Project
PRJNA375882: Comprehensive Mouse Transcriptomic BodyMap
Navigation
Downloads
Results for Obox1
Z-value: 0.95
Transcription factors associated with Obox1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Obox1
|
ENSMUSG00000054310.17 | Obox1 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Obox1 | mm39_v1_chr7_+_15122873_15122918 | 0.08 | 5.1e-01 | Click! |
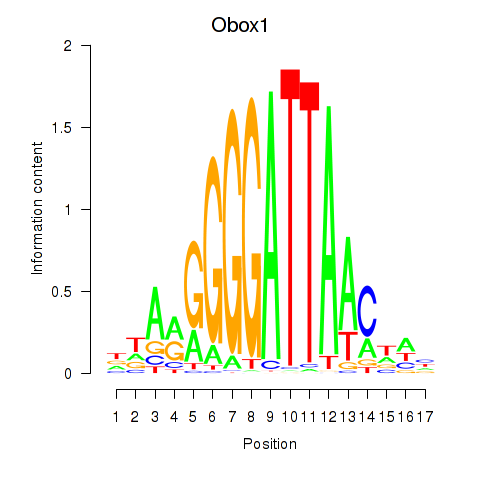
Activity profile of Obox1 motif
Sorted Z-values of Obox1 motif
Network of associatons between targets according to the STRING database.
First level regulatory network of Obox1
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr3_-_73615732 | 9.32 |
ENSMUST00000029367.6
|
Bche
|
butyrylcholinesterase |
| chr4_-_116024788 | 8.46 |
ENSMUST00000030465.10
ENSMUST00000143426.2 |
Tspan1
|
tetraspanin 1 |
| chr3_+_92123106 | 7.50 |
ENSMUST00000074449.7
ENSMUST00000090871.3 |
Sprr2a1
Sprr2a2
|
small proline-rich protein 2A1 small proline-rich protein 2A2 |
| chr3_-_73615535 | 7.13 |
ENSMUST00000138216.8
|
Bche
|
butyrylcholinesterase |
| chr11_+_3981769 | 6.85 |
ENSMUST00000019512.8
|
Sec14l4
|
SEC14-like lipid binding 4 |
| chr15_-_76501041 | 5.54 |
ENSMUST00000073428.7
|
Slc39a4
|
solute carrier family 39 (zinc transporter), member 4 |
| chr7_+_88079534 | 5.04 |
ENSMUST00000208478.2
|
Rab38
|
RAB38, member RAS oncogene family |
| chr14_+_14090981 | 4.87 |
ENSMUST00000022269.7
|
Oit1
|
oncoprotein induced transcript 1 |
| chr12_-_114621406 | 4.57 |
ENSMUST00000192077.2
|
Ighv1-15
|
immunoglobulin heavy variable 1-15 |
| chr15_-_76501525 | 4.39 |
ENSMUST00000230977.2
|
Slc39a4
|
solute carrier family 39 (zinc transporter), member 4 |
| chr9_-_21223551 | 3.98 |
ENSMUST00000003397.9
ENSMUST00000213250.2 |
Ap1m2
|
adaptor protein complex AP-1, mu 2 subunit |
| chr8_+_110717062 | 3.96 |
ENSMUST00000001720.14
ENSMUST00000143741.2 |
Tat
|
tyrosine aminotransferase |
| chr3_+_92192724 | 3.78 |
ENSMUST00000193337.2
|
Sprr2a3
|
small proline-rich protein 2A3 |
| chr7_+_88079465 | 3.62 |
ENSMUST00000107256.4
|
Rab38
|
RAB38, member RAS oncogene family |
| chr12_-_114672701 | 3.60 |
ENSMUST00000103505.3
ENSMUST00000193855.2 |
Ighv1-19
|
immunoglobulin heavy variable V1-19 |
| chr6_-_41012435 | 3.59 |
ENSMUST00000031931.6
|
2210010C04Rik
|
RIKEN cDNA 2210010C04 gene |
| chr9_-_35111172 | 3.59 |
ENSMUST00000176021.8
ENSMUST00000176531.8 ENSMUST00000176685.8 ENSMUST00000177129.8 |
Tirap
|
toll-interleukin 1 receptor (TIR) domain-containing adaptor protein |
| chr3_-_92758591 | 3.34 |
ENSMUST00000054426.5
|
Lce1l
|
late cornified envelope 1L |
| chr15_-_78739717 | 3.30 |
ENSMUST00000044584.6
|
Lgals2
|
lectin, galactose-binding, soluble 2 |
| chr6_-_5193816 | 3.18 |
ENSMUST00000002663.12
|
Pon1
|
paraoxonase 1 |
| chr6_-_5193757 | 3.18 |
ENSMUST00000177159.9
ENSMUST00000176945.2 |
Pon1
|
paraoxonase 1 |
| chr15_+_76579885 | 3.15 |
ENSMUST00000231028.2
|
Gpt
|
glutamic pyruvic transaminase, soluble |
| chr9_-_21223631 | 3.15 |
ENSMUST00000115433.11
|
Ap1m2
|
adaptor protein complex AP-1, mu 2 subunit |
| chr15_+_76579960 | 3.01 |
ENSMUST00000229679.2
|
Gpt
|
glutamic pyruvic transaminase, soluble |
| chr13_-_58758691 | 2.94 |
ENSMUST00000022036.14
|
Slc28a3
|
solute carrier family 28 (sodium-coupled nucleoside transporter), member 3 |
| chr17_+_87943401 | 2.93 |
ENSMUST00000235125.2
ENSMUST00000053577.9 ENSMUST00000234009.2 |
Epcam
|
epithelial cell adhesion molecule |
| chr12_-_114901026 | 2.88 |
ENSMUST00000103516.2
ENSMUST00000191868.2 |
Ighv1-42
|
immunoglobulin heavy variable V1-42 |
| chr7_-_110581652 | 2.86 |
ENSMUST00000005751.13
|
Irag1
|
inositol 1,4,5-triphosphate receptor associated 1 |
| chr6_+_70675416 | 2.84 |
ENSMUST00000103403.3
|
Igkv3-2
|
immunoglobulin kappa variable 3-2 |
| chr4_-_137157824 | 2.80 |
ENSMUST00000102522.5
|
Cela3b
|
chymotrypsin-like elastase family, member 3B |
| chr7_-_110581376 | 2.79 |
ENSMUST00000154466.2
|
Irag1
|
inositol 1,4,5-triphosphate receptor associated 1 |
| chr6_-_69282389 | 2.76 |
ENSMUST00000103350.3
|
Igkv4-68
|
immunoglobulin kappa variable 4-68 |
| chr1_-_139708906 | 2.71 |
ENSMUST00000111986.8
ENSMUST00000027612.11 ENSMUST00000111989.9 |
Cfhr4
|
complement factor H-related 4 |
| chr16_+_33614378 | 2.69 |
ENSMUST00000115044.8
|
Muc13
|
mucin 13, epithelial transmembrane |
| chr6_+_41155309 | 2.68 |
ENSMUST00000103276.3
|
Trbv19
|
T cell receptor beta, variable 19 |
| chr14_+_14091030 | 2.67 |
ENSMUST00000224529.2
|
Oit1
|
oncoprotein induced transcript 1 |
| chr8_+_114860375 | 2.56 |
ENSMUST00000147605.8
ENSMUST00000134593.2 |
Nudt7
|
nudix (nucleoside diphosphate linked moiety X)-type motif 7 |
| chr5_+_91287448 | 2.48 |
ENSMUST00000031325.6
|
Areg
|
amphiregulin |
| chr6_+_70348416 | 2.47 |
ENSMUST00000103391.4
|
Igkv6-17
|
immunoglobulin kappa variable 6-17 |
| chr14_-_30665232 | 2.46 |
ENSMUST00000006704.17
ENSMUST00000163118.2 |
Itih1
|
inter-alpha trypsin inhibitor, heavy chain 1 |
| chr2_-_34803988 | 2.38 |
ENSMUST00000028232.7
ENSMUST00000202907.2 |
Phf19
|
PHD finger protein 19 |
| chr6_-_59403264 | 2.38 |
ENSMUST00000051065.6
|
Gprin3
|
GPRIN family member 3 |
| chr3_+_106389732 | 2.36 |
ENSMUST00000029508.11
|
Dennd2d
|
DENN/MADD domain containing 2D |
| chr6_-_69204417 | 2.34 |
ENSMUST00000103346.3
|
Igkv4-72
|
immunoglobulin kappa chain variable 4-72 |
| chr8_+_114860342 | 2.34 |
ENSMUST00000109109.8
|
Nudt7
|
nudix (nucleoside diphosphate linked moiety X)-type motif 7 |
| chr12_-_114646685 | 2.33 |
ENSMUST00000194350.6
ENSMUST00000103504.3 |
Ighv1-18
|
immunoglobulin heavy variable V1-18 |
| chr5_-_105491795 | 2.32 |
ENSMUST00000171587.2
|
Gbp11
|
guanylate binding protein 11 |
| chr1_-_106980033 | 2.30 |
ENSMUST00000112717.3
|
Serpinb3a
|
serine (or cysteine) peptidase inhibitor, clade B (ovalbumin), member 3A |
| chr5_-_105198913 | 2.25 |
ENSMUST00000112718.5
|
Gbp8
|
guanylate-binding protein 8 |
| chr6_+_129385816 | 2.22 |
ENSMUST00000058352.15
ENSMUST00000088075.8 |
Clec9a
|
C-type lectin domain family 9, member a |
| chr10_+_127478844 | 2.19 |
ENSMUST00000092074.12
ENSMUST00000120279.2 |
Stat6
|
signal transducer and activator of transcription 6 |
| chrX_+_55825033 | 2.15 |
ENSMUST00000114772.9
ENSMUST00000114768.10 ENSMUST00000155882.8 |
Fhl1
|
four and a half LIM domains 1 |
| chr19_-_40062174 | 2.15 |
ENSMUST00000048959.5
|
Cyp2c54
|
cytochrome P450, family 2, subfamily c, polypeptide 54 |
| chr16_+_23109213 | 2.12 |
ENSMUST00000115335.2
|
St6gal1
|
beta galactoside alpha 2,6 sialyltransferase 1 |
| chr1_+_163889713 | 2.10 |
ENSMUST00000097491.10
|
Sell
|
selectin, lymphocyte |
| chr19_-_10859087 | 2.09 |
ENSMUST00000144681.2
|
Tmem109
|
transmembrane protein 109 |
| chr3_+_93227047 | 2.07 |
ENSMUST00000090856.10
ENSMUST00000093774.4 |
Hrnr
|
hornerin |
| chr15_+_54975713 | 2.05 |
ENSMUST00000096433.10
|
Deptor
|
DEP domain containing MTOR-interacting protein |
| chr19_+_33985684 | 2.05 |
ENSMUST00000225505.2
|
Lipk
|
lipase, family member K |
| chr16_+_31241085 | 2.00 |
ENSMUST00000089759.9
|
Bdh1
|
3-hydroxybutyrate dehydrogenase, type 1 |
| chr2_-_62313981 | 1.97 |
ENSMUST00000136686.2
ENSMUST00000102733.10 |
Gcg
|
glucagon |
| chr14_+_14210932 | 1.97 |
ENSMUST00000022271.14
|
Acox2
|
acyl-Coenzyme A oxidase 2, branched chain |
| chr6_-_69553484 | 1.97 |
ENSMUST00000103357.4
|
Igkv4-57
|
immunoglobulin kappa variable 4-57 |
| chr6_-_69245427 | 1.96 |
ENSMUST00000103348.3
|
Igkv4-70
|
immunoglobulin kappa chain variable 4-70 |
| chr7_+_140547941 | 1.95 |
ENSMUST00000106040.8
ENSMUST00000026564.9 |
Ifitm1
|
interferon induced transmembrane protein 1 |
| chr10_-_86843878 | 1.94 |
ENSMUST00000035288.17
|
Stab2
|
stabilin 2 |
| chr7_+_46496929 | 1.93 |
ENSMUST00000132157.2
ENSMUST00000210631.2 |
Ldha
|
lactate dehydrogenase A |
| chr13_-_74918745 | 1.91 |
ENSMUST00000223033.2
ENSMUST00000222588.2 |
Cast
|
calpastatin |
| chr1_+_178014983 | 1.91 |
ENSMUST00000161075.8
ENSMUST00000027783.14 |
Desi2
|
desumoylating isopeptidase 2 |
| chr10_+_97400990 | 1.90 |
ENSMUST00000038160.6
|
Lum
|
lumican |
| chr19_+_33985701 | 1.90 |
ENSMUST00000054260.8
|
Lipk
|
lipase, family member K |
| chr6_+_41928559 | 1.87 |
ENSMUST00000031898.5
|
Sval1
|
seminal vesicle antigen-like 1 |
| chr6_+_129385893 | 1.87 |
ENSMUST00000204860.3
ENSMUST00000164513.8 |
Clec9a
|
C-type lectin domain family 9, member a |
| chr3_+_92223927 | 1.87 |
ENSMUST00000061038.4
|
Sprr2b
|
small proline-rich protein 2B |
| chr13_-_64514830 | 1.84 |
ENSMUST00000222971.2
|
Ctsl
|
cathepsin L |
| chr1_-_139487951 | 1.84 |
ENSMUST00000023965.8
|
Cfhr1
|
complement factor H-related 1 |
| chr9_-_14815228 | 1.82 |
ENSMUST00000034409.14
ENSMUST00000117620.8 |
Izumo1r
|
IZUMO1 receptor, JUNO |
| chr7_-_12732067 | 1.82 |
ENSMUST00000032539.14
ENSMUST00000120903.8 |
Slc27a5
|
solute carrier family 27 (fatty acid transporter), member 5 |
| chr10_+_76284907 | 1.80 |
ENSMUST00000092406.12
|
2610028H24Rik
|
RIKEN cDNA 2610028H24 gene |
| chr10_-_76949762 | 1.78 |
ENSMUST00000072755.12
|
Col18a1
|
collagen, type XVIII, alpha 1 |
| chr10_-_12744025 | 1.78 |
ENSMUST00000219660.2
|
Utrn
|
utrophin |
| chr11_-_101998648 | 1.76 |
ENSMUST00000177304.8
ENSMUST00000017455.15 |
Pyy
|
peptide YY |
| chr11_-_32150222 | 1.76 |
ENSMUST00000145401.8
ENSMUST00000142396.2 ENSMUST00000128311.8 |
Il9r
|
interleukin 9 receptor |
| chr13_-_74918701 | 1.73 |
ENSMUST00000223126.2
|
Cast
|
calpastatin |
| chr2_-_34990689 | 1.73 |
ENSMUST00000226631.2
ENSMUST00000045776.5 ENSMUST00000226972.2 |
AI182371
|
expressed sequence AI182371 |
| chr14_+_40826970 | 1.73 |
ENSMUST00000225720.2
|
Mat1a
|
methionine adenosyltransferase I, alpha |
| chr14_-_40907106 | 1.71 |
ENSMUST00000077136.5
|
Sftpd
|
surfactant associated protein D |
| chr1_+_107456731 | 1.71 |
ENSMUST00000182198.8
|
Serpinb10
|
serine (or cysteine) peptidase inhibitor, clade B (ovalbumin), member 10 |
| chr14_-_25902898 | 1.69 |
ENSMUST00000183431.8
ENSMUST00000183725.2 |
Plac9a
|
placenta specific 9a |
| chr6_-_129599645 | 1.67 |
ENSMUST00000032252.8
|
Klrk1
|
killer cell lectin-like receptor subfamily K, member 1 |
| chr6_+_41139948 | 1.67 |
ENSMUST00000103275.4
|
Trbv17
|
T cell receptor beta, variable 17 |
| chr19_-_10859046 | 1.66 |
ENSMUST00000128835.8
|
Tmem109
|
transmembrane protein 109 |
| chr1_-_136888118 | 1.64 |
ENSMUST00000192357.6
ENSMUST00000027649.14 |
Nr5a2
|
nuclear receptor subfamily 5, group A, member 2 |
| chr7_+_101871623 | 1.63 |
ENSMUST00000143541.8
|
Pgap2
|
post-GPI attachment to proteins 2 |
| chr9_-_50528530 | 1.63 |
ENSMUST00000147671.2
ENSMUST00000145139.8 ENSMUST00000155435.8 |
Nkapd1
|
NKAP domain containing 1 |
| chr11_-_59937302 | 1.63 |
ENSMUST00000000310.14
ENSMUST00000102693.9 ENSMUST00000148512.2 |
Pemt
|
phosphatidylethanolamine N-methyltransferase |
| chr1_+_163889551 | 1.62 |
ENSMUST00000192047.6
ENSMUST00000027871.13 |
Sell
|
selectin, lymphocyte |
| chr12_-_113552322 | 1.60 |
ENSMUST00000103443.3
|
Ighv2-2
|
immunoglobulin heavy variable 2-2 |
| chrX_-_101232978 | 1.59 |
ENSMUST00000033683.8
|
Rps4x
|
ribosomal protein S4, X-linked |
| chr13_+_21995906 | 1.57 |
ENSMUST00000104941.4
|
H4c17
|
H4 clustered histone 17 |
| chr1_-_107206091 | 1.57 |
ENSMUST00000166100.2
ENSMUST00000027565.5 |
Serpinb3b
Serpinb3c
|
serine (or cysteine) peptidase inhibitor, clade B (ovalbumin), member 3B serine (or cysteine) peptidase inhibitor, clade B, member 3C |
| chr1_+_178015287 | 1.56 |
ENSMUST00000159284.2
|
Desi2
|
desumoylating isopeptidase 2 |
| chr19_+_32573182 | 1.55 |
ENSMUST00000235594.2
|
Papss2
|
3'-phosphoadenosine 5'-phosphosulfate synthase 2 |
| chr16_+_33614715 | 1.54 |
ENSMUST00000023520.7
|
Muc13
|
mucin 13, epithelial transmembrane |
| chr6_+_70680515 | 1.53 |
ENSMUST00000103404.2
|
Igkv3-1
|
immunoglobulin kappa variable 3-1 |
| chr10_-_12743915 | 1.52 |
ENSMUST00000219584.2
|
Utrn
|
utrophin |
| chr16_+_35861554 | 1.51 |
ENSMUST00000042203.10
|
Wdr5b
|
WD repeat domain 5B |
| chr9_-_50528727 | 1.50 |
ENSMUST00000131351.8
ENSMUST00000171462.8 |
Nkapd1
|
NKAP domain containing 1 |
| chr3_-_10366229 | 1.50 |
ENSMUST00000119761.2
ENSMUST00000029043.13 |
Fabp12
|
fatty acid binding protein 12 |
| chr1_+_40123858 | 1.49 |
ENSMUST00000027243.13
|
Il1r2
|
interleukin 1 receptor, type II |
| chr14_-_30645503 | 1.49 |
ENSMUST00000227995.2
|
Itih3
|
inter-alpha trypsin inhibitor, heavy chain 3 |
| chr6_-_69584812 | 1.48 |
ENSMUST00000103359.3
|
Igkv4-55
|
immunoglobulin kappa variable 4-55 |
| chr6_-_68887957 | 1.48 |
ENSMUST00000200454.2
|
Igkv4-86
|
immunoglobulin kappa variable 4-86 |
| chr5_+_117258185 | 1.46 |
ENSMUST00000111978.8
|
Taok3
|
TAO kinase 3 |
| chr8_+_95472218 | 1.46 |
ENSMUST00000034231.4
|
Ccl22
|
chemokine (C-C motif) ligand 22 |
| chr3_-_136031799 | 1.44 |
ENSMUST00000196159.5
ENSMUST00000041577.13 |
Bank1
|
B cell scaffold protein with ankyrin repeats 1 |
| chr14_+_59438658 | 1.44 |
ENSMUST00000173547.8
ENSMUST00000043227.13 ENSMUST00000022551.14 |
Rcbtb1
|
regulator of chromosome condensation (RCC1) and BTB (POZ) domain containing protein 1 |
| chr19_+_46120327 | 1.44 |
ENSMUST00000043739.6
ENSMUST00000237098.2 |
Elovl3
|
elongation of very long chain fatty acids (FEN1/Elo2, SUR4/Elo3, yeast)-like 3 |
| chr2_-_62476487 | 1.44 |
ENSMUST00000112459.4
ENSMUST00000028259.12 |
Ifih1
|
interferon induced with helicase C domain 1 |
| chr11_-_70350783 | 1.42 |
ENSMUST00000019064.9
|
Cxcl16
|
chemokine (C-X-C motif) ligand 16 |
| chr14_-_31362835 | 1.42 |
ENSMUST00000167066.8
ENSMUST00000127204.9 |
Hacl1
|
2-hydroxyacyl-CoA lyase 1 |
| chr18_+_67338437 | 1.39 |
ENSMUST00000210564.3
|
Chmp1b
|
charged multivesicular body protein 1B |
| chr3_+_92325386 | 1.38 |
ENSMUST00000029533.3
|
Sprr2j-ps
|
small proline-rich protein 2J, pseudogene |
| chr6_-_130044234 | 1.37 |
ENSMUST00000119096.2
|
Klra4
|
killer cell lectin-like receptor, subfamily A, member 4 |
| chr7_+_27198740 | 1.36 |
ENSMUST00000098644.9
ENSMUST00000108355.2 ENSMUST00000238936.2 |
Prx
|
periaxin |
| chr8_+_79235946 | 1.35 |
ENSMUST00000209490.2
ENSMUST00000034111.10 |
Slc10a7
|
solute carrier family 10 (sodium/bile acid cotransporter family), member 7 |
| chr14_+_40853734 | 1.34 |
ENSMUST00000022314.10
ENSMUST00000170719.2 |
Sftpa1
|
surfactant associated protein A1 |
| chr3_+_116653113 | 1.32 |
ENSMUST00000040260.11
|
Frrs1
|
ferric-chelate reductase 1 |
| chr10_-_128640232 | 1.30 |
ENSMUST00000051011.14
|
Tmem198b
|
transmembrane protein 198b |
| chr6_+_65502344 | 1.28 |
ENSMUST00000212375.2
|
Tnip3
|
TNFAIP3 interacting protein 3 |
| chr3_-_16060491 | 1.26 |
ENSMUST00000108347.3
|
Gm5150
|
predicted gene 5150 |
| chr16_+_38383154 | 1.26 |
ENSMUST00000171687.8
ENSMUST00000002924.15 |
Tmem39a
|
transmembrane protein 39a |
| chr2_-_126342551 | 1.26 |
ENSMUST00000129187.2
|
Atp8b4
|
ATPase, class I, type 8B, member 4 |
| chr3_-_16060545 | 1.25 |
ENSMUST00000194367.6
|
Gm5150
|
predicted gene 5150 |
| chr18_-_66155651 | 1.25 |
ENSMUST00000143990.2
|
Lman1
|
lectin, mannose-binding, 1 |
| chr9_+_108880158 | 1.23 |
ENSMUST00000198708.5
|
Shisa5
|
shisa family member 5 |
| chr14_-_30645711 | 1.22 |
ENSMUST00000006697.17
|
Itih3
|
inter-alpha trypsin inhibitor, heavy chain 3 |
| chr1_+_173248104 | 1.21 |
ENSMUST00000173023.2
|
Aim2
|
absent in melanoma 2 |
| chr11_+_67061837 | 1.21 |
ENSMUST00000170159.8
|
Myh2
|
myosin, heavy polypeptide 2, skeletal muscle, adult |
| chr6_-_124710084 | 1.20 |
ENSMUST00000112484.10
|
Ptpn6
|
protein tyrosine phosphatase, non-receptor type 6 |
| chr3_+_142406827 | 1.20 |
ENSMUST00000044392.11
ENSMUST00000199519.5 |
Kyat3
|
kynurenine aminotransferase 3 |
| chr4_+_62581857 | 1.20 |
ENSMUST00000126338.2
|
Rgs3
|
regulator of G-protein signaling 3 |
| chrX_-_107877909 | 1.20 |
ENSMUST00000101283.4
ENSMUST00000150434.8 |
Brwd3
|
bromodomain and WD repeat domain containing 3 |
| chr11_+_95275458 | 1.19 |
ENSMUST00000021243.16
ENSMUST00000146556.2 |
Slc35b1
|
solute carrier family 35, member B1 |
| chr19_+_44980565 | 1.19 |
ENSMUST00000179305.2
|
Sema4g
|
sema domain, immunoglobulin domain (Ig), transmembrane domain (TM) and short cytoplasmic domain, (semaphorin) 4G |
| chr3_+_142406787 | 1.18 |
ENSMUST00000106218.8
|
Kyat3
|
kynurenine aminotransferase 3 |
| chr9_+_57604895 | 1.17 |
ENSMUST00000034865.6
|
Cyp1a1
|
cytochrome P450, family 1, subfamily a, polypeptide 1 |
| chr13_+_112604037 | 1.17 |
ENSMUST00000183868.8
|
Il6st
|
interleukin 6 signal transducer |
| chr14_+_54421231 | 1.15 |
ENSMUST00000103706.2
|
Traj35
|
T cell receptor alpha joining 35 |
| chr11_+_67061908 | 1.14 |
ENSMUST00000018641.8
|
Myh2
|
myosin, heavy polypeptide 2, skeletal muscle, adult |
| chr5_-_65248962 | 1.11 |
ENSMUST00000212080.2
|
Tmem156
|
transmembrane protein 156 |
| chr9_-_48516447 | 1.11 |
ENSMUST00000034808.12
ENSMUST00000119426.2 |
Nnmt
|
nicotinamide N-methyltransferase |
| chr9_-_14815163 | 1.10 |
ENSMUST00000069408.10
|
Izumo1r
|
IZUMO1 receptor, JUNO |
| chr7_+_27147403 | 1.10 |
ENSMUST00000037399.16
ENSMUST00000108358.8 |
Blvrb
|
biliverdin reductase B (flavin reductase (NADPH)) |
| chr6_-_68887922 | 1.09 |
ENSMUST00000103337.3
|
Igkv4-86
|
immunoglobulin kappa variable 4-86 |
| chr18_-_78100610 | 1.09 |
ENSMUST00000170760.3
|
Siglec15
|
sialic acid binding Ig-like lectin 15 |
| chr17_+_24768808 | 1.09 |
ENSMUST00000228550.2
ENSMUST00000035565.5 |
Pkd1
|
polycystin 1, transient receptor poteintial channel interacting |
| chr6_-_113717689 | 1.09 |
ENSMUST00000032440.6
|
Sec13
|
SEC13 homolog, nuclear pore and COPII coat complex component |
| chr3_-_108053396 | 1.09 |
ENSMUST00000000001.5
|
Gnai3
|
guanine nucleotide binding protein (G protein), alpha inhibiting 3 |
| chr12_-_36206780 | 1.09 |
ENSMUST00000223382.2
ENSMUST00000020856.6 |
Bzw2
|
basic leucine zipper and W2 domains 2 |
| chr7_+_27147475 | 1.09 |
ENSMUST00000133750.8
|
Blvrb
|
biliverdin reductase B (flavin reductase (NADPH)) |
| chr6_+_65502292 | 1.08 |
ENSMUST00000212402.2
|
Tnip3
|
TNFAIP3 interacting protein 3 |
| chr9_+_38630317 | 1.07 |
ENSMUST00000129598.2
|
Vwa5a
|
von Willebrand factor A domain containing 5A |
| chr6_+_139564196 | 1.06 |
ENSMUST00000188066.2
ENSMUST00000190962.7 |
Pik3c2g
|
phosphatidylinositol-4-phosphate 3-kinase catalytic subunit type 2 gamma |
| chr14_-_87378641 | 1.05 |
ENSMUST00000168889.3
ENSMUST00000022599.14 |
Diaph3
|
diaphanous related formin 3 |
| chr6_+_15727798 | 1.05 |
ENSMUST00000128849.3
|
Mdfic
|
MyoD family inhibitor domain containing |
| chr4_-_117013396 | 1.05 |
ENSMUST00000102696.5
|
Rps8
|
ribosomal protein S8 |
| chr6_-_69020489 | 1.05 |
ENSMUST00000103342.4
|
Igkv4-79
|
immunoglobulin kappa variable 4-79 |
| chr12_+_21366386 | 1.04 |
ENSMUST00000076813.8
ENSMUST00000221693.2 ENSMUST00000223345.2 ENSMUST00000222344.2 |
Iah1
|
isoamyl acetate-hydrolyzing esterase 1 homolog |
| chr11_-_114825089 | 1.04 |
ENSMUST00000149663.4
ENSMUST00000239005.2 ENSMUST00000106581.5 |
Cd300lb
|
CD300 molecule like family member B |
| chr8_+_79236051 | 0.99 |
ENSMUST00000209992.2
|
Slc10a7
|
solute carrier family 10 (sodium/bile acid cotransporter family), member 7 |
| chr14_+_54431947 | 0.98 |
ENSMUST00000103715.2
|
Traj26
|
T cell receptor alpha joining 26 |
| chr11_+_115044966 | 0.98 |
ENSMUST00000021076.6
|
Rab37
|
RAB37, member RAS oncogene family |
| chr13_+_55862437 | 0.98 |
ENSMUST00000021959.11
|
Txndc15
|
thioredoxin domain containing 15 |
| chr16_+_33201227 | 0.98 |
ENSMUST00000232023.2
|
Zfp148
|
zinc finger protein 148 |
| chrX_-_133012457 | 0.97 |
ENSMUST00000159259.3
ENSMUST00000113275.10 |
Nox1
|
NADPH oxidase 1 |
| chr4_-_137137088 | 0.97 |
ENSMUST00000024200.7
|
Cela3a
|
chymotrypsin-like elastase family, member 3A |
| chr9_+_62249730 | 0.97 |
ENSMUST00000156461.2
|
Anp32a
|
acidic (leucine-rich) nuclear phosphoprotein 32 family, member A |
| chr9_+_62249341 | 0.94 |
ENSMUST00000135395.8
|
Anp32a
|
acidic (leucine-rich) nuclear phosphoprotein 32 family, member A |
| chr5_-_65248927 | 0.94 |
ENSMUST00000043352.8
|
Tmem156
|
transmembrane protein 156 |
| chr7_+_19738020 | 0.94 |
ENSMUST00000137183.9
|
Nlrp9b
|
NLR family, pyrin domain containing 9B |
| chr1_-_165830187 | 0.94 |
ENSMUST00000184643.8
ENSMUST00000160908.8 ENSMUST00000027850.15 ENSMUST00000160260.9 |
Pou2f1
|
POU domain, class 2, transcription factor 1 |
| chr3_-_136031723 | 0.94 |
ENSMUST00000198206.2
|
Bank1
|
B cell scaffold protein with ankyrin repeats 1 |
| chr10_+_79986988 | 0.94 |
ENSMUST00000146516.8
ENSMUST00000144526.2 |
Midn
|
midnolin |
| chrX_+_13527592 | 0.93 |
ENSMUST00000053659.2
|
Gpr82
|
G protein-coupled receptor 82 |
| chr8_-_70975813 | 0.92 |
ENSMUST00000121623.8
ENSMUST00000093456.12 ENSMUST00000118850.8 |
Kxd1
|
KxDL motif containing 1 |
| chr5_-_137869969 | 0.92 |
ENSMUST00000196162.5
|
Pilrb2
|
paired immunoglobin-like type 2 receptor beta 2 |
| chr16_+_48692976 | 0.91 |
ENSMUST00000065666.6
|
Retnlg
|
resistin like gamma |
| chr19_-_41790458 | 0.91 |
ENSMUST00000026150.15
ENSMUST00000163265.9 ENSMUST00000177495.2 |
Arhgap19
|
Rho GTPase activating protein 19 |
| chr11_+_96355475 | 0.91 |
ENSMUST00000107663.4
|
Skap1
|
src family associated phosphoprotein 1 |
| chr3_-_144555062 | 0.90 |
ENSMUST00000159989.2
|
Clca3b
|
chloride channel accessory 3B |
| chr16_+_59292138 | 0.90 |
ENSMUST00000023407.12
ENSMUST00000120667.2 ENSMUST00000120674.8 |
Riox2
|
ribosomal oxygenase 2 |
| chr7_-_80053063 | 0.89 |
ENSMUST00000147150.2
|
Furin
|
furin (paired basic amino acid cleaving enzyme) |
| chr8_-_70963202 | 0.89 |
ENSMUST00000125184.8
|
Uba52
|
ubiquitin A-52 residue ribosomal protein fusion product 1 |
| chr10_-_75616788 | 0.89 |
ENSMUST00000173512.2
ENSMUST00000173537.2 |
Gm20441
Gstt3
|
predicted gene 20441 glutathione S-transferase, theta 3 |
| chr5_-_137529465 | 0.88 |
ENSMUST00000150063.9
|
Gnb2
|
guanine nucleotide binding protein (G protein), beta 2 |
| chr6_+_30541581 | 0.88 |
ENSMUST00000096066.5
|
Cpa2
|
carboxypeptidase A2, pancreatic |
| chr1_-_85239791 | 0.87 |
ENSMUST00000162421.2
|
C130026I21Rik
|
RIKEN cDNA C130026I21 gene |
| chr18_+_4993795 | 0.87 |
ENSMUST00000153016.8
|
Svil
|
supervillin |
| chr2_+_122146153 | 0.86 |
ENSMUST00000099461.4
|
Duox1
|
dual oxidase 1 |
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.7 | 16.5 | GO:0014016 | neuroblast differentiation(GO:0014016) |
| 2.1 | 6.4 | GO:1902617 | response to fluoride(GO:1902617) |
| 2.0 | 9.9 | GO:0034224 | cellular response to zinc ion starvation(GO:0034224) |
| 1.3 | 5.0 | GO:0046356 | acetyl-CoA catabolic process(GO:0046356) |
| 1.2 | 8.7 | GO:1903232 | platelet dense granule organization(GO:0060155) phagosome acidification(GO:0090383) melanosome assembly(GO:1903232) |
| 1.2 | 3.6 | GO:0032618 | interleukin-15 production(GO:0032618) |
| 1.0 | 2.9 | GO:0015860 | purine nucleoside transmembrane transport(GO:0015860) nucleoside transmembrane transport(GO:1901642) |
| 0.6 | 2.5 | GO:0060598 | dichotomous subdivision of terminal units involved in mammary gland duct morphogenesis(GO:0060598) |
| 0.6 | 2.9 | GO:2000048 | negative regulation of cell-cell adhesion mediated by cadherin(GO:2000048) |
| 0.6 | 1.2 | GO:0070104 | negative regulation of interleukin-6-mediated signaling pathway(GO:0070104) |
| 0.6 | 1.7 | GO:0009087 | methionine catabolic process(GO:0009087) |
| 0.6 | 1.7 | GO:0030887 | positive regulation of myeloid dendritic cell activation(GO:0030887) |
| 0.5 | 2.2 | GO:0002296 | T-helper 1 cell lineage commitment(GO:0002296) interleukin-4-mediated signaling pathway(GO:0035771) |
| 0.5 | 3.3 | GO:0007527 | adult somatic muscle development(GO:0007527) |
| 0.5 | 1.8 | GO:0046951 | ketone body biosynthetic process(GO:0046951) |
| 0.4 | 4.0 | GO:0006572 | tyrosine catabolic process(GO:0006572) |
| 0.4 | 1.7 | GO:0043152 | induction of bacterial agglutination(GO:0043152) |
| 0.4 | 2.1 | GO:1990743 | protein sialylation(GO:1990743) |
| 0.4 | 1.2 | GO:0090481 | pyrimidine nucleotide-sugar transmembrane transport(GO:0090481) |
| 0.4 | 1.6 | GO:0050428 | purine ribonucleoside bisphosphate biosynthetic process(GO:0034036) 3'-phosphoadenosine 5'-phosphosulfate biosynthetic process(GO:0050428) |
| 0.4 | 1.5 | GO:0032690 | negative regulation of interleukin-1 alpha production(GO:0032690) negative regulation of interleukin-1 alpha secretion(GO:0050712) |
| 0.4 | 1.1 | GO:0060854 | patterning of lymph vessels(GO:0060854) |
| 0.3 | 2.0 | GO:0033540 | fatty acid beta-oxidation using acyl-CoA oxidase(GO:0033540) |
| 0.3 | 2.0 | GO:0035549 | interferon-beta secretion(GO:0035546) regulation of interferon-beta secretion(GO:0035547) positive regulation of interferon-beta secretion(GO:0035549) |
| 0.3 | 0.9 | GO:0030961 | peptidyl-arginine hydroxylation(GO:0030961) |
| 0.3 | 1.2 | GO:0018894 | dibenzo-p-dioxin metabolic process(GO:0018894) |
| 0.3 | 2.3 | GO:0042270 | protection from natural killer cell mediated cytotoxicity(GO:0042270) |
| 0.3 | 0.8 | GO:0006583 | melanin biosynthetic process from tyrosine(GO:0006583) |
| 0.3 | 0.8 | GO:0018199 | peptidyl-glutamine modification(GO:0018199) |
| 0.3 | 2.2 | GO:0006787 | porphyrin-containing compound catabolic process(GO:0006787) tetrapyrrole catabolic process(GO:0033015) heme catabolic process(GO:0042167) pigment catabolic process(GO:0046149) |
| 0.3 | 2.4 | GO:0061087 | positive regulation of histone H3-K27 methylation(GO:0061087) |
| 0.2 | 4.2 | GO:0030277 | maintenance of gastrointestinal epithelium(GO:0030277) |
| 0.2 | 2.0 | GO:0035948 | positive regulation of gluconeogenesis by positive regulation of transcription from RNA polymerase II promoter(GO:0035948) |
| 0.2 | 1.9 | GO:0032914 | positive regulation of transforming growth factor beta1 production(GO:0032914) |
| 0.2 | 1.6 | GO:0050747 | positive regulation of lipoprotein metabolic process(GO:0050747) |
| 0.2 | 0.9 | GO:0002408 | myeloid dendritic cell chemotaxis(GO:0002408) |
| 0.2 | 1.6 | GO:0001561 | fatty acid alpha-oxidation(GO:0001561) |
| 0.2 | 1.4 | GO:0032290 | peripheral nervous system myelin formation(GO:0032290) |
| 0.2 | 0.7 | GO:0032416 | negative regulation of sodium:proton antiporter activity(GO:0032416) |
| 0.2 | 0.9 | GO:0090472 | dibasic protein processing(GO:0090472) |
| 0.2 | 1.5 | GO:0072344 | rescue of stalled ribosome(GO:0072344) |
| 0.2 | 0.9 | GO:0042335 | cuticle development(GO:0042335) |
| 0.2 | 0.9 | GO:0035522 | monoubiquitinated histone deubiquitination(GO:0035521) monoubiquitinated histone H2A deubiquitination(GO:0035522) histone H2A K63-linked deubiquitination(GO:0070537) |
| 0.2 | 14.6 | GO:0018149 | peptide cross-linking(GO:0018149) |
| 0.2 | 1.8 | GO:0045919 | positive regulation of cytolysis(GO:0045919) |
| 0.2 | 3.6 | GO:2000675 | egg activation(GO:0007343) negative regulation of type B pancreatic cell apoptotic process(GO:2000675) |
| 0.2 | 1.6 | GO:1901097 | negative regulation of autophagosome maturation(GO:1901097) |
| 0.2 | 0.6 | GO:1904211 | membrane protein proteolysis involved in retrograde protein transport, ER to cytosol(GO:1904211) |
| 0.2 | 0.6 | GO:0014732 | skeletal muscle atrophy(GO:0014732) |
| 0.2 | 1.0 | GO:0045726 | positive regulation of integrin biosynthetic process(GO:0045726) |
| 0.2 | 0.6 | GO:0002014 | vasoconstriction of artery involved in ischemic response to lowering of systemic arterial blood pressure(GO:0002014) |
| 0.2 | 1.9 | GO:0019660 | glucose catabolic process to lactate(GO:0019659) glycolytic fermentation(GO:0019660) glucose catabolic process to lactate via pyruvate(GO:0019661) |
| 0.2 | 2.4 | GO:1900165 | negative regulation of interleukin-6 secretion(GO:1900165) |
| 0.2 | 2.1 | GO:0043651 | linoleic acid metabolic process(GO:0043651) |
| 0.2 | 0.9 | GO:0006566 | threonine metabolic process(GO:0006566) |
| 0.2 | 2.9 | GO:0007342 | fusion of sperm to egg plasma membrane(GO:0007342) |
| 0.2 | 0.5 | GO:0009258 | 10-formyltetrahydrofolate catabolic process(GO:0009258) folic acid-containing compound catabolic process(GO:0009397) pteridine-containing compound catabolic process(GO:0042560) |
| 0.2 | 1.8 | GO:0042637 | catagen(GO:0042637) |
| 0.2 | 0.5 | GO:0034769 | basement membrane disassembly(GO:0034769) |
| 0.2 | 0.6 | GO:0006529 | asparagine biosynthetic process(GO:0006529) |
| 0.2 | 1.2 | GO:0033277 | abortive mitotic cell cycle(GO:0033277) |
| 0.1 | 2.4 | GO:0070189 | kynurenine metabolic process(GO:0070189) |
| 0.1 | 1.3 | GO:0008228 | opsonization(GO:0008228) |
| 0.1 | 2.4 | GO:0002756 | MyD88-independent toll-like receptor signaling pathway(GO:0002756) |
| 0.1 | 2.4 | GO:0001778 | plasma membrane repair(GO:0001778) |
| 0.1 | 0.6 | GO:0031204 | posttranslational protein targeting to membrane, translocation(GO:0031204) |
| 0.1 | 1.4 | GO:0031119 | tRNA pseudouridine synthesis(GO:0031119) |
| 0.1 | 1.1 | GO:0097646 | calcitonin family receptor signaling pathway(GO:0097646) amylin receptor signaling pathway(GO:0097647) |
| 0.1 | 1.6 | GO:0061113 | pancreas morphogenesis(GO:0061113) |
| 0.1 | 0.9 | GO:0030910 | olfactory placode formation(GO:0030910) olfactory placode development(GO:0071698) olfactory placode morphogenesis(GO:0071699) |
| 0.1 | 0.5 | GO:2000110 | negative regulation of macrophage apoptotic process(GO:2000110) |
| 0.1 | 3.7 | GO:0090023 | positive regulation of neutrophil chemotaxis(GO:0090023) |
| 0.1 | 1.4 | GO:0034626 | fatty acid elongation, saturated fatty acid(GO:0019367) fatty acid elongation, unsaturated fatty acid(GO:0019368) fatty acid elongation, monounsaturated fatty acid(GO:0034625) fatty acid elongation, polyunsaturated fatty acid(GO:0034626) |
| 0.1 | 0.7 | GO:0042776 | mitochondrial ATP synthesis coupled proton transport(GO:0042776) |
| 0.1 | 5.2 | GO:0030212 | hyaluronan metabolic process(GO:0030212) |
| 0.1 | 0.9 | GO:0033132 | negative regulation of glucokinase activity(GO:0033132) negative regulation of hexokinase activity(GO:1903300) |
| 0.1 | 1.4 | GO:0034773 | histone H4-K20 trimethylation(GO:0034773) |
| 0.1 | 16.3 | GO:0006910 | phagocytosis, recognition(GO:0006910) |
| 0.1 | 0.4 | GO:1904430 | negative regulation of t-circle formation(GO:1904430) |
| 0.1 | 1.2 | GO:0018230 | peptidyl-L-cysteine S-palmitoylation(GO:0018230) peptidyl-S-diacylglycerol-L-cysteine biosynthetic process from peptidyl-cysteine(GO:0018231) |
| 0.1 | 0.5 | GO:1905068 | positive regulation of canonical Wnt signaling pathway involved in cardiac muscle cell fate commitment(GO:1901297) positive regulation of canonical Wnt signaling pathway involved in heart development(GO:1905068) |
| 0.1 | 7.3 | GO:0046676 | negative regulation of insulin secretion(GO:0046676) |
| 0.1 | 2.0 | GO:0045792 | negative regulation of cell size(GO:0045792) |
| 0.1 | 0.6 | GO:0060398 | regulation of growth hormone receptor signaling pathway(GO:0060398) |
| 0.1 | 0.6 | GO:0048496 | determination of pancreatic left/right asymmetry(GO:0035469) maintenance of organ identity(GO:0048496) |
| 0.1 | 6.6 | GO:0042771 | intrinsic apoptotic signaling pathway in response to DNA damage by p53 class mediator(GO:0042771) |
| 0.1 | 1.4 | GO:1901673 | regulation of mitotic spindle assembly(GO:1901673) |
| 0.1 | 1.2 | GO:0070269 | pyroptosis(GO:0070269) |
| 0.1 | 21.1 | GO:0002377 | immunoglobulin production(GO:0002377) |
| 0.1 | 1.8 | GO:0031424 | keratinization(GO:0031424) |
| 0.1 | 1.1 | GO:2001204 | regulation of osteoclast development(GO:2001204) |
| 0.1 | 0.9 | GO:0033504 | floor plate development(GO:0033504) |
| 0.1 | 0.4 | GO:0051572 | negative regulation of histone H3-K4 methylation(GO:0051572) |
| 0.1 | 3.3 | GO:0010972 | negative regulation of G2/M transition of mitotic cell cycle(GO:0010972) |
| 0.1 | 1.5 | GO:0046597 | negative regulation of viral entry into host cell(GO:0046597) |
| 0.1 | 0.4 | GO:0050917 | sensory perception of sweet taste(GO:0050916) sensory perception of umami taste(GO:0050917) |
| 0.1 | 0.3 | GO:0046167 | glycerol-3-phosphate biosynthetic process(GO:0046167) |
| 0.1 | 0.6 | GO:0061101 | neuroendocrine cell differentiation(GO:0061101) |
| 0.1 | 0.3 | GO:2000721 | positive regulation of transcription from RNA polymerase II promoter involved in smooth muscle cell differentiation(GO:2000721) |
| 0.1 | 1.8 | GO:0001886 | endothelial cell morphogenesis(GO:0001886) |
| 0.1 | 1.1 | GO:0090110 | cargo loading into COPII-coated vesicle(GO:0090110) |
| 0.1 | 1.0 | GO:0002862 | negative regulation of inflammatory response to antigenic stimulus(GO:0002862) |
| 0.1 | 5.9 | GO:0034341 | response to interferon-gamma(GO:0034341) |
| 0.1 | 0.5 | GO:0098535 | de novo centriole assembly(GO:0098535) |
| 0.0 | 0.4 | GO:2001268 | negative regulation of cysteine-type endopeptidase activity involved in apoptotic signaling pathway(GO:2001268) |
| 0.0 | 0.1 | GO:0042822 | pyridoxal phosphate metabolic process(GO:0042822) |
| 0.0 | 0.8 | GO:0045717 | negative regulation of fatty acid biosynthetic process(GO:0045717) |
| 0.0 | 0.7 | GO:0001731 | formation of translation preinitiation complex(GO:0001731) |
| 0.0 | 0.4 | GO:0010757 | negative regulation of plasminogen activation(GO:0010757) |
| 0.0 | 0.5 | GO:1900113 | negative regulation of histone H3-K9 trimethylation(GO:1900113) |
| 0.0 | 1.0 | GO:0002446 | neutrophil mediated immunity(GO:0002446) |
| 0.0 | 0.1 | GO:1900135 | positive regulation of renin secretion into blood stream(GO:1900135) |
| 0.0 | 8.6 | GO:0045807 | positive regulation of endocytosis(GO:0045807) |
| 0.0 | 1.0 | GO:0050434 | positive regulation of viral transcription(GO:0050434) |
| 0.0 | 1.1 | GO:0039703 | viral RNA genome replication(GO:0039694) RNA replication(GO:0039703) |
| 0.0 | 0.6 | GO:0050908 | detection of light stimulus involved in visual perception(GO:0050908) detection of light stimulus involved in sensory perception(GO:0050962) |
| 0.0 | 0.5 | GO:0090527 | actin filament reorganization(GO:0090527) |
| 0.0 | 0.6 | GO:0070816 | phosphorylation of RNA polymerase II C-terminal domain(GO:0070816) |
| 0.0 | 1.6 | GO:0042755 | eating behavior(GO:0042755) |
| 0.0 | 0.1 | GO:0018343 | protein farnesylation(GO:0018343) |
| 0.0 | 0.8 | GO:0006783 | heme biosynthetic process(GO:0006783) |
| 0.0 | 3.8 | GO:0008203 | cholesterol metabolic process(GO:0008203) |
| 0.0 | 0.9 | GO:0051560 | mitochondrial calcium ion homeostasis(GO:0051560) |
| 0.0 | 0.4 | GO:0033539 | fatty acid beta-oxidation using acyl-CoA dehydrogenase(GO:0033539) |
| 0.0 | 2.2 | GO:1900046 | regulation of blood coagulation(GO:0030193) regulation of hemostasis(GO:1900046) |
| 0.0 | 1.6 | GO:0007029 | endoplasmic reticulum organization(GO:0007029) |
| 0.0 | 0.6 | GO:0015986 | energy coupled proton transport, down electrochemical gradient(GO:0015985) ATP synthesis coupled proton transport(GO:0015986) |
| 0.0 | 0.2 | GO:0006614 | SRP-dependent cotranslational protein targeting to membrane(GO:0006614) |
| 0.0 | 1.1 | GO:0046039 | GTP metabolic process(GO:0046039) |
| 0.0 | 1.2 | GO:0050919 | negative chemotaxis(GO:0050919) |
| 0.0 | 2.8 | GO:0050715 | positive regulation of cytokine secretion(GO:0050715) |
| 0.0 | 0.4 | GO:0021554 | optic nerve development(GO:0021554) |
| 0.0 | 0.2 | GO:1900364 | negative regulation of mRNA polyadenylation(GO:1900364) |
| 0.0 | 2.0 | GO:0050852 | T cell receptor signaling pathway(GO:0050852) |
| 0.0 | 0.3 | GO:0006465 | signal peptide processing(GO:0006465) |
| 0.0 | 0.1 | GO:0035745 | CD4-positive, alpha-beta T cell cytokine production(GO:0035743) T-helper 2 cell cytokine production(GO:0035745) |
| 0.0 | 0.3 | GO:0006622 | protein targeting to lysosome(GO:0006622) |
| 0.0 | 0.1 | GO:0097475 | motor neuron migration(GO:0097475) |
| 0.0 | 0.2 | GO:0007000 | nucleolus organization(GO:0007000) |
| 0.0 | 1.0 | GO:0045454 | cell redox homeostasis(GO:0045454) |
| 0.0 | 0.3 | GO:0043968 | histone H2A acetylation(GO:0043968) |
| 0.0 | 0.9 | GO:0032418 | lysosome localization(GO:0032418) |
| 0.0 | 0.9 | GO:0006749 | glutathione metabolic process(GO:0006749) |
| 0.0 | 0.1 | GO:0001514 | selenocysteine incorporation(GO:0001514) translational readthrough(GO:0006451) |
| 0.0 | 0.3 | GO:0006379 | mRNA cleavage(GO:0006379) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.1 | 8.7 | GO:0033162 | melanosome membrane(GO:0033162) chitosome(GO:0045009) |
| 1.0 | 16.5 | GO:0005641 | nuclear envelope lumen(GO:0005641) |
| 0.4 | 6.5 | GO:0034366 | spherical high-density lipoprotein particle(GO:0034366) |
| 0.4 | 1.2 | GO:0005900 | oncostatin-M receptor complex(GO:0005900) |
| 0.3 | 18.6 | GO:0001533 | cornified envelope(GO:0001533) |
| 0.3 | 1.2 | GO:0097169 | AIM2 inflammasome complex(GO:0097169) |
| 0.3 | 2.4 | GO:0005826 | actomyosin contractile ring(GO:0005826) |
| 0.2 | 1.1 | GO:1903439 | calcitonin family receptor complex(GO:1903439) amylin receptor complex(GO:1903440) |
| 0.2 | 3.3 | GO:0070938 | contractile ring(GO:0070938) |
| 0.2 | 0.6 | GO:0097543 | ciliary inversin compartment(GO:0097543) |
| 0.2 | 2.1 | GO:0000138 | Golgi trans cisterna(GO:0000138) |
| 0.1 | 1.9 | GO:0005583 | fibrillar collagen trimer(GO:0005583) banded collagen fibril(GO:0098643) |
| 0.1 | 7.1 | GO:0030131 | clathrin adaptor complex(GO:0030131) |
| 0.1 | 0.6 | GO:0000438 | core TFIIH complex portion of holo TFIIH complex(GO:0000438) |
| 0.1 | 16.3 | GO:0042571 | immunoglobulin complex, circulating(GO:0042571) |
| 0.1 | 1.0 | GO:0071438 | invadopodium membrane(GO:0071438) |
| 0.1 | 1.4 | GO:0000815 | ESCRT III complex(GO:0000815) |
| 0.1 | 1.2 | GO:0042105 | alpha-beta T cell receptor complex(GO:0042105) |
| 0.1 | 2.0 | GO:0042101 | T cell receptor complex(GO:0042101) |
| 0.1 | 1.1 | GO:0061700 | GATOR2 complex(GO:0061700) |
| 0.1 | 0.5 | GO:0070937 | CRD-mediated mRNA stability complex(GO:0070937) |
| 0.1 | 0.6 | GO:0071556 | integral component of cytoplasmic side of endoplasmic reticulum membrane(GO:0071458) integral component of lumenal side of endoplasmic reticulum membrane(GO:0071556) lumenal side of endoplasmic reticulum membrane(GO:0098553) |
| 0.1 | 0.6 | GO:0031205 | endoplasmic reticulum Sec complex(GO:0031205) |
| 0.1 | 2.4 | GO:0035098 | ESC/E(Z) complex(GO:0035098) |
| 0.1 | 0.5 | GO:0098536 | deuterosome(GO:0098536) |
| 0.1 | 0.4 | GO:0043564 | Ku70:Ku80 complex(GO:0043564) |
| 0.1 | 0.9 | GO:0012510 | trans-Golgi network transport vesicle membrane(GO:0012510) |
| 0.1 | 0.6 | GO:0045261 | mitochondrial proton-transporting ATP synthase complex, catalytic core F(1)(GO:0000275) proton-transporting ATP synthase complex, catalytic core F(1)(GO:0045261) |
| 0.1 | 1.9 | GO:0035686 | sperm fibrous sheath(GO:0035686) |
| 0.1 | 3.0 | GO:0005771 | multivesicular body(GO:0005771) |
| 0.1 | 0.6 | GO:0061689 | tricellular tight junction(GO:0061689) |
| 0.1 | 1.2 | GO:0033655 | host cell cytoplasm(GO:0030430) host cell cytoplasm part(GO:0033655) |
| 0.1 | 0.5 | GO:0002193 | MAML1-RBP-Jkappa- ICN1 complex(GO:0002193) |
| 0.1 | 1.3 | GO:0031083 | BLOC-1 complex(GO:0031083) |
| 0.1 | 0.5 | GO:0033116 | endoplasmic reticulum-Golgi intermediate compartment membrane(GO:0033116) |
| 0.1 | 3.3 | GO:0022627 | cytosolic small ribosomal subunit(GO:0022627) |
| 0.0 | 0.8 | GO:0061702 | inflammasome complex(GO:0061702) |
| 0.0 | 0.3 | GO:0042629 | mast cell granule(GO:0042629) |
| 0.0 | 0.4 | GO:0005577 | fibrinogen complex(GO:0005577) |
| 0.0 | 1.4 | GO:0000780 | condensed nuclear chromosome, centromeric region(GO:0000780) |
| 0.0 | 3.7 | GO:0016529 | sarcoplasmic reticulum(GO:0016529) |
| 0.0 | 0.8 | GO:0042470 | melanosome(GO:0042470) pigment granule(GO:0048770) |
| 0.0 | 1.1 | GO:0042588 | zymogen granule(GO:0042588) |
| 0.0 | 1.1 | GO:0005942 | phosphatidylinositol 3-kinase complex(GO:0005942) |
| 0.0 | 1.8 | GO:0009925 | basal plasma membrane(GO:0009925) |
| 0.0 | 0.6 | GO:0019908 | nuclear cyclin-dependent protein kinase holoenzyme complex(GO:0019908) |
| 0.0 | 4.1 | GO:0072562 | blood microparticle(GO:0072562) |
| 0.0 | 2.4 | GO:0016328 | lateral plasma membrane(GO:0016328) |
| 0.0 | 0.3 | GO:0005787 | signal peptidase complex(GO:0005787) |
| 0.0 | 0.2 | GO:0005786 | signal recognition particle, endoplasmic reticulum targeting(GO:0005786) |
| 0.0 | 0.1 | GO:0031084 | BLOC-2 complex(GO:0031084) |
| 0.0 | 3.6 | GO:0032587 | ruffle membrane(GO:0032587) |
| 0.0 | 0.3 | GO:0032591 | dendritic spine membrane(GO:0032591) |
| 0.0 | 0.2 | GO:0019773 | proteasome core complex, alpha-subunit complex(GO:0019773) |
| 0.0 | 0.3 | GO:0044754 | autolysosome(GO:0044754) |
| 0.0 | 3.3 | GO:0016363 | nuclear matrix(GO:0016363) |
| 0.0 | 43.1 | GO:0005615 | extracellular space(GO:0005615) |
| 0.0 | 0.9 | GO:0043034 | costamere(GO:0043034) |
| 0.0 | 6.6 | GO:0009897 | external side of plasma membrane(GO:0009897) |
| 0.0 | 0.4 | GO:0070822 | Sin3-type complex(GO:0070822) |
| 0.0 | 1.0 | GO:0030667 | secretory granule membrane(GO:0030667) |
| 0.0 | 0.5 | GO:0005721 | pericentric heterochromatin(GO:0005721) |
| 0.0 | 0.3 | GO:0031011 | Ino80 complex(GO:0031011) |
| 0.0 | 0.6 | GO:0016235 | aggresome(GO:0016235) |
| 0.0 | 1.6 | GO:0031526 | brush border membrane(GO:0031526) |
| 0.0 | 0.9 | GO:0005604 | basement membrane(GO:0005604) |
| 0.0 | 2.0 | GO:0031225 | anchored component of membrane(GO:0031225) |
| 0.0 | 0.5 | GO:0000314 | organellar small ribosomal subunit(GO:0000314) mitochondrial small ribosomal subunit(GO:0005763) |
| 0.0 | 0.2 | GO:0090543 | Flemming body(GO:0090543) |
| 0.0 | 2.6 | GO:0090575 | RNA polymerase II transcription factor complex(GO:0090575) |
| 0.0 | 0.4 | GO:0035145 | exon-exon junction complex(GO:0035145) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 5.5 | 16.5 | GO:0003990 | acetylcholinesterase activity(GO:0003990) |
| 2.9 | 8.7 | GO:0036461 | BLOC-2 complex binding(GO:0036461) |
| 1.6 | 6.4 | GO:0004063 | aryldialkylphosphatase activity(GO:0004063) |
| 1.3 | 4.0 | GO:0080130 | L-phenylalanine:2-oxoglutarate aminotransferase activity(GO:0080130) |
| 1.2 | 6.2 | GO:0047635 | L-alanine:2-oxoglutarate aminotransferase activity(GO:0004021) alanine-oxo-acid transaminase activity(GO:0047635) |
| 1.0 | 4.9 | GO:0003986 | acetyl-CoA hydrolase activity(GO:0003986) |
| 1.0 | 2.9 | GO:0015389 | pyrimidine- and adenine-specific:sodium symporter activity(GO:0015389) |
| 0.9 | 3.6 | GO:0035663 | Toll-like receptor 2 binding(GO:0035663) |
| 0.7 | 2.2 | GO:0042602 | riboflavin reductase (NADPH) activity(GO:0042602) |
| 0.7 | 2.0 | GO:0003858 | 3-hydroxybutyrate dehydrogenase activity(GO:0003858) |
| 0.6 | 1.7 | GO:0032394 | MHC class Ib receptor activity(GO:0032394) |
| 0.5 | 2.1 | GO:0071614 | linoleic acid epoxygenase activity(GO:0071614) |
| 0.5 | 3.6 | GO:0010859 | calcium-dependent cysteine-type endopeptidase inhibitor activity(GO:0010859) |
| 0.5 | 2.4 | GO:0047804 | cysteine-S-conjugate beta-lyase activity(GO:0047804) |
| 0.4 | 1.7 | GO:0004478 | methionine adenosyltransferase activity(GO:0004478) |
| 0.4 | 1.2 | GO:0019981 | interleukin-6 receptor activity(GO:0004915) interleukin-6 binding(GO:0019981) |
| 0.4 | 1.6 | GO:0004781 | adenylylsulfate kinase activity(GO:0004020) sulfate adenylyltransferase activity(GO:0004779) sulfate adenylyltransferase (ATP) activity(GO:0004781) |
| 0.4 | 1.5 | GO:0004910 | interleukin-1, Type II, blocking receptor activity(GO:0004910) |
| 0.4 | 1.8 | GO:0030984 | kininogen binding(GO:0030984) |
| 0.4 | 3.3 | GO:0016936 | galactoside binding(GO:0016936) |
| 0.4 | 2.1 | GO:0003835 | beta-galactoside alpha-2,6-sialyltransferase activity(GO:0003835) |
| 0.3 | 1.2 | GO:0005459 | UDP-galactose transmembrane transporter activity(GO:0005459) |
| 0.2 | 9.9 | GO:0005385 | zinc ion transmembrane transporter activity(GO:0005385) |
| 0.2 | 0.9 | GO:0070905 | serine binding(GO:0070905) |
| 0.2 | 1.4 | GO:0042799 | histone methyltransferase activity (H4-K20 specific)(GO:0042799) |
| 0.2 | 1.1 | GO:0097643 | amylin receptor activity(GO:0097643) |
| 0.2 | 0.5 | GO:0033680 | ATP-dependent DNA/RNA helicase activity(GO:0033680) triplex DNA binding(GO:0045142) |
| 0.2 | 3.4 | GO:0005041 | low-density lipoprotein receptor activity(GO:0005041) |
| 0.2 | 1.9 | GO:0004459 | L-lactate dehydrogenase activity(GO:0004459) |
| 0.2 | 0.5 | GO:0016155 | formyltetrahydrofolate dehydrogenase activity(GO:0016155) |
| 0.2 | 1.3 | GO:0000293 | ferric-chelate reductase activity(GO:0000293) |
| 0.2 | 2.0 | GO:0003997 | acyl-CoA oxidase activity(GO:0003997) |
| 0.2 | 1.0 | GO:0030171 | voltage-gated proton channel activity(GO:0030171) |
| 0.2 | 0.6 | GO:0004066 | asparagine synthase (glutamine-hydrolyzing) activity(GO:0004066) |
| 0.1 | 3.7 | GO:0051861 | glycolipid binding(GO:0051861) |
| 0.1 | 1.6 | GO:0008429 | phosphatidylethanolamine binding(GO:0008429) |
| 0.1 | 0.4 | GO:0019959 | interleukin-8 binding(GO:0019959) |
| 0.1 | 0.6 | GO:0070506 | high-density lipoprotein particle receptor activity(GO:0070506) |
| 0.1 | 1.1 | GO:0031821 | G-protein coupled serotonin receptor binding(GO:0031821) |
| 0.1 | 16.3 | GO:0034987 | immunoglobulin receptor binding(GO:0034987) |
| 0.1 | 1.8 | GO:0031957 | very long-chain fatty acid-CoA ligase activity(GO:0031957) |
| 0.1 | 1.4 | GO:0102336 | fatty acid elongase activity(GO:0009922) 3-oxo-arachidoyl-CoA synthase activity(GO:0102336) 3-oxo-cerotoyl-CoA synthase activity(GO:0102337) 3-oxo-lignoceronyl-CoA synthase activity(GO:0102338) |
| 0.1 | 2.9 | GO:0098641 | cadherin binding involved in cell-cell adhesion(GO:0098641) |
| 0.1 | 0.7 | GO:0001591 | dopamine neurotransmitter receptor activity, coupled via Gi/Go(GO:0001591) |
| 0.1 | 3.3 | GO:0017166 | vinculin binding(GO:0017166) |
| 0.1 | 1.2 | GO:0005001 | transmembrane receptor protein tyrosine phosphatase activity(GO:0005001) transmembrane receptor protein phosphatase activity(GO:0019198) |
| 0.1 | 2.9 | GO:0005542 | folic acid binding(GO:0005542) |
| 0.1 | 1.4 | GO:0009982 | pseudouridine synthase activity(GO:0009982) |
| 0.1 | 0.8 | GO:0005000 | vasopressin receptor activity(GO:0005000) |
| 0.1 | 0.7 | GO:0001594 | trace-amine receptor activity(GO:0001594) |
| 0.1 | 1.4 | GO:0051864 | histone demethylase activity (H3-K36 specific)(GO:0051864) |
| 0.1 | 0.3 | GO:0004370 | glycerol kinase activity(GO:0004370) |
| 0.1 | 1.9 | GO:0001530 | lipopolysaccharide binding(GO:0001530) |
| 0.1 | 1.1 | GO:0035005 | 1-phosphatidylinositol-4-phosphate 3-kinase activity(GO:0035005) |
| 0.1 | 10.0 | GO:0004867 | serine-type endopeptidase inhibitor activity(GO:0004867) |
| 0.1 | 0.6 | GO:0042500 | aspartic endopeptidase activity, intramembrane cleaving(GO:0042500) |
| 0.1 | 1.9 | GO:0004709 | MAP kinase kinase kinase activity(GO:0004709) |
| 0.1 | 0.6 | GO:0008454 | alpha-1,3-mannosylglycoprotein 4-beta-N-acetylglucosaminyltransferase activity(GO:0008454) |
| 0.1 | 1.0 | GO:0030957 | Tat protein binding(GO:0030957) |
| 0.1 | 1.2 | GO:0005537 | mannose binding(GO:0005537) |
| 0.1 | 0.6 | GO:0004499 | N,N-dimethylaniline monooxygenase activity(GO:0004499) |
| 0.0 | 0.9 | GO:0042288 | MHC class I protein binding(GO:0042288) |
| 0.0 | 0.4 | GO:0051575 | 5'-deoxyribose-5-phosphate lyase activity(GO:0051575) |
| 0.0 | 6.8 | GO:0005200 | structural constituent of cytoskeleton(GO:0005200) |
| 0.0 | 0.7 | GO:0070628 | proteasome binding(GO:0070628) |
| 0.0 | 2.3 | GO:0042169 | SH2 domain binding(GO:0042169) |
| 0.0 | 0.8 | GO:0055106 | ubiquitin-protein transferase regulator activity(GO:0055106) |
| 0.0 | 2.4 | GO:0017112 | Rab guanyl-nucleotide exchange factor activity(GO:0017112) |
| 0.0 | 0.8 | GO:0016863 | intramolecular oxidoreductase activity, transposing C=C bonds(GO:0016863) |
| 0.0 | 2.3 | GO:0005154 | epidermal growth factor receptor binding(GO:0005154) |
| 0.0 | 0.2 | GO:0061575 | cyclin-dependent protein serine/threonine kinase activator activity(GO:0061575) |
| 0.0 | 0.2 | GO:0005047 | signal recognition particle binding(GO:0005047) endoplasmic reticulum signal peptide binding(GO:0030942) |
| 0.0 | 1.2 | GO:0070330 | aromatase activity(GO:0070330) |
| 0.0 | 0.6 | GO:0046933 | proton-transporting ATP synthase activity, rotational mechanism(GO:0046933) |
| 0.0 | 1.3 | GO:0004012 | phospholipid-translocating ATPase activity(GO:0004012) |
| 0.0 | 1.4 | GO:0048020 | CCR chemokine receptor binding(GO:0048020) |
| 0.0 | 0.5 | GO:0019957 | C-C chemokine binding(GO:0019957) |
| 0.0 | 0.5 | GO:0071933 | Arp2/3 complex binding(GO:0071933) |
| 0.0 | 1.2 | GO:0019706 | protein-cysteine S-palmitoyltransferase activity(GO:0019706) protein-cysteine S-acyltransferase activity(GO:0019707) |
| 0.0 | 0.4 | GO:0030976 | thiamine pyrophosphate binding(GO:0030976) |
| 0.0 | 0.8 | GO:0016755 | transferase activity, transferring amino-acyl groups(GO:0016755) |
| 0.0 | 0.4 | GO:0003995 | acyl-CoA dehydrogenase activity(GO:0003995) |
| 0.0 | 2.4 | GO:0031593 | polyubiquitin binding(GO:0031593) |
| 0.0 | 0.1 | GO:0016453 | acetyl-CoA C-acetyltransferase activity(GO:0003985) C-acetyltransferase activity(GO:0016453) |
| 0.0 | 0.9 | GO:0004181 | metallocarboxypeptidase activity(GO:0004181) |
| 0.0 | 0.9 | GO:0005229 | intracellular calcium activated chloride channel activity(GO:0005229) |
| 0.0 | 3.6 | GO:0000979 | RNA polymerase II core promoter sequence-specific DNA binding(GO:0000979) |
| 0.0 | 0.6 | GO:0042608 | T cell receptor binding(GO:0042608) |
| 0.0 | 2.2 | GO:0035064 | methylated histone binding(GO:0035064) |
| 0.0 | 0.6 | GO:0008353 | RNA polymerase II carboxy-terminal domain kinase activity(GO:0008353) |
| 0.0 | 1.2 | GO:0050699 | WW domain binding(GO:0050699) |
| 0.0 | 1.5 | GO:0043022 | ribosome binding(GO:0043022) |
| 0.0 | 0.1 | GO:0001042 | RNA polymerase I core binding(GO:0001042) |
| 0.0 | 0.3 | GO:0008376 | acetylgalactosaminyltransferase activity(GO:0008376) |
| 0.0 | 0.2 | GO:0000268 | peroxisome targeting sequence binding(GO:0000268) |
| 0.0 | 1.6 | GO:0004879 | RNA polymerase II transcription factor activity, ligand-activated sequence-specific DNA binding(GO:0004879) transcription factor activity, direct ligand regulated sequence-specific DNA binding(GO:0098531) |
| 0.0 | 4.3 | GO:0004252 | serine-type endopeptidase activity(GO:0004252) |
| 0.0 | 5.9 | GO:0030246 | carbohydrate binding(GO:0030246) |
| 0.0 | 2.7 | GO:0005179 | hormone activity(GO:0005179) |
| 0.0 | 1.7 | GO:0005518 | collagen binding(GO:0005518) |
| 0.0 | 0.7 | GO:0017134 | fibroblast growth factor binding(GO:0017134) |
| 0.0 | 2.4 | GO:0042393 | histone binding(GO:0042393) |
| 0.0 | 1.8 | GO:0004896 | cytokine receptor activity(GO:0004896) |
| 0.0 | 0.1 | GO:0035368 | selenocysteine insertion sequence binding(GO:0035368) |
| 0.0 | 0.9 | GO:0004364 | glutathione transferase activity(GO:0004364) |
| 0.0 | 1.2 | GO:0003823 | antigen binding(GO:0003823) |
| 0.0 | 0.1 | GO:0017150 | tRNA dihydrouridine synthase activity(GO:0017150) |
| 0.0 | 0.8 | GO:0005246 | calcium channel regulator activity(GO:0005246) |
| 0.0 | 10.4 | GO:0005198 | structural molecule activity(GO:0005198) |
| 0.0 | 0.4 | GO:0022829 | wide pore channel activity(GO:0022829) |
| 0.0 | 0.3 | GO:0031492 | nucleosomal DNA binding(GO:0031492) |
| 0.0 | 0.5 | GO:0005160 | transforming growth factor beta receptor binding(GO:0005160) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 3.7 | SA MMP CYTOKINE CONNECTION | Cytokines can induce activation of matrix metalloproteinases, which degrade extracellular matrix. |
| 0.1 | 4.9 | PID TOLL ENDOGENOUS PATHWAY | Endogenous TLR signaling |
| 0.1 | 8.7 | PID DELTA NP63 PATHWAY | Validated transcriptional targets of deltaNp63 isoforms |
| 0.1 | 2.5 | PID ERBB NETWORK PATHWAY | ErbB receptor signaling network |
| 0.1 | 0.4 | PID IL8 CXCR1 PATHWAY | IL8- and CXCR1-mediated signaling events |
| 0.1 | 6.4 | PID HNF3A PATHWAY | FOXA1 transcription factor network |
| 0.1 | 6.0 | PID HNF3B PATHWAY | FOXA2 and FOXA3 transcription factor networks |
| 0.1 | 14.3 | NABA ECM AFFILIATED | Genes encoding proteins affiliated structurally or functionally to extracellular matrix proteins |
| 0.1 | 3.9 | SIG REGULATION OF THE ACTIN CYTOSKELETON BY RHO GTPASES | Genes related to regulation of the actin cytoskeleton |
| 0.1 | 1.1 | PID S1P S1P4 PATHWAY | S1P4 pathway |
| 0.1 | 2.2 | ST INTERLEUKIN 4 PATHWAY | Interleukin 4 (IL-4) Pathway |
| 0.0 | 1.9 | PID P38 MKK3 6PATHWAY | p38 MAPK signaling pathway |
| 0.0 | 1.5 | PID IL1 PATHWAY | IL1-mediated signaling events |
| 0.0 | 10.6 | NABA ECM REGULATORS | Genes encoding enzymes and their regulators involved in the remodeling of the extracellular matrix |
| 0.0 | 1.2 | PID IL27 PATHWAY | IL27-mediated signaling events |
| 0.0 | 0.8 | PID HEDGEHOG 2PATHWAY | Signaling events mediated by the Hedgehog family |
| 0.0 | 1.2 | PID ERBB1 RECEPTOR PROXIMAL PATHWAY | EGF receptor (ErbB1) signaling pathway |
| 0.0 | 1.2 | PID EPHRINB REV PATHWAY | Ephrin B reverse signaling |
| 0.0 | 2.5 | PID HIF1 TFPATHWAY | HIF-1-alpha transcription factor network |
| 0.0 | 1.1 | PID CDC42 PATHWAY | CDC42 signaling events |
| 0.0 | 2.0 | PID MET PATHWAY | Signaling events mediated by Hepatocyte Growth Factor Receptor (c-Met) |
| 0.0 | 1.9 | PID ERA GENOMIC PATHWAY | Validated nuclear estrogen receptor alpha network |
| 0.0 | 1.1 | ST DIFFERENTIATION PATHWAY IN PC12 CELLS | Differentiation Pathway in PC12 Cells; this is a specific case of PAC1 Receptor Pathway. |
| 0.0 | 1.0 | PID RAC1 PATHWAY | RAC1 signaling pathway |
| 0.0 | 2.1 | PID AR PATHWAY | Coregulation of Androgen receptor activity |
| 0.0 | 0.3 | PID MYC PATHWAY | C-MYC pathway |
| 0.0 | 0.5 | PID IL12 2PATHWAY | IL12-mediated signaling events |
| 0.0 | 0.5 | PID PS1 PATHWAY | Presenilin action in Notch and Wnt signaling |
| 0.0 | 1.0 | PID BETA CATENIN NUC PATHWAY | Regulation of nuclear beta catenin signaling and target gene transcription |
| 0.0 | 0.1 | SA PTEN PATHWAY | PTEN is a tumor suppressor that dephosphorylates the lipid messenger phosphatidylinositol triphosphate. |
| 0.0 | 0.3 | PID INTEGRIN CS PATHWAY | Integrin family cell surface interactions |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.8 | 18.7 | REACTOME SYNTHESIS SECRETION AND DEACYLATION OF GHRELIN | Genes involved in Synthesis, Secretion, and Deacylation of Ghrelin |
| 0.5 | 7.1 | REACTOME NEF MEDIATED DOWNREGULATION OF MHC CLASS I COMPLEX CELL SURFACE EXPRESSION | Genes involved in Nef mediated downregulation of MHC class I complex cell surface expression |
| 0.4 | 1.2 | REACTOME XENOBIOTICS | Genes involved in Xenobiotics |
| 0.4 | 9.9 | REACTOME ZINC TRANSPORTERS | Genes involved in Zinc transporters |
| 0.2 | 0.5 | REACTOME ENDOSOMAL VACUOLAR PATHWAY | Genes involved in Endosomal/Vacuolar pathway |
| 0.2 | 6.3 | REACTOME TERMINATION OF O GLYCAN BIOSYNTHESIS | Genes involved in Termination of O-glycan biosynthesis |
| 0.2 | 1.9 | REACTOME KERATAN SULFATE DEGRADATION | Genes involved in Keratan sulfate degradation |
| 0.2 | 1.8 | REACTOME RECYCLING OF BILE ACIDS AND SALTS | Genes involved in Recycling of bile acids and salts |
| 0.2 | 1.9 | REACTOME HYALURONAN UPTAKE AND DEGRADATION | Genes involved in Hyaluronan uptake and degradation |
| 0.1 | 3.8 | REACTOME AMINO ACID SYNTHESIS AND INTERCONVERSION TRANSAMINATION | Genes involved in Amino acid synthesis and interconversion (transamination) |
| 0.1 | 0.5 | REACTOME BINDING AND ENTRY OF HIV VIRION | Genes involved in Binding and entry of HIV virion |
| 0.1 | 7.0 | REACTOME IMMUNOREGULATORY INTERACTIONS BETWEEN A LYMPHOID AND A NON LYMPHOID CELL | Genes involved in Immunoregulatory interactions between a Lymphoid and a non-Lymphoid cell |
| 0.1 | 1.6 | REACTOME CYTOSOLIC SULFONATION OF SMALL MOLECULES | Genes involved in Cytosolic sulfonation of small molecules |
| 0.1 | 2.0 | REACTOME SYNTHESIS OF BILE ACIDS AND BILE SALTS VIA 7ALPHA HYDROXYCHOLESTEROL | Genes involved in Synthesis of bile acids and bile salts via 7alpha-hydroxycholesterol |
| 0.1 | 2.2 | REACTOME METABOLISM OF PORPHYRINS | Genes involved in Metabolism of porphyrins |
| 0.1 | 0.9 | REACTOME IRAK2 MEDIATED ACTIVATION OF TAK1 COMPLEX UPON TLR7 8 OR 9 STIMULATION | Genes involved in IRAK2 mediated activation of TAK1 complex upon TLR7/8 or 9 stimulation |
| 0.1 | 1.4 | REACTOME NFKB ACTIVATION THROUGH FADD RIP1 PATHWAY MEDIATED BY CASPASE 8 AND10 | Genes involved in NF-kB activation through FADD/RIP-1 pathway mediated by caspase-8 and -10 |
| 0.1 | 1.2 | REACTOME PD1 SIGNALING | Genes involved in PD-1 signaling |
| 0.1 | 1.2 | REACTOME IL 6 SIGNALING | Genes involved in Interleukin-6 signaling |
| 0.1 | 3.3 | REACTOME CHEMOKINE RECEPTORS BIND CHEMOKINES | Genes involved in Chemokine receptors bind chemokines |
| 0.1 | 0.9 | REACTOME GAMMA CARBOXYLATION TRANSPORT AND AMINO TERMINAL CLEAVAGE OF PROTEINS | Genes involved in Gamma-carboxylation, transport, and amino-terminal cleavage of proteins |
| 0.1 | 1.1 | REACTOME ANTIGEN PRESENTATION FOLDING ASSEMBLY AND PEPTIDE LOADING OF CLASS I MHC | Genes involved in Antigen Presentation: Folding, assembly and peptide loading of class I MHC |
| 0.1 | 0.8 | REACTOME NEGATIVE REGULATION OF THE PI3K AKT NETWORK | Genes involved in Negative regulation of the PI3K/AKT network |
| 0.1 | 3.0 | REACTOME TRANSPORT OF VITAMINS NUCLEOSIDES AND RELATED MOLECULES | Genes involved in Transport of vitamins, nucleosides, and related molecules |
| 0.1 | 1.4 | REACTOME PEROXISOMAL LIPID METABOLISM | Genes involved in Peroxisomal lipid metabolism |
| 0.1 | 1.2 | REACTOME INFLAMMASOMES | Genes involved in Inflammasomes |
| 0.1 | 1.7 | REACTOME SULFUR AMINO ACID METABOLISM | Genes involved in Sulfur amino acid metabolism |
| 0.0 | 1.0 | REACTOME CLASS C 3 METABOTROPIC GLUTAMATE PHEROMONE RECEPTORS | Genes involved in Class C/3 (Metabotropic glutamate/pheromone receptors) |
| 0.0 | 2.1 | REACTOME REGULATION OF BETA CELL DEVELOPMENT | Genes involved in Regulation of beta-cell development |
| 0.0 | 1.6 | REACTOME SYNTHESIS OF PC | Genes involved in Synthesis of PC |
| 0.0 | 1.1 | REACTOME SYNTHESIS OF PIPS AT THE GOLGI MEMBRANE | Genes involved in Synthesis of PIPs at the Golgi membrane |
| 0.0 | 1.8 | REACTOME TRANSPORT TO THE GOLGI AND SUBSEQUENT MODIFICATION | Genes involved in Transport to the Golgi and subsequent modification |
| 0.0 | 1.4 | REACTOME SYNTHESIS OF VERY LONG CHAIN FATTY ACYL COAS | Genes involved in Synthesis of very long-chain fatty acyl-CoAs |
| 0.0 | 2.3 | REACTOME FORMATION OF THE TERNARY COMPLEX AND SUBSEQUENTLY THE 43S COMPLEX | Genes involved in Formation of the ternary complex, and subsequently, the 43S complex |
| 0.0 | 3.6 | REACTOME MYD88 MAL CASCADE INITIATED ON PLASMA MEMBRANE | Genes involved in MyD88:Mal cascade initiated on plasma membrane |
| 0.0 | 1.2 | REACTOME ENOS ACTIVATION AND REGULATION | Genes involved in eNOS activation and regulation |
| 0.0 | 1.5 | REACTOME IL1 SIGNALING | Genes involved in Interleukin-1 signaling |
| 0.0 | 0.6 | REACTOME FORMATION OF ATP BY CHEMIOSMOTIC COUPLING | Genes involved in Formation of ATP by chemiosmotic coupling |
| 0.0 | 0.4 | REACTOME INTEGRATION OF PROVIRUS | Genes involved in Integration of provirus |
| 0.0 | 0.6 | REACTOME FORMATION OF INCISION COMPLEX IN GG NER | Genes involved in Formation of incision complex in GG-NER |
| 0.0 | 1.3 | REACTOME ION TRANSPORT BY P TYPE ATPASES | Genes involved in Ion transport by P-type ATPases |
| 0.0 | 2.1 | REACTOME REGULATION OF MRNA STABILITY BY PROTEINS THAT BIND AU RICH ELEMENTS | Genes involved in Regulation of mRNA Stability by Proteins that Bind AU-rich Elements |
| 0.0 | 0.4 | REACTOME ADENYLATE CYCLASE INHIBITORY PATHWAY | Genes involved in Adenylate cyclase inhibitory pathway |
| 0.0 | 1.1 | REACTOME PHASE II CONJUGATION | Genes involved in Phase II conjugation |
| 0.0 | 0.7 | REACTOME AMINE LIGAND BINDING RECEPTORS | Genes involved in Amine ligand-binding receptors |
| 0.0 | 0.6 | REACTOME MEIOTIC SYNAPSIS | Genes involved in Meiotic Synapsis |
| 0.0 | 0.4 | REACTOME RNA POL III TRANSCRIPTION INITIATION FROM TYPE 3 PROMOTER | Genes involved in RNA Polymerase III Transcription Initiation From Type 3 Promoter |