Project
PRJNA375882: Comprehensive Mouse Transcriptomic BodyMap
Navigation
Downloads
Results for Osr2_Osr1
Z-value: 0.97
Transcription factors associated with Osr2_Osr1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
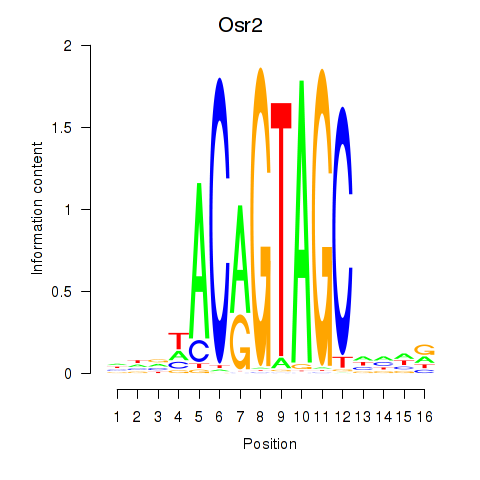
Osr2
|
ENSMUSG00000022330.6 | Osr2 |
|
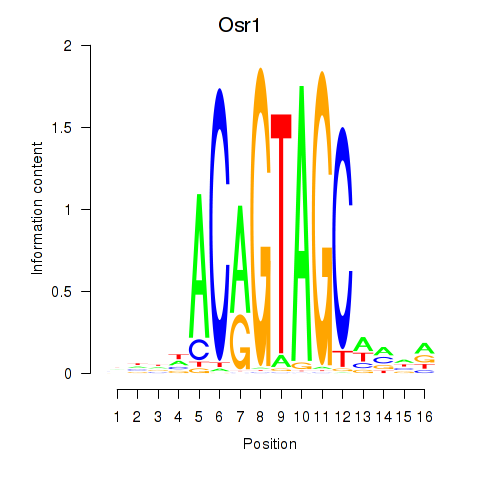
Osr1
|
ENSMUSG00000048387.9 | Osr1 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Osr1 | mm39_v1_chr12_+_9624437_9624448 | -0.38 | 9.5e-04 | Click! |
| Osr2 | mm39_v1_chr15_+_35296237_35296250 | 0.15 | 2.2e-01 | Click! |
Activity profile of Osr2_Osr1 motif
Sorted Z-values of Osr2_Osr1 motif
Network of associatons between targets according to the STRING database.
First level regulatory network of Osr2_Osr1
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr3_-_98670369 | 9.90 |
ENSMUST00000107019.8
ENSMUST00000107018.8 |
Hsd3b3
|
hydroxy-delta-5-steroid dehydrogenase, 3 beta- and steroid delta-isomerase 3 |
| chr4_-_115504907 | 8.99 |
ENSMUST00000102707.10
|
Cyp4b1
|
cytochrome P450, family 4, subfamily b, polypeptide 1 |
| chr8_+_21652293 | 6.71 |
ENSMUST00000098897.2
|
Defa22
|
defensin, alpha, 22 |
| chr8_+_21881827 | 6.60 |
ENSMUST00000120874.5
|
Defa33
|
defensin, alpha, 33 |
| chr8_+_21868531 | 6.38 |
ENSMUST00000170275.4
|
Defa2
|
defensin, alpha, 2 |
| chr8_+_21999274 | 6.22 |
ENSMUST00000084042.4
|
Defa20
|
defensin, alpha, 20 |
| chr7_-_24459736 | 6.11 |
ENSMUST00000063956.7
|
Cd177
|
CD177 antigen |
| chr8_+_22019048 | 5.97 |
ENSMUST00000084041.4
|
Defa32
|
defensin, alpha, 32 |
| chr8_+_21515561 | 5.87 |
ENSMUST00000076754.3
|
Defa21
|
defensin, alpha, 21 |
| chr1_+_88066086 | 5.52 |
ENSMUST00000014263.6
|
Ugt1a6a
|
UDP glucuronosyltransferase 1 family, polypeptide A6A |
| chr13_-_100589357 | 5.50 |
ENSMUST00000222155.2
ENSMUST00000221727.2 |
Naip1
|
NLR family, apoptosis inhibitory protein 1 |
| chr2_+_34764408 | 4.63 |
ENSMUST00000113068.9
ENSMUST00000047447.13 |
Cutal
|
cutA divalent cation tolerance homolog-like |
| chr11_+_58269862 | 4.59 |
ENSMUST00000013787.11
ENSMUST00000108826.3 |
Lypd8
|
LY6/PLAUR domain containing 8 |
| chr4_+_135691030 | 4.54 |
ENSMUST00000102541.10
ENSMUST00000149636.2 ENSMUST00000143304.2 |
Gale
|
galactose-4-epimerase, UDP |
| chr13_-_4329421 | 4.40 |
ENSMUST00000021632.5
|
Akr1c12
|
aldo-keto reductase family 1, member C12 |
| chr13_+_4241149 | 4.27 |
ENSMUST00000021634.4
|
Akr1c13
|
aldo-keto reductase family 1, member C13 |
| chr18_+_70058533 | 4.26 |
ENSMUST00000043929.11
|
Ccdc68
|
coiled-coil domain containing 68 |
| chr2_+_34764496 | 4.13 |
ENSMUST00000028228.6
|
Cutal
|
cutA divalent cation tolerance homolog-like |
| chr6_+_67586695 | 3.89 |
ENSMUST00000103303.3
|
Igkv1-135
|
immunoglobulin kappa variable 1-135 |
| chr6_-_69741999 | 3.88 |
ENSMUST00000103365.3
|
Igkv12-46
|
immunoglobulin kappa variable 12-46 |
| chr18_+_70058613 | 3.83 |
ENSMUST00000080050.6
|
Ccdc68
|
coiled-coil domain containing 68 |
| chr6_-_68907718 | 3.73 |
ENSMUST00000114212.4
|
Igkv13-85
|
immunoglobulin kappa chain variable 13-85 |
| chr12_-_108668520 | 3.40 |
ENSMUST00000167978.9
ENSMUST00000021691.6 |
Degs2
|
delta(4)-desaturase, sphingolipid 2 |
| chr7_+_19699291 | 3.40 |
ENSMUST00000094753.6
|
Ceacam20
|
carcinoembryonic antigen-related cell adhesion molecule 20 |
| chr11_+_72851989 | 3.39 |
ENSMUST00000163326.8
ENSMUST00000108485.9 ENSMUST00000021142.8 ENSMUST00000108486.8 ENSMUST00000108484.8 |
Atp2a3
|
ATPase, Ca++ transporting, ubiquitous |
| chr8_+_22108199 | 3.34 |
ENSMUST00000074343.6
|
Defa26
|
defensin, alpha, 26 |
| chr5_-_66238313 | 3.30 |
ENSMUST00000202700.4
ENSMUST00000094757.9 ENSMUST00000113724.6 |
Rbm47
|
RNA binding motif protein 47 |
| chr3_+_63148887 | 3.24 |
ENSMUST00000194324.6
|
Mme
|
membrane metallo endopeptidase |
| chr13_+_55547498 | 3.07 |
ENSMUST00000057167.9
|
Slc34a1
|
solute carrier family 34 (sodium phosphate), member 1 |
| chr9_-_51240201 | 2.89 |
ENSMUST00000039959.11
ENSMUST00000238450.3 |
1810046K07Rik
|
RIKEN cDNA 1810046K07 gene |
| chr11_-_105347500 | 2.74 |
ENSMUST00000049995.10
ENSMUST00000100332.4 |
Marchf10
|
membrane associated ring-CH-type finger 10 |
| chr9_+_51124983 | 2.72 |
ENSMUST00000034554.9
|
Pou2af1
|
POU domain, class 2, associating factor 1 |
| chr19_-_32080496 | 2.61 |
ENSMUST00000235213.2
ENSMUST00000236504.2 |
Asah2
|
N-acylsphingosine amidohydrolase 2 |
| chr3_-_98669720 | 2.60 |
ENSMUST00000146196.4
|
Hsd3b3
|
hydroxy-delta-5-steroid dehydrogenase, 3 beta- and steroid delta-isomerase 3 |
| chr6_+_68414401 | 2.59 |
ENSMUST00000103324.3
|
Igkv15-103
|
immunoglobulin kappa chain variable 15-103 |
| chr3_+_90520408 | 2.56 |
ENSMUST00000198128.2
|
S100a6
|
S100 calcium binding protein A6 (calcyclin) |
| chr6_+_70700858 | 2.55 |
ENSMUST00000199459.2
ENSMUST00000103409.2 |
Igkj5
|
immunoglobulin kappa joining 5 |
| chr19_+_5927876 | 2.50 |
ENSMUST00000235340.2
|
Slc25a45
|
solute carrier family 25, member 45 |
| chr17_-_28779678 | 2.50 |
ENSMUST00000114785.3
ENSMUST00000025062.5 |
Clps
|
colipase, pancreatic |
| chr8_-_25085654 | 2.48 |
ENSMUST00000110667.8
|
Ido1
|
indoleamine 2,3-dioxygenase 1 |
| chr9_-_50256268 | 2.31 |
ENSMUST00000076364.6
|
Rpl10-ps3
|
ribosomal protein L10, pseudogene 3 |
| chr19_+_17371401 | 2.31 |
ENSMUST00000025617.4
|
Rfk
|
riboflavin kinase |
| chr6_+_70700207 | 2.23 |
ENSMUST00000103407.3
ENSMUST00000199487.2 |
Igkj3
|
immunoglobulin kappa joining 3 |
| chr3_+_90520176 | 2.21 |
ENSMUST00000001051.9
|
S100a6
|
S100 calcium binding protein A6 (calcyclin) |
| chr9_+_45817795 | 2.15 |
ENSMUST00000039059.8
|
Pcsk7
|
proprotein convertase subtilisin/kexin type 7 |
| chr11_+_69882410 | 2.14 |
ENSMUST00000108592.2
|
Gabarap
|
gamma-aminobutyric acid receptor associated protein |
| chr6_+_70699822 | 2.12 |
ENSMUST00000198234.2
ENSMUST00000103406.2 |
Igkj2
|
immunoglobulin kappa joining 2 |
| chr17_-_57535003 | 2.11 |
ENSMUST00000177046.2
ENSMUST00000024988.15 |
C3
|
complement component 3 |
| chr14_+_30271088 | 2.06 |
ENSMUST00000022529.8
|
Tkt
|
transketolase |
| chr12_-_113700190 | 2.05 |
ENSMUST00000103452.3
ENSMUST00000192264.2 |
Ighv5-9-1
|
immunoglobulin heavy variable 5-9-1 |
| chr12_+_112645237 | 2.04 |
ENSMUST00000174780.2
ENSMUST00000169593.2 ENSMUST00000173942.2 |
Zbtb42
|
zinc finger and BTB domain containing 42 |
| chr9_+_45818250 | 1.91 |
ENSMUST00000216672.2
|
Pcsk7
|
proprotein convertase subtilisin/kexin type 7 |
| chr16_+_29884153 | 1.88 |
ENSMUST00000023171.8
|
Hes1
|
hes family bHLH transcription factor 1 |
| chrX_+_138464065 | 1.82 |
ENSMUST00000113027.8
|
Rnf128
|
ring finger protein 128 |
| chr13_+_23966524 | 1.81 |
ENSMUST00000074067.4
|
Trim38
|
tripartite motif-containing 38 |
| chr3_+_118355778 | 1.81 |
ENSMUST00000039177.12
|
Dpyd
|
dihydropyrimidine dehydrogenase |
| chr10_-_23985432 | 1.77 |
ENSMUST00000041180.7
|
Taar9
|
trace amine-associated receptor 9 |
| chr6_-_142517029 | 1.76 |
ENSMUST00000032374.9
|
Kcnj8
|
potassium inwardly-rectifying channel, subfamily J, member 8 |
| chr13_-_74498320 | 1.69 |
ENSMUST00000221594.2
ENSMUST00000022062.8 |
Sdha
|
succinate dehydrogenase complex, subunit A, flavoprotein (Fp) |
| chr3_-_144514386 | 1.67 |
ENSMUST00000197013.2
|
Clca3a2
|
chloride channel accessory 3A2 |
| chr19_+_37423198 | 1.65 |
ENSMUST00000025944.9
|
Hhex
|
hematopoietically expressed homeobox |
| chr9_-_110571645 | 1.62 |
ENSMUST00000006005.12
|
Pth1r
|
parathyroid hormone 1 receptor |
| chr7_+_119160922 | 1.61 |
ENSMUST00000130583.2
ENSMUST00000084647.13 |
Acsm2
|
acyl-CoA synthetase medium-chain family member 2 |
| chr14_-_24054352 | 1.61 |
ENSMUST00000190339.2
|
Kcnma1
|
potassium large conductance calcium-activated channel, subfamily M, alpha member 1 |
| chr3_+_118355811 | 1.56 |
ENSMUST00000149101.3
|
Dpyd
|
dihydropyrimidine dehydrogenase |
| chr17_-_7639378 | 1.56 |
ENSMUST00000231397.2
|
Gm49630
|
predicted gene, 49630 |
| chr6_+_117883783 | 1.53 |
ENSMUST00000177918.8
ENSMUST00000163168.9 |
Hnrnpf
|
heterogeneous nuclear ribonucleoprotein F |
| chr4_+_3940747 | 1.51 |
ENSMUST00000119403.2
|
Chchd7
|
coiled-coil-helix-coiled-coil-helix domain containing 7 |
| chr4_+_139350152 | 1.48 |
ENSMUST00000039818.10
|
Aldh4a1
|
aldehyde dehydrogenase 4 family, member A1 |
| chr6_+_117883732 | 1.48 |
ENSMUST00000179224.8
ENSMUST00000035493.14 |
Hnrnpf
|
heterogeneous nuclear ribonucleoprotein F |
| chr17_+_48096628 | 1.47 |
ENSMUST00000024786.14
|
Tfeb
|
transcription factor EB |
| chr13_+_20978283 | 1.46 |
ENSMUST00000021757.5
ENSMUST00000221982.2 |
Aoah
|
acyloxyacyl hydrolase |
| chr13_-_40887244 | 1.38 |
ENSMUST00000110193.9
|
Tfap2a
|
transcription factor AP-2, alpha |
| chrX_+_162873183 | 1.34 |
ENSMUST00000015545.10
|
Cltrn
|
collectrin, amino acid transport regulator |
| chr12_+_85157607 | 1.33 |
ENSMUST00000053811.10
|
Dlst
|
dihydrolipoamide S-succinyltransferase (E2 component of 2-oxo-glutarate complex) |
| chr12_-_114321838 | 1.32 |
ENSMUST00000125484.3
|
Ighv13-2
|
immunoglobulin heavy variable 13-2 |
| chr10_+_120044650 | 1.32 |
ENSMUST00000020446.11
ENSMUST00000134797.8 |
Tmbim4
|
transmembrane BAX inhibitor motif containing 4 |
| chr2_-_9888804 | 1.29 |
ENSMUST00000114915.3
|
9230102O04Rik
|
RIKEN cDNA 9230102O04 gene |
| chr8_-_94739469 | 1.22 |
ENSMUST00000053766.14
|
Amfr
|
autocrine motility factor receptor |
| chr15_+_58761057 | 1.19 |
ENSMUST00000036904.7
|
Rnf139
|
ring finger protein 139 |
| chr12_-_115031622 | 1.14 |
ENSMUST00000194257.2
|
Ighv8-5
|
immunoglobulin heavy variable V8-5 |
| chr9_+_48406706 | 1.12 |
ENSMUST00000048824.9
|
Gm5617
|
predicted gene 5617 |
| chr5_-_89583469 | 1.12 |
ENSMUST00000200534.2
|
Gc
|
vitamin D binding protein |
| chr2_+_28730418 | 1.11 |
ENSMUST00000113853.3
|
Ddx31
|
DEAD/H box helicase 31 |
| chr14_-_51295099 | 1.11 |
ENSMUST00000227764.2
|
Rnase12
|
ribonuclease, RNase A family, 12 (non-active) |
| chr1_-_85888729 | 1.11 |
ENSMUST00000086975.7
|
Gpr55
|
G protein-coupled receptor 55 |
| chr6_-_113717689 | 1.08 |
ENSMUST00000032440.6
|
Sec13
|
SEC13 homolog, nuclear pore and COPII coat complex component |
| chr2_-_164063525 | 1.07 |
ENSMUST00000018355.11
ENSMUST00000109376.9 |
Wfdc15b
|
WAP four-disulfide core domain 15B |
| chr8_+_3671599 | 1.06 |
ENSMUST00000207389.2
|
Pet100
|
PET100 homolog |
| chr8_+_3671528 | 1.05 |
ENSMUST00000156380.4
|
Pet100
|
PET100 homolog |
| chr17_+_26048010 | 1.03 |
ENSMUST00000026832.14
|
Jmjd8
|
jumonji domain containing 8 |
| chr14_+_44340111 | 1.01 |
ENSMUST00000074839.7
|
Ear2
|
eosinophil-associated, ribonuclease A family, member 2 |
| chr6_-_142517340 | 1.01 |
ENSMUST00000203945.3
|
Kcnj8
|
potassium inwardly-rectifying channel, subfamily J, member 8 |
| chr3_+_87343127 | 0.98 |
ENSMUST00000166297.7
ENSMUST00000049926.15 ENSMUST00000178261.3 |
Fcrl5
|
Fc receptor-like 5 |
| chr2_+_121859025 | 0.93 |
ENSMUST00000028668.8
|
Eif3j1
|
eukaryotic translation initiation factor 3, subunit J1 |
| chr16_-_10608766 | 0.93 |
ENSMUST00000050864.7
|
Prm3
|
protamine 3 |
| chr1_-_192880260 | 0.92 |
ENSMUST00000161367.2
|
Traf3ip3
|
TRAF3 interacting protein 3 |
| chr16_+_91188609 | 0.92 |
ENSMUST00000160764.2
|
Gm21970
|
predicted gene 21970 |
| chr19_-_18609118 | 0.91 |
ENSMUST00000025631.7
ENSMUST00000236615.2 |
Ostf1
|
osteoclast stimulating factor 1 |
| chr11_-_32150222 | 0.89 |
ENSMUST00000145401.8
ENSMUST00000142396.2 ENSMUST00000128311.8 |
Il9r
|
interleukin 9 receptor |
| chr3_+_129630380 | 0.88 |
ENSMUST00000077918.7
|
Cfi
|
complement component factor i |
| chr19_-_5416339 | 0.88 |
ENSMUST00000170010.3
|
Banf1
|
BAF nuclear assembly factor 1 |
| chr14_-_24054186 | 0.86 |
ENSMUST00000188991.7
ENSMUST00000224468.2 |
Kcnma1
|
potassium large conductance calcium-activated channel, subfamily M, alpha member 1 |
| chr16_+_45430743 | 0.86 |
ENSMUST00000161347.9
ENSMUST00000023339.5 |
Gcsam
|
germinal center associated, signaling and motility |
| chr13_+_92442488 | 0.85 |
ENSMUST00000188317.2
|
Gm20379
|
predicted gene, 20379 |
| chr2_+_153583194 | 0.85 |
ENSMUST00000028981.9
|
Mapre1
|
microtubule-associated protein, RP/EB family, member 1 |
| chr3_+_87343071 | 0.84 |
ENSMUST00000194102.6
|
Fcrl5
|
Fc receptor-like 5 |
| chr9_-_104140099 | 0.83 |
ENSMUST00000035170.13
|
Dnajc13
|
DnaJ heat shock protein family (Hsp40) member C13 |
| chr12_-_113393106 | 0.83 |
ENSMUST00000192746.2
ENSMUST00000103429.2 |
Ighj2
|
immunoglobulin heavy joining 2 |
| chr4_+_45890303 | 0.83 |
ENSMUST00000178561.8
|
Ccdc180
|
coiled-coil domain containing 180 |
| chr18_+_60515755 | 0.82 |
ENSMUST00000237185.2
|
Iigp1
|
interferon inducible GTPase 1 |
| chr1_+_90880830 | 0.81 |
ENSMUST00000166281.3
|
Prlh
|
prolactin releasing hormone |
| chr16_-_58712421 | 0.78 |
ENSMUST00000206523.3
ENSMUST00000215032.2 |
Olfr178
|
olfactory receptor 178 |
| chr6_+_91450677 | 0.76 |
ENSMUST00000032183.6
|
Tmem43
|
transmembrane protein 43 |
| chr6_+_70700522 | 0.74 |
ENSMUST00000103408.2
|
Igkj4
|
immunoglobulin kappa joining 4 |
| chr16_-_18232202 | 0.74 |
ENSMUST00000165430.8
ENSMUST00000147720.3 |
Comt
|
catechol-O-methyltransferase |
| chr17_+_6926452 | 0.73 |
ENSMUST00000097430.10
|
Sytl3
|
synaptotagmin-like 3 |
| chr9_+_108539296 | 0.72 |
ENSMUST00000035222.6
|
Slc25a20
|
solute carrier family 25 (mitochondrial carnitine/acylcarnitine translocase), member 20 |
| chr7_+_126895423 | 0.72 |
ENSMUST00000117762.8
|
Itgal
|
integrin alpha L |
| chr6_-_42574595 | 0.71 |
ENSMUST00000134707.2
|
Tcaf3
|
TRPM8 channel-associated factor 3 |
| chr5_-_145406533 | 0.70 |
ENSMUST00000031633.5
|
Cyp3a16
|
cytochrome P450, family 3, subfamily a, polypeptide 16 |
| chr2_-_5719302 | 0.69 |
ENSMUST00000044009.14
|
Camk1d
|
calcium/calmodulin-dependent protein kinase ID |
| chr9_-_39465349 | 0.69 |
ENSMUST00000215505.2
ENSMUST00000217227.2 |
Olfr958
|
olfactory receptor 958 |
| chr3_-_19682833 | 0.69 |
ENSMUST00000119133.2
|
1700064H15Rik
|
RIKEN cDNA 1700064H15 gene |
| chr3_+_87343105 | 0.68 |
ENSMUST00000193229.6
|
Fcrl5
|
Fc receptor-like 5 |
| chr9_+_49014020 | 0.68 |
ENSMUST00000070390.12
ENSMUST00000167095.8 |
Tmprss5
|
transmembrane protease, serine 5 (spinesin) |
| chr9_+_7347369 | 0.68 |
ENSMUST00000005950.12
ENSMUST00000065079.6 |
Mmp12
|
matrix metallopeptidase 12 |
| chr6_+_122929627 | 0.68 |
ENSMUST00000204427.2
|
Clec4a3
|
C-type lectin domain family 4, member a3 |
| chr10_+_43777777 | 0.67 |
ENSMUST00000054418.12
|
Rtn4ip1
|
reticulon 4 interacting protein 1 |
| chr2_+_172090412 | 0.67 |
ENSMUST00000038532.2
|
Mc3r
|
melanocortin 3 receptor |
| chr1_+_20801127 | 0.67 |
ENSMUST00000027061.5
|
Il17a
|
interleukin 17A |
| chr3_-_92393193 | 0.66 |
ENSMUST00000054599.8
|
Sprr1a
|
small proline-rich protein 1A |
| chr16_-_30129591 | 0.66 |
ENSMUST00000061190.8
|
Gp5
|
glycoprotein 5 (platelet) |
| chr17_+_8454939 | 0.65 |
ENSMUST00000164411.10
|
Ccr6
|
chemokine (C-C motif) receptor 6 |
| chr6_-_42574306 | 0.65 |
ENSMUST00000069023.4
|
Tcaf3
|
TRPM8 channel-associated factor 3 |
| chr19_-_16851169 | 0.64 |
ENSMUST00000072915.4
|
Foxb2
|
forkhead box B2 |
| chr7_+_126895463 | 0.64 |
ENSMUST00000106306.9
ENSMUST00000120857.8 |
Itgal
|
integrin alpha L |
| chr6_-_59001455 | 0.63 |
ENSMUST00000089860.12
|
Fam13a
|
family with sequence similarity 13, member A |
| chr9_-_99592116 | 0.63 |
ENSMUST00000035048.12
|
Cldn18
|
claudin 18 |
| chr3_-_92496293 | 0.62 |
ENSMUST00000098888.7
|
Smcp
|
sperm mitochondria-associated cysteine-rich protein |
| chr2_-_89583170 | 0.61 |
ENSMUST00000099765.2
|
Olfr1253
|
olfactory receptor 1253 |
| chrX_-_7185424 | 0.60 |
ENSMUST00000115746.8
|
Clcn5
|
chloride channel, voltage-sensitive 5 |
| chr17_+_8454862 | 0.59 |
ENSMUST00000231340.2
|
Ccr6
|
chemokine (C-C motif) receptor 6 |
| chr11_-_99884818 | 0.58 |
ENSMUST00000105049.2
|
Krtap17-1
|
keratin associated protein 17-1 |
| chrM_+_8603 | 0.57 |
ENSMUST00000082409.1
|
mt-Co3
|
mitochondrially encoded cytochrome c oxidase III |
| chr11_-_102209767 | 0.57 |
ENSMUST00000174302.8
ENSMUST00000178839.8 ENSMUST00000006754.14 |
Ubtf
|
upstream binding transcription factor, RNA polymerase I |
| chr5_-_87240405 | 0.56 |
ENSMUST00000132667.2
ENSMUST00000145617.8 ENSMUST00000094649.11 |
Ugt2b36
|
UDP glucuronosyltransferase 2 family, polypeptide B36 |
| chr15_-_60793115 | 0.56 |
ENSMUST00000096418.5
|
A1bg
|
alpha-1-B glycoprotein |
| chrX_-_52298601 | 0.55 |
ENSMUST00000174390.2
|
Plac1
|
placental specific protein 1 |
| chr7_+_44358144 | 0.54 |
ENSMUST00000085422.4
|
Izumo2
|
IZUMO family member 2 |
| chr2_-_19558719 | 0.54 |
ENSMUST00000062060.5
|
4921504E06Rik
|
RIKEN cDNA 4921504E06 gene |
| chr7_+_127802769 | 0.54 |
ENSMUST00000177383.8
|
Itgad
|
integrin, alpha D |
| chrX_-_7185529 | 0.53 |
ENSMUST00000128319.2
|
Clcn5
|
chloride channel, voltage-sensitive 5 |
| chrY_+_6268725 | 0.53 |
ENSMUST00000179410.2
|
Gm20873
|
predicted gene, 20873 |
| chr9_-_35179042 | 0.53 |
ENSMUST00000217306.2
ENSMUST00000125087.2 ENSMUST00000121564.8 ENSMUST00000063782.12 ENSMUST00000059057.14 |
Fam118b
|
family with sequence similarity 118, member B |
| chrY_-_6681243 | 0.53 |
ENSMUST00000115940.2
|
Gm21719
|
predicted gene, 21719 |
| chr8_+_46428551 | 0.51 |
ENSMUST00000034051.7
ENSMUST00000150943.2 |
Ufsp2
|
UFM1-specific peptidase 2 |
| chr12_+_52144511 | 0.51 |
ENSMUST00000040090.16
|
Nubpl
|
nucleotide binding protein-like |
| chr9_-_99592058 | 0.51 |
ENSMUST00000136429.8
|
Cldn18
|
claudin 18 |
| chr17_-_37511885 | 0.50 |
ENSMUST00000222190.2
|
Olfr94
|
olfactory receptor 94 |
| chr8_-_46533321 | 0.50 |
ENSMUST00000177186.2
|
Snx25
|
sorting nexin 25 |
| chr4_+_129355374 | 0.50 |
ENSMUST00000048162.10
ENSMUST00000138013.3 |
Bsdc1
|
BSD domain containing 1 |
| chr8_+_70583971 | 0.49 |
ENSMUST00000211898.2
ENSMUST00000095273.7 |
Nr2c2ap
|
nuclear receptor 2C2-associated protein |
| chr7_+_126895531 | 0.49 |
ENSMUST00000170971.8
|
Itgal
|
integrin alpha L |
| chr8_+_9914251 | 0.49 |
ENSMUST00000088080.3
|
Gm10217
|
predicted gene 10217 |
| chrY_+_5944438 | 0.49 |
ENSMUST00000178779.2
|
Gm21778
|
predicted gene, 21778 |
| chrX_-_37131983 | 0.48 |
ENSMUST00000089060.5
|
Btg1c
|
BTG anti-proliferation factor 1C |
| chr14_+_55842002 | 0.48 |
ENSMUST00000138037.2
|
Irf9
|
interferon regulatory factor 9 |
| chrX_+_106711562 | 0.48 |
ENSMUST00000168174.9
|
Tbx22
|
T-box 22 |
| chr6_-_90013823 | 0.47 |
ENSMUST00000073415.2
|
Vmn1r48
|
vomeronasal 1 receptor 48 |
| chr19_+_13211616 | 0.47 |
ENSMUST00000064102.2
|
Olfr1463
|
olfactory receptor 1463 |
| chr10_+_78261503 | 0.47 |
ENSMUST00000005185.8
|
Cstb
|
cystatin B |
| chr7_+_119803181 | 0.46 |
ENSMUST00000084640.3
|
Abca14
|
ATP-binding cassette, sub-family A (ABC1), member 14 |
| chrX_-_37107692 | 0.46 |
ENSMUST00000184432.3
|
Btg1b
|
BTG anti-proliferation factor 1B |
| chr6_+_57380765 | 0.45 |
ENSMUST00000089830.4
|
Vmn1r19
|
vomeronasal 1 receptor 19 |
| chr16_+_88555732 | 0.45 |
ENSMUST00000089111.5
|
2310034C09Rik
|
RIKEN cDNA 2310034C09 gene |
| chr3_+_93107517 | 0.44 |
ENSMUST00000098884.4
|
Flg2
|
filaggrin family member 2 |
| chr7_-_80994933 | 0.44 |
ENSMUST00000080813.5
|
Rps17
|
ribosomal protein S17 |
| chrY_+_13163582 | 0.44 |
ENSMUST00000180318.2
|
Gm21425
|
predicted gene, 21425 |
| chr19_-_6002210 | 0.43 |
ENSMUST00000236013.2
|
Pola2
|
polymerase (DNA directed), alpha 2 |
| chr10_-_129948657 | 0.42 |
ENSMUST00000081469.2
|
Olfr823
|
olfactory receptor 823 |
| chr4_+_125997788 | 0.42 |
ENSMUST00000139524.2
|
Stk40
|
serine/threonine kinase 40 |
| chrX_-_166165743 | 0.42 |
ENSMUST00000026839.5
|
Prps2
|
phosphoribosyl pyrophosphate synthetase 2 |
| chrY_+_5658985 | 0.42 |
ENSMUST00000179901.2
|
Gm21854
|
predicted gene, 21854 |
| chr1_+_161222980 | 0.42 |
ENSMUST00000028024.5
|
Tnfsf4
|
tumor necrosis factor (ligand) superfamily, member 4 |
| chr9_-_110483210 | 0.41 |
ENSMUST00000196488.5
ENSMUST00000133191.8 ENSMUST00000167320.8 |
Nbeal2
|
neurobeachin-like 2 |
| chrY_-_10145559 | 0.41 |
ENSMUST00000180130.2
|
Gm21310
|
predicted gene, 21310 |
| chr11_+_67689094 | 0.40 |
ENSMUST00000168612.8
|
Dhrs7c
|
dehydrogenase/reductase (SDR family) member 7C |
| chr19_+_7534838 | 0.40 |
ENSMUST00000141887.8
ENSMUST00000136756.2 |
Plaat3
|
phospholipase A and acyltransferase 3 |
| chrY_-_10534428 | 0.40 |
ENSMUST00000179665.2
|
Gm20737
|
predicted gene, 20737 |
| chr14_-_24054273 | 0.40 |
ENSMUST00000188285.7
ENSMUST00000190044.7 |
Kcnma1
|
potassium large conductance calcium-activated channel, subfamily M, alpha member 1 |
| chr1_+_187995096 | 0.39 |
ENSMUST00000060479.14
|
Ush2a
|
usherin |
| chrY_+_18090734 | 0.39 |
ENSMUST00000180056.2
|
Ssty1
|
spermiogenesis specific transcript on the Y 1 |
| chr7_-_142253247 | 0.38 |
ENSMUST00000105934.8
|
Ins2
|
insulin II |
| chrX_-_166047289 | 0.38 |
ENSMUST00000133722.2
|
Tlr8
|
toll-like receptor 8 |
| chr19_+_18609343 | 0.38 |
ENSMUST00000159572.9
|
Nmrk1
|
nicotinamide riboside kinase 1 |
| chr19_-_12067806 | 0.38 |
ENSMUST00000219996.2
|
Olfr1426
|
olfactory receptor 1426 |
| chr4_-_137493785 | 0.38 |
ENSMUST00000139951.8
|
Alpl
|
alkaline phosphatase, liver/bone/kidney |
| chr14_+_54405047 | 0.38 |
ENSMUST00000103693.2
|
Traj50
|
T cell receptor alpha joining 50 |
| chr6_-_59001325 | 0.38 |
ENSMUST00000173193.2
|
Fam13a
|
family with sequence similarity 13, member A |
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 3.0 | 9.0 | GO:0018879 | biphenyl metabolic process(GO:0018879) |
| 1.5 | 4.5 | GO:0061623 | glycolytic process from galactose(GO:0061623) |
| 1.1 | 3.4 | GO:0006667 | sphinganine metabolic process(GO:0006667) |
| 1.1 | 3.4 | GO:0019860 | uracil catabolic process(GO:0006212) uracil metabolic process(GO:0019860) |
| 1.0 | 3.1 | GO:2000118 | regulation of sodium-dependent phosphate transport(GO:2000118) |
| 0.8 | 2.5 | GO:0036269 | swimming behavior(GO:0036269) |
| 0.8 | 2.3 | GO:0042726 | flavin-containing compound metabolic process(GO:0042726) |
| 0.7 | 2.1 | GO:0046166 | glyceraldehyde-3-phosphate biosynthetic process(GO:0046166) |
| 0.6 | 1.9 | GO:0042668 | trochlear nerve development(GO:0021558) auditory receptor cell fate determination(GO:0042668) negative regulation of forebrain neuron differentiation(GO:2000978) |
| 0.5 | 2.1 | GO:0001970 | positive regulation of activation of membrane attack complex(GO:0001970) |
| 0.5 | 5.5 | GO:0052697 | flavonoid glucuronidation(GO:0052696) xenobiotic glucuronidation(GO:0052697) |
| 0.5 | 1.5 | GO:0010133 | proline catabolic process to glutamate(GO:0010133) |
| 0.5 | 3.2 | GO:0071492 | cellular response to UV-A(GO:0071492) |
| 0.4 | 1.2 | GO:1904155 | DN2 thymocyte differentiation(GO:1904155) DN3 thymocyte differentiation(GO:1904156) |
| 0.4 | 1.5 | GO:1902477 | defense response to bacterium, incompatible interaction(GO:0009816) regulation of defense response to bacterium, incompatible interaction(GO:1902477) |
| 0.3 | 1.7 | GO:0006121 | mitochondrial electron transport, succinate to ubiquinone(GO:0006121) |
| 0.3 | 0.9 | GO:0019043 | establishment of viral latency(GO:0019043) |
| 0.3 | 2.8 | GO:1990573 | potassium ion import across plasma membrane(GO:1990573) |
| 0.3 | 1.4 | GO:0003409 | optic cup structural organization(GO:0003409) oculomotor nerve development(GO:0021557) oculomotor nerve morphogenesis(GO:0021622) oculomotor nerve formation(GO:0021623) |
| 0.3 | 1.7 | GO:0061017 | hepatoblast differentiation(GO:0061017) |
| 0.3 | 1.6 | GO:0036112 | medium-chain fatty-acyl-CoA metabolic process(GO:0036112) |
| 0.3 | 5.5 | GO:1990001 | inhibition of cysteine-type endopeptidase activity involved in apoptotic process(GO:1990001) |
| 0.3 | 1.6 | GO:0034465 | response to carbon monoxide(GO:0034465) eye blink reflex(GO:0060082) |
| 0.3 | 3.3 | GO:0016554 | cytidine to uridine editing(GO:0016554) |
| 0.2 | 1.1 | GO:0060267 | positive regulation of respiratory burst(GO:0060267) |
| 0.2 | 0.8 | GO:1903031 | regulation of microtubule plus-end binding(GO:1903031) positive regulation of microtubule plus-end binding(GO:1903033) |
| 0.2 | 0.4 | GO:0043382 | positive regulation of memory T cell differentiation(GO:0043382) |
| 0.2 | 1.8 | GO:0002291 | T cell activation via T cell receptor contact with antigen bound to MHC molecule on antigen presenting cell(GO:0002291) |
| 0.2 | 2.5 | GO:0050859 | negative regulation of B cell receptor signaling pathway(GO:0050859) |
| 0.2 | 6.2 | GO:0019731 | antibacterial humoral response(GO:0019731) |
| 0.2 | 1.8 | GO:0046598 | positive regulation of viral entry into host cell(GO:0046598) |
| 0.2 | 1.1 | GO:2001205 | negative regulation of osteoclast development(GO:2001205) |
| 0.2 | 11.8 | GO:0050829 | defense response to Gram-negative bacterium(GO:0050829) |
| 0.1 | 0.7 | GO:0031335 | regulation of sulfur amino acid metabolic process(GO:0031335) negative regulation of catecholamine metabolic process(GO:0045914) negative regulation of dopamine metabolic process(GO:0045963) |
| 0.1 | 0.8 | GO:0002023 | reduction of food intake in response to dietary excess(GO:0002023) |
| 0.1 | 2.1 | GO:0006995 | cellular response to nitrogen starvation(GO:0006995) cellular response to nitrogen levels(GO:0043562) |
| 0.1 | 8.7 | GO:0006805 | xenobiotic metabolic process(GO:0006805) |
| 0.1 | 0.9 | GO:2000402 | negative regulation of lymphocyte migration(GO:2000402) |
| 0.1 | 0.7 | GO:0032747 | positive regulation of interleukin-23 production(GO:0032747) |
| 0.1 | 0.3 | GO:2000729 | positive regulation of mesenchymal cell proliferation involved in ureter development(GO:2000729) |
| 0.1 | 0.5 | GO:0045356 | positive regulation of interferon-alpha biosynthetic process(GO:0045356) |
| 0.1 | 0.5 | GO:0030576 | Cajal body organization(GO:0030576) |
| 0.1 | 0.4 | GO:0006015 | 5-phosphoribose 1-diphosphate biosynthetic process(GO:0006015) 5-phosphoribose 1-diphosphate metabolic process(GO:0046391) |
| 0.1 | 0.6 | GO:0090214 | spongiotrophoblast layer developmental growth(GO:0090214) |
| 0.1 | 0.3 | GO:0044206 | CMP salvage(GO:0006238) CMP biosynthetic process(GO:0009224) pyrimidine ribonucleotide salvage(GO:0010138) pyrimidine nucleotide salvage(GO:0032262) UMP salvage(GO:0044206) CMP metabolic process(GO:0046035) |
| 0.1 | 1.6 | GO:0060732 | positive regulation of inositol phosphate biosynthetic process(GO:0060732) |
| 0.1 | 1.1 | GO:0090110 | cargo loading into COPII-coated vesicle(GO:0090110) |
| 0.1 | 1.4 | GO:0033617 | mitochondrial respiratory chain complex IV assembly(GO:0033617) |
| 0.1 | 0.6 | GO:0007341 | penetration of zona pellucida(GO:0007341) |
| 0.1 | 0.8 | GO:0071763 | nuclear membrane organization(GO:0071763) |
| 0.1 | 0.7 | GO:0060054 | positive regulation of epithelial cell proliferation involved in wound healing(GO:0060054) |
| 0.1 | 0.6 | GO:0061502 | early endosome to recycling endosome transport(GO:0061502) |
| 0.1 | 0.5 | GO:0045351 | type I interferon biosynthetic process(GO:0045351) |
| 0.1 | 1.2 | GO:0060628 | regulation of ER to Golgi vesicle-mediated transport(GO:0060628) |
| 0.1 | 0.7 | GO:0015879 | carnitine transport(GO:0015879) |
| 0.1 | 20.7 | GO:0042742 | defense response to bacterium(GO:0042742) |
| 0.1 | 0.2 | GO:0060734 | regulation of endoplasmic reticulum stress-induced eIF2 alpha phosphorylation(GO:0060734) |
| 0.1 | 1.1 | GO:0042359 | vitamin D metabolic process(GO:0042359) |
| 0.1 | 0.4 | GO:0070889 | platelet alpha granule organization(GO:0070889) |
| 0.1 | 0.6 | GO:0006362 | transcription elongation from RNA polymerase I promoter(GO:0006362) |
| 0.1 | 0.4 | GO:0048496 | maintenance of organ identity(GO:0048496) |
| 0.1 | 0.3 | GO:0000738 | DNA catabolic process, exonucleolytic(GO:0000738) |
| 0.1 | 1.8 | GO:0042036 | negative regulation of cytokine biosynthetic process(GO:0042036) |
| 0.1 | 0.3 | GO:0009992 | cellular water homeostasis(GO:0009992) |
| 0.0 | 2.5 | GO:0032094 | response to food(GO:0032094) |
| 0.0 | 11.8 | GO:0006694 | steroid biosynthetic process(GO:0006694) |
| 0.0 | 1.3 | GO:0006103 | 2-oxoglutarate metabolic process(GO:0006103) |
| 0.0 | 0.1 | GO:1990167 | protein K27-linked deubiquitination(GO:1990167) |
| 0.0 | 0.1 | GO:0006550 | isoleucine metabolic process(GO:0006549) isoleucine catabolic process(GO:0006550) |
| 0.0 | 1.1 | GO:0045671 | negative regulation of osteoclast differentiation(GO:0045671) |
| 0.0 | 2.8 | GO:1902476 | chloride transmembrane transport(GO:1902476) |
| 0.0 | 0.7 | GO:0010544 | negative regulation of platelet activation(GO:0010544) |
| 0.0 | 0.3 | GO:0048743 | positive regulation of skeletal muscle fiber development(GO:0048743) |
| 0.0 | 0.2 | GO:2000434 | regulation of protein neddylation(GO:2000434) negative regulation of protein neddylation(GO:2000435) |
| 0.0 | 0.9 | GO:0006958 | complement activation, classical pathway(GO:0006958) |
| 0.0 | 0.2 | GO:0071985 | multivesicular body sorting pathway(GO:0071985) |
| 0.0 | 0.5 | GO:0060394 | negative regulation of pathway-restricted SMAD protein phosphorylation(GO:0060394) |
| 0.0 | 0.7 | GO:0045475 | locomotor rhythm(GO:0045475) |
| 0.0 | 0.3 | GO:0045617 | negative regulation of keratinocyte differentiation(GO:0045617) |
| 0.0 | 0.1 | GO:0001712 | ectodermal cell fate commitment(GO:0001712) |
| 0.0 | 0.9 | GO:0030261 | chromosome condensation(GO:0030261) |
| 0.0 | 0.6 | GO:0019363 | pyridine nucleotide biosynthetic process(GO:0019363) |
| 0.0 | 1.1 | GO:0010501 | RNA secondary structure unwinding(GO:0010501) |
| 0.0 | 0.3 | GO:0001778 | plasma membrane repair(GO:0001778) |
| 0.0 | 0.2 | GO:0042904 | 9-cis-retinoic acid biosynthetic process(GO:0042904) 9-cis-retinoic acid metabolic process(GO:0042905) |
| 0.0 | 0.7 | GO:0031424 | keratinization(GO:0031424) |
| 0.0 | 0.1 | GO:0048631 | regulation of skeletal muscle tissue growth(GO:0048631) |
| 0.0 | 0.2 | GO:0006646 | phosphatidylethanolamine biosynthetic process(GO:0006646) |
| 0.0 | 0.4 | GO:0000028 | ribosomal small subunit assembly(GO:0000028) |
| 0.0 | 0.7 | GO:0048791 | calcium ion-regulated exocytosis of neurotransmitter(GO:0048791) |
| 0.0 | 0.5 | GO:0070584 | mitochondrion morphogenesis(GO:0070584) |
| 0.0 | 3.8 | GO:0002377 | immunoglobulin production(GO:0002377) |
| 0.0 | 1.2 | GO:0051865 | protein autoubiquitination(GO:0051865) |
| 0.0 | 0.5 | GO:0033146 | regulation of intracellular estrogen receptor signaling pathway(GO:0033146) |
| 0.0 | 0.1 | GO:0006421 | asparaginyl-tRNA aminoacylation(GO:0006421) |
| 0.0 | 1.5 | GO:0050728 | negative regulation of inflammatory response(GO:0050728) |
| 0.0 | 0.5 | GO:0050798 | activated T cell proliferation(GO:0050798) |
| 0.0 | 0.2 | GO:0030259 | lipid glycosylation(GO:0030259) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 1.7 | GO:0045283 | mitochondrial respiratory chain complex II, succinate dehydrogenase complex (ubiquinone)(GO:0005749) succinate dehydrogenase complex (ubiquinone)(GO:0045257) respiratory chain complex II(GO:0045273) succinate dehydrogenase complex(GO:0045281) fumarate reductase complex(GO:0045283) |
| 0.4 | 1.8 | GO:0034687 | integrin alphaL-beta2 complex(GO:0034687) |
| 0.3 | 2.8 | GO:0008282 | ATP-sensitive potassium channel complex(GO:0008282) |
| 0.3 | 2.5 | GO:0030485 | smooth muscle contractile fiber(GO:0030485) |
| 0.1 | 2.4 | GO:0036513 | Derlin-1 retrotranslocation complex(GO:0036513) |
| 0.1 | 0.4 | GO:0014801 | longitudinal sarcoplasmic reticulum(GO:0014801) |
| 0.1 | 1.3 | GO:0045252 | oxoglutarate dehydrogenase complex(GO:0045252) |
| 0.1 | 12.5 | GO:0005758 | mitochondrial intermembrane space(GO:0005758) |
| 0.1 | 3.4 | GO:0033017 | sarcoplasmic reticulum membrane(GO:0033017) |
| 0.1 | 3.4 | GO:0031528 | microvillus membrane(GO:0031528) |
| 0.1 | 3.4 | GO:0097225 | sperm midpiece(GO:0097225) |
| 0.1 | 0.4 | GO:0002189 | ribose phosphate diphosphokinase complex(GO:0002189) |
| 0.1 | 1.1 | GO:0061700 | GATOR2 complex(GO:0061700) |
| 0.1 | 0.4 | GO:0060171 | stereocilium membrane(GO:0060171) |
| 0.1 | 1.6 | GO:0048787 | presynaptic active zone membrane(GO:0048787) |
| 0.1 | 0.8 | GO:0030981 | cortical microtubule cytoskeleton(GO:0030981) |
| 0.1 | 0.3 | GO:0042584 | chromaffin granule membrane(GO:0042584) |
| 0.1 | 4.1 | GO:0030173 | integral component of Golgi membrane(GO:0030173) |
| 0.1 | 0.4 | GO:0005658 | alpha DNA polymerase:primase complex(GO:0005658) |
| 0.1 | 0.8 | GO:0020005 | symbiont-containing vacuole membrane(GO:0020005) |
| 0.1 | 0.8 | GO:0071203 | WASH complex(GO:0071203) |
| 0.0 | 0.9 | GO:0005852 | eukaryotic translation initiation factor 3 complex(GO:0005852) |
| 0.0 | 0.4 | GO:0065010 | extracellular membrane-bounded organelle(GO:0065010) |
| 0.0 | 4.7 | GO:0031526 | brush border membrane(GO:0031526) |
| 0.0 | 0.8 | GO:0005639 | integral component of nuclear inner membrane(GO:0005639) intrinsic component of nuclear inner membrane(GO:0031229) |
| 0.0 | 6.9 | GO:0031225 | anchored component of membrane(GO:0031225) |
| 0.0 | 0.3 | GO:0000127 | transcription factor TFIIIC complex(GO:0000127) |
| 0.0 | 3.7 | GO:0042571 | immunoglobulin complex, circulating(GO:0042571) |
| 0.0 | 0.9 | GO:0000786 | nucleosome(GO:0000786) |
| 0.0 | 3.2 | GO:0005903 | brush border(GO:0005903) |
| 0.0 | 0.2 | GO:0032585 | multivesicular body membrane(GO:0032585) |
| 0.0 | 0.3 | GO:0042788 | polysomal ribosome(GO:0042788) |
| 0.0 | 2.9 | GO:0071013 | catalytic step 2 spliceosome(GO:0071013) |
| 0.0 | 2.9 | GO:0072562 | blood microparticle(GO:0072562) |
| 0.0 | 3.5 | GO:0031234 | extrinsic component of cytoplasmic side of plasma membrane(GO:0031234) |
| 0.0 | 5.2 | GO:0016323 | basolateral plasma membrane(GO:0016323) |
| 0.0 | 0.7 | GO:0019897 | extrinsic component of plasma membrane(GO:0019897) |
| 0.0 | 0.1 | GO:0008623 | CHRAC(GO:0008623) |
| 0.0 | 7.4 | GO:0005743 | mitochondrial inner membrane(GO:0005743) |
| 0.0 | 0.4 | GO:0005732 | small nucleolar ribonucleoprotein complex(GO:0005732) |
| 0.0 | 0.2 | GO:0000815 | ESCRT III complex(GO:0000815) |
| 0.0 | 0.7 | GO:0001533 | cornified envelope(GO:0001533) |
| 0.0 | 1.3 | GO:0005795 | Golgi stack(GO:0005795) |
| 0.0 | 2.7 | GO:0090575 | RNA polymerase II transcription factor complex(GO:0090575) |
| 0.0 | 24.7 | GO:0005615 | extracellular space(GO:0005615) |
| 0.0 | 0.2 | GO:0000164 | protein phosphatase type 1 complex(GO:0000164) |
| 0.0 | 2.1 | GO:0043209 | myelin sheath(GO:0043209) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.8 | 12.5 | GO:0004769 | steroid delta-isomerase activity(GO:0004769) |
| 1.1 | 3.4 | GO:0002061 | uracil binding(GO:0002058) pyrimidine nucleobase binding(GO:0002061) |
| 0.8 | 3.4 | GO:0042284 | sphingolipid delta-4 desaturase activity(GO:0042284) |
| 0.8 | 2.5 | GO:0033754 | indoleamine 2,3-dioxygenase activity(GO:0033754) |
| 0.7 | 2.1 | GO:0004802 | transketolase activity(GO:0004802) |
| 0.6 | 1.9 | GO:0071820 | N-box binding(GO:0071820) |
| 0.6 | 3.1 | GO:0015321 | sodium-dependent phosphate transmembrane transporter activity(GO:0015321) |
| 0.6 | 1.7 | GO:0008177 | succinate dehydrogenase (ubiquinone) activity(GO:0008177) |
| 0.4 | 9.0 | GO:0015643 | toxic substance binding(GO:0015643) |
| 0.4 | 1.8 | GO:0030369 | ICAM-3 receptor activity(GO:0030369) |
| 0.3 | 2.8 | GO:0015272 | ATP-activated inward rectifier potassium channel activity(GO:0015272) |
| 0.3 | 1.3 | GO:0016748 | succinyltransferase activity(GO:0016748) |
| 0.3 | 2.6 | GO:0017040 | ceramidase activity(GO:0017040) |
| 0.3 | 8.7 | GO:0004033 | aldo-keto reductase (NADP) activity(GO:0004033) |
| 0.3 | 1.1 | GO:1902271 | D3 vitamins binding(GO:1902271) |
| 0.3 | 1.1 | GO:0004949 | cannabinoid receptor activity(GO:0004949) |
| 0.3 | 4.5 | GO:0016857 | racemase and epimerase activity, acting on carbohydrates and derivatives(GO:0016857) |
| 0.2 | 0.7 | GO:0016206 | catechol O-methyltransferase activity(GO:0016206) |
| 0.2 | 1.2 | GO:1904288 | BAT3 complex binding(GO:1904288) |
| 0.2 | 1.6 | GO:0004991 | parathyroid hormone receptor activity(GO:0004991) |
| 0.2 | 0.7 | GO:0004980 | melanocyte-stimulating hormone receptor activity(GO:0004980) |
| 0.2 | 4.8 | GO:0044548 | S100 protein binding(GO:0044548) |
| 0.2 | 0.9 | GO:0045159 | myosin II binding(GO:0045159) |
| 0.2 | 1.6 | GO:0003996 | acyl-CoA ligase activity(GO:0003996) |
| 0.2 | 0.6 | GO:0050262 | ribosylnicotinamide kinase activity(GO:0050262) ribosylnicotinate kinase activity(GO:0061769) |
| 0.2 | 1.6 | GO:0060072 | large conductance calcium-activated potassium channel activity(GO:0060072) |
| 0.2 | 1.8 | GO:0001594 | trace-amine receptor activity(GO:0001594) |
| 0.1 | 6.1 | GO:0015020 | glucuronosyltransferase activity(GO:0015020) |
| 0.1 | 3.4 | GO:0005388 | calcium-transporting ATPase activity(GO:0005388) |
| 0.1 | 0.9 | GO:0004920 | interleukin-10 receptor activity(GO:0004920) |
| 0.1 | 0.7 | GO:0015226 | amino-acid betaine transmembrane transporter activity(GO:0015199) carnitine transmembrane transporter activity(GO:0015226) |
| 0.1 | 0.4 | GO:0004749 | ribose phosphate diphosphokinase activity(GO:0004749) |
| 0.1 | 1.7 | GO:0008190 | eukaryotic initiation factor 4E binding(GO:0008190) |
| 0.1 | 8.8 | GO:0005507 | copper ion binding(GO:0005507) |
| 0.1 | 0.6 | GO:0001165 | RNA polymerase I upstream control element sequence-specific DNA binding(GO:0001165) |
| 0.1 | 5.5 | GO:0043027 | cysteine-type endopeptidase inhibitor activity involved in apoptotic process(GO:0043027) |
| 0.1 | 0.5 | GO:0030280 | structural constituent of epidermis(GO:0030280) |
| 0.1 | 1.2 | GO:0019957 | C-C chemokine binding(GO:0019957) |
| 0.1 | 1.1 | GO:0005247 | voltage-gated chloride channel activity(GO:0005247) |
| 0.1 | 0.3 | GO:0008859 | exoribonuclease II activity(GO:0008859) |
| 0.1 | 1.6 | GO:0004029 | aldehyde dehydrogenase (NAD) activity(GO:0004029) |
| 0.1 | 0.4 | GO:0004035 | alkaline phosphatase activity(GO:0004035) |
| 0.1 | 4.1 | GO:0001105 | RNA polymerase II transcription coactivator activity(GO:0001105) |
| 0.1 | 1.7 | GO:0005229 | intracellular calcium activated chloride channel activity(GO:0005229) |
| 0.0 | 0.2 | GO:0004103 | choline kinase activity(GO:0004103) |
| 0.0 | 0.3 | GO:0004849 | uridine kinase activity(GO:0004849) |
| 0.0 | 0.8 | GO:0051010 | microtubule plus-end binding(GO:0051010) |
| 0.0 | 2.1 | GO:0048487 | beta-tubulin binding(GO:0048487) |
| 0.0 | 0.7 | GO:0042043 | neurexin family protein binding(GO:0042043) |
| 0.0 | 4.1 | GO:0004222 | metalloendopeptidase activity(GO:0004222) |
| 0.0 | 3.7 | GO:0034987 | immunoglobulin receptor binding(GO:0034987) |
| 0.0 | 0.5 | GO:0034713 | type I transforming growth factor beta receptor binding(GO:0034713) |
| 0.0 | 0.2 | GO:0050649 | testosterone 6-beta-hydroxylase activity(GO:0050649) |
| 0.0 | 0.8 | GO:0005184 | neuropeptide hormone activity(GO:0005184) |
| 0.0 | 3.0 | GO:0003727 | single-stranded RNA binding(GO:0003727) |
| 0.0 | 0.4 | GO:0004745 | retinol dehydrogenase activity(GO:0004745) |
| 0.0 | 0.6 | GO:0016676 | cytochrome-c oxidase activity(GO:0004129) heme-copper terminal oxidase activity(GO:0015002) oxidoreductase activity, acting on a heme group of donors, oxygen as acceptor(GO:0016676) |
| 0.0 | 1.0 | GO:0005044 | scavenger receptor activity(GO:0005044) |
| 0.0 | 2.3 | GO:0004540 | ribonuclease activity(GO:0004540) |
| 0.0 | 0.1 | GO:0003985 | acetyl-CoA C-acetyltransferase activity(GO:0003985) C-acetyltransferase activity(GO:0016453) |
| 0.0 | 0.1 | GO:0004816 | asparagine-tRNA ligase activity(GO:0004816) |
| 0.0 | 2.1 | GO:0051082 | unfolded protein binding(GO:0051082) |
| 0.0 | 0.4 | GO:0003841 | 1-acylglycerol-3-phosphate O-acyltransferase activity(GO:0003841) |
| 0.0 | 1.1 | GO:0004004 | RNA helicase activity(GO:0003724) ATP-dependent RNA helicase activity(GO:0004004) RNA-dependent ATPase activity(GO:0008186) |
| 0.0 | 0.9 | GO:0003743 | translation initiation factor activity(GO:0003743) |
| 0.0 | 1.0 | GO:0019003 | GDP binding(GO:0019003) |
| 0.0 | 0.7 | GO:0004683 | calmodulin-dependent protein kinase activity(GO:0004683) |
| 0.0 | 0.5 | GO:0051539 | 4 iron, 4 sulfur cluster binding(GO:0051539) |
| 0.0 | 2.3 | GO:0004252 | serine-type endopeptidase activity(GO:0004252) |
| 0.0 | 0.3 | GO:0003887 | DNA-directed DNA polymerase activity(GO:0003887) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 4.5 | PID INTEGRIN2 PATHWAY | Beta2 integrin cell surface interactions |
| 0.0 | 0.5 | ST TYPE I INTERFERON PATHWAY | Type I Interferon (alpha/beta IFN) Pathway |
| 0.0 | 1.5 | PID FANCONI PATHWAY | Fanconi anemia pathway |
| 0.0 | 0.7 | PID IL27 PATHWAY | IL27-mediated signaling events |
| 0.0 | 1.5 | PID NFAT TFPATHWAY | Calcineurin-regulated NFAT-dependent transcription in lymphocytes |
| 0.0 | 1.4 | PID CASPASE PATHWAY | Caspase cascade in apoptosis |
| 0.0 | 0.8 | PID NCADHERIN PATHWAY | N-cadherin signaling events |
| 0.0 | 0.7 | PID UPA UPAR PATHWAY | Urokinase-type plasminogen activator (uPA) and uPAR-mediated signaling |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 8.8 | REACTOME CYTOCHROME P450 ARRANGED BY SUBSTRATE TYPE | Genes involved in Cytochrome P450 - arranged by substrate type |
| 0.2 | 3.0 | REACTOME REGULATION OF COMPLEMENT CASCADE | Genes involved in Regulation of Complement cascade |
| 0.2 | 2.5 | REACTOME TRYPTOPHAN CATABOLISM | Genes involved in Tryptophan catabolism |
| 0.2 | 3.4 | REACTOME PYRIMIDINE CATABOLISM | Genes involved in Pyrimidine catabolism |
| 0.1 | 3.4 | REACTOME PLATELET CALCIUM HOMEOSTASIS | Genes involved in Platelet calcium homeostasis |
| 0.1 | 1.2 | REACTOME BETA DEFENSINS | Genes involved in Beta defensins |
| 0.1 | 0.9 | REACTOME APOBEC3G MEDIATED RESISTANCE TO HIV1 INFECTION | Genes involved in APOBEC3G mediated resistance to HIV-1 infection |
| 0.1 | 3.0 | REACTOME CITRIC ACID CYCLE TCA CYCLE | Genes involved in Citric acid cycle (TCA cycle) |
| 0.1 | 3.6 | REACTOME METABOLISM OF STEROID HORMONES AND VITAMINS A AND D | Genes involved in Metabolism of steroid hormones and vitamins A and D |
| 0.1 | 3.4 | REACTOME SPHINGOLIPID DE NOVO BIOSYNTHESIS | Genes involved in Sphingolipid de novo biosynthesis |
| 0.1 | 2.6 | REACTOME SPHINGOLIPID METABOLISM | Genes involved in Sphingolipid metabolism |
| 0.1 | 1.1 | REACTOME ANTIGEN PRESENTATION FOLDING ASSEMBLY AND PEPTIDE LOADING OF CLASS I MHC | Genes involved in Antigen Presentation: Folding, assembly and peptide loading of class I MHC |
| 0.0 | 0.7 | REACTOME PLATELET ADHESION TO EXPOSED COLLAGEN | Genes involved in Platelet Adhesion to exposed collagen |
| 0.0 | 0.5 | REACTOME TRAFFICKING AND PROCESSING OF ENDOSOMAL TLR | Genes involved in Trafficking and processing of endosomal TLR |
| 0.0 | 2.8 | REACTOME INWARDLY RECTIFYING K CHANNELS | Genes involved in Inwardly rectifying K+ channels |
| 0.0 | 0.6 | REACTOME RNA POL I PROMOTER OPENING | Genes involved in RNA Polymerase I Promoter Opening |
| 0.0 | 1.9 | REACTOME REGULATION OF BETA CELL DEVELOPMENT | Genes involved in Regulation of beta-cell development |
| 0.0 | 1.9 | REACTOME IMMUNOREGULATORY INTERACTIONS BETWEEN A LYMPHOID AND A NON LYMPHOID CELL | Genes involved in Immunoregulatory interactions between a Lymphoid and a non-Lymphoid cell |
| 0.0 | 0.4 | REACTOME INHIBITION OF REPLICATION INITIATION OF DAMAGED DNA BY RB1 E2F1 | Genes involved in Inhibition of replication initiation of damaged DNA by RB1/E2F1 |
| 0.0 | 1.1 | REACTOME TIGHT JUNCTION INTERACTIONS | Genes involved in Tight junction interactions |
| 0.0 | 2.3 | REACTOME METABOLISM OF VITAMINS AND COFACTORS | Genes involved in Metabolism of vitamins and cofactors |
| 0.0 | 0.7 | REACTOME ACTIVATED AMPK STIMULATES FATTY ACID OXIDATION IN MUSCLE | Genes involved in Activated AMPK stimulates fatty-acid oxidation in muscle |
| 0.0 | 3.1 | REACTOME TRANSPORT OF INORGANIC CATIONS ANIONS AND AMINO ACIDS OLIGOPEPTIDES | Genes involved in Transport of inorganic cations/anions and amino acids/oligopeptides |
| 0.0 | 3.3 | REACTOME MRNA SPLICING | Genes involved in mRNA Splicing |
| 0.0 | 1.6 | REACTOME CLASS B 2 SECRETIN FAMILY RECEPTORS | Genes involved in Class B/2 (Secretin family receptors) |
| 0.0 | 0.3 | REACTOME PYRIMIDINE METABOLISM | Genes involved in Pyrimidine metabolism |
| 0.0 | 0.2 | REACTOME ETHANOL OXIDATION | Genes involved in Ethanol oxidation |
| 0.0 | 0.3 | REACTOME RNA POL III TRANSCRIPTION INITIATION FROM TYPE 2 PROMOTER | Genes involved in RNA Polymerase III Transcription Initiation From Type 2 Promoter |
| 0.0 | 1.7 | REACTOME INTEGRATION OF ENERGY METABOLISM | Genes involved in Integration of energy metabolism |