Project
PRJNA375882: Comprehensive Mouse Transcriptomic BodyMap
Navigation
Downloads
Results for Nr3c2
Z-value: 0.45
Transcription factors associated with Nr3c2
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Nr3c2
|
ENSMUSG00000031618.14 | Nr3c2 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Nr3c2 | mm39_v1_chr8_+_77628916_77629104 | 0.15 | 2.2e-01 | Click! |
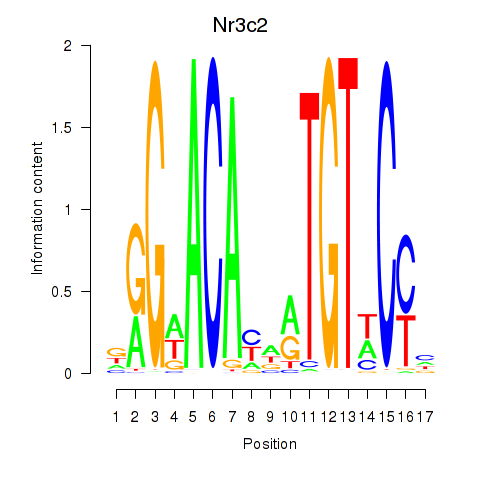
Activity profile of Nr3c2 motif
Sorted Z-values of Nr3c2 motif
Network of associatons between targets according to the STRING database.
First level regulatory network of Nr3c2
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr14_+_70768257 | 2.57 |
ENSMUST00000047331.8
|
Lgi3
|
leucine-rich repeat LGI family, member 3 |
| chr14_+_70768289 | 2.29 |
ENSMUST00000226548.2
|
Lgi3
|
leucine-rich repeat LGI family, member 3 |
| chr1_-_74974707 | 2.25 |
ENSMUST00000094844.4
|
Cfap65
|
cilia and flagella associated protein 65 |
| chr10_-_117118226 | 1.51 |
ENSMUST00000092163.9
|
Lyz2
|
lysozyme 2 |
| chr15_+_102011415 | 1.48 |
ENSMUST00000046144.10
|
Tns2
|
tensin 2 |
| chr15_+_102011352 | 1.46 |
ENSMUST00000169627.9
|
Tns2
|
tensin 2 |
| chr15_+_102010632 | 1.33 |
ENSMUST00000229592.2
|
Tns2
|
tensin 2 |
| chr6_-_69584812 | 1.17 |
ENSMUST00000103359.3
|
Igkv4-55
|
immunoglobulin kappa variable 4-55 |
| chr14_+_14159978 | 1.10 |
ENSMUST00000137133.2
ENSMUST00000036070.15 ENSMUST00000121887.8 |
Fam107a
|
family with sequence similarity 107, member A |
| chr5_+_112436599 | 1.10 |
ENSMUST00000151947.3
|
Tpst2
|
protein-tyrosine sulfotransferase 2 |
| chr11_+_96822213 | 0.98 |
ENSMUST00000107633.2
|
Prr15l
|
proline rich 15-like |
| chr3_-_95811993 | 0.92 |
ENSMUST00000147962.3
ENSMUST00000036181.15 |
Car14
|
carbonic anhydrase 14 |
| chr11_+_68989763 | 0.89 |
ENSMUST00000021271.14
|
Per1
|
period circadian clock 1 |
| chr11_+_98277276 | 0.87 |
ENSMUST00000041301.8
|
Pnmt
|
phenylethanolamine-N-methyltransferase |
| chr6_-_69282389 | 0.81 |
ENSMUST00000103350.3
|
Igkv4-68
|
immunoglobulin kappa variable 4-68 |
| chr5_-_135494775 | 0.79 |
ENSMUST00000212301.2
|
Hip1
|
huntingtin interacting protein 1 |
| chr6_+_34840057 | 0.79 |
ENSMUST00000074949.4
|
Tmem140
|
transmembrane protein 140 |
| chr1_-_157084252 | 0.75 |
ENSMUST00000134543.8
|
Rasal2
|
RAS protein activator like 2 |
| chr6_+_34840151 | 0.68 |
ENSMUST00000202010.2
|
Tmem140
|
transmembrane protein 140 |
| chr6_-_69394425 | 0.68 |
ENSMUST00000199160.2
|
Igkv4-61
|
immunoglobulin kappa chain variable 4-61 |
| chr6_-_69245427 | 0.62 |
ENSMUST00000103348.3
|
Igkv4-70
|
immunoglobulin kappa chain variable 4-70 |
| chr7_-_25089522 | 0.61 |
ENSMUST00000054301.14
|
Lipe
|
lipase, hormone sensitive |
| chr10_+_26698556 | 0.57 |
ENSMUST00000135866.2
|
Arhgap18
|
Rho GTPase activating protein 18 |
| chr14_-_68771138 | 0.55 |
ENSMUST00000022640.8
|
Adam7
|
a disintegrin and metallopeptidase domain 7 |
| chr6_-_69678271 | 0.55 |
ENSMUST00000103363.2
|
Igkv4-50
|
immunoglobulin kappa variable 4-50 |
| chr2_-_164435508 | 0.54 |
ENSMUST00000103100.2
|
Eppin
|
epididymal peptidase inhibitor |
| chr9_-_75504926 | 0.43 |
ENSMUST00000164100.2
|
Tmod2
|
tropomodulin 2 |
| chr6_-_69377328 | 0.41 |
ENSMUST00000198345.2
|
Igkv4-62
|
immunoglobulin kappa variable 4-62 |
| chr11_-_71092282 | 0.36 |
ENSMUST00000108515.9
|
Nlrp1b
|
NLR family, pyrin domain containing 1B |
| chr16_-_95993420 | 0.36 |
ENSMUST00000113804.8
ENSMUST00000054855.14 |
Lca5l
|
Leber congenital amaurosis 5-like |
| chr6_-_69020489 | 0.34 |
ENSMUST00000103342.4
|
Igkv4-79
|
immunoglobulin kappa variable 4-79 |
| chr5_-_31854942 | 0.34 |
ENSMUST00000031018.10
|
Rbks
|
ribokinase |
| chr6_-_69037208 | 0.34 |
ENSMUST00000103343.4
|
Igkv4-78
|
immunoglobulin kappa variable 4-78 |
| chr8_+_94905710 | 0.33 |
ENSMUST00000034215.8
ENSMUST00000212291.2 ENSMUST00000211807.2 |
Mt1
|
metallothionein 1 |
| chr5_+_146428173 | 0.32 |
ENSMUST00000110611.8
ENSMUST00000198912.2 |
Gm6370
|
predicted gene 6370 |
| chr6_-_69204417 | 0.29 |
ENSMUST00000103346.3
|
Igkv4-72
|
immunoglobulin kappa chain variable 4-72 |
| chr15_-_85705935 | 0.28 |
ENSMUST00000064370.6
|
Pkdrej
|
polycystin (PKD) family receptor for egg jelly |
| chr5_-_146107531 | 0.28 |
ENSMUST00000174320.2
|
Gm6309
|
predicted gene 6309 |
| chr5_+_146439209 | 0.27 |
ENSMUST00000110598.3
|
4930449I24Rik
|
RIKEN cDNA 4930449I24 gene |
| chr6_-_69377081 | 0.27 |
ENSMUST00000177795.2
|
Igkv4-62
|
immunoglobulin kappa variable 4-62 |
| chr5_+_146474457 | 0.26 |
ENSMUST00000199142.5
ENSMUST00000110597.4 ENSMUST00000110599.9 |
Gm3409
|
predicted gene 3409 |
| chr10_+_21869776 | 0.25 |
ENSMUST00000092673.11
|
Sgk1
|
serum/glucocorticoid regulated kinase 1 |
| chr5_+_146450933 | 0.25 |
ENSMUST00000200228.5
ENSMUST00000036715.16 ENSMUST00000077133.7 |
Gm3402
|
predicted gene 3402 |
| chrX_-_126644166 | 0.24 |
ENSMUST00000052500.6
|
Cldn34c4
|
claudin 34C4 |
| chr16_+_38167352 | 0.21 |
ENSMUST00000050273.9
ENSMUST00000120495.2 ENSMUST00000119704.2 |
Cox17
Gm21987
|
cytochrome c oxidase assembly protein 17, copper chaperone predicted gene 21987 |
| chr5_+_146492946 | 0.20 |
ENSMUST00000179214.8
ENSMUST00000110595.3 |
Gm3415
|
predicted gene 3415 |
| chr11_-_71092124 | 0.20 |
ENSMUST00000108514.10
|
Nlrp1b
|
NLR family, pyrin domain containing 1B |
| chr14_+_35816874 | 0.18 |
ENSMUST00000226305.2
|
4930474N05Rik
|
RIKEN cDNA 4930474N05 gene |
| chr5_-_146097347 | 0.17 |
ENSMUST00000199463.2
|
Gm5565
|
predicted gene 5565 |
| chr9_+_123195986 | 0.16 |
ENSMUST00000038863.9
ENSMUST00000216843.2 |
Lars2
|
leucyl-tRNA synthetase, mitochondrial |
| chr5_+_146418775 | 0.16 |
ENSMUST00000179032.3
|
Gm6408
|
predicted gene 6408 |
| chr6_-_126512375 | 0.15 |
ENSMUST00000060972.5
|
Kcna5
|
potassium voltage-gated channel, shaker-related subfamily, member 5 |
| chr5_+_31855009 | 0.12 |
ENSMUST00000201352.4
ENSMUST00000202815.4 |
Babam2
|
BRISC and BRCA1 A complex member 2 |
| chr6_+_52691204 | 0.10 |
ENSMUST00000138040.8
ENSMUST00000129660.2 |
Tax1bp1
|
Tax1 (human T cell leukemia virus type I) binding protein 1 |
| chr8_-_25730878 | 0.08 |
ENSMUST00000210488.2
ENSMUST00000210933.2 |
Tacc1
|
transforming, acidic coiled-coil containing protein 1 |
| chr7_+_118199375 | 0.07 |
ENSMUST00000121744.9
|
Tmc5
|
transmembrane channel-like gene family 5 |
| chrX_+_101846569 | 0.04 |
ENSMUST00000118218.8
|
Dmrtc1c1
|
DMRT-like family C1c1 |
| chrX_-_101898222 | 0.04 |
ENSMUST00000120314.8
|
Dmrtc1c2
|
DMRT-like family C1c2 |
| chr6_+_125297596 | 0.04 |
ENSMUST00000176655.8
ENSMUST00000176110.8 |
Scnn1a
|
sodium channel, nonvoltage-gated 1 alpha |
| chr17_-_23531402 | 0.04 |
ENSMUST00000168033.3
|
Vmn2r114
|
vomeronasal 2, receptor 114 |
| chr6_-_69220672 | 0.03 |
ENSMUST00000196201.2
|
Igkv4-71
|
immunoglobulin kappa chain variable 4-71 |
| chr14_+_44822935 | 0.03 |
ENSMUST00000178759.2
|
Gm8247
|
predicted gene 8247 |
| chr11_+_50905063 | 0.02 |
ENSMUST00000217480.2
ENSMUST00000215409.2 |
Olfr54
|
olfactory receptor 54 |
| chr11_+_101339233 | 0.01 |
ENSMUST00000010502.13
|
Ifi35
|
interferon-induced protein 35 |
| chr14_-_42461286 | 0.01 |
ENSMUST00000163102.2
|
Gm3633
|
predicted gene 3633 |
| chr4_-_9643636 | 0.01 |
ENSMUST00000108333.8
ENSMUST00000108334.8 ENSMUST00000108335.8 ENSMUST00000152526.8 ENSMUST00000103004.10 |
Asph
|
aspartate-beta-hydroxylase |
| chr10_-_14593935 | 0.01 |
ENSMUST00000020016.5
|
Gje1
|
gap junction protein, epsilon 1 |
| chr14_-_42116736 | 0.00 |
ENSMUST00000100698.4
|
Gm9611
|
predicted gene 9611 |
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 1.1 | GO:0006478 | peptidyl-tyrosine sulfation(GO:0006478) |
| 0.2 | 0.2 | GO:1904959 | regulation of cytochrome-c oxidase activity(GO:1904959) |
| 0.2 | 0.8 | GO:2000588 | positive regulation of platelet-derived growth factor receptor-beta signaling pathway(GO:2000588) |
| 0.2 | 4.3 | GO:0014850 | response to muscle activity(GO:0014850) |
| 0.2 | 0.9 | GO:0042414 | epinephrine metabolic process(GO:0042414) |
| 0.2 | 0.6 | GO:0046340 | termination of RNA polymerase I transcription(GO:0006363) diacylglycerol catabolic process(GO:0046340) |
| 0.1 | 0.9 | GO:0097167 | circadian regulation of translation(GO:0097167) |
| 0.1 | 0.6 | GO:1904784 | NLRP1 inflammasome complex assembly(GO:1904784) |
| 0.1 | 0.3 | GO:0006014 | D-ribose metabolic process(GO:0006014) |
| 0.1 | 1.5 | GO:0016998 | cell wall macromolecule catabolic process(GO:0016998) |
| 0.1 | 0.9 | GO:0015670 | carbon dioxide transport(GO:0015670) |
| 0.1 | 0.3 | GO:0010273 | detoxification of copper ion(GO:0010273) stress response to copper ion(GO:1990169) |
| 0.1 | 0.2 | GO:0006429 | leucyl-tRNA aminoacylation(GO:0006429) |
| 0.0 | 0.4 | GO:0051694 | pointed-end actin filament capping(GO:0051694) |
| 0.0 | 5.5 | GO:0002377 | immunoglobulin production(GO:0002377) |
| 0.0 | 5.1 | GO:0017157 | regulation of exocytosis(GO:0017157) |
| 0.0 | 0.3 | GO:0043402 | glucocorticoid mediated signaling pathway(GO:0043402) |
| 0.0 | 0.1 | GO:0098914 | bundle of His cell to Purkinje myocyte signaling(GO:0086028) bundle of His cell action potential(GO:0086043) membrane repolarization during atrial cardiac muscle cell action potential(GO:0098914) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 1.5 | GO:0048237 | rough endoplasmic reticulum lumen(GO:0048237) |
| 0.2 | 0.8 | GO:0098890 | extrinsic component of postsynaptic membrane(GO:0098890) |
| 0.1 | 0.6 | GO:0072558 | NLRP1 inflammasome complex(GO:0072558) |
| 0.0 | 0.8 | GO:0031235 | intrinsic component of the cytoplasmic side of the plasma membrane(GO:0031235) |
| 0.0 | 0.9 | GO:0043196 | varicosity(GO:0043196) |
| 0.0 | 4.9 | GO:0008021 | synaptic vesicle(GO:0008021) |
| 0.0 | 0.1 | GO:0070552 | BRISC complex(GO:0070552) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 1.1 | GO:0008476 | protein-tyrosine sulfotransferase activity(GO:0008476) |
| 0.1 | 0.6 | GO:0042134 | rRNA primary transcript binding(GO:0042134) |
| 0.1 | 1.5 | GO:0003796 | lysozyme activity(GO:0003796) |
| 0.1 | 0.8 | GO:0032051 | clathrin light chain binding(GO:0032051) |
| 0.1 | 0.2 | GO:0004823 | leucine-tRNA ligase activity(GO:0004823) |
| 0.1 | 0.9 | GO:0004089 | carbonate dehydratase activity(GO:0004089) |
| 0.0 | 0.3 | GO:0043423 | 3-phosphoinositide-dependent protein kinase binding(GO:0043423) |
| 0.0 | 0.2 | GO:0016531 | copper chaperone activity(GO:0016531) |
| 0.0 | 0.1 | GO:0086089 | voltage-gated potassium channel activity involved in atrial cardiac muscle cell action potential repolarization(GO:0086089) |
| 0.0 | 0.4 | GO:0005523 | tropomyosin binding(GO:0005523) |
| 0.0 | 4.3 | GO:0004721 | phosphoprotein phosphatase activity(GO:0004721) |
| 0.0 | 0.9 | GO:0070888 | E-box binding(GO:0070888) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.9 | PID CIRCADIAN PATHWAY | Circadian rhythm pathway |
| 0.0 | 4.9 | NABA ECM GLYCOPROTEINS | Genes encoding structural ECM glycoproteins |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.9 | REACTOME AMINE DERIVED HORMONES | Genes involved in Amine-derived hormones |
| 0.0 | 0.6 | REACTOME HORMONE SENSITIVE LIPASE HSL MEDIATED TRIACYLGLYCEROL HYDROLYSIS | Genes involved in Hormone-sensitive lipase (HSL)-mediated triacylglycerol hydrolysis |
| 0.0 | 0.9 | REACTOME BMAL1 CLOCK NPAS2 ACTIVATES CIRCADIAN EXPRESSION | Genes involved in BMAL1:CLOCK/NPAS2 Activates Circadian Expression |