Project
PRJNA375882: Comprehensive Mouse Transcriptomic BodyMap
Navigation
Downloads
Results for Pax8
Z-value: 0.58
Transcription factors associated with Pax8
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Pax8
|
ENSMUSG00000026976.16 | Pax8 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Pax8 | mm39_v1_chr2_-_24365607_24365619 | -0.39 | 7.7e-04 | Click! |
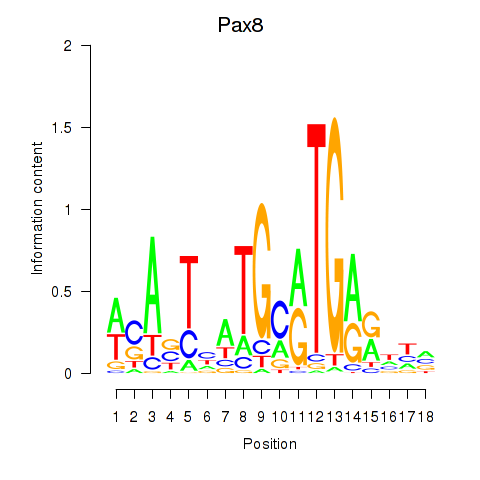
Activity profile of Pax8 motif
Sorted Z-values of Pax8 motif
Network of associatons between targets according to the STRING database.
First level regulatory network of Pax8
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr13_-_25454058 | 9.50 |
ENSMUST00000057866.13
|
Nrsn1
|
neurensin 1 |
| chr2_-_25471703 | 5.44 |
ENSMUST00000114217.3
ENSMUST00000191602.2 |
Ajm1
|
apical junction component 1 |
| chr9_-_108474757 | 4.82 |
ENSMUST00000193621.2
ENSMUST00000006853.11 |
P4htm
|
prolyl 4-hydroxylase, transmembrane (endoplasmic reticulum) |
| chr10_+_112151245 | 4.22 |
ENSMUST00000218445.2
|
Kcnc2
|
potassium voltage gated channel, Shaw-related subfamily, member 2 |
| chr1_+_149975782 | 3.21 |
ENSMUST00000035065.9
|
Ptgs2
|
prostaglandin-endoperoxide synthase 2 |
| chr14_+_60615128 | 3.14 |
ENSMUST00000022561.9
|
Amer2
|
APC membrane recruitment 2 |
| chr11_+_78389913 | 2.97 |
ENSMUST00000017488.5
|
Vtn
|
vitronectin |
| chr9_+_32027335 | 2.64 |
ENSMUST00000174641.8
|
Arhgap32
|
Rho GTPase activating protein 32 |
| chr18_-_66993567 | 2.58 |
ENSMUST00000057942.4
|
Mc4r
|
melanocortin 4 receptor |
| chr2_-_73605387 | 2.55 |
ENSMUST00000166199.9
|
Chn1
|
chimerin 1 |
| chrX_-_7439082 | 2.54 |
ENSMUST00000132788.2
|
Ppp1r3f
|
protein phosphatase 1, regulatory subunit 3F |
| chr11_+_24030663 | 2.52 |
ENSMUST00000118955.2
|
Bcl11a
|
B cell CLL/lymphoma 11A (zinc finger protein) |
| chr11_-_69692542 | 2.12 |
ENSMUST00000011285.11
ENSMUST00000102585.2 |
Fgf11
|
fibroblast growth factor 11 |
| chr18_-_60743169 | 2.05 |
ENSMUST00000115318.4
|
Synpo
|
synaptopodin |
| chr19_+_46064302 | 1.96 |
ENSMUST00000165017.2
ENSMUST00000223741.2 ENSMUST00000225780.2 |
Nolc1
|
nucleolar and coiled-body phosphoprotein 1 |
| chr8_+_61108574 | 1.83 |
ENSMUST00000034066.4
|
Mfap3l
|
microfibrillar-associated protein 3-like |
| chr9_-_103107460 | 1.75 |
ENSMUST00000165296.8
ENSMUST00000112645.8 |
Trf
|
transferrin |
| chr11_+_100900278 | 1.73 |
ENSMUST00000103110.10
ENSMUST00000044721.13 ENSMUST00000168757.9 |
Atp6v0a1
|
ATPase, H+ transporting, lysosomal V0 subunit A1 |
| chr18_+_69654900 | 1.64 |
ENSMUST00000202057.4
|
Tcf4
|
transcription factor 4 |
| chr9_-_103107495 | 1.61 |
ENSMUST00000035158.16
|
Trf
|
transferrin |
| chr15_+_99600475 | 1.60 |
ENSMUST00000228984.2
ENSMUST00000229845.2 |
Smarcd1
|
SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily d, member 1 |
| chr17_+_21910767 | 1.58 |
ENSMUST00000072133.5
|
Gm10226
|
predicted gene 10226 |
| chr1_+_6557455 | 1.50 |
ENSMUST00000140079.8
ENSMUST00000131494.8 |
St18
|
suppression of tumorigenicity 18 |
| chr7_+_114367971 | 1.49 |
ENSMUST00000117543.3
ENSMUST00000151464.2 |
Insc
|
INSC spindle orientation adaptor protein |
| chr4_+_6191084 | 1.45 |
ENSMUST00000029907.6
|
Ubxn2b
|
UBX domain protein 2B |
| chr18_+_69654992 | 1.42 |
ENSMUST00000201627.4
|
Tcf4
|
transcription factor 4 |
| chr19_+_46064409 | 1.34 |
ENSMUST00000223728.2
ENSMUST00000235620.2 |
Nolc1
|
nucleolar and coiled-body phosphoprotein 1 |
| chr1_+_74582044 | 1.21 |
ENSMUST00000113749.8
ENSMUST00000067916.13 ENSMUST00000113747.8 ENSMUST00000113750.8 |
Plcd4
|
phospholipase C, delta 4 |
| chr5_+_107655487 | 1.19 |
ENSMUST00000143074.2
|
Gm42669
|
predicted gene 42669 |
| chr19_+_46064437 | 1.13 |
ENSMUST00000223683.2
|
Nolc1
|
nucleolar and coiled-body phosphoprotein 1 |
| chr13_-_63712140 | 1.08 |
ENSMUST00000195756.6
|
Ptch1
|
patched 1 |
| chr7_-_29605504 | 1.02 |
ENSMUST00000053521.15
|
Zfp27
|
zinc finger protein 27 |
| chrX_+_81992467 | 0.80 |
ENSMUST00000114000.8
|
Dmd
|
dystrophin, muscular dystrophy |
| chr7_+_12631727 | 0.72 |
ENSMUST00000055528.11
ENSMUST00000117189.2 ENSMUST00000120809.2 ENSMUST00000119989.3 |
Zscan22
|
zinc finger and SCAN domain containing 22 |
| chr15_+_54274151 | 0.65 |
ENSMUST00000036737.4
|
Colec10
|
collectin sub-family member 10 |
| chr8_-_46577183 | 0.64 |
ENSMUST00000170416.8
|
Snx25
|
sorting nexin 25 |
| chr6_-_68784692 | 0.48 |
ENSMUST00000103334.4
|
Igkv4-90
|
immunoglobulin kappa chain variable 4-90 |
| chr13_-_62924903 | 0.48 |
ENSMUST00000167516.3
|
Gm5141
|
predicted gene 5141 |
| chr1_+_65225886 | 0.46 |
ENSMUST00000097707.5
|
Pikfyve
|
phosphoinositide kinase, FYVE type zinc finger containing |
| chr12_+_71095112 | 0.44 |
ENSMUST00000135709.2
|
Arid4a
|
AT rich interactive domain 4A (RBP1-like) |
| chr18_-_12254562 | 0.43 |
ENSMUST00000055447.14
ENSMUST00000209859.2 |
Tmem241
|
transmembrane protein 241 |
| chr14_+_53220658 | 0.36 |
ENSMUST00000200548.2
|
Trav9d-4
|
T cell receptor alpha variable 9D-4 |
| chr4_+_119397710 | 0.36 |
ENSMUST00000160219.2
|
Foxj3
|
forkhead box J3 |
| chr2_+_128809268 | 0.35 |
ENSMUST00000110320.9
ENSMUST00000110319.3 |
Zc3h6
|
zinc finger CCCH type containing 6 |
| chr4_-_57301475 | 0.35 |
ENSMUST00000130900.2
|
Ptpn3
|
protein tyrosine phosphatase, non-receptor type 3 |
| chr5_-_137305895 | 0.33 |
ENSMUST00000199243.5
ENSMUST00000197466.5 ENSMUST00000040873.12 |
Srrt
|
serrate RNA effector molecule homolog (Arabidopsis) |
| chr4_+_156300325 | 0.27 |
ENSMUST00000105572.3
|
Perm1
|
PPARGC1 and ESRR induced regulator, muscle 1 |
| chr6_+_127049865 | 0.16 |
ENSMUST00000000186.9
|
Fgf23
|
fibroblast growth factor 23 |
| chr14_+_53100756 | 0.15 |
ENSMUST00000103616.5
ENSMUST00000186370.2 |
Trav15d-1-dv6d-1
|
T cell receptor alpha variable 15D-1-DV6D-1 |
| chr14_+_50683002 | 0.15 |
ENSMUST00000214792.2
|
Olfr740
|
olfactory receptor 740 |
| chr17_+_38106337 | 0.12 |
ENSMUST00000054748.6
|
Olfr123
|
olfactory receptor 123 |
| chr14_+_50741057 | 0.11 |
ENSMUST00000217437.2
ENSMUST00000213935.2 |
Olfr742
|
olfactory receptor 742 |
| chr5_-_151051000 | 0.08 |
ENSMUST00000202111.4
|
Stard13
|
StAR-related lipid transfer (START) domain containing 13 |
| chr11_-_87788066 | 0.06 |
ENSMUST00000217095.2
ENSMUST00000215150.2 |
Olfr463
|
olfactory receptor 463 |
| chr12_-_28632514 | 0.05 |
ENSMUST00000110917.2
ENSMUST00000020965.14 |
Allc
|
allantoicase |
| chr5_+_117495337 | 0.03 |
ENSMUST00000031309.16
|
Wsb2
|
WD repeat and SOCS box-containing 2 |
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.4 | 4.2 | GO:0038060 | nitric oxide-cGMP-mediated signaling pathway(GO:0038060) |
| 1.1 | 3.2 | GO:0090362 | positive regulation of platelet-derived growth factor production(GO:0090362) |
| 0.6 | 2.5 | GO:1904800 | regulation of neuron remodeling(GO:1904799) negative regulation of neuron remodeling(GO:1904800) |
| 0.4 | 3.4 | GO:0070447 | positive regulation of oligodendrocyte progenitor proliferation(GO:0070447) |
| 0.4 | 2.1 | GO:0098886 | modification of dendritic spine(GO:0098886) |
| 0.3 | 4.4 | GO:0007000 | nucleolus organization(GO:0007000) |
| 0.3 | 0.8 | GO:0021629 | olfactory nerve morphogenesis(GO:0021627) olfactory nerve structural organization(GO:0021629) |
| 0.2 | 3.0 | GO:0061302 | smooth muscle cell-matrix adhesion(GO:0061302) liver regeneration(GO:0097421) |
| 0.2 | 1.1 | GO:0010157 | response to chlorate(GO:0010157) |
| 0.2 | 2.5 | GO:2000465 | regulation of glycogen (starch) synthase activity(GO:2000465) |
| 0.1 | 2.6 | GO:2000252 | negative regulation of feeding behavior(GO:2000252) |
| 0.1 | 1.7 | GO:0070072 | vacuolar proton-transporting V-type ATPase complex assembly(GO:0070072) |
| 0.1 | 3.1 | GO:0042118 | endothelial cell activation(GO:0042118) |
| 0.1 | 1.4 | GO:0031468 | nuclear envelope reassembly(GO:0031468) |
| 0.1 | 1.6 | GO:0006337 | nucleosome disassembly(GO:0006337) |
| 0.1 | 1.5 | GO:2001269 | positive regulation of cysteine-type endopeptidase activity involved in apoptotic signaling pathway(GO:2001269) |
| 0.1 | 4.8 | GO:0045646 | regulation of erythrocyte differentiation(GO:0045646) |
| 0.0 | 2.5 | GO:0008045 | motor neuron axon guidance(GO:0008045) |
| 0.0 | 0.4 | GO:0097368 | establishment of Sertoli cell barrier(GO:0097368) |
| 0.0 | 0.2 | GO:0010980 | regulation of vitamin D 24-hydroxylase activity(GO:0010979) positive regulation of vitamin D 24-hydroxylase activity(GO:0010980) |
| 0.0 | 0.6 | GO:0060394 | negative regulation of pathway-restricted SMAD protein phosphorylation(GO:0060394) |
| 0.0 | 0.3 | GO:0031053 | primary miRNA processing(GO:0031053) |
| 0.0 | 1.5 | GO:0000132 | establishment of mitotic spindle orientation(GO:0000132) |
| 0.0 | 0.3 | GO:0051045 | negative regulation of membrane protein ectodomain proteolysis(GO:0051045) |
| 0.0 | 1.2 | GO:0007340 | acrosome reaction(GO:0007340) |
| 0.0 | 3.1 | GO:0090090 | negative regulation of canonical Wnt signaling pathway(GO:0090090) |
| 0.0 | 0.1 | GO:0097498 | endothelial tube lumen extension(GO:0097498) |
| 0.0 | 0.3 | GO:0014850 | response to muscle activity(GO:0014850) |
| 0.0 | 0.5 | GO:0032288 | myelin assembly(GO:0032288) |
| 0.0 | 0.1 | GO:0000255 | allantoin metabolic process(GO:0000255) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 3.0 | GO:0048237 | rough endoplasmic reticulum lumen(GO:0048237) |
| 0.3 | 3.4 | GO:0097433 | dense body(GO:0097433) |
| 0.3 | 2.1 | GO:0097444 | spine apparatus(GO:0097444) |
| 0.2 | 1.7 | GO:0000220 | vacuolar proton-transporting V-type ATPase, V0 domain(GO:0000220) |
| 0.1 | 4.4 | GO:0015030 | Cajal body(GO:0015030) |
| 0.1 | 4.2 | GO:0032809 | neuronal cell body membrane(GO:0032809) |
| 0.1 | 1.1 | GO:0044294 | dendritic growth cone(GO:0044294) |
| 0.1 | 1.6 | GO:0071564 | npBAF complex(GO:0071564) |
| 0.0 | 0.8 | GO:0016010 | dystrophin-associated glycoprotein complex(GO:0016010) filopodium membrane(GO:0031527) glycoprotein complex(GO:0090665) |
| 0.0 | 9.5 | GO:0030426 | growth cone(GO:0030426) |
| 0.0 | 0.5 | GO:0032593 | insulin-responsive compartment(GO:0032593) |
| 0.0 | 4.6 | GO:0032993 | protein-DNA complex(GO:0032993) |
| 0.0 | 3.2 | GO:0005901 | caveola(GO:0005901) |
| 0.0 | 4.1 | GO:0005938 | cell cortex(GO:0005938) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.9 | 2.6 | GO:0004980 | melanocyte-stimulating hormone receptor activity(GO:0004980) |
| 0.5 | 2.5 | GO:2001069 | glycogen binding(GO:2001069) |
| 0.5 | 3.4 | GO:0015091 | ferric iron transmembrane transporter activity(GO:0015091) trivalent inorganic cation transmembrane transporter activity(GO:0072510) |
| 0.2 | 4.8 | GO:0031418 | L-ascorbic acid binding(GO:0031418) |
| 0.2 | 3.1 | GO:0001011 | transcription factor activity, sequence-specific DNA binding, RNA polymerase recruiting(GO:0001011) transcription factor activity, TFIIB-class binding(GO:0001087) |
| 0.2 | 1.1 | GO:0005119 | smoothened binding(GO:0005119) hedgehog receptor activity(GO:0008158) |
| 0.1 | 4.2 | GO:0005251 | delayed rectifier potassium channel activity(GO:0005251) |
| 0.1 | 3.0 | GO:0030247 | pattern binding(GO:0001871) polysaccharide binding(GO:0030247) |
| 0.1 | 3.2 | GO:0016702 | oxidoreductase activity, acting on single donors with incorporation of molecular oxygen, incorporation of two atoms of oxygen(GO:0016702) |
| 0.1 | 1.7 | GO:0046961 | proton-transporting ATPase activity, rotational mechanism(GO:0046961) |
| 0.1 | 0.5 | GO:0016308 | 1-phosphatidylinositol-4-phosphate 5-kinase activity(GO:0016308) |
| 0.0 | 1.2 | GO:0004435 | phosphatidylinositol phospholipase C activity(GO:0004435) |
| 0.0 | 2.5 | GO:0046875 | ephrin receptor binding(GO:0046875) |
| 0.0 | 0.6 | GO:0034713 | type I transforming growth factor beta receptor binding(GO:0034713) |
| 0.0 | 0.8 | GO:0002162 | dystroglycan binding(GO:0002162) |
| 0.0 | 3.1 | GO:0005546 | phosphatidylinositol-4,5-bisphosphate binding(GO:0005546) |
| 0.0 | 0.2 | GO:0005105 | type 1 fibroblast growth factor receptor binding(GO:0005105) |
| 0.0 | 0.7 | GO:0005537 | mannose binding(GO:0005537) |
| 0.0 | 1.4 | GO:0019888 | protein phosphatase regulator activity(GO:0019888) |
| 0.0 | 1.6 | GO:0032947 | protein complex scaffold(GO:0032947) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 3.0 | PID INTEGRIN5 PATHWAY | Beta5 beta6 beta7 and beta8 integrin cell surface interactions |
| 0.1 | 2.6 | PID SYNDECAN 3 PATHWAY | Syndecan-3-mediated signaling events |
| 0.1 | 3.2 | PID S1P S1P1 PATHWAY | S1P1 pathway |
| 0.0 | 1.1 | PID HEDGEHOG 2PATHWAY | Signaling events mediated by the Hedgehog family |
| 0.0 | 4.4 | SIG PIP3 SIGNALING IN CARDIAC MYOCTES | Genes related to PIP3 signaling in cardiac myocytes |
| 0.0 | 2.6 | PID TRKR PATHWAY | Neurotrophic factor-mediated Trk receptor signaling |
| 0.0 | 2.5 | PID RAC1 REG PATHWAY | Regulation of RAC1 activity |
| 0.0 | 3.1 | PID AR PATHWAY | Coregulation of Androgen receptor activity |
| 0.0 | 4.8 | NABA ECM REGULATORS | Genes encoding enzymes and their regulators involved in the remodeling of the extracellular matrix |
| 0.0 | 1.6 | PID REG GR PATHWAY | Glucocorticoid receptor regulatory network |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 4.2 | REACTOME VOLTAGE GATED POTASSIUM CHANNELS | Genes involved in Voltage gated Potassium channels |
| 0.1 | 3.2 | REACTOME PHASE1 FUNCTIONALIZATION OF COMPOUNDS | Genes involved in Phase 1 - Functionalization of compounds |
| 0.0 | 3.1 | REACTOME MYOGENESIS | Genes involved in Myogenesis |
| 0.0 | 0.5 | REACTOME SYNTHESIS OF PIPS AT THE LATE ENDOSOME MEMBRANE | Genes involved in Synthesis of PIPs at the late endosome membrane |
| 0.0 | 3.0 | REACTOME INTEGRIN CELL SURFACE INTERACTIONS | Genes involved in Integrin cell surface interactions |
| 0.0 | 1.1 | REACTOME CLASS B 2 SECRETIN FAMILY RECEPTORS | Genes involved in Class B/2 (Secretin family receptors) |
| 0.0 | 2.6 | REACTOME G ALPHA S SIGNALLING EVENTS | Genes involved in G alpha (s) signalling events |
| 0.0 | 0.8 | REACTOME STRIATED MUSCLE CONTRACTION | Genes involved in Striated Muscle Contraction |
| 0.0 | 2.6 | REACTOME SIGNALING BY RHO GTPASES | Genes involved in Signaling by Rho GTPases |