Project
PRJNA375882: Comprehensive Mouse Transcriptomic BodyMap
Navigation
Downloads
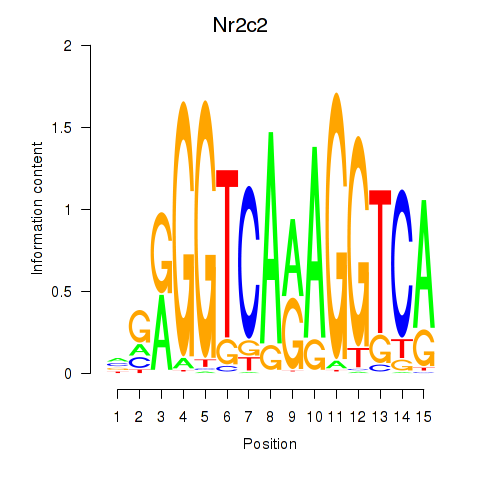
Results for Nr2c2
Z-value: 1.19
Transcription factors associated with Nr2c2
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Nr2c2
|
ENSMUSG00000005893.15 | Nr2c2 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Nr2c2 | mm39_v1_chr6_+_92069376_92069414 | -0.26 | 2.5e-02 | Click! |
Activity profile of Nr2c2 motif
Sorted Z-values of Nr2c2 motif
Network of associatons between targets according to the STRING database.
First level regulatory network of Nr2c2
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr6_-_42350188 | 9.82 |
ENSMUST00000073387.5
ENSMUST00000204357.2 |
Epha1
|
Eph receptor A1 |
| chr7_+_26821266 | 8.44 |
ENSMUST00000206552.2
|
Cyp2f2
|
cytochrome P450, family 2, subfamily f, polypeptide 2 |
| chr3_+_138121245 | 7.04 |
ENSMUST00000161312.8
ENSMUST00000013458.9 |
Adh4
|
alcohol dehydrogenase 4 (class II), pi polypeptide |
| chr16_-_38115172 | 6.44 |
ENSMUST00000023504.5
|
Nr1i2
|
nuclear receptor subfamily 1, group I, member 2 |
| chr13_+_91889626 | 5.77 |
ENSMUST00000022120.5
|
Acot12
|
acyl-CoA thioesterase 12 |
| chr7_+_43910845 | 5.70 |
ENSMUST00000124863.4
|
Gm15517
|
predicted gene 15517 |
| chr1_+_171052623 | 5.68 |
ENSMUST00000111321.8
ENSMUST00000005824.12 ENSMUST00000111320.8 ENSMUST00000111319.2 |
Apoa2
|
apolipoprotein A-II |
| chr17_-_84154173 | 5.52 |
ENSMUST00000000687.9
|
Haao
|
3-hydroxyanthranilate 3,4-dioxygenase |
| chr10_-_75658355 | 5.43 |
ENSMUST00000160211.2
|
Gstt4
|
glutathione S-transferase, theta 4 |
| chr7_-_105249308 | 5.26 |
ENSMUST00000210531.2
ENSMUST00000033185.10 |
Hpx
|
hemopexin |
| chr10_-_89342493 | 5.23 |
ENSMUST00000058126.15
ENSMUST00000105296.9 |
Nr1h4
|
nuclear receptor subfamily 1, group H, member 4 |
| chr17_-_34218301 | 5.12 |
ENSMUST00000235463.2
|
H2-K1
|
histocompatibility 2, K1, K region |
| chr11_-_89893707 | 5.11 |
ENSMUST00000020864.9
|
Pctp
|
phosphatidylcholine transfer protein |
| chr1_-_119576347 | 4.98 |
ENSMUST00000027632.14
ENSMUST00000187194.2 |
Epb41l5
|
erythrocyte membrane protein band 4.1 like 5 |
| chr10_+_128626772 | 4.93 |
ENSMUST00000219404.2
ENSMUST00000026411.8 |
Mmp19
|
matrix metallopeptidase 19 |
| chr7_-_19411866 | 4.80 |
ENSMUST00000142352.9
|
Apoc2
|
apolipoprotein C-II |
| chr16_+_4825216 | 4.78 |
ENSMUST00000185147.8
|
Smim22
|
small integral membrane protein 22 |
| chr8_+_72141858 | 4.74 |
ENSMUST00000034261.8
|
Insl3
|
insulin-like 3 |
| chr9_-_22046970 | 4.66 |
ENSMUST00000165735.9
|
Acp5
|
acid phosphatase 5, tartrate resistant |
| chr10_+_76411474 | 4.28 |
ENSMUST00000001183.8
|
Ftcd
|
formiminotransferase cyclodeaminase |
| chr5_-_139799953 | 4.26 |
ENSMUST00000044002.10
|
Tmem184a
|
transmembrane protein 184a |
| chr12_-_103796632 | 4.22 |
ENSMUST00000164454.3
|
Serpina1b
|
serine (or cysteine) preptidase inhibitor, clade A, member 1B |
| chr17_-_35351026 | 4.22 |
ENSMUST00000025249.7
|
Apom
|
apolipoprotein M |
| chr5_+_36641922 | 4.19 |
ENSMUST00000060100.3
|
Ccdc96
|
coiled-coil domain containing 96 |
| chr11_+_98337655 | 4.17 |
ENSMUST00000019456.5
|
Grb7
|
growth factor receptor bound protein 7 |
| chr19_+_38384428 | 4.16 |
ENSMUST00000054098.4
|
Slc35g1
|
solute carrier family 35, member G1 |
| chr2_-_69172944 | 4.08 |
ENSMUST00000102709.8
ENSMUST00000102710.10 ENSMUST00000180142.2 |
Abcb11
|
ATP-binding cassette, sub-family B (MDR/TAP), member 11 |
| chr2_-_164197987 | 4.02 |
ENSMUST00000165980.2
|
Slpi
|
secretory leukocyte peptidase inhibitor |
| chr1_+_191553556 | 3.98 |
ENSMUST00000027931.8
|
Nek2
|
NIMA (never in mitosis gene a)-related expressed kinase 2 |
| chr11_-_77784922 | 3.89 |
ENSMUST00000017597.5
|
Pipox
|
pipecolic acid oxidase |
| chr2_+_90613574 | 3.88 |
ENSMUST00000037206.11
|
Agbl2
|
ATP/GTP binding protein-like 2 |
| chr8_+_117822593 | 3.87 |
ENSMUST00000034308.16
ENSMUST00000176860.2 |
Bco1
|
beta-carotene oxygenase 1 |
| chr19_-_9978987 | 3.87 |
ENSMUST00000117346.2
|
Best1
|
bestrophin 1 |
| chr4_-_60457902 | 3.84 |
ENSMUST00000084548.11
ENSMUST00000103012.10 ENSMUST00000107499.4 |
Mup1
|
major urinary protein 1 |
| chr9_+_40059408 | 3.81 |
ENSMUST00000046333.9
ENSMUST00000238613.2 |
Tmem225
|
transmembrane protein 225 |
| chr10_+_81229443 | 3.80 |
ENSMUST00000118206.2
|
Smim24
|
small integral membrane protein 24 |
| chr4_-_62005498 | 3.78 |
ENSMUST00000107488.4
ENSMUST00000107472.8 ENSMUST00000084531.11 |
Mup3
|
major urinary protein 3 |
| chr7_+_43086554 | 3.78 |
ENSMUST00000206741.2
|
Nkg7
|
natural killer cell group 7 sequence |
| chr3_-_58433313 | 3.77 |
ENSMUST00000029385.9
|
Serp1
|
stress-associated endoplasmic reticulum protein 1 |
| chr11_-_106163753 | 3.75 |
ENSMUST00000021052.16
|
Smarcd2
|
SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily d, member 2 |
| chr8_-_93924426 | 3.71 |
ENSMUST00000034172.8
|
Ces1d
|
carboxylesterase 1D |
| chr16_+_48692976 | 3.70 |
ENSMUST00000065666.6
|
Retnlg
|
resistin like gamma |
| chr16_+_4825170 | 3.65 |
ENSMUST00000178155.9
|
Smim22
|
small integral membrane protein 22 |
| chr6_+_48818567 | 3.60 |
ENSMUST00000203639.3
|
Tmem176a
|
transmembrane protein 176A |
| chr4_-_61972348 | 3.58 |
ENSMUST00000074018.4
|
Mup20
|
major urinary protein 20 |
| chr7_-_14172434 | 3.58 |
ENSMUST00000210396.2
ENSMUST00000168252.9 |
Sult2a8
|
sulfotransferase family 2A, dehydroepiandrosterone (DHEA)-preferring, member 8 |
| chr1_-_64161415 | 3.56 |
ENSMUST00000135075.2
|
Klf7
|
Kruppel-like factor 7 (ubiquitous) |
| chr9_-_50657800 | 3.55 |
ENSMUST00000239417.2
ENSMUST00000034564.4 |
2310030G06Rik
|
RIKEN cDNA 2310030G06 gene |
| chr19_+_8906916 | 3.54 |
ENSMUST00000096241.6
|
Eml3
|
echinoderm microtubule associated protein like 3 |
| chr16_+_4825146 | 3.54 |
ENSMUST00000184439.8
|
Smim22
|
small integral membrane protein 22 |
| chr3_+_89043440 | 3.45 |
ENSMUST00000047111.13
|
Pklr
|
pyruvate kinase liver and red blood cell |
| chr15_-_10713621 | 3.43 |
ENSMUST00000090339.11
|
Rai14
|
retinoic acid induced 14 |
| chr7_+_44114815 | 3.42 |
ENSMUST00000035929.11
ENSMUST00000146128.8 |
Aspdh
|
aspartate dehydrogenase domain containing |
| chr6_+_108805594 | 3.39 |
ENSMUST00000089162.5
|
Edem1
|
ER degradation enhancer, mannosidase alpha-like 1 |
| chr2_+_126398048 | 3.39 |
ENSMUST00000141482.3
|
Slc27a2
|
solute carrier family 27 (fatty acid transporter), member 2 |
| chr16_-_4815645 | 3.38 |
ENSMUST00000229321.2
ENSMUST00000230362.2 ENSMUST00000170323.3 |
Septin12
|
septin 12 |
| chr3_+_135531834 | 3.33 |
ENSMUST00000029810.6
|
Slc39a8
|
solute carrier family 39 (metal ion transporter), member 8 |
| chr6_+_48818410 | 3.32 |
ENSMUST00000101426.11
ENSMUST00000168406.4 |
Tmem176a
|
transmembrane protein 176A |
| chr7_-_48531344 | 3.31 |
ENSMUST00000119223.2
|
E2f8
|
E2F transcription factor 8 |
| chr11_-_83483807 | 3.23 |
ENSMUST00000019071.4
|
Ccl6
|
chemokine (C-C motif) ligand 6 |
| chr11_-_75330302 | 3.22 |
ENSMUST00000043696.9
|
Serpinf2
|
serine (or cysteine) peptidase inhibitor, clade F, member 2 |
| chr2_+_19204076 | 3.19 |
ENSMUST00000114640.9
ENSMUST00000049255.7 |
Armc3
|
armadillo repeat containing 3 |
| chr11_-_75330415 | 3.18 |
ENSMUST00000128330.8
|
Serpinf2
|
serine (or cysteine) peptidase inhibitor, clade F, member 2 |
| chr7_+_121818692 | 3.17 |
ENSMUST00000033152.5
|
Chp2
|
calcineurin-like EF hand protein 2 |
| chr11_-_96932220 | 3.17 |
ENSMUST00000021251.7
|
Lrrc46
|
leucine rich repeat containing 46 |
| chr7_-_89914610 | 3.12 |
ENSMUST00000107221.9
ENSMUST00000107220.8 ENSMUST00000040413.2 |
Ccdc83
|
coiled-coil domain containing 83 |
| chr3_-_129126362 | 3.10 |
ENSMUST00000029658.14
|
Enpep
|
glutamyl aminopeptidase |
| chr14_-_66246652 | 3.10 |
ENSMUST00000059970.9
|
Gulo
|
gulonolactone (L-) oxidase |
| chr4_+_41569775 | 3.10 |
ENSMUST00000102963.10
|
Dnai1
|
dynein axonemal intermediate chain 1 |
| chr17_-_28299544 | 3.08 |
ENSMUST00000042692.13
ENSMUST00000114836.9 |
Tcp11
|
t-complex protein 11 |
| chr3_+_89043879 | 3.08 |
ENSMUST00000107482.10
ENSMUST00000127058.2 |
Pklr
|
pyruvate kinase liver and red blood cell |
| chr4_-_60697274 | 3.06 |
ENSMUST00000117932.2
|
Mup12
|
major urinary protein 12 |
| chr3_+_135531548 | 3.05 |
ENSMUST00000167390.8
|
Slc39a8
|
solute carrier family 39 (metal ion transporter), member 8 |
| chr2_-_164198427 | 3.03 |
ENSMUST00000109367.10
|
Slpi
|
secretory leukocyte peptidase inhibitor |
| chr2_+_172994841 | 3.02 |
ENSMUST00000029017.6
|
Pck1
|
phosphoenolpyruvate carboxykinase 1, cytosolic |
| chr1_+_93062962 | 3.00 |
ENSMUST00000027491.7
|
Agxt
|
alanine-glyoxylate aminotransferase |
| chr7_+_24310171 | 3.00 |
ENSMUST00000206422.2
|
Phldb3
|
pleckstrin homology like domain, family B, member 3 |
| chr3_-_89325594 | 3.00 |
ENSMUST00000029679.4
|
Cks1b
|
CDC28 protein kinase 1b |
| chr15_-_100976370 | 2.98 |
ENSMUST00000213610.2
|
Fignl2
|
fidgetin-like 2 |
| chr14_+_75373766 | 2.91 |
ENSMUST00000145303.8
|
Lcp1
|
lymphocyte cytosolic protein 1 |
| chr8_+_13076024 | 2.90 |
ENSMUST00000033820.4
|
F7
|
coagulation factor VII |
| chr19_+_36532061 | 2.89 |
ENSMUST00000169036.9
ENSMUST00000047247.12 |
Hectd2
|
HECT domain E3 ubiquitin protein ligase 2 |
| chr7_+_43086432 | 2.89 |
ENSMUST00000070518.4
|
Nkg7
|
natural killer cell group 7 sequence |
| chr17_-_28299569 | 2.88 |
ENSMUST00000129046.9
ENSMUST00000043925.16 |
Tcp11
|
t-complex protein 11 |
| chr12_-_57244096 | 2.87 |
ENSMUST00000044634.12
|
Slc25a21
|
solute carrier family 25 (mitochondrial oxodicarboxylate carrier), member 21 |
| chr2_-_164435508 | 2.85 |
ENSMUST00000103100.2
|
Eppin
|
epididymal peptidase inhibitor |
| chr4_+_100336003 | 2.82 |
ENSMUST00000133493.9
ENSMUST00000092730.5 |
Ube2u
|
ubiquitin-conjugating enzyme E2U (putative) |
| chr17_-_84154196 | 2.81 |
ENSMUST00000234214.2
|
Haao
|
3-hydroxyanthranilate 3,4-dioxygenase |
| chr19_-_38113249 | 2.81 |
ENSMUST00000112335.4
|
Rbp4
|
retinol binding protein 4, plasma |
| chr7_+_140658101 | 2.79 |
ENSMUST00000106039.9
|
Pkp3
|
plakophilin 3 |
| chr19_-_38113056 | 2.78 |
ENSMUST00000236283.2
|
Rbp4
|
retinol binding protein 4, plasma |
| chr4_-_92035446 | 2.77 |
ENSMUST00000107108.8
ENSMUST00000143542.2 |
Izumo3
|
IZUMO family member 3 |
| chr3_+_135531409 | 2.76 |
ENSMUST00000180196.8
|
Slc39a8
|
solute carrier family 39 (metal ion transporter), member 8 |
| chr10_+_81228877 | 2.76 |
ENSMUST00000105322.9
|
Smim24
|
small integral membrane protein 24 |
| chr11_-_118233326 | 2.74 |
ENSMUST00000103024.4
|
Cep295nl
|
CEP295 N-terminal like |
| chr3_+_55047453 | 2.74 |
ENSMUST00000118963.9
ENSMUST00000061099.14 ENSMUST00000153009.8 |
Ccdc169
|
coiled-coil domain containing 169 |
| chr11_-_51153767 | 2.74 |
ENSMUST00000065950.10
|
BC049762
|
cDNA sequence BC049762 |
| chr11_-_51153870 | 2.74 |
ENSMUST00000054226.3
|
BC049762
|
cDNA sequence BC049762 |
| chr3_+_37117779 | 2.73 |
ENSMUST00000029274.14
|
Adad1
|
adenosine deaminase domain containing 1 (testis specific) |
| chr19_-_39801188 | 2.71 |
ENSMUST00000162507.2
ENSMUST00000160476.9 ENSMUST00000239028.2 |
Cyp2c40
|
cytochrome P450, family 2, subfamily c, polypeptide 40 |
| chr19_-_4548602 | 2.71 |
ENSMUST00000048482.8
|
2010003K11Rik
|
RIKEN cDNA 2010003K11 gene |
| chr13_-_76453191 | 2.70 |
ENSMUST00000208418.2
|
Fam81b
|
family with sequence similarity 81, member B |
| chr4_+_118286898 | 2.70 |
ENSMUST00000067896.4
|
Elovl1
|
elongation of very long chain fatty acids (FEN1/Elo2, SUR4/Elo3, yeast)-like 1 |
| chr1_+_91072778 | 2.70 |
ENSMUST00000188818.2
ENSMUST00000094698.2 |
Rbm44
|
RNA binding motif protein 44 |
| chr15_-_75881289 | 2.69 |
ENSMUST00000170153.2
|
Fam83h
|
family with sequence similarity 83, member H |
| chr10_-_87982732 | 2.68 |
ENSMUST00000164121.8
ENSMUST00000164803.2 ENSMUST00000168163.8 ENSMUST00000048518.16 |
Parpbp
|
PARP1 binding protein |
| chr1_-_119576632 | 2.67 |
ENSMUST00000163147.8
ENSMUST00000052404.13 ENSMUST00000191046.7 |
Epb41l5
|
erythrocyte membrane protein band 4.1 like 5 |
| chr7_+_44114857 | 2.66 |
ENSMUST00000135624.2
|
Aspdh
|
aspartate dehydrogenase domain containing |
| chr11_-_97242842 | 2.64 |
ENSMUST00000093942.5
|
Gpr179
|
G protein-coupled receptor 179 |
| chr1_-_171854818 | 2.64 |
ENSMUST00000138714.2
ENSMUST00000027837.13 ENSMUST00000111264.8 |
Vangl2
|
VANGL planar cell polarity 2 |
| chr17_-_57137898 | 2.64 |
ENSMUST00000233000.2
ENSMUST00000002444.15 ENSMUST00000086801.7 |
Rfx2
|
regulatory factor X, 2 (influences HLA class II expression) |
| chr2_+_70339832 | 2.63 |
ENSMUST00000153121.2
|
Erich2
|
glutamate rich 2 |
| chr15_+_75881712 | 2.62 |
ENSMUST00000187868.3
|
Iqank1
|
IQ motif and ankyrin repeat containing 1 |
| chr5_-_31854942 | 2.61 |
ENSMUST00000031018.10
|
Rbks
|
ribokinase |
| chr6_-_5256282 | 2.59 |
ENSMUST00000031773.9
|
Pon3
|
paraoxonase 3 |
| chr8_+_66964401 | 2.59 |
ENSMUST00000002025.5
ENSMUST00000183187.2 |
Tktl2
|
transketolase-like 2 |
| chr10_-_75609185 | 2.56 |
ENSMUST00000001716.8
|
Ddt
|
D-dopachrome tautomerase |
| chr3_+_37117676 | 2.54 |
ENSMUST00000144629.8
|
Adad1
|
adenosine deaminase domain containing 1 (testis specific) |
| chr17_+_26332260 | 2.53 |
ENSMUST00000235821.2
ENSMUST00000025010.14 ENSMUST00000237058.2 |
Pgap6
|
post-glycosylphosphatidylinositol attachment to proteins 6 |
| chr16_-_17745999 | 2.52 |
ENSMUST00000003622.16
|
Slc25a1
|
solute carrier family 25 (mitochondrial carrier, citrate transporter), member 1 |
| chr17_+_35643818 | 2.51 |
ENSMUST00000174699.8
|
H2-Q6
|
histocompatibility 2, Q region locus 6 |
| chr1_-_162641495 | 2.51 |
ENSMUST00000144916.8
ENSMUST00000140274.2 |
Fmo4
|
flavin containing monooxygenase 4 |
| chr2_+_173364379 | 2.51 |
ENSMUST00000029023.4
|
1700021F07Rik
|
RIKEN cDNA 1700021F07 gene |
| chr2_+_158148413 | 2.49 |
ENSMUST00000109491.8
ENSMUST00000016168.9 |
Lbp
|
lipopolysaccharide binding protein |
| chr10_+_77417608 | 2.48 |
ENSMUST00000162429.8
|
Pttg1ip
|
pituitary tumor-transforming 1 interacting protein |
| chr10_+_77417554 | 2.48 |
ENSMUST00000009435.12
|
Pttg1ip
|
pituitary tumor-transforming 1 interacting protein |
| chr3_+_89325750 | 2.46 |
ENSMUST00000039110.12
ENSMUST00000125036.8 ENSMUST00000191485.7 ENSMUST00000154791.8 |
Shc1
|
src homology 2 domain-containing transforming protein C1 |
| chr18_+_50411431 | 2.43 |
ENSMUST00000039121.4
ENSMUST00000238078.2 |
Fam170a
|
family with sequence similarity 170, member A |
| chr17_+_48571298 | 2.43 |
ENSMUST00000059873.14
ENSMUST00000154335.8 ENSMUST00000136272.8 ENSMUST00000125426.8 ENSMUST00000153420.2 |
Treml4
|
triggering receptor expressed on myeloid cells-like 4 |
| chr3_-_67422821 | 2.42 |
ENSMUST00000054825.5
|
Rarres1
|
retinoic acid receptor responder (tazarotene induced) 1 |
| chr19_+_5927821 | 2.42 |
ENSMUST00000145200.8
ENSMUST00000025732.14 ENSMUST00000125114.8 ENSMUST00000155697.8 |
Slc25a45
|
solute carrier family 25, member 45 |
| chr3_+_18002574 | 2.41 |
ENSMUST00000029080.5
|
Cypt12
|
cysteine-rich perinuclear theca 12 |
| chr14_+_69409251 | 2.40 |
ENSMUST00000062437.10
|
Nkx2-6
|
NK2 homeobox 6 |
| chr15_-_82108531 | 2.38 |
ENSMUST00000109535.3
ENSMUST00000089161.10 |
Tnfrsf13c
|
tumor necrosis factor receptor superfamily, member 13c |
| chr18_+_74912268 | 2.38 |
ENSMUST00000041053.11
|
Acaa2
|
acetyl-Coenzyme A acyltransferase 2 (mitochondrial 3-oxoacyl-Coenzyme A thiolase) |
| chr19_+_46120327 | 2.37 |
ENSMUST00000043739.6
ENSMUST00000237098.2 |
Elovl3
|
elongation of very long chain fatty acids (FEN1/Elo2, SUR4/Elo3, yeast)-like 3 |
| chr19_+_4806544 | 2.34 |
ENSMUST00000182821.8
ENSMUST00000036744.8 |
Rbm4b
|
RNA binding motif protein 4B |
| chr16_+_22769822 | 2.33 |
ENSMUST00000023590.9
|
Hrg
|
histidine-rich glycoprotein |
| chr15_-_100579450 | 2.32 |
ENSMUST00000230740.2
|
Cela1
|
chymotrypsin-like elastase family, member 1 |
| chr4_+_141473983 | 2.32 |
ENSMUST00000038161.5
|
Agmat
|
agmatine ureohydrolase (agmatinase) |
| chr6_+_83031502 | 2.31 |
ENSMUST00000092618.9
|
Aup1
|
ancient ubiquitous protein 1 |
| chr19_-_39875192 | 2.31 |
ENSMUST00000168838.3
|
Cyp2c69
|
cytochrome P450, family 2, subfamily c, polypeptide 69 |
| chr6_-_48818302 | 2.30 |
ENSMUST00000203355.3
ENSMUST00000166247.8 |
Tmem176b
|
transmembrane protein 176B |
| chr5_+_31454787 | 2.30 |
ENSMUST00000201166.4
ENSMUST00000072228.9 |
Gckr
|
glucokinase regulatory protein |
| chr19_+_5927876 | 2.29 |
ENSMUST00000235340.2
|
Slc25a45
|
solute carrier family 25, member 45 |
| chr2_-_173420052 | 2.29 |
ENSMUST00000239457.2
|
Ankrd60
|
ankyrin repeat domain 60 |
| chr3_+_121761471 | 2.27 |
ENSMUST00000196479.5
ENSMUST00000197155.5 |
Arhgap29
|
Rho GTPase activating protein 29 |
| chr13_-_99121070 | 2.26 |
ENSMUST00000054425.7
|
H2bl1
|
H2B.L histone variant 1 |
| chr6_-_48818430 | 2.26 |
ENSMUST00000205147.3
|
Tmem176b
|
transmembrane protein 176B |
| chr13_-_100688949 | 2.25 |
ENSMUST00000159515.2
ENSMUST00000160859.8 ENSMUST00000069756.11 |
Ocln
|
occludin |
| chr2_+_152950388 | 2.24 |
ENSMUST00000189688.2
ENSMUST00000109799.8 ENSMUST00000003370.14 |
Hck
|
hemopoietic cell kinase |
| chr1_+_74752710 | 2.23 |
ENSMUST00000027356.7
|
Cyp27a1
|
cytochrome P450, family 27, subfamily a, polypeptide 1 |
| chr2_-_27136826 | 2.22 |
ENSMUST00000149733.8
|
Sardh
|
sarcosine dehydrogenase |
| chr14_+_41765952 | 2.22 |
ENSMUST00000168972.2
ENSMUST00000064162.11 |
1700024B05Rik
|
RIKEN cDNA 1700024B05 gene |
| chr14_+_14210932 | 2.22 |
ENSMUST00000022271.14
|
Acox2
|
acyl-Coenzyme A oxidase 2, branched chain |
| chr8_+_88290469 | 2.21 |
ENSMUST00000093342.6
|
4933402J07Rik
|
RIKEN cDNA 4933402J07 gene |
| chr5_+_145051025 | 2.20 |
ENSMUST00000085679.13
|
Arpc1b
|
actin related protein 2/3 complex, subunit 1B |
| chr14_-_67953035 | 2.19 |
ENSMUST00000163100.8
ENSMUST00000132705.8 ENSMUST00000124045.3 |
Cdca2
|
cell division cycle associated 2 |
| chr15_-_82648376 | 2.19 |
ENSMUST00000055721.6
|
Cyp2d40
|
cytochrome P450, family 2, subfamily d, polypeptide 40 |
| chr6_+_29694181 | 2.18 |
ENSMUST00000046750.14
ENSMUST00000115250.4 |
Tspan33
|
tetraspanin 33 |
| chr6_+_124807176 | 2.17 |
ENSMUST00000131847.8
ENSMUST00000151674.3 |
Cdca3
|
cell division cycle associated 3 |
| chr6_+_124639990 | 2.17 |
ENSMUST00000004381.14
|
Lpcat3
|
lysophosphatidylcholine acyltransferase 3 |
| chr2_+_58457370 | 2.17 |
ENSMUST00000071543.12
|
Upp2
|
uridine phosphorylase 2 |
| chr11_+_61575245 | 2.16 |
ENSMUST00000093019.6
|
Fam83g
|
family with sequence similarity 83, member G |
| chr11_-_106378622 | 2.15 |
ENSMUST00000001059.9
ENSMUST00000106799.2 ENSMUST00000106800.2 |
Ern1
|
endoplasmic reticulum (ER) to nucleus signalling 1 |
| chr17_-_74257164 | 2.15 |
ENSMUST00000024866.6
|
Xdh
|
xanthine dehydrogenase |
| chr5_-_36641456 | 2.14 |
ENSMUST00000119916.2
ENSMUST00000031097.8 |
Tada2b
|
transcriptional adaptor 2B |
| chr6_-_5256225 | 2.13 |
ENSMUST00000125686.8
|
Pon3
|
paraoxonase 3 |
| chr2_+_118428690 | 2.13 |
ENSMUST00000038341.8
|
Bub1b
|
BUB1B, mitotic checkpoint serine/threonine kinase |
| chr17_+_35643853 | 2.11 |
ENSMUST00000113879.4
|
H2-Q6
|
histocompatibility 2, Q region locus 6 |
| chr1_-_44140820 | 2.09 |
ENSMUST00000152239.8
|
Tex30
|
testis expressed 30 |
| chr11_+_68477812 | 2.08 |
ENSMUST00000154294.8
ENSMUST00000063006.12 |
Ccdc42
|
coiled-coil domain containing 42 |
| chr10_+_93983844 | 2.08 |
ENSMUST00000105290.9
|
Nr2c1
|
nuclear receptor subfamily 2, group C, member 1 |
| chr16_+_22769844 | 2.08 |
ENSMUST00000232422.2
|
Hrg
|
histidine-rich glycoprotein |
| chr12_-_28602302 | 2.07 |
ENSMUST00000020963.8
ENSMUST00000221349.2 |
Dcdc2c
|
doublecortin domain containing 2C |
| chr17_+_56610321 | 2.06 |
ENSMUST00000001258.15
|
Uhrf1
|
ubiquitin-like, containing PHD and RING finger domains, 1 |
| chr2_-_173420116 | 2.05 |
ENSMUST00000119453.9
|
Ankrd60
|
ankyrin repeat domain 60 |
| chr5_-_120915693 | 2.05 |
ENSMUST00000044833.9
|
Oas3
|
2'-5' oligoadenylate synthetase 3 |
| chr19_+_9979033 | 2.05 |
ENSMUST00000121418.8
|
Rab3il1
|
RAB3A interacting protein (rabin3)-like 1 |
| chr18_+_70605691 | 2.04 |
ENSMUST00000164223.8
|
Stard6
|
StAR-related lipid transfer (START) domain containing 6 |
| chr9_-_110818679 | 2.01 |
ENSMUST00000084922.6
ENSMUST00000199891.2 |
Rtp3
|
receptor transporter protein 3 |
| chr11_+_84070593 | 2.01 |
ENSMUST00000137500.9
ENSMUST00000130012.9 |
Acaca
|
acetyl-Coenzyme A carboxylase alpha |
| chr17_+_56610396 | 2.01 |
ENSMUST00000113038.8
|
Uhrf1
|
ubiquitin-like, containing PHD and RING finger domains, 1 |
| chr9_+_108216233 | 2.00 |
ENSMUST00000082429.8
|
Gpx1
|
glutathione peroxidase 1 |
| chr11_+_46295547 | 1.99 |
ENSMUST00000063166.6
|
Fam71b
|
family with sequence similarity 71, member B |
| chr7_+_103867997 | 1.99 |
ENSMUST00000098180.10
|
Trim6
|
tripartite motif-containing 6 |
| chr3_-_88241735 | 1.98 |
ENSMUST00000107552.2
|
Tmem79
|
transmembrane protein 79 |
| chr4_+_148686985 | 1.98 |
ENSMUST00000105701.9
ENSMUST00000052060.7 |
Masp2
|
mannan-binding lectin serine peptidase 2 |
| chr18_+_70605722 | 1.97 |
ENSMUST00000174118.8
|
Stard6
|
StAR-related lipid transfer (START) domain containing 6 |
| chr5_+_145063568 | 1.97 |
ENSMUST00000138922.2
|
Arpc1b
|
actin related protein 2/3 complex, subunit 1B |
| chr14_-_8123295 | 1.97 |
ENSMUST00000225775.2
ENSMUST00000224877.2 ENSMUST00000090591.4 |
Il3ra
|
interleukin 3 receptor, alpha chain |
| chr17_+_35268942 | 1.96 |
ENSMUST00000007257.10
|
Clic1
|
chloride intracellular channel 1 |
| chr10_-_62363192 | 1.95 |
ENSMUST00000160643.8
|
Srgn
|
serglycin |
| chr7_-_43906629 | 1.95 |
ENSMUST00000012921.9
|
Acp4
|
acid phosphatase 4 |
| chr16_+_43960183 | 1.94 |
ENSMUST00000159514.8
ENSMUST00000161326.8 ENSMUST00000063520.15 ENSMUST00000063542.8 |
Naa50
|
N(alpha)-acetyltransferase 50, NatE catalytic subunit |
| chr13_-_100689212 | 1.93 |
ENSMUST00000022140.12
|
Ocln
|
occludin |
| chr2_+_24226857 | 1.93 |
ENSMUST00000114487.9
ENSMUST00000142093.7 |
Il1rn
|
interleukin 1 receptor antagonist |
| chr18_+_70605630 | 1.92 |
ENSMUST00000168249.9
|
Stard6
|
StAR-related lipid transfer (START) domain containing 6 |
| chr17_+_35284315 | 1.91 |
ENSMUST00000173207.8
|
Ly6g6c
|
lymphocyte antigen 6 complex, locus G6C |
| chr15_+_81900570 | 1.91 |
ENSMUST00000069530.13
ENSMUST00000168581.8 ENSMUST00000164779.2 |
Xrcc6
|
X-ray repair complementing defective repair in Chinese hamster cells 6 |
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.8 | 8.4 | GO:0018931 | naphthalene metabolic process(GO:0018931) naphthalene-containing compound metabolic process(GO:0090420) |
| 2.5 | 7.6 | GO:0007509 | mesoderm migration involved in gastrulation(GO:0007509) |
| 1.8 | 7.0 | GO:0006069 | ethanol oxidation(GO:0006069) |
| 1.6 | 8.1 | GO:0002485 | antigen processing and presentation of endogenous peptide antigen via MHC class I via ER pathway(GO:0002484) antigen processing and presentation of endogenous peptide antigen via MHC class I via ER pathway, TAP-dependent(GO:0002485) |
| 1.6 | 4.7 | GO:0019626 | short-chain fatty acid catabolic process(GO:0019626) |
| 1.5 | 10.5 | GO:0046618 | drug export(GO:0046618) |
| 1.5 | 6.0 | GO:1902490 | regulation of sperm capacitation(GO:1902490) |
| 1.5 | 4.4 | GO:0097037 | heme export(GO:0097037) |
| 1.4 | 5.7 | GO:0010901 | regulation of very-low-density lipoprotein particle remodeling(GO:0010901) diacylglycerol catabolic process(GO:0046340) |
| 1.4 | 4.2 | GO:0034443 | negative regulation of lipoprotein oxidation(GO:0034443) |
| 1.3 | 5.3 | GO:0015886 | heme transport(GO:0015886) |
| 1.3 | 5.2 | GO:0034971 | histone H3-R17 methylation(GO:0034971) bile acid signaling pathway(GO:0038183) |
| 1.3 | 3.9 | GO:0006553 | lysine metabolic process(GO:0006553) |
| 1.2 | 4.8 | GO:0060697 | positive regulation of phospholipid catabolic process(GO:0060697) |
| 1.2 | 4.7 | GO:0046226 | coumarin catabolic process(GO:0046226) |
| 1.2 | 3.5 | GO:2000612 | regulation of thyroid-stimulating hormone secretion(GO:2000612) |
| 1.1 | 3.4 | GO:1904154 | positive regulation of retrograde protein transport, ER to cytosol(GO:1904154) |
| 1.1 | 3.4 | GO:0097089 | methyl-branched fatty acid metabolic process(GO:0097089) |
| 1.1 | 5.6 | GO:0048807 | female genitalia morphogenesis(GO:0048807) |
| 1.1 | 3.3 | GO:0032877 | positive regulation of DNA endoreduplication(GO:0032877) |
| 1.0 | 6.1 | GO:0006742 | NADP catabolic process(GO:0006742) |
| 1.0 | 3.0 | GO:0042853 | glycine biosynthetic process, by transamination of glyoxylate(GO:0019265) L-alanine catabolic process(GO:0042853) oxalic acid secretion(GO:0046724) |
| 0.9 | 8.3 | GO:0046874 | quinolinate metabolic process(GO:0046874) |
| 0.9 | 3.7 | GO:0002408 | myeloid dendritic cell chemotaxis(GO:0002408) |
| 0.9 | 5.5 | GO:0018992 | germ-line sex determination(GO:0018992) |
| 0.9 | 3.6 | GO:0042335 | cuticle development(GO:0042335) |
| 0.9 | 3.6 | GO:0033382 | protein localization to secretory granule(GO:0033366) protein localization to mast cell secretory granule(GO:0033367) protease localization to mast cell secretory granule(GO:0033368) maintenance of protein location in mast cell secretory granule(GO:0033370) T cell secretory granule organization(GO:0033371) maintenance of protease location in mast cell secretory granule(GO:0033373) protein localization to T cell secretory granule(GO:0033374) protease localization to T cell secretory granule(GO:0033375) maintenance of protein location in T cell secretory granule(GO:0033377) maintenance of protease location in T cell secretory granule(GO:0033379) granzyme B localization to T cell secretory granule(GO:0033380) maintenance of granzyme B location in T cell secretory granule(GO:0033382) |
| 0.9 | 11.5 | GO:2001199 | negative regulation of dendritic cell differentiation(GO:2001199) |
| 0.9 | 2.6 | GO:0060448 | dichotomous subdivision of terminal units involved in lung branching(GO:0060448) |
| 0.9 | 4.3 | GO:0006548 | histidine catabolic process(GO:0006548) |
| 0.8 | 2.5 | GO:0015920 | lipopolysaccharide transport(GO:0015920) |
| 0.8 | 8.9 | GO:0015691 | cadmium ion transport(GO:0015691) cadmium ion transmembrane transport(GO:0070574) |
| 0.8 | 3.2 | GO:0051325 | interphase(GO:0051325) mitotic interphase(GO:0051329) |
| 0.8 | 6.4 | GO:0002034 | regulation of blood vessel size by renin-angiotensin(GO:0002034) renal control of peripheral vascular resistance involved in regulation of systemic arterial blood pressure(GO:0003072) |
| 0.8 | 4.0 | GO:0030997 | regulation of centriole-centriole cohesion(GO:0030997) |
| 0.8 | 3.2 | GO:0038156 | interleukin-3-mediated signaling pathway(GO:0038156) |
| 0.8 | 4.7 | GO:0032929 | negative regulation of superoxide anion generation(GO:0032929) |
| 0.8 | 3.0 | GO:0061402 | glycerol biosynthetic process(GO:0006114) alditol biosynthetic process(GO:0019401) positive regulation of transcription from RNA polymerase II promoter in response to acidic pH(GO:0061402) |
| 0.8 | 5.3 | GO:0006987 | activation of signaling protein activity involved in unfolded protein response(GO:0006987) |
| 0.7 | 2.2 | GO:1901053 | sarcosine metabolic process(GO:1901052) sarcosine catabolic process(GO:1901053) |
| 0.7 | 6.5 | GO:0042866 | pyruvate biosynthetic process(GO:0042866) |
| 0.7 | 2.9 | GO:0061228 | regulation of pronephros size(GO:0035565) mesonephros morphogenesis(GO:0061206) mesonephric nephron development(GO:0061215) mesonephric nephron morphogenesis(GO:0061228) mesenchymal stem cell maintenance involved in mesonephric nephron morphogenesis(GO:0061235) regulation of mesenchymal cell apoptotic process involved in mesonephric nephron morphogenesis(GO:0061295) negative regulation of mesenchymal cell apoptotic process involved in mesonephric nephron morphogenesis(GO:0061296) mesenchymal cell apoptotic process involved in mesonephric nephron morphogenesis(GO:1901146) |
| 0.7 | 2.1 | GO:0046108 | uridine catabolic process(GO:0006218) uridine metabolic process(GO:0046108) |
| 0.7 | 4.3 | GO:1990839 | response to endothelin(GO:1990839) |
| 0.7 | 3.6 | GO:0044268 | multicellular organismal protein metabolic process(GO:0044268) |
| 0.7 | 2.8 | GO:2000297 | negative regulation of synapse maturation(GO:2000297) |
| 0.7 | 3.3 | GO:0036151 | phosphatidylcholine acyl-chain remodeling(GO:0036151) |
| 0.7 | 2.0 | GO:0046901 | tetrahydrofolylpolyglutamate biosynthetic process(GO:0046901) |
| 0.7 | 2.6 | GO:0006014 | D-ribose metabolic process(GO:0006014) |
| 0.6 | 3.8 | GO:0033140 | negative regulation of peptidyl-serine phosphorylation of STAT protein(GO:0033140) |
| 0.6 | 3.1 | GO:0019853 | L-ascorbic acid biosynthetic process(GO:0019853) |
| 0.6 | 2.4 | GO:0021526 | medial motor column neuron differentiation(GO:0021526) |
| 0.6 | 2.4 | GO:0055014 | atrial cardiac muscle cell differentiation(GO:0055011) atrial cardiac muscle cell development(GO:0055014) |
| 0.6 | 3.6 | GO:0008355 | olfactory learning(GO:0008355) |
| 0.6 | 4.2 | GO:0060309 | elastin catabolic process(GO:0060309) |
| 0.6 | 2.4 | GO:1902109 | negative regulation of mitochondrial membrane permeability involved in apoptotic process(GO:1902109) |
| 0.6 | 4.2 | GO:1990034 | calcium ion export from cell(GO:1990034) |
| 0.6 | 4.1 | GO:0031296 | B cell costimulation(GO:0031296) |
| 0.6 | 3.5 | GO:0033540 | fatty acid beta-oxidation using acyl-CoA oxidase(GO:0033540) |
| 0.6 | 1.8 | GO:0032218 | riboflavin transport(GO:0032218) |
| 0.6 | 3.9 | GO:0030321 | transepithelial chloride transport(GO:0030321) |
| 0.5 | 6.6 | GO:0019368 | fatty acid elongation, saturated fatty acid(GO:0019367) fatty acid elongation, unsaturated fatty acid(GO:0019368) fatty acid elongation, monounsaturated fatty acid(GO:0034625) fatty acid elongation, polyunsaturated fatty acid(GO:0034626) |
| 0.5 | 2.7 | GO:0042412 | taurine biosynthetic process(GO:0042412) |
| 0.5 | 1.6 | GO:0046032 | ADP catabolic process(GO:0046032) |
| 0.5 | 2.6 | GO:1901837 | negative regulation of transcription of nuclear large rRNA transcript from RNA polymerase I promoter(GO:1901837) |
| 0.5 | 2.5 | GO:0015746 | tricarboxylic acid transport(GO:0006842) citrate transport(GO:0015746) |
| 0.5 | 2.0 | GO:0009608 | response to symbiont(GO:0009608) response to symbiotic bacterium(GO:0009609) response to selenium ion(GO:0010269) |
| 0.5 | 2.9 | GO:0010641 | positive regulation of platelet-derived growth factor receptor signaling pathway(GO:0010641) |
| 0.5 | 1.4 | GO:2000474 | regulation of opioid receptor signaling pathway(GO:2000474) |
| 0.5 | 7.5 | GO:2000018 | regulation of male gonad development(GO:2000018) |
| 0.5 | 2.8 | GO:0002159 | desmosome assembly(GO:0002159) |
| 0.5 | 2.3 | GO:0009750 | response to fructose(GO:0009750) |
| 0.5 | 2.8 | GO:1903182 | regulation of SUMO transferase activity(GO:1903182) positive regulation of SUMO transferase activity(GO:1903755) |
| 0.4 | 1.8 | GO:1903237 | negative regulation of leukocyte tethering or rolling(GO:1903237) |
| 0.4 | 1.8 | GO:0035521 | monoubiquitinated histone deubiquitination(GO:0035521) monoubiquitinated histone H2A deubiquitination(GO:0035522) |
| 0.4 | 3.1 | GO:0002003 | angiotensin maturation(GO:0002003) |
| 0.4 | 2.1 | GO:0051754 | meiotic sister chromatid cohesion, centromeric(GO:0051754) |
| 0.4 | 3.4 | GO:0009446 | putrescine biosynthetic process(GO:0009446) |
| 0.4 | 1.2 | GO:0090081 | regulation of heart induction by regulation of canonical Wnt signaling pathway(GO:0090081) |
| 0.4 | 1.6 | GO:0008626 | granzyme-mediated apoptotic signaling pathway(GO:0008626) |
| 0.4 | 3.6 | GO:0010993 | regulation of ubiquitin homeostasis(GO:0010993) free ubiquitin chain polymerization(GO:0010994) |
| 0.4 | 1.9 | GO:0070601 | centromeric sister chromatid cohesion(GO:0070601) |
| 0.4 | 1.5 | GO:0003017 | lymph circulation(GO:0003017) |
| 0.4 | 1.9 | GO:2000660 | negative regulation of interleukin-1-mediated signaling pathway(GO:2000660) |
| 0.4 | 1.5 | GO:0016062 | adaptation of rhodopsin mediated signaling(GO:0016062) light adaption(GO:0036367) |
| 0.4 | 1.5 | GO:0098795 | mRNA cleavage involved in gene silencing by miRNA(GO:0035279) mRNA cleavage involved in gene silencing(GO:0098795) |
| 0.4 | 1.9 | GO:0071475 | cellular hyperosmotic salinity response(GO:0071475) |
| 0.4 | 1.1 | GO:0032487 | cerebellar cortex structural organization(GO:0021698) regulation of Rap protein signal transduction(GO:0032487) negative regulation of integrin activation(GO:0033624) |
| 0.4 | 1.1 | GO:0002277 | myeloid dendritic cell activation involved in immune response(GO:0002277) |
| 0.4 | 2.5 | GO:0042737 | drug catabolic process(GO:0042737) |
| 0.3 | 3.8 | GO:0071394 | cellular response to testosterone stimulus(GO:0071394) |
| 0.3 | 3.1 | GO:0016480 | negative regulation of transcription from RNA polymerase III promoter(GO:0016480) |
| 0.3 | 1.7 | GO:0045347 | negative regulation of MHC class II biosynthetic process(GO:0045347) |
| 0.3 | 1.4 | GO:1900060 | negative regulation of ceramide biosynthetic process(GO:1900060) |
| 0.3 | 1.7 | GO:1901252 | regulation of intracellular transport of viral material(GO:1901252) |
| 0.3 | 1.4 | GO:0090118 | receptor-mediated endocytosis of low-density lipoprotein particle involved in cholesterol transport(GO:0090118) |
| 0.3 | 1.0 | GO:0010512 | negative regulation of phosphatidylinositol biosynthetic process(GO:0010512) |
| 0.3 | 1.3 | GO:0021993 | initiation of neural tube closure(GO:0021993) |
| 0.3 | 0.9 | GO:2000670 | positive regulation of dendritic cell apoptotic process(GO:2000670) |
| 0.3 | 1.3 | GO:0042138 | meiotic DNA double-strand break formation(GO:0042138) |
| 0.3 | 1.2 | GO:0044208 | 'de novo' AMP biosynthetic process(GO:0044208) |
| 0.3 | 2.4 | GO:0002457 | T cell antigen processing and presentation(GO:0002457) |
| 0.3 | 1.2 | GO:0045659 | negative regulation of neutrophil differentiation(GO:0045659) |
| 0.3 | 1.7 | GO:0002023 | reduction of food intake in response to dietary excess(GO:0002023) positive regulation of somatostatin secretion(GO:0090274) |
| 0.3 | 0.8 | GO:1900106 | hyaluranon cable assembly(GO:0036118) regulation of hyaluranon cable assembly(GO:1900104) positive regulation of hyaluranon cable assembly(GO:1900106) |
| 0.3 | 1.4 | GO:0009115 | xanthine catabolic process(GO:0009115) |
| 0.3 | 1.4 | GO:0018101 | protein citrullination(GO:0018101) histone citrullination(GO:0036414) |
| 0.3 | 3.9 | GO:0042574 | retinal metabolic process(GO:0042574) |
| 0.3 | 2.1 | GO:0060700 | regulation of ribonuclease activity(GO:0060700) |
| 0.3 | 1.5 | GO:0021615 | glossopharyngeal nerve morphogenesis(GO:0021615) |
| 0.3 | 0.8 | GO:0051977 | lysophospholipid transport(GO:0051977) |
| 0.2 | 4.1 | GO:0070673 | response to interleukin-18(GO:0070673) |
| 0.2 | 4.1 | GO:0090308 | regulation of methylation-dependent chromatin silencing(GO:0090308) |
| 0.2 | 3.3 | GO:0016446 | somatic hypermutation of immunoglobulin genes(GO:0016446) |
| 0.2 | 4.5 | GO:0006337 | nucleosome disassembly(GO:0006337) |
| 0.2 | 1.1 | GO:2000523 | regulation of T cell costimulation(GO:2000523) positive regulation of T cell costimulation(GO:2000525) |
| 0.2 | 2.0 | GO:0008228 | opsonization(GO:0008228) |
| 0.2 | 0.9 | GO:0060133 | somatotropin secreting cell development(GO:0060133) |
| 0.2 | 1.5 | GO:0042256 | mature ribosome assembly(GO:0042256) |
| 0.2 | 1.7 | GO:1902031 | regulation of NADP metabolic process(GO:1902031) |
| 0.2 | 0.8 | GO:0071348 | cellular response to interleukin-11(GO:0071348) |
| 0.2 | 1.4 | GO:1903772 | regulation of viral budding via host ESCRT complex(GO:1903772) |
| 0.2 | 1.6 | GO:0001887 | selenium compound metabolic process(GO:0001887) |
| 0.2 | 3.6 | GO:1902916 | positive regulation of protein polyubiquitination(GO:1902916) |
| 0.2 | 0.6 | GO:0035627 | ceramide transport(GO:0035627) |
| 0.2 | 7.0 | GO:0019731 | antibacterial humoral response(GO:0019731) |
| 0.2 | 4.9 | GO:0030574 | collagen catabolic process(GO:0030574) |
| 0.2 | 2.1 | GO:0048386 | positive regulation of retinoic acid receptor signaling pathway(GO:0048386) |
| 0.2 | 1.1 | GO:0010166 | wax biosynthetic process(GO:0010025) wax metabolic process(GO:0010166) |
| 0.2 | 0.5 | GO:0002414 | immunoglobulin transcytosis in epithelial cells(GO:0002414) |
| 0.2 | 1.3 | GO:0010764 | negative regulation of fibroblast migration(GO:0010764) |
| 0.2 | 3.8 | GO:0071803 | positive regulation of podosome assembly(GO:0071803) |
| 0.2 | 0.5 | GO:0002501 | MHC protein complex assembly(GO:0002396) peptide antigen assembly with MHC protein complex(GO:0002501) |
| 0.2 | 0.4 | GO:1905167 | positive regulation of lysosomal protein catabolic process(GO:1905167) |
| 0.2 | 4.1 | GO:0001675 | acrosome assembly(GO:0001675) |
| 0.2 | 0.5 | GO:1904924 | negative regulation of mitophagy in response to mitochondrial depolarization(GO:1904924) |
| 0.2 | 2.2 | GO:0071801 | regulation of podosome assembly(GO:0071801) |
| 0.2 | 1.2 | GO:0070447 | positive regulation of oligodendrocyte progenitor proliferation(GO:0070447) |
| 0.2 | 0.5 | GO:0035965 | cardiolipin acyl-chain remodeling(GO:0035965) |
| 0.2 | 5.0 | GO:0019373 | epoxygenase P450 pathway(GO:0019373) |
| 0.2 | 2.7 | GO:0035970 | peptidyl-threonine dephosphorylation(GO:0035970) |
| 0.2 | 1.1 | GO:0046013 | regulation of T cell homeostatic proliferation(GO:0046013) |
| 0.2 | 0.5 | GO:1901994 | negative regulation of meiotic cell cycle phase transition(GO:1901994) |
| 0.2 | 1.1 | GO:2000491 | positive regulation of hepatic stellate cell activation(GO:2000491) |
| 0.2 | 1.1 | GO:0031536 | positive regulation of exit from mitosis(GO:0031536) |
| 0.2 | 0.5 | GO:0018146 | keratan sulfate biosynthetic process(GO:0018146) |
| 0.2 | 2.0 | GO:0001867 | complement activation, lectin pathway(GO:0001867) |
| 0.1 | 2.1 | GO:0050957 | equilibrioception(GO:0050957) |
| 0.1 | 2.7 | GO:0060124 | positive regulation of growth hormone secretion(GO:0060124) |
| 0.1 | 4.9 | GO:0006084 | acetyl-CoA metabolic process(GO:0006084) |
| 0.1 | 3.8 | GO:0035634 | response to stilbenoid(GO:0035634) |
| 0.1 | 0.7 | GO:0035616 | histone H2B conserved C-terminal lysine deubiquitination(GO:0035616) |
| 0.1 | 1.1 | GO:1900748 | positive regulation of vascular endothelial growth factor signaling pathway(GO:1900748) |
| 0.1 | 1.2 | GO:0006703 | estrogen biosynthetic process(GO:0006703) |
| 0.1 | 1.2 | GO:0016338 | calcium-independent cell-cell adhesion via plasma membrane cell-adhesion molecules(GO:0016338) |
| 0.1 | 3.6 | GO:1902254 | negative regulation of intrinsic apoptotic signaling pathway by p53 class mediator(GO:1902254) |
| 0.1 | 1.3 | GO:0030206 | chondroitin sulfate biosynthetic process(GO:0030206) |
| 0.1 | 4.2 | GO:0010569 | regulation of double-strand break repair via homologous recombination(GO:0010569) |
| 0.1 | 0.8 | GO:0002540 | leukotriene production involved in inflammatory response(GO:0002540) |
| 0.1 | 2.2 | GO:0001731 | formation of translation preinitiation complex(GO:0001731) |
| 0.1 | 0.6 | GO:0051885 | positive regulation of anagen(GO:0051885) |
| 0.1 | 3.2 | GO:0070886 | positive regulation of calcineurin-NFAT signaling cascade(GO:0070886) |
| 0.1 | 1.5 | GO:0009650 | UV protection(GO:0009650) elastic fiber assembly(GO:0048251) |
| 0.1 | 1.0 | GO:0051305 | chromosome movement towards spindle pole(GO:0051305) |
| 0.1 | 1.0 | GO:0051534 | negative regulation of NFAT protein import into nucleus(GO:0051534) |
| 0.1 | 1.2 | GO:0072642 | type I interferon secretion(GO:0072641) interferon-alpha secretion(GO:0072642) regulation of interferon-alpha secretion(GO:1902739) positive regulation of interferon-alpha secretion(GO:1902741) |
| 0.1 | 0.4 | GO:0051878 | lateral element assembly(GO:0051878) |
| 0.1 | 0.7 | GO:2000325 | regulation of ligand-dependent nuclear receptor transcription coactivator activity(GO:2000325) positive regulation of ligand-dependent nuclear receptor transcription coactivator activity(GO:2000327) |
| 0.1 | 8.1 | GO:0051496 | positive regulation of stress fiber assembly(GO:0051496) |
| 0.1 | 4.3 | GO:0009649 | entrainment of circadian clock(GO:0009649) |
| 0.1 | 0.4 | GO:0010725 | regulation of primitive erythrocyte differentiation(GO:0010725) eosinophil fate commitment(GO:0035854) |
| 0.1 | 1.1 | GO:1900364 | negative regulation of transcription elongation from RNA polymerase II promoter(GO:0034244) negative regulation of mRNA polyadenylation(GO:1900364) |
| 0.1 | 2.4 | GO:0034063 | stress granule assembly(GO:0034063) |
| 0.1 | 0.9 | GO:0031937 | positive regulation of chromatin silencing(GO:0031937) |
| 0.1 | 0.5 | GO:0071335 | hair follicle cell proliferation(GO:0071335) |
| 0.1 | 2.1 | GO:0019370 | leukotriene biosynthetic process(GO:0019370) |
| 0.1 | 0.2 | GO:1902174 | positive regulation of keratinocyte apoptotic process(GO:1902174) |
| 0.1 | 0.4 | GO:0046604 | positive regulation of mitotic centrosome separation(GO:0046604) |
| 0.1 | 0.6 | GO:0098734 | protein depalmitoylation(GO:0002084) macromolecule depalmitoylation(GO:0098734) |
| 0.1 | 1.1 | GO:0045075 | interleukin-12 biosynthetic process(GO:0042090) regulation of interleukin-12 biosynthetic process(GO:0045075) |
| 0.1 | 0.9 | GO:0006104 | succinyl-CoA metabolic process(GO:0006104) |
| 0.1 | 0.7 | GO:0060313 | negative regulation of neutrophil degranulation(GO:0043314) negative regulation of blood vessel remodeling(GO:0060313) |
| 0.1 | 0.9 | GO:0030854 | positive regulation of granulocyte differentiation(GO:0030854) |
| 0.1 | 0.4 | GO:0016237 | lysosomal microautophagy(GO:0016237) piecemeal microautophagy of nucleus(GO:0034727) suppression by virus of host autophagy(GO:0039521) amino acid homeostasis(GO:0080144) negative regulation of sphingolipid biosynthesis involved in cellular sphingolipid homeostasis(GO:0090157) |
| 0.1 | 0.9 | GO:0035608 | protein deglutamylation(GO:0035608) |
| 0.1 | 0.6 | GO:0006283 | transcription-coupled nucleotide-excision repair(GO:0006283) |
| 0.1 | 5.6 | GO:0043666 | regulation of phosphoprotein phosphatase activity(GO:0043666) |
| 0.1 | 1.3 | GO:0031581 | hemidesmosome assembly(GO:0031581) |
| 0.1 | 0.5 | GO:1903691 | positive regulation of wound healing, spreading of epidermal cells(GO:1903691) |
| 0.1 | 0.5 | GO:2000348 | regulation of CD40 signaling pathway(GO:2000348) |
| 0.1 | 0.4 | GO:0051790 | short-chain fatty acid biosynthetic process(GO:0051790) |
| 0.1 | 1.5 | GO:0000291 | nuclear-transcribed mRNA catabolic process, exonucleolytic(GO:0000291) |
| 0.1 | 1.3 | GO:0045899 | positive regulation of RNA polymerase II transcriptional preinitiation complex assembly(GO:0045899) |
| 0.1 | 0.7 | GO:1904668 | positive regulation of ubiquitin protein ligase activity(GO:1904668) |
| 0.1 | 1.3 | GO:0046007 | negative regulation of activated T cell proliferation(GO:0046007) |
| 0.1 | 0.9 | GO:0070423 | nucleotide-binding oligomerization domain containing signaling pathway(GO:0070423) nucleotide-binding oligomerization domain containing 2 signaling pathway(GO:0070431) |
| 0.1 | 4.2 | GO:0006953 | acute-phase response(GO:0006953) |
| 0.1 | 2.6 | GO:0042438 | melanin biosynthetic process(GO:0042438) |
| 0.1 | 1.0 | GO:0033601 | positive regulation of mammary gland epithelial cell proliferation(GO:0033601) |
| 0.1 | 0.4 | GO:0021506 | anterior neuropore closure(GO:0021506) neuropore closure(GO:0021995) |
| 0.1 | 1.6 | GO:0048312 | intracellular distribution of mitochondria(GO:0048312) |
| 0.1 | 0.3 | GO:0035964 | COPI-coated vesicle budding(GO:0035964) |
| 0.1 | 1.7 | GO:0039694 | viral RNA genome replication(GO:0039694) RNA replication(GO:0039703) |
| 0.1 | 0.6 | GO:0070327 | thyroid hormone transport(GO:0070327) |
| 0.1 | 2.3 | GO:0030970 | retrograde protein transport, ER to cytosol(GO:0030970) |
| 0.1 | 0.2 | GO:1903896 | positive regulation of IRE1-mediated unfolded protein response(GO:1903896) |
| 0.1 | 5.4 | GO:0006749 | glutathione metabolic process(GO:0006749) |
| 0.1 | 3.2 | GO:0002474 | antigen processing and presentation of peptide antigen via MHC class I(GO:0002474) |
| 0.1 | 1.0 | GO:0080009 | mRNA methylation(GO:0080009) |
| 0.1 | 3.9 | GO:0015914 | phospholipid transport(GO:0015914) |
| 0.1 | 0.9 | GO:0048096 | chromatin-mediated maintenance of transcription(GO:0048096) |
| 0.1 | 0.7 | GO:0098789 | pre-mRNA cleavage required for polyadenylation(GO:0098789) |
| 0.1 | 2.2 | GO:0019369 | arachidonic acid metabolic process(GO:0019369) |
| 0.1 | 0.2 | GO:0045004 | DNA replication proofreading(GO:0045004) |
| 0.1 | 0.4 | GO:0061051 | positive regulation of cell growth involved in cardiac muscle cell development(GO:0061051) |
| 0.1 | 3.0 | GO:0045737 | positive regulation of cyclin-dependent protein serine/threonine kinase activity(GO:0045737) |
| 0.1 | 3.1 | GO:0010862 | positive regulation of pathway-restricted SMAD protein phosphorylation(GO:0010862) |
| 0.1 | 0.6 | GO:0048680 | positive regulation of axon regeneration(GO:0048680) |
| 0.1 | 0.5 | GO:0031119 | tRNA pseudouridine synthesis(GO:0031119) |
| 0.1 | 4.8 | GO:0010923 | negative regulation of phosphatase activity(GO:0010923) |
| 0.1 | 0.4 | GO:1903797 | positive regulation of inorganic anion transmembrane transport(GO:1903797) |
| 0.1 | 2.9 | GO:0048247 | lymphocyte chemotaxis(GO:0048247) |
| 0.1 | 2.9 | GO:0044380 | protein localization to cytoskeleton(GO:0044380) |
| 0.1 | 2.0 | GO:0001580 | detection of chemical stimulus involved in sensory perception of bitter taste(GO:0001580) |
| 0.0 | 2.4 | GO:0035307 | positive regulation of protein dephosphorylation(GO:0035307) |
| 0.0 | 2.8 | GO:0016574 | histone ubiquitination(GO:0016574) |
| 0.0 | 0.2 | GO:0035660 | MyD88-dependent toll-like receptor 4 signaling pathway(GO:0035660) |
| 0.0 | 0.4 | GO:0046598 | positive regulation of viral entry into host cell(GO:0046598) |
| 0.0 | 1.8 | GO:0002089 | lens morphogenesis in camera-type eye(GO:0002089) |
| 0.0 | 2.0 | GO:0034314 | Arp2/3 complex-mediated actin nucleation(GO:0034314) |
| 0.0 | 0.2 | GO:0021691 | cerebellar Purkinje cell layer maturation(GO:0021691) |
| 0.0 | 0.4 | GO:1902583 | intracellular transport of virus(GO:0075733) multi-organism intracellular transport(GO:1902583) |
| 0.0 | 1.3 | GO:0090200 | positive regulation of release of cytochrome c from mitochondria(GO:0090200) |
| 0.0 | 0.4 | GO:0006646 | phosphatidylethanolamine biosynthetic process(GO:0006646) |
| 0.0 | 1.3 | GO:0035855 | megakaryocyte development(GO:0035855) |
| 0.0 | 1.6 | GO:0007131 | reciprocal meiotic recombination(GO:0007131) reciprocal DNA recombination(GO:0035825) |
| 0.0 | 0.3 | GO:0045618 | positive regulation of keratinocyte differentiation(GO:0045618) |
| 0.0 | 2.0 | GO:0097006 | regulation of plasma lipoprotein particle levels(GO:0097006) |
| 0.0 | 0.6 | GO:2000637 | positive regulation of gene silencing by miRNA(GO:2000637) |
| 0.0 | 1.4 | GO:0030857 | negative regulation of epithelial cell differentiation(GO:0030857) |
| 0.0 | 0.4 | GO:0006379 | mRNA cleavage(GO:0006379) |
| 0.0 | 0.5 | GO:0006488 | dolichol-linked oligosaccharide biosynthetic process(GO:0006488) |
| 0.0 | 0.2 | GO:0031120 | snRNA pseudouridine synthesis(GO:0031120) |
| 0.0 | 2.8 | GO:1903955 | positive regulation of protein targeting to mitochondrion(GO:1903955) |
| 0.0 | 0.4 | GO:0009404 | toxin metabolic process(GO:0009404) |
| 0.0 | 0.5 | GO:0046597 | negative regulation of viral entry into host cell(GO:0046597) |
| 0.0 | 0.5 | GO:0061298 | retina vasculature development in camera-type eye(GO:0061298) |
| 0.0 | 2.0 | GO:0045669 | positive regulation of osteoblast differentiation(GO:0045669) |
| 0.0 | 0.3 | GO:0035879 | lactate transport(GO:0015727) lactate transmembrane transport(GO:0035873) plasma membrane lactate transport(GO:0035879) |
| 0.0 | 0.9 | GO:0030488 | tRNA methylation(GO:0030488) |
| 0.0 | 1.1 | GO:0035066 | positive regulation of histone acetylation(GO:0035066) |
| 0.0 | 0.5 | GO:0006353 | DNA-templated transcription, termination(GO:0006353) |
| 0.0 | 0.2 | GO:0051571 | positive regulation of histone H3-K4 methylation(GO:0051571) |
| 0.0 | 0.5 | GO:0001833 | inner cell mass cell proliferation(GO:0001833) |
| 0.0 | 0.2 | GO:0090141 | positive regulation of mitochondrial fission(GO:0090141) |
| 0.0 | 0.6 | GO:0097499 | protein localization to nonmotile primary cilium(GO:0097499) |
| 0.0 | 0.4 | GO:0009083 | branched-chain amino acid catabolic process(GO:0009083) |
| 0.0 | 0.1 | GO:0060708 | spongiotrophoblast differentiation(GO:0060708) |
| 0.0 | 2.2 | GO:0006835 | dicarboxylic acid transport(GO:0006835) |
| 0.0 | 0.2 | GO:0031053 | primary miRNA processing(GO:0031053) |
| 0.0 | 0.8 | GO:1900026 | positive regulation of substrate adhesion-dependent cell spreading(GO:1900026) |
| 0.0 | 2.4 | GO:0051028 | mRNA transport(GO:0051028) |
| 0.0 | 0.9 | GO:0097120 | receptor localization to synapse(GO:0097120) |
| 0.0 | 0.1 | GO:0060685 | regulation of prostatic bud formation(GO:0060685) negative regulation of prostatic bud formation(GO:0060686) |
| 0.0 | 0.7 | GO:0030520 | intracellular estrogen receptor signaling pathway(GO:0030520) |
| 0.0 | 0.5 | GO:0050909 | sensory perception of taste(GO:0050909) |
| 0.0 | 0.2 | GO:0015747 | urate transport(GO:0015747) |
| 0.0 | 0.4 | GO:0043968 | histone H2A acetylation(GO:0043968) |
| 0.0 | 0.2 | GO:0010835 | regulation of protein ADP-ribosylation(GO:0010835) |
| 0.0 | 0.9 | GO:0016239 | positive regulation of macroautophagy(GO:0016239) |
| 0.0 | 0.6 | GO:0000184 | nuclear-transcribed mRNA catabolic process, nonsense-mediated decay(GO:0000184) |
| 0.0 | 0.8 | GO:1902099 | regulation of mitotic metaphase/anaphase transition(GO:0030071) regulation of metaphase/anaphase transition of cell cycle(GO:1902099) |
| 0.0 | 0.1 | GO:0070417 | cellular response to cold(GO:0070417) |
| 0.0 | 0.1 | GO:0006438 | valyl-tRNA aminoacylation(GO:0006438) |
| 0.0 | 0.1 | GO:0006616 | SRP-dependent cotranslational protein targeting to membrane, translocation(GO:0006616) |
| 0.0 | 0.4 | GO:0033622 | integrin activation(GO:0033622) |
| 0.0 | 0.3 | GO:0051085 | chaperone mediated protein folding requiring cofactor(GO:0051085) |
| 0.0 | 0.2 | GO:0015879 | carnitine transport(GO:0015879) |
| 0.0 | 0.6 | GO:0006414 | translational elongation(GO:0006414) |
| 0.0 | 0.5 | GO:0006284 | base-excision repair(GO:0006284) |
| 0.0 | 0.2 | GO:0032525 | somite rostral/caudal axis specification(GO:0032525) |
| 0.0 | 0.1 | GO:0031022 | nuclear migration along microfilament(GO:0031022) |
| 0.0 | 0.5 | GO:1900047 | negative regulation of blood coagulation(GO:0030195) negative regulation of hemostasis(GO:1900047) |
| 0.0 | 0.2 | GO:0001886 | endothelial cell morphogenesis(GO:0001886) |
| 0.0 | 0.7 | GO:0002181 | cytoplasmic translation(GO:0002181) |
| 0.0 | 1.0 | GO:0070301 | cellular response to hydrogen peroxide(GO:0070301) |
| 0.0 | 1.3 | GO:0042100 | B cell proliferation(GO:0042100) |
| 0.0 | 0.3 | GO:0006303 | double-strand break repair via nonhomologous end joining(GO:0006303) |
| 0.0 | 1.5 | GO:0006261 | DNA-dependent DNA replication(GO:0006261) |
| 0.0 | 0.2 | GO:1903405 | regulation of establishment of protein localization to chromosome(GO:0070202) regulation of establishment of protein localization to telomere(GO:0070203) protein localization to nuclear body(GO:1903405) positive regulation of establishment of protein localization to telomere(GO:1904851) protein localization to Cajal body(GO:1904867) regulation of protein localization to Cajal body(GO:1904869) positive regulation of protein localization to Cajal body(GO:1904871) protein localization to nucleoplasm(GO:1990173) |
| 0.0 | 0.8 | GO:0045454 | cell redox homeostasis(GO:0045454) |
| 0.0 | 0.3 | GO:0006491 | N-glycan processing(GO:0006491) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.5 | 4.4 | GO:0061474 | phagolysosome membrane(GO:0061474) |
| 1.4 | 4.3 | GO:0070435 | Shc-EGFR complex(GO:0070435) |
| 1.3 | 3.9 | GO:1990630 | IRE1-RACK1-PP2A complex(GO:1990630) |
| 1.0 | 4.2 | GO:0036284 | tubulobulbar complex(GO:0036284) |
| 1.0 | 14.7 | GO:0034366 | spherical high-density lipoprotein particle(GO:0034366) |
| 0.9 | 2.6 | GO:0060187 | cell pole(GO:0060187) |
| 0.9 | 12.3 | GO:0042612 | MHC class I protein complex(GO:0042612) |
| 0.9 | 4.3 | GO:0030868 | smooth endoplasmic reticulum membrane(GO:0030868) smooth endoplasmic reticulum part(GO:0097425) |
| 0.7 | 6.4 | GO:0005577 | fibrinogen complex(GO:0005577) |
| 0.7 | 2.1 | GO:0031477 | stereocilia tip link(GO:0002140) myosin VII complex(GO:0031477) stereocilia tip-link density(GO:1990427) upper tip-link density(GO:1990435) |
| 0.6 | 2.8 | GO:0005914 | spot adherens junction(GO:0005914) |
| 0.5 | 2.3 | GO:0000839 | Hrd1p ubiquitin ligase ERAD-L complex(GO:0000839) |
| 0.5 | 2.8 | GO:1990356 | sumoylated E2 ligase complex(GO:1990356) |
| 0.4 | 1.6 | GO:0044194 | cytolytic granule(GO:0044194) |
| 0.4 | 1.5 | GO:0071953 | elastic fiber(GO:0071953) |
| 0.4 | 5.6 | GO:0000164 | protein phosphatase type 1 complex(GO:0000164) |
| 0.4 | 1.4 | GO:0034679 | integrin alpha9-beta1 complex(GO:0034679) integrin alphav-beta3 complex(GO:0034683) |
| 0.4 | 2.8 | GO:0033503 | HULC complex(GO:0033503) |
| 0.3 | 1.3 | GO:0005608 | laminin-3 complex(GO:0005608) |
| 0.3 | 3.6 | GO:0042629 | mast cell granule(GO:0042629) |
| 0.3 | 4.8 | GO:0000940 | condensed chromosome outer kinetochore(GO:0000940) |
| 0.3 | 1.9 | GO:0043564 | Ku70:Ku80 complex(GO:0043564) |
| 0.3 | 3.9 | GO:0031983 | vesicle lumen(GO:0031983) |
| 0.3 | 1.4 | GO:0035339 | SPOTS complex(GO:0035339) |
| 0.3 | 1.4 | GO:0030121 | AP-1 adaptor complex(GO:0030121) |
| 0.3 | 2.0 | GO:0097413 | Lewy body(GO:0097413) |
| 0.2 | 1.6 | GO:0030289 | protein phosphatase 4 complex(GO:0030289) |
| 0.2 | 4.1 | GO:0046581 | intercellular canaliculus(GO:0046581) |
| 0.2 | 1.1 | GO:0045160 | myosin I complex(GO:0045160) |
| 0.2 | 1.1 | GO:0031251 | PAN complex(GO:0031251) |
| 0.2 | 5.9 | GO:0016327 | apicolateral plasma membrane(GO:0016327) |
| 0.2 | 2.5 | GO:0031080 | nuclear pore outer ring(GO:0031080) |
| 0.2 | 2.2 | GO:0097197 | tetraspanin-enriched microdomain(GO:0097197) |
| 0.2 | 0.9 | GO:0071797 | LUBAC complex(GO:0071797) |
| 0.2 | 1.7 | GO:0061574 | ASAP complex(GO:0061574) |
| 0.2 | 4.5 | GO:0016514 | SWI/SNF complex(GO:0016514) |
| 0.2 | 0.7 | GO:0008623 | CHRAC(GO:0008623) |
| 0.2 | 0.5 | GO:0055087 | Ski complex(GO:0055087) |
| 0.1 | 1.5 | GO:0019815 | B cell receptor complex(GO:0019815) |
| 0.1 | 4.8 | GO:0097225 | sperm midpiece(GO:0097225) |
| 0.1 | 2.8 | GO:0042622 | photoreceptor outer segment membrane(GO:0042622) |
| 0.1 | 1.0 | GO:0036396 | MIS complex(GO:0036396) |
| 0.1 | 0.5 | GO:0045251 | mitochondrial electron transfer flavoprotein complex(GO:0017133) electron transfer flavoprotein complex(GO:0045251) |
| 0.1 | 3.8 | GO:0001891 | phagocytic cup(GO:0001891) |
| 0.1 | 7.2 | GO:0005788 | endoplasmic reticulum lumen(GO:0005788) |
| 0.1 | 0.7 | GO:0005683 | U7 snRNP(GO:0005683) |
| 0.1 | 1.8 | GO:0035631 | CD40 receptor complex(GO:0035631) |
| 0.1 | 1.2 | GO:0097433 | dense body(GO:0097433) |
| 0.1 | 2.1 | GO:0030914 | STAGA complex(GO:0030914) |
| 0.1 | 1.1 | GO:0033116 | endoplasmic reticulum-Golgi intermediate compartment membrane(GO:0033116) |
| 0.1 | 3.3 | GO:0097038 | perinuclear endoplasmic reticulum(GO:0097038) |
| 0.1 | 1.4 | GO:0000813 | ESCRT I complex(GO:0000813) |
| 0.1 | 5.2 | GO:0005719 | nuclear euchromatin(GO:0005719) |
| 0.1 | 0.9 | GO:0070652 | HAUS complex(GO:0070652) |
| 0.1 | 2.6 | GO:0044292 | dendrite terminus(GO:0044292) |
| 0.1 | 1.4 | GO:0033179 | proton-transporting V-type ATPase, V0 domain(GO:0033179) |
| 0.1 | 5.5 | GO:0031901 | early endosome membrane(GO:0031901) |
| 0.1 | 1.2 | GO:0071203 | WASH complex(GO:0071203) |
| 0.1 | 3.3 | GO:0030687 | preribosome, large subunit precursor(GO:0030687) |
| 0.1 | 0.6 | GO:0005853 | eukaryotic translation elongation factor 1 complex(GO:0005853) |
| 0.1 | 0.9 | GO:0030015 | CCR4-NOT core complex(GO:0030015) |
| 0.1 | 6.3 | GO:0034707 | chloride channel complex(GO:0034707) |
| 0.1 | 2.0 | GO:0031907 | peroxisomal matrix(GO:0005782) microbody lumen(GO:0031907) |
| 0.1 | 0.4 | GO:0000802 | transverse filament(GO:0000802) |
| 0.1 | 0.1 | GO:0005687 | U4 snRNP(GO:0005687) |
| 0.1 | 2.1 | GO:0035145 | exon-exon junction complex(GO:0035145) |
| 0.1 | 2.3 | GO:0032588 | trans-Golgi network membrane(GO:0032588) |
| 0.1 | 1.6 | GO:0010369 | chromocenter(GO:0010369) |
| 0.1 | 0.3 | GO:0017177 | glucosidase II complex(GO:0017177) |
| 0.1 | 0.9 | GO:0005721 | pericentric heterochromatin(GO:0005721) |
| 0.1 | 1.3 | GO:0005686 | U2 snRNP(GO:0005686) |
| 0.1 | 0.5 | GO:0030062 | mitochondrial tricarboxylic acid cycle enzyme complex(GO:0030062) |
| 0.1 | 1.3 | GO:0000791 | euchromatin(GO:0000791) |
| 0.1 | 2.7 | GO:0045095 | keratin filament(GO:0045095) |
| 0.1 | 1.0 | GO:0030061 | mitochondrial crista(GO:0030061) |
| 0.1 | 0.3 | GO:0032807 | DNA ligase IV complex(GO:0032807) |
| 0.1 | 0.5 | GO:0045098 | type III intermediate filament(GO:0045098) |
| 0.1 | 0.7 | GO:0031209 | SCAR complex(GO:0031209) |
| 0.0 | 1.0 | GO:0002199 | zona pellucida receptor complex(GO:0002199) |
| 0.0 | 0.2 | GO:0090661 | box H/ACA telomerase RNP complex(GO:0090661) |
| 0.0 | 0.6 | GO:0070765 | gamma-secretase complex(GO:0070765) |
| 0.0 | 0.4 | GO:0042382 | paraspeckles(GO:0042382) |
| 0.0 | 3.0 | GO:0000307 | cyclin-dependent protein kinase holoenzyme complex(GO:0000307) |
| 0.0 | 0.9 | GO:0005665 | DNA-directed RNA polymerase II, core complex(GO:0005665) |
| 0.0 | 3.1 | GO:0031519 | PcG protein complex(GO:0031519) |
| 0.0 | 3.6 | GO:0005881 | cytoplasmic microtubule(GO:0005881) |
| 0.0 | 0.4 | GO:0000445 | THO complex(GO:0000347) THO complex part of transcription export complex(GO:0000445) |
| 0.0 | 2.7 | GO:0045171 | intercellular bridge(GO:0045171) |
| 0.0 | 1.8 | GO:0016235 | aggresome(GO:0016235) |
| 0.0 | 7.2 | GO:0001650 | fibrillar center(GO:0001650) |
| 0.0 | 3.6 | GO:0005811 | lipid particle(GO:0005811) |
| 0.0 | 0.7 | GO:0005847 | mRNA cleavage and polyadenylation specificity factor complex(GO:0005847) |
| 0.0 | 8.4 | GO:0000794 | condensed nuclear chromosome(GO:0000794) |
| 0.0 | 0.3 | GO:0001673 | male germ cell nucleus(GO:0001673) |
| 0.0 | 0.2 | GO:0072357 | PTW/PP1 phosphatase complex(GO:0072357) |
| 0.0 | 0.1 | GO:0032777 | Piccolo NuA4 histone acetyltransferase complex(GO:0032777) |
| 0.0 | 0.3 | GO:0035068 | micro-ribonucleoprotein complex(GO:0035068) RISC-loading complex(GO:0070578) |
| 0.0 | 2.4 | GO:0010494 | cytoplasmic stress granule(GO:0010494) |
| 0.0 | 0.5 | GO:0043240 | Fanconi anaemia nuclear complex(GO:0043240) |
| 0.0 | 0.9 | GO:0060077 | inhibitory synapse(GO:0060077) |
| 0.0 | 2.2 | GO:0000932 | cytoplasmic mRNA processing body(GO:0000932) |
| 0.0 | 14.9 | GO:0005667 | transcription factor complex(GO:0005667) |
| 0.0 | 4.7 | GO:0030176 | integral component of endoplasmic reticulum membrane(GO:0030176) |
| 0.0 | 4.3 | GO:0001669 | acrosomal vesicle(GO:0001669) |
| 0.0 | 16.5 | GO:0031012 | extracellular matrix(GO:0031012) |
| 0.0 | 0.2 | GO:0000322 | storage vacuole(GO:0000322) |
| 0.0 | 2.4 | GO:0032587 | ruffle membrane(GO:0032587) |
| 0.0 | 4.9 | GO:0072562 | blood microparticle(GO:0072562) |
| 0.0 | 0.4 | GO:1904115 | axon cytoplasm(GO:1904115) |
| 0.0 | 0.4 | GO:0016581 | NuRD complex(GO:0016581) CHD-type complex(GO:0090545) |
| 0.0 | 0.3 | GO:0031045 | dense core granule(GO:0031045) |
| 0.0 | 0.6 | GO:0043034 | costamere(GO:0043034) |
| 0.0 | 1.6 | GO:0016605 | PML body(GO:0016605) |
| 0.0 | 1.3 | GO:0005791 | rough endoplasmic reticulum(GO:0005791) |
| 0.0 | 0.1 | GO:1990075 | periciliary membrane compartment(GO:1990075) |
| 0.0 | 1.3 | GO:0031903 | peroxisomal membrane(GO:0005778) microbody membrane(GO:0031903) |
| 0.0 | 2.6 | GO:0005884 | actin filament(GO:0005884) |
| 0.0 | 1.0 | GO:0016328 | lateral plasma membrane(GO:0016328) |
| 0.0 | 0.6 | GO:0000407 | pre-autophagosomal structure(GO:0000407) |
| 0.0 | 0.2 | GO:0035267 | NuA4 histone acetyltransferase complex(GO:0035267) H4/H2A histone acetyltransferase complex(GO:0043189) H4 histone acetyltransferase complex(GO:1902562) |
| 0.0 | 1.6 | GO:0034399 | nuclear periphery(GO:0034399) |
| 0.0 | 0.2 | GO:0030126 | COPI vesicle coat(GO:0030126) |
| 0.0 | 0.1 | GO:0031466 | Cul5-RING ubiquitin ligase complex(GO:0031466) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.3 | 7.0 | GO:0004024 | alcohol dehydrogenase activity, zinc-dependent(GO:0004024) |
| 1.9 | 5.6 | GO:0034632 | retinol transporter activity(GO:0034632) |
| 1.4 | 5.7 | GO:0070653 | high-density lipoprotein particle receptor binding(GO:0070653) |
| 1.4 | 9.8 | GO:0005005 | transmembrane-ephrin receptor activity(GO:0005005) |
| 1.3 | 6.5 | GO:0004743 | pyruvate kinase activity(GO:0004743) |
| 1.3 | 3.8 | GO:0005009 | insulin-activated receptor activity(GO:0005009) |
| 1.2 | 3.7 | GO:0004771 | sterol esterase activity(GO:0004771) |
| 1.2 | 4.7 | GO:0004063 | aryldialkylphosphatase activity(GO:0004063) |
| 1.2 | 5.8 | GO:0003986 | acetyl-CoA hydrolase activity(GO:0003986) |
| 1.1 | 3.2 | GO:0019978 | interleukin-3 receptor activity(GO:0004912) interleukin-3 binding(GO:0019978) |
| 1.0 | 5.2 | GO:0038181 | bile acid receptor activity(GO:0038181) |
| 1.0 | 3.0 | GO:0004911 | interleukin-2 receptor activity(GO:0004911) |
| 1.0 | 3.0 | GO:0004613 | phosphoenolpyruvate carboxykinase activity(GO:0004611) phosphoenolpyruvate carboxykinase (GTP) activity(GO:0004613) |
| 0.9 | 5.3 | GO:0015232 | heme transporter activity(GO:0015232) |
| 0.9 | 2.6 | GO:0004802 | transketolase activity(GO:0004802) |
| 0.8 | 3.3 | GO:0030156 | benzodiazepine receptor binding(GO:0030156) |
| 0.8 | 4.8 | GO:0060230 | lipoprotein lipase activator activity(GO:0060230) |
| 0.8 | 3.1 | GO:0016899 | oxidoreductase activity, acting on the CH-OH group of donors, oxygen as acceptor(GO:0016899) |
| 0.8 | 12.9 | GO:0030881 | beta-2-microglobulin binding(GO:0030881) |
| 0.7 | 2.2 | GO:0008480 | sarcosine dehydrogenase activity(GO:0008480) |
| 0.7 | 2.9 | GO:0004464 | leukotriene-C4 synthase activity(GO:0004464) |
| 0.7 | 4.3 | GO:0004850 | uridine phosphorylase activity(GO:0004850) |
| 0.7 | 4.3 | GO:0048408 | epidermal growth factor binding(GO:0048408) |
| 0.7 | 2.0 | GO:0004326 | tetrahydrofolylpolyglutamate synthase activity(GO:0004326) |
| 0.6 | 3.9 | GO:0016647 | oxidoreductase activity, acting on the CH-NH group of donors, oxygen as acceptor(GO:0016647) |
| 0.6 | 16.8 | GO:0019825 | oxygen binding(GO:0019825) |
| 0.6 | 4.3 | GO:0016841 | ammonia-lyase activity(GO:0016841) |
| 0.6 | 1.8 | GO:0032217 | riboflavin transporter activity(GO:0032217) |
| 0.6 | 4.1 | GO:0044729 | hemi-methylated DNA-binding(GO:0044729) |
| 0.6 | 1.7 | GO:0004470 | malic enzyme activity(GO:0004470) malate dehydrogenase (decarboxylating) (NAD+) activity(GO:0004471) malate dehydrogenase (decarboxylating) (NADP+) activity(GO:0004473) |
| 0.5 | 6.6 | GO:0102338 | fatty acid elongase activity(GO:0009922) 3-oxo-arachidoyl-CoA synthase activity(GO:0102336) 3-oxo-cerotoyl-CoA synthase activity(GO:0102337) 3-oxo-lignoceronyl-CoA synthase activity(GO:0102338) |
| 0.5 | 2.7 | GO:0004724 | magnesium-dependent protein serine/threonine phosphatase activity(GO:0004724) |
| 0.5 | 2.7 | GO:0003989 | acetyl-CoA carboxylase activity(GO:0003989) |
| 0.5 | 2.5 | GO:0015137 | citrate transmembrane transporter activity(GO:0015137) tricarboxylic acid transmembrane transporter activity(GO:0015142) |
| 0.5 | 3.0 | GO:0008453 | alanine-glyoxylate transaminase activity(GO:0008453) |
| 0.5 | 3.0 | GO:0061575 | cyclin-dependent protein serine/threonine kinase activator activity(GO:0061575) |
| 0.5 | 1.9 | GO:0005152 | interleukin-1 receptor antagonist activity(GO:0005152) |
| 0.5 | 2.3 | GO:0008309 | double-stranded DNA exodeoxyribonuclease activity(GO:0008309) |
| 0.5 | 2.8 | GO:0061656 | SUMO conjugating enzyme activity(GO:0061656) |
| 0.4 | 3.6 | GO:0005186 | pheromone activity(GO:0005186) |
| 0.4 | 2.1 | GO:0043546 | molybdopterin cofactor binding(GO:0043546) |
| 0.4 | 5.1 | GO:0008525 | phosphatidylcholine transporter activity(GO:0008525) |
| 0.4 | 2.5 | GO:0071723 | lipopeptide binding(GO:0071723) |
| 0.4 | 1.2 | GO:0004019 | adenylosuccinate synthase activity(GO:0004019) |
| 0.4 | 2.7 | GO:0004782 | sulfinoalanine decarboxylase activity(GO:0004782) |
| 0.4 | 2.7 | GO:1990254 | keratin filament binding(GO:1990254) |
| 0.4 | 6.9 | GO:0003993 | acid phosphatase activity(GO:0003993) |
| 0.4 | 5.3 | GO:0004000 | adenosine deaminase activity(GO:0004000) |
| 0.3 | 3.7 | GO:0016813 | hydrolase activity, acting on carbon-nitrogen (but not peptide) bonds, in linear amidines(GO:0016813) |
| 0.3 | 1.3 | GO:0047961 | glycine N-acyltransferase activity(GO:0047961) |
| 0.3 | 0.9 | GO:0004774 | succinate-CoA ligase activity(GO:0004774) |
| 0.3 | 2.2 | GO:0047184 | 1-acylglycerophosphocholine O-acyltransferase activity(GO:0047184) |
| 0.3 | 2.4 | GO:0003988 | acetyl-CoA C-acyltransferase activity(GO:0003988) |
| 0.3 | 3.5 | GO:0003997 | acyl-CoA oxidase activity(GO:0003997) |
| 0.3 | 1.4 | GO:0070051 | fibrinogen binding(GO:0070051) |
| 0.3 | 1.1 | GO:0050252 | retinol O-fatty-acyltransferase activity(GO:0050252) |
| 0.3 | 2.2 | GO:0071074 | eukaryotic initiation factor eIF2 binding(GO:0071074) |
| 0.3 | 1.1 | GO:0042978 | ornithine decarboxylase activator activity(GO:0042978) |
| 0.3 | 3.4 | GO:0004571 | mannosyl-oligosaccharide 1,2-alpha-mannosidase activity(GO:0004571) |
| 0.2 | 2.3 | GO:0070095 | fructose-6-phosphate binding(GO:0070095) |
| 0.2 | 3.4 | GO:0031957 | very long-chain fatty acid-CoA ligase activity(GO:0031957) |
| 0.2 | 2.0 | GO:0031849 | olfactory receptor binding(GO:0031849) |
| 0.2 | 1.5 | GO:0052650 | NADP-retinol dehydrogenase activity(GO:0052650) |
| 0.2 | 1.7 | GO:0004046 | aminoacylase activity(GO:0004046) |
| 0.2 | 8.8 | GO:0005385 | zinc ion transmembrane transporter activity(GO:0005385) |
| 0.2 | 1.9 | GO:0051575 | 5'-deoxyribose-5-phosphate lyase activity(GO:0051575) |
| 0.2 | 2.5 | GO:0004499 | N,N-dimethylaniline monooxygenase activity(GO:0004499) |
| 0.2 | 0.8 | GO:0050501 | hyaluronan synthase activity(GO:0050501) |
| 0.2 | 1.2 | GO:0008384 | IkappaB kinase activity(GO:0008384) |
| 0.2 | 1.6 | GO:0047631 | ADP-ribose diphosphatase activity(GO:0047631) |
| 0.2 | 1.2 | GO:0051425 | PTB domain binding(GO:0051425) |
| 0.2 | 2.3 | GO:0097027 | ubiquitin-protein transferase activator activity(GO:0097027) |
| 0.2 | 5.4 | GO:0016638 | oxidoreductase activity, acting on the CH-NH2 group of donors(GO:0016638) |
| 0.2 | 1.2 | GO:0003857 | 3-hydroxyacyl-CoA dehydrogenase activity(GO:0003857) NADH binding(GO:0070404) |
| 0.2 | 2.1 | GO:0001730 | 2'-5'-oligoadenylate synthetase activity(GO:0001730) |
| 0.2 | 1.2 | GO:0072510 | ferric iron transmembrane transporter activity(GO:0015091) trivalent inorganic cation transmembrane transporter activity(GO:0072510) |
| 0.2 | 4.6 | GO:0042975 | peroxisome proliferator activated receptor binding(GO:0042975) |
| 0.2 | 0.6 | GO:0033883 | pyridoxal phosphatase activity(GO:0033883) |
| 0.2 | 0.8 | GO:0004051 | arachidonate 5-lipoxygenase activity(GO:0004051) |
| 0.2 | 0.9 | GO:0019966 | interleukin-1 binding(GO:0019966) |
| 0.2 | 2.0 | GO:0004602 | glutathione peroxidase activity(GO:0004602) |
| 0.2 | 3.8 | GO:0016702 | oxidoreductase activity, acting on single donors with incorporation of molecular oxygen, incorporation of two atoms of oxygen(GO:0016702) |
| 0.2 | 4.2 | GO:0005161 | platelet-derived growth factor receptor binding(GO:0005161) |
| 0.1 | 24.8 | GO:0004867 | serine-type endopeptidase inhibitor activity(GO:0004867) |
| 0.1 | 0.4 | GO:0003826 | alpha-ketoacid dehydrogenase activity(GO:0003826) 3-methyl-2-oxobutanoate dehydrogenase (2-methylpropanoyl-transferring) activity(GO:0003863) |
| 0.1 | 0.6 | GO:0046624 | sphingolipid transporter activity(GO:0046624) |
| 0.1 | 1.3 | GO:0015165 | pyrimidine nucleotide-sugar transmembrane transporter activity(GO:0015165) |
| 0.1 | 5.0 | GO:0008392 | arachidonic acid epoxygenase activity(GO:0008392) |
| 0.1 | 1.0 | GO:0032027 | myosin light chain binding(GO:0032027) |
| 0.1 | 2.2 | GO:0045294 | alpha-catenin binding(GO:0045294) |
| 0.1 | 4.1 | GO:0015238 | drug transmembrane transporter activity(GO:0015238) |
| 0.1 | 0.9 | GO:0047179 | platelet-activating factor acetyltransferase activity(GO:0047179) |
| 0.1 | 1.8 | GO:0045236 | CXCR chemokine receptor binding(GO:0045236) |
| 0.1 | 3.2 | GO:0030898 | actin-dependent ATPase activity(GO:0030898) |
| 0.1 | 3.1 | GO:0070006 | metalloaminopeptidase activity(GO:0070006) |
| 0.1 | 1.6 | GO:0050692 | DBD domain binding(GO:0050692) |
| 0.1 | 4.4 | GO:0008395 | steroid hydroxylase activity(GO:0008395) |
| 0.1 | 1.3 | GO:0001162 | RNA polymerase II intronic transcription regulatory region sequence-specific DNA binding(GO:0001162) |
| 0.1 | 4.5 | GO:0031492 | nucleosomal DNA binding(GO:0031492) |
| 0.1 | 2.0 | GO:0001846 | opsonin binding(GO:0001846) |
| 0.1 | 5.4 | GO:0004364 | glutathione transferase activity(GO:0004364) |
| 0.1 | 0.4 | GO:0033797 | selenate reductase activity(GO:0033797) |
| 0.1 | 1.2 | GO:0051525 | NFAT protein binding(GO:0051525) |
| 0.1 | 6.6 | GO:0015485 | cholesterol binding(GO:0015485) |
| 0.1 | 2.9 | GO:0008009 | chemokine activity(GO:0008009) |
| 0.1 | 5.3 | GO:0004864 | protein phosphatase inhibitor activity(GO:0004864) |
| 0.1 | 0.4 | GO:0019779 | Atg12 activating enzyme activity(GO:0019778) Atg8 activating enzyme activity(GO:0019779) |
| 0.1 | 0.6 | GO:0047288 | monosialoganglioside sialyltransferase activity(GO:0047288) |
| 0.1 | 1.0 | GO:0008494 | translation activator activity(GO:0008494) |
| 0.1 | 0.4 | GO:0004307 | ethanolaminephosphotransferase activity(GO:0004307) |
| 0.1 | 7.8 | GO:0004879 | RNA polymerase II transcription factor activity, ligand-activated sequence-specific DNA binding(GO:0004879) transcription factor activity, direct ligand regulated sequence-specific DNA binding(GO:0098531) |
| 0.1 | 1.7 | GO:0008191 | metalloendopeptidase inhibitor activity(GO:0008191) |
| 0.1 | 1.2 | GO:0008430 | selenium binding(GO:0008430) |
| 0.1 | 0.4 | GO:0004315 | 3-oxoacyl-[acyl-carrier-protein] synthase activity(GO:0004315) |
| 0.1 | 1.1 | GO:0042608 | T cell receptor binding(GO:0042608) |
| 0.1 | 1.5 | GO:0003796 | lysozyme activity(GO:0003796) |
| 0.1 | 5.2 | GO:0005070 | SH3/SH2 adaptor activity(GO:0005070) |
| 0.1 | 0.8 | GO:0015245 | fatty acid transporter activity(GO:0015245) |
| 0.1 | 1.3 | GO:0038036 | sphingosine-1-phosphate receptor activity(GO:0038036) |
| 0.1 | 11.8 | GO:0005200 | structural constituent of cytoskeleton(GO:0005200) |
| 0.1 | 0.8 | GO:0070053 | thrombospondin receptor activity(GO:0070053) |
| 0.1 | 0.6 | GO:0033592 | RNA strand annealing activity(GO:0033592) |
| 0.1 | 1.1 | GO:0005003 | ephrin receptor activity(GO:0005003) |
| 0.1 | 0.6 | GO:0015349 | thyroid hormone transmembrane transporter activity(GO:0015349) |
| 0.1 | 1.0 | GO:0030346 | protein phosphatase 2B binding(GO:0030346) |
| 0.1 | 1.4 | GO:0098748 | clathrin adaptor activity(GO:0035615) endocytic adaptor activity(GO:0098748) |
| 0.1 | 2.6 | GO:0019200 | carbohydrate kinase activity(GO:0019200) |
| 0.1 | 1.7 | GO:0003746 | translation elongation factor activity(GO:0003746) |
| 0.1 | 3.9 | GO:0043027 | cysteine-type endopeptidase inhibitor activity involved in apoptotic process(GO:0043027) |
| 0.1 | 0.7 | GO:0000774 | adenyl-nucleotide exchange factor activity(GO:0000774) |
| 0.1 | 0.5 | GO:0001517 | N-acetylglucosamine 6-O-sulfotransferase activity(GO:0001517) |
| 0.1 | 0.7 | GO:1990825 | sequence-specific mRNA binding(GO:1990825) |
| 0.1 | 2.9 | GO:0005310 | dicarboxylic acid transmembrane transporter activity(GO:0005310) |
| 0.1 | 1.3 | GO:0051864 | histone demethylase activity (H3-K36 specific)(GO:0051864) |
| 0.1 | 0.2 | GO:0051022 | Rho GDP-dissociation inhibitor binding(GO:0051022) |
| 0.1 | 0.4 | GO:0004030 | aldehyde dehydrogenase [NAD(P)+] activity(GO:0004030) |
| 0.1 | 0.6 | GO:0098599 | palmitoyl-(protein) hydrolase activity(GO:0008474) palmitoyl hydrolase activity(GO:0098599) |
| 0.1 | 0.3 | GO:0004525 | ribonuclease III activity(GO:0004525) double-stranded RNA-specific ribonuclease activity(GO:0032296) |
| 0.1 | 1.3 | GO:0004535 | poly(A)-specific ribonuclease activity(GO:0004535) |
| 0.1 | 3.0 | GO:0016209 | antioxidant activity(GO:0016209) |
| 0.1 | 0.8 | GO:0004438 | phosphatidylinositol-3-phosphatase activity(GO:0004438) |
| 0.1 | 6.3 | GO:0005254 | chloride channel activity(GO:0005254) |
| 0.1 | 5.4 | GO:0005518 | collagen binding(GO:0005518) |
| 0.1 | 4.1 | GO:0004715 | non-membrane spanning protein tyrosine kinase activity(GO:0004715) |
| 0.0 | 0.7 | GO:1990829 | C-rich single-stranded DNA binding(GO:1990829) |
| 0.0 | 3.1 | GO:0005160 | transforming growth factor beta receptor binding(GO:0005160) |
| 0.0 | 0.3 | GO:0004673 | protein histidine kinase activity(GO:0004673) |
| 0.0 | 0.9 | GO:0016864 | protein disulfide isomerase activity(GO:0003756) intramolecular oxidoreductase activity, transposing S-S bonds(GO:0016864) |
| 0.0 | 1.3 | GO:0017025 | TBP-class protein binding(GO:0017025) |
| 0.0 | 0.9 | GO:0016423 | tRNA (guanine) methyltransferase activity(GO:0016423) |
| 0.0 | 2.7 | GO:0030414 | peptidase inhibitor activity(GO:0030414) |
| 0.0 | 1.2 | GO:0001871 | pattern binding(GO:0001871) polysaccharide binding(GO:0030247) |
| 0.0 | 1.1 | GO:0008574 | ATP-dependent microtubule motor activity, plus-end-directed(GO:0008574) |
| 0.0 | 0.3 | GO:0043023 | ribosomal large subunit binding(GO:0043023) |
| 0.0 | 0.3 | GO:0015129 | lactate transmembrane transporter activity(GO:0015129) |
| 0.0 | 2.0 | GO:0017112 | Rab guanyl-nucleotide exchange factor activity(GO:0017112) |
| 0.0 | 0.2 | GO:0034513 | box H/ACA snoRNA binding(GO:0034513) |
| 0.0 | 4.4 | GO:0051082 | unfolded protein binding(GO:0051082) |
| 0.0 | 0.5 | GO:0003995 | acyl-CoA dehydrogenase activity(GO:0003995) |
| 0.0 | 0.7 | GO:0016922 | ligand-dependent nuclear receptor binding(GO:0016922) |
| 0.0 | 1.1 | GO:0003887 | DNA-directed DNA polymerase activity(GO:0003887) |
| 0.0 | 3.4 | GO:1990782 | protein tyrosine kinase binding(GO:1990782) |
| 0.0 | 0.6 | GO:0030169 | low-density lipoprotein particle binding(GO:0030169) |
| 0.0 | 2.3 | GO:0004402 | histone acetyltransferase activity(GO:0004402) |
| 0.0 | 3.6 | GO:0004222 | metalloendopeptidase activity(GO:0004222) |
| 0.0 | 1.5 | GO:0008235 | metalloexopeptidase activity(GO:0008235) |
| 0.0 | 0.5 | GO:0019865 | immunoglobulin binding(GO:0019865) |
| 0.0 | 0.8 | GO:0008143 | poly(A) binding(GO:0008143) |
| 0.0 | 1.4 | GO:0008536 | Ran GTPase binding(GO:0008536) |
| 0.0 | 0.5 | GO:0016500 | protein-hormone receptor activity(GO:0016500) |
| 0.0 | 0.5 | GO:0009982 | pseudouridine synthase activity(GO:0009982) |
| 0.0 | 0.1 | GO:0030621 | U4 snRNA binding(GO:0030621) |
| 0.0 | 0.2 | GO:0061665 | SUMO ligase activity(GO:0061665) |
| 0.0 | 1.8 | GO:0008146 | sulfotransferase activity(GO:0008146) |
| 0.0 | 0.4 | GO:0070742 | C2H2 zinc finger domain binding(GO:0070742) |
| 0.0 | 6.0 | GO:0004252 | serine-type endopeptidase activity(GO:0004252) |
| 0.0 | 0.2 | GO:0015199 | amino-acid betaine transmembrane transporter activity(GO:0015199) carnitine transmembrane transporter activity(GO:0015226) |
| 0.0 | 3.0 | GO:0005550 | pheromone binding(GO:0005550) |
| 0.0 | 17.7 | GO:0001159 | core promoter proximal region DNA binding(GO:0001159) |
| 0.0 | 0.5 | GO:0043395 | heparan sulfate proteoglycan binding(GO:0043395) |
| 0.0 | 0.4 | GO:0034185 | apolipoprotein binding(GO:0034185) |
| 0.0 | 0.1 | GO:0004174 | electron-transferring-flavoprotein dehydrogenase activity(GO:0004174) oxidoreductase activity, acting on the CH-NH group of donors, quinone or similar compound as acceptor(GO:0016649) |
| 0.0 | 0.4 | GO:0016290 | palmitoyl-CoA hydrolase activity(GO:0016290) |
| 0.0 | 3.0 | GO:0005179 | hormone activity(GO:0005179) |
| 0.0 | 1.1 | GO:0001078 | transcriptional repressor activity, RNA polymerase II core promoter proximal region sequence-specific binding(GO:0001078) |
| 0.0 | 0.1 | GO:0072345 | NAADP-sensitive calcium-release channel activity(GO:0072345) |
| 0.0 | 0.2 | GO:0001055 | RNA polymerase II activity(GO:0001055) |
| 0.0 | 0.1 | GO:0003839 | gamma-glutamylcyclotransferase activity(GO:0003839) |
| 0.0 | 1.8 | GO:0042826 | histone deacetylase binding(GO:0042826) |
| 0.0 | 0.1 | GO:0004832 | valine-tRNA ligase activity(GO:0004832) |
| 0.0 | 1.2 | GO:0002039 | p53 binding(GO:0002039) |
| 0.0 | 0.1 | GO:0030911 | TPR domain binding(GO:0030911) |
| 0.0 | 0.1 | GO:0070290 | N-acylphosphatidylethanolamine-specific phospholipase D activity(GO:0070290) |
| 0.0 | 0.3 | GO:0001222 | transcription corepressor binding(GO:0001222) |
| 0.0 | 0.2 | GO:0015143 | urate transmembrane transporter activity(GO:0015143) salt transmembrane transporter activity(GO:1901702) |
| 0.0 | 0.4 | GO:0016780 | phosphotransferase activity, for other substituted phosphate groups(GO:0016780) |
| 0.0 | 0.2 | GO:0004089 | carbonate dehydratase activity(GO:0004089) |
| 0.0 | 0.2 | GO:0002162 | dystroglycan binding(GO:0002162) |
| 0.0 | 0.7 | GO:0008138 | protein tyrosine/serine/threonine phosphatase activity(GO:0008138) |
| 0.0 | 0.1 | GO:0017040 | ceramidase activity(GO:0017040) |
| 0.0 | 2.6 | GO:0001228 | transcriptional activator activity, RNA polymerase II transcription regulatory region sequence-specific binding(GO:0001228) |
| 0.0 | 0.5 | GO:0005201 | extracellular matrix structural constituent(GO:0005201) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 2.5 | PID IL6 7 PATHWAY | IL6-mediated signaling events |
| 0.2 | 2.8 | PID ERBB2 ERBB3 PATHWAY | ErbB2/ErbB3 signaling events |
| 0.2 | 9.7 | PID EPHA FWDPATHWAY | EPHA forward signaling |
| 0.2 | 4.0 | PID SMAD2 3PATHWAY | Regulation of cytoplasmic and nuclear SMAD2/3 signaling |
| 0.1 | 6.1 | SA B CELL RECEPTOR COMPLEXES | Antigen binding to B cell receptors activates protein tyrosine kinases, such as the Src family, which ultimate activate MAP kinases. |
| 0.1 | 7.0 | PID FOXM1 PATHWAY | FOXM1 transcription factor network |
| 0.1 | 10.0 | PID HNF3B PATHWAY | FOXA2 and FOXA3 transcription factor networks |
| 0.1 | 4.2 | SA CASPASE CASCADE | Apoptosis is mediated by caspases, cysteine proteases arranged in a proteolytic cascade. |
| 0.1 | 1.6 | PID S1P S1P4 PATHWAY | S1P4 pathway |
| 0.1 | 30.9 | NABA ECM REGULATORS | Genes encoding enzymes and their regulators involved in the remodeling of the extracellular matrix |
| 0.1 | 2.7 | PID AVB3 OPN PATHWAY | Osteopontin-mediated events |
| 0.1 | 5.3 | PID EPHB FWD PATHWAY | EPHB forward signaling |
| 0.1 | 5.2 | PID RXR VDR PATHWAY | RXR and RAR heterodimerization with other nuclear receptor |
| 0.1 | 3.5 | PID IL2 PI3K PATHWAY | IL2 signaling events mediated by PI3K |
| 0.1 | 3.2 | PID IL3 PATHWAY | IL3-mediated signaling events |
| 0.1 | 2.2 | PID DNA PK PATHWAY | DNA-PK pathway in nonhomologous end joining |
| 0.1 | 3.8 | NABA PROTEOGLYCANS | Genes encoding proteoglycans |
| 0.1 | 1.3 | PID INTEGRIN4 PATHWAY | Alpha6 beta4 integrin-ligand interactions |
| 0.1 | 5.4 | PID RAC1 PATHWAY | RAC1 signaling pathway |
| 0.1 | 5.7 | PID TGFBR PATHWAY | TGF-beta receptor signaling |
| 0.1 | 1.3 | PID HNF3A PATHWAY | FOXA1 transcription factor network |
| 0.1 | 1.9 | PID IL1 PATHWAY | IL1-mediated signaling events |
| 0.0 | 2.8 | PID PLK1 PATHWAY | PLK1 signaling events |
| 0.0 | 0.8 | PID INTEGRIN3 PATHWAY | Beta3 integrin cell surface interactions |
| 0.0 | 2.5 | PID AR TF PATHWAY | Regulation of Androgen receptor activity |
| 0.0 | 2.2 | PID BCR 5PATHWAY | BCR signaling pathway |
| 0.0 | 1.0 | PID IL8 CXCR2 PATHWAY | IL8- and CXCR2-mediated signaling events |
| 0.0 | 1.8 | PID CXCR4 PATHWAY | CXCR4-mediated signaling events |
| 0.0 | 1.0 | SIG CHEMOTAXIS | Genes related to chemotaxis |
| 0.0 | 1.5 | PID TAP63 PATHWAY | Validated transcriptional targets of TAp63 isoforms |
| 0.0 | 0.6 | ST PHOSPHOINOSITIDE 3 KINASE PATHWAY | PI3K Pathway |
| 0.0 | 1.2 | PID CDC42 REG PATHWAY | Regulation of CDC42 activity |
| 0.0 | 1.0 | PID CERAMIDE PATHWAY | Ceramide signaling pathway |
| 0.0 | 0.3 | PID INSULIN GLUCOSE PATHWAY | Insulin-mediated glucose transport |
| 0.0 | 0.5 | PID BARD1 PATHWAY | BARD1 signaling events |
| 0.0 | 1.1 | PID TELOMERASE PATHWAY | Regulation of Telomerase |
| 0.0 | 0.2 | PID EPHRINB REV PATHWAY | Ephrin B reverse signaling |
| 0.0 | 0.5 | PID LIS1 PATHWAY | Lissencephaly gene (LIS1) in neuronal migration and development |
| 0.0 | 0.4 | PID UPA UPAR PATHWAY | Urokinase-type plasminogen activator (uPA) and uPAR-mediated signaling |
| 0.0 | 0.8 | PID P53 REGULATION PATHWAY | p53 pathway |
| 0.0 | 1.3 | PID ERA GENOMIC PATHWAY | Validated nuclear estrogen receptor alpha network |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.6 | 7.0 | REACTOME ETHANOL OXIDATION | Genes involved in Ethanol oxidation |
| 0.6 | 8.3 | REACTOME TRYPTOPHAN CATABOLISM | Genes involved in Tryptophan catabolism |
| 0.5 | 11.8 | REACTOME SYNTHESIS OF BILE ACIDS AND BILE SALTS VIA 7ALPHA HYDROXYCHOLESTEROL | Genes involved in Synthesis of bile acids and bile salts via 7alpha-hydroxycholesterol |
| 0.4 | 12.0 | REACTOME CHYLOMICRON MEDIATED LIPID TRANSPORT | Genes involved in Chylomicron-mediated lipid transport |
| 0.3 | 9.1 | REACTOME ZINC TRANSPORTERS | Genes involved in Zinc transporters |
| 0.3 | 1.0 | REACTOME TRANSPORT OF RIBONUCLEOPROTEINS INTO THE HOST NUCLEUS | Genes involved in Transport of Ribonucleoproteins into the Host Nucleus |
| 0.3 | 8.5 | REACTOME TIE2 SIGNALING | Genes involved in Tie2 Signaling |
| 0.3 | 5.9 | REACTOME APOPTOTIC CLEAVAGE OF CELL ADHESION PROTEINS | Genes involved in Apoptotic cleavage of cell adhesion proteins |
| 0.3 | 4.3 | REACTOME CALNEXIN CALRETICULIN CYCLE | Genes involved in Calnexin/calreticulin cycle |
| 0.2 | 2.0 | REACTOME CREATION OF C4 AND C2 ACTIVATORS | Genes involved in Creation of C4 and C2 activators |
| 0.2 | 1.9 | REACTOME GLYCOPROTEIN HORMONES | Genes involved in Glycoprotein hormones |
| 0.2 | 4.3 | REACTOME PYRIMIDINE CATABOLISM | Genes involved in Pyrimidine catabolism |
| 0.2 | 6.1 | REACTOME APC CDC20 MEDIATED DEGRADATION OF NEK2A | Genes involved in APC-Cdc20 mediated degradation of Nek2A |
| 0.2 | 2.9 | REACTOME GAMMA CARBOXYLATION TRANSPORT AND AMINO TERMINAL CLEAVAGE OF PROTEINS | Genes involved in Gamma-carboxylation, transport, and amino-terminal cleavage of proteins |
| 0.2 | 3.0 | REACTOME ABACAVIR TRANSPORT AND METABOLISM | Genes involved in Abacavir transport and metabolism |
| 0.2 | 7.7 | REACTOME FATTY ACYL COA BIOSYNTHESIS | Genes involved in Fatty Acyl-CoA Biosynthesis |
| 0.2 | 8.5 | REACTOME METABOLISM OF STEROID HORMONES AND VITAMINS A AND D | Genes involved in Metabolism of steroid hormones and vitamins A and D |
| 0.1 | 1.3 | REACTOME ACTIVATION OF CHAPERONE GENES BY ATF6 ALPHA | Genes involved in Activation of Chaperone Genes by ATF6-alpha |
| 0.1 | 1.3 | REACTOME ALPHA LINOLENIC ACID ALA METABOLISM | Genes involved in alpha-linolenic acid (ALA) metabolism |
| 0.1 | 7.5 | REACTOME REGULATION OF BETA CELL DEVELOPMENT | Genes involved in Regulation of beta-cell development |
| 0.1 | 7.1 | REACTOME MEIOTIC SYNAPSIS | Genes involved in Meiotic Synapsis |
| 0.1 | 2.3 | REACTOME METABOLISM OF POLYAMINES | Genes involved in Metabolism of polyamines |
| 0.1 | 4.8 | REACTOME REGULATION OF GLUCOKINASE BY GLUCOKINASE REGULATORY PROTEIN | Genes involved in Regulation of Glucokinase by Glucokinase Regulatory Protein |
| 0.1 | 2.2 | REACTOME ACYL CHAIN REMODELLING OF PS | Genes involved in Acyl chain remodelling of PS |
| 0.1 | 1.3 | REACTOME GLUCURONIDATION | Genes involved in Glucuronidation |
| 0.1 | 1.9 | REACTOME INTEGRATION OF PROVIRUS | Genes involved in Integration of provirus |
| 0.1 | 1.5 | REACTOME REGULATION OF PYRUVATE DEHYDROGENASE PDH COMPLEX | Genes involved in Regulation of pyruvate dehydrogenase (PDH) complex |
| 0.1 | 2.0 | REACTOME BILE SALT AND ORGANIC ANION SLC TRANSPORTERS | Genes involved in Bile salt and organic anion SLC transporters |
| 0.1 | 1.8 | REACTOME ASSOCIATION OF LICENSING FACTORS WITH THE PRE REPLICATIVE COMPLEX | Genes involved in Association of licensing factors with the pre-replicative complex |
| 0.1 | 3.5 | REACTOME IL RECEPTOR SHC SIGNALING | Genes involved in Interleukin receptor SHC signaling |
| 0.1 | 1.4 | REACTOME PECAM1 INTERACTIONS | Genes involved in PECAM1 interactions |
| 0.1 | 7.7 | REACTOME UNFOLDED PROTEIN RESPONSE | Genes involved in Unfolded Protein Response |
| 0.1 | 1.2 | REACTOME IRAK1 RECRUITS IKK COMPLEX | Genes involved in IRAK1 recruits IKK complex |
| 0.1 | 2.2 | REACTOME REGULATION OF SIGNALING BY CBL | Genes involved in Regulation of signaling by CBL |
| 0.1 | 0.8 | REACTOME PURINE CATABOLISM | Genes involved in Purine catabolism |
| 0.1 | 1.2 | REACTOME PURINE RIBONUCLEOSIDE MONOPHOSPHATE BIOSYNTHESIS | Genes involved in Purine ribonucleoside monophosphate biosynthesis |
| 0.1 | 0.8 | REACTOME HYALURONAN UPTAKE AND DEGRADATION | Genes involved in Hyaluronan uptake and degradation |
| 0.1 | 1.8 | REACTOME SULFUR AMINO ACID METABOLISM | Genes involved in Sulfur amino acid metabolism |
| 0.1 | 2.9 | REACTOME SEMA4D IN SEMAPHORIN SIGNALING | Genes involved in Sema4D in semaphorin signaling |
| 0.1 | 1.0 | REACTOME CYCLIN A B1 ASSOCIATED EVENTS DURING G2 M TRANSITION | Genes involved in Cyclin A/B1 associated events during G2/M transition |
| 0.1 | 5.7 | REACTOME NUCLEAR RECEPTOR TRANSCRIPTION PATHWAY | Genes involved in Nuclear Receptor transcription pathway |
| 0.1 | 1.5 | REACTOME PRE NOTCH TRANSCRIPTION AND TRANSLATION | Genes involved in Pre-NOTCH Transcription and Translation |
| 0.1 | 2.7 | REACTOME G1 PHASE | Genes involved in G1 Phase |
| 0.1 | 0.7 | REACTOME SLBP DEPENDENT PROCESSING OF REPLICATION DEPENDENT HISTONE PRE MRNAS | Genes involved in SLBP Dependent Processing of Replication-Dependent Histone Pre-mRNAs |
| 0.0 | 3.4 | REACTOME PHASE II CONJUGATION | Genes involved in Phase II conjugation |
| 0.0 | 1.9 | REACTOME IL1 SIGNALING | Genes involved in Interleukin-1 signaling |
| 0.0 | 8.7 | REACTOME METABOLISM OF AMINO ACIDS AND DERIVATIVES | Genes involved in Metabolism of amino acids and derivatives |
| 0.0 | 0.9 | REACTOME VIRAL MESSENGER RNA SYNTHESIS | Genes involved in Viral Messenger RNA Synthesis |
| 0.0 | 1.8 | REACTOME CHEMOKINE RECEPTORS BIND CHEMOKINES | Genes involved in Chemokine receptors bind chemokines |
| 0.0 | 1.6 | REACTOME INTRINSIC PATHWAY FOR APOPTOSIS | Genes involved in Intrinsic Pathway for Apoptosis |
| 0.0 | 0.7 | REACTOME PEROXISOMAL LIPID METABOLISM | Genes involved in Peroxisomal lipid metabolism |
| 0.0 | 0.3 | REACTOME MICRORNA MIRNA BIOGENESIS | Genes involved in MicroRNA (miRNA) Biogenesis |
| 0.0 | 2.1 | REACTOME COLLAGEN FORMATION | Genes involved in Collagen formation |
| 0.0 | 2.3 | REACTOME MITOTIC PROMETAPHASE | Genes involved in Mitotic Prometaphase |
| 0.0 | 3.5 | REACTOME RESPONSE TO ELEVATED PLATELET CYTOSOLIC CA2 | Genes involved in Response to elevated platelet cytosolic Ca2+ |
| 0.0 | 1.2 | REACTOME TIGHT JUNCTION INTERACTIONS | Genes involved in Tight junction interactions |
| 0.0 | 1.7 | REACTOME INTERFERON ALPHA BETA SIGNALING | Genes involved in Interferon alpha/beta signaling |
| 0.0 | 3.8 | REACTOME PEPTIDE LIGAND BINDING RECEPTORS | Genes involved in Peptide ligand-binding receptors |
| 0.0 | 0.7 | REACTOME SIGNALING BY FGFR1 FUSION MUTANTS | Genes involved in Signaling by FGFR1 fusion mutants |
| 0.0 | 0.5 | REACTOME FANCONI ANEMIA PATHWAY | Genes involved in Fanconi Anemia pathway |
| 0.0 | 0.7 | REACTOME SYNTHESIS AND INTERCONVERSION OF NUCLEOTIDE DI AND TRIPHOSPHATES | Genes involved in Synthesis and interconversion of nucleotide di- and triphosphates |
| 0.0 | 1.8 | REACTOME CELL JUNCTION ORGANIZATION | Genes involved in Cell junction organization |
| 0.0 | 1.1 | REACTOME CLEAVAGE OF GROWING TRANSCRIPT IN THE TERMINATION REGION | Genes involved in Cleavage of Growing Transcript in the Termination Region |
| 0.0 | 1.1 | REACTOME CREB PHOSPHORYLATION THROUGH THE ACTIVATION OF RAS | Genes involved in CREB phosphorylation through the activation of Ras |
| 0.0 | 1.4 | REACTOME METABOLISM OF VITAMINS AND COFACTORS | Genes involved in Metabolism of vitamins and cofactors |
| 0.0 | 2.1 | REACTOME PEPTIDE CHAIN ELONGATION | Genes involved in Peptide chain elongation |
| 0.0 | 1.6 | REACTOME MRNA SPLICING | Genes involved in mRNA Splicing |
| 0.0 | 0.5 | REACTOME BIOSYNTHESIS OF THE N GLYCAN PRECURSOR DOLICHOL LIPID LINKED OLIGOSACCHARIDE LLO AND TRANSFER TO A NASCENT PROTEIN | Genes involved in Biosynthesis of the N-glycan precursor (dolichol lipid-linked oligosaccharide, LLO) and transfer to a nascent protein |
| 0.0 | 0.2 | REACTOME REMOVAL OF THE FLAP INTERMEDIATE FROM THE C STRAND | Genes involved in Removal of the Flap Intermediate from the C-strand |
| 0.0 | 0.4 | REACTOME SYNTHESIS OF GLYCOSYLPHOSPHATIDYLINOSITOL GPI | Genes involved in Synthesis of glycosylphosphatidylinositol (GPI) |
| 0.0 | 2.8 | REACTOME SIGNALING BY RHO GTPASES | Genes involved in Signaling by Rho GTPases |
| 0.0 | 1.1 | REACTOME TRANSLATION | Genes involved in Translation |
| 0.0 | 0.2 | REACTOME DEADENYLATION OF MRNA | Genes involved in Deadenylation of mRNA |
| 0.0 | 1.4 | REACTOME FACTORS INVOLVED IN MEGAKARYOCYTE DEVELOPMENT AND PLATELET PRODUCTION | Genes involved in Factors involved in megakaryocyte development and platelet production |