Project
PRJNA375882: Comprehensive Mouse Transcriptomic BodyMap
Navigation
Downloads
Results for Pax7
Z-value: 0.66
Transcription factors associated with Pax7
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Pax7
|
ENSMUSG00000028736.14 | Pax7 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Pax7 | mm39_v1_chr4_-_139560229_139560318 | -0.19 | 1.2e-01 | Click! |
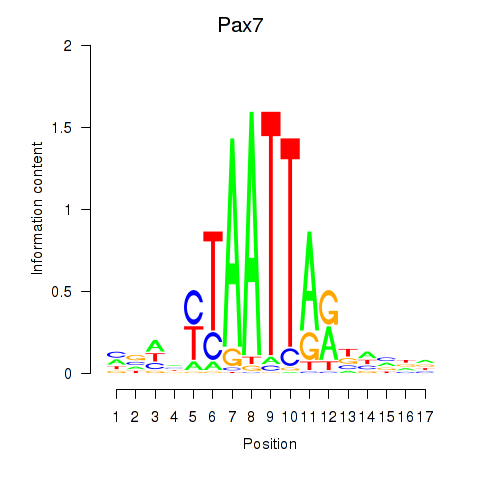
Activity profile of Pax7 motif
Sorted Z-values of Pax7 motif
Network of associatons between targets according to the STRING database.
First level regulatory network of Pax7
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr3_+_93301003 | 5.07 |
ENSMUST00000045912.3
|
Rptn
|
repetin |
| chrX_+_9751861 | 3.68 |
ENSMUST00000067529.9
ENSMUST00000086165.4 |
Sytl5
|
synaptotagmin-like 5 |
| chr19_-_15902292 | 2.45 |
ENSMUST00000025542.10
|
Psat1
|
phosphoserine aminotransferase 1 |
| chr13_-_78344492 | 2.38 |
ENSMUST00000125176.3
|
Nr2f1
|
nuclear receptor subfamily 2, group F, member 1 |
| chr19_-_15901919 | 2.31 |
ENSMUST00000162053.8
|
Psat1
|
phosphoserine aminotransferase 1 |
| chr9_-_20871081 | 2.31 |
ENSMUST00000177754.9
|
Dnmt1
|
DNA methyltransferase (cytosine-5) 1 |
| chr2_+_27055245 | 2.16 |
ENSMUST00000000910.7
|
Dbh
|
dopamine beta hydroxylase |
| chr11_-_115167775 | 2.11 |
ENSMUST00000021078.3
|
Fdxr
|
ferredoxin reductase |
| chr10_+_29074950 | 2.10 |
ENSMUST00000217011.2
|
Gm49353
|
predicted gene, 49353 |
| chr3_+_62327089 | 2.05 |
ENSMUST00000161057.2
|
Arhgef26
|
Rho guanine nucleotide exchange factor (GEF) 26 |
| chr11_-_87249837 | 1.98 |
ENSMUST00000055438.5
|
Ppm1e
|
protein phosphatase 1E (PP2C domain containing) |
| chr10_-_62215631 | 1.97 |
ENSMUST00000143236.8
ENSMUST00000133429.8 ENSMUST00000132926.8 ENSMUST00000116238.9 |
Hk1
|
hexokinase 1 |
| chr9_-_71070506 | 1.96 |
ENSMUST00000074465.9
|
Aqp9
|
aquaporin 9 |
| chr13_+_76727787 | 1.73 |
ENSMUST00000126960.8
ENSMUST00000109583.9 |
Mctp1
|
multiple C2 domains, transmembrane 1 |
| chr6_-_87510200 | 1.70 |
ENSMUST00000113637.9
ENSMUST00000071024.7 |
Arhgap25
|
Rho GTPase activating protein 25 |
| chr3_+_55369149 | 1.64 |
ENSMUST00000199585.5
ENSMUST00000070418.9 |
Dclk1
|
doublecortin-like kinase 1 |
| chr15_-_64794139 | 1.60 |
ENSMUST00000023007.7
ENSMUST00000228014.2 |
Adcy8
|
adenylate cyclase 8 |
| chr1_-_158183894 | 1.58 |
ENSMUST00000004133.11
|
Brinp2
|
bone morphogenic protein/retinoic acid inducible neural-specific 2 |
| chr1_-_55265925 | 1.52 |
ENSMUST00000027121.15
ENSMUST00000114428.3 |
Rftn2
|
raftlin family member 2 |
| chr2_+_65451100 | 1.50 |
ENSMUST00000144254.6
ENSMUST00000028377.14 |
Scn2a
|
sodium channel, voltage-gated, type II, alpha |
| chr2_+_124910037 | 1.37 |
ENSMUST00000070353.4
|
Slc24a5
|
solute carrier family 24, member 5 |
| chr16_+_22737128 | 1.34 |
ENSMUST00000170805.9
|
Fetub
|
fetuin beta |
| chr6_+_123239076 | 1.32 |
ENSMUST00000032240.4
|
Clec4d
|
C-type lectin domain family 4, member d |
| chr16_+_22737050 | 1.30 |
ENSMUST00000231768.2
|
Fetub
|
fetuin beta |
| chr15_+_9436114 | 1.29 |
ENSMUST00000042360.5
ENSMUST00000226688.2 |
Capsl
|
calcyphosine-like |
| chr1_-_182110303 | 1.25 |
ENSMUST00000035295.6
|
Degs1
|
delta(4)-desaturase, sphingolipid 1 |
| chr14_+_32507920 | 1.21 |
ENSMUST00000039191.8
ENSMUST00000227060.2 ENSMUST00000228481.2 |
Tmem273
|
transmembrane protein 273 |
| chr4_-_14621805 | 1.18 |
ENSMUST00000042221.14
|
Slc26a7
|
solute carrier family 26, member 7 |
| chr5_-_138169253 | 1.14 |
ENSMUST00000139983.8
|
Mcm7
|
minichromosome maintenance complex component 7 |
| chr16_+_22737227 | 1.06 |
ENSMUST00000231880.2
|
Fetub
|
fetuin beta |
| chr1_-_131441962 | 1.06 |
ENSMUST00000185445.3
|
Srgap2
|
SLIT-ROBO Rho GTPase activating protein 2 |
| chr17_-_43053057 | 1.04 |
ENSMUST00000239223.2
ENSMUST00000113614.3 |
Adgrf2
|
adhesion G protein-coupled receptor F2 |
| chr3_-_106697459 | 1.01 |
ENSMUST00000038845.10
|
Cd53
|
CD53 antigen |
| chr10_+_85222677 | 1.00 |
ENSMUST00000105307.8
ENSMUST00000020231.10 |
Btbd11
|
BTB (POZ) domain containing 11 |
| chr13_-_103042554 | 0.99 |
ENSMUST00000171791.8
|
Mast4
|
microtubule associated serine/threonine kinase family member 4 |
| chr19_+_8779903 | 0.95 |
ENSMUST00000172175.3
|
Zbtb3
|
zinc finger and BTB domain containing 3 |
| chr13_-_103042294 | 0.93 |
ENSMUST00000167462.8
|
Mast4
|
microtubule associated serine/threonine kinase family member 4 |
| chr12_+_117807224 | 0.91 |
ENSMUST00000021592.16
|
Cdca7l
|
cell division cycle associated 7 like |
| chr15_-_66985760 | 0.90 |
ENSMUST00000092640.6
|
St3gal1
|
ST3 beta-galactoside alpha-2,3-sialyltransferase 1 |
| chr17_+_79919267 | 0.90 |
ENSMUST00000223924.2
|
Rmdn2
|
regulator of microtubule dynamics 2 |
| chr14_+_65612788 | 0.89 |
ENSMUST00000224687.2
|
Zfp395
|
zinc finger protein 395 |
| chr11_+_6366259 | 0.88 |
ENSMUST00000213200.2
|
Ppia
|
peptidylprolyl isomerase A |
| chr14_+_65504067 | 0.87 |
ENSMUST00000224629.2
|
Fbxo16
|
F-box protein 16 |
| chr12_+_37930169 | 0.85 |
ENSMUST00000221176.2
|
Dgkb
|
diacylglycerol kinase, beta |
| chr11_-_29197222 | 0.83 |
ENSMUST00000020754.10
|
Cfap36
|
cilia and flagella associated protein 36 |
| chr2_+_124978518 | 0.80 |
ENSMUST00000238754.2
|
Ctxn2
|
cortexin 2 |
| chr19_-_46033353 | 0.79 |
ENSMUST00000026252.14
ENSMUST00000156585.9 ENSMUST00000185355.7 ENSMUST00000152946.8 |
Ldb1
|
LIM domain binding 1 |
| chr8_-_49008305 | 0.78 |
ENSMUST00000110346.9
ENSMUST00000211976.2 |
Tenm3
|
teneurin transmembrane protein 3 |
| chr13_-_24118139 | 0.77 |
ENSMUST00000052776.4
|
H2bc1
|
H2B clustered histone 1 |
| chr6_-_129449739 | 0.74 |
ENSMUST00000112076.9
ENSMUST00000184581.3 |
Clec7a
|
C-type lectin domain family 7, member a |
| chr2_-_34990689 | 0.74 |
ENSMUST00000226631.2
ENSMUST00000045776.5 ENSMUST00000226972.2 |
AI182371
|
expressed sequence AI182371 |
| chr15_-_79658608 | 0.72 |
ENSMUST00000229644.2
ENSMUST00000023055.8 |
Dnal4
|
dynein, axonemal, light chain 4 |
| chr15_-_79658584 | 0.67 |
ENSMUST00000069877.12
|
Dnal4
|
dynein, axonemal, light chain 4 |
| chr2_+_124978612 | 0.65 |
ENSMUST00000099452.3
ENSMUST00000238377.2 |
Ctxn2
|
cortexin 2 |
| chr18_-_77134980 | 0.63 |
ENSMUST00000154665.2
ENSMUST00000026486.13 ENSMUST00000123650.2 ENSMUST00000126153.8 |
Katnal2
|
katanin p60 subunit A-like 2 |
| chr9_+_21634779 | 0.63 |
ENSMUST00000034713.9
|
Ldlr
|
low density lipoprotein receptor |
| chr11_+_43572825 | 0.62 |
ENSMUST00000061070.6
ENSMUST00000094294.5 |
Pwwp2a
|
PWWP domain containing 2A |
| chr14_-_76248274 | 0.59 |
ENSMUST00000088922.5
|
Gtf2f2
|
general transcription factor IIF, polypeptide 2 |
| chr1_+_100025486 | 0.58 |
ENSMUST00000188735.2
|
Cntnap5b
|
contactin associated protein-like 5B |
| chr9_-_96601574 | 0.52 |
ENSMUST00000128269.8
|
Zbtb38
|
zinc finger and BTB domain containing 38 |
| chr13_+_76727799 | 0.51 |
ENSMUST00000109589.3
|
Mctp1
|
multiple C2 domains, transmembrane 1 |
| chr18_-_77134939 | 0.50 |
ENSMUST00000137354.8
ENSMUST00000137498.8 |
Katnal2
|
katanin p60 subunit A-like 2 |
| chr1_-_144427302 | 0.49 |
ENSMUST00000184189.3
|
Rgs21
|
regulator of G-protein signalling 21 |
| chr4_+_136038263 | 0.49 |
ENSMUST00000105850.8
ENSMUST00000148843.10 |
Hnrnpr
|
heterogeneous nuclear ribonucleoprotein R |
| chr9_+_21867043 | 0.48 |
ENSMUST00000053583.7
|
Swsap1
|
SWIM type zinc finger 7 associated protein 1 |
| chr7_+_45271229 | 0.45 |
ENSMUST00000033100.5
|
Izumo1
|
izumo sperm-egg fusion 1 |
| chrX_-_156381652 | 0.45 |
ENSMUST00000149249.2
ENSMUST00000058098.15 |
Mbtps2
|
membrane-bound transcription factor peptidase, site 2 |
| chr3_+_32490525 | 0.44 |
ENSMUST00000108242.2
|
Pik3ca
|
phosphatidylinositol-4,5-bisphosphate 3-kinase catalytic subunit alpha |
| chr5_+_104350475 | 0.44 |
ENSMUST00000066708.7
|
Dmp1
|
dentin matrix protein 1 |
| chr3_+_32490300 | 0.43 |
ENSMUST00000029201.14
|
Pik3ca
|
phosphatidylinositol-4,5-bisphosphate 3-kinase catalytic subunit alpha |
| chr4_-_14621497 | 0.43 |
ENSMUST00000149633.2
|
Slc26a7
|
solute carrier family 26, member 7 |
| chr9_+_102597486 | 0.42 |
ENSMUST00000130602.2
|
Amotl2
|
angiomotin-like 2 |
| chr4_+_136038301 | 0.40 |
ENSMUST00000084219.12
|
Hnrnpr
|
heterogeneous nuclear ribonucleoprotein R |
| chr2_-_111965322 | 0.39 |
ENSMUST00000213696.2
|
Olfr1316
|
olfactory receptor 1316 |
| chr4_-_43710231 | 0.38 |
ENSMUST00000217544.2
ENSMUST00000107862.3 |
Olfr71
|
olfactory receptor 71 |
| chr10_+_129493563 | 0.36 |
ENSMUST00000217094.2
|
Olfr800
|
olfactory receptor 800 |
| chr4_-_154721288 | 0.36 |
ENSMUST00000030902.13
ENSMUST00000105637.8 ENSMUST00000070313.14 ENSMUST00000105636.8 ENSMUST00000105638.9 ENSMUST00000097759.9 ENSMUST00000124771.2 |
Prdm16
|
PR domain containing 16 |
| chr17_-_35827676 | 0.36 |
ENSMUST00000160885.2
ENSMUST00000159009.2 ENSMUST00000161012.8 |
Tcf19
|
transcription factor 19 |
| chr18_+_39126325 | 0.35 |
ENSMUST00000137497.9
|
Arhgap26
|
Rho GTPase activating protein 26 |
| chr4_+_136038243 | 0.35 |
ENSMUST00000131671.8
|
Hnrnpr
|
heterogeneous nuclear ribonucleoprotein R |
| chr18_+_39126178 | 0.34 |
ENSMUST00000097593.9
|
Arhgap26
|
Rho GTPase activating protein 26 |
| chr6_-_8180174 | 0.33 |
ENSMUST00000213284.2
|
Col28a1
|
collagen, type XXVIII, alpha 1 |
| chr19_-_41921676 | 0.32 |
ENSMUST00000075280.12
ENSMUST00000112123.4 |
Exosc1
|
exosome component 1 |
| chr14_-_26184767 | 0.31 |
ENSMUST00000146438.2
|
Slmap
|
sarcolemma associated protein |
| chr3_-_75177378 | 0.30 |
ENSMUST00000039047.5
|
Serpini2
|
serine (or cysteine) peptidase inhibitor, clade I, member 2 |
| chr12_+_72488625 | 0.28 |
ENSMUST00000161284.3
ENSMUST00000162159.8 |
Lrrc9
|
leucine rich repeat containing 9 |
| chr14_-_76348179 | 0.28 |
ENSMUST00000022585.5
|
Gpalpp1
|
GPALPP motifs containing 1 |
| chr3_+_96175970 | 0.25 |
ENSMUST00000098843.3
|
H3c13
|
H3 clustered histone 13 |
| chr9_+_50686647 | 0.24 |
ENSMUST00000159576.2
|
Alg9
|
asparagine-linked glycosylation 9 (alpha 1,2 mannosyltransferase) |
| chr7_+_126832399 | 0.24 |
ENSMUST00000056232.7
|
Zfp553
|
zinc finger protein 553 |
| chr7_-_98305737 | 0.23 |
ENSMUST00000205911.2
ENSMUST00000038359.6 ENSMUST00000206611.2 ENSMUST00000206619.2 |
Emsy
|
EMSY, BRCA2-interacting transcriptional repressor |
| chr2_-_37249247 | 0.22 |
ENSMUST00000112940.8
ENSMUST00000009174.15 |
Pdcl
|
phosducin-like |
| chr7_-_98305986 | 0.21 |
ENSMUST00000205276.2
|
Emsy
|
EMSY, BRCA2-interacting transcriptional repressor |
| chr2_-_88157559 | 0.18 |
ENSMUST00000214207.2
|
Olfr1175
|
olfactory receptor 1175 |
| chr2_+_87686206 | 0.16 |
ENSMUST00000217376.2
|
Olfr1151
|
olfactory receptor 1151 |
| chr6_-_57512355 | 0.16 |
ENSMUST00000042766.6
|
Ppm1k
|
protein phosphatase 1K (PP2C domain containing) |
| chr14_+_76348312 | 0.14 |
ENSMUST00000022586.2
|
Nufip1
|
nuclear fragile X mental retardation protein interacting protein 1 |
| chr2_+_87362140 | 0.13 |
ENSMUST00000217113.2
|
Olfr153
|
olfactory receptor 153 |
| chrX_+_159551009 | 0.11 |
ENSMUST00000033650.14
|
Rs1
|
retinoschisis (X-linked, juvenile) 1 (human) |
| chr4_+_102446883 | 0.08 |
ENSMUST00000097949.11
ENSMUST00000106901.2 |
Pde4b
|
phosphodiesterase 4B, cAMP specific |
| chr2_-_86109346 | 0.08 |
ENSMUST00000217294.2
ENSMUST00000217245.2 ENSMUST00000216432.2 |
Olfr1051
|
olfactory receptor 1051 |
| chr11_-_99134885 | 0.06 |
ENSMUST00000103132.10
ENSMUST00000038214.7 |
Krt222
|
keratin 222 |
| chr10_+_127226180 | 0.05 |
ENSMUST00000077046.12
ENSMUST00000105250.9 |
R3hdm2
|
R3H domain containing 2 |
| chr2_+_87687521 | 0.02 |
ENSMUST00000061081.2
|
Olfr1151
|
olfactory receptor 1151 |
| chrX_+_159551171 | 0.02 |
ENSMUST00000112368.3
|
Rs1
|
retinoschisis (X-linked, juvenile) 1 (human) |
| chr2_-_111843053 | 0.02 |
ENSMUST00000213559.3
|
Olfr1310
|
olfactory receptor 1310 |
| chr4_-_14621669 | 0.02 |
ENSMUST00000143105.2
|
Slc26a7
|
solute carrier family 26, member 7 |
| chr8_+_107757847 | 0.01 |
ENSMUST00000034388.10
|
Vps4a
|
vacuolar protein sorting 4A |
| chr4_+_136013372 | 0.00 |
ENSMUST00000069195.5
ENSMUST00000130658.2 |
Zfp46
|
zinc finger protein 46 |
| chr8_-_35432783 | 0.00 |
ENSMUST00000033929.6
|
Tnks
|
tankyrase, TRF1-interacting ankyrin-related ADP-ribose polymerase |
| chr7_-_19449319 | 0.00 |
ENSMUST00000032555.10
ENSMUST00000093552.12 |
Tomm40
|
translocase of outer mitochondrial membrane 40 |
| chr6_+_57133904 | 0.00 |
ENSMUST00000226866.2
ENSMUST00000227581.2 |
Vmn1r12
|
vomeronasal 1 receptor 12 |
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.7 | 2.2 | GO:0046333 | octopamine biosynthetic process(GO:0006589) octopamine metabolic process(GO:0046333) |
| 0.6 | 4.8 | GO:0006564 | L-serine biosynthetic process(GO:0006564) |
| 0.6 | 2.3 | GO:0090309 | C-5 methylation of cytosine(GO:0090116) positive regulation of methylation-dependent chromatin silencing(GO:0090309) |
| 0.5 | 2.0 | GO:0015855 | nucleobase transport(GO:0015851) pyrimidine nucleobase transport(GO:0015855) |
| 0.3 | 1.4 | GO:0048022 | negative regulation of melanin biosynthetic process(GO:0048022) negative regulation of secondary metabolite biosynthetic process(GO:1900377) |
| 0.3 | 0.9 | GO:0044028 | DNA hypomethylation(GO:0044028) hypomethylation of CpG island(GO:0044029) |
| 0.3 | 0.8 | GO:0043973 | histone H3-K4 acetylation(GO:0043973) |
| 0.2 | 2.0 | GO:0061621 | NADH regeneration(GO:0006735) canonical glycolysis(GO:0061621) glucose catabolic process to pyruvate(GO:0061718) |
| 0.2 | 1.6 | GO:0019532 | oxalate transport(GO:0019532) |
| 0.2 | 1.5 | GO:0032596 | protein transport into membrane raft(GO:0032596) |
| 0.2 | 1.3 | GO:0038094 | Fc-gamma receptor signaling pathway(GO:0038094) |
| 0.2 | 0.6 | GO:1905167 | regulation of phosphatidylcholine catabolic process(GO:0010899) receptor-mediated endocytosis of low-density lipoprotein particle involved in cholesterol transport(GO:0090118) positive regulation of lysosomal protein catabolic process(GO:1905167) |
| 0.2 | 0.5 | GO:0036500 | ATF6-mediated unfolded protein response(GO:0036500) |
| 0.1 | 1.5 | GO:0008627 | intrinsic apoptotic signaling pathway in response to osmotic stress(GO:0008627) |
| 0.1 | 1.4 | GO:2000580 | regulation of ATP-dependent microtubule motor activity, plus-end-directed(GO:2000580) positive regulation of ATP-dependent microtubule motor activity, plus-end-directed(GO:2000582) |
| 0.1 | 2.0 | GO:0035970 | peptidyl-threonine dephosphorylation(GO:0035970) |
| 0.1 | 1.1 | GO:0021815 | modulation of microtubule cytoskeleton involved in cerebral cortex radial glia guided migration(GO:0021815) |
| 0.1 | 2.1 | GO:0070995 | NADPH oxidation(GO:0070995) |
| 0.1 | 1.1 | GO:0006268 | DNA unwinding involved in DNA replication(GO:0006268) |
| 0.1 | 2.4 | GO:0021796 | cerebral cortex regionalization(GO:0021796) |
| 0.1 | 0.4 | GO:0070173 | regulation of enamel mineralization(GO:0070173) |
| 0.1 | 2.0 | GO:0001886 | endothelial cell morphogenesis(GO:0001886) |
| 0.1 | 3.7 | GO:0048791 | calcium ion-regulated exocytosis of neurotransmitter(GO:0048791) |
| 0.1 | 4.1 | GO:0035036 | sperm-egg recognition(GO:0035036) |
| 0.1 | 0.3 | GO:1905150 | regulation of voltage-gated sodium channel activity(GO:1905150) |
| 0.0 | 0.8 | GO:0097264 | self proteolysis(GO:0097264) |
| 0.0 | 0.9 | GO:0034389 | lipid particle organization(GO:0034389) |
| 0.0 | 1.2 | GO:0061157 | mRNA destabilization(GO:0061157) |
| 0.0 | 0.4 | GO:0090336 | positive regulation of brown fat cell differentiation(GO:0090336) |
| 0.0 | 0.2 | GO:0061084 | queuosine biosynthetic process(GO:0008616) queuosine metabolic process(GO:0046116) negative regulation of protein refolding(GO:0061084) |
| 0.0 | 0.1 | GO:0036367 | adaptation of rhodopsin mediated signaling(GO:0016062) light adaption(GO:0036367) |
| 0.0 | 1.6 | GO:0021952 | central nervous system projection neuron axonogenesis(GO:0021952) |
| 0.0 | 1.6 | GO:0007616 | long-term memory(GO:0007616) |
| 0.0 | 1.0 | GO:1901741 | positive regulation of myoblast fusion(GO:1901741) |
| 0.0 | 0.6 | GO:0045899 | positive regulation of RNA polymerase II transcriptional preinitiation complex assembly(GO:0045899) |
| 0.0 | 0.8 | GO:0007205 | protein kinase C-activating G-protein coupled receptor signaling pathway(GO:0007205) |
| 0.0 | 1.6 | GO:0071300 | cellular response to retinoic acid(GO:0071300) |
| 0.0 | 1.3 | GO:0046513 | ceramide biosynthetic process(GO:0046513) |
| 0.0 | 0.9 | GO:0018279 | protein N-linked glycosylation via asparagine(GO:0018279) |
| 0.0 | 0.1 | GO:0000492 | box C/D snoRNP assembly(GO:0000492) |
| 0.0 | 0.4 | GO:0035329 | hippo signaling(GO:0035329) |
| 0.0 | 1.0 | GO:0060395 | SMAD protein signal transduction(GO:0060395) |
| 0.0 | 0.4 | GO:0044342 | type B pancreatic cell proliferation(GO:0044342) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.7 | 2.2 | GO:0034774 | secretory granule lumen(GO:0034774) cytoplasmic membrane-bounded vesicle lumen(GO:0060205) |
| 0.3 | 0.9 | GO:0005943 | phosphatidylinositol 3-kinase complex, class IA(GO:0005943) |
| 0.2 | 0.6 | GO:1990666 | PCSK9-LDLR complex(GO:1990666) |
| 0.1 | 5.1 | GO:0001533 | cornified envelope(GO:0001533) |
| 0.1 | 0.6 | GO:0097550 | transcriptional preinitiation complex(GO:0097550) |
| 0.1 | 1.1 | GO:0042555 | MCM complex(GO:0042555) |
| 0.1 | 1.1 | GO:0044327 | dendritic spine head(GO:0044327) |
| 0.1 | 1.6 | GO:0008074 | guanylate cyclase complex, soluble(GO:0008074) |
| 0.1 | 1.5 | GO:0001518 | voltage-gated sodium channel complex(GO:0001518) |
| 0.1 | 2.3 | GO:0005721 | pericentric heterochromatin(GO:0005721) |
| 0.0 | 2.0 | GO:0097228 | sperm principal piece(GO:0097228) |
| 0.0 | 0.3 | GO:0000176 | nuclear exosome (RNase complex)(GO:0000176) |
| 0.0 | 1.4 | GO:0030286 | dynein complex(GO:0030286) |
| 0.0 | 1.0 | GO:0001772 | immunological synapse(GO:0001772) |
| 0.0 | 0.5 | GO:0002080 | acrosomal membrane(GO:0002080) |
| 0.0 | 0.1 | GO:0070761 | pre-snoRNP complex(GO:0070761) |
| 0.0 | 3.8 | GO:0070382 | exocytic vesicle(GO:0070382) |
| 0.0 | 1.0 | GO:0019005 | SCF ubiquitin ligase complex(GO:0019005) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.7 | 2.2 | GO:0004500 | dopamine beta-monooxygenase activity(GO:0004500) |
| 0.4 | 2.0 | GO:0005350 | purine nucleobase transmembrane transporter activity(GO:0005345) pyrimidine nucleobase transmembrane transporter activity(GO:0005350) nucleobase transmembrane transporter activity(GO:0015205) glycerol channel activity(GO:0015254) |
| 0.3 | 1.3 | GO:0042284 | sphingolipid delta-4 desaturase activity(GO:0042284) |
| 0.2 | 2.0 | GO:0008865 | fructokinase activity(GO:0008865) mannokinase activity(GO:0019158) |
| 0.2 | 1.6 | GO:0008294 | calcium- and calmodulin-responsive adenylate cyclase activity(GO:0008294) |
| 0.2 | 1.4 | GO:0008273 | calcium, potassium:sodium antiporter activity(GO:0008273) |
| 0.2 | 2.3 | GO:0003886 | DNA (cytosine-5-)-methyltransferase activity(GO:0003886) |
| 0.2 | 1.1 | GO:0008568 | microtubule-severing ATPase activity(GO:0008568) |
| 0.2 | 3.7 | GO:0008191 | metalloendopeptidase inhibitor activity(GO:0008191) |
| 0.1 | 2.4 | GO:0044323 | retinoic acid-responsive element binding(GO:0044323) |
| 0.1 | 0.9 | GO:0016018 | cyclosporin A binding(GO:0016018) |
| 0.1 | 4.8 | GO:0008483 | transaminase activity(GO:0008483) |
| 0.1 | 0.9 | GO:0003836 | beta-galactoside (CMP) alpha-2,3-sialyltransferase activity(GO:0003836) |
| 0.1 | 1.6 | GO:0019531 | oxalate transmembrane transporter activity(GO:0019531) |
| 0.1 | 1.5 | GO:0043522 | leucine zipper domain binding(GO:0043522) |
| 0.1 | 2.1 | GO:0070402 | NADPH binding(GO:0070402) |
| 0.1 | 0.6 | GO:0030229 | very-low-density lipoprotein particle receptor activity(GO:0030229) |
| 0.1 | 0.9 | GO:0052813 | phosphatidylinositol-4,5-bisphosphate 3-kinase activity(GO:0046934) phosphatidylinositol bisphosphate kinase activity(GO:0052813) |
| 0.1 | 0.2 | GO:0000026 | alpha-1,2-mannosyltransferase activity(GO:0000026) |
| 0.1 | 0.6 | GO:0000990 | transcription factor activity, core RNA polymerase binding(GO:0000990) |
| 0.1 | 0.8 | GO:0030274 | LIM domain binding(GO:0030274) |
| 0.0 | 0.8 | GO:0004143 | diacylglycerol kinase activity(GO:0004143) |
| 0.0 | 3.7 | GO:0005544 | calcium-dependent phospholipid binding(GO:0005544) |
| 0.0 | 1.4 | GO:0008574 | ATP-dependent microtubule motor activity, plus-end-directed(GO:0008574) |
| 0.0 | 0.5 | GO:0086080 | protein binding involved in heterotypic cell-cell adhesion(GO:0086080) |
| 0.0 | 1.1 | GO:0004003 | ATP-dependent DNA helicase activity(GO:0004003) |
| 0.0 | 2.1 | GO:0004722 | protein serine/threonine phosphatase activity(GO:0004722) |
| 0.0 | 0.2 | GO:0008479 | queuine tRNA-ribosyltransferase activity(GO:0008479) |
| 0.0 | 0.5 | GO:0008327 | methyl-CpG binding(GO:0008327) |
| 0.0 | 0.1 | GO:0008429 | phosphatidylethanolamine binding(GO:0008429) |
| 0.0 | 1.1 | GO:0048365 | Rac GTPase binding(GO:0048365) |
| 0.0 | 1.2 | GO:0003730 | mRNA 3'-UTR binding(GO:0003730) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 1.6 | PID LPA4 PATHWAY | LPA4-mediated signaling events |
| 0.0 | 0.9 | PID IL5 PATHWAY | IL5-mediated signaling events |
| 0.0 | 2.1 | PID TAP63 PATHWAY | Validated transcriptional targets of TAp63 isoforms |
| 0.0 | 1.1 | PID ATR PATHWAY | ATR signaling pathway |
| 0.0 | 2.3 | PID RB 1PATHWAY | Regulation of retinoblastoma protein |
| 0.0 | 2.0 | PID HIF1 TFPATHWAY | HIF-1-alpha transcription factor network |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.9 | REACTOME BINDING AND ENTRY OF HIV VIRION | Genes involved in Binding and entry of HIV virion |
| 0.2 | 4.8 | REACTOME AMINO ACID SYNTHESIS AND INTERCONVERSION TRANSAMINATION | Genes involved in Amino acid synthesis and interconversion (transamination) |
| 0.1 | 2.0 | REACTOME PASSIVE TRANSPORT BY AQUAPORINS | Genes involved in Passive Transport by Aquaporins |
| 0.1 | 1.6 | REACTOME ADENYLATE CYCLASE ACTIVATING PATHWAY | Genes involved in Adenylate cyclase activating pathway |
| 0.1 | 1.4 | REACTOME RETROGRADE NEUROTROPHIN SIGNALLING | Genes involved in Retrograde neurotrophin signalling |
| 0.1 | 1.1 | REACTOME UNWINDING OF DNA | Genes involved in Unwinding of DNA |
| 0.0 | 0.9 | REACTOME TERMINATION OF O GLYCAN BIOSYNTHESIS | Genes involved in Termination of O-glycan biosynthesis |
| 0.0 | 0.5 | REACTOME ACTIVATION OF CHAPERONES BY ATF6 ALPHA | Genes involved in Activation of Chaperones by ATF6-alpha |
| 0.0 | 2.0 | REACTOME GLUCOSE TRANSPORT | Genes involved in Glucose transport |
| 0.0 | 0.7 | REACTOME AMINE DERIVED HORMONES | Genes involved in Amine-derived hormones |
| 0.0 | 0.6 | REACTOME VIRAL MESSENGER RNA SYNTHESIS | Genes involved in Viral Messenger RNA Synthesis |
| 0.0 | 0.3 | REACTOME MRNA DECAY BY 3 TO 5 EXORIBONUCLEASE | Genes involved in mRNA Decay by 3' to 5' Exoribonuclease |
| 0.0 | 0.9 | REACTOME TIE2 SIGNALING | Genes involved in Tie2 Signaling |
| 0.0 | 1.5 | REACTOME INTERACTION BETWEEN L1 AND ANKYRINS | Genes involved in Interaction between L1 and Ankyrins |
| 0.0 | 1.3 | REACTOME SPHINGOLIPID DE NOVO BIOSYNTHESIS | Genes involved in Sphingolipid de novo biosynthesis |
| 0.0 | 2.4 | REACTOME NUCLEAR RECEPTOR TRANSCRIPTION PATHWAY | Genes involved in Nuclear Receptor transcription pathway |
| 0.0 | 0.6 | REACTOME CHYLOMICRON MEDIATED LIPID TRANSPORT | Genes involved in Chylomicron-mediated lipid transport |
| 0.0 | 1.1 | REACTOME SIGNALING BY ROBO RECEPTOR | Genes involved in Signaling by Robo receptor |
| 0.0 | 0.8 | REACTOME EFFECTS OF PIP2 HYDROLYSIS | Genes involved in Effects of PIP2 hydrolysis |
| 0.0 | 3.0 | REACTOME TRANSPORT OF INORGANIC CATIONS ANIONS AND AMINO ACIDS OLIGOPEPTIDES | Genes involved in Transport of inorganic cations/anions and amino acids/oligopeptides |
| 0.0 | 0.4 | REACTOME SIGNALING BY HIPPO | Genes involved in Signaling by Hippo |