Project
PRJNA375882: Comprehensive Mouse Transcriptomic BodyMap
Navigation
Downloads
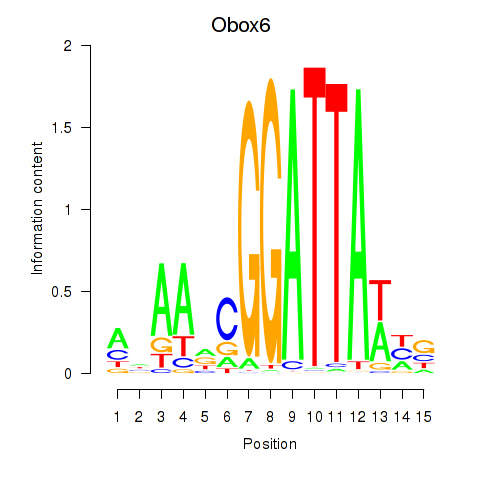
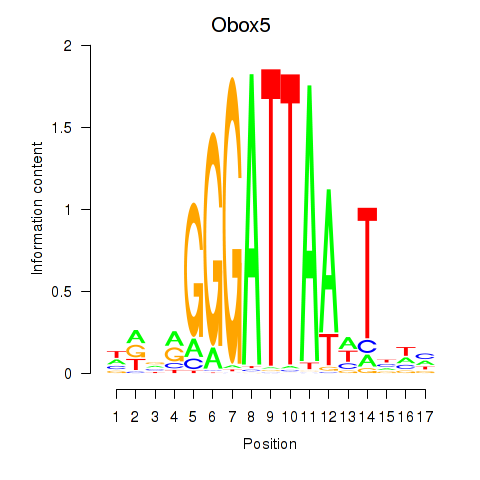
Results for Obox6_Obox5
Z-value: 1.37
Transcription factors associated with Obox6_Obox5
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Obox6
|
ENSMUSG00000041583.9 | Obox6 |
|
Obox5
|
ENSMUSG00000074366.10 | Obox5 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Obox5 | mm39_v1_chr7_+_15484056_15484056 | -0.02 | 8.5e-01 | Click! |
Activity profile of Obox6_Obox5 motif
Sorted Z-values of Obox6_Obox5 motif
Network of associatons between targets according to the STRING database.
First level regulatory network of Obox6_Obox5
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr5_-_87572060 | 24.59 |
ENSMUST00000072818.6
|
Ugt2b38
|
UDP glucuronosyltransferase 2 family, polypeptide B38 |
| chrX_+_20416019 | 11.57 |
ENSMUST00000023832.7
|
Rgn
|
regucalcin |
| chr1_-_139708906 | 10.49 |
ENSMUST00000111986.8
ENSMUST00000027612.11 ENSMUST00000111989.9 |
Cfhr4
|
complement factor H-related 4 |
| chr7_+_26534730 | 10.11 |
ENSMUST00000005685.15
|
Cyp2a5
|
cytochrome P450, family 2, subfamily a, polypeptide 5 |
| chr4_-_130116119 | 10.03 |
ENSMUST00000023884.7
|
Ldc1
|
leucine decarboxylase 1 |
| chr8_+_114860375 | 8.70 |
ENSMUST00000147605.8
ENSMUST00000134593.2 |
Nudt7
|
nudix (nucleoside diphosphate linked moiety X)-type motif 7 |
| chr7_-_119078472 | 7.66 |
ENSMUST00000209095.2
ENSMUST00000033263.6 ENSMUST00000207261.2 |
Umod
|
uromodulin |
| chr19_-_8382424 | 7.46 |
ENSMUST00000064507.12
ENSMUST00000120540.2 ENSMUST00000096269.11 |
Slc22a30
|
solute carrier family 22, member 30 |
| chr19_-_7688628 | 7.43 |
ENSMUST00000025666.8
|
Slc22a19
|
solute carrier family 22 (organic anion transporter), member 19 |
| chr11_+_104122216 | 7.35 |
ENSMUST00000106992.10
|
Mapt
|
microtubule-associated protein tau |
| chr10_-_76949762 | 6.55 |
ENSMUST00000072755.12
|
Col18a1
|
collagen, type XVIII, alpha 1 |
| chr1_-_139487951 | 6.45 |
ENSMUST00000023965.8
|
Cfhr1
|
complement factor H-related 1 |
| chr4_+_115420876 | 6.39 |
ENSMUST00000126645.8
ENSMUST00000030480.4 |
Cyp4a31
|
cytochrome P450, family 4, subfamily a, polypeptide 31 |
| chr13_+_55547498 | 6.29 |
ENSMUST00000057167.9
|
Slc34a1
|
solute carrier family 34 (sodium phosphate), member 1 |
| chr8_+_114860342 | 6.25 |
ENSMUST00000109109.8
|
Nudt7
|
nudix (nucleoside diphosphate linked moiety X)-type motif 7 |
| chr4_+_115420817 | 6.24 |
ENSMUST00000141033.8
ENSMUST00000030486.15 |
Cyp4a31
|
cytochrome P450, family 4, subfamily a, polypeptide 31 |
| chr13_+_4624074 | 6.08 |
ENSMUST00000021628.4
|
Akr1c21
|
aldo-keto reductase family 1, member C21 |
| chr13_-_4659120 | 6.03 |
ENSMUST00000091848.7
ENSMUST00000110691.10 |
Akr1e1
|
aldo-keto reductase family 1, member E1 |
| chr11_-_59937302 | 5.85 |
ENSMUST00000000310.14
ENSMUST00000102693.9 ENSMUST00000148512.2 |
Pemt
|
phosphatidylethanolamine N-methyltransferase |
| chr7_-_119078330 | 5.80 |
ENSMUST00000207460.2
|
Umod
|
uromodulin |
| chr13_-_41373638 | 5.74 |
ENSMUST00000117096.2
|
Elovl2
|
elongation of very long chain fatty acids (FEN1/Elo2, SUR4/Elo3, yeast)-like 2 |
| chr5_-_87288177 | 5.74 |
ENSMUST00000067790.7
|
Ugt2b5
|
UDP glucuronosyltransferase 2 family, polypeptide B5 |
| chr10_+_127637015 | 5.58 |
ENSMUST00000071646.2
|
Rdh16
|
retinol dehydrogenase 16 |
| chr8_+_114860297 | 5.57 |
ENSMUST00000073521.12
ENSMUST00000066514.13 |
Nudt7
|
nudix (nucleoside diphosphate linked moiety X)-type motif 7 |
| chr3_+_87704258 | 5.56 |
ENSMUST00000029711.9
ENSMUST00000107582.3 |
Insrr
|
insulin receptor-related receptor |
| chr9_+_119186447 | 5.38 |
ENSMUST00000039610.10
|
Xylb
|
xylulokinase homolog (H. influenzae) |
| chr9_+_32283779 | 5.26 |
ENSMUST00000047334.10
|
Kcnj1
|
potassium inwardly-rectifying channel, subfamily J, member 1 |
| chr9_-_53521585 | 5.09 |
ENSMUST00000034547.6
|
Acat1
|
acetyl-Coenzyme A acetyltransferase 1 |
| chr3_+_151143557 | 5.06 |
ENSMUST00000196970.3
|
Adgrl4
|
adhesion G protein-coupled receptor L4 |
| chr7_+_119206233 | 5.00 |
ENSMUST00000126367.8
|
Acsm1
|
acyl-CoA synthetase medium-chain family member 1 |
| chr11_-_62172164 | 4.98 |
ENSMUST00000072916.5
|
Zswim7
|
zinc finger SWIM-type containing 7 |
| chr3_-_73615732 | 4.97 |
ENSMUST00000029367.6
|
Bche
|
butyrylcholinesterase |
| chr7_+_26006594 | 4.97 |
ENSMUST00000098657.5
|
Cyp2a4
|
cytochrome P450, family 2, subfamily a, polypeptide 4 |
| chr7_+_51530060 | 4.87 |
ENSMUST00000145049.2
|
Gas2
|
growth arrest specific 2 |
| chr19_-_8109346 | 4.69 |
ENSMUST00000065651.5
|
Slc22a28
|
solute carrier family 22, member 28 |
| chr1_-_139786421 | 4.64 |
ENSMUST00000194186.6
ENSMUST00000094489.5 ENSMUST00000239380.2 |
Cfhr2
|
complement factor H-related 2 |
| chr6_-_71121324 | 4.62 |
ENSMUST00000074241.9
|
Thnsl2
|
threonine synthase-like 2 (bacterial) |
| chr1_-_119576632 | 4.62 |
ENSMUST00000163147.8
ENSMUST00000052404.13 ENSMUST00000191046.7 |
Epb41l5
|
erythrocyte membrane protein band 4.1 like 5 |
| chr19_-_6593049 | 4.60 |
ENSMUST00000113451.9
|
Slc22a12
|
solute carrier family 22 (organic anion/cation transporter), member 12 |
| chr7_-_12732067 | 4.47 |
ENSMUST00000032539.14
ENSMUST00000120903.8 |
Slc27a5
|
solute carrier family 27 (fatty acid transporter), member 5 |
| chr6_-_71121347 | 4.39 |
ENSMUST00000160918.8
|
Thnsl2
|
threonine synthase-like 2 (bacterial) |
| chr6_-_21851827 | 4.30 |
ENSMUST00000202353.2
ENSMUST00000134635.2 ENSMUST00000123116.8 ENSMUST00000120965.8 ENSMUST00000143531.2 |
Tspan12
|
tetraspanin 12 |
| chr4_+_101276474 | 4.26 |
ENSMUST00000102780.8
ENSMUST00000106946.8 ENSMUST00000106945.8 |
Ak4
|
adenylate kinase 4 |
| chr7_+_119160922 | 4.25 |
ENSMUST00000130583.2
ENSMUST00000084647.13 |
Acsm2
|
acyl-CoA synthetase medium-chain family member 2 |
| chr13_+_4486105 | 4.23 |
ENSMUST00000156277.2
|
Akr1c6
|
aldo-keto reductase family 1, member C6 |
| chr19_+_8595369 | 3.97 |
ENSMUST00000010250.4
|
Slc22a6
|
solute carrier family 22 (organic anion transporter), member 6 |
| chr11_-_109613040 | 3.83 |
ENSMUST00000020938.8
|
Fam20a
|
FAM20A, golgi associated secretory pathway pseudokinase |
| chr1_+_21310821 | 3.83 |
ENSMUST00000121676.8
ENSMUST00000124990.3 |
Gsta3
|
glutathione S-transferase, alpha 3 |
| chr2_-_27138347 | 3.83 |
ENSMUST00000139312.8
|
Sardh
|
sarcosine dehydrogenase |
| chr15_+_6329278 | 3.82 |
ENSMUST00000159046.2
ENSMUST00000161040.8 |
Dab2
|
disabled 2, mitogen-responsive phosphoprotein |
| chr15_+_6329263 | 3.72 |
ENSMUST00000078019.13
|
Dab2
|
disabled 2, mitogen-responsive phosphoprotein |
| chr13_+_4109566 | 3.71 |
ENSMUST00000041768.7
|
Akr1c14
|
aldo-keto reductase family 1, member C14 |
| chr9_+_65536892 | 3.67 |
ENSMUST00000169003.8
|
Rbpms2
|
RNA binding protein with multiple splicing 2 |
| chr1_+_21310803 | 3.64 |
ENSMUST00000027067.15
|
Gsta3
|
glutathione S-transferase, alpha 3 |
| chr14_+_66205932 | 3.63 |
ENSMUST00000022616.14
|
Clu
|
clusterin |
| chr10_+_79977291 | 3.63 |
ENSMUST00000105367.8
|
Atp5d
|
ATP synthase, H+ transporting, mitochondrial F1 complex, delta subunit |
| chr7_+_140500848 | 3.59 |
ENSMUST00000184560.2
|
Nlrp6
|
NLR family, pyrin domain containing 6 |
| chr1_+_88093726 | 3.54 |
ENSMUST00000097659.5
|
Ugt1a5
|
UDP glucuronosyltransferase 1 family, polypeptide A5 |
| chr11_+_98932586 | 3.51 |
ENSMUST00000177092.8
|
Igfbp4
|
insulin-like growth factor binding protein 4 |
| chr11_+_3981769 | 3.51 |
ENSMUST00000019512.8
|
Sec14l4
|
SEC14-like lipid binding 4 |
| chr9_-_48516447 | 3.47 |
ENSMUST00000034808.12
ENSMUST00000119426.2 |
Nnmt
|
nicotinamide N-methyltransferase |
| chr11_-_30599510 | 3.41 |
ENSMUST00000074613.4
|
Acyp2
|
acylphosphatase 2, muscle type |
| chr5_-_87240405 | 3.39 |
ENSMUST00000132667.2
ENSMUST00000145617.8 ENSMUST00000094649.11 |
Ugt2b36
|
UDP glucuronosyltransferase 2 family, polypeptide B36 |
| chr19_+_39980868 | 3.36 |
ENSMUST00000049178.3
|
Cyp2c37
|
cytochrome P450, family 2. subfamily c, polypeptide 37 |
| chr2_+_19376447 | 3.34 |
ENSMUST00000023856.9
|
Msrb2
|
methionine sulfoxide reductase B2 |
| chr3_-_73615535 | 3.33 |
ENSMUST00000138216.8
|
Bche
|
butyrylcholinesterase |
| chr6_-_85739462 | 3.29 |
ENSMUST00000168531.2
|
Nat8f3
|
N-acetyltransferase 8 (GCN5-related) family member 3 |
| chr10_+_94034817 | 3.22 |
ENSMUST00000020209.16
ENSMUST00000179990.8 |
Ndufa12
|
NADH:ubiquinone oxidoreductase subunit A12 |
| chr14_+_40826970 | 3.18 |
ENSMUST00000225720.2
|
Mat1a
|
methionine adenosyltransferase I, alpha |
| chr8_+_123920233 | 3.17 |
ENSMUST00000212773.2
|
Dpep1
|
dipeptidase 1 |
| chr19_-_7943365 | 3.16 |
ENSMUST00000182102.8
ENSMUST00000075619.5 |
Slc22a27
|
solute carrier family 22, member 27 |
| chr1_-_140111138 | 3.10 |
ENSMUST00000111976.9
ENSMUST00000066859.13 |
Cfh
|
complement component factor h |
| chr14_-_31362835 | 3.08 |
ENSMUST00000167066.8
ENSMUST00000127204.9 |
Hacl1
|
2-hydroxyacyl-CoA lyase 1 |
| chr7_-_86425016 | 3.06 |
ENSMUST00000107271.10
|
Folh1
|
folate hydrolase 1 |
| chr13_-_4200627 | 3.06 |
ENSMUST00000110704.9
ENSMUST00000021635.9 |
Akr1c18
|
aldo-keto reductase family 1, member C18 |
| chr2_+_144435974 | 3.03 |
ENSMUST00000136628.2
|
Smim26
|
small integral membrane protein 26 |
| chr8_+_127790772 | 3.02 |
ENSMUST00000079777.12
ENSMUST00000160272.8 ENSMUST00000162907.8 ENSMUST00000162536.8 ENSMUST00000026921.13 ENSMUST00000162665.8 ENSMUST00000162602.8 ENSMUST00000160581.8 ENSMUST00000161355.8 ENSMUST00000162531.8 ENSMUST00000160766.8 ENSMUST00000159537.8 |
Pard3
|
par-3 family cell polarity regulator |
| chr12_-_103660946 | 3.00 |
ENSMUST00000118101.2
|
Serpina1f
|
serine (or cysteine) peptidase inhibitor, clade A, member 1F |
| chr4_-_138546564 | 2.99 |
ENSMUST00000102512.11
|
Pla2g5
|
phospholipase A2, group V |
| chr10_-_76895779 | 2.93 |
ENSMUST00000218407.2
|
Col18a1
|
collagen, type XVIII, alpha 1 |
| chr7_-_119122681 | 2.90 |
ENSMUST00000033267.4
|
Pdilt
|
protein disulfide isomerase-like, testis expressed |
| chr14_-_64654397 | 2.88 |
ENSMUST00000210428.2
|
Msra
|
methionine sulfoxide reductase A |
| chr1_-_140111018 | 2.88 |
ENSMUST00000192880.6
ENSMUST00000111977.8 |
Cfh
|
complement component factor h |
| chr9_+_110782861 | 2.86 |
ENSMUST00000196834.2
|
Lrrc2
|
leucine rich repeat containing 2 |
| chr13_+_4278681 | 2.85 |
ENSMUST00000118663.9
|
Akr1c19
|
aldo-keto reductase family 1, member C19 |
| chr16_+_31241085 | 2.85 |
ENSMUST00000089759.9
|
Bdh1
|
3-hydroxybutyrate dehydrogenase, type 1 |
| chr19_-_10079091 | 2.82 |
ENSMUST00000025567.9
|
Fads2
|
fatty acid desaturase 2 |
| chr11_-_116080361 | 2.82 |
ENSMUST00000148601.2
|
Acox1
|
acyl-Coenzyme A oxidase 1, palmitoyl |
| chr3_+_118355811 | 2.82 |
ENSMUST00000149101.3
|
Dpyd
|
dihydropyrimidine dehydrogenase |
| chr3_-_88162012 | 2.80 |
ENSMUST00000171887.4
|
Rhbg
|
Rhesus blood group-associated B glycoprotein |
| chr7_-_101518484 | 2.77 |
ENSMUST00000140584.2
ENSMUST00000134145.8 |
Folr1
|
folate receptor 1 (adult) |
| chrX_-_84820250 | 2.74 |
ENSMUST00000113978.9
|
Gk
|
glycerol kinase |
| chr9_+_108356935 | 2.71 |
ENSMUST00000194147.2
ENSMUST00000065014.10 ENSMUST00000195483.6 ENSMUST00000195058.2 |
Lamb2
|
laminin, beta 2 |
| chr9_+_68561042 | 2.70 |
ENSMUST00000034766.14
|
Rora
|
RAR-related orphan receptor alpha |
| chr5_+_87148697 | 2.69 |
ENSMUST00000031186.9
|
Ugt2b35
|
UDP glucuronosyltransferase 2 family, polypeptide B35 |
| chr15_+_76579885 | 2.67 |
ENSMUST00000231028.2
|
Gpt
|
glutamic pyruvic transaminase, soluble |
| chr9_-_103107495 | 2.65 |
ENSMUST00000035158.16
|
Trf
|
transferrin |
| chr13_-_64514830 | 2.59 |
ENSMUST00000222971.2
|
Ctsl
|
cathepsin L |
| chr5_+_25427860 | 2.56 |
ENSMUST00000045737.14
|
Galnt11
|
polypeptide N-acetylgalactosaminyltransferase 11 |
| chr5_-_121798541 | 2.56 |
ENSMUST00000031412.12
ENSMUST00000111770.2 |
Acad10
|
acyl-Coenzyme A dehydrogenase family, member 10 |
| chr1_-_186947651 | 2.55 |
ENSMUST00000183819.8
|
Spata17
|
spermatogenesis associated 17 |
| chrX_+_141009756 | 2.54 |
ENSMUST00000112916.9
|
Nxt2
|
nuclear transport factor 2-like export factor 2 |
| chr4_-_138590764 | 2.53 |
ENSMUST00000030524.14
ENSMUST00000102513.8 |
Pla2g5
|
phospholipase A2, group V |
| chr4_+_134042423 | 2.53 |
ENSMUST00000105875.8
ENSMUST00000030638.7 |
Trim63
|
tripartite motif-containing 63 |
| chr15_-_76544308 | 2.47 |
ENSMUST00000066677.10
ENSMUST00000177359.2 |
Cyhr1
|
cysteine and histidine rich 1 |
| chr6_-_85797917 | 2.46 |
ENSMUST00000174143.2
|
Nat8f6
|
N-acetyltransferase 8 (GCN5-related) family member 6 |
| chr11_-_53321242 | 2.43 |
ENSMUST00000109019.8
|
Uqcrq
|
ubiquinol-cytochrome c reductase, complex III subunit VII |
| chr5_-_18093739 | 2.43 |
ENSMUST00000169095.6
ENSMUST00000197574.2 |
Cd36
|
CD36 molecule |
| chr12_-_102390000 | 2.39 |
ENSMUST00000110020.8
|
Lgmn
|
legumain |
| chr4_-_137512682 | 2.38 |
ENSMUST00000133473.2
|
Alpl
|
alkaline phosphatase, liver/bone/kidney |
| chrX_-_74423647 | 2.38 |
ENSMUST00000114085.9
|
F8
|
coagulation factor VIII |
| chr14_+_40827317 | 2.37 |
ENSMUST00000047286.7
|
Mat1a
|
methionine adenosyltransferase I, alpha |
| chr13_-_90237631 | 2.35 |
ENSMUST00000160232.8
|
Xrcc4
|
X-ray repair complementing defective repair in Chinese hamster cells 4 |
| chr2_-_13496624 | 2.35 |
ENSMUST00000091436.7
|
Cubn
|
cubilin (intrinsic factor-cobalamin receptor) |
| chr15_+_76579960 | 2.35 |
ENSMUST00000229679.2
|
Gpt
|
glutamic pyruvic transaminase, soluble |
| chr19_+_28812474 | 2.34 |
ENSMUST00000025875.5
|
Slc1a1
|
solute carrier family 1 (neuronal/epithelial high affinity glutamate transporter, system Xag), member 1 |
| chr2_+_67276338 | 2.33 |
ENSMUST00000239060.2
ENSMUST00000028410.4 ENSMUST00000112347.8 ENSMUST00000238878.2 |
Xirp2
|
xin actin-binding repeat containing 2 |
| chr14_-_34225281 | 2.31 |
ENSMUST00000171343.9
|
Bmpr1a
|
bone morphogenetic protein receptor, type 1A |
| chr14_-_30645711 | 2.29 |
ENSMUST00000006697.17
|
Itih3
|
inter-alpha trypsin inhibitor, heavy chain 3 |
| chr3_-_63872189 | 2.28 |
ENSMUST00000029402.15
|
Slc33a1
|
solute carrier family 33 (acetyl-CoA transporter), member 1 |
| chr1_-_121255503 | 2.27 |
ENSMUST00000160688.2
|
Insig2
|
insulin induced gene 2 |
| chr7_+_133311062 | 2.26 |
ENSMUST00000033282.5
|
Bccip
|
BRCA2 and CDKN1A interacting protein |
| chr16_+_22739028 | 2.25 |
ENSMUST00000232097.2
|
Fetub
|
fetuin beta |
| chrY_+_1010543 | 2.25 |
ENSMUST00000091197.4
|
Eif2s3y
|
eukaryotic translation initiation factor 2, subunit 3, structural gene Y-linked |
| chr15_+_100202021 | 2.23 |
ENSMUST00000230472.2
|
Mettl7a1
|
methyltransferase like 7A1 |
| chr10_+_120044650 | 2.22 |
ENSMUST00000020446.11
ENSMUST00000134797.8 |
Tmbim4
|
transmembrane BAX inhibitor motif containing 4 |
| chr9_-_103107460 | 2.21 |
ENSMUST00000165296.8
ENSMUST00000112645.8 |
Trf
|
transferrin |
| chr9_+_64028783 | 2.19 |
ENSMUST00000118215.3
|
Lctl
|
lactase-like |
| chr12_+_52144511 | 2.16 |
ENSMUST00000040090.16
|
Nubpl
|
nucleotide binding protein-like |
| chr1_+_182591425 | 2.13 |
ENSMUST00000155229.7
ENSMUST00000153348.8 |
Susd4
|
sushi domain containing 4 |
| chr1_-_121255448 | 2.08 |
ENSMUST00000186915.2
ENSMUST00000160968.8 ENSMUST00000162582.2 |
Insig2
|
insulin induced gene 2 |
| chr13_+_99481283 | 2.06 |
ENSMUST00000052249.7
ENSMUST00000224660.3 |
Mrps27
|
mitochondrial ribosomal protein S27 |
| chr11_+_60956624 | 2.06 |
ENSMUST00000041944.9
ENSMUST00000108717.3 |
Kcnj12
|
potassium inwardly-rectifying channel, subfamily J, member 12 |
| chr17_+_56935118 | 2.04 |
ENSMUST00000112979.4
|
Catsperd
|
cation channel sperm associated auxiliary subunit delta |
| chr6_-_85797946 | 2.04 |
ENSMUST00000032074.5
|
Nat8f5
|
N-acetyltransferase 8 (GCN5-related) family member 5 |
| chr5_-_91550853 | 2.01 |
ENSMUST00000121044.6
|
Btc
|
betacellulin, epidermal growth factor family member |
| chr3_+_94217396 | 2.01 |
ENSMUST00000049822.10
|
Them4
|
thioesterase superfamily member 4 |
| chr10_-_86843878 | 2.00 |
ENSMUST00000035288.17
|
Stab2
|
stabilin 2 |
| chr1_-_121255400 | 1.99 |
ENSMUST00000159085.8
ENSMUST00000159125.2 ENSMUST00000161818.2 |
Insig2
|
insulin induced gene 2 |
| chr6_+_137387718 | 1.99 |
ENSMUST00000167002.4
|
Ptpro
|
protein tyrosine phosphatase, receptor type, O |
| chr11_+_6242555 | 1.98 |
ENSMUST00000081894.5
|
Ogdh
|
oxoglutarate (alpha-ketoglutarate) dehydrogenase (lipoamide) |
| chr5_+_105848598 | 1.98 |
ENSMUST00000120847.8
|
Lrrc8d
|
leucine rich repeat containing 8D |
| chr3_+_55023594 | 1.98 |
ENSMUST00000146109.2
|
Spg20
|
spastic paraplegia 20, spartin (Troyer syndrome) homolog (human) |
| chr11_+_6242119 | 1.97 |
ENSMUST00000135124.8
|
Ogdh
|
oxoglutarate (alpha-ketoglutarate) dehydrogenase (lipoamide) |
| chr1_-_121255753 | 1.95 |
ENSMUST00000003818.14
|
Insig2
|
insulin induced gene 2 |
| chr11_+_96920751 | 1.93 |
ENSMUST00000021249.11
|
Scrn2
|
secernin 2 |
| chr19_+_3373285 | 1.90 |
ENSMUST00000025835.6
|
Cpt1a
|
carnitine palmitoyltransferase 1a, liver |
| chr8_-_65489791 | 1.90 |
ENSMUST00000124790.8
|
Apela
|
apelin receptor early endogenous ligand |
| chr10_-_75617245 | 1.88 |
ENSMUST00000001715.10
|
Gstt3
|
glutathione S-transferase, theta 3 |
| chr11_-_53871121 | 1.84 |
ENSMUST00000076493.11
|
Slc22a21
|
solute carrier family 22 (organic cation transporter), member 21 |
| chr3_+_107137924 | 1.83 |
ENSMUST00000179399.3
|
A630076J17Rik
|
RIKEN cDNA A630076J17 gene |
| chr19_-_17316906 | 1.82 |
ENSMUST00000169897.2
|
Gcnt1
|
glucosaminyl (N-acetyl) transferase 1, core 2 |
| chr14_-_30645503 | 1.82 |
ENSMUST00000227995.2
|
Itih3
|
inter-alpha trypsin inhibitor, heavy chain 3 |
| chr1_-_10108325 | 1.79 |
ENSMUST00000027050.10
ENSMUST00000188619.2 |
Cops5
|
COP9 signalosome subunit 5 |
| chr9_-_64248570 | 1.73 |
ENSMUST00000068367.14
|
Dis3l
|
DIS3 like exosome 3'-5' exoribonuclease |
| chr2_-_23939401 | 1.73 |
ENSMUST00000051416.12
|
Hnmt
|
histamine N-methyltransferase |
| chr11_-_70350783 | 1.70 |
ENSMUST00000019064.9
|
Cxcl16
|
chemokine (C-X-C motif) ligand 16 |
| chr9_-_71075939 | 1.68 |
ENSMUST00000113570.8
|
Aqp9
|
aquaporin 9 |
| chr16_+_22739191 | 1.68 |
ENSMUST00000116625.10
|
Fetub
|
fetuin beta |
| chr8_-_65489834 | 1.67 |
ENSMUST00000142822.4
|
Apela
|
apelin receptor early endogenous ligand |
| chr9_+_77661808 | 1.67 |
ENSMUST00000034905.9
|
Gclc
|
glutamate-cysteine ligase, catalytic subunit |
| chr11_-_88754543 | 1.65 |
ENSMUST00000107904.3
|
Akap1
|
A kinase (PRKA) anchor protein 1 |
| chr11_-_120618052 | 1.64 |
ENSMUST00000106148.10
ENSMUST00000026144.5 |
Dcxr
|
dicarbonyl L-xylulose reductase |
| chr5_-_5799315 | 1.61 |
ENSMUST00000015796.9
|
Steap1
|
six transmembrane epithelial antigen of the prostate 1 |
| chr16_-_18161746 | 1.59 |
ENSMUST00000231372.2
ENSMUST00000130752.2 ENSMUST00000231605.2 ENSMUST00000115628.10 |
Tango2
|
transport and golgi organization 2 |
| chr9_+_50528608 | 1.59 |
ENSMUST00000000171.15
ENSMUST00000151197.8 |
Pih1d2
|
PIH1 domain containing 2 |
| chr10_-_75673175 | 1.58 |
ENSMUST00000220440.2
|
Gstt2
|
glutathione S-transferase, theta 2 |
| chr16_-_45664591 | 1.55 |
ENSMUST00000076333.12
|
Phldb2
|
pleckstrin homology like domain, family B, member 2 |
| chr16_-_45664664 | 1.55 |
ENSMUST00000036355.13
|
Phldb2
|
pleckstrin homology like domain, family B, member 2 |
| chr11_-_103640640 | 1.55 |
ENSMUST00000018630.3
|
Wnt9b
|
wingless-type MMTV integration site family, member 9B |
| chr8_+_22996233 | 1.55 |
ENSMUST00000210854.2
|
Slc20a2
|
solute carrier family 20, member 2 |
| chr11_-_115167775 | 1.54 |
ENSMUST00000021078.3
|
Fdxr
|
ferredoxin reductase |
| chr5_+_129924619 | 1.53 |
ENSMUST00000077320.3
|
Zbed5
|
zinc finger, BED type containing 5 |
| chrM_+_14138 | 1.53 |
ENSMUST00000082421.1
|
mt-Cytb
|
mitochondrially encoded cytochrome b |
| chr16_+_22738987 | 1.53 |
ENSMUST00000023587.12
|
Fetub
|
fetuin beta |
| chr8_-_71085097 | 1.53 |
ENSMUST00000110103.2
|
Gdf15
|
growth differentiation factor 15 |
| chr3_-_63872079 | 1.51 |
ENSMUST00000161659.8
|
Slc33a1
|
solute carrier family 33 (acetyl-CoA transporter), member 1 |
| chr2_+_164404499 | 1.49 |
ENSMUST00000017867.10
ENSMUST00000109344.9 ENSMUST00000109345.9 |
Wfdc2
|
WAP four-disulfide core domain 2 |
| chr3_-_79760067 | 1.49 |
ENSMUST00000192341.2
ENSMUST00000193410.3 |
Tmem144
|
transmembrane protein 144 |
| chr11_+_96173355 | 1.48 |
ENSMUST00000125410.2
|
Hoxb8
|
homeobox B8 |
| chrX_+_141010919 | 1.48 |
ENSMUST00000042329.12
|
Nxt2
|
nuclear transport factor 2-like export factor 2 |
| chr8_-_13304096 | 1.48 |
ENSMUST00000171619.2
|
Adprhl1
|
ADP-ribosylhydrolase like 1 |
| chr4_-_88562696 | 1.47 |
ENSMUST00000105149.3
|
Ifna13
|
interferon alpha 13 |
| chrM_-_14061 | 1.44 |
ENSMUST00000082419.1
|
mt-Nd6
|
mitochondrially encoded NADH dehydrogenase 6 |
| chr2_-_101459274 | 1.44 |
ENSMUST00000099682.9
|
Iftap
|
intraflagellar transport associated protein |
| chr19_+_44980565 | 1.43 |
ENSMUST00000179305.2
|
Sema4g
|
sema domain, immunoglobulin domain (Ig), transmembrane domain (TM) and short cytoplasmic domain, (semaphorin) 4G |
| chr16_+_84312009 | 1.43 |
ENSMUST00000209558.2
|
A730009L09Rik
|
RIKEN cDNA A730009L09 gene |
| chr18_-_68562385 | 1.42 |
ENSMUST00000052347.8
|
Mc2r
|
melanocortin 2 receptor |
| chr19_+_13742643 | 1.42 |
ENSMUST00000215930.2
|
Olfr1495
|
olfactory receptor 1495 |
| chr11_-_29197222 | 1.41 |
ENSMUST00000020754.10
|
Cfap36
|
cilia and flagella associated protein 36 |
| chr14_-_20231871 | 1.41 |
ENSMUST00000024011.10
|
Kcnk5
|
potassium channel, subfamily K, member 5 |
| chr3_-_57202301 | 1.40 |
ENSMUST00000171384.8
|
Tm4sf1
|
transmembrane 4 superfamily member 1 |
| chr8_+_124173678 | 1.39 |
ENSMUST00000212391.2
ENSMUST00000212827.2 |
Def8
|
differentially expressed in FDCP 8 |
| chr9_-_105022272 | 1.39 |
ENSMUST00000190661.2
ENSMUST00000035180.5 |
Nudt16l2
|
nudix (nucleoside diphosphate linked moiety X)-type motif 16-like 2 |
| chr1_+_167136217 | 1.39 |
ENSMUST00000193446.6
|
Tmco1
|
transmembrane and coiled-coil domains 1 |
| chr3_-_107952146 | 1.37 |
ENSMUST00000178808.8
ENSMUST00000106670.2 ENSMUST00000029489.15 |
Gstm4
|
glutathione S-transferase, mu 4 |
| chr6_+_34389269 | 1.36 |
ENSMUST00000007449.9
|
Akr1b7
|
aldo-keto reductase family 1, member B7 |
| chr7_-_132178101 | 1.35 |
ENSMUST00000084500.8
|
Oat
|
ornithine aminotransferase |
| chr1_+_128171859 | 1.34 |
ENSMUST00000027592.6
|
Ubxn4
|
UBX domain protein 4 |
| chr16_+_33650002 | 1.34 |
ENSMUST00000115028.11
ENSMUST00000069345.6 |
Itgb5
|
integrin beta 5 |
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 6.4 | 25.6 | GO:0046356 | acetyl-CoA catabolic process(GO:0046356) |
| 3.4 | 13.5 | GO:0072021 | ascending thin limb development(GO:0072021) thick ascending limb development(GO:0072023) metanephric ascending thin limb development(GO:0072218) metanephric thick ascending limb development(GO:0072233) |
| 2.9 | 11.6 | GO:0051344 | negative regulation of cyclic-nucleotide phosphodiesterase activity(GO:0051344) regulation of aminoacyl-tRNA ligase activity(GO:1903630) |
| 2.5 | 7.5 | GO:0046223 | mycotoxin catabolic process(GO:0043387) aflatoxin catabolic process(GO:0046223) organic heteropentacyclic compound catabolic process(GO:1901377) |
| 2.4 | 7.3 | GO:0009753 | response to jasmonic acid(GO:0009753) cellular response to jasmonic acid stimulus(GO:0071395) |
| 2.3 | 7.0 | GO:0042732 | D-xylose metabolic process(GO:0042732) |
| 2.1 | 6.3 | GO:2000118 | regulation of sodium-dependent phosphate transport(GO:2000118) |
| 2.1 | 31.3 | GO:0015747 | urate transport(GO:0015747) |
| 1.9 | 5.6 | GO:0009087 | methionine catabolic process(GO:0009087) |
| 1.8 | 9.0 | GO:0006566 | threonine metabolic process(GO:0006566) |
| 1.5 | 4.6 | GO:0007509 | mesoderm migration involved in gastrulation(GO:0007509) |
| 1.4 | 5.6 | GO:0071469 | cellular response to alkaline pH(GO:0071469) |
| 1.4 | 8.3 | GO:0014016 | neuroblast differentiation(GO:0014016) |
| 1.3 | 11.7 | GO:0045919 | positive regulation of cytolysis(GO:0045919) |
| 1.3 | 3.8 | GO:1901052 | sarcosine metabolic process(GO:1901052) sarcosine catabolic process(GO:1901053) |
| 1.3 | 7.5 | GO:0035026 | leading edge cell differentiation(GO:0035026) |
| 1.1 | 4.5 | GO:0046951 | ketone body biosynthetic process(GO:0046951) |
| 0.9 | 2.8 | GO:0006212 | uracil catabolic process(GO:0006212) uracil metabolic process(GO:0019860) |
| 0.9 | 5.5 | GO:0006663 | platelet activating factor biosynthetic process(GO:0006663) |
| 0.9 | 2.7 | GO:0072248 | metanephric glomerular visceral epithelial cell differentiation(GO:0072248) metanephric glomerular visceral epithelial cell development(GO:0072249) metanephric glomerular epithelial cell differentiation(GO:0072312) metanephric glomerular epithelial cell development(GO:0072313) |
| 0.9 | 4.3 | GO:0003383 | apical constriction(GO:0003383) |
| 0.8 | 2.5 | GO:0014878 | response to electrical stimulus involved in regulation of muscle adaptation(GO:0014878) |
| 0.8 | 5.8 | GO:0050747 | positive regulation of lipoprotein metabolic process(GO:0050747) |
| 0.8 | 3.3 | GO:0030091 | protein repair(GO:0030091) |
| 0.8 | 3.2 | GO:0016999 | antibiotic metabolic process(GO:0016999) |
| 0.8 | 2.3 | GO:0021998 | neural plate mediolateral regionalization(GO:0021998) mesoderm structural organization(GO:0048338) paraxial mesoderm structural organization(GO:0048352) positive regulation of cardiac ventricle development(GO:1904414) |
| 0.7 | 7.4 | GO:1900034 | regulation of cellular response to heat(GO:1900034) |
| 0.7 | 4.2 | GO:0036112 | medium-chain fatty-acyl-CoA metabolic process(GO:0036112) |
| 0.7 | 2.8 | GO:0070634 | transepithelial ammonium transport(GO:0070634) |
| 0.7 | 2.0 | GO:0048698 | negative regulation of collateral sprouting in absence of injury(GO:0048698) |
| 0.7 | 5.3 | GO:0070294 | renal sodium ion absorption(GO:0070294) |
| 0.6 | 5.6 | GO:0042904 | 9-cis-retinoic acid biosynthetic process(GO:0042904) 9-cis-retinoic acid metabolic process(GO:0042905) |
| 0.6 | 4.9 | GO:0070447 | positive regulation of oligodendrocyte progenitor proliferation(GO:0070447) |
| 0.6 | 3.6 | GO:1902847 | regulation of neuronal signal transduction(GO:1902847) positive regulation of neurofibrillary tangle assembly(GO:1902998) |
| 0.6 | 2.4 | GO:0006624 | vacuolar protein processing(GO:0006624) |
| 0.6 | 7.1 | GO:0034626 | fatty acid elongation, saturated fatty acid(GO:0019367) fatty acid elongation, unsaturated fatty acid(GO:0019368) fatty acid elongation, monounsaturated fatty acid(GO:0034625) fatty acid elongation, polyunsaturated fatty acid(GO:0034626) |
| 0.6 | 18.4 | GO:0019373 | epoxygenase P450 pathway(GO:0019373) |
| 0.5 | 2.7 | GO:0046167 | glycerol-3-phosphate biosynthetic process(GO:0046167) |
| 0.5 | 3.8 | GO:0051725 | protein de-ADP-ribosylation(GO:0051725) |
| 0.5 | 2.1 | GO:0030450 | regulation of complement activation, classical pathway(GO:0030450) |
| 0.5 | 2.0 | GO:0036060 | filtration diaphragm assembly(GO:0036058) slit diaphragm assembly(GO:0036060) |
| 0.5 | 2.4 | GO:0070543 | response to linoleic acid(GO:0070543) |
| 0.5 | 2.8 | GO:0033540 | fatty acid beta-oxidation using acyl-CoA oxidase(GO:0033540) |
| 0.5 | 2.3 | GO:0000101 | sulfur amino acid transport(GO:0000101) |
| 0.5 | 8.3 | GO:0060363 | cranial suture morphogenesis(GO:0060363) |
| 0.4 | 3.1 | GO:0001561 | fatty acid alpha-oxidation(GO:0001561) |
| 0.4 | 1.7 | GO:0015851 | nucleobase transport(GO:0015851) pyrimidine nucleobase transport(GO:0015855) |
| 0.4 | 1.2 | GO:1904733 | negative regulation of electron carrier activity(GO:1904733) regulation of fatty acid beta-oxidation using acyl-CoA dehydrogenase(GO:1904735) negative regulation of fatty acid beta-oxidation using acyl-CoA dehydrogenase(GO:1904736) |
| 0.4 | 9.5 | GO:2000353 | positive regulation of endothelial cell apoptotic process(GO:2000353) |
| 0.4 | 3.9 | GO:0006104 | succinyl-CoA metabolic process(GO:0006104) olfactory bulb mitral cell layer development(GO:0061034) |
| 0.4 | 2.3 | GO:0015889 | cobalamin transport(GO:0015889) |
| 0.4 | 1.5 | GO:0009786 | regulation of asymmetric cell division(GO:0009786) |
| 0.4 | 4.7 | GO:0006122 | mitochondrial electron transport, ubiquinol to cytochrome c(GO:0006122) |
| 0.3 | 1.7 | GO:0001692 | histamine metabolic process(GO:0001692) |
| 0.3 | 2.6 | GO:0018243 | protein O-linked glycosylation via threonine(GO:0018243) |
| 0.3 | 3.1 | GO:1903690 | negative regulation of wound healing, spreading of epidermal cells(GO:1903690) |
| 0.3 | 2.8 | GO:0003147 | neural crest cell migration involved in heart formation(GO:0003147) anterior neural tube closure(GO:0061713) cellular response to folic acid(GO:0071231) |
| 0.3 | 3.7 | GO:0051151 | negative regulation of smooth muscle cell differentiation(GO:0051151) |
| 0.3 | 0.9 | GO:0002501 | MHC protein complex assembly(GO:0002396) peptide antigen assembly with MHC protein complex(GO:0002501) |
| 0.3 | 2.4 | GO:0007597 | blood coagulation, intrinsic pathway(GO:0007597) |
| 0.3 | 3.5 | GO:0052697 | flavonoid glucuronidation(GO:0052696) xenobiotic glucuronidation(GO:0052697) |
| 0.3 | 0.8 | GO:0070676 | intralumenal vesicle formation(GO:0070676) negative regulation of early endosome to late endosome transport(GO:2000642) |
| 0.3 | 0.3 | GO:0006711 | estrogen catabolic process(GO:0006711) |
| 0.3 | 1.0 | GO:0045338 | farnesyl diphosphate metabolic process(GO:0045338) |
| 0.3 | 1.0 | GO:0060800 | regulation of cell differentiation involved in embryonic placenta development(GO:0060800) |
| 0.2 | 0.7 | GO:0070104 | negative regulation of interleukin-6-mediated signaling pathway(GO:0070104) |
| 0.2 | 4.1 | GO:0070166 | enamel mineralization(GO:0070166) |
| 0.2 | 0.9 | GO:0008594 | photoreceptor cell morphogenesis(GO:0008594) |
| 0.2 | 3.5 | GO:0051103 | DNA ligation involved in DNA repair(GO:0051103) |
| 0.2 | 1.1 | GO:0006041 | glucosamine metabolic process(GO:0006041) |
| 0.2 | 3.7 | GO:0009437 | carnitine metabolic process(GO:0009437) |
| 0.2 | 2.6 | GO:0042637 | catagen(GO:0042637) |
| 0.2 | 2.3 | GO:0061101 | neuroendocrine cell differentiation(GO:0061101) |
| 0.2 | 2.6 | GO:0033539 | fatty acid beta-oxidation using acyl-CoA dehydrogenase(GO:0033539) |
| 0.2 | 1.4 | GO:0018916 | nitrobenzene metabolic process(GO:0018916) |
| 0.2 | 3.5 | GO:0043568 | positive regulation of insulin-like growth factor receptor signaling pathway(GO:0043568) |
| 0.2 | 1.7 | GO:0051409 | response to nitrosative stress(GO:0051409) |
| 0.2 | 1.7 | GO:0051534 | negative regulation of NFAT protein import into nucleus(GO:0051534) |
| 0.2 | 0.7 | GO:1904430 | negative regulation of t-circle formation(GO:1904430) |
| 0.2 | 0.4 | GO:0061643 | chemorepulsion of axon(GO:0061643) |
| 0.2 | 4.3 | GO:0046033 | AMP metabolic process(GO:0046033) |
| 0.2 | 0.2 | GO:1903722 | regulation of centriole elongation(GO:1903722) |
| 0.2 | 0.5 | GO:0070682 | proteasome regulatory particle assembly(GO:0070682) |
| 0.2 | 0.7 | GO:0034552 | respiratory chain complex II assembly(GO:0034552) mitochondrial respiratory chain complex II assembly(GO:0034553) mitochondrial respiratory chain complex II biogenesis(GO:0097032) |
| 0.2 | 1.8 | GO:1903894 | regulation of IRE1-mediated unfolded protein response(GO:1903894) |
| 0.2 | 1.6 | GO:0097461 | ferric iron import into cell(GO:0097461) ferric iron import across plasma membrane(GO:0098706) |
| 0.2 | 0.6 | GO:1901491 | negative regulation of lymphangiogenesis(GO:1901491) |
| 0.2 | 1.1 | GO:0072344 | rescue of stalled ribosome(GO:0072344) |
| 0.2 | 0.6 | GO:2000048 | negative regulation of cell-cell adhesion mediated by cadherin(GO:2000048) |
| 0.2 | 0.8 | GO:1901526 | positive regulation of macromitophagy(GO:1901526) positive regulation of mitophagy in response to mitochondrial depolarization(GO:1904925) |
| 0.2 | 1.2 | GO:0031179 | peptide modification(GO:0031179) leukotriene D4 metabolic process(GO:1901748) leukotriene D4 biosynthetic process(GO:1901750) |
| 0.2 | 2.1 | GO:0006348 | chromatin silencing at telomere(GO:0006348) |
| 0.1 | 1.3 | GO:0034214 | protein hexamerization(GO:0034214) |
| 0.1 | 2.7 | GO:0036315 | cellular response to sterol(GO:0036315) |
| 0.1 | 1.3 | GO:0006102 | isocitrate metabolic process(GO:0006102) |
| 0.1 | 1.1 | GO:0090074 | negative regulation of protein homodimerization activity(GO:0090074) |
| 0.1 | 0.6 | GO:0071873 | response to norepinephrine(GO:0071873) |
| 0.1 | 4.5 | GO:0015986 | energy coupled proton transport, down electrochemical gradient(GO:0015985) ATP synthesis coupled proton transport(GO:0015986) |
| 0.1 | 0.8 | GO:0021615 | glossopharyngeal nerve morphogenesis(GO:0021615) |
| 0.1 | 0.5 | GO:0033686 | positive regulation of luteinizing hormone secretion(GO:0033686) generation of ovulation cycle rhythm(GO:0060112) |
| 0.1 | 0.7 | GO:0034227 | tRNA thio-modification(GO:0034227) |
| 0.1 | 0.9 | GO:0019262 | N-acetylneuraminate catabolic process(GO:0019262) |
| 0.1 | 2.6 | GO:0070255 | regulation of mucus secretion(GO:0070255) |
| 0.1 | 0.5 | GO:0000239 | pachytene(GO:0000239) |
| 0.1 | 9.1 | GO:0060395 | SMAD protein signal transduction(GO:0060395) |
| 0.1 | 1.1 | GO:0061732 | mitochondrial acetyl-CoA biosynthetic process from pyruvate(GO:0061732) |
| 0.1 | 4.9 | GO:0010842 | retina layer formation(GO:0010842) |
| 0.1 | 1.3 | GO:0045945 | positive regulation of transcription from RNA polymerase III promoter(GO:0045945) |
| 0.1 | 2.0 | GO:0035810 | positive regulation of urine volume(GO:0035810) |
| 0.1 | 0.8 | GO:0070127 | tRNA aminoacylation for mitochondrial protein translation(GO:0070127) |
| 0.1 | 0.5 | GO:1902045 | regulation of Fas signaling pathway(GO:1902044) negative regulation of Fas signaling pathway(GO:1902045) |
| 0.1 | 0.5 | GO:0010273 | detoxification of copper ion(GO:0010273) stress response to copper ion(GO:1990169) |
| 0.1 | 0.6 | GO:0086046 | membrane depolarization during SA node cell action potential(GO:0086046) |
| 0.1 | 1.4 | GO:0006983 | ER overload response(GO:0006983) |
| 0.1 | 1.6 | GO:0006388 | tRNA splicing, via endonucleolytic cleavage and ligation(GO:0006388) |
| 0.1 | 0.4 | GO:1902774 | late endosome to lysosome transport(GO:1902774) |
| 0.1 | 5.3 | GO:0001702 | gastrulation with mouth forming second(GO:0001702) |
| 0.1 | 1.2 | GO:0008063 | Toll signaling pathway(GO:0008063) |
| 0.1 | 1.3 | GO:0090503 | RNA phosphodiester bond hydrolysis, exonucleolytic(GO:0090503) |
| 0.1 | 1.1 | GO:0032049 | cardiolipin biosynthetic process(GO:0032049) |
| 0.1 | 0.5 | GO:0060295 | regulation of cilium movement involved in cell motility(GO:0060295) regulation of cilium beat frequency involved in ciliary motility(GO:0060296) regulation of cilium-dependent cell motility(GO:1902019) |
| 0.1 | 4.9 | GO:0010257 | NADH dehydrogenase complex assembly(GO:0010257) mitochondrial respiratory chain complex I assembly(GO:0032981) mitochondrial respiratory chain complex I biogenesis(GO:0097031) |
| 0.1 | 0.5 | GO:0003241 | growth involved in heart morphogenesis(GO:0003241) |
| 0.1 | 0.4 | GO:0060697 | positive regulation of phospholipid catabolic process(GO:0060697) |
| 0.1 | 0.5 | GO:2000360 | negative regulation of binding of sperm to zona pellucida(GO:2000360) |
| 0.1 | 5.0 | GO:0007339 | binding of sperm to zona pellucida(GO:0007339) |
| 0.1 | 1.0 | GO:0061727 | methylglyoxal catabolic process to D-lactate via S-lactoyl-glutathione(GO:0019243) methylglyoxal catabolic process(GO:0051596) methylglyoxal catabolic process to lactate(GO:0061727) |
| 0.1 | 1.1 | GO:0035845 | photoreceptor cell outer segment organization(GO:0035845) |
| 0.1 | 0.5 | GO:1904426 | positive regulation of GTP binding(GO:1904426) |
| 0.1 | 0.3 | GO:0009258 | 10-formyltetrahydrofolate catabolic process(GO:0009258) folic acid-containing compound catabolic process(GO:0009397) pteridine-containing compound catabolic process(GO:0042560) |
| 0.1 | 1.8 | GO:0060352 | cell adhesion molecule production(GO:0060352) |
| 0.1 | 0.5 | GO:0070858 | negative regulation of bile acid biosynthetic process(GO:0070858) negative regulation of bile acid metabolic process(GO:1904252) |
| 0.1 | 0.6 | GO:1990416 | cellular response to brain-derived neurotrophic factor stimulus(GO:1990416) |
| 0.1 | 0.4 | GO:0001830 | trophectodermal cell fate commitment(GO:0001830) |
| 0.1 | 0.3 | GO:2000562 | negative regulation of CD4-positive, alpha-beta T cell proliferation(GO:2000562) |
| 0.1 | 1.4 | GO:0030322 | stabilization of membrane potential(GO:0030322) |
| 0.1 | 1.5 | GO:0070995 | NADPH oxidation(GO:0070995) |
| 0.1 | 2.9 | GO:1902175 | regulation of oxidative stress-induced intrinsic apoptotic signaling pathway(GO:1902175) |
| 0.1 | 3.2 | GO:0042775 | mitochondrial ATP synthesis coupled electron transport(GO:0042775) |
| 0.1 | 1.2 | GO:0048312 | intracellular distribution of mitochondria(GO:0048312) |
| 0.1 | 0.9 | GO:0035269 | protein O-linked mannosylation(GO:0035269) |
| 0.1 | 0.6 | GO:0040032 | post-embryonic body morphogenesis(GO:0040032) |
| 0.1 | 0.4 | GO:0032917 | polyamine acetylation(GO:0032917) spermidine acetylation(GO:0032918) |
| 0.1 | 4.0 | GO:0006749 | glutathione metabolic process(GO:0006749) |
| 0.1 | 1.9 | GO:0006817 | phosphate ion transport(GO:0006817) |
| 0.1 | 1.1 | GO:0046415 | urate metabolic process(GO:0046415) |
| 0.1 | 0.9 | GO:0042761 | fatty acid elongation(GO:0030497) very long-chain fatty acid biosynthetic process(GO:0042761) |
| 0.1 | 0.8 | GO:0006390 | transcription from mitochondrial promoter(GO:0006390) |
| 0.1 | 0.7 | GO:0002318 | myeloid progenitor cell differentiation(GO:0002318) |
| 0.1 | 1.7 | GO:0006516 | glycoprotein catabolic process(GO:0006516) |
| 0.1 | 1.7 | GO:0010818 | T cell chemotaxis(GO:0010818) |
| 0.1 | 0.5 | GO:0032510 | endosome to lysosome transport via multivesicular body sorting pathway(GO:0032510) |
| 0.1 | 2.6 | GO:0030212 | hyaluronan metabolic process(GO:0030212) |
| 0.1 | 1.3 | GO:0031340 | positive regulation of vesicle fusion(GO:0031340) |
| 0.1 | 1.3 | GO:0090136 | epithelial cell-cell adhesion(GO:0090136) |
| 0.1 | 0.8 | GO:0008300 | isoprenoid catabolic process(GO:0008300) |
| 0.1 | 1.2 | GO:0061179 | negative regulation of insulin secretion involved in cellular response to glucose stimulus(GO:0061179) |
| 0.1 | 5.0 | GO:0035383 | acyl-CoA metabolic process(GO:0006637) thioester metabolic process(GO:0035383) |
| 0.1 | 1.5 | GO:0034219 | carbohydrate transmembrane transport(GO:0034219) |
| 0.1 | 0.4 | GO:0038109 | response to stem cell factor(GO:0036215) cellular response to stem cell factor stimulus(GO:0036216) Kit signaling pathway(GO:0038109) |
| 0.1 | 0.2 | GO:2000474 | regulation of opioid receptor signaling pathway(GO:2000474) |
| 0.1 | 1.9 | GO:0006783 | heme biosynthetic process(GO:0006783) |
| 0.1 | 0.4 | GO:0019532 | oxalate transport(GO:0019532) |
| 0.1 | 1.0 | GO:0030150 | protein import into mitochondrial matrix(GO:0030150) |
| 0.1 | 0.3 | GO:0060356 | leucine import(GO:0060356) |
| 0.1 | 0.8 | GO:0048280 | vesicle fusion with Golgi apparatus(GO:0048280) |
| 0.1 | 0.5 | GO:0098909 | regulation of cardiac muscle cell action potential involved in regulation of contraction(GO:0098909) |
| 0.1 | 0.6 | GO:0048630 | skeletal muscle tissue growth(GO:0048630) |
| 0.1 | 2.2 | GO:0048240 | sperm capacitation(GO:0048240) |
| 0.1 | 2.5 | GO:0010107 | potassium ion import(GO:0010107) |
| 0.1 | 0.2 | GO:0001998 | angiotensin mediated vasoconstriction involved in regulation of systemic arterial blood pressure(GO:0001998) |
| 0.1 | 0.6 | GO:0008272 | sulfate transport(GO:0008272) |
| 0.1 | 0.9 | GO:0055070 | copper ion homeostasis(GO:0055070) |
| 0.1 | 1.5 | GO:0033141 | positive regulation of peptidyl-serine phosphorylation of STAT protein(GO:0033141) |
| 0.1 | 0.7 | GO:0051177 | meiotic sister chromatid cohesion(GO:0051177) |
| 0.0 | 1.6 | GO:0040023 | nuclear migration(GO:0007097) establishment of nucleus localization(GO:0040023) |
| 0.0 | 0.3 | GO:0030916 | otic vesicle formation(GO:0030916) |
| 0.0 | 0.3 | GO:0000707 | meiotic DNA recombinase assembly(GO:0000707) |
| 0.0 | 0.4 | GO:0044351 | macropinocytosis(GO:0044351) |
| 0.0 | 0.5 | GO:0000963 | mitochondrial RNA processing(GO:0000963) |
| 0.0 | 5.0 | GO:0000724 | double-strand break repair via homologous recombination(GO:0000724) recombinational repair(GO:0000725) |
| 0.0 | 0.9 | GO:0016558 | protein import into peroxisome matrix(GO:0016558) |
| 0.0 | 1.1 | GO:0097499 | protein localization to nonmotile primary cilium(GO:0097499) |
| 0.0 | 2.0 | GO:1902108 | regulation of mitochondrial membrane permeability involved in apoptotic process(GO:1902108) |
| 0.0 | 1.2 | GO:0021516 | dorsal spinal cord development(GO:0021516) |
| 0.0 | 0.3 | GO:0015886 | heme transport(GO:0015886) |
| 0.0 | 0.9 | GO:0034389 | lipid particle organization(GO:0034389) |
| 0.0 | 0.3 | GO:0007258 | JUN phosphorylation(GO:0007258) |
| 0.0 | 0.4 | GO:0071763 | nuclear membrane organization(GO:0071763) |
| 0.0 | 0.3 | GO:2000623 | regulation of nuclear-transcribed mRNA catabolic process, nonsense-mediated decay(GO:2000622) negative regulation of nuclear-transcribed mRNA catabolic process, nonsense-mediated decay(GO:2000623) |
| 0.0 | 0.3 | GO:0006561 | proline biosynthetic process(GO:0006561) |
| 0.0 | 1.1 | GO:0007202 | activation of phospholipase C activity(GO:0007202) |
| 0.0 | 3.4 | GO:0055008 | cardiac muscle tissue morphogenesis(GO:0055008) |
| 0.0 | 0.4 | GO:0015937 | coenzyme A biosynthetic process(GO:0015937) |
| 0.0 | 0.1 | GO:0018343 | protein farnesylation(GO:0018343) |
| 0.0 | 0.4 | GO:0045741 | positive regulation of epidermal growth factor-activated receptor activity(GO:0045741) |
| 0.0 | 1.2 | GO:0060384 | innervation(GO:0060384) |
| 0.0 | 1.0 | GO:0006760 | folic acid-containing compound metabolic process(GO:0006760) |
| 0.0 | 0.3 | GO:2000467 | positive regulation of glycogen (starch) synthase activity(GO:2000467) |
| 0.0 | 1.5 | GO:0036075 | endochondral ossification(GO:0001958) replacement ossification(GO:0036075) |
| 0.0 | 5.5 | GO:0006694 | steroid biosynthetic process(GO:0006694) |
| 0.0 | 0.4 | GO:0043248 | proteasome assembly(GO:0043248) |
| 0.0 | 1.1 | GO:0033962 | cytoplasmic mRNA processing body assembly(GO:0033962) |
| 0.0 | 0.2 | GO:0071639 | positive regulation of monocyte chemotactic protein-1 production(GO:0071639) |
| 0.0 | 0.6 | GO:0021796 | cerebral cortex regionalization(GO:0021796) |
| 0.0 | 0.5 | GO:0036159 | inner dynein arm assembly(GO:0036159) |
| 0.0 | 1.2 | GO:0001580 | detection of chemical stimulus involved in sensory perception of bitter taste(GO:0001580) |
| 0.0 | 1.2 | GO:0009410 | response to xenobiotic stimulus(GO:0009410) |
| 0.0 | 0.2 | GO:0035902 | response to immobilization stress(GO:0035902) |
| 0.0 | 0.5 | GO:0090435 | protein localization to nuclear envelope(GO:0090435) |
| 0.0 | 1.3 | GO:0050919 | negative chemotaxis(GO:0050919) |
| 0.0 | 1.9 | GO:0030433 | ER-associated ubiquitin-dependent protein catabolic process(GO:0030433) |
| 0.0 | 0.2 | GO:0070417 | cellular response to cold(GO:0070417) |
| 0.0 | 0.3 | GO:0051967 | negative regulation of synaptic transmission, glutamatergic(GO:0051967) |
| 0.0 | 0.0 | GO:1902065 | response to L-glutamate(GO:1902065) |
| 0.0 | 1.2 | GO:0051496 | positive regulation of stress fiber assembly(GO:0051496) |
| 0.0 | 0.1 | GO:0006788 | heme oxidation(GO:0006788) |
| 0.0 | 0.1 | GO:0034093 | positive regulation of maintenance of sister chromatid cohesion(GO:0034093) positive regulation of maintenance of mitotic sister chromatid cohesion(GO:0034184) protein localization to site of double-strand break(GO:1990166) |
| 0.0 | 0.2 | GO:0046784 | viral mRNA export from host cell nucleus(GO:0046784) |
| 0.0 | 0.3 | GO:0090336 | positive regulation of brown fat cell differentiation(GO:0090336) |
| 0.0 | 0.6 | GO:0071108 | protein K48-linked deubiquitination(GO:0071108) |
| 0.0 | 1.5 | GO:0046324 | regulation of glucose import(GO:0046324) |
| 0.0 | 0.2 | GO:0003215 | cardiac right ventricle morphogenesis(GO:0003215) |
| 0.0 | 0.1 | GO:1904158 | axonemal central apparatus assembly(GO:1904158) |
| 0.0 | 0.3 | GO:0060285 | cilium or flagellum-dependent cell motility(GO:0001539) cilium-dependent cell motility(GO:0060285) |
| 0.0 | 17.9 | GO:0007608 | sensory perception of smell(GO:0007608) |
| 0.0 | 2.1 | GO:0006413 | translational initiation(GO:0006413) |
| 0.0 | 0.5 | GO:0016339 | calcium-dependent cell-cell adhesion via plasma membrane cell adhesion molecules(GO:0016339) |
| 0.0 | 0.5 | GO:0030199 | collagen fibril organization(GO:0030199) |
| 0.0 | 0.2 | GO:0048875 | G-protein coupled glutamate receptor signaling pathway(GO:0007216) chemical homeostasis within a tissue(GO:0048875) |
| 0.0 | 0.9 | GO:1902807 | negative regulation of cell cycle G1/S phase transition(GO:1902807) |
| 0.0 | 0.8 | GO:0045454 | cell redox homeostasis(GO:0045454) |
| 0.0 | 0.9 | GO:0055013 | cardiac muscle cell development(GO:0055013) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.5 | 7.5 | GO:0070022 | transforming growth factor beta receptor homodimeric complex(GO:0070022) |
| 1.0 | 8.3 | GO:0032937 | SREBP-SCAP-Insig complex(GO:0032937) |
| 0.8 | 7.4 | GO:0045298 | tubulin complex(GO:0045298) |
| 0.7 | 2.7 | GO:0005608 | laminin-3 complex(GO:0005608) |
| 0.6 | 3.5 | GO:0032807 | DNA ligase IV complex(GO:0032807) |
| 0.6 | 1.7 | GO:0017109 | glutamate-cysteine ligase complex(GO:0017109) |
| 0.5 | 4.3 | GO:0033269 | internode region of axon(GO:0033269) |
| 0.5 | 5.3 | GO:0097433 | dense body(GO:0097433) |
| 0.5 | 2.3 | GO:0043202 | lysosomal lumen(GO:0043202) |
| 0.4 | 1.3 | GO:0036488 | CHOP-C/EBP complex(GO:0036488) |
| 0.4 | 3.6 | GO:0045261 | mitochondrial proton-transporting ATP synthase complex, catalytic core F(1)(GO:0000275) proton-transporting ATP synthase complex, catalytic core F(1)(GO:0045261) |
| 0.4 | 3.9 | GO:0045252 | oxoglutarate dehydrogenase complex(GO:0045252) |
| 0.4 | 4.7 | GO:0045275 | respiratory chain complex III(GO:0045275) |
| 0.4 | 1.5 | GO:1990851 | Wnt-Frizzled-LRP5/6 complex(GO:1990851) |
| 0.3 | 1.2 | GO:0071920 | cleavage body(GO:0071920) |
| 0.3 | 4.0 | GO:0034366 | spherical high-density lipoprotein particle(GO:0034366) |
| 0.3 | 1.3 | GO:0034684 | integrin alphav-beta5 complex(GO:0034684) |
| 0.2 | 0.7 | GO:0005900 | oncostatin-M receptor complex(GO:0005900) |
| 0.2 | 15.2 | GO:0005788 | endoplasmic reticulum lumen(GO:0005788) |
| 0.2 | 0.9 | GO:1990429 | Pex17p-Pex14p docking complex(GO:1990415) peroxisomal importomer complex(GO:1990429) |
| 0.2 | 1.7 | GO:0016012 | sarcoglycan complex(GO:0016012) |
| 0.2 | 0.5 | GO:0034751 | aryl hydrocarbon receptor complex(GO:0034751) |
| 0.2 | 22.2 | GO:0031526 | brush border membrane(GO:0031526) |
| 0.2 | 3.1 | GO:0045180 | basal cortex(GO:0045180) |
| 0.2 | 0.5 | GO:0060473 | cortical granule(GO:0060473) |
| 0.2 | 0.6 | GO:0016035 | zeta DNA polymerase complex(GO:0016035) |
| 0.2 | 2.2 | GO:0036128 | CatSper complex(GO:0036128) |
| 0.2 | 1.1 | GO:0045254 | pyruvate dehydrogenase complex(GO:0045254) |
| 0.1 | 12.9 | GO:0005881 | cytoplasmic microtubule(GO:0005881) |
| 0.1 | 0.6 | GO:0005742 | mitochondrial outer membrane translocase complex(GO:0005742) |
| 0.1 | 1.7 | GO:0044322 | endoplasmic reticulum quality control compartment(GO:0044322) |
| 0.1 | 6.9 | GO:0045271 | mitochondrial respiratory chain complex I(GO:0005747) NADH dehydrogenase complex(GO:0030964) respiratory chain complex I(GO:0045271) |
| 0.1 | 31.3 | GO:0005777 | peroxisome(GO:0005777) microbody(GO:0042579) |
| 0.1 | 2.7 | GO:0065010 | extracellular membrane-bounded organelle(GO:0065010) |
| 0.1 | 1.1 | GO:0008282 | ATP-sensitive potassium channel complex(GO:0008282) |
| 0.1 | 1.0 | GO:0005744 | mitochondrial inner membrane presequence translocase complex(GO:0005744) |
| 0.1 | 0.7 | GO:0043564 | Ku70:Ku80 complex(GO:0043564) |
| 0.1 | 1.7 | GO:0000177 | cytoplasmic exosome (RNase complex)(GO:0000177) |
| 0.1 | 4.5 | GO:1990391 | DNA repair complex(GO:1990391) |
| 0.1 | 0.6 | GO:0097057 | TRAF2-GSTP1 complex(GO:0097057) |
| 0.1 | 2.3 | GO:0019908 | nuclear cyclin-dependent protein kinase holoenzyme complex(GO:0019908) |
| 0.1 | 2.8 | GO:0014731 | spectrin-associated cytoskeleton(GO:0014731) |
| 0.1 | 2.1 | GO:0048188 | Set1C/COMPASS complex(GO:0048188) |
| 0.1 | 1.9 | GO:0061702 | inflammasome complex(GO:0061702) |
| 0.1 | 0.4 | GO:0031084 | BLOC-2 complex(GO:0031084) |
| 0.1 | 0.3 | GO:0070939 | Dsl1p complex(GO:0070939) |
| 0.1 | 1.4 | GO:0070469 | respiratory chain(GO:0070469) |
| 0.1 | 0.9 | GO:0034098 | VCP-NPL4-UFD1 AAA ATPase complex(GO:0034098) |
| 0.1 | 0.5 | GO:0005775 | vacuolar lumen(GO:0005775) |
| 0.1 | 22.7 | GO:0005759 | mitochondrial matrix(GO:0005759) |
| 0.1 | 8.4 | GO:0005581 | collagen trimer(GO:0005581) |
| 0.1 | 15.5 | GO:0031225 | anchored component of membrane(GO:0031225) |
| 0.1 | 4.3 | GO:0009925 | basal plasma membrane(GO:0009925) |
| 0.1 | 0.8 | GO:0032009 | early phagosome(GO:0032009) |
| 0.1 | 0.7 | GO:0097542 | ciliary tip(GO:0097542) |
| 0.1 | 1.9 | GO:0031307 | integral component of mitochondrial outer membrane(GO:0031307) |
| 0.1 | 0.4 | GO:0000214 | tRNA-intron endonuclease complex(GO:0000214) |
| 0.1 | 0.9 | GO:0000276 | mitochondrial proton-transporting ATP synthase complex, coupling factor F(o)(GO:0000276) |
| 0.1 | 0.4 | GO:0033263 | CORVET complex(GO:0033263) |
| 0.1 | 5.9 | GO:0005901 | caveola(GO:0005901) |
| 0.1 | 0.7 | GO:0034464 | BBSome(GO:0034464) |
| 0.1 | 0.5 | GO:0000127 | transcription factor TFIIIC complex(GO:0000127) |
| 0.0 | 0.3 | GO:0033063 | Rad51B-Rad51C-Rad51D-XRCC2 complex(GO:0033063) |
| 0.0 | 1.0 | GO:0070069 | cytochrome complex(GO:0070069) |
| 0.0 | 17.0 | GO:0016324 | apical plasma membrane(GO:0016324) |
| 0.0 | 4.2 | GO:0005811 | lipid particle(GO:0005811) |
| 0.0 | 1.1 | GO:0030014 | CCR4-NOT complex(GO:0030014) |
| 0.0 | 6.1 | GO:0045177 | apical part of cell(GO:0045177) |
| 0.0 | 0.4 | GO:0032541 | cortical endoplasmic reticulum(GO:0032541) |
| 0.0 | 7.2 | GO:0030176 | integral component of endoplasmic reticulum membrane(GO:0030176) |
| 0.0 | 0.3 | GO:0033588 | Elongator holoenzyme complex(GO:0033588) |
| 0.0 | 0.1 | GO:0014802 | terminal cisterna(GO:0014802) |
| 0.0 | 1.1 | GO:1902711 | GABA-A receptor complex(GO:1902711) |
| 0.0 | 0.2 | GO:0036449 | microtubule minus-end(GO:0036449) |
| 0.0 | 0.5 | GO:0036156 | inner dynein arm(GO:0036156) |
| 0.0 | 0.5 | GO:0008540 | proteasome regulatory particle, base subcomplex(GO:0008540) |
| 0.0 | 0.4 | GO:0008541 | proteasome regulatory particle, lid subcomplex(GO:0008541) |
| 0.0 | 3.5 | GO:0005795 | Golgi stack(GO:0005795) |
| 0.0 | 1.8 | GO:0008180 | COP9 signalosome(GO:0008180) |
| 0.0 | 0.1 | GO:0030689 | Noc complex(GO:0030689) |
| 0.0 | 0.2 | GO:0000178 | exosome (RNase complex)(GO:0000178) |
| 0.0 | 1.8 | GO:0031201 | SNARE complex(GO:0031201) |
| 0.0 | 0.3 | GO:0061574 | ASAP complex(GO:0061574) |
| 0.0 | 0.4 | GO:0071006 | U2-type catalytic step 1 spliceosome(GO:0071006) |
| 0.0 | 2.0 | GO:0005793 | endoplasmic reticulum-Golgi intermediate compartment(GO:0005793) |
| 0.0 | 0.6 | GO:0005892 | acetylcholine-gated channel complex(GO:0005892) |
| 0.0 | 2.9 | GO:0005741 | mitochondrial outer membrane(GO:0005741) |
| 0.0 | 0.4 | GO:0036038 | MKS complex(GO:0036038) |
| 0.0 | 0.2 | GO:0000347 | THO complex(GO:0000347) THO complex part of transcription export complex(GO:0000445) |
| 0.0 | 40.2 | GO:0005783 | endoplasmic reticulum(GO:0005783) |
| 0.0 | 0.6 | GO:0032588 | trans-Golgi network membrane(GO:0032588) |
| 0.0 | 0.2 | GO:0097255 | R2TP complex(GO:0097255) |
| 0.0 | 0.7 | GO:0097546 | ciliary base(GO:0097546) |
| 0.0 | 0.4 | GO:0001673 | male germ cell nucleus(GO:0001673) |
| 0.0 | 30.1 | GO:0005739 | mitochondrion(GO:0005739) |
| 0.0 | 1.3 | GO:0031941 | filamentous actin(GO:0031941) |
| 0.0 | 0.5 | GO:0060170 | ciliary membrane(GO:0060170) |
| 0.0 | 0.2 | GO:0030285 | integral component of synaptic vesicle membrane(GO:0030285) intrinsic component of synaptic vesicle membrane(GO:0098563) |
| 0.0 | 0.1 | GO:0090537 | CERF complex(GO:0090537) |
| 0.0 | 7.9 | GO:0048471 | perinuclear region of cytoplasm(GO:0048471) |
| 0.0 | 0.5 | GO:0001917 | photoreceptor inner segment(GO:0001917) |
| 0.0 | 0.1 | GO:1990716 | axonemal central apparatus(GO:1990716) |
| 0.0 | 0.5 | GO:0031901 | early endosome membrane(GO:0031901) |
| 0.0 | 0.4 | GO:0030173 | integral component of Golgi membrane(GO:0030173) intrinsic component of Golgi membrane(GO:0031228) |
| 0.0 | 0.2 | GO:0000815 | ESCRT III complex(GO:0000815) |
| 0.0 | 0.4 | GO:0005921 | gap junction(GO:0005921) |
| 0.0 | 0.4 | GO:0016235 | aggresome(GO:0016235) |
| 0.0 | 20.7 | GO:0005887 | integral component of plasma membrane(GO:0005887) |
| 0.0 | 0.0 | GO:0032133 | chromosome passenger complex(GO:0032133) |
| 0.0 | 0.6 | GO:0045171 | intercellular bridge(GO:0045171) |
| 0.0 | 29.2 | GO:0070062 | extracellular exosome(GO:0070062) |
| 0.0 | 0.4 | GO:0031226 | intrinsic component of plasma membrane(GO:0031226) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 4.1 | 20.5 | GO:0003986 | acetyl-CoA hydrolase activity(GO:0003986) |
| 2.8 | 17.1 | GO:0047023 | androsterone dehydrogenase activity(GO:0047023) |
| 2.8 | 8.3 | GO:0003990 | acetylcholinesterase activity(GO:0003990) |
| 2.3 | 9.0 | GO:0070905 | serine binding(GO:0070905) |
| 1.9 | 32.4 | GO:1901702 | urate transmembrane transporter activity(GO:0015143) salt transmembrane transporter activity(GO:1901702) |
| 1.8 | 12.6 | GO:0050051 | alkane 1-monooxygenase activity(GO:0018685) leukotriene-B4 20-monooxygenase activity(GO:0050051) |
| 1.5 | 7.4 | GO:0099609 | microtubule lateral binding(GO:0099609) |
| 1.4 | 4.3 | GO:0046899 | nucleoside triphosphate adenylate kinase activity(GO:0046899) |
| 1.4 | 5.6 | GO:0004478 | methionine adenosyltransferase activity(GO:0004478) |
| 1.3 | 3.8 | GO:0008480 | sarcosine dehydrogenase activity(GO:0008480) |
| 1.3 | 6.3 | GO:0015321 | sodium-dependent phosphate transmembrane transporter activity(GO:0015321) |
| 1.2 | 9.3 | GO:0003996 | acyl-CoA ligase activity(GO:0003996) |
| 1.1 | 13.5 | GO:0019864 | IgG binding(GO:0019864) |
| 1.0 | 5.0 | GO:0047635 | L-alanine:2-oxoglutarate aminotransferase activity(GO:0004021) alanine-oxo-acid transaminase activity(GO:0047635) |
| 1.0 | 40.8 | GO:0015020 | glucuronosyltransferase activity(GO:0015020) |
| 0.9 | 2.8 | GO:0003858 | 3-hydroxybutyrate dehydrogenase activity(GO:0003858) |
| 0.9 | 2.8 | GO:0002061 | uracil binding(GO:0002058) pyrimidine nucleobase binding(GO:0002061) |
| 0.9 | 3.8 | GO:0003875 | ADP-ribosylarginine hydrolase activity(GO:0003875) |
| 0.9 | 6.0 | GO:0001851 | complement component C3b binding(GO:0001851) |
| 0.8 | 3.3 | GO:0033743 | peptide-methionine (R)-S-oxide reductase activity(GO:0033743) |
| 0.8 | 3.2 | GO:0003998 | acylphosphatase activity(GO:0003998) |
| 0.8 | 6.4 | GO:0015272 | ATP-activated inward rectifier potassium channel activity(GO:0015272) |
| 0.8 | 3.9 | GO:0034602 | oxoglutarate dehydrogenase (NAD+) activity(GO:0034602) |
| 0.8 | 3.8 | GO:0051185 | coenzyme transporter activity(GO:0051185) |
| 0.7 | 5.1 | GO:0003985 | acetyl-CoA C-acetyltransferase activity(GO:0003985) C-acetyltransferase activity(GO:0016453) |
| 0.7 | 2.9 | GO:0008113 | peptide-methionine (S)-S-oxide reductase activity(GO:0008113) |
| 0.7 | 2.8 | GO:0016401 | palmitoyl-CoA oxidase activity(GO:0016401) |
| 0.7 | 4.9 | GO:0015091 | ferric iron transmembrane transporter activity(GO:0015091) trivalent inorganic cation transmembrane transporter activity(GO:0072510) |
| 0.6 | 1.9 | GO:1990698 | palmitoleoyltransferase activity(GO:1990698) |
| 0.6 | 5.5 | GO:0047498 | calcium-dependent phospholipase A2 activity(GO:0047498) |
| 0.6 | 7.1 | GO:0102336 | fatty acid elongase activity(GO:0009922) 3-oxo-arachidoyl-CoA synthase activity(GO:0102336) 3-oxo-cerotoyl-CoA synthase activity(GO:0102337) 3-oxo-lignoceronyl-CoA synthase activity(GO:0102338) |
| 0.6 | 18.4 | GO:0008392 | arachidonic acid epoxygenase activity(GO:0008392) |
| 0.6 | 3.5 | GO:1990599 | 3' overhang single-stranded DNA endodeoxyribonuclease activity(GO:1990599) |
| 0.6 | 1.7 | GO:0004357 | glutamate-cysteine ligase activity(GO:0004357) |
| 0.5 | 2.7 | GO:0004370 | glycerol kinase activity(GO:0004370) |
| 0.5 | 5.8 | GO:0008429 | phosphatidylethanolamine binding(GO:0008429) |
| 0.5 | 2.6 | GO:0030984 | kininogen binding(GO:0030984) |
| 0.5 | 4.0 | GO:0016681 | ubiquinol-cytochrome-c reductase activity(GO:0008121) oxidoreductase activity, acting on diphenols and related substances as donors, cytochrome as acceptor(GO:0016681) |
| 0.5 | 2.4 | GO:0070892 | lipoteichoic acid receptor activity(GO:0070892) |
| 0.5 | 3.2 | GO:0070573 | metallodipeptidase activity(GO:0070573) |
| 0.4 | 2.7 | GO:0008142 | oxysterol binding(GO:0008142) |
| 0.4 | 3.1 | GO:0030976 | thiamine pyrophosphate binding(GO:0030976) |
| 0.4 | 8.7 | GO:0035612 | AP-2 adaptor complex binding(GO:0035612) |
| 0.4 | 2.3 | GO:0015501 | glutamate:sodium symporter activity(GO:0015501) |
| 0.4 | 1.5 | GO:0015319 | sodium:inorganic phosphate symporter activity(GO:0015319) |
| 0.4 | 1.5 | GO:0019828 | aspartic-type endopeptidase inhibitor activity(GO:0019828) |
| 0.4 | 1.8 | GO:0003829 | beta-1,3-galactosyl-O-glycosyl-glycoprotein beta-1,6-N-acetylglucosaminyltransferase activity(GO:0003829) |
| 0.4 | 1.1 | GO:0004739 | pyruvate dehydrogenase (acetyl-transferring) activity(GO:0004739) |
| 0.4 | 3.6 | GO:0005000 | vasopressin receptor activity(GO:0005000) |
| 0.4 | 3.5 | GO:0031995 | insulin-like growth factor II binding(GO:0031995) |
| 0.3 | 2.4 | GO:0004035 | alkaline phosphatase activity(GO:0004035) |
| 0.3 | 1.7 | GO:0015254 | glycerol channel activity(GO:0015254) |
| 0.3 | 2.3 | GO:0030492 | hemoglobin binding(GO:0030492) |
| 0.3 | 2.8 | GO:0004768 | stearoyl-CoA 9-desaturase activity(GO:0004768) |
| 0.3 | 5.0 | GO:0016805 | dipeptidase activity(GO:0016805) |
| 0.3 | 1.8 | GO:0015226 | amino-acid betaine transmembrane transporter activity(GO:0015199) carnitine transmembrane transporter activity(GO:0015226) |
| 0.3 | 4.5 | GO:0031957 | very long-chain fatty acid-CoA ligase activity(GO:0031957) |
| 0.3 | 0.9 | GO:0080023 | 3R-hydroxyacyl-CoA dehydratase activity(GO:0080023) |
| 0.3 | 5.6 | GO:0043560 | insulin receptor substrate binding(GO:0043560) |
| 0.3 | 0.8 | GO:0030337 | DNA polymerase processivity factor activity(GO:0030337) |
| 0.3 | 3.7 | GO:0005041 | low-density lipoprotein receptor activity(GO:0005041) |
| 0.3 | 1.3 | GO:0004449 | isocitrate dehydrogenase (NAD+) activity(GO:0004449) |
| 0.3 | 1.5 | GO:0008422 | beta-glucosidase activity(GO:0008422) |
| 0.3 | 1.0 | GO:0019166 | trans-2-enoyl-CoA reductase (NADPH) activity(GO:0019166) |
| 0.3 | 2.8 | GO:0051870 | methotrexate binding(GO:0051870) |
| 0.2 | 5.5 | GO:0008191 | metalloendopeptidase inhibitor activity(GO:0008191) |
| 0.2 | 0.7 | GO:0019981 | interleukin-6 receptor activity(GO:0004915) interleukin-6 binding(GO:0019981) |
| 0.2 | 5.6 | GO:0004745 | retinol dehydrogenase activity(GO:0004745) |
| 0.2 | 2.9 | GO:0031545 | peptidyl-proline 4-dioxygenase activity(GO:0031545) |
| 0.2 | 4.5 | GO:0046933 | proton-transporting ATP synthase activity, rotational mechanism(GO:0046933) |
| 0.2 | 2.3 | GO:0098821 | BMP receptor activity(GO:0098821) |
| 0.2 | 1.4 | GO:0004032 | alditol:NADP+ 1-oxidoreductase activity(GO:0004032) |
| 0.2 | 0.9 | GO:0017150 | tRNA dihydrouridine synthase activity(GO:0017150) |
| 0.2 | 12.9 | GO:0004364 | glutathione transferase activity(GO:0004364) |
| 0.2 | 0.9 | GO:0016833 | oxo-acid-lyase activity(GO:0016833) |
| 0.2 | 0.9 | GO:0033883 | pyridoxal phosphatase activity(GO:0033883) |
| 0.2 | 1.4 | GO:0004977 | melanocortin receptor activity(GO:0004977) |
| 0.2 | 1.6 | GO:0052851 | cupric reductase activity(GO:0008823) ferric-chelate reductase (NADPH) activity(GO:0052851) |
| 0.2 | 5.9 | GO:0008137 | NADH dehydrogenase (ubiquinone) activity(GO:0008137) NADH dehydrogenase (quinone) activity(GO:0050136) |
| 0.2 | 0.6 | GO:0086059 | voltage-gated calcium channel activity involved SA node cell action potential(GO:0086059) |
| 0.2 | 4.3 | GO:0042813 | Wnt-activated receptor activity(GO:0042813) |
| 0.2 | 0.7 | GO:0004161 | dimethylallyltranstransferase activity(GO:0004161) geranyltranstransferase activity(GO:0004337) |
| 0.2 | 2.5 | GO:0031432 | titin binding(GO:0031432) |
| 0.2 | 0.5 | GO:0070002 | glutamic-type peptidase activity(GO:0070002) |
| 0.2 | 4.1 | GO:0051787 | misfolded protein binding(GO:0051787) |
| 0.2 | 0.5 | GO:0001962 | alpha-1,3-galactosyltransferase activity(GO:0001962) |
| 0.1 | 0.4 | GO:0004140 | dephospho-CoA kinase activity(GO:0004140) |
| 0.1 | 2.0 | GO:0005225 | volume-sensitive anion channel activity(GO:0005225) |
| 0.1 | 2.6 | GO:0003995 | acyl-CoA dehydrogenase activity(GO:0003995) |
| 0.1 | 2.0 | GO:0017147 | Wnt-protein binding(GO:0017147) |
| 0.1 | 1.5 | GO:0039706 | co-receptor binding(GO:0039706) |
| 0.1 | 1.0 | GO:0004416 | hydroxyacylglutathione hydrolase activity(GO:0004416) |
| 0.1 | 1.1 | GO:0034046 | poly(G) binding(GO:0034046) |
| 0.1 | 2.6 | GO:0004653 | polypeptide N-acetylgalactosaminyltransferase activity(GO:0004653) |
| 0.1 | 3.3 | GO:0010485 | H4 histone acetyltransferase activity(GO:0010485) |
| 0.1 | 1.7 | GO:1904264 | ubiquitin protein ligase activity involved in ERAD pathway(GO:1904264) |
| 0.1 | 2.0 | GO:0016290 | palmitoyl-CoA hydrolase activity(GO:0016290) |
| 0.1 | 1.2 | GO:0036374 | glutathione hydrolase activity(GO:0036374) |
| 0.1 | 1.2 | GO:0016655 | oxidoreductase activity, acting on NAD(P)H, quinone or similar compound as acceptor(GO:0016655) |
| 0.1 | 2.8 | GO:0008519 | ammonium transmembrane transporter activity(GO:0008519) |
| 0.1 | 0.5 | GO:0034584 | piRNA binding(GO:0034584) |
| 0.1 | 0.4 | GO:0060230 | lipoprotein lipase activator activity(GO:0060230) |
| 0.1 | 0.4 | GO:0000822 | inositol hexakisphosphate binding(GO:0000822) |
| 0.1 | 2.9 | GO:0005242 | inward rectifier potassium channel activity(GO:0005242) |
| 0.1 | 0.3 | GO:0016155 | formyltetrahydrofolate dehydrogenase activity(GO:0016155) |
| 0.1 | 0.6 | GO:0019784 | NEDD8-specific protease activity(GO:0019784) |
| 0.1 | 0.4 | GO:0019809 | spermidine binding(GO:0019809) |
| 0.1 | 2.1 | GO:0042800 | histone methyltransferase activity (H3-K4 specific)(GO:0042800) |
| 0.1 | 1.0 | GO:0033691 | sialic acid binding(GO:0033691) |
| 0.1 | 4.9 | GO:0032266 | phosphatidylinositol-3-phosphate binding(GO:0032266) |
| 0.1 | 1.5 | GO:0070402 | NADPH binding(GO:0070402) |
| 0.1 | 3.8 | GO:0019200 | carbohydrate kinase activity(GO:0019200) |
| 0.1 | 1.2 | GO:0017162 | aryl hydrocarbon receptor binding(GO:0017162) |
| 0.1 | 1.0 | GO:0015266 | protein channel activity(GO:0015266) |
| 0.1 | 0.4 | GO:0004045 | aminoacyl-tRNA hydrolase activity(GO:0004045) |
| 0.1 | 0.3 | GO:0004427 | inorganic diphosphatase activity(GO:0004427) |
| 0.1 | 0.7 | GO:0051575 | 5'-deoxyribose-5-phosphate lyase activity(GO:0051575) |
| 0.1 | 0.3 | GO:0004311 | farnesyltranstransferase activity(GO:0004311) |
| 0.1 | 0.7 | GO:0008889 | glycerophosphodiester phosphodiesterase activity(GO:0008889) |
| 0.1 | 1.4 | GO:0022841 | potassium ion leak channel activity(GO:0022841) |
| 0.1 | 1.3 | GO:0001013 | RNA polymerase I regulatory region DNA binding(GO:0001013) |
| 0.1 | 0.2 | GO:0052594 | tryptamine:oxygen oxidoreductase (deaminating) activity(GO:0052593) aminoacetone:oxygen oxidoreductase(deaminating) activity(GO:0052594) aliphatic-amine oxidase activity(GO:0052595) phenethylamine:oxygen oxidoreductase (deaminating) activity(GO:0052596) |
| 0.1 | 0.9 | GO:0035473 | lipase binding(GO:0035473) |
| 0.1 | 6.3 | GO:0008170 | N-methyltransferase activity(GO:0008170) |
| 0.1 | 1.9 | GO:0004129 | cytochrome-c oxidase activity(GO:0004129) heme-copper terminal oxidase activity(GO:0015002) oxidoreductase activity, acting on a heme group of donors, oxygen as acceptor(GO:0016676) |
| 0.1 | 11.4 | GO:0052689 | carboxylic ester hydrolase activity(GO:0052689) |
| 0.1 | 0.1 | GO:0004027 | alcohol sulfotransferase activity(GO:0004027) |
| 0.1 | 1.5 | GO:0005132 | type I interferon receptor binding(GO:0005132) |
| 0.1 | 1.1 | GO:0005078 | MAP-kinase scaffold activity(GO:0005078) |
| 0.1 | 1.7 | GO:0030247 | pattern binding(GO:0001871) polysaccharide binding(GO:0030247) |
| 0.1 | 0.3 | GO:0004735 | pyrroline-5-carboxylate reductase activity(GO:0004735) |
| 0.1 | 1.2 | GO:0033038 | bitter taste receptor activity(GO:0033038) |
| 0.1 | 0.4 | GO:0000213 | tRNA-intron endonuclease activity(GO:0000213) |
| 0.1 | 0.8 | GO:0019869 | chloride channel inhibitor activity(GO:0019869) |
| 0.1 | 0.6 | GO:0031852 | mu-type opioid receptor binding(GO:0031852) |
| 0.1 | 0.3 | GO:0004705 | JUN kinase activity(GO:0004705) SAP kinase activity(GO:0016909) |
| 0.1 | 2.8 | GO:0005154 | epidermal growth factor receptor binding(GO:0005154) |
| 0.1 | 0.4 | GO:0001094 | TFIID-class transcription factor binding(GO:0001094) |
| 0.1 | 1.4 | GO:0017160 | Ral GTPase binding(GO:0017160) |
| 0.0 | 0.3 | GO:0015232 | heme transporter activity(GO:0015232) |
| 0.0 | 0.3 | GO:0089720 | caspase binding(GO:0089720) |
| 0.0 | 1.7 | GO:0034237 | protein kinase A regulatory subunit binding(GO:0034237) |
| 0.0 | 1.3 | GO:0008483 | transaminase activity(GO:0008483) |
| 0.0 | 0.6 | GO:0044323 | retinoic acid-responsive element binding(GO:0044323) |
| 0.0 | 0.3 | GO:0000150 | recombinase activity(GO:0000150) |
| 0.0 | 3.5 | GO:0008080 | N-acetyltransferase activity(GO:0008080) |
| 0.0 | 0.3 | GO:0050294 | steroid sulfotransferase activity(GO:0050294) |
| 0.0 | 0.2 | GO:0030160 | GKAP/Homer scaffold activity(GO:0030160) |
| 0.0 | 3.0 | GO:0005507 | copper ion binding(GO:0005507) |
| 0.0 | 1.1 | GO:0016780 | phosphotransferase activity, for other substituted phosphate groups(GO:0016780) |
| 0.0 | 4.0 | GO:0016616 | oxidoreductase activity, acting on the CH-OH group of donors, NAD or NADP as acceptor(GO:0016616) |
| 0.0 | 5.6 | GO:0004867 | serine-type endopeptidase inhibitor activity(GO:0004867) |
| 0.0 | 0.6 | GO:0042166 | acetylcholine binding(GO:0042166) |
| 0.0 | 1.2 | GO:0017166 | vinculin binding(GO:0017166) |
| 0.0 | 0.1 | GO:0001607 | neuromedin U receptor activity(GO:0001607) |
| 0.0 | 0.4 | GO:0004499 | N,N-dimethylaniline monooxygenase activity(GO:0004499) |
| 0.0 | 1.2 | GO:0044183 | protein binding involved in protein folding(GO:0044183) |
| 0.0 | 2.3 | GO:0003743 | translation initiation factor activity(GO:0003743) |
| 0.0 | 0.8 | GO:0005521 | lamin binding(GO:0005521) |
| 0.0 | 0.6 | GO:0008432 | JUN kinase binding(GO:0008432) |
| 0.0 | 1.8 | GO:0001540 | beta-amyloid binding(GO:0001540) |
| 0.0 | 0.5 | GO:0034450 | ubiquitin-ubiquitin ligase activity(GO:0034450) |
| 0.0 | 4.2 | GO:0005178 | integrin binding(GO:0005178) |
| 0.0 | 0.5 | GO:0017017 | MAP kinase tyrosine/serine/threonine phosphatase activity(GO:0017017) |
| 0.0 | 0.4 | GO:0009931 | calcium-dependent protein serine/threonine kinase activity(GO:0009931) |
| 0.0 | 1.5 | GO:0015144 | carbohydrate transmembrane transporter activity(GO:0015144) carbohydrate transporter activity(GO:1901476) |
| 0.0 | 1.3 | GO:0001786 | phosphatidylserine binding(GO:0001786) |
| 0.0 | 0.2 | GO:0004991 | parathyroid hormone receptor activity(GO:0004991) |
| 0.0 | 0.3 | GO:0016783 | sulfurtransferase activity(GO:0016783) |
| 0.0 | 0.9 | GO:0000175 | 3'-5'-exoribonuclease activity(GO:0000175) |
| 0.0 | 0.1 | GO:0004392 | heme oxygenase (decyclizing) activity(GO:0004392) |
| 0.0 | 2.6 | GO:0036459 | thiol-dependent ubiquitinyl hydrolase activity(GO:0036459) ubiquitinyl hydrolase activity(GO:0101005) |
| 0.0 | 0.9 | GO:0008138 | protein tyrosine/serine/threonine phosphatase activity(GO:0008138) |
| 0.0 | 0.4 | GO:0019531 | oxalate transmembrane transporter activity(GO:0019531) |
| 0.0 | 0.7 | GO:0009055 | electron carrier activity(GO:0009055) |
| 0.0 | 0.5 | GO:0005123 | death receptor binding(GO:0005123) |
| 0.0 | 0.2 | GO:0001594 | trace-amine receptor activity(GO:0001594) |
| 0.0 | 0.2 | GO:0051022 | Rho GDP-dissociation inhibitor binding(GO:0051022) |
| 0.0 | 0.8 | GO:0000049 | tRNA binding(GO:0000049) |
| 0.0 | 0.5 | GO:0071855 | neuropeptide receptor binding(GO:0071855) |
| 0.0 | 18.8 | GO:0004984 | olfactory receptor activity(GO:0004984) |
| 0.0 | 0.2 | GO:0019783 | ubiquitin-like protein-specific protease activity(GO:0019783) |
| 0.0 | 0.3 | GO:0050431 | transforming growth factor beta binding(GO:0050431) |
| 0.0 | 0.4 | GO:0022829 | wide pore channel activity(GO:0022829) |
| 0.0 | 0.2 | GO:0097027 | ubiquitin-protein transferase activator activity(GO:0097027) |
| 0.0 | 0.2 | GO:0031210 | phosphatidylcholine binding(GO:0031210) |
| 0.0 | 1.9 | GO:0005200 | structural constituent of cytoskeleton(GO:0005200) |
| 0.0 | 0.5 | GO:0001671 | ATPase activator activity(GO:0001671) |
| 0.0 | 0.4 | GO:0003887 | DNA-directed DNA polymerase activity(GO:0003887) |
| 0.0 | 0.1 | GO:0030957 | Tat protein binding(GO:0030957) |
| 0.0 | 1.7 | GO:0008083 | growth factor activity(GO:0008083) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 7.4 | PID P38 GAMMA DELTA PATHWAY | Signaling mediated by p38-gamma and p38-delta |
| 0.2 | 12.3 | NABA BASEMENT MEMBRANES | Genes encoding structural components of basement membranes |
| 0.1 | 3.7 | PID DNA PK PATHWAY | DNA-PK pathway in nonhomologous end joining |
| 0.1 | 3.5 | PID WNT SIGNALING PATHWAY | Wnt signaling network |
| 0.1 | 7.2 | PID HNF3B PATHWAY | FOXA2 and FOXA3 transcription factor networks |
| 0.1 | 1.8 | PID HIF1A PATHWAY | Hypoxic and oxygen homeostasis regulation of HIF-1-alpha |
| 0.1 | 1.5 | PID CDC42 PATHWAY | CDC42 signaling events |
| 0.1 | 2.4 | PID ERBB NETWORK PATHWAY | ErbB receptor signaling network |
| 0.1 | 1.3 | PID INTEGRIN5 PATHWAY | Beta5 beta6 beta7 and beta8 integrin cell surface interactions |
| 0.1 | 6.5 | PID TGFBR PATHWAY | TGF-beta receptor signaling |
| 0.1 | 4.3 | SIG INSULIN RECEPTOR PATHWAY IN CARDIAC MYOCYTES | Genes related to the insulin receptor pathway |
| 0.1 | 4.9 | PID CASPASE PATHWAY | Caspase cascade in apoptosis |
| 0.1 | 2.3 | PID BMP PATHWAY | BMP receptor signaling |
| 0.0 | 3.6 | PID MYC REPRESS PATHWAY | Validated targets of C-MYC transcriptional repression |
| 0.0 | 2.6 | PID TAP63 PATHWAY | Validated transcriptional targets of TAp63 isoforms |
| 0.0 | 0.6 | PID NECTIN PATHWAY | Nectin adhesion pathway |
| 0.0 | 1.2 | PID MYC ACTIV PATHWAY | Validated targets of C-MYC transcriptional activation |
| 0.0 | 0.9 | PID ARF6 TRAFFICKING PATHWAY | Arf6 trafficking events |
| 0.0 | 0.4 | PID P38 MK2 PATHWAY | p38 signaling mediated by MAPKAP kinases |
| 0.0 | 0.6 | PID RHOA PATHWAY | RhoA signaling pathway |
| 0.0 | 0.4 | PID VEGFR1 2 PATHWAY | Signaling events mediated by VEGFR1 and VEGFR2 |
| 0.0 | 1.4 | PID REG GR PATHWAY | Glucocorticoid receptor regulatory network |
| 0.0 | 0.4 | PID NCADHERIN PATHWAY | N-cadherin signaling events |
| 0.0 | 0.7 | PID FAK PATHWAY | Signaling events mediated by focal adhesion kinase |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.3 | 1.3 | REACTOME CIRCADIAN REPRESSION OF EXPRESSION BY REV ERBA | Genes involved in Circadian Repression of Expression by REV-ERBA |
| 0.5 | 8.6 | REACTOME ORGANIC CATION ANION ZWITTERION TRANSPORT | Genes involved in Organic cation/anion/zwitterion transport |
| 0.5 | 11.4 | REACTOME ALPHA LINOLENIC ACID ALA METABOLISM | Genes involved in alpha-linolenic acid (ALA) metabolism |
| 0.4 | 6.0 | REACTOME REGULATION OF COMPLEMENT CASCADE | Genes involved in Regulation of Complement cascade |
| 0.4 | 4.5 | REACTOME RECYCLING OF BILE ACIDS AND SALTS | Genes involved in Recycling of bile acids and salts |
| 0.4 | 8.3 | REACTOME SYNTHESIS SECRETION AND DEACYLATION OF GHRELIN | Genes involved in Synthesis, Secretion, and Deacylation of Ghrelin |
| 0.3 | 6.9 | REACTOME GAP JUNCTION DEGRADATION | Genes involved in Gap junction degradation |
| 0.3 | 12.2 | REACTOME CASPASE MEDIATED CLEAVAGE OF CYTOSKELETAL PROTEINS | Genes involved in Caspase-mediated cleavage of cytoskeletal proteins |
| 0.3 | 3.5 | REACTOME GLUCURONIDATION | Genes involved in Glucuronidation |
| 0.3 | 5.5 | REACTOME ACYL CHAIN REMODELLING OF PS | Genes involved in Acyl chain remodelling of PS |
| 0.3 | 26.0 | REACTOME PHASE II CONJUGATION | Genes involved in Phase II conjugation |
| 0.3 | 4.2 | REACTOME INTEGRATION OF PROVIRUS | Genes involved in Integration of provirus |
| 0.2 | 6.4 | REACTOME AMINO ACID SYNTHESIS AND INTERCONVERSION TRANSAMINATION | Genes involved in Amino acid synthesis and interconversion (transamination) |
| 0.2 | 5.1 | REACTOME BRANCHED CHAIN AMINO ACID CATABOLISM | Genes involved in Branched-chain amino acid catabolism |
| 0.2 | 5.3 | REACTOME CITRIC ACID CYCLE TCA CYCLE | Genes involved in Citric acid cycle (TCA cycle) |
| 0.2 | 3.6 | REACTOME FORMATION OF ATP BY CHEMIOSMOTIC COUPLING | Genes involved in Formation of ATP by chemiosmotic coupling |
| 0.2 | 5.8 | REACTOME SYNTHESIS OF PC | Genes involved in Synthesis of PC |
| 0.2 | 2.0 | REACTOME HYALURONAN UPTAKE AND DEGRADATION | Genes involved in Hyaluronan uptake and degradation |
| 0.2 | 3.5 | REACTOME REGULATION OF INSULIN LIKE GROWTH FACTOR IGF ACTIVITY BY INSULIN LIKE GROWTH FACTOR BINDING PROTEINS IGFBPS | Genes involved in Regulation of Insulin-like Growth Factor (IGF) Activity by Insulin-like Growth Factor Binding Proteins (IGFBPs) |
| 0.2 | 0.9 | REACTOME DOPAMINE NEUROTRANSMITTER RELEASE CYCLE | Genes involved in Dopamine Neurotransmitter Release Cycle |
| 0.2 | 1.1 | REACTOME ACYL CHAIN REMODELLING OF PG | Genes involved in Acyl chain remodelling of PG |
| 0.1 | 2.7 | REACTOME HDL MEDIATED LIPID TRANSPORT | Genes involved in HDL-mediated lipid transport |
| 0.1 | 10.2 | REACTOME RESPIRATORY ELECTRON TRANSPORT | Genes involved in Respiratory electron transport |
| 0.1 | 2.8 | REACTOME PYRIMIDINE CATABOLISM | Genes involved in Pyrimidine catabolism |
| 0.1 | 2.0 | REACTOME NEGATIVE REGULATION OF THE PI3K AKT NETWORK | Genes involved in Negative regulation of the PI3K/AKT network |
| 0.1 | 2.3 | REACTOME GLUTAMATE NEUROTRANSMITTER RELEASE CYCLE | Genes involved in Glutamate Neurotransmitter Release Cycle |
| 0.1 | 1.7 | REACTOME PASSIVE TRANSPORT BY AQUAPORINS | Genes involved in Passive Transport by Aquaporins |
| 0.1 | 3.1 | REACTOME PEROXISOMAL LIPID METABOLISM | Genes involved in Peroxisomal lipid metabolism |
| 0.1 | 3.2 | REACTOME TGF BETA RECEPTOR SIGNALING IN EMT EPITHELIAL TO MESENCHYMAL TRANSITION | Genes involved in TGF-beta receptor signaling in EMT (epithelial to mesenchymal transition) |
| 0.1 | 2.4 | REACTOME INTRINSIC PATHWAY | Genes involved in Intrinsic Pathway |
| 0.1 | 4.1 | REACTOME RORA ACTIVATES CIRCADIAN EXPRESSION | Genes involved in RORA Activates Circadian Expression |
| 0.1 | 1.2 | REACTOME IRAK1 RECRUITS IKK COMPLEX | Genes involved in IRAK1 recruits IKK complex |
| 0.1 | 1.1 | REACTOME REGULATION OF PYRUVATE DEHYDROGENASE PDH COMPLEX | Genes involved in Regulation of pyruvate dehydrogenase (PDH) complex |
| 0.1 | 0.9 | REACTOME ACTIVATION OF CHAPERONE GENES BY ATF6 ALPHA | Genes involved in Activation of Chaperone Genes by ATF6-alpha |
| 0.1 | 2.3 | REACTOME SIGNALING BY BMP | Genes involved in Signaling by BMP |
| 0.1 | 4.7 | REACTOME INWARDLY RECTIFYING K CHANNELS | Genes involved in Inwardly rectifying K+ channels |
| 0.1 | 2.8 | REACTOME STEROID HORMONES | Genes involved in Steroid hormones |
| 0.1 | 1.1 | REACTOME FACILITATIVE NA INDEPENDENT GLUCOSE TRANSPORTERS | Genes involved in Facilitative Na+-independent glucose transporters |
| 0.1 | 1.1 | REACTOME SYNTHESIS OF SUBSTRATES IN N GLYCAN BIOSYTHESIS | Genes involved in Synthesis of substrates in N-glycan biosythesis |
| 0.0 | 1.2 | REACTOME GABA A RECEPTOR ACTIVATION | Genes involved in GABA A receptor activation |
| 0.0 | 2.5 | REACTOME SMOOTH MUSCLE CONTRACTION | Genes involved in Smooth Muscle Contraction |
| 0.0 | 2.0 | REACTOME SHC1 EVENTS IN ERBB4 SIGNALING | Genes involved in SHC1 events in ERBB4 signaling |
| 0.0 | 2.7 | REACTOME TRIGLYCERIDE BIOSYNTHESIS | Genes involved in Triglyceride Biosynthesis |
| 0.0 | 0.7 | REACTOME PROSTACYCLIN SIGNALLING THROUGH PROSTACYCLIN RECEPTOR | Genes involved in Prostacyclin signalling through prostacyclin receptor |
| 0.0 | 0.5 | REACTOME KERATAN SULFATE DEGRADATION | Genes involved in Keratan sulfate degradation |
| 0.0 | 5.1 | REACTOME RESPONSE TO ELEVATED PLATELET CYTOSOLIC CA2 | Genes involved in Response to elevated platelet cytosolic Ca2+ |
| 0.0 | 0.5 | REACTOME RETROGRADE NEUROTROPHIN SIGNALLING | Genes involved in Retrograde neurotrophin signalling |
| 0.0 | 0.6 | REACTOME PRESYNAPTIC NICOTINIC ACETYLCHOLINE RECEPTORS | Genes involved in Presynaptic nicotinic acetylcholine receptors |
| 0.0 | 0.8 | REACTOME MITOCHONDRIAL TRNA AMINOACYLATION | Genes involved in Mitochondrial tRNA aminoacylation |
| 0.0 | 0.1 | REACTOME SHC MEDIATED SIGNALLING | Genes involved in SHC-mediated signalling |
| 0.0 | 1.7 | REACTOME TRANSPORT OF VITAMINS NUCLEOSIDES AND RELATED MOLECULES | Genes involved in Transport of vitamins, nucleosides, and related molecules |
| 0.0 | 1.0 | REACTOME CHOLESTEROL BIOSYNTHESIS | Genes involved in Cholesterol biosynthesis |
| 0.0 | 0.3 | REACTOME ACTIVATION OF THE AP1 FAMILY OF TRANSCRIPTION FACTORS | Genes involved in Activation of the AP-1 family of transcription factors |
| 0.0 | 0.4 | REACTOME SIGNALING BY SCF KIT | Genes involved in Signaling by SCF-KIT |
| 0.0 | 0.4 | REACTOME SIGNALLING TO RAS | Genes involved in Signalling to RAS |
| 0.0 | 1.0 | REACTOME MITOCHONDRIAL PROTEIN IMPORT | Genes involved in Mitochondrial Protein Import |
| 0.0 | 0.4 | REACTOME YAP1 AND WWTR1 TAZ STIMULATED GENE EXPRESSION | Genes involved in YAP1- and WWTR1 (TAZ)-stimulated gene expression |
| 0.0 | 1.7 | REACTOME CLASS B 2 SECRETIN FAMILY RECEPTORS | Genes involved in Class B/2 (Secretin family receptors) |
| 0.0 | 2.3 | REACTOME FATTY ACID TRIACYLGLYCEROL AND KETONE BODY METABOLISM | Genes involved in Fatty acid, triacylglycerol, and ketone body metabolism |
| 0.0 | 2.8 | REACTOME TRANSPORT OF INORGANIC CATIONS ANIONS AND AMINO ACIDS OLIGOPEPTIDES | Genes involved in Transport of inorganic cations/anions and amino acids/oligopeptides |
| 0.0 | 0.8 | REACTOME MEIOTIC SYNAPSIS | Genes involved in Meiotic Synapsis |
| 0.0 | 0.7 | REACTOME ASSOCIATION OF TRIC CCT WITH TARGET PROTEINS DURING BIOSYNTHESIS | Genes involved in Association of TriC/CCT with target proteins during biosynthesis |
| 0.0 | 1.0 | REACTOME O LINKED GLYCOSYLATION OF MUCINS | Genes involved in O-linked glycosylation of mucins |
| 0.0 | 1.0 | REACTOME NUCLEAR RECEPTOR TRANSCRIPTION PATHWAY | Genes involved in Nuclear Receptor transcription pathway |