Project
PRJNA375882: Comprehensive Mouse Transcriptomic BodyMap
Navigation
Downloads
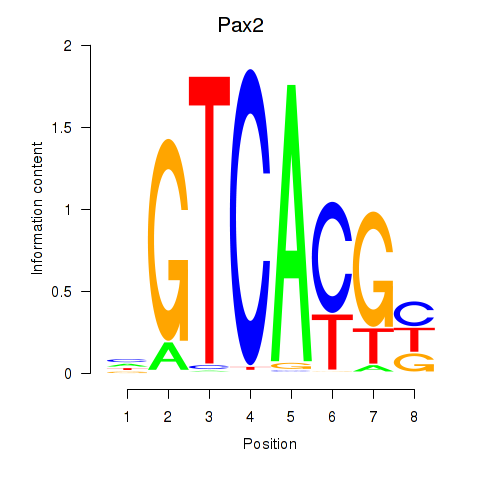
Results for Pax2
Z-value: 0.72
Transcription factors associated with Pax2
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Pax2
|
ENSMUSG00000004231.16 | Pax2 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Pax2 | mm39_v1_chr19_+_44745833_44745833 | -0.14 | 2.5e-01 | Click! |
Activity profile of Pax2 motif
Sorted Z-values of Pax2 motif
Network of associatons between targets according to the STRING database.
First level regulatory network of Pax2
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr17_+_44499451 | 3.70 |
ENSMUST00000024755.7
|
Clic5
|
chloride intracellular channel 5 |
| chr3_+_104545974 | 3.66 |
ENSMUST00000046212.2
|
Slc16a1
|
solute carrier family 16 (monocarboxylic acid transporters), member 1 |
| chr14_+_55797468 | 3.64 |
ENSMUST00000147981.2
ENSMUST00000133256.8 |
Dcaf11
|
DDB1 and CUL4 associated factor 11 |
| chr1_+_13738967 | 3.43 |
ENSMUST00000088542.4
|
Xkr9
|
X-linked Kx blood group related 9 |
| chr10_-_17823736 | 3.18 |
ENSMUST00000037879.8
|
Heca
|
hdc homolog, cell cycle regulator |
| chr4_+_99544536 | 3.16 |
ENSMUST00000087285.5
|
Foxd3
|
forkhead box D3 |
| chr1_+_88139678 | 2.92 |
ENSMUST00000073049.7
|
Ugt1a1
|
UDP glucuronosyltransferase 1 family, polypeptide A1 |
| chr19_+_43600738 | 2.89 |
ENSMUST00000057178.11
|
Nkx2-3
|
NK2 homeobox 3 |
| chr7_+_46700349 | 2.84 |
ENSMUST00000010451.8
|
Tmem86a
|
transmembrane protein 86A |
| chr14_-_34310602 | 2.79 |
ENSMUST00000064098.14
ENSMUST00000090040.12 ENSMUST00000022330.9 ENSMUST00000022327.13 |
Ldb3
|
LIM domain binding 3 |
| chr17_-_12988492 | 2.74 |
ENSMUST00000024599.14
|
Igf2r
|
insulin-like growth factor 2 receptor |
| chr18_-_64794338 | 2.73 |
ENSMUST00000025482.10
|
Atp8b1
|
ATPase, class I, type 8B, member 1 |
| chr2_+_67004178 | 2.70 |
ENSMUST00000239009.2
ENSMUST00000238912.2 |
Xirp2
|
xin actin-binding repeat containing 2 |
| chr16_-_97763780 | 2.70 |
ENSMUST00000232187.2
ENSMUST00000231263.2 ENSMUST00000052089.9 ENSMUST00000063605.15 ENSMUST00000113734.9 ENSMUST00000231560.2 ENSMUST00000232165.2 |
Zbtb21
C2cd2
|
zinc finger and BTB domain containing 21 C2 calcium-dependent domain containing 2 |
| chr10_+_126899468 | 2.69 |
ENSMUST00000120226.8
ENSMUST00000133115.8 |
Cdk4
|
cyclin-dependent kinase 4 |
| chr5_-_66309244 | 2.67 |
ENSMUST00000167950.8
|
Rbm47
|
RNA binding motif protein 47 |
| chr14_-_34310438 | 2.67 |
ENSMUST00000228044.2
ENSMUST00000022328.14 |
Ldb3
|
LIM domain binding 3 |
| chr14_+_14475188 | 2.64 |
ENSMUST00000026315.8
|
Dnase1l3
|
deoxyribonuclease 1-like 3 |
| chr14_-_34310637 | 2.63 |
ENSMUST00000227819.2
|
Ldb3
|
LIM domain binding 3 |
| chr10_+_126899396 | 2.59 |
ENSMUST00000006911.12
|
Cdk4
|
cyclin-dependent kinase 4 |
| chr4_-_89229256 | 2.57 |
ENSMUST00000097981.6
|
Cdkn2b
|
cyclin dependent kinase inhibitor 2B |
| chr6_-_52203146 | 2.47 |
ENSMUST00000114425.3
|
Hoxa9
|
homeobox A9 |
| chr6_+_17463925 | 2.47 |
ENSMUST00000115442.8
|
Met
|
met proto-oncogene |
| chr2_+_164675697 | 2.41 |
ENSMUST00000143780.9
|
Ctsa
|
cathepsin A |
| chr6_+_17463748 | 2.37 |
ENSMUST00000115443.8
|
Met
|
met proto-oncogene |
| chr6_-_33037107 | 2.29 |
ENSMUST00000115091.2
ENSMUST00000127666.8 |
Chchd3
|
coiled-coil-helix-coiled-coil-helix domain containing 3 |
| chr19_-_56378309 | 2.22 |
ENSMUST00000166203.2
ENSMUST00000167239.8 |
Nrap
|
nebulin-related anchoring protein |
| chr6_-_6217126 | 2.14 |
ENSMUST00000188414.4
|
Slc25a13
|
solute carrier family 25 (mitochondrial carrier, adenine nucleotide translocator), member 13 |
| chr6_-_6217021 | 2.10 |
ENSMUST00000015256.15
|
Slc25a13
|
solute carrier family 25 (mitochondrial carrier, adenine nucleotide translocator), member 13 |
| chr17_-_46956920 | 2.09 |
ENSMUST00000233974.2
|
Klc4
|
kinesin light chain 4 |
| chr7_-_19363280 | 2.09 |
ENSMUST00000094762.10
ENSMUST00000049912.15 ENSMUST00000098754.5 |
Relb
|
avian reticuloendotheliosis viral (v-rel) oncogene related B |
| chr6_-_33037191 | 2.07 |
ENSMUST00000066379.11
|
Chchd3
|
coiled-coil-helix-coiled-coil-helix domain containing 3 |
| chr1_+_61017057 | 2.04 |
ENSMUST00000027162.12
ENSMUST00000102827.4 |
Icos
|
inducible T cell co-stimulator |
| chr14_-_47426863 | 1.99 |
ENSMUST00000089959.7
|
Gch1
|
GTP cyclohydrolase 1 |
| chr4_+_62278932 | 1.95 |
ENSMUST00000084526.12
|
Slc31a1
|
solute carrier family 31, member 1 |
| chr6_+_17463819 | 1.93 |
ENSMUST00000140070.8
|
Met
|
met proto-oncogene |
| chr11_-_69563133 | 1.91 |
ENSMUST00000163666.3
|
Eif4a1
|
eukaryotic translation initiation factor 4A1 |
| chr1_-_128256048 | 1.88 |
ENSMUST00000073490.7
|
Lct
|
lactase |
| chr5_-_66308421 | 1.87 |
ENSMUST00000200775.4
ENSMUST00000094756.11 |
Rbm47
|
RNA binding motif protein 47 |
| chr10_+_41395870 | 1.84 |
ENSMUST00000189300.2
|
Cd164
|
CD164 antigen |
| chr14_+_55797934 | 1.83 |
ENSMUST00000121622.8
ENSMUST00000143431.2 ENSMUST00000150481.8 |
Dcaf11
|
DDB1 and CUL4 associated factor 11 |
| chr1_-_136888118 | 1.82 |
ENSMUST00000192357.6
ENSMUST00000027649.14 |
Nr5a2
|
nuclear receptor subfamily 5, group A, member 2 |
| chr3_-_123029745 | 1.82 |
ENSMUST00000106426.8
|
Synpo2
|
synaptopodin 2 |
| chr15_+_59520493 | 1.82 |
ENSMUST00000118228.2
|
Trib1
|
tribbles pseudokinase 1 |
| chr5_-_124563611 | 1.81 |
ENSMUST00000198420.5
|
Sbno1
|
strawberry notch 1 |
| chr2_+_121279842 | 1.80 |
ENSMUST00000110615.8
ENSMUST00000099475.12 |
Serf2
|
small EDRK-rich factor 2 |
| chr14_-_30740946 | 1.79 |
ENSMUST00000228341.2
|
Gnl3
|
guanine nucleotide binding protein-like 3 (nucleolar) |
| chr3_-_123029782 | 1.78 |
ENSMUST00000106427.8
ENSMUST00000198584.2 |
Synpo2
|
synaptopodin 2 |
| chr15_+_59520199 | 1.75 |
ENSMUST00000067543.8
|
Trib1
|
tribbles pseudokinase 1 |
| chr18_-_61344644 | 1.74 |
ENSMUST00000146409.8
|
Slc26a2
|
solute carrier family 26 (sulfate transporter), member 2 |
| chr17_-_67939702 | 1.72 |
ENSMUST00000097290.4
|
Lrrc30
|
leucine rich repeat containing 30 |
| chr5_-_24745436 | 1.70 |
ENSMUST00000048302.13
ENSMUST00000119657.2 |
Asb10
|
ankyrin repeat and SOCS box-containing 10 |
| chr7_+_89779564 | 1.70 |
ENSMUST00000208742.2
ENSMUST00000049537.9 |
Picalm
|
phosphatidylinositol binding clathrin assembly protein |
| chr7_+_89779493 | 1.67 |
ENSMUST00000208730.2
|
Picalm
|
phosphatidylinositol binding clathrin assembly protein |
| chr5_+_119970733 | 1.62 |
ENSMUST00000202723.4
|
Tbx5
|
T-box 5 |
| chrX_-_161671421 | 1.59 |
ENSMUST00000033723.4
|
Syap1
|
synapse associated protein 1 |
| chr10_+_41395410 | 1.59 |
ENSMUST00000019962.15
|
Cd164
|
CD164 antigen |
| chr2_+_147206910 | 1.56 |
ENSMUST00000109968.3
|
Pax1
|
paired box 1 |
| chr11_+_70453666 | 1.55 |
ENSMUST00000072237.13
ENSMUST00000072873.14 |
Mink1
|
misshapen-like kinase 1 (zebrafish) |
| chr2_-_28806639 | 1.54 |
ENSMUST00000113847.3
|
Barhl1
|
BarH like homeobox 1 |
| chr5_-_46013838 | 1.53 |
ENSMUST00000087164.10
ENSMUST00000121573.8 |
Lcorl
|
ligand dependent nuclear receptor corepressor-like |
| chr11_-_101062111 | 1.52 |
ENSMUST00000164474.8
ENSMUST00000043397.14 |
Plekhh3
|
pleckstrin homology domain containing, family H (with MyTH4 domain) member 3 |
| chr13_+_75855695 | 1.52 |
ENSMUST00000222194.2
ENSMUST00000223535.2 ENSMUST00000222853.2 |
Ell2
|
elongation factor for RNA polymerase II 2 |
| chr2_+_18677195 | 1.52 |
ENSMUST00000171845.8
ENSMUST00000061158.5 |
Commd3
|
COMM domain containing 3 |
| chr3_-_89959917 | 1.52 |
ENSMUST00000197903.5
|
Ubap2l
|
ubiquitin-associated protein 2-like |
| chr14_+_55798362 | 1.47 |
ENSMUST00000072530.11
ENSMUST00000128490.9 |
Dcaf11
|
DDB1 and CUL4 associated factor 11 |
| chr11_+_70453724 | 1.47 |
ENSMUST00000102559.11
|
Mink1
|
misshapen-like kinase 1 (zebrafish) |
| chr9_+_108765701 | 1.45 |
ENSMUST00000026743.14
ENSMUST00000194047.3 |
Uqcrc1
|
ubiquinol-cytochrome c reductase core protein 1 |
| chr11_+_35660288 | 1.43 |
ENSMUST00000018990.8
|
Pank3
|
pantothenate kinase 3 |
| chr19_+_46293210 | 1.42 |
ENSMUST00000236591.2
|
Nfkb2
|
nuclear factor of kappa light polypeptide gene enhancer in B cells 2, p49/p100 |
| chr3_-_142587419 | 1.41 |
ENSMUST00000174422.8
ENSMUST00000173830.8 |
Pkn2
|
protein kinase N2 |
| chr19_+_46293160 | 1.40 |
ENSMUST00000073116.13
|
Nfkb2
|
nuclear factor of kappa light polypeptide gene enhancer in B cells 2, p49/p100 |
| chr14_+_55797443 | 1.40 |
ENSMUST00000117236.8
|
Dcaf11
|
DDB1 and CUL4 associated factor 11 |
| chr6_+_121320339 | 1.39 |
ENSMUST00000168295.2
|
Slc6a12
|
solute carrier family 6 (neurotransmitter transporter, betaine/GABA), member 12 |
| chr10_+_43355113 | 1.39 |
ENSMUST00000040147.8
|
Bend3
|
BEN domain containing 3 |
| chr16_-_94171556 | 1.37 |
ENSMUST00000113906.9
|
Pigp
|
phosphatidylinositol glycan anchor biosynthesis, class P |
| chr15_-_53209513 | 1.37 |
ENSMUST00000077273.9
|
Ext1
|
exostosin glycosyltransferase 1 |
| chr2_+_84669739 | 1.36 |
ENSMUST00000146816.8
ENSMUST00000028469.14 |
Slc43a1
|
solute carrier family 43, member 1 |
| chr11_+_88890202 | 1.36 |
ENSMUST00000100627.9
ENSMUST00000107896.10 ENSMUST00000000284.7 |
Trim25
|
tripartite motif-containing 25 |
| chr11_-_101061153 | 1.34 |
ENSMUST00000123864.2
|
Plekhh3
|
pleckstrin homology domain containing, family H (with MyTH4 domain) member 3 |
| chr4_-_129083251 | 1.34 |
ENSMUST00000117965.8
|
S100pbp
|
S100P binding protein |
| chr7_+_89779421 | 1.33 |
ENSMUST00000207225.2
ENSMUST00000207484.2 ENSMUST00000209068.2 |
Picalm
|
phosphatidylinositol binding clathrin assembly protein |
| chr5_-_65855199 | 1.33 |
ENSMUST00000031104.7
|
Pds5a
|
PDS5 cohesin associated factor A |
| chr5_-_24745970 | 1.32 |
ENSMUST00000117900.8
|
Asb10
|
ankyrin repeat and SOCS box-containing 10 |
| chr5_-_113285852 | 1.31 |
ENSMUST00000212276.2
|
2900026A02Rik
|
RIKEN cDNA 2900026A02 gene |
| chr12_-_84497718 | 1.31 |
ENSMUST00000085192.7
ENSMUST00000220491.2 |
Aldh6a1
|
aldehyde dehydrogenase family 6, subfamily A1 |
| chr12_-_85421467 | 1.30 |
ENSMUST00000040766.9
|
Tmed10
|
transmembrane p24 trafficking protein 10 |
| chr6_-_52135261 | 1.29 |
ENSMUST00000000964.6
ENSMUST00000120363.2 |
Hoxa1
|
homeobox A1 |
| chr18_-_31742946 | 1.28 |
ENSMUST00000060396.7
|
Slc25a46
|
solute carrier family 25, member 46 |
| chr19_-_7218363 | 1.26 |
ENSMUST00000236769.2
|
Naa40
|
N(alpha)-acetyltransferase 40, NatD catalytic subunit |
| chr2_+_74512638 | 1.25 |
ENSMUST00000142312.3
|
Hoxd11
|
homeobox D11 |
| chr18_+_31742565 | 1.24 |
ENSMUST00000164667.2
|
B930094E09Rik
|
RIKEN cDNA B930094E09 gene |
| chr3_+_95226093 | 1.23 |
ENSMUST00000139866.2
|
Cers2
|
ceramide synthase 2 |
| chr5_+_28370687 | 1.22 |
ENSMUST00000036177.9
|
En2
|
engrailed 2 |
| chr3_+_134918298 | 1.21 |
ENSMUST00000062893.12
|
Cenpe
|
centromere protein E |
| chr17_-_56916771 | 1.20 |
ENSMUST00000052832.6
|
Micos13
|
mitochondrial contact site and cristae organizing system subunit 13 |
| chr3_+_67490068 | 1.18 |
ENSMUST00000029344.10
|
Mfsd1
|
major facilitator superfamily domain containing 1 |
| chrX_+_105230706 | 1.17 |
ENSMUST00000081593.13
|
Pgk1
|
phosphoglycerate kinase 1 |
| chr7_-_37472979 | 1.16 |
ENSMUST00000176534.8
|
Zfp536
|
zinc finger protein 536 |
| chr5_-_106606032 | 1.15 |
ENSMUST00000086795.8
|
Barhl2
|
BarH like homeobox 2 |
| chr15_-_76540916 | 1.15 |
ENSMUST00000229524.2
|
Cyhr1
|
cysteine and histidine rich 1 |
| chr1_+_131935342 | 1.13 |
ENSMUST00000086556.12
|
Elk4
|
ELK4, member of ETS oncogene family |
| chr9_+_35179153 | 1.12 |
ENSMUST00000034543.5
|
Rpusd4
|
RNA pseudouridylate synthase domain containing 4 |
| chr4_-_119279551 | 1.11 |
ENSMUST00000106316.2
ENSMUST00000030385.13 |
Ppcs
|
phosphopantothenoylcysteine synthetase |
| chr13_-_3995349 | 1.11 |
ENSMUST00000058610.8
|
Ucn3
|
urocortin 3 |
| chr15_-_76541105 | 1.11 |
ENSMUST00000176274.2
|
Cyhr1
|
cysteine and histidine rich 1 |
| chr10_-_63174801 | 1.10 |
ENSMUST00000020257.13
ENSMUST00000177694.8 ENSMUST00000105442.3 |
Sirt1
|
sirtuin 1 |
| chr13_-_12355604 | 1.09 |
ENSMUST00000168193.8
ENSMUST00000064204.14 |
Actn2
|
actinin alpha 2 |
| chr15_-_81283795 | 1.08 |
ENSMUST00000023039.15
|
St13
|
suppression of tumorigenicity 13 |
| chr4_-_126096376 | 1.07 |
ENSMUST00000106142.8
ENSMUST00000169403.8 ENSMUST00000130334.2 |
Thrap3
|
thyroid hormone receptor associated protein 3 |
| chr2_-_164675357 | 1.07 |
ENSMUST00000042775.5
|
Neurl2
|
neuralized E3 ubiquitin protein ligase 2 |
| chr10_+_43354807 | 1.07 |
ENSMUST00000167488.9
|
Bend3
|
BEN domain containing 3 |
| chr2_+_75489596 | 1.06 |
ENSMUST00000111964.8
ENSMUST00000111962.8 ENSMUST00000111961.8 ENSMUST00000164947.9 ENSMUST00000090792.11 |
Hnrnpa3
|
heterogeneous nuclear ribonucleoprotein A3 |
| chr16_-_94171533 | 1.06 |
ENSMUST00000113910.8
|
Pigp
|
phosphatidylinositol glycan anchor biosynthesis, class P |
| chr1_-_75119277 | 1.05 |
ENSMUST00000168720.8
ENSMUST00000041213.12 ENSMUST00000189809.2 |
Cnppd1
|
cyclin Pas1/PHO80 domain containing 1 |
| chr12_+_79177523 | 1.05 |
ENSMUST00000021550.7
|
Arg2
|
arginase type II |
| chr1_-_10302895 | 1.03 |
ENSMUST00000088615.11
ENSMUST00000131556.2 |
Arfgef1
|
ADP-ribosylation factor guanine nucleotide-exchange factor 1(brefeldin A-inhibited) |
| chr4_-_129083392 | 1.02 |
ENSMUST00000117497.8
ENSMUST00000117350.2 |
S100pbp
|
S100P binding protein |
| chr13_-_64422775 | 0.98 |
ENSMUST00000221634.2
ENSMUST00000039318.16 |
Cdc14b
|
CDC14 cell division cycle 14B |
| chr19_-_7218512 | 0.97 |
ENSMUST00000025675.11
|
Naa40
|
N(alpha)-acetyltransferase 40, NatD catalytic subunit |
| chr4_-_126096112 | 0.97 |
ENSMUST00000142125.2
ENSMUST00000106141.3 |
Thrap3
|
thyroid hormone receptor associated protein 3 |
| chr17_+_45874800 | 0.95 |
ENSMUST00000224905.2
ENSMUST00000226086.2 ENSMUST00000041353.7 |
Slc35b2
|
solute carrier family 35, member B2 |
| chr14_-_30741012 | 0.94 |
ENSMUST00000037739.8
|
Gnl3
|
guanine nucleotide binding protein-like 3 (nucleolar) |
| chr5_-_124563636 | 0.94 |
ENSMUST00000196711.5
ENSMUST00000200474.5 ENSMUST00000199808.5 |
Sbno1
|
strawberry notch 1 |
| chr6_+_90439596 | 0.93 |
ENSMUST00000203039.3
|
Klf15
|
Kruppel-like factor 15 |
| chr10_-_18891095 | 0.93 |
ENSMUST00000019997.11
|
Tnfaip3
|
tumor necrosis factor, alpha-induced protein 3 |
| chr10_-_62628008 | 0.93 |
ENSMUST00000217768.2
ENSMUST00000020268.7 ENSMUST00000218946.2 ENSMUST00000219527.2 |
Ccar1
|
cell division cycle and apoptosis regulator 1 |
| chr6_+_90439544 | 0.93 |
ENSMUST00000032174.12
|
Klf15
|
Kruppel-like factor 15 |
| chr10_+_79852487 | 0.92 |
ENSMUST00000099501.10
|
Arhgap45
|
Rho GTPase activating protein 45 |
| chr6_-_39702127 | 0.90 |
ENSMUST00000101497.4
|
Braf
|
Braf transforming gene |
| chr16_+_44979086 | 0.90 |
ENSMUST00000023343.4
|
Atg3
|
autophagy related 3 |
| chr1_+_191638854 | 0.89 |
ENSMUST00000044954.7
|
Slc30a1
|
solute carrier family 30 (zinc transporter), member 1 |
| chr11_+_69891398 | 0.89 |
ENSMUST00000019362.15
ENSMUST00000190940.2 |
Dvl2
|
dishevelled segment polarity protein 2 |
| chr11_-_51891575 | 0.88 |
ENSMUST00000109086.8
|
Ube2b
|
ubiquitin-conjugating enzyme E2B |
| chr17_-_65920481 | 0.88 |
ENSMUST00000024897.10
|
Vapa
|
vesicle-associated membrane protein, associated protein A |
| chr5_+_97145533 | 0.87 |
ENSMUST00000112974.6
ENSMUST00000035635.10 |
Bmp2k
|
BMP2 inducible kinase |
| chr18_+_65021974 | 0.87 |
ENSMUST00000225261.3
|
Nedd4l
|
neural precursor cell expressed, developmentally down-regulated gene 4-like |
| chr15_+_99291100 | 0.86 |
ENSMUST00000159209.8
|
Tmbim6
|
transmembrane BAX inhibitor motif containing 6 |
| chr4_+_99184137 | 0.86 |
ENSMUST00000094955.3
|
Gm12689
|
predicted gene 12689 |
| chr13_-_64518112 | 0.85 |
ENSMUST00000021933.8
ENSMUST00000222462.2 |
Ctsl
|
cathepsin L |
| chr2_-_37593856 | 0.85 |
ENSMUST00000155237.2
|
Strbp
|
spermatid perinuclear RNA binding protein |
| chr3_+_137570334 | 0.84 |
ENSMUST00000174561.8
ENSMUST00000173790.8 |
H2az1
|
H2A.Z variant histone 1 |
| chr3_-_89959770 | 0.83 |
ENSMUST00000029553.16
ENSMUST00000195995.5 ENSMUST00000064639.15 ENSMUST00000199834.5 |
Ubap2l
|
ubiquitin-associated protein 2-like |
| chr8_+_13209141 | 0.83 |
ENSMUST00000033824.8
|
Lamp1
|
lysosomal-associated membrane protein 1 |
| chr7_+_100997268 | 0.82 |
ENSMUST00000107010.9
|
Arap1
|
ArfGAP with RhoGAP domain, ankyrin repeat and PH domain 1 |
| chr2_+_110427643 | 0.82 |
ENSMUST00000045972.13
ENSMUST00000111026.3 |
Slc5a12
|
solute carrier family 5 (sodium/glucose cotransporter), member 12 |
| chr5_-_100648487 | 0.82 |
ENSMUST00000239512.1
|
LIN54
|
lin-54 homolog (C. elegans) |
| chr17_+_46957151 | 0.82 |
ENSMUST00000002844.14
ENSMUST00000113429.8 ENSMUST00000113430.2 |
Mrpl2
|
mitochondrial ribosomal protein L2 |
| chr3_+_159201077 | 0.81 |
ENSMUST00000029825.14
|
Depdc1a
|
DEP domain containing 1a |
| chr1_-_155617773 | 0.81 |
ENSMUST00000027740.14
|
Lhx4
|
LIM homeobox protein 4 |
| chr16_+_4867876 | 0.81 |
ENSMUST00000230703.2
ENSMUST00000052449.6 |
Ubn1
|
ubinuclein 1 |
| chr9_-_35179042 | 0.80 |
ENSMUST00000217306.2
ENSMUST00000125087.2 ENSMUST00000121564.8 ENSMUST00000063782.12 ENSMUST00000059057.14 |
Fam118b
|
family with sequence similarity 118, member B |
| chr11_-_51891259 | 0.80 |
ENSMUST00000020657.13
|
Ube2b
|
ubiquitin-conjugating enzyme E2B |
| chr14_+_30741115 | 0.80 |
ENSMUST00000112094.8
ENSMUST00000144009.2 |
Pbrm1
|
polybromo 1 |
| chr2_+_155223728 | 0.79 |
ENSMUST00000043237.14
ENSMUST00000174685.8 |
Trp53inp2
|
transformation related protein 53 inducible nuclear protein 2 |
| chr7_-_46445085 | 0.79 |
ENSMUST00000123725.2
|
Hps5
|
HPS5, biogenesis of lysosomal organelles complex 2 subunit 2 |
| chr4_-_129083439 | 0.78 |
ENSMUST00000106059.8
|
S100pbp
|
S100P binding protein |
| chr13_-_64422693 | 0.78 |
ENSMUST00000109770.2
|
Cdc14b
|
CDC14 cell division cycle 14B |
| chr4_-_126096551 | 0.78 |
ENSMUST00000080919.12
|
Thrap3
|
thyroid hormone receptor associated protein 3 |
| chr10_+_51356728 | 0.78 |
ENSMUST00000102894.6
ENSMUST00000219661.2 ENSMUST00000219696.2 ENSMUST00000217706.2 |
Lilr4b
Gm49339
|
leukocyte immunoglobulin-like receptor, subfamily B, member 4B predicted gene, 49339 |
| chr5_-_124564014 | 0.76 |
ENSMUST00000196329.5
ENSMUST00000196644.5 |
Sbno1
|
strawberry notch 1 |
| chrX_-_93166964 | 0.76 |
ENSMUST00000137853.8
|
Zfx
|
zinc finger protein X-linked |
| chr11_+_53241561 | 0.75 |
ENSMUST00000060945.12
|
Aff4
|
AF4/FMR2 family, member 4 |
| chr10_-_128759331 | 0.74 |
ENSMUST00000153731.8
ENSMUST00000026405.10 |
Bloc1s1
|
biogenesis of lysosomal organelles complex-1, subunit 1 |
| chr15_+_99291455 | 0.74 |
ENSMUST00000162624.8
|
Tmbim6
|
transmembrane BAX inhibitor motif containing 6 |
| chr17_+_35235552 | 0.73 |
ENSMUST00000007245.8
ENSMUST00000172499.2 |
Vwa7
|
von Willebrand factor A domain containing 7 |
| chr2_-_30176324 | 0.72 |
ENSMUST00000100219.5
|
Dolk
|
dolichol kinase |
| chr1_+_87254729 | 0.72 |
ENSMUST00000172794.8
ENSMUST00000164992.9 ENSMUST00000173173.8 |
Gigyf2
|
GRB10 interacting GYF protein 2 |
| chr2_-_39116044 | 0.72 |
ENSMUST00000204368.2
|
Ppp6c
|
protein phosphatase 6, catalytic subunit |
| chr5_-_65855511 | 0.71 |
ENSMUST00000201948.4
|
Pds5a
|
PDS5 cohesin associated factor A |
| chr5_-_124563969 | 0.71 |
ENSMUST00000065263.12
|
Sbno1
|
strawberry notch 1 |
| chr11_+_69881885 | 0.71 |
ENSMUST00000018711.15
|
Gabarap
|
gamma-aminobutyric acid receptor associated protein |
| chrX_-_93166693 | 0.70 |
ENSMUST00000113925.8
|
Zfx
|
zinc finger protein X-linked |
| chr11_+_70453806 | 0.69 |
ENSMUST00000079244.12
ENSMUST00000102558.11 |
Mink1
|
misshapen-like kinase 1 (zebrafish) |
| chr10_-_29238009 | 0.69 |
ENSMUST00000161508.3
|
Rnf146
|
ring finger protein 146 |
| chr10_-_62723103 | 0.69 |
ENSMUST00000218438.2
|
Tet1
|
tet methylcytosine dioxygenase 1 |
| chr19_+_37674029 | 0.68 |
ENSMUST00000073391.5
|
Cyp26c1
|
cytochrome P450, family 26, subfamily c, polypeptide 1 |
| chr4_-_41275091 | 0.68 |
ENSMUST00000030143.13
ENSMUST00000108068.8 |
Ubap2
|
ubiquitin-associated protein 2 |
| chr2_+_119727689 | 0.68 |
ENSMUST00000046717.13
ENSMUST00000079934.12 ENSMUST00000110774.8 ENSMUST00000110773.9 ENSMUST00000156510.2 |
Mga
|
MAX gene associated |
| chr18_+_53596570 | 0.67 |
ENSMUST00000236096.2
|
Prdm6
|
PR domain containing 6 |
| chrX_-_93166992 | 0.67 |
ENSMUST00000088102.12
ENSMUST00000113927.8 |
Zfx
|
zinc finger protein X-linked |
| chr3_+_159201092 | 0.67 |
ENSMUST00000106041.3
|
Depdc1a
|
DEP domain containing 1a |
| chr14_+_8508484 | 0.67 |
ENSMUST00000065865.10
ENSMUST00000225891.2 |
Thoc7
|
THO complex 7 |
| chr7_-_45109262 | 0.67 |
ENSMUST00000094434.13
|
Ftl1
|
ferritin light polypeptide 1 |
| chr10_-_29238028 | 0.67 |
ENSMUST00000160372.9
ENSMUST00000214896.2 |
Rnf146
|
ring finger protein 146 |
| chr11_+_115671523 | 0.65 |
ENSMUST00000239299.2
|
Tmem94
|
transmembrane protein 94 |
| chr7_+_28580847 | 0.65 |
ENSMUST00000066880.6
|
Capn12
|
calpain 12 |
| chr2_+_160573604 | 0.64 |
ENSMUST00000174885.2
ENSMUST00000109462.8 |
Plcg1
|
phospholipase C, gamma 1 |
| chr16_-_5021843 | 0.64 |
ENSMUST00000147567.2
ENSMUST00000023911.11 |
Nagpa
|
N-acetylglucosamine-1-phosphodiester alpha-N-acetylglucosaminidase |
| chr14_-_72840373 | 0.63 |
ENSMUST00000162825.8
|
Fndc3a
|
fibronectin type III domain containing 3A |
| chr3_-_89959739 | 0.63 |
ENSMUST00000199929.2
ENSMUST00000090908.11 ENSMUST00000198322.5 ENSMUST00000196843.5 |
Ubap2l
|
ubiquitin-associated protein 2-like |
| chr2_-_37593287 | 0.63 |
ENSMUST00000072186.12
|
Strbp
|
spermatid perinuclear RNA binding protein |
| chr7_+_27878894 | 0.62 |
ENSMUST00000085901.13
ENSMUST00000172761.8 |
Dyrk1b
|
dual-specificity tyrosine-(Y)-phosphorylation regulated kinase 1b |
| chr15_+_99291491 | 0.61 |
ENSMUST00000159531.3
|
Tmbim6
|
transmembrane BAX inhibitor motif containing 6 |
| chr3_+_138233004 | 0.59 |
ENSMUST00000196990.5
ENSMUST00000200239.5 ENSMUST00000200100.2 |
Eif4e
|
eukaryotic translation initiation factor 4E |
| chr4_-_129083335 | 0.59 |
ENSMUST00000106061.9
ENSMUST00000072431.13 |
S100pbp
|
S100P binding protein |
| chr7_-_46445305 | 0.59 |
ENSMUST00000107653.8
ENSMUST00000107654.8 ENSMUST00000014562.14 ENSMUST00000152759.8 |
Hps5
|
HPS5, biogenesis of lysosomal organelles complex 2 subunit 2 |
| chr9_+_45924120 | 0.59 |
ENSMUST00000120463.9
ENSMUST00000120247.8 |
Sik3
|
SIK family kinase 3 |
| chr3_+_89960121 | 0.58 |
ENSMUST00000160640.8
ENSMUST00000029552.13 ENSMUST00000162114.8 ENSMUST00000068798.13 |
4933434E20Rik
|
RIKEN cDNA 4933434E20 gene |
| chr16_-_94171340 | 0.58 |
ENSMUST00000138514.2
|
Pigp
|
phosphatidylinositol glycan anchor biosynthesis, class P |
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.2 | 3.6 | GO:0045643 | regulation of eosinophil differentiation(GO:0045643) positive regulation of eosinophil differentiation(GO:0045645) |
| 1.0 | 7.2 | GO:0010288 | response to lead ion(GO:0010288) |
| 1.0 | 2.9 | GO:0018879 | biphenyl metabolic process(GO:0018879) |
| 1.0 | 6.8 | GO:0070495 | regulation of thrombin receptor signaling pathway(GO:0070494) negative regulation of thrombin receptor signaling pathway(GO:0070495) |
| 0.8 | 4.2 | GO:0043490 | malate-aspartate shuttle(GO:0043490) |
| 0.8 | 2.4 | GO:1904715 | negative regulation of chaperone-mediated autophagy(GO:1904715) |
| 0.6 | 1.7 | GO:0010845 | positive regulation of reciprocal meiotic recombination(GO:0010845) negative regulation of histone phosphorylation(GO:0033128) |
| 0.5 | 1.6 | GO:0003218 | cardiac left ventricle formation(GO:0003218) |
| 0.5 | 4.7 | GO:1902961 | positive regulation of aspartic-type endopeptidase activity involved in amyloid precursor protein catabolic process(GO:1902961) positive regulation of aspartic-type peptidase activity(GO:1905247) |
| 0.5 | 2.0 | GO:0014916 | regulation of lung blood pressure(GO:0014916) |
| 0.4 | 2.2 | GO:1904721 | regulation of mRNA cleavage(GO:0031437) negative regulation of mRNA cleavage(GO:0031438) regulation of mRNA endonucleolytic cleavage involved in unfolded protein response(GO:1904720) negative regulation of mRNA endonucleolytic cleavage involved in unfolded protein response(GO:1904721) |
| 0.4 | 1.3 | GO:0000720 | pyrimidine dimer repair by nucleotide-excision repair(GO:0000720) |
| 0.4 | 1.3 | GO:0021570 | rhombomere 4 development(GO:0021570) |
| 0.4 | 1.3 | GO:0090149 | mitochondrial membrane fission(GO:0090149) |
| 0.4 | 2.9 | GO:0002317 | plasma cell differentiation(GO:0002317) |
| 0.4 | 5.1 | GO:0016554 | cytidine to uridine editing(GO:0016554) |
| 0.4 | 1.9 | GO:0003349 | epicardium-derived cardiac endothelial cell differentiation(GO:0003349) |
| 0.4 | 1.1 | GO:0051695 | actin filament uncapping(GO:0051695) phospholipase C-activating angiotensin-activated signaling pathway(GO:0086097) |
| 0.3 | 2.7 | GO:0021650 | vestibulocochlear nerve formation(GO:0021650) |
| 0.3 | 1.3 | GO:0006208 | pyrimidine nucleobase catabolic process(GO:0006208) thymine catabolic process(GO:0006210) thymine metabolic process(GO:0019859) |
| 0.3 | 1.3 | GO:0035964 | COPI-coated vesicle budding(GO:0035964) |
| 0.3 | 0.9 | GO:0034147 | regulation of granuloma formation(GO:0002631) negative regulation of granuloma formation(GO:0002632) regulation of toll-like receptor 5 signaling pathway(GO:0034147) negative regulation of toll-like receptor 5 signaling pathway(GO:0034148) negative regulation of nucleotide-binding oligomerization domain containing 1 signaling pathway(GO:0070429) |
| 0.3 | 2.5 | GO:0098532 | histone H3-K27 trimethylation(GO:0098532) |
| 0.3 | 1.2 | GO:1903976 | negative regulation of glial cell migration(GO:1903976) |
| 0.3 | 3.7 | GO:0035873 | lactate transport(GO:0015727) lactate transmembrane transport(GO:0035873) plasma membrane lactate transport(GO:0035879) |
| 0.3 | 1.2 | GO:0090310 | negative regulation of methylation-dependent chromatin silencing(GO:0090310) |
| 0.3 | 0.9 | GO:0071579 | regulation of zinc ion transport(GO:0071579) |
| 0.3 | 1.4 | GO:0002266 | follicular dendritic cell activation(GO:0002266) follicular dendritic cell differentiation(GO:0002268) |
| 0.3 | 1.9 | GO:0098705 | cellular response to cisplatin(GO:0072719) copper ion import across plasma membrane(GO:0098705) copper ion import into cell(GO:1902861) |
| 0.3 | 2.8 | GO:0061484 | hematopoietic stem cell homeostasis(GO:0061484) |
| 0.3 | 0.8 | GO:0043323 | positive regulation of natural killer cell degranulation(GO:0043323) |
| 0.3 | 1.6 | GO:0061056 | sclerotome development(GO:0061056) |
| 0.3 | 3.6 | GO:2000298 | regulation of Rho-dependent protein serine/threonine kinase activity(GO:2000298) |
| 0.3 | 1.0 | GO:0090283 | regulation of protein glycosylation in Golgi(GO:0090283) |
| 0.3 | 4.4 | GO:0042407 | cristae formation(GO:0042407) |
| 0.2 | 1.4 | GO:0009992 | cellular water homeostasis(GO:0009992) |
| 0.2 | 0.9 | GO:0022007 | neural plate elongation(GO:0014022) convergent extension involved in neural plate elongation(GO:0022007) |
| 0.2 | 1.1 | GO:0010958 | regulation of amino acid import(GO:0010958) L-arginine import(GO:0043091) arginine import(GO:0090467) |
| 0.2 | 0.8 | GO:0021526 | medial motor column neuron differentiation(GO:0021526) |
| 0.2 | 1.4 | GO:0061084 | negative regulation of protein refolding(GO:0061084) |
| 0.2 | 3.7 | GO:0045060 | negative thymic T cell selection(GO:0045060) |
| 0.2 | 3.7 | GO:0002024 | diet induced thermogenesis(GO:0002024) |
| 0.2 | 3.9 | GO:1904816 | positive regulation of protein localization to chromosome, telomeric region(GO:1904816) |
| 0.2 | 1.5 | GO:0042795 | snRNA transcription from RNA polymerase II promoter(GO:0042795) |
| 0.2 | 0.8 | GO:0030576 | Cajal body organization(GO:0030576) |
| 0.2 | 1.8 | GO:1904668 | mitotic spindle midzone assembly(GO:0051256) positive regulation of ubiquitin protein ligase activity(GO:1904668) |
| 0.2 | 1.4 | GO:0015014 | heparan sulfate proteoglycan biosynthetic process, polysaccharide chain biosynthetic process(GO:0015014) |
| 0.2 | 1.8 | GO:0061113 | pancreas morphogenesis(GO:0061113) |
| 0.1 | 0.4 | GO:0010536 | positive regulation of activation of Janus kinase activity(GO:0010536) |
| 0.1 | 0.4 | GO:0002501 | MHC protein complex assembly(GO:0002396) peptide antigen assembly with MHC protein complex(GO:0002501) |
| 0.1 | 1.2 | GO:0051987 | positive regulation of attachment of spindle microtubules to kinetochore(GO:0051987) |
| 0.1 | 3.0 | GO:0006309 | apoptotic DNA fragmentation(GO:0006309) |
| 0.1 | 2.5 | GO:0015937 | coenzyme A biosynthetic process(GO:0015937) |
| 0.1 | 1.4 | GO:0006122 | mitochondrial electron transport, ubiquinol to cytochrome c(GO:0006122) |
| 0.1 | 0.6 | GO:0044861 | protein transport into plasma membrane raft(GO:0044861) |
| 0.1 | 1.1 | GO:0031119 | tRNA pseudouridine synthesis(GO:0031119) |
| 0.1 | 0.9 | GO:0044828 | negative regulation by host of viral genome replication(GO:0044828) |
| 0.1 | 9.2 | GO:0045214 | sarcomere organization(GO:0045214) |
| 0.1 | 0.7 | GO:0016103 | diterpenoid catabolic process(GO:0016103) retinoic acid catabolic process(GO:0034653) |
| 0.1 | 0.9 | GO:0051503 | adenine nucleotide transport(GO:0051503) |
| 0.1 | 1.6 | GO:0048387 | negative regulation of retinoic acid receptor signaling pathway(GO:0048387) |
| 0.1 | 4.5 | GO:0009954 | proximal/distal pattern formation(GO:0009954) |
| 0.1 | 0.8 | GO:0042637 | catagen(GO:0042637) |
| 0.1 | 1.1 | GO:0070932 | histone H3 deacetylation(GO:0070932) |
| 0.1 | 0.4 | GO:2000584 | regulation of platelet-derived growth factor receptor-alpha signaling pathway(GO:2000583) negative regulation of platelet-derived growth factor receptor-alpha signaling pathway(GO:2000584) |
| 0.1 | 2.1 | GO:0032688 | negative regulation of interferon-beta production(GO:0032688) |
| 0.1 | 1.7 | GO:0008272 | sulfate transport(GO:0008272) |
| 0.1 | 2.2 | GO:0006474 | N-terminal protein amino acid acetylation(GO:0006474) |
| 0.1 | 1.2 | GO:1990403 | embryonic brain development(GO:1990403) |
| 0.1 | 3.2 | GO:0001829 | trophectodermal cell differentiation(GO:0001829) |
| 0.1 | 0.9 | GO:0010764 | negative regulation of fibroblast migration(GO:0010764) |
| 0.1 | 2.8 | GO:0048026 | positive regulation of mRNA splicing, via spliceosome(GO:0048026) |
| 0.1 | 0.3 | GO:1903297 | regulation of hypoxia-induced intrinsic apoptotic signaling pathway(GO:1903297) negative regulation of hypoxia-induced intrinsic apoptotic signaling pathway(GO:1903298) |
| 0.1 | 2.6 | GO:0031670 | cellular response to nutrient(GO:0031670) |
| 0.1 | 2.1 | GO:0060746 | parental behavior(GO:0060746) |
| 0.1 | 2.0 | GO:0031295 | T cell costimulation(GO:0031295) |
| 0.1 | 0.3 | GO:1901228 | positive regulation of transcription from RNA polymerase II promoter involved in heart development(GO:1901228) |
| 0.1 | 0.9 | GO:0043653 | mitochondrial fragmentation involved in apoptotic process(GO:0043653) |
| 0.1 | 0.7 | GO:0046784 | viral mRNA export from host cell nucleus(GO:0046784) |
| 0.1 | 0.7 | GO:0097577 | intracellular sequestering of iron ion(GO:0006880) sequestering of iron ion(GO:0097577) |
| 0.1 | 1.4 | GO:1902187 | negative regulation of viral release from host cell(GO:1902187) |
| 0.1 | 0.8 | GO:2001288 | positive regulation of caveolin-mediated endocytosis(GO:2001288) |
| 0.1 | 2.0 | GO:0007064 | mitotic sister chromatid cohesion(GO:0007064) |
| 0.1 | 1.9 | GO:0072311 | renal filtration cell differentiation(GO:0061318) glomerular visceral epithelial cell differentiation(GO:0072112) glomerular epithelial cell differentiation(GO:0072311) |
| 0.1 | 0.3 | GO:0097498 | endothelial tube lumen extension(GO:0097498) |
| 0.1 | 0.4 | GO:0070127 | tRNA aminoacylation for mitochondrial protein translation(GO:0070127) |
| 0.1 | 3.0 | GO:0006506 | GPI anchor biosynthetic process(GO:0006506) |
| 0.1 | 0.4 | GO:1900740 | regulation of protein insertion into mitochondrial membrane involved in apoptotic signaling pathway(GO:1900739) positive regulation of protein insertion into mitochondrial membrane involved in apoptotic signaling pathway(GO:1900740) |
| 0.1 | 0.8 | GO:1901339 | regulation of store-operated calcium channel activity(GO:1901339) |
| 0.1 | 0.2 | GO:1901979 | regulation of inward rectifier potassium channel activity(GO:1901979) |
| 0.0 | 0.9 | GO:0048642 | negative regulation of skeletal muscle tissue development(GO:0048642) |
| 0.0 | 0.7 | GO:0051151 | negative regulation of smooth muscle cell differentiation(GO:0051151) |
| 0.0 | 0.8 | GO:0006336 | DNA replication-independent nucleosome assembly(GO:0006336) |
| 0.0 | 1.2 | GO:0031571 | mitotic G1 DNA damage checkpoint(GO:0031571) |
| 0.0 | 1.4 | GO:0032467 | positive regulation of cytokinesis(GO:0032467) |
| 0.0 | 1.2 | GO:0031639 | plasminogen activation(GO:0031639) |
| 0.0 | 1.9 | GO:0010501 | RNA secondary structure unwinding(GO:0010501) |
| 0.0 | 0.3 | GO:1990034 | calcium ion export from cell(GO:1990034) |
| 0.0 | 0.2 | GO:1903347 | negative regulation of bicellular tight junction assembly(GO:1903347) |
| 0.0 | 0.4 | GO:0007549 | dosage compensation(GO:0007549) dosage compensation by inactivation of X chromosome(GO:0009048) |
| 0.0 | 0.2 | GO:0010587 | miRNA catabolic process(GO:0010587) |
| 0.0 | 1.5 | GO:0030901 | midbrain development(GO:0030901) |
| 0.0 | 0.5 | GO:2001014 | regulation of skeletal muscle cell differentiation(GO:2001014) |
| 0.0 | 0.6 | GO:0006895 | Golgi to endosome transport(GO:0006895) |
| 0.0 | 0.3 | GO:0033133 | positive regulation of glucokinase activity(GO:0033133) |
| 0.0 | 0.2 | GO:1900107 | regulation of nodal signaling pathway(GO:1900107) |
| 0.0 | 1.4 | GO:0015804 | neutral amino acid transport(GO:0015804) |
| 0.0 | 0.6 | GO:0060009 | Sertoli cell development(GO:0060009) |
| 0.0 | 0.4 | GO:0090190 | positive regulation of branching involved in ureteric bud morphogenesis(GO:0090190) |
| 0.0 | 2.7 | GO:0032526 | response to retinoic acid(GO:0032526) |
| 0.0 | 0.8 | GO:0001921 | positive regulation of receptor recycling(GO:0001921) |
| 0.0 | 0.3 | GO:0006268 | DNA unwinding involved in DNA replication(GO:0006268) |
| 0.0 | 0.9 | GO:2000369 | regulation of clathrin-mediated endocytosis(GO:2000369) |
| 0.0 | 1.2 | GO:0001709 | cell fate determination(GO:0001709) |
| 0.0 | 0.5 | GO:0016973 | poly(A)+ mRNA export from nucleus(GO:0016973) |
| 0.0 | 1.1 | GO:0045838 | positive regulation of membrane potential(GO:0045838) |
| 0.0 | 1.0 | GO:0036075 | endochondral ossification(GO:0001958) replacement ossification(GO:0036075) |
| 0.0 | 3.3 | GO:0045930 | negative regulation of mitotic cell cycle(GO:0045930) |
| 0.0 | 0.0 | GO:0046005 | positive regulation of circadian sleep/wake cycle, REM sleep(GO:0046005) |
| 0.0 | 0.5 | GO:0032967 | positive regulation of collagen biosynthetic process(GO:0032967) |
| 0.0 | 1.4 | GO:0051865 | protein autoubiquitination(GO:0051865) |
| 0.0 | 0.6 | GO:0035115 | embryonic forelimb morphogenesis(GO:0035115) |
| 0.0 | 0.3 | GO:0071281 | cellular response to iron ion(GO:0071281) |
| 0.0 | 0.9 | GO:0032543 | mitochondrial translation(GO:0032543) |
| 0.0 | 0.3 | GO:0071542 | dopaminergic neuron differentiation(GO:0071542) |
| 0.0 | 0.1 | GO:0045423 | granulocyte macrophage colony-stimulating factor biosynthetic process(GO:0042253) regulation of granulocyte macrophage colony-stimulating factor biosynthetic process(GO:0045423) |
| 0.0 | 1.1 | GO:0000079 | regulation of cyclin-dependent protein serine/threonine kinase activity(GO:0000079) |
| 0.0 | 0.1 | GO:0034058 | endosomal vesicle fusion(GO:0034058) |
| 0.0 | 1.0 | GO:0055008 | cardiac muscle tissue morphogenesis(GO:0055008) |
| 0.0 | 0.3 | GO:0034508 | centromere complex assembly(GO:0034508) |
| 0.0 | 0.4 | GO:0046622 | positive regulation of organ growth(GO:0046622) |
| 0.0 | 0.2 | GO:0003094 | glomerular filtration(GO:0003094) |
| 0.0 | 0.7 | GO:0009060 | aerobic respiration(GO:0009060) |
| 0.0 | 0.6 | GO:0007520 | myoblast fusion(GO:0007520) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.7 | 4.7 | GO:0070381 | endosome to plasma membrane transport vesicle(GO:0070381) |
| 0.7 | 5.3 | GO:0097129 | cyclin D2-CDK4 complex(GO:0097129) |
| 0.5 | 4.4 | GO:0061617 | MICOS complex(GO:0061617) |
| 0.4 | 3.3 | GO:0005927 | muscle tendon junction(GO:0005927) |
| 0.3 | 1.4 | GO:0031084 | BLOC-2 complex(GO:0031084) |
| 0.3 | 1.4 | GO:0033257 | Bcl3/NF-kappaB2 complex(GO:0033257) |
| 0.3 | 0.8 | GO:0061474 | phagolysosome membrane(GO:0061474) |
| 0.3 | 2.1 | GO:0033256 | I-kappaB/NF-kappaB complex(GO:0033256) |
| 0.2 | 1.7 | GO:0033503 | HULC complex(GO:0033503) |
| 0.2 | 8.3 | GO:0080008 | Cul4-RING E3 ubiquitin ligase complex(GO:0080008) |
| 0.2 | 2.9 | GO:0034663 | endoplasmic reticulum chaperone complex(GO:0034663) |
| 0.2 | 1.1 | GO:0033553 | rDNA heterochromatin(GO:0033553) |
| 0.2 | 3.0 | GO:0000506 | glycosylphosphatidylinositol-N-acetylglucosaminyltransferase (GPI-GnT) complex(GO:0000506) |
| 0.1 | 1.4 | GO:0005750 | mitochondrial respiratory chain complex III(GO:0005750) |
| 0.1 | 2.0 | GO:0005641 | nuclear envelope lumen(GO:0005641) |
| 0.1 | 0.3 | GO:0005854 | nascent polypeptide-associated complex(GO:0005854) |
| 0.1 | 0.4 | GO:0005896 | interleukin-6 receptor complex(GO:0005896) |
| 0.1 | 1.2 | GO:0000778 | condensed nuclear chromosome kinetochore(GO:0000778) |
| 0.1 | 0.5 | GO:0000346 | transcription export complex(GO:0000346) |
| 0.1 | 0.7 | GO:0000445 | THO complex(GO:0000347) THO complex part of transcription export complex(GO:0000445) |
| 0.1 | 6.8 | GO:0009925 | basal plasma membrane(GO:0009925) |
| 0.1 | 6.4 | GO:0032420 | stereocilium(GO:0032420) |
| 0.1 | 2.8 | GO:0035145 | exon-exon junction complex(GO:0035145) |
| 0.1 | 1.1 | GO:0005832 | chaperonin-containing T-complex(GO:0005832) |
| 0.1 | 1.3 | GO:0042589 | zymogen granule membrane(GO:0042589) |
| 0.1 | 0.6 | GO:0044352 | pinosome(GO:0044352) macropinosome(GO:0044354) |
| 0.1 | 0.9 | GO:0000153 | cytoplasmic ubiquitin ligase complex(GO:0000153) |
| 0.1 | 12.9 | GO:0030018 | Z disc(GO:0030018) |
| 0.1 | 0.2 | GO:0034457 | Mpp10 complex(GO:0034457) |
| 0.0 | 0.5 | GO:0008250 | oligosaccharyltransferase complex(GO:0008250) |
| 0.0 | 0.4 | GO:0042824 | MHC class I peptide loading complex(GO:0042824) |
| 0.0 | 1.9 | GO:0090544 | BAF-type complex(GO:0090544) |
| 0.0 | 1.1 | GO:0043196 | varicosity(GO:0043196) |
| 0.0 | 0.5 | GO:0035102 | PRC1 complex(GO:0035102) |
| 0.0 | 0.7 | GO:0035327 | transcriptionally active chromatin(GO:0035327) |
| 0.0 | 0.3 | GO:1990712 | HFE-transferrin receptor complex(GO:1990712) |
| 0.0 | 0.7 | GO:0031083 | BLOC-1 complex(GO:0031083) |
| 0.0 | 2.5 | GO:0005720 | nuclear heterochromatin(GO:0005720) |
| 0.0 | 0.1 | GO:0030906 | retromer, cargo-selective complex(GO:0030906) |
| 0.0 | 1.4 | GO:0008023 | transcription elongation factor complex(GO:0008023) |
| 0.0 | 0.4 | GO:0044754 | autolysosome(GO:0044754) |
| 0.0 | 0.7 | GO:0097225 | sperm midpiece(GO:0097225) |
| 0.0 | 2.1 | GO:0005871 | kinesin complex(GO:0005871) |
| 0.0 | 0.3 | GO:0042555 | MCM complex(GO:0042555) |
| 0.0 | 1.4 | GO:0032154 | cleavage furrow(GO:0032154) cell surface furrow(GO:0097610) |
| 0.0 | 0.2 | GO:0030877 | beta-catenin destruction complex(GO:0030877) |
| 0.0 | 0.3 | GO:0032591 | dendritic spine membrane(GO:0032591) |
| 0.0 | 0.9 | GO:0016235 | aggresome(GO:0016235) |
| 0.0 | 0.9 | GO:0030173 | integral component of Golgi membrane(GO:0030173) |
| 0.0 | 0.1 | GO:0070695 | FHF complex(GO:0070695) |
| 0.0 | 0.7 | GO:0044665 | MLL1/2 complex(GO:0044665) MLL1 complex(GO:0071339) |
| 0.0 | 0.1 | GO:0033263 | CORVET complex(GO:0033263) |
| 0.0 | 0.1 | GO:0005827 | polar microtubule(GO:0005827) |
| 0.0 | 0.8 | GO:0008180 | COP9 signalosome(GO:0008180) |
| 0.0 | 0.6 | GO:0030131 | clathrin adaptor complex(GO:0030131) |
| 0.0 | 0.2 | GO:0016600 | flotillin complex(GO:0016600) |
| 0.0 | 0.3 | GO:0032045 | guanyl-nucleotide exchange factor complex(GO:0032045) |
| 0.0 | 0.2 | GO:0031616 | spindle pole centrosome(GO:0031616) |
| 0.0 | 0.8 | GO:0000315 | organellar large ribosomal subunit(GO:0000315) mitochondrial large ribosomal subunit(GO:0005762) |
| 0.0 | 1.8 | GO:0030176 | integral component of endoplasmic reticulum membrane(GO:0030176) |
| 0.0 | 1.0 | GO:0030532 | small nuclear ribonucleoprotein complex(GO:0030532) |
| 0.0 | 2.8 | GO:0005770 | late endosome(GO:0005770) |
| 0.0 | 6.5 | GO:0005667 | transcription factor complex(GO:0005667) |
| 0.0 | 0.8 | GO:0005902 | microvillus(GO:0005902) |
| 0.0 | 1.0 | GO:0072686 | mitotic spindle(GO:0072686) |
| 0.0 | 1.9 | GO:0005903 | brush border(GO:0005903) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.7 | 6.8 | GO:0005008 | hepatocyte growth factor-activated receptor activity(GO:0005008) |
| 0.8 | 4.2 | GO:0015183 | L-aspartate transmembrane transporter activity(GO:0015183) |
| 0.5 | 4.5 | GO:0015129 | lactate transmembrane transporter activity(GO:0015129) |
| 0.5 | 13.9 | GO:0051371 | muscle alpha-actinin binding(GO:0051371) |
| 0.5 | 1.4 | GO:0050508 | glucuronosyl-N-acetylglucosaminyl-proteoglycan 4-alpha-N-acetylglucosaminyltransferase activity(GO:0050508) |
| 0.4 | 2.2 | GO:0060698 | endoribonuclease inhibitor activity(GO:0060698) |
| 0.4 | 1.1 | GO:0046969 | histone deacetylase activity (H3-K9 specific)(GO:0032129) NAD-dependent histone deacetylase activity (H3-K9 specific)(GO:0046969) |
| 0.3 | 0.9 | GO:0046964 | 3'-phosphoadenosine 5'-phosphosulfate transmembrane transporter activity(GO:0046964) |
| 0.3 | 1.9 | GO:0008422 | beta-glucosidase activity(GO:0008422) |
| 0.3 | 4.7 | GO:0032050 | clathrin heavy chain binding(GO:0032050) |
| 0.3 | 0.9 | GO:0019776 | Atg8 ligase activity(GO:0019776) |
| 0.3 | 0.9 | GO:0033149 | FFAT motif binding(GO:0033149) |
| 0.3 | 1.1 | GO:0051431 | corticotropin-releasing hormone receptor 2 binding(GO:0051431) |
| 0.3 | 2.7 | GO:0031995 | insulin-like growth factor II binding(GO:0031995) |
| 0.3 | 2.7 | GO:1901612 | cardiolipin binding(GO:1901612) |
| 0.3 | 3.1 | GO:0004185 | serine-type carboxypeptidase activity(GO:0004185) |
| 0.2 | 1.4 | GO:0004594 | pantothenate kinase activity(GO:0004594) |
| 0.2 | 1.2 | GO:0016774 | phosphotransferase activity, carboxyl group as acceptor(GO:0016774) |
| 0.2 | 2.2 | GO:1990189 | peptide-serine-N-acetyltransferase activity(GO:1990189) |
| 0.2 | 2.0 | GO:0019238 | cyclohydrolase activity(GO:0019238) |
| 0.2 | 3.5 | GO:0008349 | MAP kinase kinase kinase kinase activity(GO:0008349) |
| 0.2 | 1.1 | GO:0051373 | FATZ binding(GO:0051373) |
| 0.2 | 3.0 | GO:0017176 | phosphatidylinositol N-acetylglucosaminyltransferase activity(GO:0017176) |
| 0.2 | 4.0 | GO:0055106 | ubiquitin-protein transferase regulator activity(GO:0055106) |
| 0.2 | 1.4 | GO:0005332 | gamma-aminobutyric acid:sodium symporter activity(GO:0005332) |
| 0.2 | 1.1 | GO:0051032 | nucleic acid transmembrane transporter activity(GO:0051032) RNA transmembrane transporter activity(GO:0051033) |
| 0.2 | 1.2 | GO:0043515 | kinetochore binding(GO:0043515) |
| 0.2 | 2.2 | GO:0000182 | rDNA binding(GO:0000182) |
| 0.2 | 0.8 | GO:0030984 | kininogen binding(GO:0030984) |
| 0.2 | 3.6 | GO:0001972 | retinoic acid binding(GO:0001972) |
| 0.2 | 1.4 | GO:0032564 | dATP binding(GO:0032564) |
| 0.1 | 0.4 | GO:0004915 | interleukin-6 receptor activity(GO:0004915) interleukin-6 binding(GO:0019981) |
| 0.1 | 2.6 | GO:0004861 | cyclin-dependent protein serine/threonine kinase inhibitor activity(GO:0004861) |
| 0.1 | 6.2 | GO:0030332 | cyclin binding(GO:0030332) |
| 0.1 | 1.9 | GO:0005375 | copper ion transmembrane transporter activity(GO:0005375) |
| 0.1 | 1.2 | GO:0050291 | sphingosine N-acyltransferase activity(GO:0050291) |
| 0.1 | 0.9 | GO:0061578 | Lys63-specific deubiquitinase activity(GO:0061578) |
| 0.1 | 1.2 | GO:0044323 | retinoic acid-responsive element binding(GO:0044323) |
| 0.1 | 0.6 | GO:0004531 | deoxyribonuclease II activity(GO:0004531) |
| 0.1 | 1.0 | GO:0070579 | methylcytosine dioxygenase activity(GO:0070579) |
| 0.1 | 0.5 | GO:0072345 | NAADP-sensitive calcium-release channel activity(GO:0072345) |
| 0.1 | 1.7 | GO:0019531 | oxalate transmembrane transporter activity(GO:0019531) |
| 0.1 | 2.7 | GO:0008327 | methyl-CpG binding(GO:0008327) |
| 0.1 | 1.1 | GO:0016813 | hydrolase activity, acting on carbon-nitrogen (but not peptide) bonds, in linear amidines(GO:0016813) |
| 0.1 | 0.4 | GO:0005459 | UDP-galactose transmembrane transporter activity(GO:0005459) |
| 0.1 | 0.7 | GO:0047134 | protein-disulfide reductase activity(GO:0047134) |
| 0.1 | 0.6 | GO:0008449 | N-acetylglucosamine-6-sulfatase activity(GO:0008449) |
| 0.1 | 1.0 | GO:0004887 | thyroid hormone receptor activity(GO:0004887) |
| 0.1 | 0.2 | GO:0030519 | snoRNP binding(GO:0030519) |
| 0.1 | 1.4 | GO:1904264 | ubiquitin protein ligase activity involved in ERAD pathway(GO:1904264) |
| 0.1 | 2.7 | GO:0051393 | alpha-actinin binding(GO:0051393) |
| 0.1 | 0.2 | GO:0004946 | bombesin receptor activity(GO:0004946) |
| 0.1 | 1.1 | GO:0009982 | pseudouridine synthase activity(GO:0009982) |
| 0.1 | 2.8 | GO:0046966 | thyroid hormone receptor binding(GO:0046966) |
| 0.1 | 0.3 | GO:0004998 | transferrin receptor activity(GO:0004998) |
| 0.1 | 0.9 | GO:0019855 | calcium channel inhibitor activity(GO:0019855) |
| 0.1 | 0.8 | GO:0005168 | neurotrophin TRKA receptor binding(GO:0005168) |
| 0.0 | 0.9 | GO:0035612 | AP-2 adaptor complex binding(GO:0035612) |
| 0.0 | 0.1 | GO:0004616 | phosphogluconate dehydrogenase (decarboxylating) activity(GO:0004616) |
| 0.0 | 0.8 | GO:0031702 | type 1 angiotensin receptor binding(GO:0031702) |
| 0.0 | 2.4 | GO:0004520 | endodeoxyribonuclease activity(GO:0004520) |
| 0.0 | 0.7 | GO:0015929 | hexosaminidase activity(GO:0015929) |
| 0.0 | 1.2 | GO:0070064 | proline-rich region binding(GO:0070064) |
| 0.0 | 1.7 | GO:0061631 | ubiquitin conjugating enzyme activity(GO:0061631) |
| 0.0 | 0.8 | GO:0019871 | sodium channel inhibitor activity(GO:0019871) |
| 0.0 | 3.8 | GO:0032947 | protein complex scaffold(GO:0032947) |
| 0.0 | 0.9 | GO:0031434 | mitogen-activated protein kinase kinase binding(GO:0031434) |
| 0.0 | 0.3 | GO:0003873 | 6-phosphofructo-2-kinase activity(GO:0003873) |
| 0.0 | 1.1 | GO:0016881 | acid-amino acid ligase activity(GO:0016881) |
| 0.0 | 1.8 | GO:0008138 | protein tyrosine/serine/threonine phosphatase activity(GO:0008138) |
| 0.0 | 0.5 | GO:0051959 | dynein light intermediate chain binding(GO:0051959) |
| 0.0 | 1.9 | GO:0003743 | translation initiation factor activity(GO:0003743) |
| 0.0 | 0.6 | GO:0051010 | microtubule plus-end binding(GO:0051010) |
| 0.0 | 0.6 | GO:0004198 | calcium-dependent cysteine-type endopeptidase activity(GO:0004198) |
| 0.0 | 1.4 | GO:0015175 | neutral amino acid transmembrane transporter activity(GO:0015175) |
| 0.0 | 0.2 | GO:0070097 | delta-catenin binding(GO:0070097) |
| 0.0 | 0.3 | GO:0004983 | neuropeptide Y receptor activity(GO:0004983) |
| 0.0 | 1.4 | GO:0070063 | RNA polymerase binding(GO:0070063) |
| 0.0 | 1.3 | GO:0016790 | thiolester hydrolase activity(GO:0016790) |
| 0.0 | 0.5 | GO:0008432 | JUN kinase binding(GO:0008432) |
| 0.0 | 2.1 | GO:0003777 | microtubule motor activity(GO:0003777) |
| 0.0 | 0.2 | GO:0031492 | nucleosomal DNA binding(GO:0031492) |
| 0.0 | 0.2 | GO:0015272 | ATP-activated inward rectifier potassium channel activity(GO:0015272) |
| 0.0 | 0.9 | GO:0005109 | frizzled binding(GO:0005109) |
| 0.0 | 0.9 | GO:0030374 | ligand-dependent nuclear receptor transcription coactivator activity(GO:0030374) |
| 0.0 | 0.3 | GO:0004708 | MAP kinase kinase activity(GO:0004708) |
| 0.0 | 0.4 | GO:0050681 | androgen receptor binding(GO:0050681) |
| 0.0 | 1.8 | GO:0000980 | RNA polymerase II distal enhancer sequence-specific DNA binding(GO:0000980) |
| 0.0 | 6.3 | GO:0000978 | RNA polymerase II core promoter proximal region sequence-specific DNA binding(GO:0000978) |
| 0.0 | 0.3 | GO:0017025 | TBP-class protein binding(GO:0017025) |
| 0.0 | 0.2 | GO:0042813 | Wnt-activated receptor activity(GO:0042813) |
| 0.0 | 3.9 | GO:0001227 | transcriptional repressor activity, RNA polymerase II transcription regulatory region sequence-specific binding(GO:0001227) |
| 0.0 | 0.7 | GO:0018024 | histone-lysine N-methyltransferase activity(GO:0018024) |
| 0.0 | 0.7 | GO:0048487 | beta-tubulin binding(GO:0048487) |
| 0.0 | 0.3 | GO:0034237 | protein kinase A regulatory subunit binding(GO:0034237) |
| 0.0 | 0.3 | GO:0048027 | mRNA 5'-UTR binding(GO:0048027) |
| 0.0 | 0.4 | GO:0044183 | protein binding involved in protein folding(GO:0044183) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 5.3 | SA REG CASCADE OF CYCLIN EXPR | Expression of cyclins regulates progression through the cell cycle by activating cyclin-dependent kinases. |
| 0.1 | 6.8 | PID ARF6 PATHWAY | Arf6 signaling events |
| 0.1 | 1.2 | PID TCR RAS PATHWAY | Ras signaling in the CD4+ TCR pathway |
| 0.0 | 2.3 | PID HIF2PATHWAY | HIF-2-alpha transcription factor network |
| 0.0 | 0.8 | PID S1P S1P4 PATHWAY | S1P4 pathway |
| 0.0 | 2.4 | ST TUMOR NECROSIS FACTOR PATHWAY | Tumor Necrosis Factor Pathway. |
| 0.0 | 3.2 | PID PLK1 PATHWAY | PLK1 signaling events |
| 0.0 | 0.9 | PID BETA CATENIN DEG PATHWAY | Degradation of beta catenin |
| 0.0 | 2.1 | PID IL12 2PATHWAY | IL12-mediated signaling events |
| 0.0 | 2.9 | PID SMAD2 3NUCLEAR PATHWAY | Regulation of nuclear SMAD2/3 signaling |
| 0.0 | 0.9 | PID P38 ALPHA BETA DOWNSTREAM PATHWAY | Signaling mediated by p38-alpha and p38-beta |
| 0.0 | 3.1 | NABA ECM REGULATORS | Genes encoding enzymes and their regulators involved in the remodeling of the extracellular matrix |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 1.9 | REACTOME DIGESTION OF DIETARY CARBOHYDRATE | Genes involved in Digestion of dietary carbohydrate |
| 0.3 | 2.9 | REACTOME GLUCURONIDATION | Genes involved in Glucuronidation |
| 0.2 | 3.7 | REACTOME BILE SALT AND ORGANIC ANION SLC TRANSPORTERS | Genes involved in Bile salt and organic anion SLC transporters |
| 0.2 | 0.6 | REACTOME LYSOSOME VESICLE BIOGENESIS | Genes involved in Lysosome Vesicle Biogenesis |
| 0.2 | 7.0 | REACTOME SEMA4D IN SEMAPHORIN SIGNALING | Genes involved in Sema4D in semaphorin signaling |
| 0.1 | 2.5 | REACTOME VITAMIN B5 PANTOTHENATE METABOLISM | Genes involved in Vitamin B5 (pantothenate) metabolism |
| 0.1 | 5.3 | REACTOME MEIOTIC RECOMBINATION | Genes involved in Meiotic Recombination |
| 0.1 | 3.0 | REACTOME SYNTHESIS OF GLYCOSYLPHOSPHATIDYLINOSITOL GPI | Genes involved in Synthesis of glycosylphosphatidylinositol (GPI) |
| 0.1 | 7.8 | REACTOME GOLGI ASSOCIATED VESICLE BIOGENESIS | Genes involved in Golgi Associated Vesicle Biogenesis |
| 0.1 | 2.8 | REACTOME TRAF6 MEDIATED NFKB ACTIVATION | Genes involved in TRAF6 mediated NF-kB activation |
| 0.1 | 2.0 | REACTOME TETRAHYDROBIOPTERIN BH4 SYNTHESIS RECYCLING SALVAGE AND REGULATION | Genes involved in Tetrahydrobiopterin (BH4) synthesis, recycling, salvage and regulation |
| 0.1 | 4.8 | REACTOME GLUCONEOGENESIS | Genes involved in Gluconeogenesis |
| 0.1 | 2.8 | REACTOME METAL ION SLC TRANSPORTERS | Genes involved in Metal ion SLC transporters |
| 0.1 | 3.0 | REACTOME SMAD2 SMAD3 SMAD4 HETEROTRIMER REGULATES TRANSCRIPTION | Genes involved in SMAD2/SMAD3:SMAD4 heterotrimer regulates transcription |
| 0.1 | 2.1 | REACTOME KINESINS | Genes involved in Kinesins |
| 0.1 | 2.0 | REACTOME PROLONGED ERK ACTIVATION EVENTS | Genes involved in Prolonged ERK activation events |
| 0.1 | 1.4 | REACTOME NA CL DEPENDENT NEUROTRANSMITTER TRANSPORTERS | Genes involved in Na+/Cl- dependent neurotransmitter transporters |
| 0.1 | 1.3 | REACTOME BRANCHED CHAIN AMINO ACID CATABOLISM | Genes involved in Branched-chain amino acid catabolism |
| 0.1 | 2.4 | REACTOME GLYCOSPHINGOLIPID METABOLISM | Genes involved in Glycosphingolipid metabolism |
| 0.0 | 1.7 | REACTOME DEADENYLATION OF MRNA | Genes involved in Deadenylation of mRNA |
| 0.0 | 2.7 | REACTOME ION TRANSPORT BY P TYPE ATPASES | Genes involved in Ion transport by P-type ATPases |
| 0.0 | 2.1 | REACTOME SPHINGOLIPID DE NOVO BIOSYNTHESIS | Genes involved in Sphingolipid de novo biosynthesis |
| 0.0 | 0.4 | REACTOME ACTIVATION OF CHAPERONE GENES BY ATF6 ALPHA | Genes involved in Activation of Chaperone Genes by ATF6-alpha |
| 0.0 | 1.6 | REACTOME YAP1 AND WWTR1 TAZ STIMULATED GENE EXPRESSION | Genes involved in YAP1- and WWTR1 (TAZ)-stimulated gene expression |
| 0.0 | 1.1 | REACTOME FORMATION OF TUBULIN FOLDING INTERMEDIATES BY CCT TRIC | Genes involved in Formation of tubulin folding intermediates by CCT/TriC |
| 0.0 | 0.7 | REACTOME SYNTHESIS OF SUBSTRATES IN N GLYCAN BIOSYTHESIS | Genes involved in Synthesis of substrates in N-glycan biosythesis |
| 0.0 | 1.1 | REACTOME CREB PHOSPHORYLATION THROUGH THE ACTIVATION OF CAMKII | Genes involved in CREB phosphorylation through the activation of CaMKII |
| 0.0 | 1.9 | REACTOME REGULATION OF BETA CELL DEVELOPMENT | Genes involved in Regulation of beta-cell development |
| 0.0 | 0.9 | REACTOME SIGNALING BY HIPPO | Genes involved in Signaling by Hippo |
| 0.0 | 1.3 | REACTOME TRANSPORT OF VITAMINS NUCLEOSIDES AND RELATED MOLECULES | Genes involved in Transport of vitamins, nucleosides, and related molecules |
| 0.0 | 0.3 | REACTOME GLYCOLYSIS | Genes involved in Glycolysis |
| 0.0 | 1.4 | REACTOME AMINO ACID TRANSPORT ACROSS THE PLASMA MEMBRANE | Genes involved in Amino acid transport across the plasma membrane |
| 0.0 | 2.0 | REACTOME COSTIMULATION BY THE CD28 FAMILY | Genes involved in Costimulation by the CD28 family |
| 0.0 | 0.9 | REACTOME NOD1 2 SIGNALING PATHWAY | Genes involved in NOD1/2 Signaling Pathway |
| 0.0 | 0.9 | REACTOME HS GAG BIOSYNTHESIS | Genes involved in HS-GAG biosynthesis |
| 0.0 | 1.5 | REACTOME RESPIRATORY ELECTRON TRANSPORT | Genes involved in Respiratory electron transport |
| 0.0 | 0.3 | REACTOME UNWINDING OF DNA | Genes involved in Unwinding of DNA |
| 0.0 | 0.7 | REACTOME CYTOCHROME P450 ARRANGED BY SUBSTRATE TYPE | Genes involved in Cytochrome P450 - arranged by substrate type |
| 0.0 | 0.4 | REACTOME ACTIVATION OF BH3 ONLY PROTEINS | Genes involved in Activation of BH3-only proteins |
| 0.0 | 0.4 | REACTOME MITOCHONDRIAL TRNA AMINOACYLATION | Genes involved in Mitochondrial tRNA aminoacylation |
| 0.0 | 1.5 | REACTOME TRANSPORT OF INORGANIC CATIONS ANIONS AND AMINO ACIDS OLIGOPEPTIDES | Genes involved in Transport of inorganic cations/anions and amino acids/oligopeptides |
| 0.0 | 0.3 | REACTOME DEPOSITION OF NEW CENPA CONTAINING NUCLEOSOMES AT THE CENTROMERE | Genes involved in Deposition of New CENPA-containing Nucleosomes at the Centromere |
| 0.0 | 0.3 | REACTOME DCC MEDIATED ATTRACTIVE SIGNALING | Genes involved in DCC mediated attractive signaling |