Project
PRJNA375882: Comprehensive Mouse Transcriptomic BodyMap
Navigation
Downloads
Results for Nr2e1
Z-value: 1.84
Transcription factors associated with Nr2e1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Nr2e1
|
ENSMUSG00000019803.12 | Nr2e1 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Nr2e1 | mm39_v1_chr10_-_42459624_42459635 | -0.12 | 3.1e-01 | Click! |
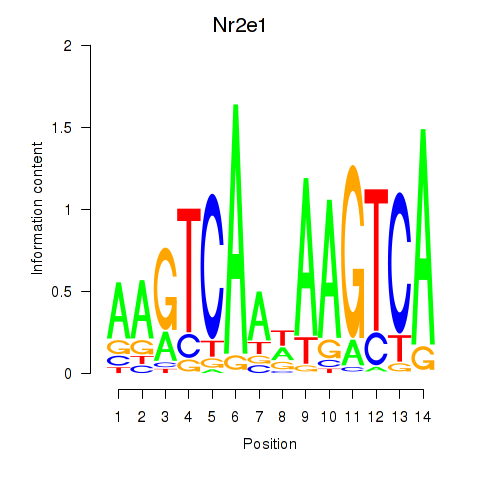
Activity profile of Nr2e1 motif
Sorted Z-values of Nr2e1 motif
Network of associatons between targets according to the STRING database.
First level regulatory network of Nr2e1
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr4_-_60457902 | 38.06 |
ENSMUST00000084548.11
ENSMUST00000103012.10 ENSMUST00000107499.4 |
Mup1
|
major urinary protein 1 |
| chr4_-_62005498 | 34.84 |
ENSMUST00000107488.4
ENSMUST00000107472.8 ENSMUST00000084531.11 |
Mup3
|
major urinary protein 3 |
| chr4_-_60777462 | 30.91 |
ENSMUST00000211875.2
|
Mup22
|
major urinary protein 22 |
| chr4_-_61437704 | 29.88 |
ENSMUST00000095051.6
ENSMUST00000107483.8 |
Mup16
|
major urinary protein 16 |
| chr4_-_60697274 | 27.04 |
ENSMUST00000117932.2
|
Mup12
|
major urinary protein 12 |
| chr3_+_96432479 | 25.55 |
ENSMUST00000049208.11
|
Hjv
|
hemojuvelin BMP co-receptor |
| chr4_-_61259997 | 22.94 |
ENSMUST00000071005.9
ENSMUST00000075206.12 |
Mup14
|
major urinary protein 14 |
| chr4_-_61259801 | 22.49 |
ENSMUST00000125461.8
|
Mup14
|
major urinary protein 14 |
| chr6_-_141892517 | 18.79 |
ENSMUST00000168119.8
|
Slco1a1
|
solute carrier organic anion transporter family, member 1a1 |
| chr3_+_130411294 | 18.10 |
ENSMUST00000163620.8
|
Etnppl
|
ethanolamine phosphate phospholyase |
| chr9_-_71070506 | 18.05 |
ENSMUST00000074465.9
|
Aqp9
|
aquaporin 9 |
| chr4_-_62069046 | 17.74 |
ENSMUST00000077719.4
|
Mup21
|
major urinary protein 21 |
| chr5_-_87288177 | 16.96 |
ENSMUST00000067790.7
|
Ugt2b5
|
UDP glucuronosyltransferase 2 family, polypeptide B5 |
| chr17_+_32904629 | 15.48 |
ENSMUST00000008801.7
|
Cyp4f15
|
cytochrome P450, family 4, subfamily f, polypeptide 15 |
| chr17_+_32904601 | 14.94 |
ENSMUST00000168171.8
|
Cyp4f15
|
cytochrome P450, family 4, subfamily f, polypeptide 15 |
| chr5_-_87054796 | 14.84 |
ENSMUST00000031181.16
ENSMUST00000113333.2 |
Ugt2b34
|
UDP glucuronosyltransferase 2 family, polypeptide B34 |
| chr13_+_24023428 | 13.59 |
ENSMUST00000091698.12
ENSMUST00000110422.3 ENSMUST00000166467.9 |
Slc17a3
|
solute carrier family 17 (sodium phosphate), member 3 |
| chr7_+_114367971 | 12.11 |
ENSMUST00000117543.3
ENSMUST00000151464.2 |
Insc
|
INSC spindle orientation adaptor protein |
| chr13_+_24023386 | 9.14 |
ENSMUST00000039721.14
|
Slc17a3
|
solute carrier family 17 (sodium phosphate), member 3 |
| chr12_+_86781141 | 8.96 |
ENSMUST00000223308.2
|
Lrrc74a
|
leucine rich repeat containing 74A |
| chr12_+_86781154 | 8.07 |
ENSMUST00000095527.6
|
Lrrc74a
|
leucine rich repeat containing 74A |
| chr19_-_11261177 | 6.19 |
ENSMUST00000186937.7
ENSMUST00000067673.13 |
Ms4a5
|
membrane-spanning 4-domains, subfamily A, member 5 |
| chr14_-_33737131 | 5.99 |
ENSMUST00000227795.2
|
Gm30083
|
predicted gene, 30083 |
| chr4_+_60003438 | 5.85 |
ENSMUST00000107517.8
ENSMUST00000107520.2 |
Mup6
|
major urinary protein 6 |
| chr14_+_28740162 | 5.76 |
ENSMUST00000055662.4
|
Lrtm1
|
leucine-rich repeats and transmembrane domains 1 |
| chr8_+_71156071 | 5.42 |
ENSMUST00000212436.2
|
Iqcn
|
IQ motif containing N |
| chr18_-_60881405 | 4.89 |
ENSMUST00000237070.2
|
Ndst1
|
N-deacetylase/N-sulfotransferase (heparan glucosaminyl) 1 |
| chr1_-_75196496 | 4.13 |
ENSMUST00000186758.7
|
Tuba4a
|
tubulin, alpha 4A |
| chr14_+_33775423 | 3.87 |
ENSMUST00000058725.5
|
Antxrl
|
anthrax toxin receptor-like |
| chrX_+_30080456 | 3.79 |
ENSMUST00000177917.2
|
Gm14632
|
predicted gene 14632 |
| chrX_-_30540953 | 3.79 |
ENSMUST00000177566.2
|
Gm10487
|
predicted gene 10487 |
| chrX_+_34669447 | 3.75 |
ENSMUST00000169396.3
|
Gm14819
|
predicted gene 14819 |
| chrX_-_27378797 | 3.75 |
ENSMUST00000178745.2
|
Gm10230
|
predicted gene 10230 |
| chrX_-_28386687 | 3.75 |
ENSMUST00000178966.2
|
Gm10147
|
predicted gene 10147 |
| chr14_+_33739013 | 3.59 |
ENSMUST00000226499.2
|
Gm5460
|
predicted gene 5460 |
| chrX_-_27725713 | 3.55 |
ENSMUST00000178152.2
|
Gm10058
|
predicted gene 10058 |
| chrX_-_28733497 | 3.55 |
ENSMUST00000178672.2
|
Gm10096
|
predicted gene 10096 |
| chr17_+_48037758 | 3.25 |
ENSMUST00000024782.12
ENSMUST00000144955.2 |
Pgc
|
progastricsin (pepsinogen C) |
| chrY_-_79161056 | 3.22 |
ENSMUST00000179922.2
|
Gm20917
|
predicted gene, 20917 |
| chrY_-_78847812 | 3.18 |
ENSMUST00000180324.2
|
Gm20806
|
predicted gene, 20806 |
| chr14_+_33775115 | 3.14 |
ENSMUST00000227979.2
|
Antxrl
|
anthrax toxin receptor-like |
| chrY_+_21166083 | 3.13 |
ENSMUST00000178234.2
|
Gm20909
|
predicted gene, 20909 |
| chrY_-_53413354 | 3.13 |
ENSMUST00000179137.2
|
Gm20747
|
predicted gene, 20747 |
| chrY_+_24411927 | 3.09 |
ENSMUST00000179663.2
|
Gm20809
|
predicted gene, 20809 |
| chrY_-_84574663 | 3.08 |
ENSMUST00000179523.2
|
Gm21394
|
predicted gene, 21394 |
| chrY_-_66751879 | 3.06 |
ENSMUST00000178761.2
|
Gm20852
|
predicted gene, 20852 |
| chrY_-_85540787 | 3.01 |
ENSMUST00000178889.2
|
Gm20854
|
predicted gene, 20854 |
| chrY_-_67057835 | 2.91 |
ENSMUST00000179136.2
|
Ssty2
|
spermiogenesis specific transcript on the Y 2 |
| chrX_-_24639814 | 2.91 |
ENSMUST00000178986.2
|
Gm5935
|
predicted gene 5935 |
| chrY_-_77972959 | 2.85 |
ENSMUST00000178900.2
|
Gm20867
|
predicted gene, 20867 |
| chrY_+_68550741 | 2.83 |
ENSMUST00000177765.2
|
Gm20816
|
predicted gene, 20816 |
| chr4_-_35845204 | 2.80 |
ENSMUST00000164772.8
ENSMUST00000065173.9 |
Lingo2
|
leucine rich repeat and Ig domain containing 2 |
| chrY_-_65214536 | 2.79 |
ENSMUST00000177663.2
|
Gm20924
|
predicted gene, 20924 |
| chrX_-_26004758 | 2.77 |
ENSMUST00000168002.3
|
Gm5168
|
predicted gene 5168 |
| chrY_-_70283393 | 2.76 |
ENSMUST00000180179.2
|
Gm21118
|
predicted gene, 21118 |
| chr14_+_26414422 | 2.75 |
ENSMUST00000022433.12
|
Dnah12
|
dynein, axonemal, heavy chain 12 |
| chrX_-_105528972 | 2.65 |
ENSMUST00000140707.2
|
Fndc3c1
|
fibronectin type III domain containing 3C1 |
| chr4_+_135455427 | 2.57 |
ENSMUST00000102546.4
|
Il22ra1
|
interleukin 22 receptor, alpha 1 |
| chr7_-_80055168 | 2.55 |
ENSMUST00000107362.10
ENSMUST00000135306.3 |
Furin
|
furin (paired basic amino acid cleaving enzyme) |
| chrY_+_9783380 | 2.50 |
ENSMUST00000180352.2
|
Gm20821
|
predicted gene, 20821 |
| chr3_-_24837772 | 2.48 |
ENSMUST00000203414.2
|
Naaladl2
|
N-acetylated alpha-linked acidic dipeptidase-like 2 |
| chrY_-_9345654 | 2.42 |
ENSMUST00000179590.2
|
Gm21812
|
predicted gene, 21812 |
| chr11_-_99482165 | 2.39 |
ENSMUST00000104930.2
|
Krtap1-3
|
keratin associated protein 1-3 |
| chr18_-_60881679 | 2.38 |
ENSMUST00000237783.2
|
Ndst1
|
N-deacetylase/N-sulfotransferase (heparan glucosaminyl) 1 |
| chr4_-_59960659 | 2.35 |
ENSMUST00000075973.3
|
Mup4
|
major urinary protein 4 |
| chr5_-_123185073 | 2.22 |
ENSMUST00000031437.14
|
Morn3
|
MORN repeat containing 3 |
| chr2_-_85632888 | 2.22 |
ENSMUST00000217410.3
ENSMUST00000216425.3 |
Olfr1016
|
olfactory receptor 1016 |
| chrX_-_142610371 | 2.06 |
ENSMUST00000087316.6
|
Capn6
|
calpain 6 |
| chrY_-_9132561 | 2.05 |
ENSMUST00000171947.3
|
Gm21292
|
predicted gene, 21292 |
| chrY_+_21242966 | 2.04 |
ENSMUST00000179095.2
|
Gm20865
|
predicted gene, 20865 |
| chr2_+_51517523 | 1.96 |
ENSMUST00000070028.3
|
Tas2r134
|
taste receptor, type 2, member 134 |
| chr7_+_131144596 | 1.92 |
ENSMUST00000046093.6
|
Hmx3
|
H6 homeobox 3 |
| chrY_-_6917117 | 1.81 |
ENSMUST00000178016.2
|
Gm20830
|
predicted gene, 20830 |
| chrX_-_105528503 | 1.81 |
ENSMUST00000138724.8
ENSMUST00000149331.2 |
Fndc3c1
|
fibronectin type III domain containing 3C1 |
| chrY_-_9200643 | 1.79 |
ENSMUST00000179595.2
|
Gm21721
|
predicted gene, 21721 |
| chr5_+_16996808 | 1.77 |
ENSMUST00000211738.2
|
Gm28710
|
predicted gene 28710 |
| chrY_+_5727496 | 1.73 |
ENSMUST00000180372.2
|
Gm20914
|
predicted gene, 20914 |
| chrY_-_7636973 | 1.65 |
ENSMUST00000179256.2
|
Gm20826
|
predicted gene, 20826 |
| chr12_-_75596441 | 1.63 |
ENSMUST00000218716.2
|
Ppp2r5e
|
protein phosphatase 2, regulatory subunit B', epsilon |
| chr11_-_58521327 | 1.54 |
ENSMUST00000214132.2
|
Olfr323
|
olfactory receptor 323 |
| chrY_-_7167777 | 1.52 |
ENSMUST00000178447.2
|
Gm21244
|
predicted gene, 21244 |
| chr6_+_42377172 | 1.47 |
ENSMUST00000057398.4
|
Tas2r143
|
taste receptor, type 2, member 143 |
| chr5_-_123185029 | 1.44 |
ENSMUST00000045843.15
|
Morn3
|
MORN repeat containing 3 |
| chrX_+_48552803 | 1.30 |
ENSMUST00000130558.8
|
Arhgap36
|
Rho GTPase activating protein 36 |
| chr4_-_119272640 | 1.15 |
ENSMUST00000238293.2
|
Ccdc30
|
coiled-coil domain containing 30 |
| chr3_+_14643669 | 1.02 |
ENSMUST00000029069.13
ENSMUST00000165922.3 |
E2f5
|
E2F transcription factor 5 |
| chrY_-_15883245 | 0.96 |
ENSMUST00000165756.2
|
Gm20822
|
predicted gene, 20822 |
| chrY_-_10323434 | 0.91 |
ENSMUST00000179558.2
|
Gm20834
|
predicted gene, 20834 |
| chrY_-_11197848 | 0.81 |
ENSMUST00000170889.2
|
Gm20828
|
predicted gene, 20828 |
| chrY_+_12383184 | 0.76 |
ENSMUST00000179220.2
|
Gm20812
|
predicted gene, 20812 |
| chr8_-_62355690 | 0.64 |
ENSMUST00000121785.9
ENSMUST00000034057.14 |
Palld
|
palladin, cytoskeletal associated protein |
| chr4_-_119272667 | 0.60 |
ENSMUST00000238609.2
|
Ccdc30
|
coiled-coil domain containing 30 |
| chr2_-_111820618 | 0.56 |
ENSMUST00000216948.2
ENSMUST00000214935.2 ENSMUST00000217452.2 ENSMUST00000215045.2 |
Olfr1309
|
olfactory receptor 1309 |
| chr18_+_77369654 | 0.31 |
ENSMUST00000096547.11
ENSMUST00000148341.9 ENSMUST00000123410.9 |
Loxhd1
|
lipoxygenase homology domains 1 |
| chr4_-_96029375 | 0.29 |
ENSMUST00000097972.5
|
Cyp2j12
|
cytochrome P450, family 2, subfamily j, polypeptide 12 |
| chr4_+_28813125 | 0.21 |
ENSMUST00000080934.11
ENSMUST00000029964.12 |
Epha7
|
Eph receptor A7 |
| chr12_+_111132779 | 0.18 |
ENSMUST00000117269.8
|
Traf3
|
TNF receptor-associated factor 3 |
| chr7_+_104713682 | 0.16 |
ENSMUST00000213622.2
|
Olfr678
|
olfactory receptor 678 |
| chrX_-_133012457 | 0.12 |
ENSMUST00000159259.3
ENSMUST00000113275.10 |
Nox1
|
NADPH oxidase 1 |
| chr2_-_22930149 | 0.10 |
ENSMUST00000091394.13
ENSMUST00000093171.13 |
Abi1
|
abl interactor 1 |
| chr2_-_111251294 | 0.06 |
ENSMUST00000099617.2
|
Olfr1286
|
olfactory receptor 1286 |
| chr8_+_36956345 | 0.03 |
ENSMUST00000171777.2
|
Trmt9b
|
tRNA methyltransferase 9B |
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 4.5 | 18.0 | GO:0015851 | nucleobase transport(GO:0015851) pyrimidine nucleobase transport(GO:0015855) |
| 3.2 | 38.1 | GO:0071394 | cellular response to testosterone stimulus(GO:0071394) |
| 1.7 | 53.6 | GO:0035634 | response to stilbenoid(GO:0035634) |
| 1.5 | 22.7 | GO:0015747 | urate transport(GO:0015747) |
| 0.8 | 7.3 | GO:0015014 | heparan sulfate proteoglycan biosynthetic process, polysaccharide chain biosynthetic process(GO:0015014) |
| 0.6 | 3.2 | GO:0002225 | positive regulation of antimicrobial peptide production(GO:0002225) positive regulation of antibacterial peptide production(GO:0002803) |
| 0.6 | 2.5 | GO:0090472 | dibasic protein processing(GO:0090472) |
| 0.3 | 12.1 | GO:0000132 | establishment of mitotic spindle orientation(GO:0000132) |
| 0.2 | 2.7 | GO:0036159 | inner dynein arm assembly(GO:0036159) |
| 0.1 | 3.4 | GO:0001580 | detection of chemical stimulus involved in sensory perception of bitter taste(GO:0001580) |
| 0.1 | 5.8 | GO:0046426 | negative regulation of JAK-STAT cascade(GO:0046426) negative regulation of STAT cascade(GO:1904893) |
| 0.0 | 21.1 | GO:0051321 | meiotic cell cycle(GO:0051321) |
| 0.0 | 2.6 | GO:0050829 | defense response to Gram-negative bacterium(GO:0050829) |
| 0.0 | 0.1 | GO:0045726 | positive regulation of integrin biosynthetic process(GO:0045726) |
| 0.0 | 0.6 | GO:0071803 | positive regulation of podosome assembly(GO:0071803) |
| 0.0 | 1.9 | GO:0060135 | maternal process involved in female pregnancy(GO:0060135) |
| 0.0 | 2.8 | GO:0051965 | positive regulation of synapse assembly(GO:0051965) |
| 0.0 | 2.1 | GO:0001578 | microtubule bundle formation(GO:0001578) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 31.6 | GO:0000800 | lateral element(GO:0000800) |
| 0.2 | 2.7 | GO:0036156 | inner dynein arm(GO:0036156) |
| 0.1 | 2.5 | GO:0012510 | trans-Golgi network transport vesicle membrane(GO:0012510) |
| 0.1 | 36.8 | GO:0016323 | basolateral plasma membrane(GO:0016323) |
| 0.1 | 34.8 | GO:0045177 | apical part of cell(GO:0045177) |
| 0.0 | 1.6 | GO:0000159 | protein phosphatase type 2A complex(GO:0000159) |
| 0.0 | 2.1 | GO:0005876 | spindle microtubule(GO:0005876) |
| 0.0 | 0.1 | GO:0071438 | invadopodium membrane(GO:0071438) |
| 0.0 | 15.5 | GO:0005743 | mitochondrial inner membrane(GO:0005743) |
| 0.0 | 58.6 | GO:0005615 | extracellular space(GO:0005615) |
| 0.0 | 0.6 | GO:0002102 | podosome(GO:0002102) |
| 0.0 | 1.0 | GO:0016528 | sarcoplasm(GO:0016528) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 12.7 | 38.1 | GO:0005009 | insulin-activated receptor activity(GO:0005009) |
| 6.0 | 18.1 | GO:0016838 | carbon-oxygen lyase activity, acting on phosphates(GO:0016838) |
| 3.6 | 18.0 | GO:0005345 | purine nucleobase transmembrane transporter activity(GO:0005345) pyrimidine nucleobase transmembrane transporter activity(GO:0005350) nucleobase transmembrane transporter activity(GO:0015205) glycerol channel activity(GO:0015254) |
| 2.3 | 22.7 | GO:0015562 | efflux transmembrane transporter activity(GO:0015562) |
| 1.0 | 7.3 | GO:0042328 | heparan sulfate N-acetylglucosaminyltransferase activity(GO:0042328) |
| 0.9 | 2.6 | GO:0042015 | interleukin-20 binding(GO:0042015) |
| 0.8 | 31.8 | GO:0015020 | glucuronosyltransferase activity(GO:0015020) |
| 0.4 | 18.6 | GO:0015347 | sodium-independent organic anion transmembrane transporter activity(GO:0015347) |
| 0.3 | 36.2 | GO:0005550 | pheromone binding(GO:0005550) |
| 0.2 | 30.4 | GO:0020037 | heme binding(GO:0020037) |
| 0.2 | 3.4 | GO:0033038 | bitter taste receptor activity(GO:0033038) |
| 0.1 | 2.5 | GO:0048406 | nerve growth factor binding(GO:0048406) |
| 0.1 | 3.2 | GO:0004190 | aspartic-type endopeptidase activity(GO:0004190) |
| 0.1 | 2.7 | GO:0008569 | ATP-dependent microtubule motor activity, minus-end-directed(GO:0008569) |
| 0.1 | 2.1 | GO:0004198 | calcium-dependent cysteine-type endopeptidase activity(GO:0004198) |
| 0.0 | 5.8 | GO:0004860 | protein kinase inhibitor activity(GO:0004860) |
| 0.0 | 4.1 | GO:0005200 | structural constituent of cytoskeleton(GO:0005200) |
| 0.0 | 0.1 | GO:0030171 | voltage-gated proton channel activity(GO:0030171) |
| 0.0 | 1.6 | GO:0019888 | protein phosphatase regulator activity(GO:0019888) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 2.5 | PID NOTCH PATHWAY | Notch signaling pathway |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.3 | 18.0 | REACTOME PASSIVE TRANSPORT BY AQUAPORINS | Genes involved in Passive Transport by Aquaporins |
| 0.2 | 4.1 | REACTOME POST CHAPERONIN TUBULIN FOLDING PATHWAY | Genes involved in Post-chaperonin tubulin folding pathway |
| 0.2 | 2.5 | REACTOME GAMMA CARBOXYLATION TRANSPORT AND AMINO TERMINAL CLEAVAGE OF PROTEINS | Genes involved in Gamma-carboxylation, transport, and amino-terminal cleavage of proteins |
| 0.2 | 7.3 | REACTOME HS GAG BIOSYNTHESIS | Genes involved in HS-GAG biosynthesis |
| 0.1 | 1.6 | REACTOME CTNNB1 PHOSPHORYLATION CASCADE | Genes involved in Beta-catenin phosphorylation cascade |
| 0.0 | 1.0 | REACTOME G0 AND EARLY G1 | Genes involved in G0 and Early G1 |