Project
PRJNA375882: Comprehensive Mouse Transcriptomic BodyMap
Navigation
Downloads
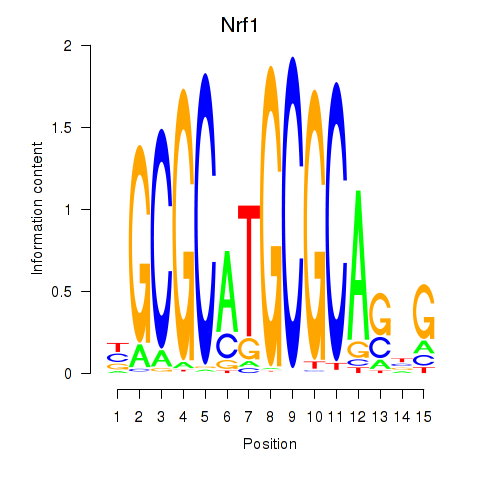
Results for Nrf1
Z-value: 3.80
Transcription factors associated with Nrf1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Nrf1
|
ENSMUSG00000058440.15 | Nrf1 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Nrf1 | mm39_v1_chr6_+_30048061_30048150 | 0.74 | 1.3e-13 | Click! |
Activity profile of Nrf1 motif
Sorted Z-values of Nrf1 motif
Network of associatons between targets according to the STRING database.
First level regulatory network of Nrf1
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 5.9 | 17.7 | GO:1990918 | double-strand break repair involved in meiotic recombination(GO:1990918) |
| 4.6 | 13.7 | GO:1902630 | regulation of membrane hyperpolarization(GO:1902630) |
| 4.1 | 12.4 | GO:0008358 | oocyte construction(GO:0007308) oocyte axis specification(GO:0007309) oocyte anterior/posterior axis specification(GO:0007314) pole plasm assembly(GO:0007315) maternal determination of anterior/posterior axis, embryo(GO:0008358) P granule organization(GO:0030719) |
| 3.9 | 11.6 | GO:0072429 | response to intra-S DNA damage checkpoint signaling(GO:0072429) positive regulation of t-circle formation(GO:1904431) |
| 3.4 | 17.1 | GO:0000727 | double-strand break repair via break-induced replication(GO:0000727) |
| 3.4 | 10.3 | GO:2000769 | regulation of establishment or maintenance of cell polarity regulating cell shape(GO:2000769) positive regulation of establishment or maintenance of cell polarity regulating cell shape(GO:2000771) |
| 3.1 | 24.8 | GO:0046600 | negative regulation of centriole replication(GO:0046600) |
| 3.0 | 12.0 | GO:0051316 | attachment of spindle microtubules to kinetochore involved in meiotic chromosome segregation(GO:0051316) |
| 2.8 | 25.5 | GO:1904781 | positive regulation of protein localization to centrosome(GO:1904781) |
| 2.8 | 2.8 | GO:0071163 | DNA replication preinitiation complex assembly(GO:0071163) |
| 2.4 | 4.9 | GO:1904779 | regulation of protein localization to centrosome(GO:1904779) |
| 2.4 | 7.1 | GO:0051436 | negative regulation of ubiquitin-protein ligase activity involved in mitotic cell cycle(GO:0051436) regulation of ubiquitin-protein ligase activity involved in mitotic cell cycle(GO:0051439) |
| 2.1 | 14.8 | GO:0045583 | regulation of cytotoxic T cell differentiation(GO:0045583) positive regulation of cytotoxic T cell differentiation(GO:0045585) |
| 2.0 | 8.1 | GO:2000312 | regulation of kainate selective glutamate receptor activity(GO:2000312) |
| 2.0 | 13.7 | GO:0006290 | pyrimidine dimer repair(GO:0006290) |
| 2.0 | 5.9 | GO:1904172 | positive regulation of bleb assembly(GO:1904172) |
| 1.9 | 9.6 | GO:1904975 | response to bleomycin(GO:1904975) cellular response to bleomycin(GO:1904976) |
| 1.9 | 17.1 | GO:0090267 | positive regulation of mitotic cell cycle spindle assembly checkpoint(GO:0090267) |
| 1.7 | 7.0 | GO:0048388 | endosomal lumen acidification(GO:0048388) |
| 1.7 | 22.7 | GO:0000012 | single strand break repair(GO:0000012) |
| 1.7 | 6.9 | GO:0090309 | positive regulation of methylation-dependent chromatin silencing(GO:0090309) |
| 1.7 | 5.2 | GO:1900368 | regulation of RNA interference(GO:1900368) |
| 1.7 | 10.1 | GO:1904751 | positive regulation of protein localization to nucleolus(GO:1904751) |
| 1.5 | 16.5 | GO:1903608 | protein localization to cytoplasmic stress granule(GO:1903608) |
| 1.5 | 7.3 | GO:0034164 | negative regulation of toll-like receptor 9 signaling pathway(GO:0034164) |
| 1.4 | 15.4 | GO:2000051 | negative regulation of non-canonical Wnt signaling pathway(GO:2000051) |
| 1.4 | 4.2 | GO:0006530 | asparagine catabolic process(GO:0006530) |
| 1.4 | 1.4 | GO:0016078 | tRNA catabolic process(GO:0016078) |
| 1.4 | 5.5 | GO:0010424 | DNA methylation on cytosine within a CG sequence(GO:0010424) |
| 1.4 | 4.2 | GO:0042822 | pyridoxal phosphate metabolic process(GO:0042822) |
| 1.4 | 9.7 | GO:0010032 | meiotic chromosome condensation(GO:0010032) |
| 1.3 | 1.3 | GO:0021508 | floor plate formation(GO:0021508) |
| 1.3 | 5.2 | GO:0071544 | diphosphoinositol polyphosphate catabolic process(GO:0071544) |
| 1.3 | 6.5 | GO:0035470 | positive regulation of vascular wound healing(GO:0035470) |
| 1.3 | 1.3 | GO:1902990 | mitotic telomere maintenance via semi-conservative replication(GO:1902990) |
| 1.3 | 24.4 | GO:0010603 | regulation of cytoplasmic mRNA processing body assembly(GO:0010603) |
| 1.3 | 3.8 | GO:0034476 | U1 snRNA 3'-end processing(GO:0034473) U5 snRNA 3'-end processing(GO:0034476) |
| 1.2 | 2.4 | GO:0000715 | nucleotide-excision repair, DNA damage recognition(GO:0000715) |
| 1.2 | 19.1 | GO:0019985 | translesion synthesis(GO:0019985) |
| 1.2 | 3.6 | GO:0070844 | misfolded protein transport(GO:0070843) polyubiquitinated protein transport(GO:0070844) polyubiquitinated misfolded protein transport(GO:0070845) Hsp90 deacetylation(GO:0070846) |
| 1.2 | 3.6 | GO:0007181 | transforming growth factor beta receptor complex assembly(GO:0007181) |
| 1.2 | 3.5 | GO:0019389 | glucuronoside metabolic process(GO:0019389) |
| 1.2 | 3.5 | GO:0051892 | negative regulation of cardioblast differentiation(GO:0051892) |
| 1.2 | 3.5 | GO:1900195 | positive regulation of oocyte maturation(GO:1900195) |
| 1.1 | 3.4 | GO:0061511 | centriole elongation(GO:0061511) |
| 1.1 | 4.5 | GO:0060382 | regulation of DNA strand elongation(GO:0060382) |
| 1.1 | 5.6 | GO:0046601 | positive regulation of centriole replication(GO:0046601) |
| 1.1 | 2.2 | GO:1904742 | regulation of telomeric DNA binding(GO:1904742) |
| 1.1 | 3.3 | GO:1904924 | negative regulation of mitophagy in response to mitochondrial depolarization(GO:1904924) |
| 1.1 | 7.6 | GO:0006933 | negative regulation of cell adhesion involved in substrate-bound cell migration(GO:0006933) |
| 1.1 | 3.3 | GO:0009629 | response to gravity(GO:0009629) |
| 1.1 | 16.2 | GO:0036297 | interstrand cross-link repair(GO:0036297) |
| 1.1 | 2.2 | GO:0060623 | regulation of chromosome condensation(GO:0060623) |
| 1.1 | 3.2 | GO:0036509 | trimming of terminal mannose on B branch(GO:0036509) |
| 1.1 | 2.1 | GO:0061055 | myotome development(GO:0061055) |
| 1.0 | 3.1 | GO:2000670 | positive regulation of dendritic cell apoptotic process(GO:2000670) |
| 1.0 | 5.2 | GO:0035609 | C-terminal protein deglutamylation(GO:0035609) |
| 1.0 | 5.2 | GO:0007066 | female meiosis sister chromatid cohesion(GO:0007066) |
| 1.0 | 4.0 | GO:0070100 | negative regulation of chemokine-mediated signaling pathway(GO:0070100) |
| 1.0 | 5.0 | GO:0070829 | heterochromatin maintenance(GO:0070829) |
| 1.0 | 6.0 | GO:0070458 | detoxification of nitrogen compound(GO:0051410) cellular detoxification of nitrogen compound(GO:0070458) |
| 1.0 | 13.8 | GO:0035562 | negative regulation of chromatin binding(GO:0035562) |
| 1.0 | 7.8 | GO:0036506 | maintenance of unfolded protein(GO:0036506) maintenance of unfolded protein involved in ERAD pathway(GO:1904378) |
| 1.0 | 6.8 | GO:0030910 | olfactory placode formation(GO:0030910) olfactory placode development(GO:0071698) olfactory placode morphogenesis(GO:0071699) |
| 1.0 | 11.6 | GO:0000463 | maturation of LSU-rRNA from tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000463) |
| 1.0 | 6.7 | GO:0000722 | telomere maintenance via recombination(GO:0000722) |
| 1.0 | 5.8 | GO:0050916 | sensory perception of sweet taste(GO:0050916) |
| 1.0 | 4.8 | GO:0061086 | negative regulation of histone H3-K27 methylation(GO:0061086) |
| 1.0 | 8.6 | GO:1903333 | negative regulation of protein folding(GO:1903333) |
| 0.9 | 0.9 | GO:0072710 | response to hydroxyurea(GO:0072710) cellular response to hydroxyurea(GO:0072711) |
| 0.9 | 7.5 | GO:0070294 | renal sodium ion absorption(GO:0070294) |
| 0.9 | 3.6 | GO:0071169 | establishment of protein localization to chromatin(GO:0071169) |
| 0.9 | 2.7 | GO:0031848 | protection from non-homologous end joining at telomere(GO:0031848) |
| 0.9 | 3.5 | GO:0033615 | mitochondrial proton-transporting ATP synthase complex assembly(GO:0033615) |
| 0.9 | 3.5 | GO:0044837 | assembly of actomyosin apparatus involved in cytokinesis(GO:0000912) actomyosin contractile ring assembly(GO:0000915) actomyosin contractile ring organization(GO:0044837) |
| 0.9 | 9.6 | GO:0032226 | positive regulation of synaptic transmission, dopaminergic(GO:0032226) |
| 0.9 | 5.2 | GO:0006384 | transcription initiation from RNA polymerase III promoter(GO:0006384) |
| 0.9 | 2.6 | GO:2000299 | negative regulation of Rho-dependent protein serine/threonine kinase activity(GO:2000299) |
| 0.9 | 3.5 | GO:0043418 | homocysteine catabolic process(GO:0043418) |
| 0.9 | 2.6 | GO:0032241 | positive regulation of nucleobase-containing compound transport(GO:0032241) |
| 0.9 | 3.4 | GO:0051325 | interphase(GO:0051325) mitotic interphase(GO:0051329) |
| 0.8 | 2.5 | GO:0006500 | N-terminal protein palmitoylation(GO:0006500) |
| 0.8 | 2.5 | GO:0016131 | brassinosteroid metabolic process(GO:0016131) brassinosteroid biosynthetic process(GO:0016132) |
| 0.8 | 4.1 | GO:0030576 | Cajal body organization(GO:0030576) |
| 0.8 | 2.4 | GO:0070376 | regulation of ERK5 cascade(GO:0070376) |
| 0.8 | 2.4 | GO:0031125 | rRNA 3'-end processing(GO:0031125) |
| 0.8 | 7.9 | GO:0051596 | methylglyoxal catabolic process to D-lactate via S-lactoyl-glutathione(GO:0019243) methylglyoxal catabolic process(GO:0051596) methylglyoxal catabolic process to lactate(GO:0061727) |
| 0.8 | 2.4 | GO:0031554 | regulation of DNA-templated transcription, termination(GO:0031554) |
| 0.8 | 1.6 | GO:0009438 | methylglyoxal metabolic process(GO:0009438) |
| 0.8 | 6.3 | GO:0043928 | exonucleolytic nuclear-transcribed mRNA catabolic process involved in deadenylation-dependent decay(GO:0043928) |
| 0.8 | 2.3 | GO:0043988 | histone H3-S28 phosphorylation(GO:0043988) histone H2A phosphorylation(GO:1990164) |
| 0.8 | 3.1 | GO:0035425 | autocrine signaling(GO:0035425) |
| 0.8 | 6.9 | GO:2000234 | positive regulation of ribosome biogenesis(GO:0090070) positive regulation of rRNA processing(GO:2000234) |
| 0.8 | 19.0 | GO:0043968 | histone H2A acetylation(GO:0043968) |
| 0.8 | 5.3 | GO:2000144 | positive regulation of DNA-templated transcription, initiation(GO:2000144) |
| 0.7 | 4.5 | GO:0061641 | CENP-A containing nucleosome assembly(GO:0034080) CENP-A containing chromatin organization(GO:0061641) |
| 0.7 | 3.7 | GO:0007084 | mitotic nuclear envelope reassembly(GO:0007084) |
| 0.7 | 2.2 | GO:0000019 | regulation of mitotic recombination(GO:0000019) negative regulation of mitotic recombination(GO:0045950) |
| 0.7 | 8.1 | GO:0036123 | histone H3-K9 dimethylation(GO:0036123) |
| 0.7 | 2.2 | GO:1901074 | regulation of engulfment of apoptotic cell(GO:1901074) |
| 0.7 | 2.2 | GO:0098884 | postsynaptic neurotransmitter receptor internalization(GO:0098884) |
| 0.7 | 2.9 | GO:0072717 | cellular response to actinomycin D(GO:0072717) |
| 0.7 | 5.1 | GO:0007144 | female meiosis I(GO:0007144) |
| 0.7 | 13.6 | GO:0071578 | zinc II ion transmembrane import(GO:0071578) |
| 0.7 | 2.8 | GO:0046606 | negative regulation of centrosome duplication(GO:0010826) negative regulation of centrosome cycle(GO:0046606) |
| 0.7 | 2.1 | GO:1990180 | mitochondrial tRNA 3'-end processing(GO:1990180) |
| 0.7 | 8.5 | GO:0021785 | branchiomotor neuron axon guidance(GO:0021785) |
| 0.7 | 2.1 | GO:0097045 | activation of blood coagulation via clotting cascade(GO:0002543) phosphatidylserine exposure on blood platelet(GO:0097045) |
| 0.7 | 11.2 | GO:0034501 | protein localization to kinetochore(GO:0034501) |
| 0.7 | 3.4 | GO:0046125 | pyrimidine deoxyribonucleoside metabolic process(GO:0046125) |
| 0.7 | 7.5 | GO:0098789 | pre-mRNA cleavage required for polyadenylation(GO:0098789) |
| 0.7 | 10.9 | GO:0016180 | snRNA processing(GO:0016180) |
| 0.7 | 2.7 | GO:0046725 | negative regulation by virus of viral protein levels in host cell(GO:0046725) |
| 0.7 | 2.7 | GO:0000707 | meiotic DNA recombinase assembly(GO:0000707) |
| 0.7 | 2.7 | GO:0000973 | posttranscriptional tethering of RNA polymerase II gene DNA at nuclear periphery(GO:0000973) |
| 0.7 | 4.6 | GO:0018171 | peptidyl-cysteine oxidation(GO:0018171) |
| 0.6 | 1.9 | GO:0071459 | protein localization to chromosome, centromeric region(GO:0071459) |
| 0.6 | 1.3 | GO:0046726 | positive regulation by virus of viral protein levels in host cell(GO:0046726) |
| 0.6 | 2.6 | GO:0031660 | regulation of cyclin-dependent protein serine/threonine kinase activity involved in G2/M transition of mitotic cell cycle(GO:0031660) positive regulation of cyclin-dependent protein serine/threonine kinase activity involved in G2/M transition of mitotic cell cycle(GO:0031662) |
| 0.6 | 7.1 | GO:0000479 | endonucleolytic cleavage of tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000479) |
| 0.6 | 4.5 | GO:0021776 | smoothened signaling pathway involved in ventral spinal cord interneuron specification(GO:0021775) smoothened signaling pathway involved in spinal cord motor neuron cell fate specification(GO:0021776) |
| 0.6 | 1.9 | GO:0042138 | meiotic DNA double-strand break formation(GO:0042138) |
| 0.6 | 1.9 | GO:1900062 | regulation of replicative cell aging(GO:1900062) |
| 0.6 | 3.7 | GO:0046599 | regulation of centriole replication(GO:0046599) |
| 0.6 | 8.1 | GO:0000244 | spliceosomal tri-snRNP complex assembly(GO:0000244) |
| 0.6 | 2.4 | GO:1902268 | negative regulation of polyamine transmembrane transport(GO:1902268) |
| 0.6 | 2.4 | GO:0010457 | centriole-centriole cohesion(GO:0010457) |
| 0.6 | 2.9 | GO:0033364 | mast cell secretory granule organization(GO:0033364) |
| 0.6 | 5.3 | GO:0017196 | N-terminal peptidyl-methionine acetylation(GO:0017196) |
| 0.6 | 4.1 | GO:0042796 | snRNA transcription from RNA polymerase III promoter(GO:0042796) |
| 0.6 | 11.1 | GO:0051299 | centrosome separation(GO:0051299) |
| 0.6 | 7.6 | GO:0048304 | positive regulation of isotype switching to IgG isotypes(GO:0048304) |
| 0.6 | 4.0 | GO:1904100 | regulation of protein O-linked glycosylation(GO:1904098) positive regulation of protein O-linked glycosylation(GO:1904100) |
| 0.6 | 2.3 | GO:0090063 | positive regulation of microtubule nucleation(GO:0090063) |
| 0.6 | 1.7 | GO:1903347 | negative regulation of bicellular tight junction assembly(GO:1903347) |
| 0.6 | 7.3 | GO:0036158 | outer dynein arm assembly(GO:0036158) |
| 0.6 | 1.7 | GO:0002396 | MHC protein complex assembly(GO:0002396) peptide antigen assembly with MHC protein complex(GO:0002501) |
| 0.5 | 1.6 | GO:0070375 | ERK5 cascade(GO:0070375) |
| 0.5 | 6.0 | GO:1902166 | negative regulation of intrinsic apoptotic signaling pathway in response to DNA damage by p53 class mediator(GO:1902166) |
| 0.5 | 0.5 | GO:0072385 | minus-end-directed organelle transport along microtubule(GO:0072385) |
| 0.5 | 1.6 | GO:2000845 | positive regulation of testosterone secretion(GO:2000845) |
| 0.5 | 10.2 | GO:0034587 | piRNA metabolic process(GO:0034587) |
| 0.5 | 3.2 | GO:0097501 | stress response to metal ion(GO:0097501) |
| 0.5 | 1.6 | GO:0015825 | L-serine transport(GO:0015825) |
| 0.5 | 1.6 | GO:0030472 | mitotic spindle organization in nucleus(GO:0030472) |
| 0.5 | 1.0 | GO:2000825 | positive regulation of androgen receptor activity(GO:2000825) |
| 0.5 | 6.8 | GO:0018095 | protein polyglutamylation(GO:0018095) |
| 0.5 | 5.2 | GO:0045162 | clustering of voltage-gated sodium channels(GO:0045162) |
| 0.5 | 7.3 | GO:0098764 | meiotic prophase I(GO:0007128) prophase(GO:0051324) meiotic cell cycle phase(GO:0098762) meiosis I cell cycle phase(GO:0098764) |
| 0.5 | 6.7 | GO:0070131 | positive regulation of mitochondrial translation(GO:0070131) |
| 0.5 | 11.3 | GO:0035518 | histone H2A monoubiquitination(GO:0035518) |
| 0.5 | 19.0 | GO:0006270 | DNA replication initiation(GO:0006270) |
| 0.5 | 2.6 | GO:0046167 | glycerol-3-phosphate biosynthetic process(GO:0046167) |
| 0.5 | 5.6 | GO:0051574 | positive regulation of histone H3-K9 methylation(GO:0051574) |
| 0.5 | 1.5 | GO:0033566 | gamma-tubulin complex localization(GO:0033566) |
| 0.5 | 2.0 | GO:1903371 | regulation of endoplasmic reticulum tubular network organization(GO:1903371) |
| 0.5 | 2.0 | GO:1905150 | regulation of voltage-gated sodium channel activity(GO:1905150) |
| 0.5 | 3.9 | GO:0030263 | apoptotic chromosome condensation(GO:0030263) |
| 0.5 | 1.5 | GO:1902523 | positive regulation of protein K63-linked ubiquitination(GO:1902523) positive regulation of protein K48-linked ubiquitination(GO:1902524) positive regulation of protein monoubiquitination(GO:1902527) |
| 0.5 | 1.5 | GO:1902174 | positive regulation of keratinocyte apoptotic process(GO:1902174) |
| 0.5 | 1.9 | GO:1904354 | negative regulation of telomere capping(GO:1904354) |
| 0.5 | 5.3 | GO:0043553 | negative regulation of phosphatidylinositol 3-kinase activity(GO:0043553) |
| 0.5 | 1.4 | GO:2000847 | negative regulation of corticosteroid hormone secretion(GO:2000847) negative regulation of glucocorticoid secretion(GO:2000850) |
| 0.5 | 1.4 | GO:2000537 | regulation of cGMP-mediated signaling(GO:0010752) regulation of B cell chemotaxis(GO:2000537) positive regulation of B cell chemotaxis(GO:2000538) |
| 0.5 | 11.9 | GO:0010569 | regulation of double-strand break repair via homologous recombination(GO:0010569) |
| 0.5 | 6.7 | GO:1901673 | regulation of mitotic spindle assembly(GO:1901673) |
| 0.5 | 4.8 | GO:0034244 | negative regulation of transcription elongation from RNA polymerase II promoter(GO:0034244) |
| 0.5 | 2.4 | GO:0097309 | cap1 mRNA methylation(GO:0097309) |
| 0.5 | 4.2 | GO:0071787 | endoplasmic reticulum tubular network assembly(GO:0071787) |
| 0.5 | 2.3 | GO:0033563 | dorsal/ventral axon guidance(GO:0033563) |
| 0.5 | 2.3 | GO:0006481 | C-terminal protein methylation(GO:0006481) |
| 0.5 | 1.4 | GO:0006713 | glucocorticoid catabolic process(GO:0006713) |
| 0.5 | 9.5 | GO:0046597 | negative regulation of viral entry into host cell(GO:0046597) |
| 0.5 | 3.2 | GO:1903208 | neuron death in response to hydrogen peroxide(GO:0036476) regulation of hydrogen peroxide-induced neuron death(GO:1903207) negative regulation of hydrogen peroxide-induced neuron death(GO:1903208) |
| 0.5 | 1.4 | GO:0098501 | polynucleotide dephosphorylation(GO:0098501) |
| 0.4 | 1.3 | GO:0046035 | CMP salvage(GO:0006238) CMP biosynthetic process(GO:0009224) pyrimidine ribonucleotide salvage(GO:0010138) pyrimidine nucleotide salvage(GO:0032262) UMP salvage(GO:0044206) CMP metabolic process(GO:0046035) |
| 0.4 | 2.2 | GO:0035617 | stress granule disassembly(GO:0035617) |
| 0.4 | 6.1 | GO:0051823 | regulation of synapse structural plasticity(GO:0051823) |
| 0.4 | 0.9 | GO:0018312 | peptidyl-serine ADP-ribosylation(GO:0018312) |
| 0.4 | 0.4 | GO:0033127 | regulation of histone phosphorylation(GO:0033127) |
| 0.4 | 2.2 | GO:0090669 | telomerase RNA stabilization(GO:0090669) |
| 0.4 | 2.1 | GO:0014886 | transition between slow and fast fiber(GO:0014886) |
| 0.4 | 9.8 | GO:0034508 | centromere complex assembly(GO:0034508) |
| 0.4 | 3.4 | GO:0000712 | resolution of meiotic recombination intermediates(GO:0000712) |
| 0.4 | 3.3 | GO:0070345 | negative regulation of fat cell proliferation(GO:0070345) |
| 0.4 | 2.5 | GO:0032053 | ciliary basal body organization(GO:0032053) positive regulation of protein localization to cilium(GO:1903566) |
| 0.4 | 5.0 | GO:0050684 | regulation of mRNA processing(GO:0050684) |
| 0.4 | 2.5 | GO:0006438 | valyl-tRNA aminoacylation(GO:0006438) |
| 0.4 | 1.2 | GO:0016256 | N-glycan processing to lysosome(GO:0016256) |
| 0.4 | 2.5 | GO:0060339 | negative regulation of type I interferon-mediated signaling pathway(GO:0060339) |
| 0.4 | 1.2 | GO:0061357 | positive regulation of Wnt protein secretion(GO:0061357) |
| 0.4 | 2.0 | GO:0000972 | transcription-dependent tethering of RNA polymerase II gene DNA at nuclear periphery(GO:0000972) |
| 0.4 | 2.8 | GO:0051533 | positive regulation of NFAT protein import into nucleus(GO:0051533) |
| 0.4 | 1.6 | GO:0006244 | pyrimidine nucleotide catabolic process(GO:0006244) pyrimidine deoxyribonucleotide catabolic process(GO:0009223) |
| 0.4 | 2.0 | GO:0098886 | modification of dendritic spine(GO:0098886) |
| 0.4 | 2.0 | GO:1903422 | negative regulation of synaptic vesicle recycling(GO:1903422) regulation of spontaneous neurotransmitter secretion(GO:1904048) |
| 0.4 | 12.8 | GO:0007020 | microtubule nucleation(GO:0007020) |
| 0.4 | 6.0 | GO:0030261 | chromosome condensation(GO:0030261) |
| 0.4 | 8.3 | GO:0007064 | mitotic sister chromatid cohesion(GO:0007064) |
| 0.4 | 2.8 | GO:0098532 | histone H3-K27 trimethylation(GO:0098532) |
| 0.4 | 1.6 | GO:0034421 | post-translational protein acetylation(GO:0034421) |
| 0.4 | 1.2 | GO:0036515 | serotonergic neuron axon guidance(GO:0036515) |
| 0.4 | 10.6 | GO:1904353 | regulation of telomere capping(GO:1904353) |
| 0.4 | 1.6 | GO:0036116 | medium-chain fatty-acyl-CoA catabolic process(GO:0036114) long-chain fatty-acyl-CoA catabolic process(GO:0036116) palmitic acid metabolic process(GO:1900533) palmitic acid biosynthetic process(GO:1900535) |
| 0.4 | 1.9 | GO:1902172 | keratinocyte apoptotic process(GO:0097283) regulation of keratinocyte apoptotic process(GO:1902172) |
| 0.4 | 1.9 | GO:0051310 | metaphase plate congression(GO:0051310) |
| 0.4 | 1.5 | GO:2001245 | regulation of phosphatidylcholine biosynthetic process(GO:2001245) |
| 0.4 | 4.3 | GO:0051103 | DNA ligation(GO:0006266) DNA ligation involved in DNA repair(GO:0051103) |
| 0.4 | 1.2 | GO:0006344 | maintenance of chromatin silencing(GO:0006344) |
| 0.4 | 1.1 | GO:0030397 | membrane disassembly(GO:0030397) nuclear envelope disassembly(GO:0051081) |
| 0.4 | 2.3 | GO:0071484 | cellular response to light intensity(GO:0071484) cellular response to high light intensity(GO:0071486) retinal rod cell apoptotic process(GO:0097473) |
| 0.4 | 1.9 | GO:0031145 | anaphase-promoting complex-dependent catabolic process(GO:0031145) |
| 0.4 | 3.0 | GO:0006283 | transcription-coupled nucleotide-excision repair(GO:0006283) |
| 0.4 | 9.5 | GO:0016578 | histone deubiquitination(GO:0016578) |
| 0.4 | 3.7 | GO:0030579 | ubiquitin-dependent SMAD protein catabolic process(GO:0030579) |
| 0.4 | 1.5 | GO:0016080 | synaptic vesicle targeting(GO:0016080) |
| 0.4 | 1.9 | GO:0032849 | positive regulation of cellular pH reduction(GO:0032849) |
| 0.4 | 0.7 | GO:2000111 | positive regulation of macrophage apoptotic process(GO:2000111) |
| 0.4 | 1.1 | GO:0000350 | generation of catalytic spliceosome for second transesterification step(GO:0000350) spliceosomal conformational changes to generate catalytic conformation(GO:0000393) |
| 0.4 | 1.8 | GO:0008298 | intracellular mRNA localization(GO:0008298) |
| 0.4 | 1.8 | GO:0042271 | susceptibility to natural killer cell mediated cytotoxicity(GO:0042271) |
| 0.4 | 5.5 | GO:0035871 | protein K11-linked deubiquitination(GO:0035871) |
| 0.4 | 8.1 | GO:0016973 | poly(A)+ mRNA export from nucleus(GO:0016973) |
| 0.4 | 2.9 | GO:1900264 | regulation of DNA-directed DNA polymerase activity(GO:1900262) positive regulation of DNA-directed DNA polymerase activity(GO:1900264) |
| 0.4 | 9.4 | GO:0008608 | attachment of spindle microtubules to kinetochore(GO:0008608) |
| 0.4 | 1.8 | GO:0007023 | post-chaperonin tubulin folding pathway(GO:0007023) |
| 0.4 | 1.1 | GO:0032066 | nucleolus to nucleoplasm transport(GO:0032066) |
| 0.4 | 1.4 | GO:0051031 | tRNA transport(GO:0051031) |
| 0.4 | 2.2 | GO:0035864 | response to potassium ion(GO:0035864) cellular response to potassium ion(GO:0035865) |
| 0.4 | 3.6 | GO:1900364 | negative regulation of mRNA polyadenylation(GO:1900364) |
| 0.4 | 6.8 | GO:1902857 | positive regulation of nonmotile primary cilium assembly(GO:1902857) |
| 0.4 | 1.8 | GO:0010825 | positive regulation of centrosome duplication(GO:0010825) |
| 0.4 | 2.1 | GO:0006265 | DNA topological change(GO:0006265) |
| 0.4 | 3.2 | GO:0033210 | leptin-mediated signaling pathway(GO:0033210) |
| 0.3 | 3.8 | GO:0006013 | mannose metabolic process(GO:0006013) |
| 0.3 | 1.0 | GO:0030961 | peptidyl-arginine hydroxylation(GO:0030961) |
| 0.3 | 4.2 | GO:0018231 | peptidyl-L-cysteine S-palmitoylation(GO:0018230) peptidyl-S-diacylglycerol-L-cysteine biosynthetic process from peptidyl-cysteine(GO:0018231) |
| 0.3 | 1.4 | GO:0000414 | regulation of histone H3-K36 methylation(GO:0000414) |
| 0.3 | 1.4 | GO:0045897 | regulation of transcription during mitosis(GO:0045896) positive regulation of transcription during mitosis(GO:0045897) |
| 0.3 | 1.4 | GO:1903575 | cornified envelope assembly(GO:1903575) |
| 0.3 | 1.4 | GO:2000467 | positive regulation of glycogen (starch) synthase activity(GO:2000467) |
| 0.3 | 18.2 | GO:0042073 | intraciliary transport(GO:0042073) |
| 0.3 | 2.3 | GO:0051725 | protein de-ADP-ribosylation(GO:0051725) |
| 0.3 | 3.3 | GO:0031293 | membrane protein intracellular domain proteolysis(GO:0031293) |
| 0.3 | 3.0 | GO:0097112 | gamma-aminobutyric acid receptor clustering(GO:0097112) |
| 0.3 | 2.6 | GO:0016233 | telomere capping(GO:0016233) |
| 0.3 | 4.9 | GO:0000083 | regulation of transcription involved in G1/S transition of mitotic cell cycle(GO:0000083) |
| 0.3 | 13.4 | GO:0000184 | nuclear-transcribed mRNA catabolic process, nonsense-mediated decay(GO:0000184) |
| 0.3 | 1.3 | GO:0015827 | aromatic amino acid transport(GO:0015801) tryptophan transport(GO:0015827) |
| 0.3 | 1.6 | GO:0043144 | snoRNA processing(GO:0043144) |
| 0.3 | 4.2 | GO:0070544 | histone H3-K36 demethylation(GO:0070544) |
| 0.3 | 0.6 | GO:0010387 | COP9 signalosome assembly(GO:0010387) |
| 0.3 | 3.5 | GO:0006610 | ribosomal protein import into nucleus(GO:0006610) |
| 0.3 | 2.9 | GO:0046784 | viral mRNA export from host cell nucleus(GO:0046784) |
| 0.3 | 0.6 | GO:0018364 | peptidyl-glutamine methylation(GO:0018364) |
| 0.3 | 4.8 | GO:0070203 | establishment of protein localization to chromosome(GO:0070199) establishment of protein localization to telomere(GO:0070200) regulation of establishment of protein localization to chromosome(GO:0070202) regulation of establishment of protein localization to telomere(GO:0070203) positive regulation of establishment of protein localization to telomere(GO:1904851) |
| 0.3 | 3.8 | GO:2000288 | positive regulation of myoblast proliferation(GO:2000288) |
| 0.3 | 5.0 | GO:0035970 | peptidyl-threonine dephosphorylation(GO:0035970) |
| 0.3 | 1.9 | GO:0048096 | chromatin-mediated maintenance of transcription(GO:0048096) |
| 0.3 | 9.4 | GO:0046827 | positive regulation of protein export from nucleus(GO:0046827) |
| 0.3 | 0.9 | GO:0000451 | rRNA 2'-O-methylation(GO:0000451) |
| 0.3 | 1.6 | GO:2000327 | regulation of ligand-dependent nuclear receptor transcription coactivator activity(GO:2000325) positive regulation of ligand-dependent nuclear receptor transcription coactivator activity(GO:2000327) |
| 0.3 | 9.3 | GO:0043631 | mRNA polyadenylation(GO:0006378) RNA polyadenylation(GO:0043631) |
| 0.3 | 1.2 | GO:2000016 | negative regulation of determination of dorsal identity(GO:2000016) |
| 0.3 | 3.4 | GO:1902036 | regulation of hematopoietic stem cell differentiation(GO:1902036) |
| 0.3 | 2.5 | GO:0010792 | DNA double-strand break processing involved in repair via single-strand annealing(GO:0010792) |
| 0.3 | 0.9 | GO:0060385 | axonogenesis involved in innervation(GO:0060385) |
| 0.3 | 2.4 | GO:0016926 | protein desumoylation(GO:0016926) |
| 0.3 | 17.5 | GO:0046677 | response to antibiotic(GO:0046677) |
| 0.3 | 2.1 | GO:0000185 | activation of MAPKKK activity(GO:0000185) |
| 0.3 | 5.4 | GO:0090435 | protein localization to nuclear envelope(GO:0090435) |
| 0.3 | 7.2 | GO:0061157 | mRNA destabilization(GO:0061157) |
| 0.3 | 1.2 | GO:1901491 | negative regulation of lymphangiogenesis(GO:1901491) |
| 0.3 | 0.9 | GO:0060722 | spongiotrophoblast cell proliferation(GO:0060720) cell proliferation involved in embryonic placenta development(GO:0060722) |
| 0.3 | 1.2 | GO:0060434 | bronchus morphogenesis(GO:0060434) |
| 0.3 | 1.2 | GO:0045876 | positive regulation of sister chromatid cohesion(GO:0045876) |
| 0.3 | 0.3 | GO:0090365 | regulation of mRNA modification(GO:0090365) negative regulation of mRNA modification(GO:0090367) |
| 0.3 | 10.3 | GO:0040015 | negative regulation of multicellular organism growth(GO:0040015) |
| 0.3 | 7.4 | GO:0033235 | positive regulation of protein sumoylation(GO:0033235) |
| 0.3 | 5.1 | GO:0001675 | acrosome assembly(GO:0001675) |
| 0.3 | 12.2 | GO:0010501 | RNA secondary structure unwinding(GO:0010501) |
| 0.3 | 5.7 | GO:0045947 | negative regulation of translational initiation(GO:0045947) |
| 0.3 | 1.1 | GO:0033314 | mitotic DNA replication checkpoint(GO:0033314) |
| 0.3 | 3.7 | GO:0046498 | S-adenosylhomocysteine metabolic process(GO:0046498) |
| 0.3 | 6.2 | GO:0006353 | DNA-templated transcription, termination(GO:0006353) |
| 0.3 | 1.1 | GO:0072365 | regulation of cellular ketone metabolic process by negative regulation of transcription from RNA polymerase II promoter(GO:0072365) |
| 0.3 | 11.1 | GO:0051123 | RNA polymerase II transcriptional preinitiation complex assembly(GO:0051123) |
| 0.3 | 13.9 | GO:0048246 | macrophage chemotaxis(GO:0048246) |
| 0.3 | 1.1 | GO:0051661 | maintenance of centrosome location(GO:0051661) |
| 0.3 | 1.9 | GO:1903527 | positive regulation of membrane tubulation(GO:1903527) |
| 0.3 | 1.6 | GO:0033140 | negative regulation of peptidyl-serine phosphorylation of STAT protein(GO:0033140) |
| 0.3 | 3.2 | GO:0002051 | osteoblast fate commitment(GO:0002051) |
| 0.3 | 20.2 | GO:0000245 | spliceosomal complex assembly(GO:0000245) |
| 0.3 | 3.1 | GO:0006622 | protein targeting to lysosome(GO:0006622) |
| 0.3 | 1.6 | GO:1990403 | embryonic brain development(GO:1990403) |
| 0.3 | 1.3 | GO:1903799 | negative regulation of production of miRNAs involved in gene silencing by miRNA(GO:1903799) |
| 0.3 | 2.3 | GO:0006108 | malate metabolic process(GO:0006108) |
| 0.3 | 7.3 | GO:2000369 | regulation of clathrin-mediated endocytosis(GO:2000369) |
| 0.3 | 2.0 | GO:0001833 | inner cell mass cell proliferation(GO:0001833) |
| 0.3 | 1.3 | GO:0016185 | synaptic vesicle budding from presynaptic endocytic zone membrane(GO:0016185) |
| 0.3 | 0.5 | GO:0045650 | negative regulation of macrophage differentiation(GO:0045650) |
| 0.2 | 1.5 | GO:0036265 | RNA (guanine-N7)-methylation(GO:0036265) |
| 0.2 | 2.7 | GO:0007099 | centriole replication(GO:0007099) |
| 0.2 | 4.7 | GO:0048026 | positive regulation of mRNA splicing, via spliceosome(GO:0048026) |
| 0.2 | 1.0 | GO:2000210 | positive regulation of anoikis(GO:2000210) |
| 0.2 | 2.4 | GO:0016191 | synaptic vesicle uncoating(GO:0016191) |
| 0.2 | 1.2 | GO:0099558 | maintenance of synapse structure(GO:0099558) |
| 0.2 | 0.2 | GO:0034454 | microtubule anchoring at centrosome(GO:0034454) |
| 0.2 | 2.7 | GO:0034063 | stress granule assembly(GO:0034063) |
| 0.2 | 1.1 | GO:1903898 | negative regulation of PERK-mediated unfolded protein response(GO:1903898) |
| 0.2 | 2.3 | GO:0032485 | regulation of Ral protein signal transduction(GO:0032485) |
| 0.2 | 1.8 | GO:1904263 | positive regulation of TORC1 signaling(GO:1904263) |
| 0.2 | 0.7 | GO:0042128 | nitrate assimilation(GO:0042128) |
| 0.2 | 2.7 | GO:0010216 | maintenance of DNA methylation(GO:0010216) |
| 0.2 | 1.1 | GO:2000584 | regulation of platelet-derived growth factor receptor-alpha signaling pathway(GO:2000583) negative regulation of platelet-derived growth factor receptor-alpha signaling pathway(GO:2000584) |
| 0.2 | 1.1 | GO:0090148 | membrane fission(GO:0090148) |
| 0.2 | 15.8 | GO:0000070 | mitotic sister chromatid segregation(GO:0000070) |
| 0.2 | 1.7 | GO:0035542 | regulation of SNARE complex assembly(GO:0035542) |
| 0.2 | 5.4 | GO:0010566 | regulation of ketone biosynthetic process(GO:0010566) |
| 0.2 | 2.0 | GO:0035234 | ectopic germ cell programmed cell death(GO:0035234) |
| 0.2 | 1.1 | GO:0001832 | blastocyst growth(GO:0001832) |
| 0.2 | 2.6 | GO:0090385 | phagosome-lysosome fusion(GO:0090385) |
| 0.2 | 3.7 | GO:0032897 | negative regulation of viral transcription(GO:0032897) |
| 0.2 | 14.3 | GO:0035176 | social behavior(GO:0035176) intraspecies interaction between organisms(GO:0051703) |
| 0.2 | 0.6 | GO:0000469 | cleavage involved in rRNA processing(GO:0000469) |
| 0.2 | 1.7 | GO:0031642 | negative regulation of myelination(GO:0031642) |
| 0.2 | 3.0 | GO:0000460 | maturation of 5.8S rRNA(GO:0000460) |
| 0.2 | 3.9 | GO:0018345 | protein palmitoylation(GO:0018345) |
| 0.2 | 0.4 | GO:0060287 | epithelial cilium movement involved in determination of left/right asymmetry(GO:0060287) |
| 0.2 | 3.4 | GO:0006614 | SRP-dependent cotranslational protein targeting to membrane(GO:0006614) |
| 0.2 | 0.6 | GO:2000156 | regulation of constitutive secretory pathway(GO:1903433) regulation of retrograde vesicle-mediated transport, Golgi to ER(GO:2000156) |
| 0.2 | 1.3 | GO:1901096 | regulation of autophagosome maturation(GO:1901096) |
| 0.2 | 15.8 | GO:0007088 | regulation of mitotic nuclear division(GO:0007088) |
| 0.2 | 2.9 | GO:0046036 | CTP biosynthetic process(GO:0006241) CTP metabolic process(GO:0046036) |
| 0.2 | 12.6 | GO:0006367 | transcription initiation from RNA polymerase II promoter(GO:0006367) |
| 0.2 | 6.1 | GO:0071549 | cellular response to dexamethasone stimulus(GO:0071549) |
| 0.2 | 2.5 | GO:1902018 | negative regulation of cilium assembly(GO:1902018) |
| 0.2 | 1.9 | GO:0007141 | male meiosis I(GO:0007141) |
| 0.2 | 2.7 | GO:0045955 | negative regulation of calcium ion-dependent exocytosis(GO:0045955) |
| 0.2 | 3.7 | GO:0006346 | methylation-dependent chromatin silencing(GO:0006346) |
| 0.2 | 0.2 | GO:1901300 | positive regulation of hydrogen peroxide-mediated programmed cell death(GO:1901300) |
| 0.2 | 1.0 | GO:0033313 | meiotic cell cycle checkpoint(GO:0033313) |
| 0.2 | 4.8 | GO:0038202 | TORC1 signaling(GO:0038202) |
| 0.2 | 4.0 | GO:0006044 | N-acetylglucosamine metabolic process(GO:0006044) |
| 0.2 | 7.1 | GO:0048266 | behavioral response to pain(GO:0048266) |
| 0.2 | 1.0 | GO:1902570 | protein localization to nucleolus(GO:1902570) |
| 0.2 | 65.3 | GO:0008380 | RNA splicing(GO:0008380) |
| 0.2 | 0.9 | GO:0046341 | CDP-diacylglycerol metabolic process(GO:0046341) |
| 0.2 | 1.3 | GO:0031118 | rRNA pseudouridine synthesis(GO:0031118) |
| 0.2 | 0.6 | GO:0006428 | isoleucyl-tRNA aminoacylation(GO:0006428) |
| 0.2 | 2.1 | GO:1904357 | negative regulation of telomere maintenance via telomere lengthening(GO:1904357) |
| 0.2 | 2.6 | GO:0001522 | pseudouridine synthesis(GO:0001522) |
| 0.2 | 1.5 | GO:0061525 | hindgut development(GO:0061525) |
| 0.2 | 2.8 | GO:0060736 | prostate gland growth(GO:0060736) |
| 0.2 | 5.3 | GO:0050860 | negative regulation of T cell receptor signaling pathway(GO:0050860) |
| 0.2 | 0.7 | GO:0036124 | histone H3-K9 trimethylation(GO:0036124) |
| 0.2 | 5.3 | GO:0070884 | regulation of calcineurin-NFAT signaling cascade(GO:0070884) |
| 0.2 | 0.5 | GO:0034970 | histone H3-R2 methylation(GO:0034970) |
| 0.2 | 2.8 | GO:0040001 | establishment of mitotic spindle localization(GO:0040001) |
| 0.2 | 1.0 | GO:0016557 | peroxisome membrane biogenesis(GO:0016557) |
| 0.2 | 0.7 | GO:0070475 | rRNA base methylation(GO:0070475) |
| 0.2 | 3.3 | GO:0030317 | sperm motility(GO:0030317) |
| 0.2 | 0.9 | GO:0006432 | phenylalanyl-tRNA aminoacylation(GO:0006432) |
| 0.2 | 0.3 | GO:1905064 | cardiac fibroblast cell differentiation(GO:0060935) cardiac fibroblast cell development(GO:0060936) epicardium-derived cardiac fibroblast cell differentiation(GO:0060938) epicardium-derived cardiac fibroblast cell development(GO:0060939) negative regulation of vascular smooth muscle cell differentiation(GO:1905064) |
| 0.2 | 1.9 | GO:0061635 | regulation of protein complex stability(GO:0061635) |
| 0.2 | 0.3 | GO:0097167 | circadian regulation of translation(GO:0097167) |
| 0.2 | 2.3 | GO:0006337 | nucleosome disassembly(GO:0006337) |
| 0.2 | 0.8 | GO:0018343 | protein farnesylation(GO:0018343) |
| 0.2 | 0.5 | GO:0043181 | vacuolar sequestering(GO:0043181) |
| 0.2 | 4.7 | GO:0000462 | maturation of SSU-rRNA from tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000462) |
| 0.2 | 1.4 | GO:0018344 | protein geranylgeranylation(GO:0018344) |
| 0.2 | 3.0 | GO:0043982 | histone H4-K5 acetylation(GO:0043981) histone H4-K8 acetylation(GO:0043982) |
| 0.2 | 1.8 | GO:1990118 | sodium ion import across plasma membrane(GO:0098719) sodium ion import into cell(GO:1990118) |
| 0.2 | 2.9 | GO:0035090 | maintenance of apical/basal cell polarity(GO:0035090) |
| 0.2 | 1.7 | GO:0070862 | negative regulation of protein exit from endoplasmic reticulum(GO:0070862) negative regulation of retrograde protein transport, ER to cytosol(GO:1904153) |
| 0.2 | 0.6 | GO:0043097 | pyrimidine-containing compound salvage(GO:0008655) pyrimidine nucleoside salvage(GO:0043097) |
| 0.2 | 1.2 | GO:0044821 | telomere tethering at nuclear periphery(GO:0034398) meiotic telomere tethering at nuclear periphery(GO:0044821) meiotic attachment of telomere to nuclear envelope(GO:0070197) chromosome attachment to the nuclear envelope(GO:0097240) |
| 0.1 | 1.9 | GO:0006303 | double-strand break repair via nonhomologous end joining(GO:0006303) |
| 0.1 | 0.9 | GO:0042985 | negative regulation of amyloid precursor protein biosynthetic process(GO:0042985) |
| 0.1 | 2.8 | GO:0032204 | regulation of telomere maintenance(GO:0032204) |
| 0.1 | 5.0 | GO:0050901 | leukocyte tethering or rolling(GO:0050901) |
| 0.1 | 7.8 | GO:0007019 | microtubule depolymerization(GO:0007019) |
| 0.1 | 5.0 | GO:0051225 | spindle assembly(GO:0051225) |
| 0.1 | 1.1 | GO:0070431 | nucleotide-binding oligomerization domain containing signaling pathway(GO:0070423) nucleotide-binding oligomerization domain containing 2 signaling pathway(GO:0070431) |
| 0.1 | 1.7 | GO:0006020 | inositol metabolic process(GO:0006020) |
| 0.1 | 0.7 | GO:0031296 | B cell costimulation(GO:0031296) |
| 0.1 | 0.7 | GO:1903003 | positive regulation of protein deubiquitination(GO:1903003) |
| 0.1 | 0.5 | GO:0061534 | negative regulation of neurotransmitter secretion(GO:0046929) gamma-aminobutyric acid secretion, neurotransmission(GO:0061534) |
| 0.1 | 1.1 | GO:0071712 | ER-associated misfolded protein catabolic process(GO:0071712) |
| 0.1 | 1.2 | GO:2000601 | positive regulation of Arp2/3 complex-mediated actin nucleation(GO:2000601) |
| 0.1 | 0.7 | GO:0007296 | vitellogenesis(GO:0007296) |
| 0.1 | 1.4 | GO:0007143 | female meiotic division(GO:0007143) |
| 0.1 | 0.4 | GO:0035519 | protein K29-linked ubiquitination(GO:0035519) |
| 0.1 | 9.7 | GO:0031497 | chromatin assembly(GO:0031497) |
| 0.1 | 1.0 | GO:0006348 | chromatin silencing at telomere(GO:0006348) |
| 0.1 | 3.5 | GO:0031572 | G2 DNA damage checkpoint(GO:0031572) |
| 0.1 | 0.5 | GO:0034553 | respiratory chain complex II assembly(GO:0034552) mitochondrial respiratory chain complex II assembly(GO:0034553) mitochondrial respiratory chain complex II biogenesis(GO:0097032) |
| 0.1 | 1.4 | GO:0043084 | penile erection(GO:0043084) |
| 0.1 | 1.4 | GO:0043117 | positive regulation of vascular permeability(GO:0043117) |
| 0.1 | 0.1 | GO:0072368 | regulation of lipid transport by negative regulation of transcription from RNA polymerase II promoter(GO:0072368) |
| 0.1 | 1.8 | GO:0048757 | endosome to melanosome transport(GO:0035646) endosome to pigment granule transport(GO:0043485) pigment granule maturation(GO:0048757) |
| 0.1 | 0.8 | GO:0051388 | positive regulation of neurotrophin TRK receptor signaling pathway(GO:0051388) |
| 0.1 | 2.3 | GO:0036295 | cellular response to increased oxygen levels(GO:0036295) |
| 0.1 | 0.6 | GO:0051096 | regulation of helicase activity(GO:0051095) positive regulation of helicase activity(GO:0051096) |
| 0.1 | 0.2 | GO:0010459 | negative regulation of heart rate(GO:0010459) |
| 0.1 | 3.0 | GO:0016925 | protein sumoylation(GO:0016925) |
| 0.1 | 10.1 | GO:0050982 | detection of mechanical stimulus(GO:0050982) |
| 0.1 | 2.3 | GO:0060285 | cilium or flagellum-dependent cell motility(GO:0001539) cilium-dependent cell motility(GO:0060285) |
| 0.1 | 3.4 | GO:0046856 | phosphatidylinositol dephosphorylation(GO:0046856) |
| 0.1 | 1.8 | GO:0040037 | negative regulation of fibroblast growth factor receptor signaling pathway(GO:0040037) |
| 0.1 | 0.3 | GO:2000474 | regulation of opioid receptor signaling pathway(GO:2000474) |
| 0.1 | 2.1 | GO:0032780 | negative regulation of ATPase activity(GO:0032780) |
| 0.1 | 1.5 | GO:0006054 | N-acetylneuraminate metabolic process(GO:0006054) |
| 0.1 | 1.0 | GO:0065004 | protein-DNA complex assembly(GO:0065004) |
| 0.1 | 6.3 | GO:0031109 | microtubule polymerization or depolymerization(GO:0031109) |
| 0.1 | 0.4 | GO:0006529 | asparagine biosynthetic process(GO:0006529) |
| 0.1 | 3.2 | GO:0000724 | double-strand break repair via homologous recombination(GO:0000724) recombinational repair(GO:0000725) |
| 0.1 | 5.5 | GO:0010596 | negative regulation of endothelial cell migration(GO:0010596) |
| 0.1 | 5.9 | GO:0048146 | positive regulation of fibroblast proliferation(GO:0048146) |
| 0.1 | 0.6 | GO:0008090 | retrograde axonal transport(GO:0008090) |
| 0.1 | 1.9 | GO:0071392 | cellular response to estradiol stimulus(GO:0071392) |
| 0.1 | 3.2 | GO:0009651 | response to salt stress(GO:0009651) |
| 0.1 | 2.0 | GO:0099514 | anterograde synaptic vesicle transport(GO:0048490) synaptic vesicle cytoskeletal transport(GO:0099514) synaptic vesicle transport along microtubule(GO:0099517) |
| 0.1 | 1.1 | GO:0051151 | negative regulation of smooth muscle cell differentiation(GO:0051151) |
| 0.1 | 0.6 | GO:0010993 | regulation of ubiquitin homeostasis(GO:0010993) free ubiquitin chain polymerization(GO:0010994) |
| 0.1 | 0.1 | GO:0044340 | canonical Wnt signaling pathway involved in regulation of cell proliferation(GO:0044340) |
| 0.1 | 1.2 | GO:1901407 | regulation of phosphorylation of RNA polymerase II C-terminal domain(GO:1901407) |
| 0.1 | 1.8 | GO:1902236 | negative regulation of endoplasmic reticulum stress-induced intrinsic apoptotic signaling pathway(GO:1902236) |
| 0.1 | 0.7 | GO:0006352 | DNA-templated transcription, initiation(GO:0006352) |
| 0.1 | 0.8 | GO:0001682 | tRNA 5'-leader removal(GO:0001682) |
| 0.1 | 1.5 | GO:0007052 | mitotic spindle organization(GO:0007052) |
| 0.1 | 4.1 | GO:0048240 | sperm capacitation(GO:0048240) |
| 0.1 | 0.7 | GO:0000729 | DNA double-strand break processing(GO:0000729) |
| 0.1 | 1.3 | GO:0042989 | sequestering of actin monomers(GO:0042989) |
| 0.1 | 1.3 | GO:0046007 | negative regulation of activated T cell proliferation(GO:0046007) |
| 0.1 | 3.6 | GO:0007339 | binding of sperm to zona pellucida(GO:0007339) |
| 0.1 | 0.5 | GO:0007182 | common-partner SMAD protein phosphorylation(GO:0007182) |
| 0.1 | 1.1 | GO:0015074 | DNA integration(GO:0015074) |
| 0.1 | 2.1 | GO:0001702 | gastrulation with mouth forming second(GO:0001702) |
| 0.1 | 1.6 | GO:0007190 | activation of adenylate cyclase activity(GO:0007190) |
| 0.1 | 2.4 | GO:0030488 | tRNA methylation(GO:0030488) |
| 0.1 | 0.2 | GO:0046909 | intermembrane transport(GO:0046909) protein transport from ciliary membrane to plasma membrane(GO:1903445) |
| 0.1 | 0.9 | GO:1902514 | regulation of calcium ion transmembrane transport via high voltage-gated calcium channel(GO:1902514) |
| 0.1 | 3.0 | GO:0051281 | positive regulation of release of sequestered calcium ion into cytosol(GO:0051281) |
| 0.1 | 3.0 | GO:0002548 | monocyte chemotaxis(GO:0002548) |
| 0.1 | 2.7 | GO:0060444 | branching involved in mammary gland duct morphogenesis(GO:0060444) |
| 0.1 | 0.6 | GO:0051967 | negative regulation of synaptic transmission, glutamatergic(GO:0051967) |
| 0.1 | 1.0 | GO:0048681 | negative regulation of axon regeneration(GO:0048681) |
| 0.1 | 1.2 | GO:0070979 | protein K11-linked ubiquitination(GO:0070979) |
| 0.1 | 0.1 | GO:0021861 | forebrain radial glial cell differentiation(GO:0021861) |
| 0.1 | 1.8 | GO:0048172 | regulation of short-term neuronal synaptic plasticity(GO:0048172) |
| 0.1 | 6.9 | GO:0007218 | neuropeptide signaling pathway(GO:0007218) |
| 0.1 | 0.6 | GO:0035457 | cellular response to interferon-alpha(GO:0035457) |
| 0.1 | 4.5 | GO:0070936 | protein K48-linked ubiquitination(GO:0070936) |
| 0.1 | 2.2 | GO:0045987 | positive regulation of smooth muscle contraction(GO:0045987) |
| 0.1 | 0.5 | GO:0042754 | negative regulation of circadian rhythm(GO:0042754) |
| 0.1 | 6.3 | GO:0006310 | DNA recombination(GO:0006310) |
| 0.1 | 8.2 | GO:0008213 | protein methylation(GO:0006479) protein alkylation(GO:0008213) |
| 0.1 | 1.2 | GO:0031034 | myosin filament assembly(GO:0031034) |
| 0.1 | 3.4 | GO:0009214 | cyclic nucleotide catabolic process(GO:0009214) |
| 0.1 | 0.6 | GO:0033262 | regulation of nuclear cell cycle DNA replication(GO:0033262) |
| 0.1 | 1.7 | GO:0040019 | positive regulation of embryonic development(GO:0040019) |
| 0.1 | 2.1 | GO:0035774 | positive regulation of insulin secretion involved in cellular response to glucose stimulus(GO:0035774) |
| 0.1 | 1.1 | GO:0007343 | egg activation(GO:0007343) |
| 0.1 | 0.5 | GO:0051204 | protein insertion into mitochondrial membrane(GO:0051204) |
| 0.1 | 1.2 | GO:0048714 | positive regulation of oligodendrocyte differentiation(GO:0048714) |
| 0.1 | 0.9 | GO:0051457 | maintenance of protein location in nucleus(GO:0051457) |
| 0.1 | 1.2 | GO:0015809 | arginine transport(GO:0015809) |
| 0.1 | 2.1 | GO:0060216 | definitive hemopoiesis(GO:0060216) |
| 0.1 | 1.8 | GO:0010172 | embryonic body morphogenesis(GO:0010172) |
| 0.1 | 0.6 | GO:0033147 | negative regulation of intracellular estrogen receptor signaling pathway(GO:0033147) |
| 0.1 | 0.9 | GO:0014049 | positive regulation of glutamate secretion(GO:0014049) |
| 0.1 | 1.0 | GO:0030889 | negative regulation of B cell proliferation(GO:0030889) |
| 0.1 | 1.4 | GO:0032026 | response to magnesium ion(GO:0032026) |
| 0.1 | 0.7 | GO:0007289 | spermatid nucleus differentiation(GO:0007289) |
| 0.1 | 2.5 | GO:0032012 | ARF protein signal transduction(GO:0032011) regulation of ARF protein signal transduction(GO:0032012) |
| 0.1 | 0.7 | GO:0051683 | establishment of Golgi localization(GO:0051683) |
| 0.1 | 1.3 | GO:0043306 | positive regulation of mast cell activation involved in immune response(GO:0033008) positive regulation of mast cell degranulation(GO:0043306) |
| 0.1 | 1.2 | GO:0051443 | positive regulation of ubiquitin-protein transferase activity(GO:0051443) |
| 0.1 | 2.6 | GO:0043966 | histone H3 acetylation(GO:0043966) |
| 0.1 | 5.5 | GO:0032259 | methylation(GO:0032259) |
| 0.1 | 1.9 | GO:0061512 | protein localization to cilium(GO:0061512) |
| 0.1 | 0.7 | GO:0030490 | maturation of SSU-rRNA(GO:0030490) |
| 0.1 | 2.1 | GO:0001578 | microtubule bundle formation(GO:0001578) |
| 0.1 | 0.5 | GO:0006686 | sphingomyelin biosynthetic process(GO:0006686) |
| 0.1 | 0.6 | GO:0031124 | mRNA 3'-end processing(GO:0031124) |
| 0.1 | 0.5 | GO:0034058 | endosomal vesicle fusion(GO:0034058) |
| 0.1 | 0.4 | GO:0021800 | cerebral cortex tangential migration(GO:0021800) |
| 0.1 | 0.2 | GO:0007217 | tachykinin receptor signaling pathway(GO:0007217) |
| 0.1 | 0.4 | GO:0006271 | DNA strand elongation involved in DNA replication(GO:0006271) |
| 0.1 | 0.7 | GO:0061213 | positive regulation of mesonephros development(GO:0061213) positive regulation of branching involved in ureteric bud morphogenesis(GO:0090190) |
| 0.1 | 0.5 | GO:0006003 | fructose 2,6-bisphosphate metabolic process(GO:0006003) |
| 0.1 | 0.1 | GO:1901874 | regulation of post-translational protein modification(GO:1901873) negative regulation of post-translational protein modification(GO:1901874) |
| 0.1 | 0.4 | GO:0035455 | response to interferon-alpha(GO:0035455) |
| 0.1 | 0.2 | GO:0061668 | mitochondrial ribosome assembly(GO:0061668) mitochondrial large ribosomal subunit assembly(GO:1902775) |
| 0.1 | 4.3 | GO:0050868 | negative regulation of T cell activation(GO:0050868) |
| 0.1 | 20.8 | GO:0048232 | spermatogenesis(GO:0007283) male gamete generation(GO:0048232) |
| 0.1 | 1.5 | GO:0097035 | regulation of membrane lipid distribution(GO:0097035) |
| 0.1 | 1.0 | GO:0015701 | bicarbonate transport(GO:0015701) |
| 0.1 | 2.3 | GO:0060612 | adipose tissue development(GO:0060612) |
| 0.1 | 1.6 | GO:0042755 | eating behavior(GO:0042755) |
| 0.0 | 0.6 | GO:0006968 | cellular defense response(GO:0006968) |
| 0.0 | 0.6 | GO:0097264 | self proteolysis(GO:0097264) |
| 0.0 | 0.4 | GO:0048280 | vesicle fusion with Golgi apparatus(GO:0048280) |
| 0.0 | 0.2 | GO:1903715 | regulation of aerobic respiration(GO:1903715) |
| 0.0 | 0.8 | GO:0034776 | response to histamine(GO:0034776) cellular response to histamine(GO:0071420) |
| 0.0 | 1.0 | GO:0042761 | fatty acid elongation(GO:0030497) very long-chain fatty acid biosynthetic process(GO:0042761) |
| 0.0 | 0.6 | GO:0014047 | glutamate secretion(GO:0014047) |
| 0.0 | 1.1 | GO:0032007 | negative regulation of TOR signaling(GO:0032007) |
| 0.0 | 0.2 | GO:0001732 | formation of cytoplasmic translation initiation complex(GO:0001732) |
| 0.0 | 0.4 | GO:0061088 | sequestering of zinc ion(GO:0032119) regulation of sequestering of zinc ion(GO:0061088) |
| 0.0 | 0.3 | GO:0016266 | O-glycan processing(GO:0016266) |
| 0.0 | 0.7 | GO:0006302 | double-strand break repair(GO:0006302) |
| 0.0 | 1.1 | GO:0014003 | oligodendrocyte development(GO:0014003) |
| 0.0 | 1.2 | GO:0016573 | histone acetylation(GO:0016573) |
| 0.0 | 0.6 | GO:0046716 | muscle cell cellular homeostasis(GO:0046716) |
| 0.0 | 1.4 | GO:0006891 | intra-Golgi vesicle-mediated transport(GO:0006891) |
| 0.0 | 3.0 | GO:0090090 | negative regulation of canonical Wnt signaling pathway(GO:0090090) |
| 0.0 | 0.3 | GO:0030970 | retrograde protein transport, ER to cytosol(GO:0030970) |
| 0.0 | 1.1 | GO:1902042 | negative regulation of extrinsic apoptotic signaling pathway via death domain receptors(GO:1902042) |
| 0.0 | 0.9 | GO:0000186 | activation of MAPKK activity(GO:0000186) |
| 0.0 | 1.0 | GO:0006471 | protein ADP-ribosylation(GO:0006471) |
| 0.0 | 0.5 | GO:1903779 | regulation of cardiac conduction(GO:1903779) |
| 0.0 | 2.9 | GO:0007188 | adenylate cyclase-modulating G-protein coupled receptor signaling pathway(GO:0007188) |
| 0.0 | 0.9 | GO:0007274 | neuromuscular synaptic transmission(GO:0007274) |
| 0.0 | 0.3 | GO:2001183 | negative regulation of interleukin-12 secretion(GO:2001183) |
| 0.0 | 3.5 | GO:0000209 | protein polyubiquitination(GO:0000209) |
| 0.0 | 0.4 | GO:0043248 | proteasome assembly(GO:0043248) |
| 0.0 | 0.8 | GO:0071353 | cellular response to interleukin-4(GO:0071353) |
| 0.0 | 2.9 | GO:0048706 | embryonic skeletal system development(GO:0048706) |
| 0.0 | 0.7 | GO:0006400 | tRNA modification(GO:0006400) |
| 0.0 | 0.8 | GO:0048709 | oligodendrocyte differentiation(GO:0048709) |
| 0.0 | 2.5 | GO:0007224 | smoothened signaling pathway(GO:0007224) |
| 0.0 | 1.8 | GO:0033138 | positive regulation of peptidyl-serine phosphorylation(GO:0033138) |
| 0.0 | 1.4 | GO:0003014 | renal system process(GO:0003014) |
| 0.0 | 0.7 | GO:0032330 | regulation of chondrocyte differentiation(GO:0032330) |
| 0.0 | 2.2 | GO:0016579 | protein deubiquitination(GO:0016579) |
| 0.0 | 0.5 | GO:0016339 | calcium-dependent cell-cell adhesion via plasma membrane cell adhesion molecules(GO:0016339) |
| 0.0 | 0.1 | GO:0035511 | oxidative DNA demethylation(GO:0035511) |
| 0.0 | 0.4 | GO:0006513 | protein monoubiquitination(GO:0006513) |
| 0.0 | 0.4 | GO:0019933 | cAMP-mediated signaling(GO:0019933) |
| 0.0 | 0.0 | GO:0070272 | proton-transporting ATP synthase complex assembly(GO:0043461) proton-transporting ATP synthase complex biogenesis(GO:0070272) |
| 0.0 | 0.1 | GO:0035507 | regulation of myosin-light-chain-phosphatase activity(GO:0035507) |
| 0.0 | 0.2 | GO:2001241 | positive regulation of extrinsic apoptotic signaling pathway in absence of ligand(GO:2001241) |
| 0.0 | 0.5 | GO:0032728 | positive regulation of interferon-beta production(GO:0032728) |
| 0.0 | 0.4 | GO:0032232 | negative regulation of actin filament bundle assembly(GO:0032232) negative regulation of stress fiber assembly(GO:0051497) |
| 0.0 | 0.1 | GO:0097428 | protein maturation by iron-sulfur cluster transfer(GO:0097428) |
| 0.0 | 0.1 | GO:0001964 | startle response(GO:0001964) |
| 0.0 | 0.1 | GO:0016322 | neuron remodeling(GO:0016322) |
| 0.0 | 0.1 | GO:0048386 | positive regulation of retinoic acid receptor signaling pathway(GO:0048386) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 3.9 | 11.7 | GO:0016939 | kinesin II complex(GO:0016939) |
| 3.3 | 9.8 | GO:0000818 | nuclear MIS12/MIND complex(GO:0000818) |
| 2.6 | 7.7 | GO:0097135 | cyclin E2-CDK2 complex(GO:0097135) |
| 2.4 | 7.1 | GO:0031074 | nucleocytoplasmic shuttling complex(GO:0031074) |
| 2.4 | 14.2 | GO:0097226 | sperm mitochondrial sheath(GO:0097226) |
| 2.3 | 11.6 | GO:0048476 | Holliday junction resolvase complex(GO:0048476) |
| 2.1 | 6.4 | GO:0005673 | transcription factor TFIIE complex(GO:0005673) |
| 1.9 | 9.7 | GO:0000799 | nuclear condensin complex(GO:0000799) |
| 1.9 | 5.7 | GO:0005712 | chiasma(GO:0005712) |
| 1.8 | 7.4 | GO:0071821 | FANCM-MHF complex(GO:0071821) |
| 1.7 | 6.8 | GO:0000942 | condensed nuclear chromosome outer kinetochore(GO:0000942) |
| 1.6 | 9.7 | GO:0005958 | DNA-dependent protein kinase-DNA ligase 4 complex(GO:0005958) |
| 1.6 | 17.7 | GO:0071546 | pi-body(GO:0071546) |
| 1.5 | 5.9 | GO:0035061 | interchromatin granule(GO:0035061) |
| 1.5 | 13.2 | GO:0097165 | nuclear stress granule(GO:0097165) |
| 1.5 | 1.5 | GO:0032807 | DNA ligase IV complex(GO:0032807) |
| 1.4 | 4.3 | GO:0031510 | SUMO activating enzyme complex(GO:0031510) |
| 1.4 | 9.6 | GO:0000796 | condensin complex(GO:0000796) |
| 1.4 | 1.4 | GO:0044094 | viral replication complex(GO:0019034) host cell nucleus(GO:0042025) host cell nuclear part(GO:0044094) |
| 1.4 | 5.4 | GO:0033597 | mitotic checkpoint complex(GO:0033597) bub1-bub3 complex(GO:1990298) |
| 1.2 | 2.4 | GO:0071942 | XPC complex(GO:0071942) |
| 1.2 | 13.4 | GO:0044613 | nuclear pore central transport channel(GO:0044613) |
| 1.1 | 3.4 | GO:0070522 | ERCC4-ERCC1 complex(GO:0070522) |
| 1.1 | 10.0 | GO:0070545 | PeBoW complex(GO:0070545) |
| 1.1 | 7.7 | GO:0005658 | alpha DNA polymerase:primase complex(GO:0005658) |
| 1.0 | 11.5 | GO:0000940 | condensed chromosome outer kinetochore(GO:0000940) |
| 1.0 | 5.2 | GO:0000802 | transverse filament(GO:0000802) |
| 1.0 | 35.4 | GO:0030992 | intraciliary transport particle B(GO:0030992) |
| 0.9 | 4.6 | GO:0005672 | transcription factor TFIIA complex(GO:0005672) |
| 0.9 | 6.4 | GO:0005964 | phosphorylase kinase complex(GO:0005964) |
| 0.9 | 3.5 | GO:0042585 | germinal vesicle(GO:0042585) |
| 0.9 | 2.6 | GO:1990879 | CST complex(GO:1990879) |
| 0.9 | 7.8 | GO:0071818 | BAT3 complex(GO:0071818) ER membrane insertion complex(GO:0072379) |
| 0.9 | 7.7 | GO:1990726 | Lsm1-7-Pat1 complex(GO:1990726) |
| 0.8 | 9.3 | GO:0000782 | telomere cap complex(GO:0000782) nuclear telomere cap complex(GO:0000783) |
| 0.8 | 8.0 | GO:0031390 | Ctf18 RFC-like complex(GO:0031390) |
| 0.8 | 17.6 | GO:0031011 | Ino80 complex(GO:0031011) |
| 0.8 | 5.6 | GO:0098536 | deuterosome(GO:0098536) |
| 0.8 | 3.2 | GO:1990037 | Lewy body core(GO:1990037) |
| 0.8 | 101.7 | GO:0005814 | centriole(GO:0005814) |
| 0.8 | 11.7 | GO:0001939 | female pronucleus(GO:0001939) |
| 0.8 | 3.8 | GO:0031251 | PAN complex(GO:0031251) |
| 0.7 | 3.7 | GO:0036449 | microtubule minus-end(GO:0036449) |
| 0.7 | 5.9 | GO:0005826 | actomyosin contractile ring(GO:0005826) |
| 0.7 | 2.2 | GO:0098843 | postsynaptic endocytic zone(GO:0098843) |
| 0.7 | 21.7 | GO:0035098 | ESC/E(Z) complex(GO:0035098) |
| 0.7 | 4.8 | GO:0071556 | integral component of cytoplasmic side of endoplasmic reticulum membrane(GO:0071458) integral component of lumenal side of endoplasmic reticulum membrane(GO:0071556) lumenal side of endoplasmic reticulum membrane(GO:0098553) |
| 0.7 | 3.4 | GO:1990130 | Iml1 complex(GO:1990130) |
| 0.7 | 7.4 | GO:0005664 | origin recognition complex(GO:0000808) nuclear origin of replication recognition complex(GO:0005664) |
| 0.7 | 7.9 | GO:0000176 | nuclear exosome (RNase complex)(GO:0000176) |
| 0.7 | 3.9 | GO:0071008 | U2-type post-mRNA release spliceosomal complex(GO:0071008) |
| 0.7 | 3.3 | GO:0032389 | MutLalpha complex(GO:0032389) |
| 0.7 | 3.3 | GO:0070033 | synaptobrevin 2-SNAP-25-syntaxin-1a-complexin I complex(GO:0070032) synaptobrevin 2-SNAP-25-syntaxin-1a-complexin II complex(GO:0070033) |
| 0.6 | 6.5 | GO:0030991 | intraciliary transport particle A(GO:0030991) |
| 0.6 | 1.9 | GO:0005863 | striated muscle myosin thick filament(GO:0005863) |
| 0.6 | 0.6 | GO:1990257 | piccolo-bassoon transport vesicle(GO:1990257) |
| 0.6 | 11.6 | GO:0000242 | pericentriolar material(GO:0000242) |
| 0.6 | 1.9 | GO:0034657 | GID complex(GO:0034657) |
| 0.6 | 4.5 | GO:0031314 | extrinsic component of mitochondrial inner membrane(GO:0031314) |
| 0.6 | 3.9 | GO:1990393 | 3M complex(GO:1990393) |
| 0.6 | 3.8 | GO:0097149 | centralspindlin complex(GO:0097149) |
| 0.6 | 10.9 | GO:0000346 | transcription export complex(GO:0000346) |
| 0.6 | 3.4 | GO:0043564 | Ku70:Ku80 complex(GO:0043564) |
| 0.6 | 10.9 | GO:0032039 | integrator complex(GO:0032039) |
| 0.6 | 2.9 | GO:0035363 | histone locus body(GO:0035363) |
| 0.6 | 11.8 | GO:0005847 | mRNA cleavage and polyadenylation specificity factor complex(GO:0005847) |
| 0.6 | 16.8 | GO:0005868 | cytoplasmic dynein complex(GO:0005868) |
| 0.6 | 4.4 | GO:0097504 | Gemini of coiled bodies(GO:0097504) |
| 0.6 | 3.9 | GO:0030915 | Smc5-Smc6 complex(GO:0030915) |
| 0.5 | 4.9 | GO:0070436 | Grb2-EGFR complex(GO:0070436) |
| 0.5 | 5.5 | GO:0042382 | paraspeckles(GO:0042382) |
| 0.5 | 2.2 | GO:0000778 | condensed nuclear chromosome kinetochore(GO:0000778) |
| 0.5 | 4.9 | GO:0070652 | HAUS complex(GO:0070652) |
| 0.5 | 6.0 | GO:0000777 | condensed chromosome kinetochore(GO:0000777) |
| 0.5 | 13.9 | GO:0043189 | NuA4 histone acetyltransferase complex(GO:0035267) H4/H2A histone acetyltransferase complex(GO:0043189) H4 histone acetyltransferase complex(GO:1902562) |
| 0.5 | 2.1 | GO:0032021 | NELF complex(GO:0032021) |
| 0.5 | 4.2 | GO:0001652 | granular component(GO:0001652) |
| 0.5 | 4.2 | GO:0005845 | mRNA cap binding complex(GO:0005845) RNA cap binding complex(GO:0034518) |
| 0.5 | 6.3 | GO:0031080 | nuclear pore outer ring(GO:0031080) |
| 0.5 | 3.6 | GO:0031465 | Cul4B-RING E3 ubiquitin ligase complex(GO:0031465) |
| 0.5 | 1.5 | GO:0043614 | multi-eIF complex(GO:0043614) |
| 0.5 | 5.1 | GO:0000127 | transcription factor TFIIIC complex(GO:0000127) |
| 0.5 | 9.7 | GO:0002199 | zona pellucida receptor complex(GO:0002199) |
| 0.5 | 1.5 | GO:0097361 | CIA complex(GO:0097361) |
| 0.5 | 1.5 | GO:0044317 | rod spherule(GO:0044317) |
| 0.5 | 2.0 | GO:1990769 | proximal neuron projection(GO:1990769) |
| 0.5 | 7.3 | GO:0005689 | U12-type spliceosomal complex(GO:0005689) |
| 0.5 | 2.4 | GO:0071014 | post-mRNA release spliceosomal complex(GO:0071014) |
| 0.5 | 2.4 | GO:0097255 | R2TP complex(GO:0097255) |
| 0.5 | 6.1 | GO:0000974 | Prp19 complex(GO:0000974) |
| 0.5 | 0.9 | GO:0071817 | MMXD complex(GO:0071817) |
| 0.5 | 5.1 | GO:0030015 | CCR4-NOT core complex(GO:0030015) |
| 0.5 | 6.0 | GO:0000124 | SAGA complex(GO:0000124) |
| 0.5 | 7.8 | GO:0031258 | lamellipodium membrane(GO:0031258) |
| 0.5 | 5.9 | GO:0044666 | MLL3/4 complex(GO:0044666) |
| 0.4 | 2.7 | GO:0030981 | cortical microtubule cytoskeleton(GO:0030981) |
| 0.4 | 2.7 | GO:0033063 | Rad51B-Rad51C-Rad51D-XRCC2 complex(GO:0033063) |
| 0.4 | 4.0 | GO:0072669 | tRNA-splicing ligase complex(GO:0072669) |
| 0.4 | 9.8 | GO:0071564 | npBAF complex(GO:0071564) |
| 0.4 | 11.1 | GO:0034451 | centriolar satellite(GO:0034451) |
| 0.4 | 4.0 | GO:0000801 | central element(GO:0000801) |
| 0.4 | 4.4 | GO:0005869 | dynactin complex(GO:0005869) |
| 0.4 | 4.4 | GO:0000235 | astral microtubule(GO:0000235) |
| 0.4 | 0.4 | GO:0097422 | tubular endosome(GO:0097422) |
| 0.4 | 1.3 | GO:0005760 | gamma DNA polymerase complex(GO:0005760) |
| 0.4 | 7.3 | GO:0031932 | TORC2 complex(GO:0031932) |
| 0.4 | 9.3 | GO:0010369 | chromocenter(GO:0010369) |
| 0.4 | 37.0 | GO:0072686 | mitotic spindle(GO:0072686) |
| 0.4 | 6.2 | GO:0036038 | MKS complex(GO:0036038) |
| 0.4 | 33.9 | GO:0010494 | cytoplasmic stress granule(GO:0010494) |
| 0.4 | 1.2 | GO:1990622 | CHOP-ATF3 complex(GO:1990622) |
| 0.4 | 12.8 | GO:0071011 | precatalytic spliceosome(GO:0071011) |
| 0.4 | 4.4 | GO:0016272 | prefoldin complex(GO:0016272) |
| 0.4 | 2.4 | GO:0071204 | histone pre-mRNA 3'end processing complex(GO:0071204) |
| 0.4 | 3.6 | GO:0005686 | U2 snRNP(GO:0005686) |
| 0.4 | 3.5 | GO:0005786 | signal recognition particle, endoplasmic reticulum targeting(GO:0005786) |
| 0.4 | 5.5 | GO:0090544 | BAF-type complex(GO:0090544) |
| 0.4 | 5.1 | GO:0030688 | preribosome, small subunit precursor(GO:0030688) |
| 0.4 | 2.7 | GO:0036157 | outer dynein arm(GO:0036157) |
| 0.4 | 1.1 | GO:0031262 | Ndc80 complex(GO:0031262) |
| 0.4 | 5.7 | GO:0035748 | myelin sheath abaxonal region(GO:0035748) |
| 0.4 | 4.5 | GO:0035327 | transcriptionally active chromatin(GO:0035327) |
| 0.4 | 10.1 | GO:0071339 | MLL1/2 complex(GO:0044665) MLL1 complex(GO:0071339) |
| 0.4 | 2.6 | GO:0036396 | MIS complex(GO:0036396) |
| 0.4 | 4.5 | GO:0033180 | proton-transporting V-type ATPase, V1 domain(GO:0033180) |
| 0.4 | 1.8 | GO:0034719 | SMN-Sm protein complex(GO:0034719) |
| 0.4 | 3.9 | GO:0000923 | equatorial microtubule organizing center(GO:0000923) |
| 0.4 | 3.2 | GO:0008278 | cohesin complex(GO:0008278) |
| 0.3 | 1.0 | GO:0071001 | U4/U6 snRNP(GO:0071001) |
| 0.3 | 4.7 | GO:0000015 | phosphopyruvate hydratase complex(GO:0000015) |
| 0.3 | 0.3 | GO:0000110 | nucleotide-excision repair factor 1 complex(GO:0000110) |
| 0.3 | 1.6 | GO:0031501 | mannosyltransferase complex(GO:0031501) |
| 0.3 | 25.3 | GO:0000776 | kinetochore(GO:0000776) |
| 0.3 | 3.2 | GO:0071004 | U2-type prespliceosome(GO:0071004) |
| 0.3 | 3.8 | GO:0005671 | Ada2/Gcn5/Ada3 transcription activator complex(GO:0005671) |
| 0.3 | 4.4 | GO:0036128 | CatSper complex(GO:0036128) |
| 0.3 | 1.9 | GO:0000172 | ribonuclease MRP complex(GO:0000172) |
| 0.3 | 7.8 | GO:0005665 | DNA-directed RNA polymerase II, core complex(GO:0005665) |
| 0.3 | 1.2 | GO:0090661 | box H/ACA telomerase RNP complex(GO:0090661) |
| 0.3 | 3.7 | GO:0035869 | ciliary transition zone(GO:0035869) |
| 0.3 | 5.2 | GO:0070938 | contractile ring(GO:0070938) |
| 0.3 | 2.1 | GO:0031298 | replication fork protection complex(GO:0031298) |
| 0.3 | 7.9 | GO:0032391 | photoreceptor connecting cilium(GO:0032391) |
| 0.3 | 3.3 | GO:0070531 | BRCA1-A complex(GO:0070531) |
| 0.3 | 3.6 | GO:0016580 | Sin3 complex(GO:0016580) |
| 0.3 | 3.6 | GO:0000930 | gamma-tubulin complex(GO:0000930) |
| 0.3 | 0.6 | GO:0032300 | mismatch repair complex(GO:0032300) |
| 0.3 | 13.4 | GO:0031519 | PcG protein complex(GO:0031519) |
| 0.3 | 1.5 | GO:0035189 | Rb-E2F complex(GO:0035189) |
| 0.3 | 0.9 | GO:0009328 | phenylalanine-tRNA ligase complex(GO:0009328) |
| 0.3 | 1.1 | GO:0097550 | transcriptional preinitiation complex(GO:0097550) |
| 0.3 | 9.3 | GO:0015030 | Cajal body(GO:0015030) |
| 0.3 | 0.8 | GO:0005965 | protein farnesyltransferase complex(GO:0005965) |
| 0.3 | 8.1 | GO:0001741 | XY body(GO:0001741) |
| 0.3 | 1.6 | GO:0005684 | U2-type spliceosomal complex(GO:0005684) |
| 0.3 | 7.2 | GO:0033176 | proton-transporting V-type ATPase complex(GO:0033176) |
| 0.3 | 42.0 | GO:0098687 | chromosomal region(GO:0098687) |
| 0.3 | 1.8 | GO:0005853 | eukaryotic translation elongation factor 1 complex(GO:0005853) |
| 0.2 | 2.2 | GO:0034464 | BBSome(GO:0034464) |
| 0.2 | 1.5 | GO:0043527 | tRNA methyltransferase complex(GO:0043527) |
| 0.2 | 4.2 | GO:0034098 | VCP-NPL4-UFD1 AAA ATPase complex(GO:0034098) |
| 0.2 | 1.4 | GO:0005968 | Rab-protein geranylgeranyltransferase complex(GO:0005968) |
| 0.2 | 1.7 | GO:0002177 | manchette(GO:0002177) |
| 0.2 | 1.7 | GO:0033588 | Elongator holoenzyme complex(GO:0033588) |
| 0.2 | 3.6 | GO:0017119 | Golgi transport complex(GO:0017119) |
| 0.2 | 0.7 | GO:0031372 | UBC13-MMS2 complex(GO:0031372) |
| 0.2 | 1.6 | GO:0034388 | Pwp2p-containing subcomplex of 90S preribosome(GO:0034388) |
| 0.2 | 1.2 | GO:0044301 | climbing fiber(GO:0044301) |
| 0.2 | 3.0 | GO:0044322 | endoplasmic reticulum quality control compartment(GO:0044322) |
| 0.2 | 1.1 | GO:0035859 | Seh1-associated complex(GO:0035859) |
| 0.2 | 2.2 | GO:0048188 | Set1C/COMPASS complex(GO:0048188) |
| 0.2 | 1.1 | GO:0071797 | LUBAC complex(GO:0071797) |
| 0.2 | 5.5 | GO:0030686 | 90S preribosome(GO:0030686) |
| 0.2 | 2.0 | GO:0001673 | male germ cell nucleus(GO:0001673) |
| 0.2 | 2.4 | GO:0032541 | cortical endoplasmic reticulum(GO:0032541) |
| 0.2 | 1.5 | GO:0033391 | chromatoid body(GO:0033391) |
| 0.2 | 1.8 | GO:0032584 | growth cone membrane(GO:0032584) |
| 0.2 | 3.6 | GO:0016600 | flotillin complex(GO:0016600) |
| 0.2 | 0.6 | GO:0036020 | endolysosome membrane(GO:0036020) |
| 0.2 | 9.6 | GO:0016235 | aggresome(GO:0016235) |
| 0.2 | 1.0 | GO:1990923 | PET complex(GO:1990923) |
| 0.2 | 4.6 | GO:0005876 | spindle microtubule(GO:0005876) |
| 0.2 | 1.0 | GO:0046540 | U4/U6 x U5 tri-snRNP complex(GO:0046540) |
| 0.2 | 15.0 | GO:0071013 | catalytic step 2 spliceosome(GO:0071013) |
| 0.2 | 30.1 | GO:0001669 | acrosomal vesicle(GO:0001669) |
| 0.2 | 0.9 | GO:0000120 | RNA polymerase I transcription factor complex(GO:0000120) |
| 0.2 | 1.5 | GO:0033263 | CORVET complex(GO:0033263) |
| 0.2 | 0.9 | GO:0072487 | MSL complex(GO:0072487) |
| 0.2 | 11.4 | GO:0000785 | chromatin(GO:0000785) |
| 0.2 | 13.6 | GO:0099501 | synaptic vesicle membrane(GO:0030672) exocytic vesicle membrane(GO:0099501) |
| 0.2 | 3.7 | GO:0090545 | NuRD complex(GO:0016581) CHD-type complex(GO:0090545) |
| 0.2 | 2.5 | GO:0005640 | nuclear outer membrane(GO:0005640) |
| 0.2 | 8.4 | GO:0016592 | mediator complex(GO:0016592) |
| 0.2 | 0.5 | GO:0042721 | mitochondrial inner membrane protein insertion complex(GO:0042721) |
| 0.2 | 3.8 | GO:0005892 | acetylcholine-gated channel complex(GO:0005892) |
| 0.2 | 25.5 | GO:0031514 | motile cilium(GO:0031514) |
| 0.2 | 6.5 | GO:0008023 | transcription elongation factor complex(GO:0008023) |
| 0.2 | 3.4 | GO:0005669 | transcription factor TFIID complex(GO:0005669) |
| 0.2 | 1.1 | GO:0005851 | eukaryotic translation initiation factor 2B complex(GO:0005851) |
| 0.2 | 3.2 | GO:0030687 | preribosome, large subunit precursor(GO:0030687) |
| 0.2 | 1.8 | GO:0044327 | dendritic spine head(GO:0044327) |
| 0.2 | 8.5 | GO:0005844 | polysome(GO:0005844) |
| 0.2 | 6.0 | GO:0000791 | euchromatin(GO:0000791) |
| 0.2 | 2.7 | GO:0097546 | ciliary base(GO:0097546) |
| 0.1 | 6.0 | GO:0030904 | retromer complex(GO:0030904) |
| 0.1 | 1.4 | GO:0005742 | mitochondrial outer membrane translocase complex(GO:0005742) |
| 0.1 | 2.0 | GO:0048787 | presynaptic active zone membrane(GO:0048787) |
| 0.1 | 1.0 | GO:0030289 | protein phosphatase 4 complex(GO:0030289) |
| 0.1 | 2.4 | GO:0031414 | N-terminal protein acetyltransferase complex(GO:0031414) |
| 0.1 | 0.7 | GO:0030877 | beta-catenin destruction complex(GO:0030877) |
| 0.1 | 1.7 | GO:0042613 | MHC class II protein complex(GO:0042613) |
| 0.1 | 0.3 | GO:0033061 | DNA recombinase mediator complex(GO:0033061) |
| 0.1 | 1.4 | GO:0031595 | nuclear proteasome complex(GO:0031595) |
| 0.1 | 1.2 | GO:0005871 | kinesin complex(GO:0005871) |
| 0.1 | 2.4 | GO:0031235 | intrinsic component of the cytoplasmic side of the plasma membrane(GO:0031235) |
| 0.1 | 2.9 | GO:0044815 | DNA packaging complex(GO:0044815) |
| 0.1 | 1.9 | GO:0005666 | DNA-directed RNA polymerase III complex(GO:0005666) |
| 0.1 | 3.2 | GO:0005680 | anaphase-promoting complex(GO:0005680) |
| 0.1 | 10.9 | GO:0017053 | transcriptional repressor complex(GO:0017053) |
| 0.1 | 3.2 | GO:0000159 | protein phosphatase type 2A complex(GO:0000159) |
| 0.1 | 0.8 | GO:0036194 | muscle cell projection(GO:0036194) muscle cell projection membrane(GO:0036195) |
| 0.1 | 7.9 | GO:0016363 | nuclear matrix(GO:0016363) |
| 0.1 | 0.4 | GO:0089701 | U2AF(GO:0089701) |
| 0.1 | 9.9 | GO:0000932 | cytoplasmic mRNA processing body(GO:0000932) |
| 0.1 | 2.0 | GO:0031045 | dense core granule(GO:0031045) |
| 0.1 | 5.2 | GO:0043194 | axon initial segment(GO:0043194) |
| 0.1 | 42.5 | GO:0005874 | microtubule(GO:0005874) |
| 0.1 | 1.6 | GO:0044300 | cerebellar mossy fiber(GO:0044300) |
| 0.1 | 4.0 | GO:0016234 | inclusion body(GO:0016234) |
| 0.1 | 8.3 | GO:0016605 | PML body(GO:0016605) |
| 0.1 | 8.5 | GO:0031463 | Cul3-RING ubiquitin ligase complex(GO:0031463) |
| 0.1 | 1.2 | GO:0005641 | nuclear envelope lumen(GO:0005641) |
| 0.1 | 0.7 | GO:0045120 | pronucleus(GO:0045120) |
| 0.1 | 1.3 | GO:0035097 | histone methyltransferase complex(GO:0035097) |
| 0.1 | 28.8 | GO:0016607 | nuclear speck(GO:0016607) |
| 0.1 | 0.4 | GO:0043625 | delta DNA polymerase complex(GO:0043625) |
| 0.1 | 3.3 | GO:0032588 | trans-Golgi network membrane(GO:0032588) |
| 0.1 | 3.9 | GO:0008180 | COP9 signalosome(GO:0008180) |
| 0.1 | 2.2 | GO:0016010 | dystrophin-associated glycoprotein complex(GO:0016010) glycoprotein complex(GO:0090665) |
| 0.1 | 1.3 | GO:0043240 | Fanconi anaemia nuclear complex(GO:0043240) |
| 0.1 | 1.2 | GO:0031209 | SCAR complex(GO:0031209) |
| 0.1 | 0.5 | GO:0008091 | spectrin(GO:0008091) |
| 0.1 | 3.1 | GO:0048786 | presynaptic active zone(GO:0048786) |
| 0.1 | 3.1 | GO:0005681 | spliceosomal complex(GO:0005681) |
| 0.1 | 0.4 | GO:0044194 | cytolytic granule(GO:0044194) |
| 0.1 | 3.1 | GO:0032281 | AMPA glutamate receptor complex(GO:0032281) |
| 0.1 | 2.8 | GO:0030131 | clathrin adaptor complex(GO:0030131) |
| 0.1 | 1.1 | GO:0032040 | small-subunit processome(GO:0032040) |
| 0.1 | 2.1 | GO:0032993 | protein-DNA complex(GO:0032993) |
| 0.1 | 7.3 | GO:0090575 | RNA polymerase II transcription factor complex(GO:0090575) |
| 0.1 | 13.7 | GO:0016604 | nuclear body(GO:0016604) |
| 0.1 | 9.7 | GO:0000794 | condensed nuclear chromosome(GO:0000794) |
| 0.1 | 4.3 | GO:0019005 | SCF ubiquitin ligase complex(GO:0019005) |
| 0.1 | 1.6 | GO:0097038 | perinuclear endoplasmic reticulum(GO:0097038) |
| 0.1 | 2.0 | GO:0005637 | nuclear inner membrane(GO:0005637) |
| 0.1 | 17.6 | GO:0014069 | postsynaptic density(GO:0014069) |
| 0.0 | 0.4 | GO:0008541 | proteasome regulatory particle, lid subcomplex(GO:0008541) |
| 0.0 | 10.4 | GO:0000228 | nuclear chromosome(GO:0000228) |
| 0.0 | 0.3 | GO:0071203 | WASH complex(GO:0071203) |
| 0.0 | 0.2 | GO:0033010 | paranodal junction(GO:0033010) |
| 0.0 | 0.2 | GO:0030868 | smooth endoplasmic reticulum membrane(GO:0030868) smooth endoplasmic reticulum part(GO:0097425) |
| 0.0 | 0.3 | GO:1990316 | ATG1/ULK1 kinase complex(GO:1990316) |
| 0.0 | 0.6 | GO:0005852 | eukaryotic translation initiation factor 3 complex(GO:0005852) |
| 0.0 | 0.3 | GO:0000408 | EKC/KEOPS complex(GO:0000408) |
| 0.0 | 0.2 | GO:0005849 | mRNA cleavage factor complex(GO:0005849) |
| 0.0 | 0.3 | GO:0010009 | cytoplasmic side of endosome membrane(GO:0010009) |
| 0.0 | 0.8 | GO:0005797 | Golgi medial cisterna(GO:0005797) |
| 0.0 | 2.2 | GO:0032420 | stereocilium(GO:0032420) |
| 0.0 | 0.8 | GO:1902711 | GABA-A receptor complex(GO:1902711) |
| 0.0 | 0.3 | GO:1904115 | axon cytoplasm(GO:1904115) |
| 0.0 | 1.7 | GO:0005834 | heterotrimeric G-protein complex(GO:0005834) |
| 0.0 | 0.3 | GO:0017146 | NMDA selective glutamate receptor complex(GO:0017146) |
| 0.0 | 0.0 | GO:0097632 | extrinsic component of pre-autophagosomal structure membrane(GO:0097632) |
| 0.0 | 0.9 | GO:0034707 | chloride channel complex(GO:0034707) |
| 0.0 | 0.2 | GO:0031254 | uropod(GO:0001931) cell trailing edge(GO:0031254) |
| 0.0 | 0.7 | GO:0034704 | calcium channel complex(GO:0034704) |
| 0.0 | 0.2 | GO:0044798 | nuclear transcription factor complex(GO:0044798) |
| 0.0 | 2.3 | GO:0031225 | anchored component of membrane(GO:0031225) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 4.1 | 16.3 | GO:0061505 | DNA topoisomerase type II (ATP-hydrolyzing) activity(GO:0003918) DNA topoisomerase II activity(GO:0061505) |
| 2.6 | 12.8 | GO:0017108 | 5'-flap endonuclease activity(GO:0017108) |
| 2.1 | 8.5 | GO:0019237 | centromeric DNA binding(GO:0019237) |
| 2.1 | 6.4 | GO:0045142 | triplex DNA binding(GO:0045142) |
| 2.0 | 16.3 | GO:0010521 | telomerase inhibitor activity(GO:0010521) |
| 2.0 | 14.2 | GO:0002135 | CTP binding(GO:0002135) |
| 1.8 | 5.5 | GO:0001716 | L-amino-acid oxidase activity(GO:0001716) |
| 1.7 | 17.3 | GO:0008140 | cAMP response element binding protein binding(GO:0008140) |
| 1.6 | 6.2 | GO:0033883 | pyridoxal phosphatase activity(GO:0033883) |
| 1.5 | 4.5 | GO:0035851 | Krueppel-associated box domain binding(GO:0035851) |
| 1.5 | 4.4 | GO:0010698 | acetyltransferase activator activity(GO:0010698) |
| 1.4 | 4.3 | GO:0019948 | SUMO activating enzyme activity(GO:0019948) |
| 1.4 | 4.2 | GO:0004067 | asparaginase activity(GO:0004067) |
| 1.4 | 4.1 | GO:0070138 | isopeptidase activity(GO:0070122) ubiquitin-like protein-specific isopeptidase activity(GO:0070138) SUMO-specific isopeptidase activity(GO:0070140) |
| 1.4 | 5.5 | GO:0001884 | pyrimidine nucleoside binding(GO:0001884) |
| 1.4 | 1.4 | GO:0034739 | histone deacetylase activity (H4-K16 specific)(GO:0034739) |
| 1.4 | 6.8 | GO:0071987 | WD40-repeat domain binding(GO:0071987) |
| 1.4 | 4.1 | GO:0004342 | glucosamine-6-phosphate deaminase activity(GO:0004342) |
| 1.3 | 3.8 | GO:0004572 | mannosyl-oligosaccharide 1,3-1,6-alpha-mannosidase activity(GO:0004572) |
| 1.2 | 8.2 | GO:0016972 | thiol oxidase activity(GO:0016972) |
| 1.2 | 7.0 | GO:0008545 | JUN kinase kinase activity(GO:0008545) |
| 1.1 | 4.5 | GO:0036313 | phosphatidylinositol 3-kinase catalytic subunit binding(GO:0036313) |
| 1.1 | 3.4 | GO:0003963 | RNA-3'-phosphate cyclase activity(GO:0003963) |
| 1.1 | 11.2 | GO:0003909 | DNA ligase activity(GO:0003909) DNA ligase (ATP) activity(GO:0003910) |
| 1.1 | 7.8 | GO:0051425 | PTB domain binding(GO:0051425) |
| 1.1 | 4.4 | GO:0070139 | ubiquitin-like protein-specific endopeptidase activity(GO:0070137) SUMO-specific endopeptidase activity(GO:0070139) |
| 1.1 | 2.2 | GO:0032138 | single base insertion or deletion binding(GO:0032138) |
| 1.1 | 4.2 | GO:0000992 | polymerase III regulatory region sequence-specific DNA binding(GO:0000992) RNA polymerase III type 1 promoter sequence-specific DNA binding(GO:0001002) RNA polymerase III type 2 promoter sequence-specific DNA binding(GO:0001003) |
| 1.0 | 7.9 | GO:0004416 | hydroxyacylglutathione hydrolase activity(GO:0004416) |
| 1.0 | 4.8 | GO:0004719 | protein-L-isoaspartate (D-aspartate) O-methyltransferase activity(GO:0004719) |
| 0.9 | 4.6 | GO:0005150 | interleukin-1, Type I receptor binding(GO:0005150) |
| 0.9 | 6.5 | GO:0032027 | myosin light chain binding(GO:0032027) |
| 0.9 | 6.4 | GO:0004689 | phosphorylase kinase activity(GO:0004689) |
| 0.9 | 2.7 | GO:0004677 | DNA-dependent protein kinase activity(GO:0004677) |
| 0.8 | 3.4 | GO:0032356 | oxidized DNA binding(GO:0032356) |
| 0.8 | 2.5 | GO:0004802 | transketolase activity(GO:0004802) |
| 0.8 | 2.5 | GO:0047598 | sterol delta7 reductase activity(GO:0009918) 7-dehydrocholesterol reductase activity(GO:0047598) |
| 0.8 | 5.8 | GO:0043515 | kinetochore binding(GO:0043515) |
| 0.8 | 3.2 | GO:0003998 | acylphosphatase activity(GO:0003998) |
| 0.8 | 4.0 | GO:0004668 | protein-arginine deiminase activity(GO:0004668) |
| 0.8 | 2.4 | GO:0001147 | transcription termination site sequence-specific DNA binding(GO:0001147) transcription termination site DNA binding(GO:0001160) |
| 0.8 | 2.4 | GO:0016426 | tRNA (adenine) methyltransferase activity(GO:0016426) |
| 0.8 | 6.3 | GO:0070883 | pre-miRNA binding(GO:0070883) |
| 0.7 | 4.5 | GO:0008568 | microtubule-severing ATPase activity(GO:0008568) |
| 0.7 | 0.7 | GO:0004828 | serine-tRNA ligase activity(GO:0004828) |
| 0.7 | 2.9 | GO:0003883 | CTP synthase activity(GO:0003883) |
| 0.7 | 2.2 | GO:0031798 | type 1 metabotropic glutamate receptor binding(GO:0031798) |
| 0.7 | 3.6 | GO:1990188 | euchromatin binding(GO:1990188) |
| 0.7 | 9.4 | GO:0001055 | RNA polymerase II activity(GO:0001055) |
| 0.7 | 7.3 | GO:0035374 | chondroitin sulfate binding(GO:0035374) |
| 0.7 | 1.4 | GO:0043423 | 3-phosphoinositide-dependent protein kinase binding(GO:0043423) |
| 0.7 | 3.6 | GO:0042903 | tubulin deacetylase activity(GO:0042903) |
| 0.7 | 3.6 | GO:0005483 | soluble NSF attachment protein activity(GO:0005483) |
| 0.7 | 2.1 | GO:0097100 | supercoiled DNA binding(GO:0097100) |
| 0.7 | 0.7 | GO:0009383 | rRNA (cytosine-C5-)-methyltransferase activity(GO:0009383) |
| 0.7 | 2.1 | GO:0031491 | nucleosome binding(GO:0031491) |
| 0.7 | 2.1 | GO:0097677 | STAT family protein binding(GO:0097677) |
| 0.7 | 23.3 | GO:0003887 | DNA-directed DNA polymerase activity(GO:0003887) |
| 0.7 | 0.7 | GO:0016886 | ligase activity, forming phosphoric ester bonds(GO:0016886) |
| 0.6 | 5.8 | GO:0055104 | ligase inhibitor activity(GO:0055104) ubiquitin ligase inhibitor activity(GO:1990948) |
| 0.6 | 3.8 | GO:1904047 | S-adenosyl-L-methionine binding(GO:1904047) |
| 0.6 | 3.8 | GO:0061575 | cyclin-dependent protein serine/threonine kinase activator activity(GO:0061575) |
| 0.6 | 3.7 | GO:0032184 | SUMO polymer binding(GO:0032184) |
| 0.6 | 1.9 | GO:0035614 | snRNA stem-loop binding(GO:0035614) |
| 0.6 | 4.3 | GO:0061665 | SUMO ligase activity(GO:0061665) |
| 0.6 | 2.4 | GO:0008073 | ornithine decarboxylase inhibitor activity(GO:0008073) |
| 0.6 | 0.6 | GO:0004741 | [pyruvate dehydrogenase (lipoamide)] phosphatase activity(GO:0004741) |
| 0.6 | 5.4 | GO:0001162 | RNA polymerase II intronic transcription regulatory region sequence-specific DNA binding(GO:0001162) |
| 0.6 | 4.2 | GO:0000298 | endopolyphosphatase activity(GO:0000298) diphosphoinositol-polyphosphate diphosphatase activity(GO:0008486) bis(5'-adenosyl)-hexaphosphatase activity(GO:0034431) bis(5'-adenosyl)-pentaphosphatase activity(GO:0034432) |
| 0.6 | 2.4 | GO:0071207 | histone pre-mRNA stem-loop binding(GO:0071207) |
| 0.6 | 3.5 | GO:0030942 | endoplasmic reticulum signal peptide binding(GO:0030942) |
| 0.6 | 4.0 | GO:0004169 | dolichyl-phosphate-mannose-protein mannosyltransferase activity(GO:0004169) |
| 0.6 | 3.4 | GO:0008821 | crossover junction endodeoxyribonuclease activity(GO:0008821) |
| 0.6 | 2.8 | GO:0010340 | carboxyl-O-methyltransferase activity(GO:0010340) protein carboxyl O-methyltransferase activity(GO:0051998) |
| 0.6 | 24.2 | GO:0017091 | AU-rich element binding(GO:0017091) |
| 0.6 | 2.2 | GO:0031727 | CCR2 chemokine receptor binding(GO:0031727) |
| 0.6 | 2.2 | GO:0004052 | arachidonate 12-lipoxygenase activity(GO:0004052) |
| 0.5 | 1.6 | GO:0031826 | type 2A serotonin receptor binding(GO:0031826) |
| 0.5 | 1.1 | GO:0070336 | flap-structured DNA binding(GO:0070336) |
| 0.5 | 3.7 | GO:0044547 | DNA topoisomerase binding(GO:0044547) |
| 0.5 | 2.7 | GO:0004724 | magnesium-dependent protein serine/threonine phosphatase activity(GO:0004724) |
| 0.5 | 3.2 | GO:0004095 | carnitine O-palmitoyltransferase activity(GO:0004095) O-palmitoyltransferase activity(GO:0016416) |
| 0.5 | 3.2 | GO:0033142 | progesterone receptor binding(GO:0033142) |
| 0.5 | 5.1 | GO:0000340 | RNA 7-methylguanosine cap binding(GO:0000340) |
| 0.5 | 2.6 | GO:0004370 | glycerol kinase activity(GO:0004370) |
| 0.5 | 5.0 | GO:1990189 | peptide-serine-N-acetyltransferase activity(GO:1990189) |
| 0.5 | 2.5 | GO:0016435 | rRNA (guanine) methyltransferase activity(GO:0016435) |
| 0.5 | 2.5 | GO:0030060 | L-malate dehydrogenase activity(GO:0030060) |
| 0.5 | 3.4 | GO:0035613 | RNA stem-loop binding(GO:0035613) |
| 0.5 | 1.5 | GO:0047192 | 1-alkylglycerophosphocholine O-acetyltransferase activity(GO:0047192) |
| 0.5 | 4.8 | GO:0042500 | aspartic endopeptidase activity, intramembrane cleaving(GO:0042500) |
| 0.5 | 1.4 | GO:0030943 | mitochondrion targeting sequence binding(GO:0030943) |
| 0.5 | 1.4 | GO:0061649 | ubiquitinated histone binding(GO:0061649) |
| 0.5 | 7.5 | GO:0051011 | microtubule minus-end binding(GO:0051011) |
| 0.5 | 1.9 | GO:0034511 | U3 snoRNA binding(GO:0034511) |
| 0.5 | 4.6 | GO:1990825 | sequence-specific mRNA binding(GO:1990825) |
| 0.5 | 1.4 | GO:0000171 | ribonuclease MRP activity(GO:0000171) |
| 0.5 | 1.9 | GO:0034648 | histone demethylase activity (H3-dimethyl-K4 specific)(GO:0034648) |
| 0.5 | 2.7 | GO:0098519 | nucleotide phosphatase activity, acting on free nucleotides(GO:0098519) |
| 0.5 | 3.7 | GO:0031493 | nucleosomal histone binding(GO:0031493) |
| 0.5 | 7.8 | GO:0001163 | RNA polymerase I regulatory region sequence-specific DNA binding(GO:0001163) RNA polymerase I CORE element sequence-specific DNA binding(GO:0001164) |
| 0.4 | 1.3 | GO:0016422 | mRNA (2'-O-methyladenosine-N6-)-methyltransferase activity(GO:0016422) |
| 0.4 | 10.7 | GO:0008253 | 5'-nucleotidase activity(GO:0008253) |
| 0.4 | 6.2 | GO:0009008 | DNA-methyltransferase activity(GO:0009008) |
| 0.4 | 3.0 | GO:0016933 | extracellular-glycine-gated ion channel activity(GO:0016933) extracellular-glycine-gated chloride channel activity(GO:0016934) |
| 0.4 | 1.7 | GO:0061628 | H3K27me3 modified histone binding(GO:0061628) |
| 0.4 | 32.2 | GO:0035064 | methylated histone binding(GO:0035064) |
| 0.4 | 12.2 | GO:0070840 | dynein complex binding(GO:0070840) |
| 0.4 | 11.8 | GO:0070530 | K63-linked polyubiquitin binding(GO:0070530) |
| 0.4 | 14.5 | GO:0005487 | nucleocytoplasmic transporter activity(GO:0005487) |
| 0.4 | 2.5 | GO:0004832 | valine-tRNA ligase activity(GO:0004832) |
| 0.4 | 1.7 | GO:0070740 | tubulin-glutamic acid ligase activity(GO:0070740) |
| 0.4 | 2.9 | GO:0015616 | DNA translocase activity(GO:0015616) |
| 0.4 | 15.7 | GO:0008009 | chemokine activity(GO:0008009) |
| 0.4 | 5.6 | GO:0043008 | ATP-dependent protein binding(GO:0043008) |
| 0.4 | 2.4 | GO:0033192 | calmodulin-dependent protein phosphatase activity(GO:0033192) |
| 0.4 | 2.8 | GO:0005087 | Ran guanyl-nucleotide exchange factor activity(GO:0005087) |
| 0.4 | 7.8 | GO:0015643 | toxic substance binding(GO:0015643) |
| 0.4 | 2.3 | GO:0048495 | Roundabout binding(GO:0048495) |
| 0.4 | 7.0 | GO:0015250 | water channel activity(GO:0015250) |
| 0.4 | 2.3 | GO:0052724 | inositol hexakisphosphate kinase activity(GO:0000828) inositol hexakisphosphate 5-kinase activity(GO:0000832) inositol hexakisphosphate 1-kinase activity(GO:0052723) inositol hexakisphosphate 3-kinase activity(GO:0052724) |
| 0.4 | 2.7 | GO:0000150 | recombinase activity(GO:0000150) |
| 0.4 | 3.4 | GO:0032795 | heterotrimeric G-protein binding(GO:0032795) |
| 0.4 | 4.2 | GO:0086080 | protein binding involved in heterotypic cell-cell adhesion(GO:0086080) |
| 0.4 | 1.5 | GO:0004534 | 5'-3' exoribonuclease activity(GO:0004534) |
| 0.4 | 1.5 | GO:0008176 | tRNA (guanine-N7-)-methyltransferase activity(GO:0008176) |
| 0.4 | 11.6 | GO:0008143 | poly(A) binding(GO:0008143) |
| 0.4 | 4.4 | GO:0001223 | transcription coactivator binding(GO:0001223) |
| 0.4 | 1.5 | GO:0098770 | FBXO family protein binding(GO:0098770) |
| 0.4 | 3.3 | GO:0097157 | pre-mRNA intronic binding(GO:0097157) |
| 0.4 | 16.6 | GO:0043015 | gamma-tubulin binding(GO:0043015) |
| 0.4 | 8.3 | GO:0004535 | poly(A)-specific ribonuclease activity(GO:0004535) |
| 0.4 | 1.8 | GO:0070012 | oligopeptidase activity(GO:0070012) |
| 0.4 | 18.0 | GO:0005164 | tumor necrosis factor receptor binding(GO:0005164) |
| 0.4 | 1.4 | GO:0004161 | dimethylallyltranstransferase activity(GO:0004161) geranyltranstransferase activity(GO:0004337) |
| 0.3 | 2.4 | GO:0030619 | U1 snRNA binding(GO:0030619) |
| 0.3 | 8.6 | GO:0005092 | GDP-dissociation inhibitor activity(GO:0005092) |
| 0.3 | 0.7 | GO:0098809 | nitrite reductase activity(GO:0098809) |
| 0.3 | 7.5 | GO:0036312 | phosphatidylinositol 3-kinase regulatory subunit binding(GO:0036312) |
| 0.3 | 1.0 | GO:0004771 | sterol esterase activity(GO:0004771) |
| 0.3 | 1.3 | GO:0004711 | ribosomal protein S6 kinase activity(GO:0004711) |
| 0.3 | 1.0 | GO:0046974 | histone methyltransferase activity (H3-K9 specific)(GO:0046974) |
| 0.3 | 1.0 | GO:0004723 | calcium-dependent protein serine/threonine phosphatase activity(GO:0004723) |
| 0.3 | 4.7 | GO:0004634 | phosphopyruvate hydratase activity(GO:0004634) |
| 0.3 | 8.0 | GO:0017049 | GTP-Rho binding(GO:0017049) |
| 0.3 | 3.6 | GO:0070181 | small ribosomal subunit rRNA binding(GO:0070181) |
| 0.3 | 1.3 | GO:1990460 | leptin receptor binding(GO:1990460) |
| 0.3 | 4.9 | GO:0008171 | O-methyltransferase activity(GO:0008171) |
| 0.3 | 4.2 | GO:0051864 | histone demethylase activity (H3-K36 specific)(GO:0051864) |
| 0.3 | 0.9 | GO:0004605 | phosphatidate cytidylyltransferase activity(GO:0004605) |
| 0.3 | 12.8 | GO:0005385 | zinc ion transmembrane transporter activity(GO:0005385) |
| 0.3 | 1.2 | GO:0003976 | UDP-N-acetylglucosamine-lysosomal-enzyme N-acetylglucosaminephosphotransferase activity(GO:0003976) |
| 0.3 | 2.2 | GO:0050733 | RS domain binding(GO:0050733) |
| 0.3 | 2.5 | GO:0000014 | single-stranded DNA endodeoxyribonuclease activity(GO:0000014) |
| 0.3 | 3.7 | GO:0008097 | 5S rRNA binding(GO:0008097) |
| 0.3 | 5.1 | GO:0009982 | pseudouridine synthase activity(GO:0009982) |
| 0.3 | 4.3 | GO:0005068 | transmembrane receptor protein tyrosine kinase adaptor activity(GO:0005068) |
| 0.3 | 2.0 | GO:0008158 | hedgehog receptor activity(GO:0008158) |
| 0.3 | 2.8 | GO:0045504 | dynein heavy chain binding(GO:0045504) |
| 0.3 | 4.5 | GO:0070403 | NAD+ binding(GO:0070403) |
| 0.3 | 1.1 | GO:0070728 | leucine binding(GO:0070728) |
| 0.3 | 2.0 | GO:0004849 | uridine kinase activity(GO:0004849) |
| 0.3 | 0.8 | GO:0031531 | thyrotropin-releasing hormone receptor binding(GO:0031531) |
| 0.3 | 29.1 | GO:0003697 | single-stranded DNA binding(GO:0003697) |
| 0.3 | 2.4 | GO:0016308 | 1-phosphatidylinositol-4-phosphate 5-kinase activity(GO:0016308) |
| 0.3 | 4.3 | GO:0003688 | DNA replication origin binding(GO:0003688) |
| 0.3 | 0.8 | GO:0004660 | protein farnesyltransferase activity(GO:0004660) |
| 0.3 | 0.8 | GO:0010484 | H3 histone acetyltransferase activity(GO:0010484) |
| 0.3 | 2.9 | GO:0033170 | DNA clamp loader activity(GO:0003689) protein-DNA loading ATPase activity(GO:0033170) |
| 0.3 | 6.8 | GO:0031681 | G-protein beta-subunit binding(GO:0031681) |
| 0.3 | 1.3 | GO:0015173 | aromatic amino acid transmembrane transporter activity(GO:0015173) |
| 0.3 | 2.8 | GO:0008409 | 5'-3' exonuclease activity(GO:0008409) |
| 0.3 | 2.8 | GO:0004565 | beta-galactosidase activity(GO:0004565) |
| 0.3 | 6.8 | GO:0005540 | hyaluronic acid binding(GO:0005540) |
| 0.2 | 5.2 | GO:0042162 | telomeric DNA binding(GO:0042162) |
| 0.2 | 2.0 | GO:0097200 | cysteine-type endopeptidase activity involved in execution phase of apoptosis(GO:0097200) |
| 0.2 | 2.2 | GO:0043996 | histone acetyltransferase activity (H4-K5 specific)(GO:0043995) histone acetyltransferase activity (H4-K8 specific)(GO:0043996) histone acetyltransferase activity (H4-K16 specific)(GO:0046972) |
| 0.2 | 5.4 | GO:0030296 | protein tyrosine kinase activator activity(GO:0030296) |
| 0.2 | 13.4 | GO:0097472 | cyclin-dependent protein kinase activity(GO:0097472) |
| 0.2 | 1.6 | GO:0043208 | glycosphingolipid binding(GO:0043208) |
| 0.2 | 2.8 | GO:0017151 | DEAD/H-box RNA helicase binding(GO:0017151) |
| 0.2 | 10.3 | GO:0030331 | estrogen receptor binding(GO:0030331) |
| 0.2 | 1.4 | GO:0010485 | H4 histone acetyltransferase activity(GO:0010485) |
| 0.2 | 1.6 | GO:0008294 | calcium- and calmodulin-responsive adenylate cyclase activity(GO:0008294) |
| 0.2 | 7.1 | GO:0031492 | nucleosomal DNA binding(GO:0031492) |
| 0.2 | 1.6 | GO:0031812 | G-protein coupled nucleotide receptor binding(GO:0031811) P2Y1 nucleotide receptor binding(GO:0031812) |
| 0.2 | 15.7 | GO:0003724 | RNA helicase activity(GO:0003724) ATP-dependent RNA helicase activity(GO:0004004) RNA-dependent ATPase activity(GO:0008186) |
| 0.2 | 1.8 | GO:0052629 | phosphatidylinositol-3,5-bisphosphate 3-phosphatase activity(GO:0052629) |
| 0.2 | 2.7 | GO:0004596 | peptide alpha-N-acetyltransferase activity(GO:0004596) |
| 0.2 | 22.9 | GO:0043130 | ubiquitin binding(GO:0043130) |
| 0.2 | 3.5 | GO:0016723 | oxidoreductase activity, oxidizing metal ions, NAD or NADP as acceptor(GO:0016723) |
| 0.2 | 13.6 | GO:0004712 | protein serine/threonine/tyrosine kinase activity(GO:0004712) |
| 0.2 | 0.9 | GO:0016433 | rRNA (adenine) methyltransferase activity(GO:0016433) |
| 0.2 | 1.7 | GO:0001190 | transcriptional activator activity, RNA polymerase II transcription factor binding(GO:0001190) transcriptional repressor activity, RNA polymerase II activating transcription factor binding(GO:0098811) |
| 0.2 | 3.6 | GO:0046332 | SMAD binding(GO:0046332) |
| 0.2 | 4.2 | GO:0003950 | NAD+ ADP-ribosyltransferase activity(GO:0003950) |
| 0.2 | 5.2 | GO:0034062 | DNA-directed RNA polymerase activity(GO:0003899) RNA polymerase activity(GO:0034062) |
| 0.2 | 3.3 | GO:0004143 | diacylglycerol kinase activity(GO:0004143) |
| 0.2 | 0.6 | GO:0004637 | phosphoribosylamine-glycine ligase activity(GO:0004637) |
| 0.2 | 0.4 | GO:0046403 | polynucleotide 3'-phosphatase activity(GO:0046403) |
| 0.2 | 5.5 | GO:0016538 | cyclin-dependent protein serine/threonine kinase regulator activity(GO:0016538) |
| 0.2 | 1.6 | GO:0070290 | N-acylphosphatidylethanolamine-specific phospholipase D activity(GO:0070290) |
| 0.2 | 0.4 | GO:0045236 | CXCR chemokine receptor binding(GO:0045236) |
| 0.2 | 1.7 | GO:0046976 | histone methyltransferase activity (H3-K27 specific)(GO:0046976) |
| 0.2 | 6.1 | GO:0008536 | Ran GTPase binding(GO:0008536) |
| 0.2 | 1.1 | GO:1990405 | protein antigen binding(GO:1990405) |
| 0.2 | 0.6 | GO:0004822 | isoleucine-tRNA ligase activity(GO:0004822) |
| 0.2 | 0.9 | GO:0043139 | 5'-3' DNA helicase activity(GO:0043139) |
| 0.2 | 1.7 | GO:0071535 | RING-like zinc finger domain binding(GO:0071535) |
| 0.2 | 2.2 | GO:0010857 | calcium-dependent protein kinase activity(GO:0010857) |
| 0.2 | 0.7 | GO:0017150 | tRNA dihydrouridine synthase activity(GO:0017150) |
| 0.2 | 1.8 | GO:0015386 | potassium:proton antiporter activity(GO:0015386) |
| 0.2 | 2.0 | GO:0004003 | ATP-dependent DNA helicase activity(GO:0004003) |
| 0.2 | 1.3 | GO:0031013 | troponin I binding(GO:0031013) |
| 0.2 | 13.7 | GO:0019894 | kinesin binding(GO:0019894) |
| 0.2 | 5.9 | GO:0019707 | protein-cysteine S-palmitoyltransferase activity(GO:0019706) protein-cysteine S-acyltransferase activity(GO:0019707) |
| 0.2 | 7.9 | GO:0008173 | RNA methyltransferase activity(GO:0008173) |
| 0.2 | 1.4 | GO:0004865 | protein serine/threonine phosphatase inhibitor activity(GO:0004865) |
| 0.2 | 1.2 | GO:0061676 | importin-alpha family protein binding(GO:0061676) |
| 0.2 | 0.7 | GO:0030627 | pre-mRNA 5'-splice site binding(GO:0030627) |
| 0.2 | 7.6 | GO:0001965 | G-protein alpha-subunit binding(GO:0001965) |
| 0.2 | 5.2 | GO:0004181 | metallocarboxypeptidase activity(GO:0004181) |
| 0.2 | 1.0 | GO:0000182 | rDNA binding(GO:0000182) |
| 0.2 | 1.1 | GO:0000990 | transcription factor activity, core RNA polymerase binding(GO:0000990) |
| 0.2 | 0.3 | GO:0045322 | unmethylated CpG binding(GO:0045322) |
| 0.2 | 1.6 | GO:0017162 | aryl hydrocarbon receptor binding(GO:0017162) |
| 0.2 | 0.6 | GO:0001222 | transcription corepressor binding(GO:0001222) |
| 0.2 | 1.4 | GO:0016929 | SUMO-specific protease activity(GO:0016929) |
| 0.2 | 3.9 | GO:0017160 | Ral GTPase binding(GO:0017160) |
| 0.2 | 3.9 | GO:0031434 | mitogen-activated protein kinase kinase binding(GO:0031434) |
| 0.2 | 0.9 | GO:0010997 | anaphase-promoting complex binding(GO:0010997) |
| 0.1 | 2.7 | GO:0035612 | AP-2 adaptor complex binding(GO:0035612) |
| 0.1 | 1.6 | GO:0004117 | calmodulin-dependent cyclic-nucleotide phosphodiesterase activity(GO:0004117) calcium- and calmodulin-regulated 3',5'-cyclic-GMP phosphodiesterase activity(GO:0048101) |
| 0.1 | 1.0 | GO:0016861 | intramolecular oxidoreductase activity, interconverting aldoses and ketoses(GO:0016861) |
| 0.1 | 7.1 | GO:0070063 | RNA polymerase binding(GO:0070063) |
| 0.1 | 0.6 | GO:0045505 | dynein intermediate chain binding(GO:0045505) |
| 0.1 | 0.9 | GO:0004826 | phenylalanine-tRNA ligase activity(GO:0004826) |
| 0.1 | 3.0 | GO:0070577 | lysine-acetylated histone binding(GO:0070577) |
| 0.1 | 4.1 | GO:0034237 | protein kinase A regulatory subunit binding(GO:0034237) |
| 0.1 | 5.3 | GO:0030742 | GTP-dependent protein binding(GO:0030742) |
| 0.1 | 5.5 | GO:0051019 | mitogen-activated protein kinase binding(GO:0051019) |
| 0.1 | 2.2 | GO:1990226 | histone methyltransferase binding(GO:1990226) |
| 0.1 | 2.6 | GO:0042166 | acetylcholine binding(GO:0042166) |
| 0.1 | 1.2 | GO:0015181 | arginine transmembrane transporter activity(GO:0015181) |
| 0.1 | 2.7 | GO:0061733 | peptide-lysine-N-acetyltransferase activity(GO:0061733) |
| 0.1 | 4.0 | GO:0017128 | phospholipid scramblase activity(GO:0017128) |
| 0.1 | 0.5 | GO:0035241 | protein-arginine omega-N monomethyltransferase activity(GO:0035241) histone methyltransferase activity (H4-R3 specific)(GO:0044020) |
| 0.1 | 1.4 | GO:0009881 | photoreceptor activity(GO:0009881) |
| 0.1 | 8.6 | GO:0008276 | protein methyltransferase activity(GO:0008276) |
| 0.1 | 6.6 | GO:0035035 | histone acetyltransferase binding(GO:0035035) |
| 0.1 | 3.0 | GO:0004629 | phosphatidylinositol phospholipase C activity(GO:0004435) phospholipase C activity(GO:0004629) |
| 0.1 | 0.4 | GO:0019784 | NEDD8-specific protease activity(GO:0019784) |
| 0.1 | 11.2 | GO:0042393 | histone binding(GO:0042393) |
| 0.1 | 35.4 | GO:0008017 | microtubule binding(GO:0008017) |
| 0.1 | 1.6 | GO:0015194 | L-serine transmembrane transporter activity(GO:0015194) serine transmembrane transporter activity(GO:0022889) |
| 0.1 | 0.5 | GO:0004574 | oligo-1,6-glucosidase activity(GO:0004574) |
| 0.1 | 4.3 | GO:0000983 | transcription factor activity, RNA polymerase II core promoter sequence-specific(GO:0000983) |
| 0.1 | 3.5 | GO:0043395 | heparan sulfate proteoglycan binding(GO:0043395) |
| 0.1 | 1.7 | GO:0032794 | GTPase activating protein binding(GO:0032794) |
| 0.1 | 3.2 | GO:0052866 | phosphatidylinositol phosphate phosphatase activity(GO:0052866) |
| 0.1 | 0.2 | GO:0008934 | inositol monophosphate 1-phosphatase activity(GO:0008934) inositol monophosphate 3-phosphatase activity(GO:0052832) inositol monophosphate 4-phosphatase activity(GO:0052833) inositol monophosphate phosphatase activity(GO:0052834) |
| 0.1 | 0.4 | GO:0008263 | pyrimidine-specific mismatch base pair DNA N-glycosylase activity(GO:0008263) |
| 0.1 | 0.6 | GO:0070016 | armadillo repeat domain binding(GO:0070016) |
| 0.1 | 0.2 | GO:0005237 | inhibitory extracellular ligand-gated ion channel activity(GO:0005237) |
| 0.1 | 2.4 | GO:0022841 | potassium ion leak channel activity(GO:0022841) |
| 0.1 | 0.4 | GO:0004066 | asparagine synthase (glutamine-hydrolyzing) activity(GO:0004066) |
| 0.1 | 2.5 | GO:0035259 | glucocorticoid receptor binding(GO:0035259) |
| 0.1 | 1.9 | GO:0004089 | carbonate dehydratase activity(GO:0004089) |
| 0.1 | 0.3 | GO:0000033 | alpha-1,3-mannosyltransferase activity(GO:0000033) |
| 0.1 | 2.9 | GO:0048487 | beta-tubulin binding(GO:0048487) |
| 0.1 | 0.8 | GO:0004366 | glycerol-3-phosphate O-acyltransferase activity(GO:0004366) |
| 0.1 | 0.3 | GO:0061711 | N(6)-L-threonylcarbamoyladenine synthase(GO:0061711) |
| 0.1 | 0.7 | GO:0030628 | pre-mRNA 3'-splice site binding(GO:0030628) |
| 0.1 | 1.0 | GO:0030306 | ADP-ribosylation factor binding(GO:0030306) |
| 0.1 | 3.3 | GO:0046961 | proton-transporting ATPase activity, rotational mechanism(GO:0046961) |
| 0.1 | 1.5 | GO:0005522 | profilin binding(GO:0005522) |
| 0.1 | 1.0 | GO:0015252 | hydrogen ion channel activity(GO:0015252) |
| 0.1 | 1.9 | GO:0005161 | platelet-derived growth factor receptor binding(GO:0005161) |
| 0.1 | 4.3 | GO:0004112 | cyclic-nucleotide phosphodiesterase activity(GO:0004112) |
| 0.1 | 0.9 | GO:0008494 | translation activator activity(GO:0008494) |
| 0.1 | 0.8 | GO:0008499 | UDP-galactose:beta-N-acetylglucosamine beta-1,3-galactosyltransferase activity(GO:0008499) |
| 0.1 | 0.8 | GO:0071933 | Arp2/3 complex binding(GO:0071933) |
| 0.1 | 0.7 | GO:0016742 | hydroxymethyl-, formyl- and related transferase activity(GO:0016742) |
| 0.1 | 0.6 | GO:0022851 | GABA-gated chloride ion channel activity(GO:0022851) |
| 0.1 | 0.5 | GO:0036402 | proteasome-activating ATPase activity(GO:0036402) |
| 0.1 | 2.4 | GO:1900750 | glutathione binding(GO:0043295) oligopeptide binding(GO:1900750) |
| 0.1 | 3.6 | GO:0035255 | ionotropic glutamate receptor binding(GO:0035255) |
| 0.1 | 1.5 | GO:0008484 | sulfuric ester hydrolase activity(GO:0008484) |
| 0.1 | 1.6 | GO:0070410 | co-SMAD binding(GO:0070410) |
| 0.1 | 6.4 | GO:0004843 | thiol-dependent ubiquitin-specific protease activity(GO:0004843) |
| 0.1 | 1.0 | GO:0004386 | helicase activity(GO:0004386) |
| 0.1 | 1.4 | GO:0003684 | damaged DNA binding(GO:0003684) |
| 0.1 | 0.3 | GO:0031208 | POZ domain binding(GO:0031208) |
| 0.1 | 0.5 | GO:0033188 | sphingomyelin synthase activity(GO:0033188) ceramide cholinephosphotransferase activity(GO:0047493) |
| 0.1 | 1.2 | GO:0042165 | neurotransmitter binding(GO:0042165) |
| 0.1 | 0.4 | GO:0102345 | 3-hydroxy-behenoyl-CoA dehydratase activity(GO:0102344) 3-hydroxy-lignoceroyl-CoA dehydratase activity(GO:0102345) |
| 0.1 | 1.0 | GO:0017154 | semaphorin receptor activity(GO:0017154) |
| 0.1 | 0.5 | GO:0001515 | opioid peptide activity(GO:0001515) |
| 0.1 | 18.2 | GO:0003714 | transcription corepressor activity(GO:0003714) |
| 0.1 | 1.2 | GO:0000030 | mannosyltransferase activity(GO:0000030) |
| 0.1 | 1.4 | GO:0019783 | ubiquitin-like protein-specific protease activity(GO:0019783) |
| 0.1 | 4.3 | GO:0008138 | protein tyrosine/serine/threonine phosphatase activity(GO:0008138) |
| 0.1 | 1.0 | GO:0004954 | prostanoid receptor activity(GO:0004954) |
| 0.1 | 1.0 | GO:0001104 | RNA polymerase II transcription cofactor activity(GO:0001104) |
| 0.1 | 0.6 | GO:1990446 | U1 snRNP binding(GO:1990446) |
| 0.1 | 43.2 | GO:0004842 | ubiquitin-protein transferase activity(GO:0004842) |
| 0.1 | 1.2 | GO:0033558 | histone deacetylase activity(GO:0004407) protein deacetylase activity(GO:0033558) |
| 0.1 | 1.6 | GO:0042826 | histone deacetylase binding(GO:0042826) |
| 0.1 | 0.7 | GO:0031419 | cobalamin binding(GO:0031419) |
| 0.1 | 0.5 | GO:0002161 | aminoacyl-tRNA editing activity(GO:0002161) |
| 0.1 | 1.5 | GO:0017025 | TBP-class protein binding(GO:0017025) |
| 0.1 | 1.3 | GO:0016290 | palmitoyl-CoA hydrolase activity(GO:0016290) |
| 0.1 | 0.3 | GO:0004704 | NF-kappaB-inducing kinase activity(GO:0004704) |
| 0.1 | 0.2 | GO:0051959 | dynein light intermediate chain binding(GO:0051959) |
| 0.1 | 1.4 | GO:0017075 | syntaxin-1 binding(GO:0017075) |
| 0.1 | 21.3 | GO:0005096 | GTPase activator activity(GO:0005096) |
| 0.1 | 2.5 | GO:0005086 | ARF guanyl-nucleotide exchange factor activity(GO:0005086) |
| 0.1 | 1.2 | GO:0097602 | cullin family protein binding(GO:0097602) |
| 0.1 | 0.6 | GO:0001640 | adenylate cyclase inhibiting G-protein coupled glutamate receptor activity(GO:0001640) |
| 0.1 | 1.3 | GO:0019825 | oxygen binding(GO:0019825) |
| 0.1 | 1.2 | GO:0004190 | aspartic-type endopeptidase activity(GO:0004190) |
| 0.1 | 0.6 | GO:0042975 | peroxisome proliferator activated receptor binding(GO:0042975) |
| 0.1 | 3.2 | GO:0001540 | beta-amyloid binding(GO:0001540) |
| 0.1 | 0.6 | GO:0004332 | fructose-bisphosphate aldolase activity(GO:0004332) |
| 0.1 | 4.3 | GO:0008234 | cysteine-type peptidase activity(GO:0008234) |
| 0.1 | 0.2 | GO:0051734 | ATP-dependent polydeoxyribonucleotide 5'-hydroxyl-kinase activity(GO:0046404) polydeoxyribonucleotide kinase activity(GO:0051733) ATP-dependent polynucleotide kinase activity(GO:0051734) |
| 0.1 | 1.7 | GO:0043539 | protein serine/threonine kinase activator activity(GO:0043539) |
| 0.1 | 0.5 | GO:0001665 | alpha-N-acetylgalactosaminide alpha-2,6-sialyltransferase activity(GO:0001665) |
| 0.1 | 3.6 | GO:0003682 | chromatin binding(GO:0003682) |
| 0.1 | 10.5 | GO:0001078 | transcriptional repressor activity, RNA polymerase II core promoter proximal region sequence-specific binding(GO:0001078) |
| 0.1 | 7.0 | GO:0005262 | calcium channel activity(GO:0005262) |
| 0.1 | 0.3 | GO:0031748 | D1 dopamine receptor binding(GO:0031748) |
| 0.0 | 1.7 | GO:0004012 | phospholipid-translocating ATPase activity(GO:0004012) |
| 0.0 | 0.9 | GO:0008430 | selenium binding(GO:0008430) |
| 0.0 | 0.9 | GO:0070300 | phosphatidic acid binding(GO:0070300) |
| 0.0 | 0.2 | GO:1990247 | N6-methyladenosine-containing RNA binding(GO:1990247) |
| 0.0 | 1.4 | GO:0005184 | neuropeptide hormone activity(GO:0005184) |
| 0.0 | 0.3 | GO:0035252 | UDP-xylosyltransferase activity(GO:0035252) |
| 0.0 | 0.7 | GO:0043522 | leucine zipper domain binding(GO:0043522) |
| 0.0 | 0.2 | GO:0047696 | beta-adrenergic receptor kinase activity(GO:0047696) |
| 0.0 | 82.3 | GO:0003677 | DNA binding(GO:0003677) |
| 0.0 | 2.2 | GO:0051393 | alpha-actinin binding(GO:0051393) |
| 0.0 | 1.3 | GO:0000049 | tRNA binding(GO:0000049) |
| 0.0 | 3.1 | GO:0035591 | signaling adaptor activity(GO:0035591) |
| 0.0 | 1.0 | GO:0019209 | kinase activator activity(GO:0019209) |
| 0.0 | 6.2 | GO:0004721 | phosphoprotein phosphatase activity(GO:0004721) |
| 0.0 | 0.1 | GO:0019797 | procollagen-proline 3-dioxygenase activity(GO:0019797) |
| 0.0 | 16.2 | GO:0004674 | protein serine/threonine kinase activity(GO:0004674) |
| 0.0 | 0.3 | GO:0047288 | monosialoganglioside sialyltransferase activity(GO:0047288) |
| 0.0 | 1.4 | GO:0003777 | microtubule motor activity(GO:0003777) |
| 0.0 | 0.7 | GO:0010181 | FMN binding(GO:0010181) |
| 0.0 | 0.1 | GO:0047391 | alkylglycerophosphoethanolamine phosphodiesterase activity(GO:0047391) |
| 0.0 | 0.7 | GO:0030374 | ligand-dependent nuclear receptor transcription coactivator activity(GO:0030374) |
| 0.0 | 0.5 | GO:0023026 | MHC class II protein complex binding(GO:0023026) |
| 0.0 | 1.2 | GO:0005104 | fibroblast growth factor receptor binding(GO:0005104) |
| 0.0 | 0.9 | GO:0031369 | translation initiation factor binding(GO:0031369) |
| 0.0 | 0.2 | GO:0043023 | ribosomal large subunit binding(GO:0043023) |
| 0.0 | 0.9 | GO:0004527 | exonuclease activity(GO:0004527) |
| 0.0 | 0.8 | GO:0050431 | transforming growth factor beta binding(GO:0050431) |
| 0.0 | 0.6 | GO:0005545 | 1-phosphatidylinositol binding(GO:0005545) |
| 0.0 | 2.7 | GO:0030674 | protein binding, bridging(GO:0030674) |
| 0.0 | 1.0 | GO:0008168 | methyltransferase activity(GO:0008168) |
| 0.0 | 1.1 | GO:0043531 | ADP binding(GO:0043531) |
| 0.0 | 0.3 | GO:0008641 | small protein activating enzyme activity(GO:0008641) |
| 0.0 | 5.7 | GO:0003700 | nucleic acid binding transcription factor activity(GO:0001071) transcription factor activity, sequence-specific DNA binding(GO:0003700) |
| 0.0 | 0.2 | GO:0033613 | activating transcription factor binding(GO:0033613) |
| 0.0 | 0.2 | GO:0015165 | pyrimidine nucleotide-sugar transmembrane transporter activity(GO:0015165) |
| 0.0 | 0.2 | GO:0019855 | calcium channel inhibitor activity(GO:0019855) |
| 0.0 | 0.1 | GO:0036442 | hydrogen-exporting ATPase activity(GO:0036442) |
| 0.0 | 6.2 | GO:0008270 | zinc ion binding(GO:0008270) |
| 0.0 | 0.2 | GO:0017056 | structural constituent of nuclear pore(GO:0017056) |
| 0.0 | 0.5 | GO:0030506 | ankyrin binding(GO:0030506) |
| 0.0 | 3.2 | GO:0008237 | metallopeptidase activity(GO:0008237) |
| 0.0 | 0.1 | GO:0070628 | proteasome binding(GO:0070628) |
| 0.0 | 0.1 | GO:0047710 | bis(5'-adenosyl)-triphosphatase activity(GO:0047710) |
| 0.0 | 0.0 | GO:0008107 | galactoside 2-alpha-L-fucosyltransferase activity(GO:0008107) alpha-(1,2)-fucosyltransferase activity(GO:0031127) |
| 0.0 | 0.6 | GO:0032947 | protein complex scaffold(GO:0032947) |
| 0.0 | 0.2 | GO:0004862 | cAMP-dependent protein kinase inhibitor activity(GO:0004862) |
| 0.0 | 1.4 | GO:0044325 | ion channel binding(GO:0044325) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.8 | 6.8 | SA G1 AND S PHASES | Cdk2, 4, and 6 bind cyclin D in G1, while cdk2/cyclin E promotes the G1/S transition. |
| 0.6 | 10.1 | SA REG CASCADE OF CYCLIN EXPR | Expression of cyclins regulates progression through the cell cycle by activating cyclin-dependent kinases. |
| 0.6 | 14.7 | PID HIF1A PATHWAY | Hypoxic and oxygen homeostasis regulation of HIF-1-alpha |
| 0.5 | 1.6 | PID NFKAPPAB ATYPICAL PATHWAY | Atypical NF-kappaB pathway |
| 0.5 | 4.0 | PID INTEGRIN A4B1 PATHWAY | Alpha4 beta1 integrin signaling events |
| 0.5 | 12.7 | PID DNA PK PATHWAY | DNA-PK pathway in nonhomologous end joining |
| 0.4 | 6.0 | ST JAK STAT PATHWAY | Jak-STAT Pathway |
| 0.4 | 23.3 | PID AURORA B PATHWAY | Aurora B signaling |
| 0.4 | 7.4 | PID ERB GENOMIC PATHWAY | Validated nuclear estrogen receptor beta network |
| 0.3 | 2.3 | ST INTERFERON GAMMA PATHWAY | Interferon gamma pathway. |
| 0.3 | 8.0 | PID ATM PATHWAY | ATM pathway |
| 0.3 | 9.4 | PID HDAC CLASSI PATHWAY | Signaling events mediated by HDAC Class I |
| 0.3 | 1.4 | PID AVB3 INTEGRIN PATHWAY | Integrins in angiogenesis |
| 0.2 | 8.7 | PID BARD1 PATHWAY | BARD1 signaling events |
| 0.2 | 9.7 | ST GRANULE CELL SURVIVAL PATHWAY | Granule Cell Survival Pathway is a specific case of more general PAC1 Receptor Pathway. |
| 0.2 | 14.4 | PID HES HEY PATHWAY | Notch-mediated HES/HEY network |
| 0.2 | 11.8 | PID PLK1 PATHWAY | PLK1 signaling events |
| 0.2 | 9.2 | PID EPHA FWDPATHWAY | EPHA forward signaling |
| 0.2 | 9.4 | PID TCR CALCIUM PATHWAY | Calcium signaling in the CD4+ TCR pathway |
| 0.2 | 6.0 | PID ATR PATHWAY | ATR signaling pathway |
| 0.2 | 6.5 | PID S1P S1P3 PATHWAY | S1P3 pathway |
| 0.2 | 2.4 | PID FAK PATHWAY | Signaling events mediated by focal adhesion kinase |
| 0.2 | 7.4 | PID HDAC CLASSII PATHWAY | Signaling events mediated by HDAC Class II |
| 0.2 | 10.8 | PID BCR 5PATHWAY | BCR signaling pathway |
| 0.2 | 12.0 | PID AP1 PATHWAY | AP-1 transcription factor network |
| 0.2 | 9.3 | PID INSULIN PATHWAY | Insulin Pathway |
| 0.2 | 4.0 | ST P38 MAPK PATHWAY | p38 MAPK Pathway |
| 0.1 | 4.1 | ST WNT CA2 CYCLIC GMP PATHWAY | Wnt/Ca2+/cyclic GMP signaling. |
| 0.1 | 3.5 | PID ARF 3PATHWAY | Arf1 pathway |
| 0.1 | 3.7 | PID P38 MKK3 6PATHWAY | p38 MAPK signaling pathway |
| 0.1 | 8.2 | PID P73PATHWAY | p73 transcription factor network |
| 0.1 | 2.5 | PID LPA4 PATHWAY | LPA4-mediated signaling events |
| 0.1 | 13.5 | PID AR PATHWAY | Coregulation of Androgen receptor activity |
| 0.1 | 1.7 | PID FANCONI PATHWAY | Fanconi anemia pathway |
| 0.1 | 12.2 | PID TRKR PATHWAY | Neurotrophic factor-mediated Trk receptor signaling |
| 0.1 | 7.7 | PID ILK PATHWAY | Integrin-linked kinase signaling |
| 0.1 | 7.3 | PID RB 1PATHWAY | Regulation of retinoblastoma protein |
| 0.1 | 2.8 | PID TELOMERASE PATHWAY | Regulation of Telomerase |
| 0.1 | 0.6 | ST TYPE I INTERFERON PATHWAY | Type I Interferon (alpha/beta IFN) Pathway |
| 0.1 | 0.7 | ST G ALPHA I PATHWAY | G alpha i Pathway |
| 0.1 | 1.7 | PID MAPK TRK PATHWAY | Trk receptor signaling mediated by the MAPK pathway |
| 0.1 | 2.9 | PID LIS1 PATHWAY | Lissencephaly gene (LIS1) in neuronal migration and development |
| 0.1 | 1.8 | PID MYC PATHWAY | C-MYC pathway |
| 0.1 | 2.4 | PID CD40 PATHWAY | CD40/CD40L signaling |
| 0.1 | 1.2 | PID RAC1 PATHWAY | RAC1 signaling pathway |
| 0.1 | 0.9 | PID IFNG PATHWAY | IFN-gamma pathway |
| 0.1 | 1.5 | PID P38 ALPHA BETA DOWNSTREAM PATHWAY | Signaling mediated by p38-alpha and p38-beta |
| 0.1 | 1.6 | ST MYOCYTE AD PATHWAY | Myocyte Adrenergic Pathway is a specific case of the generalized Adrenergic Pathway. |
| 0.1 | 1.1 | PID P38 MK2 PATHWAY | p38 signaling mediated by MAPKAP kinases |
| 0.1 | 2.3 | PID IL4 2PATHWAY | IL4-mediated signaling events |
| 0.0 | 0.5 | PID ATF2 PATHWAY | ATF-2 transcription factor network |
| 0.0 | 2.2 | PID E2F PATHWAY | E2F transcription factor network |
| 0.0 | 3.0 | PID BETA CATENIN NUC PATHWAY | Regulation of nuclear beta catenin signaling and target gene transcription |
| 0.0 | 0.6 | PID HDAC CLASSIII PATHWAY | Signaling events mediated by HDAC Class III |
| 0.0 | 1.1 | PID PI3KCI PATHWAY | Class I PI3K signaling events |
| 0.0 | 1.1 | PID INTEGRIN A9B1 PATHWAY | Alpha9 beta1 integrin signaling events |
| 0.0 | 1.0 | PID NCADHERIN PATHWAY | N-cadherin signaling events |
| 0.0 | 1.5 | PID P53 REGULATION PATHWAY | p53 pathway |
| 0.0 | 0.4 | ST GAQ PATHWAY | G alpha q Pathway |
| 0.0 | 5.5 | NABA ECM AFFILIATED | Genes encoding proteins affiliated structurally or functionally to extracellular matrix proteins |
| 0.0 | 0.5 | PID TXA2PATHWAY | Thromboxane A2 receptor signaling |
| 0.0 | 0.3 | PID CD8 TCR DOWNSTREAM PATHWAY | Downstream signaling in naïve CD8+ T cells |
| 0.0 | 0.6 | PID AJDISS 2PATHWAY | Posttranslational regulation of adherens junction stability and dissassembly |
| 0.0 | 0.6 | PID HIF1 TFPATHWAY | HIF-1-alpha transcription factor network |
| 0.0 | 0.2 | PID MET PATHWAY | Signaling events mediated by Hepatocyte Growth Factor Receptor (c-Met) |
| 0.0 | 0.2 | PID ERBB4 PATHWAY | ErbB4 signaling events |
| 0.0 | 0.2 | PID IL2 PI3K PATHWAY | IL2 signaling events mediated by PI3K |
| 0.0 | 0.3 | PID HIV NEF PATHWAY | HIV-1 Nef: Negative effector of Fas and TNF-alpha |
| 0.0 | 0.5 | PID WNT SIGNALING PATHWAY | Wnt signaling network |
| 0.0 | 0.1 | PID WNT CANONICAL PATHWAY | Canonical Wnt signaling pathway |
| 0.0 | 0.1 | ST DIFFERENTIATION PATHWAY IN PC12 CELLS | Differentiation Pathway in PC12 Cells; this is a specific case of PAC1 Receptor Pathway. |
| 0.0 | 3.6 | NABA MATRISOME ASSOCIATED | Ensemble of genes encoding ECM-associated proteins including ECM-affilaited proteins, ECM regulators and secreted factors |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.6 | 6.5 | REACTOME BINDING AND ENTRY OF HIV VIRION | Genes involved in Binding and entry of HIV virion |
| 1.3 | 1.3 | REACTOME REMOVAL OF THE FLAP INTERMEDIATE FROM THE C STRAND | Genes involved in Removal of the Flap Intermediate from the C-strand |
| 1.2 | 1.2 | REACTOME E2F MEDIATED REGULATION OF DNA REPLICATION | Genes involved in E2F mediated regulation of DNA replication |
| 0.9 | 6.3 | REACTOME G1 S SPECIFIC TRANSCRIPTION | Genes involved in G1/S-Specific Transcription |
| 0.9 | 16.0 | REACTOME HYALURONAN METABOLISM | Genes involved in Hyaluronan metabolism |
| 0.9 | 42.2 | REACTOME TRANSPORT OF MATURE MRNA DERIVED FROM AN INTRONLESS TRANSCRIPT | Genes involved in Transport of Mature mRNA Derived from an Intronless Transcript |
| 0.9 | 2.6 | REACTOME E2F ENABLED INHIBITION OF PRE REPLICATION COMPLEX FORMATION | Genes involved in E2F-enabled inhibition of pre-replication complex formation |
| 0.9 | 15.3 | REACTOME DOUBLE STRAND BREAK REPAIR | Genes involved in Double-Strand Break Repair |
| 0.8 | 1.6 | REACTOME VIRAL MESSENGER RNA SYNTHESIS | Genes involved in Viral Messenger RNA Synthesis |
| 0.8 | 32.0 | REACTOME ACTIVATION OF THE PRE REPLICATIVE COMPLEX | Genes involved in Activation of the pre-replicative complex |
| 0.7 | 0.7 | REACTOME SCFSKP2 MEDIATED DEGRADATION OF P27 P21 | Genes involved in SCF(Skp2)-mediated degradation of p27/p21 |
| 0.7 | 16.9 | REACTOME INHIBITION OF THE PROTEOLYTIC ACTIVITY OF APC C REQUIRED FOR THE ONSET OF ANAPHASE BY MITOTIC SPINDLE CHECKPOINT COMPONENTS | Genes involved in Inhibition of the proteolytic activity of APC/C required for the onset of anaphase by mitotic spindle checkpoint components |
| 0.7 | 23.9 | REACTOME MEIOTIC RECOMBINATION | Genes involved in Meiotic Recombination |
| 0.7 | 19.1 | REACTOME SEMA3A PAK DEPENDENT AXON REPULSION | Genes involved in Sema3A PAK dependent Axon repulsion |
| 0.6 | 5.1 | REACTOME FORMATION OF TRANSCRIPTION COUPLED NER TC NER REPAIR COMPLEX | Genes involved in Formation of transcription-coupled NER (TC-NER) repair complex |
| 0.6 | 16.9 | REACTOME G0 AND EARLY G1 | Genes involved in G0 and Early G1 |
| 0.6 | 16.7 | REACTOME MICRORNA MIRNA BIOGENESIS | Genes involved in MicroRNA (miRNA) Biogenesis |
| 0.6 | 2.9 | REACTOME P53 DEPENDENT G1 DNA DAMAGE RESPONSE | Genes involved in p53-Dependent G1 DNA Damage Response |
| 0.6 | 6.3 | REACTOME MRNA DECAY BY 3 TO 5 EXORIBONUCLEASE | Genes involved in mRNA Decay by 3' to 5' Exoribonuclease |
| 0.5 | 14.6 | REACTOME ZINC TRANSPORTERS | Genes involved in Zinc transporters |
| 0.5 | 8.9 | REACTOME IL 6 SIGNALING | Genes involved in Interleukin-6 signaling |
| 0.5 | 41.8 | REACTOME LOSS OF NLP FROM MITOTIC CENTROSOMES | Genes involved in Loss of Nlp from mitotic centrosomes |
| 0.5 | 10.2 | REACTOME DEPOSITION OF NEW CENPA CONTAINING NUCLEOSOMES AT THE CENTROMERE | Genes involved in Deposition of New CENPA-containing Nucleosomes at the Centromere |
| 0.5 | 2.7 | REACTOME RNA POL III TRANSCRIPTION TERMINATION | Genes involved in RNA Polymerase III Transcription Termination |
| 0.4 | 10.0 | REACTOME RNA POL III TRANSCRIPTION INITIATION FROM TYPE 3 PROMOTER | Genes involved in RNA Polymerase III Transcription Initiation From Type 3 Promoter |
| 0.4 | 5.8 | REACTOME RESOLUTION OF AP SITES VIA THE SINGLE NUCLEOTIDE REPLACEMENT PATHWAY | Genes involved in Resolution of AP sites via the single-nucleotide replacement pathway |
| 0.4 | 3.3 | REACTOME CYCLIN A B1 ASSOCIATED EVENTS DURING G2 M TRANSITION | Genes involved in Cyclin A/B1 associated events during G2/M transition |
| 0.4 | 12.9 | REACTOME RNA POL II TRANSCRIPTION PRE INITIATION AND PROMOTER OPENING | Genes involved in RNA Polymerase II Transcription Pre-Initiation And Promoter Opening |
| 0.4 | 2.8 | REACTOME FORMATION OF INCISION COMPLEX IN GG NER | Genes involved in Formation of incision complex in GG-NER |
| 0.4 | 12.5 | REACTOME KINESINS | Genes involved in Kinesins |
| 0.4 | 3.2 | REACTOME THE ACTIVATION OF ARYLSULFATASES | Genes involved in The activation of arylsulfatases |
| 0.3 | 2.4 | REACTOME PACKAGING OF TELOMERE ENDS | Genes involved in Packaging Of Telomere Ends |
| 0.3 | 9.2 | REACTOME PROLACTIN RECEPTOR SIGNALING | Genes involved in Prolactin receptor signaling |
| 0.3 | 7.1 | REACTOME MITOCHONDRIAL TRNA AMINOACYLATION | Genes involved in Mitochondrial tRNA aminoacylation |
| 0.3 | 6.3 | REACTOME GLYCOGEN BREAKDOWN GLYCOGENOLYSIS | Genes involved in Glycogen breakdown (glycogenolysis) |
| 0.3 | 22.4 | REACTOME MITOTIC PROMETAPHASE | Genes involved in Mitotic Prometaphase |
| 0.3 | 3.9 | REACTOME RECRUITMENT OF MITOTIC CENTROSOME PROTEINS AND COMPLEXES | Genes involved in Recruitment of mitotic centrosome proteins and complexes |
| 0.3 | 7.6 | REACTOME JNK C JUN KINASES PHOSPHORYLATION AND ACTIVATION MEDIATED BY ACTIVATED HUMAN TAK1 | Genes involved in JNK (c-Jun kinases) phosphorylation and activation mediated by activated human TAK1 |
| 0.3 | 13.0 | REACTOME DNA REPAIR | Genes involved in DNA Repair |
| 0.3 | 3.8 | REACTOME PHOSPHORYLATION OF CD3 AND TCR ZETA CHAINS | Genes involved in Phosphorylation of CD3 and TCR zeta chains |
| 0.3 | 0.5 | REACTOME FRS2 MEDIATED CASCADE | Genes involved in FRS2-mediated cascade |
| 0.3 | 7.8 | REACTOME TRANSPORT OF MATURE TRANSCRIPT TO CYTOPLASM | Genes involved in Transport of Mature Transcript to Cytoplasm |
| 0.3 | 0.5 | REACTOME SIGNALLING TO RAS | Genes involved in Signalling to RAS |
| 0.2 | 5.5 | REACTOME MRNA DECAY BY 5 TO 3 EXORIBONUCLEASE | Genes involved in mRNA Decay by 5' to 3' Exoribonuclease |
| 0.2 | 2.4 | REACTOME CS DS DEGRADATION | Genes involved in CS/DS degradation |
| 0.2 | 3.8 | REACTOME MTORC1 MEDIATED SIGNALLING | Genes involved in mTORC1-mediated signalling |
| 0.2 | 3.3 | REACTOME NOREPINEPHRINE NEUROTRANSMITTER RELEASE CYCLE | Genes involved in Norepinephrine Neurotransmitter Release Cycle |
| 0.2 | 1.8 | REACTOME SIGNALING BY WNT | Genes involved in Signaling by Wnt |
| 0.2 | 2.2 | REACTOME SPRY REGULATION OF FGF SIGNALING | Genes involved in Spry regulation of FGF signaling |
| 0.2 | 3.7 | REACTOME REGULATION OF IFNG SIGNALING | Genes involved in Regulation of IFNG signaling |
| 0.2 | 3.4 | REACTOME METABOLISM OF NON CODING RNA | Genes involved in Metabolism of non-coding RNA |
| 0.2 | 7.0 | REACTOME MEIOSIS | Genes involved in Meiosis |
| 0.2 | 10.8 | REACTOME ASSOCIATION OF TRIC CCT WITH TARGET PROTEINS DURING BIOSYNTHESIS | Genes involved in Association of TriC/CCT with target proteins during biosynthesis |
| 0.2 | 3.1 | REACTOME NEF MEDIATED DOWNREGULATION OF MHC CLASS I COMPLEX CELL SURFACE EXPRESSION | Genes involved in Nef mediated downregulation of MHC class I complex cell surface expression |
| 0.2 | 7.0 | REACTOME PYRIMIDINE METABOLISM | Genes involved in Pyrimidine metabolism |
| 0.2 | 21.8 | REACTOME MRNA SPLICING | Genes involved in mRNA Splicing |
| 0.2 | 2.4 | REACTOME ERKS ARE INACTIVATED | Genes involved in ERKs are inactivated |
| 0.2 | 2.0 | REACTOME OPSINS | Genes involved in Opsins |
| 0.2 | 3.1 | REACTOME IL 7 SIGNALING | Genes involved in Interleukin-7 signaling |
| 0.2 | 4.6 | REACTOME DEADENYLATION OF MRNA | Genes involved in Deadenylation of mRNA |
| 0.2 | 4.6 | REACTOME A TETRASACCHARIDE LINKER SEQUENCE IS REQUIRED FOR GAG SYNTHESIS | Genes involved in A tetrasaccharide linker sequence is required for GAG synthesis |
| 0.2 | 5.8 | REACTOME INSULIN RECEPTOR RECYCLING | Genes involved in Insulin receptor recycling |
| 0.2 | 2.6 | REACTOME PRESYNAPTIC NICOTINIC ACETYLCHOLINE RECEPTORS | Genes involved in Presynaptic nicotinic acetylcholine receptors |
| 0.2 | 2.8 | REACTOME SYNTHESIS OF PIPS AT THE LATE ENDOSOME MEMBRANE | Genes involved in Synthesis of PIPs at the late endosome membrane |
| 0.2 | 3.8 | REACTOME SIGNALING BY FGFR1 FUSION MUTANTS | Genes involved in Signaling by FGFR1 fusion mutants |
| 0.1 | 6.0 | REACTOME STEROID HORMONES | Genes involved in Steroid hormones |
| 0.1 | 1.2 | REACTOME TELOMERE MAINTENANCE | Genes involved in Telomere Maintenance |
| 0.1 | 4.1 | REACTOME RNA POL I RNA POL III AND MITOCHONDRIAL TRANSCRIPTION | Genes involved in RNA Polymerase I, RNA Polymerase III, and Mitochondrial Transcription |
| 0.1 | 2.0 | REACTOME NEGATIVE REGULATION OF THE PI3K AKT NETWORK | Genes involved in Negative regulation of the PI3K/AKT network |
| 0.1 | 1.1 | REACTOME ACTIVATION OF RAC | Genes involved in Activation of Rac |
| 0.1 | 3.4 | REACTOME NOD1 2 SIGNALING PATHWAY | Genes involved in NOD1/2 Signaling Pathway |
| 0.1 | 1.8 | REACTOME ROLE OF DCC IN REGULATING APOPTOSIS | Genes involved in Role of DCC in regulating apoptosis |
| 0.1 | 1.5 | REACTOME P38MAPK EVENTS | Genes involved in p38MAPK events |
| 0.1 | 3.8 | REACTOME LIGAND GATED ION CHANNEL TRANSPORT | Genes involved in Ligand-gated ion channel transport |
| 0.1 | 1.6 | REACTOME THROMBIN SIGNALLING THROUGH PROTEINASE ACTIVATED RECEPTORS PARS | Genes involved in Thrombin signalling through proteinase activated receptors (PARs) |
| 0.1 | 1.0 | REACTOME PROSTANOID LIGAND RECEPTORS | Genes involved in Prostanoid ligand receptors |
| 0.1 | 7.2 | REACTOME NOTCH1 INTRACELLULAR DOMAIN REGULATES TRANSCRIPTION | Genes involved in NOTCH1 Intracellular Domain Regulates Transcription |
| 0.1 | 1.4 | REACTOME TRAFFICKING OF GLUR2 CONTAINING AMPA RECEPTORS | Genes involved in Trafficking of GluR2-containing AMPA receptors |
| 0.1 | 0.9 | REACTOME REGULATION OF ORNITHINE DECARBOXYLASE ODC | Genes involved in Regulation of ornithine decarboxylase (ODC) |
| 0.1 | 4.9 | REACTOME INTERACTION BETWEEN L1 AND ANKYRINS | Genes involved in Interaction between L1 and Ankyrins |
| 0.1 | 1.5 | REACTOME TANDEM PORE DOMAIN POTASSIUM CHANNELS | Genes involved in Tandem pore domain potassium channels |
| 0.1 | 2.4 | REACTOME ERK MAPK TARGETS | Genes involved in ERK/MAPK targets |
| 0.1 | 1.1 | REACTOME POST CHAPERONIN TUBULIN FOLDING PATHWAY | Genes involved in Post-chaperonin tubulin folding pathway |
| 0.1 | 0.9 | REACTOME CRMPS IN SEMA3A SIGNALING | Genes involved in CRMPs in Sema3A signaling |
| 0.1 | 2.0 | REACTOME LATE PHASE OF HIV LIFE CYCLE | Genes involved in Late Phase of HIV Life Cycle |
| 0.1 | 0.6 | REACTOME REGULATION OF IFNA SIGNALING | Genes involved in Regulation of IFNA signaling |
| 0.1 | 14.8 | REACTOME ANTIGEN PROCESSING UBIQUITINATION PROTEASOME DEGRADATION | Genes involved in Antigen processing: Ubiquitination & Proteasome degradation |
| 0.1 | 1.3 | REACTOME RETROGRADE NEUROTROPHIN SIGNALLING | Genes involved in Retrograde neurotrophin signalling |
| 0.1 | 1.2 | REACTOME TRAF3 DEPENDENT IRF ACTIVATION PATHWAY | Genes involved in TRAF3-dependent IRF activation pathway |
| 0.1 | 1.1 | REACTOME ADENYLATE CYCLASE ACTIVATING PATHWAY | Genes involved in Adenylate cyclase activating pathway |
| 0.1 | 3.7 | REACTOME APC C CDH1 MEDIATED DEGRADATION OF CDC20 AND OTHER APC C CDH1 TARGETED PROTEINS IN LATE MITOSIS EARLY G1 | Genes involved in APC/C:Cdh1 mediated degradation of Cdc20 and other APC/C:Cdh1 targeted proteins in late mitosis/early G1 |
| 0.1 | 3.0 | REACTOME CHEMOKINE RECEPTORS BIND CHEMOKINES | Genes involved in Chemokine receptors bind chemokines |
| 0.1 | 3.9 | REACTOME G ALPHA Z SIGNALLING EVENTS | Genes involved in G alpha (z) signalling events |
| 0.0 | 1.4 | REACTOME SYNTHESIS AND INTERCONVERSION OF NUCLEOTIDE DI AND TRIPHOSPHATES | Genes involved in Synthesis and interconversion of nucleotide di- and triphosphates |
| 0.0 | 0.3 | REACTOME OXYGEN DEPENDENT PROLINE HYDROXYLATION OF HYPOXIA INDUCIBLE FACTOR ALPHA | Genes involved in Oxygen-dependent Proline Hydroxylation of Hypoxia-inducible Factor Alpha |
| 0.0 | 0.7 | REACTOME PERK REGULATED GENE EXPRESSION | Genes involved in PERK regulated gene expression |
| 0.0 | 0.9 | REACTOME SIGNALING BY EGFR IN CANCER | Genes involved in Signaling by EGFR in Cancer |
| 0.0 | 0.3 | REACTOME GABA SYNTHESIS RELEASE REUPTAKE AND DEGRADATION | Genes involved in GABA synthesis, release, reuptake and degradation |
| 0.0 | 1.8 | REACTOME IMMUNOREGULATORY INTERACTIONS BETWEEN A LYMPHOID AND A NON LYMPHOID CELL | Genes involved in Immunoregulatory interactions between a Lymphoid and a non-Lymphoid cell |
| 0.0 | 0.7 | REACTOME ACYL CHAIN REMODELLING OF PG | Genes involved in Acyl chain remodelling of PG |
| 0.0 | 1.6 | REACTOME MHC CLASS II ANTIGEN PRESENTATION | Genes involved in MHC class II antigen presentation |
| 0.0 | 1.0 | REACTOME SYNTHESIS OF VERY LONG CHAIN FATTY ACYL COAS | Genes involved in Synthesis of very long-chain fatty acyl-CoAs |
| 0.0 | 0.6 | REACTOME CLASS C 3 METABOTROPIC GLUTAMATE PHEROMONE RECEPTORS | Genes involved in Class C/3 (Metabotropic glutamate/pheromone receptors) |
| 0.0 | 0.1 | REACTOME NFKB IS ACTIVATED AND SIGNALS SURVIVAL | Genes involved in NF-kB is activated and signals survival |
| 0.0 | 0.7 | REACTOME AMINE COMPOUND SLC TRANSPORTERS | Genes involved in Amine compound SLC transporters |
| 0.0 | 0.5 | REACTOME TERMINATION OF O GLYCAN BIOSYNTHESIS | Genes involved in Termination of O-glycan biosynthesis |
| 0.0 | 0.2 | REACTOME GLUCURONIDATION | Genes involved in Glucuronidation |
| 0.0 | 0.2 | REACTOME CD28 DEPENDENT PI3K AKT SIGNALING | Genes involved in CD28 dependent PI3K/Akt signaling |
| 0.0 | 0.5 | REACTOME GLYCOSPHINGOLIPID METABOLISM | Genes involved in Glycosphingolipid metabolism |
| 0.0 | 0.4 | REACTOME NUCLEAR SIGNALING BY ERBB4 | Genes involved in Nuclear signaling by ERBB4 |
| 0.0 | 0.5 | REACTOME SIGNALING BY NOTCH1 | Genes involved in Signaling by NOTCH1 |
| 0.0 | 0.3 | REACTOME DOWNREGULATION OF SMAD2 3 SMAD4 TRANSCRIPTIONAL ACTIVITY | Genes involved in Downregulation of SMAD2/3:SMAD4 transcriptional activity |
| 0.0 | 0.3 | REACTOME UNBLOCKING OF NMDA RECEPTOR GLUTAMATE BINDING AND ACTIVATION | Genes involved in Unblocking of NMDA receptor, glutamate binding and activation |