Project
PRJNA375882: Comprehensive Mouse Transcriptomic BodyMap
Navigation
Downloads
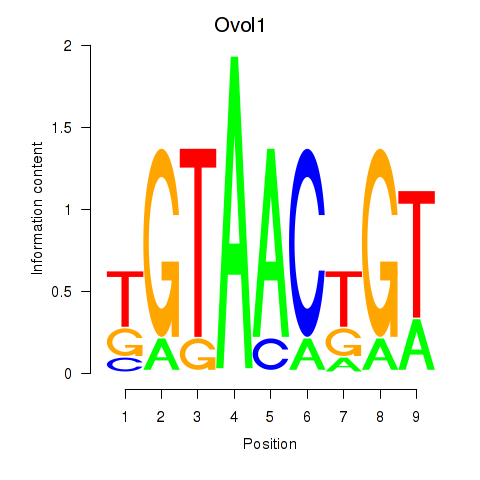
Results for Ovol1
Z-value: 0.31
Transcription factors associated with Ovol1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Ovol1
|
ENSMUSG00000024922.3 | Ovol1 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Ovol1 | mm39_v1_chr19_-_5610628_5610674 | -0.60 | 3.2e-08 | Click! |
Activity profile of Ovol1 motif
Sorted Z-values of Ovol1 motif
Network of associatons between targets according to the STRING database.
First level regulatory network of Ovol1
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr6_+_21215472 | 3.84 |
ENSMUST00000081542.6
|
Kcnd2
|
potassium voltage-gated channel, Shal-related family, member 2 |
| chr11_-_42070517 | 2.78 |
ENSMUST00000206105.2
ENSMUST00000153147.3 |
Gabra1
|
gamma-aminobutyric acid (GABA) A receptor, subunit alpha 1 |
| chr18_+_37153149 | 1.73 |
ENSMUST00000047614.4
|
Pcdha12
|
protocadherin alpha 12 |
| chr18_+_37822865 | 1.49 |
ENSMUST00000195112.2
|
Pcdhgb2
|
protocadherin gamma subfamily B, 2 |
| chr7_+_18810167 | 1.35 |
ENSMUST00000108479.2
|
Dmwd
|
dystrophia myotonica-containing WD repeat motif |
| chr2_-_79959178 | 1.35 |
ENSMUST00000102654.11
ENSMUST00000102655.10 |
Pde1a
|
phosphodiesterase 1A, calmodulin-dependent |
| chrX_-_23151771 | 0.80 |
ENSMUST00000115319.9
|
Klhl13
|
kelch-like 13 |
| chr5_+_65505657 | 0.77 |
ENSMUST00000031096.11
|
Klb
|
klotho beta |
| chr14_-_34032311 | 0.74 |
ENSMUST00000111917.3
ENSMUST00000228704.2 |
Shld2
|
shieldin complex subunit 2 |
| chr11_-_32217547 | 0.60 |
ENSMUST00000109389.9
ENSMUST00000129010.2 ENSMUST00000020530.12 |
Nprl3
|
nitrogen permease regulator-like 3 |
| chr17_+_72076728 | 0.59 |
ENSMUST00000230305.2
|
Clip4
|
CAP-GLY domain containing linker protein family, member 4 |
| chr14_-_34032450 | 0.58 |
ENSMUST00000227375.2
|
Shld2
|
shieldin complex subunit 2 |
| chr17_+_72076678 | 0.57 |
ENSMUST00000230427.2
ENSMUST00000229952.2 ENSMUST00000230333.2 |
Clip4
|
CAP-GLY domain containing linker protein family, member 4 |
| chr14_-_75829389 | 0.47 |
ENSMUST00000165569.3
ENSMUST00000035243.5 |
Cby2
|
chibby family member 2 |
| chr8_-_106052884 | 0.44 |
ENSMUST00000210412.2
ENSMUST00000210801.2 ENSMUST00000070508.8 |
Lrrc29
|
leucine rich repeat containing 29 |
| chr2_-_44912927 | 0.30 |
ENSMUST00000202432.4
|
Zeb2
|
zinc finger E-box binding homeobox 2 |
| chr2_+_172090412 | 0.22 |
ENSMUST00000038532.2
|
Mc3r
|
melanocortin 3 receptor |
| chr2_-_89156522 | 0.20 |
ENSMUST00000099785.2
|
Olfr1232
|
olfactory receptor 1232 |
| chr10_+_73782857 | 0.11 |
ENSMUST00000191709.6
ENSMUST00000193739.6 ENSMUST00000195531.6 |
Pcdh15
|
protocadherin 15 |
| chr8_+_106052970 | 0.05 |
ENSMUST00000015000.12
ENSMUST00000098453.9 |
Tmem208
|
transmembrane protein 208 |
| chr19_+_12186932 | 0.05 |
ENSMUST00000072316.5
|
Olfr1431
|
olfactory receptor 1431 |
| chr2_-_111100733 | 0.04 |
ENSMUST00000099619.6
|
Olfr1277
|
olfactory receptor 1277 |
| chr14_-_51493569 | 0.02 |
ENSMUST00000061936.8
|
Rnase2a
|
ribonuclease, RNase A family, 2A (liver, eosinophil-derived neurotoxin) |
| chr2_-_111104451 | 0.01 |
ENSMUST00000214760.2
|
Olfr1277
|
olfactory receptor 1277 |
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 2.8 | GO:0071420 | cellular response to histamine(GO:0071420) |
| 0.1 | 4.1 | GO:0045475 | locomotor rhythm(GO:0045475) |
| 0.1 | 0.8 | GO:0090080 | positive regulation of MAPKKK cascade by fibroblast growth factor receptor signaling pathway(GO:0090080) |
| 0.1 | 1.3 | GO:0046069 | cGMP catabolic process(GO:0046069) |
| 0.0 | 0.3 | GO:1902748 | positive regulation of lens fiber cell differentiation(GO:1902748) |
| 0.0 | 0.1 | GO:0050973 | detection of mechanical stimulus involved in equilibrioception(GO:0050973) |
| 0.0 | 0.6 | GO:2000785 | TORC1 signaling(GO:0038202) regulation of autophagosome assembly(GO:2000785) |
| 0.0 | 0.1 | GO:0006624 | vacuolar protein processing(GO:0006624) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.6 | GO:1990130 | Iml1 complex(GO:1990130) |
| 0.1 | 3.8 | GO:0097038 | perinuclear endoplasmic reticulum(GO:0097038) |
| 0.1 | 2.8 | GO:1902711 | GABA-A receptor complex(GO:1902711) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.8 | 3.8 | GO:0005250 | A-type (transient outward) potassium channel activity(GO:0005250) |
| 0.3 | 2.8 | GO:1904315 | transmitter-gated ion channel activity involved in regulation of postsynaptic membrane potential(GO:1904315) |
| 0.1 | 1.3 | GO:0004117 | calmodulin-dependent cyclic-nucleotide phosphodiesterase activity(GO:0004117) calcium- and calmodulin-regulated 3',5'-cyclic-GMP phosphodiesterase activity(GO:0048101) |
| 0.1 | 0.2 | GO:0004980 | melanocyte-stimulating hormone receptor activity(GO:0004980) |
| 0.0 | 0.8 | GO:0017134 | fibroblast growth factor binding(GO:0017134) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.8 | PID AURORA B PATHWAY | Aurora B signaling |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 2.8 | REACTOME GABA A RECEPTOR ACTIVATION | Genes involved in GABA A receptor activation |
| 0.1 | 3.8 | REACTOME VOLTAGE GATED POTASSIUM CHANNELS | Genes involved in Voltage gated Potassium channels |
| 0.0 | 1.3 | REACTOME CGMP EFFECTS | Genes involved in cGMP effects |
| 0.0 | 0.8 | REACTOME FGFR LIGAND BINDING AND ACTIVATION | Genes involved in FGFR ligand binding and activation |