Project
PRJNA375882: Comprehensive Mouse Transcriptomic BodyMap
Navigation
Downloads
Results for Pax4
Z-value: 0.92
Transcription factors associated with Pax4
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Pax4
|
ENSMUSG00000029706.16 | Pax4 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Pax4 | mm39_v1_chr6_-_28447179_28447179 | -0.16 | 1.9e-01 | Click! |
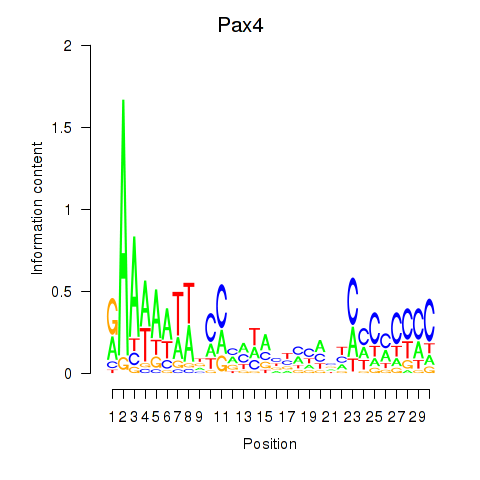
Activity profile of Pax4 motif
Sorted Z-values of Pax4 motif
Network of associatons between targets according to the STRING database.
First level regulatory network of Pax4
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr9_+_59564482 | 6.90 |
ENSMUST00000216620.2
ENSMUST00000217038.2 |
Pkm
|
pyruvate kinase, muscle |
| chr9_+_59563838 | 6.58 |
ENSMUST00000163694.4
|
Pkm
|
pyruvate kinase, muscle |
| chr1_-_79838897 | 6.49 |
ENSMUST00000190724.2
|
Serpine2
|
serine (or cysteine) peptidase inhibitor, clade E, member 2 |
| chr9_+_59563872 | 5.99 |
ENSMUST00000215623.2
ENSMUST00000215660.2 ENSMUST00000217353.2 |
Pkm
|
pyruvate kinase, muscle |
| chr5_+_66833434 | 5.87 |
ENSMUST00000031131.11
|
Uchl1
|
ubiquitin carboxy-terminal hydrolase L1 |
| chr9_-_58066484 | 5.81 |
ENSMUST00000041477.15
|
Islr
|
immunoglobulin superfamily containing leucine-rich repeat |
| chr9_+_58395850 | 5.41 |
ENSMUST00000085658.5
|
Insyn1
|
inhibitory synaptic factor 1 |
| chr17_+_35278011 | 4.90 |
ENSMUST00000007255.13
ENSMUST00000174493.8 |
Ddah2
|
dimethylarginine dimethylaminohydrolase 2 |
| chr9_+_36743980 | 4.84 |
ENSMUST00000034630.15
|
Fez1
|
fasciculation and elongation protein zeta 1 (zygin I) |
| chr17_+_27160356 | 4.73 |
ENSMUST00000229490.2
ENSMUST00000201702.5 ENSMUST00000177932.7 ENSMUST00000201349.6 |
Syngap1
|
synaptic Ras GTPase activating protein 1 homolog (rat) |
| chr1_-_65071897 | 4.27 |
ENSMUST00000162800.8
ENSMUST00000069142.12 |
Akr1cl
|
aldo-keto reductase family 1, member C-like |
| chr16_+_22965286 | 4.23 |
ENSMUST00000023593.6
|
Adipoq
|
adiponectin, C1Q and collagen domain containing |
| chr16_+_22965330 | 3.95 |
ENSMUST00000171309.2
|
Adipoq
|
adiponectin, C1Q and collagen domain containing |
| chr12_-_90705212 | 3.88 |
ENSMUST00000082432.6
|
Dio2
|
deiodinase, iodothyronine, type II |
| chr17_+_17622934 | 3.87 |
ENSMUST00000115576.3
|
Lix1
|
limb and CNS expressed 1 |
| chr7_+_58307930 | 3.66 |
ENSMUST00000168747.3
|
Atp10a
|
ATPase, class V, type 10A |
| chr17_+_27160203 | 3.56 |
ENSMUST00000194598.6
|
Syngap1
|
synaptic Ras GTPase activating protein 1 homolog (rat) |
| chr9_-_37058590 | 3.49 |
ENSMUST00000080754.12
ENSMUST00000188057.7 ENSMUST00000039674.13 |
Pknox2
|
Pbx/knotted 1 homeobox 2 |
| chr13_-_110416637 | 3.48 |
ENSMUST00000167824.3
ENSMUST00000224180.2 |
Rab3c
|
RAB3C, member RAS oncogene family |
| chr18_-_43925932 | 3.19 |
ENSMUST00000237926.2
ENSMUST00000096570.4 |
Gm94
|
predicted gene 94 |
| chr19_+_4022322 | 3.12 |
ENSMUST00000143380.3
|
Aldh3b2
|
aldehyde dehydrogenase 3 family, member B2 |
| chr15_-_88863210 | 3.12 |
ENSMUST00000042594.13
ENSMUST00000109368.2 |
Mlc1
|
megalencephalic leukoencephalopathy with subcortical cysts 1 homolog (human) |
| chr11_+_62465304 | 3.05 |
ENSMUST00000018651.14
|
Trpv2
|
transient receptor potential cation channel, subfamily V, member 2 |
| chr2_+_181322077 | 2.92 |
ENSMUST00000103042.10
|
Tcea2
|
transcription elongation factor A (SII), 2 |
| chr5_-_103247920 | 2.78 |
ENSMUST00000112848.8
|
Mapk10
|
mitogen-activated protein kinase 10 |
| chr7_-_144292288 | 2.68 |
ENSMUST00000238848.2
ENSMUST00000118556.9 ENSMUST00000033393.15 |
Ano1
|
anoctamin 1, calcium activated chloride channel |
| chr2_-_125348305 | 2.60 |
ENSMUST00000028633.13
|
Fbn1
|
fibrillin 1 |
| chrX_-_74918709 | 2.56 |
ENSMUST00000114059.10
|
Pls3
|
plastin 3 (T-isoform) |
| chr17_+_47604995 | 2.54 |
ENSMUST00000190020.4
|
Trerf1
|
transcriptional regulating factor 1 |
| chr1_-_169938298 | 2.53 |
ENSMUST00000192312.6
|
Ddr2
|
discoidin domain receptor family, member 2 |
| chr9_+_36744016 | 2.36 |
ENSMUST00000214772.2
|
Fez1
|
fasciculation and elongation protein zeta 1 (zygin I) |
| chr2_-_32976378 | 2.31 |
ENSMUST00000049618.9
|
Garnl3
|
GTPase activating RANGAP domain-like 3 |
| chr14_+_9646630 | 2.30 |
ENSMUST00000112658.8
ENSMUST00000112657.9 ENSMUST00000177814.2 ENSMUST00000067491.14 |
Cadps
|
Ca2+-dependent secretion activator |
| chrX_+_72235828 | 2.29 |
ENSMUST00000101486.5
|
Xlr3b
|
X-linked lymphocyte-regulated 3B |
| chrX_-_8072714 | 2.23 |
ENSMUST00000089403.10
ENSMUST00000077595.12 ENSMUST00000089402.10 ENSMUST00000082320.12 |
Porcn
|
porcupine O-acyltransferase |
| chrX_+_142301572 | 2.23 |
ENSMUST00000033640.14
|
Pak3
|
p21 (RAC1) activated kinase 3 |
| chr16_-_14109219 | 2.17 |
ENSMUST00000230397.2
ENSMUST00000231567.2 ENSMUST00000090287.5 |
Myh11
|
myosin, heavy polypeptide 11, smooth muscle |
| chr4_+_136011969 | 2.12 |
ENSMUST00000144217.8
|
Zfp46
|
zinc finger protein 46 |
| chr7_+_24561616 | 2.04 |
ENSMUST00000170837.3
|
Gm9844
|
predicted pseudogene 9844 |
| chr15_-_79048674 | 1.95 |
ENSMUST00000230261.2
ENSMUST00000040019.5 |
Sox10
|
SRY (sex determining region Y)-box 10 |
| chrX_+_21350783 | 1.95 |
ENSMUST00000089188.9
|
Agtr2
|
angiotensin II receptor, type 2 |
| chr2_-_114485421 | 1.89 |
ENSMUST00000028640.14
ENSMUST00000102542.10 |
Dph6
|
diphthamine biosynthesis 6 |
| chr19_+_53128861 | 1.87 |
ENSMUST00000111741.10
|
Add3
|
adducin 3 (gamma) |
| chr15_+_78481247 | 1.78 |
ENSMUST00000043069.6
ENSMUST00000231180.2 ENSMUST00000229796.2 ENSMUST00000229295.2 |
Cyth4
|
cytohesin 4 |
| chr15_-_50753061 | 1.73 |
ENSMUST00000165201.9
ENSMUST00000184458.8 |
Trps1
|
transcriptional repressor GATA binding 1 |
| chr3_+_95532282 | 1.73 |
ENSMUST00000058230.13
ENSMUST00000037983.6 |
Ensa
|
endosulfine alpha |
| chr13_+_75237939 | 1.72 |
ENSMUST00000022075.6
|
Pcsk1
|
proprotein convertase subtilisin/kexin type 1 |
| chr9_-_101076198 | 1.71 |
ENSMUST00000066773.9
|
Ppp2r3a
|
protein phosphatase 2, regulatory subunit B'', alpha |
| chr1_-_74974707 | 1.71 |
ENSMUST00000094844.4
|
Cfap65
|
cilia and flagella associated protein 65 |
| chr11_-_18968955 | 1.57 |
ENSMUST00000068264.14
ENSMUST00000185131.8 |
Meis1
|
Meis homeobox 1 |
| chr1_-_155910546 | 1.56 |
ENSMUST00000169241.8
|
Tor1aip1
|
torsin A interacting protein 1 |
| chr7_-_144292257 | 1.55 |
ENSMUST00000121758.8
|
Ano1
|
anoctamin 1, calcium activated chloride channel |
| chr13_+_109769294 | 1.52 |
ENSMUST00000135275.8
|
Pde4d
|
phosphodiesterase 4D, cAMP specific |
| chr4_+_147216495 | 1.42 |
ENSMUST00000084149.10
|
Zfp991
|
zinc finger protein 991 |
| chr6_+_136495784 | 1.39 |
ENSMUST00000032335.13
ENSMUST00000185724.7 |
Atf7ip
|
activating transcription factor 7 interacting protein |
| chr2_+_158509039 | 1.37 |
ENSMUST00000045503.11
|
Ppp1r16b
|
protein phosphatase 1, regulatory subunit 16B |
| chr15_+_80861966 | 1.37 |
ENSMUST00000139517.9
ENSMUST00000137255.3 ENSMUST00000137004.2 |
Sgsm3
|
small G protein signaling modulator 3 |
| chr5_-_121641461 | 1.37 |
ENSMUST00000079368.5
|
Adam1b
|
a disintegrin and metallopeptidase domain 1b |
| chr4_+_136012019 | 1.36 |
ENSMUST00000130223.8
|
Zfp46
|
zinc finger protein 46 |
| chr10_-_63926044 | 1.35 |
ENSMUST00000105439.2
|
Lrrtm3
|
leucine rich repeat transmembrane neuronal 3 |
| chr7_-_64575308 | 1.34 |
ENSMUST00000149851.8
|
Fam189a1
|
family with sequence similarity 189, member A1 |
| chr1_-_84912810 | 1.32 |
ENSMUST00000027422.7
|
Slc16a14
|
solute carrier family 16 (monocarboxylic acid transporters), member 14 |
| chr10_+_119655294 | 1.26 |
ENSMUST00000105262.9
ENSMUST00000147454.8 ENSMUST00000138410.8 ENSMUST00000144825.8 ENSMUST00000148954.8 ENSMUST00000144959.8 |
Grip1
|
glutamate receptor interacting protein 1 |
| chr2_-_114485342 | 1.23 |
ENSMUST00000055144.8
|
Dph6
|
diphthamine biosynthesis 6 |
| chr2_-_131170902 | 1.23 |
ENSMUST00000110194.8
|
Rnf24
|
ring finger protein 24 |
| chr2_+_22785534 | 1.22 |
ENSMUST00000053729.14
|
Pdss1
|
prenyl (solanesyl) diphosphate synthase, subunit 1 |
| chrX_+_9066105 | 1.22 |
ENSMUST00000069763.3
|
Lancl3
|
LanC lantibiotic synthetase component C-like 3 (bacterial) |
| chrM_+_3906 | 1.13 |
ENSMUST00000082396.1
|
mt-Nd2
|
mitochondrially encoded NADH dehydrogenase 2 |
| chr4_+_102427247 | 1.13 |
ENSMUST00000097950.9
|
Pde4b
|
phosphodiesterase 4B, cAMP specific |
| chr4_+_118522716 | 1.10 |
ENSMUST00000102666.5
|
Olfr62
|
olfactory receptor 62 |
| chr18_-_84104574 | 1.08 |
ENSMUST00000175783.3
|
Tshz1
|
teashirt zinc finger family member 1 |
| chr5_+_63957805 | 1.07 |
ENSMUST00000162757.4
|
Nwd2
|
NACHT and WD repeat domain containing 2 |
| chr5_+_124690908 | 1.06 |
ENSMUST00000071057.14
ENSMUST00000111438.2 |
Ddx55
|
DEAD box helicase 55 |
| chr2_+_158508609 | 1.00 |
ENSMUST00000103116.10
|
Ppp1r16b
|
protein phosphatase 1, regulatory subunit 16B |
| chr8_-_85500010 | 0.94 |
ENSMUST00000109764.8
|
Nfix
|
nuclear factor I/X |
| chr11_+_87017878 | 0.93 |
ENSMUST00000041282.13
|
Trim37
|
tripartite motif-containing 37 |
| chrM_+_11735 | 0.92 |
ENSMUST00000082418.1
|
mt-Nd5
|
mitochondrially encoded NADH dehydrogenase 5 |
| chrX_+_142301666 | 0.89 |
ENSMUST00000134402.8
|
Pak3
|
p21 (RAC1) activated kinase 3 |
| chr11_-_18968714 | 0.85 |
ENSMUST00000177417.8
|
Meis1
|
Meis homeobox 1 |
| chr6_+_136495818 | 0.84 |
ENSMUST00000186577.7
|
Atf7ip
|
activating transcription factor 7 interacting protein |
| chr7_+_78563513 | 0.80 |
ENSMUST00000038142.15
|
Isg20
|
interferon-stimulated protein |
| chr15_+_80862074 | 0.73 |
ENSMUST00000229727.2
|
Sgsm3
|
small G protein signaling modulator 3 |
| chr8_-_83129134 | 0.70 |
ENSMUST00000209363.2
|
Il15
|
interleukin 15 |
| chr4_+_146695418 | 0.70 |
ENSMUST00000130825.8
|
Zfp993
|
zinc finger protein 993 |
| chr7_+_78563184 | 0.69 |
ENSMUST00000121645.8
|
Isg20
|
interferon-stimulated protein |
| chr18_+_58969739 | 0.69 |
ENSMUST00000052907.7
|
Adamts19
|
a disintegrin-like and metallopeptidase (reprolysin type) with thrombospondin type 1 motif, 19 |
| chr2_+_22785593 | 0.68 |
ENSMUST00000152170.8
|
Pdss1
|
prenyl (solanesyl) diphosphate synthase, subunit 1 |
| chr18_-_84104507 | 0.64 |
ENSMUST00000060303.10
|
Tshz1
|
teashirt zinc finger family member 1 |
| chr15_-_59245998 | 0.60 |
ENSMUST00000022976.6
|
Washc5
|
WASH complex subunit 5 |
| chr16_-_45664591 | 0.59 |
ENSMUST00000076333.12
|
Phldb2
|
pleckstrin homology like domain, family B, member 2 |
| chr6_+_113460258 | 0.59 |
ENSMUST00000032422.6
|
Creld1
|
cysteine-rich with EGF-like domains 1 |
| chr1_+_153628598 | 0.53 |
ENSMUST00000182538.3
|
Rnasel
|
ribonuclease L (2', 5'-oligoisoadenylate synthetase-dependent) |
| chr6_+_47854138 | 0.51 |
ENSMUST00000061890.8
|
Zfp282
|
zinc finger protein 282 |
| chr10_+_21870565 | 0.50 |
ENSMUST00000020145.12
|
Sgk1
|
serum/glucocorticoid regulated kinase 1 |
| chr7_+_7174315 | 0.49 |
ENSMUST00000051435.8
|
Zfp418
|
zinc finger protein 418 |
| chr7_+_78563150 | 0.48 |
ENSMUST00000118867.8
|
Isg20
|
interferon-stimulated protein |
| chr4_+_155915729 | 0.48 |
ENSMUST00000139651.8
ENSMUST00000084097.12 |
Aurkaip1
|
aurora kinase A interacting protein 1 |
| chr4_+_150366028 | 0.47 |
ENSMUST00000105682.9
|
Rere
|
arginine glutamic acid dipeptide (RE) repeats |
| chr7_+_102246977 | 0.44 |
ENSMUST00000215712.2
|
Olfr552
|
olfactory receptor 552 |
| chr13_+_23922783 | 0.42 |
ENSMUST00000040914.3
|
H1f2
|
H1.2 linker histone, cluster member |
| chr10_+_127256993 | 0.41 |
ENSMUST00000170336.8
|
R3hdm2
|
R3H domain containing 2 |
| chr10_-_129509659 | 0.40 |
ENSMUST00000213294.2
ENSMUST00000216067.3 ENSMUST00000203424.2 |
Olfr801
|
olfactory receptor 801 |
| chr9_-_48747232 | 0.40 |
ENSMUST00000093852.5
|
Zbtb16
|
zinc finger and BTB domain containing 16 |
| chr10_-_67384898 | 0.40 |
ENSMUST00000075686.7
|
Ado
|
2-aminoethanethiol (cysteamine) dioxygenase |
| chr13_-_55676334 | 0.39 |
ENSMUST00000047877.5
|
Dok3
|
docking protein 3 |
| chr7_+_43057611 | 0.34 |
ENSMUST00000005592.7
|
Siglecg
|
sialic acid binding Ig-like lectin G |
| chr17_+_35327255 | 0.29 |
ENSMUST00000037849.3
|
Ly6g5c
|
lymphocyte antigen 6 complex, locus G5C |
| chrX_-_9529189 | 0.29 |
ENSMUST00000033519.3
|
Dynlt3
|
dynein light chain Tctex-type 3 |
| chr1_+_21419819 | 0.27 |
ENSMUST00000088407.4
|
Khdc1a
|
KH domain containing 1A |
| chr19_+_55169148 | 0.27 |
ENSMUST00000154886.8
ENSMUST00000120936.8 ENSMUST00000025936.12 |
Tectb
|
tectorin beta |
| chr8_-_22966831 | 0.24 |
ENSMUST00000163774.3
ENSMUST00000033935.16 |
Smim19
|
small integral membrane protein 19 |
| chr6_-_130170075 | 0.24 |
ENSMUST00000112032.8
ENSMUST00000071554.3 |
Klra9
|
killer cell lectin-like receptor subfamily A, member 9 |
| chr11_-_58346806 | 0.23 |
ENSMUST00000055204.6
|
Olfr30
|
olfactory receptor 30 |
| chr17_+_34457868 | 0.21 |
ENSMUST00000095342.11
ENSMUST00000167280.8 ENSMUST00000236838.2 |
H2-Ob
|
histocompatibility 2, O region beta locus |
| chr7_-_103928939 | 0.21 |
ENSMUST00000051795.10
|
Trim5
|
tripartite motif-containing 5 |
| chr13_+_21901791 | 0.21 |
ENSMUST00000188775.2
|
H3c10
|
H3 clustered histone 10 |
| chr10_+_129153986 | 0.20 |
ENSMUST00000215503.2
|
Olfr780
|
olfactory receptor 780 |
| chr11_+_106916430 | 0.16 |
ENSMUST00000140447.4
|
1810010H24Rik
|
RIKEN cDNA 1810010H24 gene |
| chrX_-_153911405 | 0.15 |
ENSMUST00000076671.4
|
Cldn34b2
|
claudin 34B2 |
| chr6_+_65567373 | 0.14 |
ENSMUST00000114236.2
|
Tnip3
|
TNFAIP3 interacting protein 3 |
| chr16_-_45664664 | 0.12 |
ENSMUST00000036355.13
|
Phldb2
|
pleckstrin homology like domain, family B, member 2 |
| chr9_-_57065572 | 0.11 |
ENSMUST00000065358.9
|
Commd4
|
COMM domain containing 4 |
| chr19_-_10460238 | 0.10 |
ENSMUST00000235392.2
ENSMUST00000237522.2 ENSMUST00000038842.5 |
Ppp1r32
|
protein phosphatase 1, regulatory subunit 32 |
| chr7_-_102591782 | 0.10 |
ENSMUST00000210571.2
ENSMUST00000064830.6 |
Olfr573-ps1
|
olfactory receptor 573, pseudogene 1 |
| chr9_+_96141299 | 0.09 |
ENSMUST00000179065.8
|
Tfdp2
|
transcription factor Dp 2 |
| chr9_+_96141317 | 0.09 |
ENSMUST00000165768.4
|
Tfdp2
|
transcription factor Dp 2 |
| chr4_+_6365694 | 0.08 |
ENSMUST00000175769.8
ENSMUST00000140830.8 ENSMUST00000108374.8 |
Sdcbp
|
syndecan binding protein |
| chr5_-_116427003 | 0.07 |
ENSMUST00000086483.4
ENSMUST00000050178.13 |
Ccdc60
|
coiled-coil domain containing 60 |
| chr4_+_6365650 | 0.07 |
ENSMUST00000029912.11
ENSMUST00000103008.12 |
Sdcbp
|
syndecan binding protein |
| chr7_-_104246386 | 0.07 |
ENSMUST00000057385.5
|
Olfr655
|
olfactory receptor 655 |
| chr2_-_89491560 | 0.05 |
ENSMUST00000111527.4
|
Olfr1250
|
olfactory receptor 1250 |
| chr9_+_18848418 | 0.05 |
ENSMUST00000218385.2
|
Olfr832
|
olfactory receptor 832 |
| chr16_-_58747023 | 0.04 |
ENSMUST00000205742.2
|
Olfr181
|
olfactory receptor 181 |
| chr10_+_127257077 | 0.03 |
ENSMUST00000168780.8
|
R3hdm2
|
R3H domain containing 2 |
| chr3_+_20111958 | 0.02 |
ENSMUST00000002502.12
|
Hltf
|
helicase-like transcription factor |
| chr2_+_73102269 | 0.01 |
ENSMUST00000090813.6
|
Sp9
|
trans-acting transcription factor 9 |
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.7 | 8.2 | GO:0072313 | metanephric glomerular visceral epithelial cell differentiation(GO:0072248) metanephric glomerular visceral epithelial cell development(GO:0072249) metanephric glomerular epithelial cell differentiation(GO:0072312) metanephric glomerular epithelial cell development(GO:0072313) |
| 2.2 | 6.5 | GO:0061107 | seminal vesicle development(GO:0061107) |
| 1.9 | 19.5 | GO:0042866 | pyruvate biosynthetic process(GO:0042866) |
| 1.0 | 5.9 | GO:0007412 | axon target recognition(GO:0007412) |
| 1.0 | 3.9 | GO:0070460 | thyroid-stimulating hormone secretion(GO:0070460) |
| 0.6 | 2.6 | GO:0035582 | sequestering of BMP in extracellular matrix(GO:0035582) |
| 0.6 | 2.4 | GO:1902309 | negative regulation of peptidyl-serine dephosphorylation(GO:1902309) |
| 0.6 | 2.3 | GO:0099525 | presynaptic dense core granule exocytosis(GO:0099525) |
| 0.5 | 1.9 | GO:0035566 | regulation of metanephros size(GO:0035566) |
| 0.4 | 7.2 | GO:1902902 | negative regulation of autophagosome assembly(GO:1902902) |
| 0.4 | 2.8 | GO:0007258 | JUN phosphorylation(GO:0007258) |
| 0.4 | 4.2 | GO:0015705 | iodide transport(GO:0015705) |
| 0.4 | 4.9 | GO:0006527 | arginine catabolic process(GO:0006527) |
| 0.3 | 2.0 | GO:0000738 | DNA catabolic process, exonucleolytic(GO:0000738) |
| 0.3 | 2.1 | GO:0048227 | plasma membrane to endosome transport(GO:0048227) |
| 0.3 | 3.1 | GO:0047484 | regulation of response to osmotic stress(GO:0047484) |
| 0.2 | 1.7 | GO:0060023 | soft palate development(GO:0060023) |
| 0.2 | 1.7 | GO:0090245 | axis elongation involved in somitogenesis(GO:0090245) |
| 0.2 | 1.9 | GO:0010626 | negative regulation of Schwann cell proliferation(GO:0010626) |
| 0.2 | 2.6 | GO:1901898 | negative regulation of relaxation of cardiac muscle(GO:1901898) |
| 0.2 | 2.2 | GO:0048251 | elastic fiber assembly(GO:0048251) |
| 0.2 | 2.5 | GO:0090091 | positive regulation of extracellular matrix disassembly(GO:0090091) |
| 0.1 | 0.7 | GO:1903690 | negative regulation of wound healing, spreading of epidermal cells(GO:1903690) |
| 0.1 | 1.6 | GO:0071763 | nuclear membrane organization(GO:0071763) |
| 0.1 | 0.9 | GO:0036353 | histone H2A-K119 monoubiquitination(GO:0036353) |
| 0.1 | 8.3 | GO:0048169 | regulation of long-term neuronal synaptic plasticity(GO:0048169) |
| 0.1 | 0.5 | GO:0021691 | cerebellar Purkinje cell layer maturation(GO:0021691) |
| 0.1 | 0.7 | GO:0035723 | interleukin-15-mediated signaling pathway(GO:0035723) extrathymic T cell selection(GO:0045062) cellular response to interleukin-15(GO:0071350) |
| 0.1 | 5.4 | GO:0061003 | positive regulation of dendritic spine morphogenesis(GO:0061003) |
| 0.1 | 3.9 | GO:0097352 | autophagosome maturation(GO:0097352) |
| 0.1 | 2.4 | GO:0060044 | negative regulation of cardiac muscle cell proliferation(GO:0060044) |
| 0.1 | 2.6 | GO:0051764 | actin crosslink formation(GO:0051764) |
| 0.1 | 0.4 | GO:0051138 | positive regulation of NK T cell differentiation(GO:0051138) |
| 0.1 | 1.9 | GO:0006744 | ubiquinone biosynthetic process(GO:0006744) |
| 0.1 | 0.6 | GO:0040038 | meiotic cytokinesis(GO:0033206) polar body extrusion after meiotic divisions(GO:0040038) spindle assembly involved in meiosis(GO:0090306) |
| 0.1 | 2.2 | GO:0045898 | regulation of RNA polymerase II transcriptional preinitiation complex assembly(GO:0045898) |
| 0.1 | 0.2 | GO:0002581 | negative regulation of antigen processing and presentation of peptide or polysaccharide antigen via MHC class II(GO:0002581) |
| 0.1 | 1.4 | GO:1902004 | positive regulation of beta-amyloid formation(GO:1902004) |
| 0.1 | 1.1 | GO:0006120 | mitochondrial electron transport, NADH to ubiquinone(GO:0006120) |
| 0.1 | 1.7 | GO:0016486 | peptide hormone processing(GO:0016486) |
| 0.1 | 1.5 | GO:1902043 | positive regulation of extrinsic apoptotic signaling pathway via death domain receptors(GO:1902043) |
| 0.0 | 1.8 | GO:0032011 | ARF protein signal transduction(GO:0032011) regulation of ARF protein signal transduction(GO:0032012) |
| 0.0 | 0.9 | GO:0021684 | cerebellar granular layer formation(GO:0021684) cerebellar granule cell differentiation(GO:0021707) |
| 0.0 | 3.1 | GO:0090280 | positive regulation of calcium ion import(GO:0090280) |
| 0.0 | 2.7 | GO:0015914 | phospholipid transport(GO:0015914) |
| 0.0 | 0.5 | GO:0043402 | glucocorticoid mediated signaling pathway(GO:0043402) |
| 0.0 | 3.1 | GO:0006081 | cellular aldehyde metabolic process(GO:0006081) |
| 0.0 | 3.5 | GO:0019882 | antigen processing and presentation(GO:0019882) |
| 0.0 | 0.2 | GO:1903553 | positive regulation of extracellular exosome assembly(GO:1903553) |
| 0.0 | 0.5 | GO:2001275 | positive regulation of glucose import in response to insulin stimulus(GO:2001275) |
| 0.0 | 2.2 | GO:1903955 | positive regulation of protein targeting to mitochondrion(GO:1903955) |
| 0.0 | 1.1 | GO:0010501 | RNA secondary structure unwinding(GO:0010501) |
| 0.0 | 2.2 | GO:0006497 | protein lipidation(GO:0006497) |
| 0.0 | 1.7 | GO:0035308 | negative regulation of protein dephosphorylation(GO:0035308) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 6.5 | 19.5 | GO:1902912 | pyruvate kinase complex(GO:1902912) |
| 0.4 | 8.3 | GO:0031235 | intrinsic component of the cytoplasmic side of the plasma membrane(GO:0031235) |
| 0.2 | 2.2 | GO:0030485 | smooth muscle contractile fiber(GO:0030485) |
| 0.2 | 3.1 | GO:0032584 | growth cone membrane(GO:0032584) |
| 0.2 | 3.2 | GO:0031232 | extrinsic component of external side of plasma membrane(GO:0031232) |
| 0.2 | 2.6 | GO:0043205 | microfibril(GO:0001527) fibril(GO:0043205) |
| 0.1 | 1.9 | GO:0031315 | extrinsic component of mitochondrial outer membrane(GO:0031315) |
| 0.1 | 2.3 | GO:0031045 | dense core granule(GO:0031045) |
| 0.1 | 8.2 | GO:0005581 | collagen trimer(GO:0005581) |
| 0.1 | 4.2 | GO:0034707 | chloride channel complex(GO:0034707) |
| 0.1 | 1.7 | GO:0000159 | protein phosphatase type 2A complex(GO:0000159) |
| 0.0 | 3.5 | GO:0045171 | intercellular bridge(GO:0045171) |
| 0.0 | 2.1 | GO:0005921 | gap junction(GO:0005921) |
| 0.0 | 2.2 | GO:0032281 | AMPA glutamate receptor complex(GO:0032281) |
| 0.0 | 2.6 | GO:0005891 | voltage-gated calcium channel complex(GO:0005891) |
| 0.0 | 0.6 | GO:0071203 | WASH complex(GO:0071203) |
| 0.0 | 0.7 | GO:0045180 | basal cortex(GO:0045180) |
| 0.0 | 2.0 | GO:0015030 | Cajal body(GO:0015030) |
| 0.0 | 0.9 | GO:0035098 | ESC/E(Z) complex(GO:0035098) |
| 0.0 | 2.6 | GO:0032420 | stereocilium(GO:0032420) |
| 0.0 | 3.1 | GO:0005811 | lipid particle(GO:0005811) |
| 0.0 | 3.0 | GO:0000118 | histone deacetylase complex(GO:0000118) |
| 0.0 | 4.1 | GO:0055037 | recycling endosome(GO:0055037) |
| 0.0 | 0.2 | GO:1990462 | omegasome(GO:1990462) |
| 0.0 | 5.9 | GO:0044306 | neuron projection terminus(GO:0044306) |
| 0.0 | 3.5 | GO:0008021 | synaptic vesicle(GO:0008021) |
| 0.0 | 2.8 | GO:0043204 | perikaryon(GO:0043204) |
| 0.0 | 0.2 | GO:0042613 | MHC class II protein complex(GO:0042613) |
| 0.0 | 7.5 | GO:0005874 | microtubule(GO:0005874) |
| 0.0 | 2.6 | GO:0000794 | condensed nuclear chromosome(GO:0000794) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 3.9 | 19.5 | GO:0004743 | pyruvate kinase activity(GO:0004743) |
| 1.5 | 5.9 | GO:0031694 | alpha-2A adrenergic receptor binding(GO:0031694) |
| 1.0 | 3.9 | GO:0004800 | thyroxine 5'-deiodinase activity(GO:0004800) |
| 0.7 | 8.2 | GO:0033691 | sialic acid binding(GO:0033691) |
| 0.7 | 2.2 | GO:1990698 | palmitoleoyltransferase activity(GO:1990698) |
| 0.7 | 4.2 | GO:0015111 | iodide transmembrane transporter activity(GO:0015111) |
| 0.6 | 2.8 | GO:0004705 | JUN kinase activity(GO:0004705) SAP kinase activity(GO:0016909) |
| 0.5 | 1.9 | GO:0050347 | trans-hexaprenyltranstransferase activity(GO:0000010) trans-octaprenyltranstransferase activity(GO:0050347) |
| 0.4 | 4.9 | GO:0016813 | hydrolase activity, acting on carbon-nitrogen (but not peptide) bonds, in linear amidines(GO:0016813) |
| 0.4 | 2.0 | GO:0008859 | exoribonuclease II activity(GO:0008859) |
| 0.4 | 1.9 | GO:0004945 | angiotensin type II receptor activity(GO:0004945) |
| 0.3 | 3.1 | GO:0004030 | aldehyde dehydrogenase [NAD(P)+] activity(GO:0004030) |
| 0.2 | 2.5 | GO:0038062 | protein tyrosine kinase collagen receptor activity(GO:0038062) |
| 0.1 | 8.3 | GO:0043015 | gamma-tubulin binding(GO:0043015) |
| 0.1 | 3.1 | GO:0004708 | MAP kinase kinase activity(GO:0004708) |
| 0.1 | 3.5 | GO:0031489 | myosin V binding(GO:0031489) |
| 0.1 | 3.7 | GO:0004012 | phospholipid-translocating ATPase activity(GO:0004012) |
| 0.1 | 2.1 | GO:0050136 | NADH dehydrogenase (ubiquinone) activity(GO:0008137) NADH dehydrogenase (quinone) activity(GO:0050136) |
| 0.1 | 1.7 | GO:0019870 | potassium channel inhibitor activity(GO:0019870) |
| 0.1 | 0.5 | GO:0043423 | 3-phosphoinositide-dependent protein kinase binding(GO:0043423) |
| 0.1 | 2.9 | GO:0005201 | extracellular matrix structural constituent(GO:0005201) |
| 0.1 | 1.6 | GO:0005521 | lamin binding(GO:0005521) |
| 0.1 | 0.2 | GO:0005137 | interleukin-5 receptor binding(GO:0005137) |
| 0.0 | 1.5 | GO:0004115 | 3',5'-cyclic-AMP phosphodiesterase activity(GO:0004115) |
| 0.0 | 1.8 | GO:0005086 | ARF guanyl-nucleotide exchange factor activity(GO:0005086) |
| 0.0 | 1.9 | GO:1990841 | promoter-specific chromatin binding(GO:1990841) |
| 0.0 | 6.5 | GO:0004867 | serine-type endopeptidase inhibitor activity(GO:0004867) |
| 0.0 | 2.2 | GO:0008307 | structural constituent of muscle(GO:0008307) |
| 0.0 | 7.6 | GO:0017124 | SH3 domain binding(GO:0017124) |
| 0.0 | 0.9 | GO:0030159 | receptor signaling complex scaffold activity(GO:0030159) |
| 0.0 | 2.3 | GO:0005546 | phosphatidylinositol-4,5-bisphosphate binding(GO:0005546) |
| 0.0 | 6.1 | GO:0051015 | actin filament binding(GO:0051015) |
| 0.0 | 0.4 | GO:0001206 | transcriptional repressor activity, RNA polymerase II distal enhancer sequence-specific binding(GO:0001206) |
| 0.0 | 3.1 | GO:0008565 | protein transporter activity(GO:0008565) |
| 0.0 | 1.9 | GO:0005080 | protein kinase C binding(GO:0005080) |
| 0.0 | 3.1 | GO:0005262 | calcium channel activity(GO:0005262) |
| 0.0 | 2.4 | GO:0019888 | protein phosphatase regulator activity(GO:0019888) |
| 0.0 | 1.3 | GO:0008028 | monocarboxylic acid transmembrane transporter activity(GO:0008028) |
| 0.0 | 0.9 | GO:0005164 | tumor necrosis factor receptor binding(GO:0005164) |
| 0.0 | 1.1 | GO:0004004 | RNA helicase activity(GO:0003724) ATP-dependent RNA helicase activity(GO:0004004) |
| 0.0 | 0.4 | GO:0016702 | oxidoreductase activity, acting on single donors with incorporation of molecular oxygen, incorporation of two atoms of oxygen(GO:0016702) |
| 0.0 | 1.9 | GO:0030674 | protein binding, bridging(GO:0030674) |
| 0.0 | 1.4 | GO:0004860 | protein kinase inhibitor activity(GO:0004860) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 8.3 | PID RAS PATHWAY | Regulation of Ras family activation |
| 0.1 | 5.9 | PID ALPHA SYNUCLEIN PATHWAY | Alpha-synuclein signaling |
| 0.1 | 3.1 | PID P38 ALPHA BETA PATHWAY | Regulation of p38-alpha and p38-beta |
| 0.1 | 2.8 | ST GRANULE CELL SURVIVAL PATHWAY | Granule Cell Survival Pathway is a specific case of more general PAC1 Receptor Pathway. |
| 0.0 | 8.3 | NABA ECM GLYCOPROTEINS | Genes encoding structural ECM glycoproteins |
| 0.0 | 5.3 | NABA ECM REGULATORS | Genes encoding enzymes and their regulators involved in the remodeling of the extracellular matrix |
| 0.0 | 2.2 | WNT SIGNALING | Genes related to Wnt-mediated signal transduction |
| 0.0 | 0.5 | PID AURORA A PATHWAY | Aurora A signaling |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 3.9 | REACTOME AMINE DERIVED HORMONES | Genes involved in Amine-derived hormones |
| 0.2 | 1.7 | REACTOME PEPTIDE HORMONE BIOSYNTHESIS | Genes involved in Peptide hormone biosynthesis |
| 0.1 | 2.8 | REACTOME ACTIVATION OF THE AP1 FAMILY OF TRANSCRIPTION FACTORS | Genes involved in Activation of the AP-1 family of transcription factors |
| 0.1 | 8.2 | REACTOME TRANSCRIPTIONAL REGULATION OF WHITE ADIPOCYTE DIFFERENTIATION | Genes involved in Transcriptional Regulation of White Adipocyte Differentiation |
| 0.1 | 3.7 | REACTOME ION TRANSPORT BY P TYPE ATPASES | Genes involved in Ion transport by P-type ATPases |
| 0.1 | 2.2 | REACTOME SEMA4D INDUCED CELL MIGRATION AND GROWTH CONE COLLAPSE | Genes involved in Sema4D induced cell migration and growth-cone collapse |
| 0.1 | 2.6 | REACTOME DARPP 32 EVENTS | Genes involved in DARPP-32 events |
| 0.0 | 2.5 | REACTOME INTERFERON ALPHA BETA SIGNALING | Genes involved in Interferon alpha/beta signaling |