Project
PRJNA375882: Comprehensive Mouse Transcriptomic BodyMap
Navigation
Downloads
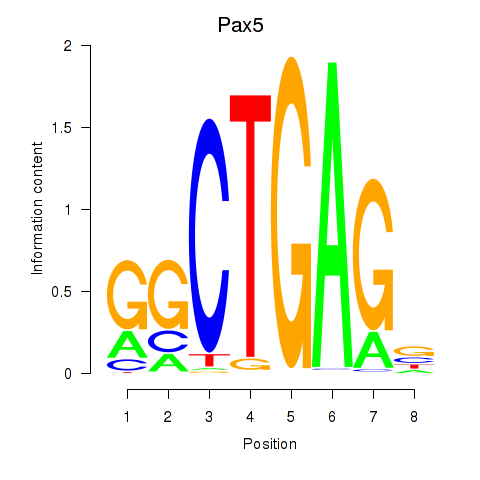
Results for Pax5
Z-value: 2.04
Transcription factors associated with Pax5
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Pax5
|
ENSMUSG00000014030.16 | Pax5 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Pax5 | mm39_v1_chr4_-_44710408_44710487 | -0.19 | 1.0e-01 | Click! |
Activity profile of Pax5 motif
Sorted Z-values of Pax5 motif
Network of associatons between targets according to the STRING database.
First level regulatory network of Pax5
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr17_+_35455532 | 9.33 |
ENSMUST00000068261.9
|
Atp6v1g2
|
ATPase, H+ transporting, lysosomal V1 subunit G2 |
| chr11_+_31950452 | 8.90 |
ENSMUST00000109409.8
ENSMUST00000020537.9 |
Nsg2
|
neuron specific gene family member 2 |
| chr13_+_58956495 | 8.88 |
ENSMUST00000225950.2
ENSMUST00000225583.2 |
Ntrk2
|
neurotrophic tyrosine kinase, receptor, type 2 |
| chr1_-_79417732 | 8.75 |
ENSMUST00000185234.2
ENSMUST00000049972.6 |
Scg2
|
secretogranin II |
| chr13_+_58956077 | 8.07 |
ENSMUST00000109838.10
|
Ntrk2
|
neurotrophic tyrosine kinase, receptor, type 2 |
| chr3_-_152687877 | 7.92 |
ENSMUST00000044278.6
|
St6galnac5
|
ST6 (alpha-N-acetyl-neuraminyl-2,3-beta-galactosyl-1,3)-N-acetylgalactosaminide alpha-2,6-sialyltransferase 5 |
| chr11_-_102787950 | 7.85 |
ENSMUST00000067444.10
|
Gfap
|
glial fibrillary acidic protein |
| chr2_-_85027041 | 7.69 |
ENSMUST00000099930.9
ENSMUST00000111601.2 |
Lrrc55
|
leucine rich repeat containing 55 |
| chr16_+_20513658 | 7.68 |
ENSMUST00000056518.13
|
Fam131a
|
family with sequence similarity 131, member A |
| chr9_-_29323500 | 7.63 |
ENSMUST00000115237.8
|
Ntm
|
neurotrimin |
| chr12_+_16703383 | 7.32 |
ENSMUST00000221596.2
ENSMUST00000111064.3 ENSMUST00000220892.2 |
Ntsr2
|
neurotensin receptor 2 |
| chr14_+_55173696 | 7.30 |
ENSMUST00000037814.8
|
Cmtm5
|
CKLF-like MARVEL transmembrane domain containing 5 |
| chr11_-_69451012 | 7.28 |
ENSMUST00000004036.6
|
Efnb3
|
ephrin B3 |
| chr17_+_26036893 | 7.27 |
ENSMUST00000235694.2
|
Fbxl16
|
F-box and leucine-rich repeat protein 16 |
| chr1_-_171359228 | 7.09 |
ENSMUST00000168184.2
|
Itln1
|
intelectin 1 (galactofuranose binding) |
| chr15_-_79048674 | 7.06 |
ENSMUST00000230261.2
ENSMUST00000040019.5 |
Sox10
|
SRY (sex determining region Y)-box 10 |
| chr14_-_68362284 | 7.05 |
ENSMUST00000111089.8
ENSMUST00000022638.6 |
Nefm
|
neurofilament, medium polypeptide |
| chr5_-_137246852 | 6.98 |
ENSMUST00000179412.2
|
Muc3a
|
mucin 3A, cell surface associated |
| chr12_-_76869510 | 6.77 |
ENSMUST00000154765.8
|
Rab15
|
RAB15, member RAS oncogene family |
| chr15_+_103411461 | 6.76 |
ENSMUST00000023132.5
|
Pde1b
|
phosphodiesterase 1B, Ca2+-calmodulin dependent |
| chr1_-_135095344 | 6.69 |
ENSMUST00000027682.9
|
Gpr37l1
|
G protein-coupled receptor 37-like 1 |
| chr2_-_180798785 | 6.53 |
ENSMUST00000055990.8
|
Eef1a2
|
eukaryotic translation elongation factor 1 alpha 2 |
| chr11_-_102787972 | 6.53 |
ENSMUST00000077902.5
|
Gfap
|
glial fibrillary acidic protein |
| chr6_-_124746510 | 6.42 |
ENSMUST00000149652.2
ENSMUST00000112476.8 ENSMUST00000004378.15 |
Eno2
|
enolase 2, gamma neuronal |
| chr7_-_79036691 | 6.36 |
ENSMUST00000053718.15
ENSMUST00000179243.3 |
Rlbp1
|
retinaldehyde binding protein 1 |
| chr17_+_27171834 | 6.29 |
ENSMUST00000231853.2
|
Syngap1
|
synaptic Ras GTPase activating protein 1 homolog (rat) |
| chr9_+_119978773 | 6.26 |
ENSMUST00000068698.15
ENSMUST00000215512.2 ENSMUST00000111627.3 ENSMUST00000093773.8 |
Mobp
|
myelin-associated oligodendrocytic basic protein |
| chr10_-_67748461 | 6.21 |
ENSMUST00000064656.8
|
Zfp365
|
zinc finger protein 365 |
| chr15_+_82140224 | 6.21 |
ENSMUST00000143238.2
|
1500009C09Rik
|
RIKEN cDNA 1500009C09 gene |
| chr3_+_45332831 | 6.17 |
ENSMUST00000193252.2
ENSMUST00000171554.8 ENSMUST00000166126.7 ENSMUST00000170695.4 |
Pcdh10
|
protocadherin 10 |
| chr12_-_112477536 | 6.07 |
ENSMUST00000066791.7
|
Tmem179
|
transmembrane protein 179 |
| chr2_+_61876956 | 6.06 |
ENSMUST00000112480.3
|
Slc4a10
|
solute carrier family 4, sodium bicarbonate cotransporter-like, member 10 |
| chr8_+_114932312 | 6.02 |
ENSMUST00000049509.7
ENSMUST00000150963.2 |
Vat1l
|
vesicle amine transport protein 1 like |
| chr7_-_109092834 | 6.02 |
ENSMUST00000106739.8
|
Trim66
|
tripartite motif-containing 66 |
| chr15_+_38740784 | 5.95 |
ENSMUST00000226440.3
ENSMUST00000239553.1 |
Baalc
|
brain and acute leukemia, cytoplasmic |
| chr2_+_61876923 | 5.93 |
ENSMUST00000054484.15
|
Slc4a10
|
solute carrier family 4, sodium bicarbonate cotransporter-like, member 10 |
| chr2_+_119572770 | 5.92 |
ENSMUST00000028758.8
|
Itpka
|
inositol 1,4,5-trisphosphate 3-kinase A |
| chr15_+_99590098 | 5.92 |
ENSMUST00000228185.2
|
Asic1
|
acid-sensing (proton-gated) ion channel 1 |
| chr11_+_78215026 | 5.91 |
ENSMUST00000102478.4
|
Aldoc
|
aldolase C, fructose-bisphosphate |
| chr2_+_61876806 | 5.89 |
ENSMUST00000102735.10
|
Slc4a10
|
solute carrier family 4, sodium bicarbonate cotransporter-like, member 10 |
| chr6_-_68681962 | 5.87 |
ENSMUST00000103330.2
|
Igkv10-94
|
immunoglobulin kappa variable 10-94 |
| chr6_-_124746468 | 5.66 |
ENSMUST00000204896.3
|
Eno2
|
enolase 2, gamma neuronal |
| chr9_+_36743980 | 5.64 |
ENSMUST00000034630.15
|
Fez1
|
fasciculation and elongation protein zeta 1 (zygin I) |
| chr19_+_8641369 | 5.62 |
ENSMUST00000035444.10
ENSMUST00000163785.2 |
Chrm1
|
cholinergic receptor, muscarinic 1, CNS |
| chr15_+_103411689 | 5.53 |
ENSMUST00000226493.2
|
Pde1b
|
phosphodiesterase 1B, Ca2+-calmodulin dependent |
| chr13_+_83721696 | 5.50 |
ENSMUST00000197146.5
ENSMUST00000185052.6 ENSMUST00000195984.5 |
Mef2c
|
myocyte enhancer factor 2C |
| chr13_+_75237939 | 5.47 |
ENSMUST00000022075.6
|
Pcsk1
|
proprotein convertase subtilisin/kexin type 1 |
| chr9_-_75591274 | 5.45 |
ENSMUST00000214244.2
ENSMUST00000213324.2 ENSMUST00000034699.8 |
Scg3
|
secretogranin III |
| chr9_-_112046329 | 5.37 |
ENSMUST00000159451.8
ENSMUST00000162796.8 ENSMUST00000161097.8 |
Arpp21
|
cyclic AMP-regulated phosphoprotein, 21 |
| chr6_-_127746390 | 5.36 |
ENSMUST00000032500.9
|
Prmt8
|
protein arginine N-methyltransferase 8 |
| chr3_-_33137209 | 5.33 |
ENSMUST00000194016.6
ENSMUST00000193681.6 ENSMUST00000192093.6 ENSMUST00000193289.6 |
Pex5l
|
peroxisomal biogenesis factor 5-like |
| chr7_+_105074938 | 5.33 |
ENSMUST00000033189.6
ENSMUST00000181339.8 |
Cckbr
|
cholecystokinin B receptor |
| chr13_+_109769294 | 5.32 |
ENSMUST00000135275.8
|
Pde4d
|
phosphodiesterase 4D, cAMP specific |
| chr15_-_79688910 | 5.30 |
ENSMUST00000175858.10
ENSMUST00000023057.10 |
Nptxr
|
neuronal pentraxin receptor |
| chr14_+_115329676 | 5.28 |
ENSMUST00000176912.8
ENSMUST00000175665.8 |
Gpc5
|
glypican 5 |
| chr12_+_16703709 | 5.25 |
ENSMUST00000221049.2
|
Ntsr2
|
neurotensin receptor 2 |
| chr4_-_126647156 | 5.20 |
ENSMUST00000030637.14
ENSMUST00000106116.2 |
Ncdn
|
neurochondrin |
| chr12_-_76869282 | 5.18 |
ENSMUST00000021459.14
|
Rab15
|
RAB15, member RAS oncogene family |
| chr7_-_109380745 | 5.10 |
ENSMUST00000207400.2
ENSMUST00000033331.7 |
Nrip3
|
nuclear receptor interacting protein 3 |
| chr19_-_8816530 | 5.07 |
ENSMUST00000096259.6
|
Gng3
|
guanine nucleotide binding protein (G protein), gamma 3 |
| chr13_-_51723473 | 5.05 |
ENSMUST00000239056.2
ENSMUST00000223543.3 |
Shc3
|
src homology 2 domain-containing transforming protein C3 |
| chr10_-_5144699 | 4.99 |
ENSMUST00000215467.2
|
Syne1
|
spectrin repeat containing, nuclear envelope 1 |
| chr14_+_55173936 | 4.97 |
ENSMUST00000227441.2
|
Cmtm5
|
CKLF-like MARVEL transmembrane domain containing 5 |
| chr1_-_172125555 | 4.88 |
ENSMUST00000085913.11
ENSMUST00000097464.4 |
Atp1a2
|
ATPase, Na+/K+ transporting, alpha 2 polypeptide |
| chr6_-_138399896 | 4.88 |
ENSMUST00000161450.8
ENSMUST00000163024.8 ENSMUST00000162185.8 |
Lmo3
|
LIM domain only 3 |
| chr10_+_90412570 | 4.85 |
ENSMUST00000182430.8
ENSMUST00000182960.8 ENSMUST00000182045.2 ENSMUST00000182083.2 |
Anks1b
|
ankyrin repeat and sterile alpha motif domain containing 1B |
| chr9_-_52590686 | 4.83 |
ENSMUST00000098768.3
ENSMUST00000213843.2 |
AI593442
|
expressed sequence AI593442 |
| chr10_+_90412638 | 4.82 |
ENSMUST00000183136.8
ENSMUST00000182595.8 |
Anks1b
|
ankyrin repeat and sterile alpha motif domain containing 1B |
| chr11_+_70396070 | 4.80 |
ENSMUST00000019063.3
|
Tm4sf5
|
transmembrane 4 superfamily member 5 |
| chr16_-_23709564 | 4.76 |
ENSMUST00000004480.5
|
Sst
|
somatostatin |
| chr5_+_22951015 | 4.74 |
ENSMUST00000197992.2
|
Lhfpl3
|
lipoma HMGIC fusion partner-like 3 |
| chr9_+_110074574 | 4.74 |
ENSMUST00000197850.5
|
Cspg5
|
chondroitin sulfate proteoglycan 5 |
| chr7_+_130633776 | 4.73 |
ENSMUST00000084509.7
ENSMUST00000213064.3 ENSMUST00000208311.4 |
Dmbt1
|
deleted in malignant brain tumors 1 |
| chr10_-_63926044 | 4.67 |
ENSMUST00000105439.2
|
Lrrtm3
|
leucine rich repeat transmembrane neuronal 3 |
| chr3_-_33137165 | 4.65 |
ENSMUST00000078226.10
ENSMUST00000108224.8 |
Pex5l
|
peroxisomal biogenesis factor 5-like |
| chr4_-_68872585 | 4.61 |
ENSMUST00000030036.6
|
Brinp1
|
bone morphogenic protein/retinoic acid inducible neural specific 1 |
| chrX_-_94209913 | 4.61 |
ENSMUST00000113873.9
ENSMUST00000113876.9 ENSMUST00000199920.5 ENSMUST00000113885.8 ENSMUST00000113883.8 ENSMUST00000196012.2 ENSMUST00000182001.8 ENSMUST00000113878.8 ENSMUST00000113882.8 ENSMUST00000182562.2 |
Arhgef9
|
CDC42 guanine nucleotide exchange factor (GEF) 9 |
| chr17_+_70276068 | 4.60 |
ENSMUST00000133983.8
|
Dlgap1
|
DLG associated protein 1 |
| chr9_+_26645024 | 4.57 |
ENSMUST00000160899.8
ENSMUST00000161431.3 ENSMUST00000159799.8 |
B3gat1
|
beta-1,3-glucuronyltransferase 1 (glucuronosyltransferase P) |
| chr10_-_127098932 | 4.52 |
ENSMUST00000217895.2
|
Kif5a
|
kinesin family member 5A |
| chr10_+_13841819 | 4.52 |
ENSMUST00000187083.7
|
Hivep2
|
human immunodeficiency virus type I enhancer binding protein 2 |
| chr9_-_86762467 | 4.50 |
ENSMUST00000074501.12
ENSMUST00000239074.2 ENSMUST00000098495.10 ENSMUST00000036347.13 ENSMUST00000074468.13 |
Snap91
|
synaptosomal-associated protein 91 |
| chr19_-_5148506 | 4.47 |
ENSMUST00000025805.8
|
Cnih2
|
cornichon family AMPA receptor auxiliary protein 2 |
| chr11_+_87651359 | 4.47 |
ENSMUST00000039627.12
ENSMUST00000100644.10 |
Tspoap1
|
TSPO associated protein 1 |
| chr4_+_42917228 | 4.45 |
ENSMUST00000107976.9
ENSMUST00000069184.9 |
Phf24
|
PHD finger protein 24 |
| chr1_+_50966670 | 4.45 |
ENSMUST00000081851.4
|
Tmeff2
|
transmembrane protein with EGF-like and two follistatin-like domains 2 |
| chr16_+_32480040 | 4.44 |
ENSMUST00000238806.2
ENSMUST00000238856.2 |
Tnk2
|
tyrosine kinase, non-receptor, 2 |
| chr2_+_121125918 | 4.36 |
ENSMUST00000110639.8
|
Map1a
|
microtubule-associated protein 1 A |
| chr17_+_70276210 | 4.34 |
ENSMUST00000060072.12
|
Dlgap1
|
DLG associated protein 1 |
| chr11_+_101066867 | 4.33 |
ENSMUST00000103109.4
|
Cntnap1
|
contactin associated protein-like 1 |
| chr9_+_58395850 | 4.32 |
ENSMUST00000085658.5
|
Insyn1
|
inhibitory synaptic factor 1 |
| chr10_-_62258195 | 4.30 |
ENSMUST00000020277.9
|
Hkdc1
|
hexokinase domain containing 1 |
| chr3_+_55154486 | 4.23 |
ENSMUST00000200348.2
|
Dclk1
|
doublecortin-like kinase 1 |
| chr7_-_105218472 | 4.19 |
ENSMUST00000187683.7
ENSMUST00000210079.2 ENSMUST00000187051.7 ENSMUST00000189265.7 ENSMUST00000190369.7 |
Apbb1
|
amyloid beta (A4) precursor protein-binding, family B, member 1 |
| chrX_-_42256694 | 4.19 |
ENSMUST00000115058.8
ENSMUST00000115059.8 |
Tenm1
|
teneurin transmembrane protein 1 |
| chr2_+_49509288 | 4.17 |
ENSMUST00000028102.14
|
Kif5c
|
kinesin family member 5C |
| chr6_-_13838423 | 4.16 |
ENSMUST00000115492.2
|
Gpr85
|
G protein-coupled receptor 85 |
| chr6_-_136150076 | 4.14 |
ENSMUST00000053880.13
|
Grin2b
|
glutamate receptor, ionotropic, NMDA2B (epsilon 2) |
| chr10_+_90412827 | 4.12 |
ENSMUST00000182550.8
ENSMUST00000099364.12 |
Anks1b
|
ankyrin repeat and sterile alpha motif domain containing 1B |
| chr15_+_99122742 | 4.12 |
ENSMUST00000041415.5
|
Kcnh3
|
potassium voltage-gated channel, subfamily H (eag-related), member 3 |
| chr10_+_90412432 | 4.10 |
ENSMUST00000182786.8
ENSMUST00000182600.8 |
Anks1b
|
ankyrin repeat and sterile alpha motif domain containing 1B |
| chr15_+_82159094 | 4.08 |
ENSMUST00000116423.3
ENSMUST00000230418.2 |
Septin3
|
septin 3 |
| chr5_+_35971767 | 4.07 |
ENSMUST00000114203.8
ENSMUST00000150146.2 |
Ablim2
|
actin-binding LIM protein 2 |
| chr6_+_38639945 | 4.04 |
ENSMUST00000114874.5
|
Clec2l
|
C-type lectin domain family 2, member L |
| chr3_+_94386385 | 3.99 |
ENSMUST00000199775.5
|
Celf3
|
CUGBP, Elav-like family member 3 |
| chr8_-_4309257 | 3.90 |
ENSMUST00000053252.9
|
Ctxn1
|
cortexin 1 |
| chr7_+_131568167 | 3.90 |
ENSMUST00000045840.5
|
Gpr26
|
G protein-coupled receptor 26 |
| chr1_+_75377616 | 3.88 |
ENSMUST00000122266.3
|
Speg
|
SPEG complex locus |
| chr5_+_57876401 | 3.87 |
ENSMUST00000094783.7
|
Pcdh7
|
protocadherin 7 |
| chr12_+_88689638 | 3.87 |
ENSMUST00000190626.7
ENSMUST00000167103.8 |
Nrxn3
|
neurexin III |
| chr1_-_132470672 | 3.85 |
ENSMUST00000086521.11
|
Cntn2
|
contactin 2 |
| chr16_+_7011580 | 3.83 |
ENSMUST00000231194.2
|
Rbfox1
|
RNA binding protein, fox-1 homolog (C. elegans) 1 |
| chr4_+_103476777 | 3.82 |
ENSMUST00000106827.8
|
Dab1
|
disabled 1 |
| chr16_-_9812787 | 3.82 |
ENSMUST00000199708.5
|
Grin2a
|
glutamate receptor, ionotropic, NMDA2A (epsilon 1) |
| chr9_-_52591030 | 3.82 |
ENSMUST00000213937.2
|
AI593442
|
expressed sequence AI593442 |
| chr17_+_70276382 | 3.82 |
ENSMUST00000146730.9
|
Dlgap1
|
DLG associated protein 1 |
| chr5_+_129661233 | 3.81 |
ENSMUST00000031390.10
|
Mmp17
|
matrix metallopeptidase 17 |
| chr7_-_79036654 | 3.80 |
ENSMUST00000206695.2
|
Rlbp1
|
retinaldehyde binding protein 1 |
| chr5_-_49682150 | 3.79 |
ENSMUST00000087395.11
|
Kcnip4
|
Kv channel interacting protein 4 |
| chr2_+_32465730 | 3.78 |
ENSMUST00000055304.14
|
Pip5kl1
|
phosphatidylinositol-4-phosphate 5-kinase-like 1 |
| chrX_+_139243012 | 3.77 |
ENSMUST00000208130.2
|
Frmpd3
|
FERM and PDZ domain containing 3 |
| chr2_-_129541753 | 3.77 |
ENSMUST00000028883.12
|
Pdyn
|
prodynorphin |
| chr8_+_73325871 | 3.77 |
ENSMUST00000212763.2
|
Tmem38a
|
transmembrane protein 38A |
| chr12_-_114263874 | 3.76 |
ENSMUST00000103482.2
ENSMUST00000194159.2 |
Ighv9-4
|
immunoglobulin heavy variable 9-4 |
| chr8_-_68427217 | 3.75 |
ENSMUST00000098696.10
ENSMUST00000038959.16 ENSMUST00000093469.11 |
Psd3
|
pleckstrin and Sec7 domain containing 3 |
| chr5_+_35971697 | 3.72 |
ENSMUST00000130233.8
|
Ablim2
|
actin-binding LIM protein 2 |
| chr15_-_72418189 | 3.70 |
ENSMUST00000044624.8
|
Kcnk9
|
potassium channel, subfamily K, member 9 |
| chr4_+_124779592 | 3.69 |
ENSMUST00000149146.2
|
Epha10
|
Eph receptor A10 |
| chr9_+_37278647 | 3.68 |
ENSMUST00000051839.9
|
Hepacam
|
hepatocyte cell adhesion molecule |
| chr6_-_138398376 | 3.68 |
ENSMUST00000163065.8
|
Lmo3
|
LIM domain only 3 |
| chr4_+_73931679 | 3.68 |
ENSMUST00000098006.9
ENSMUST00000084474.6 |
Frmd3
|
FERM domain containing 3 |
| chr3_-_36575955 | 3.68 |
ENSMUST00000124606.3
|
Smim43
|
small integral membrane protein 43 |
| chr8_+_71207326 | 3.67 |
ENSMUST00000110093.9
ENSMUST00000143118.3 ENSMUST00000034301.12 ENSMUST00000110090.8 |
Rab3a
|
RAB3A, member RAS oncogene family |
| chr7_-_25374472 | 3.67 |
ENSMUST00000108404.8
ENSMUST00000108405.2 ENSMUST00000079439.10 |
Tmem91
|
transmembrane protein 91 |
| chr6_-_136150491 | 3.67 |
ENSMUST00000111905.8
ENSMUST00000152012.8 ENSMUST00000143943.8 ENSMUST00000125905.2 |
Grin2b
|
glutamate receptor, ionotropic, NMDA2B (epsilon 2) |
| chr9_-_91247831 | 3.64 |
ENSMUST00000065360.5
|
Zic1
|
zinc finger protein of the cerebellum 1 |
| chrX_+_149981074 | 3.63 |
ENSMUST00000184730.8
ENSMUST00000184392.8 ENSMUST00000096285.5 |
Wnk3
|
WNK lysine deficient protein kinase 3 |
| chr12_-_114878652 | 3.58 |
ENSMUST00000103515.2
|
Ighv1-39
|
immunoglobulin heavy variable 1-39 |
| chr9_-_112046411 | 3.57 |
ENSMUST00000161412.8
|
Arpp21
|
cyclic AMP-regulated phosphoprotein, 21 |
| chr9_+_32135540 | 3.55 |
ENSMUST00000168954.9
|
Arhgap32
|
Rho GTPase activating protein 32 |
| chr11_+_56902658 | 3.53 |
ENSMUST00000094179.11
|
Gria1
|
glutamate receptor, ionotropic, AMPA1 (alpha 1) |
| chr10_+_90412539 | 3.53 |
ENSMUST00000182284.8
|
Anks1b
|
ankyrin repeat and sterile alpha motif domain containing 1B |
| chr15_+_83676140 | 3.52 |
ENSMUST00000172115.8
ENSMUST00000172398.2 |
Mpped1
|
metallophosphoesterase domain containing 1 |
| chr8_+_73325912 | 3.51 |
ENSMUST00000034244.9
|
Tmem38a
|
transmembrane protein 38A |
| chr9_-_91247809 | 3.50 |
ENSMUST00000034927.13
|
Zic1
|
zinc finger protein of the cerebellum 1 |
| chr7_-_19338349 | 3.49 |
ENSMUST00000086041.7
|
Clasrp
|
CLK4-associating serine/arginine rich protein |
| chr13_-_49301407 | 3.47 |
ENSMUST00000162581.8
ENSMUST00000110097.9 ENSMUST00000049265.15 ENSMUST00000035538.13 ENSMUST00000110096.8 ENSMUST00000091623.10 |
Wnk2
|
WNK lysine deficient protein kinase 2 |
| chr6_-_28830344 | 3.47 |
ENSMUST00000171353.2
|
Lrrc4
|
leucine rich repeat containing 4 |
| chr3_+_94391676 | 3.46 |
ENSMUST00000198384.3
|
Celf3
|
CUGBP, Elav-like family member 3 |
| chr7_-_79036222 | 3.46 |
ENSMUST00000205638.2
ENSMUST00000206320.3 ENSMUST00000205442.2 |
Rlbp1
|
retinaldehyde binding protein 1 |
| chrX_+_99773784 | 3.45 |
ENSMUST00000113744.2
|
Gdpd2
|
glycerophosphodiester phosphodiesterase domain containing 2 |
| chr2_+_32496990 | 3.44 |
ENSMUST00000095045.9
ENSMUST00000095044.10 ENSMUST00000126636.8 |
St6galnac6
|
ST6 (alpha-N-acetyl-neuraminyl-2,3-beta-galactosyl-1,3)-N-acetylgalactosaminide alpha-2,6-sialyltransferase 6 |
| chr16_-_94798509 | 3.43 |
ENSMUST00000095873.12
ENSMUST00000099508.4 |
Kcnj6
|
potassium inwardly-rectifying channel, subfamily J, member 6 |
| chr1_-_22845124 | 3.43 |
ENSMUST00000115273.10
|
Rims1
|
regulating synaptic membrane exocytosis 1 |
| chrX_-_132589727 | 3.40 |
ENSMUST00000149154.8
|
Pcdh19
|
protocadherin 19 |
| chr15_+_82159398 | 3.40 |
ENSMUST00000023095.14
ENSMUST00000230365.2 |
Septin3
|
septin 3 |
| chr5_+_137059127 | 3.40 |
ENSMUST00000041543.9
ENSMUST00000186451.2 |
Vgf
|
VGF nerve growth factor inducible |
| chr17_+_70829050 | 3.39 |
ENSMUST00000133717.9
ENSMUST00000148486.8 |
Dlgap1
|
DLG associated protein 1 |
| chr3_+_13536696 | 3.38 |
ENSMUST00000191806.3
ENSMUST00000193117.3 |
Ralyl
|
RALY RNA binding protein-like |
| chr12_-_115706126 | 3.38 |
ENSMUST00000166645.2
ENSMUST00000196690.2 |
Ighv1-71
|
immunoglobulin heavy variable 1-71 |
| chr11_+_69217078 | 3.38 |
ENSMUST00000018614.3
|
Kcnab3
|
potassium voltage-gated channel, shaker-related subfamily, beta member 3 |
| chr9_+_36744016 | 3.37 |
ENSMUST00000214772.2
|
Fez1
|
fasciculation and elongation protein zeta 1 (zygin I) |
| chr11_-_101973288 | 3.35 |
ENSMUST00000100398.5
|
Mpp2
|
membrane protein, palmitoylated 2 (MAGUK p55 subfamily member 2) |
| chr2_+_177783713 | 3.35 |
ENSMUST00000103066.10
|
Phactr3
|
phosphatase and actin regulator 3 |
| chr5_+_14564932 | 3.35 |
ENSMUST00000182407.8
ENSMUST00000030691.17 |
Pclo
|
piccolo (presynaptic cytomatrix protein) |
| chrX_+_99773523 | 3.35 |
ENSMUST00000019503.14
|
Gdpd2
|
glycerophosphodiester phosphodiesterase domain containing 2 |
| chr7_+_101060093 | 3.33 |
ENSMUST00000084894.15
|
Gm45837
|
predicted gene 45837 |
| chr14_-_24053994 | 3.31 |
ENSMUST00000225431.2
ENSMUST00000188210.8 ENSMUST00000224787.2 ENSMUST00000225315.2 ENSMUST00000225556.2 ENSMUST00000223727.2 ENSMUST00000223655.2 ENSMUST00000224077.2 ENSMUST00000224812.2 ENSMUST00000224285.2 ENSMUST00000225471.2 ENSMUST00000224232.2 ENSMUST00000223749.2 ENSMUST00000224025.2 |
Kcnma1
|
potassium large conductance calcium-activated channel, subfamily M, alpha member 1 |
| chr7_+_25005510 | 3.29 |
ENSMUST00000119703.8
ENSMUST00000205639.3 ENSMUST00000108409.2 |
Tmem145
|
transmembrane protein 145 |
| chr10_-_10958031 | 3.29 |
ENSMUST00000105561.9
ENSMUST00000044306.13 |
Grm1
|
glutamate receptor, metabotropic 1 |
| chr3_-_88669551 | 3.28 |
ENSMUST00000183267.2
|
Syt11
|
synaptotagmin XI |
| chr13_-_34314328 | 3.28 |
ENSMUST00000075774.5
|
Tubb2b
|
tubulin, beta 2B class IIB |
| chr8_+_94763826 | 3.27 |
ENSMUST00000109556.9
ENSMUST00000093301.9 ENSMUST00000060632.8 |
Ogfod1
|
2-oxoglutarate and iron-dependent oxygenase domain containing 1 |
| chr3_+_94385602 | 3.26 |
ENSMUST00000199884.5
ENSMUST00000198316.5 ENSMUST00000197558.5 |
Celf3
|
CUGBP, Elav-like family member 3 |
| chr11_-_33793461 | 3.26 |
ENSMUST00000101368.9
ENSMUST00000065970.6 ENSMUST00000109340.9 |
Kcnip1
|
Kv channel-interacting protein 1 |
| chr5_-_113428407 | 3.25 |
ENSMUST00000112324.2
ENSMUST00000057209.12 |
Sgsm1
|
small G protein signaling modulator 1 |
| chr14_+_20732804 | 3.24 |
ENSMUST00000228545.2
|
Sec24c
|
Sec24 related gene family, member C (S. cerevisiae) |
| chr7_+_90739904 | 3.24 |
ENSMUST00000107196.10
ENSMUST00000074273.10 |
Dlg2
|
discs large MAGUK scaffold protein 2 |
| chr3_-_89230190 | 3.21 |
ENSMUST00000200436.2
ENSMUST00000029673.10 |
Efna3
|
ephrin A3 |
| chr5_-_8672951 | 3.19 |
ENSMUST00000047485.15
ENSMUST00000115378.2 |
Rundc3b
|
RUN domain containing 3B |
| chrX_+_133305291 | 3.18 |
ENSMUST00000113228.8
ENSMUST00000153424.8 |
Drp2
|
dystrophin related protein 2 |
| chr15_-_77417512 | 3.18 |
ENSMUST00000062562.7
ENSMUST00000230863.2 |
Apol7c
|
apolipoprotein L 7c |
| chr4_-_134099840 | 3.17 |
ENSMUST00000030643.3
|
Extl1
|
exostosin-like glycosyltransferase 1 |
| chr11_+_56902624 | 3.16 |
ENSMUST00000036315.16
|
Gria1
|
glutamate receptor, ionotropic, AMPA1 (alpha 1) |
| chr12_-_115410489 | 3.13 |
ENSMUST00000194581.2
|
Ighv1-62-2
|
immunoglobulin heavy variable 1-62-2 |
| chr7_+_49624612 | 3.13 |
ENSMUST00000151721.8
ENSMUST00000081872.13 |
Nell1
|
NEL-like 1 |
| chr1_-_128256048 | 3.09 |
ENSMUST00000073490.7
|
Lct
|
lactase |
| chr4_+_124550600 | 3.07 |
ENSMUST00000053491.9
|
Pou3f1
|
POU domain, class 3, transcription factor 1 |
| chr8_+_63404395 | 3.07 |
ENSMUST00000119068.8
|
Spock3
|
sparc/osteonectin, cwcv and kazal-like domains proteoglycan 3 |
| chr6_-_116050081 | 3.07 |
ENSMUST00000173548.3
|
Tmcc1
|
transmembrane and coiled coil domains 1 |
| chr12_+_102915709 | 3.07 |
ENSMUST00000179002.8
|
Unc79
|
unc-79 homolog |
| chr10_+_101994841 | 3.05 |
ENSMUST00000020039.13
|
Mgat4c
|
MGAT4 family, member C |
| chr6_-_69415741 | 3.05 |
ENSMUST00000103354.3
|
Igkv4-59
|
immunoglobulin kappa variable 4-59 |
| chr6_-_124746445 | 3.05 |
ENSMUST00000156033.2
|
Eno2
|
enolase 2, gamma neuronal |
| chr4_-_155870321 | 3.04 |
ENSMUST00000097742.3
|
Tmem88b
|
transmembrane protein 88B |
| chr7_+_101070897 | 3.04 |
ENSMUST00000163751.10
ENSMUST00000211368.2 ENSMUST00000166652.2 |
Pde2a
|
phosphodiesterase 2A, cGMP-stimulated |
| chr1_-_133681419 | 3.02 |
ENSMUST00000125659.8
ENSMUST00000048953.14 ENSMUST00000165602.9 |
Atp2b4
|
ATPase, Ca++ transporting, plasma membrane 4 |
| chr10_-_110292053 | 3.01 |
ENSMUST00000239301.2
|
Nav3
|
neuron navigator 3 |
| chr18_+_86413077 | 3.01 |
ENSMUST00000058829.4
|
Neto1
|
neuropilin (NRP) and tolloid (TLL)-like 1 |
| chr17_+_70829144 | 3.00 |
ENSMUST00000140728.8
|
Dlgap1
|
DLG associated protein 1 |
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.8 | 17.0 | GO:0099538 | synaptic signaling via neuropeptide(GO:0099538) trans-synaptic signaling by neuropeptide(GO:0099540) trans-synaptic signaling by neuropeptide, modulating synaptic transmission(GO:0099551) |
| 2.4 | 7.1 | GO:0033693 | neurofilament bundle assembly(GO:0033693) |
| 2.2 | 11.1 | GO:0099566 | regulation of postsynaptic cytosolic calcium ion concentration(GO:0099566) |
| 2.2 | 21.5 | GO:0010624 | regulation of Schwann cell proliferation(GO:0010624) |
| 1.9 | 21.4 | GO:1900383 | regulation of synaptic plasticity by receptor localization to synapse(GO:1900383) |
| 1.7 | 10.0 | GO:0099527 | postsynapse to nucleus signaling pathway(GO:0099527) |
| 1.6 | 6.2 | GO:0033566 | gamma-tubulin complex localization(GO:0033566) |
| 1.5 | 19.3 | GO:0070842 | aggresome assembly(GO:0070842) |
| 1.5 | 5.9 | GO:0017055 | negative regulation of RNA polymerase II transcriptional preinitiation complex assembly(GO:0017055) |
| 1.4 | 5.8 | GO:0032470 | positive regulation of endoplasmic reticulum calcium ion concentration(GO:0032470) |
| 1.4 | 10.0 | GO:0045163 | clustering of voltage-gated potassium channels(GO:0045163) |
| 1.4 | 8.3 | GO:0016185 | synaptic vesicle budding from presynaptic endocytic zone membrane(GO:0016185) |
| 1.4 | 10.9 | GO:0016560 | protein import into peroxisome matrix, docking(GO:0016560) |
| 1.3 | 3.8 | GO:0021589 | hindbrain structural organization(GO:0021577) cerebellum structural organization(GO:0021589) |
| 1.2 | 4.9 | GO:1903279 | regulation of calcium:sodium antiporter activity(GO:1903279) |
| 1.2 | 3.6 | GO:2000686 | regulation of rubidium ion transmembrane transporter activity(GO:2000686) |
| 1.2 | 7.2 | GO:0050758 | thymidylate synthase biosynthetic process(GO:0050757) regulation of thymidylate synthase biosynthetic process(GO:0050758) negative regulation of thymidylate synthase biosynthetic process(GO:0050760) |
| 1.2 | 5.9 | GO:0099558 | maintenance of synapse structure(GO:0099558) |
| 1.2 | 7.1 | GO:0050915 | sensory perception of sour taste(GO:0050915) |
| 1.2 | 7.1 | GO:0009624 | response to nematode(GO:0009624) |
| 1.2 | 3.5 | GO:0042668 | auditory receptor cell fate determination(GO:0042668) |
| 1.1 | 5.6 | GO:0007197 | adenylate cyclase-inhibiting G-protein coupled acetylcholine receptor signaling pathway(GO:0007197) phospholipase C-activating G-protein coupled acetylcholine receptor signaling pathway(GO:0007207) |
| 1.1 | 15.7 | GO:0036005 | response to macrophage colony-stimulating factor(GO:0036005) cellular response to macrophage colony-stimulating factor stimulus(GO:0036006) |
| 1.1 | 4.5 | GO:1902683 | regulation of receptor localization to synapse(GO:1902683) |
| 1.1 | 3.3 | GO:0098928 | presynaptic signal transduction(GO:0098928) presynapse to nucleus signaling pathway(GO:0099526) |
| 1.1 | 6.5 | GO:0090274 | positive regulation of somatostatin secretion(GO:0090274) |
| 1.1 | 5.4 | GO:0019919 | peptidyl-arginine methylation, to asymmetrical-dimethyl arginine(GO:0019919) |
| 1.0 | 4.2 | GO:0098963 | dendritic transport of ribonucleoprotein complex(GO:0098961) dendritic transport of messenger ribonucleoprotein complex(GO:0098963) anterograde dendritic transport of messenger ribonucleoprotein complex(GO:0098964) |
| 1.0 | 3.0 | GO:0045763 | negative regulation of cellular amino acid metabolic process(GO:0045763) |
| 1.0 | 5.0 | GO:0018125 | peptidyl-cysteine methylation(GO:0018125) |
| 0.9 | 1.8 | GO:0035606 | peptidyl-cysteine S-trans-nitrosylation(GO:0035606) |
| 0.9 | 2.7 | GO:0045212 | neurotransmitter receptor biosynthetic process(GO:0045212) |
| 0.9 | 4.5 | GO:0098971 | anterograde dendritic transport of neurotransmitter receptor complex(GO:0098971) |
| 0.9 | 2.7 | GO:0002777 | antimicrobial peptide biosynthetic process(GO:0002777) antibacterial peptide biosynthetic process(GO:0002780) |
| 0.8 | 3.4 | GO:1903033 | regulation of microtubule plus-end binding(GO:1903031) positive regulation of microtubule plus-end binding(GO:1903033) |
| 0.8 | 3.4 | GO:0015801 | aromatic amino acid transport(GO:0015801) tryptophan transport(GO:0015827) |
| 0.8 | 3.3 | GO:0050428 | purine ribonucleoside bisphosphate biosynthetic process(GO:0034036) 3'-phosphoadenosine 5'-phosphosulfate biosynthetic process(GO:0050428) |
| 0.8 | 3.3 | GO:1990927 | negative regulation of membrane invagination(GO:1905154) calcium ion regulated lysosome exocytosis(GO:1990927) |
| 0.8 | 4.5 | GO:0000160 | phosphorelay signal transduction system(GO:0000160) |
| 0.8 | 3.0 | GO:2000312 | regulation of kainate selective glutamate receptor activity(GO:2000312) |
| 0.7 | 2.2 | GO:0002184 | cytoplasmic translational termination(GO:0002184) |
| 0.7 | 2.2 | GO:0048162 | preantral ovarian follicle growth(GO:0001546) multi-layer follicle stage(GO:0048162) |
| 0.7 | 5.1 | GO:0001757 | somite specification(GO:0001757) |
| 0.7 | 4.4 | GO:1990504 | dense core granule exocytosis(GO:1990504) |
| 0.7 | 4.4 | GO:0060082 | response to carbon monoxide(GO:0034465) eye blink reflex(GO:0060082) |
| 0.7 | 5.0 | GO:0090292 | nuclear matrix organization(GO:0043578) nuclear matrix anchoring at nuclear membrane(GO:0090292) |
| 0.7 | 2.1 | GO:0008628 | hormone-mediated apoptotic signaling pathway(GO:0008628) |
| 0.7 | 17.9 | GO:0021860 | pyramidal neuron development(GO:0021860) |
| 0.7 | 3.4 | GO:0010808 | positive regulation of synaptic vesicle priming(GO:0010808) |
| 0.7 | 6.8 | GO:0016198 | axon choice point recognition(GO:0016198) |
| 0.7 | 2.0 | GO:0061743 | motor learning(GO:0061743) |
| 0.7 | 2.7 | GO:2000297 | negative regulation of synapse maturation(GO:2000297) |
| 0.7 | 9.8 | GO:0048245 | eosinophil chemotaxis(GO:0048245) |
| 0.6 | 7.8 | GO:0019800 | peptide cross-linking via chondroitin 4-sulfate glycosaminoglycan(GO:0019800) |
| 0.6 | 4.4 | GO:0045113 | regulation of integrin biosynthetic process(GO:0045113) |
| 0.6 | 3.1 | GO:0010040 | response to iron(II) ion(GO:0010040) |
| 0.6 | 1.8 | GO:1903244 | positive regulation of cardiac muscle adaptation(GO:0010615) positive regulation of cardiac muscle hypertrophy in response to stress(GO:1903244) |
| 0.6 | 4.0 | GO:0032899 | regulation of neurotrophin production(GO:0032899) |
| 0.6 | 1.7 | GO:0071929 | alpha-tubulin acetylation(GO:0071929) |
| 0.6 | 11.4 | GO:0009312 | oligosaccharide biosynthetic process(GO:0009312) |
| 0.6 | 6.8 | GO:0030050 | vesicle transport along actin filament(GO:0030050) |
| 0.6 | 2.8 | GO:0098886 | modification of dendritic spine(GO:0098886) |
| 0.5 | 2.2 | GO:0070173 | regulation of enamel mineralization(GO:0070173) |
| 0.5 | 1.6 | GO:2000538 | regulation of cGMP-mediated signaling(GO:0010752) regulation of B cell chemotaxis(GO:2000537) positive regulation of B cell chemotaxis(GO:2000538) |
| 0.5 | 8.4 | GO:0030388 | fructose 1,6-bisphosphate metabolic process(GO:0030388) |
| 0.5 | 4.7 | GO:0038026 | reelin-mediated signaling pathway(GO:0038026) |
| 0.5 | 16.1 | GO:0030575 | nuclear body organization(GO:0030575) |
| 0.5 | 5.2 | GO:1903753 | MHC class I biosynthetic process(GO:0045341) regulation of MHC class I biosynthetic process(GO:0045343) positive regulation of MHC class I biosynthetic process(GO:0045345) negative regulation of p38MAPK cascade(GO:1903753) |
| 0.5 | 3.1 | GO:0034184 | positive regulation of maintenance of mitotic sister chromatid cohesion(GO:0034184) |
| 0.5 | 8.0 | GO:1902902 | negative regulation of autophagosome assembly(GO:1902902) |
| 0.5 | 1.5 | GO:0006867 | asparagine transport(GO:0006867) positive regulation of glutamine transport(GO:2000487) |
| 0.5 | 2.0 | GO:0070649 | formin-nucleated actin cable assembly(GO:0070649) |
| 0.5 | 7.9 | GO:1902993 | positive regulation of amyloid precursor protein catabolic process(GO:1902993) |
| 0.5 | 2.9 | GO:0071233 | response to leucine(GO:0043201) cellular response to leucine(GO:0071233) |
| 0.5 | 4.3 | GO:0071205 | protein localization to juxtaparanode region of axon(GO:0071205) |
| 0.5 | 2.8 | GO:0097118 | neuroligin clustering involved in postsynaptic membrane assembly(GO:0097118) |
| 0.5 | 8.0 | GO:0090527 | actin filament reorganization(GO:0090527) |
| 0.5 | 1.4 | GO:0090063 | positive regulation of microtubule nucleation(GO:0090063) |
| 0.5 | 4.2 | GO:1901552 | positive regulation of endothelial cell development(GO:1901552) positive regulation of establishment of endothelial barrier(GO:1903142) |
| 0.5 | 5.5 | GO:0003138 | primary heart field specification(GO:0003138) sinoatrial valve development(GO:0003172) sinoatrial valve morphogenesis(GO:0003185) |
| 0.5 | 68.7 | GO:0006910 | phagocytosis, recognition(GO:0006910) |
| 0.5 | 5.9 | GO:0006020 | inositol metabolic process(GO:0006020) |
| 0.5 | 5.4 | GO:0023041 | neuronal signal transduction(GO:0023041) |
| 0.5 | 5.4 | GO:0033689 | negative regulation of osteoblast proliferation(GO:0033689) |
| 0.5 | 2.7 | GO:0006382 | adenosine to inosine editing(GO:0006382) |
| 0.4 | 1.3 | GO:0002940 | tRNA N2-guanine methylation(GO:0002940) |
| 0.4 | 2.2 | GO:1905150 | regulation of voltage-gated sodium channel activity(GO:1905150) |
| 0.4 | 2.2 | GO:0060633 | negative regulation of transcription initiation from RNA polymerase II promoter(GO:0060633) |
| 0.4 | 2.2 | GO:1900738 | positive regulation of phospholipase C-activating G-protein coupled receptor signaling pathway(GO:1900738) |
| 0.4 | 2.9 | GO:1904222 | regulation of CDP-diacylglycerol-serine O-phosphatidyltransferase activity(GO:1904217) positive regulation of CDP-diacylglycerol-serine O-phosphatidyltransferase activity(GO:1904219) positive regulation of serine C-palmitoyltransferase activity(GO:1904222) |
| 0.4 | 2.9 | GO:0007256 | activation of JNKK activity(GO:0007256) |
| 0.4 | 2.1 | GO:1903378 | positive regulation of oxidative stress-induced neuron intrinsic apoptotic signaling pathway(GO:1903378) |
| 0.4 | 5.3 | GO:1901898 | negative regulation of relaxation of cardiac muscle(GO:1901898) |
| 0.4 | 2.0 | GO:0014042 | positive regulation of mitochondrial fusion(GO:0010636) positive regulation of neuron maturation(GO:0014042) |
| 0.4 | 4.0 | GO:0019243 | methylglyoxal catabolic process to D-lactate via S-lactoyl-glutathione(GO:0019243) methylglyoxal catabolic process(GO:0051596) methylglyoxal catabolic process to lactate(GO:0061727) |
| 0.4 | 2.0 | GO:0034224 | cellular response to zinc ion starvation(GO:0034224) |
| 0.4 | 2.4 | GO:0015015 | heparan sulfate proteoglycan biosynthetic process, enzymatic modification(GO:0015015) |
| 0.4 | 1.2 | GO:1903237 | negative regulation of leukocyte tethering or rolling(GO:1903237) |
| 0.4 | 1.2 | GO:1900126 | negative regulation of hyaluronan biosynthetic process(GO:1900126) |
| 0.4 | 3.5 | GO:2000651 | positive regulation of sodium ion transmembrane transporter activity(GO:2000651) |
| 0.4 | 1.5 | GO:0061026 | cardiac muscle tissue regeneration(GO:0061026) |
| 0.4 | 6.4 | GO:0043084 | penile erection(GO:0043084) |
| 0.4 | 1.5 | GO:0071596 | ubiquitin-dependent protein catabolic process via the N-end rule pathway(GO:0071596) |
| 0.4 | 3.0 | GO:0033058 | directional locomotion(GO:0033058) |
| 0.4 | 3.3 | GO:0006449 | regulation of translational termination(GO:0006449) |
| 0.4 | 1.4 | GO:0009608 | response to symbiont(GO:0009608) response to symbiotic bacterium(GO:0009609) |
| 0.4 | 1.4 | GO:0010609 | mRNA localization resulting in posttranscriptional regulation of gene expression(GO:0010609) positive regulation of DNA damage response, signal transduction by p53 class mediator resulting in transcription of p21 class mediator(GO:1902164) |
| 0.4 | 6.7 | GO:0048712 | negative regulation of astrocyte differentiation(GO:0048712) |
| 0.3 | 1.4 | GO:1901491 | negative regulation of lymphangiogenesis(GO:1901491) |
| 0.3 | 2.8 | GO:0061669 | spontaneous neurotransmitter secretion(GO:0061669) spontaneous synaptic transmission(GO:0098814) |
| 0.3 | 8.2 | GO:0007614 | short-term memory(GO:0007614) |
| 0.3 | 2.6 | GO:0098712 | L-glutamate import across plasma membrane(GO:0098712) |
| 0.3 | 2.6 | GO:0007217 | tachykinin receptor signaling pathway(GO:0007217) |
| 0.3 | 2.6 | GO:1902564 | negative regulation of neutrophil degranulation(GO:0043314) negative regulation of blood vessel remodeling(GO:0060313) negative regulation of neutrophil activation(GO:1902564) |
| 0.3 | 1.6 | GO:0051835 | positive regulation of synapse structural plasticity(GO:0051835) |
| 0.3 | 1.2 | GO:0003360 | brainstem development(GO:0003360) |
| 0.3 | 1.5 | GO:0086048 | membrane depolarization during bundle of His cell action potential(GO:0086048) |
| 0.3 | 1.2 | GO:0051182 | coenzyme transport(GO:0051182) |
| 0.3 | 2.1 | GO:1901897 | regulation of relaxation of cardiac muscle(GO:1901897) |
| 0.3 | 4.9 | GO:1903818 | positive regulation of voltage-gated potassium channel activity(GO:1903818) |
| 0.3 | 3.3 | GO:1902669 | positive regulation of axon guidance(GO:1902669) |
| 0.3 | 3.9 | GO:0090129 | regulation of synapse maturation(GO:0090128) positive regulation of synapse maturation(GO:0090129) |
| 0.3 | 1.8 | GO:0060907 | positive regulation of macrophage cytokine production(GO:0060907) |
| 0.3 | 1.5 | GO:0035095 | behavioral response to nicotine(GO:0035095) |
| 0.3 | 3.5 | GO:0097119 | postsynaptic density protein 95 clustering(GO:0097119) |
| 0.3 | 2.8 | GO:0021840 | directional guidance of interneurons involved in migration from the subpallium to the cortex(GO:0021840) chemorepulsion involved in interneuron migration from the subpallium to the cortex(GO:0021842) |
| 0.3 | 3.4 | GO:0051045 | negative regulation of membrane protein ectodomain proteolysis(GO:0051045) |
| 0.3 | 0.6 | GO:0031446 | regulation of fast-twitch skeletal muscle fiber contraction(GO:0031446) positive regulation of fast-twitch skeletal muscle fiber contraction(GO:0031448) |
| 0.3 | 3.3 | GO:0045161 | neuronal ion channel clustering(GO:0045161) |
| 0.3 | 3.0 | GO:0061577 | calcium ion transmembrane transport via high voltage-gated calcium channel(GO:0061577) |
| 0.3 | 0.8 | GO:0061762 | CAMKK-AMPK signaling cascade(GO:0061762) |
| 0.3 | 9.0 | GO:2000369 | regulation of clathrin-mediated endocytosis(GO:2000369) |
| 0.3 | 6.2 | GO:0050884 | neuromuscular process controlling posture(GO:0050884) |
| 0.3 | 0.5 | GO:0070973 | protein localization to endoplasmic reticulum exit site(GO:0070973) |
| 0.3 | 4.2 | GO:0048149 | behavioral response to ethanol(GO:0048149) |
| 0.3 | 0.5 | GO:0071799 | response to prostaglandin D(GO:0071798) cellular response to prostaglandin D stimulus(GO:0071799) |
| 0.3 | 0.8 | GO:0002631 | regulation of granuloma formation(GO:0002631) negative regulation of granuloma formation(GO:0002632) regulation of toll-like receptor 5 signaling pathway(GO:0034147) negative regulation of toll-like receptor 5 signaling pathway(GO:0034148) negative regulation of nucleotide-binding oligomerization domain containing 1 signaling pathway(GO:0070429) |
| 0.3 | 0.8 | GO:1900135 | positive regulation of renin secretion into blood stream(GO:1900135) |
| 0.3 | 2.1 | GO:0072321 | chaperone-mediated protein transport(GO:0072321) |
| 0.3 | 13.2 | GO:0048169 | regulation of long-term neuronal synaptic plasticity(GO:0048169) |
| 0.3 | 12.6 | GO:0006904 | vesicle docking involved in exocytosis(GO:0006904) |
| 0.3 | 0.5 | GO:0031587 | positive regulation of inositol 1,4,5-trisphosphate-sensitive calcium-release channel activity(GO:0031587) |
| 0.3 | 1.0 | GO:0007185 | transmembrane receptor protein tyrosine phosphatase signaling pathway(GO:0007185) |
| 0.3 | 1.8 | GO:0043553 | negative regulation of phosphatidylinositol 3-kinase activity(GO:0043553) |
| 0.3 | 5.8 | GO:0048268 | clathrin coat assembly(GO:0048268) |
| 0.3 | 2.3 | GO:0033600 | negative regulation of mammary gland epithelial cell proliferation(GO:0033600) regulation of branching involved in mammary gland duct morphogenesis(GO:0060762) |
| 0.2 | 1.5 | GO:1903347 | negative regulation of bicellular tight junction assembly(GO:1903347) |
| 0.2 | 2.5 | GO:0007213 | G-protein coupled acetylcholine receptor signaling pathway(GO:0007213) |
| 0.2 | 2.0 | GO:0033314 | mitotic DNA replication checkpoint(GO:0033314) |
| 0.2 | 1.0 | GO:1905053 | regulation of base-excision repair(GO:1905051) positive regulation of base-excision repair(GO:1905053) |
| 0.2 | 1.0 | GO:0010807 | regulation of synaptic vesicle priming(GO:0010807) |
| 0.2 | 1.9 | GO:0071802 | negative regulation of podosome assembly(GO:0071802) |
| 0.2 | 1.0 | GO:1904395 | positive regulation of skeletal muscle acetylcholine-gated channel clustering(GO:1904395) |
| 0.2 | 0.7 | GO:0071286 | cellular response to magnesium ion(GO:0071286) |
| 0.2 | 1.1 | GO:0010935 | regulation of macrophage cytokine production(GO:0010935) negative regulation of macrophage cytokine production(GO:0010936) |
| 0.2 | 2.0 | GO:0005513 | detection of calcium ion(GO:0005513) |
| 0.2 | 5.2 | GO:0001833 | inner cell mass cell proliferation(GO:0001833) |
| 0.2 | 4.5 | GO:0032228 | regulation of synaptic transmission, GABAergic(GO:0032228) |
| 0.2 | 2.2 | GO:0021785 | branchiomotor neuron axon guidance(GO:0021785) |
| 0.2 | 2.7 | GO:0034380 | high-density lipoprotein particle assembly(GO:0034380) |
| 0.2 | 0.9 | GO:0042539 | hypotonic salinity response(GO:0042539) cellular hypotonic salinity response(GO:0071477) |
| 0.2 | 2.0 | GO:0048852 | diencephalon morphogenesis(GO:0048852) |
| 0.2 | 1.5 | GO:0006689 | ganglioside catabolic process(GO:0006689) |
| 0.2 | 2.2 | GO:0035630 | bone mineralization involved in bone maturation(GO:0035630) |
| 0.2 | 2.8 | GO:0042756 | drinking behavior(GO:0042756) |
| 0.2 | 1.5 | GO:0043652 | engulfment of apoptotic cell(GO:0043652) |
| 0.2 | 1.3 | GO:0003065 | positive regulation of heart rate by epinephrine(GO:0003065) |
| 0.2 | 1.1 | GO:0018343 | protein farnesylation(GO:0018343) |
| 0.2 | 1.1 | GO:1902162 | regulation of DNA damage response, signal transduction by p53 class mediator resulting in transcription of p21 class mediator(GO:1902162) negative regulation of DNA damage response, signal transduction by p53 class mediator resulting in transcription of p21 class mediator(GO:1902163) |
| 0.2 | 2.1 | GO:0031915 | positive regulation of synaptic plasticity(GO:0031915) |
| 0.2 | 0.8 | GO:0051823 | regulation of synapse structural plasticity(GO:0051823) |
| 0.2 | 1.0 | GO:2000587 | regulation of platelet-derived growth factor receptor-beta signaling pathway(GO:2000586) negative regulation of platelet-derived growth factor receptor-beta signaling pathway(GO:2000587) |
| 0.2 | 0.8 | GO:0035279 | mRNA cleavage involved in gene silencing by miRNA(GO:0035279) mRNA cleavage involved in gene silencing(GO:0098795) |
| 0.2 | 2.9 | GO:0035584 | calcium-mediated signaling using intracellular calcium source(GO:0035584) |
| 0.2 | 3.7 | GO:0071625 | vocalization behavior(GO:0071625) |
| 0.2 | 1.4 | GO:0030950 | establishment or maintenance of actin cytoskeleton polarity(GO:0030950) |
| 0.2 | 0.6 | GO:0018312 | peptidyl-serine ADP-ribosylation(GO:0018312) |
| 0.2 | 2.2 | GO:0019372 | lipoxygenase pathway(GO:0019372) |
| 0.2 | 0.8 | GO:1990091 | sodium-dependent self proteolysis(GO:1990091) |
| 0.2 | 2.1 | GO:2000300 | regulation of synaptic vesicle exocytosis(GO:2000300) |
| 0.2 | 7.2 | GO:0060292 | long term synaptic depression(GO:0060292) |
| 0.2 | 0.6 | GO:0000389 | mRNA 3'-splice site recognition(GO:0000389) |
| 0.2 | 0.7 | GO:0072021 | ascending thin limb development(GO:0072021) thick ascending limb development(GO:0072023) metanephric ascending thin limb development(GO:0072218) metanephric thick ascending limb development(GO:0072233) |
| 0.2 | 4.0 | GO:0034063 | stress granule assembly(GO:0034063) |
| 0.2 | 5.1 | GO:0048168 | regulation of neuronal synaptic plasticity(GO:0048168) |
| 0.2 | 6.3 | GO:0032012 | ARF protein signal transduction(GO:0032011) regulation of ARF protein signal transduction(GO:0032012) |
| 0.2 | 3.2 | GO:0090110 | cargo loading into COPII-coated vesicle(GO:0090110) |
| 0.2 | 1.1 | GO:0060478 | acrosomal vesicle exocytosis(GO:0060478) |
| 0.2 | 1.1 | GO:0021914 | negative regulation of smoothened signaling pathway involved in ventral spinal cord patterning(GO:0021914) |
| 0.2 | 4.0 | GO:0045760 | positive regulation of action potential(GO:0045760) |
| 0.2 | 0.3 | GO:0061642 | chemoattraction of axon(GO:0061642) |
| 0.2 | 1.2 | GO:0046826 | negative regulation of protein export from nucleus(GO:0046826) |
| 0.2 | 0.3 | GO:0055096 | lipoprotein particle mediated signaling(GO:0055095) low-density lipoprotein particle mediated signaling(GO:0055096) |
| 0.2 | 1.8 | GO:0015868 | purine ribonucleotide transport(GO:0015868) |
| 0.2 | 4.9 | GO:0030252 | growth hormone secretion(GO:0030252) |
| 0.2 | 1.3 | GO:0090073 | positive regulation of protein homodimerization activity(GO:0090073) |
| 0.2 | 6.2 | GO:0010107 | potassium ion import(GO:0010107) |
| 0.2 | 1.3 | GO:0021999 | neural plate anterior/posterior regionalization(GO:0021999) |
| 0.2 | 1.1 | GO:0071787 | endoplasmic reticulum tubular network assembly(GO:0071787) |
| 0.2 | 0.8 | GO:0003376 | sphingosine-1-phosphate signaling pathway(GO:0003376) |
| 0.2 | 1.2 | GO:0014059 | dopamine secretion(GO:0014046) regulation of dopamine secretion(GO:0014059) |
| 0.1 | 0.6 | GO:0042335 | cuticle development(GO:0042335) |
| 0.1 | 1.5 | GO:1904259 | regulation of basement membrane assembly involved in embryonic body morphogenesis(GO:1904259) positive regulation of basement membrane assembly involved in embryonic body morphogenesis(GO:1904261) basement membrane assembly involved in embryonic body morphogenesis(GO:2001197) |
| 0.1 | 0.6 | GO:0015910 | peroxisomal long-chain fatty acid import(GO:0015910) |
| 0.1 | 1.6 | GO:0001682 | tRNA 5'-leader removal(GO:0001682) |
| 0.1 | 1.0 | GO:2000348 | regulation of CD40 signaling pathway(GO:2000348) |
| 0.1 | 0.6 | GO:0006114 | glycerol biosynthetic process(GO:0006114) |
| 0.1 | 2.2 | GO:0033603 | positive regulation of dopamine secretion(GO:0033603) |
| 0.1 | 0.4 | GO:0061090 | positive regulation of sequestering of zinc ion(GO:0061090) |
| 0.1 | 0.7 | GO:0006481 | C-terminal protein methylation(GO:0006481) |
| 0.1 | 0.7 | GO:0035507 | regulation of myosin-light-chain-phosphatase activity(GO:0035507) |
| 0.1 | 1.0 | GO:0001574 | ganglioside biosynthetic process(GO:0001574) |
| 0.1 | 1.0 | GO:0060480 | lung goblet cell differentiation(GO:0060480) |
| 0.1 | 0.4 | GO:0061537 | glycine secretion(GO:0061536) glycine secretion, neurotransmission(GO:0061537) |
| 0.1 | 1.0 | GO:0032237 | activation of store-operated calcium channel activity(GO:0032237) |
| 0.1 | 5.7 | GO:0007026 | negative regulation of microtubule depolymerization(GO:0007026) |
| 0.1 | 2.8 | GO:0033169 | histone H3-K9 demethylation(GO:0033169) |
| 0.1 | 1.2 | GO:0014850 | response to muscle activity(GO:0014850) |
| 0.1 | 0.4 | GO:0021979 | hypothalamus cell differentiation(GO:0021979) |
| 0.1 | 0.8 | GO:0060005 | vestibular reflex(GO:0060005) |
| 0.1 | 1.2 | GO:1900118 | negative regulation of execution phase of apoptosis(GO:1900118) |
| 0.1 | 2.6 | GO:0051481 | negative regulation of cytosolic calcium ion concentration(GO:0051481) |
| 0.1 | 1.0 | GO:1903003 | positive regulation of protein deubiquitination(GO:1903003) |
| 0.1 | 0.9 | GO:0072092 | ureteric bud invasion(GO:0072092) |
| 0.1 | 3.5 | GO:0061003 | positive regulation of dendritic spine morphogenesis(GO:0061003) |
| 0.1 | 1.3 | GO:0042866 | pyruvate biosynthetic process(GO:0042866) |
| 0.1 | 1.4 | GO:0061635 | regulation of protein complex stability(GO:0061635) |
| 0.1 | 1.2 | GO:0034983 | peptidyl-lysine deacetylation(GO:0034983) |
| 0.1 | 2.3 | GO:0034315 | regulation of Arp2/3 complex-mediated actin nucleation(GO:0034315) |
| 0.1 | 1.9 | GO:0045542 | positive regulation of cholesterol biosynthetic process(GO:0045542) positive regulation of cholesterol metabolic process(GO:0090205) |
| 0.1 | 2.1 | GO:0046069 | cGMP catabolic process(GO:0046069) |
| 0.1 | 2.6 | GO:0060081 | membrane hyperpolarization(GO:0060081) |
| 0.1 | 3.3 | GO:0045475 | locomotor rhythm(GO:0045475) |
| 0.1 | 9.4 | GO:0007218 | neuropeptide signaling pathway(GO:0007218) |
| 0.1 | 5.3 | GO:0007628 | adult walking behavior(GO:0007628) |
| 0.1 | 2.2 | GO:0030204 | chondroitin sulfate metabolic process(GO:0030204) |
| 0.1 | 0.2 | GO:0072194 | kidney smooth muscle tissue development(GO:0072194) |
| 0.1 | 1.5 | GO:2000465 | regulation of glycogen (starch) synthase activity(GO:2000465) |
| 0.1 | 1.4 | GO:0048681 | negative regulation of axon regeneration(GO:0048681) |
| 0.1 | 1.9 | GO:0034643 | establishment of mitochondrion localization, microtubule-mediated(GO:0034643) mitochondrion transport along microtubule(GO:0047497) |
| 0.1 | 1.2 | GO:0039536 | negative regulation of RIG-I signaling pathway(GO:0039536) |
| 0.1 | 6.2 | GO:0006414 | translational elongation(GO:0006414) |
| 0.1 | 7.7 | GO:0034605 | cellular response to heat(GO:0034605) |
| 0.1 | 4.5 | GO:0021952 | central nervous system projection neuron axonogenesis(GO:0021952) |
| 0.1 | 1.0 | GO:0097120 | receptor localization to synapse(GO:0097120) |
| 0.1 | 2.1 | GO:0006054 | N-acetylneuraminate metabolic process(GO:0006054) |
| 0.1 | 11.2 | GO:0006096 | glycolytic process(GO:0006096) |
| 0.1 | 0.4 | GO:2000774 | positive regulation of cellular senescence(GO:2000774) |
| 0.1 | 1.4 | GO:0008090 | retrograde axonal transport(GO:0008090) |
| 0.1 | 0.8 | GO:0071569 | protein ufmylation(GO:0071569) |
| 0.1 | 3.1 | GO:0018345 | protein palmitoylation(GO:0018345) |
| 0.1 | 0.9 | GO:0042905 | 9-cis-retinoic acid biosynthetic process(GO:0042904) 9-cis-retinoic acid metabolic process(GO:0042905) |
| 0.1 | 0.3 | GO:0046061 | dATP catabolic process(GO:0046061) |
| 0.1 | 1.1 | GO:0001778 | plasma membrane repair(GO:0001778) |
| 0.1 | 0.5 | GO:0090037 | positive regulation of protein kinase C signaling(GO:0090037) |
| 0.1 | 1.5 | GO:0021942 | radial glia guided migration of Purkinje cell(GO:0021942) |
| 0.1 | 0.5 | GO:0060423 | foregut regionalization(GO:0060423) lung field specification(GO:0060424) lung induction(GO:0060492) |
| 0.1 | 1.4 | GO:0071397 | cellular response to cholesterol(GO:0071397) |
| 0.1 | 0.3 | GO:0021660 | rhombomere formation(GO:0021594) rhombomere 3 formation(GO:0021660) rhombomere 5 morphogenesis(GO:0021664) rhombomere 5 formation(GO:0021666) |
| 0.1 | 1.2 | GO:0009301 | snRNA transcription(GO:0009301) |
| 0.1 | 0.6 | GO:0098532 | histone H3-K27 trimethylation(GO:0098532) |
| 0.1 | 0.4 | GO:1902044 | regulation of Fas signaling pathway(GO:1902044) negative regulation of Fas signaling pathway(GO:1902045) |
| 0.1 | 2.8 | GO:0045780 | positive regulation of bone resorption(GO:0045780) positive regulation of bone remodeling(GO:0046852) |
| 0.1 | 1.4 | GO:0010172 | embryonic body morphogenesis(GO:0010172) |
| 0.1 | 2.4 | GO:0006107 | oxaloacetate metabolic process(GO:0006107) |
| 0.1 | 0.5 | GO:0016557 | peroxisomal membrane transport(GO:0015919) peroxisome membrane biogenesis(GO:0016557) protein import into peroxisome membrane(GO:0045046) |
| 0.1 | 0.1 | GO:0070885 | negative regulation of calcineurin-NFAT signaling cascade(GO:0070885) |
| 0.1 | 0.3 | GO:0032227 | negative regulation of synaptic transmission, dopaminergic(GO:0032227) |
| 0.1 | 1.3 | GO:0002031 | G-protein coupled receptor internalization(GO:0002031) |
| 0.1 | 1.2 | GO:0033572 | transferrin transport(GO:0033572) |
| 0.1 | 0.5 | GO:0030321 | transepithelial chloride transport(GO:0030321) |
| 0.1 | 3.3 | GO:0001782 | B cell homeostasis(GO:0001782) |
| 0.1 | 0.8 | GO:0051196 | regulation of coenzyme metabolic process(GO:0051196) |
| 0.1 | 1.3 | GO:1904262 | negative regulation of TORC1 signaling(GO:1904262) |
| 0.1 | 0.8 | GO:0006527 | arginine catabolic process(GO:0006527) |
| 0.1 | 0.3 | GO:0014707 | branchiomeric skeletal muscle development(GO:0014707) |
| 0.1 | 0.7 | GO:0070255 | regulation of mucus secretion(GO:0070255) |
| 0.1 | 1.6 | GO:0030322 | stabilization of membrane potential(GO:0030322) |
| 0.1 | 3.6 | GO:0035249 | synaptic transmission, glutamatergic(GO:0035249) |
| 0.1 | 0.4 | GO:0060290 | transdifferentiation(GO:0060290) |
| 0.1 | 0.8 | GO:0010032 | meiotic chromosome condensation(GO:0010032) |
| 0.1 | 20.9 | GO:0002377 | immunoglobulin production(GO:0002377) |
| 0.1 | 0.3 | GO:0046951 | ketone body biosynthetic process(GO:0046951) |
| 0.1 | 0.5 | GO:0051140 | regulation of NK T cell proliferation(GO:0051140) positive regulation of NK T cell proliferation(GO:0051142) |
| 0.1 | 3.3 | GO:0060291 | long-term synaptic potentiation(GO:0060291) |
| 0.1 | 0.1 | GO:2000642 | negative regulation of early endosome to late endosome transport(GO:2000642) |
| 0.1 | 0.4 | GO:0045872 | positive regulation of rhodopsin gene expression(GO:0045872) |
| 0.1 | 2.5 | GO:0033344 | cholesterol efflux(GO:0033344) |
| 0.1 | 2.5 | GO:0043113 | receptor clustering(GO:0043113) |
| 0.1 | 0.9 | GO:0070131 | positive regulation of mitochondrial translation(GO:0070131) |
| 0.1 | 0.5 | GO:0097167 | circadian regulation of translation(GO:0097167) |
| 0.1 | 0.2 | GO:2000110 | negative regulation of macrophage apoptotic process(GO:2000110) |
| 0.1 | 1.1 | GO:2001223 | negative regulation of neuron migration(GO:2001223) |
| 0.1 | 0.5 | GO:0045086 | positive regulation of interleukin-2 biosynthetic process(GO:0045086) |
| 0.1 | 2.4 | GO:0006376 | mRNA splice site selection(GO:0006376) |
| 0.1 | 4.7 | GO:0051965 | positive regulation of synapse assembly(GO:0051965) |
| 0.1 | 0.7 | GO:2000507 | positive regulation of energy homeostasis(GO:2000507) |
| 0.1 | 0.4 | GO:1900245 | positive regulation of MDA-5 signaling pathway(GO:1900245) |
| 0.1 | 0.1 | GO:0006788 | heme oxidation(GO:0006788) |
| 0.1 | 0.3 | GO:0070445 | oligodendrocyte progenitor proliferation(GO:0070444) regulation of oligodendrocyte progenitor proliferation(GO:0070445) |
| 0.1 | 2.9 | GO:0030212 | hyaluronan metabolic process(GO:0030212) |
| 0.1 | 1.0 | GO:0097264 | self proteolysis(GO:0097264) |
| 0.1 | 1.6 | GO:0061098 | positive regulation of protein tyrosine kinase activity(GO:0061098) |
| 0.1 | 0.6 | GO:0071044 | histone mRNA catabolic process(GO:0071044) |
| 0.1 | 0.3 | GO:0006682 | galactosylceramide biosynthetic process(GO:0006682) galactolipid biosynthetic process(GO:0019375) |
| 0.1 | 0.2 | GO:0019918 | peptidyl-arginine methylation, to symmetrical-dimethyl arginine(GO:0019918) |
| 0.1 | 1.7 | GO:0030497 | fatty acid elongation(GO:0030497) very long-chain fatty acid biosynthetic process(GO:0042761) |
| 0.1 | 1.8 | GO:1902043 | positive regulation of extrinsic apoptotic signaling pathway via death domain receptors(GO:1902043) |
| 0.1 | 4.2 | GO:0000381 | regulation of alternative mRNA splicing, via spliceosome(GO:0000381) |
| 0.1 | 1.3 | GO:0046475 | glycerophospholipid catabolic process(GO:0046475) |
| 0.1 | 0.7 | GO:2000582 | regulation of ATP-dependent microtubule motor activity, plus-end-directed(GO:2000580) positive regulation of ATP-dependent microtubule motor activity, plus-end-directed(GO:2000582) |
| 0.1 | 2.3 | GO:0090662 | ATP hydrolysis coupled proton transport(GO:0015991) ATP hydrolysis coupled transmembrane transport(GO:0090662) |
| 0.1 | 4.9 | GO:0031338 | regulation of vesicle fusion(GO:0031338) |
| 0.1 | 1.7 | GO:0035024 | negative regulation of Rho protein signal transduction(GO:0035024) |
| 0.1 | 1.0 | GO:0001502 | cartilage condensation(GO:0001502) |
| 0.1 | 1.1 | GO:0045725 | positive regulation of glycogen biosynthetic process(GO:0045725) |
| 0.1 | 0.4 | GO:0070245 | positive regulation of thymocyte apoptotic process(GO:0070245) |
| 0.1 | 0.2 | GO:1901026 | ripoptosome assembly(GO:0097343) ripoptosome assembly involved in necroptotic process(GO:1901026) |
| 0.1 | 0.2 | GO:1990859 | cellular response to endothelin(GO:1990859) |
| 0.1 | 0.5 | GO:0032532 | regulation of microvillus length(GO:0032532) |
| 0.1 | 0.9 | GO:0032776 | DNA methylation on cytosine(GO:0032776) |
| 0.1 | 1.0 | GO:0007274 | neuromuscular synaptic transmission(GO:0007274) |
| 0.1 | 0.6 | GO:0034088 | maintenance of mitotic sister chromatid cohesion(GO:0034088) |
| 0.1 | 1.5 | GO:0017004 | cytochrome complex assembly(GO:0017004) |
| 0.1 | 2.2 | GO:0015804 | neutral amino acid transport(GO:0015804) |
| 0.1 | 0.2 | GO:0035522 | monoubiquitinated histone deubiquitination(GO:0035521) monoubiquitinated histone H2A deubiquitination(GO:0035522) |
| 0.1 | 2.1 | GO:0045197 | establishment or maintenance of epithelial cell apical/basal polarity(GO:0045197) |
| 0.1 | 1.3 | GO:0042118 | endothelial cell activation(GO:0042118) |
| 0.1 | 1.7 | GO:0014003 | oligodendrocyte development(GO:0014003) |
| 0.1 | 0.2 | GO:0034729 | histone H3-K79 methylation(GO:0034729) |
| 0.1 | 0.4 | GO:0033184 | positive regulation of histone ubiquitination(GO:0033184) |
| 0.1 | 0.6 | GO:0000492 | box C/D snoRNP assembly(GO:0000492) |
| 0.1 | 0.7 | GO:0048280 | vesicle fusion with Golgi apparatus(GO:0048280) |
| 0.0 | 0.2 | GO:0043418 | homocysteine catabolic process(GO:0043418) |
| 0.0 | 0.1 | GO:0030200 | heparan sulfate proteoglycan catabolic process(GO:0030200) |
| 0.0 | 0.7 | GO:0003334 | keratinocyte development(GO:0003334) |
| 0.0 | 0.8 | GO:0048505 | regulation of timing of cell differentiation(GO:0048505) |
| 0.0 | 0.6 | GO:0048733 | sebaceous gland development(GO:0048733) |
| 0.0 | 0.0 | GO:0014057 | acetylcholine secretion, neurotransmission(GO:0014055) regulation of acetylcholine secretion, neurotransmission(GO:0014056) positive regulation of acetylcholine secretion, neurotransmission(GO:0014057) |
| 0.0 | 3.4 | GO:0050804 | modulation of synaptic transmission(GO:0050804) |
| 0.0 | 0.2 | GO:0001920 | negative regulation of receptor recycling(GO:0001920) |
| 0.0 | 5.0 | GO:0071804 | cellular potassium ion transport(GO:0071804) potassium ion transmembrane transport(GO:0071805) |
| 0.0 | 0.5 | GO:0045217 | cell-cell junction maintenance(GO:0045217) |
| 0.0 | 6.7 | GO:0010977 | negative regulation of neuron projection development(GO:0010977) |
| 0.0 | 1.4 | GO:0042501 | serine phosphorylation of STAT protein(GO:0042501) |
| 0.0 | 1.2 | GO:0046854 | phosphatidylinositol phosphorylation(GO:0046854) |
| 0.0 | 1.1 | GO:1903859 | regulation of dendrite extension(GO:1903859) |
| 0.0 | 0.7 | GO:0000338 | protein deneddylation(GO:0000338) |
| 0.0 | 5.0 | GO:0050808 | synapse organization(GO:0050808) |
| 0.0 | 5.9 | GO:0007601 | visual perception(GO:0007601) |
| 0.0 | 4.5 | GO:0051291 | protein heterooligomerization(GO:0051291) |
| 0.0 | 1.3 | GO:0008206 | bile acid metabolic process(GO:0008206) |
| 0.0 | 0.5 | GO:0002523 | leukocyte migration involved in inflammatory response(GO:0002523) |
| 0.0 | 2.2 | GO:0015992 | proton transport(GO:0015992) |
| 0.0 | 0.2 | GO:0048207 | vesicle targeting, rough ER to cis-Golgi(GO:0048207) COPII vesicle coating(GO:0048208) |
| 0.0 | 1.0 | GO:0032528 | microvillus assembly(GO:0030033) microvillus organization(GO:0032528) |
| 0.0 | 0.0 | GO:0018307 | enzyme active site formation(GO:0018307) |
| 0.0 | 0.3 | GO:0097428 | protein maturation by iron-sulfur cluster transfer(GO:0097428) |
| 0.0 | 0.4 | GO:2000671 | regulation of motor neuron apoptotic process(GO:2000671) |
| 0.0 | 0.2 | GO:0019659 | glucose catabolic process to lactate(GO:0019659) glycolytic fermentation(GO:0019660) glucose catabolic process to lactate via pyruvate(GO:0019661) |
| 0.0 | 0.4 | GO:0034205 | beta-amyloid formation(GO:0034205) |
| 0.0 | 3.9 | GO:0046488 | phosphatidylinositol metabolic process(GO:0046488) |
| 0.0 | 1.5 | GO:0006813 | potassium ion transport(GO:0006813) |
| 0.0 | 1.5 | GO:0031572 | G2 DNA damage checkpoint(GO:0031572) |
| 0.0 | 3.1 | GO:1903955 | positive regulation of protein targeting to mitochondrion(GO:1903955) |
| 0.0 | 0.3 | GO:0048642 | negative regulation of skeletal muscle tissue development(GO:0048642) |
| 0.0 | 0.3 | GO:0006610 | ribosomal protein import into nucleus(GO:0006610) |
| 0.0 | 1.0 | GO:0006182 | cGMP biosynthetic process(GO:0006182) |
| 0.0 | 3.0 | GO:0031398 | positive regulation of protein ubiquitination(GO:0031398) |
| 0.0 | 0.3 | GO:0032968 | positive regulation of transcription elongation from RNA polymerase II promoter(GO:0032968) |
| 0.0 | 0.6 | GO:0006999 | nuclear pore organization(GO:0006999) |
| 0.0 | 0.1 | GO:0030860 | regulation of polarized epithelial cell differentiation(GO:0030860) |
| 0.0 | 0.4 | GO:0061302 | smooth muscle cell-matrix adhesion(GO:0061302) |
| 0.0 | 0.5 | GO:0042462 | eye photoreceptor cell development(GO:0042462) |
| 0.0 | 0.2 | GO:0038031 | non-canonical Wnt signaling pathway via JNK cascade(GO:0038031) |
| 0.0 | 0.6 | GO:0071526 | semaphorin-plexin signaling pathway(GO:0071526) |
| 0.0 | 0.8 | GO:0015721 | bile acid and bile salt transport(GO:0015721) |
| 0.0 | 0.3 | GO:0032264 | purine nucleotide salvage(GO:0032261) IMP salvage(GO:0032264) |
| 0.0 | 0.4 | GO:0070935 | 3'-UTR-mediated mRNA stabilization(GO:0070935) |
| 0.0 | 0.2 | GO:1903300 | negative regulation of glucokinase activity(GO:0033132) negative regulation of hexokinase activity(GO:1903300) |
| 0.0 | 0.5 | GO:0046686 | response to cadmium ion(GO:0046686) |
| 0.0 | 3.3 | GO:0019236 | response to pheromone(GO:0019236) |
| 0.0 | 0.4 | GO:0098543 | detection of bacterium(GO:0016045) detection of other organism(GO:0098543) |
| 0.0 | 0.5 | GO:0006620 | posttranslational protein targeting to membrane(GO:0006620) |
| 0.0 | 0.8 | GO:0042149 | cellular response to glucose starvation(GO:0042149) |
| 0.0 | 0.5 | GO:0032094 | response to food(GO:0032094) |
| 0.0 | 0.2 | GO:0046490 | isopentenyl diphosphate biosynthetic process(GO:0009240) isopentenyl diphosphate metabolic process(GO:0046490) |
| 0.0 | 0.2 | GO:0051418 | interphase microtubule nucleation by interphase microtubule organizing center(GO:0051415) microtubule nucleation by microtubule organizing center(GO:0051418) |
| 0.0 | 0.2 | GO:1990126 | retrograde transport, endosome to plasma membrane(GO:1990126) |
| 0.0 | 0.4 | GO:0070816 | phosphorylation of RNA polymerase II C-terminal domain(GO:0070816) |
| 0.0 | 0.2 | GO:0031179 | peptide modification(GO:0031179) |
| 0.0 | 0.2 | GO:0071578 | zinc II ion transmembrane import(GO:0071578) |
| 0.0 | 0.3 | GO:1900017 | positive regulation of cytokine production involved in inflammatory response(GO:1900017) |
| 0.0 | 0.4 | GO:0010259 | multicellular organism aging(GO:0010259) |
| 0.0 | 0.2 | GO:0032815 | negative regulation of natural killer cell activation(GO:0032815) |
| 0.0 | 0.6 | GO:0007613 | memory(GO:0007613) |
| 0.0 | 0.4 | GO:0045948 | positive regulation of translational initiation(GO:0045948) |
| 0.0 | 0.1 | GO:0015904 | tetracycline transport(GO:0015904) |
| 0.0 | 0.3 | GO:0006744 | ubiquinone biosynthetic process(GO:0006744) |
| 0.0 | 0.8 | GO:0003170 | heart valve development(GO:0003170) |
| 0.0 | 0.0 | GO:0002541 | activation of plasma proteins involved in acute inflammatory response(GO:0002541) |
| 0.0 | 0.5 | GO:0070979 | protein K11-linked ubiquitination(GO:0070979) |
| 0.0 | 0.1 | GO:0002082 | regulation of oxidative phosphorylation(GO:0002082) |
| 0.0 | 0.1 | GO:0000480 | endonucleolytic cleavage in 5'-ETS of tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000480) |
| 0.0 | 0.3 | GO:0034314 | Arp2/3 complex-mediated actin nucleation(GO:0034314) |
| 0.0 | 0.1 | GO:0006561 | proline biosynthetic process(GO:0006561) |
| 0.0 | 0.2 | GO:0035871 | protein K11-linked deubiquitination(GO:0035871) |
| 0.0 | 0.3 | GO:0000413 | protein peptidyl-prolyl isomerization(GO:0000413) |
| 0.0 | 0.5 | GO:0001523 | retinoid metabolic process(GO:0001523) |
| 0.0 | 0.1 | GO:0006663 | platelet activating factor biosynthetic process(GO:0006663) |
| 0.0 | 0.0 | GO:0070493 | thrombin receptor signaling pathway(GO:0070493) |
| 0.0 | 0.0 | GO:0098749 | cerebellar neuron development(GO:0098749) |
| 0.0 | 0.0 | GO:0046013 | T cell homeostatic proliferation(GO:0001777) regulation of T cell homeostatic proliferation(GO:0046013) |
| 0.0 | 0.1 | GO:0007016 | cytoskeletal anchoring at plasma membrane(GO:0007016) |
| 0.0 | 0.5 | GO:0007200 | phospholipase C-activating G-protein coupled receptor signaling pathway(GO:0007200) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.6 | 17.9 | GO:0097441 | basilar dendrite(GO:0097441) |
| 2.3 | 7.0 | GO:0098830 | presynaptic endosome(GO:0098830) |
| 1.8 | 14.4 | GO:0098574 | cytoplasmic side of lysosomal membrane(GO:0098574) |
| 1.3 | 6.7 | GO:0044308 | axonal spine(GO:0044308) |
| 1.2 | 10.0 | GO:0017071 | intracellular cyclic nucleotide activated cation channel complex(GO:0017071) |
| 1.2 | 11.2 | GO:0070436 | Grb2-EGFR complex(GO:0070436) |
| 1.1 | 6.8 | GO:0098831 | presynaptic active zone cytoplasmic component(GO:0098831) |
| 1.1 | 15.1 | GO:0000015 | phosphopyruvate hydratase complex(GO:0000015) |
| 1.0 | 3.0 | GO:0044302 | dentate gyrus mossy fiber(GO:0044302) |
| 0.9 | 6.5 | GO:0005853 | eukaryotic translation elongation factor 1 complex(GO:0005853) |
| 0.8 | 2.5 | GO:0014801 | longitudinal sarcoplasmic reticulum(GO:0014801) |
| 0.7 | 2.2 | GO:0018444 | translation release factor complex(GO:0018444) |
| 0.7 | 2.1 | GO:0070033 | synaptobrevin 2-SNAP-25-syntaxin-1a-complexin II complex(GO:0070033) |
| 0.7 | 2.8 | GO:0097444 | spine apparatus(GO:0097444) |
| 0.7 | 7.2 | GO:1990812 | growth cone lamellipodium(GO:1990761) growth cone filopodium(GO:1990812) |
| 0.6 | 3.2 | GO:0044301 | climbing fiber(GO:0044301) |
| 0.6 | 4.7 | GO:0031673 | H zone(GO:0031673) |
| 0.6 | 5.1 | GO:1990357 | terminal web(GO:1990357) |
| 0.6 | 69.9 | GO:0042571 | immunoglobulin complex, circulating(GO:0042571) |
| 0.5 | 13.7 | GO:0098839 | postsynaptic density membrane(GO:0098839) |
| 0.5 | 31.9 | GO:0048786 | presynaptic active zone(GO:0048786) |
| 0.5 | 2.1 | GO:1990452 | Parkin-FBXW7-Cul1 ubiquitin ligase complex(GO:1990452) |
| 0.5 | 7.4 | GO:0005883 | neurofilament(GO:0005883) |
| 0.5 | 1.0 | GO:0070044 | synaptobrevin 2-SNAP-25-syntaxin-1a complex(GO:0070044) |
| 0.5 | 8.7 | GO:0031235 | intrinsic component of the cytoplasmic side of the plasma membrane(GO:0031235) |
| 0.5 | 3.8 | GO:0032983 | kainate selective glutamate receptor complex(GO:0032983) |
| 0.5 | 13.3 | GO:0044224 | juxtaparanode region of axon(GO:0044224) |
| 0.5 | 5.6 | GO:0032279 | asymmetric synapse(GO:0032279) |
| 0.5 | 6.1 | GO:0031315 | extrinsic component of mitochondrial outer membrane(GO:0031315) |
| 0.5 | 4.5 | GO:0005890 | sodium:potassium-exchanging ATPase complex(GO:0005890) |
| 0.4 | 9.3 | GO:0098563 | integral component of synaptic vesicle membrane(GO:0030285) intrinsic component of synaptic vesicle membrane(GO:0098563) |
| 0.4 | 8.7 | GO:0031045 | dense core granule(GO:0031045) |
| 0.4 | 1.3 | GO:1902912 | pyruvate kinase complex(GO:1902912) |
| 0.4 | 10.1 | GO:0035253 | ciliary rootlet(GO:0035253) |
| 0.4 | 4.8 | GO:0034992 | microtubule organizing center attachment site(GO:0034992) LINC complex(GO:0034993) |
| 0.4 | 1.6 | GO:0017146 | NMDA selective glutamate receptor complex(GO:0017146) |
| 0.4 | 1.2 | GO:0031523 | Myb complex(GO:0031523) |
| 0.4 | 1.2 | GO:0036501 | UFD1-NPL4 complex(GO:0036501) |
| 0.4 | 12.5 | GO:0032281 | AMPA glutamate receptor complex(GO:0032281) |
| 0.4 | 4.7 | GO:0030670 | phagocytic vesicle membrane(GO:0030670) |
| 0.4 | 1.1 | GO:0005965 | protein farnesyltransferase complex(GO:0005965) |
| 0.3 | 4.4 | GO:0097427 | microtubule bundle(GO:0097427) |
| 0.3 | 2.4 | GO:0070381 | endosome to plasma membrane transport vesicle(GO:0070381) |
| 0.3 | 0.3 | GO:0034681 | integrin alpha11-beta1 complex(GO:0034681) |
| 0.3 | 4.5 | GO:1990454 | L-type voltage-gated calcium channel complex(GO:1990454) |
| 0.3 | 2.0 | GO:0031314 | extrinsic component of mitochondrial inner membrane(GO:0031314) |
| 0.3 | 1.6 | GO:0000172 | ribonuclease MRP complex(GO:0000172) |
| 0.3 | 2.9 | GO:0031931 | TORC1 complex(GO:0031931) |
| 0.3 | 2.6 | GO:0071203 | WASH complex(GO:0071203) |
| 0.3 | 4.2 | GO:0008290 | F-actin capping protein complex(GO:0008290) |
| 0.3 | 18.2 | GO:0043198 | dendritic shaft(GO:0043198) |
| 0.3 | 1.8 | GO:0005955 | calcineurin complex(GO:0005955) |
| 0.2 | 1.0 | GO:0060053 | neurofilament cytoskeleton(GO:0060053) |
| 0.2 | 26.5 | GO:0008076 | voltage-gated potassium channel complex(GO:0008076) |
| 0.2 | 1.2 | GO:0097059 | CNTFR-CLCF1 complex(GO:0097059) |
| 0.2 | 4.4 | GO:0036056 | filtration diaphragm(GO:0036056) slit diaphragm(GO:0036057) |
| 0.2 | 86.4 | GO:0045211 | postsynaptic membrane(GO:0045211) |
| 0.2 | 1.4 | GO:0097452 | GAIT complex(GO:0097452) |
| 0.2 | 1.0 | GO:1990429 | Pex17p-Pex14p docking complex(GO:1990415) peroxisomal importomer complex(GO:1990429) |
| 0.2 | 6.8 | GO:0032809 | neuronal cell body membrane(GO:0032809) |
| 0.2 | 1.1 | GO:0005899 | insulin receptor complex(GO:0005899) |
| 0.2 | 1.5 | GO:0045180 | basal cortex(GO:0045180) |
| 0.2 | 2.1 | GO:0043196 | varicosity(GO:0043196) |
| 0.2 | 6.4 | GO:0000930 | gamma-tubulin complex(GO:0000930) |
| 0.2 | 2.0 | GO:0030915 | Smc5-Smc6 complex(GO:0030915) |
| 0.2 | 2.2 | GO:0030877 | beta-catenin destruction complex(GO:0030877) |
| 0.2 | 5.5 | GO:0010369 | chromocenter(GO:0010369) |
| 0.2 | 0.9 | GO:0034271 | phosphatidylinositol 3-kinase complex, class III, type I(GO:0034271) phosphatidylinositol 3-kinase complex, class III, type II(GO:0034272) |
| 0.2 | 3.2 | GO:0030127 | COPII vesicle coat(GO:0030127) |
| 0.2 | 1.2 | GO:0031095 | platelet dense tubular network membrane(GO:0031095) |
| 0.2 | 0.8 | GO:0000799 | nuclear condensin complex(GO:0000799) |
| 0.2 | 0.5 | GO:0034066 | RIC1-RGP1 guanyl-nucleotide exchange factor complex(GO:0034066) |
| 0.1 | 3.9 | GO:0005922 | connexon complex(GO:0005922) |
| 0.1 | 0.4 | GO:0034673 | inhibin-betaglycan-ActRII complex(GO:0034673) |
| 0.1 | 2.1 | GO:0017101 | aminoacyl-tRNA synthetase multienzyme complex(GO:0017101) |
| 0.1 | 1.4 | GO:0030122 | AP-2 adaptor complex(GO:0030122) |
| 0.1 | 2.0 | GO:0005869 | dynactin complex(GO:0005869) |
| 0.1 | 3.9 | GO:0042734 | presynaptic membrane(GO:0042734) |
| 0.1 | 1.3 | GO:0061700 | GATOR2 complex(GO:0061700) |
| 0.1 | 3.2 | GO:0097038 | perinuclear endoplasmic reticulum(GO:0097038) |
| 0.1 | 0.5 | GO:0033010 | paranodal junction(GO:0033010) |
| 0.1 | 0.6 | GO:0000839 | Hrd1p ubiquitin ligase ERAD-L complex(GO:0000839) |
| 0.1 | 30.4 | GO:0060076 | excitatory synapse(GO:0060076) |
| 0.1 | 1.0 | GO:0072357 | PTW/PP1 phosphatase complex(GO:0072357) |
| 0.1 | 9.8 | GO:0044309 | dendritic spine(GO:0043197) neuron spine(GO:0044309) |
| 0.1 | 0.4 | GO:0031084 | BLOC-2 complex(GO:0031084) |
| 0.1 | 0.5 | GO:0044614 | nuclear pore cytoplasmic filaments(GO:0044614) |
| 0.1 | 2.0 | GO:0042599 | lamellar body(GO:0042599) |
| 0.1 | 2.2 | GO:0065010 | extracellular membrane-bounded organelle(GO:0065010) |
| 0.1 | 0.6 | GO:0000322 | storage vacuole(GO:0000322) |
| 0.1 | 3.6 | GO:0032588 | trans-Golgi network membrane(GO:0032588) |
| 0.1 | 1.2 | GO:0016281 | eukaryotic translation initiation factor 4F complex(GO:0016281) |
| 0.1 | 0.7 | GO:0012507 | ER to Golgi transport vesicle membrane(GO:0012507) |
| 0.1 | 6.8 | GO:0016529 | sarcoplasmic reticulum(GO:0016529) |
| 0.1 | 1.9 | GO:0033268 | node of Ranvier(GO:0033268) |
| 0.1 | 2.2 | GO:0033178 | proton-transporting two-sector ATPase complex, catalytic domain(GO:0033178) |
| 0.1 | 2.3 | GO:0008074 | guanylate cyclase complex, soluble(GO:0008074) |
| 0.1 | 4.1 | GO:0031672 | A band(GO:0031672) |
| 0.1 | 0.7 | GO:0000439 | core TFIIH complex(GO:0000439) |
| 0.1 | 0.5 | GO:0033553 | rDNA heterochromatin(GO:0033553) |
| 0.1 | 16.8 | GO:0031225 | anchored component of membrane(GO:0031225) |
| 0.1 | 2.8 | GO:0005891 | voltage-gated calcium channel complex(GO:0005891) |
| 0.1 | 1.4 | GO:0032593 | insulin-responsive compartment(GO:0032593) |
| 0.1 | 0.8 | GO:0031209 | SCAR complex(GO:0031209) |
| 0.1 | 2.7 | GO:0034364 | high-density lipoprotein particle(GO:0034364) |
| 0.1 | 0.5 | GO:0032585 | multivesicular body membrane(GO:0032585) |
| 0.1 | 0.7 | GO:0005685 | U1 snRNP(GO:0005685) |
| 0.1 | 3.0 | GO:0016328 | lateral plasma membrane(GO:0016328) |
| 0.1 | 0.2 | GO:0030906 | retromer, cargo-selective complex(GO:0030906) |
| 0.1 | 1.4 | GO:0071004 | U2-type prespliceosome(GO:0071004) |
| 0.1 | 0.9 | GO:0030126 | COPI vesicle coat(GO:0030126) |
| 0.1 | 13.5 | GO:0030016 | myofibril(GO:0030016) |
| 0.1 | 0.7 | GO:0016461 | unconventional myosin complex(GO:0016461) |
| 0.1 | 8.9 | GO:0008021 | synaptic vesicle(GO:0008021) |
| 0.1 | 1.7 | GO:0046540 | U4/U6 x U5 tri-snRNP complex(GO:0046540) |
| 0.1 | 0.3 | GO:0070449 | elongin complex(GO:0070449) |
| 0.1 | 0.6 | GO:1990712 | HFE-transferrin receptor complex(GO:1990712) |
| 0.1 | 7.0 | GO:0030659 | cytoplasmic vesicle membrane(GO:0030659) |
| 0.0 | 3.2 | GO:0005871 | kinesin complex(GO:0005871) |
| 0.0 | 0.8 | GO:0097440 | apical dendrite(GO:0097440) |
| 0.0 | 0.6 | GO:0016442 | RISC complex(GO:0016442) RNAi effector complex(GO:0031332) |
| 0.0 | 15.8 | GO:0030425 | dendrite(GO:0030425) |
| 0.0 | 0.5 | GO:0031464 | Cul4A-RING E3 ubiquitin ligase complex(GO:0031464) |
| 0.0 | 0.1 | GO:0046691 | intracellular canaliculus(GO:0046691) |
| 0.0 | 0.3 | GO:0097165 | nuclear stress granule(GO:0097165) |
| 0.0 | 0.4 | GO:0070652 | HAUS complex(GO:0070652) |
| 0.0 | 1.1 | GO:0005779 | integral component of peroxisomal membrane(GO:0005779) |
| 0.0 | 0.5 | GO:0008540 | proteasome regulatory particle, base subcomplex(GO:0008540) cytosolic proteasome complex(GO:0031597) |
| 0.0 | 0.4 | GO:0070765 | gamma-secretase complex(GO:0070765) |
| 0.0 | 0.2 | GO:0045298 | tubulin complex(GO:0045298) |
| 0.0 | 3.0 | GO:0031463 | Cul3-RING ubiquitin ligase complex(GO:0031463) |
| 0.0 | 0.2 | GO:0046696 | lipopolysaccharide receptor complex(GO:0046696) |
| 0.0 | 0.7 | GO:0061702 | inflammasome complex(GO:0061702) |
| 0.0 | 1.8 | GO:0005801 | cis-Golgi network(GO:0005801) |
| 0.0 | 0.6 | GO:0005689 | U12-type spliceosomal complex(GO:0005689) |
| 0.0 | 1.6 | GO:0043596 | nuclear replication fork(GO:0043596) |
| 0.0 | 1.7 | GO:0005881 | cytoplasmic microtubule(GO:0005881) |
| 0.0 | 1.6 | GO:0010494 | cytoplasmic stress granule(GO:0010494) |
| 0.0 | 0.5 | GO:0031616 | spindle pole centrosome(GO:0031616) |
| 0.0 | 0.3 | GO:0005885 | Arp2/3 protein complex(GO:0005885) |
| 0.0 | 1.1 | GO:0005876 | spindle microtubule(GO:0005876) |
| 0.0 | 0.9 | GO:0001741 | XY body(GO:0001741) |
| 0.0 | 0.5 | GO:0051233 | spindle midzone(GO:0051233) |
| 0.0 | 2.9 | GO:0005884 | actin filament(GO:0005884) |
| 0.0 | 0.2 | GO:0001739 | sex chromatin(GO:0001739) |
| 0.0 | 0.8 | GO:0031305 | intrinsic component of mitochondrial inner membrane(GO:0031304) integral component of mitochondrial inner membrane(GO:0031305) |
| 0.0 | 0.6 | GO:0035098 | ESC/E(Z) complex(GO:0035098) |
| 0.0 | 0.1 | GO:0097422 | tubular endosome(GO:0097422) |
| 0.0 | 0.8 | GO:0009925 | basal plasma membrane(GO:0009925) |
| 0.0 | 3.5 | GO:0043209 | myelin sheath(GO:0043209) |
| 0.0 | 0.2 | GO:0042470 | melanosome(GO:0042470) pigment granule(GO:0048770) |
| 0.0 | 0.6 | GO:0012506 | vesicle membrane(GO:0012506) |
| 0.0 | 0.1 | GO:0031428 | box C/D snoRNP complex(GO:0031428) |
| 0.0 | 0.2 | GO:0042405 | nuclear inclusion body(GO:0042405) |
| 0.0 | 0.9 | GO:0022625 | cytosolic large ribosomal subunit(GO:0022625) |
| 0.0 | 0.9 | GO:0019005 | SCF ubiquitin ligase complex(GO:0019005) |
| 0.0 | 1.0 | GO:0030426 | growth cone(GO:0030426) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 4.2 | 12.6 | GO:0016492 | G-protein coupled neurotensin receptor activity(GO:0016492) |
| 2.8 | 17.0 | GO:0060175 | brain-derived neurotrophic factor-activated receptor activity(GO:0060175) |
| 2.7 | 13.6 | GO:0005502 | 11-cis retinal binding(GO:0005502) |
| 2.2 | 17.8 | GO:0099583 | neurotransmitter receptor activity involved in regulation of postsynaptic cytosolic calcium ion concentration(GO:0099583) |
| 1.8 | 5.3 | GO:0001847 | opsonin receptor activity(GO:0001847) |
| 1.7 | 10.0 | GO:0005052 | peroxisome matrix targeting signal-1 binding(GO:0005052) |
| 1.6 | 4.7 | GO:0035375 | zymogen binding(GO:0035375) |
| 1.5 | 5.9 | GO:0002153 | steroid receptor RNA activator RNA binding(GO:0002153) |
| 1.3 | 16.2 | GO:0097322 | 7SK snRNA binding(GO:0097322) |
| 1.3 | 14.4 | GO:0048101 | calmodulin-dependent cyclic-nucleotide phosphodiesterase activity(GO:0004117) calcium- and calmodulin-regulated 3',5'-cyclic-GMP phosphodiesterase activity(GO:0048101) |
| 1.2 | 7.4 | GO:0015018 | galactosylgalactosylxylosylprotein 3-beta-glucuronosyltransferase activity(GO:0015018) |
| 1.1 | 11.4 | GO:0001665 | alpha-N-acetylgalactosaminide alpha-2,6-sialyltransferase activity(GO:0001665) |
| 1.1 | 5.6 | GO:0016907 | G-protein coupled acetylcholine receptor activity(GO:0016907) |
| 1.1 | 4.5 | GO:0030156 | benzodiazepine receptor binding(GO:0030156) |
| 1.1 | 15.1 | GO:0004634 | phosphopyruvate hydratase activity(GO:0004634) |
| 1.1 | 3.2 | GO:0050508 | glucuronosyl-N-acetylglucosaminyl-proteoglycan 4-alpha-N-acetylglucosaminyltransferase activity(GO:0050508) |
| 1.0 | 3.0 | GO:0036004 | GAF domain binding(GO:0036004) |
| 1.0 | 2.9 | GO:0001156 | TFIIIC-class transcription factor binding(GO:0001156) |
| 0.9 | 3.8 | GO:0038025 | reelin receptor activity(GO:0038025) |
| 0.9 | 3.7 | GO:0051381 | histamine binding(GO:0051381) |
| 0.9 | 2.7 | GO:0003990 | acetylcholinesterase activity(GO:0003990) |
| 0.9 | 5.4 | GO:0035241 | protein-arginine omega-N monomethyltransferase activity(GO:0035241) |
| 0.8 | 8.4 | GO:0004332 | fructose-bisphosphate aldolase activity(GO:0004332) |
| 0.8 | 10.1 | GO:0032051 | clathrin light chain binding(GO:0032051) |
| 0.8 | 3.3 | GO:0004781 | adenylylsulfate kinase activity(GO:0004020) sulfate adenylyltransferase activity(GO:0004779) sulfate adenylyltransferase (ATP) activity(GO:0004781) |
| 0.8 | 5.7 | GO:0005005 | transmembrane-ephrin receptor activity(GO:0005005) |
| 0.8 | 7.8 | GO:0051766 | inositol trisphosphate kinase activity(GO:0051766) |
| 0.8 | 4.6 | GO:0034190 | apolipoprotein receptor binding(GO:0034190) |
| 0.8 | 3.1 | GO:0005219 | ryanodine-sensitive calcium-release channel activity(GO:0005219) |
| 0.8 | 2.3 | GO:0070540 | stearic acid binding(GO:0070540) |
| 0.8 | 4.5 | GO:0000155 | phosphorelay sensor kinase activity(GO:0000155) |
| 0.7 | 15.3 | GO:0008510 | sodium:bicarbonate symporter activity(GO:0008510) |
| 0.7 | 2.0 | GO:0015275 | stretch-activated, cation-selective, calcium channel activity(GO:0015275) |
| 0.6 | 1.9 | GO:0035870 | dITP diphosphatase activity(GO:0035870) |
| 0.6 | 3.8 | GO:0015277 | kainate selective glutamate receptor activity(GO:0015277) |
| 0.6 | 6.3 | GO:0008889 | glycerophosphodiester phosphodiesterase activity(GO:0008889) |
| 0.6 | 7.1 | GO:0015280 | ligand-gated sodium channel activity(GO:0015280) |
| 0.6 | 2.4 | GO:0017095 | heparan sulfate 6-O-sulfotransferase activity(GO:0017095) |
| 0.6 | 2.3 | GO:0001588 | dopamine neurotransmitter receptor activity, coupled via Gs(GO:0001588) |
| 0.6 | 3.4 | GO:0008241 | peptidyl-dipeptidase activity(GO:0008241) |
| 0.6 | 11.6 | GO:0008553 | hydrogen-exporting ATPase activity, phosphorylative mechanism(GO:0008553) |
| 0.5 | 4.4 | GO:0060072 | large conductance calcium-activated potassium channel activity(GO:0060072) |
| 0.5 | 2.6 | GO:0005250 | A-type (transient outward) potassium channel activity(GO:0005250) |
| 0.5 | 3.1 | GO:0008422 | beta-glucosidase activity(GO:0008422) |
| 0.5 | 1.5 | GO:0086057 | voltage-gated calcium channel activity involved in bundle of His cell action potential(GO:0086057) |
| 0.5 | 2.5 | GO:0034041 | sterol-transporting ATPase activity(GO:0034041) |
| 0.5 | 4.0 | GO:0004416 | hydroxyacylglutathione hydrolase activity(GO:0004416) |
| 0.5 | 4.0 | GO:0019158 | fructokinase activity(GO:0008865) mannokinase activity(GO:0019158) |
| 0.5 | 5.4 | GO:0004972 | NMDA glutamate receptor activity(GO:0004972) |
| 0.5 | 5.4 | GO:0008454 | alpha-1,3-mannosylglycoprotein 4-beta-N-acetylglucosaminyltransferase activity(GO:0008454) |
| 0.5 | 5.2 | GO:0000774 | adenyl-nucleotide exchange factor activity(GO:0000774) |
| 0.5 | 7.1 | GO:0070492 | oligosaccharide binding(GO:0070492) |
| 0.5 | 61.8 | GO:0034987 | immunoglobulin receptor binding(GO:0034987) |
| 0.5 | 3.8 | GO:0001515 | opioid peptide activity(GO:0001515) |
| 0.5 | 7.0 | GO:0032050 | clathrin heavy chain binding(GO:0032050) |
| 0.5 | 1.8 | GO:0019828 | aspartic-type endopeptidase inhibitor activity(GO:0019828) |
| 0.5 | 31.4 | GO:0046875 | ephrin receptor binding(GO:0046875) |
| 0.4 | 1.3 | GO:0004819 | glutamine-tRNA ligase activity(GO:0004819) |
| 0.4 | 3.9 | GO:0016308 | 1-phosphatidylinositol-4-phosphate 5-kinase activity(GO:0016308) |
| 0.4 | 3.0 | GO:0030160 | GKAP/Homer scaffold activity(GO:0030160) |
| 0.4 | 2.5 | GO:0030294 | receptor signaling protein tyrosine kinase inhibitor activity(GO:0030294) |
| 0.4 | 2.5 | GO:0003835 | beta-galactoside alpha-2,6-sialyltransferase activity(GO:0003835) |
| 0.4 | 2.5 | GO:0004614 | phosphoglucomutase activity(GO:0004614) |
| 0.4 | 6.3 | GO:0019911 | structural constituent of myelin sheath(GO:0019911) |
| 0.4 | 4.9 | GO:0005391 | sodium:potassium-exchanging ATPase activity(GO:0005391) steroid hormone binding(GO:1990239) |
| 0.4 | 1.6 | GO:0004105 | choline-phosphate cytidylyltransferase activity(GO:0004105) |
| 0.4 | 2.8 | GO:0048403 | brain-derived neurotrophic factor binding(GO:0048403) |
| 0.4 | 3.6 | GO:0008597 | calcium-dependent protein serine/threonine phosphatase regulator activity(GO:0008597) |
| 0.4 | 2.0 | GO:0099609 | microtubule lateral binding(GO:0099609) |
| 0.4 | 2.0 | GO:0097001 | ceramide binding(GO:0097001) |
| 0.4 | 3.4 | GO:0015467 | G-protein activated inward rectifier potassium channel activity(GO:0015467) |
| 0.4 | 2.3 | GO:0034186 | apolipoprotein A-I binding(GO:0034186) |
| 0.4 | 1.5 | GO:0015182 | L-asparagine transmembrane transporter activity(GO:0015182) |
| 0.4 | 3.3 | GO:0005519 | cytoskeletal regulatory protein binding(GO:0005519) |
| 0.4 | 11.0 | GO:0005184 | neuropeptide hormone activity(GO:0005184) |
| 0.4 | 1.1 | GO:0004660 | protein farnesyltransferase activity(GO:0004660) |
| 0.4 | 2.8 | GO:0070699 | type II activin receptor binding(GO:0070699) |
| 0.3 | 2.4 | GO:0046912 | transferase activity, transferring acyl groups, acyl groups converted into alkyl on transfer(GO:0046912) |
| 0.3 | 1.4 | GO:0031544 | peptidyl-proline 3-dioxygenase activity(GO:0031544) |
| 0.3 | 9.2 | GO:0031489 | myosin V binding(GO:0031489) |
| 0.3 | 2.0 | GO:0010997 | anaphase-promoting complex binding(GO:0010997) |
| 0.3 | 1.3 | GO:0004711 | ribosomal protein S6 kinase activity(GO:0004711) |
| 0.3 | 1.5 | GO:2001069 | glycogen binding(GO:2001069) |
| 0.3 | 1.5 | GO:0004308 | exo-alpha-sialidase activity(GO:0004308) alpha-sialidase activity(GO:0016997) exo-alpha-(2->3)-sialidase activity(GO:0052794) exo-alpha-(2->6)-sialidase activity(GO:0052795) exo-alpha-(2->8)-sialidase activity(GO:0052796) |
| 0.3 | 1.8 | GO:0033192 | calmodulin-dependent protein phosphatase activity(GO:0033192) |
| 0.3 | 1.2 | GO:0031735 | CCR10 chemokine receptor binding(GO:0031735) |
| 0.3 | 6.4 | GO:0008191 | metalloendopeptidase inhibitor activity(GO:0008191) |
| 0.3 | 2.2 | GO:0003747 | translation release factor activity(GO:0003747) translation termination factor activity(GO:0008079) |
| 0.3 | 6.7 | GO:0015269 | calcium-activated potassium channel activity(GO:0015269) |
| 0.3 | 7.3 | GO:0005388 | calcium-transporting ATPase activity(GO:0005388) |
| 0.3 | 1.4 | GO:0016499 | orexin receptor activity(GO:0016499) |
| 0.3 | 1.3 | GO:0004809 | tRNA (guanine-N2-)-methyltransferase activity(GO:0004809) |
| 0.3 | 2.1 | GO:1903763 | gap junction channel activity involved in cell communication by electrical coupling(GO:1903763) |
| 0.3 | 1.3 | GO:0004743 | pyruvate kinase activity(GO:0004743) |
| 0.3 | 4.8 | GO:0045499 | chemorepellent activity(GO:0045499) |
| 0.2 | 6.2 | GO:0003746 | translation elongation factor activity(GO:0003746) |
| 0.2 | 4.7 | GO:0015643 | toxic substance binding(GO:0015643) |
| 0.2 | 7.5 | GO:0048156 | tau protein binding(GO:0048156) |
| 0.2 | 2.3 | GO:0097027 | ubiquitin-protein transferase activator activity(GO:0097027) |
| 0.2 | 1.3 | GO:0070728 | leucine binding(GO:0070728) |
| 0.2 | 5.5 | GO:0004385 | guanylate kinase activity(GO:0004385) |
| 0.2 | 0.9 | GO:0035827 | rubidium ion transmembrane transporter activity(GO:0035827) |
| 0.2 | 11.5 | GO:0050699 | WW domain binding(GO:0050699) |
| 0.2 | 2.5 | GO:0070097 | delta-catenin binding(GO:0070097) |
| 0.2 | 5.2 | GO:0008574 | ATP-dependent microtubule motor activity, plus-end-directed(GO:0008574) |
| 0.2 | 2.0 | GO:1901612 | cardiolipin binding(GO:1901612) |
| 0.2 | 0.8 | GO:0008169 | C-methyltransferase activity(GO:0008169) |
| 0.2 | 0.8 | GO:0036313 | phosphatidylinositol 3-kinase catalytic subunit binding(GO:0036313) |
| 0.2 | 0.6 | GO:0008967 | phosphoglycolate phosphatase activity(GO:0008967) |
| 0.2 | 0.8 | GO:0034617 | tetrahydrobiopterin binding(GO:0034617) |
| 0.2 | 1.0 | GO:0005483 | soluble NSF attachment protein activity(GO:0005483) |
| 0.2 | 1.7 | GO:0004716 | receptor signaling protein tyrosine kinase activity(GO:0004716) |
| 0.2 | 0.8 | GO:0003726 | double-stranded RNA adenosine deaminase activity(GO:0003726) |
| 0.2 | 1.3 | GO:0034711 | inhibin binding(GO:0034711) |
| 0.2 | 5.1 | GO:0003680 | AT DNA binding(GO:0003680) |
| 0.2 | 25.7 | GO:0005267 | potassium channel activity(GO:0005267) |
| 0.2 | 1.8 | GO:0005243 | gap junction channel activity(GO:0005243) |
| 0.2 | 2.9 | GO:0008349 | MAP kinase kinase kinase kinase activity(GO:0008349) |
| 0.2 | 3.1 | GO:0050321 | tau-protein kinase activity(GO:0050321) |
| 0.2 | 0.5 | GO:0045030 | UTP-activated nucleotide receptor activity(GO:0045030) |
| 0.2 | 9.8 | GO:0004864 | protein phosphatase inhibitor activity(GO:0004864) |
| 0.2 | 5.0 | GO:0005521 | lamin binding(GO:0005521) |
| 0.2 | 2.7 | GO:0031078 | histone deacetylase activity (H3-K14 specific)(GO:0031078) NAD-dependent histone deacetylase activity (H3-K14 specific)(GO:0032041) |
| 0.2 | 1.0 | GO:0001025 | RNA polymerase III transcription factor binding(GO:0001025) |
| 0.2 | 5.7 | GO:0004190 | aspartic-type endopeptidase activity(GO:0004190) |
| 0.2 | 6.0 | GO:0070273 | phosphatidylinositol-4-phosphate binding(GO:0070273) |
| 0.2 | 1.3 | GO:0008508 | bile acid:sodium symporter activity(GO:0008508) |
| 0.2 | 1.6 | GO:0004526 | ribonuclease P activity(GO:0004526) |
| 0.2 | 2.6 | GO:0005314 | high-affinity glutamate transmembrane transporter activity(GO:0005314) |
| 0.2 | 6.9 | GO:0005086 | ARF guanyl-nucleotide exchange factor activity(GO:0005086) |
| 0.2 | 0.8 | GO:0046624 | sphingolipid transporter activity(GO:0046624) |
| 0.2 | 1.4 | GO:0047498 | calcium-dependent phospholipase A2 activity(GO:0047498) |
| 0.1 | 0.9 | GO:0030280 | structural constituent of epidermis(GO:0030280) |
| 0.1 | 0.7 | GO:0003880 | protein C-terminal carboxyl O-methyltransferase activity(GO:0003880) |
| 0.1 | 1.0 | GO:0008499 | UDP-galactose:beta-N-acetylglucosamine beta-1,3-galactosyltransferase activity(GO:0008499) |
| 0.1 | 0.6 | GO:0051022 | Rho GDP-dissociation inhibitor binding(GO:0051022) |
| 0.1 | 3.5 | GO:0042608 | T cell receptor binding(GO:0042608) |
| 0.1 | 0.7 | GO:0047635 | L-alanine:2-oxoglutarate aminotransferase activity(GO:0004021) alanine-oxo-acid transaminase activity(GO:0047635) |
| 0.1 | 0.6 | GO:0035402 | histone kinase activity (H3-T11 specific)(GO:0035402) |
| 0.1 | 1.2 | GO:0004897 | ciliary neurotrophic factor receptor activity(GO:0004897) |
| 0.1 | 1.2 | GO:0032795 | heterotrimeric G-protein binding(GO:0032795) |
| 0.1 | 2.0 | GO:0005522 | profilin binding(GO:0005522) |
| 0.1 | 0.5 | GO:0055105 | ubiquitin-protein transferase inhibitor activity(GO:0055105) |
| 0.1 | 20.6 | GO:0005200 | structural constituent of cytoskeleton(GO:0005200) |
| 0.1 | 4.2 | GO:0005057 | receptor signaling protein activity(GO:0005057) |
| 0.1 | 5.5 | GO:0043015 | gamma-tubulin binding(GO:0043015) |
| 0.1 | 1.0 | GO:0061578 | Lys63-specific deubiquitinase activity(GO:0061578) |
| 0.1 | 2.3 | GO:0017049 | GTP-Rho binding(GO:0017049) |
| 0.1 | 0.6 | GO:0004534 | 5'-3' exoribonuclease activity(GO:0004534) |
| 0.1 | 5.8 | GO:0003785 | actin monomer binding(GO:0003785) |
| 0.1 | 5.1 | GO:0042169 | SH2 domain binding(GO:0042169) |
| 0.1 | 2.9 | GO:0004708 | MAP kinase kinase activity(GO:0004708) |
| 0.1 | 1.4 | GO:0022889 | L-serine transmembrane transporter activity(GO:0015194) serine transmembrane transporter activity(GO:0022889) |
| 0.1 | 2.9 | GO:0004115 | 3',5'-cyclic-AMP phosphodiesterase activity(GO:0004115) |
| 0.1 | 3.0 | GO:0008331 | high voltage-gated calcium channel activity(GO:0008331) |
| 0.1 | 1.0 | GO:0015038 | glutathione disulfide oxidoreductase activity(GO:0015038) |
| 0.1 | 0.6 | GO:0005275 | amine transmembrane transporter activity(GO:0005275) |
| 0.1 | 1.9 | GO:0042166 | acetylcholine binding(GO:0042166) |
| 0.1 | 6.0 | GO:0031369 | translation initiation factor binding(GO:0031369) |
| 0.1 | 1.8 | GO:0001206 | transcriptional repressor activity, RNA polymerase II distal enhancer sequence-specific binding(GO:0001206) |
| 0.1 | 1.1 | GO:0035374 | chondroitin sulfate binding(GO:0035374) |
| 0.1 | 0.3 | GO:0035539 | 8-oxo-7,8-dihydroguanosine triphosphate pyrophosphatase activity(GO:0008413) 8-oxo-7,8-dihydrodeoxyguanosine triphosphate pyrophosphatase activity(GO:0035539) |
| 0.1 | 0.7 | GO:0019784 | NEDD8-specific protease activity(GO:0019784) |
| 0.1 | 1.2 | GO:0090482 | vitamin transmembrane transporter activity(GO:0090482) |
| 0.1 | 3.4 | GO:0019707 | protein-cysteine S-palmitoyltransferase activity(GO:0019706) protein-cysteine S-acyltransferase activity(GO:0019707) |
| 0.1 | 2.7 | GO:0031402 | sodium ion binding(GO:0031402) |
| 0.1 | 2.2 | GO:0070016 | armadillo repeat domain binding(GO:0070016) |
| 0.1 | 1.2 | GO:0036435 | K48-linked polyubiquitin binding(GO:0036435) |
| 0.1 | 3.0 | GO:0071855 | neuropeptide receptor binding(GO:0071855) |
| 0.1 | 2.7 | GO:0043395 | heparan sulfate proteoglycan binding(GO:0043395) |
| 0.1 | 2.5 | GO:0034237 | protein kinase A regulatory subunit binding(GO:0034237) |
| 0.1 | 17.4 | GO:0017124 | SH3 domain binding(GO:0017124) |
| 0.1 | 0.3 | GO:1902379 | chemoattractant activity involved in axon guidance(GO:1902379) |
| 0.1 | 0.3 | GO:0003945 | N-acetyllactosamine synthase activity(GO:0003945) |
| 0.1 | 0.6 | GO:0005324 | long-chain fatty acid transporter activity(GO:0005324) |
| 0.1 | 2.2 | GO:0015026 | coreceptor activity(GO:0015026) |
| 0.1 | 2.2 | GO:0043014 | alpha-tubulin binding(GO:0043014) |
| 0.1 | 3.8 | GO:0005246 | calcium channel regulator activity(GO:0005246) |
| 0.1 | 0.8 | GO:0019869 | chloride channel inhibitor activity(GO:0019869) |
| 0.1 | 0.8 | GO:0030306 | ADP-ribosylation factor binding(GO:0030306) |
| 0.1 | 8.8 | GO:0048306 | calcium-dependent protein binding(GO:0048306) |
| 0.1 | 0.3 | GO:0045131 | pre-mRNA branch point binding(GO:0045131) |
| 0.1 | 3.1 | GO:0008139 | nuclear localization sequence binding(GO:0008139) |
| 0.1 | 4.3 | GO:0004693 | cyclin-dependent protein serine/threonine kinase activity(GO:0004693) |
| 0.1 | 0.6 | GO:0032027 | myosin light chain binding(GO:0032027) |
| 0.1 | 1.3 | GO:0005048 | signal sequence binding(GO:0005048) |
| 0.1 | 0.9 | GO:0003886 | DNA (cytosine-5-)-methyltransferase activity(GO:0003886) |
| 0.1 | 0.6 | GO:0070290 | N-acylphosphatidylethanolamine-specific phospholipase D activity(GO:0070290) |
| 0.1 | 0.5 | GO:0004957 | prostaglandin E receptor activity(GO:0004957) |
| 0.1 | 1.8 | GO:0004745 | retinol dehydrogenase activity(GO:0004745) |
| 0.1 | 11.4 | GO:0005089 | Rho guanyl-nucleotide exchange factor activity(GO:0005089) |
| 0.1 | 1.4 | GO:0048365 | Rac GTPase binding(GO:0048365) |
| 0.1 | 0.6 | GO:0015379 | potassium:chloride symporter activity(GO:0015379) potassium ion symporter activity(GO:0022820) |
| 0.1 | 0.7 | GO:0005000 | vasopressin receptor activity(GO:0005000) |
| 0.1 | 0.2 | GO:0035243 | protein-arginine omega-N symmetric methyltransferase activity(GO:0035243) |
| 0.1 | 2.7 | GO:0005385 | zinc ion transmembrane transporter activity(GO:0005385) |
| 0.1 | 2.0 | GO:0035255 | ionotropic glutamate receptor binding(GO:0035255) |
| 0.1 | 1.9 | GO:0042923 | neuropeptide binding(GO:0042923) |
| 0.1 | 0.6 | GO:0008093 | cytoskeletal adaptor activity(GO:0008093) |
| 0.1 | 1.1 | GO:0030215 | semaphorin receptor binding(GO:0030215) |
| 0.1 | 0.4 | GO:0046976 | histone methyltransferase activity (H3-K27 specific)(GO:0046976) |
| 0.1 | 2.9 | GO:0004683 | calmodulin-dependent protein kinase activity(GO:0004683) |
| 0.1 | 0.7 | GO:0008353 | RNA polymerase II carboxy-terminal domain kinase activity(GO:0008353) |
| 0.1 | 0.3 | GO:0015091 | ferric iron transmembrane transporter activity(GO:0015091) trivalent inorganic cation transmembrane transporter activity(GO:0072510) |
| 0.1 | 2.5 | GO:0001784 | phosphotyrosine binding(GO:0001784) |
| 0.1 | 0.9 | GO:0016846 | carbon-sulfur lyase activity(GO:0016846) |
| 0.1 | 2.2 | GO:1990841 | promoter-specific chromatin binding(GO:1990841) |
| 0.1 | 0.3 | GO:0055100 | adiponectin binding(GO:0055100) |
| 0.1 | 1.6 | GO:0005212 | structural constituent of eye lens(GO:0005212) |
| 0.1 | 0.5 | GO:0015129 | lactate transmembrane transporter activity(GO:0015129) |
| 0.1 | 0.7 | GO:0017154 | semaphorin receptor activity(GO:0017154) |
| 0.1 | 0.5 | GO:0032052 | bile acid binding(GO:0032052) |
| 0.1 | 0.3 | GO:0098639 | collagen binding involved in cell-matrix adhesion(GO:0098639) |
| 0.1 | 0.2 | GO:0060228 | phosphatidylcholine-sterol O-acyltransferase activator activity(GO:0060228) |
| 0.1 | 2.8 | GO:0016247 | channel regulator activity(GO:0016247) |
| 0.1 | 0.3 | GO:0004998 | transferrin receptor activity(GO:0004998) |
| 0.1 | 1.0 | GO:0031702 | type 1 angiotensin receptor binding(GO:0031702) |
| 0.1 | 0.4 | GO:0039706 | co-receptor binding(GO:0039706) |
| 0.0 | 1.7 | GO:0042056 | chemoattractant activity(GO:0042056) |
| 0.0 | 0.1 | GO:0004392 | heme oxygenase (decyclizing) activity(GO:0004392) |
| 0.0 | 1.4 | GO:0070840 | dynein complex binding(GO:0070840) |
| 0.0 | 3.1 | GO:0001105 | RNA polymerase II transcription coactivator activity(GO:0001105) |
| 0.0 | 4.8 | GO:0008528 | G-protein coupled peptide receptor activity(GO:0008528) |
| 0.0 | 0.2 | GO:0019212 | phosphatase inhibitor activity(GO:0019212) |
| 0.0 | 16.4 | GO:0030695 | GTPase regulator activity(GO:0030695) |
| 0.0 | 2.7 | GO:0015485 | cholesterol binding(GO:0015485) |
| 0.0 | 1.0 | GO:0030332 | cyclin binding(GO:0030332) |
| 0.0 | 2.4 | GO:0005080 | protein kinase C binding(GO:0005080) |
| 0.0 | 0.2 | GO:0004487 | methylenetetrahydrofolate dehydrogenase (NAD+) activity(GO:0004487) |
| 0.0 | 0.2 | GO:0016842 | amidine-lyase activity(GO:0016842) |
| 0.0 | 0.2 | GO:0004452 | isopentenyl-diphosphate delta-isomerase activity(GO:0004452) |
| 0.0 | 2.0 | GO:0001540 | beta-amyloid binding(GO:0001540) |
| 0.0 | 0.7 | GO:0008239 | dipeptidyl-peptidase activity(GO:0008239) |
| 0.0 | 0.4 | GO:0005347 | ATP transmembrane transporter activity(GO:0005347) ADP transmembrane transporter activity(GO:0015217) |
| 0.0 | 0.4 | GO:0050733 | RS domain binding(GO:0050733) |
| 0.0 | 0.3 | GO:0043024 | ribosomal small subunit binding(GO:0043024) |
| 0.0 | 0.5 | GO:0001087 | transcription factor activity, sequence-specific DNA binding, RNA polymerase recruiting(GO:0001011) transcription factor activity, TFIIB-class binding(GO:0001087) |
| 0.0 | 8.9 | GO:0003924 | GTPase activity(GO:0003924) |
| 0.0 | 0.3 | GO:0030368 | interleukin-17 receptor activity(GO:0030368) |
| 0.0 | 0.3 | GO:0015245 | fatty acid transporter activity(GO:0015245) |
| 0.0 | 0.4 | GO:0015187 | glycine transmembrane transporter activity(GO:0015187) |
| 0.0 | 0.6 | GO:0030247 | pattern binding(GO:0001871) polysaccharide binding(GO:0030247) |
| 0.0 | 0.8 | GO:0070182 | DNA polymerase binding(GO:0070182) |
| 0.0 | 1.0 | GO:0071889 | 14-3-3 protein binding(GO:0071889) |
| 0.0 | 4.4 | GO:0005516 | calmodulin binding(GO:0005516) |
| 0.0 | 1.5 | GO:0019894 | kinesin binding(GO:0019894) |
| 0.0 | 2.8 | GO:0005179 | hormone activity(GO:0005179) |
| 0.0 | 6.5 | GO:0008017 | microtubule binding(GO:0008017) |
| 0.0 | 0.3 | GO:0004602 | glutathione peroxidase activity(GO:0004602) |
| 0.0 | 2.3 | GO:0016765 | transferase activity, transferring alkyl or aryl (other than methyl) groups(GO:0016765) |
| 0.0 | 0.3 | GO:0003876 | AMP deaminase activity(GO:0003876) adenosine-phosphate deaminase activity(GO:0047623) |
| 0.0 | 2.7 | GO:0004860 | protein kinase inhibitor activity(GO:0004860) |
| 0.0 | 11.4 | GO:0004674 | protein serine/threonine kinase activity(GO:0004674) |
| 0.0 | 0.4 | GO:0045295 | gamma-catenin binding(GO:0045295) |
| 0.0 | 0.1 | GO:0003963 | RNA-3'-phosphate cyclase activity(GO:0003963) |
| 0.0 | 0.1 | GO:0045545 | syndecan binding(GO:0045545) |
| 0.0 | 0.1 | GO:0005135 | interleukin-3 receptor binding(GO:0005135) |
| 0.0 | 0.6 | GO:0008307 | structural constituent of muscle(GO:0008307) |
| 0.0 | 0.1 | GO:0004735 | pyrroline-5-carboxylate reductase activity(GO:0004735) |
| 0.0 | 0.7 | GO:0051537 | 2 iron, 2 sulfur cluster binding(GO:0051537) |
| 0.0 | 0.1 | GO:0009019 | tRNA (guanine-N1-)-methyltransferase activity(GO:0009019) |
| 0.0 | 0.2 | GO:0004459 | L-lactate dehydrogenase activity(GO:0004459) |
| 0.0 | 0.1 | GO:0008493 | tetracycline transporter activity(GO:0008493) |
| 0.0 | 2.3 | GO:0002020 | protease binding(GO:0002020) |
| 0.0 | 3.2 | GO:0004252 | serine-type endopeptidase activity(GO:0004252) |
| 0.0 | 0.1 | GO:0034452 | dynactin binding(GO:0034452) |
| 0.0 | 1.5 | GO:0044325 | ion channel binding(GO:0044325) |
| 0.0 | 0.1 | GO:0046974 | histone methyltransferase activity (H3-K9 specific)(GO:0046974) |
| 0.0 | 0.3 | GO:0005245 | voltage-gated calcium channel activity(GO:0005245) |
| 0.0 | 0.1 | GO:0045322 | unmethylated CpG binding(GO:0045322) |
| 0.0 | 0.1 | GO:0045294 | alpha-catenin binding(GO:0045294) |
| 0.0 | 0.8 | GO:0035064 | methylated histone binding(GO:0035064) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.7 | 5.9 | ST PAC1 RECEPTOR PATHWAY | PAC1 Receptor Pathway |
| 0.3 | 21.2 | PID REELIN PATHWAY | Reelin signaling pathway |
| 0.3 | 7.7 | PID ARF 3PATHWAY | Arf1 pathway |
| 0.2 | 8.8 | PID RAS PATHWAY | Regulation of Ras family activation |
| 0.2 | 18.9 | PID TRKR PATHWAY | Neurotrophic factor-mediated Trk receptor signaling |
| 0.2 | 14.2 | PID EPHB FWD PATHWAY | EPHB forward signaling |
| 0.2 | 9.1 | PID CDC42 REG PATHWAY | Regulation of CDC42 activity |
| 0.2 | 4.6 | PID ERBB NETWORK PATHWAY | ErbB receptor signaling network |
| 0.1 | 5.5 | PID MAPK TRK PATHWAY | Trk receptor signaling mediated by the MAPK pathway |
| 0.1 | 11.3 | PID CDC42 PATHWAY | CDC42 signaling events |
| 0.1 | 5.9 | PID NCADHERIN PATHWAY | N-cadherin signaling events |
| 0.1 | 3.1 | PID AVB3 OPN PATHWAY | Osteopontin-mediated events |
| 0.1 | 4.8 | ST T CELL SIGNAL TRANSDUCTION | T Cell Signal Transduction |
| 0.1 | 5.6 | PID RXR VDR PATHWAY | RXR and RAR heterodimerization with other nuclear receptor |
| 0.1 | 1.2 | PID FAS PATHWAY | FAS (CD95) signaling pathway |
| 0.1 | 2.3 | PID NETRIN PATHWAY | Netrin-mediated signaling events |
| 0.1 | 2.6 | PID AR TF PATHWAY | Regulation of Androgen receptor activity |
| 0.1 | 4.9 | PID LKB1 PATHWAY | LKB1 signaling events |
| 0.1 | 1.2 | SA CASPASE CASCADE | Apoptosis is mediated by caspases, cysteine proteases arranged in a proteolytic cascade. |
| 0.1 | 4.2 | PID HDAC CLASSII PATHWAY | Signaling events mediated by HDAC Class II |
| 0.1 | 0.3 | PID A6B1 A6B4 INTEGRIN PATHWAY | a6b1 and a6b4 Integrin signaling |
| 0.1 | 1.6 | PID PI3KCI AKT PATHWAY | Class I PI3K signaling events mediated by Akt |
| 0.1 | 0.5 | PID TXA2PATHWAY | Thromboxane A2 receptor signaling |
| 0.1 | 2.5 | PID TAP63 PATHWAY | Validated transcriptional targets of TAp63 isoforms |
| 0.1 | 1.9 | ST ERK1 ERK2 MAPK PATHWAY | ERK1/ERK2 MAPK Pathway |
| 0.1 | 0.6 | PID P38 GAMMA DELTA PATHWAY | Signaling mediated by p38-gamma and p38-delta |
| 0.1 | 11.6 | NABA ECM AFFILIATED | Genes encoding proteins affiliated structurally or functionally to extracellular matrix proteins |
| 0.1 | 1.0 | PID IL6 7 PATHWAY | IL6-mediated signaling events |
| 0.1 | 0.9 | PID ECADHERIN KERATINOCYTE PATHWAY | E-cadherin signaling in keratinocytes |
| 0.1 | 1.8 | PID IL12 STAT4 PATHWAY | IL12 signaling mediated by STAT4 |
| 0.0 | 2.9 | PID ERBB1 DOWNSTREAM PATHWAY | ErbB1 downstream signaling |
| 0.0 | 3.3 | PID P75 NTR PATHWAY | p75(NTR)-mediated signaling |
| 0.0 | 1.0 | PID LIS1 PATHWAY | Lissencephaly gene (LIS1) in neuronal migration and development |
| 0.0 | 1.9 | PID ATR PATHWAY | ATR signaling pathway |
| 0.0 | 1.1 | SIG INSULIN RECEPTOR PATHWAY IN CARDIAC MYOCYTES | Genes related to the insulin receptor pathway |
| 0.0 | 0.5 | PID AMB2 NEUTROPHILS PATHWAY | amb2 Integrin signaling |
| 0.0 | 1.4 | NABA PROTEOGLYCANS | Genes encoding proteoglycans |
| 0.0 | 2.1 | PID BETA CATENIN NUC PATHWAY | Regulation of nuclear beta catenin signaling and target gene transcription |
| 0.0 | 0.5 | PID CIRCADIAN PATHWAY | Circadian rhythm pathway |
| 0.0 | 0.5 | PID BETA CATENIN DEG PATHWAY | Degradation of beta catenin |
| 0.0 | 0.3 | ST DIFFERENTIATION PATHWAY IN PC12 CELLS | Differentiation Pathway in PC12 Cells; this is a specific case of PAC1 Receptor Pathway. |
| 0.0 | 1.1 | PID ATF2 PATHWAY | ATF-2 transcription factor network |
| 0.0 | 0.5 | PID RET PATHWAY | Signaling events regulated by Ret tyrosine kinase |
| 0.0 | 0.4 | PID ALK1 PATHWAY | ALK1 signaling events |
| 0.0 | 0.2 | PID IL12 2PATHWAY | IL12-mediated signaling events |
| 0.0 | 0.4 | PID BARD1 PATHWAY | BARD1 signaling events |
| 0.0 | 0.6 | PID ARF6 TRAFFICKING PATHWAY | Arf6 trafficking events |
| 0.0 | 0.2 | PID NFKAPPAB CANONICAL PATHWAY | Canonical NF-kappaB pathway |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.8 | 3.1 | REACTOME DIGESTION OF DIETARY CARBOHYDRATE | Genes involved in Digestion of dietary carbohydrate |
| 0.6 | 18.6 | REACTOME CGMP EFFECTS | Genes involved in cGMP effects |
| 0.5 | 18.3 | REACTOME UNBLOCKING OF NMDA RECEPTOR GLUTAMATE BINDING AND ACTIVATION | Genes involved in Unblocking of NMDA receptor, glutamate binding and activation |
| 0.5 | 7.1 | REACTOME ACETYLCHOLINE NEUROTRANSMITTER RELEASE CYCLE | Genes involved in Acetylcholine Neurotransmitter Release Cycle |
| 0.5 | 23.8 | REACTOME GLYCOLYSIS | Genes involved in Glycolysis |
| 0.4 | 5.5 | REACTOME PEPTIDE HORMONE BIOSYNTHESIS | Genes involved in Peptide hormone biosynthesis |
| 0.3 | 3.0 | REACTOME INHIBITION OF INSULIN SECRETION BY ADRENALINE NORADRENALINE | Genes involved in Inhibition of Insulin Secretion by Adrenaline/Noradrenaline |
| 0.3 | 6.1 | REACTOME IONOTROPIC ACTIVITY OF KAINATE RECEPTORS | Genes involved in Ionotropic activity of Kainate Receptors |
| 0.3 | 7.3 | REACTOME SIGNAL ATTENUATION | Genes involved in Signal attenuation |
| 0.3 | 10.1 | REACTOME A TETRASACCHARIDE LINKER SEQUENCE IS REQUIRED FOR GAG SYNTHESIS | Genes involved in A tetrasaccharide linker sequence is required for GAG synthesis |
| 0.3 | 2.3 | REACTOME ROLE OF SECOND MESSENGERS IN NETRIN1 SIGNALING | Genes involved in Role of second messengers in netrin-1 signaling |
| 0.3 | 2.9 | REACTOME RECYCLING OF BILE ACIDS AND SALTS | Genes involved in Recycling of bile acids and salts |
| 0.2 | 3.7 | REACTOME PRESYNAPTIC NICOTINIC ACETYLCHOLINE RECEPTORS | Genes involved in Presynaptic nicotinic acetylcholine receptors |
| 0.2 | 3.3 | REACTOME CYTOSOLIC SULFONATION OF SMALL MOLECULES | Genes involved in Cytosolic sulfonation of small molecules |
| 0.2 | 2.6 | REACTOME GLUTAMATE NEUROTRANSMITTER RELEASE CYCLE | Genes involved in Glutamate Neurotransmitter Release Cycle |
| 0.2 | 7.1 | REACTOME SYNTHESIS OF PC | Genes involved in Synthesis of PC |
| 0.2 | 5.7 | REACTOME N GLYCAN ANTENNAE ELONGATION | Genes involved in N-Glycan antennae elongation |
| 0.2 | 13.4 | REACTOME VOLTAGE GATED POTASSIUM CHANNELS | Genes involved in Voltage gated Potassium channels |
| 0.2 | 3.9 | REACTOME MTORC1 MEDIATED SIGNALLING | Genes involved in mTORC1-mediated signalling |
| 0.2 | 3.3 | REACTOME CLASS C 3 METABOTROPIC GLUTAMATE PHEROMONE RECEPTORS | Genes involved in Class C/3 (Metabotropic glutamate/pheromone receptors) |
| 0.2 | 7.7 | REACTOME DARPP 32 EVENTS | Genes involved in DARPP-32 events |
| 0.2 | 8.3 | REACTOME AMINE LIGAND BINDING RECEPTORS | Genes involved in Amine ligand-binding receptors |
| 0.2 | 4.6 | REACTOME GABA A RECEPTOR ACTIVATION | Genes involved in GABA A receptor activation |
| 0.2 | 6.3 | REACTOME KINESINS | Genes involved in Kinesins |
| 0.2 | 5.3 | REACTOME OTHER SEMAPHORIN INTERACTIONS | Genes involved in Other semaphorin interactions |
| 0.2 | 4.3 | REACTOME PLATELET CALCIUM HOMEOSTASIS | Genes involved in Platelet calcium homeostasis |
| 0.2 | 3.5 | REACTOME POST CHAPERONIN TUBULIN FOLDING PATHWAY | Genes involved in Post-chaperonin tubulin folding pathway |
| 0.2 | 3.9 | REACTOME GAP JUNCTION ASSEMBLY | Genes involved in Gap junction assembly |
| 0.2 | 5.6 | REACTOME INSULIN RECEPTOR RECYCLING | Genes involved in Insulin receptor recycling |
| 0.2 | 9.1 | REACTOME INHIBITION OF VOLTAGE GATED CA2 CHANNELS VIA GBETA GAMMA SUBUNITS | Genes involved in Inhibition of voltage gated Ca2+ channels via Gbeta/gamma subunits |
| 0.2 | 2.8 | REACTOME G PROTEIN ACTIVATION | Genes involved in G-protein activation |
| 0.1 | 2.5 | REACTOME GAP JUNCTION TRAFFICKING | Genes involved in Gap junction trafficking |
| 0.1 | 2.3 | REACTOME TANDEM PORE DOMAIN POTASSIUM CHANNELS | Genes involved in Tandem pore domain potassium channels |
| 0.1 | 3.6 | REACTOME HS GAG BIOSYNTHESIS | Genes involved in HS-GAG biosynthesis |
| 0.1 | 5.5 | REACTOME ERK MAPK TARGETS | Genes involved in ERK/MAPK targets |
| 0.1 | 0.8 | REACTOME NITRIC OXIDE STIMULATES GUANYLATE CYCLASE | Genes involved in Nitric oxide stimulates guanylate cyclase |
| 0.1 | 4.0 | REACTOME DEGRADATION OF THE EXTRACELLULAR MATRIX | Genes involved in Degradation of the extracellular matrix |
| 0.1 | 10.2 | REACTOME NUCLEAR SIGNALING BY ERBB4 | Genes involved in Nuclear signaling by ERBB4 |
| 0.1 | 2.0 | REACTOME HDL MEDIATED LIPID TRANSPORT | Genes involved in HDL-mediated lipid transport |
| 0.1 | 3.1 | REACTOME ABCA TRANSPORTERS IN LIPID HOMEOSTASIS | Genes involved in ABCA transporters in lipid homeostasis |
| 0.1 | 4.9 | REACTOME NEPHRIN INTERACTIONS | Genes involved in Nephrin interactions |
| 0.1 | 1.2 | REACTOME COPI MEDIATED TRANSPORT | Genes involved in COPI Mediated Transport |
| 0.1 | 1.9 | REACTOME ANTIGEN PRESENTATION FOLDING ASSEMBLY AND PEPTIDE LOADING OF CLASS I MHC | Genes involved in Antigen Presentation: Folding, assembly and peptide loading of class I MHC |
| 0.1 | 1.2 | REACTOME ACTIVATION OF RAC | Genes involved in Activation of Rac |
| 0.1 | 14.5 | REACTOME NGF SIGNALLING VIA TRKA FROM THE PLASMA MEMBRANE | Genes involved in NGF signalling via TRKA from the plasma membrane |
| 0.1 | 5.3 | REACTOME ACTIVATION OF CHAPERONE GENES BY XBP1S | Genes involved in Activation of Chaperone Genes by XBP1(S) |
| 0.1 | 2.4 | REACTOME CITRIC ACID CYCLE TCA CYCLE | Genes involved in Citric acid cycle (TCA cycle) |
| 0.1 | 2.2 | REACTOME ACYL CHAIN REMODELLING OF PG | Genes involved in Acyl chain remodelling of PG |
| 0.1 | 2.5 | REACTOME ZINC TRANSPORTERS | Genes involved in Zinc transporters |
| 0.1 | 16.5 | REACTOME PEPTIDE LIGAND BINDING RECEPTORS | Genes involved in Peptide ligand-binding receptors |
| 0.1 | 4.7 | REACTOME NCAM1 INTERACTIONS | Genes involved in NCAM1 interactions |
| 0.1 | 3.3 | REACTOME TRANSPORT OF RIBONUCLEOPROTEINS INTO THE HOST NUCLEUS | Genes involved in Transport of Ribonucleoproteins into the Host Nucleus |
| 0.1 | 1.0 | REACTOME REGULATION OF THE FANCONI ANEMIA PATHWAY | Genes involved in Regulation of the Fanconi anemia pathway |
| 0.1 | 1.8 | REACTOME NUCLEOTIDE LIKE PURINERGIC RECEPTORS | Genes involved in Nucleotide-like (purinergic) receptors |
| 0.1 | 2.5 | REACTOME SEMA4D INDUCED CELL MIGRATION AND GROWTH CONE COLLAPSE | Genes involved in Sema4D induced cell migration and growth-cone collapse |
| 0.1 | 2.1 | REACTOME CYTOSOLIC TRNA AMINOACYLATION | Genes involved in Cytosolic tRNA aminoacylation |
| 0.1 | 2.6 | REACTOME NETRIN1 SIGNALING | Genes involved in Netrin-1 signaling |
| 0.1 | 1.8 | REACTOME SIGNALING BY ROBO RECEPTOR | Genes involved in Signaling by Robo receptor |
| 0.1 | 0.7 | REACTOME MRNA CAPPING | Genes involved in mRNA Capping |
| 0.1 | 0.5 | REACTOME PROSTANOID LIGAND RECEPTORS | Genes involved in Prostanoid ligand receptors |
| 0.1 | 0.4 | REACTOME NA CL DEPENDENT NEUROTRANSMITTER TRANSPORTERS | Genes involved in Na+/Cl- dependent neurotransmitter transporters |
| 0.1 | 0.2 | REACTOME DOWNREGULATION OF ERBB2 ERBB3 SIGNALING | Genes involved in Downregulation of ERBB2:ERBB3 signaling |
| 0.0 | 0.7 | REACTOME METAL ION SLC TRANSPORTERS | Genes involved in Metal ion SLC transporters |
| 0.0 | 0.6 | REACTOME LIPOPROTEIN METABOLISM | Genes involved in Lipoprotein metabolism |
| 0.0 | 0.4 | REACTOME SIGNALING BY SCF KIT | Genes involved in Signaling by SCF-KIT |
| 0.0 | 0.4 | REACTOME JNK C JUN KINASES PHOSPHORYLATION AND ACTIVATION MEDIATED BY ACTIVATED HUMAN TAK1 | Genes involved in JNK (c-Jun kinases) phosphorylation and activation mediated by activated human TAK1 |
| 0.0 | 3.3 | REACTOME NUCLEAR RECEPTOR TRANSCRIPTION PATHWAY | Genes involved in Nuclear Receptor transcription pathway |
| 0.0 | 1.5 | REACTOME GLYCOSPHINGOLIPID METABOLISM | Genes involved in Glycosphingolipid metabolism |
| 0.0 | 0.6 | REACTOME MRNA DECAY BY 5 TO 3 EXORIBONUCLEASE | Genes involved in mRNA Decay by 5' to 3' Exoribonuclease |
| 0.0 | 2.6 | REACTOME FACTORS INVOLVED IN MEGAKARYOCYTE DEVELOPMENT AND PLATELET PRODUCTION | Genes involved in Factors involved in megakaryocyte development and platelet production |
| 0.0 | 1.0 | REACTOME NOD1 2 SIGNALING PATHWAY | Genes involved in NOD1/2 Signaling Pathway |
| 0.0 | 1.6 | REACTOME MRNA SPLICING MINOR PATHWAY | Genes involved in mRNA Splicing - Minor Pathway |
| 0.0 | 0.7 | REACTOME AMINO ACID SYNTHESIS AND INTERCONVERSION TRANSAMINATION | Genes involved in Amino acid synthesis and interconversion (transamination) |
| 0.0 | 0.5 | REACTOME BILE SALT AND ORGANIC ANION SLC TRANSPORTERS | Genes involved in Bile salt and organic anion SLC transporters |
| 0.0 | 1.5 | REACTOME ASSOCIATION OF TRIC CCT WITH TARGET PROTEINS DURING BIOSYNTHESIS | Genes involved in Association of TriC/CCT with target proteins during biosynthesis |
| 0.0 | 0.7 | REACTOME RNA POL III TRANSCRIPTION INITIATION FROM TYPE 3 PROMOTER | Genes involved in RNA Polymerase III Transcription Initiation From Type 3 Promoter |
| 0.0 | 0.6 | REACTOME PROCESSING OF INTRONLESS PRE MRNAS | Genes involved in Processing of Intronless Pre-mRNAs |
| 0.0 | 0.5 | REACTOME BMAL1 CLOCK NPAS2 ACTIVATES CIRCADIAN EXPRESSION | Genes involved in BMAL1:CLOCK/NPAS2 Activates Circadian Expression |
| 0.0 | 0.5 | REACTOME PHOSPHORYLATION OF THE APC C | Genes involved in Phosphorylation of the APC/C |
| 0.0 | 0.1 | REACTOME HEPARAN SULFATE HEPARIN HS GAG METABOLISM | Genes involved in Heparan sulfate/heparin (HS-GAG) metabolism |
| 0.0 | 0.8 | REACTOME MYOGENESIS | Genes involved in Myogenesis |
| 0.0 | 1.3 | REACTOME NRAGE SIGNALS DEATH THROUGH JNK | Genes involved in NRAGE signals death through JNK |
| 0.0 | 0.5 | REACTOME SYNTHESIS AND INTERCONVERSION OF NUCLEOTIDE DI AND TRIPHOSPHATES | Genes involved in Synthesis and interconversion of nucleotide di- and triphosphates |
| 0.0 | 0.2 | REACTOME PLATELET HOMEOSTASIS | Genes involved in Platelet homeostasis |
| 0.0 | 0.3 | REACTOME GLUCAGON TYPE LIGAND RECEPTORS | Genes involved in Glucagon-type ligand receptors |
| 0.0 | 1.1 | REACTOME L1CAM INTERACTIONS | Genes involved in L1CAM interactions |
| 0.0 | 0.2 | REACTOME PYRUVATE METABOLISM | Genes involved in Pyruvate metabolism |