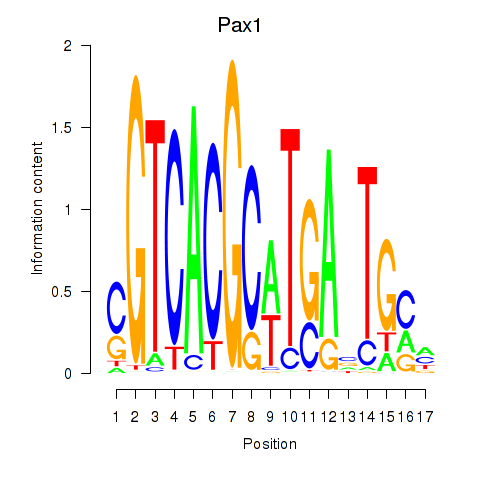
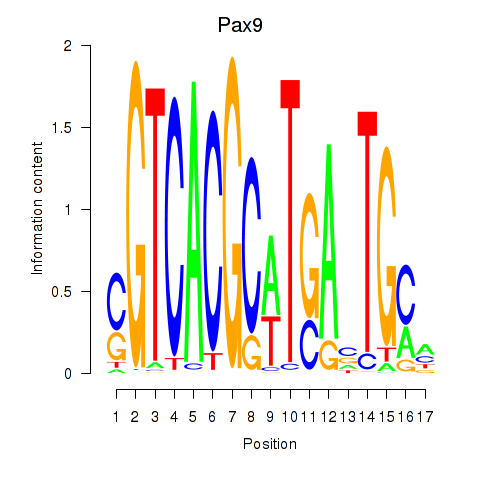
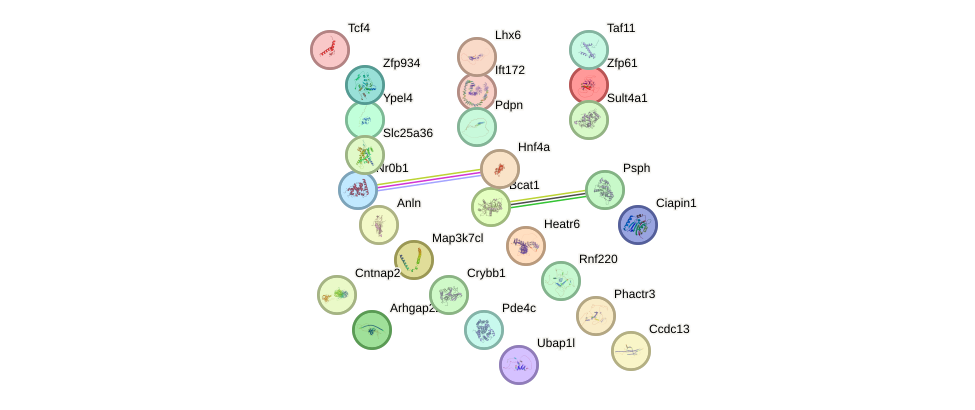
|
chr2_+_177834868
Show fit
|
6.91 |
ENSMUST00000103065.2
|
Phactr3
|
phosphatase and actin regulator 3
|
|
chr6_-_144994534
Show fit
|
6.54 |
ENSMUST00000032402.12
|
Bcat1
|
branched chain aminotransferase 1, cytosolic
|
|
chr5_+_112403678
Show fit
|
5.78 |
ENSMUST00000031286.13
ENSMUST00000131673.8
ENSMUST00000112375.2
|
Crybb1
|
crystallin, beta B1
|
|
chr6_-_144994327
Show fit
|
5.59 |
ENSMUST00000204138.3
|
Bcat1
|
branched chain aminotransferase 1, cytosolic
|
|
chr15_-_83989801
Show fit
|
5.26 |
ENSMUST00000229826.2
ENSMUST00000082365.6
|
Sult4a1
|
sulfotransferase family 4A, member 1
|
|
chr18_+_69477541
Show fit
|
2.19 |
ENSMUST00000114985.10
ENSMUST00000128706.8
ENSMUST00000201781.4
ENSMUST00000202674.4
|
Tcf4
|
transcription factor 4
|
|
chr9_-_22300409
Show fit
|
1.72 |
ENSMUST00000040912.9
|
Anln
|
anillin, actin binding protein
|
|
chr2_+_84564394
Show fit
|
1.51 |
ENSMUST00000238573.2
ENSMUST00000090729.9
|
Ypel4
|
yippee like 4
|
|
chr5_-_129864202
Show fit
|
1.51 |
ENSMUST00000136507.4
|
Psph
|
phosphoserine phosphatase
|
|
chr9_-_121668527
Show fit
|
1.43 |
ENSMUST00000135986.9
|
Ccdc13
|
coiled-coil domain containing 13
|
|
chr17_-_24424456
Show fit
|
1.40 |
ENSMUST00000201583.2
ENSMUST00000202925.4
ENSMUST00000167791.9
ENSMUST00000201960.4
ENSMUST00000040474.11
ENSMUST00000201089.4
ENSMUST00000201301.4
ENSMUST00000201805.4
ENSMUST00000168410.9
ENSMUST00000097376.10
|
Tbc1d24
|
TBC1 domain family, member 24
|
|
chr9_+_65268304
Show fit
|
1.38 |
ENSMUST00000147185.3
|
Ubap1l
|
ubiquitin-associated protein 1-like
|
|
chr4_-_117232650
Show fit
|
1.37 |
ENSMUST00000094853.9
|
Rnf220
|
ring finger protein 220
|
|
chr6_+_47021979
Show fit
|
1.20 |
ENSMUST00000150737.2
|
Cntnap2
|
contactin associated protein-like 2
|
|
chr4_-_143026068
Show fit
|
1.09 |
ENSMUST00000030317.14
|
Pdpn
|
podoplanin
|
|
chr9_-_96993169
Show fit
|
1.01 |
ENSMUST00000085206.11
|
Slc25a36
|
solute carrier family 25, member 36
|
|
chr10_+_77814358
Show fit
|
1.00 |
ENSMUST00000105397.10
ENSMUST00000105398.2
|
Cfap410
|
cilia and flagella associated protein 410
|
|
chr18_-_10706701
Show fit
|
0.87 |
ENSMUST00000002549.9
ENSMUST00000117726.9
ENSMUST00000117828.9
|
Abhd3
|
abhydrolase domain containing 3
|
|
chr5_-_31448370
Show fit
|
0.85 |
ENSMUST00000041565.11
ENSMUST00000201809.2
|
Ift172
|
intraflagellar transport 172
|
|
chr5_+_129864044
Show fit
|
0.77 |
ENSMUST00000201414.5
|
Cct6a
|
chaperonin containing Tcp1, subunit 6a (zeta)
|
|
chr2_-_35995283
Show fit
|
0.77 |
ENSMUST00000112960.8
ENSMUST00000112967.12
ENSMUST00000112963.8
|
Lhx6
|
LIM homeobox protein 6
|
|
chr7_-_23998735
Show fit
|
0.76 |
ENSMUST00000145131.8
|
Zfp61
|
zinc finger protein 61
|
|
chr4_-_143026033
Show fit
|
0.73 |
ENSMUST00000119654.2
|
Pdpn
|
podoplanin
|
|
chr14_+_32938774
Show fit
|
0.68 |
ENSMUST00000111956.9
|
Arhgap22
|
Rho GTPase activating protein 22
|
|
chr8_+_31640332
Show fit
|
0.65 |
ENSMUST00000209851.2
ENSMUST00000098842.3
ENSMUST00000210129.2
|
Tti2
|
TELO2 interacting protein 2
|
|
chr17_-_28128180
Show fit
|
0.58 |
ENSMUST00000114848.8
|
Taf11
|
TATA-box binding protein associated factor 11
|
|
chr13_-_62924903
Show fit
|
0.42 |
ENSMUST00000167516.3
|
Gm5141
|
predicted gene 5141
|
|
chr16_+_87350202
Show fit
|
0.39 |
ENSMUST00000026700.8
|
Map3k7cl
|
Map3k7 C-terminal like
|
|
chr8_+_71176713
Show fit
|
0.24 |
ENSMUST00000034307.14
ENSMUST00000239435.2
ENSMUST00000239487.2
ENSMUST00000110095.3
|
Pde4c
|
phosphodiesterase 4C, cAMP specific
|
|
chr1_+_87683592
Show fit
|
0.19 |
ENSMUST00000144047.8
ENSMUST00000027512.13
ENSMUST00000113186.8
ENSMUST00000113190.3
|
Atg16l1
|
autophagy related 16-like 1 (S. cerevisiae)
|
|
chr18_-_78640066
Show fit
|
0.19 |
ENSMUST00000235389.2
ENSMUST00000237674.2
|
Slc14a2
|
solute carrier family 14 (urea transporter), member 2
|
|
chr8_-_95564881
Show fit
|
0.16 |
ENSMUST00000034233.15
ENSMUST00000162538.9
|
Ciapin1
|
cytokine induced apoptosis inhibitor 1
|
|
chrX_+_85235370
Show fit
|
0.12 |
ENSMUST00000026036.5
|
Nr0b1
|
nuclear receptor subfamily 0, group B, member 1
|
|
chr11_+_83644514
Show fit
|
0.01 |
ENSMUST00000001002.8
|
Heatr6
|
HEAT repeat containing 6
|
|
chr13_-_62668265
Show fit
|
0.00 |
ENSMUST00000082203.7
|
Zfp934
|
zinc finger protein 934
|