Project
PRJNA375882: Comprehensive Mouse Transcriptomic BodyMap
Navigation
Downloads
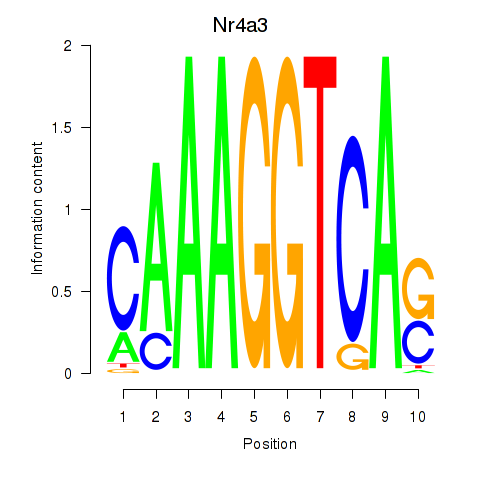
Results for Nr4a3
Z-value: 0.79
Transcription factors associated with Nr4a3
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Nr4a3
|
ENSMUSG00000028341.10 | Nr4a3 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Nr4a3 | mm39_v1_chr4_+_48045143_48045160 | -0.17 | 1.4e-01 | Click! |
Activity profile of Nr4a3 motif
Sorted Z-values of Nr4a3 motif
Network of associatons between targets according to the STRING database.
First level regulatory network of Nr4a3
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr7_+_125202653 | 4.64 |
ENSMUST00000206103.2
ENSMUST00000033000.8 |
Il21r
|
interleukin 21 receptor |
| chr7_-_127805518 | 4.37 |
ENSMUST00000033049.9
|
Cox6a2
|
cytochrome c oxidase subunit 6A2 |
| chr6_+_41523664 | 4.00 |
ENSMUST00000103299.3
|
Trbc2
|
T cell receptor beta, constant 2 |
| chr3_-_101195213 | 3.84 |
ENSMUST00000029456.5
|
Cd2
|
CD2 antigen |
| chr11_+_70548022 | 3.60 |
ENSMUST00000157027.8
ENSMUST00000072841.12 ENSMUST00000108548.8 ENSMUST00000126241.8 |
Eno3
|
enolase 3, beta muscle |
| chr15_-_78449172 | 3.42 |
ENSMUST00000230952.2
|
Rac2
|
Rac family small GTPase 2 |
| chr11_+_70548622 | 3.29 |
ENSMUST00000170716.8
|
Eno3
|
enolase 3, beta muscle |
| chr4_+_131600918 | 3.14 |
ENSMUST00000053819.6
|
Srsf4
|
serine and arginine-rich splicing factor 4 |
| chr14_-_70414236 | 2.82 |
ENSMUST00000153735.8
|
Pdlim2
|
PDZ and LIM domain 2 |
| chr2_-_13496624 | 2.68 |
ENSMUST00000091436.7
|
Cubn
|
cubilin (intrinsic factor-cobalamin receptor) |
| chr8_-_124621483 | 2.54 |
ENSMUST00000034453.6
ENSMUST00000212584.2 |
Acta1
|
actin, alpha 1, skeletal muscle |
| chr5_+_34527230 | 2.44 |
ENSMUST00000180376.8
|
Fam193a
|
family with sequence homology 193, member A |
| chr15_-_5273659 | 2.37 |
ENSMUST00000047379.15
|
Ptger4
|
prostaglandin E receptor 4 (subtype EP4) |
| chr6_+_113508636 | 2.37 |
ENSMUST00000036340.12
ENSMUST00000204827.3 |
Fancd2
|
Fanconi anemia, complementation group D2 |
| chr15_-_5273645 | 2.30 |
ENSMUST00000120563.2
|
Ptger4
|
prostaglandin E receptor 4 (subtype EP4) |
| chr5_+_67125759 | 2.30 |
ENSMUST00000238993.2
ENSMUST00000038188.14 |
Limch1
|
LIM and calponin homology domains 1 |
| chr15_-_63932288 | 2.26 |
ENSMUST00000063838.11
ENSMUST00000228908.2 |
Cyrib
|
CYFIP related Rac1 interactor B |
| chr17_-_74257164 | 2.20 |
ENSMUST00000024866.6
|
Xdh
|
xanthine dehydrogenase |
| chr3_-_104725853 | 2.17 |
ENSMUST00000106775.8
ENSMUST00000166979.8 |
Mov10
|
Mov10 RISC complex RNA helicase |
| chrX_-_149372840 | 2.16 |
ENSMUST00000112725.8
ENSMUST00000112720.8 |
Apex2
|
apurinic/apyrimidinic endonuclease 2 |
| chr7_-_79492091 | 2.09 |
ENSMUST00000049004.8
|
Anpep
|
alanyl (membrane) aminopeptidase |
| chr19_-_7218363 | 2.05 |
ENSMUST00000236769.2
|
Naa40
|
N(alpha)-acetyltransferase 40, NatD catalytic subunit |
| chr16_-_4340920 | 2.04 |
ENSMUST00000090500.10
ENSMUST00000023161.8 |
Srl
|
sarcalumenin |
| chr3_-_104725581 | 1.98 |
ENSMUST00000168015.8
|
Mov10
|
Mov10 RISC complex RNA helicase |
| chr14_-_54655079 | 1.94 |
ENSMUST00000226753.2
ENSMUST00000197440.5 |
Slc7a7
|
solute carrier family 7 (cationic amino acid transporter, y+ system), member 7 |
| chr2_+_69500444 | 1.88 |
ENSMUST00000100050.4
|
Klhl41
|
kelch-like 41 |
| chr6_-_116693849 | 1.72 |
ENSMUST00000056623.13
|
Tmem72
|
transmembrane protein 72 |
| chr10_-_23226684 | 1.71 |
ENSMUST00000220299.2
|
Eya4
|
EYA transcriptional coactivator and phosphatase 4 |
| chr12_-_110945415 | 1.66 |
ENSMUST00000135131.2
ENSMUST00000043459.13 ENSMUST00000128353.8 |
Ankrd9
|
ankyrin repeat domain 9 |
| chr11_-_61384998 | 1.63 |
ENSMUST00000101085.9
ENSMUST00000079080.13 ENSMUST00000108714.2 |
Mapk7
|
mitogen-activated protein kinase 7 |
| chr7_+_43910845 | 1.61 |
ENSMUST00000124863.4
|
Gm15517
|
predicted gene 15517 |
| chr2_+_164647002 | 1.56 |
ENSMUST00000052107.5
|
Zswim3
|
zinc finger SWIM-type containing 3 |
| chr10_-_80184238 | 1.56 |
ENSMUST00000095446.10
ENSMUST00000105352.2 |
Adamtsl5
|
ADAMTS-like 5 |
| chr12_-_110945052 | 1.54 |
ENSMUST00000140788.8
|
Ankrd9
|
ankyrin repeat domain 9 |
| chr15_+_84553801 | 1.54 |
ENSMUST00000171460.8
|
Prr5
|
proline rich 5 (renal) |
| chr12_-_110945376 | 1.50 |
ENSMUST00000142012.2
|
Ankrd9
|
ankyrin repeat domain 9 |
| chr3_-_104725535 | 1.48 |
ENSMUST00000002297.12
|
Mov10
|
Mov10 RISC complex RNA helicase |
| chr4_-_126096551 | 1.47 |
ENSMUST00000080919.12
|
Thrap3
|
thyroid hormone receptor associated protein 3 |
| chr2_+_153850177 | 1.44 |
ENSMUST00000048103.9
ENSMUST00000145388.2 |
Bpifa2
|
BPI fold containing family A, member 2 |
| chr7_-_79115915 | 1.43 |
ENSMUST00000073889.14
|
Polg
|
polymerase (DNA directed), gamma |
| chr3_-_95041246 | 1.43 |
ENSMUST00000172572.9
ENSMUST00000173462.3 |
Scnm1
|
sodium channel modifier 1 |
| chr6_-_137548004 | 1.41 |
ENSMUST00000100841.9
|
Eps8
|
epidermal growth factor receptor pathway substrate 8 |
| chr17_+_29579882 | 1.41 |
ENSMUST00000024810.8
|
Fgd2
|
FYVE, RhoGEF and PH domain containing 2 |
| chr14_-_8798841 | 1.38 |
ENSMUST00000061045.3
|
Sntn
|
sentan, cilia apical structure protein |
| chr15_-_76084776 | 1.33 |
ENSMUST00000169108.8
ENSMUST00000170728.8 |
Plec
|
plectin |
| chr11_-_100713348 | 1.33 |
ENSMUST00000107358.9
|
Stat5b
|
signal transducer and activator of transcription 5B |
| chr17_-_36220518 | 1.32 |
ENSMUST00000141132.2
|
Atat1
|
alpha tubulin acetyltransferase 1 |
| chr17_-_27842237 | 1.31 |
ENSMUST00000062397.13
ENSMUST00000176876.8 ENSMUST00000146321.3 |
Nudt3
|
nudix (nucleotide diphosphate linked moiety X)-type motif 3 |
| chr14_-_52252003 | 1.30 |
ENSMUST00000226522.2
|
Zfp219
|
zinc finger protein 219 |
| chr16_-_50411484 | 1.30 |
ENSMUST00000062439.6
|
Ccdc54
|
coiled-coil domain containing 54 |
| chr15_+_100659622 | 1.28 |
ENSMUST00000023776.13
|
Slc4a8
|
solute carrier family 4 (anion exchanger), member 8 |
| chr11_-_105346120 | 1.27 |
ENSMUST00000138977.8
|
Marchf10
|
membrane associated ring-CH-type finger 10 |
| chr9_-_21913896 | 1.25 |
ENSMUST00000044926.6
|
Odad3
|
outer dynein arm docking complex subunit 3 |
| chr15_-_63932176 | 1.24 |
ENSMUST00000226675.2
ENSMUST00000228226.2 ENSMUST00000227024.2 |
Cyrib
|
CYFIP related Rac1 interactor B |
| chr1_-_160079007 | 1.23 |
ENSMUST00000191909.6
|
Rabgap1l
|
RAB GTPase activating protein 1-like |
| chrX_+_55493325 | 1.17 |
ENSMUST00000079663.7
|
Gm2174
|
predicted gene 2174 |
| chr3_+_101993787 | 1.16 |
ENSMUST00000165540.9
ENSMUST00000164123.2 |
Casq2
|
calsequestrin 2 |
| chr15_-_102425241 | 1.14 |
ENSMUST00000169162.8
ENSMUST00000023812.10 ENSMUST00000165174.8 ENSMUST00000169367.8 ENSMUST00000169377.8 |
Map3k12
|
mitogen-activated protein kinase kinase kinase 12 |
| chr11_+_115366470 | 1.12 |
ENSMUST00000035240.7
|
Armc7
|
armadillo repeat containing 7 |
| chr2_+_79538124 | 1.12 |
ENSMUST00000090760.9
ENSMUST00000040863.11 ENSMUST00000111780.3 |
Ppp1r1c
|
protein phosphatase 1, regulatory inhibitor subunit 1C |
| chr4_-_126096376 | 1.11 |
ENSMUST00000106142.8
ENSMUST00000169403.8 ENSMUST00000130334.2 |
Thrap3
|
thyroid hormone receptor associated protein 3 |
| chr7_-_79115760 | 1.11 |
ENSMUST00000125562.2
|
Polg
|
polymerase (DNA directed), gamma |
| chr3_-_137837117 | 1.10 |
ENSMUST00000029805.13
|
Mttp
|
microsomal triglyceride transfer protein |
| chr2_-_157121440 | 1.05 |
ENSMUST00000143663.2
|
Mroh8
|
maestro heat-like repeat family member 8 |
| chr5_+_67125902 | 1.04 |
ENSMUST00000127184.8
|
Limch1
|
LIM and calponin homology domains 1 |
| chr14_-_52252318 | 1.03 |
ENSMUST00000228051.2
|
Zfp219
|
zinc finger protein 219 |
| chr9_-_21913833 | 1.01 |
ENSMUST00000115336.10
|
Odad3
|
outer dynein arm docking complex subunit 3 |
| chr8_+_120588977 | 0.99 |
ENSMUST00000034287.10
|
Klhl36
|
kelch-like 36 |
| chr17_+_47905553 | 0.99 |
ENSMUST00000182846.3
|
Ccnd3
|
cyclin D3 |
| chr2_+_158636727 | 0.99 |
ENSMUST00000029186.14
ENSMUST00000109478.9 ENSMUST00000156893.2 |
Dhx35
|
DEAH (Asp-Glu-Ala-His) box polypeptide 35 |
| chr2_-_167334746 | 0.98 |
ENSMUST00000109211.9
ENSMUST00000057627.16 |
Spata2
|
spermatogenesis associated 2 |
| chr2_-_163259012 | 0.94 |
ENSMUST00000127038.2
|
Oser1
|
oxidative stress responsive serine rich 1 |
| chr6_-_76474767 | 0.91 |
ENSMUST00000097218.7
|
Gm9008
|
predicted pseudogene 9008 |
| chr18_-_60757361 | 0.90 |
ENSMUST00000097566.10
|
Synpo
|
synaptopodin |
| chr12_-_84240781 | 0.88 |
ENSMUST00000110294.2
|
Mideas
|
mitotic deacetylase associated SANT domain protein |
| chr19_+_46140942 | 0.87 |
ENSMUST00000026254.14
|
Gbf1
|
golgi-specific brefeldin A-resistance factor 1 |
| chr3_-_59038634 | 0.84 |
ENSMUST00000200358.2
ENSMUST00000197220.2 |
P2ry14
|
purinergic receptor P2Y, G-protein coupled, 14 |
| chr3_-_59038539 | 0.82 |
ENSMUST00000198838.2
|
P2ry14
|
purinergic receptor P2Y, G-protein coupled, 14 |
| chr7_-_109215754 | 0.80 |
ENSMUST00000084738.5
|
Denn2b
|
DENN domain containing 2B |
| chr8_-_121394742 | 0.80 |
ENSMUST00000181334.2
ENSMUST00000181950.2 ENSMUST00000181333.2 |
Emc8
Gm27021
|
ER membrane protein complex subunit 8 predicted gene, 27021 |
| chr14_+_53007210 | 0.80 |
ENSMUST00000178768.4
|
Trav7d-4
|
T cell receptor alpha variable 7D-4 |
| chr3_-_89121147 | 0.79 |
ENSMUST00000173477.8
ENSMUST00000119222.9 |
Mtx1
|
metaxin 1 |
| chr14_-_52252432 | 0.77 |
ENSMUST00000226527.2
|
Zfp219
|
zinc finger protein 219 |
| chr3_+_5815863 | 0.75 |
ENSMUST00000192045.2
|
Gm8797
|
predicted pseudogene 8797 |
| chr19_+_37184927 | 0.73 |
ENSMUST00000024078.15
ENSMUST00000112391.8 |
Marchf5
|
membrane associated ring-CH-type finger 5 |
| chr16_+_84631789 | 0.72 |
ENSMUST00000114184.8
|
Gabpa
|
GA repeat binding protein, alpha |
| chr1_-_152262425 | 0.70 |
ENSMUST00000015124.15
|
Tsen15
|
tRNA splicing endonuclease subunit 15 |
| chr15_+_100659729 | 0.70 |
ENSMUST00000161564.2
|
Slc4a8
|
solute carrier family 4 (anion exchanger), member 8 |
| chr8_-_105350533 | 0.68 |
ENSMUST00000212662.2
|
Cdh16
|
cadherin 16 |
| chr2_-_76698725 | 0.68 |
ENSMUST00000149616.8
ENSMUST00000152185.8 ENSMUST00000130915.8 ENSMUST00000155365.8 ENSMUST00000128071.8 |
Ttn
|
titin |
| chr17_-_36220924 | 0.67 |
ENSMUST00000141662.8
ENSMUST00000056034.13 ENSMUST00000077494.13 ENSMUST00000149277.8 ENSMUST00000061052.12 |
Atat1
|
alpha tubulin acetyltransferase 1 |
| chr9_-_18585826 | 0.66 |
ENSMUST00000208663.2
|
Muc16
|
mucin 16 |
| chr2_-_165125519 | 0.62 |
ENSMUST00000155289.8
|
Slc35c2
|
solute carrier family 35, member C2 |
| chr11_-_72157450 | 0.61 |
ENSMUST00000021161.14
|
Slc13a5
|
solute carrier family 13 (sodium-dependent citrate transporter), member 5 |
| chr19_-_37184692 | 0.61 |
ENSMUST00000132580.8
ENSMUST00000079754.11 ENSMUST00000136286.8 ENSMUST00000126188.8 ENSMUST00000126781.2 |
Cpeb3
|
cytoplasmic polyadenylation element binding protein 3 |
| chr8_-_105350898 | 0.60 |
ENSMUST00000212882.2
ENSMUST00000163783.4 |
Cdh16
|
cadherin 16 |
| chr5_-_23881353 | 0.58 |
ENSMUST00000198661.5
|
Srpk2
|
serine/arginine-rich protein specific kinase 2 |
| chr7_-_126074256 | 0.57 |
ENSMUST00000205440.2
ENSMUST00000032978.8 ENSMUST00000205340.2 |
Sh2b1
|
SH2B adaptor protein 1 |
| chr19_-_40371016 | 0.56 |
ENSMUST00000225766.3
|
Sorbs1
|
sorbin and SH3 domain containing 1 |
| chr7_+_110368037 | 0.54 |
ENSMUST00000213373.2
|
Ampd3
|
adenosine monophosphate deaminase 3 |
| chr5_+_16758777 | 0.54 |
ENSMUST00000030683.8
|
Hgf
|
hepatocyte growth factor |
| chr13_+_73476629 | 0.52 |
ENSMUST00000221730.2
|
Mrpl36
|
mitochondrial ribosomal protein L36 |
| chr4_+_150999019 | 0.51 |
ENSMUST00000135169.8
|
Tnfrsf9
|
tumor necrosis factor receptor superfamily, member 9 |
| chr8_-_105350881 | 0.50 |
ENSMUST00000211903.2
|
Cdh16
|
cadherin 16 |
| chr6_+_124986627 | 0.48 |
ENSMUST00000046064.17
ENSMUST00000152752.8 ENSMUST00000088308.10 ENSMUST00000112425.8 ENSMUST00000084275.12 |
Zfp384
|
zinc finger protein 384 |
| chr19_-_45986919 | 0.47 |
ENSMUST00000045396.9
|
Armh3
|
armadillo-like helical domain containing 3 |
| chr3_+_63883527 | 0.47 |
ENSMUST00000029405.8
|
Gmps
|
guanine monophosphate synthetase |
| chr11_-_72157419 | 0.46 |
ENSMUST00000208056.2
ENSMUST00000208912.2 ENSMUST00000140167.3 ENSMUST00000137701.3 |
Slc13a5
|
solute carrier family 13 (sodium-dependent citrate transporter), member 5 |
| chr3_+_122298308 | 0.43 |
ENSMUST00000199358.2
|
Bcar3
|
breast cancer anti-estrogen resistance 3 |
| chr8_-_105350816 | 0.43 |
ENSMUST00000212447.2
|
Cdh16
|
cadherin 16 |
| chr1_+_75412574 | 0.42 |
ENSMUST00000037796.14
ENSMUST00000113584.8 ENSMUST00000145166.8 ENSMUST00000143730.8 ENSMUST00000133418.8 ENSMUST00000144874.8 ENSMUST00000140287.8 |
Gmppa
|
GDP-mannose pyrophosphorylase A |
| chr7_-_126074222 | 0.41 |
ENSMUST00000205497.2
|
Sh2b1
|
SH2B adaptor protein 1 |
| chr4_-_152213315 | 0.40 |
ENSMUST00000049305.14
|
Espn
|
espin |
| chr11_+_116547932 | 0.39 |
ENSMUST00000116318.3
|
Prcd
|
photoreceptor disc component |
| chrX_+_149372903 | 0.39 |
ENSMUST00000080884.11
|
Pfkfb1
|
6-phosphofructo-2-kinase/fructose-2,6-biphosphatase 1 |
| chr4_+_125952349 | 0.38 |
ENSMUST00000035497.5
ENSMUST00000143712.3 |
Oscp1
|
organic solute carrier partner 1 |
| chr8_-_105350842 | 0.37 |
ENSMUST00000212324.2
|
Cdh16
|
cadherin 16 |
| chr16_+_10363222 | 0.37 |
ENSMUST00000155633.8
ENSMUST00000066345.15 |
Clec16a
|
C-type lectin domain family 16, member A |
| chr14_+_54453748 | 0.36 |
ENSMUST00000103736.2
|
Traj4
|
T cell receptor alpha joining 4 |
| chr18_-_60757272 | 0.34 |
ENSMUST00000155195.3
|
Synpo
|
synaptopodin |
| chrX_+_74425990 | 0.34 |
ENSMUST00000033541.5
|
Fundc2
|
FUN14 domain containing 2 |
| chr11_-_4654303 | 0.33 |
ENSMUST00000058407.6
|
Uqcr10
|
ubiquinol-cytochrome c reductase, complex III subunit X |
| chr14_+_53073315 | 0.29 |
ENSMUST00000197128.2
|
Trav7d-5
|
T cell receptor alpha variable 7D-5 |
| chr16_+_4704103 | 0.29 |
ENSMUST00000023159.10
ENSMUST00000070658.16 ENSMUST00000229038.2 |
Mgrn1
|
mahogunin, ring finger 1 |
| chr4_+_115932466 | 0.29 |
ENSMUST00000030471.9
|
Lrrc41
|
leucine rich repeat containing 41 |
| chr9_+_21914083 | 0.29 |
ENSMUST00000216344.2
|
Prkcsh
|
protein kinase C substrate 80K-H |
| chr5_-_66308666 | 0.27 |
ENSMUST00000201561.4
|
Rbm47
|
RNA binding motif protein 47 |
| chr8_-_22550035 | 0.26 |
ENSMUST00000110738.3
|
Atp7b
|
ATPase, Cu++ transporting, beta polypeptide |
| chr1_-_152262339 | 0.25 |
ENSMUST00000162371.2
|
Tsen15
|
tRNA splicing endonuclease subunit 15 |
| chr16_+_10363203 | 0.24 |
ENSMUST00000115824.10
|
Clec16a
|
C-type lectin domain family 16, member A |
| chr9_+_21914296 | 0.24 |
ENSMUST00000003493.9
|
Prkcsh
|
protein kinase C substrate 80K-H |
| chr7_-_109215960 | 0.23 |
ENSMUST00000077909.9
|
Denn2b
|
DENN domain containing 2B |
| chr9_+_21914513 | 0.23 |
ENSMUST00000215795.2
|
Prkcsh
|
protein kinase C substrate 80K-H |
| chr9_+_21914334 | 0.22 |
ENSMUST00000115331.10
|
Prkcsh
|
protein kinase C substrate 80K-H |
| chr13_-_73476561 | 0.22 |
ENSMUST00000222930.2
ENSMUST00000223293.2 ENSMUST00000022097.6 |
Ndufs6
|
NADH:ubiquinone oxidoreductase core subunit S6 |
| chr5_+_16758897 | 0.20 |
ENSMUST00000196645.2
|
Hgf
|
hepatocyte growth factor |
| chr16_+_10363254 | 0.20 |
ENSMUST00000115827.8
ENSMUST00000150894.2 ENSMUST00000038145.13 |
Clec16a
|
C-type lectin domain family 16, member A |
| chr4_-_43578823 | 0.19 |
ENSMUST00000030189.14
|
Gba2
|
glucosidase beta 2 |
| chr8_+_95722289 | 0.19 |
ENSMUST00000211984.2
|
Adgrg1
|
adhesion G protein-coupled receptor G1 |
| chr15_-_79025387 | 0.17 |
ENSMUST00000187550.7
ENSMUST00000188562.7 ENSMUST00000190509.7 ENSMUST00000190730.7 ENSMUST00000190959.7 ENSMUST00000169604.8 ENSMUST00000186053.7 |
1700088E04Rik
|
RIKEN cDNA 1700088E04 gene |
| chr12_-_112640626 | 0.15 |
ENSMUST00000001780.10
|
Akt1
|
thymoma viral proto-oncogene 1 |
| chr19_-_56996617 | 0.15 |
ENSMUST00000118800.8
ENSMUST00000111584.9 ENSMUST00000122359.8 ENSMUST00000148049.8 |
Afap1l2
|
actin filament associated protein 1-like 2 |
| chr16_+_4704166 | 0.06 |
ENSMUST00000230990.2
|
Mgrn1
|
mahogunin, ring finger 1 |
| chr8_+_121394961 | 0.05 |
ENSMUST00000034276.13
ENSMUST00000181586.8 |
Cox4i1
|
cytochrome c oxidase subunit 4I1 |
| chr7_-_126944754 | 0.03 |
ENSMUST00000205266.2
|
Zfp768
|
zinc finger protein 768 |
| chr8_-_121394860 | 0.02 |
ENSMUST00000034277.14
|
Emc8
|
ER membrane protein complex subunit 8 |
| chr14_-_21898992 | 0.02 |
ENSMUST00000124549.9
|
Comtd1
|
catechol-O-methyltransferase domain containing 1 |
| chr7_+_99659121 | 0.00 |
ENSMUST00000107084.8
|
Chrdl2
|
chordin-like 2 |
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.6 | 4.7 | GO:1904464 | negative regulation of integrin activation(GO:0033624) regulation of matrix metallopeptidase secretion(GO:1904464) matrix metallopeptidase secretion(GO:1990773) positive regulation of ovarian follicle development(GO:2000386) regulation of antral ovarian follicle growth(GO:2000387) positive regulation of antral ovarian follicle growth(GO:2000388) negative regulation of eosinophil migration(GO:2000417) |
| 1.4 | 5.6 | GO:0098795 | mRNA cleavage involved in gene silencing by miRNA(GO:0035279) mRNA cleavage involved in gene silencing(GO:0098795) |
| 0.7 | 2.0 | GO:0071929 | alpha-tubulin acetylation(GO:0071929) |
| 0.5 | 1.6 | GO:0070376 | regulation of ERK5 cascade(GO:0070376) |
| 0.5 | 2.0 | GO:0014873 | response to muscle activity involved in regulation of muscle adaptation(GO:0014873) |
| 0.4 | 2.7 | GO:0015889 | cobalamin transport(GO:0015889) |
| 0.4 | 2.2 | GO:0009115 | xanthine catabolic process(GO:0009115) |
| 0.4 | 1.4 | GO:0070358 | actin polymerization-dependent cell motility(GO:0070358) |
| 0.3 | 3.4 | GO:0060753 | regulation of mast cell chemotaxis(GO:0060753) |
| 0.3 | 1.3 | GO:0071544 | diphosphoinositol polyphosphate catabolic process(GO:0071544) |
| 0.3 | 3.2 | GO:0030240 | skeletal muscle thin filament assembly(GO:0030240) |
| 0.3 | 0.9 | GO:0070973 | protein localization to endoplasmic reticulum exit site(GO:0070973) |
| 0.3 | 1.1 | GO:0015744 | succinate transport(GO:0015744) |
| 0.3 | 0.8 | GO:1904766 | negative regulation of macroautophagy by TORC1 signaling(GO:1904766) |
| 0.3 | 2.4 | GO:2000348 | regulation of CD40 signaling pathway(GO:2000348) |
| 0.3 | 3.8 | GO:1902715 | positive regulation of interferon-gamma secretion(GO:1902715) |
| 0.2 | 1.2 | GO:0098886 | modification of dendritic spine(GO:0098886) |
| 0.2 | 1.9 | GO:0000821 | regulation of arginine metabolic process(GO:0000821) |
| 0.2 | 0.6 | GO:1900248 | cytoplasmic translational elongation(GO:0002182) regulation of cytoplasmic translational elongation(GO:1900247) negative regulation of cytoplasmic translational elongation(GO:1900248) |
| 0.2 | 1.3 | GO:0046544 | development of secondary male sexual characteristics(GO:0046544) |
| 0.2 | 1.7 | GO:0035589 | G-protein coupled purinergic nucleotide receptor signaling pathway(GO:0035589) |
| 0.2 | 1.5 | GO:0038203 | TORC2 signaling(GO:0038203) |
| 0.2 | 0.6 | GO:0015786 | UDP-glucose transport(GO:0015786) |
| 0.1 | 1.2 | GO:0071313 | cellular response to caffeine(GO:0071313) |
| 0.1 | 1.1 | GO:0034196 | acylglycerol transport(GO:0034196) triglyceride transport(GO:0034197) |
| 0.1 | 2.5 | GO:0006264 | mitochondrial DNA replication(GO:0006264) |
| 0.1 | 1.1 | GO:2000672 | negative regulation of motor neuron apoptotic process(GO:2000672) |
| 0.1 | 1.7 | GO:0016576 | histone dephosphorylation(GO:0016576) |
| 0.1 | 1.9 | GO:0031272 | regulation of pseudopodium assembly(GO:0031272) |
| 0.1 | 2.1 | GO:0035814 | negative regulation of renal sodium excretion(GO:0035814) |
| 0.1 | 0.6 | GO:0035063 | nuclear speck organization(GO:0035063) |
| 0.1 | 1.3 | GO:0031581 | hemidesmosome assembly(GO:0031581) |
| 0.1 | 0.3 | GO:0015680 | intracellular copper ion transport(GO:0015680) |
| 0.1 | 0.7 | GO:0045653 | negative regulation of megakaryocyte differentiation(GO:0045653) |
| 0.1 | 2.6 | GO:0048026 | positive regulation of mRNA splicing, via spliceosome(GO:0048026) |
| 0.1 | 2.1 | GO:0043968 | histone H2A acetylation(GO:0043968) |
| 0.1 | 3.1 | GO:0048025 | negative regulation of mRNA splicing, via spliceosome(GO:0048025) |
| 0.1 | 2.6 | GO:0016339 | calcium-dependent cell-cell adhesion via plasma membrane cell adhesion molecules(GO:0016339) |
| 0.1 | 1.0 | GO:0006388 | tRNA splicing, via endonucleolytic cleavage and ligation(GO:0006388) |
| 0.1 | 0.5 | GO:0006177 | GMP biosynthetic process(GO:0006177) |
| 0.1 | 0.7 | GO:0045409 | negative regulation of interleukin-6 biosynthetic process(GO:0045409) |
| 0.1 | 0.5 | GO:2001183 | negative regulation of interleukin-12 secretion(GO:2001183) |
| 0.1 | 4.2 | GO:0009060 | aerobic respiration(GO:0009060) |
| 0.1 | 0.4 | GO:0006000 | fructose metabolic process(GO:0006000) fructose 2,6-bisphosphate metabolic process(GO:0006003) |
| 0.1 | 0.5 | GO:0032264 | IMP salvage(GO:0032264) |
| 0.1 | 0.7 | GO:0060665 | regulation of branching involved in salivary gland morphogenesis by mesenchymal-epithelial signaling(GO:0060665) |
| 0.1 | 6.9 | GO:0006096 | glycolytic process(GO:0006096) |
| 0.1 | 0.4 | GO:0030046 | parallel actin filament bundle assembly(GO:0030046) |
| 0.0 | 2.2 | GO:0006284 | base-excision repair(GO:0006284) |
| 0.0 | 1.0 | GO:0030213 | hyaluronan biosynthetic process(GO:0030213) |
| 0.0 | 0.2 | GO:0006680 | glucosylceramide catabolic process(GO:0006680) |
| 0.0 | 1.0 | GO:0006491 | N-glycan processing(GO:0006491) |
| 0.0 | 1.2 | GO:0035855 | megakaryocyte development(GO:0035855) |
| 0.0 | 4.0 | GO:0006910 | phagocytosis, recognition(GO:0006910) |
| 0.0 | 0.4 | GO:0043951 | negative regulation of cAMP-mediated signaling(GO:0043951) |
| 0.0 | 1.1 | GO:1900087 | positive regulation of G1/S transition of mitotic cell cycle(GO:1900087) |
| 0.0 | 2.0 | GO:0051453 | regulation of intracellular pH(GO:0051453) |
| 0.0 | 0.4 | GO:0050962 | detection of light stimulus involved in visual perception(GO:0050908) detection of light stimulus involved in sensory perception(GO:0050962) |
| 0.0 | 1.0 | GO:0045840 | positive regulation of mitotic nuclear division(GO:0045840) |
| 0.0 | 1.4 | GO:0043507 | positive regulation of JUN kinase activity(GO:0043507) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.8 | 2.5 | GO:0005760 | gamma DNA polymerase complex(GO:0005760) |
| 0.5 | 2.7 | GO:0043202 | lysosomal lumen(GO:0043202) |
| 0.5 | 6.9 | GO:0000015 | phosphopyruvate hydratase complex(GO:0000015) |
| 0.4 | 1.2 | GO:0033018 | sarcoplasmic reticulum lumen(GO:0033018) |
| 0.3 | 0.8 | GO:0036020 | endolysosome membrane(GO:0036020) |
| 0.2 | 1.0 | GO:0017177 | glucosidase II complex(GO:0017177) |
| 0.2 | 1.2 | GO:0097444 | spine apparatus(GO:0097444) |
| 0.2 | 2.0 | GO:0097427 | microtubule bundle(GO:0097427) |
| 0.1 | 2.1 | GO:0031983 | vesicle lumen(GO:0031983) |
| 0.1 | 0.4 | GO:0043540 | 6-phosphofructo-2-kinase/fructose-2,6-biphosphatase complex(GO:0043540) |
| 0.1 | 1.6 | GO:0043205 | microfibril(GO:0001527) fibril(GO:0043205) |
| 0.1 | 1.5 | GO:0031932 | TORC2 complex(GO:0031932) |
| 0.1 | 1.9 | GO:0031143 | pseudopodium(GO:0031143) |
| 0.1 | 2.6 | GO:0035145 | exon-exon junction complex(GO:0035145) |
| 0.1 | 3.2 | GO:0005865 | striated muscle thin filament(GO:0005865) |
| 0.1 | 3.8 | GO:0046658 | anchored component of plasma membrane(GO:0046658) |
| 0.1 | 1.3 | GO:0017146 | NMDA selective glutamate receptor complex(GO:0017146) |
| 0.0 | 0.6 | GO:0030014 | CCR4-NOT complex(GO:0030014) |
| 0.0 | 5.6 | GO:0016528 | sarcoplasm(GO:0016528) |
| 0.0 | 1.1 | GO:0031528 | microvillus membrane(GO:0031528) |
| 0.0 | 2.0 | GO:0032809 | neuronal cell body membrane(GO:0032809) |
| 0.0 | 5.4 | GO:0000932 | cytoplasmic mRNA processing body(GO:0000932) |
| 0.0 | 0.7 | GO:0005751 | mitochondrial respiratory chain complex IV(GO:0005751) |
| 0.0 | 4.0 | GO:0042571 | immunoglobulin complex, circulating(GO:0042571) |
| 0.0 | 0.4 | GO:0032426 | stereocilium tip(GO:0032426) |
| 0.0 | 2.8 | GO:0030864 | cortical actin cytoskeleton(GO:0030864) |
| 0.0 | 2.9 | GO:0005884 | actin filament(GO:0005884) |
| 0.0 | 1.5 | GO:0005793 | endoplasmic reticulum-Golgi intermediate compartment(GO:0005793) |
| 0.0 | 0.2 | GO:0097451 | glial limiting end-foot(GO:0097451) |
| 0.0 | 4.7 | GO:0031965 | nuclear membrane(GO:0031965) |
| 0.0 | 2.8 | GO:0001650 | fibrillar center(GO:0001650) |
| 0.0 | 1.0 | GO:0000307 | cyclin-dependent protein kinase holoenzyme complex(GO:0000307) |
| 0.0 | 3.9 | GO:0016323 | basolateral plasma membrane(GO:0016323) |
| 0.0 | 0.3 | GO:0031307 | integral component of mitochondrial outer membrane(GO:0031307) |
| 0.0 | 1.0 | GO:0031463 | Cul3-RING ubiquitin ligase complex(GO:0031463) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.7 | 4.7 | GO:0004957 | prostaglandin E receptor activity(GO:0004957) |
| 0.5 | 2.2 | GO:0008311 | double-stranded DNA 3'-5' exodeoxyribonuclease activity(GO:0008311) |
| 0.5 | 6.9 | GO:0004634 | phosphopyruvate hydratase activity(GO:0004634) |
| 0.4 | 2.2 | GO:0004854 | xanthine dehydrogenase activity(GO:0004854) oxidoreductase activity, acting on CH or CH2 groups, NAD or NADP as acceptor(GO:0016726) molybdenum ion binding(GO:0030151) molybdopterin cofactor binding(GO:0043546) |
| 0.4 | 2.7 | GO:0030492 | hemoglobin binding(GO:0030492) |
| 0.4 | 1.1 | GO:0017153 | sodium:dicarboxylate symporter activity(GO:0017153) |
| 0.2 | 3.1 | GO:1990825 | sequence-specific mRNA binding(GO:1990825) |
| 0.2 | 2.1 | GO:1990189 | peptide-serine-N-acetyltransferase activity(GO:1990189) |
| 0.2 | 1.3 | GO:0034432 | endopolyphosphatase activity(GO:0000298) diphosphoinositol-polyphosphate diphosphatase activity(GO:0008486) bis(5'-adenosyl)-hexaphosphatase activity(GO:0034431) bis(5'-adenosyl)-pentaphosphatase activity(GO:0034432) |
| 0.2 | 2.0 | GO:0004468 | lysine N-acetyltransferase activity, acting on acetyl phosphate as donor(GO:0004468) |
| 0.2 | 1.0 | GO:0000213 | tRNA-intron endonuclease activity(GO:0000213) |
| 0.1 | 1.1 | GO:0004865 | protein serine/threonine phosphatase inhibitor activity(GO:0004865) |
| 0.1 | 2.8 | GO:0032036 | myosin heavy chain binding(GO:0032036) |
| 0.1 | 1.9 | GO:0015174 | basic amino acid transmembrane transporter activity(GO:0015174) |
| 0.1 | 2.0 | GO:0008510 | sodium:bicarbonate symporter activity(GO:0008510) |
| 0.1 | 1.7 | GO:0045028 | G-protein coupled nucleotide receptor activity(GO:0001608) G-protein coupled purinergic nucleotide receptor activity(GO:0045028) |
| 0.1 | 2.1 | GO:0070006 | metalloaminopeptidase activity(GO:0070006) |
| 0.1 | 2.5 | GO:0003887 | DNA-directed DNA polymerase activity(GO:0003887) |
| 0.1 | 2.4 | GO:0070182 | DNA polymerase binding(GO:0070182) |
| 0.1 | 1.1 | GO:0045294 | alpha-catenin binding(GO:0045294) |
| 0.1 | 0.5 | GO:0003876 | AMP deaminase activity(GO:0003876) adenosine-phosphate deaminase activity(GO:0047623) |
| 0.1 | 0.3 | GO:0004008 | copper-exporting ATPase activity(GO:0004008) copper-transporting ATPase activity(GO:0043682) |
| 0.1 | 1.6 | GO:0004707 | MAP kinase activity(GO:0004707) |
| 0.1 | 0.2 | GO:0004348 | glucosylceramidase activity(GO:0004348) |
| 0.1 | 0.7 | GO:0031433 | telethonin binding(GO:0031433) |
| 0.1 | 2.6 | GO:0046966 | thyroid hormone receptor binding(GO:0046966) |
| 0.0 | 1.4 | GO:0001530 | lipopolysaccharide binding(GO:0001530) |
| 0.0 | 0.4 | GO:0003873 | 6-phosphofructo-2-kinase activity(GO:0003873) |
| 0.0 | 1.3 | GO:0035259 | glucocorticoid receptor binding(GO:0035259) |
| 0.0 | 0.6 | GO:0035925 | mRNA 3'-UTR AU-rich region binding(GO:0035925) |
| 0.0 | 7.8 | GO:0003823 | antigen binding(GO:0003823) |
| 0.0 | 0.5 | GO:0016884 | carbon-nitrogen ligase activity, with glutamine as amido-N-donor(GO:0016884) |
| 0.0 | 1.3 | GO:0030506 | ankyrin binding(GO:0030506) |
| 0.0 | 1.1 | GO:0004709 | MAP kinase kinase kinase activity(GO:0004709) |
| 0.0 | 2.6 | GO:0035591 | signaling adaptor activity(GO:0035591) |
| 0.0 | 0.7 | GO:0004129 | cytochrome-c oxidase activity(GO:0004129) heme-copper terminal oxidase activity(GO:0015002) oxidoreductase activity, acting on a heme group of donors, oxygen as acceptor(GO:0016676) |
| 0.0 | 0.9 | GO:0080025 | phosphatidylinositol-3,5-bisphosphate binding(GO:0080025) |
| 0.0 | 3.0 | GO:0004896 | cytokine receptor activity(GO:0004896) |
| 0.0 | 4.0 | GO:0004386 | helicase activity(GO:0004386) |
| 0.0 | 1.0 | GO:0004693 | cyclin-dependent protein serine/threonine kinase activity(GO:0004693) |
| 0.0 | 0.6 | GO:0071889 | 14-3-3 protein binding(GO:0071889) |
| 0.0 | 0.7 | GO:0042056 | chemoattractant activity(GO:0042056) |
| 0.0 | 0.5 | GO:0005035 | tumor necrosis factor-activated receptor activity(GO:0005031) death receptor activity(GO:0005035) |
| 0.0 | 3.4 | GO:0019887 | protein kinase regulator activity(GO:0019887) |
| 0.0 | 3.1 | GO:0001078 | transcriptional repressor activity, RNA polymerase II core promoter proximal region sequence-specific binding(GO:0001078) |
| 0.0 | 1.1 | GO:0005548 | phospholipid transporter activity(GO:0005548) |
| 0.0 | 1.1 | GO:0005080 | protein kinase C binding(GO:0005080) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 2.4 | PID ATM PATHWAY | ATM pathway |
| 0.1 | 3.2 | PID IL8 CXCR2 PATHWAY | IL8- and CXCR2-mediated signaling events |
| 0.0 | 2.8 | ST JNK MAPK PATHWAY | JNK MAPK Pathway |
| 0.0 | 2.5 | PID RHOA PATHWAY | RhoA signaling pathway |
| 0.0 | 1.3 | PID IL5 PATHWAY | IL5-mediated signaling events |
| 0.0 | 0.9 | PID ARF 3PATHWAY | Arf1 pathway |
| 0.0 | 1.0 | PID IL2 STAT5 PATHWAY | IL2 signaling events mediated by STAT5 |
| 0.0 | 0.7 | PID ARF6 PATHWAY | Arf6 signaling events |
| 0.0 | 1.4 | PID RAC1 REG PATHWAY | Regulation of RAC1 activity |
| 0.0 | 1.8 | PID CMYB PATHWAY | C-MYB transcription factor network |
| 0.0 | 1.5 | PID MTOR 4PATHWAY | mTOR signaling pathway |
| 0.0 | 0.4 | PID CDC42 REG PATHWAY | Regulation of CDC42 activity |
| 0.0 | 1.0 | PID TRKR PATHWAY | Neurotrophic factor-mediated Trk receptor signaling |
| 0.0 | 0.5 | PID CD8 TCR DOWNSTREAM PATHWAY | Downstream signaling in naïve CD8+ T cells |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 4.7 | REACTOME PROSTANOID LIGAND RECEPTORS | Genes involved in Prostanoid ligand receptors |
| 0.2 | 2.4 | REACTOME REGULATION OF THE FANCONI ANEMIA PATHWAY | Genes involved in Regulation of the Fanconi anemia pathway |
| 0.2 | 5.6 | REACTOME PRE NOTCH TRANSCRIPTION AND TRANSLATION | Genes involved in Pre-NOTCH Transcription and Translation |
| 0.1 | 7.3 | REACTOME GLYCOLYSIS | Genes involved in Glycolysis |
| 0.1 | 1.6 | REACTOME ERKS ARE INACTIVATED | Genes involved in ERKs are inactivated |
| 0.1 | 2.2 | REACTOME PURINE CATABOLISM | Genes involved in Purine catabolism |
| 0.1 | 3.4 | REACTOME SEMA4D INDUCED CELL MIGRATION AND GROWTH CONE COLLAPSE | Genes involved in Sema4D induced cell migration and growth-cone collapse |
| 0.1 | 1.7 | REACTOME P2Y RECEPTORS | Genes involved in P2Y receptors |
| 0.1 | 2.0 | REACTOME IL 7 SIGNALING | Genes involved in Interleukin-7 signaling |
| 0.1 | 0.9 | REACTOME COPI MEDIATED TRANSPORT | Genes involved in COPI Mediated Transport |
| 0.1 | 2.7 | REACTOME STEROID HORMONES | Genes involved in Steroid hormones |
| 0.1 | 1.1 | REACTOME BILE SALT AND ORGANIC ANION SLC TRANSPORTERS | Genes involved in Bile salt and organic anion SLC transporters |
| 0.1 | 3.1 | REACTOME MRNA 3 END PROCESSING | Genes involved in mRNA 3'-end processing |
| 0.1 | 1.0 | REACTOME CALNEXIN CALRETICULIN CYCLE | Genes involved in Calnexin/calreticulin cycle |
| 0.0 | 1.9 | REACTOME BASIGIN INTERACTIONS | Genes involved in Basigin interactions |
| 0.0 | 1.0 | REACTOME PROLACTIN RECEPTOR SIGNALING | Genes involved in Prolactin receptor signaling |
| 0.0 | 3.8 | REACTOME CELL SURFACE INTERACTIONS AT THE VASCULAR WALL | Genes involved in Cell surface interactions at the vascular wall |
| 0.0 | 0.5 | REACTOME PURINE RIBONUCLEOSIDE MONOPHOSPHATE BIOSYNTHESIS | Genes involved in Purine ribonucleoside monophosphate biosynthesis |
| 0.0 | 0.7 | REACTOME TERMINATION OF O GLYCAN BIOSYNTHESIS | Genes involved in Termination of O-glycan biosynthesis |
| 0.0 | 0.5 | REACTOME PURINE METABOLISM | Genes involved in Purine metabolism |
| 0.0 | 1.0 | REACTOME G1 PHASE | Genes involved in G1 Phase |
| 0.0 | 0.4 | REACTOME SYNTHESIS OF SUBSTRATES IN N GLYCAN BIOSYTHESIS | Genes involved in Synthesis of substrates in N-glycan biosythesis |
| 0.0 | 1.4 | REACTOME NRAGE SIGNALS DEATH THROUGH JNK | Genes involved in NRAGE signals death through JNK |
| 0.0 | 2.0 | REACTOME TRANSPORT OF INORGANIC CATIONS ANIONS AND AMINO ACIDS OLIGOPEPTIDES | Genes involved in Transport of inorganic cations/anions and amino acids/oligopeptides |
| 0.0 | 0.6 | REACTOME MITOCHONDRIAL PROTEIN IMPORT | Genes involved in Mitochondrial Protein Import |