Project
PRJNA375882: Comprehensive Mouse Transcriptomic BodyMap
Navigation
Downloads
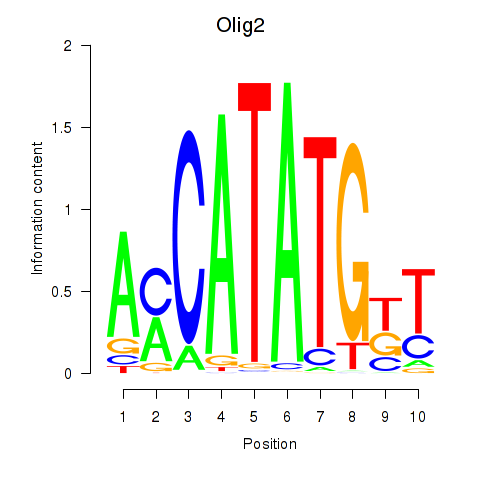
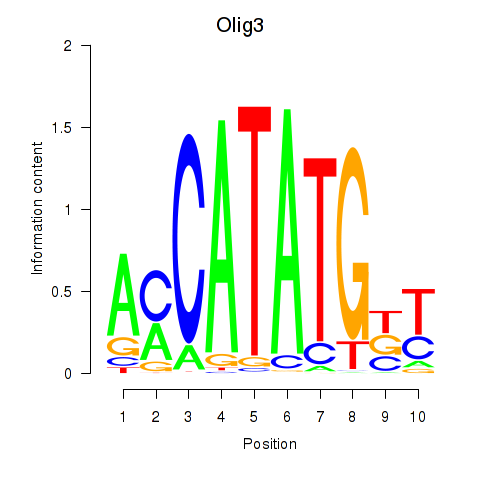
Results for Olig2_Olig3
Z-value: 1.05
Transcription factors associated with Olig2_Olig3
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Olig2
|
ENSMUSG00000039830.10 | Olig2 |
|
Olig3
|
ENSMUSG00000045591.7 | Olig3 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Olig3 | mm39_v1_chr10_+_19232281_19232312 | -0.36 | 1.9e-03 | Click! |
| Olig2 | mm39_v1_chr16_+_91022300_91022345 | -0.15 | 1.9e-01 | Click! |
Activity profile of Olig2_Olig3 motif
Sorted Z-values of Olig2_Olig3 motif
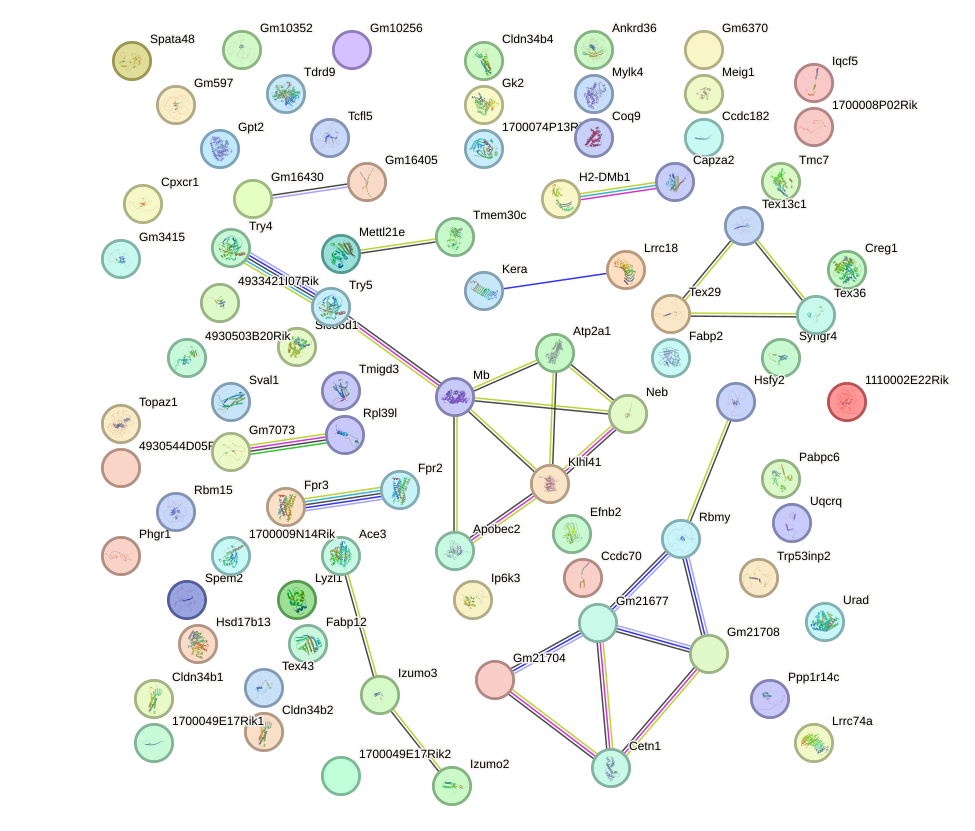
Network of associatons between targets according to the STRING database.
First level regulatory network of Olig2_Olig3
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr7_-_126062272 | 8.16 |
ENSMUST00000032974.13
|
Atp2a1
|
ATPase, Ca++ transporting, cardiac muscle, fast twitch 1 |
| chr2_+_118603247 | 7.35 |
ENSMUST00000061360.4
ENSMUST00000130293.8 |
Phgr1
|
proline/histidine/glycine-rich 1 |
| chr5_-_5529119 | 6.56 |
ENSMUST00000115447.2
|
Pttg1ip2
|
PTTG1IP family member 2 |
| chr2_+_69500444 | 5.89 |
ENSMUST00000100050.4
|
Klhl41
|
kelch-like 41 |
| chr18_+_4165856 | 4.78 |
ENSMUST00000120837.2
|
Lyzl1
|
lysozyme-like 1 |
| chr11_-_69709283 | 4.78 |
ENSMUST00000056941.3
|
Spem2
|
SPEM family member 2 |
| chr18_+_56721413 | 4.41 |
ENSMUST00000035640.13
ENSMUST00000127591.2 ENSMUST00000147775.2 |
Tex43
|
testis expressed 43 |
| chr7_+_46636562 | 4.33 |
ENSMUST00000185832.2
|
Gm9999
|
predicted gene 9999 |
| chr13_-_32960379 | 4.16 |
ENSMUST00000230119.2
|
Mylk4
|
myosin light chain kinase family, member 4 |
| chr15_-_76906150 | 4.14 |
ENSMUST00000230031.2
|
Mb
|
myoglobin |
| chrX_+_42680037 | 4.10 |
ENSMUST00000105113.4
|
Tex13c1
|
TEX13 family member C1 |
| chr3_+_137770813 | 4.02 |
ENSMUST00000163080.3
|
1110002E22Rik
|
RIKEN cDNA 1110002E22 gene |
| chr14_+_32713349 | 3.81 |
ENSMUST00000120866.8
ENSMUST00000120588.8 |
Lrrc18
|
leucine rich repeat containing 18 |
| chr2_-_52225146 | 3.81 |
ENSMUST00000075301.10
|
Neb
|
nebulin |
| chr16_-_57113207 | 3.75 |
ENSMUST00000023434.15
ENSMUST00000120112.2 ENSMUST00000119407.8 |
Tmem30c
|
transmembrane protein 30C |
| chr7_+_99827886 | 3.59 |
ENSMUST00000207358.2
ENSMUST00000207995.2 ENSMUST00000049333.13 ENSMUST00000170954.10 ENSMUST00000179842.3 ENSMUST00000208260.2 |
Kcne3
|
potassium voltage-gated channel, Isk-related subfamily, gene 3 |
| chr14_+_32713387 | 3.56 |
ENSMUST00000123822.8
ENSMUST00000120951.2 |
Lrrc18
|
leucine rich repeat containing 18 |
| chr15_-_76906832 | 3.53 |
ENSMUST00000019037.10
ENSMUST00000169226.9 |
Mb
|
myoglobin |
| chr6_+_41928559 | 3.47 |
ENSMUST00000031898.5
|
Sval1
|
seminal vesicle antigen-like 1 |
| chr8_+_11890474 | 3.45 |
ENSMUST00000033909.14
ENSMUST00000209692.2 |
Tex29
|
testis expressed 29 |
| chr3_+_105821450 | 3.45 |
ENSMUST00000198080.5
ENSMUST00000199977.2 |
Tmigd3
|
transmembrane and immunoglobulin domain containing 3 |
| chr6_-_40906665 | 3.41 |
ENSMUST00000136499.2
|
1700074P13Rik
|
RIKEN cDNA 1700074P13 gene |
| chr7_-_133203838 | 3.32 |
ENSMUST00000033275.4
|
Tex36
|
testis expressed 36 |
| chr17_-_48739874 | 3.24 |
ENSMUST00000046549.5
|
Apobec2
|
apolipoprotein B mRNA editing enzyme, catalytic polypeptide 2 |
| chr5_+_146450933 | 3.21 |
ENSMUST00000200228.5
ENSMUST00000036715.16 ENSMUST00000077133.7 |
Gm3402
|
predicted gene 3402 |
| chrX_+_115358631 | 3.18 |
ENSMUST00000101269.2
|
Cpxcr1
|
CPX chromosome region, candidate 1 |
| chr6_+_91881193 | 3.13 |
ENSMUST00000205686.2
|
4930590J08Rik
|
RIKEN cDNA 4930590J08 gene |
| chr1_+_98348817 | 3.10 |
ENSMUST00000027575.13
ENSMUST00000160796.2 |
Slco6d1
|
solute carrier organic anion transporter family, member 6d1 |
| chr4_-_25242858 | 3.07 |
ENSMUST00000029922.14
ENSMUST00000108204.2 |
Fhl5
|
four and a half LIM domains 5 |
| chrX_+_53625767 | 3.01 |
ENSMUST00000169006.3
|
Gm16430
|
predicted gene 16430 |
| chr3_+_122688721 | 2.98 |
ENSMUST00000023820.6
|
Fabp2
|
fatty acid binding protein 2, intestinal |
| chr14_+_32713336 | 2.97 |
ENSMUST00000038956.12
|
Lrrc18
|
leucine rich repeat containing 18 |
| chrX_+_53534764 | 2.97 |
ENSMUST00000169247.3
|
Gm16405
|
predicted gene 16405 |
| chr7_+_130633776 | 2.86 |
ENSMUST00000084509.7
ENSMUST00000213064.3 ENSMUST00000208311.4 |
Dmbt1
|
deleted in malignant brain tumors 1 |
| chr11_+_94858417 | 2.86 |
ENSMUST00000038928.7
|
H1f9
|
H1.9 linker histone |
| chr1_-_44258112 | 2.81 |
ENSMUST00000054801.4
|
Mettl21e
|
methyltransferase like 21E |
| chr7_-_126014027 | 2.77 |
ENSMUST00000032968.7
ENSMUST00000206325.2 |
Cd19
|
CD19 antigen |
| chr17_-_15198955 | 2.66 |
ENSMUST00000231584.2
ENSMUST00000097399.6 ENSMUST00000232173.2 |
Dynlt2a3
|
dynein light chain Tctex-type 2A3 |
| chr12_+_86781141 | 2.65 |
ENSMUST00000223308.2
|
Lrrc74a
|
leucine rich repeat containing 74A |
| chr12_+_86781154 | 2.62 |
ENSMUST00000095527.6
|
Lrrc74a
|
leucine rich repeat containing 74A |
| chr6_-_41291634 | 2.61 |
ENSMUST00000064324.12
|
Try5
|
trypsin 5 |
| chr7_-_45546081 | 2.60 |
ENSMUST00000120299.4
ENSMUST00000039049.16 |
Syngr4
|
synaptogyrin 4 |
| chr1_-_28819331 | 2.51 |
ENSMUST00000059937.5
|
Gm597
|
predicted gene 597 |
| chr19_+_20579322 | 2.48 |
ENSMUST00000087638.4
|
Aldh1a1
|
aldehyde dehydrogenase family 1, subfamily A1 |
| chr3_-_146357059 | 2.46 |
ENSMUST00000149825.2
ENSMUST00000049703.6 |
4930503B20Rik
|
RIKEN cDNA 4930503B20 gene |
| chr10_+_3316057 | 2.45 |
ENSMUST00000043374.7
|
Ppp1r14c
|
protein phosphatase 1, regulatory inhibitor subunit 14C |
| chr17_-_27386763 | 2.39 |
ENSMUST00000025046.4
|
Ip6k3
|
inositol hexaphosphate kinase 3 |
| chr2_-_180284468 | 2.38 |
ENSMUST00000037877.11
|
Tcfl5
|
transcription factor-like 5 (basic helix-loop-helix) |
| chr10_+_97442727 | 2.38 |
ENSMUST00000105286.4
|
Kera
|
keratocan |
| chrX_+_75436956 | 2.35 |
ENSMUST00000101419.2
ENSMUST00000178974.2 |
Cldn34b4
|
claudin 34B4 |
| chr2_+_67276338 | 2.35 |
ENSMUST00000239060.2
ENSMUST00000028410.4 ENSMUST00000112347.8 ENSMUST00000238878.2 |
Xirp2
|
xin actin-binding repeat containing 2 |
| chr3_-_51184895 | 2.35 |
ENSMUST00000108051.8
ENSMUST00000108053.9 |
Elf2
|
E74-like factor 2 |
| chr6_-_68907718 | 2.31 |
ENSMUST00000114212.4
|
Igkv13-85
|
immunoglobulin kappa chain variable 13-85 |
| chr1_-_56676589 | 2.30 |
ENSMUST00000062085.6
|
Hsfy2
|
heat shock transcription factor, Y-linked 2 |
| chr16_-_10609959 | 2.28 |
ENSMUST00000037996.7
|
Prm2
|
protamine 2 |
| chr7_+_123061497 | 2.28 |
ENSMUST00000033023.10
|
Aqp8
|
aquaporin 8 |
| chr11_+_105885461 | 2.28 |
ENSMUST00000190995.2
|
Ace3
|
angiotensin I converting enzyme (peptidyl-dipeptidase A) 3 |
| chr11_+_48729499 | 2.23 |
ENSMUST00000129674.2
|
Trim7
|
tripartite motif-containing 7 |
| chr5_+_146428173 | 2.23 |
ENSMUST00000110611.8
ENSMUST00000198912.2 |
Gm6370
|
predicted gene 6370 |
| chr14_+_43156329 | 2.20 |
ENSMUST00000228117.2
|
Gm9732
|
predicted gene 9732 |
| chr8_+_95564949 | 2.19 |
ENSMUST00000034234.15
ENSMUST00000159871.4 |
Coq9
|
coenzyme Q9 |
| chr18_+_43897354 | 2.16 |
ENSMUST00000187157.7
ENSMUST00000043803.13 ENSMUST00000189750.2 |
Scgb3a2
|
secretoglobin, family 3A, member 2 |
| chr5_-_104125192 | 2.15 |
ENSMUST00000120320.8
|
Hsd17b13
|
hydroxysteroid (17-beta) dehydrogenase 13 |
| chrY_-_3410148 | 2.13 |
ENSMUST00000190283.7
ENSMUST00000188091.7 ENSMUST00000169382.3 |
Gm21708
Gm21704
|
predicted gene, 21708 predicted gene, 21704 |
| chr5_-_147259245 | 2.10 |
ENSMUST00000100433.5
|
Urad
|
ureidoimidazoline (2-oxo-4-hydroxy-4-carboxy-5) decarboxylase |
| chrY_+_2830680 | 2.10 |
ENSMUST00000171534.8
ENSMUST00000100360.5 ENSMUST00000179404.8 |
Rbmy
Gm10256
|
RNA binding motif protein, Y chromosome predicted gene 10256 |
| chr18_-_9619460 | 2.09 |
ENSMUST00000234003.2
ENSMUST00000062769.7 |
Cetn1
|
centrin 1 |
| chr7_+_44358144 | 2.09 |
ENSMUST00000085422.4
|
Izumo2
|
IZUMO family member 2 |
| chr5_-_104125270 | 2.07 |
ENSMUST00000112803.3
|
Hsd17b13
|
hydroxysteroid (17-beta) dehydrogenase 13 |
| chr9_+_106391771 | 2.04 |
ENSMUST00000085113.5
|
Iqcf5
|
IQ motif containing F5 |
| chr16_+_9988080 | 2.03 |
ENSMUST00000121292.8
ENSMUST00000044103.6 |
Rpl39l
|
ribosomal protein L39-like |
| chr6_-_70318437 | 2.00 |
ENSMUST00000196599.2
|
Igkv8-19
|
immunoglobulin kappa variable 8-19 |
| chr11_+_88184872 | 1.98 |
ENSMUST00000037268.6
|
Ccdc182
|
coiled-coil domain containing 182 |
| chrX_-_153673263 | 1.93 |
ENSMUST00000096852.5
|
Cldn34b1
|
claudin 34B1 |
| chrX_-_59582855 | 1.92 |
ENSMUST00000166562.3
|
Gm7073
|
predicted gene 7073 |
| chr7_-_42097503 | 1.86 |
ENSMUST00000032648.5
|
4933421I07Rik
|
RIKEN cDNA 4933421I07 gene |
| chr1_+_165591315 | 1.86 |
ENSMUST00000111432.10
|
Creg1
|
cellular repressor of E1A-stimulated genes 1 |
| chr8_+_86219191 | 1.86 |
ENSMUST00000034136.12
|
Gpt2
|
glutamic pyruvate transaminase (alanine aminotransferase) 2 |
| chr12_+_111937978 | 1.85 |
ENSMUST00000079009.11
|
Tdrd9
|
tudor domain containing 9 |
| chr3_-_6685492 | 1.85 |
ENSMUST00000091364.4
|
1700008P02Rik
|
RIKEN cDNA 1700008P02 gene |
| chr6_-_68609426 | 1.84 |
ENSMUST00000103328.3
|
Igkv10-96
|
immunoglobulin kappa variable 10-96 |
| chr5_-_104125226 | 1.83 |
ENSMUST00000048118.15
|
Hsd17b13
|
hydroxysteroid (17-beta) dehydrogenase 13 |
| chrY_+_2862139 | 1.80 |
ENSMUST00000189964.7
ENSMUST00000188114.2 |
Gm10256
|
predicted gene 10256 |
| chr7_-_118183892 | 1.80 |
ENSMUST00000044195.6
|
Tmc7
|
transmembrane channel-like gene family 7 |
| chr8_+_22460606 | 1.76 |
ENSMUST00000017193.2
|
Ccdc70
|
coiled-coil domain containing 70 |
| chr5_-_97604830 | 1.76 |
ENSMUST00000059657.4
|
Gk2
|
glycerol kinase 2 |
| chr4_+_39450265 | 1.75 |
ENSMUST00000029955.5
|
1700009N14Rik
|
RIKEN cDNA 1700009N14 gene |
| chrX_-_153911405 | 1.74 |
ENSMUST00000076671.4
|
Cldn34b2
|
claudin 34B2 |
| chr2_-_3423646 | 1.71 |
ENSMUST00000064685.14
|
Meig1
|
meiosis expressed gene 1 |
| chr6_-_67919524 | 1.71 |
ENSMUST00000196768.2
|
Igkv9-124
|
immunoglobulin kappa chain variable 9-124 |
| chr2_+_155224105 | 1.68 |
ENSMUST00000134218.2
|
Trp53inp2
|
transformation related protein 53 inducible nuclear protein 2 |
| chr3_+_132335575 | 1.68 |
ENSMUST00000212804.2
ENSMUST00000212852.2 |
Gimd1
|
GIMAP family P-loop NTPase domain containing 1 |
| chr12_-_114104740 | 1.67 |
ENSMUST00000103473.2
|
Ighv9-3
|
immunoglobulin heavy variable V9-3 |
| chr6_+_17636979 | 1.67 |
ENSMUST00000015877.14
ENSMUST00000152005.3 |
Capza2
|
capping protein (actin filament) muscle Z-line, alpha 2 |
| chr3_-_10366229 | 1.67 |
ENSMUST00000119761.2
ENSMUST00000029043.13 |
Fabp12
|
fatty acid binding protein 12 |
| chr17_-_9888639 | 1.67 |
ENSMUST00000057190.4
|
Pabpc6
|
poly(A) binding protein, cytoplasmic 6 |
| chr7_+_123061535 | 1.66 |
ENSMUST00000098056.6
|
Aqp8
|
aquaporin 8 |
| chr10_+_70040483 | 1.65 |
ENSMUST00000020090.8
|
Mrln
|
myoregulin |
| chr17_+_18108086 | 1.61 |
ENSMUST00000149944.2
|
Fpr2
|
formyl peptide receptor 2 |
| chr2_-_3423613 | 1.60 |
ENSMUST00000144584.2
|
Meig1
|
meiosis expressed gene 1 |
| chr12_-_114815280 | 1.60 |
ENSMUST00000103512.3
|
Ighv1-34
|
immunoglobulin heavy variable 1-34 |
| chr6_+_42957599 | 1.60 |
ENSMUST00000167638.2
|
Gm17472
|
predicted gene, 17472 |
| chr17_+_38234397 | 1.59 |
ENSMUST00000080231.5
|
Olfr128
|
olfactory receptor 128 |
| chr11_+_5578738 | 1.58 |
ENSMUST00000137933.2
|
Ankrd36
|
ankyrin repeat domain 36 |
| chr6_-_69877961 | 1.58 |
ENSMUST00000197290.2
|
Igkv5-39
|
immunoglobulin kappa variable 5-39 |
| chr13_+_83723743 | 1.57 |
ENSMUST00000198217.5
ENSMUST00000199210.5 |
Mef2c
|
myocyte enhancer factor 2C |
| chr11_+_11414256 | 1.56 |
ENSMUST00000020410.11
|
Spata48
|
spermatogenesis associated 48 |
| chr9_+_122576325 | 1.56 |
ENSMUST00000178679.3
|
Topaz1
|
testis and ovary specific PAZ domain containing 1 |
| chr11_-_53321242 | 1.55 |
ENSMUST00000109019.8
|
Uqcrq
|
ubiquinol-cytochrome c reductase, complex III subunit VII |
| chr3_-_107240989 | 1.55 |
ENSMUST00000061772.11
|
Rbm15
|
RNA binding motif protein 15 |
| chr11_+_70506716 | 1.53 |
ENSMUST00000144960.2
|
4930544D05Rik
|
RIKEN cDNA 4930544D05 gene |
| chr4_-_92035446 | 1.53 |
ENSMUST00000107108.8
ENSMUST00000143542.2 |
Izumo3
|
IZUMO family member 3 |
| chrY_-_3306449 | 1.53 |
ENSMUST00000189592.7
|
Gm21677
|
predicted gene, 21677 |
| chrY_-_3378783 | 1.53 |
ENSMUST00000187277.7
|
Gm21704
|
predicted gene, 21704 |
| chrY_+_2932582 | 1.52 |
ENSMUST00000188358.2
|
Gm29289
|
predicted gene 29289 |
| chr3_-_124219673 | 1.51 |
ENSMUST00000029598.10
|
1700006A11Rik
|
RIKEN cDNA 1700006A11 gene |
| chr11_-_30421792 | 1.49 |
ENSMUST00000041763.14
|
4930505A04Rik
|
RIKEN cDNA 4930505A04 gene |
| chr6_+_68026941 | 1.47 |
ENSMUST00000103316.2
|
Igkv9-120
|
immunoglobulin kappa chain variable 9-120 |
| chr6_-_69877642 | 1.46 |
ENSMUST00000103370.3
|
Igkv5-39
|
immunoglobulin kappa variable 5-39 |
| chrY_-_3345329 | 1.46 |
ENSMUST00000186047.7
|
Gm21693
|
predicted gene, 21693 |
| chr3_+_3573084 | 1.46 |
ENSMUST00000108393.8
|
Hnf4g
|
hepatocyte nuclear factor 4, gamma |
| chr11_+_14549252 | 1.46 |
ENSMUST00000117584.3
|
Pom121l12
|
POM121 membrane glycoprotein-like 12 |
| chr5_+_90916370 | 1.46 |
ENSMUST00000031319.8
|
Ppbp
|
pro-platelet basic protein |
| chr11_+_70506674 | 1.45 |
ENSMUST00000180052.8
|
4930544D05Rik
|
RIKEN cDNA 4930544D05 gene |
| chr11_+_96025045 | 1.45 |
ENSMUST00000107680.2
|
Ttll6
|
tubulin tyrosine ligase-like family, member 6 |
| chr5_+_77163869 | 1.44 |
ENSMUST00000031161.11
ENSMUST00000117880.8 |
Thegl
|
theg spermatid protein like |
| chr2_-_79287095 | 1.44 |
ENSMUST00000041099.5
|
Neurod1
|
neurogenic differentiation 1 |
| chrY_-_68304748 | 1.44 |
ENSMUST00000180329.2
|
Gm20937
|
predicted gene, 20937 |
| chrY_+_3771673 | 1.44 |
ENSMUST00000186140.7
|
Gm3376
|
predicted gene 3376 |
| chrY_+_49569158 | 1.43 |
ENSMUST00000178556.2
|
Gm21209
|
predicted gene, 21209 |
| chrY_+_62200817 | 1.43 |
ENSMUST00000178115.2
|
Gm21518
|
predicted gene, 21518 |
| chr9_+_110663656 | 1.43 |
ENSMUST00000011391.12
ENSMUST00000146794.4 |
Prss45
|
protease, serine 45 |
| chr11_+_102036356 | 1.43 |
ENSMUST00000055409.6
|
Nags
|
N-acetylglutamate synthase |
| chr3_+_99147677 | 1.43 |
ENSMUST00000151606.8
|
Tbx15
|
T-box 15 |
| chr5_+_123648523 | 1.43 |
ENSMUST00000031384.6
|
B3gnt4
|
UDP-GlcNAc:betaGal beta-1,3-N-acetylglucosaminyltransferase 4 |
| chr1_-_161807205 | 1.43 |
ENSMUST00000162676.2
|
4930558K02Rik
|
RIKEN cDNA 4930558K02 gene |
| chrY_+_77705501 | 1.42 |
ENSMUST00000179073.2
|
Gm21650
|
predicted gene, 21650 |
| chrY_-_72256967 | 1.42 |
ENSMUST00000178505.2
|
Gm20843
|
predicted gene, 20843 |
| chrY_+_2900989 | 1.41 |
ENSMUST00000187842.7
|
Gm10352
|
predicted gene 10352 |
| chrX_+_134274033 | 1.41 |
ENSMUST00000113173.8
ENSMUST00000180198.2 |
Gm5128
|
predicted gene 5128 |
| chrY_-_2796205 | 1.40 |
ENSMUST00000187482.2
|
Gm4064
|
predicted gene 4064 |
| chr3_+_26691767 | 1.40 |
ENSMUST00000047005.11
|
Spata16
|
spermatogenesis associated 16 |
| chr2_-_164315634 | 1.40 |
ENSMUST00000017864.3
|
Trp53tg5
|
transformation related protein 53 target 5 |
| chr12_-_114901026 | 1.39 |
ENSMUST00000103516.2
ENSMUST00000191868.2 |
Ighv1-42
|
immunoglobulin heavy variable V1-42 |
| chr11_+_58808830 | 1.39 |
ENSMUST00000020792.12
ENSMUST00000108818.4 |
Btnl10
|
butyrophilin-like 10 |
| chr2_-_163239865 | 1.37 |
ENSMUST00000017961.11
ENSMUST00000109425.3 |
Jph2
|
junctophilin 2 |
| chr1_+_153300874 | 1.37 |
ENSMUST00000042373.12
|
Shcbp1l
|
Shc SH2-domain binding protein 1-like |
| chr18_-_32595515 | 1.36 |
ENSMUST00000207669.2
|
Gm35060
|
predicted gene, 35060 |
| chr2_-_76700830 | 1.34 |
ENSMUST00000138542.2
|
Ttn
|
titin |
| chr1_-_53391778 | 1.34 |
ENSMUST00000236737.2
ENSMUST00000027264.10 ENSMUST00000123519.9 |
Gm50478
Asnsd1
|
predicted gene, 50478 asparagine synthetase domain containing 1 |
| chr10_-_53252210 | 1.34 |
ENSMUST00000095691.7
|
Cep85l
|
centrosomal protein 85-like |
| chrX_+_64091244 | 1.33 |
ENSMUST00000033524.3
|
Ctag2
|
cancer/testis antigen 2 |
| chr9_-_72892617 | 1.31 |
ENSMUST00000124565.3
|
Ccpg1os
|
cell cycle progression 1, opposite strand |
| chr12_-_40249314 | 1.31 |
ENSMUST00000095760.3
|
Lsmem1
|
leucine-rich single-pass membrane protein 1 |
| chrY_+_51123234 | 1.30 |
ENSMUST00000180133.2
|
Gm21117
|
predicted gene, 21117 |
| chr2_+_136733421 | 1.30 |
ENSMUST00000141463.8
|
Slx4ip
|
SLX4 interacting protein |
| chr4_+_43441939 | 1.30 |
ENSMUST00000060864.13
|
Tesk1
|
testis specific protein kinase 1 |
| chr5_-_146107531 | 1.29 |
ENSMUST00000174320.2
|
Gm6309
|
predicted gene 6309 |
| chr1_-_180027151 | 1.28 |
ENSMUST00000161743.3
|
Coq8a
|
coenzyme Q8A |
| chr2_+_136374186 | 1.28 |
ENSMUST00000121717.2
|
Ankef1
|
ankyrin repeat and EF-hand domain containing 1 |
| chr17_-_65946817 | 1.28 |
ENSMUST00000233702.2
|
Txndc2
|
thioredoxin domain containing 2 (spermatozoa) |
| chr16_-_64422716 | 1.27 |
ENSMUST00000209382.3
|
Csnka2ip
|
casein kinase 2, alpha prime interacting protein |
| chr7_-_6014009 | 1.25 |
ENSMUST00000086338.2
|
Vmn1r65
|
vomeronasal 1 receptor 65 |
| chr11_+_100211363 | 1.25 |
ENSMUST00000152521.2
|
Eif1
|
eukaryotic translation initiation factor 1 |
| chr6_-_24515036 | 1.25 |
ENSMUST00000052277.5
|
Iqub
|
IQ motif and ubiquitin domain containing |
| chrY_+_70454574 | 1.25 |
ENSMUST00000178934.2
|
Gm20888
|
predicted gene, 20888 |
| chr11_+_87684299 | 1.25 |
ENSMUST00000020779.11
|
Mpo
|
myeloperoxidase |
| chr3_-_27764571 | 1.25 |
ENSMUST00000046157.10
|
Fndc3b
|
fibronectin type III domain containing 3B |
| chrY_+_65387652 | 1.23 |
ENSMUST00000178198.2
|
Gm20736
|
predicted gene, 20736 |
| chr12_-_114186874 | 1.23 |
ENSMUST00000103477.4
ENSMUST00000192499.3 |
Ighv7-4
|
immunoglobulin heavy variable 7-4 |
| chrX_+_68988904 | 1.23 |
ENSMUST00000075654.2
|
1700020N15Rik
|
RIKEN cDNA 1700020N15 gene |
| chr11_-_103511966 | 1.22 |
ENSMUST00000059279.13
|
Gm884
|
predicted gene 884 |
| chr17_+_18108102 | 1.21 |
ENSMUST00000054871.12
ENSMUST00000064068.5 |
Fpr3
Fpr2
|
formyl peptide receptor 3 formyl peptide receptor 2 |
| chr3_+_20039775 | 1.21 |
ENSMUST00000172860.2
|
Cp
|
ceruloplasmin |
| chr6_-_52203146 | 1.19 |
ENSMUST00000114425.3
|
Hoxa9
|
homeobox A9 |
| chr12_-_114878652 | 1.19 |
ENSMUST00000103515.2
|
Ighv1-39
|
immunoglobulin heavy variable 1-39 |
| chr15_-_66372704 | 1.18 |
ENSMUST00000023006.7
|
Lrrc6
|
leucine rich repeat containing 6 (testis) |
| chrY_+_50093855 | 1.16 |
ENSMUST00000187033.3
|
Gm28870
|
predicted gene 28870 |
| chrY_+_79332266 | 1.15 |
ENSMUST00000178063.2
|
Gm20916
|
predicted gene, 20916 |
| chr9_-_106421834 | 1.14 |
ENSMUST00010181660.1
ENSMUST00000215525.2 |
ENSMUSG00001074846.1
ENSMUSG00000118396.3
|
IQ motif containing F3 IQ motif containing F3 |
| chr1_+_167445815 | 1.13 |
ENSMUST00000111380.2
|
Rxrg
|
retinoid X receptor gamma |
| chr10_+_70040504 | 1.12 |
ENSMUST00000190199.2
|
Mrln
|
myoregulin |
| chr7_-_119694400 | 1.12 |
ENSMUST00000209154.3
ENSMUST00000046993.4 |
Dnah3
|
dynein, axonemal, heavy chain 3 |
| chr9_-_121745354 | 1.11 |
ENSMUST00000062474.5
|
Cyp8b1
|
cytochrome P450, family 8, subfamily b, polypeptide 1 |
| chr17_-_35285146 | 1.11 |
ENSMUST00000174190.2
ENSMUST00000097337.8 |
Mpig6b
|
megakaryocyte and platelet inhibitory receptor G6b |
| chr4_-_116024788 | 1.11 |
ENSMUST00000030465.10
ENSMUST00000143426.2 |
Tspan1
|
tetraspanin 1 |
| chrY_-_35085749 | 1.11 |
ENSMUST00000180170.2
|
Gm20855
|
predicted gene, 20855 |
| chrX_+_45417884 | 1.10 |
ENSMUST00000059466.3
|
Actrt1
|
actin-related protein T1 |
| chrY_+_55211732 | 1.10 |
ENSMUST00000180249.2
|
Gm20931
|
predicted gene, 20931 |
| chrY_+_84109980 | 1.09 |
ENSMUST00000177775.2
|
Gm21095
|
predicted gene, 21095 |
| chr3_-_144738526 | 1.08 |
ENSMUST00000029919.7
|
Clca1
|
chloride channel accessory 1 |
| chr11_-_55042529 | 1.08 |
ENSMUST00000020502.9
ENSMUST00000069816.6 |
Slc36a3
|
solute carrier family 36 (proton/amino acid symporter), member 3 |
| chr2_-_34951443 | 1.08 |
ENSMUST00000028233.7
|
Hc
|
hemolytic complement |
| chr1_-_139487951 | 1.07 |
ENSMUST00000023965.8
|
Cfhr1
|
complement factor H-related 1 |
| chr18_-_46444804 | 1.06 |
ENSMUST00000236934.2
|
Ccdc112
|
coiled-coil domain containing 112 |
| chr6_-_5496261 | 1.05 |
ENSMUST00000203347.3
ENSMUST00000019721.7 |
Pdk4
|
pyruvate dehydrogenase kinase, isoenzyme 4 |
| chrX_+_143253677 | 1.05 |
ENSMUST00000178233.2
|
Trpc5os
|
transient receptor potential cation channel, subfamily C, member 5, opposite strand |
| chr2_+_152873772 | 1.04 |
ENSMUST00000037235.7
|
Xkr7
|
X-linked Kx blood group related 7 |
| chr15_-_76010736 | 1.04 |
ENSMUST00000054022.12
ENSMUST00000089654.4 |
BC024139
|
cDNA sequence BC024139 |
| chr9_+_111011327 | 1.03 |
ENSMUST00000216430.2
|
Lrrfip2
|
leucine rich repeat (in FLII) interacting protein 2 |
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.8 | 8.4 | GO:0051659 | maintenance of mitochondrion location(GO:0051659) |
| 0.9 | 3.6 | GO:1902303 | negative regulation of potassium ion export(GO:1902303) |
| 0.9 | 7.7 | GO:0015671 | oxygen transport(GO:0015671) |
| 0.7 | 2.8 | GO:1902080 | regulation of calcium ion import into sarcoplasmic reticulum(GO:1902080) negative regulation of calcium ion import into sarcoplasmic reticulum(GO:1902081) |
| 0.6 | 1.9 | GO:0035702 | monocyte homeostasis(GO:0035702) |
| 0.5 | 3.9 | GO:0015722 | canalicular bile acid transport(GO:0015722) |
| 0.5 | 1.9 | GO:0042851 | L-alanine metabolic process(GO:0042851) |
| 0.4 | 1.2 | GO:0002149 | hypochlorous acid metabolic process(GO:0002148) hypochlorous acid biosynthetic process(GO:0002149) |
| 0.4 | 2.8 | GO:0002430 | complement receptor mediated signaling pathway(GO:0002430) |
| 0.4 | 2.0 | GO:0035660 | MyD88-dependent toll-like receptor 4 signaling pathway(GO:0035660) |
| 0.4 | 1.6 | GO:0000239 | pachytene(GO:0000239) meiotic cell cycle phase transition(GO:0044771) regulation of meiotic cell cycle phase transition(GO:1901993) |
| 0.4 | 1.4 | GO:0030309 | poly-N-acetyllactosamine metabolic process(GO:0030309) poly-N-acetyllactosamine biosynthetic process(GO:0030311) |
| 0.4 | 1.8 | GO:0046167 | glycerol-3-phosphate biosynthetic process(GO:0046167) |
| 0.3 | 1.3 | GO:0006529 | asparagine biosynthetic process(GO:0006529) |
| 0.3 | 5.4 | GO:0031272 | regulation of pseudopodium assembly(GO:0031272) |
| 0.3 | 1.5 | GO:0034224 | cellular response to zinc ion starvation(GO:0034224) |
| 0.3 | 2.5 | GO:0042905 | 9-cis-retinoic acid biosynthetic process(GO:0042904) 9-cis-retinoic acid metabolic process(GO:0042905) |
| 0.3 | 1.3 | GO:0043056 | forward locomotion(GO:0043056) |
| 0.3 | 4.8 | GO:0016998 | cell wall macromolecule catabolic process(GO:0016998) |
| 0.3 | 1.1 | GO:0010510 | regulation of acetyl-CoA biosynthetic process from pyruvate(GO:0010510) |
| 0.2 | 3.2 | GO:0016554 | cytidine to uridine editing(GO:0016554) |
| 0.2 | 1.3 | GO:0042776 | mitochondrial ATP synthesis coupled proton transport(GO:0042776) |
| 0.2 | 1.5 | GO:0021539 | subthalamus development(GO:0021539) |
| 0.2 | 1.5 | GO:1990009 | retinal cell apoptotic process(GO:1990009) |
| 0.2 | 0.8 | GO:0014734 | skeletal muscle hypertrophy(GO:0014734) |
| 0.2 | 1.1 | GO:0060729 | intestinal epithelial structure maintenance(GO:0060729) |
| 0.2 | 1.4 | GO:0051013 | microtubule severing(GO:0051013) |
| 0.2 | 1.0 | GO:0006311 | meiotic gene conversion(GO:0006311) gene conversion(GO:0035822) |
| 0.2 | 0.8 | GO:0002767 | immune response-inhibiting cell surface receptor signaling pathway(GO:0002767) |
| 0.2 | 2.1 | GO:0000255 | allantoin metabolic process(GO:0000255) |
| 0.2 | 0.6 | GO:0072347 | response to anesthetic(GO:0072347) |
| 0.2 | 1.4 | GO:0006526 | arginine biosynthetic process(GO:0006526) |
| 0.2 | 1.4 | GO:0060316 | positive regulation of ryanodine-sensitive calcium-release channel activity(GO:0060316) |
| 0.2 | 1.2 | GO:0060287 | epithelial cilium movement involved in determination of left/right asymmetry(GO:0060287) |
| 0.2 | 2.4 | GO:0061303 | cornea development in camera-type eye(GO:0061303) |
| 0.1 | 3.5 | GO:0006744 | ubiquinone biosynthetic process(GO:0006744) |
| 0.1 | 0.6 | GO:1902174 | positive regulation of keratinocyte apoptotic process(GO:1902174) |
| 0.1 | 0.7 | GO:2001274 | immunoglobulin biosynthetic process(GO:0002378) negative regulation of glucose import in response to insulin stimulus(GO:2001274) |
| 0.1 | 1.1 | GO:0045919 | positive regulation of cytolysis(GO:0045919) |
| 0.1 | 1.6 | GO:0003172 | primary heart field specification(GO:0003138) sinoatrial valve development(GO:0003172) sinoatrial valve morphogenesis(GO:0003185) |
| 0.1 | 0.4 | GO:0016999 | antibiotic metabolic process(GO:0016999) |
| 0.1 | 1.5 | GO:0007221 | positive regulation of transcription of Notch receptor target(GO:0007221) |
| 0.1 | 2.2 | GO:0006662 | glycerol ether metabolic process(GO:0006662) |
| 0.1 | 1.3 | GO:0007549 | dosage compensation(GO:0007549) dosage compensation by inactivation of X chromosome(GO:0009048) |
| 0.1 | 0.8 | GO:0072092 | ureteric bud invasion(GO:0072092) |
| 0.1 | 17.9 | GO:0006958 | complement activation, classical pathway(GO:0006958) |
| 0.1 | 0.7 | GO:0034334 | desmosome assembly(GO:0002159) adherens junction maintenance(GO:0034334) |
| 0.1 | 0.3 | GO:0061033 | secretion by lung epithelial cell involved in lung growth(GO:0061033) |
| 0.1 | 0.4 | GO:0030046 | parallel actin filament bundle assembly(GO:0030046) |
| 0.1 | 0.3 | GO:0072429 | response to intra-S DNA damage checkpoint signaling(GO:0072429) |
| 0.1 | 1.9 | GO:0034587 | piRNA metabolic process(GO:0034587) |
| 0.1 | 1.6 | GO:0060510 | Type II pneumocyte differentiation(GO:0060510) |
| 0.1 | 1.7 | GO:0000054 | ribosomal subunit export from nucleus(GO:0000054) ribosome localization(GO:0033750) establishment of ribosome localization(GO:0033753) |
| 0.1 | 1.6 | GO:0006968 | cellular defense response(GO:0006968) |
| 0.1 | 1.1 | GO:0036159 | inner dynein arm assembly(GO:0036159) |
| 0.1 | 2.1 | GO:0001833 | inner cell mass cell proliferation(GO:0001833) |
| 0.1 | 0.5 | GO:1904098 | regulation of protein O-linked glycosylation(GO:1904098) positive regulation of protein O-linked glycosylation(GO:1904100) |
| 0.1 | 0.2 | GO:0021912 | regulation of transcription from RNA polymerase II promoter involved in spinal cord motor neuron fate specification(GO:0021912) |
| 0.1 | 0.7 | GO:0035754 | B cell chemotaxis(GO:0035754) |
| 0.1 | 2.3 | GO:0030261 | chromosome condensation(GO:0030261) |
| 0.1 | 0.9 | GO:2000344 | positive regulation of acrosome reaction(GO:2000344) |
| 0.1 | 0.7 | GO:0032000 | positive regulation of fatty acid beta-oxidation(GO:0032000) very long-chain fatty acid catabolic process(GO:0042760) |
| 0.1 | 0.3 | GO:1904000 | positive regulation of eating behavior(GO:1904000) regulation of gastric mucosal blood circulation(GO:1904344) positive regulation of gastric mucosal blood circulation(GO:1904346) positive regulation of small intestine smooth muscle contraction(GO:1904349) gastric mucosal blood circulation(GO:1990768) negative regulation of energy homeostasis(GO:2000506) |
| 0.1 | 0.6 | GO:0006572 | tyrosine catabolic process(GO:0006572) |
| 0.1 | 0.4 | GO:2000325 | regulation of ligand-dependent nuclear receptor transcription coactivator activity(GO:2000325) positive regulation of ligand-dependent nuclear receptor transcription coactivator activity(GO:2000327) |
| 0.1 | 0.2 | GO:0043382 | defense response to nematode(GO:0002215) positive regulation of memory T cell differentiation(GO:0043382) |
| 0.1 | 3.2 | GO:0043252 | sodium-independent organic anion transport(GO:0043252) |
| 0.1 | 0.2 | GO:0051030 | snRNA transport(GO:0051030) |
| 0.1 | 3.0 | GO:0050892 | intestinal absorption(GO:0050892) |
| 0.1 | 0.3 | GO:0070966 | nuclear-transcribed mRNA catabolic process, no-go decay(GO:0070966) |
| 0.1 | 0.6 | GO:0047484 | regulation of response to osmotic stress(GO:0047484) |
| 0.1 | 1.1 | GO:0015816 | glycine transport(GO:0015816) |
| 0.1 | 0.4 | GO:0002023 | reduction of food intake in response to dietary excess(GO:0002023) |
| 0.1 | 0.2 | GO:0002940 | tRNA N2-guanine methylation(GO:0002940) |
| 0.1 | 1.0 | GO:0097500 | receptor localization to nonmotile primary cilium(GO:0097500) |
| 0.1 | 1.5 | GO:0090023 | positive regulation of neutrophil chemotaxis(GO:0090023) |
| 0.1 | 23.5 | GO:0051321 | meiotic cell cycle(GO:0051321) |
| 0.1 | 4.1 | GO:0045214 | sarcomere organization(GO:0045214) |
| 0.1 | 0.5 | GO:0006122 | mitochondrial electron transport, ubiquinol to cytochrome c(GO:0006122) |
| 0.1 | 0.9 | GO:0015721 | bile acid and bile salt transport(GO:0015721) |
| 0.1 | 0.4 | GO:0016560 | protein import into peroxisome matrix, docking(GO:0016560) |
| 0.1 | 0.4 | GO:0010898 | positive regulation of triglyceride catabolic process(GO:0010898) |
| 0.1 | 0.7 | GO:0006123 | mitochondrial electron transport, cytochrome c to oxygen(GO:0006123) |
| 0.0 | 1.2 | GO:0042118 | endothelial cell activation(GO:0042118) |
| 0.0 | 0.3 | GO:1903265 | positive regulation of tumor necrosis factor-mediated signaling pathway(GO:1903265) |
| 0.0 | 1.7 | GO:0010614 | negative regulation of cardiac muscle hypertrophy(GO:0010614) |
| 0.0 | 0.3 | GO:0048861 | oncostatin-M-mediated signaling pathway(GO:0038165) leukemia inhibitory factor signaling pathway(GO:0048861) |
| 0.0 | 1.7 | GO:0051016 | barbed-end actin filament capping(GO:0051016) |
| 0.0 | 0.2 | GO:0071544 | diphosphoinositol polyphosphate catabolic process(GO:0071544) |
| 0.0 | 9.6 | GO:0002377 | immunoglobulin production(GO:0002377) |
| 0.0 | 0.4 | GO:0006102 | isocitrate metabolic process(GO:0006102) |
| 0.0 | 0.5 | GO:0050957 | equilibrioception(GO:0050957) |
| 0.0 | 1.0 | GO:0014898 | muscle hypertrophy in response to stress(GO:0003299) cardiac muscle adaptation(GO:0014887) cardiac muscle hypertrophy in response to stress(GO:0014898) |
| 0.0 | 0.3 | GO:0015014 | heparan sulfate proteoglycan biosynthetic process, polysaccharide chain biosynthetic process(GO:0015014) |
| 0.0 | 0.5 | GO:0042573 | retinoic acid metabolic process(GO:0042573) |
| 0.0 | 1.2 | GO:0046688 | response to copper ion(GO:0046688) |
| 0.0 | 4.5 | GO:0046889 | positive regulation of lipid biosynthetic process(GO:0046889) |
| 0.0 | 1.0 | GO:0048240 | sperm capacitation(GO:0048240) |
| 0.0 | 0.5 | GO:0060088 | auditory receptor cell stereocilium organization(GO:0060088) |
| 0.0 | 0.8 | GO:0030574 | collagen catabolic process(GO:0030574) |
| 0.0 | 1.1 | GO:1901522 | positive regulation of transcription from RNA polymerase II promoter involved in cellular response to chemical stimulus(GO:1901522) |
| 0.0 | 0.2 | GO:0001830 | trophectodermal cell fate commitment(GO:0001830) |
| 0.0 | 1.0 | GO:0006978 | DNA damage response, signal transduction by p53 class mediator resulting in transcription of p21 class mediator(GO:0006978) |
| 0.0 | 0.4 | GO:0035871 | protein K11-linked deubiquitination(GO:0035871) |
| 0.0 | 0.4 | GO:0018206 | peptidyl-methionine modification(GO:0018206) |
| 0.0 | 2.4 | GO:0055008 | cardiac muscle tissue morphogenesis(GO:0055008) |
| 0.0 | 0.6 | GO:0000737 | DNA catabolic process, endonucleolytic(GO:0000737) apoptotic DNA fragmentation(GO:0006309) |
| 0.0 | 0.2 | GO:0002553 | histamine production involved in inflammatory response(GO:0002349) histamine secretion involved in inflammatory response(GO:0002441) histamine secretion by mast cell(GO:0002553) |
| 0.0 | 0.3 | GO:2000507 | positive regulation of energy homeostasis(GO:2000507) |
| 0.0 | 0.5 | GO:0045953 | negative regulation of natural killer cell mediated cytotoxicity(GO:0045953) |
| 0.0 | 0.6 | GO:2001014 | regulation of skeletal muscle cell differentiation(GO:2001014) |
| 0.0 | 0.3 | GO:0021891 | olfactory bulb interneuron development(GO:0021891) |
| 0.0 | 0.2 | GO:0006686 | sphingomyelin biosynthetic process(GO:0006686) |
| 0.0 | 0.1 | GO:0006363 | termination of RNA polymerase I transcription(GO:0006363) |
| 0.0 | 0.1 | GO:2000138 | positive regulation of cell proliferation involved in heart morphogenesis(GO:2000138) |
| 0.0 | 0.1 | GO:1902261 | positive regulation of delayed rectifier potassium channel activity(GO:1902261) |
| 0.0 | 0.3 | GO:0048386 | positive regulation of retinoic acid receptor signaling pathway(GO:0048386) |
| 0.0 | 0.7 | GO:0050482 | arachidonic acid secretion(GO:0050482) arachidonate transport(GO:1903963) |
| 0.0 | 1.1 | GO:0048701 | embryonic cranial skeleton morphogenesis(GO:0048701) |
| 0.0 | 0.2 | GO:1902570 | protein localization to nucleolus(GO:1902570) |
| 0.0 | 0.1 | GO:0002606 | positive regulation of dendritic cell antigen processing and presentation(GO:0002606) |
| 0.0 | 0.1 | GO:0002457 | T cell antigen processing and presentation(GO:0002457) |
| 0.0 | 0.2 | GO:2000659 | regulation of interleukin-1-mediated signaling pathway(GO:2000659) |
| 0.0 | 0.2 | GO:0045792 | negative regulation of cell size(GO:0045792) |
| 0.0 | 0.8 | GO:0002474 | antigen processing and presentation of peptide antigen via MHC class I(GO:0002474) |
| 0.0 | 0.1 | GO:0072268 | pattern specification involved in metanephros development(GO:0072268) |
| 0.0 | 0.1 | GO:0046684 | response to pyrethroid(GO:0046684) |
| 0.0 | 0.5 | GO:0032288 | myelin assembly(GO:0032288) |
| 0.0 | 0.6 | GO:0032801 | receptor catabolic process(GO:0032801) |
| 0.0 | 1.0 | GO:0034605 | cellular response to heat(GO:0034605) |
| 0.0 | 0.4 | GO:2000678 | negative regulation of transcription regulatory region DNA binding(GO:2000678) |
| 0.0 | 2.5 | GO:0046488 | phosphatidylinositol metabolic process(GO:0046488) |
| 0.0 | 0.2 | GO:0001682 | tRNA 5'-leader removal(GO:0001682) |
| 0.0 | 0.5 | GO:0006270 | DNA replication initiation(GO:0006270) |
| 0.0 | 0.5 | GO:0051290 | protein heterotetramerization(GO:0051290) |
| 0.0 | 0.8 | GO:0018279 | protein N-linked glycosylation via asparagine(GO:0018279) |
| 0.0 | 0.1 | GO:0010578 | regulation of adenylate cyclase activity involved in G-protein coupled receptor signaling pathway(GO:0010578) positive regulation of adenylate cyclase activity involved in G-protein coupled receptor signaling pathway(GO:0010579) |
| 0.0 | 0.4 | GO:1904355 | positive regulation of telomere capping(GO:1904355) |
| 0.0 | 1.7 | GO:0018107 | peptidyl-threonine phosphorylation(GO:0018107) |
| 0.0 | 0.2 | GO:0070389 | chaperone cofactor-dependent protein refolding(GO:0070389) |
| 0.0 | 0.2 | GO:0052695 | cellular glucuronidation(GO:0052695) |
| 0.0 | 0.1 | GO:0009249 | protein lipoylation(GO:0009249) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.0 | 3.9 | GO:0046691 | intracellular canaliculus(GO:0046691) |
| 0.9 | 7.5 | GO:0031673 | H zone(GO:0031673) |
| 0.4 | 1.4 | GO:0070722 | Tle3-Aes complex(GO:0070722) |
| 0.3 | 1.9 | GO:0071547 | piP-body(GO:0071547) |
| 0.3 | 5.9 | GO:0031143 | pseudopodium(GO:0031143) |
| 0.3 | 3.0 | GO:0045179 | apical cortex(GO:0045179) |
| 0.2 | 2.9 | GO:0030670 | phagocytic vesicle membrane(GO:0030670) |
| 0.2 | 2.1 | GO:0005750 | mitochondrial respiratory chain complex III(GO:0005750) |
| 0.2 | 26.7 | GO:0000800 | lateral element(GO:0000800) |
| 0.2 | 1.4 | GO:0072687 | meiotic spindle(GO:0072687) |
| 0.2 | 1.4 | GO:0005579 | membrane attack complex(GO:0005579) |
| 0.2 | 2.2 | GO:0001520 | outer dense fiber(GO:0001520) |
| 0.1 | 2.4 | GO:0001673 | male germ cell nucleus(GO:0001673) |
| 0.1 | 1.4 | GO:0030314 | junctional membrane complex(GO:0030314) |
| 0.1 | 14.9 | GO:0042571 | immunoglobulin complex, circulating(GO:0042571) |
| 0.1 | 2.8 | GO:0033017 | sarcoplasmic reticulum membrane(GO:0033017) |
| 0.1 | 1.0 | GO:0000801 | central element(GO:0000801) |
| 0.1 | 0.9 | GO:0017071 | intracellular cyclic nucleotide activated cation channel complex(GO:0017071) |
| 0.1 | 1.7 | GO:0008290 | F-actin capping protein complex(GO:0008290) |
| 0.1 | 1.5 | GO:0042582 | primary lysosome(GO:0005766) azurophil granule(GO:0042582) |
| 0.1 | 5.1 | GO:0005865 | striated muscle thin filament(GO:0005865) |
| 0.1 | 1.6 | GO:0042589 | zymogen granule membrane(GO:0042589) |
| 0.1 | 1.1 | GO:0036156 | inner dynein arm(GO:0036156) |
| 0.1 | 1.0 | GO:0036128 | CatSper complex(GO:0036128) |
| 0.1 | 1.3 | GO:0000276 | mitochondrial proton-transporting ATP synthase complex, coupling factor F(o)(GO:0000276) |
| 0.1 | 0.7 | GO:0044299 | C-fiber(GO:0044299) |
| 0.1 | 0.3 | GO:0097169 | AIM2 inflammasome complex(GO:0097169) |
| 0.1 | 0.8 | GO:0035748 | myelin sheath abaxonal region(GO:0035748) |
| 0.1 | 2.3 | GO:0000786 | nucleosome(GO:0000786) |
| 0.1 | 3.6 | GO:0032809 | neuronal cell body membrane(GO:0032809) |
| 0.1 | 0.8 | GO:0000243 | commitment complex(GO:0000243) |
| 0.1 | 0.8 | GO:0034663 | endoplasmic reticulum chaperone complex(GO:0034663) |
| 0.0 | 0.9 | GO:0032593 | insulin-responsive compartment(GO:0032593) |
| 0.0 | 0.7 | GO:0005751 | mitochondrial respiratory chain complex IV(GO:0005751) |
| 0.0 | 0.7 | GO:0005852 | eukaryotic translation initiation factor 3 complex(GO:0005852) |
| 0.0 | 2.1 | GO:0031941 | filamentous actin(GO:0031941) |
| 0.0 | 0.4 | GO:0030015 | CCR4-NOT core complex(GO:0030015) |
| 0.0 | 0.6 | GO:0033391 | chromatoid body(GO:0033391) |
| 0.0 | 0.1 | GO:0005833 | hemoglobin complex(GO:0005833) |
| 0.0 | 3.0 | GO:0005811 | lipid particle(GO:0005811) |
| 0.0 | 0.2 | GO:0005786 | signal recognition particle, endoplasmic reticulum targeting(GO:0005786) |
| 0.0 | 0.3 | GO:0016281 | eukaryotic translation initiation factor 4F complex(GO:0016281) |
| 0.0 | 3.9 | GO:0072562 | blood microparticle(GO:0072562) |
| 0.0 | 0.2 | GO:0042567 | insulin-like growth factor ternary complex(GO:0042567) |
| 0.0 | 0.2 | GO:0030289 | protein phosphatase 4 complex(GO:0030289) |
| 0.0 | 0.2 | GO:0031931 | TORC1 complex(GO:0031931) |
| 0.0 | 0.2 | GO:0005655 | nucleolar ribonuclease P complex(GO:0005655) |
| 0.0 | 0.2 | GO:0070369 | beta-catenin-TCF7L2 complex(GO:0070369) |
| 0.0 | 0.4 | GO:0090543 | Flemming body(GO:0090543) |
| 0.0 | 1.7 | GO:0030672 | synaptic vesicle membrane(GO:0030672) exocytic vesicle membrane(GO:0099501) |
| 0.0 | 3.2 | GO:0001669 | acrosomal vesicle(GO:0001669) |
| 0.0 | 0.2 | GO:0019773 | proteasome core complex, alpha-subunit complex(GO:0019773) |
| 0.0 | 0.2 | GO:0000138 | Golgi trans cisterna(GO:0000138) |
| 0.0 | 0.9 | GO:0005844 | polysome(GO:0005844) |
| 0.0 | 1.2 | GO:0022625 | cytosolic large ribosomal subunit(GO:0022625) |
| 0.0 | 0.8 | GO:0032391 | photoreceptor connecting cilium(GO:0032391) |
| 0.0 | 1.2 | GO:0000502 | proteasome complex(GO:0000502) |
| 0.0 | 0.3 | GO:1902711 | GABA-A receptor complex(GO:1902711) |
| 0.0 | 1.1 | GO:0005776 | autophagosome(GO:0005776) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.0 | 2.9 | GO:0035375 | zymogen binding(GO:0035375) |
| 0.9 | 2.8 | GO:0004982 | N-formyl peptide receptor activity(GO:0004982) |
| 0.9 | 7.7 | GO:0005344 | oxygen transporter activity(GO:0005344) |
| 0.6 | 8.2 | GO:0005338 | nucleotide-sugar transmembrane transporter activity(GO:0005338) |
| 0.6 | 2.9 | GO:0018479 | benzaldehyde dehydrogenase (NAD+) activity(GO:0018479) |
| 0.5 | 1.4 | GO:0008532 | N-acetyllactosaminide beta-1,3-N-acetylglucosaminyltransferase activity(GO:0008532) |
| 0.5 | 4.2 | GO:0004687 | myosin light chain kinase activity(GO:0004687) |
| 0.4 | 3.5 | GO:0004865 | protein serine/threonine phosphatase inhibitor activity(GO:0004865) |
| 0.4 | 2.4 | GO:0000832 | inositol hexakisphosphate kinase activity(GO:0000828) inositol hexakisphosphate 5-kinase activity(GO:0000832) inositol hexakisphosphate 1-kinase activity(GO:0052723) inositol hexakisphosphate 3-kinase activity(GO:0052724) |
| 0.4 | 2.3 | GO:0008241 | peptidyl-dipeptidase activity(GO:0008241) |
| 0.4 | 1.9 | GO:0047635 | L-alanine:2-oxoglutarate aminotransferase activity(GO:0004021) alanine-oxo-acid transaminase activity(GO:0047635) |
| 0.4 | 3.2 | GO:0004126 | cytidine deaminase activity(GO:0004126) |
| 0.4 | 1.8 | GO:0004370 | glycerol kinase activity(GO:0004370) |
| 0.3 | 1.3 | GO:0004066 | asparagine synthase (glutamine-hydrolyzing) activity(GO:0004066) |
| 0.3 | 3.0 | GO:0005324 | long-chain fatty acid transporter activity(GO:0005324) |
| 0.3 | 0.9 | GO:0050253 | retinyl-palmitate esterase activity(GO:0050253) |
| 0.3 | 4.8 | GO:0003796 | lysozyme activity(GO:0003796) |
| 0.3 | 1.1 | GO:0004740 | pyruvate dehydrogenase (acetyl-transferring) kinase activity(GO:0004740) |
| 0.2 | 2.2 | GO:0047134 | protein-disulfide reductase activity(GO:0047134) |
| 0.2 | 1.4 | GO:0034618 | arginine binding(GO:0034618) |
| 0.2 | 2.1 | GO:0032795 | heterotrimeric G-protein binding(GO:0032795) |
| 0.2 | 3.9 | GO:0015250 | water channel activity(GO:0015250) |
| 0.2 | 1.1 | GO:0005280 | hydrogen:amino acid symporter activity(GO:0005280) |
| 0.2 | 0.9 | GO:0004574 | oligo-1,6-glucosidase activity(GO:0004574) |
| 0.2 | 0.6 | GO:0016034 | maleylacetoacetate isomerase activity(GO:0016034) |
| 0.2 | 1.5 | GO:0008121 | ubiquinol-cytochrome-c reductase activity(GO:0008121) oxidoreductase activity, acting on diphenols and related substances as donors, cytochrome as acceptor(GO:0016681) |
| 0.2 | 0.7 | GO:0047016 | cholest-5-ene-3-beta,7-alpha-diol 3-beta-dehydrogenase activity(GO:0047016) |
| 0.1 | 0.8 | GO:0032027 | myosin light chain binding(GO:0032027) |
| 0.1 | 1.7 | GO:0031433 | telethonin binding(GO:0031433) |
| 0.1 | 0.4 | GO:0004558 | alpha-1,4-glucosidase activity(GO:0004558) maltose alpha-glucosidase activity(GO:0032450) |
| 0.1 | 1.4 | GO:0070739 | protein-glutamic acid ligase activity(GO:0070739) |
| 0.1 | 1.7 | GO:0045236 | CXCR chemokine receptor binding(GO:0045236) |
| 0.1 | 0.4 | GO:0051538 | 3 iron, 4 sulfur cluster binding(GO:0051538) |
| 0.1 | 0.6 | GO:0004531 | deoxyribonuclease II activity(GO:0004531) |
| 0.1 | 1.1 | GO:0004886 | 9-cis retinoic acid receptor activity(GO:0004886) |
| 0.1 | 14.9 | GO:0034987 | immunoglobulin receptor binding(GO:0034987) |
| 0.1 | 0.8 | GO:0047288 | monosialoganglioside sialyltransferase activity(GO:0047288) |
| 0.1 | 0.9 | GO:0015016 | [heparan sulfate]-glucosamine N-sulfotransferase activity(GO:0015016) |
| 0.1 | 1.2 | GO:0016724 | ferroxidase activity(GO:0004322) oxidoreductase activity, oxidizing metal ions, oxygen as acceptor(GO:0016724) |
| 0.1 | 0.3 | GO:0031768 | ghrelin receptor binding(GO:0031768) |
| 0.1 | 0.7 | GO:0060072 | large conductance calcium-activated potassium channel activity(GO:0060072) |
| 0.1 | 0.5 | GO:0005223 | intracellular cGMP activated cation channel activity(GO:0005223) |
| 0.1 | 0.6 | GO:0016160 | alpha-amylase activity(GO:0004556) amylase activity(GO:0016160) |
| 0.1 | 0.6 | GO:0086089 | voltage-gated potassium channel activity involved in atrial cardiac muscle cell action potential repolarization(GO:0086089) |
| 0.1 | 0.6 | GO:0005166 | neurotrophin p75 receptor binding(GO:0005166) |
| 0.1 | 0.5 | GO:0004169 | dolichyl-phosphate-mannose-protein mannosyltransferase activity(GO:0004169) |
| 0.1 | 0.4 | GO:0070573 | metallodipeptidase activity(GO:0070573) |
| 0.1 | 3.2 | GO:0015347 | sodium-independent organic anion transmembrane transporter activity(GO:0015347) |
| 0.1 | 1.3 | GO:0046933 | proton-transporting ATP synthase activity, rotational mechanism(GO:0046933) |
| 0.1 | 0.4 | GO:0005052 | peroxisome matrix targeting signal-1 binding(GO:0005052) |
| 0.1 | 1.4 | GO:0010314 | phosphatidylinositol-5-phosphate binding(GO:0010314) |
| 0.1 | 0.7 | GO:0008494 | translation activator activity(GO:0008494) |
| 0.1 | 0.8 | GO:0005001 | transmembrane receptor protein tyrosine phosphatase activity(GO:0005001) transmembrane receptor protein phosphatase activity(GO:0019198) |
| 0.1 | 1.6 | GO:0003680 | AT DNA binding(GO:0003680) |
| 0.1 | 0.3 | GO:0004923 | leukemia inhibitory factor receptor activity(GO:0004923) |
| 0.0 | 0.3 | GO:0005111 | type 2 fibroblast growth factor receptor binding(GO:0005111) |
| 0.0 | 2.3 | GO:0001106 | RNA polymerase II transcription corepressor activity(GO:0001106) |
| 0.0 | 0.8 | GO:0008430 | selenium binding(GO:0008430) |
| 0.0 | 0.7 | GO:0045294 | alpha-catenin binding(GO:0045294) |
| 0.0 | 0.2 | GO:0030942 | endoplasmic reticulum signal peptide binding(GO:0030942) |
| 0.0 | 0.2 | GO:0033188 | sphingomyelin synthase activity(GO:0033188) ceramide cholinephosphotransferase activity(GO:0047493) |
| 0.0 | 2.1 | GO:0016831 | carboxy-lyase activity(GO:0016831) |
| 0.0 | 1.5 | GO:0005385 | zinc ion transmembrane transporter activity(GO:0005385) |
| 0.0 | 1.1 | GO:0008143 | poly(A) binding(GO:0008143) |
| 0.0 | 1.1 | GO:0008569 | ATP-dependent microtubule motor activity, minus-end-directed(GO:0008569) |
| 0.0 | 0.2 | GO:0004809 | tRNA (guanine-N2-)-methyltransferase activity(GO:0004809) |
| 0.0 | 0.2 | GO:0048256 | flap endonuclease activity(GO:0048256) |
| 0.0 | 0.4 | GO:0050700 | CARD domain binding(GO:0050700) |
| 0.0 | 2.1 | GO:0004004 | RNA helicase activity(GO:0003724) ATP-dependent RNA helicase activity(GO:0004004) |
| 0.0 | 2.2 | GO:0003743 | translation initiation factor activity(GO:0003743) |
| 0.0 | 0.2 | GO:0047035 | testosterone dehydrogenase (NAD+) activity(GO:0047035) |
| 0.0 | 0.8 | GO:0004993 | G-protein coupled serotonin receptor activity(GO:0004993) |
| 0.0 | 0.1 | GO:0016979 | lipoate-protein ligase activity(GO:0016979) |
| 0.0 | 3.5 | GO:0001191 | transcriptional repressor activity, RNA polymerase II transcription factor binding(GO:0001191) |
| 0.0 | 5.7 | GO:0004252 | serine-type endopeptidase activity(GO:0004252) |
| 0.0 | 0.5 | GO:0070273 | mannose binding(GO:0005537) phosphatidylinositol-4-phosphate binding(GO:0070273) |
| 0.0 | 0.7 | GO:0016676 | cytochrome-c oxidase activity(GO:0004129) heme-copper terminal oxidase activity(GO:0015002) oxidoreductase activity, acting on a heme group of donors, oxygen as acceptor(GO:0016676) |
| 0.0 | 0.2 | GO:0000298 | endopolyphosphatase activity(GO:0000298) diphosphoinositol-polyphosphate diphosphatase activity(GO:0008486) bis(5'-adenosyl)-hexaphosphatase activity(GO:0034431) bis(5'-adenosyl)-pentaphosphatase activity(GO:0034432) |
| 0.0 | 1.3 | GO:0004693 | cyclin-dependent protein serine/threonine kinase activity(GO:0004693) |
| 0.0 | 2.1 | GO:0015459 | potassium channel regulator activity(GO:0015459) |
| 0.0 | 0.4 | GO:0004707 | MAP kinase activity(GO:0004707) |
| 0.0 | 0.4 | GO:0004806 | triglyceride lipase activity(GO:0004806) |
| 0.0 | 1.4 | GO:0008395 | steroid hydroxylase activity(GO:0008395) |
| 0.0 | 0.4 | GO:0019841 | retinol binding(GO:0019841) |
| 0.0 | 0.8 | GO:0042605 | peptide antigen binding(GO:0042605) |
| 0.0 | 0.1 | GO:0042134 | rRNA primary transcript binding(GO:0042134) |
| 0.0 | 1.3 | GO:0004712 | protein serine/threonine/tyrosine kinase activity(GO:0004712) |
| 0.0 | 1.5 | GO:0004879 | RNA polymerase II transcription factor activity, ligand-activated sequence-specific DNA binding(GO:0004879) transcription factor activity, direct ligand regulated sequence-specific DNA binding(GO:0098531) |
| 0.0 | 0.6 | GO:0008236 | serine-type peptidase activity(GO:0008236) |
| 0.0 | 0.6 | GO:0032266 | phosphatidylinositol-3-phosphate binding(GO:0032266) |
| 0.0 | 0.2 | GO:0042288 | MHC class I protein binding(GO:0042288) |
| 0.0 | 0.3 | GO:0070628 | proteasome binding(GO:0070628) |
| 0.0 | 0.5 | GO:0050136 | NADH dehydrogenase (ubiquinone) activity(GO:0008137) NADH dehydrogenase (quinone) activity(GO:0050136) |
| 0.0 | 0.2 | GO:0004526 | ribonuclease P activity(GO:0004526) |
| 0.0 | 0.0 | GO:0030345 | extracellular matrix structural constituent conferring compression resistance(GO:0030021) structural constituent of tooth enamel(GO:0030345) |
| 0.0 | 0.3 | GO:0015643 | toxic substance binding(GO:0015643) |
| 0.0 | 0.1 | GO:0004566 | beta-glucuronidase activity(GO:0004566) |
| 0.0 | 0.2 | GO:0005243 | gap junction channel activity(GO:0005243) |
| 0.0 | 1.2 | GO:0020037 | heme binding(GO:0020037) |
| 0.0 | 0.1 | GO:0004128 | cytochrome-b5 reductase activity, acting on NAD(P)H(GO:0004128) |
| 0.0 | 0.1 | GO:0043024 | ribosomal small subunit binding(GO:0043024) |
| 0.0 | 0.7 | GO:0004364 | glutathione transferase activity(GO:0004364) |
| 0.0 | 0.1 | GO:0008190 | eukaryotic initiation factor 4E binding(GO:0008190) |
| 0.0 | 0.3 | GO:0042923 | neuropeptide binding(GO:0042923) |
| 0.0 | 0.4 | GO:0005184 | neuropeptide hormone activity(GO:0005184) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 2.7 | NABA PROTEOGLYCANS | Genes encoding proteoglycans |
| 0.1 | 3.6 | SIG PIP3 SIGNALING IN B LYMPHOCYTES | Genes related to PIP3 signaling in B lymphocytes |
| 0.0 | 2.8 | PID UPA UPAR PATHWAY | Urokinase-type plasminogen activator (uPA) and uPAR-mediated signaling |
| 0.0 | 0.2 | PID IL2 PI3K PATHWAY | IL2 signaling events mediated by PI3K |
| 0.0 | 0.8 | PID ECADHERIN KERATINOCYTE PATHWAY | E-cadherin signaling in keratinocytes |
| 0.0 | 3.5 | PID ILK PATHWAY | Integrin-linked kinase signaling |
| 0.0 | 1.6 | PID MAPK TRK PATHWAY | Trk receptor signaling mediated by the MAPK pathway |
| 0.0 | 1.6 | PID A6B1 A6B4 INTEGRIN PATHWAY | a6b1 and a6b4 Integrin signaling |
| 0.0 | 2.2 | PID ANGIOPOIETIN RECEPTOR PATHWAY | Angiopoietin receptor Tie2-mediated signaling |
| 0.0 | 0.6 | PID SMAD2 3PATHWAY | Regulation of cytoplasmic and nuclear SMAD2/3 signaling |
| 0.0 | 0.3 | PID ANTHRAX PATHWAY | Cellular roles of Anthrax toxin |
| 0.0 | 0.9 | PID IL23 PATHWAY | IL23-mediated signaling events |
| 0.0 | 0.1 | ST PHOSPHOINOSITIDE 3 KINASE PATHWAY | PI3K Pathway |
| 0.0 | 0.4 | PID PRL SIGNALING EVENTS PATHWAY | Signaling events mediated by PRL |
| 0.0 | 2.5 | NABA ECM GLYCOPROTEINS | Genes encoding structural ECM glycoproteins |
| 0.0 | 0.4 | SIG BCR SIGNALING PATHWAY | Members of the BCR signaling pathway |
| 0.0 | 0.2 | PID IGF1 PATHWAY | IGF1 pathway |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 8.6 | REACTOME PLATELET CALCIUM HOMEOSTASIS | Genes involved in Platelet calcium homeostasis |
| 0.3 | 3.9 | REACTOME PASSIVE TRANSPORT BY AQUAPORINS | Genes involved in Passive Transport by Aquaporins |
| 0.2 | 2.5 | REACTOME ETHANOL OXIDATION | Genes involved in Ethanol oxidation |
| 0.2 | 2.7 | REACTOME KERATAN SULFATE DEGRADATION | Genes involved in Keratan sulfate degradation |
| 0.1 | 1.1 | REACTOME RECYCLING OF BILE ACIDS AND SALTS | Genes involved in Recycling of bile acids and salts |
| 0.1 | 1.8 | REACTOME SYNTHESIS OF BILE ACIDS AND BILE SALTS VIA 24 HYDROXYCHOLESTEROL | Genes involved in Synthesis of bile acids and bile salts via 24-hydroxycholesterol |
| 0.1 | 3.8 | REACTOME REGULATION OF GENE EXPRESSION IN BETA CELLS | Genes involved in Regulation of gene expression in beta cells |
| 0.1 | 0.4 | REACTOME DIGESTION OF DIETARY CARBOHYDRATE | Genes involved in Digestion of dietary carbohydrate |
| 0.1 | 1.1 | REACTOME REGULATION OF PYRUVATE DEHYDROGENASE PDH COMPLEX | Genes involved in Regulation of pyruvate dehydrogenase (PDH) complex |
| 0.1 | 1.7 | REACTOME ADVANCED GLYCOSYLATION ENDPRODUCT RECEPTOR SIGNALING | Genes involved in Advanced glycosylation endproduct receptor signaling |
| 0.1 | 2.7 | REACTOME METAL ION SLC TRANSPORTERS | Genes involved in Metal ion SLC transporters |
| 0.1 | 1.3 | REACTOME FORMATION OF ATP BY CHEMIOSMOTIC COUPLING | Genes involved in Formation of ATP by chemiosmotic coupling |
| 0.1 | 3.8 | REACTOME STRIATED MUSCLE CONTRACTION | Genes involved in Striated Muscle Contraction |
| 0.1 | 1.4 | REACTOME KERATAN SULFATE KERATIN METABOLISM | Genes involved in Keratan sulfate/keratin metabolism |
| 0.0 | 0.6 | REACTOME SEROTONIN RECEPTORS | Genes involved in Serotonin receptors |
| 0.0 | 1.3 | REACTOME CELL EXTRACELLULAR MATRIX INTERACTIONS | Genes involved in Cell-extracellular matrix interactions |
| 0.0 | 2.3 | REACTOME IMMUNOREGULATORY INTERACTIONS BETWEEN A LYMPHOID AND A NON LYMPHOID CELL | Genes involved in Immunoregulatory interactions between a Lymphoid and a non-Lymphoid cell |
| 0.0 | 1.6 | REACTOME ERK MAPK TARGETS | Genes involved in ERK/MAPK targets |
| 0.0 | 2.6 | REACTOME RESPIRATORY ELECTRON TRANSPORT | Genes involved in Respiratory electron transport |
| 0.0 | 1.0 | REACTOME DEGRADATION OF THE EXTRACELLULAR MATRIX | Genes involved in Degradation of the extracellular matrix |
| 0.0 | 1.9 | REACTOME PLATELET AGGREGATION PLUG FORMATION | Genes involved in Platelet Aggregation (Plug Formation) |
| 0.0 | 0.7 | REACTOME ABCA TRANSPORTERS IN LIPID HOMEOSTASIS | Genes involved in ABCA transporters in lipid homeostasis |
| 0.0 | 1.5 | REACTOME CHEMOKINE RECEPTORS BIND CHEMOKINES | Genes involved in Chemokine receptors bind chemokines |
| 0.0 | 0.7 | REACTOME AMINO ACID SYNTHESIS AND INTERCONVERSION TRANSAMINATION | Genes involved in Amino acid synthesis and interconversion (transamination) |
| 0.0 | 0.9 | REACTOME HS GAG BIOSYNTHESIS | Genes involved in HS-GAG biosynthesis |
| 0.0 | 0.4 | REACTOME CITRIC ACID CYCLE TCA CYCLE | Genes involved in Citric acid cycle (TCA cycle) |
| 0.0 | 0.2 | REACTOME SRP DEPENDENT COTRANSLATIONAL PROTEIN TARGETING TO MEMBRANE | Genes involved in SRP-dependent cotranslational protein targeting to membrane |
| 0.0 | 0.4 | REACTOME HORMONE SENSITIVE LIPASE HSL MEDIATED TRIACYLGLYCEROL HYDROLYSIS | Genes involved in Hormone-sensitive lipase (HSL)-mediated triacylglycerol hydrolysis |
| 0.0 | 0.3 | REACTOME AMYLOIDS | Genes involved in Amyloids |
| 0.0 | 0.2 | REACTOME AKT PHOSPHORYLATES TARGETS IN THE CYTOSOL | Genes involved in AKT phosphorylates targets in the cytosol |
| 0.0 | 0.3 | REACTOME MTORC1 MEDIATED SIGNALLING | Genes involved in mTORC1-mediated signalling |
| 0.0 | 0.3 | REACTOME SYNTHESIS SECRETION AND DEACYLATION OF GHRELIN | Genes involved in Synthesis, Secretion, and Deacylation of Ghrelin |
| 0.0 | 0.3 | REACTOME TERMINATION OF O GLYCAN BIOSYNTHESIS | Genes involved in Termination of O-glycan biosynthesis |
| 0.0 | 0.4 | REACTOME COMPLEMENT CASCADE | Genes involved in Complement cascade |
| 0.0 | 1.1 | REACTOME NUCLEAR RECEPTOR TRANSCRIPTION PATHWAY | Genes involved in Nuclear Receptor transcription pathway |
| 0.0 | 0.2 | REACTOME GAP JUNCTION ASSEMBLY | Genes involved in Gap junction assembly |
| 0.0 | 0.6 | REACTOME CROSS PRESENTATION OF SOLUBLE EXOGENOUS ANTIGENS ENDOSOMES | Genes involved in Cross-presentation of soluble exogenous antigens (endosomes) |
| 0.0 | 0.3 | REACTOME ACTIVATED POINT MUTANTS OF FGFR2 | Genes involved in Activated point mutants of FGFR2 |