prostate gland development(GO:0060742) |
| 0.1 | 0.2 | GO:2000395 | regulation of ubiquitin-dependent endocytosis(GO:2000395) positive regulation of ubiquitin-dependent endocytosis(GO:2000397) |
| 0.1 | 1.0 | GO:1903818 | positive regulation of voltage-gated potassium channel activity(GO:1903818) |
| 0.1 | 1.1 | GO:0021796 | cerebral cortex regionalization(GO:0021796) |
| 0.1 | 0.9 | GO:0018298 | protein-chromophore linkage(GO:0018298) |
| 0.1 | 2.5 | GO:0007026 | negative regulation of microtubule depolymerization(GO:0007026) |
| 0.1 | 1.7 | GO:0045475 | locomotor rhythm(GO:0045475) |
| 0.1 | 0.3 | GO:0060767 | epithelial cell proliferation involved in prostate gland development(GO:0060767) regulation of epithelial cell proliferation involved in prostate gland development(GO:0060768) negative regulation of epithelial cell proliferation involved in prostate gland development(GO:0060770) |
| 0.1 | 0.8 | GO:0051451 | myoblast migration(GO:0051451) |
| 0.1 | 1.1 | GO:0007220 | Notch receptor processing(GO:0007220) |
| 0.1 | 1.4 | GO:0097499 | protein localization to nonmotile primary cilium(GO:0097499) |
| 0.1 | 1.1 | GO:0043586 | tongue development(GO:0043586) |
| 0.1 | 0.3 | GO:0009946 | proximal/distal axis specification(GO:0009946) |
| 0.1 | 0.2 | GO:0006311 | meiotic gene conversion(GO:0006311) gene conversion(GO:0035822) |
| 0.0 | 0.9 | GO:0006120 | mitochondrial electron transport, NADH to ubiquinone(GO:0006120) |
| 0.0 | 0.2 | GO:1902167 | positive regulation of intrinsic apoptotic signaling pathway in response to DNA damage by p53 class mediator(GO:1902167) |
| 0.0 | 0.3 | GO:2000327 | regulation of ligand-dependent nuclear receptor transcription coactivator activity(GO:2000325) positive regulation of ligand-dependent nuclear receptor transcription coactivator activity(GO:2000327) |
| 0.0 | 0.2 | GO:0033313 | meiotic cell cycle checkpoint(GO:0033313) |
| 0.0 | 0.6 | GO:0034551 | respiratory chain complex III assembly(GO:0017062) mitochondrial respiratory chain complex III assembly(GO:0034551) |
| 0.0 | 2.1 | GO:0022011 | myelination in peripheral nervous system(GO:0022011) peripheral nervous system axon ensheathment(GO:0032292) |
| 0.0 | 0.1 | GO:2001160 | regulation of histone H3-K79 methylation(GO:2001160) |
| 0.0 | 1.7 | GO:0007214 | gamma-aminobutyric acid signaling pathway(GO:0007214) |
| 0.0 | 2.1 | GO:0009409 | response to cold(GO:0009409) |
| 0.0 | 2.4 | GO:0071385 | cellular response to glucocorticoid stimulus(GO:0071385) |
| 0.0 | 1.6 | GO:1903846 | positive regulation of transforming growth factor beta receptor signaling pathway(GO:0030511) positive regulation of cellular response to transforming growth factor beta stimulus(GO:1903846) |
| 0.0 | 0.2 | GO:0018343 | protein farnesylation(GO:0018343) |
| 0.0 | 0.6 | GO:0042541 | hemoglobin biosynthetic process(GO:0042541) |
| 0.0 | 3.5 | GO:0051865 | protein autoubiquitination(GO:0051865) |
| 0.0 | 1.4 | GO:0000289 | nuclear-transcribed mRNA poly(A) tail shortening(GO:0000289) |
| 0.0 | 0.5 | GO:0060136 | embryonic process involved in female pregnancy(GO:0060136) |
| 0.0 | 0.5 | GO:0021542 | dentate gyrus development(GO:0021542) |
| 0.0 | 0.2 | GO:0031442 | positive regulation of mRNA 3'-end processing(GO:0031442) |
| 0.0 | 1.8 | GO:0048488 | synaptic vesicle endocytosis(GO:0048488) |
| 0.0 | 4.8 | GO:0019236 | response to pheromone(GO:0019236) |
| 0.0 | 0.8 | GO:0045724 | positive regulation of cilium assembly(GO:0045724) |
| 0.0 | 0.2 | GO:0034975 | protein folding in endoplasmic reticulum(GO:0034975) |
| 0.0 | 0.3 | GO:0007525 | somatic muscle development(GO:0007525) |
| 0.0 | 0.3 | GO:0033600 | negative regulation of mammary gland epithelial cell proliferation(GO:0033600) |
| 0.0 | 0.5 | GO:0072189 | ureter development(GO:0072189) |
| 0.0 | 0.3 | GO:0048739 | cardiac muscle fiber development(GO:0048739) |
| 0.0 | 0.5 | GO:0032094 | response to food(GO:0032094) |
| 0.0 | 0.5 | GO:0033033 | negative regulation of myeloid cell apoptotic process(GO:0033033) |
| 0.0 | 0.8 | GO:0048268 | clathrin coat assembly(GO:0048268) |
| 0.0 | 0.5 | GO:0034374 | low-density lipoprotein particle remodeling(GO:0034374) |
| 0.0 | 2.1 | GO:0007566 | embryo implantation(GO:0007566) |
| 0.0 | 0.3 | GO:0010457 | centriole-centriole cohesion(GO:0010457) |
| 0.0 | 0.4 | GO:0034116 | positive regulation of heterotypic cell-cell adhesion(GO:0034116) |
| 0.0 | 0.5 | GO:0048671 | negative regulation of collateral sprouting(GO:0048671) |
| 0.0 | 1.4 | GO:0048013 | ephrin receptor signaling pathway(GO:0048013) |
| 0.0 | 0.5 | GO:0043508 | negative regulation of JUN kinase activity(GO:0043508) |
| 0.0 | 0.2 | GO:0019371 | cyclooxygenase pathway(GO:0019371) |
| 0.0 | 0.1 | GO:1903778 | protein localization to vacuolar membrane(GO:1903778) |
| 0.0 | 0.7 | GO:0035094 | response to nicotine(GO:0035094) |
| 0.0 | 0.4 | GO:0045779 | negative regulation of bone resorption(GO:0045779) |
| 0.0 | 2.6 | GO:1903955 | positive regulation of protein targeting to mitochondrion(GO:1903955) |
| 0.0 | 0.4 | GO:0086027 | AV node cell action potential(GO:0086016) AV node cell to bundle of His cell signaling(GO:0086027) |
| 0.0 | 2.4 | GO:0048709 | oligodendrocyte differentiation(GO:0048709) |
| 0.0 | 0.4 | GO:0048934 | peripheral nervous system neuron differentiation(GO:0048934) peripheral nervous system neuron development(GO:0048935) |
| 0.0 | 0.3 | GO:0070986 | left/right axis specification(GO:0070986) |
| 0.0 | 0.6 | GO:0050650 | chondroitin sulfate proteoglycan biosynthetic process(GO:0050650) |
| 0.0 | 1.6 | GO:0007029 | endoplasmic reticulum organization(GO:0007029) |
| 0.0 | 0.2 | GO:0051703 | social behavior(GO:0035176) intraspecies interaction between organisms(GO:0051703) |
| 0.0 | 0.2 | GO:0035735 | intraciliary transport involved in cilium morphogenesis(GO:0035735) |
| 0.0 | 0.6 | GO:0008272 | sulfate transport(GO:0008272) |
| 0.0 | 0.3 | GO:0051418 | interphase microtubule nucleation by interphase microtubule organizing center(GO:0051415) microtubule nucleation by microtubule organizing center(GO:0051418) |
| 0.0 | 2.4 | GO:0051965 | positive regulation of synapse assembly(GO:0051965) |
| 0.0 | 0.1 | GO:0060406 | positive regulation of penile erection(GO:0060406) |
| 0.0 | 0.6 | GO:0050910 | detection of mechanical stimulus involved in sensory perception of sound(GO:0050910) |
| 0.0 | 0.4 | GO:2000096 | positive regulation of Wnt signaling pathway, planar cell polarity pathway(GO:2000096) |
| 0.0 | 1.4 | GO:1904893 | negative regulation of JAK-STAT cascade(GO:0046426) negative regulation of STAT cascade(GO:1904893) |
| 0.0 | 0.5 | GO:0009954 | proximal/distal pattern formation(GO:0009954) |
| 0.0 | 0.9 | GO:0007212 | dopamine receptor signaling pathway(GO:0007212) |
| 0.0 | 0.0 | GO:0003331 | regulation of extracellular matrix constituent secretion(GO:0003330) positive regulation of extracellular matrix constituent secretion(GO:0003331) |
| 0.0 | 0.9 | GO:0030514 | negative regulation of BMP signaling pathway(GO:0030514) |
| 0.0 | 1.3 | GO:0016572 | histone phosphorylation(GO:0016572) |
| 0.0 | 0.4 | GO:0036159 | inner dynein arm assembly(GO:0036159) |
| 0.0 | 0.1 | GO:0014707 | branchiomeric skeletal muscle development(GO:0014707) |
| 0.0 | 1.1 | GO:0097009 | energy homeostasis(GO:0097009) |
| 0.0 | 0.1 | GO:1900108 | negative regulation of nodal signaling pathway(GO:1900108) |
| 0.0 | 2.4 | GO:0007156 | homophilic cell adhesion via plasma membrane adhesion molecules(GO:0007156) |
| 0.0 | 0.1 | GO:2000983 | regulation of ATP citrate synthase activity(GO:2000983) negative regulation of ATP citrate synthase activity(GO:2000984) |
| 0.0 | 0.9 | GO:0000266 | mitochondrial fission(GO:0000266) |
| 0.0 | 5.5 | GO:0010466 | negative regulation of peptidase activity(GO:0010466) |
| 0.0 | 0.1 | GO:0014816 | skeletal muscle satellite cell differentiation(GO:0014816) |
| 0.0 | 0.7 | GO:0048791 | calcium ion-regulated exocytosis of neurotransmitter(GO:0048791) |
| 0.0 | 0.8 | GO:0048025 | negative regulation of mRNA splicing, via spliceosome(GO:0048025) |
| 0.0 | 0.6 | GO:0019373 | epoxygenase P450 pathway(GO:0019373) |
| 0.0 | 0.8 | GO:0045599 | negative regulation of fat cell differentiation(GO:0045599) |
| 0.0 | 1.2 | GO:0000381 | regulation of alternative mRNA splicing, via spliceosome(GO:0000381) |
| 0.0 | 0.3 | GO:0043496 | regulation of protein homodimerization activity(GO:0043496) |
| 0.0 | 1.1 | GO:0007218 | neuropeptide signaling pathway(GO:0007218) |
| 0.0 | 0.4 | GO:0007095 | mitotic G2 DNA damage checkpoint(GO:0007095) |
| 0.0 | 0.6 | GO:0060997 | dendritic spine morphogenesis(GO:0060997) |
| 0.0 | 0.5 | GO:1990089 | response to nerve growth factor(GO:1990089) cellular response to nerve growth factor stimulus(GO:1990090) |
| 0.0 | 0.1 | GO:0045292 | mRNA cis splicing, via spliceosome(GO:0045292) |
| 0.0 | 1.3 | GO:0051453 | regulation of intracellular pH(GO:0051453) |
| 0.0 | 0.5 | GO:0043001 | Golgi to plasma membrane protein transport(GO:0043001) |
| 0.0 | 0.1 | GO:0090210 | regulation of establishment of blood-brain barrier(GO:0090210) |
| 0.0 | 3.0 | GO:0071805 | potassium ion transmembrane transport(GO:0071805) |
| 0.0 | 0.3 | GO:0070536 | protein K63-linked deubiquitination(GO:0070536) |
| 0.0 | 0.5 | GO:0050911 | detection of chemical stimulus involved in sensory perception of smell(GO:0050911) |
| 0.0 | 3.4 | GO:0050890 | cognition(GO:0050890) |
| 0.0 | 0.3 | GO:0060218 | hematopoietic stem cell differentiation(GO:0060218) |
| 0.0 | 1.7 | GO:0006906 | vesicle fusion(GO:0006906) |
| 0.0 | 0.2 | GO:0045899 | positive regulation of RNA polymerase II transcriptional preinitiation complex assembly(GO:0045899) |
| 0.0 | 0.1 | GO:0031665 | negative regulation of lipopolysaccharide-mediated signaling pathway(GO:0031665) |
| 0.0 | 0.8 | GO:0007200 | phospholipase C-activating G-protein coupled receptor signaling pathway(GO:0007200) |
| 0.0 | 2.2 | GO:0030198 | extracellular matrix organization(GO:0030198) |
| 0.0 | 0.1 | GO:0019348 | dolichol metabolic process(GO:0019348) |
| 0.0 | 0.5 | GO:0006378 | mRNA polyadenylation(GO:0006378) RNA polyadenylation(GO:0043631) |
| 0.0 | 0.2 | GO:0006620 | posttranslational protein targeting to membrane(GO:0006620) |
| 0.0 | 0.7 | GO:0032648 | regulation of interferon-beta production(GO:0032648) |
| 0.0 | 0.6 | GO:0000245 | spliceosomal complex assembly(GO:0000245) |
| 0.0 | 0.4 | GO:0015893 | drug transport(GO:0015893) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.7 | 2.8 | GO:0031092 | platelet alpha granule membrane(GO:0031092) |
| 0.7 | 2.7 | GO:0032280 | symmetric synapse(GO:0032280) |
| 0.6 | 1.9 | GO:0098843 | postsynaptic endocytic zone(GO:0098843) |
| 0.6 | 2.4 | GO:0044316 | cone cell pedicle(GO:0044316) |
| 0.6 | 3.0 | GO:0097209 | epidermal lamellar body(GO:0097209) |
| 0.5 | 3.0 | GO:0098831 | presynaptic active zone cytoplasmic component(GO:0098831) |
| 0.5 | 6.5 | GO:0031232 | extrinsic component of external side of plasma membrane(GO:0031232) |
| 0.4 | 2.7 | GO:1990584 | cardiac Troponin complex(GO:1990584) |
| 0.4 | 3.6 | GO:0033116 | endoplasmic reticulum-Golgi intermediate compartment membrane(GO:0033116) |
| 0.4 | 2.5 | GO:0031095 | platelet dense tubular network membrane(GO:0031095) |
| 0.3 | 4.1 | GO:0034993 | microtubule organizing center attachment site(GO:0034992) LINC complex(GO:0034993) |
| 0.3 | 1.5 | GO:0005914 | spot adherens junction(GO:0005914) |
| 0.3 | 16.1 | GO:0001533 | cornified envelope(GO:0001533) |
| 0.2 | 3.9 | GO:0043083 | synaptic cleft(GO:0043083) |
| 0.2 | 2.8 | GO:0097427 | microtubule bundle(GO:0097427) |
| 0.2 | 1.6 | GO:0031673 | H zone(GO:0031673) |
| 0.2 | 0.6 | GO:0055028 | cortical microtubule(GO:0055028) cortical microtubule plus-end(GO:1903754) cytoplasmic microtubule plus-end(GO:1904511) |
| 0.2 | 2.3 | GO:0001520 | outer dense fiber(GO:0001520) |
| 0.2 | 8.0 | GO:0031430 | M band(GO:0031430) |
| 0.2 | 1.9 | GO:0005861 | troponin complex(GO:0005861) |
| 0.1 | 0.7 | GO:1990696 | USH2 complex(GO:1990696) |
| 0.1 | 3.6 | GO:0031672 | A band(GO:0031672) |
| 0.1 | 2.3 | GO:0030123 | AP-3 adaptor complex(GO:0030123) |
| 0.1 | 0.3 | GO:0008282 | ATP-sensitive potassium channel complex(GO:0008282) |
| 0.1 | 4.4 | GO:0031307 | integral component of mitochondrial outer membrane(GO:0031307) |
| 0.1 | 0.6 | GO:0097512 | cardiac myofibril(GO:0097512) |
| 0.1 | 0.4 | GO:0071007 | U2-type catalytic step 2 spliceosome(GO:0071007) |
| 0.1 | 0.8 | GO:0036195 | muscle cell projection(GO:0036194) muscle cell projection membrane(GO:0036195) |
| 0.1 | 1.6 | GO:0000137 | Golgi cis cisterna(GO:0000137) |
| 0.1 | 1.8 | GO:0043203 | axon hillock(GO:0043203) |
| 0.1 | 2.0 | GO:0036056 | filtration diaphragm(GO:0036056) slit diaphragm(GO:0036057) |
| 0.1 | 1.4 | GO:0060091 | kinocilium(GO:0060091) |
| 0.1 | 4.6 | GO:0032590 | dendrite membrane(GO:0032590) |
| 0.1 | 1.8 | GO:0098984 | neuron to neuron synapse(GO:0098984) |
| 0.1 | 1.9 | GO:0032045 | guanyl-nucleotide exchange factor complex(GO:0032045) |
| 0.1 | 1.2 | GO:0031315 | extrinsic component of mitochondrial outer membrane(GO:0031315) |
| 0.1 | 0.3 | GO:0036398 | TCR signalosome(GO:0036398) |
| 0.1 | 2.1 | GO:1902711 | GABA-A receptor complex(GO:1902711) |
| 0.1 | 0.7 | GO:0031258 | lamellipodium membrane(GO:0031258) |
| 0.1 | 0.3 | GO:0097598 | sperm cytoplasmic droplet(GO:0097598) |
| 0.1 | 3.1 | GO:0032809 | neuronal cell body membrane(GO:0032809) |
| 0.1 | 0.9 | GO:0032584 | growth cone membrane(GO:0032584) |
| 0.1 | 1.0 | GO:0005883 | neurofilament(GO:0005883) |
| 0.1 | 0.2 | GO:1990879 | CST complex(GO:1990879) |
| 0.1 | 3.3 | GO:0043034 | costamere(GO:0043034) |
| 0.1 | 0.1 | GO:0034680 | integrin alpha10-beta1 complex(GO:0034680) |
| 0.1 | 8.5 | GO:0005791 | rough endoplasmic reticulum(GO:0005791) |
| 0.1 | 1.4 | GO:0017146 | NMDA selective glutamate receptor complex(GO:0017146) |
| 0.1 | 0.9 | GO:0030126 | COPI vesicle coat(GO:0030126) COPI-coated vesicle membrane(GO:0030663) |
| 0.1 | 2.8 | GO:0030131 | clathrin adaptor complex(GO:0030131) |
| 0.1 | 0.3 | GO:0034272 | phosphatidylinositol 3-kinase complex, class III, type I(GO:0034271) phosphatidylinositol 3-kinase complex, class III, type II(GO:0034272) |
| 0.1 | 8.0 | GO:0043195 | terminal bouton(GO:0043195) |
| 0.1 | 7.1 | GO:0008076 | voltage-gated potassium channel complex(GO:0008076) |
| 0.1 | 1.9 | GO:0005868 | cytoplasmic dynein complex(GO:0005868) |
| 0.1 | 7.6 | GO:0005814 | centriole(GO:0005814) |
| 0.1 | 1.8 | GO:0005865 | striated muscle thin filament(GO:0005865) |
| 0.1 | 1.5 | GO:0033017 | sarcoplasmic reticulum membrane(GO:0033017) |
| 0.1 | 1.9 | GO:0032391 | photoreceptor connecting cilium(GO:0032391) |
| 0.1 | 0.2 | GO:0070722 | Tle3-Aes complex(GO:0070722) |
| 0.1 | 0.5 | GO:0042382 | paraspeckles(GO:0042382) |
| 0.1 | 0.3 | GO:0008274 | gamma-tubulin large complex(GO:0000931) gamma-tubulin ring complex(GO:0008274) |
| 0.0 | 0.5 | GO:0034464 | BBSome(GO:0034464) |
| 0.0 | 0.2 | GO:0030896 | checkpoint clamp complex(GO:0030896) |
| 0.0 | 0.1 | GO:0071438 | invadopodium membrane(GO:0071438) |
| 0.0 | 0.4 | GO:0016593 | Cdc73/Paf1 complex(GO:0016593) |
| 0.0 | 0.7 | GO:0005892 | acetylcholine-gated channel complex(GO:0005892) |
| 0.0 | 0.3 | GO:0044300 | cerebellar mossy fiber(GO:0044300) |
| 0.0 | 3.6 | GO:0005581 | collagen trimer(GO:0005581) |
| 0.0 | 1.3 | GO:0098878 | ionotropic glutamate receptor complex(GO:0008328) neurotransmitter receptor complex(GO:0098878) |
| 0.0 | 1.7 | GO:0043679 | axon terminus(GO:0043679) |
| 0.0 | 0.3 | GO:0070552 | BRISC complex(GO:0070552) |
| 0.0 | 10.6 | GO:0097060 | synaptic membrane(GO:0097060) |
| 0.0 | 0.3 | GO:0071141 | SMAD protein complex(GO:0071141) |
| 0.0 | 0.1 | GO:0005927 | muscle tendon junction(GO:0005927) |
| 0.0 | 0.3 | GO:0030015 | CCR4-NOT core complex(GO:0030015) |
| 0.0 | 0.2 | GO:0005851 | eukaryotic translation initiation factor 2B complex(GO:0005851) |
| 0.0 | 0.6 | GO:0044295 | axonal growth cone(GO:0044295) |
| 0.0 | 0.6 | GO:0030057 | desmosome(GO:0030057) |
| 0.0 | 0.9 | GO:0051233 | spindle midzone(GO:0051233) |
| 0.0 | 0.3 | GO:0034045 | pre-autophagosomal structure membrane(GO:0034045) |
| 0.0 | 5.7 | GO:0001726 | ruffle(GO:0001726) |
| 0.0 | 0.6 | GO:0005682 | U5 snRNP(GO:0005682) |
| 0.0 | 0.7 | GO:0097440 | apical dendrite(GO:0097440) |
| 0.0 | 3.0 | GO:0005875 | microtubule associated complex(GO:0005875) |
| 0.0 | 0.5 | GO:0008074 | guanylate cyclase complex, soluble(GO:0008074) |
| 0.0 | 0.6 | GO:0005614 | interstitial matrix(GO:0005614) |
| 0.0 | 0.1 | GO:0033185 | dolichol-phosphate-mannose synthase complex(GO:0033185) |
| 0.0 | 0.2 | GO:0097504 | Gemini of coiled bodies(GO:0097504) |
| 0.0 | 0.2 | GO:0000815 | ESCRT III complex(GO:0000815) |
| 0.0 | 0.5 | GO:0001518 | voltage-gated sodium channel complex(GO:0001518) |
| 0.0 | 1.7 | GO:0031463 | Cul3-RING ubiquitin ligase complex(GO:0031463) |
| 0.0 | 0.2 | GO:0000813 | ESCRT I complex(GO:0000813) |
| 0.0 | 0.2 | GO:0000801 | central element(GO:0000801) |
| 0.0 | 0.2 | GO:0072546 | ER membrane protein complex(GO:0072546) |
| 0.0 | 2.0 | GO:0016605 | PML body(GO:0016605) |
| 0.0 | 3.4 | GO:0031225 | anchored component of membrane(GO:0031225) |
| 0.0 | 3.3 | GO:0043209 | myelin sheath(GO:0043209) |
| 0.0 | 2.3 | GO:0030176 | integral component of endoplasmic reticulum membrane(GO:0030176) |
| 0.0 | 1.6 | GO:0016363 | nuclear matrix(GO:0016363) |
| 0.0 | 0.1 | GO:0019815 | B cell receptor complex(GO:0019815) |
| 0.0 | 0.3 | GO:0005666 | DNA-directed RNA polymerase III complex(GO:0005666) |
| 0.0 | 0.2 | GO:0034518 | mRNA cap binding complex(GO:0005845) RNA cap binding complex(GO:0034518) |
| 0.0 | 0.5 | GO:0031901 | early endosome membrane(GO:0031901) |
| 0.0 | 0.5 | GO:0034399 | nuclear periphery(GO:0034399) |
| 0.0 | 3.5 | GO:0030424 | axon(GO:0030424) |
| 0.0 | 0.1 | GO:0001891 | phagocytic cup(GO:0001891) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.8 | 11.3 | GO:0004667 | prostaglandin-D synthase activity(GO:0004667) |
| 1.9 | 5.8 | GO:0004698 | calcium-dependent protein kinase C activity(GO:0004698) |
| 0.9 | 2.6 | GO:0050479 | glyceryl-ether monooxygenase activity(GO:0050479) |
| 0.7 | 4.0 | GO:0003835 | beta-galactoside alpha-2,6-sialyltransferase activity(GO:0003835) |
| 0.6 | 1.9 | GO:0031798 | type 1 metabotropic glutamate receptor binding(GO:0031798) |
| 0.6 | 4.2 | GO:0015091 | ferric iron transmembrane transporter activity(GO:0015091) trivalent inorganic cation transmembrane transporter activity(GO:0072510) |
| 0.6 | 5.0 | GO:0008597 | calcium-dependent protein serine/threonine phosphatase regulator activity(GO:0008597) |
| 0.5 | 3.1 | GO:0030280 | structural constituent of epidermis(GO:0030280) |
| 0.5 | 1.5 | GO:0005169 | neurotrophin TRKB receptor binding(GO:0005169) |
| 0.5 | 1.5 | GO:0005009 | insulin-activated receptor activity(GO:0005009) |
| 0.5 | 1.4 | GO:0004980 | melanocyte-stimulating hormone receptor activity(GO:0004980) |
| 0.5 | 1.4 | GO:0070996 | type 1 melanocortin receptor binding(GO:0070996) |
| 0.5 | 1.4 | GO:0050508 | glucuronosyl-N-acetylglucosaminyl-proteoglycan 4-alpha-N-acetylglucosaminyltransferase activity(GO:0050508) |
| 0.4 | 1.8 | GO:0004740 | pyruvate dehydrogenase (acetyl-transferring) kinase activity(GO:0004740) |
| 0.4 | 3.3 | GO:0001515 | opioid peptide activity(GO:0001515) |
| 0.4 | 4.1 | GO:0030550 | acetylcholine receptor inhibitor activity(GO:0030550) |
| 0.4 | 2.8 | GO:0019784 | NEDD8-specific protease activity(GO:0019784) |
| 0.4 | 2.7 | GO:0030172 | troponin C binding(GO:0030172) |
| 0.4 | 2.1 | GO:0004359 | glutaminase activity(GO:0004359) |
| 0.4 | 3.2 | GO:0031995 | insulin-like growth factor II binding(GO:0031995) |
| 0.3 | 2.4 | GO:0005005 | transmembrane-ephrin receptor activity(GO:0005005) |
| 0.3 | 1.2 | GO:0019862 | IgA binding(GO:0019862) |
| 0.3 | 1.8 | GO:0015277 | kainate selective glutamate receptor activity(GO:0015277) |
| 0.3 | 1.8 | GO:0004994 | somatostatin receptor activity(GO:0004994) |
| 0.3 | 1.7 | GO:0015232 | heme transporter activity(GO:0015232) |
| 0.3 | 2.8 | GO:0015193 | L-proline transmembrane transporter activity(GO:0015193) |
| 0.3 | 1.3 | GO:0005250 | A-type (transient outward) potassium channel activity(GO:0005250) |
| 0.3 | 2.8 | GO:1903136 | cuprous ion binding(GO:1903136) |
| 0.3 | 1.0 | GO:0008073 | ornithine decarboxylase inhibitor activity(GO:0008073) |
| 0.3 | 2.8 | GO:0015379 | potassium:chloride symporter activity(GO:0015379) potassium ion symporter activity(GO:0022820) |
| 0.3 | 2.8 | GO:0003810 | protein-glutamine gamma-glutamyltransferase activity(GO:0003810) |
| 0.3 | 1.0 | GO:0004711 | ribosomal protein S6 kinase activity(GO:0004711) |
| 0.2 | 4.7 | GO:0038191 | neuropilin binding(GO:0038191) |
| 0.2 | 1.7 | GO:0008294 | calcium- and calmodulin-responsive adenylate cyclase activity(GO:0008294) |
| 0.2 | 2.1 | GO:0005237 | inhibitory extracellular ligand-gated ion channel activity(GO:0005237) |
| 0.2 | 1.8 | GO:0099583 | neurotransmitter receptor activity involved in regulation of postsynaptic cytosolic calcium ion concentration(GO:0099583) |
| 0.2 | 1.5 | GO:0015265 | urea channel activity(GO:0015265) |
| 0.2 | 3.2 | GO:0019911 | structural constituent of myelin sheath(GO:0019911) |
| 0.2 | 0.8 | GO:0008332 | low voltage-gated calcium channel activity(GO:0008332) |
| 0.2 | 1.9 | GO:0004687 | myosin light chain kinase activity(GO:0004687) |
| 0.2 | 2.3 | GO:0004468 | lysine N-acetyltransferase activity, acting on acetyl phosphate as donor(GO:0004468) |
| 0.2 | 1.0 | GO:0072590 | N-acetyl-L-aspartate-L-glutamate ligase activity(GO:0072590) |
| 0.2 | 0.6 | GO:0003870 | 5-aminolevulinate synthase activity(GO:0003870) N-succinyltransferase activity(GO:0016749) |
| 0.2 | 1.2 | GO:0051033 | nucleic acid transmembrane transporter activity(GO:0051032) RNA transmembrane transporter activity(GO:0051033) |
| 0.2 | 1.9 | GO:0045504 | dynein heavy chain binding(GO:0045504) |
| 0.2 | 1.2 | GO:0019770 | IgG receptor activity(GO:0019770) |
| 0.2 | 0.7 | GO:0086089 | voltage-gated potassium channel activity involved in atrial cardiac muscle cell action potential repolarization(GO:0086089) |
| 0.2 | 2.3 | GO:0004017 | adenylate kinase activity(GO:0004017) |
| 0.2 | 1.9 | GO:0102337 | fatty acid elongase activity(GO:0009922) 3-oxo-arachidoyl-CoA synthase activity(GO:0102336) 3-oxo-cerotoyl-CoA synthase activity(GO:0102337) 3-oxo-lignoceronyl-CoA synthase activity(GO:0102338) |
| 0.2 | 3.9 | GO:0050811 | GABA receptor binding(GO:0050811) |
| 0.2 | 0.6 | GO:0034714 | type III transforming growth factor beta receptor binding(GO:0034714) |
| 0.1 | 4.3 | GO:0005184 | neuropeptide hormone activity(GO:0005184) |
| 0.1 | 4.6 | GO:0004181 | metallocarboxypeptidase activity(GO:0004181) |
| 0.1 | 1.3 | GO:0015198 | oligopeptide transporter activity(GO:0015198) |
| 0.1 | 4.1 | GO:0005521 | lamin binding(GO:0005521) |
| 0.1 | 0.9 | GO:0004966 | galanin receptor activity(GO:0004966) |
| 0.1 | 2.0 | GO:0097109 | neuroligin family protein binding(GO:0097109) |
| 0.1 | 1.0 | GO:0019808 | polyamine binding(GO:0019808) |
| 0.1 | 2.0 | GO:0051378 | serotonin binding(GO:0051378) |
| 0.1 | 1.5 | GO:0031802 | type 5 metabotropic glutamate receptor binding(GO:0031802) |
| 0.1 | 5.8 | GO:0008157 | protein phosphatase 1 binding(GO:0008157) |
| 0.1 | 0.9 | GO:0005095 | GTPase inhibitor activity(GO:0005095) |
| 0.1 | 0.4 | GO:0005026 | transforming growth factor beta receptor activity, type II(GO:0005026) |
| 0.1 | 3.2 | GO:0005540 | hyaluronic acid binding(GO:0005540) |
| 0.1 | 0.5 | GO:0019136 | deoxynucleoside kinase activity(GO:0019136) |
| 0.1 | 0.3 | GO:0008330 | protein tyrosine/threonine phosphatase activity(GO:0008330) |
| 0.1 | 1.8 | GO:0001087 | transcription factor activity, sequence-specific DNA binding, RNA polymerase recruiting(GO:0001011) transcription factor activity, TFIIB-class binding(GO:0001087) |
| 0.1 | 1.8 | GO:0004445 | inositol-polyphosphate 5-phosphatase activity(GO:0004445) inositol trisphosphate phosphatase activity(GO:0046030) |
| 0.1 | 1.5 | GO:0019855 | calcium channel inhibitor activity(GO:0019855) |
| 0.1 | 3.0 | GO:0005388 | calcium-transporting ATPase activity(GO:0005388) |
| 0.1 | 0.4 | GO:0008493 | tetracycline transporter activity(GO:0008493) |
| 0.1 | 0.5 | GO:0035478 | chylomicron binding(GO:0035478) |
| 0.1 | 1.4 | GO:0044323 | retinoic acid-responsive element binding(GO:0044323) |
| 0.1 | 1.4 | GO:0042043 | neurexin family protein binding(GO:0042043) |
| 0.1 | 1.4 | GO:0031957 | very long-chain fatty acid-CoA ligase activity(GO:0031957) |
| 0.1 | 0.4 | GO:0008109 | N-acetyllactosaminide beta-1,6-N-acetylglucosaminyltransferase activity(GO:0008109) |
| 0.1 | 1.3 | GO:0030274 | LIM domain binding(GO:0030274) |
| 0.1 | 0.8 | GO:0034711 | inhibin binding(GO:0034711) |
| 0.1 | 1.8 | GO:0097493 | structural molecule activity conferring elasticity(GO:0097493) |
| 0.1 | 0.2 | GO:0004802 | transketolase activity(GO:0004802) |
| 0.1 | 2.9 | GO:0005112 | Notch binding(GO:0005112) |
| 0.1 | 0.7 | GO:0001609 | G-protein coupled adenosine receptor activity(GO:0001609) |
| 0.1 | 1.3 | GO:0035014 | phosphatidylinositol 3-kinase regulator activity(GO:0035014) |
| 0.1 | 3.2 | GO:0005086 | ARF guanyl-nucleotide exchange factor activity(GO:0005086) |
| 0.1 | 0.5 | GO:0061629 | RNA polymerase II sequence-specific DNA binding transcription factor binding(GO:0061629) |
| 0.1 | 0.6 | GO:0005520 | insulin-like growth factor binding(GO:0005520) |
| 0.1 | 4.4 | GO:0046875 | ephrin receptor binding(GO:0046875) |
| 0.1 | 0.8 | GO:1990825 | sequence-specific mRNA binding(GO:1990825) |
| 0.1 | 1.9 | GO:0043274 | phospholipase binding(GO:0043274) |
| 0.1 | 1.2 | GO:0004385 | guanylate kinase activity(GO:0004385) |
| 0.1 | 1.7 | GO:0004890 | GABA-A receptor activity(GO:0004890) |
| 0.1 | 0.8 | GO:0048403 | brain-derived neurotrophic factor binding(GO:0048403) |
| 0.1 | 0.2 | GO:0031370 | eukaryotic initiation factor 4G binding(GO:0031370) |
| 0.1 | 0.3 | GO:0002046 | opsin binding(GO:0002046) |
| 0.1 | 0.5 | GO:0019158 | fructokinase activity(GO:0008865) mannokinase activity(GO:0019158) |
| 0.1 | 9.4 | GO:0004867 | serine-type endopeptidase inhibitor activity(GO:0004867) |
| 0.1 | 1.3 | GO:0008510 | sodium:bicarbonate symporter activity(GO:0008510) |
| 0.1 | 0.3 | GO:0004614 | phosphoglucomutase activity(GO:0004614) |
| 0.1 | 1.7 | GO:0005212 | structural constituent of eye lens(GO:0005212) |
| 0.1 | 0.3 | GO:1902282 | voltage-gated potassium channel activity involved in ventricular cardiac muscle cell action potential repolarization(GO:1902282) |
| 0.1 | 0.4 | GO:0086006 | voltage-gated sodium channel activity involved in cardiac muscle cell action potential(GO:0086006) |
| 0.0 | 0.7 | GO:0017017 | MAP kinase tyrosine/serine/threonine phosphatase activity(GO:0017017) |
| 0.0 | 2.4 | GO:0030552 | cAMP binding(GO:0030552) |
| 0.0 | 0.2 | GO:0004449 | isocitrate dehydrogenase (NAD+) activity(GO:0004449) |
| 0.0 | 1.0 | GO:0008143 | poly(A) binding(GO:0008143) |
| 0.0 | 0.4 | GO:0004366 | glycerol-3-phosphate O-acyltransferase activity(GO:0004366) |
| 0.0 | 0.2 | GO:0005502 | 11-cis retinal binding(GO:0005502) |
| 0.0 | 1.1 | GO:0016503 | pheromone receptor activity(GO:0016503) |
| 0.0 | 1.1 | GO:0030371 | translation repressor activity(GO:0030371) |
| 0.0 | 0.3 | GO:0008607 | cAMP-dependent protein kinase regulator activity(GO:0008603) phosphorylase kinase regulator activity(GO:0008607) |
| 0.0 | 1.2 | GO:0008266 | poly(U) RNA binding(GO:0008266) |
| 0.0 | 0.9 | GO:0019825 | oxygen binding(GO:0019825) |
| 0.0 | 5.2 | GO:0005249 | voltage-gated potassium channel activity(GO:0005249) |
| 0.0 | 0.5 | GO:0019992 | diacylglycerol binding(GO:0019992) |
| 0.0 | 3.3 | GO:0015459 | potassium channel regulator activity(GO:0015459) |
| 0.0 | 0.1 | GO:0045030 | A1 adenosine receptor binding(GO:0031686) UTP-activated nucleotide receptor activity(GO:0045030) |
| 0.0 | 0.6 | GO:0045028 | G-protein coupled nucleotide receptor activity(GO:0001608) G-protein coupled purinergic nucleotide receptor activity(GO:0045028) |
| 0.0 | 1.9 | GO:0003785 | actin monomer binding(GO:0003785) |
| 0.0 | 1.3 | GO:0016702 | oxidoreductase activity, acting on single donors with incorporation of molecular oxygen, incorporation of two atoms of oxygen(GO:0016702) |
| 0.0 | 0.4 | GO:0001162 | RNA polymerase II intronic transcription regulatory region sequence-specific DNA binding(GO:0001162) |
| 0.0 | 4.3 | GO:0005550 | pheromone binding(GO:0005550) |
| 0.0 | 0.2 | GO:1990269 | RNA polymerase II C-terminal domain phosphoserine binding(GO:1990269) |
| 0.0 | 0.7 | GO:0042166 | acetylcholine binding(GO:0042166) |
| 0.0 | 3.1 | GO:0043621 | protein self-association(GO:0043621) |
| 0.0 | 0.4 | GO:0031432 | titin binding(GO:0031432) |
| 0.0 | 1.2 | GO:0005159 | insulin-like growth factor receptor binding(GO:0005159) |
| 0.0 | 0.4 | GO:0004887 | thyroid hormone receptor activity(GO:0004887) |
| 0.0 | 1.2 | GO:0035255 | ionotropic glutamate receptor binding(GO:0035255) |
| 0.0 | 0.2 | GO:0005087 | Ran guanyl-nucleotide exchange factor activity(GO:0005087) |
| 0.0 | 1.5 | GO:0005109 | frizzled binding(GO:0005109) |
| 0.0 | 0.6 | GO:0019531 | oxalate transmembrane transporter activity(GO:0019531) |
| 0.0 | 0.4 | GO:0005049 | nuclear export signal receptor activity(GO:0005049) |
| 0.0 | 1.2 | GO:0043236 | laminin binding(GO:0043236) |
| 0.0 | 0.6 | GO:0002162 | dystroglycan binding(GO:0002162) |
| 0.0 | 0.6 | GO:0043422 | protein kinase B binding(GO:0043422) |
| 0.0 | 1.5 | GO:0005160 | transforming growth factor beta receptor binding(GO:0005160) |
| 0.0 | 1.9 | GO:0030276 | clathrin binding(GO:0030276) |
| 0.0 | 0.8 | GO:0050840 | extracellular matrix binding(GO:0050840) |
| 0.0 | 0.6 | GO:0097602 | cullin family protein binding(GO:0097602) |
| 0.0 | 0.5 | GO:0001618 | virus receptor activity(GO:0001618) |
| 0.0 | 0.2 | GO:0097371 | MDM2/MDM4 family protein binding(GO:0097371) |
| 0.0 | 4.1 | GO:0001191 | transcriptional repressor activity, RNA polymerase II transcription factor binding(GO:0001191) |
| 0.0 | 0.9 | GO:0005201 | extracellular matrix structural constituent(GO:0005201) |
| 0.0 | 0.2 | GO:0004311 | farnesyltranstransferase activity(GO:0004311) |
| 0.0 | 5.5 | GO:0004252 | serine-type endopeptidase activity(GO:0004252) |
| 0.0 | 0.1 | GO:0098639 | collagen binding involved in cell-matrix adhesion(GO:0098639) |
| 0.0 | 1.6 | GO:0030544 | Hsp70 protein binding(GO:0030544) |
| 0.0 | 1.8 | GO:0005546 | phosphatidylinositol-4,5-bisphosphate binding(GO:0005546) |
| 0.0 | 2.3 | GO:0004860 | protein kinase inhibitor activity(GO:0004860) |
| 0.0 | 0.5 | GO:0017056 | structural constituent of nuclear pore(GO:0017056) |
| 0.0 | 0.6 | GO:0070182 | DNA polymerase binding(GO:0070182) |
| 0.0 | 0.3 | GO:0005078 | MAP-kinase scaffold activity(GO:0005078) |
| 0.0 | 0.5 | GO:0001786 | phosphatidylserine binding(GO:0001786) |
| 0.0 | 0.4 | GO:0017070 | U6 snRNA binding(GO:0017070) |
| 0.0 | 0.1 | GO:0004582 | dolichyl-phosphate beta-D-mannosyltransferase activity(GO:0004582) |
| 0.0 | 0.8 | GO:0008307 | structural constituent of muscle(GO:0008307) |
| 0.0 | 0.0 | GO:0031013 | troponin I binding(GO:0031013) |
| 0.0 | 0.2 | GO:0008481 | sphinganine kinase activity(GO:0008481) D-erythro-sphingosine kinase activity(GO:0017050) |
| 0.0 | 0.7 | GO:0044183 | protein binding involved in protein folding(GO:0044183) |
| 0.0 | 1.1 | GO:0001540 | beta-amyloid binding(GO:0001540) |
| 0.0 | 0.2 | GO:0019869 | chloride channel inhibitor activity(GO:0019869) |
| 0.0 | 0.9 | GO:0004364 | glutathione transferase activity(GO:0004364) |
| 0.0 | 1.7 | GO:0004843 | thiol-dependent ubiquitin-specific protease activity(GO:0004843) |
| 0.0 | 0.7 | GO:0003899 | DNA-directed RNA polymerase activity(GO:0003899) |
| 0.0 | 0.6 | GO:0051393 | alpha-actinin binding(GO:0051393) |
| 0.0 | 0.4 | GO:0004198 | calcium-dependent cysteine-type endopeptidase activity(GO:0004198) |
| 0.0 | 0.6 | GO:0017080 | sodium channel regulator activity(GO:0017080) |
| 0.0 | 0.3 | GO:0051011 | microtubule minus-end binding(GO:0051011) |
| 0.0 | 0.3 | GO:0070530 | K63-linked polyubiquitin binding(GO:0070530) |
| 0.0 | 0.3 | GO:0004535 | poly(A)-specific ribonuclease activity(GO:0004535) |
| 0.0 | 0.1 | GO:0050649 | testosterone 6-beta-hydroxylase activity(GO:0050649) |
| 0.0 | 0.4 | GO:0051059 | NF-kappaB binding(GO:0051059) |
| 0.0 | 1.2 | GO:0048306 | calcium-dependent protein binding(GO:0048306) |
| 0.0 | 0.7 | GO:0016814 | hydrolase activity, acting on carbon-nitrogen (but not peptide) bonds, in cyclic amidines(GO:0016814) |
| 0.0 | 0.2 | GO:0030553 | cGMP binding(GO:0030553) |
| 0.0 | 0.2 | GO:0005234 | extracellular-glutamate-gated ion channel activity(GO:0005234) |
| 0.0 | 0.4 | GO:0008137 | NADH dehydrogenase (ubiquinone) activity(GO:0008137) NADH dehydrogenase (quinone) activity(GO:0050136) |
| 0.0 | 0.3 | GO:0030275 | LRR domain binding(GO:0030275) |
| 0.0 | 0.5 | GO:0032266 | phosphatidylinositol-3-phosphate binding(GO:0032266) |
| 0.0 | 2.4 | GO:0017137 | Rab GTPase binding(GO:0017137) |
| 0.0 | 1.3 | GO:0004222 | metalloendopeptidase activity(GO:0004222) |
| 0.0 | 0.1 | GO:0070053 | thrombospondin receptor activity(GO:0070053) |
| 0.0 | 0.3 | GO:0031435 | mitogen-activated protein kinase kinase kinase binding(GO:0031435) |
| 0.0 | 0.3 | GO:0017160 | Ral GTPase binding(GO:0017160) |
| 0.0 | 0.6 | GO:0005518 | collagen binding(GO:0005518) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.4 | PID ALK2 PATHWAY | ALK2 signaling events |
| 0.1 | 5.1 | PID ALPHA SYNUCLEIN PATHWAY | Alpha-synuclein signaling |
| 0.1 | 2.6 | PID SYNDECAN 3 PATHWAY | Syndecan-3-mediated signaling events |
| 0.1 | 10.6 | PID NFAT 3PATHWAY | Role of Calcineurin-dependent NFAT signaling in lymphocytes |
| 0.1 | 1.7 | PID LPA4 PATHWAY | LPA4-mediated signaling events |
| 0.1 | 1.4 | PID EPHA2 FWD PATHWAY | EPHA2 forward signaling |
| 0.1 | 2.9 | PID PI3K PLC TRK PATHWAY | Trk receptor signaling mediated by PI3K and PLC-gamma |
| 0.0 | 12.9 | NABA ECM REGULATORS | Genes encoding enzymes and their regulators involved in the remodeling of the extracellular matrix |
| 0.0 | 1.8 | PID NCADHERIN PATHWAY | N-cadherin signaling events |
| 0.0 | 0.6 | SA MMP CYTOKINE CONNECTION | Cytokines can induce activation of matrix metalloproteinases, which degrade extracellular matrix. |
| 0.0 | 2.2 | NABA COLLAGENS | Genes encoding collagen proteins |
| 0.0 | 2.2 | PID ERBB1 INTERNALIZATION PATHWAY | Internalization of ErbB1 |
| 0.0 | 1.4 | PID REELIN PATHWAY | Reelin signaling pathway |
| 0.0 | 1.1 | PID IFNG PATHWAY | IFN-gamma pathway |
| 0.0 | 0.7 | PID ANTHRAX PATHWAY | Cellular roles of Anthrax toxin |
| 0.0 | 2.7 | PID RAC1 REG PATHWAY | Regulation of RAC1 activity |
| 0.0 | 0.7 | PID CIRCADIAN PATHWAY | Circadian rhythm pathway |
| 0.0 | 0.2 | ST INTERFERON GAMMA PATHWAY | Interferon gamma pathway. |
| 0.0 | 1.2 | ST ERK1 ERK2 MAPK PATHWAY | ERK1/ERK2 MAPK Pathway |
| 0.0 | 2.5 | PID NOTCH PATHWAY | Notch signaling pathway |
| 0.0 | 0.5 | PID IL23 PATHWAY | IL23-mediated signaling events |
| 0.0 | 1.5 | PID P75 NTR PATHWAY | p75(NTR)-mediated signaling |
| 0.0 | 1.6 | PID AP1 PATHWAY | AP-1 transcription factor network |
| 0.0 | 0.9 | PID ATR PATHWAY | ATR signaling pathway |
| 0.0 | 2.2 | PID BETA CATENIN NUC PATHWAY | Regulation of nuclear beta catenin signaling and target gene transcription |
| 0.0 | 0.4 | PID P38 MK2 PATHWAY | p38 signaling mediated by MAPKAP kinases |
| 0.0 | 0.5 | PID AMB2 NEUTROPHILS PATHWAY | amb2 Integrin signaling |
| 0.0 | 2.6 | NABA ECM AFFILIATED | Genes encoding proteins affiliated structurally or functionally to extracellular matrix proteins |
| 0.0 | 0.8 | PID HIF1 TFPATHWAY | HIF-1-alpha transcription factor network |
| 0.0 | 0.5 | PID HNF3A PATHWAY | FOXA1 transcription factor network |
| 0.0 | 0.5 | PID LKB1 PATHWAY | LKB1 signaling events |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 5.7 | REACTOME DOPAMINE NEUROTRANSMITTER RELEASE CYCLE | Genes involved in Dopamine Neurotransmitter Release Cycle |
| 0.3 | 7.6 | REACTOME TRAFFICKING OF GLUR2 CONTAINING AMPA RECEPTORS | Genes involved in Trafficking of GluR2-containing AMPA receptors |
| 0.2 | 0.2 | REACTOME OPSINS | Genes involved in Opsins |
| 0.2 | 4.4 | REACTOME REGULATION OF INSULIN LIKE GROWTH FACTOR IGF ACTIVITY BY INSULIN LIKE GROWTH FACTOR BINDING PROTEINS IGFBPS | Genes involved in Regulation of Insulin-like Growth Factor (IGF) Activity by Insulin-like Growth Factor Binding Proteins (IGFBPs) |
| 0.2 | 2.1 | REACTOME GLUTAMATE NEUROTRANSMITTER RELEASE CYCLE | Genes involved in Glutamate Neurotransmitter Release Cycle |
| 0.2 | 2.0 | REACTOME SEROTONIN RECEPTORS | Genes involved in Serotonin receptors |
| 0.2 | 3.8 | REACTOME GABA A RECEPTOR ACTIVATION | Genes involved in GABA A receptor activation |
| 0.2 | 6.1 | REACTOME AMINE COMPOUND SLC TRANSPORTERS | Genes involved in Amine compound SLC transporters |
| 0.1 | 1.8 | REACTOME REGULATION OF PYRUVATE DEHYDROGENASE PDH COMPLEX | Genes involved in Regulation of pyruvate dehydrogenase (PDH) complex |
| 0.1 | 1.8 | REACTOME CS DS DEGRADATION | Genes involved in CS/DS degradation |
| 0.1 | 2.5 | REACTOME SEMA3A PLEXIN REPULSION SIGNALING BY INHIBITING INTEGRIN ADHESION | Genes involved in SEMA3A-Plexin repulsion signaling by inhibiting Integrin adhesion |
| 0.1 | 8.1 | REACTOME STRIATED MUSCLE CONTRACTION | Genes involved in Striated Muscle Contraction |
| 0.1 | 2.6 | REACTOME OTHER SEMAPHORIN INTERACTIONS | Genes involved in Other semaphorin interactions |
| 0.1 | 0.8 | REACTOME CREATION OF C4 AND C2 ACTIVATORS | Genes involved in Creation of C4 and C2 activators |
| 0.1 | 6.4 | REACTOME VOLTAGE GATED POTASSIUM CHANNELS | Genes involved in Voltage gated Potassium channels |
| 0.1 | 2.3 | REACTOME PLATELET CALCIUM HOMEOSTASIS | Genes involved in Platelet calcium homeostasis |
| 0.1 | 1.7 | REACTOME ADENYLATE CYCLASE ACTIVATING PATHWAY | Genes involved in Adenylate cyclase activating pathway |
| 0.1 | 2.2 | REACTOME UNBLOCKING OF NMDA RECEPTOR GLUTAMATE BINDING AND ACTIVATION | Genes involved in Unblocking of NMDA receptor, glutamate binding and activation |
| 0.1 | 2.6 | REACTOME AMYLOIDS | Genes involved in Amyloids |
| 0.1 | 1.3 | REACTOME NEF MEDIATED DOWNREGULATION OF MHC CLASS I COMPLEX CELL SURFACE EXPRESSION | Genes involved in Nef mediated downregulation of MHC class I complex cell surface expression |
| 0.1 | 2.2 | REACTOME RAP1 SIGNALLING | Genes involved in Rap1 signalling |
| 0.1 | 4.6 | REACTOME O LINKED GLYCOSYLATION OF MUCINS | Genes involved in O-linked glycosylation of mucins |
| 0.1 | 2.1 | REACTOME ADHERENS JUNCTIONS INTERACTIONS | Genes involved in Adherens junctions interactions |
| 0.1 | 3.2 | REACTOME MEIOTIC SYNAPSIS | Genes involved in Meiotic Synapsis |
| 0.1 | 1.9 | REACTOME SYNTHESIS OF VERY LONG CHAIN FATTY ACYL COAS | Genes involved in Synthesis of very long-chain fatty acyl-CoAs |
| 0.1 | 1.9 | REACTOME EFFECTS OF PIP2 HYDROLYSIS | Genes involved in Effects of PIP2 hydrolysis |
| 0.1 | 1.4 | REACTOME NUCLEOTIDE LIKE PURINERGIC RECEPTORS | Genes involved in Nucleotide-like (purinergic) receptors |
| 0.1 | 0.6 | REACTOME REGULATION OF THE FANCONI ANEMIA PATHWAY | Genes involved in Regulation of the Fanconi anemia pathway |
| 0.0 | 0.7 | REACTOME PRESYNAPTIC NICOTINIC ACETYLCHOLINE RECEPTORS | Genes involved in Presynaptic nicotinic acetylcholine receptors |
| 0.0 | 9.2 | REACTOME PEPTIDE LIGAND BINDING RECEPTORS | Genes involved in Peptide ligand-binding receptors |
| 0.0 | 0.7 | REACTOME ERKS ARE INACTIVATED | Genes involved in ERKs are inactivated |
| 0.0 | 0.5 | REACTOME LIPID DIGESTION MOBILIZATION AND TRANSPORT | Genes involved in Lipid digestion, mobilization, and transport |
| 0.0 | 0.4 | REACTOME RECEPTOR LIGAND BINDING INITIATES THE SECOND PROTEOLYTIC CLEAVAGE OF NOTCH RECEPTOR | Genes involved in Receptor-ligand binding initiates the second proteolytic cleavage of Notch receptor |
| 0.0 | 1.1 | REACTOME ACTIVATED NOTCH1 TRANSMITS SIGNAL TO THE NUCLEUS | Genes involved in Activated NOTCH1 Transmits Signal to the Nucleus |
| 0.0 | 4.7 | REACTOME TRANSPORT OF INORGANIC CATIONS ANIONS AND AMINO ACIDS OLIGOPEPTIDES | Genes involved in Transport of inorganic cations/anions and amino acids/oligopeptides |
| 0.0 | 1.0 | REACTOME CREB PHOSPHORYLATION THROUGH THE ACTIVATION OF RAS | Genes involved in CREB phosphorylation through the activation of Ras |
| 0.0 | 0.7 | REACTOME GABA B RECEPTOR ACTIVATION | Genes involved in GABA B receptor activation |
| 0.0 | 0.3 | REACTOME G2 M DNA DAMAGE CHECKPOINT | Genes involved in G2/M DNA damage checkpoint |
| 0.0 | 0.5 | REACTOME REGULATION OF AMPK ACTIVITY VIA LKB1 | Genes involved in Regulation of AMPK activity via LKB1 |
| 0.0 | 0.5 | REACTOME APOPTOTIC CLEAVAGE OF CELL ADHESION PROTEINS | Genes involved in Apoptotic cleavage of cell adhesion proteins |
| 0.0 | 0.8 | REACTOME SYNTHESIS AND INTERCONVERSION OF NUCLEOTIDE DI AND TRIPHOSPHATES | Genes involved in Synthesis and interconversion of nucleotide di- and triphosphates |
| 0.0 | 2.0 | REACTOME LOSS OF NLP FROM MITOTIC CENTROSOMES | Genes involved in Loss of Nlp from mitotic centrosomes |
| 0.0 | 0.2 | REACTOME GROWTH HORMONE RECEPTOR SIGNALING | Genes involved in Growth hormone receptor signaling |
| 0.0 | 0.6 | REACTOME METABOLISM OF PORPHYRINS | Genes involved in Metabolism of porphyrins |
| 0.0 | 0.3 | REACTOME HYALURONAN METABOLISM | Genes involved in Hyaluronan metabolism |
| 0.0 | 0.6 | REACTOME RNA POL III TRANSCRIPTION TERMINATION | Genes involved in RNA Polymerase III Transcription Termination |
| 0.0 | 0.3 | REACTOME RECRUITMENT OF MITOTIC CENTROSOME PROTEINS AND COMPLEXES | Genes involved in Recruitment of mitotic centrosome proteins and complexes |
| 0.0 | 0.2 | REACTOME MEMBRANE BINDING AND TARGETTING OF GAG PROTEINS | Genes involved in Membrane binding and targetting of GAG proteins |
| 0.0 | 0.5 | REACTOME GLUCOSE TRANSPORT | Genes involved in Glucose transport |
| 0.0 | 0.5 | REACTOME PURINE SALVAGE | Genes involved in Purine salvage |
| 0.0 | 1.5 | REACTOME TRANSPORT OF MATURE TRANSCRIPT TO CYTOPLASM | Genes involved in Transport of Mature Transcript to Cytoplasm |
| 0.0 | 0.9 | REACTOME ASSOCIATION OF TRIC CCT WITH TARGET PROTEINS DURING BIOSYNTHESIS | Genes involved in Association of TriC/CCT with target proteins during biosynthesis |
| 0.0 | 1.6 | REACTOME NRAGE SIGNALS DEATH THROUGH JNK | Genes involved in NRAGE signals death through JNK |
| 0.0 | 2.2 | REACTOME SIGNALING BY RHO GTPASES | Genes involved in Signaling by Rho GTPases |
| 0.0 | 0.8 | REACTOME NCAM1 INTERACTIONS | Genes involved in NCAM1 interactions |
| 0.0 | 0.2 | REACTOME PROTEOLYTIC CLEAVAGE OF SNARE COMPLEX PROTEINS | Genes involved in Proteolytic cleavage of SNARE complex proteins |
| 0.0 | 0.3 | REACTOME INSULIN SYNTHESIS AND PROCESSING | Genes involved in Insulin Synthesis and Processing |
| 0.0 | 0.8 | REACTOME REGULATION OF ORNITHINE DECARBOXYLASE ODC | Genes involved in Regulation of ornithine decarboxylase (ODC) |
| 0.0 | 0.2 | REACTOME ENDOSOMAL SORTING COMPLEX REQUIRED FOR TRANSPORT ESCRT | Genes involved in Endosomal Sorting Complex Required For Transport (ESCRT) |