Project
PRJNA375882: Comprehensive Mouse Transcriptomic BodyMap
Navigation
Downloads
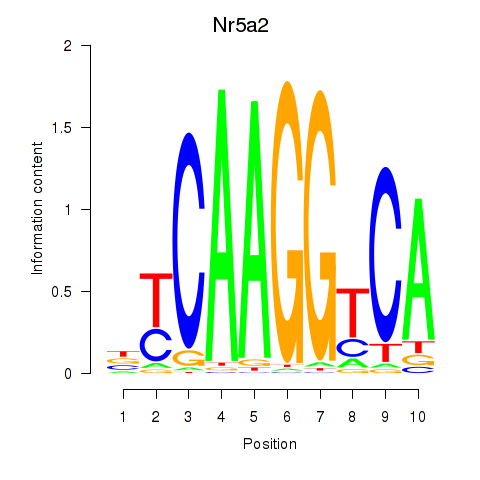
Results for Nr5a2
Z-value: 1.28
Transcription factors associated with Nr5a2
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Nr5a2
|
ENSMUSG00000026398.15 | Nr5a2 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Nr5a2 | mm39_v1_chr1_-_136876902_136876929 | 0.04 | 7.6e-01 | Click! |
Activity profile of Nr5a2 motif
Sorted Z-values of Nr5a2 motif
Network of associatons between targets according to the STRING database.
First level regulatory network of Nr5a2
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr9_-_54568950 | 10.15 |
ENSMUST00000128624.2
|
Acsbg1
|
acyl-CoA synthetase bubblegum family member 1 |
| chr9_+_57921954 | 8.45 |
ENSMUST00000034874.14
|
Cyp11a1
|
cytochrome P450, family 11, subfamily a, polypeptide 1 |
| chr9_-_54569128 | 8.08 |
ENSMUST00000034822.12
|
Acsbg1
|
acyl-CoA synthetase bubblegum family member 1 |
| chr6_-_54543446 | 7.75 |
ENSMUST00000019268.11
|
Scrn1
|
secernin 1 |
| chr1_-_134163102 | 7.72 |
ENSMUST00000187631.2
ENSMUST00000038191.8 ENSMUST00000086465.6 |
Adora1
|
adenosine A1 receptor |
| chr19_-_43512929 | 7.14 |
ENSMUST00000026196.14
|
Got1
|
glutamic-oxaloacetic transaminase 1, soluble |
| chr18_+_45402018 | 6.52 |
ENSMUST00000183850.8
ENSMUST00000066890.14 |
Kcnn2
|
potassium intermediate/small conductance calcium-activated channel, subfamily N, member 2 |
| chr1_+_75526225 | 6.20 |
ENSMUST00000154101.8
|
Slc4a3
|
solute carrier family 4 (anion exchanger), member 3 |
| chr3_-_107366868 | 6.04 |
ENSMUST00000009617.10
ENSMUST00000238670.2 |
Kcnc4
|
potassium voltage gated channel, Shaw-related subfamily, member 4 |
| chr1_+_75522902 | 5.90 |
ENSMUST00000124341.8
|
Slc4a3
|
solute carrier family 4 (anion exchanger), member 3 |
| chr1_-_43235914 | 5.85 |
ENSMUST00000187357.2
|
Fhl2
|
four and a half LIM domains 2 |
| chrX_+_156482116 | 5.48 |
ENSMUST00000112521.8
|
Smpx
|
small muscle protein, X-linked |
| chr5_+_118165808 | 5.46 |
ENSMUST00000031304.14
|
Tesc
|
tescalcin |
| chr8_+_94879235 | 5.13 |
ENSMUST00000034211.10
ENSMUST00000211930.2 ENSMUST00000211915.2 |
Mt3
|
metallothionein 3 |
| chr17_-_26087696 | 4.92 |
ENSMUST00000236479.2
ENSMUST00000235806.2 ENSMUST00000026828.7 |
Mcrip2
|
MAPK regulated corepressor interacting protein 2 |
| chr2_+_32518402 | 4.80 |
ENSMUST00000156578.8
|
Ak1
|
adenylate kinase 1 |
| chr1_-_175319842 | 4.71 |
ENSMUST00000195324.6
ENSMUST00000192227.6 ENSMUST00000194555.6 |
Rgs7
|
regulator of G protein signaling 7 |
| chr12_+_102521225 | 4.65 |
ENSMUST00000021610.7
|
Chga
|
chromogranin A |
| chr9_+_110595224 | 4.65 |
ENSMUST00000136695.3
|
Myl3
|
myosin, light polypeptide 3 |
| chr17_-_56424577 | 4.62 |
ENSMUST00000019808.12
|
Plin5
|
perilipin 5 |
| chr9_-_107546166 | 4.54 |
ENSMUST00000177567.8
|
Slc38a3
|
solute carrier family 38, member 3 |
| chr17_-_56424265 | 4.46 |
ENSMUST00000113072.3
|
Plin5
|
perilipin 5 |
| chr5_+_122239007 | 4.45 |
ENSMUST00000014080.13
ENSMUST00000111750.8 |
Myl2
|
myosin, light polypeptide 2, regulatory, cardiac, slow |
| chr13_-_92024994 | 4.36 |
ENSMUST00000022122.4
|
Ckmt2
|
creatine kinase, mitochondrial 2 |
| chr3_+_27237114 | 4.36 |
ENSMUST00000046515.15
|
Nceh1
|
neutral cholesterol ester hydrolase 1 |
| chr9_-_43027809 | 4.26 |
ENSMUST00000216126.2
ENSMUST00000213544.2 ENSMUST00000061833.6 |
Tlcd5
|
TLC domain containing 5 |
| chr10_+_79552421 | 4.20 |
ENSMUST00000099513.8
ENSMUST00000020581.3 |
Hcn2
|
hyperpolarization-activated, cyclic nucleotide-gated K+ 2 |
| chr5_+_122239030 | 4.10 |
ENSMUST00000139213.8
ENSMUST00000111751.8 ENSMUST00000155612.8 |
Myl2
|
myosin, light polypeptide 2, regulatory, cardiac, slow |
| chr3_+_27237143 | 4.10 |
ENSMUST00000091284.5
|
Nceh1
|
neutral cholesterol ester hydrolase 1 |
| chrX_+_74425990 | 3.85 |
ENSMUST00000033541.5
|
Fundc2
|
FUN14 domain containing 2 |
| chr7_+_29883611 | 3.80 |
ENSMUST00000208441.2
|
Cox7a1
|
cytochrome c oxidase subunit 7A1 |
| chr17_+_44263890 | 3.79 |
ENSMUST00000177857.9
ENSMUST00000044792.6 |
Rcan2
|
regulator of calcineurin 2 |
| chr17_-_35023521 | 3.78 |
ENSMUST00000025223.9
|
Cyp21a1
|
cytochrome P450, family 21, subfamily a, polypeptide 1 |
| chr8_+_46944000 | 3.78 |
ENSMUST00000110372.9
ENSMUST00000130563.2 |
Acsl1
|
acyl-CoA synthetase long-chain family member 1 |
| chr13_-_99653045 | 3.76 |
ENSMUST00000064762.6
|
Map1b
|
microtubule-associated protein 1B |
| chr15_+_98006346 | 3.75 |
ENSMUST00000051226.8
|
Pfkm
|
phosphofructokinase, muscle |
| chr1_+_75523092 | 3.62 |
ENSMUST00000150142.8
|
Slc4a3
|
solute carrier family 4 (anion exchanger), member 3 |
| chr4_-_141143313 | 3.61 |
ENSMUST00000006378.9
ENSMUST00000105788.2 |
Clcnkb
|
chloride channel, voltage-sensitive Kb |
| chr19_-_60862964 | 3.57 |
ENSMUST00000025961.7
|
Prdx3
|
peroxiredoxin 3 |
| chr5_+_129802127 | 3.48 |
ENSMUST00000086046.10
ENSMUST00000186265.6 |
Nipsnap2
|
nipsnap homolog 2 |
| chr7_-_4525793 | 3.45 |
ENSMUST00000140424.8
|
Tnni3
|
troponin I, cardiac 3 |
| chr15_-_83989801 | 3.44 |
ENSMUST00000229826.2
ENSMUST00000082365.6 |
Sult4a1
|
sulfotransferase family 4A, member 1 |
| chr15_+_89218601 | 3.41 |
ENSMUST00000023282.9
|
Miox
|
myo-inositol oxygenase |
| chr10_+_94034817 | 3.39 |
ENSMUST00000020209.16
ENSMUST00000179990.8 |
Ndufa12
|
NADH:ubiquinone oxidoreductase subunit A12 |
| chr9_-_107546195 | 3.36 |
ENSMUST00000192990.6
|
Slc38a3
|
solute carrier family 38, member 3 |
| chr19_+_8568618 | 3.31 |
ENSMUST00000170817.2
ENSMUST00000010251.11 |
Slc22a8
|
solute carrier family 22 (organic anion transporter), member 8 |
| chr1_+_75523166 | 3.30 |
ENSMUST00000138814.2
|
Slc4a3
|
solute carrier family 4 (anion exchanger), member 3 |
| chr15_-_77813123 | 3.28 |
ENSMUST00000109748.9
ENSMUST00000109747.9 ENSMUST00000100486.6 ENSMUST00000005487.12 |
Txn2
|
thioredoxin 2 |
| chr7_-_4525426 | 3.26 |
ENSMUST00000209148.2
ENSMUST00000098859.10 |
Tnni3
|
troponin I, cardiac 3 |
| chr11_-_3454766 | 3.25 |
ENSMUST00000044507.12
|
Inpp5j
|
inositol polyphosphate 5-phosphatase J |
| chr19_+_10366753 | 3.20 |
ENSMUST00000169121.9
ENSMUST00000076968.11 ENSMUST00000235479.2 ENSMUST00000223586.2 ENSMUST00000235784.2 ENSMUST00000224135.3 ENSMUST00000225452.3 ENSMUST00000237366.2 |
Syt7
|
synaptotagmin VII |
| chr9_+_65008735 | 3.14 |
ENSMUST00000213533.2
ENSMUST00000035499.5 ENSMUST00000077696.13 ENSMUST00000166273.2 |
Igdcc4
|
immunoglobulin superfamily, DCC subclass, member 4 |
| chr13_-_53083494 | 3.09 |
ENSMUST00000123599.8
|
Auh
|
AU RNA binding protein/enoyl-coenzyme A hydratase |
| chr4_+_130297132 | 3.05 |
ENSMUST00000105993.4
|
Nkain1
|
Na+/K+ transporting ATPase interacting 1 |
| chr9_+_65538352 | 3.02 |
ENSMUST00000216342.2
ENSMUST00000216382.2 |
Rbpms2
|
RNA binding protein with multiple splicing 2 |
| chr15_+_68800261 | 2.97 |
ENSMUST00000022954.7
|
Khdrbs3
|
KH domain containing, RNA binding, signal transduction associated 3 |
| chr17_+_17669082 | 2.91 |
ENSMUST00000140134.2
|
Lix1
|
limb and CNS expressed 1 |
| chr6_+_90596123 | 2.89 |
ENSMUST00000032177.10
|
Slc41a3
|
solute carrier family 41, member 3 |
| chr9_-_121621544 | 2.85 |
ENSMUST00000035110.11
|
Hhatl
|
hedgehog acyltransferase-like |
| chr11_+_116547932 | 2.84 |
ENSMUST00000116318.3
|
Prcd
|
photoreceptor disc component |
| chr10_-_89093441 | 2.83 |
ENSMUST00000182341.8
ENSMUST00000182613.8 |
Ano4
|
anoctamin 4 |
| chr9_+_107217786 | 2.82 |
ENSMUST00000042581.4
|
6430571L13Rik
|
RIKEN cDNA 6430571L13 gene |
| chr1_+_36730530 | 2.81 |
ENSMUST00000081180.7
ENSMUST00000193210.6 ENSMUST00000195151.6 |
Cox5b
|
cytochrome c oxidase subunit 5B |
| chr14_+_122419116 | 2.80 |
ENSMUST00000026625.7
|
Clybl
|
citrate lyase beta like |
| chr15_+_74435587 | 2.66 |
ENSMUST00000185682.7
ENSMUST00000170845.8 ENSMUST00000187599.2 |
Adgrb1
|
adhesion G protein-coupled receptor B1 |
| chr1_-_155120190 | 2.64 |
ENSMUST00000186156.7
|
BC034090
|
cDNA sequence BC034090 |
| chr9_+_65536892 | 2.62 |
ENSMUST00000169003.8
|
Rbpms2
|
RNA binding protein with multiple splicing 2 |
| chr2_-_64806106 | 2.56 |
ENSMUST00000156765.2
|
Grb14
|
growth factor receptor bound protein 14 |
| chr9_-_70048766 | 2.52 |
ENSMUST00000034749.16
|
Fam81a
|
family with sequence similarity 81, member A |
| chrX_-_70536198 | 2.52 |
ENSMUST00000080035.11
|
Cd99l2
|
CD99 antigen-like 2 |
| chr11_+_98632631 | 2.51 |
ENSMUST00000064187.12
|
Thra
|
thyroid hormone receptor alpha |
| chrX_-_70536449 | 2.50 |
ENSMUST00000037391.12
ENSMUST00000114586.9 ENSMUST00000114587.3 |
Cd99l2
|
CD99 antigen-like 2 |
| chr1_+_75377616 | 2.49 |
ENSMUST00000122266.3
|
Speg
|
SPEG complex locus |
| chr9_+_54493618 | 2.46 |
ENSMUST00000217484.2
|
Idh3a
|
isocitrate dehydrogenase 3 (NAD+) alpha |
| chr13_-_25121568 | 2.43 |
ENSMUST00000037615.7
|
Aldh5a1
|
aldhehyde dehydrogenase family 5, subfamily A1 |
| chr9_+_54493784 | 2.42 |
ENSMUST00000167866.2
|
Idh3a
|
isocitrate dehydrogenase 3 (NAD+) alpha |
| chr2_+_102536701 | 2.41 |
ENSMUST00000123759.8
ENSMUST00000005220.11 ENSMUST00000111212.8 |
Slc1a2
|
solute carrier family 1 (glial high affinity glutamate transporter), member 2 |
| chr9_-_53882530 | 2.38 |
ENSMUST00000048409.14
|
Elmod1
|
ELMO/CED-12 domain containing 1 |
| chr14_+_58310143 | 2.37 |
ENSMUST00000022545.14
|
Fgf9
|
fibroblast growth factor 9 |
| chr5_-_137609691 | 2.35 |
ENSMUST00000031731.14
|
Pcolce
|
procollagen C-endopeptidase enhancer protein |
| chr2_-_17465410 | 2.32 |
ENSMUST00000145492.2
|
Nebl
|
nebulette |
| chr5_-_137609634 | 2.32 |
ENSMUST00000054564.13
|
Pcolce
|
procollagen C-endopeptidase enhancer protein |
| chr18_+_77273510 | 2.26 |
ENSMUST00000075290.8
ENSMUST00000079618.11 |
St8sia5
|
ST8 alpha-N-acetyl-neuraminide alpha-2,8-sialyltransferase 5 |
| chr5_-_137608886 | 2.25 |
ENSMUST00000142675.8
|
Pcolce
|
procollagen C-endopeptidase enhancer protein |
| chr6_+_38639945 | 2.19 |
ENSMUST00000114874.5
|
Clec2l
|
C-type lectin domain family 2, member L |
| chr2_+_103396638 | 2.19 |
ENSMUST00000076212.4
|
Abtb2
|
ankyrin repeat and BTB (POZ) domain containing 2 |
| chr5_+_24569802 | 2.19 |
ENSMUST00000115090.6
ENSMUST00000030834.7 |
Nos3
|
nitric oxide synthase 3, endothelial cell |
| chr11_-_97464755 | 2.18 |
ENSMUST00000126287.2
ENSMUST00000107590.9 |
Srcin1
|
SRC kinase signaling inhibitor 1 |
| chr7_-_126398165 | 2.17 |
ENSMUST00000205890.2
ENSMUST00000205336.2 ENSMUST00000087566.11 |
Aldoa
|
aldolase A, fructose-bisphosphate |
| chr10_-_79973210 | 2.15 |
ENSMUST00000170219.9
ENSMUST00000169546.9 |
Cbarp
|
calcium channel, voltage-dependent, beta subunit associated regulatory protein |
| chr2_-_102903680 | 2.13 |
ENSMUST00000132449.8
ENSMUST00000111183.2 ENSMUST00000011058.9 |
Pdhx
|
pyruvate dehydrogenase complex, component X |
| chr15_-_79688910 | 2.13 |
ENSMUST00000175858.10
ENSMUST00000023057.10 |
Nptxr
|
neuronal pentraxin receptor |
| chr11_-_69451012 | 2.11 |
ENSMUST00000004036.6
|
Efnb3
|
ephrin B3 |
| chr13_+_104246259 | 2.06 |
ENSMUST00000160322.8
ENSMUST00000159574.2 |
Sgtb
|
small glutamine-rich tetratricopeptide repeat (TPR)-containing, beta |
| chr15_+_81756671 | 2.06 |
ENSMUST00000135198.2
ENSMUST00000157003.8 ENSMUST00000229068.2 |
Aco2
|
aconitase 2, mitochondrial |
| chr3_-_107992662 | 2.03 |
ENSMUST00000078912.7
|
Ampd2
|
adenosine monophosphate deaminase 2 |
| chr4_-_73869071 | 2.01 |
ENSMUST00000095023.2
ENSMUST00000030101.4 |
2310002L09Rik
|
RIKEN cDNA 2310002L09 gene |
| chr7_+_25016492 | 1.97 |
ENSMUST00000128119.2
|
Megf8
|
multiple EGF-like-domains 8 |
| chr2_+_155453103 | 1.96 |
ENSMUST00000092995.6
|
Myh7b
|
myosin, heavy chain 7B, cardiac muscle, beta |
| chr5_-_24650164 | 1.94 |
ENSMUST00000115043.8
ENSMUST00000115041.2 ENSMUST00000030800.13 |
Fastk
|
Fas-activated serine/threonine kinase |
| chr4_-_61437704 | 1.94 |
ENSMUST00000095051.6
ENSMUST00000107483.8 |
Mup16
|
major urinary protein 16 |
| chr5_+_31274064 | 1.92 |
ENSMUST00000202769.2
|
Trim54
|
tripartite motif-containing 54 |
| chr14_+_79086665 | 1.92 |
ENSMUST00000227255.2
|
Vwa8
|
von Willebrand factor A domain containing 8 |
| chr3_-_51316347 | 1.89 |
ENSMUST00000193279.2
ENSMUST00000038108.12 |
Ndufc1
|
NADH:ubiquinone oxidoreductase subunit C1 |
| chr13_-_53083674 | 1.89 |
ENSMUST00000120535.8
ENSMUST00000119311.8 ENSMUST00000021913.16 ENSMUST00000110031.4 |
Auh
|
AU RNA binding protein/enoyl-coenzyme A hydratase |
| chr16_-_28383615 | 1.87 |
ENSMUST00000231399.2
|
Fgf12
|
fibroblast growth factor 12 |
| chr5_+_31274046 | 1.86 |
ENSMUST00000013771.15
|
Trim54
|
tripartite motif-containing 54 |
| chr11_-_96720309 | 1.86 |
ENSMUST00000167149.8
|
Nfe2l1
|
nuclear factor, erythroid derived 2,-like 1 |
| chr2_+_140012560 | 1.85 |
ENSMUST00000044825.5
|
Ndufaf5
|
NADH:ubiquinone oxidoreductase complex assembly factor 5 |
| chr15_-_98193995 | 1.85 |
ENSMUST00000023722.12
ENSMUST00000169721.3 |
Zfp641
|
zinc finger protein 641 |
| chr8_+_95564949 | 1.84 |
ENSMUST00000034234.15
ENSMUST00000159871.4 |
Coq9
|
coenzyme Q9 |
| chr13_+_104246245 | 1.83 |
ENSMUST00000044385.14
|
Sgtb
|
small glutamine-rich tetratricopeptide repeat (TPR)-containing, beta |
| chr5_-_5744559 | 1.78 |
ENSMUST00000115426.9
|
Steap2
|
six transmembrane epithelial antigen of prostate 2 |
| chr10_-_10958031 | 1.78 |
ENSMUST00000105561.9
ENSMUST00000044306.13 |
Grm1
|
glutamate receptor, metabotropic 1 |
| chr4_-_148244028 | 1.77 |
ENSMUST00000167160.8
ENSMUST00000151246.8 |
Fbxo44
|
F-box protein 44 |
| chr13_-_95170755 | 1.77 |
ENSMUST00000162670.8
|
Pde8b
|
phosphodiesterase 8B |
| chr18_-_60757272 | 1.76 |
ENSMUST00000155195.3
|
Synpo
|
synaptopodin |
| chr4_+_140688514 | 1.74 |
ENSMUST00000010007.9
|
Sdhb
|
succinate dehydrogenase complex, subunit B, iron sulfur (Ip) |
| chr14_+_79086492 | 1.74 |
ENSMUST00000040990.7
|
Vwa8
|
von Willebrand factor A domain containing 8 |
| chr2_+_26518456 | 1.69 |
ENSMUST00000074240.4
|
Dipk1b
|
divergent protein kinase domain 1B |
| chr6_-_125142539 | 1.67 |
ENSMUST00000183272.2
ENSMUST00000182052.8 ENSMUST00000182277.2 |
Gapdh
|
glyceraldehyde-3-phosphate dehydrogenase |
| chr11_-_115158062 | 1.66 |
ENSMUST00000106554.2
|
Grin2c
|
glutamate receptor, ionotropic, NMDA2C (epsilon 3) |
| chr5_-_5744024 | 1.65 |
ENSMUST00000115425.9
ENSMUST00000115427.8 ENSMUST00000115424.9 ENSMUST00000015797.11 |
Steap2
|
six transmembrane epithelial antigen of prostate 2 |
| chr1_-_16589425 | 1.61 |
ENSMUST00000159558.8
ENSMUST00000054668.13 ENSMUST00000162627.8 ENSMUST00000162007.8 ENSMUST00000128957.9 ENSMUST00000115359.10 ENSMUST00000151888.8 |
Stau2
|
staufen double-stranded RNA binding protein 2 |
| chr13_-_21685588 | 1.60 |
ENSMUST00000044043.3
|
Cox5b-ps
|
cytochrome c oxidase subunit 5B, pseudogene |
| chr7_-_109380745 | 1.60 |
ENSMUST00000207400.2
ENSMUST00000033331.7 |
Nrip3
|
nuclear receptor interacting protein 3 |
| chr2_-_157408239 | 1.59 |
ENSMUST00000109528.9
ENSMUST00000088494.3 |
Blcap
|
bladder cancer associated protein |
| chr5_+_150042092 | 1.59 |
ENSMUST00000200960.4
ENSMUST00000202530.4 |
Fry
|
FRY microtubule binding protein |
| chr11_+_42310557 | 1.59 |
ENSMUST00000007797.10
|
Gabrb2
|
gamma-aminobutyric acid (GABA) A receptor, subunit beta 2 |
| chrX_-_73412078 | 1.58 |
ENSMUST00000155676.8
|
Ubl4a
|
ubiquitin-like 4A |
| chr13_-_74498320 | 1.58 |
ENSMUST00000221594.2
ENSMUST00000022062.8 |
Sdha
|
succinate dehydrogenase complex, subunit A, flavoprotein (Fp) |
| chr15_-_78428865 | 1.57 |
ENSMUST00000053239.4
|
Sstr3
|
somatostatin receptor 3 |
| chr7_+_141503719 | 1.57 |
ENSMUST00000105989.9
ENSMUST00000075528.12 ENSMUST00000174499.8 |
Brsk2
|
BR serine/threonine kinase 2 |
| chrM_+_8603 | 1.56 |
ENSMUST00000082409.1
|
mt-Co3
|
mitochondrially encoded cytochrome c oxidase III |
| chr12_-_31401432 | 1.55 |
ENSMUST00000110857.5
|
Dld
|
dihydrolipoamide dehydrogenase |
| chr1_-_135241429 | 1.55 |
ENSMUST00000134088.3
ENSMUST00000081104.10 |
Timm17a
|
translocase of inner mitochondrial membrane 17a |
| chr19_-_3962733 | 1.54 |
ENSMUST00000075092.8
ENSMUST00000235847.2 ENSMUST00000235301.2 ENSMUST00000237341.2 |
Ndufs8
|
NADH:ubiquinone oxidoreductase core subunit S8 |
| chr9_+_45311000 | 1.54 |
ENSMUST00000216289.2
|
Fxyd2
|
FXYD domain-containing ion transport regulator 2 |
| chr11_+_68979308 | 1.54 |
ENSMUST00000021273.13
|
Vamp2
|
vesicle-associated membrane protein 2 |
| chr8_+_4298189 | 1.53 |
ENSMUST00000110993.2
|
Tgfbr3l
|
transforming growth factor, beta receptor III-like |
| chr2_+_90865958 | 1.53 |
ENSMUST00000111445.10
ENSMUST00000111446.10 ENSMUST00000050323.6 |
Rapsn
|
receptor-associated protein of the synapse |
| chr11_+_74540284 | 1.53 |
ENSMUST00000117818.2
ENSMUST00000092915.12 |
Cluh
|
clustered mitochondria (cluA/CLU1) homolog |
| chr6_+_113259262 | 1.51 |
ENSMUST00000041203.6
|
Cpne9
|
copine family member IX |
| chr6_+_145692439 | 1.51 |
ENSMUST00000111704.8
|
Rassf8
|
Ras association (RalGDS/AF-6) domain family (N-terminal) member 8 |
| chr6_+_48604869 | 1.51 |
ENSMUST00000154570.2
|
AI854703
|
expressed sequence AI854703 |
| chr4_+_99786611 | 1.50 |
ENSMUST00000058351.16
|
Pgm1
|
phosphoglucomutase 1 |
| chr14_-_104081119 | 1.50 |
ENSMUST00000227824.2
ENSMUST00000172237.2 |
Ednrb
|
endothelin receptor type B |
| chr3_+_96128427 | 1.49 |
ENSMUST00000090781.8
|
H2bc21
|
H2B clustered histone 21 |
| chr7_+_78432867 | 1.48 |
ENSMUST00000032840.5
|
Mrps11
|
mitochondrial ribosomal protein S11 |
| chr1_+_131898325 | 1.46 |
ENSMUST00000027695.8
|
Slc45a3
|
solute carrier family 45, member 3 |
| chr4_-_68872585 | 1.46 |
ENSMUST00000030036.6
|
Brinp1
|
bone morphogenic protein/retinoic acid inducible neural specific 1 |
| chr11_+_67477347 | 1.46 |
ENSMUST00000108682.9
|
Gas7
|
growth arrest specific 7 |
| chr15_-_35938155 | 1.38 |
ENSMUST00000156915.3
|
Cox6c
|
cytochrome c oxidase subunit 6C |
| chr7_+_110368037 | 1.37 |
ENSMUST00000213373.2
|
Ampd3
|
adenosine monophosphate deaminase 3 |
| chr15_+_76227695 | 1.37 |
ENSMUST00000023210.8
ENSMUST00000231045.2 |
Cyc1
|
cytochrome c-1 |
| chr8_-_70573465 | 1.36 |
ENSMUST00000002412.9
|
Ncan
|
neurocan |
| chr13_-_67053384 | 1.35 |
ENSMUST00000021993.5
|
Uqcrb
|
ubiquinol-cytochrome c reductase binding protein |
| chr8_-_106022676 | 1.33 |
ENSMUST00000057855.4
|
Exoc3l
|
exocyst complex component 3-like |
| chr13_+_97208069 | 1.33 |
ENSMUST00000042517.8
|
Fam169a
|
family with sequence similarity 169, member A |
| chr4_+_123127349 | 1.31 |
ENSMUST00000040821.5
|
Heyl
|
hairy/enhancer-of-split related with YRPW motif-like |
| chr6_+_91133755 | 1.31 |
ENSMUST00000143621.8
|
Hdac11
|
histone deacetylase 11 |
| chr5_+_117501557 | 1.31 |
ENSMUST00000111959.2
|
Wsb2
|
WD repeat and SOCS box-containing 2 |
| chr16_-_91728162 | 1.30 |
ENSMUST00000139277.8
ENSMUST00000154661.8 |
Atp5o
|
ATP synthase, H+ transporting, mitochondrial F1 complex, O subunit |
| chr1_-_171050004 | 1.29 |
ENSMUST00000147246.2
ENSMUST00000111326.8 ENSMUST00000138184.8 |
Tomm40l
|
translocase of outer mitochondrial membrane 40-like |
| chr7_-_141009264 | 1.29 |
ENSMUST00000164387.2
ENSMUST00000137488.2 ENSMUST00000084436.10 |
Cend1
|
cell cycle exit and neuronal differentiation 1 |
| chr13_-_30170031 | 1.29 |
ENSMUST00000102948.11
|
E2f3
|
E2F transcription factor 3 |
| chr4_-_82423985 | 1.28 |
ENSMUST00000107245.9
ENSMUST00000107246.2 |
Nfib
|
nuclear factor I/B |
| chr8_+_121395047 | 1.27 |
ENSMUST00000181795.2
|
Cox4i1
|
cytochrome c oxidase subunit 4I1 |
| chr2_-_84508385 | 1.27 |
ENSMUST00000189772.2
ENSMUST00000053664.9 ENSMUST00000111664.8 |
Gm28635
Tmx2
|
predicted gene 28635 thioredoxin-related transmembrane protein 2 |
| chr11_+_67477501 | 1.27 |
ENSMUST00000108680.2
|
Gas7
|
growth arrest specific 7 |
| chr18_-_35087355 | 1.27 |
ENSMUST00000025217.11
|
Hspa9
|
heat shock protein 9 |
| chr14_-_20844034 | 1.27 |
ENSMUST00000226630.2
|
Camk2g
|
calcium/calmodulin-dependent protein kinase II gamma |
| chr4_-_138858340 | 1.26 |
ENSMUST00000143971.2
|
Micos10
|
mitochondrial contact site and cristae organizing system subunit 10 |
| chr13_+_21971631 | 1.26 |
ENSMUST00000110473.3
ENSMUST00000102982.2 |
H2bc22
|
H2B clustered histone 22 |
| chr6_+_83326424 | 1.25 |
ENSMUST00000136501.2
|
Bola3
|
bolA-like 3 (E. coli) |
| chr11_+_6241964 | 1.25 |
ENSMUST00000140765.8
|
Ogdh
|
oxoglutarate (alpha-ketoglutarate) dehydrogenase (lipoamide) |
| chr1_+_104696235 | 1.25 |
ENSMUST00000062528.9
|
Cdh20
|
cadherin 20 |
| chr6_-_114018982 | 1.25 |
ENSMUST00000101045.10
|
Atp2b2
|
ATPase, Ca++ transporting, plasma membrane 2 |
| chr4_-_132939790 | 1.24 |
ENSMUST00000052090.9
|
Gpr3
|
G-protein coupled receptor 3 |
| chr8_-_105350898 | 1.22 |
ENSMUST00000212882.2
ENSMUST00000163783.4 |
Cdh16
|
cadherin 16 |
| chr16_+_4501934 | 1.22 |
ENSMUST00000060067.12
ENSMUST00000115854.4 ENSMUST00000229529.2 |
Dnaja3
|
DnaJ heat shock protein family (Hsp40) member A3 |
| chr19_-_46661321 | 1.22 |
ENSMUST00000026012.8
|
Cyp17a1
|
cytochrome P450, family 17, subfamily a, polypeptide 1 |
| chr11_+_6241660 | 1.21 |
ENSMUST00000101554.9
ENSMUST00000093350.10 |
Ogdh
|
oxoglutarate (alpha-ketoglutarate) dehydrogenase (lipoamide) |
| chr19_-_46661501 | 1.21 |
ENSMUST00000236174.2
|
Cyp17a1
|
cytochrome P450, family 17, subfamily a, polypeptide 1 |
| chr11_-_53321606 | 1.20 |
ENSMUST00000061326.5
ENSMUST00000109021.4 |
Uqcrq
|
ubiquinol-cytochrome c reductase, complex III subunit VII |
| chr3_+_121243395 | 1.19 |
ENSMUST00000198393.2
|
Cnn3
|
calponin 3, acidic |
| chr2_+_121125918 | 1.18 |
ENSMUST00000110639.8
|
Map1a
|
microtubule-associated protein 1 A |
| chr3_+_96484294 | 1.17 |
ENSMUST00000148290.2
|
Gm16253
|
predicted gene 16253 |
| chr15_+_74435217 | 1.16 |
ENSMUST00000190524.7
|
Adgrb1
|
adhesion G protein-coupled receptor B1 |
| chr2_-_30305472 | 1.15 |
ENSMUST00000134120.2
ENSMUST00000102854.10 |
Crat
|
carnitine acetyltransferase |
| chr11_+_70735751 | 1.15 |
ENSMUST00000177731.8
ENSMUST00000108533.10 ENSMUST00000081362.13 ENSMUST00000178245.2 |
Rabep1
|
rabaptin, RAB GTPase binding effector protein 1 |
| chr4_-_82423944 | 1.12 |
ENSMUST00000107248.8
ENSMUST00000107247.8 |
Nfib
|
nuclear factor I/B |
| chr15_+_25843225 | 1.11 |
ENSMUST00000022881.15
|
Retreg1
|
reticulophagy regulator 1 |
| chr4_-_155845500 | 1.11 |
ENSMUST00000030903.12
|
Atad3a
|
ATPase family, AAA domain containing 3A |
| chr8_-_71315902 | 1.10 |
ENSMUST00000212611.2
|
Kcnn1
|
potassium intermediate/small conductance calcium-activated channel, subfamily N, member 1 |
| chr12_-_16639756 | 1.08 |
ENSMUST00000222989.2
ENSMUST00000067124.6 ENSMUST00000221230.2 |
Lpin1
|
lipin 1 |
| chr2_-_30305779 | 1.08 |
ENSMUST00000102855.8
ENSMUST00000028207.13 |
Crat
|
carnitine acetyltransferase |
| chr8_+_121394961 | 1.08 |
ENSMUST00000034276.13
ENSMUST00000181586.8 |
Cox4i1
|
cytochrome c oxidase subunit 4I1 |
| chr11_-_120521382 | 1.07 |
ENSMUST00000106181.8
|
Mafg
|
v-maf musculoaponeurotic fibrosarcoma oncogene family, protein G (avian) |
| chr9_+_4376556 | 1.05 |
ENSMUST00000212075.2
|
Msantd4
|
Myb/SANT-like DNA-binding domain containing 4 with coiled-coils |
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.8 | 8.5 | GO:0042694 | muscle cell fate specification(GO:0042694) |
| 2.7 | 8.1 | GO:0006106 | fumarate metabolic process(GO:0006106) |
| 2.6 | 7.9 | GO:2000487 | asparagine transport(GO:0006867) positive regulation of glutamine transport(GO:2000487) |
| 2.6 | 7.7 | GO:0032242 | positive regulation of nucleobase-containing compound transport(GO:0032241) regulation of nucleoside transport(GO:0032242) negative regulation of circadian sleep/wake cycle, non-REM sleep(GO:0042323) negative regulation of mucus secretion(GO:0070256) |
| 2.0 | 6.0 | GO:1904456 | negative regulation of neuronal action potential(GO:1904456) |
| 1.8 | 9.1 | GO:0035359 | negative regulation of peroxisome proliferator activated receptor signaling pathway(GO:0035359) |
| 1.5 | 5.9 | GO:0055011 | atrial cardiac muscle cell differentiation(GO:0055011) atrial cardiac muscle cell development(GO:0055014) |
| 1.4 | 5.5 | GO:0032417 | positive regulation of sodium:proton antiporter activity(GO:0032417) |
| 1.2 | 3.7 | GO:0097212 | lysosomal membrane organization(GO:0097212) |
| 1.2 | 4.6 | GO:2000705 | positive regulation of relaxation of muscle(GO:1901079) regulation of dense core granule biogenesis(GO:2000705) |
| 1.1 | 6.5 | GO:0042776 | mitochondrial ATP synthesis coupled proton transport(GO:0042776) |
| 1.1 | 3.2 | GO:1990926 | short-term synaptic potentiation(GO:1990926) |
| 1.1 | 2.1 | GO:0021941 | negative regulation of cerebellar granule cell precursor proliferation(GO:0021941) |
| 1.0 | 2.9 | GO:0006500 | N-terminal protein palmitoylation(GO:0006500) |
| 0.9 | 3.5 | GO:2000984 | regulation of ATP citrate synthase activity(GO:2000983) negative regulation of ATP citrate synthase activity(GO:2000984) |
| 0.9 | 2.6 | GO:1903537 | meiotic cell cycle process involved in oocyte maturation(GO:1903537) regulation of meiotic cell cycle process involved in oocyte maturation(GO:1903538) |
| 0.8 | 3.4 | GO:2000795 | negative regulation of epithelial cell proliferation involved in lung morphogenesis(GO:2000795) |
| 0.8 | 3.3 | GO:0042939 | glutathione transport(GO:0034635) tripeptide transport(GO:0042939) |
| 0.8 | 2.4 | GO:0006538 | glutamate catabolic process(GO:0006538) |
| 0.8 | 4.8 | GO:0001980 | regulation of systemic arterial blood pressure by ischemic conditions(GO:0001980) |
| 0.8 | 4.8 | GO:0046103 | inosine biosynthetic process(GO:0046103) |
| 0.8 | 6.9 | GO:0006102 | isocitrate metabolic process(GO:0006102) |
| 0.7 | 3.7 | GO:0061622 | glycolytic process through glucose-1-phosphate(GO:0061622) |
| 0.7 | 2.2 | GO:0014805 | smooth muscle adaptation(GO:0014805) |
| 0.6 | 2.5 | GO:0017055 | negative regulation of RNA polymerase II transcriptional preinitiation complex assembly(GO:0017055) |
| 0.6 | 1.8 | GO:0030961 | peptidyl-arginine hydroxylation(GO:0030961) |
| 0.6 | 4.2 | GO:0071321 | cellular response to cGMP(GO:0071321) |
| 0.6 | 1.7 | GO:0035606 | peptidyl-cysteine S-trans-nitrosylation(GO:0035606) |
| 0.5 | 3.6 | GO:0018171 | peptidyl-cysteine oxidation(GO:0018171) |
| 0.5 | 2.0 | GO:0097155 | embryonic heart tube left/right pattern formation(GO:0060971) fasciculation of sensory neuron axon(GO:0097155) |
| 0.5 | 5.3 | GO:0098914 | membrane repolarization during atrial cardiac muscle cell action potential(GO:0098914) |
| 0.5 | 2.8 | GO:0061589 | calcium activated phosphatidylserine scrambling(GO:0061589) |
| 0.4 | 12.2 | GO:0006700 | C21-steroid hormone biosynthetic process(GO:0006700) |
| 0.4 | 4.3 | GO:0097461 | ferric iron import into cell(GO:0097461) ferric iron import across plasma membrane(GO:0098706) |
| 0.4 | 3.7 | GO:0061732 | mitochondrial acetyl-CoA biosynthetic process from pyruvate(GO:0061732) |
| 0.4 | 1.6 | GO:0098963 | dendritic transport of ribonucleoprotein complex(GO:0098961) dendritic transport of messenger ribonucleoprotein complex(GO:0098963) anterograde dendritic transport of messenger ribonucleoprotein complex(GO:0098964) |
| 0.4 | 1.6 | GO:1904428 | negative regulation of tubulin deacetylation(GO:1904428) |
| 0.4 | 1.9 | GO:0098886 | modification of dendritic spine(GO:0098886) |
| 0.4 | 4.6 | GO:0006122 | mitochondrial electron transport, ubiquinol to cytochrome c(GO:0006122) |
| 0.4 | 5.6 | GO:0051151 | negative regulation of smooth muscle cell differentiation(GO:0051151) |
| 0.4 | 1.9 | GO:1905150 | regulation of voltage-gated sodium channel activity(GO:1905150) |
| 0.4 | 3.3 | GO:0006104 | succinyl-CoA metabolic process(GO:0006104) |
| 0.4 | 1.5 | GO:0015770 | disaccharide transport(GO:0015766) sucrose transport(GO:0015770) oligosaccharide transport(GO:0015772) |
| 0.4 | 1.8 | GO:0099566 | regulation of postsynaptic cytosolic calcium ion concentration(GO:0099566) |
| 0.4 | 1.8 | GO:0090032 | negative regulation of steroid hormone biosynthetic process(GO:0090032) |
| 0.3 | 2.4 | GO:0070779 | D-aspartate transport(GO:0070777) D-aspartate import(GO:0070779) |
| 0.3 | 3.4 | GO:0032261 | purine nucleotide salvage(GO:0032261) IMP salvage(GO:0032264) |
| 0.3 | 18.2 | GO:0000038 | very long-chain fatty acid metabolic process(GO:0000038) |
| 0.3 | 1.3 | GO:2000823 | regulation of androgen receptor activity(GO:2000823) |
| 0.3 | 1.6 | GO:0006121 | mitochondrial electron transport, succinate to ubiquinone(GO:0006121) |
| 0.3 | 2.2 | GO:0019254 | carnitine metabolic process, CoA-linked(GO:0019254) |
| 0.3 | 1.3 | GO:1902037 | negative regulation of hematopoietic stem cell differentiation(GO:1902037) |
| 0.3 | 3.8 | GO:0044539 | long-chain fatty acid import(GO:0044539) |
| 0.3 | 1.0 | GO:1901003 | negative regulation of fermentation(GO:1901003) |
| 0.3 | 1.5 | GO:0034334 | adherens junction maintenance(GO:0034334) |
| 0.2 | 1.5 | GO:0014826 | vein smooth muscle contraction(GO:0014826) |
| 0.2 | 5.0 | GO:0009083 | branched-chain amino acid catabolic process(GO:0009083) |
| 0.2 | 0.7 | GO:0042560 | 10-formyltetrahydrofolate catabolic process(GO:0009258) folic acid-containing compound catabolic process(GO:0009397) pteridine-containing compound catabolic process(GO:0042560) |
| 0.2 | 1.8 | GO:0007597 | blood coagulation, intrinsic pathway(GO:0007597) |
| 0.2 | 0.7 | GO:1900248 | cytoplasmic translational elongation(GO:0002182) regulation of cytoplasmic translational elongation(GO:1900247) negative regulation of cytoplasmic translational elongation(GO:1900248) |
| 0.2 | 2.4 | GO:0006704 | glucocorticoid biosynthetic process(GO:0006704) |
| 0.2 | 2.6 | GO:0006020 | inositol metabolic process(GO:0006020) |
| 0.2 | 2.8 | GO:0006123 | mitochondrial electron transport, cytochrome c to oxygen(GO:0006123) |
| 0.2 | 0.4 | GO:0061110 | dense core granule biogenesis(GO:0061110) |
| 0.2 | 2.1 | GO:0016198 | axon choice point recognition(GO:0016198) |
| 0.2 | 5.2 | GO:0007614 | short-term memory(GO:0007614) |
| 0.2 | 1.7 | GO:0033058 | directional locomotion(GO:0033058) |
| 0.2 | 1.2 | GO:0060331 | negative regulation of response to interferon-gamma(GO:0060331) negative regulation of interferon-gamma-mediated signaling pathway(GO:0060336) |
| 0.2 | 3.6 | GO:0030150 | protein import into mitochondrial matrix(GO:0030150) |
| 0.2 | 5.1 | GO:0034643 | establishment of mitochondrion localization, microtubule-mediated(GO:0034643) mitochondrion transport along microtubule(GO:0047497) |
| 0.2 | 0.9 | GO:0050902 | leukocyte adhesive activation(GO:0050902) |
| 0.2 | 0.7 | GO:1901475 | pyruvate transmembrane transport(GO:1901475) |
| 0.2 | 1.2 | GO:0048840 | otolith development(GO:0048840) |
| 0.2 | 0.5 | GO:0072363 | regulation of glycolytic process by positive regulation of transcription from RNA polymerase II promoter(GO:0072363) |
| 0.2 | 1.6 | GO:0071816 | tail-anchored membrane protein insertion into ER membrane(GO:0071816) |
| 0.2 | 5.2 | GO:0042775 | mitochondrial ATP synthesis coupled electron transport(GO:0042775) |
| 0.2 | 3.3 | GO:0006662 | glycerol ether metabolic process(GO:0006662) |
| 0.2 | 0.7 | GO:0000105 | histidine biosynthetic process(GO:0000105) |
| 0.2 | 0.5 | GO:0019626 | short-chain fatty acid catabolic process(GO:0019626) |
| 0.2 | 1.7 | GO:0006105 | succinate metabolic process(GO:0006105) |
| 0.2 | 0.8 | GO:1904253 | positive regulation of bile acid biosynthetic process(GO:0070859) positive regulation of bile acid metabolic process(GO:1904253) |
| 0.2 | 0.5 | GO:0046684 | response to pyrethroid(GO:0046684) |
| 0.2 | 0.3 | GO:1901856 | negative regulation of cellular respiration(GO:1901856) |
| 0.2 | 0.5 | GO:0048162 | preantral ovarian follicle growth(GO:0001546) multi-layer follicle stage(GO:0048162) |
| 0.2 | 0.3 | GO:0050923 | regulation of negative chemotaxis(GO:0050923) |
| 0.2 | 0.5 | GO:1903215 | negative regulation of protein targeting to mitochondrion(GO:1903215) |
| 0.1 | 2.5 | GO:0061615 | glycolytic process through fructose-6-phosphate(GO:0061615) |
| 0.1 | 0.4 | GO:1903281 | positive regulation of calcium:sodium antiporter activity(GO:1903281) |
| 0.1 | 3.2 | GO:0031115 | negative regulation of microtubule polymerization(GO:0031115) |
| 0.1 | 1.8 | GO:0002536 | respiratory burst involved in inflammatory response(GO:0002536) |
| 0.1 | 0.7 | GO:0010747 | positive regulation of plasma membrane long-chain fatty acid transport(GO:0010747) |
| 0.1 | 5.6 | GO:0010257 | NADH dehydrogenase complex assembly(GO:0010257) mitochondrial respiratory chain complex I assembly(GO:0032981) mitochondrial respiratory chain complex I biogenesis(GO:0097031) |
| 0.1 | 0.8 | GO:0045578 | negative regulation of B cell differentiation(GO:0045578) |
| 0.1 | 0.6 | GO:0015744 | succinate transport(GO:0015744) |
| 0.1 | 0.4 | GO:0003130 | BMP signaling pathway involved in heart induction(GO:0003130) endodermal-mesodermal cell signaling(GO:0003133) endodermal-mesodermal cell signaling involved in heart induction(GO:0003134) regulation of heart induction by regulation of canonical Wnt signaling pathway(GO:0090081) |
| 0.1 | 2.4 | GO:0032927 | positive regulation of activin receptor signaling pathway(GO:0032927) |
| 0.1 | 0.4 | GO:0045645 | regulation of eosinophil differentiation(GO:0045643) positive regulation of eosinophil differentiation(GO:0045645) |
| 0.1 | 2.3 | GO:0051823 | regulation of synapse structural plasticity(GO:0051823) |
| 0.1 | 0.5 | GO:0086047 | membrane depolarization during Purkinje myocyte cell action potential(GO:0086047) |
| 0.1 | 1.1 | GO:0030242 | pexophagy(GO:0030242) |
| 0.1 | 14.8 | GO:0051453 | regulation of intracellular pH(GO:0051453) |
| 0.1 | 1.4 | GO:0034982 | mitochondrial protein processing(GO:0034982) |
| 0.1 | 0.9 | GO:0034551 | respiratory chain complex III assembly(GO:0017062) mitochondrial respiratory chain complex III assembly(GO:0034551) |
| 0.1 | 1.1 | GO:0061709 | reticulophagy(GO:0061709) |
| 0.1 | 0.7 | GO:1904781 | positive regulation of protein localization to centrosome(GO:1904781) |
| 0.1 | 1.0 | GO:0098532 | histone H3-K27 trimethylation(GO:0098532) |
| 0.1 | 2.1 | GO:1901386 | negative regulation of voltage-gated calcium channel activity(GO:1901386) |
| 0.1 | 0.8 | GO:0043308 | eosinophil activation involved in immune response(GO:0002278) eosinophil mediated immunity(GO:0002447) eosinophil degranulation(GO:0043308) |
| 0.1 | 1.5 | GO:0071340 | skeletal muscle acetylcholine-gated channel clustering(GO:0071340) |
| 0.1 | 0.8 | GO:1903527 | positive regulation of membrane tubulation(GO:1903527) |
| 0.1 | 0.8 | GO:0050747 | positive regulation of lipoprotein metabolic process(GO:0050747) |
| 0.1 | 0.5 | GO:0045039 | protein import into mitochondrial inner membrane(GO:0045039) |
| 0.1 | 2.1 | GO:0015986 | energy coupled proton transport, down electrochemical gradient(GO:0015985) ATP synthesis coupled proton transport(GO:0015986) |
| 0.1 | 0.9 | GO:0010756 | positive regulation of plasminogen activation(GO:0010756) |
| 0.1 | 1.3 | GO:0006642 | triglyceride mobilization(GO:0006642) |
| 0.1 | 0.8 | GO:0015803 | branched-chain amino acid transport(GO:0015803) leucine transport(GO:0015820) |
| 0.1 | 1.5 | GO:0048312 | intracellular distribution of mitochondria(GO:0048312) |
| 0.1 | 5.0 | GO:0031111 | negative regulation of microtubule depolymerization(GO:0007026) negative regulation of microtubule polymerization or depolymerization(GO:0031111) |
| 0.1 | 4.6 | GO:0002026 | regulation of the force of heart contraction(GO:0002026) |
| 0.1 | 2.8 | GO:0050962 | detection of light stimulus involved in visual perception(GO:0050908) detection of light stimulus involved in sensory perception(GO:0050962) |
| 0.1 | 0.7 | GO:0035469 | determination of pancreatic left/right asymmetry(GO:0035469) |
| 0.1 | 5.2 | GO:1902476 | chloride transmembrane transport(GO:1902476) |
| 0.1 | 1.4 | GO:2000465 | regulation of glycogen (starch) synthase activity(GO:2000465) |
| 0.1 | 1.0 | GO:0035507 | regulation of myosin-light-chain-phosphatase activity(GO:0035507) |
| 0.1 | 0.8 | GO:0009249 | protein lipoylation(GO:0009249) |
| 0.1 | 7.2 | GO:0006805 | xenobiotic metabolic process(GO:0006805) |
| 0.1 | 1.9 | GO:1901897 | regulation of relaxation of cardiac muscle(GO:1901897) |
| 0.1 | 0.5 | GO:0034975 | protein folding in endoplasmic reticulum(GO:0034975) |
| 0.1 | 0.5 | GO:0014846 | esophagus smooth muscle contraction(GO:0014846) |
| 0.1 | 0.3 | GO:0006393 | termination of mitochondrial transcription(GO:0006393) |
| 0.1 | 0.5 | GO:0003383 | apical constriction(GO:0003383) |
| 0.1 | 0.6 | GO:0090074 | negative regulation of protein homodimerization activity(GO:0090074) |
| 0.1 | 1.3 | GO:0051601 | exocyst localization(GO:0051601) |
| 0.1 | 0.7 | GO:0006572 | tyrosine catabolic process(GO:0006572) |
| 0.1 | 2.1 | GO:0097237 | cellular response to toxic substance(GO:0097237) |
| 0.1 | 2.7 | GO:0016339 | calcium-dependent cell-cell adhesion via plasma membrane cell adhesion molecules(GO:0016339) |
| 0.1 | 1.6 | GO:0022904 | respiratory electron transport chain(GO:0022904) |
| 0.1 | 0.2 | GO:1902202 | regulation of hepatocyte growth factor receptor signaling pathway(GO:1902202) |
| 0.1 | 2.3 | GO:0097352 | autophagosome maturation(GO:0097352) |
| 0.1 | 0.4 | GO:0035063 | nuclear speck organization(GO:0035063) |
| 0.1 | 0.5 | GO:0046013 | regulation of T cell homeostatic proliferation(GO:0046013) |
| 0.1 | 0.3 | GO:2001287 | negative regulation of caveolin-mediated endocytosis(GO:2001287) |
| 0.1 | 1.6 | GO:0021796 | cerebral cortex regionalization(GO:0021796) |
| 0.1 | 0.9 | GO:0032780 | negative regulation of ATPase activity(GO:0032780) |
| 0.1 | 0.6 | GO:0030007 | cellular potassium ion homeostasis(GO:0030007) |
| 0.1 | 1.5 | GO:0000028 | ribosomal small subunit assembly(GO:0000028) |
| 0.1 | 0.1 | GO:0009629 | response to gravity(GO:0009629) |
| 0.1 | 2.7 | GO:0014003 | oligodendrocyte development(GO:0014003) |
| 0.1 | 1.5 | GO:1903861 | positive regulation of dendrite extension(GO:1903861) |
| 0.0 | 0.4 | GO:0032790 | ribosome disassembly(GO:0032790) |
| 0.0 | 1.1 | GO:0048875 | chemical homeostasis within a tissue(GO:0048875) |
| 0.0 | 1.0 | GO:2000060 | positive regulation of protein ubiquitination involved in ubiquitin-dependent protein catabolic process(GO:2000060) |
| 0.0 | 1.0 | GO:0048490 | anterograde synaptic vesicle transport(GO:0048490) synaptic vesicle cytoskeletal transport(GO:0099514) synaptic vesicle transport along microtubule(GO:0099517) |
| 0.0 | 1.6 | GO:0030262 | apoptotic nuclear changes(GO:0030262) |
| 0.0 | 2.0 | GO:0061098 | positive regulation of protein tyrosine kinase activity(GO:0061098) |
| 0.0 | 0.8 | GO:0007019 | microtubule depolymerization(GO:0007019) |
| 0.0 | 0.7 | GO:2000311 | regulation of alpha-amino-3-hydroxy-5-methyl-4-isoxazole propionate selective glutamate receptor activity(GO:2000311) |
| 0.0 | 3.9 | GO:0061726 | mitophagy(GO:0000422) mitochondrion disassembly(GO:0061726) |
| 0.0 | 1.4 | GO:0000266 | mitochondrial fission(GO:0000266) |
| 0.0 | 4.4 | GO:0051965 | positive regulation of synapse assembly(GO:0051965) |
| 0.0 | 1.0 | GO:0090201 | negative regulation of release of cytochrome c from mitochondria(GO:0090201) |
| 0.0 | 0.5 | GO:0006228 | UTP biosynthetic process(GO:0006228) |
| 0.0 | 4.6 | GO:0002028 | regulation of sodium ion transport(GO:0002028) |
| 0.0 | 5.5 | GO:0008277 | regulation of G-protein coupled receptor protein signaling pathway(GO:0008277) |
| 0.0 | 0.3 | GO:0031282 | regulation of guanylate cyclase activity(GO:0031282) |
| 0.0 | 5.0 | GO:0006941 | striated muscle contraction(GO:0006941) |
| 0.0 | 1.1 | GO:0040020 | regulation of meiotic nuclear division(GO:0040020) |
| 0.0 | 0.3 | GO:0021842 | directional guidance of interneurons involved in migration from the subpallium to the cortex(GO:0021840) chemorepulsion involved in interneuron migration from the subpallium to the cortex(GO:0021842) |
| 0.0 | 0.6 | GO:0043457 | regulation of cellular respiration(GO:0043457) |
| 0.0 | 0.3 | GO:0030641 | regulation of cellular pH(GO:0030641) |
| 0.0 | 0.4 | GO:0006388 | tRNA splicing, via endonucleolytic cleavage and ligation(GO:0006388) |
| 0.0 | 2.5 | GO:0055013 | cardiac muscle cell development(GO:0055013) |
| 0.0 | 0.3 | GO:0009437 | carnitine metabolic process(GO:0009437) |
| 0.0 | 0.1 | GO:2000360 | negative regulation of binding of sperm to zona pellucida(GO:2000360) |
| 0.0 | 0.6 | GO:0007190 | activation of adenylate cyclase activity(GO:0007190) |
| 0.0 | 0.3 | GO:0002087 | regulation of respiratory gaseous exchange by neurological system process(GO:0002087) |
| 0.0 | 0.2 | GO:1903142 | positive regulation of endothelial cell development(GO:1901552) positive regulation of establishment of endothelial barrier(GO:1903142) |
| 0.0 | 0.3 | GO:0042637 | catagen(GO:0042637) |
| 0.0 | 0.2 | GO:0006390 | transcription from mitochondrial promoter(GO:0006390) |
| 0.0 | 0.4 | GO:0000338 | protein deneddylation(GO:0000338) |
| 0.0 | 0.2 | GO:0061113 | pancreas morphogenesis(GO:0061113) |
| 0.0 | 8.2 | GO:0006887 | exocytosis(GO:0006887) |
| 0.0 | 0.7 | GO:0019432 | triglyceride biosynthetic process(GO:0019432) |
| 0.0 | 1.3 | GO:0045454 | cell redox homeostasis(GO:0045454) |
| 0.0 | 0.8 | GO:0006953 | acute-phase response(GO:0006953) |
| 0.0 | 0.5 | GO:0045187 | regulation of circadian sleep/wake cycle, sleep(GO:0045187) |
| 0.0 | 0.8 | GO:0006458 | 'de novo' protein folding(GO:0006458) |
| 0.0 | 0.8 | GO:0006821 | chloride transport(GO:0006821) |
| 0.0 | 0.8 | GO:0051489 | regulation of filopodium assembly(GO:0051489) |
| 0.0 | 0.7 | GO:0046627 | negative regulation of insulin receptor signaling pathway(GO:0046627) |
| 0.0 | 0.6 | GO:0034394 | protein localization to cell surface(GO:0034394) |
| 0.0 | 0.7 | GO:0015844 | monoamine transport(GO:0015844) |
| 0.0 | 0.2 | GO:0021520 | spinal cord motor neuron cell fate specification(GO:0021520) |
| 0.0 | 2.0 | GO:0006813 | potassium ion transport(GO:0006813) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.4 | 4.2 | GO:0098855 | HCN channel complex(GO:0098855) |
| 1.0 | 6.7 | GO:1990584 | cardiac Troponin complex(GO:1990584) |
| 0.8 | 3.4 | GO:0045283 | mitochondrial respiratory chain complex II, succinate dehydrogenase complex (ubiquinone)(GO:0005749) succinate dehydrogenase complex (ubiquinone)(GO:0045257) respiratory chain complex II(GO:0045273) succinate dehydrogenase complex(GO:0045281) fumarate reductase complex(GO:0045283) |
| 0.8 | 10.5 | GO:0097512 | cardiac myofibril(GO:0097512) |
| 0.8 | 4.6 | GO:0042583 | chromaffin granule(GO:0042583) |
| 0.7 | 3.7 | GO:0005945 | 6-phosphofructokinase complex(GO:0005945) |
| 0.7 | 5.5 | GO:0005927 | muscle tendon junction(GO:0005927) |
| 0.5 | 5.8 | GO:0005750 | mitochondrial respiratory chain complex III(GO:0005750) |
| 0.5 | 4.9 | GO:0045252 | oxoglutarate dehydrogenase complex(GO:0045252) |
| 0.4 | 8.5 | GO:0030061 | mitochondrial crista(GO:0030061) |
| 0.4 | 8.4 | GO:0032279 | asymmetric synapse(GO:0032279) |
| 0.4 | 1.8 | GO:0031314 | extrinsic component of mitochondrial inner membrane(GO:0031314) |
| 0.3 | 4.8 | GO:0001520 | outer dense fiber(GO:0001520) |
| 0.3 | 5.6 | GO:0005751 | mitochondrial respiratory chain complex IV(GO:0005751) |
| 0.3 | 2.1 | GO:0045254 | pyruvate dehydrogenase complex(GO:0045254) |
| 0.3 | 4.3 | GO:0008385 | IkappaB kinase complex(GO:0008385) |
| 0.3 | 1.7 | GO:0097452 | GAIT complex(GO:0097452) |
| 0.3 | 1.9 | GO:0097444 | spine apparatus(GO:0097444) |
| 0.3 | 1.0 | GO:1990769 | proximal neuron projection(GO:1990769) |
| 0.2 | 4.0 | GO:0000276 | mitochondrial proton-transporting ATP synthase complex, coupling factor F(o)(GO:0000276) |
| 0.2 | 3.4 | GO:0044300 | cerebellar mossy fiber(GO:0044300) |
| 0.2 | 2.8 | GO:0005742 | mitochondrial outer membrane translocase complex(GO:0005742) |
| 0.2 | 12.5 | GO:0005746 | mitochondrial respiratory chain(GO:0005746) |
| 0.2 | 1.5 | GO:0005744 | mitochondrial inner membrane presequence translocase complex(GO:0005744) |
| 0.2 | 3.2 | GO:0032009 | early phagosome(GO:0032009) |
| 0.2 | 4.6 | GO:0099634 | postsynaptic specialization membrane(GO:0099634) |
| 0.2 | 4.7 | GO:0044292 | dendrite terminus(GO:0044292) |
| 0.2 | 1.6 | GO:0072379 | BAT3 complex(GO:0071818) ER membrane insertion complex(GO:0072379) |
| 0.2 | 0.8 | GO:0070033 | synaptobrevin 2-SNAP-25-syntaxin-1a-complexin I complex(GO:0070032) synaptobrevin 2-SNAP-25-syntaxin-1a-complexin II complex(GO:0070033) |
| 0.2 | 6.5 | GO:0005790 | smooth endoplasmic reticulum(GO:0005790) |
| 0.2 | 0.5 | GO:0042721 | mitochondrial inner membrane protein insertion complex(GO:0042721) |
| 0.1 | 2.8 | GO:0042622 | photoreceptor outer segment membrane(GO:0042622) |
| 0.1 | 5.9 | GO:0031430 | M band(GO:0031430) |
| 0.1 | 3.8 | GO:0043196 | varicosity(GO:0043196) |
| 0.1 | 4.4 | GO:0031672 | A band(GO:0031672) |
| 0.1 | 0.5 | GO:0005745 | m-AAA complex(GO:0005745) |
| 0.1 | 1.1 | GO:0000275 | mitochondrial proton-transporting ATP synthase complex, catalytic core F(1)(GO:0000275) proton-transporting ATP synthase complex, catalytic core F(1)(GO:0045261) |
| 0.1 | 0.3 | GO:0071914 | prominosome(GO:0071914) |
| 0.1 | 0.9 | GO:0001652 | granular component(GO:0001652) |
| 0.1 | 0.9 | GO:0045239 | tricarboxylic acid cycle enzyme complex(GO:0045239) |
| 0.1 | 0.6 | GO:0098560 | cytoplasmic side of late endosome membrane(GO:0098560) |
| 0.1 | 4.3 | GO:0030140 | trans-Golgi network transport vesicle(GO:0030140) |
| 0.1 | 1.0 | GO:0070469 | respiratory chain(GO:0070469) |
| 0.1 | 1.7 | GO:0017146 | NMDA selective glutamate receptor complex(GO:0017146) |
| 0.1 | 20.4 | GO:0042579 | peroxisome(GO:0005777) microbody(GO:0042579) |
| 0.1 | 2.4 | GO:1902711 | GABA-A receptor complex(GO:1902711) |
| 0.1 | 9.1 | GO:0005811 | lipid particle(GO:0005811) |
| 0.1 | 6.5 | GO:0010494 | cytoplasmic stress granule(GO:0010494) |
| 0.1 | 2.5 | GO:0005763 | organellar small ribosomal subunit(GO:0000314) mitochondrial small ribosomal subunit(GO:0005763) |
| 0.1 | 3.6 | GO:0034707 | chloride channel complex(GO:0034707) |
| 0.1 | 0.7 | GO:0097451 | glial limiting end-foot(GO:0097451) |
| 0.1 | 1.5 | GO:0031307 | integral component of mitochondrial outer membrane(GO:0031307) |
| 0.1 | 0.5 | GO:0033269 | internode region of axon(GO:0033269) |
| 0.1 | 2.7 | GO:0030673 | axolemma(GO:0030673) |
| 0.1 | 7.0 | GO:0008076 | voltage-gated potassium channel complex(GO:0008076) |
| 0.1 | 1.3 | GO:0000145 | exocyst(GO:0000145) |
| 0.1 | 0.9 | GO:0005832 | chaperonin-containing T-complex(GO:0005832) |
| 0.0 | 0.3 | GO:0019907 | cyclin-dependent protein kinase activating kinase holoenzyme complex(GO:0019907) |
| 0.0 | 4.3 | GO:0019005 | SCF ubiquitin ligase complex(GO:0019005) |
| 0.0 | 0.1 | GO:0060473 | cortical granule(GO:0060473) |
| 0.0 | 1.1 | GO:0032426 | stereocilium tip(GO:0032426) |
| 0.0 | 1.0 | GO:0001518 | voltage-gated sodium channel complex(GO:0001518) |
| 0.0 | 0.6 | GO:0005916 | fascia adherens(GO:0005916) |
| 0.0 | 0.1 | GO:1990851 | Wnt-Frizzled-LRP5/6 complex(GO:1990851) |
| 0.0 | 1.0 | GO:0016581 | NuRD complex(GO:0016581) CHD-type complex(GO:0090545) |
| 0.0 | 0.4 | GO:0090533 | cation-transporting ATPase complex(GO:0090533) |
| 0.0 | 0.6 | GO:0005852 | eukaryotic translation initiation factor 3 complex(GO:0005852) |
| 0.0 | 3.4 | GO:0043679 | axon terminus(GO:0043679) |
| 0.0 | 0.4 | GO:0031232 | extrinsic component of external side of plasma membrane(GO:0031232) |
| 0.0 | 2.7 | GO:0000118 | histone deacetylase complex(GO:0000118) |
| 0.0 | 2.7 | GO:0043198 | dendritic shaft(GO:0043198) |
| 0.0 | 0.7 | GO:0034451 | centriolar satellite(GO:0034451) |
| 0.0 | 5.9 | GO:0008021 | synaptic vesicle(GO:0008021) |
| 0.0 | 8.5 | GO:0031965 | nuclear membrane(GO:0031965) |
| 0.0 | 0.6 | GO:0008074 | guanylate cyclase complex, soluble(GO:0008074) |
| 0.0 | 0.8 | GO:0002102 | podosome(GO:0002102) |
| 0.0 | 3.4 | GO:0005604 | basement membrane(GO:0005604) |
| 0.0 | 1.9 | GO:0016529 | sarcoplasmic reticulum(GO:0016529) |
| 0.0 | 2.5 | GO:0005901 | caveola(GO:0005901) |
| 0.0 | 0.8 | GO:0035371 | microtubule plus-end(GO:0035371) |
| 0.0 | 1.8 | GO:0001750 | photoreceptor outer segment(GO:0001750) |
| 0.0 | 4.0 | GO:0043209 | myelin sheath(GO:0043209) |
| 0.0 | 0.7 | GO:0097440 | apical dendrite(GO:0097440) |
| 0.0 | 2.2 | GO:0005884 | actin filament(GO:0005884) |
| 0.0 | 4.9 | GO:0030027 | lamellipodium(GO:0030027) |
| 0.0 | 1.1 | GO:0005801 | cis-Golgi network(GO:0005801) |
| 0.0 | 4.2 | GO:0016323 | basolateral plasma membrane(GO:0016323) |
| 0.0 | 7.5 | GO:0031012 | extracellular matrix(GO:0031012) |
| 0.0 | 6.5 | GO:0031966 | mitochondrial membrane(GO:0031966) |
| 0.0 | 0.8 | GO:0031526 | brush border membrane(GO:0031526) |
| 0.0 | 0.4 | GO:0030992 | intraciliary transport particle B(GO:0030992) |
| 0.0 | 0.2 | GO:0030867 | rough endoplasmic reticulum membrane(GO:0030867) |
| 0.0 | 0.4 | GO:0008180 | COP9 signalosome(GO:0008180) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.4 | 7.1 | GO:0080130 | L-phenylalanine:2-oxoglutarate aminotransferase activity(GO:0080130) |
| 2.1 | 8.5 | GO:0008386 | cholesterol monooxygenase (side-chain-cleaving) activity(GO:0008386) |
| 2.0 | 7.9 | GO:0015182 | L-asparagine transmembrane transporter activity(GO:0015182) |
| 1.3 | 3.8 | GO:0051538 | 3 iron, 4 sulfur cluster binding(GO:0051538) |
| 1.2 | 18.2 | GO:0031957 | very long-chain fatty acid-CoA ligase activity(GO:0031957) |
| 1.1 | 3.2 | GO:0052658 | inositol-1,4,5-trisphosphate 5-phosphatase activity(GO:0052658) |
| 1.0 | 4.9 | GO:0004449 | isocitrate dehydrogenase (NAD+) activity(GO:0004449) |
| 1.0 | 6.7 | GO:0030172 | troponin C binding(GO:0030172) |
| 0.9 | 7.7 | GO:0032795 | heterotrimeric G-protein binding(GO:0032795) |
| 0.8 | 7.6 | GO:0016286 | small conductance calcium-activated potassium channel activity(GO:0016286) |
| 0.8 | 18.1 | GO:0008510 | sodium:bicarbonate symporter activity(GO:0008510) |
| 0.8 | 3.3 | GO:0008113 | peptide-methionine (S)-S-oxide reductase activity(GO:0008113) |
| 0.8 | 1.6 | GO:0008177 | succinate dehydrogenase (ubiquinone) activity(GO:0008177) |
| 0.8 | 3.8 | GO:0043758 | acetate-CoA ligase (ADP-forming) activity(GO:0043758) |
| 0.7 | 3.7 | GO:0003872 | 6-phosphofructokinase activity(GO:0003872) |
| 0.7 | 2.1 | GO:0001847 | opsonin receptor activity(GO:0001847) |
| 0.7 | 4.2 | GO:0005222 | intracellular cAMP activated cation channel activity(GO:0005222) |
| 0.7 | 9.1 | GO:0035473 | lipase binding(GO:0035473) |
| 0.7 | 3.3 | GO:0034602 | oxoglutarate dehydrogenase (NAD+) activity(GO:0034602) |
| 0.6 | 2.5 | GO:0002153 | steroid receptor RNA activator RNA binding(GO:0002153) |
| 0.6 | 3.7 | GO:0004738 | pyruvate dehydrogenase activity(GO:0004738) pyruvate dehydrogenase [NAD(P)+] activity(GO:0034603) pyruvate dehydrogenase (NAD+) activity(GO:0034604) |
| 0.6 | 1.8 | GO:0099530 | PLC activating G-protein coupled glutamate receptor activity(GO:0001639) G-protein coupled receptor activity involved in regulation of postsynaptic membrane potential(GO:0099530) |
| 0.6 | 2.2 | GO:0004092 | carnitine O-acetyltransferase activity(GO:0004092) |
| 0.5 | 2.2 | GO:0034617 | tetrahydrobiopterin binding(GO:0034617) |
| 0.5 | 4.3 | GO:0052851 | cupric reductase activity(GO:0008823) ferric-chelate reductase (NADPH) activity(GO:0052851) |
| 0.5 | 1.5 | GO:0004962 | endothelin receptor activity(GO:0004962) |
| 0.5 | 8.5 | GO:0042301 | phosphate ion binding(GO:0042301) |
| 0.5 | 13.4 | GO:0015002 | cytochrome-c oxidase activity(GO:0004129) heme-copper terminal oxidase activity(GO:0015002) oxidoreductase activity, acting on a heme group of donors, oxygen as acceptor(GO:0016676) |
| 0.4 | 3.6 | GO:0016681 | ubiquinol-cytochrome-c reductase activity(GO:0008121) oxidoreductase activity, acting on diphenols and related substances as donors, cytochrome as acceptor(GO:0016681) |
| 0.4 | 1.3 | GO:0035939 | microsatellite binding(GO:0035939) |
| 0.4 | 4.4 | GO:1901612 | cardiolipin binding(GO:1901612) |
| 0.4 | 3.8 | GO:0008597 | calcium-dependent protein serine/threonine phosphatase regulator activity(GO:0008597) |
| 0.4 | 1.7 | GO:0019828 | aspartic-type endopeptidase inhibitor activity(GO:0019828) |
| 0.4 | 3.8 | GO:0005519 | cytoskeletal regulatory protein binding(GO:0005519) |
| 0.4 | 5.0 | GO:0004300 | enoyl-CoA hydratase activity(GO:0004300) |
| 0.4 | 2.4 | GO:0015501 | glutamate:sodium symporter activity(GO:0015501) |
| 0.4 | 2.8 | GO:0046912 | transferase activity, transferring acyl groups, acyl groups converted into alkyl on transfer(GO:0046912) |
| 0.4 | 2.3 | GO:0003828 | alpha-N-acetylneuraminate alpha-2,8-sialyltransferase activity(GO:0003828) |
| 0.4 | 1.5 | GO:0008506 | sucrose:proton symporter activity(GO:0008506) sucrose transmembrane transporter activity(GO:0008515) disaccharide transmembrane transporter activity(GO:0015154) oligosaccharide transmembrane transporter activity(GO:0015157) |
| 0.4 | 5.3 | GO:0008379 | thioredoxin peroxidase activity(GO:0008379) |
| 0.3 | 3.4 | GO:0003876 | AMP deaminase activity(GO:0003876) adenosine-phosphate deaminase activity(GO:0047623) |
| 0.3 | 7.8 | GO:0016805 | dipeptidase activity(GO:0016805) |
| 0.3 | 3.7 | GO:0015266 | protein channel activity(GO:0015266) |
| 0.3 | 3.3 | GO:0015651 | quaternary ammonium group transmembrane transporter activity(GO:0015651) |
| 0.3 | 1.1 | GO:0043532 | angiostatin binding(GO:0043532) |
| 0.3 | 2.2 | GO:0070061 | fructose binding(GO:0070061) |
| 0.3 | 13.2 | GO:0003785 | actin monomer binding(GO:0003785) |
| 0.3 | 3.4 | GO:0008199 | ferric iron binding(GO:0008199) |
| 0.3 | 1.6 | GO:0004994 | somatostatin receptor activity(GO:0004994) |
| 0.3 | 3.5 | GO:0004017 | adenylate kinase activity(GO:0004017) |
| 0.3 | 1.5 | GO:0004614 | phosphoglucomutase activity(GO:0004614) |
| 0.2 | 1.0 | GO:0061628 | H3K27me3 modified histone binding(GO:0061628) |
| 0.2 | 0.7 | GO:0016155 | formyltetrahydrofolate dehydrogenase activity(GO:0016155) |
| 0.2 | 4.0 | GO:0046933 | proton-transporting ATP synthase activity, rotational mechanism(GO:0046933) |
| 0.2 | 0.7 | GO:0008127 | 4-hydroxyphenylpyruvate dioxygenase activity(GO:0003868) quercetin 2,3-dioxygenase activity(GO:0008127) |
| 0.2 | 1.2 | GO:0030899 | calcium-dependent ATPase activity(GO:0030899) |
| 0.2 | 0.6 | GO:0017153 | sodium:dicarboxylate symporter activity(GO:0017153) |
| 0.2 | 4.7 | GO:0031681 | G-protein beta-subunit binding(GO:0031681) |
| 0.2 | 1.6 | GO:0005237 | inhibitory extracellular ligand-gated ion channel activity(GO:0005237) |
| 0.2 | 2.4 | GO:0016832 | aldehyde-lyase activity(GO:0016832) |
| 0.2 | 0.9 | GO:2001069 | glycogen binding(GO:2001069) |
| 0.2 | 6.6 | GO:0009055 | electron carrier activity(GO:0009055) |
| 0.2 | 0.5 | GO:0086062 | voltage-gated sodium channel activity involved in Purkinje myocyte action potential(GO:0086062) |
| 0.2 | 0.7 | GO:0004487 | methylenetetrahydrofolate dehydrogenase (NAD+) activity(GO:0004487) |
| 0.2 | 3.6 | GO:0005247 | voltage-gated chloride channel activity(GO:0005247) |
| 0.2 | 1.1 | GO:0030160 | GKAP/Homer scaffold activity(GO:0030160) |
| 0.1 | 0.4 | GO:0005137 | interleukin-5 receptor binding(GO:0005137) |
| 0.1 | 6.9 | GO:0016504 | peptidase activator activity(GO:0016504) |
| 0.1 | 1.5 | GO:0070181 | small ribosomal subunit rRNA binding(GO:0070181) |
| 0.1 | 2.8 | GO:0017128 | phospholipid scramblase activity(GO:0017128) |
| 0.1 | 1.7 | GO:0004972 | NMDA glutamate receptor activity(GO:0004972) |
| 0.1 | 7.1 | GO:0019212 | phosphatase inhibitor activity(GO:0019212) |
| 0.1 | 2.7 | GO:0032041 | histone deacetylase activity (H3-K14 specific)(GO:0031078) NAD-dependent histone deacetylase activity (H3-K14 specific)(GO:0032041) |
| 0.1 | 0.3 | GO:0016649 | electron-transferring-flavoprotein dehydrogenase activity(GO:0004174) oxidoreductase activity, acting on the CH-NH group of donors, quinone or similar compound as acceptor(GO:0016649) |
| 0.1 | 0.5 | GO:0016499 | orexin receptor activity(GO:0016499) |
| 0.1 | 0.7 | GO:0050833 | pyruvate transmembrane transporter activity(GO:0050833) |
| 0.1 | 0.3 | GO:0008048 | calcium sensitive guanylate cyclase activator activity(GO:0008048) |
| 0.1 | 4.2 | GO:0005104 | fibroblast growth factor receptor binding(GO:0005104) |
| 0.1 | 0.5 | GO:0086006 | voltage-gated sodium channel activity involved in cardiac muscle cell action potential(GO:0086006) |
| 0.1 | 0.7 | GO:0004467 | long-chain fatty acid-CoA ligase activity(GO:0004467) decanoate--CoA ligase activity(GO:0102391) |
| 0.1 | 0.3 | GO:1990698 | palmitoleoyltransferase activity(GO:1990698) |
| 0.1 | 0.6 | GO:0008294 | calcium- and calmodulin-responsive adenylate cyclase activity(GO:0008294) |
| 0.1 | 0.7 | GO:0004321 | acyl-CoA ligase activity(GO:0003996) fatty-acyl-CoA synthase activity(GO:0004321) |
| 0.1 | 0.7 | GO:0008504 | monoamine transmembrane transporter activity(GO:0008504) |
| 0.1 | 1.6 | GO:0050321 | tau-protein kinase activity(GO:0050321) |
| 0.1 | 1.9 | GO:0008195 | phosphatidate phosphatase activity(GO:0008195) |
| 0.1 | 0.5 | GO:0008449 | N-acetylglucosamine-6-sulfatase activity(GO:0008449) |
| 0.1 | 3.4 | GO:0001106 | RNA polymerase II transcription corepressor activity(GO:0001106) |
| 0.1 | 0.3 | GO:0030984 | kininogen binding(GO:0030984) |
| 0.1 | 0.2 | GO:0005171 | hepatocyte growth factor receptor binding(GO:0005171) |
| 0.1 | 4.0 | GO:0008395 | steroid hydroxylase activity(GO:0008395) |
| 0.1 | 1.1 | GO:0051537 | 2 iron, 2 sulfur cluster binding(GO:0051537) |
| 0.1 | 1.5 | GO:0050136 | NADH dehydrogenase (ubiquinone) activity(GO:0008137) NADH dehydrogenase (quinone) activity(GO:0050136) |
| 0.1 | 1.4 | GO:0005540 | hyaluronic acid binding(GO:0005540) |
| 0.1 | 7.0 | GO:0005249 | voltage-gated potassium channel activity(GO:0005249) |
| 0.0 | 0.5 | GO:0051525 | NFAT protein binding(GO:0051525) |
| 0.0 | 2.4 | GO:0016620 | oxidoreductase activity, acting on the aldehyde or oxo group of donors, NAD or NADP as acceptor(GO:0016620) |
| 0.0 | 1.5 | GO:0033130 | acetylcholine receptor binding(GO:0033130) |
| 0.0 | 1.6 | GO:0000062 | fatty-acyl-CoA binding(GO:0000062) |
| 0.0 | 1.2 | GO:0098641 | cadherin binding involved in cell-cell adhesion(GO:0098641) |
| 0.0 | 0.4 | GO:0015386 | potassium:proton antiporter activity(GO:0015386) |
| 0.0 | 0.1 | GO:0070002 | glutamic-type peptidase activity(GO:0070002) |
| 0.0 | 0.4 | GO:0043023 | ribosomal large subunit binding(GO:0043023) |
| 0.0 | 0.6 | GO:0045294 | alpha-catenin binding(GO:0045294) |
| 0.0 | 3.3 | GO:0005544 | calcium-dependent phospholipid binding(GO:0005544) |
| 0.0 | 0.7 | GO:0035925 | mRNA 3'-UTR AU-rich region binding(GO:0035925) |
| 0.0 | 1.8 | GO:0004115 | 3',5'-cyclic-AMP phosphodiesterase activity(GO:0004115) |
| 0.0 | 2.6 | GO:0005070 | SH3/SH2 adaptor activity(GO:0005070) |
| 0.0 | 1.9 | GO:0004683 | calmodulin-dependent protein kinase activity(GO:0004683) |
| 0.0 | 0.2 | GO:0004839 | ubiquitin activating enzyme activity(GO:0004839) |
| 0.0 | 2.2 | GO:0051019 | mitogen-activated protein kinase binding(GO:0051019) |
| 0.0 | 0.5 | GO:0004550 | nucleoside diphosphate kinase activity(GO:0004550) |
| 0.0 | 0.4 | GO:0016861 | intramolecular oxidoreductase activity, interconverting aldoses and ketoses(GO:0016861) |
| 0.0 | 2.1 | GO:0046875 | ephrin receptor binding(GO:0046875) |
| 0.0 | 1.2 | GO:0051059 | NF-kappaB binding(GO:0051059) |
| 0.0 | 0.4 | GO:0001162 | RNA polymerase II intronic transcription regulatory region sequence-specific DNA binding(GO:0001162) |
| 0.0 | 1.3 | GO:0017134 | fibroblast growth factor binding(GO:0017134) |
| 0.0 | 0.8 | GO:0017075 | syntaxin-1 binding(GO:0017075) |
| 0.0 | 0.6 | GO:0050811 | GABA receptor binding(GO:0050811) |
| 0.0 | 2.2 | GO:0008146 | sulfotransferase activity(GO:0008146) |
| 0.0 | 1.0 | GO:0004602 | glutathione peroxidase activity(GO:0004602) |
| 0.0 | 0.8 | GO:0004890 | GABA-A receptor activity(GO:0004890) |
| 0.0 | 0.9 | GO:0044183 | protein binding involved in protein folding(GO:0044183) |
| 0.0 | 0.6 | GO:0017081 | chloride channel regulator activity(GO:0017081) |
| 0.0 | 0.3 | GO:0005176 | ErbB-2 class receptor binding(GO:0005176) |
| 0.0 | 0.3 | GO:0017154 | semaphorin receptor activity(GO:0017154) |
| 0.0 | 0.9 | GO:0048020 | CCR chemokine receptor binding(GO:0048020) |
| 0.0 | 1.5 | GO:0017080 | sodium channel regulator activity(GO:0017080) |
| 0.0 | 0.6 | GO:0005328 | neurotransmitter:sodium symporter activity(GO:0005328) |
| 0.0 | 3.1 | GO:0051117 | ATPase binding(GO:0051117) |
| 0.0 | 0.7 | GO:0030742 | GTP-dependent protein binding(GO:0030742) |
| 0.0 | 1.4 | GO:0001205 | transcriptional activator activity, RNA polymerase II distal enhancer sequence-specific binding(GO:0001205) |
| 0.0 | 1.6 | GO:0005507 | copper ion binding(GO:0005507) |
| 0.0 | 0.6 | GO:0005212 | structural constituent of eye lens(GO:0005212) |
| 0.0 | 2.0 | GO:0008201 | heparin binding(GO:0008201) |
| 0.0 | 0.4 | GO:0004181 | metallocarboxypeptidase activity(GO:0004181) |
| 0.0 | 0.1 | GO:0004359 | glutaminase activity(GO:0004359) |
| 0.0 | 0.4 | GO:0071889 | 14-3-3 protein binding(GO:0071889) |
| 0.0 | 0.1 | GO:0044547 | DNA topoisomerase binding(GO:0044547) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 8.5 | PID THROMBIN PAR4 PATHWAY | PAR4-mediated thrombin signaling events |
| 0.2 | 5.9 | PID HDAC CLASSIII PATHWAY | Signaling events mediated by HDAC Class III |
| 0.1 | 3.8 | PID LIS1 PATHWAY | Lissencephaly gene (LIS1) in neuronal migration and development |
| 0.1 | 3.7 | PID HIF1 TFPATHWAY | HIF-1-alpha transcription factor network |
| 0.1 | 3.8 | PID TCR CALCIUM PATHWAY | Calcium signaling in the CD4+ TCR pathway |
| 0.1 | 2.2 | PID FRA PATHWAY | Validated transcriptional targets of AP1 family members Fra1 and Fra2 |
| 0.1 | 1.4 | PID SYNDECAN 3 PATHWAY | Syndecan-3-mediated signaling events |
| 0.1 | 2.1 | PID EPHB FWD PATHWAY | EPHB forward signaling |
| 0.1 | 3.4 | PID HNF3A PATHWAY | FOXA1 transcription factor network |
| 0.1 | 3.0 | PID RXR VDR PATHWAY | RXR and RAR heterodimerization with other nuclear receptor |
| 0.1 | 0.7 | ST INTERFERON GAMMA PATHWAY | Interferon gamma pathway. |
| 0.0 | 2.6 | PID ENDOTHELIN PATHWAY | Endothelins |
| 0.0 | 1.9 | PID NCADHERIN PATHWAY | N-cadherin signaling events |
| 0.0 | 2.9 | PID ANGIOPOIETIN RECEPTOR PATHWAY | Angiopoietin receptor Tie2-mediated signaling |
| 0.0 | 0.4 | PID IL5 PATHWAY | IL5-mediated signaling events |
| 0.0 | 0.4 | PID ALK2 PATHWAY | ALK2 signaling events |
| 0.0 | 1.4 | PID FAK PATHWAY | Signaling events mediated by focal adhesion kinase |
| 0.0 | 6.7 | NABA ECM GLYCOPROTEINS | Genes encoding structural ECM glycoproteins |
| 0.0 | 0.8 | PID NETRIN PATHWAY | Netrin-mediated signaling events |
| 0.0 | 1.9 | PID FGF PATHWAY | FGF signaling pathway |
| 0.0 | 1.2 | SIG INSULIN RECEPTOR PATHWAY IN CARDIAC MYOCYTES | Genes related to the insulin receptor pathway |
| 0.0 | 0.8 | PID INSULIN GLUCOSE PATHWAY | Insulin-mediated glucose transport |
| 0.0 | 1.4 | PID CERAMIDE PATHWAY | Ceramide signaling pathway |
| 0.0 | 1.5 | PID LKB1 PATHWAY | LKB1 signaling events |
| 0.0 | 1.1 | PID AP1 PATHWAY | AP-1 transcription factor network |
| 0.0 | 0.9 | ST JNK MAPK PATHWAY | JNK MAPK Pathway |
| 0.0 | 0.6 | PID RHODOPSIN PATHWAY | Visual signal transduction: Rods |
| 0.0 | 3.4 | NABA ECM AFFILIATED | Genes encoding proteins affiliated structurally or functionally to extracellular matrix proteins |
| 0.0 | 1.2 | PID TRKR PATHWAY | Neurotrophic factor-mediated Trk receptor signaling |
| 0.0 | 0.3 | PID ERBB NETWORK PATHWAY | ErbB receptor signaling network |
| 0.0 | 0.4 | PID HIF2PATHWAY | HIF-2-alpha transcription factor network |
| 0.0 | 3.7 | NABA SECRETED FACTORS | Genes encoding secreted soluble factors |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.6 | 11.7 | REACTOME ENDOGENOUS STEROLS | Genes involved in Endogenous sterols |
| 0.6 | 15.2 | REACTOME CITRIC ACID CYCLE TCA CYCLE | Genes involved in Citric acid cycle (TCA cycle) |
| 0.4 | 7.1 | REACTOME METABOLISM OF POLYAMINES | Genes involved in Metabolism of polyamines |
| 0.3 | 22.5 | REACTOME RESPIRATORY ELECTRON TRANSPORT | Genes involved in Respiratory electron transport |
| 0.3 | 19.9 | REACTOME STRIATED MUSCLE CONTRACTION | Genes involved in Striated Muscle Contraction |
| 0.3 | 7.7 | REACTOME NUCLEOTIDE LIKE PURINERGIC RECEPTORS | Genes involved in Nucleotide-like (purinergic) receptors |
| 0.2 | 3.4 | REACTOME CYTOSOLIC SULFONATION OF SMALL MOLECULES | Genes involved in Cytosolic sulfonation of small molecules |
| 0.2 | 5.0 | REACTOME BRANCHED CHAIN AMINO ACID CATABOLISM | Genes involved in Branched-chain amino acid catabolism |
| 0.2 | 3.3 | REACTOME ORGANIC CATION ANION ZWITTERION TRANSPORT | Genes involved in Organic cation/anion/zwitterion transport |
| 0.2 | 4.3 | REACTOME FORMATION OF ATP BY CHEMIOSMOTIC COUPLING | Genes involved in Formation of ATP by chemiosmotic coupling |
| 0.2 | 28.9 | REACTOME TRANSPORT OF INORGANIC CATIONS ANIONS AND AMINO ACIDS OLIGOPEPTIDES | Genes involved in Transport of inorganic cations/anions and amino acids/oligopeptides |
| 0.2 | 2.1 | REACTOME REGULATION OF PYRUVATE DEHYDROGENASE PDH COMPLEX | Genes involved in Regulation of pyruvate dehydrogenase (PDH) complex |
| 0.2 | 7.6 | REACTOME GLYCOLYSIS | Genes involved in Glycolysis |
| 0.2 | 3.8 | REACTOME ALPHA LINOLENIC ACID ALA METABOLISM | Genes involved in alpha-linolenic acid (ALA) metabolism |
| 0.1 | 3.3 | REACTOME GABA SYNTHESIS RELEASE REUPTAKE AND DEGRADATION | Genes involved in GABA synthesis, release, reuptake and degradation |
| 0.1 | 3.4 | REACTOME PURINE SALVAGE | Genes involved in Purine salvage |
| 0.1 | 3.4 | REACTOME RNA POL III TRANSCRIPTION TERMINATION | Genes involved in RNA Polymerase III Transcription Termination |
| 0.1 | 2.4 | REACTOME FGFR4 LIGAND BINDING AND ACTIVATION | Genes involved in FGFR4 ligand binding and activation |
| 0.1 | 1.8 | REACTOME CLASS C 3 METABOTROPIC GLUTAMATE PHEROMONE RECEPTORS | Genes involved in Class C/3 (Metabotropic glutamate/pheromone receptors) |
| 0.1 | 7.0 | REACTOME VOLTAGE GATED POTASSIUM CHANNELS | Genes involved in Voltage gated Potassium channels |
| 0.1 | 3.5 | REACTOME SYNTHESIS AND INTERCONVERSION OF NUCLEOTIDE DI AND TRIPHOSPHATES | Genes involved in Synthesis and interconversion of nucleotide di- and triphosphates |
| 0.1 | 7.4 | REACTOME COLLAGEN FORMATION | Genes involved in Collagen formation |
| 0.1 | 2.4 | REACTOME GABA A RECEPTOR ACTIVATION | Genes involved in GABA A receptor activation |
| 0.1 | 2.2 | REACTOME TETRAHYDROBIOPTERIN BH4 SYNTHESIS RECYCLING SALVAGE AND REGULATION | Genes involved in Tetrahydrobiopterin (BH4) synthesis, recycling, salvage and regulation |
| 0.1 | 10.1 | REACTOME POTASSIUM CHANNELS | Genes involved in Potassium Channels |
| 0.1 | 1.9 | REACTOME SYNTHESIS OF PE | Genes involved in Synthesis of PE |
| 0.1 | 1.4 | REACTOME CS DS DEGRADATION | Genes involved in CS/DS degradation |
| 0.1 | 1.5 | REACTOME GLYCOGEN BREAKDOWN GLYCOGENOLYSIS | Genes involved in Glycogen breakdown (glycogenolysis) |
| 0.1 | 2.2 | REACTOME PEROXISOMAL LIPID METABOLISM | Genes involved in Peroxisomal lipid metabolism |
| 0.1 | 2.6 | REACTOME TIE2 SIGNALING | Genes involved in Tie2 Signaling |
| 0.1 | 1.8 | REACTOME INTRINSIC PATHWAY | Genes involved in Intrinsic Pathway |
| 0.1 | 0.9 | REACTOME CHEMOKINE RECEPTORS BIND CHEMOKINES | Genes involved in Chemokine receptors bind chemokines |
| 0.1 | 3.2 | REACTOME SYNTHESIS OF PIPS AT THE PLASMA MEMBRANE | Genes involved in Synthesis of PIPs at the plasma membrane |
| 0.1 | 3.4 | REACTOME MITOCHONDRIAL PROTEIN IMPORT | Genes involved in Mitochondrial Protein Import |
| 0.1 | 1.2 | REACTOME PLATELET CALCIUM HOMEOSTASIS | Genes involved in Platelet calcium homeostasis |
| 0.1 | 1.7 | REACTOME RAS ACTIVATION UOPN CA2 INFUX THROUGH NMDA RECEPTOR | Genes involved in Ras activation uopn Ca2+ infux through NMDA receptor |
| 0.1 | 1.2 | REACTOME TGF BETA RECEPTOR SIGNALING IN EMT EPITHELIAL TO MESENCHYMAL TRANSITION | Genes involved in TGF-beta receptor signaling in EMT (epithelial to mesenchymal transition) |
| 0.0 | 0.7 | REACTOME HYALURONAN METABOLISM | Genes involved in Hyaluronan metabolism |
| 0.0 | 4.0 | REACTOME NOTCH1 INTRACELLULAR DOMAIN REGULATES TRANSCRIPTION | Genes involved in NOTCH1 Intracellular Domain Regulates Transcription |
| 0.0 | 0.8 | REACTOME SYNTHESIS OF PC | Genes involved in Synthesis of PC |
| 0.0 | 0.6 | REACTOME ADENYLATE CYCLASE ACTIVATING PATHWAY | Genes involved in Adenylate cyclase activating pathway |
| 0.0 | 0.7 | REACTOME SYNTHESIS OF VERY LONG CHAIN FATTY ACYL COAS | Genes involved in Synthesis of very long-chain fatty acyl-CoAs |
| 0.0 | 1.5 | REACTOME ION TRANSPORT BY P TYPE ATPASES | Genes involved in Ion transport by P-type ATPases |
| 0.0 | 0.5 | REACTOME MITOCHONDRIAL FATTY ACID BETA OXIDATION | Genes involved in Mitochondrial Fatty Acid Beta-Oxidation |
| 0.0 | 0.8 | REACTOME RORA ACTIVATES CIRCADIAN EXPRESSION | Genes involved in RORA Activates Circadian Expression |
| 0.0 | 0.9 | REACTOME ADHERENS JUNCTIONS INTERACTIONS | Genes involved in Adherens junctions interactions |
| 0.0 | 0.3 | REACTOME CDC6 ASSOCIATION WITH THE ORC ORIGIN COMPLEX | Genes involved in CDC6 association with the ORC:origin complex |
| 0.0 | 4.2 | REACTOME G ALPHA I SIGNALLING EVENTS | Genes involved in G alpha (i) signalling events |
| 0.0 | 1.7 | REACTOME PHASE II CONJUGATION | Genes involved in Phase II conjugation |
| 0.0 | 1.0 | REACTOME INTERACTION BETWEEN L1 AND ANKYRINS | Genes involved in Interaction between L1 and Ankyrins |
| 0.0 | 0.4 | REACTOME POST CHAPERONIN TUBULIN FOLDING PATHWAY | Genes involved in Post-chaperonin tubulin folding pathway |
| 0.0 | 1.3 | REACTOME PEPTIDE LIGAND BINDING RECEPTORS | Genes involved in Peptide ligand-binding receptors |
| 0.0 | 3.3 | REACTOME METABOLISM OF AMINO ACIDS AND DERIVATIVES | Genes involved in Metabolism of amino acids and derivatives |
| 0.0 | 1.0 | REACTOME NETRIN1 SIGNALING | Genes involved in Netrin-1 signaling |
| 0.0 | 1.4 | REACTOME NUCLEAR RECEPTOR TRANSCRIPTION PATHWAY | Genes involved in Nuclear Receptor transcription pathway |
| 0.0 | 0.7 | REACTOME TRANSPORT OF VITAMINS NUCLEOSIDES AND RELATED MOLECULES | Genes involved in Transport of vitamins, nucleosides, and related molecules |
| 0.0 | 0.6 | REACTOME ASSOCIATION OF TRIC CCT WITH TARGET PROTEINS DURING BIOSYNTHESIS | Genes involved in Association of TriC/CCT with target proteins during biosynthesis |
| 0.0 | 0.3 | REACTOME DOWNREGULATION OF ERBB2 ERBB3 SIGNALING | Genes involved in Downregulation of ERBB2:ERBB3 signaling |
| 0.0 | 0.4 | REACTOME IL RECEPTOR SHC SIGNALING | Genes involved in Interleukin receptor SHC signaling |