Project
PRJNA375882: Comprehensive Mouse Transcriptomic BodyMap
Navigation
Downloads
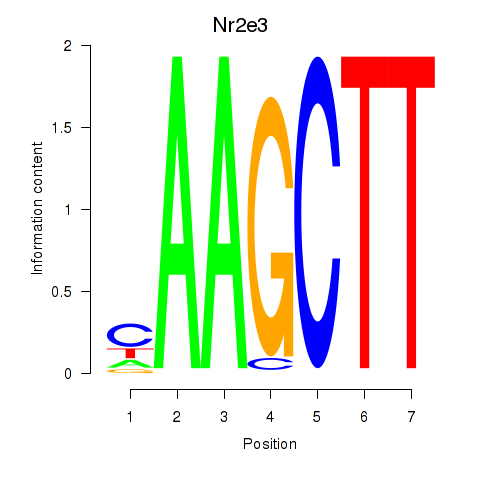
Results for Nr2e3
Z-value: 1.86
Transcription factors associated with Nr2e3
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Nr2e3
|
ENSMUSG00000032292.9 | Nr2e3 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Nr2e3 | mm39_v1_chr9_-_59857355_59857407 | -0.07 | 5.4e-01 | Click! |
Activity profile of Nr2e3 motif
Sorted Z-values of Nr2e3 motif
Network of associatons between targets according to the STRING database.
First level regulatory network of Nr2e3
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr18_-_35348049 | 27.06 |
ENSMUST00000091636.5
ENSMUST00000236680.2 |
Lrrtm2
|
leucine rich repeat transmembrane neuronal 2 |
| chr2_-_73490746 | 22.95 |
ENSMUST00000102677.11
|
Chn1
|
chimerin 1 |
| chr2_-_73490719 | 22.08 |
ENSMUST00000154258.8
|
Chn1
|
chimerin 1 |
| chr3_+_75981577 | 20.44 |
ENSMUST00000038364.15
|
Fstl5
|
follistatin-like 5 |
| chr5_-_71815318 | 20.35 |
ENSMUST00000199357.2
|
Gabra4
|
gamma-aminobutyric acid (GABA) A receptor, subunit alpha 4 |
| chr14_+_14159978 | 18.79 |
ENSMUST00000137133.2
ENSMUST00000036070.15 ENSMUST00000121887.8 |
Fam107a
|
family with sequence similarity 107, member A |
| chr6_-_36787096 | 18.35 |
ENSMUST00000201321.2
ENSMUST00000101534.5 |
Ptn
|
pleiotrophin |
| chr1_+_143516402 | 16.62 |
ENSMUST00000038252.4
|
B3galt2
|
UDP-Gal:betaGlcNAc beta 1,3-galactosyltransferase, polypeptide 2 |
| chr10_+_90412432 | 16.53 |
ENSMUST00000182786.8
ENSMUST00000182600.8 |
Anks1b
|
ankyrin repeat and sterile alpha motif domain containing 1B |
| chr11_-_42073737 | 15.93 |
ENSMUST00000206085.2
ENSMUST00000020707.12 ENSMUST00000132971.3 |
Gabra1
|
gamma-aminobutyric acid (GABA) A receptor, subunit alpha 1 |
| chr10_-_108846816 | 15.59 |
ENSMUST00000105276.8
ENSMUST00000064054.14 |
Syt1
|
synaptotagmin I |
| chr11_-_42072990 | 15.54 |
ENSMUST00000205546.2
|
Gabra1
|
gamma-aminobutyric acid (GABA) A receptor, subunit alpha 1 |
| chr12_+_88920169 | 15.12 |
ENSMUST00000057634.14
|
Nrxn3
|
neurexin III |
| chr6_+_141470105 | 15.01 |
ENSMUST00000032362.12
ENSMUST00000205214.3 |
Slco1c1
|
solute carrier organic anion transporter family, member 1c1 |
| chr3_+_68375495 | 14.83 |
ENSMUST00000182532.8
|
Schip1
|
schwannomin interacting protein 1 |
| chr11_-_65053710 | 14.67 |
ENSMUST00000093002.12
ENSMUST00000047463.15 |
Arhgap44
|
Rho GTPase activating protein 44 |
| chr10_+_90412114 | 14.65 |
ENSMUST00000182427.8
ENSMUST00000182053.8 ENSMUST00000182113.8 |
Anks1b
|
ankyrin repeat and sterile alpha motif domain containing 1B |
| chr10_+_90412539 | 14.61 |
ENSMUST00000182284.8
|
Anks1b
|
ankyrin repeat and sterile alpha motif domain containing 1B |
| chr2_+_65451100 | 14.40 |
ENSMUST00000144254.6
ENSMUST00000028377.14 |
Scn2a
|
sodium channel, voltage-gated, type II, alpha |
| chr14_-_9015639 | 13.80 |
ENSMUST00000112656.4
|
Synpr
|
synaptoporin |
| chr7_-_105217851 | 13.34 |
ENSMUST00000188368.7
ENSMUST00000187057.7 |
Apbb1
|
amyloid beta (A4) precursor protein-binding, family B, member 1 |
| chr2_-_25471703 | 13.34 |
ENSMUST00000114217.3
ENSMUST00000191602.2 |
Ajm1
|
apical junction component 1 |
| chr2_+_143388062 | 13.32 |
ENSMUST00000028905.10
|
Pcsk2
|
proprotein convertase subtilisin/kexin type 2 |
| chr6_+_141470261 | 12.82 |
ENSMUST00000203140.2
|
Slco1c1
|
solute carrier organic anion transporter family, member 1c1 |
| chr10_+_90412570 | 12.60 |
ENSMUST00000182430.8
ENSMUST00000182960.8 ENSMUST00000182045.2 ENSMUST00000182083.2 |
Anks1b
|
ankyrin repeat and sterile alpha motif domain containing 1B |
| chrX_+_165127688 | 12.38 |
ENSMUST00000112223.8
ENSMUST00000112224.8 ENSMUST00000112229.9 ENSMUST00000112228.8 ENSMUST00000112227.9 ENSMUST00000112226.3 |
Gpm6b
|
glycoprotein m6b |
| chr13_-_78345932 | 12.19 |
ENSMUST00000127137.3
|
Nr2f1
|
nuclear receptor subfamily 2, group F, member 1 |
| chr10_+_90412638 | 12.12 |
ENSMUST00000183136.8
ENSMUST00000182595.8 |
Anks1b
|
ankyrin repeat and sterile alpha motif domain containing 1B |
| chr6_-_13839914 | 12.05 |
ENSMUST00000060442.14
|
Gpr85
|
G protein-coupled receptor 85 |
| chrX_-_58211440 | 11.53 |
ENSMUST00000119306.2
|
Fgf13
|
fibroblast growth factor 13 |
| chr5_+_71815382 | 11.37 |
ENSMUST00000199967.5
|
Gabrb1
|
gamma-aminobutyric acid (GABA) A receptor, subunit beta 1 |
| chr18_+_37952556 | 11.23 |
ENSMUST00000055935.11
|
Pcdhgc5
|
protocadherin gamma subfamily C, 5 |
| chr3_+_54063459 | 11.20 |
ENSMUST00000029311.11
ENSMUST00000200048.5 |
Trpc4
|
transient receptor potential cation channel, subfamily C, member 4 |
| chr7_-_105218472 | 11.11 |
ENSMUST00000187683.7
ENSMUST00000210079.2 ENSMUST00000187051.7 ENSMUST00000189265.7 ENSMUST00000190369.7 |
Apbb1
|
amyloid beta (A4) precursor protein-binding, family B, member 1 |
| chr9_-_29323032 | 10.52 |
ENSMUST00000115236.2
|
Ntm
|
neurotrimin |
| chr4_-_151946155 | 10.32 |
ENSMUST00000049790.14
|
Camta1
|
calmodulin binding transcription activator 1 |
| chr9_+_113641615 | 10.08 |
ENSMUST00000111838.10
ENSMUST00000166734.10 ENSMUST00000214522.2 ENSMUST00000163895.3 |
Clasp2
|
CLIP associating protein 2 |
| chr4_-_138123700 | 9.61 |
ENSMUST00000105032.4
|
Fam43b
|
family with sequence similarity 43, member B |
| chr6_+_104469751 | 9.58 |
ENSMUST00000161446.2
ENSMUST00000161070.8 ENSMUST00000089215.12 |
Cntn6
|
contactin 6 |
| chrX_+_119199956 | 9.22 |
ENSMUST00000113364.10
ENSMUST00000050239.16 ENSMUST00000113358.10 |
Pcdh11x
|
protocadherin 11 X-linked |
| chr4_+_102617495 | 9.14 |
ENSMUST00000072481.12
ENSMUST00000156596.8 ENSMUST00000080728.13 ENSMUST00000106882.9 |
Sgip1
|
SH3-domain GRB2-like (endophilin) interacting protein 1 |
| chr5_+_88731386 | 8.88 |
ENSMUST00000031229.11
|
Rufy3
|
RUN and FYVE domain containing 3 |
| chr5_-_71815218 | 8.84 |
ENSMUST00000198138.2
|
Gabra4
|
gamma-aminobutyric acid (GABA) A receptor, subunit alpha 4 |
| chr2_-_136229849 | 8.74 |
ENSMUST00000035264.9
ENSMUST00000077200.4 |
Pak5
|
p21 (RAC1) activated kinase 5 |
| chr9_-_77255099 | 8.53 |
ENSMUST00000184138.8
ENSMUST00000184006.8 ENSMUST00000185144.8 ENSMUST00000034910.16 |
Mlip
|
muscular LMNA-interacting protein |
| chr18_+_37952596 | 8.51 |
ENSMUST00000193890.2
ENSMUST00000193941.2 |
Pcdhgc5
|
protocadherin gamma subfamily C, 5 |
| chr5_+_19431976 | 8.48 |
ENSMUST00000197354.5
ENSMUST00000088516.10 ENSMUST00000197443.5 |
Magi2
|
membrane associated guanylate kinase, WW and PDZ domain containing 2 |
| chr8_-_55171699 | 8.25 |
ENSMUST00000144711.9
|
Wdr17
|
WD repeat domain 17 |
| chr2_+_96148418 | 8.07 |
ENSMUST00000135431.8
ENSMUST00000162807.9 |
Lrrc4c
|
leucine rich repeat containing 4C |
| chr9_+_65268304 | 7.96 |
ENSMUST00000147185.3
|
Ubap1l
|
ubiquitin-associated protein 1-like |
| chr14_-_9015757 | 7.94 |
ENSMUST00000153954.8
|
Synpr
|
synaptoporin |
| chr5_-_5315968 | 7.90 |
ENSMUST00000115451.8
ENSMUST00000115452.8 ENSMUST00000131392.8 |
Cdk14
|
cyclin-dependent kinase 14 |
| chr9_-_77255069 | 7.86 |
ENSMUST00000184848.8
ENSMUST00000184415.8 |
Mlip
|
muscular LMNA-interacting protein |
| chr13_+_43070127 | 7.85 |
ENSMUST00000239286.2
|
Phactr1
|
phosphatase and actin regulator 1 |
| chr18_-_22983794 | 7.73 |
ENSMUST00000092015.11
ENSMUST00000069215.13 |
Nol4
|
nucleolar protein 4 |
| chr2_-_26917921 | 7.51 |
ENSMUST00000102890.11
ENSMUST00000153388.2 ENSMUST00000045702.6 |
Slc2a6
|
solute carrier family 2 (facilitated glucose transporter), member 6 |
| chr11_-_73217633 | 7.36 |
ENSMUST00000134079.2
|
Aspa
|
aspartoacylase |
| chr4_-_97472844 | 7.26 |
ENSMUST00000107067.8
ENSMUST00000107068.9 |
E130114P18Rik
|
RIKEN cDNA E130114P18 gene |
| chr4_-_151946124 | 7.17 |
ENSMUST00000169423.9
|
Camta1
|
calmodulin binding transcription activator 1 |
| chr4_+_102617332 | 7.10 |
ENSMUST00000066824.14
|
Sgip1
|
SH3-domain GRB2-like (endophilin) interacting protein 1 |
| chr4_-_151946219 | 7.06 |
ENSMUST00000097774.9
|
Camta1
|
calmodulin binding transcription activator 1 |
| chrX_-_142716200 | 7.05 |
ENSMUST00000112851.8
ENSMUST00000112856.3 ENSMUST00000033642.10 |
Dcx
|
doublecortin |
| chr1_+_179938904 | 6.66 |
ENSMUST00000145181.2
|
Cdc42bpa
|
CDC42 binding protein kinase alpha |
| chr18_+_33072194 | 6.60 |
ENSMUST00000042868.6
|
Camk4
|
calcium/calmodulin-dependent protein kinase IV |
| chr11_+_69909659 | 6.39 |
ENSMUST00000232002.2
ENSMUST00000134376.10 ENSMUST00000231221.2 |
Dlg4
|
discs large MAGUK scaffold protein 4 |
| chr5_-_28672091 | 6.30 |
ENSMUST00000002708.5
|
Shh
|
sonic hedgehog |
| chrX_+_7656225 | 6.11 |
ENSMUST00000136930.8
ENSMUST00000115675.9 ENSMUST00000101694.10 |
Gripap1
|
GRIP1 associated protein 1 |
| chr19_-_40365318 | 5.77 |
ENSMUST00000239304.2
|
Sorbs1
|
sorbin and SH3 domain containing 1 |
| chr8_+_105267431 | 5.75 |
ENSMUST00000056051.11
|
Car7
|
carbonic anhydrase 7 |
| chr9_-_77255171 | 5.62 |
ENSMUST00000185039.8
|
Mlip
|
muscular LMNA-interacting protein |
| chr18_-_34712123 | 5.62 |
ENSMUST00000079287.12
|
Nme5
|
NME/NM23 family member 5 |
| chr11_+_110888313 | 5.60 |
ENSMUST00000106635.2
|
Kcnj16
|
potassium inwardly-rectifying channel, subfamily J, member 16 |
| chr2_-_17465410 | 5.55 |
ENSMUST00000145492.2
|
Nebl
|
nebulette |
| chr13_+_21663077 | 5.54 |
ENSMUST00000062609.6
ENSMUST00000225845.2 |
Zkscan4
|
zinc finger with KRAB and SCAN domains 4 |
| chr4_-_123458465 | 5.20 |
ENSMUST00000238731.2
|
Macf1
|
microtubule-actin crosslinking factor 1 |
| chr3_-_79053182 | 5.16 |
ENSMUST00000118340.7
|
Rapgef2
|
Rap guanine nucleotide exchange factor (GEF) 2 |
| chr10_+_119655294 | 5.09 |
ENSMUST00000105262.9
ENSMUST00000147454.8 ENSMUST00000138410.8 ENSMUST00000144825.8 ENSMUST00000148954.8 ENSMUST00000144959.8 |
Grip1
|
glutamate receptor interacting protein 1 |
| chr11_-_3724706 | 5.06 |
ENSMUST00000155197.2
|
Osbp2
|
oxysterol binding protein 2 |
| chr6_+_149310471 | 4.96 |
ENSMUST00000086829.11
ENSMUST00000111513.9 |
Bicd1
|
BICD cargo adaptor 1 |
| chr14_-_78866714 | 4.96 |
ENSMUST00000228362.2
ENSMUST00000227767.2 |
Dgkh
|
diacylglycerol kinase, eta |
| chrX_+_7656251 | 4.91 |
ENSMUST00000140540.2
|
Gripap1
|
GRIP1 associated protein 1 |
| chr15_+_22549108 | 4.75 |
ENSMUST00000163361.8
|
Cdh18
|
cadherin 18 |
| chrX_-_133012600 | 4.73 |
ENSMUST00000033610.13
|
Nox1
|
NADPH oxidase 1 |
| chr10_-_10956700 | 4.70 |
ENSMUST00000105560.2
|
Grm1
|
glutamate receptor, metabotropic 1 |
| chrX_-_93585668 | 4.60 |
ENSMUST00000026142.8
|
Maged1
|
MAGE family member D1 |
| chr2_-_45001141 | 4.59 |
ENSMUST00000201969.4
ENSMUST00000201623.4 |
Zeb2
|
zinc finger E-box binding homeobox 2 |
| chr19_+_22670134 | 4.50 |
ENSMUST00000237470.2
ENSMUST00000099564.10 ENSMUST00000099569.10 ENSMUST00000099566.5 ENSMUST00000235712.2 |
Trpm3
|
transient receptor potential cation channel, subfamily M, member 3 |
| chr7_+_83234118 | 4.32 |
ENSMUST00000039317.14
ENSMUST00000164944.2 |
Tmc3
|
transmembrane channel-like gene family 3 |
| chr12_+_95662124 | 4.15 |
ENSMUST00000110117.2
|
Flrt2
|
fibronectin leucine rich transmembrane protein 2 |
| chr1_+_185187000 | 4.14 |
ENSMUST00000061093.7
|
Slc30a10
|
solute carrier family 30, member 10 |
| chr1_+_167445815 | 4.14 |
ENSMUST00000111380.2
|
Rxrg
|
retinoid X receptor gamma |
| chr7_+_30450896 | 4.10 |
ENSMUST00000182229.8
ENSMUST00000080518.14 ENSMUST00000182227.8 ENSMUST00000182721.8 |
Sbsn
|
suprabasin |
| chr11_+_19874403 | 3.94 |
ENSMUST00000093298.12
|
Spred2
|
sprouty-related EVH1 domain containing 2 |
| chr15_+_81756671 | 3.88 |
ENSMUST00000135198.2
ENSMUST00000157003.8 ENSMUST00000229068.2 |
Aco2
|
aconitase 2, mitochondrial |
| chr4_-_140780972 | 3.80 |
ENSMUST00000040222.14
|
Crocc
|
ciliary rootlet coiled-coil, rootletin |
| chr18_+_35695736 | 3.79 |
ENSMUST00000235851.2
ENSMUST00000235581.2 |
Matr3
|
matrin 3 |
| chr9_-_16289527 | 3.71 |
ENSMUST00000082170.6
|
Fat3
|
FAT atypical cadherin 3 |
| chr4_-_154721288 | 3.66 |
ENSMUST00000030902.13
ENSMUST00000105637.8 ENSMUST00000070313.14 ENSMUST00000105636.8 ENSMUST00000105638.9 ENSMUST00000097759.9 ENSMUST00000124771.2 |
Prdm16
|
PR domain containing 16 |
| chr1_-_97904958 | 3.60 |
ENSMUST00000161567.8
|
Pam
|
peptidylglycine alpha-amidating monooxygenase |
| chr5_+_76957812 | 3.56 |
ENSMUST00000120818.8
|
Cracd
|
capping protein inhibiting regulator of actin |
| chr5_+_20112500 | 3.45 |
ENSMUST00000101558.10
|
Magi2
|
membrane associated guanylate kinase, WW and PDZ domain containing 2 |
| chr2_-_140513320 | 3.44 |
ENSMUST00000056760.4
|
Flrt3
|
fibronectin leucine rich transmembrane protein 3 |
| chr13_-_92931317 | 3.27 |
ENSMUST00000022213.8
|
Thbs4
|
thrombospondin 4 |
| chr3_-_143908060 | 3.26 |
ENSMUST00000121112.6
|
Lmo4
|
LIM domain only 4 |
| chr11_-_73217298 | 3.24 |
ENSMUST00000155630.9
|
Aspa
|
aspartoacylase |
| chr5_+_20112704 | 3.13 |
ENSMUST00000115267.7
|
Magi2
|
membrane associated guanylate kinase, WW and PDZ domain containing 2 |
| chr14_+_123897383 | 3.13 |
ENSMUST00000049681.14
|
Itgbl1
|
integrin, beta-like 1 |
| chr3_-_143908111 | 3.09 |
ENSMUST00000121796.8
|
Lmo4
|
LIM domain only 4 |
| chr19_-_45619559 | 3.01 |
ENSMUST00000160718.9
|
Fbxw4
|
F-box and WD-40 domain protein 4 |
| chr16_-_22258469 | 3.01 |
ENSMUST00000079601.13
|
Etv5
|
ets variant 5 |
| chr9_+_106080307 | 2.95 |
ENSMUST00000024047.12
ENSMUST00000216348.2 |
Twf2
|
twinfilin actin binding protein 2 |
| chr16_+_91343936 | 2.94 |
ENSMUST00000023687.9
|
Ifngr2
|
interferon gamma receptor 2 |
| chr6_-_93890520 | 2.93 |
ENSMUST00000203688.3
|
Magi1
|
membrane associated guanylate kinase, WW and PDZ domain containing 1 |
| chr15_-_80989200 | 2.92 |
ENSMUST00000109579.9
|
Mrtfa
|
myocardin related transcription factor A |
| chr14_+_102077937 | 2.84 |
ENSMUST00000159026.8
|
Lmo7
|
LIM domain only 7 |
| chr14_+_20724378 | 2.80 |
ENSMUST00000224492.2
ENSMUST00000223751.2 ENSMUST00000225108.2 ENSMUST00000224754.2 |
Sec24c
|
Sec24 related gene family, member C (S. cerevisiae) |
| chr18_+_65054188 | 2.72 |
ENSMUST00000236898.2
ENSMUST00000237410.2 |
Nedd4l
|
neural precursor cell expressed, developmentally down-regulated gene 4-like |
| chr12_-_110669076 | 2.66 |
ENSMUST00000155242.8
|
Hsp90aa1
|
heat shock protein 90, alpha (cytosolic), class A member 1 |
| chr13_+_25127127 | 2.66 |
ENSMUST00000021773.13
|
Gpld1
|
glycosylphosphatidylinositol specific phospholipase D1 |
| chr16_+_31241085 | 2.66 |
ENSMUST00000089759.9
|
Bdh1
|
3-hydroxybutyrate dehydrogenase, type 1 |
| chr7_+_27430823 | 2.65 |
ENSMUST00000130997.8
ENSMUST00000042641.14 |
Zfp60
|
zinc finger protein 60 |
| chr8_-_39128662 | 2.63 |
ENSMUST00000118896.2
|
Sgcz
|
sarcoglycan zeta |
| chr14_+_102078038 | 2.61 |
ENSMUST00000159314.8
|
Lmo7
|
LIM domain only 7 |
| chr16_-_22258320 | 2.56 |
ENSMUST00000170393.2
|
Etv5
|
ets variant 5 |
| chr18_+_65022035 | 2.54 |
ENSMUST00000224385.3
ENSMUST00000163516.9 |
Nedd4l
|
neural precursor cell expressed, developmentally down-regulated gene 4-like |
| chr5_+_35156454 | 2.47 |
ENSMUST00000114283.8
|
Rgs12
|
regulator of G-protein signaling 12 |
| chr7_+_27430796 | 2.43 |
ENSMUST00000108336.8
|
Zfp60
|
zinc finger protein 60 |
| chr11_+_19874354 | 2.40 |
ENSMUST00000093299.13
|
Spred2
|
sprouty-related EVH1 domain containing 2 |
| chr14_-_34310438 | 2.35 |
ENSMUST00000228044.2
ENSMUST00000022328.14 |
Ldb3
|
LIM domain binding 3 |
| chr5_-_51711204 | 2.35 |
ENSMUST00000196968.5
|
Ppargc1a
|
peroxisome proliferative activated receptor, gamma, coactivator 1 alpha |
| chr4_+_143139490 | 2.33 |
ENSMUST00000132915.2
ENSMUST00000037356.8 |
Pramel12
|
PRAME like 12 |
| chr1_-_37575313 | 2.32 |
ENSMUST00000042161.15
|
Mgat4a
|
mannoside acetylglucosaminyltransferase 4, isoenzyme A |
| chr4_+_143139571 | 2.31 |
ENSMUST00000155157.2
|
Pramel12
|
PRAME like 12 |
| chr14_+_20724366 | 2.25 |
ENSMUST00000048657.10
|
Sec24c
|
Sec24 related gene family, member C (S. cerevisiae) |
| chr10_+_115854118 | 2.25 |
ENSMUST00000063470.11
|
Ptprr
|
protein tyrosine phosphatase, receptor type, R |
| chr2_-_140513382 | 2.17 |
ENSMUST00000110057.3
|
Flrt3
|
fibronectin leucine rich transmembrane protein 3 |
| chr19_-_7318798 | 2.15 |
ENSMUST00000165965.8
ENSMUST00000051711.16 ENSMUST00000169541.8 ENSMUST00000165989.2 |
Mark2
|
MAP/microtubule affinity regulating kinase 2 |
| chr6_-_122317484 | 2.12 |
ENSMUST00000112600.9
|
Phc1
|
polyhomeotic 1 |
| chr19_+_39102342 | 2.06 |
ENSMUST00000087234.3
|
Cyp2c66
|
cytochrome P450, family 2, subfamily c, polypeptide 66 |
| chr14_+_76741625 | 2.02 |
ENSMUST00000177207.2
|
Tsc22d1
|
TSC22 domain family, member 1 |
| chr18_+_35695485 | 2.01 |
ENSMUST00000235199.2
ENSMUST00000237744.2 ENSMUST00000236276.2 |
Matr3
|
matrin 3 |
| chr14_+_76741918 | 2.00 |
ENSMUST00000022587.10
ENSMUST00000134109.2 |
Tsc22d1
|
TSC22 domain family, member 1 |
| chr10_+_29020055 | 1.92 |
ENSMUST00000216757.2
|
Soga3
|
SOGA family member 3 |
| chr11_+_115656246 | 1.88 |
ENSMUST00000093912.11
ENSMUST00000136720.8 ENSMUST00000103034.10 ENSMUST00000141871.8 |
Tmem94
|
transmembrane protein 94 |
| chrX_-_142716085 | 1.86 |
ENSMUST00000087313.10
|
Dcx
|
doublecortin |
| chr1_-_75293447 | 1.79 |
ENSMUST00000189551.7
|
Dnpep
|
aspartyl aminopeptidase |
| chr2_-_42543069 | 1.77 |
ENSMUST00000203080.3
|
Lrp1b
|
low density lipoprotein-related protein 1B |
| chr6_+_97906760 | 1.73 |
ENSMUST00000101123.10
|
Mitf
|
melanogenesis associated transcription factor |
| chr1_-_40829801 | 1.64 |
ENSMUST00000039672.6
|
Mfsd9
|
major facilitator superfamily domain containing 9 |
| chr10_-_128204545 | 1.61 |
ENSMUST00000220027.2
|
Coq10a
|
coenzyme Q10A |
| chr6_-_93890659 | 1.61 |
ENSMUST00000093769.8
|
Magi1
|
membrane associated guanylate kinase, WW and PDZ domain containing 1 |
| chr2_+_124452194 | 1.58 |
ENSMUST00000051419.15
ENSMUST00000076335.12 ENSMUST00000078621.12 ENSMUST00000077847.12 |
Sema6d
|
sema domain, transmembrane domain (TM), and cytoplasmic domain, (semaphorin) 6D |
| chrX_+_168662592 | 1.54 |
ENSMUST00000112105.8
ENSMUST00000078947.12 |
Mid1
|
midline 1 |
| chr5_-_113285852 | 1.50 |
ENSMUST00000212276.2
|
2900026A02Rik
|
RIKEN cDNA 2900026A02 gene |
| chr2_-_160950936 | 1.41 |
ENSMUST00000039782.14
ENSMUST00000134178.8 |
Chd6
|
chromodomain helicase DNA binding protein 6 |
| chr13_+_112425322 | 1.38 |
ENSMUST00000022275.14
|
Ankrd55
|
ankyrin repeat domain 55 |
| chr2_+_83554741 | 1.38 |
ENSMUST00000028499.11
|
Itgav
|
integrin alpha V |
| chr8_+_79754980 | 1.32 |
ENSMUST00000087927.11
ENSMUST00000098614.9 |
Zfp827
|
zinc finger protein 827 |
| chr16_+_32090286 | 1.32 |
ENSMUST00000093183.5
|
Smco1
|
single-pass membrane protein with coiled-coil domains 1 |
| chr18_+_61688378 | 1.31 |
ENSMUST00000165721.8
ENSMUST00000115246.9 ENSMUST00000166990.8 ENSMUST00000163205.8 ENSMUST00000170862.8 |
Csnk1a1
|
casein kinase 1, alpha 1 |
| chr10_-_120815232 | 1.30 |
ENSMUST00000119944.8
ENSMUST00000119093.2 |
Lemd3
|
LEM domain containing 3 |
| chr5_-_51711237 | 1.27 |
ENSMUST00000132734.8
|
Ppargc1a
|
peroxisome proliferative activated receptor, gamma, coactivator 1 alpha |
| chr9_-_20085353 | 1.25 |
ENSMUST00000215984.3
|
Olfr870
|
olfactory receptor 870 |
| chr6_+_82379768 | 1.24 |
ENSMUST00000203775.2
|
Tacr1
|
tachykinin receptor 1 |
| chr3_+_40594899 | 1.24 |
ENSMUST00000091186.7
|
Intu
|
inturned planar cell polarity protein |
| chr1_+_90131702 | 1.20 |
ENSMUST00000065587.5
ENSMUST00000159654.2 |
Ackr3
|
atypical chemokine receptor 3 |
| chr3_+_69129745 | 1.17 |
ENSMUST00000183126.2
|
Arl14
|
ADP-ribosylation factor-like 14 |
| chr6_+_15185399 | 1.15 |
ENSMUST00000115474.8
ENSMUST00000115472.8 ENSMUST00000031545.14 |
Foxp2
|
forkhead box P2 |
| chr7_+_92426772 | 1.08 |
ENSMUST00000208945.2
|
Rab30
|
RAB30, member RAS oncogene family |
| chrX_-_133012457 | 1.00 |
ENSMUST00000159259.3
ENSMUST00000113275.10 |
Nox1
|
NADPH oxidase 1 |
| chr11_+_93934940 | 1.00 |
ENSMUST00000132079.8
|
Spag9
|
sperm associated antigen 9 |
| chr7_+_101714692 | 0.96 |
ENSMUST00000106950.8
ENSMUST00000146450.8 |
Xndc1
|
Xrcc1 N-terminal domain containing 1 |
| chr14_+_67470884 | 0.96 |
ENSMUST00000176161.8
|
Ebf2
|
early B cell factor 2 |
| chr1_-_160040286 | 0.95 |
ENSMUST00000195654.2
ENSMUST00000014370.11 |
Cacybp
|
calcyclin binding protein |
| chrM_+_11735 | 0.94 |
ENSMUST00000082418.1
|
mt-Nd5
|
mitochondrially encoded NADH dehydrogenase 5 |
| chr6_+_82379456 | 0.94 |
ENSMUST00000032122.11
|
Tacr1
|
tachykinin receptor 1 |
| chr19_+_8748347 | 0.94 |
ENSMUST00000010248.4
|
Tmem223
|
transmembrane protein 223 |
| chr7_+_36397426 | 0.91 |
ENSMUST00000021641.8
|
Tshz3
|
teashirt zinc finger family member 3 |
| chr6_+_30568366 | 0.91 |
ENSMUST00000049251.6
|
Cpa4
|
carboxypeptidase A4 |
| chr7_+_120634834 | 0.82 |
ENSMUST00000207351.2
|
Mettl9
|
methyltransferase like 9 |
| chr11_+_34264757 | 0.81 |
ENSMUST00000165963.9
ENSMUST00000093192.4 |
Insyn2b
|
inhibitory synaptic factor family member 2B |
| chr6_+_136509922 | 0.75 |
ENSMUST00000187429.4
|
Atf7ip
|
activating transcription factor 7 interacting protein |
| chr17_-_21134625 | 0.72 |
ENSMUST00000077001.3
|
Vmn1r232
|
vomeronasal 1 receptor 232 |
| chr1_-_75293942 | 0.70 |
ENSMUST00000187075.7
|
Dnpep
|
aspartyl aminopeptidase |
| chr16_+_84312009 | 0.70 |
ENSMUST00000209558.2
|
A730009L09Rik
|
RIKEN cDNA A730009L09 gene |
| chr5_-_65492940 | 0.70 |
ENSMUST00000203471.3
ENSMUST00000172732.8 ENSMUST00000204965.3 |
Rfc1
|
replication factor C (activator 1) 1 |
| chr3_-_90685187 | 0.69 |
ENSMUST00000180151.3
|
Gm5849
|
predicted gene 5849 |
| chr12_+_86999366 | 0.68 |
ENSMUST00000191463.2
|
Cipc
|
CLOCK interacting protein, circadian |
| chr7_-_101714251 | 0.66 |
ENSMUST00000130074.2
ENSMUST00000131104.3 ENSMUST00000096639.12 |
Rnf121
|
ring finger protein 121 |
| chr16_-_63684477 | 0.61 |
ENSMUST00000232654.2
ENSMUST00000064405.8 |
Epha3
|
Eph receptor A3 |
| chrX_+_37689503 | 0.59 |
ENSMUST00000000365.3
|
Mcts1
|
malignant T cell amplified sequence 1 |
| chr16_+_22926504 | 0.58 |
ENSMUST00000187168.7
ENSMUST00000232287.2 ENSMUST00000077605.12 |
Eif4a2
|
eukaryotic translation initiation factor 4A2 |
| chr11_-_102470287 | 0.55 |
ENSMUST00000107081.8
|
Gm11627
|
predicted gene 11627 |
| chr14_+_67470735 | 0.47 |
ENSMUST00000022637.14
|
Ebf2
|
early B cell factor 2 |
| chr1_+_177273226 | 0.45 |
ENSMUST00000077225.8
|
Zbtb18
|
zinc finger and BTB domain containing 18 |
| chr2_+_24839758 | 0.45 |
ENSMUST00000028350.9
|
Zmynd19
|
zinc finger, MYND domain containing 19 |
| chr2_+_83554770 | 0.41 |
ENSMUST00000141725.3
|
Itgav
|
integrin alpha V |
| chr19_-_4843860 | 0.38 |
ENSMUST00000178615.8
ENSMUST00000179189.9 ENSMUST00000237760.2 ENSMUST00000164376.3 ENSMUST00000164209.9 ENSMUST00000180248.8 |
Rbm4
|
RNA binding motif protein 4 |
| chr12_-_34578842 | 0.31 |
ENSMUST00000110819.4
|
Hdac9
|
histone deacetylase 9 |
| chr16_-_63684425 | 0.29 |
ENSMUST00000232049.2
|
Epha3
|
Eph receptor A3 |
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 6.4 | 70.5 | GO:1900383 | regulation of synaptic plasticity by receptor localization to synapse(GO:1900383) |
| 4.6 | 18.3 | GO:1904395 | positive regulation of skeletal muscle acetylcholine-gated channel clustering(GO:1904395) |
| 4.4 | 13.3 | GO:0030070 | insulin processing(GO:0030070) |
| 4.2 | 29.2 | GO:2001023 | regulation of response to drug(GO:2001023) |
| 4.1 | 24.4 | GO:0050758 | thymidylate synthase biosynthetic process(GO:0050757) regulation of thymidylate synthase biosynthetic process(GO:0050758) negative regulation of thymidylate synthase biosynthetic process(GO:0050760) |
| 2.9 | 14.7 | GO:0098886 | modification of dendritic spine(GO:0098886) |
| 2.4 | 7.2 | GO:0072194 | kidney smooth muscle tissue development(GO:0072194) |
| 2.1 | 4.1 | GO:0071421 | manganese ion transmembrane transport(GO:0071421) |
| 1.9 | 15.3 | GO:0005513 | detection of calcium ion(GO:0005513) |
| 1.9 | 42.8 | GO:0071420 | cellular response to histamine(GO:0071420) |
| 1.8 | 16.6 | GO:0006682 | galactosylceramide biosynthetic process(GO:0006682) galactolipid biosynthetic process(GO:0019375) |
| 1.7 | 5.2 | GO:2000670 | positive regulation of dendritic cell apoptotic process(GO:2000670) |
| 1.7 | 33.4 | GO:0002091 | negative regulation of receptor internalization(GO:0002091) |
| 1.7 | 5.0 | GO:1900275 | negative regulation of phospholipase C activity(GO:1900275) |
| 1.6 | 11.5 | GO:0045196 | establishment or maintenance of neuroblast polarity(GO:0045196) establishment of neuroblast polarity(GO:0045200) |
| 1.6 | 11.0 | GO:1905244 | regulation of modification of synaptic structure(GO:1905244) |
| 1.5 | 12.4 | GO:0051612 | negative regulation of neurotransmitter uptake(GO:0051581) negative regulation of serotonin uptake(GO:0051612) |
| 1.5 | 10.6 | GO:0006083 | acetate metabolic process(GO:0006083) |
| 1.3 | 14.4 | GO:0008627 | intrinsic apoptotic signaling pathway in response to osmotic stress(GO:0008627) |
| 1.2 | 19.9 | GO:2000507 | positive regulation of energy homeostasis(GO:2000507) |
| 1.1 | 5.7 | GO:0032849 | positive regulation of cellular pH reduction(GO:0032849) |
| 1.1 | 5.7 | GO:0045726 | positive regulation of integrin biosynthetic process(GO:0045726) |
| 1.1 | 3.3 | GO:0071603 | endothelial cell-cell adhesion(GO:0071603) |
| 1.0 | 10.1 | GO:1903690 | negative regulation of wound healing, spreading of epidermal cells(GO:1903690) |
| 0.9 | 4.7 | GO:0099566 | regulation of postsynaptic cytosolic calcium ion concentration(GO:0099566) |
| 0.9 | 2.7 | GO:0010040 | response to iron(II) ion(GO:0010040) |
| 0.9 | 45.9 | GO:0008045 | motor neuron axon guidance(GO:0008045) |
| 0.8 | 27.8 | GO:0015721 | bile acid and bile salt transport(GO:0015721) |
| 0.8 | 9.4 | GO:0097118 | neuroligin clustering involved in postsynaptic membrane assembly(GO:0097118) |
| 0.6 | 3.8 | GO:1903566 | ciliary basal body organization(GO:0032053) positive regulation of protein localization to cilium(GO:1903566) |
| 0.6 | 5.6 | GO:0042078 | germ-line stem cell division(GO:0042078) male germ-line stem cell asymmetric division(GO:0048133) regulation of branching involved in mammary gland duct morphogenesis(GO:0060762) germline stem cell asymmetric division(GO:0098728) |
| 0.6 | 15.1 | GO:0090129 | positive regulation of synapse maturation(GO:0090129) |
| 0.6 | 5.6 | GO:0003344 | pericardium morphogenesis(GO:0003344) |
| 0.6 | 22.0 | GO:0010614 | negative regulation of cardiac muscle hypertrophy(GO:0010614) |
| 0.5 | 14.8 | GO:0001553 | luteinization(GO:0001553) |
| 0.5 | 3.6 | GO:1902748 | positive regulation of lens fiber cell differentiation(GO:1902748) |
| 0.5 | 8.9 | GO:0048672 | positive regulation of collateral sprouting(GO:0048672) |
| 0.5 | 2.5 | GO:0038032 | termination of G-protein coupled receptor signaling pathway(GO:0038032) |
| 0.5 | 12.2 | GO:0021796 | cerebral cortex regionalization(GO:0021796) |
| 0.5 | 11.2 | GO:0006828 | manganese ion transport(GO:0006828) gamma-aminobutyric acid secretion(GO:0014051) |
| 0.4 | 3.6 | GO:0031179 | peptide modification(GO:0031179) |
| 0.4 | 5.6 | GO:0006228 | UTP biosynthetic process(GO:0006228) |
| 0.4 | 3.9 | GO:0006102 | isocitrate metabolic process(GO:0006102) |
| 0.4 | 4.1 | GO:0061343 | cell adhesion involved in heart morphogenesis(GO:0061343) |
| 0.4 | 7.5 | GO:0035428 | hexose transmembrane transport(GO:0035428) |
| 0.4 | 1.1 | GO:2000536 | negative regulation of entry of bacterium into host cell(GO:2000536) |
| 0.4 | 2.7 | GO:0045585 | regulation of cytotoxic T cell differentiation(GO:0045583) positive regulation of cytotoxic T cell differentiation(GO:0045585) |
| 0.4 | 8.0 | GO:0043162 | ubiquitin-dependent protein catabolic process via the multivesicular body sorting pathway(GO:0043162) |
| 0.4 | 2.2 | GO:0038128 | ERBB2 signaling pathway(GO:0038128) |
| 0.4 | 6.3 | GO:0021527 | spinal cord association neuron differentiation(GO:0021527) |
| 0.4 | 2.2 | GO:0003051 | angiotensin-mediated drinking behavior(GO:0003051) positive regulation of saliva secretion(GO:0046878) |
| 0.4 | 1.4 | GO:0036091 | positive regulation of transcription from RNA polymerase II promoter in response to oxidative stress(GO:0036091) |
| 0.3 | 6.6 | GO:0043011 | myeloid dendritic cell differentiation(GO:0043011) |
| 0.3 | 6.3 | GO:0043517 | positive regulation of DNA damage response, signal transduction by p53 class mediator(GO:0043517) |
| 0.3 | 2.9 | GO:0032532 | regulation of microvillus length(GO:0032532) |
| 0.3 | 5.0 | GO:0090110 | cargo loading into COPII-coated vesicle(GO:0090110) |
| 0.3 | 8.9 | GO:2000114 | regulation of establishment of cell polarity(GO:2000114) |
| 0.3 | 3.7 | GO:0030853 | negative regulation of granulocyte differentiation(GO:0030853) |
| 0.2 | 5.2 | GO:0006620 | posttranslational protein targeting to membrane(GO:0006620) |
| 0.2 | 2.7 | GO:2001288 | positive regulation of caveolin-mediated endocytosis(GO:2001288) |
| 0.2 | 5.0 | GO:0046473 | phosphatidic acid metabolic process(GO:0046473) |
| 0.2 | 9.6 | GO:0045747 | positive regulation of Notch signaling pathway(GO:0045747) |
| 0.2 | 6.7 | GO:0040023 | nuclear migration(GO:0007097) establishment of nucleus localization(GO:0040023) |
| 0.2 | 24.6 | GO:0050885 | neuromuscular process controlling balance(GO:0050885) |
| 0.2 | 1.5 | GO:0035372 | protein localization to microtubule(GO:0035372) |
| 0.2 | 1.3 | GO:0044821 | meiotic telomere tethering at nuclear periphery(GO:0044821) meiotic attachment of telomere to nuclear envelope(GO:0070197) chromosome attachment to the nuclear envelope(GO:0097240) |
| 0.2 | 4.6 | GO:0090190 | positive regulation of branching involved in ureteric bud morphogenesis(GO:0090190) |
| 0.1 | 5.8 | GO:0045725 | positive regulation of glycogen biosynthetic process(GO:0045725) |
| 0.1 | 1.0 | GO:0090074 | negative regulation of protein homodimerization activity(GO:0090074) |
| 0.1 | 5.6 | GO:0010107 | potassium ion import(GO:0010107) |
| 0.1 | 1.7 | GO:0044336 | canonical Wnt signaling pathway involved in negative regulation of apoptotic process(GO:0044336) |
| 0.1 | 0.6 | GO:0002188 | translation reinitiation(GO:0002188) |
| 0.1 | 0.4 | GO:0032055 | negative regulation of translation in response to stress(GO:0032055) |
| 0.1 | 3.7 | GO:0010842 | retina layer formation(GO:0010842) |
| 0.1 | 9.2 | GO:0010923 | negative regulation of phosphatase activity(GO:0010923) |
| 0.1 | 1.4 | GO:0035563 | positive regulation of chromatin binding(GO:0035563) |
| 0.1 | 1.2 | GO:0010839 | negative regulation of keratinocyte proliferation(GO:0010839) |
| 0.1 | 3.0 | GO:0031146 | SCF-dependent proteasomal ubiquitin-dependent protein catabolic process(GO:0031146) |
| 0.1 | 0.7 | GO:1904424 | regulation of GTP binding(GO:1904424) |
| 0.1 | 1.6 | GO:0006744 | ubiquinone biosynthetic process(GO:0006744) |
| 0.1 | 4.1 | GO:0048384 | retinoic acid receptor signaling pathway(GO:0048384) |
| 0.1 | 2.1 | GO:0071300 | cellular response to retinoic acid(GO:0071300) |
| 0.1 | 5.1 | GO:1904893 | negative regulation of JAK-STAT cascade(GO:0046426) negative regulation of STAT cascade(GO:1904893) |
| 0.1 | 0.9 | GO:0060416 | response to growth hormone(GO:0060416) |
| 0.1 | 7.8 | GO:0043149 | contractile actin filament bundle assembly(GO:0030038) stress fiber assembly(GO:0043149) |
| 0.1 | 19.7 | GO:0050808 | synapse organization(GO:0050808) |
| 0.1 | 7.9 | GO:0000086 | G2/M transition of mitotic cell cycle(GO:0000086) |
| 0.1 | 2.1 | GO:0045197 | establishment or maintenance of epithelial cell apical/basal polarity(GO:0045197) |
| 0.0 | 1.1 | GO:0014912 | negative regulation of smooth muscle cell migration(GO:0014912) |
| 0.0 | 0.7 | GO:0042754 | negative regulation of circadian rhythm(GO:0042754) |
| 0.0 | 1.2 | GO:1902230 | negative regulation of intrinsic apoptotic signaling pathway in response to DNA damage(GO:1902230) |
| 0.0 | 2.4 | GO:0045214 | sarcomere organization(GO:0045214) |
| 0.0 | 0.3 | GO:0034983 | peptidyl-lysine deacetylation(GO:0034983) |
| 0.0 | 1.3 | GO:0050911 | detection of chemical stimulus involved in sensory perception of smell(GO:0050911) |
| 0.0 | 4.7 | GO:0010977 | negative regulation of neuron projection development(GO:0010977) |
| 0.0 | 0.8 | GO:0045898 | regulation of RNA polymerase II transcriptional preinitiation complex assembly(GO:0045898) |
| 0.0 | 4.4 | GO:0007286 | spermatid development(GO:0007286) |
| 0.0 | 0.3 | GO:0043353 | enucleate erythrocyte differentiation(GO:0043353) |
| 0.0 | 0.9 | GO:0042773 | ATP synthesis coupled electron transport(GO:0042773) |
| 0.0 | 3.6 | GO:0016358 | dendrite development(GO:0016358) |
| 0.0 | 2.6 | GO:0060047 | heart contraction(GO:0060047) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 3.4 | 10.1 | GO:1903754 | cortical microtubule plus-end(GO:1903754) cytoplasmic microtubule plus-end(GO:1904511) |
| 2.6 | 72.0 | GO:1902711 | GABA-A receptor complex(GO:1902711) |
| 2.2 | 24.4 | GO:1990812 | growth cone lamellipodium(GO:1990761) growth cone filopodium(GO:1990812) |
| 1.6 | 7.9 | GO:0000308 | cytoplasmic cyclin-dependent protein kinase holoenzyme complex(GO:0000308) |
| 1.6 | 11.0 | GO:0098837 | postsynaptic recycling endosome(GO:0098837) |
| 1.4 | 37.3 | GO:0030285 | integral component of synaptic vesicle membrane(GO:0030285) intrinsic component of synaptic vesicle membrane(GO:0098563) |
| 1.2 | 3.6 | GO:1990843 | subsarcolemmal mitochondrion(GO:1990843) interfibrillar mitochondrion(GO:1990844) |
| 1.0 | 5.0 | GO:0033647 | host intracellular organelle(GO:0033647) host intracellular membrane-bounded organelle(GO:0033648) |
| 1.0 | 5.8 | GO:0005899 | insulin receptor complex(GO:0005899) |
| 0.8 | 16.2 | GO:0030122 | AP-2 adaptor complex(GO:0030122) |
| 0.7 | 5.7 | GO:0071438 | invadopodium membrane(GO:0071438) |
| 0.6 | 8.0 | GO:0000813 | ESCRT I complex(GO:0000813) |
| 0.6 | 14.4 | GO:0001518 | voltage-gated sodium channel complex(GO:0001518) |
| 0.5 | 8.9 | GO:0071437 | invadopodium(GO:0071437) |
| 0.5 | 9.4 | GO:0036056 | filtration diaphragm(GO:0036056) slit diaphragm(GO:0036057) |
| 0.4 | 15.1 | GO:0042734 | presynaptic membrane(GO:0042734) |
| 0.4 | 2.7 | GO:0097226 | sperm mitochondrial sheath(GO:0097226) |
| 0.4 | 11.1 | GO:0098839 | postsynaptic density membrane(GO:0098839) |
| 0.4 | 14.7 | GO:0048786 | presynaptic active zone(GO:0048786) |
| 0.3 | 2.6 | GO:0016012 | sarcoglycan complex(GO:0016012) |
| 0.3 | 1.1 | GO:0034686 | integrin alphav-beta3 complex(GO:0034683) integrin alphav-beta8 complex(GO:0034686) |
| 0.3 | 5.0 | GO:0030127 | COPII vesicle coat(GO:0030127) |
| 0.3 | 108.7 | GO:0045211 | postsynaptic membrane(GO:0045211) |
| 0.3 | 21.6 | GO:0031594 | neuromuscular junction(GO:0031594) |
| 0.2 | 3.8 | GO:0035253 | ciliary rootlet(GO:0035253) |
| 0.2 | 7.5 | GO:0097440 | apical dendrite(GO:0097440) |
| 0.2 | 1.6 | GO:0030877 | beta-catenin destruction complex(GO:0030877) |
| 0.2 | 2.1 | GO:0001739 | sex chromatin(GO:0001739) |
| 0.2 | 2.1 | GO:0097427 | microtubule bundle(GO:0097427) |
| 0.2 | 22.0 | GO:0016605 | PML body(GO:0016605) |
| 0.1 | 11.7 | GO:0016328 | lateral plasma membrane(GO:0016328) |
| 0.1 | 11.2 | GO:0034704 | calcium channel complex(GO:0034704) |
| 0.1 | 5.6 | GO:0044295 | axonal growth cone(GO:0044295) |
| 0.1 | 16.9 | GO:0043204 | perikaryon(GO:0043204) |
| 0.1 | 3.7 | GO:0016235 | aggresome(GO:0016235) |
| 0.1 | 20.1 | GO:0031225 | anchored component of membrane(GO:0031225) |
| 0.1 | 2.7 | GO:0034364 | high-density lipoprotein particle(GO:0034364) |
| 0.1 | 1.3 | GO:0031229 | integral component of nuclear inner membrane(GO:0005639) intrinsic component of nuclear inner membrane(GO:0031229) |
| 0.1 | 12.5 | GO:0030426 | growth cone(GO:0030426) |
| 0.1 | 1.4 | GO:0005665 | DNA-directed RNA polymerase II, core complex(GO:0005665) |
| 0.0 | 4.2 | GO:0016363 | nuclear matrix(GO:0016363) |
| 0.0 | 4.7 | GO:0042641 | actomyosin(GO:0042641) |
| 0.0 | 6.6 | GO:0001650 | fibrillar center(GO:0001650) |
| 0.0 | 1.2 | GO:0005905 | clathrin-coated pit(GO:0005905) |
| 0.0 | 7.6 | GO:0045121 | membrane raft(GO:0045121) membrane microdomain(GO:0098857) |
| 0.0 | 27.6 | GO:0045202 | synapse(GO:0045202) |
| 0.0 | 22.4 | GO:0043005 | neuron projection(GO:0043005) |
| 0.0 | 4.1 | GO:0055037 | recycling endosome(GO:0055037) |
| 0.0 | 39.0 | GO:0005887 | integral component of plasma membrane(GO:0005887) |
| 0.0 | 25.5 | GO:0005730 | nucleolus(GO:0005730) |
| 0.0 | 1.5 | GO:0005881 | cytoplasmic microtubule(GO:0005881) |
| 0.0 | 2.4 | GO:0030018 | Z disc(GO:0030018) |
| 0.0 | 3.5 | GO:0016324 | apical plasma membrane(GO:0016324) |
| 0.0 | 2.8 | GO:0043209 | myelin sheath(GO:0043209) |
| 0.0 | 1.1 | GO:0005801 | cis-Golgi network(GO:0005801) |
| 0.0 | 0.2 | GO:0032045 | guanyl-nucleotide exchange factor complex(GO:0032045) |
| 0.0 | 1.3 | GO:0019005 | SCF ubiquitin ligase complex(GO:0019005) |
| 0.0 | 0.6 | GO:0022627 | cytosolic small ribosomal subunit(GO:0022627) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 5.2 | 15.6 | GO:0030348 | syntaxin-3 binding(GO:0030348) |
| 4.6 | 18.3 | GO:0035373 | chondroitin sulfate proteoglycan binding(GO:0035373) |
| 4.3 | 42.8 | GO:0022851 | GABA-gated chloride ion channel activity(GO:0022851) |
| 4.2 | 16.6 | GO:0047275 | glucosaminylgalactosylglucosylceramide beta-galactosyltransferase activity(GO:0047275) |
| 3.5 | 10.6 | GO:0019807 | aspartoacylase activity(GO:0019807) |
| 3.5 | 27.8 | GO:0015349 | thyroid hormone transmembrane transporter activity(GO:0015349) |
| 2.4 | 14.6 | GO:0031697 | beta-1 adrenergic receptor binding(GO:0031697) |
| 1.7 | 33.4 | GO:0042043 | neurexin family protein binding(GO:0042043) |
| 1.7 | 5.0 | GO:0031871 | proteinase activated receptor binding(GO:0031871) |
| 1.7 | 115.5 | GO:0046875 | ephrin receptor binding(GO:0046875) |
| 1.6 | 4.7 | GO:0099530 | PLC activating G-protein coupled glutamate receptor activity(GO:0001639) G-protein coupled receptor activity involved in regulation of postsynaptic membrane potential(GO:0099530) |
| 1.3 | 29.2 | GO:0004890 | GABA-A receptor activity(GO:0004890) |
| 1.3 | 3.9 | GO:0051538 | 3 iron, 4 sulfur cluster binding(GO:0051538) |
| 1.3 | 6.3 | GO:0005173 | stem cell factor receptor binding(GO:0005173) |
| 1.2 | 9.6 | GO:0043237 | laminin-1 binding(GO:0043237) |
| 1.0 | 5.7 | GO:0030171 | voltage-gated proton channel activity(GO:0030171) |
| 0.9 | 3.6 | GO:0016842 | amidine-lyase activity(GO:0016842) |
| 0.9 | 2.7 | GO:0003858 | 3-hydroxybutyrate dehydrogenase activity(GO:0003858) |
| 0.8 | 14.4 | GO:0043522 | leucine zipper domain binding(GO:0043522) |
| 0.8 | 27.1 | GO:0048156 | tau protein binding(GO:0048156) |
| 0.7 | 11.2 | GO:0015279 | store-operated calcium channel activity(GO:0015279) |
| 0.7 | 12.2 | GO:0044323 | retinoic acid-responsive element binding(GO:0044323) |
| 0.7 | 4.1 | GO:0004886 | 9-cis retinoic acid receptor activity(GO:0004886) |
| 0.5 | 2.2 | GO:0004995 | tachykinin receptor activity(GO:0004995) |
| 0.5 | 9.8 | GO:0045499 | chemorepellent activity(GO:0045499) |
| 0.5 | 10.1 | GO:0002162 | dystroglycan binding(GO:0002162) |
| 0.4 | 2.9 | GO:0004920 | interleukin-10 receptor activity(GO:0004920) |
| 0.4 | 2.1 | GO:0008390 | testosterone 16-alpha-hydroxylase activity(GO:0008390) |
| 0.4 | 6.6 | GO:0009931 | calcium-dependent protein serine/threonine kinase activity(GO:0009931) |
| 0.3 | 5.7 | GO:0004089 | carbonate dehydratase activity(GO:0004089) |
| 0.3 | 2.7 | GO:0004630 | phospholipase D activity(GO:0004630) |
| 0.3 | 5.0 | GO:0004143 | diacylglycerol kinase activity(GO:0004143) |
| 0.3 | 7.5 | GO:0005351 | sugar:proton symporter activity(GO:0005351) |
| 0.3 | 15.1 | GO:0005246 | calcium channel regulator activity(GO:0005246) |
| 0.3 | 9.6 | GO:0005112 | Notch binding(GO:0005112) |
| 0.2 | 1.6 | GO:0048039 | ubiquinone binding(GO:0048039) |
| 0.2 | 5.6 | GO:0004550 | nucleoside diphosphate kinase activity(GO:0004550) |
| 0.2 | 2.3 | GO:0008454 | alpha-1,3-mannosylglycoprotein 4-beta-N-acetylglucosaminyltransferase activity(GO:0008454) |
| 0.2 | 6.1 | GO:0030159 | receptor signaling complex scaffold activity(GO:0030159) |
| 0.2 | 11.5 | GO:0048487 | beta-tubulin binding(GO:0048487) |
| 0.2 | 1.2 | GO:0019958 | C-X-C chemokine binding(GO:0019958) |
| 0.2 | 14.7 | GO:0048365 | Rac GTPase binding(GO:0048365) |
| 0.2 | 7.9 | GO:0030332 | cyclin binding(GO:0030332) |
| 0.2 | 6.2 | GO:0008157 | protein phosphatase 1 binding(GO:0008157) |
| 0.1 | 5.6 | GO:0005242 | inward rectifier potassium channel activity(GO:0005242) |
| 0.1 | 2.7 | GO:0019870 | potassium channel inhibitor activity(GO:0019870) |
| 0.1 | 0.9 | GO:0005004 | GPI-linked ephrin receptor activity(GO:0005004) |
| 0.1 | 1.1 | GO:0019960 | C-X3-C chemokine binding(GO:0019960) |
| 0.1 | 2.1 | GO:0050321 | tau-protein kinase activity(GO:0050321) |
| 0.1 | 3.6 | GO:0043014 | alpha-tubulin binding(GO:0043014) |
| 0.1 | 4.1 | GO:0005385 | zinc ion transmembrane transporter activity(GO:0005385) |
| 0.1 | 5.8 | GO:0005070 | SH3/SH2 adaptor activity(GO:0005070) |
| 0.1 | 8.2 | GO:0042805 | actinin binding(GO:0042805) |
| 0.1 | 2.4 | GO:0070006 | metalloaminopeptidase activity(GO:0070006) |
| 0.1 | 3.6 | GO:0070412 | R-SMAD binding(GO:0070412) |
| 0.1 | 14.5 | GO:0005057 | receptor signaling protein activity(GO:0005057) |
| 0.1 | 12.9 | GO:0001191 | transcriptional repressor activity, RNA polymerase II transcription factor binding(GO:0001191) |
| 0.1 | 3.8 | GO:0019894 | kinesin binding(GO:0019894) |
| 0.1 | 5.1 | GO:0015485 | cholesterol binding(GO:0015485) |
| 0.1 | 22.2 | GO:0008017 | microtubule binding(GO:0008017) |
| 0.1 | 1.1 | GO:0030215 | semaphorin receptor binding(GO:0030215) |
| 0.1 | 2.9 | GO:0003785 | actin monomer binding(GO:0003785) |
| 0.1 | 13.3 | GO:0004252 | serine-type endopeptidase activity(GO:0004252) |
| 0.1 | 8.0 | GO:0043130 | ubiquitin binding(GO:0043130) |
| 0.0 | 3.7 | GO:0033613 | activating transcription factor binding(GO:0033613) |
| 0.0 | 0.4 | GO:0097157 | pre-mRNA intronic binding(GO:0097157) |
| 0.0 | 0.3 | GO:0032041 | histone deacetylase activity (H3-K14 specific)(GO:0031078) NAD-dependent histone deacetylase activity (H3-K14 specific)(GO:0032041) |
| 0.0 | 0.9 | GO:0004181 | metallocarboxypeptidase activity(GO:0004181) |
| 0.0 | 4.9 | GO:0000149 | SNARE binding(GO:0000149) |
| 0.0 | 1.7 | GO:0070888 | E-box binding(GO:0070888) |
| 0.0 | 1.4 | GO:0001221 | transcription cofactor binding(GO:0001221) |
| 0.0 | 2.2 | GO:0004860 | protein kinase inhibitor activity(GO:0004860) |
| 0.0 | 12.4 | GO:0005509 | calcium ion binding(GO:0005509) |
| 0.0 | 0.1 | GO:0005344 | oxygen transporter activity(GO:0005344) |
| 0.0 | 4.2 | GO:0005261 | cation channel activity(GO:0005261) |
| 0.0 | 1.2 | GO:0001158 | enhancer sequence-specific DNA binding(GO:0001158) |
| 0.0 | 1.3 | GO:0031490 | chromatin DNA binding(GO:0031490) |
| 0.0 | 2.2 | GO:0004725 | protein tyrosine phosphatase activity(GO:0004725) |
| 0.0 | 4.5 | GO:0001077 | transcriptional activator activity, RNA polymerase II core promoter proximal region sequence-specific binding(GO:0001077) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.8 | 18.3 | PID SYNDECAN 3 PATHWAY | Syndecan-3-mediated signaling events |
| 0.5 | 45.0 | PID RAC1 REG PATHWAY | Regulation of RAC1 activity |
| 0.2 | 30.8 | PID ERA GENOMIC PATHWAY | Validated nuclear estrogen receptor alpha network |
| 0.2 | 15.6 | SIG CD40PATHWAYMAP | Genes related to CD40 signaling |
| 0.2 | 8.9 | PID LIS1 PATHWAY | Lissencephaly gene (LIS1) in neuronal migration and development |
| 0.2 | 6.3 | PID HEDGEHOG 2PATHWAY | Signaling events mediated by the Hedgehog family |
| 0.2 | 5.7 | PID RAC1 PATHWAY | RAC1 signaling pathway |
| 0.2 | 6.4 | ST GA12 PATHWAY | G alpha 12 Pathway |
| 0.1 | 8.1 | PID KIT PATHWAY | Signaling events mediated by Stem cell factor receptor (c-Kit) |
| 0.1 | 5.6 | PID IL12 STAT4 PATHWAY | IL12 signaling mediated by STAT4 |
| 0.1 | 4.1 | PID RETINOIC ACID PATHWAY | Retinoic acid receptors-mediated signaling |
| 0.1 | 11.7 | PID NOTCH PATHWAY | Notch signaling pathway |
| 0.1 | 6.6 | PID PI3K PLC TRK PATHWAY | Trk receptor signaling mediated by PI3K and PLC-gamma |
| 0.1 | 3.6 | PID HDAC CLASSIII PATHWAY | Signaling events mediated by HDAC Class III |
| 0.1 | 2.7 | PID HIF1A PATHWAY | Hypoxic and oxygen homeostasis regulation of HIF-1-alpha |
| 0.1 | 5.8 | SIG INSULIN RECEPTOR PATHWAY IN CARDIAC MYOCYTES | Genes related to the insulin receptor pathway |
| 0.1 | 7.3 | PID TRKR PATHWAY | Neurotrophic factor-mediated Trk receptor signaling |
| 0.1 | 3.8 | PID UPA UPAR PATHWAY | Urokinase-type plasminogen activator (uPA) and uPAR-mediated signaling |
| 0.0 | 3.6 | PID CDC42 PATHWAY | CDC42 signaling events |
| 0.0 | 0.7 | PID BETA CATENIN DEG PATHWAY | Degradation of beta catenin |
| 0.0 | 0.9 | PID EPHA FWDPATHWAY | EPHA forward signaling |
| 0.0 | 1.0 | PID ARF6 TRAFFICKING PATHWAY | Arf6 trafficking events |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 3.0 | 72.0 | REACTOME GABA A RECEPTOR ACTIVATION | Genes involved in GABA A receptor activation |
| 2.0 | 27.8 | REACTOME TRANSPORT OF ORGANIC ANIONS | Genes involved in Transport of organic anions |
| 1.1 | 15.6 | REACTOME NOREPINEPHRINE NEUROTRANSMITTER RELEASE CYCLE | Genes involved in Norepinephrine Neurotransmitter Release Cycle |
| 0.8 | 11.2 | REACTOME ROLE OF SECOND MESSENGERS IN NETRIN1 SIGNALING | Genes involved in Role of second messengers in netrin-1 signaling |
| 0.6 | 13.3 | REACTOME INSULIN SYNTHESIS AND PROCESSING | Genes involved in Insulin Synthesis and Processing |
| 0.4 | 7.5 | REACTOME FACILITATIVE NA INDEPENDENT GLUCOSE TRANSPORTERS | Genes involved in Facilitative Na+-independent glucose transporters |
| 0.3 | 4.6 | REACTOME ROLE OF DCC IN REGULATING APOPTOSIS | Genes involved in Role of DCC in regulating apoptosis |
| 0.3 | 14.4 | REACTOME INTERACTION BETWEEN L1 AND ANKYRINS | Genes involved in Interaction between L1 and Ankyrins |
| 0.3 | 9.4 | REACTOME NEPHRIN INTERACTIONS | Genes involved in Nephrin interactions |
| 0.3 | 5.0 | REACTOME ANTIGEN PRESENTATION FOLDING ASSEMBLY AND PEPTIDE LOADING OF CLASS I MHC | Genes involved in Antigen Presentation: Folding, assembly and peptide loading of class I MHC |
| 0.2 | 55.8 | REACTOME SIGNALING BY RHO GTPASES | Genes involved in Signaling by Rho GTPases |
| 0.2 | 4.7 | REACTOME CLASS C 3 METABOTROPIC GLUTAMATE PHEROMONE RECEPTORS | Genes involved in Class C/3 (Metabotropic glutamate/pheromone receptors) |
| 0.2 | 19.6 | REACTOME L1CAM INTERACTIONS | Genes involved in L1CAM interactions |
| 0.2 | 16.3 | REACTOME NUCLEAR RECEPTOR TRANSCRIPTION PATHWAY | Genes involved in Nuclear Receptor transcription pathway |
| 0.2 | 3.9 | REACTOME CITRIC ACID CYCLE TCA CYCLE | Genes involved in Citric acid cycle (TCA cycle) |
| 0.1 | 2.7 | REACTOME TETRAHYDROBIOPTERIN BH4 SYNTHESIS RECYCLING SALVAGE AND REGULATION | Genes involved in Tetrahydrobiopterin (BH4) synthesis, recycling, salvage and regulation |
| 0.1 | 5.0 | REACTOME EFFECTS OF PIP2 HYDROLYSIS | Genes involved in Effects of PIP2 hydrolysis |
| 0.1 | 6.6 | REACTOME POST NMDA RECEPTOR ACTIVATION EVENTS | Genes involved in Post NMDA receptor activation events |
| 0.1 | 3.6 | REACTOME CIRCADIAN REPRESSION OF EXPRESSION BY REV ERBA | Genes involved in Circadian Repression of Expression by REV-ERBA |
| 0.1 | 5.8 | REACTOME SMOOTH MUSCLE CONTRACTION | Genes involved in Smooth Muscle Contraction |
| 0.1 | 2.9 | REACTOME REGULATION OF IFNG SIGNALING | Genes involved in Regulation of IFNG signaling |
| 0.1 | 5.6 | REACTOME INHIBITION OF VOLTAGE GATED CA2 CHANNELS VIA GBETA GAMMA SUBUNITS | Genes involved in Inhibition of voltage gated Ca2+ channels via Gbeta/gamma subunits |
| 0.1 | 2.3 | REACTOME N GLYCAN ANTENNAE ELONGATION | Genes involved in N-Glycan antennae elongation |
| 0.1 | 3.0 | REACTOME ASSOCIATION OF TRIC CCT WITH TARGET PROTEINS DURING BIOSYNTHESIS | Genes involved in Association of TriC/CCT with target proteins during biosynthesis |
| 0.0 | 1.1 | REACTOME OTHER SEMAPHORIN INTERACTIONS | Genes involved in Other semaphorin interactions |
| 0.0 | 0.7 | REACTOME CTNNB1 PHOSPHORYLATION CASCADE | Genes involved in Beta-catenin phosphorylation cascade |
| 0.0 | 2.2 | REACTOME G ALPHA I SIGNALLING EVENTS | Genes involved in G alpha (i) signalling events |
| 0.0 | 2.5 | REACTOME GENERIC TRANSCRIPTION PATHWAY | Genes involved in Generic Transcription Pathway |
| 0.0 | 0.6 | REACTOME DEADENYLATION OF MRNA | Genes involved in Deadenylation of mRNA |
| 0.0 | 2.5 | REACTOME FATTY ACID TRIACYLGLYCEROL AND KETONE BODY METABOLISM | Genes involved in Fatty acid, triacylglycerol, and ketone body metabolism |
| 0.0 | 2.2 | REACTOME PEPTIDE LIGAND BINDING RECEPTORS | Genes involved in Peptide ligand-binding receptors |