Project
PRJNA375882: Comprehensive Mouse Transcriptomic BodyMap
Navigation
Downloads
Results for Onecut2_Onecut3
Z-value: 1.22
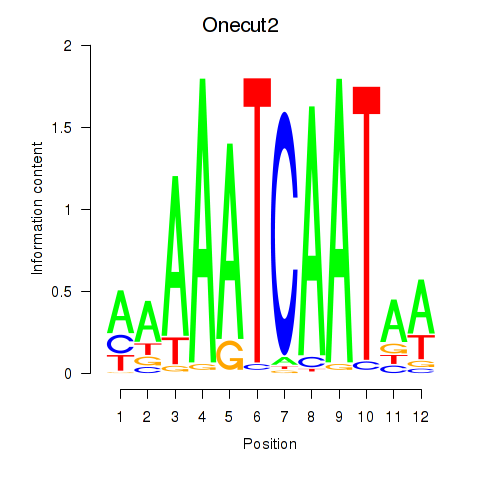
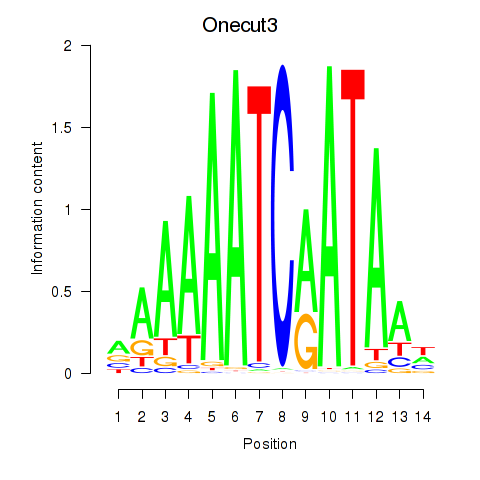
Transcription factors associated with Onecut2_Onecut3
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Onecut2
|
ENSMUSG00000045991.20 | Onecut2 |
|
Onecut3
|
ENSMUSG00000045518.9 | Onecut3 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Onecut2 | mm39_v1_chr18_+_64473091_64473114 | 0.22 | 5.8e-02 | Click! |
| Onecut3 | mm39_v1_chr10_+_80330669_80330714 | 0.10 | 3.9e-01 | Click! |
Activity profile of Onecut2_Onecut3 motif
Sorted Z-values of Onecut2_Onecut3 motif
Network of associatons between targets according to the STRING database.
First level regulatory network of Onecut2_Onecut3
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr6_-_128503666 | 12.86 |
ENSMUST00000143664.2
ENSMUST00000112132.8 |
Pzp
|
PZP, alpha-2-macroglobulin like |
| chr14_-_9015639 | 10.99 |
ENSMUST00000112656.4
|
Synpr
|
synaptoporin |
| chrX_-_149440388 | 8.99 |
ENSMUST00000151403.9
ENSMUST00000087253.11 ENSMUST00000112709.8 ENSMUST00000163969.8 ENSMUST00000087258.10 |
Tro
|
trophinin |
| chr3_+_20011201 | 8.92 |
ENSMUST00000091309.12
ENSMUST00000108329.8 ENSMUST00000003714.13 |
Cp
|
ceruloplasmin |
| chr11_+_78389913 | 8.77 |
ENSMUST00000017488.5
|
Vtn
|
vitronectin |
| chr3_+_20011405 | 8.02 |
ENSMUST00000108325.9
|
Cp
|
ceruloplasmin |
| chr19_+_38252984 | 7.16 |
ENSMUST00000198518.5
ENSMUST00000199812.5 |
Lgi1
|
leucine-rich repeat LGI family, member 1 |
| chr3_+_20011251 | 7.15 |
ENSMUST00000108328.8
|
Cp
|
ceruloplasmin |
| chr6_-_127746390 | 6.87 |
ENSMUST00000032500.9
|
Prmt8
|
protein arginine N-methyltransferase 8 |
| chr17_-_35023521 | 6.52 |
ENSMUST00000025223.9
|
Cyp21a1
|
cytochrome P450, family 21, subfamily a, polypeptide 1 |
| chr6_-_21852508 | 6.28 |
ENSMUST00000031678.10
|
Tspan12
|
tetraspanin 12 |
| chr3_+_68479578 | 6.07 |
ENSMUST00000170788.9
|
Schip1
|
schwannomin interacting protein 1 |
| chrX_-_149440362 | 5.70 |
ENSMUST00000148604.2
|
Tro
|
trophinin |
| chr6_-_92458324 | 5.70 |
ENSMUST00000113445.2
|
Prickle2
|
prickle planar cell polarity protein 2 |
| chr19_+_38253077 | 5.58 |
ENSMUST00000198045.5
|
Lgi1
|
leucine-rich repeat LGI family, member 1 |
| chr14_-_9015757 | 5.56 |
ENSMUST00000153954.8
|
Synpr
|
synaptoporin |
| chr19_-_30152814 | 5.28 |
ENSMUST00000025778.9
|
Gldc
|
glycine decarboxylase |
| chr16_+_7011580 | 5.13 |
ENSMUST00000231194.2
|
Rbfox1
|
RNA binding protein, fox-1 homolog (C. elegans) 1 |
| chr14_-_4488167 | 5.05 |
ENSMUST00000022304.12
|
Thrb
|
thyroid hormone receptor beta |
| chr1_+_11063678 | 4.77 |
ENSMUST00000027056.12
|
Prex2
|
phosphatidylinositol-3,4,5-trisphosphate-dependent Rac exchange factor 2 |
| chr1_+_172525613 | 4.76 |
ENSMUST00000038495.5
|
Crp
|
C-reactive protein, pentraxin-related |
| chr13_+_60749735 | 4.57 |
ENSMUST00000226059.2
ENSMUST00000077453.13 |
Dapk1
|
death associated protein kinase 1 |
| chr19_+_38253105 | 4.53 |
ENSMUST00000196090.2
|
Lgi1
|
leucine-rich repeat LGI family, member 1 |
| chr4_-_61972348 | 4.36 |
ENSMUST00000074018.4
|
Mup20
|
major urinary protein 20 |
| chr3_-_81883509 | 4.27 |
ENSMUST00000029645.14
ENSMUST00000193879.2 |
Tdo2
|
tryptophan 2,3-dioxygenase |
| chr3_+_64884839 | 4.27 |
ENSMUST00000239069.2
|
Kcnab1
|
potassium voltage-gated channel, shaker-related subfamily, beta member 1 |
| chr9_-_86577940 | 4.04 |
ENSMUST00000034989.15
|
Me1
|
malic enzyme 1, NADP(+)-dependent, cytosolic |
| chr4_+_43384320 | 4.00 |
ENSMUST00000136360.2
|
Rusc2
|
RUN and SH3 domain containing 2 |
| chr10_+_69369632 | 3.78 |
ENSMUST00000182155.8
ENSMUST00000183169.8 ENSMUST00000183148.8 |
Ank3
|
ankyrin 3, epithelial |
| chr15_-_96929086 | 3.75 |
ENSMUST00000230086.2
|
Slc38a4
|
solute carrier family 38, member 4 |
| chr8_+_94537910 | 3.59 |
ENSMUST00000138659.9
|
Gnao1
|
guanine nucleotide binding protein, alpha O |
| chr7_-_27010068 | 3.57 |
ENSMUST00000125455.2
|
Ltbp4
|
latent transforming growth factor beta binding protein 4 |
| chr2_-_10135449 | 3.50 |
ENSMUST00000042290.14
|
Itih2
|
inter-alpha trypsin inhibitor, heavy chain 2 |
| chr1_+_34199333 | 3.41 |
ENSMUST00000183302.6
ENSMUST00000185897.7 ENSMUST00000185269.7 |
Dst
|
dystonin |
| chr6_+_141470105 | 3.41 |
ENSMUST00000032362.12
ENSMUST00000205214.3 |
Slco1c1
|
solute carrier organic anion transporter family, member 1c1 |
| chr1_+_171041539 | 3.30 |
ENSMUST00000005820.11
ENSMUST00000075469.12 ENSMUST00000155126.8 |
Nr1i3
|
nuclear receptor subfamily 1, group I, member 3 |
| chr10_+_128626772 | 3.23 |
ENSMUST00000219404.2
ENSMUST00000026411.8 |
Mmp19
|
matrix metallopeptidase 19 |
| chr12_-_46767619 | 3.22 |
ENSMUST00000219886.2
|
Nova1
|
NOVA alternative splicing regulator 1 |
| chr10_+_69369590 | 3.19 |
ENSMUST00000182884.8
|
Ank3
|
ankyrin 3, epithelial |
| chr13_+_60749995 | 3.08 |
ENSMUST00000044083.9
|
Dapk1
|
death associated protein kinase 1 |
| chr4_-_147726953 | 3.03 |
ENSMUST00000133006.2
ENSMUST00000037565.14 ENSMUST00000105720.8 |
Zfp979
|
zinc finger protein 979 |
| chrX_+_118836893 | 3.02 |
ENSMUST00000040961.3
ENSMUST00000113366.2 |
Pabpc5
|
poly(A) binding protein, cytoplasmic 5 |
| chr17_-_90763300 | 2.94 |
ENSMUST00000159778.8
ENSMUST00000174337.8 ENSMUST00000172466.8 |
Nrxn1
|
neurexin I |
| chr1_-_5140504 | 2.92 |
ENSMUST00000147158.2
ENSMUST00000118000.8 |
Rgs20
|
regulator of G-protein signaling 20 |
| chr6_-_42301574 | 2.91 |
ENSMUST00000031891.15
ENSMUST00000143278.8 |
Fam131b
|
family with sequence similarity 131, member B |
| chr11_-_42070517 | 2.88 |
ENSMUST00000206105.2
ENSMUST00000153147.3 |
Gabra1
|
gamma-aminobutyric acid (GABA) A receptor, subunit alpha 1 |
| chr7_-_142215595 | 2.85 |
ENSMUST00000145896.3
|
Igf2
|
insulin-like growth factor 2 |
| chr8_-_72124359 | 2.85 |
ENSMUST00000177517.8
ENSMUST00000030170.15 |
Unc13a
|
unc-13 homolog A |
| chr10_+_76089674 | 2.78 |
ENSMUST00000036387.8
|
S100b
|
S100 protein, beta polypeptide, neural |
| chr7_+_87233554 | 2.78 |
ENSMUST00000125009.9
|
Grm5
|
glutamate receptor, metabotropic 5 |
| chr7_+_19024994 | 2.77 |
ENSMUST00000108468.5
|
Rtn2
|
reticulon 2 (Z-band associated protein) |
| chr8_+_67150055 | 2.72 |
ENSMUST00000039303.7
|
Npy1r
|
neuropeptide Y receptor Y1 |
| chr12_+_103498542 | 2.66 |
ENSMUST00000021631.12
|
Ppp4r4
|
protein phosphatase 4, regulatory subunit 4 |
| chr17_+_43671314 | 2.61 |
ENSMUST00000226087.2
|
Adgrf5
|
adhesion G protein-coupled receptor F5 |
| chr10_-_18619658 | 2.61 |
ENSMUST00000215836.2
|
Arfgef3
|
ARFGEF family member 3 |
| chr4_+_43383449 | 2.53 |
ENSMUST00000135216.2
ENSMUST00000152322.8 |
Rusc2
|
RUN and SH3 domain containing 2 |
| chr8_+_67149815 | 2.52 |
ENSMUST00000212588.2
|
Npy1r
|
neuropeptide Y receptor Y1 |
| chr17_-_90395771 | 2.43 |
ENSMUST00000197268.5
ENSMUST00000173917.8 |
Nrxn1
|
neurexin I |
| chr5_+_110751362 | 2.41 |
ENSMUST00000165856.3
|
Galnt9
|
polypeptide N-acetylgalactosaminyltransferase 9 |
| chr7_+_51537645 | 2.40 |
ENSMUST00000208711.2
|
Gas2
|
growth arrest specific 2 |
| chr6_+_134617903 | 2.40 |
ENSMUST00000062755.10
|
Borcs5
|
BLOC-1 related complex subunit 5 |
| chr10_+_69369854 | 2.34 |
ENSMUST00000182557.8
|
Ank3
|
ankyrin 3, epithelial |
| chr6_-_42301488 | 2.33 |
ENSMUST00000095974.4
|
Fam131b
|
family with sequence similarity 131, member B |
| chr2_-_174188505 | 2.30 |
ENSMUST00000168292.2
|
Gm20721
|
predicted gene, 20721 |
| chr18_+_62790356 | 2.28 |
ENSMUST00000162511.3
|
Spink10
|
serine peptidase inhibitor, Kazal type 10 |
| chr8_+_94537460 | 2.24 |
ENSMUST00000034198.15
ENSMUST00000125716.8 |
Gnao1
|
guanine nucleotide binding protein, alpha O |
| chr2_+_14828903 | 2.23 |
ENSMUST00000193800.6
|
Cacnb2
|
calcium channel, voltage-dependent, beta 2 subunit |
| chr10_-_89568106 | 2.21 |
ENSMUST00000020109.5
|
Actr6
|
ARP6 actin-related protein 6 |
| chr18_-_24663260 | 2.18 |
ENSMUST00000046206.5
|
Rprd1a
|
regulation of nuclear pre-mRNA domain containing 1A |
| chr1_+_167426019 | 2.17 |
ENSMUST00000111386.8
ENSMUST00000111384.8 |
Rxrg
|
retinoid X receptor gamma |
| chr1_-_180083859 | 2.08 |
ENSMUST00000111108.10
|
Psen2
|
presenilin 2 |
| chr7_+_108549545 | 2.08 |
ENSMUST00000207583.2
|
Tub
|
tubby bipartite transcription factor |
| chr10_+_128089965 | 2.07 |
ENSMUST00000060782.5
ENSMUST00000218722.2 |
Apon
|
apolipoprotein N |
| chr8_+_48277493 | 2.06 |
ENSMUST00000038693.8
|
Cldn22
|
claudin 22 |
| chr10_+_33071964 | 2.06 |
ENSMUST00000219211.2
|
Trdn
|
triadin |
| chr6_-_67743756 | 2.06 |
ENSMUST00000103306.2
|
Igkv1-131
|
immunoglobulin kappa variable 1-131 |
| chr15_+_38940307 | 2.04 |
ENSMUST00000067072.5
|
Cthrc1
|
collagen triple helix repeat containing 1 |
| chr17_-_33252341 | 2.02 |
ENSMUST00000087654.5
|
Zfp763
|
zinc finger protein 763 |
| chr9_+_45749869 | 2.01 |
ENSMUST00000078111.11
ENSMUST00000034591.11 |
Bace1
|
beta-site APP cleaving enzyme 1 |
| chr2_-_30249202 | 2.01 |
ENSMUST00000100215.11
ENSMUST00000113620.10 ENSMUST00000163668.3 ENSMUST00000028214.15 ENSMUST00000113621.10 |
Sh3glb2
|
SH3-domain GRB2-like endophilin B2 |
| chr10_-_63926044 | 2.00 |
ENSMUST00000105439.2
|
Lrrtm3
|
leucine rich repeat transmembrane neuronal 3 |
| chr6_-_82916655 | 1.99 |
ENSMUST00000000641.15
|
Sema4f
|
sema domain, immunoglobulin domain (Ig), TM domain, and short cytoplasmic domain |
| chr3_+_75982890 | 1.97 |
ENSMUST00000160261.8
|
Fstl5
|
follistatin-like 5 |
| chr2_-_17465410 | 1.94 |
ENSMUST00000145492.2
|
Nebl
|
nebulette |
| chr7_-_141009264 | 1.93 |
ENSMUST00000164387.2
ENSMUST00000137488.2 ENSMUST00000084436.10 |
Cend1
|
cell cycle exit and neuronal differentiation 1 |
| chr8_-_68270936 | 1.89 |
ENSMUST00000120071.8
|
Psd3
|
pleckstrin and Sec7 domain containing 3 |
| chr2_-_174973657 | 1.89 |
ENSMUST00000108929.3
|
Gm14399
|
predicted gene 14399 |
| chr3_+_137046828 | 1.88 |
ENSMUST00000122064.8
ENSMUST00000119475.6 |
Emcn
|
endomucin |
| chr2_+_144435974 | 1.88 |
ENSMUST00000136628.2
|
Smim26
|
small integral membrane protein 26 |
| chr1_-_170042947 | 1.87 |
ENSMUST00000027979.14
ENSMUST00000123399.2 |
Uhmk1
|
U2AF homology motif (UHM) kinase 1 |
| chr8_+_22966736 | 1.86 |
ENSMUST00000067786.9
|
Slc20a2
|
solute carrier family 20, member 2 |
| chr3_+_115801869 | 1.81 |
ENSMUST00000106502.2
|
Extl2
|
exostosin-like glycosyltransferase 2 |
| chr14_-_56448874 | 1.77 |
ENSMUST00000022757.5
|
Gzmf
|
granzyme F |
| chr9_+_74959259 | 1.77 |
ENSMUST00000170310.2
ENSMUST00000166549.2 |
Arpp19
|
cAMP-regulated phosphoprotein 19 |
| chr11_-_30148230 | 1.76 |
ENSMUST00000102838.10
|
Sptbn1
|
spectrin beta, non-erythrocytic 1 |
| chr7_-_89590404 | 1.74 |
ENSMUST00000153470.9
|
Hikeshi
|
heat shock protein nuclear import factor |
| chr8_-_85500998 | 1.70 |
ENSMUST00000109762.8
|
Nfix
|
nuclear factor I/X |
| chr7_-_141009346 | 1.70 |
ENSMUST00000124444.2
|
Cend1
|
cell cycle exit and neuronal differentiation 1 |
| chr3_+_137046935 | 1.69 |
ENSMUST00000197511.2
|
Emcn
|
endomucin |
| chr5_-_87288177 | 1.68 |
ENSMUST00000067790.7
|
Ugt2b5
|
UDP glucuronosyltransferase 2 family, polypeptide B5 |
| chr17_-_90395568 | 1.67 |
ENSMUST00000173222.2
|
Nrxn1
|
neurexin I |
| chr8_-_85500010 | 1.66 |
ENSMUST00000109764.8
|
Nfix
|
nuclear factor I/X |
| chr15_+_38940351 | 1.66 |
ENSMUST00000226433.2
ENSMUST00000132192.3 |
Cthrc1
|
collagen triple helix repeat containing 1 |
| chr10_-_63039709 | 1.64 |
ENSMUST00000095580.3
|
Mypn
|
myopalladin |
| chr13_+_49658249 | 1.62 |
ENSMUST00000051504.8
|
Ecm2
|
extracellular matrix protein 2, female organ and adipocyte specific |
| chr9_+_55234197 | 1.61 |
ENSMUST00000085754.10
ENSMUST00000034862.5 |
Tmem266
|
transmembrane protein 266 |
| chr8_-_45715049 | 1.61 |
ENSMUST00000034064.5
|
F11
|
coagulation factor XI |
| chr7_+_21006104 | 1.60 |
ENSMUST00000098739.3
|
Vmn1r125
|
vomeronasal 1 receptor 125 |
| chr7_-_78798790 | 1.58 |
ENSMUST00000107409.5
ENSMUST00000032825.14 |
Mfge8
|
milk fat globule EGF and factor V/VIII domain containing |
| chr7_+_19927635 | 1.57 |
ENSMUST00000168984.2
|
Vmn1r95
|
vomeronasal 1 receptor, 95 |
| chr8_+_22966889 | 1.53 |
ENSMUST00000209305.2
|
Slc20a2
|
solute carrier family 20, member 2 |
| chr9_-_51874846 | 1.51 |
ENSMUST00000034552.8
ENSMUST00000214013.2 |
Fdx1
|
ferredoxin 1 |
| chr3_+_92259448 | 1.49 |
ENSMUST00000068399.2
|
Sprr2e
|
small proline-rich protein 2E |
| chr3_-_126918491 | 1.49 |
ENSMUST00000238781.2
|
Ank2
|
ankyrin 2, brain |
| chr3_+_136376440 | 1.48 |
ENSMUST00000056758.9
|
Ppp3ca
|
protein phosphatase 3, catalytic subunit, alpha isoform |
| chr6_+_89691185 | 1.45 |
ENSMUST00000075158.2
|
Vmn1r40
|
vomeronasal 1 receptor 40 |
| chr11_-_98220466 | 1.44 |
ENSMUST00000041685.7
|
Neurod2
|
neurogenic differentiation 2 |
| chr11_-_41891111 | 1.43 |
ENSMUST00000109290.2
|
Gabrg2
|
gamma-aminobutyric acid (GABA) A receptor, subunit gamma 2 |
| chr17_+_21787250 | 1.41 |
ENSMUST00000088787.7
|
Zfp948
|
zinc finger protein 948 |
| chr1_+_167425953 | 1.40 |
ENSMUST00000015987.10
|
Rxrg
|
retinoid X receptor gamma |
| chr5_+_110088292 | 1.39 |
ENSMUST00000170826.2
|
Gm15446
|
predicted gene 15446 |
| chr5_-_69699965 | 1.33 |
ENSMUST00000031045.10
|
Yipf7
|
Yip1 domain family, member 7 |
| chr4_-_97472844 | 1.31 |
ENSMUST00000107067.8
ENSMUST00000107068.9 |
E130114P18Rik
|
RIKEN cDNA E130114P18 gene |
| chrX_-_110446022 | 1.31 |
ENSMUST00000156639.2
|
Rps6ka6
|
ribosomal protein S6 kinase polypeptide 6 |
| chr7_-_89590334 | 1.31 |
ENSMUST00000207309.2
ENSMUST00000130609.3 |
Hikeshi
|
heat shock protein nuclear import factor |
| chr2_-_86722536 | 1.31 |
ENSMUST00000111576.3
ENSMUST00000217403.2 |
Olfr1097
|
olfactory receptor 1097 |
| chr13_+_112604037 | 1.28 |
ENSMUST00000183868.8
|
Il6st
|
interleukin 6 signal transducer |
| chr7_+_20505110 | 1.28 |
ENSMUST00000174538.2
|
Vmn1r112
|
vomeronasal 1 receptor 112 |
| chr5_-_52723700 | 1.27 |
ENSMUST00000039750.7
|
Lgi2
|
leucine-rich repeat LGI family, member 2 |
| chr10_-_128505096 | 1.25 |
ENSMUST00000238610.2
ENSMUST00000238712.2 |
Ikzf4
|
IKAROS family zinc finger 4 |
| chr9_-_19799300 | 1.24 |
ENSMUST00000079660.5
|
Olfr862
|
olfactory receptor 862 |
| chrX_+_105059305 | 1.24 |
ENSMUST00000033582.5
|
Cox7b
|
cytochrome c oxidase subunit 7B |
| chr17_+_35327255 | 1.23 |
ENSMUST00000037849.3
|
Ly6g5c
|
lymphocyte antigen 6 complex, locus G5C |
| chr4_+_147637714 | 1.22 |
ENSMUST00000139784.8
ENSMUST00000143885.8 ENSMUST00000081742.7 |
Zfp985
|
zinc finger protein 985 |
| chr4_-_134279433 | 1.22 |
ENSMUST00000060435.8
|
Selenon
|
selenoprotein N |
| chr6_+_97968737 | 1.20 |
ENSMUST00000043628.13
ENSMUST00000203938.2 |
Mitf
|
melanogenesis associated transcription factor |
| chr11_-_74238498 | 1.20 |
ENSMUST00000080365.6
|
Olfr411
|
olfactory receptor 411 |
| chr5_-_52723607 | 1.19 |
ENSMUST00000199942.5
|
Lgi2
|
leucine-rich repeat LGI family, member 2 |
| chr18_+_37646674 | 1.18 |
ENSMUST00000061405.6
|
Pcdhb21
|
protocadherin beta 21 |
| chr4_-_4138817 | 1.17 |
ENSMUST00000133567.2
|
Penk
|
preproenkephalin |
| chr5_-_69699932 | 1.16 |
ENSMUST00000202423.2
|
Yipf7
|
Yip1 domain family, member 7 |
| chr4_+_147576874 | 1.15 |
ENSMUST00000105721.9
|
Zfp982
|
zinc finger protein 982 |
| chr7_+_112118817 | 1.15 |
ENSMUST00000106640.2
|
Parva
|
parvin, alpha |
| chr17_+_34783242 | 1.15 |
ENSMUST00000015612.14
|
Notch4
|
notch 4 |
| chr7_-_30234481 | 1.15 |
ENSMUST00000207858.2
|
Arhgap33
|
Rho GTPase activating protein 33 |
| chr14_+_26616514 | 1.14 |
ENSMUST00000238987.2
ENSMUST00000239004.2 ENSMUST00000165929.4 ENSMUST00000090337.12 |
Asb14
|
ankyrin repeat and SOCS box-containing 14 |
| chr18_+_36431732 | 1.13 |
ENSMUST00000210490.3
|
Igip
|
IgA inducing protein |
| chrX_-_112095181 | 1.11 |
ENSMUST00000026607.15
ENSMUST00000113388.3 |
Chm
|
choroidermia (RAB escort protein 1) |
| chr9_-_21710029 | 1.11 |
ENSMUST00000034717.7
|
Kank2
|
KN motif and ankyrin repeat domains 2 |
| chr7_-_90125178 | 1.11 |
ENSMUST00000032843.9
|
Tmem126b
|
transmembrane protein 126B |
| chr16_-_10360893 | 1.10 |
ENSMUST00000184863.8
ENSMUST00000038281.6 |
Dexi
|
dexamethasone-induced transcript |
| chr8_-_22966831 | 1.10 |
ENSMUST00000163774.3
ENSMUST00000033935.16 |
Smim19
|
small integral membrane protein 19 |
| chr3_-_93600377 | 1.10 |
ENSMUST00000193565.2
|
Tdpoz6
|
TD and POZ domain containing 6 |
| chr2_-_85592444 | 1.09 |
ENSMUST00000214255.2
|
Olfr1012
|
olfactory receptor 1012 |
| chr3_-_98496123 | 1.09 |
ENSMUST00000178221.4
|
Gm10681
|
predicted gene 10681 |
| chr2_-_94236991 | 1.08 |
ENSMUST00000111237.9
ENSMUST00000094801.5 ENSMUST00000111238.8 |
Ttc17
|
tetratricopeptide repeat domain 17 |
| chrX_+_9751861 | 1.07 |
ENSMUST00000067529.9
ENSMUST00000086165.4 |
Sytl5
|
synaptotagmin-like 5 |
| chr10_+_69370038 | 1.05 |
ENSMUST00000182439.8
ENSMUST00000092434.12 ENSMUST00000047061.13 ENSMUST00000092432.12 ENSMUST00000092431.12 ENSMUST00000054167.15 |
Ank3
|
ankyrin 3, epithelial |
| chr2_+_81883566 | 1.03 |
ENSMUST00000047527.8
|
Zfp804a
|
zinc finger protein 804A |
| chr7_-_10009278 | 1.03 |
ENSMUST00000060374.5
|
Vmn1r66
|
vomeronasal 1 receptor 66 |
| chr8_-_105350898 | 1.02 |
ENSMUST00000212882.2
ENSMUST00000163783.4 |
Cdh16
|
cadherin 16 |
| chr3_+_136375839 | 1.02 |
ENSMUST00000070198.14
|
Ppp3ca
|
protein phosphatase 3, catalytic subunit, alpha isoform |
| chr9_+_109865810 | 1.01 |
ENSMUST00000163190.8
|
Map4
|
microtubule-associated protein 4 |
| chr12_+_29578354 | 1.01 |
ENSMUST00000218583.2
ENSMUST00000049784.17 |
Myt1l
|
myelin transcription factor 1-like |
| chr7_-_30234422 | 1.01 |
ENSMUST00000208522.2
ENSMUST00000207860.2 ENSMUST00000208538.2 |
Arhgap33
|
Rho GTPase activating protein 33 |
| chr5_+_29940686 | 1.00 |
ENSMUST00000008733.15
|
Dnajb6
|
DnaJ heat shock protein family (Hsp40) member B6 |
| chr16_-_28748410 | 1.00 |
ENSMUST00000100023.3
|
Mb21d2
|
Mab-21 domain containing 2 |
| chr2_-_23986544 | 0.97 |
ENSMUST00000153338.2
ENSMUST00000080453.8 |
Tbpl2
|
TATA box binding protein like 2 |
| chrX_-_76968668 | 0.97 |
ENSMUST00000063127.4
|
5430402E10Rik
|
RIKEN cDNA 5430402E10 gene |
| chr6_-_122833109 | 0.96 |
ENSMUST00000042081.9
|
C3ar1
|
complement component 3a receptor 1 |
| chr6_+_116185077 | 0.96 |
ENSMUST00000204051.2
|
Washc2
|
WASH complex subunit 2` |
| chr15_+_54975814 | 0.96 |
ENSMUST00000100660.11
|
Deptor
|
DEP domain containing MTOR-interacting protein |
| chr2_+_38229270 | 0.95 |
ENSMUST00000143783.9
|
Lhx2
|
LIM homeobox protein 2 |
| chr2_+_111329683 | 0.95 |
ENSMUST00000219064.3
|
Olfr1291-ps1
|
olfactory receptor 1291, pseudogene 1 |
| chr11_-_58346806 | 0.92 |
ENSMUST00000055204.6
|
Olfr30
|
olfactory receptor 30 |
| chr2_-_160169414 | 0.91 |
ENSMUST00000099127.3
|
Gm826
|
predicted gene 826 |
| chr15_+_92495007 | 0.91 |
ENSMUST00000035399.10
|
Pdzrn4
|
PDZ domain containing RING finger 4 |
| chr15_+_54975713 | 0.90 |
ENSMUST00000096433.10
|
Deptor
|
DEP domain containing MTOR-interacting protein |
| chr5_-_31684036 | 0.90 |
ENSMUST00000202421.2
ENSMUST00000201769.4 ENSMUST00000065388.11 |
Supt7l
|
SPT7-like, STAGA complex gamma subunit |
| chr2_-_93988229 | 0.90 |
ENSMUST00000028619.5
|
Hsd17b12
|
hydroxysteroid (17-beta) dehydrogenase 12 |
| chrX_+_162694397 | 0.89 |
ENSMUST00000140845.2
|
Ap1s2
|
adaptor-related protein complex 1, sigma 2 subunit |
| chr7_+_101512922 | 0.88 |
ENSMUST00000209334.2
|
Anapc15
|
anaphase promoting complex C subunit 15 |
| chr19_+_9957940 | 0.88 |
ENSMUST00000235196.2
|
Fth1
|
ferritin heavy polypeptide 1 |
| chr6_+_106095726 | 0.87 |
ENSMUST00000113258.8
ENSMUST00000079416.6 |
Cntn4
|
contactin 4 |
| chr4_+_145237329 | 0.87 |
ENSMUST00000105742.8
ENSMUST00000136309.8 |
Zfp990
|
zinc finger protein 990 |
| chr13_+_14787819 | 0.86 |
ENSMUST00000170836.4
|
Psma2
|
proteasome subunit alpha 2 |
| chr2_+_175978653 | 0.85 |
ENSMUST00000166464.2
|
2210418O10Rik
|
RIKEN cDNA 2210418O10 gene |
| chr2_-_176609311 | 0.85 |
ENSMUST00000108931.3
|
Gm14296
|
predicted gene 14296 |
| chr16_-_10131804 | 0.85 |
ENSMUST00000078357.5
|
Emp2
|
epithelial membrane protein 2 |
| chr1_+_70764874 | 0.84 |
ENSMUST00000053922.12
ENSMUST00000161937.2 ENSMUST00000162182.2 |
Vwc2l
|
von Willebrand factor C domain-containing protein 2-like |
| chr6_+_116184991 | 0.84 |
ENSMUST00000036759.11
ENSMUST00000204476.3 |
Washc2
|
WASH complex subunit 2` |
| chr9_-_39914761 | 0.83 |
ENSMUST00000215523.2
|
Olfr979
|
olfactory receptor 979 |
| chr9_+_38883609 | 0.83 |
ENSMUST00000216238.3
|
Olfr933
|
olfactory receptor 933 |
| chr13_+_35059285 | 0.83 |
ENSMUST00000077853.5
|
Prpf4b
|
pre-mRNA processing factor 4B |
| chr7_-_23508625 | 0.82 |
ENSMUST00000166141.2
|
Vmn1r175
|
vomeronasal 1 receptor 175 |
| chr7_+_23330147 | 0.82 |
ENSMUST00000227774.2
ENSMUST00000226771.2 ENSMUST00000228681.2 ENSMUST00000228559.2 ENSMUST00000228674.2 ENSMUST00000227866.2 ENSMUST00000227386.2 ENSMUST00000228484.2 ENSMUST00000226321.2 ENSMUST00000226128.2 ENSMUST00000226733.2 ENSMUST00000228228.2 |
Vmn1r171
|
vomeronasal 1 receptor 171 |
| chr8_-_68270870 | 0.81 |
ENSMUST00000059374.5
|
Psd3
|
pleckstrin and Sec7 domain containing 3 |
| chr7_-_110443557 | 0.80 |
ENSMUST00000177462.8
ENSMUST00000176716.3 ENSMUST00000176746.8 ENSMUST00000177236.8 |
Rnf141
|
ring finger protein 141 |
| chr16_-_30406355 | 0.79 |
ENSMUST00000117363.9
|
Lsg1
|
large 60S subunit nuclear export GTPase 1 |
| chr7_+_21348017 | 0.78 |
ENSMUST00000098736.5
|
Vmn1r132
|
vomeronasal 1 receptor 132 |
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.8 | 5.3 | GO:0019464 | glycine catabolic process(GO:0006546) glycine decarboxylation via glycine cleavage system(GO:0019464) |
| 1.4 | 4.3 | GO:0019442 | tryptophan catabolic process to acetyl-CoA(GO:0019442) |
| 1.4 | 6.9 | GO:0019919 | peptidyl-arginine methylation, to asymmetrical-dimethyl arginine(GO:0019919) |
| 1.2 | 7.0 | GO:0090126 | protein complex assembly involved in synapse maturation(GO:0090126) |
| 1.1 | 5.7 | GO:0001830 | trophectodermal cell fate commitment(GO:0001830) |
| 0.9 | 2.8 | GO:0099553 | trans-synaptic signaling by lipid, modulating synaptic transmission(GO:0099552) trans-synaptic signaling by endocannabinoid, modulating synaptic transmission(GO:0099553) regulation of intracellular calcium activated chloride channel activity(GO:1902938) |
| 0.9 | 3.6 | GO:0021933 | radial glia guided migration of cerebellar granule cell(GO:0021933) negative regulation of cerebellar granule cell precursor proliferation(GO:0021941) |
| 0.8 | 5.1 | GO:0007621 | negative regulation of female receptivity(GO:0007621) |
| 0.8 | 2.5 | GO:1903244 | positive regulation of cardiac muscle adaptation(GO:0010615) positive regulation of cardiac muscle hypertrophy in response to stress(GO:1903244) |
| 0.8 | 10.4 | GO:1900827 | positive regulation of cell communication by electrical coupling(GO:0010650) positive regulation of membrane depolarization during cardiac muscle cell action potential(GO:1900827) |
| 0.7 | 4.4 | GO:0008355 | olfactory learning(GO:0008355) |
| 0.7 | 8.8 | GO:0097421 | smooth muscle cell-matrix adhesion(GO:0061302) liver regeneration(GO:0097421) |
| 0.7 | 2.7 | GO:0080163 | regulation of protein serine/threonine phosphatase activity(GO:0080163) |
| 0.7 | 24.1 | GO:0006825 | copper ion transport(GO:0006825) |
| 0.6 | 4.8 | GO:0032929 | negative regulation of superoxide anion generation(GO:0032929) |
| 0.6 | 2.2 | GO:1904879 | positive regulation of calcium ion transmembrane transport via high voltage-gated calcium channel(GO:1904879) |
| 0.5 | 4.0 | GO:1902031 | regulation of NADP metabolic process(GO:1902031) |
| 0.5 | 2.8 | GO:0099525 | presynaptic dense core granule exocytosis(GO:0099525) |
| 0.4 | 1.3 | GO:0070104 | negative regulation of interleukin-6-mediated signaling pathway(GO:0070104) |
| 0.4 | 1.2 | GO:0014873 | response to muscle activity involved in regulation of muscle adaptation(GO:0014873) |
| 0.4 | 6.5 | GO:0006705 | mineralocorticoid biosynthetic process(GO:0006705) |
| 0.4 | 1.1 | GO:0070563 | negative regulation of vitamin D receptor signaling pathway(GO:0070563) |
| 0.4 | 1.4 | GO:2000297 | negative regulation of synapse maturation(GO:2000297) |
| 0.4 | 7.8 | GO:0071447 | cellular response to hydroperoxide(GO:0071447) |
| 0.3 | 2.1 | GO:0090158 | endoplasmic reticulum membrane organization(GO:0090158) |
| 0.3 | 1.0 | GO:0060715 | syncytiotrophoblast cell differentiation involved in labyrinthine layer development(GO:0060715) |
| 0.3 | 1.6 | GO:0051919 | positive regulation of fibrinolysis(GO:0051919) |
| 0.3 | 2.8 | GO:2001015 | negative regulation of skeletal muscle cell differentiation(GO:2001015) |
| 0.3 | 5.2 | GO:0030432 | peristalsis(GO:0030432) |
| 0.3 | 4.3 | GO:1902260 | negative regulation of delayed rectifier potassium channel activity(GO:1902260) |
| 0.3 | 1.8 | GO:0019276 | UDP-N-acetylgalactosamine metabolic process(GO:0019276) |
| 0.3 | 3.4 | GO:0021684 | cerebellar granular layer formation(GO:0021684) cerebellar granule cell differentiation(GO:0021707) |
| 0.2 | 2.9 | GO:2000467 | positive regulation of glycogen (starch) synthase activity(GO:2000467) |
| 0.2 | 6.1 | GO:0001553 | luteinization(GO:0001553) |
| 0.2 | 2.2 | GO:0070940 | dephosphorylation of RNA polymerase II C-terminal domain(GO:0070940) |
| 0.2 | 3.7 | GO:0033690 | positive regulation of osteoblast proliferation(GO:0033690) |
| 0.2 | 4.3 | GO:0071420 | cellular response to histamine(GO:0071420) |
| 0.2 | 2.6 | GO:0071073 | positive regulation of phospholipid biosynthetic process(GO:0071073) |
| 0.2 | 0.9 | GO:0097309 | cap1 mRNA methylation(GO:0097309) |
| 0.2 | 13.7 | GO:0007566 | embryo implantation(GO:0007566) |
| 0.2 | 0.6 | GO:0018199 | peptidyl-glutamine modification(GO:0018199) |
| 0.2 | 1.6 | GO:2000427 | positive regulation of apoptotic cell clearance(GO:2000427) |
| 0.2 | 1.2 | GO:1903367 | positive regulation of fear response(GO:1903367) positive regulation of behavioral fear response(GO:2000987) |
| 0.2 | 1.5 | GO:0036309 | protein localization to M-band(GO:0036309) protein localization to T-tubule(GO:0036371) |
| 0.2 | 1.2 | GO:1903847 | regulation of aorta morphogenesis(GO:1903847) positive regulation of aorta morphogenesis(GO:1903849) |
| 0.2 | 1.8 | GO:1901727 | positive regulation of histone deacetylase activity(GO:1901727) |
| 0.2 | 6.3 | GO:0010842 | retina layer formation(GO:0010842) |
| 0.2 | 0.6 | GO:0030210 | heparin biosynthetic process(GO:0030210) |
| 0.1 | 0.6 | GO:0010979 | regulation of vitamin D 24-hydroxylase activity(GO:0010979) positive regulation of vitamin D 24-hydroxylase activity(GO:0010980) |
| 0.1 | 1.8 | GO:1900042 | common-partner SMAD protein phosphorylation(GO:0007182) positive regulation of interleukin-2 secretion(GO:1900042) |
| 0.1 | 1.0 | GO:0051012 | microtubule sliding(GO:0051012) |
| 0.1 | 5.3 | GO:0032011 | ARF protein signal transduction(GO:0032011) regulation of ARF protein signal transduction(GO:0032012) |
| 0.1 | 3.4 | GO:0008090 | retrograde axonal transport(GO:0008090) |
| 0.1 | 5.1 | GO:2001014 | regulation of skeletal muscle cell differentiation(GO:2001014) |
| 0.1 | 1.0 | GO:0002430 | complement receptor mediated signaling pathway(GO:0002430) tolerance induction dependent upon immune response(GO:0002461) |
| 0.1 | 2.0 | GO:0043589 | skin morphogenesis(GO:0043589) |
| 0.1 | 3.2 | GO:0030574 | collagen catabolic process(GO:0030574) |
| 0.1 | 0.7 | GO:0018343 | protein farnesylation(GO:0018343) |
| 0.1 | 2.1 | GO:1903546 | protein localization to photoreceptor outer segment(GO:1903546) |
| 0.1 | 1.1 | GO:0018344 | protein geranylgeranylation(GO:0018344) |
| 0.1 | 0.7 | GO:0071484 | response to high light intensity(GO:0009644) cellular response to light intensity(GO:0071484) cellular response to high light intensity(GO:0071486) retinal rod cell apoptotic process(GO:0097473) |
| 0.1 | 0.7 | GO:0097118 | neuroligin clustering involved in postsynaptic membrane assembly(GO:0097118) |
| 0.1 | 3.4 | GO:0006817 | phosphate ion transport(GO:0006817) |
| 0.1 | 0.9 | GO:0060762 | regulation of branching involved in mammary gland duct morphogenesis(GO:0060762) |
| 0.1 | 0.6 | GO:0006685 | sphingomyelin catabolic process(GO:0006685) |
| 0.1 | 6.5 | GO:0032007 | negative regulation of TOR signaling(GO:0032007) |
| 0.1 | 2.0 | GO:1902004 | positive regulation of beta-amyloid formation(GO:1902004) |
| 0.1 | 0.7 | GO:0048743 | positive regulation of skeletal muscle fiber development(GO:0048743) |
| 0.1 | 2.8 | GO:0016339 | calcium-dependent cell-cell adhesion via plasma membrane cell adhesion molecules(GO:0016339) |
| 0.1 | 5.8 | GO:0007212 | dopamine receptor signaling pathway(GO:0007212) |
| 0.1 | 1.5 | GO:1904322 | response to forskolin(GO:1904321) cellular response to forskolin(GO:1904322) |
| 0.1 | 0.9 | GO:0097577 | intracellular sequestering of iron ion(GO:0006880) sequestering of iron ion(GO:0097577) |
| 0.1 | 1.6 | GO:0048739 | cardiac muscle fiber development(GO:0048739) |
| 0.1 | 0.4 | GO:1905035 | regulation of antifungal innate immune response(GO:1905034) negative regulation of antifungal innate immune response(GO:1905035) |
| 0.1 | 2.0 | GO:0031290 | retinal ganglion cell axon guidance(GO:0031290) |
| 0.1 | 1.2 | GO:0071670 | smooth muscle cell chemotaxis(GO:0071670) |
| 0.1 | 0.5 | GO:1903800 | positive regulation of production of miRNAs involved in gene silencing by miRNA(GO:1903800) |
| 0.1 | 1.3 | GO:2000381 | negative regulation of mesoderm development(GO:2000381) |
| 0.1 | 3.5 | GO:0030212 | hyaluronan metabolic process(GO:0030212) |
| 0.1 | 0.3 | GO:0018894 | dibenzo-p-dioxin metabolic process(GO:0018894) |
| 0.1 | 1.9 | GO:0045948 | positive regulation of translational initiation(GO:0045948) |
| 0.1 | 17.3 | GO:0050806 | positive regulation of synaptic transmission(GO:0050806) |
| 0.1 | 1.2 | GO:0044336 | canonical Wnt signaling pathway involved in negative regulation of apoptotic process(GO:0044336) |
| 0.1 | 0.6 | GO:0009249 | protein lipoylation(GO:0009249) |
| 0.1 | 3.4 | GO:0043252 | sodium-independent organic anion transport(GO:0043252) |
| 0.1 | 3.7 | GO:0003333 | amino acid transmembrane transport(GO:0003333) |
| 0.1 | 0.7 | GO:0021869 | forebrain ventricular zone progenitor cell division(GO:0021869) |
| 0.1 | 2.0 | GO:0050435 | beta-amyloid metabolic process(GO:0050435) |
| 0.1 | 3.6 | GO:0048384 | retinoic acid receptor signaling pathway(GO:0048384) |
| 0.1 | 0.1 | GO:0048549 | positive regulation of pinocytosis(GO:0048549) |
| 0.1 | 1.0 | GO:0045199 | maintenance of epithelial cell apical/basal polarity(GO:0045199) |
| 0.1 | 12.8 | GO:0030308 | negative regulation of cell growth(GO:0030308) |
| 0.1 | 0.2 | GO:0051710 | regulation of cytolysis in other organism(GO:0051710) positive regulation of cytolysis in other organism(GO:0051714) |
| 0.1 | 0.4 | GO:0006848 | pyruvate transport(GO:0006848) pyruvate transmembrane transport(GO:1901475) |
| 0.0 | 1.8 | GO:0031424 | keratinization(GO:0031424) |
| 0.0 | 0.9 | GO:0051457 | maintenance of protein location in nucleus(GO:0051457) |
| 0.0 | 0.9 | GO:0090266 | regulation of mitotic cell cycle spindle assembly checkpoint(GO:0090266) regulation of mitotic spindle checkpoint(GO:1903504) |
| 0.0 | 0.7 | GO:0001731 | formation of translation preinitiation complex(GO:0001731) |
| 0.0 | 2.8 | GO:0071806 | intracellular protein transmembrane transport(GO:0065002) protein transmembrane transport(GO:0071806) |
| 0.0 | 1.3 | GO:0050911 | detection of chemical stimulus involved in sensory perception of smell(GO:0050911) |
| 0.0 | 0.7 | GO:0009083 | branched-chain amino acid catabolic process(GO:0009083) |
| 0.0 | 0.3 | GO:0043951 | negative regulation of cAMP-mediated signaling(GO:0043951) |
| 0.0 | 0.4 | GO:0018298 | protein-chromophore linkage(GO:0018298) |
| 0.0 | 2.4 | GO:0072384 | organelle transport along microtubule(GO:0072384) |
| 0.0 | 1.1 | GO:0048791 | calcium ion-regulated exocytosis of neurotransmitter(GO:0048791) |
| 0.0 | 0.7 | GO:0006677 | glycosylceramide metabolic process(GO:0006677) |
| 0.0 | 3.2 | GO:0019236 | response to pheromone(GO:0019236) |
| 0.0 | 3.1 | GO:0007030 | Golgi organization(GO:0007030) |
| 0.0 | 0.9 | GO:0036465 | synaptic vesicle recycling(GO:0036465) |
| 0.0 | 0.1 | GO:0070245 | positive regulation of thymocyte apoptotic process(GO:0070245) |
| 0.0 | 0.6 | GO:0007214 | gamma-aminobutyric acid signaling pathway(GO:0007214) |
| 0.0 | 0.2 | GO:0006610 | ribosomal protein import into nucleus(GO:0006610) |
| 0.0 | 0.7 | GO:0006182 | cGMP biosynthetic process(GO:0006182) |
| 0.0 | 0.5 | GO:0043552 | positive regulation of phosphatidylinositol 3-kinase activity(GO:0043552) |
| 0.0 | 2.5 | GO:0010811 | positive regulation of cell-substrate adhesion(GO:0010811) |
| 0.0 | 0.2 | GO:0070475 | rRNA base methylation(GO:0070475) |
| 0.0 | 0.2 | GO:0097062 | dendritic spine maintenance(GO:0097062) |
| 0.0 | 1.8 | GO:0035308 | negative regulation of protein dephosphorylation(GO:0035308) |
| 0.0 | 0.5 | GO:0015991 | ATP hydrolysis coupled proton transport(GO:0015991) |
| 0.0 | 0.2 | GO:1900028 | negative regulation of ruffle assembly(GO:1900028) |
| 0.0 | 0.1 | GO:0002457 | T cell antigen processing and presentation(GO:0002457) histone H3-K4 demethylation(GO:0034720) |
| 0.0 | 0.5 | GO:0006346 | methylation-dependent chromatin silencing(GO:0006346) |
| 0.0 | 0.4 | GO:0019373 | epoxygenase P450 pathway(GO:0019373) |
| 0.0 | 15.0 | GO:0007606 | sensory perception of chemical stimulus(GO:0007606) |
| 0.0 | 0.3 | GO:0001958 | endochondral ossification(GO:0001958) replacement ossification(GO:0036075) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.3 | 8.8 | GO:0048237 | rough endoplasmic reticulum lumen(GO:0048237) |
| 0.7 | 2.8 | GO:0014802 | terminal cisterna(GO:0014802) |
| 0.6 | 16.5 | GO:0098563 | integral component of synaptic vesicle membrane(GO:0030285) intrinsic component of synaptic vesicle membrane(GO:0098563) |
| 0.5 | 2.1 | GO:0014701 | junctional sarcoplasmic reticulum membrane(GO:0014701) |
| 0.5 | 2.8 | GO:0098831 | calyx of Held(GO:0044305) presynaptic active zone cytoplasmic component(GO:0098831) |
| 0.4 | 3.4 | GO:0031673 | H zone(GO:0031673) |
| 0.4 | 1.3 | GO:0005900 | oncostatin-M receptor complex(GO:0005900) |
| 0.4 | 7.7 | GO:0036057 | filtration diaphragm(GO:0036056) slit diaphragm(GO:0036057) |
| 0.4 | 10.4 | GO:0014731 | spectrin-associated cytoskeleton(GO:0014731) |
| 0.4 | 2.5 | GO:0005955 | calcineurin complex(GO:0005955) |
| 0.3 | 24.0 | GO:0046658 | anchored component of plasma membrane(GO:0046658) |
| 0.3 | 2.4 | GO:0098574 | cytoplasmic side of lysosomal membrane(GO:0098574) |
| 0.3 | 1.2 | GO:0032280 | symmetric synapse(GO:0032280) |
| 0.3 | 1.8 | GO:0008091 | spectrin(GO:0008091) cuticular plate(GO:0032437) |
| 0.3 | 1.9 | GO:0089701 | U2AF(GO:0089701) |
| 0.3 | 4.3 | GO:1990635 | proximal dendrite(GO:1990635) |
| 0.2 | 0.7 | GO:0005965 | protein farnesyltransferase complex(GO:0005965) |
| 0.2 | 1.1 | GO:0005968 | Rab-protein geranylgeranyltransferase complex(GO:0005968) |
| 0.2 | 5.7 | GO:0016327 | apicolateral plasma membrane(GO:0016327) |
| 0.2 | 4.3 | GO:1902711 | GABA-A receptor complex(GO:1902711) |
| 0.1 | 2.2 | GO:1990454 | L-type voltage-gated calcium channel complex(GO:1990454) |
| 0.1 | 2.8 | GO:0097449 | astrocyte projection(GO:0097449) |
| 0.1 | 1.8 | GO:0071203 | WASH complex(GO:0071203) |
| 0.1 | 0.7 | GO:0005947 | mitochondrial alpha-ketoglutarate dehydrogenase complex(GO:0005947) |
| 0.1 | 2.1 | GO:0035253 | ciliary rootlet(GO:0035253) |
| 0.1 | 5.8 | GO:0005834 | heterotrimeric G-protein complex(GO:0005834) |
| 0.1 | 1.5 | GO:0030061 | mitochondrial crista(GO:0030061) |
| 0.1 | 0.9 | GO:0019773 | proteasome core complex, alpha-subunit complex(GO:0019773) |
| 0.1 | 1.2 | GO:0045277 | respiratory chain complex IV(GO:0045277) |
| 0.1 | 0.9 | GO:0044754 | autolysosome(GO:0044754) |
| 0.1 | 1.6 | GO:0005614 | interstitial matrix(GO:0005614) |
| 0.0 | 5.8 | GO:1990204 | oxidoreductase complex(GO:1990204) |
| 0.0 | 0.9 | GO:0030914 | STAGA complex(GO:0030914) |
| 0.0 | 2.0 | GO:0005771 | multivesicular body(GO:0005771) |
| 0.0 | 2.7 | GO:0097610 | cleavage furrow(GO:0032154) cell surface furrow(GO:0097610) |
| 0.0 | 0.7 | GO:0032593 | insulin-responsive compartment(GO:0032593) |
| 0.0 | 2.7 | GO:1903293 | protein serine/threonine phosphatase complex(GO:0008287) phosphatase complex(GO:1903293) |
| 0.0 | 1.5 | GO:0031430 | M band(GO:0031430) |
| 0.0 | 8.7 | GO:0005802 | trans-Golgi network(GO:0005802) |
| 0.0 | 0.5 | GO:0016471 | vacuolar proton-transporting V-type ATPase complex(GO:0016471) |
| 0.0 | 0.9 | GO:0005680 | anaphase-promoting complex(GO:0005680) |
| 0.0 | 0.7 | GO:0008074 | guanylate cyclase complex, soluble(GO:0008074) |
| 0.0 | 2.8 | GO:0001726 | ruffle(GO:0001726) |
| 0.0 | 6.1 | GO:0070382 | exocytic vesicle(GO:0070382) |
| 0.0 | 1.3 | GO:0001533 | cornified envelope(GO:0001533) |
| 0.0 | 0.1 | GO:0044354 | pinosome(GO:0044352) macropinosome(GO:0044354) |
| 0.0 | 0.8 | GO:0032281 | AMPA glutamate receptor complex(GO:0032281) |
| 0.0 | 0.1 | GO:0035363 | histone locus body(GO:0035363) |
| 0.0 | 2.7 | GO:0030175 | filopodium(GO:0030175) |
| 0.0 | 0.5 | GO:0032839 | dendrite cytoplasm(GO:0032839) |
| 0.0 | 2.2 | GO:0016591 | DNA-directed RNA polymerase II, holoenzyme(GO:0016591) |
| 0.0 | 19.5 | GO:0045202 | synapse(GO:0045202) |
| 0.0 | 0.7 | GO:0043679 | axon terminus(GO:0043679) |
| 0.0 | 1.1 | GO:0001750 | photoreceptor outer segment(GO:0001750) |
| 0.0 | 2.1 | GO:0005923 | bicellular tight junction(GO:0005923) |
| 0.0 | 1.7 | GO:0030018 | Z disc(GO:0030018) |
| 0.0 | 7.8 | GO:0015629 | actin cytoskeleton(GO:0015629) |
| 0.0 | 1.0 | GO:0005901 | caveola(GO:0005901) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.1 | 25.0 | GO:0016724 | ferroxidase activity(GO:0004322) oxidoreductase activity, oxidizing metal ions, oxygen as acceptor(GO:0016724) |
| 1.3 | 4.0 | GO:0004471 | malic enzyme activity(GO:0004470) malate dehydrogenase (decarboxylating) (NAD+) activity(GO:0004471) malate dehydrogenase (decarboxylating) (NADP+) activity(GO:0004473) |
| 1.1 | 6.9 | GO:0035241 | protein-arginine omega-N monomethyltransferase activity(GO:0035241) |
| 1.1 | 4.3 | GO:0004833 | tryptophan 2,3-dioxygenase activity(GO:0004833) |
| 1.0 | 12.9 | GO:0048403 | brain-derived neurotrophic factor binding(GO:0048403) |
| 0.9 | 2.8 | GO:0099530 | PLC activating G-protein coupled glutamate receptor activity(GO:0001639) G-protein coupled receptor activity involved in regulation of postsynaptic membrane potential(GO:0099530) |
| 0.7 | 5.2 | GO:0001602 | peptide YY receptor activity(GO:0001601) pancreatic polypeptide receptor activity(GO:0001602) |
| 0.7 | 5.8 | GO:0031821 | G-protein coupled serotonin receptor binding(GO:0031821) |
| 0.7 | 8.8 | GO:0004887 | thyroid hormone receptor activity(GO:0004887) |
| 0.7 | 3.4 | GO:0015319 | sodium:inorganic phosphate symporter activity(GO:0015319) |
| 0.6 | 1.8 | GO:0001888 | glucuronyl-galactosyl-proteoglycan 4-alpha-N-acetylglucosaminyltransferase activity(GO:0001888) |
| 0.6 | 4.8 | GO:0001849 | complement component C1q binding(GO:0001849) |
| 0.5 | 7.0 | GO:0097109 | neuroligin family protein binding(GO:0097109) |
| 0.4 | 1.3 | GO:0004915 | interleukin-6 receptor activity(GO:0004915) interleukin-6 binding(GO:0019981) |
| 0.4 | 3.4 | GO:0015349 | thyroid hormone transmembrane transporter activity(GO:0015349) |
| 0.4 | 4.2 | GO:0005186 | pheromone activity(GO:0005186) |
| 0.4 | 2.5 | GO:0033192 | calmodulin-dependent protein phosphatase activity(GO:0033192) |
| 0.4 | 1.6 | GO:0070052 | collagen V binding(GO:0070052) |
| 0.4 | 1.5 | GO:0008386 | cholesterol monooxygenase (side-chain-cleaving) activity(GO:0008386) |
| 0.4 | 2.9 | GO:1904315 | transmitter-gated ion channel activity involved in regulation of postsynaptic membrane potential(GO:1904315) |
| 0.4 | 3.6 | GO:0004886 | 9-cis retinoic acid receptor activity(GO:0004886) |
| 0.3 | 2.2 | GO:0086056 | voltage-gated calcium channel activity involved in AV node cell action potential(GO:0086056) |
| 0.3 | 10.5 | GO:0017075 | syntaxin-1 binding(GO:0017075) |
| 0.3 | 0.9 | GO:0016509 | long-chain-3-hydroxyacyl-CoA dehydrogenase activity(GO:0016509) |
| 0.3 | 8.8 | GO:0030247 | pattern binding(GO:0001871) polysaccharide binding(GO:0030247) |
| 0.3 | 2.8 | GO:0050786 | RAGE receptor binding(GO:0050786) |
| 0.3 | 6.3 | GO:0042813 | Wnt-activated receptor activity(GO:0042813) |
| 0.3 | 5.3 | GO:0016594 | glycine binding(GO:0016594) |
| 0.2 | 3.7 | GO:0017147 | Wnt-protein binding(GO:0017147) |
| 0.2 | 0.7 | GO:0004660 | protein farnesyltransferase activity(GO:0004660) |
| 0.2 | 2.1 | GO:0042500 | aspartic endopeptidase activity, intramembrane cleaving(GO:0042500) |
| 0.2 | 0.6 | GO:0031177 | phosphopantetheine binding(GO:0031177) |
| 0.2 | 4.3 | GO:0070402 | NADPH binding(GO:0070402) |
| 0.2 | 1.0 | GO:0004875 | complement receptor activity(GO:0004875) |
| 0.2 | 0.9 | GO:0004483 | mRNA (nucleoside-2'-O-)-methyltransferase activity(GO:0004483) |
| 0.1 | 1.2 | GO:0001515 | opioid peptide activity(GO:0001515) |
| 0.1 | 1.6 | GO:0008429 | phosphatidylethanolamine binding(GO:0008429) |
| 0.1 | 1.9 | GO:0030628 | pre-mRNA 3'-splice site binding(GO:0030628) |
| 0.1 | 11.9 | GO:0030507 | spectrin binding(GO:0030507) |
| 0.1 | 4.8 | GO:0030676 | Rac guanyl-nucleotide exchange factor activity(GO:0030676) |
| 0.1 | 5.3 | GO:0005086 | ARF guanyl-nucleotide exchange factor activity(GO:0005086) |
| 0.1 | 0.6 | GO:0005105 | type 1 fibroblast growth factor receptor binding(GO:0005105) |
| 0.1 | 0.7 | GO:0050656 | 3'-phosphoadenosine 5'-phosphosulfate binding(GO:0050656) |
| 0.1 | 0.7 | GO:0031697 | beta-1 adrenergic receptor binding(GO:0031697) |
| 0.1 | 7.2 | GO:0008395 | steroid hydroxylase activity(GO:0008395) |
| 0.1 | 1.8 | GO:0010314 | phosphatidylinositol-5-phosphate binding(GO:0010314) |
| 0.1 | 1.6 | GO:0070008 | serine-type exopeptidase activity(GO:0070008) |
| 0.1 | 0.4 | GO:0005502 | 11-cis retinal binding(GO:0005502) |
| 0.1 | 1.0 | GO:0044323 | retinoic acid-responsive element binding(GO:0044323) |
| 0.1 | 3.4 | GO:0043539 | protein serine/threonine kinase activator activity(GO:0043539) |
| 0.1 | 2.0 | GO:0004890 | GABA-A receptor activity(GO:0004890) |
| 0.1 | 0.7 | GO:0001665 | alpha-N-acetylgalactosaminide alpha-2,6-sialyltransferase activity(GO:0001665) |
| 0.1 | 2.0 | GO:0050431 | transforming growth factor beta binding(GO:0050431) |
| 0.1 | 0.6 | GO:0004767 | sphingomyelin phosphodiesterase activity(GO:0004767) |
| 0.1 | 0.4 | GO:0042030 | ATPase inhibitor activity(GO:0042030) |
| 0.1 | 1.6 | GO:0051371 | muscle alpha-actinin binding(GO:0051371) |
| 0.1 | 0.4 | GO:0050833 | pyruvate transmembrane transporter activity(GO:0050833) |
| 0.0 | 1.8 | GO:0030506 | ankyrin binding(GO:0030506) |
| 0.0 | 1.1 | GO:0005092 | GDP-dissociation inhibitor activity(GO:0005092) |
| 0.0 | 1.2 | GO:0015002 | cytochrome-c oxidase activity(GO:0004129) heme-copper terminal oxidase activity(GO:0015002) oxidoreductase activity, acting on a heme group of donors, oxygen as acceptor(GO:0016676) |
| 0.0 | 3.1 | GO:0030544 | Hsp70 protein binding(GO:0030544) |
| 0.0 | 4.4 | GO:0019888 | protein phosphatase regulator activity(GO:0019888) |
| 0.0 | 0.6 | GO:0003854 | 3-beta-hydroxy-delta5-steroid dehydrogenase activity(GO:0003854) |
| 0.0 | 7.7 | GO:0017137 | Rab GTPase binding(GO:0017137) |
| 0.0 | 2.6 | GO:0070888 | E-box binding(GO:0070888) |
| 0.0 | 1.4 | GO:0044183 | protein binding involved in protein folding(GO:0044183) |
| 0.0 | 0.9 | GO:0070003 | threonine-type endopeptidase activity(GO:0004298) threonine-type peptidase activity(GO:0070003) |
| 0.0 | 1.3 | GO:0043425 | bHLH transcription factor binding(GO:0043425) |
| 0.0 | 2.0 | GO:0001540 | beta-amyloid binding(GO:0001540) |
| 0.0 | 0.7 | GO:0015026 | coreceptor activity(GO:0015026) |
| 0.0 | 3.7 | GO:0015171 | amino acid transmembrane transporter activity(GO:0015171) |
| 0.0 | 0.7 | GO:0004383 | guanylate cyclase activity(GO:0004383) |
| 0.0 | 8.8 | GO:0044822 | mRNA binding(GO:0003729) poly(A) RNA binding(GO:0044822) |
| 0.0 | 0.5 | GO:0008553 | hydrogen-exporting ATPase activity, phosphorylative mechanism(GO:0008553) |
| 0.0 | 3.1 | GO:0016503 | pheromone receptor activity(GO:0016503) |
| 0.0 | 0.6 | GO:0016755 | transferase activity, transferring amino-acyl groups(GO:0016755) |
| 0.0 | 0.5 | GO:0008327 | methyl-CpG binding(GO:0008327) |
| 0.0 | 6.9 | GO:0005549 | odorant binding(GO:0005549) |
| 0.0 | 0.6 | GO:0005112 | Notch binding(GO:0005112) |
| 0.0 | 0.7 | GO:0031435 | mitogen-activated protein kinase kinase kinase binding(GO:0031435) |
| 0.0 | 0.1 | GO:0001729 | ceramide kinase activity(GO:0001729) |
| 0.0 | 0.1 | GO:0032453 | histone demethylase activity (H3-K4 specific)(GO:0032453) |
| 0.0 | 0.9 | GO:0004402 | histone acetyltransferase activity(GO:0004402) |
| 0.0 | 0.2 | GO:0005522 | profilin binding(GO:0005522) |
| 0.0 | 0.2 | GO:0030215 | semaphorin receptor binding(GO:0030215) |
| 0.0 | 0.4 | GO:0015020 | glucuronosyltransferase activity(GO:0015020) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 8.8 | PID INTEGRIN5 PATHWAY | Beta5 beta6 beta7 and beta8 integrin cell surface interactions |
| 0.3 | 5.8 | PID S1P S1P4 PATHWAY | S1P4 pathway |
| 0.2 | 23.2 | PID HIF1 TFPATHWAY | HIF-1-alpha transcription factor network |
| 0.1 | 8.1 | PID IFNG PATHWAY | IFN-gamma pathway |
| 0.1 | 7.2 | PID IL6 7 PATHWAY | IL6-mediated signaling events |
| 0.1 | 3.7 | PID WNT SIGNALING PATHWAY | Wnt signaling network |
| 0.1 | 5.5 | PID RXR VDR PATHWAY | RXR and RAR heterodimerization with other nuclear receptor |
| 0.1 | 2.5 | PID IL12 STAT4 PATHWAY | IL12 signaling mediated by STAT4 |
| 0.1 | 3.4 | ST G ALPHA I PATHWAY | G alpha i Pathway |
| 0.1 | 12.6 | NABA ECM GLYCOPROTEINS | Genes encoding structural ECM glycoproteins |
| 0.1 | 4.8 | PID RAC1 REG PATHWAY | Regulation of RAC1 activity |
| 0.0 | 3.8 | PID REG GR PATHWAY | Glucocorticoid receptor regulatory network |
| 0.0 | 2.8 | PID LKB1 PATHWAY | LKB1 signaling events |
| 0.0 | 9.6 | NABA ECM REGULATORS | Genes encoding enzymes and their regulators involved in the remodeling of the extracellular matrix |
| 0.0 | 0.9 | PID MYC PATHWAY | C-MYC pathway |
| 0.0 | 2.0 | PID TAP63 PATHWAY | Validated transcriptional targets of TAp63 isoforms |
| 0.0 | 0.7 | PID ARF 3PATHWAY | Arf1 pathway |
| 0.0 | 1.9 | PID MTOR 4PATHWAY | mTOR signaling pathway |
| 0.0 | 2.4 | PID CASPASE PATHWAY | Caspase cascade in apoptosis |
| 0.0 | 1.2 | PID ILK PATHWAY | Integrin-linked kinase signaling |
| 0.0 | 1.6 | PID AJDISS 2PATHWAY | Posttranslational regulation of adherens junction stability and dissassembly |
| 0.0 | 0.6 | PID AURORA A PATHWAY | Aurora A signaling |
| 0.0 | 0.6 | PID SHP2 PATHWAY | SHP2 signaling |
| 0.0 | 0.9 | PID MYC REPRESS PATHWAY | Validated targets of C-MYC transcriptional repression |
| 0.0 | 0.7 | PID BMP PATHWAY | BMP receptor signaling |
| 0.0 | 0.6 | PID FGF PATHWAY | FGF signaling pathway |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.6 | 4.8 | REACTOME CREATION OF C4 AND C2 ACTIVATORS | Genes involved in Creation of C4 and C2 activators |
| 0.6 | 24.1 | REACTOME METAL ION SLC TRANSPORTERS | Genes involved in Metal ion SLC transporters |
| 0.5 | 7.7 | REACTOME ROLE OF DCC IN REGULATING APOPTOSIS | Genes involved in Role of DCC in regulating apoptosis |
| 0.3 | 4.3 | REACTOME TRYPTOPHAN CATABOLISM | Genes involved in Tryptophan catabolism |
| 0.2 | 3.4 | REACTOME TRANSPORT OF ORGANIC ANIONS | Genes involved in Transport of organic anions |
| 0.2 | 12.1 | REACTOME INTERACTION BETWEEN L1 AND ANKYRINS | Genes involved in Interaction between L1 and Ankyrins |
| 0.2 | 4.9 | REACTOME GABA A RECEPTOR ACTIVATION | Genes involved in GABA A receptor activation |
| 0.2 | 3.2 | REACTOME SIGNALING BY NOTCH4 | Genes involved in Signaling by NOTCH4 |
| 0.2 | 8.1 | REACTOME INHIBITION OF INSULIN SECRETION BY ADRENALINE NORADRENALINE | Genes involved in Inhibition of Insulin Secretion by Adrenaline/Noradrenaline |
| 0.1 | 2.8 | REACTOME ADVANCED GLYCOSYLATION ENDPRODUCT RECEPTOR SIGNALING | Genes involved in Advanced glycosylation endproduct receptor signaling |
| 0.1 | 0.3 | REACTOME XENOBIOTICS | Genes involved in Xenobiotics |
| 0.1 | 1.9 | REACTOME REGULATION OF INSULIN LIKE GROWTH FACTOR IGF ACTIVITY BY INSULIN LIKE GROWTH FACTOR BINDING PROTEINS IGFBPS | Genes involved in Regulation of Insulin-like Growth Factor (IGF) Activity by Insulin-like Growth Factor Binding Proteins (IGFBPs) |
| 0.1 | 8.5 | REACTOME NUCLEAR RECEPTOR TRANSCRIPTION PATHWAY | Genes involved in Nuclear Receptor transcription pathway |
| 0.1 | 1.3 | REACTOME IL 6 SIGNALING | Genes involved in Interleukin-6 signaling |
| 0.1 | 4.5 | REACTOME VOLTAGE GATED POTASSIUM CHANNELS | Genes involved in Voltage gated Potassium channels |
| 0.1 | 1.6 | REACTOME INTRINSIC PATHWAY | Genes involved in Intrinsic Pathway |
| 0.1 | 2.4 | REACTOME CASPASE MEDIATED CLEAVAGE OF CYTOSKELETAL PROTEINS | Genes involved in Caspase-mediated cleavage of cytoskeletal proteins |
| 0.1 | 2.9 | REACTOME G ALPHA Z SIGNALLING EVENTS | Genes involved in G alpha (z) signalling events |
| 0.1 | 8.8 | REACTOME INTEGRIN CELL SURFACE INTERACTIONS | Genes involved in Integrin cell surface interactions |
| 0.1 | 0.7 | REACTOME NEGATIVE REGULATION OF THE PI3K AKT NETWORK | Genes involved in Negative regulation of the PI3K/AKT network |
| 0.1 | 0.7 | REACTOME COPI MEDIATED TRANSPORT | Genes involved in COPI Mediated Transport |
| 0.1 | 0.9 | REACTOME NEF MEDIATED DOWNREGULATION OF MHC CLASS I COMPLEX CELL SURFACE EXPRESSION | Genes involved in Nef mediated downregulation of MHC class I complex cell surface expression |
| 0.1 | 3.7 | REACTOME AMINO ACID TRANSPORT ACROSS THE PLASMA MEMBRANE | Genes involved in Amino acid transport across the plasma membrane |
| 0.1 | 2.5 | REACTOME DARPP 32 EVENTS | Genes involved in DARPP-32 events |
| 0.1 | 2.1 | REACTOME TIGHT JUNCTION INTERACTIONS | Genes involved in Tight junction interactions |
| 0.0 | 3.1 | REACTOME O LINKED GLYCOSYLATION OF MUCINS | Genes involved in O-linked glycosylation of mucins |
| 0.0 | 1.2 | REACTOME CELL EXTRACELLULAR MATRIX INTERACTIONS | Genes involved in Cell-extracellular matrix interactions |
| 0.0 | 0.5 | REACTOME RAF MAP KINASE CASCADE | Genes involved in RAF/MAP kinase cascade |
| 0.0 | 0.4 | REACTOME OPSINS | Genes involved in Opsins |
| 0.0 | 3.4 | REACTOME TRANSPORT OF INORGANIC CATIONS ANIONS AND AMINO ACIDS OLIGOPEPTIDES | Genes involved in Transport of inorganic cations/anions and amino acids/oligopeptides |
| 0.0 | 0.9 | REACTOME AMYLOIDS | Genes involved in Amyloids |
| 0.0 | 0.9 | REACTOME SYNTHESIS OF VERY LONG CHAIN FATTY ACYL COAS | Genes involved in Synthesis of very long-chain fatty acyl-CoAs |
| 0.0 | 1.3 | REACTOME RECYCLING PATHWAY OF L1 | Genes involved in Recycling pathway of L1 |
| 0.0 | 5.6 | REACTOME PEPTIDE LIGAND BINDING RECEPTORS | Genes involved in Peptide ligand-binding receptors |
| 0.0 | 0.6 | REACTOME FGFR4 LIGAND BINDING AND ACTIVATION | Genes involved in FGFR4 ligand binding and activation |
| 0.0 | 4.2 | REACTOME PPARA ACTIVATES GENE EXPRESSION | Genes involved in PPARA Activates Gene Expression |
| 0.0 | 1.3 | REACTOME IRON UPTAKE AND TRANSPORT | Genes involved in Iron uptake and transport |
| 0.0 | 1.8 | REACTOME RESPIRATORY ELECTRON TRANSPORT | Genes involved in Respiratory electron transport |
| 0.0 | 0.3 | REACTOME G ALPHA S SIGNALLING EVENTS | Genes involved in G alpha (s) signalling events |
| 0.0 | 0.4 | REACTOME BILE SALT AND ORGANIC ANION SLC TRANSPORTERS | Genes involved in Bile salt and organic anion SLC transporters |
| 0.0 | 0.9 | REACTOME CROSS PRESENTATION OF SOLUBLE EXOGENOUS ANTIGENS ENDOSOMES | Genes involved in Cross-presentation of soluble exogenous antigens (endosomes) |