Project
PRJNA375882: Comprehensive Mouse Transcriptomic BodyMap
Navigation
Downloads
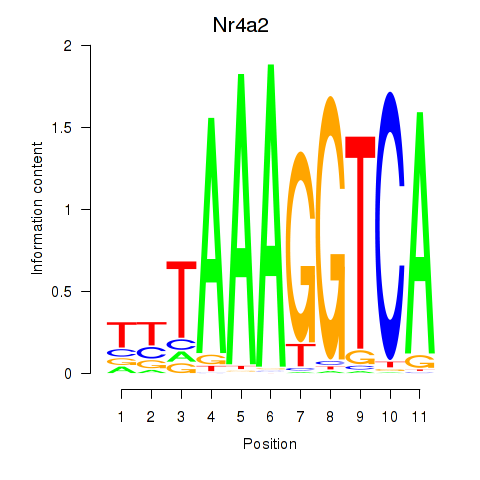
Results for Nr4a2
Z-value: 0.77
Transcription factors associated with Nr4a2
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Nr4a2
|
ENSMUSG00000026826.14 | Nr4a2 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Nr4a2 | mm39_v1_chr2_-_57004933_57005050 | 0.16 | 1.9e-01 | Click! |
Activity profile of Nr4a2 motif
Sorted Z-values of Nr4a2 motif
Network of associatons between targets according to the STRING database.
First level regulatory network of Nr4a2
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr3_-_144680801 | 9.82 |
ENSMUST00000029923.10
ENSMUST00000238960.2 |
Clca4a
|
chloride channel accessory 4A |
| chr12_+_31488208 | 9.08 |
ENSMUST00000001254.6
|
Slc26a3
|
solute carrier family 26, member 3 |
| chr3_-_144638284 | 7.89 |
ENSMUST00000098549.4
|
Clca4b
|
chloride channel accessory 4B |
| chr11_+_70548022 | 6.08 |
ENSMUST00000157027.8
ENSMUST00000072841.12 ENSMUST00000108548.8 ENSMUST00000126241.8 |
Eno3
|
enolase 3, beta muscle |
| chr13_+_74269554 | 5.88 |
ENSMUST00000036208.7
ENSMUST00000225423.2 ENSMUST00000221703.2 |
Slc9a3
|
solute carrier family 9 (sodium/hydrogen exchanger), member 3 |
| chr2_-_163239865 | 4.90 |
ENSMUST00000017961.11
ENSMUST00000109425.3 |
Jph2
|
junctophilin 2 |
| chr12_-_76842263 | 4.61 |
ENSMUST00000082431.6
|
Gpx2
|
glutathione peroxidase 2 |
| chr3_+_122688721 | 3.81 |
ENSMUST00000023820.6
|
Fabp2
|
fatty acid binding protein 2, intestinal |
| chr2_+_132623198 | 3.62 |
ENSMUST00000028826.4
|
Chgb
|
chromogranin B |
| chr8_-_46664321 | 3.48 |
ENSMUST00000034049.5
|
Slc25a4
|
solute carrier family 25 (mitochondrial carrier, adenine nucleotide translocator), member 4 |
| chr11_+_70548622 | 3.47 |
ENSMUST00000170716.8
|
Eno3
|
enolase 3, beta muscle |
| chr8_+_105573693 | 3.18 |
ENSMUST00000055052.6
|
Ces2c
|
carboxylesterase 2C |
| chr1_-_190950129 | 3.15 |
ENSMUST00000195117.2
|
Atf3
|
activating transcription factor 3 |
| chr11_-_69838971 | 2.87 |
ENSMUST00000179298.3
ENSMUST00000018710.13 ENSMUST00000135437.3 ENSMUST00000141837.9 ENSMUST00000142500.8 |
Slc2a4
|
solute carrier family 2 (facilitated glucose transporter), member 4 |
| chr11_+_70548513 | 2.70 |
ENSMUST00000134087.8
|
Eno3
|
enolase 3, beta muscle |
| chr1_+_57884693 | 2.66 |
ENSMUST00000169772.3
|
Spats2l
|
spermatogenesis associated, serine-rich 2-like |
| chr1_+_83094481 | 2.61 |
ENSMUST00000027351.13
ENSMUST00000113437.9 ENSMUST00000186832.2 |
Ccl20
|
chemokine (C-C motif) ligand 20 |
| chr6_-_124791259 | 2.43 |
ENSMUST00000172132.10
ENSMUST00000239432.2 |
Tpi1
|
triosephosphate isomerase 1 |
| chr15_+_35296237 | 2.30 |
ENSMUST00000022952.6
|
Osr2
|
odd-skipped related 2 |
| chr6_-_3487369 | 2.28 |
ENSMUST00000201607.4
|
Hepacam2
|
HEPACAM family member 2 |
| chr1_+_87501721 | 2.20 |
ENSMUST00000166259.8
ENSMUST00000172222.8 ENSMUST00000163606.8 |
Neu2
|
neuraminidase 2 |
| chr1_-_167221344 | 2.16 |
ENSMUST00000028005.3
|
Mgst3
|
microsomal glutathione S-transferase 3 |
| chr13_+_118851214 | 2.06 |
ENSMUST00000022246.9
|
Fgf10
|
fibroblast growth factor 10 |
| chr7_-_89176294 | 1.87 |
ENSMUST00000207932.2
|
Prss23
|
protease, serine 23 |
| chr4_-_141351110 | 1.74 |
ENSMUST00000038661.8
|
Slc25a34
|
solute carrier family 25, member 34 |
| chr13_-_98152768 | 1.68 |
ENSMUST00000238746.2
|
Arhgef28
|
Rho guanine nucleotide exchange factor (GEF) 28 |
| chr16_+_21644692 | 1.66 |
ENSMUST00000232240.2
|
Map3k13
|
mitogen-activated protein kinase kinase kinase 13 |
| chr2_-_13496624 | 1.62 |
ENSMUST00000091436.7
|
Cubn
|
cubilin (intrinsic factor-cobalamin receptor) |
| chr1_-_63215812 | 1.56 |
ENSMUST00000185847.2
ENSMUST00000185732.7 ENSMUST00000188370.7 ENSMUST00000168099.9 |
Ndufs1
|
NADH:ubiquinone oxidoreductase core subunit S1 |
| chr10_-_42354276 | 1.55 |
ENSMUST00000151747.8
|
Afg1l
|
AFG1 like ATPase |
| chr10_-_42354482 | 1.42 |
ENSMUST00000041024.15
|
Afg1l
|
AFG1 like ATPase |
| chr15_-_98505508 | 1.41 |
ENSMUST00000096224.6
|
Adcy6
|
adenylate cyclase 6 |
| chr9_+_118335294 | 1.38 |
ENSMUST00000084820.6
|
Golga4
|
golgi autoantigen, golgin subfamily a, 4 |
| chr18_+_86729184 | 1.36 |
ENSMUST00000068423.10
|
Cbln2
|
cerebellin 2 precursor protein |
| chr7_-_141009264 | 1.34 |
ENSMUST00000164387.2
ENSMUST00000137488.2 ENSMUST00000084436.10 |
Cend1
|
cell cycle exit and neuronal differentiation 1 |
| chr15_+_25940859 | 1.31 |
ENSMUST00000226750.2
|
Retreg1
|
reticulophagy regulator 1 |
| chr1_-_123972900 | 1.31 |
ENSMUST00000112603.4
|
Dpp10
|
dipeptidylpeptidase 10 |
| chr3_+_89344006 | 1.30 |
ENSMUST00000038942.10
ENSMUST00000130858.8 ENSMUST00000146630.8 ENSMUST00000145753.2 |
Pbxip1
|
pre B cell leukemia transcription factor interacting protein 1 |
| chr15_-_76113692 | 1.30 |
ENSMUST00000074834.12
|
Plec
|
plectin |
| chr1_-_63215952 | 1.24 |
ENSMUST00000185412.7
ENSMUST00000027111.15 ENSMUST00000189664.2 |
Ndufs1
|
NADH:ubiquinone oxidoreductase core subunit S1 |
| chr2_-_129541753 | 1.18 |
ENSMUST00000028883.12
|
Pdyn
|
prodynorphin |
| chr10_+_98943999 | 1.18 |
ENSMUST00000161240.4
|
Galnt4
|
polypeptide N-acetylgalactosaminyltransferase 4 |
| chr5_-_121665249 | 1.15 |
ENSMUST00000152270.8
|
Mapkapk5
|
MAP kinase-activated protein kinase 5 |
| chr1_-_87501548 | 1.15 |
ENSMUST00000068681.12
|
Ngef
|
neuronal guanine nucleotide exchange factor |
| chr11_-_59029996 | 1.14 |
ENSMUST00000219084.3
|
Obscn
|
obscurin, cytoskeletal calmodulin and titin-interacting RhoGEF |
| chr6_-_138399896 | 1.11 |
ENSMUST00000161450.8
ENSMUST00000163024.8 ENSMUST00000162185.8 |
Lmo3
|
LIM domain only 3 |
| chr7_+_120234399 | 1.10 |
ENSMUST00000033176.7
ENSMUST00000208400.2 |
Uqcrc2
|
ubiquinol cytochrome c reductase core protein 2 |
| chr15_+_102875229 | 1.10 |
ENSMUST00000001699.8
|
Hoxc10
|
homeobox C10 |
| chr19_-_32038838 | 1.05 |
ENSMUST00000096119.5
|
Asah2
|
N-acylsphingosine amidohydrolase 2 |
| chr11_+_121593582 | 1.04 |
ENSMUST00000125580.2
|
Metrnl
|
meteorin, glial cell differentiation regulator-like |
| chr4_+_99812912 | 1.02 |
ENSMUST00000102783.5
|
Pgm1
|
phosphoglucomutase 1 |
| chr19_+_11724913 | 1.00 |
ENSMUST00000025585.4
|
Cblif
|
cobalamin binding intrinsic factor |
| chr10_-_23226684 | 1.00 |
ENSMUST00000220299.2
|
Eya4
|
EYA transcriptional coactivator and phosphatase 4 |
| chr11_+_4207557 | 0.91 |
ENSMUST00000066283.12
|
Lif
|
leukemia inhibitory factor |
| chr15_+_25940781 | 0.87 |
ENSMUST00000227275.2
|
Retreg1
|
reticulophagy regulator 1 |
| chr5_-_66309244 | 0.85 |
ENSMUST00000167950.8
|
Rbm47
|
RNA binding motif protein 47 |
| chr2_-_120394288 | 0.85 |
ENSMUST00000055241.13
ENSMUST00000135625.8 |
Zfp106
|
zinc finger protein 106 |
| chr7_+_140500848 | 0.81 |
ENSMUST00000184560.2
|
Nlrp6
|
NLR family, pyrin domain containing 6 |
| chr8_-_65186565 | 0.80 |
ENSMUST00000141021.2
|
Msmo1
|
methylsterol monoxygenase 1 |
| chr5_+_30437579 | 0.79 |
ENSMUST00000145167.9
|
Selenoi
|
selenoprotein I |
| chrX_-_16683578 | 0.76 |
ENSMUST00000040820.13
|
Maob
|
monoamine oxidase B |
| chr7_-_141009346 | 0.73 |
ENSMUST00000124444.2
|
Cend1
|
cell cycle exit and neuronal differentiation 1 |
| chr7_+_140500808 | 0.71 |
ENSMUST00000106045.8
ENSMUST00000183845.8 |
Nlrp6
|
NLR family, pyrin domain containing 6 |
| chr5_-_67256578 | 0.71 |
ENSMUST00000012664.11
|
Phox2b
|
paired-like homeobox 2b |
| chr15_+_25940931 | 0.69 |
ENSMUST00000110438.3
|
Retreg1
|
reticulophagy regulator 1 |
| chr10_-_20600442 | 0.68 |
ENSMUST00000170265.8
|
Pde7b
|
phosphodiesterase 7B |
| chr19_-_40371016 | 0.65 |
ENSMUST00000225766.3
|
Sorbs1
|
sorbin and SH3 domain containing 1 |
| chr5_+_150183201 | 0.61 |
ENSMUST00000087204.9
|
Fry
|
FRY microtubule binding protein |
| chr6_+_96090127 | 0.60 |
ENSMUST00000122120.8
|
Tafa1
|
TAFA chemokine like family member 1 |
| chr10_-_20600797 | 0.59 |
ENSMUST00000020165.14
|
Pde7b
|
phosphodiesterase 7B |
| chr7_+_4122523 | 0.57 |
ENSMUST00000119661.8
ENSMUST00000129423.8 |
Ttyh1
|
tweety family member 1 |
| chr10_+_110756031 | 0.56 |
ENSMUST00000220409.2
ENSMUST00000219502.2 |
Csrp2
|
cysteine and glycine-rich protein 2 |
| chr10_+_27950809 | 0.56 |
ENSMUST00000166468.2
ENSMUST00000218359.2 ENSMUST00000218276.2 |
Ptprk
|
protein tyrosine phosphatase, receptor type, K |
| chr2_+_121125918 | 0.54 |
ENSMUST00000110639.8
|
Map1a
|
microtubule-associated protein 1 A |
| chr13_+_93440265 | 0.53 |
ENSMUST00000109494.8
|
Homer1
|
homer scaffolding protein 1 |
| chr15_+_25940912 | 0.51 |
ENSMUST00000226438.2
|
Retreg1
|
reticulophagy regulator 1 |
| chr2_+_105499233 | 0.49 |
ENSMUST00000111086.11
ENSMUST00000111087.10 |
Pax6
|
paired box 6 |
| chr3_-_87934772 | 0.48 |
ENSMUST00000005014.9
|
Hapln2
|
hyaluronan and proteoglycan link protein 2 |
| chr7_+_4122555 | 0.47 |
ENSMUST00000079415.12
|
Ttyh1
|
tweety family member 1 |
| chr2_+_158636727 | 0.47 |
ENSMUST00000029186.14
ENSMUST00000109478.9 ENSMUST00000156893.2 |
Dhx35
|
DEAH (Asp-Glu-Ala-His) box polypeptide 35 |
| chr1_+_167445815 | 0.46 |
ENSMUST00000111380.2
|
Rxrg
|
retinoid X receptor gamma |
| chr9_-_106533279 | 0.46 |
ENSMUST00000023959.13
ENSMUST00000201681.2 |
Grm2
|
glutamate receptor, metabotropic 2 |
| chr10_-_40178182 | 0.46 |
ENSMUST00000099945.6
ENSMUST00000238953.2 ENSMUST00000238969.2 |
Amd1
|
S-adenosylmethionine decarboxylase 1 |
| chr2_+_172994841 | 0.44 |
ENSMUST00000029017.6
|
Pck1
|
phosphoenolpyruvate carboxykinase 1, cytosolic |
| chr3_-_20329823 | 0.43 |
ENSMUST00000011607.6
|
Cpb1
|
carboxypeptidase B1 (tissue) |
| chr18_+_77861656 | 0.43 |
ENSMUST00000114748.2
|
Atp5a1
|
ATP synthase, H+ transporting, mitochondrial F1 complex, alpha subunit 1 |
| chr11_-_79394904 | 0.42 |
ENSMUST00000164465.3
|
Omg
|
oligodendrocyte myelin glycoprotein |
| chrX_-_166638057 | 0.42 |
ENSMUST00000238211.2
|
Frmpd4
|
FERM and PDZ domain containing 4 |
| chr19_-_40371242 | 0.42 |
ENSMUST00000224583.2
|
Sorbs1
|
sorbin and SH3 domain containing 1 |
| chr10_+_90665639 | 0.38 |
ENSMUST00000179337.9
|
Anks1b
|
ankyrin repeat and sterile alpha motif domain containing 1B |
| chr2_-_73741664 | 0.37 |
ENSMUST00000111996.8
ENSMUST00000018914.3 |
Atp5g3
|
ATP synthase, H+ transporting, mitochondrial F0 complex, subunit C3 (subunit 9) |
| chr16_+_8647959 | 0.37 |
ENSMUST00000023150.7
|
1810013L24Rik
|
RIKEN cDNA 1810013L24 gene |
| chr8_-_13544478 | 0.37 |
ENSMUST00000033828.7
|
Gas6
|
growth arrest specific 6 |
| chr19_-_5168251 | 0.35 |
ENSMUST00000113728.8
ENSMUST00000113727.8 ENSMUST00000025798.13 |
Klc2
|
kinesin light chain 2 |
| chr2_-_101451383 | 0.35 |
ENSMUST00000090513.11
|
Iftap
|
intraflagellar transport associated protein |
| chr3_-_95041246 | 0.35 |
ENSMUST00000172572.9
ENSMUST00000173462.3 |
Scnm1
|
sodium channel modifier 1 |
| chr2_+_4980923 | 0.35 |
ENSMUST00000167607.8
ENSMUST00000115010.9 |
Ucma
|
upper zone of growth plate and cartilage matrix associated |
| chr2_+_4980986 | 0.29 |
ENSMUST00000027978.7
ENSMUST00000195688.2 |
Ucma
|
upper zone of growth plate and cartilage matrix associated |
| chr4_+_102617495 | 0.28 |
ENSMUST00000072481.12
ENSMUST00000156596.8 ENSMUST00000080728.13 ENSMUST00000106882.9 |
Sgip1
|
SH3-domain GRB2-like (endophilin) interacting protein 1 |
| chr2_-_165125519 | 0.28 |
ENSMUST00000155289.8
|
Slc35c2
|
solute carrier family 35, member C2 |
| chr11_-_24025054 | 0.27 |
ENSMUST00000068360.2
|
A830031A19Rik
|
RIKEN cDNA A830031A19 gene |
| chr2_-_26127360 | 0.27 |
ENSMUST00000036187.9
|
Qsox2
|
quiescin Q6 sulfhydryl oxidase 2 |
| chr10_-_99595498 | 0.27 |
ENSMUST00000056085.6
|
Csl
|
citrate synthase like |
| chr7_-_65177444 | 0.27 |
ENSMUST00000206228.2
|
Tjp1
|
tight junction protein 1 |
| chr10_-_12745109 | 0.26 |
ENSMUST00000218635.2
|
Utrn
|
utrophin |
| chr19_+_46140942 | 0.24 |
ENSMUST00000026254.14
|
Gbf1
|
golgi-specific brefeldin A-resistance factor 1 |
| chr2_+_61423469 | 0.24 |
ENSMUST00000112494.2
|
Tank
|
TRAF family member-associated Nf-kappa B activator |
| chr9_+_21914513 | 0.23 |
ENSMUST00000215795.2
|
Prkcsh
|
protein kinase C substrate 80K-H |
| chr1_+_63215976 | 0.23 |
ENSMUST00000129339.8
|
Eef1b2
|
eukaryotic translation elongation factor 1 beta 2 |
| chr1_+_63216281 | 0.21 |
ENSMUST00000188524.2
|
Eef1b2
|
eukaryotic translation elongation factor 1 beta 2 |
| chr9_+_21914296 | 0.20 |
ENSMUST00000003493.9
|
Prkcsh
|
protein kinase C substrate 80K-H |
| chr15_+_76579960 | 0.20 |
ENSMUST00000229679.2
|
Gpt
|
glutamic pyruvic transaminase, soluble |
| chr2_+_128907854 | 0.20 |
ENSMUST00000035812.14
|
Ttl
|
tubulin tyrosine ligase |
| chr5_+_63806451 | 0.19 |
ENSMUST00000159584.3
|
Nwd2
|
NACHT and WD repeat domain containing 2 |
| chr2_+_127180559 | 0.17 |
ENSMUST00000059839.9
|
Astl
|
astacin-like metalloendopeptidase (M12 family) |
| chr9_+_21914083 | 0.17 |
ENSMUST00000216344.2
|
Prkcsh
|
protein kinase C substrate 80K-H |
| chr7_-_102566717 | 0.16 |
ENSMUST00000214160.2
ENSMUST00000215773.2 |
Olfr571
|
olfactory receptor 571 |
| chr16_-_20245071 | 0.16 |
ENSMUST00000115547.9
ENSMUST00000096199.5 |
Abcc5
|
ATP-binding cassette, sub-family C (CFTR/MRP), member 5 |
| chrX_-_47602395 | 0.15 |
ENSMUST00000114945.9
ENSMUST00000037349.8 |
Aifm1
|
apoptosis-inducing factor, mitochondrion-associated 1 |
| chr10_-_93425553 | 0.15 |
ENSMUST00000020203.7
|
Snrpf
|
small nuclear ribonucleoprotein polypeptide F |
| chr15_+_25774070 | 0.13 |
ENSMUST00000125667.3
|
Myo10
|
myosin X |
| chr9_+_21914334 | 0.13 |
ENSMUST00000115331.10
|
Prkcsh
|
protein kinase C substrate 80K-H |
| chr4_-_150998857 | 0.12 |
ENSMUST00000105675.8
|
Park7
|
Parkinson disease (autosomal recessive, early onset) 7 |
| chr16_-_20245138 | 0.11 |
ENSMUST00000079158.13
|
Abcc5
|
ATP-binding cassette, sub-family C (CFTR/MRP), member 5 |
| chr10_-_35587888 | 0.10 |
ENSMUST00000080898.4
|
Amd2
|
S-adenosylmethionine decarboxylase 2 |
| chr14_+_27598021 | 0.08 |
ENSMUST00000211684.2
ENSMUST00000210924.2 |
Erc2
|
ELKS/RAB6-interacting/CAST family member 2 |
| chr19_-_45986919 | 0.07 |
ENSMUST00000045396.9
|
Armh3
|
armadillo-like helical domain containing 3 |
| chr12_+_119356318 | 0.06 |
ENSMUST00000221866.2
|
Macc1
|
metastasis associated in colon cancer 1 |
| chr4_-_36136463 | 0.06 |
ENSMUST00000098151.9
|
Lingo2
|
leucine rich repeat and Ig domain containing 2 |
| chr4_+_33081505 | 0.06 |
ENSMUST00000147889.2
|
Gabrr2
|
gamma-aminobutyric acid (GABA) C receptor, subunit rho 2 |
| chr9_+_98746796 | 0.05 |
ENSMUST00000167951.3
|
Prr23a3
|
proline rich 23A, member 3 |
| chr16_-_43959993 | 0.05 |
ENSMUST00000137557.8
|
Atp6v1a
|
ATPase, H+ transporting, lysosomal V1 subunit A |
| chr2_-_73722874 | 0.05 |
ENSMUST00000136958.8
ENSMUST00000112010.9 ENSMUST00000128531.8 ENSMUST00000112017.8 |
Atf2
|
activating transcription factor 2 |
| chr17_+_70829050 | 0.03 |
ENSMUST00000133717.9
ENSMUST00000148486.8 |
Dlgap1
|
DLG associated protein 1 |
| chr2_+_75662806 | 0.02 |
ENSMUST00000175646.2
|
Agps
|
alkylglycerone phosphate synthase |
| chr2_-_25911544 | 0.02 |
ENSMUST00000136750.3
|
Ubac1
|
ubiquitin associated domain containing 1 |
| chr9_-_21996693 | 0.01 |
ENSMUST00000179422.8
ENSMUST00000098937.10 ENSMUST00000177967.2 ENSMUST00000180180.8 |
Ecsit
|
ECSIT signalling integrator |
| chr6_+_132832401 | 0.01 |
ENSMUST00000032315.3
|
Tas2r116
|
taste receptor, type 2, member 116 |
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.2 | 4.6 | GO:0009609 | response to symbiont(GO:0009608) response to symbiotic bacterium(GO:0009609) |
| 0.9 | 3.5 | GO:1902109 | negative regulation of mitochondrial membrane permeability involved in apoptotic process(GO:1902109) |
| 0.9 | 2.6 | GO:0045362 | regulation of interleukin-1 biosynthetic process(GO:0045360) positive regulation of interleukin-1 biosynthetic process(GO:0045362) |
| 0.8 | 2.4 | GO:0019405 | alditol catabolic process(GO:0019405) glyceraldehyde-3-phosphate biosynthetic process(GO:0046166) |
| 0.8 | 3.1 | GO:1903984 | positive regulation of TRAIL-activated apoptotic signaling pathway(GO:1903984) |
| 0.7 | 2.1 | GO:0060435 | bronchiole development(GO:0060435) secretion by lung epithelial cell involved in lung growth(GO:0061033) |
| 0.6 | 4.9 | GO:0060316 | positive regulation of ryanodine-sensitive calcium-release channel activity(GO:0060316) |
| 0.6 | 9.1 | GO:0045852 | pH elevation(GO:0045852) intracellular pH elevation(GO:0051454) |
| 0.6 | 3.0 | GO:0035694 | mitochondrial protein catabolic process(GO:0035694) |
| 0.5 | 2.1 | GO:0021941 | radial glia guided migration of cerebellar granule cell(GO:0021933) negative regulation of cerebellar granule cell precursor proliferation(GO:0021941) |
| 0.5 | 5.9 | GO:1990118 | sodium ion import across plasma membrane(GO:0098719) sodium ion import into cell(GO:1990118) |
| 0.5 | 1.4 | GO:1904117 | response to vasopressin(GO:1904116) cellular response to vasopressin(GO:1904117) |
| 0.5 | 2.3 | GO:0036022 | limb joint morphogenesis(GO:0036022) embryonic skeletal limb joint morphogenesis(GO:0036023) |
| 0.4 | 3.4 | GO:0061709 | reticulophagy(GO:0061709) |
| 0.4 | 0.4 | GO:1900141 | regulation of oligodendrocyte apoptotic process(GO:1900141) negative regulation of oligodendrocyte apoptotic process(GO:1900142) |
| 0.3 | 2.2 | GO:0009313 | ganglioside catabolic process(GO:0006689) oligosaccharide catabolic process(GO:0009313) |
| 0.3 | 1.6 | GO:0015889 | cobalamin transport(GO:0015889) |
| 0.2 | 0.7 | GO:0021934 | hindbrain tangential cell migration(GO:0021934) |
| 0.2 | 0.6 | GO:0006597 | spermine biosynthetic process(GO:0006597) |
| 0.2 | 1.1 | GO:0006627 | protein processing involved in protein targeting to mitochondrion(GO:0006627) |
| 0.2 | 0.6 | GO:1904428 | negative regulation of tubulin deacetylation(GO:1904428) |
| 0.2 | 0.8 | GO:0014063 | negative regulation of serotonin secretion(GO:0014063) |
| 0.2 | 0.9 | GO:0043974 | histone H3-K27 acetylation(GO:0043974) spongiotrophoblast differentiation(GO:0060708) regulation of histone H3-K27 acetylation(GO:1901674) |
| 0.1 | 18.8 | GO:0006821 | chloride transport(GO:0006821) |
| 0.1 | 0.4 | GO:0061402 | glycerol biosynthetic process(GO:0006114) positive regulation of transcription from RNA polymerase II promoter in response to acidic pH(GO:0061402) |
| 0.1 | 1.1 | GO:0090400 | stress-induced premature senescence(GO:0090400) |
| 0.1 | 1.7 | GO:0060052 | neurofilament cytoskeleton organization(GO:0060052) |
| 0.1 | 12.2 | GO:0006096 | glycolytic process(GO:0006096) |
| 0.1 | 2.9 | GO:0044381 | glucose import in response to insulin stimulus(GO:0044381) |
| 0.1 | 1.5 | GO:0002862 | negative regulation of inflammatory response to antigenic stimulus(GO:0002862) |
| 0.1 | 1.1 | GO:2000507 | positive regulation of energy homeostasis(GO:2000507) |
| 0.1 | 1.3 | GO:0031581 | hemidesmosome assembly(GO:0031581) |
| 0.1 | 0.2 | GO:0070973 | protein localization to endoplasmic reticulum exit site(GO:0070973) |
| 0.1 | 0.8 | GO:0006646 | phosphatidylethanolamine biosynthetic process(GO:0006646) |
| 0.1 | 3.8 | GO:0050892 | intestinal absorption(GO:0050892) |
| 0.1 | 1.1 | GO:0061002 | negative regulation of dendritic spine morphogenesis(GO:0061002) |
| 0.1 | 0.3 | GO:0015786 | UDP-glucose transport(GO:0015786) |
| 0.1 | 0.5 | GO:0008065 | establishment of blood-nerve barrier(GO:0008065) |
| 0.1 | 1.0 | GO:0016576 | histone dephosphorylation(GO:0016576) |
| 0.1 | 1.4 | GO:0021520 | spinal cord motor neuron cell fate specification(GO:0021520) |
| 0.1 | 1.1 | GO:0045725 | positive regulation of glycogen biosynthetic process(GO:0045725) |
| 0.0 | 2.9 | GO:0042773 | ATP synthesis coupled electron transport(GO:0042773) |
| 0.0 | 0.6 | GO:0010839 | negative regulation of keratinocyte proliferation(GO:0010839) |
| 0.0 | 0.3 | GO:0007527 | adult somatic muscle development(GO:0007527) |
| 0.0 | 0.2 | GO:2000360 | negative regulation of binding of sperm to zona pellucida(GO:2000360) |
| 0.0 | 0.4 | GO:1900383 | regulation of synaptic plasticity by receptor localization to synapse(GO:1900383) |
| 0.0 | 0.5 | GO:2001256 | regulation of store-operated calcium entry(GO:2001256) |
| 0.0 | 0.8 | GO:0015986 | energy coupled proton transport, down electrochemical gradient(GO:0015985) ATP synthesis coupled proton transport(GO:0015986) |
| 0.0 | 0.7 | GO:0006491 | N-glycan processing(GO:0006491) |
| 0.0 | 1.4 | GO:0043001 | Golgi to plasma membrane protein transport(GO:0043001) |
| 0.0 | 0.3 | GO:0090557 | establishment of endothelial intestinal barrier(GO:0090557) |
| 0.0 | 1.3 | GO:1903078 | positive regulation of protein localization to plasma membrane(GO:1903078) |
| 0.0 | 0.9 | GO:0000186 | activation of MAPKK activity(GO:0000186) |
| 0.0 | 0.4 | GO:0032291 | central nervous system myelination(GO:0022010) axon ensheathment in central nervous system(GO:0032291) |
| 0.0 | 1.1 | GO:0045600 | positive regulation of fat cell differentiation(GO:0045600) |
| 0.0 | 2.3 | GO:0007098 | centrosome cycle(GO:0007098) |
| 0.0 | 1.4 | GO:0051965 | positive regulation of synapse assembly(GO:0051965) |
| 0.0 | 0.5 | GO:1901522 | positive regulation of transcription from RNA polymerase II promoter involved in cellular response to chemical stimulus(GO:1901522) |
| 0.0 | 0.9 | GO:0007218 | neuropeptide signaling pathway(GO:0007218) |
| 0.0 | 0.2 | GO:1902510 | regulation of apoptotic DNA fragmentation(GO:1902510) |
| 0.0 | 0.5 | GO:0007026 | negative regulation of microtubule depolymerization(GO:0007026) |
| 0.0 | 0.5 | GO:0014047 | glutamate secretion(GO:0014047) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.0 | 3.1 | GO:1990622 | CHOP-ATF3 complex(GO:1990622) |
| 0.9 | 12.2 | GO:0000015 | phosphopyruvate hydratase complex(GO:0000015) |
| 0.4 | 4.9 | GO:0030314 | junctional membrane complex(GO:0030314) |
| 0.4 | 1.1 | GO:0032156 | septin cytoskeleton(GO:0032156) |
| 0.4 | 1.1 | GO:0005863 | striated muscle myosin thick filament(GO:0005863) |
| 0.3 | 3.8 | GO:0045179 | apical cortex(GO:0045179) |
| 0.3 | 1.6 | GO:0043202 | lysosomal lumen(GO:0043202) |
| 0.3 | 9.1 | GO:0097225 | sperm midpiece(GO:0097225) |
| 0.2 | 1.0 | GO:0097425 | smooth endoplasmic reticulum membrane(GO:0030868) smooth endoplasmic reticulum part(GO:0097425) |
| 0.2 | 1.1 | GO:0005899 | insulin receptor complex(GO:0005899) |
| 0.2 | 2.9 | GO:0032593 | insulin-responsive compartment(GO:0032593) |
| 0.1 | 0.7 | GO:0017177 | glucosidase II complex(GO:0017177) |
| 0.1 | 1.1 | GO:0019907 | cyclin-dependent protein kinase activating kinase holoenzyme complex(GO:0019907) |
| 0.1 | 1.7 | GO:0008385 | IkappaB kinase complex(GO:0008385) |
| 0.1 | 1.1 | GO:0005750 | mitochondrial respiratory chain complex III(GO:0005750) |
| 0.1 | 1.5 | GO:0061702 | inflammasome complex(GO:0061702) |
| 0.1 | 0.4 | GO:0005853 | eukaryotic translation elongation factor 1 complex(GO:0005853) |
| 0.1 | 0.2 | GO:0060473 | cortical granule(GO:0060473) |
| 0.1 | 2.9 | GO:0030964 | mitochondrial respiratory chain complex I(GO:0005747) NADH dehydrogenase complex(GO:0030964) respiratory chain complex I(GO:0045271) |
| 0.1 | 1.4 | GO:0008074 | guanylate cyclase complex, soluble(GO:0008074) |
| 0.1 | 3.9 | GO:0005801 | cis-Golgi network(GO:0005801) |
| 0.0 | 0.4 | GO:0045261 | mitochondrial proton-transporting ATP synthase complex, catalytic core F(1)(GO:0000275) proton-transporting ATP synthase complex, catalytic core F(1)(GO:0045261) |
| 0.0 | 1.3 | GO:0030056 | hemidesmosome(GO:0030056) |
| 0.0 | 4.4 | GO:0031526 | brush border membrane(GO:0031526) |
| 0.0 | 0.2 | GO:0005683 | U7 snRNP(GO:0005683) |
| 0.0 | 0.4 | GO:0000276 | mitochondrial proton-transporting ATP synthase complex, coupling factor F(o)(GO:0000276) |
| 0.0 | 0.3 | GO:0046581 | intercellular canaliculus(GO:0046581) |
| 0.0 | 0.4 | GO:0035253 | ciliary rootlet(GO:0035253) |
| 0.0 | 0.3 | GO:0070938 | contractile ring(GO:0070938) |
| 0.0 | 0.6 | GO:0016235 | aggresome(GO:0016235) |
| 0.0 | 2.3 | GO:0030496 | midbody(GO:0030496) |
| 0.0 | 3.1 | GO:0043209 | myelin sheath(GO:0043209) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.9 | 12.2 | GO:0004634 | phosphopyruvate hydratase activity(GO:0004634) |
| 0.6 | 5.9 | GO:0015386 | potassium:proton antiporter activity(GO:0015386) |
| 0.6 | 2.9 | GO:0055056 | D-glucose transmembrane transporter activity(GO:0055056) |
| 0.6 | 17.7 | GO:0005229 | intracellular calcium activated chloride channel activity(GO:0005229) |
| 0.5 | 9.1 | GO:0019531 | oxalate transmembrane transporter activity(GO:0019531) |
| 0.4 | 2.2 | GO:0004308 | exo-alpha-sialidase activity(GO:0004308) alpha-sialidase activity(GO:0016997) exo-alpha-(2->3)-sialidase activity(GO:0052794) exo-alpha-(2->6)-sialidase activity(GO:0052795) exo-alpha-(2->8)-sialidase activity(GO:0052796) |
| 0.4 | 3.8 | GO:0005324 | long-chain fatty acid transporter activity(GO:0005324) |
| 0.3 | 3.5 | GO:0005347 | ATP transmembrane transporter activity(GO:0005347) ADP transmembrane transporter activity(GO:0015217) |
| 0.3 | 2.1 | GO:0005111 | type 2 fibroblast growth factor receptor binding(GO:0005111) |
| 0.3 | 1.0 | GO:0072320 | volume-sensitive chloride channel activity(GO:0072320) |
| 0.3 | 2.0 | GO:0001515 | opioid peptide activity(GO:0001515) |
| 0.2 | 2.4 | GO:0016861 | intramolecular oxidoreductase activity, interconverting aldoses and ketoses(GO:0016861) |
| 0.2 | 1.6 | GO:0030492 | hemoglobin binding(GO:0030492) |
| 0.2 | 0.9 | GO:0005146 | leukemia inhibitory factor receptor binding(GO:0005146) |
| 0.2 | 4.9 | GO:0099604 | calcium-release channel activity(GO:0015278) ligand-gated calcium channel activity(GO:0099604) |
| 0.2 | 1.4 | GO:0008294 | calcium- and calmodulin-responsive adenylate cyclase activity(GO:0008294) |
| 0.2 | 6.8 | GO:0004602 | glutathione peroxidase activity(GO:0004602) |
| 0.2 | 0.8 | GO:0004307 | ethanolaminephosphotransferase activity(GO:0004307) |
| 0.2 | 1.0 | GO:0004614 | phosphoglucomutase activity(GO:0004614) |
| 0.2 | 1.5 | GO:0005000 | vasopressin receptor activity(GO:0005000) |
| 0.1 | 0.4 | GO:0004611 | phosphoenolpyruvate carboxykinase activity(GO:0004611) phosphoenolpyruvate carboxykinase (GTP) activity(GO:0004613) |
| 0.1 | 1.1 | GO:0017040 | ceramidase activity(GO:0017040) |
| 0.1 | 0.4 | GO:0043532 | angiostatin binding(GO:0043532) |
| 0.1 | 2.8 | GO:0008137 | NADH dehydrogenase (ubiquinone) activity(GO:0008137) NADH dehydrogenase (quinone) activity(GO:0050136) |
| 0.1 | 0.8 | GO:0008131 | primary amine oxidase activity(GO:0008131) |
| 0.1 | 0.6 | GO:0019808 | polyamine binding(GO:0019808) |
| 0.1 | 1.1 | GO:0031432 | titin binding(GO:0031432) |
| 0.1 | 1.1 | GO:0009931 | calcium-dependent protein serine/threonine kinase activity(GO:0009931) |
| 0.1 | 1.3 | GO:0008239 | dipeptidyl-peptidase activity(GO:0008239) |
| 0.1 | 2.6 | GO:0048020 | CCR chemokine receptor binding(GO:0048020) |
| 0.1 | 0.2 | GO:0070002 | glutamic-type peptidase activity(GO:0070002) |
| 0.1 | 0.3 | GO:0016971 | flavin-linked sulfhydryl oxidase activity(GO:0016971) |
| 0.1 | 1.2 | GO:0004653 | polypeptide N-acetylgalactosaminyltransferase activity(GO:0004653) |
| 0.0 | 0.5 | GO:0004886 | 9-cis retinoic acid receptor activity(GO:0004886) |
| 0.0 | 0.5 | GO:0001640 | adenylate cyclase inhibiting G-protein coupled glutamate receptor activity(GO:0001640) |
| 0.0 | 0.2 | GO:0047635 | L-alanine:2-oxoglutarate aminotransferase activity(GO:0004021) alanine-oxo-acid transaminase activity(GO:0047635) |
| 0.0 | 0.5 | GO:0031802 | type 5 metabotropic glutamate receptor binding(GO:0031802) |
| 0.0 | 0.1 | GO:0036478 | tyrosine 3-monooxygenase activator activity(GO:0036470) L-dopa decarboxylase activator activity(GO:0036478) cupric ion binding(GO:1903135) |
| 0.0 | 0.2 | GO:0004174 | electron-transferring-flavoprotein dehydrogenase activity(GO:0004174) oxidoreductase activity, acting on the CH-NH group of donors, quinone or similar compound as acceptor(GO:0016649) |
| 0.0 | 1.3 | GO:0004115 | 3',5'-cyclic-AMP phosphodiesterase activity(GO:0004115) |
| 0.0 | 1.3 | GO:0030506 | ankyrin binding(GO:0030506) |
| 0.0 | 0.9 | GO:0004709 | MAP kinase kinase kinase activity(GO:0004709) |
| 0.0 | 0.3 | GO:0071253 | connexin binding(GO:0071253) |
| 0.0 | 1.5 | GO:0046875 | ephrin receptor binding(GO:0046875) |
| 0.0 | 0.4 | GO:0046933 | proton-transporting ATP synthase activity, rotational mechanism(GO:0046933) |
| 0.0 | 1.1 | GO:0005070 | SH3/SH2 adaptor activity(GO:0005070) |
| 0.0 | 0.6 | GO:0045295 | gamma-catenin binding(GO:0045295) |
| 0.0 | 0.5 | GO:0005540 | hyaluronic acid binding(GO:0005540) |
| 0.0 | 0.4 | GO:0048018 | receptor agonist activity(GO:0048018) |
| 0.0 | 0.4 | GO:0003746 | translation elongation factor activity(GO:0003746) |
| 0.0 | 0.8 | GO:0016709 | oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, NAD(P)H as one donor, and incorporation of one atom of oxygen(GO:0016709) |
| 0.0 | 0.4 | GO:0004181 | metallocarboxypeptidase activity(GO:0004181) |
| 0.0 | 1.7 | GO:0005089 | Rho guanyl-nucleotide exchange factor activity(GO:0005089) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 5.8 | PID RHOA PATHWAY | RhoA signaling pathway |
| 0.1 | 2.9 | PID INSULIN GLUCOSE PATHWAY | Insulin-mediated glucose transport |
| 0.1 | 4.1 | PID DELTA NP63 PATHWAY | Validated transcriptional targets of deltaNp63 isoforms |
| 0.1 | 1.4 | PID LPA4 PATHWAY | LPA4-mediated signaling events |
| 0.0 | 3.7 | PID ATF2 PATHWAY | ATF-2 transcription factor network |
| 0.0 | 1.1 | ST P38 MAPK PATHWAY | p38 MAPK Pathway |
| 0.0 | 0.9 | PID FRA PATHWAY | Validated transcriptional targets of AP1 family members Fra1 and Fra2 |
| 0.0 | 0.8 | ST JNK MAPK PATHWAY | JNK MAPK Pathway |
| 0.0 | 1.1 | PID EPHA FWDPATHWAY | EPHA forward signaling |
| 0.0 | 1.1 | SIG INSULIN RECEPTOR PATHWAY IN CARDIAC MYOCYTES | Genes related to the insulin receptor pathway |
| 0.0 | 0.3 | PID NEPHRIN NEPH1 PATHWAY | Nephrin/Neph1 signaling in the kidney podocyte |
| 0.0 | 0.5 | PID RETINOIC ACID PATHWAY | Retinoic acid receptors-mediated signaling |
| 0.0 | 0.4 | PID ALPHA SYNUCLEIN PATHWAY | Alpha-synuclein signaling |
| 0.0 | 1.1 | PID RHOA REG PATHWAY | Regulation of RhoA activity |
| 0.0 | 1.0 | PID HIF1 TFPATHWAY | HIF-1-alpha transcription factor network |
| 0.0 | 0.5 | NABA PROTEOGLYCANS | Genes encoding proteoglycans |
| 0.0 | 0.2 | PID ARF 3PATHWAY | Arf1 pathway |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 13.2 | REACTOME GLYCOLYSIS | Genes involved in Glycolysis |
| 0.2 | 2.9 | REACTOME FACILITATIVE NA INDEPENDENT GLUCOSE TRANSPORTERS | Genes involved in Facilitative Na+-independent glucose transporters |
| 0.1 | 3.1 | REACTOME ACTIVATION OF GENES BY ATF4 | Genes involved in Activation of Genes by ATF4 |
| 0.1 | 15.0 | REACTOME TRANSPORT OF INORGANIC CATIONS ANIONS AND AMINO ACIDS OLIGOPEPTIDES | Genes involved in Transport of inorganic cations/anions and amino acids/oligopeptides |
| 0.1 | 1.4 | REACTOME ADENYLATE CYCLASE ACTIVATING PATHWAY | Genes involved in Adenylate cyclase activating pathway |
| 0.1 | 1.6 | REACTOME HDL MEDIATED LIPID TRANSPORT | Genes involved in HDL-mediated lipid transport |
| 0.1 | 3.5 | REACTOME INTERACTIONS OF VPR WITH HOST CELLULAR PROTEINS | Genes involved in Interactions of Vpr with host cellular proteins |
| 0.1 | 2.1 | REACTOME FGFR1 LIGAND BINDING AND ACTIVATION | Genes involved in FGFR1 ligand binding and activation |
| 0.1 | 3.3 | REACTOME GLYCOSPHINGOLIPID METABOLISM | Genes involved in Glycosphingolipid metabolism |
| 0.1 | 3.9 | REACTOME RESPIRATORY ELECTRON TRANSPORT | Genes involved in Respiratory electron transport |
| 0.0 | 2.6 | REACTOME CHEMOKINE RECEPTORS BIND CHEMOKINES | Genes involved in Chemokine receptors bind chemokines |
| 0.0 | 1.0 | REACTOME GLYCOGEN BREAKDOWN GLYCOGENOLYSIS | Genes involved in Glycogen breakdown (glycogenolysis) |
| 0.0 | 0.7 | REACTOME CALNEXIN CALRETICULIN CYCLE | Genes involved in Calnexin/calreticulin cycle |
| 0.0 | 2.2 | REACTOME GLUTATHIONE CONJUGATION | Genes involved in Glutathione conjugation |
| 0.0 | 0.4 | REACTOME ABACAVIR TRANSPORT AND METABOLISM | Genes involved in Abacavir transport and metabolism |
| 0.0 | 0.8 | REACTOME CHOLESTEROL BIOSYNTHESIS | Genes involved in Cholesterol biosynthesis |
| 0.0 | 0.5 | REACTOME METABOLISM OF POLYAMINES | Genes involved in Metabolism of polyamines |
| 0.0 | 2.3 | REACTOME NRAGE SIGNALS DEATH THROUGH JNK | Genes involved in NRAGE signals death through JNK |
| 0.0 | 1.1 | REACTOME SMOOTH MUSCLE CONTRACTION | Genes involved in Smooth Muscle Contraction |
| 0.0 | 0.2 | REACTOME COPI MEDIATED TRANSPORT | Genes involved in COPI Mediated Transport |
| 0.0 | 0.5 | REACTOME CLASS C 3 METABOTROPIC GLUTAMATE PHEROMONE RECEPTORS | Genes involved in Class C/3 (Metabotropic glutamate/pheromone receptors) |
| 0.0 | 0.4 | REACTOME FORMATION OF ATP BY CHEMIOSMOTIC COUPLING | Genes involved in Formation of ATP by chemiosmotic coupling |
| 0.0 | 0.3 | REACTOME HYALURONAN METABOLISM | Genes involved in Hyaluronan metabolism |
| 0.0 | 0.8 | REACTOME PHASE1 FUNCTIONALIZATION OF COMPOUNDS | Genes involved in Phase 1 - Functionalization of compounds |
| 0.0 | 0.3 | REACTOME APOPTOTIC CLEAVAGE OF CELL ADHESION PROTEINS | Genes involved in Apoptotic cleavage of cell adhesion proteins |
| 0.0 | 0.2 | REACTOME SLBP DEPENDENT PROCESSING OF REPLICATION DEPENDENT HISTONE PRE MRNAS | Genes involved in SLBP Dependent Processing of Replication-Dependent Histone Pre-mRNAs |
| 0.0 | 0.3 | REACTOME SYNTHESIS SECRETION AND INACTIVATION OF GIP | Genes involved in Synthesis, Secretion, and Inactivation of Glucose-dependent Insulinotropic Polypeptide (GIP) |
| 0.0 | 0.5 | REACTOME G PROTEIN ACTIVATION | Genes involved in G-protein activation |
| 0.0 | 0.4 | REACTOME KINESINS | Genes involved in Kinesins |
| 0.0 | 1.3 | REACTOME CELL JUNCTION ORGANIZATION | Genes involved in Cell junction organization |