Project
PRJNA375882: Comprehensive Mouse Transcriptomic BodyMap
Navigation
Downloads
Results for Nr2c1
Z-value: 0.70
Transcription factors associated with Nr2c1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Nr2c1
|
ENSMUSG00000005897.15 | Nr2c1 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Nr2c1 | mm39_v1_chr10_+_93983844_93983885 | -0.32 | 6.9e-03 | Click! |
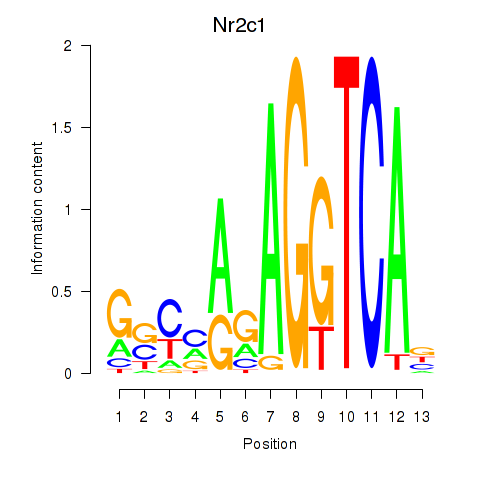
Activity profile of Nr2c1 motif
Sorted Z-values of Nr2c1 motif
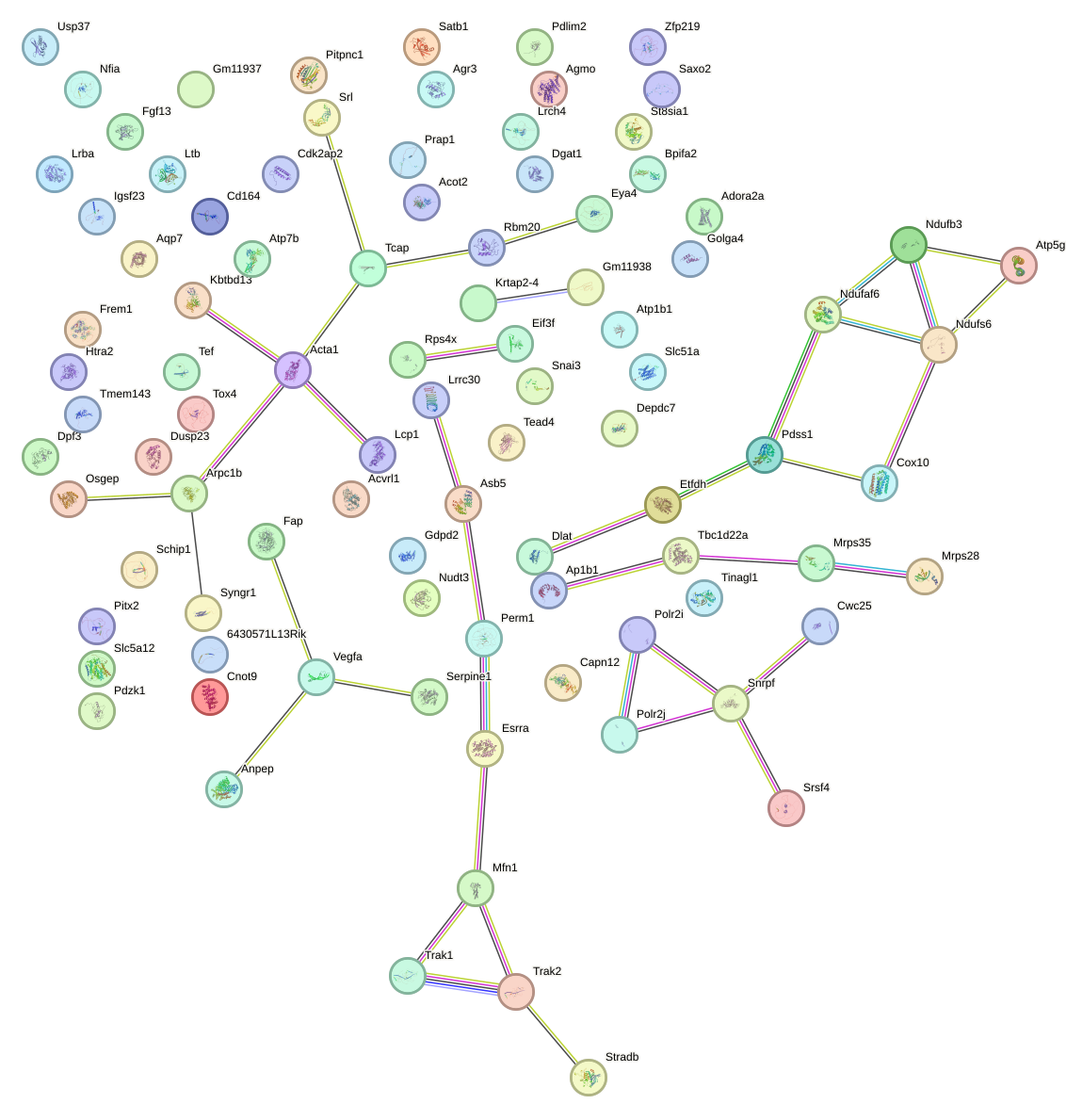
Network of associatons between targets according to the STRING database.
First level regulatory network of Nr2c1
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr8_-_124621483 | 4.09 |
ENSMUST00000034453.6
ENSMUST00000212584.2 |
Acta1
|
actin, alpha 1, skeletal muscle |
| chr11_+_98274637 | 3.79 |
ENSMUST00000008021.3
|
Tcap
|
titin-cap |
| chr16_-_4340920 | 3.74 |
ENSMUST00000090500.10
ENSMUST00000023161.8 |
Srl
|
sarcalumenin |
| chr7_+_139673300 | 3.07 |
ENSMUST00000026540.9
|
Prap1
|
proline-rich acidic protein 1 |
| chr4_+_156300325 | 2.72 |
ENSMUST00000105572.3
|
Perm1
|
PPARGC1 and ESRR induced regulator, muscle 1 |
| chr8_+_55024446 | 2.65 |
ENSMUST00000239166.2
ENSMUST00000239106.2 ENSMUST00000239152.2 |
Asb5
|
ankyrin repeat and SOCs box-containing 5 |
| chr2_+_110427643 | 2.62 |
ENSMUST00000045972.13
ENSMUST00000111026.3 |
Slc5a12
|
solute carrier family 5 (sodium/glucose cotransporter), member 12 |
| chr13_-_73476561 | 2.61 |
ENSMUST00000222930.2
ENSMUST00000223293.2 ENSMUST00000022097.6 |
Ndufs6
|
NADH:ubiquinone oxidoreductase core subunit S6 |
| chr17_-_67939702 | 2.53 |
ENSMUST00000097290.4
|
Lrrc30
|
leucine rich repeat containing 30 |
| chr19_+_53781721 | 2.24 |
ENSMUST00000162910.2
|
Rbm20
|
RNA binding motif protein 20 |
| chr4_-_41048124 | 2.24 |
ENSMUST00000030136.13
|
Aqp7
|
aquaporin 7 |
| chr16_-_32306683 | 1.94 |
ENSMUST00000042042.9
|
Slc51a
|
solute carrier family 51, alpha subunit |
| chr14_+_75373766 | 1.87 |
ENSMUST00000145303.8
|
Lcp1
|
lymphocyte cytosolic protein 1 |
| chr3_+_129007599 | 1.85 |
ENSMUST00000042587.12
|
Pitx2
|
paired-like homeodomain transcription factor 2 |
| chr8_-_123187406 | 1.85 |
ENSMUST00000006762.7
|
Snai3
|
snail family zinc finger 3 |
| chr15_+_79975520 | 1.70 |
ENSMUST00000009728.13
ENSMUST00000009727.12 |
Syngr1
|
synaptogyrin 1 |
| chr9_+_107217786 | 1.62 |
ENSMUST00000042581.4
|
6430571L13Rik
|
RIKEN cDNA 6430571L13 gene |
| chr1_-_59012579 | 1.54 |
ENSMUST00000173590.2
ENSMUST00000027186.12 |
Trak2
|
trafficking protein, kinesin binding 2 |
| chr15_+_101026454 | 1.53 |
ENSMUST00000117984.8
|
Acvrl1
|
activin A receptor, type II-like 1 |
| chr14_-_70414236 | 1.47 |
ENSMUST00000153735.8
|
Pdlim2
|
PDZ and LIM domain 2 |
| chr9_-_107483855 | 1.47 |
ENSMUST00000073448.12
ENSMUST00000194606.2 ENSMUST00000195662.6 |
Sema3b
|
sema domain, immunoglobulin domain (Ig), short basic domain, secreted, (semaphorin) 3B |
| chr7_-_79492091 | 1.43 |
ENSMUST00000049004.8
|
Anpep
|
alanyl (membrane) aminopeptidase |
| chr15_+_101026803 | 1.43 |
ENSMUST00000119063.8
ENSMUST00000121718.8 |
Acvrl1
|
activin A receptor, type II-like 1 |
| chr1_-_74544946 | 1.42 |
ENSMUST00000044260.11
ENSMUST00000186282.7 |
Usp37
|
ubiquitin specific peptidase 37 |
| chr9_+_118335294 | 1.31 |
ENSMUST00000084820.6
|
Golga4
|
golgi autoantigen, golgin subfamily a, 4 |
| chr7_-_19684654 | 1.27 |
ENSMUST00000043440.8
|
Igsf23
|
immunoglobulin superfamily, member 23 |
| chr7_+_45546365 | 1.23 |
ENSMUST00000069772.16
ENSMUST00000210503.2 ENSMUST00000209350.2 |
Tmem143
|
transmembrane protein 143 |
| chr12_-_115083839 | 1.17 |
ENSMUST00000103521.3
|
Ighv1-50
|
immunoglobulin heavy variable 1-50 |
| chr11_-_107228382 | 1.15 |
ENSMUST00000040380.13
|
Pitpnc1
|
phosphatidylinositol transfer protein, cytoplasmic 1 |
| chr3_+_129007917 | 1.15 |
ENSMUST00000174623.3
|
Pitx2
|
paired-like homeodomain transcription factor 2 |
| chr10_+_75152705 | 1.15 |
ENSMUST00000105420.3
|
Adora2a
|
adenosine A2a receptor |
| chr3_+_96737385 | 1.13 |
ENSMUST00000058865.14
|
Pdzk1
|
PDZ domain containing 1 |
| chr12_-_83534482 | 1.13 |
ENSMUST00000177959.8
ENSMUST00000178756.8 |
Dpf3
|
D4, zinc and double PHD fingers, family 3 |
| chr5_-_137101108 | 1.11 |
ENSMUST00000077523.4
ENSMUST00000041388.11 |
Serpine1
|
serine (or cysteine) peptidase inhibitor, clade E, member 1 |
| chr17_+_35413415 | 1.03 |
ENSMUST00000025262.6
ENSMUST00000173600.2 |
Ltb
|
lymphotoxin B |
| chr6_-_142910094 | 1.02 |
ENSMUST00000032421.4
|
St8sia1
|
ST8 alpha-N-acetyl-neuraminide alpha-2,8-sialyltransferase 1 |
| chr1_+_74545203 | 1.02 |
ENSMUST00000087215.7
|
Cnot9
|
CCR4-NOT transcription complex, subunit 9 |
| chrX_+_99773523 | 1.00 |
ENSMUST00000019503.14
|
Gdpd2
|
glycerophosphodiester phosphodiesterase domain containing 2 |
| chr2_-_62404195 | 0.98 |
ENSMUST00000174234.8
ENSMUST00000000402.16 ENSMUST00000174448.8 ENSMUST00000102732.10 |
Fap
|
fibroblast activation protein |
| chr14_+_75373915 | 0.97 |
ENSMUST00000122840.8
|
Lcp1
|
lymphocyte cytosolic protein 1 |
| chr10_-_23226684 | 0.96 |
ENSMUST00000220299.2
|
Eya4
|
EYA transcriptional coactivator and phosphatase 4 |
| chr17_-_13211075 | 0.94 |
ENSMUST00000159986.8
ENSMUST00000007007.14 |
Wtap
|
Wilms tumour 1-associating protein |
| chr19_-_6899173 | 0.92 |
ENSMUST00000025906.12
ENSMUST00000239322.2 |
Esrra
|
estrogen related receptor, alpha |
| chr4_+_97665992 | 0.90 |
ENSMUST00000107062.9
ENSMUST00000052018.12 ENSMUST00000107057.8 |
Nfia
|
nuclear factor I/A |
| chr17_-_46343291 | 0.89 |
ENSMUST00000071648.12
ENSMUST00000142351.9 |
Vegfa
|
vascular endothelial growth factor A |
| chr1_+_59012680 | 0.89 |
ENSMUST00000114296.8
ENSMUST00000027185.11 |
Stradb
|
STE20-related kinase adaptor beta |
| chr17_-_13211374 | 0.88 |
ENSMUST00000159551.8
ENSMUST00000160781.8 |
Wtap
|
Wilms tumour 1-associating protein |
| chr15_-_102579463 | 0.87 |
ENSMUST00000185641.7
|
Atp5g2
|
ATP synthase, H+ transporting, mitochondrial F0 complex, subunit C2 (subunit 9) |
| chr3_+_68479578 | 0.84 |
ENSMUST00000170788.9
|
Schip1
|
schwannomin interacting protein 1 |
| chr15_-_76396151 | 0.81 |
ENSMUST00000023214.11
|
Dgat1
|
diacylglycerol O-acyltransferase 1 |
| chr1_+_58625539 | 0.81 |
ENSMUST00000027193.9
|
Ndufb3
|
NADH:ubiquinone oxidoreductase subunit B3 |
| chr10_+_75152908 | 0.79 |
ENSMUST00000219044.2
|
Adora2a
|
adenosine A2a receptor |
| chr17_+_75772475 | 0.75 |
ENSMUST00000095204.6
|
Rasgrp3
|
RAS, guanyl releasing protein 3 |
| chr4_-_11076159 | 0.75 |
ENSMUST00000058183.9
|
Ndufaf6
|
NADH:ubiquinone oxidoreductase complex assembly factor 6 |
| chr4_+_97665843 | 0.74 |
ENSMUST00000075448.13
ENSMUST00000092532.13 |
Nfia
|
nuclear factor I/A |
| chr17_-_52117894 | 0.73 |
ENSMUST00000176669.8
|
Satb1
|
special AT-rich sequence binding protein 1 |
| chr1_-_164285914 | 0.72 |
ENSMUST00000027863.13
|
Atp1b1
|
ATPase, Na+/K+ transporting, beta 1 polypeptide |
| chr4_+_131600918 | 0.70 |
ENSMUST00000053819.6
|
Srsf4
|
serine and arginine-rich splicing factor 4 |
| chr12_+_84034628 | 0.70 |
ENSMUST00000021649.8
|
Acot2
|
acyl-CoA thioesterase 2 |
| chr3_-_79535966 | 0.68 |
ENSMUST00000120992.8
|
Etfdh
|
electron transferring flavoprotein, dehydrogenase |
| chr7_+_108533613 | 0.66 |
ENSMUST00000033342.7
|
Eif3f
|
eukaryotic translation initiation factor 3, subunit F |
| chr11_-_63970270 | 0.62 |
ENSMUST00000049091.9
|
Cox10
|
heme A:farnesyltransferase cytochrome c oxidase assembly factor 10 |
| chr7_-_82297676 | 0.61 |
ENSMUST00000207693.2
ENSMUST00000056728.5 ENSMUST00000156720.8 ENSMUST00000126478.2 |
Saxo2
|
stabilizer of axonemal microtubules 2 |
| chr9_-_65298934 | 0.58 |
ENSMUST00000068307.4
|
Kbtbd13
|
kelch repeat and BTB (POZ) domain containing 13 |
| chr7_+_28580847 | 0.57 |
ENSMUST00000066880.6
|
Capn12
|
calpain 12 |
| chr15_-_76093256 | 0.56 |
ENSMUST00000071869.12
ENSMUST00000170915.2 |
Plec
|
plectin |
| chrX_-_58612709 | 0.56 |
ENSMUST00000124402.2
|
Fgf13
|
fibroblast growth factor 13 |
| chr9_+_121126562 | 0.56 |
ENSMUST00000238988.2
ENSMUST00000210798.3 ENSMUST00000211439.2 |
Trak1
|
trafficking protein, kinesin binding 1 |
| chr6_-_83030759 | 0.55 |
ENSMUST00000134606.8
|
Htra2
|
HtrA serine peptidase 2 |
| chr14_-_51162083 | 0.55 |
ENSMUST00000160375.8
ENSMUST00000162177.8 |
Osgep
|
O-sialoglycoprotein endopeptidase |
| chr2_+_153850177 | 0.55 |
ENSMUST00000048103.9
ENSMUST00000145388.2 |
Bpifa2
|
BPI fold containing family A, member 2 |
| chr6_-_128252540 | 0.54 |
ENSMUST00000130454.8
|
Tead4
|
TEA domain family member 4 |
| chr19_-_6899121 | 0.54 |
ENSMUST00000173635.2
|
Esrra
|
estrogen related receptor, alpha |
| chr4_-_130068484 | 0.53 |
ENSMUST00000132545.3
ENSMUST00000175992.8 ENSMUST00000105999.9 |
Tinagl1
|
tubulointerstitial nephritis antigen-like 1 |
| chr7_+_29931735 | 0.52 |
ENSMUST00000108193.2
ENSMUST00000108192.2 |
Polr2i
|
polymerase (RNA) II (DNA directed) polypeptide I |
| chr9_-_50571080 | 0.51 |
ENSMUST00000034567.4
|
Dlat
|
dihydrolipoamide S-acetyltransferase (E2 component of pyruvate dehydrogenase complex) |
| chr4_-_106360302 | 0.50 |
ENSMUST00000177702.2
|
Tmem61
|
transmembrane protein 61 |
| chr5_+_145060489 | 0.50 |
ENSMUST00000136074.2
|
Arpc1b
|
actin related protein 2/3 complex, subunit 1B |
| chr10_-_93425553 | 0.49 |
ENSMUST00000020203.7
|
Snrpf
|
small nuclear ribonucleoprotein polypeptide F |
| chr14_-_51162346 | 0.48 |
ENSMUST00000159292.8
|
Osgep
|
O-sialoglycoprotein endopeptidase |
| chr8_-_22550035 | 0.48 |
ENSMUST00000110738.3
|
Atp7b
|
ATPase, Cu++ transporting, beta polypeptide |
| chr8_-_22550321 | 0.48 |
ENSMUST00000006742.11
|
Atp7b
|
ATPase, Cu++ transporting, beta polypeptide |
| chr7_+_29931309 | 0.46 |
ENSMUST00000019882.16
ENSMUST00000149654.8 |
Polr2i
|
polymerase (RNA) II (DNA directed) polypeptide I |
| chr3_+_32583681 | 0.45 |
ENSMUST00000147350.8
|
Mfn1
|
mitofusin 1 |
| chr2_-_104573179 | 0.44 |
ENSMUST00000028595.8
|
Depdc7
|
DEP domain containing 7 |
| chr5_+_137628377 | 0.44 |
ENSMUST00000175968.8
|
Lrch4
|
leucine-rich repeats and calponin homology (CH) domain containing 4 |
| chr14_-_52252003 | 0.41 |
ENSMUST00000226522.2
|
Zfp219
|
zinc finger protein 219 |
| chr3_+_32583602 | 0.41 |
ENSMUST00000091257.11
|
Mfn1
|
mitofusin 1 |
| chr3_-_8988854 | 0.40 |
ENSMUST00000042148.6
|
Mrps28
|
mitochondrial ribosomal protein S28 |
| chr8_+_121395047 | 0.39 |
ENSMUST00000181795.2
|
Cox4i1
|
cytochrome c oxidase subunit 4I1 |
| chr11_+_4936824 | 0.39 |
ENSMUST00000109897.8
ENSMUST00000009234.16 |
Ap1b1
|
adaptor protein complex AP-1, beta 1 subunit |
| chr12_+_35975619 | 0.38 |
ENSMUST00000042101.5
ENSMUST00000154042.2 |
Agr3
|
anterior gradient 3 |
| chr2_+_22785593 | 0.38 |
ENSMUST00000152170.8
|
Pdss1
|
prenyl (solanesyl) diphosphate synthase, subunit 1 |
| chr1_-_172460497 | 0.37 |
ENSMUST00000027826.7
|
Dusp23
|
dual specificity phosphatase 23 |
| chr11_-_99494134 | 0.36 |
ENSMUST00000072306.4
|
Gm11938
|
predicted gene 11938 |
| chr6_-_83031358 | 0.36 |
ENSMUST00000113962.8
ENSMUST00000089645.13 ENSMUST00000113963.8 |
Htra2
|
HtrA serine peptidase 2 |
| chr15_+_86098660 | 0.36 |
ENSMUST00000063414.9
|
Tbc1d22a
|
TBC1 domain family, member 22a |
| chrX_-_101232978 | 0.36 |
ENSMUST00000033683.8
|
Rps4x
|
ribosomal protein S4, X-linked |
| chr3_+_86131970 | 0.35 |
ENSMUST00000192145.6
ENSMUST00000194759.6 ENSMUST00000107635.7 |
Lrba
|
LPS-responsive beige-like anchor |
| chr17_+_57369490 | 0.34 |
ENSMUST00000163628.2
|
Crb3
|
crumbs family member 3 |
| chr15_+_79025523 | 0.34 |
ENSMUST00000040077.8
|
Polr2f
|
polymerase (RNA) II (DNA directed) polypeptide F |
| chr15_-_79025387 | 0.33 |
ENSMUST00000187550.7
ENSMUST00000188562.7 ENSMUST00000190509.7 ENSMUST00000190730.7 ENSMUST00000190959.7 ENSMUST00000169604.8 ENSMUST00000186053.7 |
1700088E04Rik
|
RIKEN cDNA 1700088E04 gene |
| chr11_-_97657327 | 0.32 |
ENSMUST00000107579.2
ENSMUST00000018685.9 |
Cwc25
|
CWC25 spliceosome-associated protein |
| chr12_+_37291728 | 0.32 |
ENSMUST00000160768.8
|
Agmo
|
alkylglycerol monooxygenase |
| chr15_+_81695615 | 0.31 |
ENSMUST00000023024.8
|
Tef
|
thyrotroph embryonic factor |
| chr10_+_41395410 | 0.30 |
ENSMUST00000019962.15
|
Cd164
|
CD164 antigen |
| chr17_-_27816151 | 0.30 |
ENSMUST00000231742.2
|
Nudt3
|
nudix (nucleotide diphosphate linked moiety X)-type motif 3 |
| chr19_+_4147391 | 0.29 |
ENSMUST00000174514.2
ENSMUST00000174149.8 |
Cdk2ap2
|
CDK2-associated protein 2 |
| chr2_+_22785534 | 0.29 |
ENSMUST00000053729.14
|
Pdss1
|
prenyl (solanesyl) diphosphate synthase, subunit 1 |
| chr4_-_82939330 | 0.29 |
ENSMUST00000071708.12
|
Frem1
|
Fras1 related extracellular matrix protein 1 |
| chr2_+_109522781 | 0.28 |
ENSMUST00000111050.10
|
Bdnf
|
brain derived neurotrophic factor |
| chr2_-_19558719 | 0.28 |
ENSMUST00000062060.5
|
4921504E06Rik
|
RIKEN cDNA 4921504E06 gene |
| chr13_+_73476629 | 0.27 |
ENSMUST00000221730.2
|
Mrpl36
|
mitochondrial ribosomal protein L36 |
| chr7_+_79836581 | 0.27 |
ENSMUST00000032754.9
|
Sema4b
|
sema domain, immunoglobulin domain (Ig), transmembrane domain (TM) and short cytoplasmic domain, (semaphorin) 4B |
| chr13_+_104424359 | 0.27 |
ENSMUST00000065766.7
|
Adamts6
|
a disintegrin-like and metallopeptidase (reprolysin type) with thrombospondin type 1 motif, 6 |
| chr17_-_34247016 | 0.27 |
ENSMUST00000236627.2
ENSMUST00000237759.2 ENSMUST00000045467.14 ENSMUST00000114303.4 |
H2-Ke6
|
H2-K region expressed gene 6 |
| chr17_+_75772567 | 0.27 |
ENSMUST00000234660.2
|
Rasgrp3
|
RAS, guanyl releasing protein 3 |
| chr5_-_121974325 | 0.26 |
ENSMUST00000137682.2
|
Sh2b3
|
SH2B adaptor protein 3 |
| chr7_+_43600038 | 0.25 |
ENSMUST00000072204.5
|
Klk1b8
|
kallikrein 1-related peptidase b8 |
| chr6_+_83031502 | 0.24 |
ENSMUST00000092618.9
|
Aup1
|
ancient ubiquitous protein 1 |
| chr1_-_155627430 | 0.23 |
ENSMUST00000195275.2
|
Lhx4
|
LIM homeobox protein 4 |
| chr10_-_4382311 | 0.23 |
ENSMUST00000126102.8
ENSMUST00000131853.2 ENSMUST00000042251.11 |
Rmnd1
|
required for meiotic nuclear division 1 homolog |
| chr7_+_43910845 | 0.23 |
ENSMUST00000124863.4
|
Gm15517
|
predicted gene 15517 |
| chr7_-_28297565 | 0.22 |
ENSMUST00000040531.9
ENSMUST00000108283.8 |
Samd4b
Pak4
|
sterile alpha motif domain containing 4B p21 (RAC1) activated kinase 4 |
| chr10_+_58207229 | 0.22 |
ENSMUST00000238939.2
|
Lims1
|
LIM and senescent cell antigen-like domains 1 |
| chr17_+_38110779 | 0.22 |
ENSMUST00000215168.2
ENSMUST00000216478.2 |
Olfr124
|
olfactory receptor 124 |
| chr17_-_35454729 | 0.21 |
ENSMUST00000048994.7
|
Nfkbil1
|
nuclear factor of kappa light polypeptide gene enhancer in B cells inhibitor like 1 |
| chr4_+_138606671 | 0.21 |
ENSMUST00000105804.2
|
Pla2g2e
|
phospholipase A2, group IIE |
| chr5_+_135216090 | 0.21 |
ENSMUST00000002825.6
|
Baz1b
|
bromodomain adjacent to zinc finger domain, 1B |
| chr19_+_5687503 | 0.20 |
ENSMUST00000025867.6
|
Rela
|
v-rel reticuloendotheliosis viral oncogene homolog A (avian) |
| chr17_+_7292967 | 0.20 |
ENSMUST00000097422.6
|
Gm1604b
|
predicted gene 1604b |
| chr6_+_125122172 | 0.20 |
ENSMUST00000119527.8
ENSMUST00000117675.8 |
Iffo1
|
intermediate filament family orphan 1 |
| chr7_-_45570674 | 0.19 |
ENSMUST00000210939.2
|
Emp3
|
epithelial membrane protein 3 |
| chr19_-_45986919 | 0.18 |
ENSMUST00000045396.9
|
Armh3
|
armadillo-like helical domain containing 3 |
| chr11_-_62172164 | 0.18 |
ENSMUST00000072916.5
|
Zswim7
|
zinc finger SWIM-type containing 7 |
| chr7_-_45570254 | 0.18 |
ENSMUST00000164119.3
|
Emp3
|
epithelial membrane protein 3 |
| chr4_+_117581609 | 0.17 |
ENSMUST00000203629.2
ENSMUST00000204276.2 |
Gm12845
|
predicted gene 12845 |
| chr11_+_116744578 | 0.17 |
ENSMUST00000021173.14
|
Mfsd11
|
major facilitator superfamily domain containing 11 |
| chr7_+_101043568 | 0.16 |
ENSMUST00000098243.4
|
Arap1
|
ArfGAP with RhoGAP domain, ankyrin repeat and PH domain 1 |
| chr14_+_58036093 | 0.16 |
ENSMUST00000111267.2
|
Sap18
|
Sin3-associated polypeptide 18 |
| chr14_-_52252318 | 0.16 |
ENSMUST00000228051.2
|
Zfp219
|
zinc finger protein 219 |
| chr7_-_45570828 | 0.16 |
ENSMUST00000038876.13
|
Emp3
|
epithelial membrane protein 3 |
| chr9_+_44398524 | 0.15 |
ENSMUST00000218183.2
|
Bcl9l
|
B cell CLL/lymphoma 9-like |
| chr9_-_35755446 | 0.15 |
ENSMUST00000054175.7
|
Pate5
|
prostate and testis expressed 5 |
| chr13_-_18118736 | 0.13 |
ENSMUST00000009003.9
|
Rala
|
v-ral simian leukemia viral oncogene A (ras related) |
| chr11_-_24025054 | 0.13 |
ENSMUST00000068360.2
|
A830031A19Rik
|
RIKEN cDNA A830031A19 gene |
| chr15_-_102274781 | 0.12 |
ENSMUST00000078508.7
|
Sp7
|
Sp7 transcription factor 7 |
| chr12_+_37292029 | 0.12 |
ENSMUST00000160390.2
|
Agmo
|
alkylglycerol monooxygenase |
| chr7_-_45570538 | 0.10 |
ENSMUST00000210297.2
|
Emp3
|
epithelial membrane protein 3 |
| chr10_-_129010923 | 0.10 |
ENSMUST00000169800.4
|
Olfr772
|
olfactory receptor 772 |
| chrX_+_10351360 | 0.09 |
ENSMUST00000076354.13
ENSMUST00000115526.2 |
Tspan7
|
tetraspanin 7 |
| chr3_+_122298308 | 0.09 |
ENSMUST00000199358.2
|
Bcar3
|
breast cancer anti-estrogen resistance 3 |
| chr16_+_3648742 | 0.08 |
ENSMUST00000214238.2
ENSMUST00000214590.2 |
Olfr15
|
olfactory receptor 15 |
| chr14_+_58036071 | 0.08 |
ENSMUST00000111268.8
|
Sap18
|
Sin3-associated polypeptide 18 |
| chr11_+_62172283 | 0.08 |
ENSMUST00000101075.11
ENSMUST00000050646.13 |
Ttc19
|
tetratricopeptide repeat domain 19 |
| chr2_+_158636727 | 0.08 |
ENSMUST00000029186.14
ENSMUST00000109478.9 ENSMUST00000156893.2 |
Dhx35
|
DEAH (Asp-Glu-Ala-His) box polypeptide 35 |
| chr1_-_58625431 | 0.07 |
ENSMUST00000161000.2
ENSMUST00000161600.8 |
Fam126b
|
family with sequence similarity 126, member B |
| chr11_-_115698894 | 0.07 |
ENSMUST00000132780.8
|
Caskin2
|
CASK-interacting protein 2 |
| chr11_-_100098333 | 0.06 |
ENSMUST00000007272.8
|
Krt14
|
keratin 14 |
| chr13_-_55718899 | 0.06 |
ENSMUST00000225240.3
ENSMUST00000021957.8 |
Fam193b
|
family with sequence similarity 193, member B |
| chr3_+_58433236 | 0.06 |
ENSMUST00000029387.15
|
Eif2a
|
eukaryotic translation initiation factor 2A |
| chr2_-_63928339 | 0.06 |
ENSMUST00000131615.9
|
Fign
|
fidgetin |
| chr16_-_58850641 | 0.05 |
ENSMUST00000216415.2
ENSMUST00000206463.3 |
Olfr186
|
olfactory receptor 186 |
| chr19_+_10914517 | 0.05 |
ENSMUST00000037261.4
|
Ptgdr2
|
prostaglandin D2 receptor 2 |
| chr4_-_117109074 | 0.03 |
ENSMUST00000165128.9
|
Armh1
|
armadillo-like helical domain containing 1 |
| chr14_+_58035640 | 0.03 |
ENSMUST00000111269.2
|
Sap18
|
Sin3-associated polypeptide 18 |
| chr4_-_137084973 | 0.01 |
ENSMUST00000030417.10
ENSMUST00000051477.13 |
Cdc42
|
cell division cycle 42 |
| chr2_-_128890092 | 0.00 |
ENSMUST00000145798.4
|
Vinac1
|
vinculin/alpha-catenin family member 1 |
| chr2_+_153187729 | 0.00 |
ENSMUST00000227428.2
ENSMUST00000109790.2 |
Asxl1
|
ASXL transcriptional regulator 1 |
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.9 | 3.7 | GO:0014873 | response to muscle activity involved in regulation of muscle adaptation(GO:0014873) |
| 0.7 | 7.9 | GO:0030240 | skeletal muscle thin filament assembly(GO:0030240) |
| 0.6 | 3.0 | GO:0021763 | subthalamic nucleus development(GO:0021763) superior vena cava morphogenesis(GO:0060578) |
| 0.4 | 2.2 | GO:0070295 | glycerol transport(GO:0015793) renal water absorption(GO:0070295) |
| 0.4 | 1.9 | GO:0014057 | positive regulation of acetylcholine secretion, neurotransmission(GO:0014057) |
| 0.4 | 1.1 | GO:2000097 | regulation of smooth muscle cell-matrix adhesion(GO:2000097) |
| 0.3 | 1.0 | GO:0097325 | regulation of collagen catabolic process(GO:0010710) melanocyte proliferation(GO:0097325) |
| 0.3 | 1.0 | GO:0015680 | intracellular copper ion transport(GO:0015680) |
| 0.3 | 0.9 | GO:1904924 | negative regulation of mitophagy in response to mitochondrial depolarization(GO:1904924) |
| 0.3 | 0.9 | GO:0038190 | neuropilin signaling pathway(GO:0038189) VEGF-activated neuropilin signaling pathway(GO:0038190) |
| 0.3 | 0.8 | GO:1901738 | regulation of vitamin A metabolic process(GO:1901738) |
| 0.3 | 1.0 | GO:0002949 | tRNA threonylcarbamoyladenosine modification(GO:0002949) |
| 0.2 | 3.0 | GO:0090500 | lymphatic endothelial cell differentiation(GO:0060836) endocardial cushion to mesenchymal transition(GO:0090500) |
| 0.2 | 0.7 | GO:1903281 | positive regulation of calcium:sodium antiporter activity(GO:1903281) |
| 0.2 | 0.9 | GO:0010636 | positive regulation of mitochondrial fusion(GO:0010636) |
| 0.2 | 1.0 | GO:2000325 | regulation of ligand-dependent nuclear receptor transcription coactivator activity(GO:2000325) positive regulation of ligand-dependent nuclear receptor transcription coactivator activity(GO:2000327) |
| 0.2 | 1.5 | GO:0098937 | dendritic transport(GO:0098935) anterograde dendritic transport(GO:0098937) |
| 0.1 | 1.2 | GO:0045084 | positive regulation of interleukin-12 biosynthetic process(GO:0045084) |
| 0.1 | 2.7 | GO:0014850 | response to muscle activity(GO:0014850) |
| 0.1 | 2.9 | GO:0006120 | mitochondrial electron transport, NADH to ubiquinone(GO:0006120) |
| 0.1 | 1.8 | GO:0080009 | mRNA methylation(GO:0080009) |
| 0.1 | 0.6 | GO:0018343 | protein farnesylation(GO:0018343) |
| 0.1 | 2.8 | GO:0071803 | positive regulation of podosome assembly(GO:0071803) |
| 0.1 | 0.6 | GO:0098957 | anterograde axonal transport of mitochondrion(GO:0098957) |
| 0.1 | 0.5 | GO:0001830 | trophectodermal cell fate commitment(GO:0001830) |
| 0.1 | 1.1 | GO:0015879 | carnitine transport(GO:0015879) |
| 0.1 | 1.0 | GO:0006283 | transcription-coupled nucleotide-excision repair(GO:0006283) |
| 0.1 | 0.6 | GO:0045200 | establishment or maintenance of neuroblast polarity(GO:0045196) establishment of neuroblast polarity(GO:0045200) |
| 0.1 | 0.3 | GO:0071544 | diphosphoinositol polyphosphate catabolic process(GO:0071544) |
| 0.1 | 1.6 | GO:0072189 | ureter development(GO:0072189) |
| 0.1 | 1.7 | GO:0048172 | regulation of short-term neuronal synaptic plasticity(GO:0048172) |
| 0.1 | 0.2 | GO:0098749 | cerebellar neuron development(GO:0098749) |
| 0.1 | 1.1 | GO:0035871 | protein K11-linked deubiquitination(GO:0035871) |
| 0.1 | 0.7 | GO:0042760 | very long-chain fatty acid catabolic process(GO:0042760) |
| 0.1 | 1.4 | GO:0035814 | negative regulation of renal sodium excretion(GO:0035814) |
| 0.1 | 1.0 | GO:0016576 | histone dephosphorylation(GO:0016576) |
| 0.1 | 0.5 | GO:0061732 | mitochondrial acetyl-CoA biosynthetic process from pyruvate(GO:0061732) |
| 0.1 | 1.9 | GO:0015721 | bile acid and bile salt transport(GO:0015721) |
| 0.1 | 1.0 | GO:0090527 | actin filament reorganization(GO:0090527) |
| 0.1 | 0.2 | GO:0021526 | medial motor column neuron differentiation(GO:0021526) |
| 0.0 | 1.1 | GO:0008210 | estrogen metabolic process(GO:0008210) |
| 0.0 | 0.6 | GO:0031581 | hemidesmosome assembly(GO:0031581) |
| 0.0 | 0.7 | GO:0043374 | CD8-positive, alpha-beta T cell differentiation(GO:0043374) |
| 0.0 | 1.6 | GO:0010257 | NADH dehydrogenase complex assembly(GO:0010257) mitochondrial respiratory chain complex I assembly(GO:0032981) mitochondrial respiratory chain complex I biogenesis(GO:0097031) |
| 0.0 | 0.4 | GO:0046485 | ether lipid metabolic process(GO:0046485) |
| 0.0 | 2.2 | GO:0033120 | positive regulation of RNA splicing(GO:0033120) |
| 0.0 | 0.7 | GO:0006744 | ubiquinone biosynthetic process(GO:0006744) |
| 0.0 | 0.7 | GO:0075522 | IRES-dependent viral translational initiation(GO:0075522) |
| 0.0 | 0.7 | GO:0033539 | fatty acid beta-oxidation using acyl-CoA dehydrogenase(GO:0033539) |
| 0.0 | 1.7 | GO:0050919 | negative chemotaxis(GO:0050919) |
| 0.0 | 1.5 | GO:1900078 | positive regulation of cellular response to insulin stimulus(GO:1900078) |
| 0.0 | 0.4 | GO:0006123 | mitochondrial electron transport, cytochrome c to oxygen(GO:0006123) |
| 0.0 | 0.9 | GO:0015985 | energy coupled proton transport, down electrochemical gradient(GO:0015985) ATP synthesis coupled proton transport(GO:0015986) |
| 0.0 | 0.4 | GO:0045199 | maintenance of epithelial cell apical/basal polarity(GO:0045199) |
| 0.0 | 0.7 | GO:0048025 | negative regulation of mRNA splicing, via spliceosome(GO:0048025) |
| 0.0 | 1.3 | GO:0043001 | Golgi to plasma membrane protein transport(GO:0043001) |
| 0.0 | 0.2 | GO:0031665 | negative regulation of lipopolysaccharide-mediated signaling pathway(GO:0031665) |
| 0.0 | 0.6 | GO:0032060 | bleb assembly(GO:0032060) |
| 0.0 | 0.9 | GO:2001240 | negative regulation of signal transduction in absence of ligand(GO:1901099) negative regulation of extrinsic apoptotic signaling pathway in absence of ligand(GO:2001240) |
| 0.0 | 0.2 | GO:2000346 | negative regulation of hepatocyte proliferation(GO:2000346) |
| 0.0 | 0.2 | GO:0070131 | positive regulation of mitochondrial translation(GO:0070131) |
| 0.0 | 0.1 | GO:0051665 | membrane raft localization(GO:0051665) |
| 0.0 | 0.3 | GO:2000035 | regulation of stem cell division(GO:2000035) |
| 0.0 | 0.1 | GO:0017062 | respiratory chain complex III assembly(GO:0017062) mitochondrial respiratory chain complex III assembly(GO:0034551) |
| 0.0 | 1.0 | GO:0034605 | cellular response to heat(GO:0034605) |
| 0.0 | 0.5 | GO:0000387 | spliceosomal snRNP assembly(GO:0000387) |
| 0.0 | 0.1 | GO:1990928 | response to amino acid starvation(GO:1990928) |
| 0.0 | 0.4 | GO:0048268 | clathrin coat assembly(GO:0048268) |
| 0.0 | 0.5 | GO:0034314 | Arp2/3 complex-mediated actin nucleation(GO:0034314) |
| 0.0 | 0.3 | GO:0097094 | craniofacial suture morphogenesis(GO:0097094) |
| 0.0 | 0.2 | GO:0034374 | low-density lipoprotein particle remodeling(GO:0034374) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 1.8 | GO:0036396 | MIS complex(GO:0036396) |
| 0.2 | 1.1 | GO:0042583 | chromaffin granule(GO:0042583) |
| 0.2 | 0.5 | GO:0005967 | mitochondrial pyruvate dehydrogenase complex(GO:0005967) |
| 0.2 | 0.7 | GO:0045251 | mitochondrial electron transfer flavoprotein complex(GO:0017133) electron transfer flavoprotein complex(GO:0045251) |
| 0.1 | 0.5 | GO:0036284 | tubulobulbar complex(GO:0036284) |
| 0.1 | 1.0 | GO:0071438 | invadopodium membrane(GO:0071438) |
| 0.1 | 1.0 | GO:0000408 | EKC/KEOPS complex(GO:0000408) |
| 0.1 | 1.4 | GO:0031983 | vesicle lumen(GO:0031983) |
| 0.1 | 1.0 | GO:0030015 | CCR4-NOT core complex(GO:0030015) |
| 0.1 | 1.9 | GO:0032279 | asymmetric synapse(GO:0032279) |
| 0.1 | 2.8 | GO:0001891 | phagocytic cup(GO:0001891) |
| 0.1 | 0.5 | GO:0005683 | U7 snRNP(GO:0005683) |
| 0.1 | 0.7 | GO:0005890 | sodium:potassium-exchanging ATPase complex(GO:0005890) |
| 0.1 | 4.1 | GO:0005865 | striated muscle thin filament(GO:0005865) |
| 0.1 | 3.4 | GO:0045271 | mitochondrial respiratory chain complex I(GO:0005747) NADH dehydrogenase complex(GO:0030964) respiratory chain complex I(GO:0045271) |
| 0.1 | 0.7 | GO:0071541 | eukaryotic translation initiation factor 3 complex, eIF3m(GO:0071541) |
| 0.1 | 0.9 | GO:0000276 | mitochondrial proton-transporting ATP synthase complex, coupling factor F(o)(GO:0000276) |
| 0.1 | 0.9 | GO:0035631 | CD40 receptor complex(GO:0035631) |
| 0.1 | 0.9 | GO:0098799 | outer mitochondrial membrane protein complex(GO:0098799) |
| 0.1 | 1.0 | GO:0032045 | guanyl-nucleotide exchange factor complex(GO:0032045) |
| 0.0 | 1.3 | GO:0005665 | DNA-directed RNA polymerase II, core complex(GO:0005665) |
| 0.0 | 0.2 | GO:0000839 | Hrd1p ubiquitin ligase ERAD-L complex(GO:0000839) |
| 0.0 | 1.1 | GO:0071565 | nBAF complex(GO:0071565) |
| 0.0 | 1.1 | GO:0031528 | microvillus membrane(GO:0031528) |
| 0.0 | 3.7 | GO:0016529 | sarcoplasmic reticulum(GO:0016529) |
| 0.0 | 1.0 | GO:0070069 | cytochrome complex(GO:0070069) |
| 0.0 | 1.2 | GO:0032839 | dendrite cytoplasm(GO:0032839) |
| 0.0 | 0.6 | GO:0016528 | sarcoplasm(GO:0016528) |
| 0.0 | 0.6 | GO:1904115 | axon cytoplasm(GO:1904115) |
| 0.0 | 1.0 | GO:0032588 | trans-Golgi network membrane(GO:0032588) |
| 0.0 | 0.2 | GO:0061574 | ASAP complex(GO:0061574) |
| 0.0 | 2.2 | GO:0031526 | brush border membrane(GO:0031526) |
| 0.0 | 3.6 | GO:0030018 | Z disc(GO:0030018) |
| 0.0 | 1.7 | GO:0031594 | neuromuscular junction(GO:0031594) |
| 0.0 | 0.3 | GO:0030061 | mitochondrial crista(GO:0030061) |
| 0.0 | 0.2 | GO:0033256 | I-kappaB/NF-kappaB complex(GO:0033256) |
| 0.0 | 0.9 | GO:0016235 | aggresome(GO:0016235) |
| 0.0 | 1.5 | GO:0001725 | stress fiber(GO:0001725) contractile actin filament bundle(GO:0097517) |
| 0.0 | 0.4 | GO:0005763 | organellar small ribosomal subunit(GO:0000314) mitochondrial small ribosomal subunit(GO:0005763) |
| 0.0 | 0.6 | GO:0014704 | intercalated disc(GO:0014704) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.8 | 3.8 | GO:0051373 | FATZ binding(GO:0051373) |
| 0.4 | 2.2 | GO:0015254 | glycerol channel activity(GO:0015254) |
| 0.4 | 3.0 | GO:0016361 | activin receptor activity, type I(GO:0016361) |
| 0.3 | 1.0 | GO:0061711 | N(6)-L-threonylcarbamoyladenine synthase(GO:0061711) |
| 0.3 | 0.9 | GO:0043183 | vascular endothelial growth factor receptor 1 binding(GO:0043183) |
| 0.3 | 2.6 | GO:0015129 | lactate transmembrane transporter activity(GO:0015129) |
| 0.2 | 1.0 | GO:0043682 | copper-exporting ATPase activity(GO:0004008) copper-transporting ATPase activity(GO:0043682) |
| 0.2 | 0.8 | GO:0050252 | retinol O-fatty-acyltransferase activity(GO:0050252) |
| 0.2 | 1.0 | GO:0003828 | alpha-N-acetylneuraminate alpha-2,8-sialyltransferase activity(GO:0003828) |
| 0.2 | 0.5 | GO:0016418 | S-acetyltransferase activity(GO:0016418) |
| 0.2 | 0.7 | GO:0050347 | trans-hexaprenyltranstransferase activity(GO:0000010) trans-octaprenyltranstransferase activity(GO:0050347) |
| 0.2 | 1.9 | GO:0001609 | G-protein coupled adenosine receptor activity(GO:0001609) |
| 0.1 | 0.4 | GO:0050479 | glyceryl-ether monooxygenase activity(GO:0050479) |
| 0.1 | 1.1 | GO:0005124 | scavenger receptor binding(GO:0005124) |
| 0.1 | 0.7 | GO:0004174 | electron-transferring-flavoprotein dehydrogenase activity(GO:0004174) oxidoreductase activity, acting on the CH-NH group of donors, quinone or similar compound as acceptor(GO:0016649) |
| 0.1 | 1.0 | GO:0008889 | glycerophosphodiester phosphodiesterase activity(GO:0008889) |
| 0.1 | 1.2 | GO:0008526 | phosphatidylinositol transporter activity(GO:0008526) |
| 0.1 | 0.3 | GO:0005169 | neurotrophin TRKB receptor binding(GO:0005169) |
| 0.1 | 1.5 | GO:0038191 | neuropilin binding(GO:0038191) |
| 0.1 | 2.1 | GO:0050811 | GABA receptor binding(GO:0050811) |
| 0.1 | 1.5 | GO:0032036 | myosin heavy chain binding(GO:0032036) |
| 0.1 | 0.6 | GO:0004311 | farnesyltranstransferase activity(GO:0004311) |
| 0.1 | 1.4 | GO:0070006 | metalloaminopeptidase activity(GO:0070006) |
| 0.1 | 0.7 | GO:1990825 | sequence-specific mRNA binding(GO:1990825) |
| 0.1 | 1.0 | GO:0008239 | dipeptidyl-peptidase activity(GO:0008239) |
| 0.0 | 0.9 | GO:0046933 | proton-transporting ATP synthase activity, rotational mechanism(GO:0046933) |
| 0.0 | 0.1 | GO:0031755 | endothelial differentiation G-protein coupled receptor binding(GO:0031753) Edg-2 lysophosphatidic acid receptor binding(GO:0031755) |
| 0.0 | 0.3 | GO:0034431 | endopolyphosphatase activity(GO:0000298) diphosphoinositol-polyphosphate diphosphatase activity(GO:0008486) bis(5'-adenosyl)-hexaphosphatase activity(GO:0034431) bis(5'-adenosyl)-pentaphosphatase activity(GO:0034432) |
| 0.0 | 0.3 | GO:0070404 | NADH binding(GO:0070404) |
| 0.0 | 3.0 | GO:0001102 | RNA polymerase II activating transcription factor binding(GO:0001102) |
| 0.0 | 0.7 | GO:0030955 | potassium ion binding(GO:0030955) |
| 0.0 | 1.3 | GO:0003899 | DNA-directed RNA polymerase activity(GO:0003899) |
| 0.0 | 0.2 | GO:0071532 | ankyrin repeat binding(GO:0071532) |
| 0.0 | 1.0 | GO:0005164 | tumor necrosis factor receptor binding(GO:0005164) |
| 0.0 | 0.2 | GO:0097027 | ubiquitin-protein transferase activator activity(GO:0097027) |
| 0.0 | 0.5 | GO:0030247 | pattern binding(GO:0001871) polysaccharide binding(GO:0030247) |
| 0.0 | 0.6 | GO:0004198 | calcium-dependent cysteine-type endopeptidase activity(GO:0004198) |
| 0.0 | 1.0 | GO:0005154 | epidermal growth factor receptor binding(GO:0005154) |
| 0.0 | 0.4 | GO:0002162 | dystroglycan binding(GO:0002162) |
| 0.0 | 0.0 | GO:0004958 | prostaglandin F receptor activity(GO:0004958) |
| 0.0 | 1.7 | GO:0004843 | thiol-dependent ubiquitin-specific protease activity(GO:0004843) |
| 0.0 | 0.4 | GO:0015002 | cytochrome-c oxidase activity(GO:0004129) heme-copper terminal oxidase activity(GO:0015002) oxidoreductase activity, acting on a heme group of donors, oxygen as acceptor(GO:0016676) |
| 0.0 | 0.6 | GO:0030506 | ankyrin binding(GO:0030506) |
| 0.0 | 0.4 | GO:0001530 | lipopolysaccharide binding(GO:0001530) |
| 0.0 | 0.1 | GO:0017151 | DEAD/H-box RNA helicase binding(GO:0017151) |
| 0.0 | 1.5 | GO:0003707 | steroid hormone receptor activity(GO:0003707) |
| 0.0 | 0.2 | GO:0016290 | palmitoyl-CoA hydrolase activity(GO:0016290) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.9 | PID VEGF VEGFR PATHWAY | VEGF and VEGFR signaling network |
| 0.1 | 3.0 | PID ALK1 PATHWAY | ALK1 signaling events |
| 0.1 | 3.2 | ST GAQ PATHWAY | G alpha q Pathway |
| 0.1 | 4.1 | PID RHOA PATHWAY | RhoA signaling pathway |
| 0.1 | 2.4 | PID HIF2PATHWAY | HIF-2-alpha transcription factor network |
| 0.0 | 2.0 | PID HNF3A PATHWAY | FOXA1 transcription factor network |
| 0.0 | 1.0 | PID RAS PATHWAY | Regulation of Ras family activation |
| 0.0 | 0.9 | PID LKB1 PATHWAY | LKB1 signaling events |
| 0.0 | 1.4 | PID CMYB PATHWAY | C-MYB transcription factor network |
| 0.0 | 0.6 | SIG REGULATION OF THE ACTIN CYTOSKELETON BY RHO GTPASES | Genes related to regulation of the actin cytoskeleton |
| 0.0 | 2.0 | NABA ECM AFFILIATED | Genes encoding proteins affiliated structurally or functionally to extracellular matrix proteins |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 2.2 | REACTOME PASSIVE TRANSPORT BY AQUAPORINS | Genes involved in Passive Transport by Aquaporins |
| 0.1 | 0.9 | REACTOME VEGF LIGAND RECEPTOR INTERACTIONS | Genes involved in VEGF ligand-receptor interactions |
| 0.1 | 1.9 | REACTOME NUCLEOTIDE LIKE PURINERGIC RECEPTORS | Genes involved in Nucleotide-like (purinergic) receptors |
| 0.1 | 1.3 | REACTOME VIRAL MESSENGER RNA SYNTHESIS | Genes involved in Viral Messenger RNA Synthesis |
| 0.1 | 4.5 | REACTOME RESPIRATORY ELECTRON TRANSPORT | Genes involved in Respiratory electron transport |
| 0.1 | 3.8 | REACTOME STRIATED MUSCLE CONTRACTION | Genes involved in Striated Muscle Contraction |
| 0.0 | 0.9 | REACTOME REGULATION OF AMPK ACTIVITY VIA LKB1 | Genes involved in Regulation of AMPK activity via LKB1 |
| 0.0 | 0.5 | REACTOME REGULATION OF PYRUVATE DEHYDROGENASE PDH COMPLEX | Genes involved in Regulation of pyruvate dehydrogenase (PDH) complex |
| 0.0 | 0.5 | REACTOME SLBP DEPENDENT PROCESSING OF REPLICATION DEPENDENT HISTONE PRE MRNAS | Genes involved in SLBP Dependent Processing of Replication-Dependent Histone Pre-mRNAs |
| 0.0 | 0.4 | REACTOME NEF MEDIATED DOWNREGULATION OF MHC CLASS I COMPLEX CELL SURFACE EXPRESSION | Genes involved in Nef mediated downregulation of MHC class I complex cell surface expression |
| 0.0 | 1.1 | REACTOME SMAD2 SMAD3 SMAD4 HETEROTRIMER REGULATES TRANSCRIPTION | Genes involved in SMAD2/SMAD3:SMAD4 heterotrimer regulates transcription |
| 0.0 | 1.1 | REACTOME ION TRANSPORT BY P TYPE ATPASES | Genes involved in Ion transport by P-type ATPases |
| 0.0 | 0.2 | REACTOME NFKB IS ACTIVATED AND SIGNALS SURVIVAL | Genes involved in NF-kB is activated and signals survival |
| 0.0 | 0.5 | REACTOME YAP1 AND WWTR1 TAZ STIMULATED GENE EXPRESSION | Genes involved in YAP1- and WWTR1 (TAZ)-stimulated gene expression |
| 0.0 | 0.6 | REACTOME CASPASE MEDIATED CLEAVAGE OF CYTOSKELETAL PROTEINS | Genes involved in Caspase-mediated cleavage of cytoskeletal proteins |
| 0.0 | 1.0 | REACTOME FORMATION OF THE TERNARY COMPLEX AND SUBSEQUENTLY THE 43S COMPLEX | Genes involved in Formation of the ternary complex, and subsequently, the 43S complex |
| 0.0 | 0.7 | REACTOME APOPTOTIC CLEAVAGE OF CELLULAR PROTEINS | Genes involved in Apoptotic cleavage of cellular proteins |
| 0.0 | 0.7 | REACTOME CLEAVAGE OF GROWING TRANSCRIPT IN THE TERMINATION REGION | Genes involved in Cleavage of Growing Transcript in the Termination Region |
| 0.0 | 1.5 | REACTOME NUCLEAR RECEPTOR TRANSCRIPTION PATHWAY | Genes involved in Nuclear Receptor transcription pathway |
| 0.0 | 0.2 | REACTOME ACTIVATION OF RAC | Genes involved in Activation of Rac |
| 0.0 | 0.2 | REACTOME ACYL CHAIN REMODELLING OF PS | Genes involved in Acyl chain remodelling of PS |