Project
PRJNA375882: Comprehensive Mouse Transcriptomic BodyMap
Navigation
Downloads
Results for Otx1
Z-value: 0.88
Transcription factors associated with Otx1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Otx1
|
ENSMUSG00000005917.16 | Otx1 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Otx1 | mm39_v1_chr11_-_21951605_21951631 | 0.14 | 2.5e-01 | Click! |
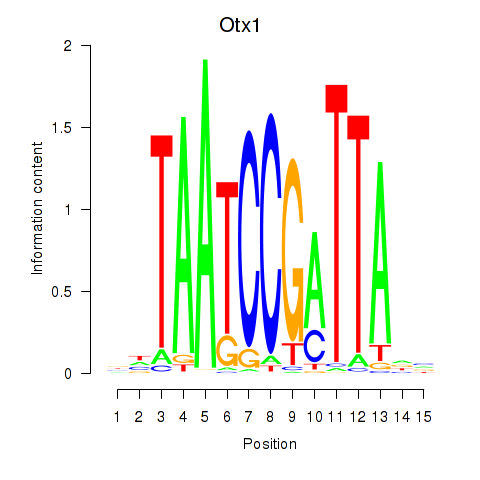
Activity profile of Otx1 motif
Sorted Z-values of Otx1 motif
Network of associatons between targets according to the STRING database.
First level regulatory network of Otx1
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr18_-_68562385 | 5.80 |
ENSMUST00000052347.8
|
Mc2r
|
melanocortin 2 receptor |
| chr8_+_55024446 | 4.99 |
ENSMUST00000239166.2
ENSMUST00000239106.2 ENSMUST00000239152.2 |
Asb5
|
ankyrin repeat and SOCs box-containing 5 |
| chr11_+_98277276 | 4.38 |
ENSMUST00000041301.8
|
Pnmt
|
phenylethanolamine-N-methyltransferase |
| chr2_+_158344532 | 3.87 |
ENSMUST00000059889.4
|
Adig
|
adipogenin |
| chr2_+_91376650 | 3.41 |
ENSMUST00000099716.11
ENSMUST00000046769.16 ENSMUST00000111337.3 |
Ckap5
|
cytoskeleton associated protein 5 |
| chr9_+_108356935 | 3.40 |
ENSMUST00000194147.2
ENSMUST00000065014.10 ENSMUST00000195483.6 ENSMUST00000195058.2 |
Lamb2
|
laminin, beta 2 |
| chr13_-_61084358 | 3.19 |
ENSMUST00000225859.2
ENSMUST00000225167.2 ENSMUST00000021880.10 |
Gm49391
Ctla2a
|
predicted gene, 49391 cytotoxic T lymphocyte-associated protein 2 alpha |
| chr19_-_10847121 | 3.13 |
ENSMUST00000120524.2
ENSMUST00000025645.14 |
Tmem132a
|
transmembrane protein 132A |
| chr2_+_158344553 | 3.09 |
ENSMUST00000109484.2
|
Adig
|
adipogenin |
| chr5_+_146392371 | 2.91 |
ENSMUST00000238592.2
|
Wasf3
|
WASP family, member 3 |
| chr4_-_11965691 | 2.71 |
ENSMUST00000108301.8
ENSMUST00000095144.10 ENSMUST00000108302.8 |
Pdp1
|
pyruvate dehyrogenase phosphatase catalytic subunit 1 |
| chr6_+_58810674 | 2.69 |
ENSMUST00000041401.11
|
Herc3
|
hect domain and RLD 3 |
| chr2_+_85715984 | 2.64 |
ENSMUST00000213441.3
|
Olfr1023
|
olfactory receptor 1023 |
| chr5_-_104261556 | 2.29 |
ENSMUST00000031249.8
|
Sparcl1
|
SPARC-like 1 |
| chr5_-_104261285 | 2.06 |
ENSMUST00000199947.2
|
Sparcl1
|
SPARC-like 1 |
| chr5_-_69748126 | 2.03 |
ENSMUST00000166298.8
|
Gnpda2
|
glucosamine-6-phosphate deaminase 2 |
| chr16_-_48592372 | 2.02 |
ENSMUST00000231701.3
|
Trat1
|
T cell receptor associated transmembrane adaptor 1 |
| chr7_-_37970734 | 1.96 |
ENSMUST00000032585.8
|
Pop4
|
processing of precursor 4, ribonuclease P/MRP family, (S. cerevisiae) |
| chr5_+_146321757 | 1.90 |
ENSMUST00000016143.9
|
Wasf3
|
WASP family, member 3 |
| chr2_+_169475436 | 1.89 |
ENSMUST00000109157.2
|
Tshz2
|
teashirt zinc finger family member 2 |
| chr4_-_138546564 | 1.89 |
ENSMUST00000102512.11
|
Pla2g5
|
phospholipase A2, group V |
| chrX_+_36390430 | 1.88 |
ENSMUST00000016553.5
|
Nkap
|
NFKB activating protein |
| chr5_+_25427860 | 1.85 |
ENSMUST00000045737.14
|
Galnt11
|
polypeptide N-acetylgalactosaminyltransferase 11 |
| chr2_-_153067297 | 1.80 |
ENSMUST00000099194.4
|
Tspyl3
|
TSPY-like 3 |
| chr9_-_20432562 | 1.79 |
ENSMUST00000215908.2
ENSMUST00000068296.8 ENSMUST00000174462.8 ENSMUST00000213418.2 |
Zfp266
|
zinc finger protein 266 |
| chr10_-_7423341 | 1.73 |
ENSMUST00000169796.4
ENSMUST00000218087.2 |
Ulbp1
|
UL16 binding protein 1 |
| chr10_+_3923086 | 1.67 |
ENSMUST00000117291.8
ENSMUST00000120585.8 ENSMUST00000043735.8 |
Mthfd1l
|
methylenetetrahydrofolate dehydrogenase (NADP+ dependent) 1-like |
| chr1_-_165535617 | 1.65 |
ENSMUST00000040357.11
|
Rcsd1
|
RCSD domain containing 1 |
| chr19_+_5074070 | 1.63 |
ENSMUST00000025826.7
ENSMUST00000237371.2 ENSMUST00000235416.2 |
Slc29a2
|
solute carrier family 29 (nucleoside transporters), member 2 |
| chr16_-_48592319 | 1.59 |
ENSMUST00000239408.2
|
Trat1
|
T cell receptor associated transmembrane adaptor 1 |
| chr17_-_21110913 | 1.59 |
ENSMUST00000061278.2
|
Vmn1r231
|
vomeronasal 1 receptor 231 |
| chr13_+_93908138 | 1.56 |
ENSMUST00000091403.6
|
Arsb
|
arylsulfatase B |
| chr5_-_65117375 | 1.53 |
ENSMUST00000062315.7
ENSMUST00000239485.2 ENSMUST00000201307.3 |
Tlr6
|
toll-like receptor 6 |
| chr5_-_115622356 | 1.52 |
ENSMUST00000112067.8
|
Sirt4
|
sirtuin 4 |
| chr16_-_59452883 | 1.47 |
ENSMUST00000118438.8
|
Arl6
|
ADP-ribosylation factor-like 6 |
| chr5_+_110071266 | 1.43 |
ENSMUST00000112544.8
|
Gm15446
|
predicted gene 15446 |
| chr18_+_24087725 | 1.39 |
ENSMUST00000225682.2
ENSMUST00000060762.6 |
Zfp397
|
zinc finger protein 397 |
| chr3_+_18108313 | 1.36 |
ENSMUST00000026120.8
|
Bhlhe22
|
basic helix-loop-helix family, member e22 |
| chr2_-_73283010 | 1.32 |
ENSMUST00000151939.2
|
Wipf1
|
WAS/WASL interacting protein family, member 1 |
| chr12_-_114012399 | 1.31 |
ENSMUST00000103468.3
|
Ighv11-2
|
immunoglobulin heavy variable V11-2 |
| chr16_+_22926504 | 1.27 |
ENSMUST00000187168.7
ENSMUST00000232287.2 ENSMUST00000077605.12 |
Eif4a2
|
eukaryotic translation initiation factor 4A2 |
| chr7_-_7250327 | 1.25 |
ENSMUST00000170922.2
|
Vmn2r29
|
vomeronasal 2, receptor 29 |
| chr7_-_29772226 | 1.18 |
ENSMUST00000183115.8
ENSMUST00000182919.8 ENSMUST00000183190.2 ENSMUST00000080834.15 |
Zfp82
|
zinc finger protein 82 |
| chr12_+_103400470 | 1.17 |
ENSMUST00000079294.12
ENSMUST00000076788.12 ENSMUST00000076702.12 ENSMUST00000066701.13 ENSMUST00000085065.12 ENSMUST00000140838.2 |
Ifi27
|
interferon, alpha-inducible protein 27 |
| chr16_+_49620883 | 1.15 |
ENSMUST00000229640.2
|
Cd47
|
CD47 antigen (Rh-related antigen, integrin-associated signal transducer) |
| chr8_-_94825556 | 1.14 |
ENSMUST00000034206.6
|
Bbs2
|
Bardet-Biedl syndrome 2 (human) |
| chr13_-_23553139 | 1.13 |
ENSMUST00000152557.8
|
Zfp322a
|
zinc finger protein 322A |
| chrX_+_106132840 | 1.13 |
ENSMUST00000118666.8
ENSMUST00000053375.4 |
P2ry10
|
purinergic receptor P2Y, G-protein coupled 10 |
| chr19_-_42768374 | 1.13 |
ENSMUST00000069298.13
ENSMUST00000160455.8 ENSMUST00000162004.8 |
Hps1
|
HPS1, biogenesis of lysosomal organelles complex 3 subunit 1 |
| chr11_+_94827050 | 1.12 |
ENSMUST00000001547.8
|
Col1a1
|
collagen, type I, alpha 1 |
| chr13_-_61045252 | 1.11 |
ENSMUST00000021884.10
|
Ctla2b
|
cytotoxic T lymphocyte-associated protein 2 beta |
| chr4_-_137512682 | 1.10 |
ENSMUST00000133473.2
|
Alpl
|
alkaline phosphatase, liver/bone/kidney |
| chr6_-_148732893 | 1.08 |
ENSMUST00000145960.2
|
Ipo8
|
importin 8 |
| chr7_-_4066154 | 1.07 |
ENSMUST00000086401.10
ENSMUST00000068865.13 |
Lair1
|
leukocyte-associated Ig-like receptor 1 |
| chr7_-_4066125 | 1.06 |
ENSMUST00000108600.9
ENSMUST00000205296.2 |
Lair1
|
leukocyte-associated Ig-like receptor 1 |
| chr7_-_4066194 | 1.05 |
ENSMUST00000086400.13
|
Lair1
|
leukocyte-associated Ig-like receptor 1 |
| chr2_-_12306722 | 1.02 |
ENSMUST00000028106.11
|
Itga8
|
integrin alpha 8 |
| chr12_-_65120674 | 1.00 |
ENSMUST00000220983.2
ENSMUST00000220730.2 ENSMUST00000021332.10 |
Fkbp3
|
FK506 binding protein 3 |
| chr17_+_24768808 | 1.00 |
ENSMUST00000228550.2
ENSMUST00000035565.5 |
Pkd1
|
polycystin 1, transient receptor poteintial channel interacting |
| chr2_-_156022054 | 0.99 |
ENSMUST00000126992.8
ENSMUST00000146288.8 ENSMUST00000029149.13 ENSMUST00000109587.9 ENSMUST00000109584.8 |
Rbm39
|
RNA binding motif protein 39 |
| chr11_+_78237492 | 0.98 |
ENSMUST00000100755.4
|
Unc119
|
unc-119 lipid binding chaperone |
| chr1_-_118239146 | 0.96 |
ENSMUST00000027623.9
|
Tsn
|
translin |
| chr5_-_5799315 | 0.96 |
ENSMUST00000015796.9
|
Steap1
|
six transmembrane epithelial antigen of the prostate 1 |
| chr18_+_57601541 | 0.95 |
ENSMUST00000091892.4
ENSMUST00000209782.2 |
Ctxn3
|
cortexin 3 |
| chr1_-_85664246 | 0.94 |
ENSMUST00000064788.14
|
A630001G21Rik
|
RIKEN cDNA A630001G21 gene |
| chr15_+_80017315 | 0.93 |
ENSMUST00000023050.9
|
Tab1
|
TGF-beta activated kinase 1/MAP3K7 binding protein 1 |
| chr3_+_66892979 | 0.92 |
ENSMUST00000162362.8
ENSMUST00000065074.14 ENSMUST00000065047.13 |
Rsrc1
|
arginine/serine-rich coiled-coil 1 |
| chr2_+_112096154 | 0.89 |
ENSMUST00000110991.9
|
Slc12a6
|
solute carrier family 12, member 6 |
| chr5_+_53713137 | 0.89 |
ENSMUST00000087360.9
|
Rbpj
|
recombination signal binding protein for immunoglobulin kappa J region |
| chr7_+_27537982 | 0.88 |
ENSMUST00000205701.2
|
Zfp59
|
zinc finger protein 59 |
| chr3_+_66893031 | 0.87 |
ENSMUST00000046542.13
ENSMUST00000162693.8 |
Rsrc1
|
arginine/serine-rich coiled-coil 1 |
| chr10_+_11025150 | 0.87 |
ENSMUST00000044053.13
|
Shprh
|
SNF2 histone linker PHD RING helicase |
| chr16_-_19341016 | 0.86 |
ENSMUST00000214315.2
|
Olfr167
|
olfactory receptor 167 |
| chr3_+_95018324 | 0.85 |
ENSMUST00000009102.9
|
Vps72
|
vacuolar protein sorting 72 |
| chr6_+_108771840 | 0.84 |
ENSMUST00000204483.2
|
Arl8b
|
ADP-ribosylation factor-like 8B |
| chr17_+_33483650 | 0.84 |
ENSMUST00000217023.3
|
Olfr63
|
olfactory receptor 63 |
| chr13_-_61045212 | 0.84 |
ENSMUST00000171347.9
|
Ctla2b
|
cytotoxic T lymphocyte-associated protein 2 beta |
| chr4_-_126094910 | 0.82 |
ENSMUST00000136157.8
|
Thrap3
|
thyroid hormone receptor associated protein 3 |
| chr4_+_74160705 | 0.79 |
ENSMUST00000077851.10
|
Kdm4c
|
lysine (K)-specific demethylase 4C |
| chr9_-_62895197 | 0.78 |
ENSMUST00000216209.2
|
Pias1
|
protein inhibitor of activated STAT 1 |
| chr12_-_98225676 | 0.76 |
ENSMUST00000021390.9
|
Galc
|
galactosylceramidase |
| chr17_+_21031817 | 0.75 |
ENSMUST00000232810.2
ENSMUST00000233712.2 ENSMUST00000232852.2 |
Vmn1r229
|
vomeronasal 1 receptor 229 |
| chrM_+_10167 | 0.74 |
ENSMUST00000082414.1
|
mt-Nd4
|
mitochondrially encoded NADH dehydrogenase 4 |
| chr6_+_48531710 | 0.74 |
ENSMUST00000114545.8
ENSMUST00000153222.2 ENSMUST00000204071.2 ENSMUST00000101436.3 ENSMUST00000203627.2 |
Lrrc61
|
leucine rich repeat containing 61 |
| chr15_-_57128522 | 0.72 |
ENSMUST00000137764.2
ENSMUST00000022995.13 |
Slc22a22
|
solute carrier family 22 (organic cation transporter), member 22 |
| chr9_+_117888124 | 0.71 |
ENSMUST00000123690.2
|
Azi2
|
5-azacytidine induced gene 2 |
| chr2_+_112114906 | 0.71 |
ENSMUST00000053666.8
|
Slc12a6
|
solute carrier family 12, member 6 |
| chr7_-_85985625 | 0.69 |
ENSMUST00000069279.5
|
Olfr307
|
olfactory receptor 307 |
| chr9_+_108367801 | 0.69 |
ENSMUST00000006854.13
|
Usp19
|
ubiquitin specific peptidase 19 |
| chr6_-_89847511 | 0.68 |
ENSMUST00000089418.5
|
Vmn1r43
|
vomeronasal 1 receptor 43 |
| chr11_+_3913970 | 0.67 |
ENSMUST00000109985.8
ENSMUST00000020705.5 |
Pes1
|
pescadillo ribosomal biogenesis factor 1 |
| chr19_-_7017295 | 0.66 |
ENSMUST00000025918.9
|
Stip1
|
stress-induced phosphoprotein 1 |
| chr13_+_18901459 | 0.65 |
ENSMUST00000072961.6
|
Vps41
|
VPS41 HOPS complex subunit |
| chr17_+_47922497 | 0.64 |
ENSMUST00000024778.3
|
Med20
|
mediator complex subunit 20 |
| chr8_-_55171699 | 0.63 |
ENSMUST00000144711.9
|
Wdr17
|
WD repeat domain 17 |
| chr13_-_63579497 | 0.61 |
ENSMUST00000160931.2
ENSMUST00000099444.10 ENSMUST00000220684.2 ENSMUST00000161977.8 ENSMUST00000163091.8 |
Fancc
|
Fanconi anemia, complementation group C |
| chr11_-_120344299 | 0.60 |
ENSMUST00000026452.3
|
Pde6g
|
phosphodiesterase 6G, cGMP-specific, rod, gamma |
| chr17_+_36172210 | 0.56 |
ENSMUST00000074259.15
ENSMUST00000174873.2 |
Nrm
|
nurim (nuclear envelope membrane protein) |
| chr19_-_5738936 | 0.56 |
ENSMUST00000113615.9
|
Pcnx3
|
pecanex homolog 3 |
| chr16_-_63684477 | 0.55 |
ENSMUST00000232654.2
ENSMUST00000064405.8 |
Epha3
|
Eph receptor A3 |
| chr13_-_21934675 | 0.54 |
ENSMUST00000102983.2
|
H4c12
|
H4 clustered histone 12 |
| chr12_-_52074846 | 0.53 |
ENSMUST00000040161.5
|
Gpr33
|
G protein-coupled receptor 33 |
| chr16_+_91444730 | 0.53 |
ENSMUST00000119368.8
ENSMUST00000114037.9 ENSMUST00000114036.9 ENSMUST00000122302.8 |
Son
|
Son DNA binding protein |
| chr8_-_96615138 | 0.53 |
ENSMUST00000034097.8
|
Got2
|
glutamatic-oxaloacetic transaminase 2, mitochondrial |
| chr6_+_70332836 | 0.52 |
ENSMUST00000103390.3
|
Igkv8-18
|
immunoglobulin kappa variable 8-18 |
| chr9_+_108911547 | 0.52 |
ENSMUST00000026735.9
|
Ccdc51
|
coiled-coil domain containing 51 |
| chr11_+_115455260 | 0.51 |
ENSMUST00000021085.11
|
Nup85
|
nucleoporin 85 |
| chr6_+_68657317 | 0.50 |
ENSMUST00000198735.2
|
Igkv10-95
|
immunoglobulin kappa variable 10-95 |
| chr6_-_133032779 | 0.49 |
ENSMUST00000095391.3
|
Tas2r140
|
taste receptor, type 2, member 140 |
| chr7_+_16186704 | 0.49 |
ENSMUST00000019302.10
|
Tmem160
|
transmembrane protein 160 |
| chr6_-_58412879 | 0.49 |
ENSMUST00000078890.5
|
Vmn1r30
|
vomeronasal 1 receptor 30 |
| chr1_+_131838220 | 0.47 |
ENSMUST00000189946.7
|
Nucks1
|
nuclear casein kinase and cyclin-dependent kinase substrate 1 |
| chr6_-_29507944 | 0.47 |
ENSMUST00000101614.10
ENSMUST00000078112.13 |
Kcp
|
kielin/chordin-like protein |
| chr15_+_78819119 | 0.46 |
ENSMUST00000138880.9
ENSMUST00000041164.4 |
Nol12
|
nucleolar protein 12 |
| chr1_+_118249558 | 0.42 |
ENSMUST00000027626.13
ENSMUST00000112688.10 |
Nifk
|
nucleolar protein interacting with the FHA domain of MKI67 |
| chr12_-_46865709 | 0.40 |
ENSMUST00000021438.8
|
Nova1
|
NOVA alternative splicing regulator 1 |
| chr9_+_106330437 | 0.40 |
ENSMUST00000185874.7
|
Pcbp4
|
poly(rC) binding protein 4 |
| chr7_-_100311522 | 0.40 |
ENSMUST00000151123.8
ENSMUST00000208812.2 ENSMUST00000107047.10 |
Plekhb1
|
pleckstrin homology domain containing, family B (evectins) member 1 |
| chr6_+_132928065 | 0.40 |
ENSMUST00000070991.5
|
Tas2r129
|
taste receptor, type 2, member 129 |
| chr14_+_52122439 | 0.39 |
ENSMUST00000167984.2
|
Mettl17
|
methyltransferase like 17 |
| chr12_-_26506422 | 0.39 |
ENSMUST00000020970.10
|
Rsad2
|
radical S-adenosyl methionine domain containing 2 |
| chr10_-_111829393 | 0.38 |
ENSMUST00000161870.3
|
Glipr1
|
GLI pathogenesis-related 1 (glioma) |
| chr10_+_123099945 | 0.37 |
ENSMUST00000238972.2
ENSMUST00000050756.8 |
Tafa2
|
TAFA chemokine like family member 2 |
| chr12_-_13299197 | 0.34 |
ENSMUST00000071103.10
|
Ddx1
|
DEAD box helicase 1 |
| chr8_-_65440298 | 0.33 |
ENSMUST00000095295.3
|
Trim75
|
tripartite motif-containing 75 |
| chr4_+_143515922 | 0.32 |
ENSMUST00000085144.11
ENSMUST00000149739.2 ENSMUST00000105770.2 |
Pramel25
|
PRAME like 25 |
| chr19_-_41921676 | 0.32 |
ENSMUST00000075280.12
ENSMUST00000112123.4 |
Exosc1
|
exosome component 1 |
| chr2_+_87576198 | 0.32 |
ENSMUST00000217572.2
|
Olfr1140
|
olfactory receptor 1140 |
| chrX_-_42256694 | 0.31 |
ENSMUST00000115058.8
ENSMUST00000115059.8 |
Tenm1
|
teneurin transmembrane protein 1 |
| chr16_-_18885809 | 0.31 |
ENSMUST00000200211.2
|
Iglj3
|
immunoglobulin lambda joining 3 |
| chr9_-_53521585 | 0.29 |
ENSMUST00000034547.6
|
Acat1
|
acetyl-Coenzyme A acetyltransferase 1 |
| chr12_-_103660946 | 0.28 |
ENSMUST00000118101.2
|
Serpina1f
|
serine (or cysteine) peptidase inhibitor, clade A, member 1F |
| chr5_-_146158235 | 0.27 |
ENSMUST00000161859.8
|
Rnf6
|
ring finger protein (C3H2C3 type) 6 |
| chr11_-_116165024 | 0.25 |
ENSMUST00000021133.16
|
Srp68
|
signal recognition particle 68 |
| chr11_+_103007054 | 0.23 |
ENSMUST00000053063.7
|
Hexim1
|
hexamethylene bis-acetamide inducible 1 |
| chr6_+_57012898 | 0.23 |
ENSMUST00000078186.4
|
Vmn1r8
|
vomeronasal 1 receptor 8 |
| chr7_-_100311621 | 0.23 |
ENSMUST00000079176.14
|
Plekhb1
|
pleckstrin homology domain containing, family B (evectins) member 1 |
| chr4_-_96552349 | 0.21 |
ENSMUST00000030299.8
|
Cyp2j5
|
cytochrome P450, family 2, subfamily j, polypeptide 5 |
| chr4_-_130116119 | 0.21 |
ENSMUST00000023884.7
|
Ldc1
|
leucine decarboxylase 1 |
| chr17_+_28075415 | 0.20 |
ENSMUST00000114849.3
|
Uhrf1bp1
|
UHRF1 (ICBP90) binding protein 1 |
| chr9_+_68561042 | 0.20 |
ENSMUST00000034766.14
|
Rora
|
RAR-related orphan receptor alpha |
| chr9_-_38577138 | 0.20 |
ENSMUST00000076542.2
|
Olfr917
|
olfactory receptor 917 |
| chr13_+_36142822 | 0.20 |
ENSMUST00000225537.2
|
Ppp1r3g
|
protein phosphatase 1, regulatory subunit 3G |
| chr9_-_109105194 | 0.18 |
ENSMUST00000198048.5
|
Fbxw14
|
F-box and WD-40 domain protein 14 |
| chr17_+_36172235 | 0.17 |
ENSMUST00000172931.2
|
Nrm
|
nurim (nuclear envelope membrane protein) |
| chr2_-_37312881 | 0.17 |
ENSMUST00000112936.4
ENSMUST00000112934.8 |
Rc3h2
|
ring finger and CCCH-type zinc finger domains 2 |
| chr8_-_65440483 | 0.16 |
ENSMUST00000210982.2
|
Trim75
|
tripartite motif-containing 75 |
| chr12_-_103660916 | 0.16 |
ENSMUST00000117053.8
|
Serpina1f
|
serine (or cysteine) peptidase inhibitor, clade A, member 1F |
| chr3_-_148696155 | 0.16 |
ENSMUST00000196526.5
ENSMUST00000200543.5 ENSMUST00000200154.5 |
Adgrl2
|
adhesion G protein-coupled receptor L2 |
| chr2_+_111145783 | 0.14 |
ENSMUST00000082167.5
|
Olfr1280
|
olfactory receptor 1280 |
| chr15_+_102391614 | 0.12 |
ENSMUST00000229432.2
|
Pcbp2
|
poly(rC) binding protein 2 |
| chr7_+_103652466 | 0.11 |
ENSMUST00000209757.3
|
Olfr638
|
olfactory receptor 638 |
| chr1_+_186947683 | 0.11 |
ENSMUST00000065573.14
ENSMUST00000110943.9 ENSMUST00000044812.12 |
Gpatch2
|
G patch domain containing 2 |
| chr11_-_116164928 | 0.10 |
ENSMUST00000106425.4
|
Srp68
|
signal recognition particle 68 |
| chr7_+_86444235 | 0.09 |
ENSMUST00000233099.2
ENSMUST00000164996.2 |
Vmn2r77
|
vomeronasal 2, receptor 77 |
| chr10_+_78848306 | 0.09 |
ENSMUST00000216030.2
|
Olfr1351
|
olfactory receptor 1351 |
| chr1_+_131838294 | 0.08 |
ENSMUST00000062264.8
|
Nucks1
|
nuclear casein kinase and cyclin-dependent kinase substrate 1 |
| chr8_+_85676489 | 0.08 |
ENSMUST00000121880.8
|
Rtbdn
|
retbindin |
| chr10_-_39009844 | 0.07 |
ENSMUST00000134279.8
ENSMUST00000139743.8 ENSMUST00000149949.8 ENSMUST00000124941.8 ENSMUST00000125042.8 ENSMUST00000063204.9 |
Fam229b
|
family with sequence similarity 229, member B |
| chr2_-_89273344 | 0.07 |
ENSMUST00000216123.2
|
Olfr1240
|
olfactory receptor 1240 |
| chr11_-_69127848 | 0.06 |
ENSMUST00000021259.9
ENSMUST00000108665.8 ENSMUST00000108664.2 |
Gucy2e
|
guanylate cyclase 2e |
| chr16_+_88966321 | 0.06 |
ENSMUST00000089098.2
|
Gm7735
|
predicted gene 7735 |
| chr13_+_21363602 | 0.04 |
ENSMUST00000222544.2
|
Trim27
|
tripartite motif-containing 27 |
| chr6_-_68609426 | 0.04 |
ENSMUST00000103328.3
|
Igkv10-96
|
immunoglobulin kappa variable 10-96 |
| chr13_-_58422647 | 0.03 |
ENSMUST00000225034.2
|
Gkap1
|
G kinase anchoring protein 1 |
| chr12_+_70499869 | 0.03 |
ENSMUST00000021471.13
|
Tmx1
|
thioredoxin-related transmembrane protein 1 |
| chr19_-_6107766 | 0.02 |
ENSMUST00000235520.2
ENSMUST00000007482.8 |
Mrpl49
|
mitochondrial ribosomal protein L49 |
| chr18_+_86729645 | 0.01 |
ENSMUST00000122464.8
|
Cbln2
|
cerebellin 2 precursor protein |
| chr9_+_19351562 | 0.00 |
ENSMUST00000217273.2
|
Olfr849
|
olfactory receptor 849 |
| chr10_+_127478844 | 0.00 |
ENSMUST00000092074.12
ENSMUST00000120279.2 |
Stat6
|
signal transducer and activator of transcription 6 |
| chr1_+_37258272 | 0.00 |
ENSMUST00000027288.10
|
Cnga3
|
cyclic nucleotide gated channel alpha 3 |
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.1 | 3.4 | GO:0072248 | metanephric glomerular visceral epithelial cell differentiation(GO:0072248) metanephric glomerular visceral epithelial cell development(GO:0072249) metanephric glomerular epithelial cell differentiation(GO:0072312) metanephric glomerular epithelial cell development(GO:0072313) |
| 0.9 | 4.4 | GO:0042414 | epinephrine metabolic process(GO:0042414) |
| 0.6 | 3.6 | GO:0001920 | negative regulation of receptor recycling(GO:0001920) |
| 0.6 | 3.4 | GO:0030951 | establishment or maintenance of microtubule cytoskeleton polarity(GO:0030951) |
| 0.4 | 1.7 | GO:0009257 | 10-formyltetrahydrofolate biosynthetic process(GO:0009257) |
| 0.4 | 1.6 | GO:0042539 | hypotonic salinity response(GO:0042539) cellular hypotonic salinity response(GO:0071477) |
| 0.4 | 1.5 | GO:0070340 | negative regulation of toll-like receptor 2 signaling pathway(GO:0034136) TRIF-dependent toll-like receptor signaling pathway(GO:0035666) detection of bacterial lipopeptide(GO:0070340) |
| 0.3 | 1.7 | GO:0042271 | susceptibility to natural killer cell mediated cytotoxicity(GO:0042271) |
| 0.3 | 1.0 | GO:0060854 | patterning of lymph vessels(GO:0060854) |
| 0.3 | 1.9 | GO:0006663 | platelet activating factor biosynthetic process(GO:0006663) |
| 0.3 | 1.5 | GO:1903215 | negative regulation of protein targeting to mitochondrion(GO:1903215) |
| 0.3 | 1.5 | GO:0046909 | intermembrane transport(GO:0046909) protein transport from ciliary membrane to plasma membrane(GO:1903445) |
| 0.3 | 1.1 | GO:0038108 | negative regulation of appetite by leptin-mediated signaling pathway(GO:0038108) |
| 0.2 | 1.0 | GO:2001287 | negative regulation of caveolin-mediated endocytosis(GO:2001287) |
| 0.2 | 1.9 | GO:0018243 | protein O-linked glycosylation via threonine(GO:0018243) |
| 0.2 | 1.0 | GO:2000721 | positive regulation of transcription from RNA polymerase II promoter involved in smooth muscle cell differentiation(GO:2000721) |
| 0.2 | 0.5 | GO:0097156 | fasciculation of motor neuron axon(GO:0097156) |
| 0.2 | 0.9 | GO:1901297 | positive regulation of canonical Wnt signaling pathway involved in cardiac muscle cell fate commitment(GO:1901297) positive regulation of canonical Wnt signaling pathway involved in heart development(GO:1905068) |
| 0.2 | 2.0 | GO:0001682 | tRNA 5'-leader removal(GO:0001682) |
| 0.2 | 0.5 | GO:0006106 | fumarate metabolic process(GO:0006106) aspartate catabolic process(GO:0006533) |
| 0.2 | 2.7 | GO:0035970 | peptidyl-threonine dephosphorylation(GO:0035970) |
| 0.2 | 1.6 | GO:0071474 | cellular hyperosmotic response(GO:0071474) |
| 0.2 | 1.1 | GO:1903232 | melanosome assembly(GO:1903232) |
| 0.1 | 4.7 | GO:0050872 | white fat cell differentiation(GO:0050872) |
| 0.1 | 0.7 | GO:0071718 | sodium-independent icosanoid transport(GO:0071718) |
| 0.1 | 0.7 | GO:0044565 | dendritic cell proliferation(GO:0044565) |
| 0.1 | 1.1 | GO:0001957 | intramembranous ossification(GO:0001957) direct ossification(GO:0036072) |
| 0.1 | 0.6 | GO:0019046 | release from viral latency(GO:0019046) regulation of DNA strand elongation(GO:0060382) |
| 0.1 | 1.4 | GO:0021960 | anterior commissure morphogenesis(GO:0021960) |
| 0.1 | 0.6 | GO:1902774 | late endosome to lysosome transport(GO:1902774) |
| 0.1 | 1.1 | GO:0008228 | opsonization(GO:0008228) |
| 0.1 | 2.0 | GO:0006044 | N-acetylglucosamine metabolic process(GO:0006044) |
| 0.1 | 3.2 | GO:0045589 | regulation of regulatory T cell differentiation(GO:0045589) |
| 0.1 | 0.4 | GO:0034165 | positive regulation of toll-like receptor 9 signaling pathway(GO:0034165) |
| 0.1 | 0.3 | GO:0006550 | isoleucine catabolic process(GO:0006550) |
| 0.1 | 1.0 | GO:0098706 | ferric iron import into cell(GO:0097461) ferric iron import across plasma membrane(GO:0098706) |
| 0.1 | 4.8 | GO:0014003 | oligodendrocyte development(GO:0014003) |
| 0.1 | 0.4 | GO:1902163 | negative regulation of DNA damage response, signal transduction by p53 class mediator resulting in transcription of p21 class mediator(GO:1902163) |
| 0.1 | 0.5 | GO:0002430 | complement receptor mediated signaling pathway(GO:0002430) |
| 0.1 | 0.2 | GO:2000863 | positive regulation of estrogen secretion(GO:2000863) |
| 0.1 | 1.6 | GO:0015858 | nucleoside transport(GO:0015858) |
| 0.1 | 0.9 | GO:0000185 | activation of MAPKKK activity(GO:0000185) |
| 0.1 | 0.8 | GO:1900113 | negative regulation of histone H3-K9 trimethylation(GO:1900113) |
| 0.1 | 0.8 | GO:0046477 | glycosylceramide catabolic process(GO:0046477) |
| 0.1 | 0.8 | GO:0051152 | positive regulation of smooth muscle cell differentiation(GO:0051152) |
| 0.1 | 5.8 | GO:0030819 | positive regulation of cAMP biosynthetic process(GO:0030819) |
| 0.1 | 0.7 | GO:0000463 | maturation of LSU-rRNA from tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000463) nucleolus organization(GO:0007000) |
| 0.0 | 2.5 | GO:0046825 | regulation of protein export from nucleus(GO:0046825) |
| 0.0 | 1.9 | GO:0071425 | hematopoietic stem cell proliferation(GO:0071425) |
| 0.0 | 2.9 | GO:0046677 | response to antibiotic(GO:0046677) |
| 0.0 | 0.7 | GO:0006120 | mitochondrial electron transport, NADH to ubiquinone(GO:0006120) |
| 0.0 | 0.3 | GO:0044314 | protein K27-linked ubiquitination(GO:0044314) |
| 0.0 | 0.3 | GO:1903608 | protein localization to cytoplasmic stress granule(GO:1903608) |
| 0.0 | 0.7 | GO:1900037 | regulation of cellular response to hypoxia(GO:1900037) |
| 0.0 | 1.1 | GO:0051482 | positive regulation of cytosolic calcium ion concentration involved in phospholipase C-activating G-protein coupled signaling pathway(GO:0051482) |
| 0.0 | 0.8 | GO:0043486 | histone exchange(GO:0043486) |
| 0.0 | 1.0 | GO:0042753 | positive regulation of circadian rhythm(GO:0042753) |
| 0.0 | 3.7 | GO:0019236 | response to pheromone(GO:0019236) |
| 0.0 | 1.3 | GO:0010501 | RNA secondary structure unwinding(GO:0010501) |
| 0.0 | 1.6 | GO:0009268 | response to pH(GO:0009268) |
| 0.0 | 0.6 | GO:0097150 | neuronal stem cell population maintenance(GO:0097150) |
| 0.0 | 0.9 | GO:0001580 | detection of chemical stimulus involved in sensory perception of bitter taste(GO:0001580) |
| 0.0 | 0.2 | GO:2000467 | positive regulation of glycogen (starch) synthase activity(GO:2000467) |
| 0.0 | 0.4 | GO:0006614 | SRP-dependent cotranslational protein targeting to membrane(GO:0006614) |
| 0.0 | 0.2 | GO:0061470 | T follicular helper cell differentiation(GO:0061470) |
| 0.0 | 0.6 | GO:0045745 | positive regulation of G-protein coupled receptor protein signaling pathway(GO:0045745) |
| 0.0 | 0.9 | GO:0006334 | nucleosome assembly(GO:0006334) |
| 0.0 | 1.0 | GO:0061077 | chaperone-mediated protein folding(GO:0061077) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.9 | 3.4 | GO:0005608 | laminin-3 complex(GO:0005608) |
| 0.4 | 1.1 | GO:0005584 | collagen type I trimer(GO:0005584) |
| 0.3 | 2.0 | GO:0000172 | ribonuclease MRP complex(GO:0000172) |
| 0.2 | 4.4 | GO:0043196 | varicosity(GO:0043196) |
| 0.1 | 3.6 | GO:0042101 | T cell receptor complex(GO:0042101) |
| 0.1 | 0.9 | GO:0002193 | MAML1-RBP-Jkappa- ICN1 complex(GO:0002193) |
| 0.1 | 3.4 | GO:0000930 | gamma-tubulin complex(GO:0000930) |
| 0.1 | 1.1 | GO:0034464 | BBSome(GO:0034464) |
| 0.1 | 0.3 | GO:0071920 | cleavage body(GO:0071920) |
| 0.1 | 0.6 | GO:0033263 | CORVET complex(GO:0033263) |
| 0.1 | 1.0 | GO:0032591 | dendritic spine membrane(GO:0032591) |
| 0.1 | 0.7 | GO:0070545 | PeBoW complex(GO:0070545) |
| 0.1 | 7.3 | GO:0005811 | lipid particle(GO:0005811) |
| 0.0 | 1.1 | GO:0065010 | extracellular membrane-bounded organelle(GO:0065010) |
| 0.0 | 1.1 | GO:0031082 | BLOC complex(GO:0031082) |
| 0.0 | 0.4 | GO:0005786 | signal recognition particle, endoplasmic reticulum targeting(GO:0005786) |
| 0.0 | 1.8 | GO:0051233 | spindle midzone(GO:0051233) |
| 0.0 | 0.5 | GO:0031080 | nuclear pore outer ring(GO:0031080) |
| 0.0 | 1.9 | GO:0005637 | nuclear inner membrane(GO:0005637) |
| 0.0 | 0.6 | GO:0043240 | Fanconi anaemia nuclear complex(GO:0043240) |
| 0.0 | 1.5 | GO:0016592 | mediator complex(GO:0016592) |
| 0.0 | 0.9 | GO:0000786 | nucleosome(GO:0000786) |
| 0.0 | 0.3 | GO:0000176 | nuclear exosome (RNase complex)(GO:0000176) |
| 0.0 | 1.7 | GO:0046658 | anchored component of plasma membrane(GO:0046658) |
| 0.0 | 0.8 | GO:0005721 | pericentric heterochromatin(GO:0005721) |
| 0.0 | 0.1 | GO:0033165 | interphotoreceptor matrix(GO:0033165) |
| 0.0 | 4.8 | GO:0030027 | lamellipodium(GO:0030027) |
| 0.0 | 1.6 | GO:0005791 | rough endoplasmic reticulum(GO:0005791) |
| 0.0 | 5.0 | GO:0016323 | basolateral plasma membrane(GO:0016323) |
| 0.0 | 4.8 | GO:0016607 | nuclear speck(GO:0016607) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.8 | 5.8 | GO:0004977 | melanocortin receptor activity(GO:0004977) |
| 0.7 | 2.7 | GO:0004741 | [pyruvate dehydrogenase (lipoamide)] phosphatase activity(GO:0004741) |
| 0.7 | 2.0 | GO:0004342 | glucosamine-6-phosphate deaminase activity(GO:0004342) |
| 0.7 | 2.0 | GO:0000171 | ribonuclease MRP activity(GO:0000171) |
| 0.4 | 1.7 | GO:0004329 | formate-tetrahydrofolate ligase activity(GO:0004329) |
| 0.4 | 1.6 | GO:0035827 | rubidium ion transmembrane transporter activity(GO:0035827) |
| 0.4 | 1.5 | GO:0035663 | Toll-like receptor 2 binding(GO:0035663) |
| 0.2 | 1.9 | GO:0047498 | calcium-dependent phospholipase A2 activity(GO:0047498) |
| 0.2 | 1.6 | GO:0004065 | arylsulfatase activity(GO:0004065) |
| 0.2 | 0.5 | GO:0080130 | L-phenylalanine:2-oxoglutarate aminotransferase activity(GO:0080130) |
| 0.2 | 1.1 | GO:0004035 | alkaline phosphatase activity(GO:0004035) |
| 0.1 | 3.4 | GO:0051010 | microtubule plus-end binding(GO:0051010) |
| 0.1 | 0.5 | GO:0031727 | CCR2 chemokine receptor binding(GO:0031727) |
| 0.1 | 1.0 | GO:0008823 | cupric reductase activity(GO:0008823) ferric-chelate reductase (NADPH) activity(GO:0052851) |
| 0.1 | 0.8 | GO:0061665 | SUMO ligase activity(GO:0061665) |
| 0.1 | 1.1 | GO:0070053 | thrombospondin receptor activity(GO:0070053) |
| 0.1 | 1.7 | GO:0046703 | natural killer cell lectin-like receptor binding(GO:0046703) |
| 0.1 | 1.9 | GO:0004653 | polypeptide N-acetylgalactosaminyltransferase activity(GO:0004653) |
| 0.1 | 1.5 | GO:0070403 | NAD+ binding(GO:0070403) |
| 0.1 | 0.3 | GO:0033677 | DNA/RNA helicase activity(GO:0033677) |
| 0.1 | 1.1 | GO:0048407 | platelet-derived growth factor binding(GO:0048407) |
| 0.1 | 0.5 | GO:0004875 | complement receptor activity(GO:0004875) |
| 0.1 | 0.4 | GO:0005047 | signal recognition particle binding(GO:0005047) endoplasmic reticulum signal peptide binding(GO:0030942) |
| 0.1 | 1.1 | GO:0045028 | G-protein coupled nucleotide receptor activity(GO:0001608) G-protein coupled purinergic nucleotide receptor activity(GO:0045028) |
| 0.1 | 0.7 | GO:0004955 | prostaglandin receptor activity(GO:0004955) |
| 0.1 | 0.5 | GO:0005004 | GPI-linked ephrin receptor activity(GO:0005004) |
| 0.1 | 1.6 | GO:0005337 | nucleoside transmembrane transporter activity(GO:0005337) |
| 0.1 | 0.7 | GO:1990380 | Lys48-specific deubiquitinase activity(GO:1990380) |
| 0.1 | 0.5 | GO:0050733 | RS domain binding(GO:0050733) |
| 0.1 | 4.4 | GO:0050840 | extracellular matrix binding(GO:0050840) |
| 0.1 | 0.9 | GO:0048273 | mitogen-activated protein kinase p38 binding(GO:0048273) |
| 0.0 | 1.0 | GO:0005528 | macrolide binding(GO:0005527) FK506 binding(GO:0005528) |
| 0.0 | 0.9 | GO:0033038 | bitter taste receptor activity(GO:0033038) |
| 0.0 | 0.3 | GO:0016453 | acetyl-CoA C-acetyltransferase activity(GO:0003985) C-acetyltransferase activity(GO:0016453) |
| 0.0 | 1.2 | GO:0005521 | lamin binding(GO:0005521) |
| 0.0 | 0.2 | GO:2001069 | glycogen binding(GO:2001069) |
| 0.0 | 0.8 | GO:0051864 | histone demethylase activity (H3-K36 specific)(GO:0051864) |
| 0.0 | 4.4 | GO:0008170 | N-methyltransferase activity(GO:0008170) |
| 0.0 | 0.2 | GO:0008142 | oxysterol binding(GO:0008142) |
| 0.0 | 0.8 | GO:0004993 | G-protein coupled serotonin receptor activity(GO:0004993) |
| 0.0 | 3.7 | GO:0016503 | pheromone receptor activity(GO:0016503) |
| 0.0 | 0.7 | GO:0050136 | NADH dehydrogenase (ubiquinone) activity(GO:0008137) NADH dehydrogenase (quinone) activity(GO:0050136) |
| 0.0 | 0.6 | GO:0030553 | cGMP binding(GO:0030553) |
| 0.0 | 1.1 | GO:0008536 | Ran GTPase binding(GO:0008536) |
| 0.0 | 0.2 | GO:0097322 | 7SK snRNA binding(GO:0097322) |
| 0.0 | 0.8 | GO:0046966 | thyroid hormone receptor binding(GO:0046966) |
| 0.0 | 1.3 | GO:0003743 | translation initiation factor activity(GO:0003743) |
| 0.0 | 2.0 | GO:0004869 | cysteine-type endopeptidase inhibitor activity(GO:0004869) |
| 0.0 | 0.7 | GO:0001103 | RNA polymerase II repressing transcription factor binding(GO:0001103) |
| 0.0 | 2.1 | GO:0003697 | single-stranded DNA binding(GO:0003697) |
| 0.0 | 1.9 | GO:0031490 | chromatin DNA binding(GO:0031490) |
| 0.0 | 2.4 | GO:0005178 | integrin binding(GO:0005178) |
| 0.0 | 0.8 | GO:0048487 | beta-tubulin binding(GO:0048487) |
| 0.0 | 0.7 | GO:0030544 | Hsp70 protein binding(GO:0030544) |
| 0.0 | 3.8 | GO:0061630 | ubiquitin protein ligase activity(GO:0061630) ubiquitin-like protein ligase activity(GO:0061659) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 3.4 | PID INTEGRIN4 PATHWAY | Alpha6 beta4 integrin-ligand interactions |
| 0.1 | 3.4 | PID AURORA A PATHWAY | Aurora A signaling |
| 0.1 | 0.8 | ST JAK STAT PATHWAY | Jak-STAT Pathway |
| 0.0 | 1.5 | PID TOLL ENDOGENOUS PATHWAY | Endogenous TLR signaling |
| 0.0 | 1.1 | PID INTEGRIN3 PATHWAY | Beta3 integrin cell surface interactions |
| 0.0 | 1.0 | PID INTEGRIN CS PATHWAY | Integrin family cell surface interactions |
| 0.0 | 0.9 | PID P38 MKK3 6PATHWAY | p38 MAPK signaling pathway |
| 0.0 | 1.1 | PID LYMPH ANGIOGENESIS PATHWAY | VEGFR3 signaling in lymphatic endothelium |
| 0.0 | 2.5 | PID HDAC CLASSI PATHWAY | Signaling events mediated by HDAC Class I |
| 0.0 | 0.6 | PID RHODOPSIN PATHWAY | Visual signal transduction: Rods |
| 0.0 | 3.5 | NABA ECM GLYCOPROTEINS | Genes encoding structural ECM glycoproteins |
| 0.0 | 0.9 | PID PS1 PATHWAY | Presenilin action in Notch and Wnt signaling |
| 0.0 | 1.3 | PID FCER1 PATHWAY | Fc-epsilon receptor I signaling in mast cells |
| 0.0 | 0.6 | PID BARD1 PATHWAY | BARD1 signaling events |
| 0.0 | 0.5 | PID EPHA FWDPATHWAY | EPHA forward signaling |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 2.7 | REACTOME REGULATION OF PYRUVATE DEHYDROGENASE PDH COMPLEX | Genes involved in Regulation of pyruvate dehydrogenase (PDH) complex |
| 0.2 | 4.4 | REACTOME AMINE DERIVED HORMONES | Genes involved in Amine-derived hormones |
| 0.2 | 1.6 | REACTOME THE ACTIVATION OF ARYLSULFATASES | Genes involved in The activation of arylsulfatases |
| 0.1 | 1.9 | REACTOME ACYL CHAIN REMODELLING OF PS | Genes involved in Acyl chain remodelling of PS |
| 0.1 | 3.6 | REACTOME DOWNSTREAM TCR SIGNALING | Genes involved in Downstream TCR signaling |
| 0.1 | 0.9 | REACTOME IRAK2 MEDIATED ACTIVATION OF TAK1 COMPLEX UPON TLR7 8 OR 9 STIMULATION | Genes involved in IRAK2 mediated activation of TAK1 complex upon TLR7/8 or 9 stimulation |
| 0.1 | 1.1 | REACTOME PLATELET ADHESION TO EXPOSED COLLAGEN | Genes involved in Platelet Adhesion to exposed collagen |
| 0.1 | 1.1 | REACTOME P2Y RECEPTORS | Genes involved in P2Y receptors |
| 0.0 | 0.9 | REACTOME NOTCH HLH TRANSCRIPTION PATHWAY | Genes involved in Notch-HLH transcription pathway |
| 0.0 | 0.8 | REACTOME REGULATION OF IFNG SIGNALING | Genes involved in Regulation of IFNG signaling |
| 0.0 | 1.7 | REACTOME IMMUNOREGULATORY INTERACTIONS BETWEEN A LYMPHOID AND A NON LYMPHOID CELL | Genes involved in Immunoregulatory interactions between a Lymphoid and a non-Lymphoid cell |
| 0.0 | 3.4 | REACTOME LOSS OF NLP FROM MITOTIC CENTROSOMES | Genes involved in Loss of Nlp from mitotic centrosomes |
| 0.0 | 1.3 | REACTOME DEADENYLATION OF MRNA | Genes involved in Deadenylation of mRNA |
| 0.0 | 1.1 | REACTOME SIGNAL REGULATORY PROTEIN SIRP FAMILY INTERACTIONS | Genes involved in Signal regulatory protein (SIRP) family interactions |
| 0.0 | 3.4 | REACTOME INTEGRIN CELL SURFACE INTERACTIONS | Genes involved in Integrin cell surface interactions |
| 0.0 | 0.6 | REACTOME CGMP EFFECTS | Genes involved in cGMP effects |
| 0.0 | 1.6 | REACTOME TRANSPORT OF VITAMINS NUCLEOSIDES AND RELATED MOLECULES | Genes involved in Transport of vitamins, nucleosides, and related molecules |
| 0.0 | 5.8 | REACTOME G ALPHA S SIGNALLING EVENTS | Genes involved in G alpha (s) signalling events |
| 0.0 | 0.3 | REACTOME MRNA DECAY BY 3 TO 5 EXORIBONUCLEASE | Genes involved in mRNA Decay by 3' to 5' Exoribonuclease |
| 0.0 | 0.6 | REACTOME FANCONI ANEMIA PATHWAY | Genes involved in Fanconi Anemia pathway |
| 0.0 | 0.8 | REACTOME GLYCOSPHINGOLIPID METABOLISM | Genes involved in Glycosphingolipid metabolism |
| 0.0 | 1.2 | REACTOME INTERFERON ALPHA BETA SIGNALING | Genes involved in Interferon alpha/beta signaling |
| 0.0 | 0.5 | REACTOME AMINO ACID SYNTHESIS AND INTERCONVERSION TRANSAMINATION | Genes involved in Amino acid synthesis and interconversion (transamination) |
| 0.0 | 5.3 | REACTOME ANTIGEN PROCESSING UBIQUITINATION PROTEASOME DEGRADATION | Genes involved in Antigen processing: Ubiquitination & Proteasome degradation |
| 0.0 | 0.5 | REACTOME TRANSPORT OF RIBONUCLEOPROTEINS INTO THE HOST NUCLEUS | Genes involved in Transport of Ribonucleoproteins into the Host Nucleus |
| 0.0 | 1.5 | REACTOME MYD88 MAL CASCADE INITIATED ON PLASMA MEMBRANE | Genes involved in MyD88:Mal cascade initiated on plasma membrane |
| 0.0 | 0.3 | REACTOME BRANCHED CHAIN AMINO ACID CATABOLISM | Genes involved in Branched-chain amino acid catabolism |
| 0.0 | 1.6 | REACTOME TRANSPORT OF INORGANIC CATIONS ANIONS AND AMINO ACIDS OLIGOPEPTIDES | Genes involved in Transport of inorganic cations/anions and amino acids/oligopeptides |