Project
PRJNA375882: Comprehensive Mouse Transcriptomic BodyMap
Navigation
Downloads
Results for Pbx1_Pbx3
Z-value: 1.55
Transcription factors associated with Pbx1_Pbx3
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
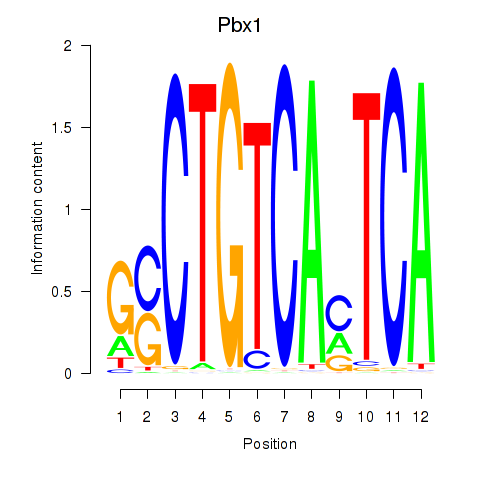
Pbx1
|
ENSMUSG00000052534.16 | Pbx1 |
|
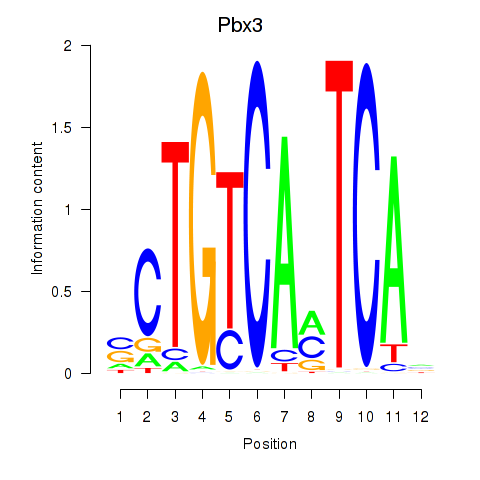
Pbx3
|
ENSMUSG00000038718.16 | Pbx3 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Pbx1 | mm39_v1_chr1_-_168259839_168259916 | 0.50 | 7.1e-06 | Click! |
| Pbx3 | mm39_v1_chr2_-_34261121_34261265 | 0.28 | 1.5e-02 | Click! |
Activity profile of Pbx1_Pbx3 motif
Sorted Z-values of Pbx1_Pbx3 motif
Network of associatons between targets according to the STRING database.
First level regulatory network of Pbx1_Pbx3
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 3.4 | 10.2 | GO:0044413 | evasion or tolerance of host defenses by virus(GO:0019049) avoidance of host defenses(GO:0044413) evasion or tolerance of host defenses(GO:0044415) avoidance of defenses of other organism involved in symbiotic interaction(GO:0051832) evasion or tolerance of defenses of other organism involved in symbiotic interaction(GO:0051834) |
| 3.0 | 15.1 | GO:0043056 | forward locomotion(GO:0043056) |
| 1.9 | 11.3 | GO:0070885 | negative regulation of calcineurin-NFAT signaling cascade(GO:0070885) |
| 1.8 | 5.5 | GO:2000820 | negative regulation of transcription from RNA polymerase II promoter involved in smooth muscle cell differentiation(GO:2000820) |
| 1.8 | 7.2 | GO:1904209 | regulation of chemokine (C-C motif) ligand 2 secretion(GO:1904207) positive regulation of chemokine (C-C motif) ligand 2 secretion(GO:1904209) |
| 1.4 | 4.3 | GO:0002649 | B cell tolerance induction(GO:0002514) regulation of tolerance induction to self antigen(GO:0002649) regulation of B cell tolerance induction(GO:0002661) positive regulation of B cell tolerance induction(GO:0002663) |
| 1.4 | 4.3 | GO:0031448 | regulation of fast-twitch skeletal muscle fiber contraction(GO:0031446) positive regulation of fast-twitch skeletal muscle fiber contraction(GO:0031448) maintenance of mitochondrion location(GO:0051659) relaxation of skeletal muscle(GO:0090076) |
| 1.3 | 4.0 | GO:0043988 | histone H3-S28 phosphorylation(GO:0043988) histone H2A phosphorylation(GO:1990164) |
| 1.2 | 5.9 | GO:0030240 | skeletal muscle thin filament assembly(GO:0030240) |
| 1.2 | 4.7 | GO:0001928 | regulation of exocyst assembly(GO:0001928) |
| 1.1 | 6.8 | GO:0043456 | regulation of pentose-phosphate shunt(GO:0043456) |
| 1.1 | 6.4 | GO:0070893 | transposon integration(GO:0070893) regulation of transposon integration(GO:0070894) negative regulation of transposon integration(GO:0070895) |
| 1.0 | 7.3 | GO:0098912 | membrane depolarization during atrial cardiac muscle cell action potential(GO:0098912) |
| 1.0 | 5.0 | GO:0048769 | sarcomerogenesis(GO:0048769) |
| 0.9 | 2.8 | GO:0038189 | neuropilin signaling pathway(GO:0038189) VEGF-activated neuropilin signaling pathway(GO:0038190) |
| 0.9 | 2.7 | GO:0036500 | ATF6-mediated unfolded protein response(GO:0036500) |
| 0.8 | 18.0 | GO:0042759 | long-chain fatty acid biosynthetic process(GO:0042759) |
| 0.8 | 4.8 | GO:0051387 | negative regulation of neurotrophin TRK receptor signaling pathway(GO:0051387) |
| 0.8 | 2.3 | GO:1904956 | regulation of midbrain dopaminergic neuron differentiation(GO:1904956) regulation of planar cell polarity pathway involved in axis elongation(GO:2000040) negative regulation of planar cell polarity pathway involved in axis elongation(GO:2000041) |
| 0.8 | 3.1 | GO:0070650 | actin filament bundle distribution(GO:0070650) |
| 0.8 | 3.1 | GO:0051344 | negative regulation of cyclic-nucleotide phosphodiesterase activity(GO:0051344) |
| 0.8 | 3.8 | GO:0070543 | response to linoleic acid(GO:0070543) |
| 0.7 | 2.9 | GO:0000379 | tRNA-type intron splice site recognition and cleavage(GO:0000379) |
| 0.7 | 5.0 | GO:0030473 | nucleokinesis involved in cell motility in cerebral cortex radial glia guided migration(GO:0021817) nuclear migration along microtubule(GO:0030473) nuclear matrix organization(GO:0043578) nuclear matrix anchoring at nuclear membrane(GO:0090292) |
| 0.7 | 6.2 | GO:1901552 | positive regulation of endothelial cell development(GO:1901552) positive regulation of establishment of endothelial barrier(GO:1903142) |
| 0.7 | 2.7 | GO:0070649 | formin-nucleated actin cable assembly(GO:0070649) |
| 0.6 | 5.2 | GO:0051964 | negative regulation of synapse assembly(GO:0051964) |
| 0.6 | 11.3 | GO:0071374 | cellular response to parathyroid hormone stimulus(GO:0071374) |
| 0.6 | 5.6 | GO:0098728 | germ-line stem cell division(GO:0042078) male germ-line stem cell asymmetric division(GO:0048133) regulation of branching involved in mammary gland duct morphogenesis(GO:0060762) germline stem cell asymmetric division(GO:0098728) |
| 0.6 | 8.7 | GO:0098953 | receptor diffusion trapping(GO:0098953) postsynaptic neurotransmitter receptor diffusion trapping(GO:0098970) neurotransmitter receptor diffusion trapping(GO:0099628) |
| 0.5 | 2.2 | GO:1903225 | negative regulation of endodermal cell differentiation(GO:1903225) |
| 0.5 | 4.9 | GO:0060346 | bone trabecula formation(GO:0060346) |
| 0.5 | 3.6 | GO:0050847 | progesterone receptor signaling pathway(GO:0050847) |
| 0.5 | 1.5 | GO:0006597 | spermine biosynthetic process(GO:0006597) |
| 0.5 | 1.5 | GO:0002543 | activation of blood coagulation via clotting cascade(GO:0002543) phosphatidylserine exposure on blood platelet(GO:0097045) |
| 0.5 | 2.9 | GO:1990166 | protein localization to site of double-strand break(GO:1990166) |
| 0.5 | 2.4 | GO:0050976 | detection of mechanical stimulus involved in sensory perception of touch(GO:0050976) |
| 0.5 | 2.8 | GO:0000160 | phosphorelay signal transduction system(GO:0000160) |
| 0.5 | 5.2 | GO:2000042 | negative regulation of double-strand break repair via homologous recombination(GO:2000042) |
| 0.5 | 1.4 | GO:0019085 | early viral transcription(GO:0019085) |
| 0.4 | 3.0 | GO:1901509 | regulation of endothelial tube morphogenesis(GO:1901509) |
| 0.4 | 5.4 | GO:0018026 | peptidyl-lysine monomethylation(GO:0018026) |
| 0.4 | 3.2 | GO:0043519 | myosin II filament organization(GO:0031038) regulation of myosin II filament organization(GO:0043519) |
| 0.4 | 1.6 | GO:0046725 | modulation by host of viral RNA genome replication(GO:0044830) negative regulation by virus of viral protein levels in host cell(GO:0046725) |
| 0.4 | 8.0 | GO:0048934 | peripheral nervous system neuron differentiation(GO:0048934) peripheral nervous system neuron development(GO:0048935) |
| 0.4 | 3.2 | GO:0071442 | positive regulation of histone H3-K14 acetylation(GO:0071442) |
| 0.4 | 1.8 | GO:0097498 | endothelial tube lumen extension(GO:0097498) |
| 0.3 | 1.0 | GO:2000729 | positive regulation of mesenchymal cell proliferation involved in ureter development(GO:2000729) |
| 0.3 | 16.7 | GO:0035335 | peptidyl-tyrosine dephosphorylation(GO:0035335) |
| 0.3 | 2.3 | GO:0045196 | establishment or maintenance of neuroblast polarity(GO:0045196) establishment of neuroblast polarity(GO:0045200) |
| 0.3 | 2.3 | GO:0007256 | activation of JNKK activity(GO:0007256) |
| 0.3 | 2.6 | GO:0097428 | protein maturation by iron-sulfur cluster transfer(GO:0097428) |
| 0.3 | 1.9 | GO:0099525 | presynaptic dense core granule exocytosis(GO:0099525) |
| 0.3 | 5.7 | GO:0015693 | magnesium ion transport(GO:0015693) |
| 0.3 | 3.1 | GO:0035635 | entry of bacterium into host cell(GO:0035635) regulation of entry of bacterium into host cell(GO:2000535) |
| 0.3 | 1.5 | GO:1901843 | positive regulation of high voltage-gated calcium channel activity(GO:1901843) |
| 0.3 | 2.3 | GO:0002457 | T cell antigen processing and presentation(GO:0002457) |
| 0.3 | 4.1 | GO:0060136 | embryonic process involved in female pregnancy(GO:0060136) |
| 0.3 | 15.7 | GO:0045022 | early endosome to late endosome transport(GO:0045022) |
| 0.3 | 4.0 | GO:1901339 | regulation of store-operated calcium channel activity(GO:1901339) |
| 0.3 | 5.0 | GO:0036010 | protein localization to endosome(GO:0036010) |
| 0.3 | 0.3 | GO:0072554 | blood vessel lumenization(GO:0072554) |
| 0.3 | 1.1 | GO:0018197 | peptidyl-aspartic acid modification(GO:0018197) |
| 0.3 | 0.8 | GO:0010808 | positive regulation of synaptic vesicle priming(GO:0010808) |
| 0.3 | 1.8 | GO:1903265 | positive regulation of tumor necrosis factor-mediated signaling pathway(GO:1903265) |
| 0.3 | 1.5 | GO:0072318 | clathrin coat disassembly(GO:0072318) |
| 0.3 | 1.3 | GO:0036414 | protein citrullination(GO:0018101) histone citrullination(GO:0036414) |
| 0.2 | 1.0 | GO:1902365 | regulation of spindle elongation(GO:0032887) regulation of mitotic spindle elongation(GO:0032888) anastral spindle assembly(GO:0055048) protein localization to spindle pole body(GO:0071988) regulation of protein localization to spindle pole body(GO:1902363) positive regulation of protein localization to spindle pole body(GO:1902365) positive regulation of mitotic spindle elongation(GO:1902846) |
| 0.2 | 1.2 | GO:0006432 | phenylalanyl-tRNA aminoacylation(GO:0006432) |
| 0.2 | 2.6 | GO:0071688 | striated muscle myosin thick filament assembly(GO:0071688) |
| 0.2 | 1.4 | GO:0048743 | positive regulation of skeletal muscle fiber development(GO:0048743) |
| 0.2 | 5.2 | GO:0032060 | bleb assembly(GO:0032060) |
| 0.2 | 0.9 | GO:0007185 | transmembrane receptor protein tyrosine phosphatase signaling pathway(GO:0007185) |
| 0.2 | 1.1 | GO:0046882 | negative regulation of follicle-stimulating hormone secretion(GO:0046882) |
| 0.2 | 1.1 | GO:0072086 | specification of loop of Henle identity(GO:0072086) |
| 0.2 | 1.3 | GO:0061668 | mitochondrial ribosome assembly(GO:0061668) |
| 0.2 | 3.9 | GO:0000188 | inactivation of MAPK activity(GO:0000188) |
| 0.2 | 17.7 | GO:0065002 | intracellular protein transmembrane transport(GO:0065002) protein transmembrane transport(GO:0071806) |
| 0.2 | 0.6 | GO:0070973 | protein localization to endoplasmic reticulum exit site(GO:0070973) |
| 0.2 | 3.9 | GO:0006337 | nucleosome disassembly(GO:0006337) |
| 0.2 | 2.3 | GO:0006002 | fructose 6-phosphate metabolic process(GO:0006002) |
| 0.2 | 0.8 | GO:0038108 | negative regulation of appetite by leptin-mediated signaling pathway(GO:0038108) |
| 0.2 | 0.7 | GO:2000016 | negative regulation of determination of dorsal identity(GO:2000016) |
| 0.2 | 0.5 | GO:0030860 | regulation of polarized epithelial cell differentiation(GO:0030860) |
| 0.2 | 1.9 | GO:0060120 | auditory receptor cell fate commitment(GO:0009912) inner ear receptor cell fate commitment(GO:0060120) |
| 0.2 | 1.1 | GO:0008355 | olfactory learning(GO:0008355) |
| 0.2 | 1.0 | GO:0009794 | regulation of mitotic cell cycle, embryonic(GO:0009794) mitotic cell cycle, embryonic(GO:0045448) |
| 0.2 | 0.9 | GO:0003376 | sphingosine-1-phosphate signaling pathway(GO:0003376) |
| 0.2 | 0.8 | GO:0070173 | regulation of enamel mineralization(GO:0070173) |
| 0.2 | 1.6 | GO:1901727 | positive regulation of histone deacetylase activity(GO:1901727) |
| 0.2 | 0.6 | GO:1904580 | regulation of intracellular mRNA localization(GO:1904580) |
| 0.1 | 0.6 | GO:0032915 | positive regulation of transforming growth factor beta2 production(GO:0032915) |
| 0.1 | 1.0 | GO:0060023 | soft palate development(GO:0060023) |
| 0.1 | 1.0 | GO:0001955 | blood vessel maturation(GO:0001955) negative regulation of metalloendopeptidase activity(GO:1904684) |
| 0.1 | 3.3 | GO:0042921 | glucocorticoid receptor signaling pathway(GO:0042921) |
| 0.1 | 1.4 | GO:1901525 | negative regulation of macromitophagy(GO:1901525) |
| 0.1 | 1.5 | GO:1901621 | negative regulation of smoothened signaling pathway involved in dorsal/ventral neural tube patterning(GO:1901621) |
| 0.1 | 2.1 | GO:0006654 | phosphatidic acid biosynthetic process(GO:0006654) |
| 0.1 | 1.9 | GO:0098870 | neuronal action potential propagation(GO:0019227) action potential propagation(GO:0098870) |
| 0.1 | 5.1 | GO:0007205 | protein kinase C-activating G-protein coupled receptor signaling pathway(GO:0007205) |
| 0.1 | 1.4 | GO:0006610 | ribosomal protein import into nucleus(GO:0006610) |
| 0.1 | 0.5 | GO:1903999 | negative regulation of eating behavior(GO:1903999) |
| 0.1 | 2.0 | GO:0071447 | cellular response to hydroperoxide(GO:0071447) |
| 0.1 | 2.0 | GO:0043084 | penile erection(GO:0043084) |
| 0.1 | 5.4 | GO:0001958 | endochondral ossification(GO:0001958) replacement ossification(GO:0036075) |
| 0.1 | 6.4 | GO:0042417 | dopamine metabolic process(GO:0042417) |
| 0.1 | 1.9 | GO:0046855 | inositol phosphate dephosphorylation(GO:0046855) |
| 0.1 | 0.5 | GO:0070900 | mitochondrial tRNA modification(GO:0070900) mitochondrial RNA modification(GO:1900864) |
| 0.1 | 1.9 | GO:1904262 | negative regulation of TORC1 signaling(GO:1904262) |
| 0.1 | 1.1 | GO:1902412 | regulation of mitotic cytokinesis(GO:1902412) |
| 0.1 | 3.4 | GO:0070536 | protein K63-linked deubiquitination(GO:0070536) |
| 0.1 | 4.0 | GO:0097352 | autophagosome maturation(GO:0097352) |
| 0.1 | 2.2 | GO:0038063 | collagen-activated tyrosine kinase receptor signaling pathway(GO:0038063) |
| 0.1 | 0.7 | GO:0072344 | rescue of stalled ribosome(GO:0072344) |
| 0.1 | 2.1 | GO:0031290 | axonal fasciculation(GO:0007413) retinal ganglion cell axon guidance(GO:0031290) |
| 0.1 | 3.0 | GO:0010971 | positive regulation of G2/M transition of mitotic cell cycle(GO:0010971) |
| 0.1 | 6.5 | GO:0009250 | glycogen biosynthetic process(GO:0005978) glucan biosynthetic process(GO:0009250) |
| 0.1 | 2.8 | GO:0035855 | megakaryocyte development(GO:0035855) |
| 0.1 | 1.0 | GO:1900113 | negative regulation of histone H3-K9 trimethylation(GO:1900113) |
| 0.1 | 9.2 | GO:0050885 | neuromuscular process controlling balance(GO:0050885) |
| 0.1 | 0.6 | GO:0005513 | detection of calcium ion(GO:0005513) |
| 0.1 | 0.8 | GO:0033147 | negative regulation of intracellular estrogen receptor signaling pathway(GO:0033147) |
| 0.1 | 1.0 | GO:0001865 | NK T cell differentiation(GO:0001865) |
| 0.1 | 0.2 | GO:0021957 | corticospinal tract morphogenesis(GO:0021957) |
| 0.1 | 2.3 | GO:0045187 | regulation of circadian sleep/wake cycle, sleep(GO:0045187) |
| 0.1 | 1.6 | GO:0046697 | decidualization(GO:0046697) |
| 0.1 | 2.4 | GO:0015986 | energy coupled proton transport, down electrochemical gradient(GO:0015985) ATP synthesis coupled proton transport(GO:0015986) |
| 0.1 | 0.8 | GO:2001288 | positive regulation of caveolin-mediated endocytosis(GO:2001288) |
| 0.1 | 1.9 | GO:0051044 | positive regulation of membrane protein ectodomain proteolysis(GO:0051044) |
| 0.1 | 3.3 | GO:0007026 | negative regulation of microtubule depolymerization(GO:0007026) |
| 0.1 | 0.7 | GO:2000270 | negative regulation of fibroblast apoptotic process(GO:2000270) |
| 0.1 | 2.1 | GO:0060135 | maternal process involved in female pregnancy(GO:0060135) |
| 0.1 | 0.5 | GO:0035405 | histone-threonine phosphorylation(GO:0035405) |
| 0.1 | 2.2 | GO:0043949 | regulation of cAMP-mediated signaling(GO:0043949) |
| 0.1 | 3.3 | GO:0042462 | eye photoreceptor cell development(GO:0042462) |
| 0.1 | 2.2 | GO:0060122 | inner ear receptor stereocilium organization(GO:0060122) |
| 0.1 | 0.4 | GO:1903691 | positive regulation of wound healing, spreading of epidermal cells(GO:1903691) |
| 0.1 | 1.0 | GO:0051131 | chaperone-mediated protein complex assembly(GO:0051131) |
| 0.1 | 1.5 | GO:0044406 | adhesion of symbiont to host(GO:0044406) |
| 0.1 | 1.5 | GO:0043153 | entrainment of circadian clock by photoperiod(GO:0043153) |
| 0.1 | 3.0 | GO:2000134 | negative regulation of G1/S transition of mitotic cell cycle(GO:2000134) |
| 0.1 | 1.2 | GO:0036297 | interstrand cross-link repair(GO:0036297) |
| 0.1 | 2.2 | GO:0071158 | positive regulation of cell cycle arrest(GO:0071158) |
| 0.1 | 0.9 | GO:0035493 | SNARE complex assembly(GO:0035493) |
| 0.1 | 2.8 | GO:0042092 | type 2 immune response(GO:0042092) |
| 0.1 | 2.7 | GO:0007257 | activation of JUN kinase activity(GO:0007257) |
| 0.1 | 0.4 | GO:0035589 | G-protein coupled purinergic nucleotide receptor signaling pathway(GO:0035589) |
| 0.1 | 2.0 | GO:0032012 | ARF protein signal transduction(GO:0032011) regulation of ARF protein signal transduction(GO:0032012) |
| 0.1 | 0.4 | GO:1902513 | regulation of organelle transport along microtubule(GO:1902513) |
| 0.0 | 2.1 | GO:0045745 | positive regulation of G-protein coupled receptor protein signaling pathway(GO:0045745) |
| 0.0 | 1.2 | GO:0016180 | snRNA processing(GO:0016180) |
| 0.0 | 2.8 | GO:0043029 | T cell homeostasis(GO:0043029) |
| 0.0 | 0.4 | GO:0042636 | negative regulation of hair cycle(GO:0042636) |
| 0.0 | 0.3 | GO:0090073 | positive regulation of protein homodimerization activity(GO:0090073) |
| 0.0 | 5.0 | GO:0051965 | positive regulation of synapse assembly(GO:0051965) |
| 0.0 | 3.0 | GO:0032543 | mitochondrial translation(GO:0032543) |
| 0.0 | 0.4 | GO:0018344 | protein geranylgeranylation(GO:0018344) |
| 0.0 | 1.8 | GO:0019835 | cytolysis(GO:0019835) |
| 0.0 | 1.9 | GO:0006779 | porphyrin-containing compound biosynthetic process(GO:0006779) |
| 0.0 | 0.3 | GO:0007042 | lysosomal lumen acidification(GO:0007042) |
| 0.0 | 0.6 | GO:2001054 | negative regulation of mesenchymal cell apoptotic process(GO:2001054) |
| 0.0 | 2.9 | GO:0031110 | regulation of microtubule polymerization or depolymerization(GO:0031110) |
| 0.0 | 4.9 | GO:0018107 | peptidyl-threonine phosphorylation(GO:0018107) |
| 0.0 | 3.5 | GO:0000086 | G2/M transition of mitotic cell cycle(GO:0000086) |
| 0.0 | 2.3 | GO:0045839 | negative regulation of mitotic nuclear division(GO:0045839) |
| 0.0 | 1.0 | GO:0031639 | plasminogen activation(GO:0031639) |
| 0.0 | 4.2 | GO:0007156 | homophilic cell adhesion via plasma membrane adhesion molecules(GO:0007156) |
| 0.0 | 1.0 | GO:0008053 | mitochondrial fusion(GO:0008053) |
| 0.0 | 0.2 | GO:1902310 | positive regulation of peptidyl-serine dephosphorylation(GO:1902310) |
| 0.0 | 2.5 | GO:0006334 | nucleosome assembly(GO:0006334) |
| 0.0 | 3.5 | GO:0046330 | positive regulation of JNK cascade(GO:0046330) |
| 0.0 | 0.2 | GO:0007207 | adenylate cyclase-inhibiting G-protein coupled acetylcholine receptor signaling pathway(GO:0007197) phospholipase C-activating G-protein coupled acetylcholine receptor signaling pathway(GO:0007207) |
| 0.0 | 0.7 | GO:0035357 | peroxisome proliferator activated receptor signaling pathway(GO:0035357) |
| 0.0 | 0.1 | GO:0071169 | establishment of protein localization to chromatin(GO:0071169) |
| 0.0 | 0.1 | GO:0021764 | amygdala development(GO:0021764) |
| 0.0 | 0.2 | GO:0060117 | auditory receptor cell development(GO:0060117) |
| 0.0 | 0.3 | GO:0051152 | positive regulation of smooth muscle cell differentiation(GO:0051152) |
| 0.0 | 0.4 | GO:2000114 | regulation of establishment of cell polarity(GO:2000114) |
| 0.0 | 1.2 | GO:0016601 | Rac protein signal transduction(GO:0016601) |
| 0.0 | 11.9 | GO:0003012 | muscle system process(GO:0003012) |
| 0.0 | 5.2 | GO:0006457 | protein folding(GO:0006457) |
| 0.0 | 3.1 | GO:1903364 | positive regulation of cellular protein catabolic process(GO:1903364) |
| 0.0 | 0.6 | GO:0042403 | thyroid hormone metabolic process(GO:0042403) |
| 0.0 | 1.9 | GO:0008625 | extrinsic apoptotic signaling pathway via death domain receptors(GO:0008625) |
| 0.0 | 0.8 | GO:0035065 | regulation of histone acetylation(GO:0035065) |
| 0.0 | 0.2 | GO:0006265 | DNA topological change(GO:0006265) |
| 0.0 | 1.1 | GO:0006801 | superoxide metabolic process(GO:0006801) |
| 0.0 | 1.6 | GO:0045931 | positive regulation of mitotic cell cycle(GO:0045931) |
| 0.0 | 0.3 | GO:0045199 | maintenance of epithelial cell apical/basal polarity(GO:0045199) |
| 0.0 | 2.6 | GO:0006821 | chloride transport(GO:0006821) |
| 0.0 | 0.3 | GO:0060670 | branching involved in labyrinthine layer morphogenesis(GO:0060670) |
| 0.0 | 0.4 | GO:0048520 | positive regulation of behavior(GO:0048520) |
| 0.0 | 1.6 | GO:0035914 | skeletal muscle cell differentiation(GO:0035914) |
| 0.0 | 0.9 | GO:0010761 | fibroblast migration(GO:0010761) |
| 0.0 | 0.1 | GO:0009816 | defense response, incompatible interaction(GO:0009814) defense response to bacterium, incompatible interaction(GO:0009816) regulation of defense response to bacterium, incompatible interaction(GO:1902477) |
| 0.0 | 0.2 | GO:0033572 | transferrin transport(GO:0033572) |
| 0.0 | 0.3 | GO:0051290 | protein heterotetramerization(GO:0051290) |
| 0.0 | 0.6 | GO:0007218 | neuropeptide signaling pathway(GO:0007218) |
| 0.0 | 0.2 | GO:0006895 | Golgi to endosome transport(GO:0006895) |
| 0.0 | 0.2 | GO:0045671 | negative regulation of osteoclast differentiation(GO:0045671) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 4.2 | 16.7 | GO:0014802 | terminal cisterna(GO:0014802) |
| 2.2 | 8.7 | GO:0099573 | glutamatergic postsynaptic density(GO:0099573) |
| 2.1 | 15.0 | GO:0070695 | FHF complex(GO:0070695) |
| 2.1 | 6.2 | GO:0016014 | dystrobrevin complex(GO:0016014) |
| 2.0 | 10.2 | GO:0071144 | SMAD2-SMAD3 protein complex(GO:0071144) |
| 2.0 | 5.9 | GO:0042643 | actomyosin, actin portion(GO:0042643) |
| 1.7 | 5.0 | GO:0002095 | caveolar macromolecular signaling complex(GO:0002095) |
| 0.9 | 7.2 | GO:0033269 | internode region of axon(GO:0033269) |
| 0.7 | 6.6 | GO:0016012 | sarcoglycan complex(GO:0016012) |
| 0.7 | 2.2 | GO:0005588 | collagen type V trimer(GO:0005588) |
| 0.7 | 19.5 | GO:0005859 | muscle myosin complex(GO:0005859) |
| 0.6 | 3.1 | GO:0034751 | aryl hydrocarbon receptor complex(GO:0034751) |
| 0.6 | 8.3 | GO:0005862 | muscle thin filament tropomyosin(GO:0005862) |
| 0.5 | 4.3 | GO:0031673 | H zone(GO:0031673) |
| 0.5 | 2.9 | GO:0000214 | tRNA-intron endonuclease complex(GO:0000214) |
| 0.5 | 2.3 | GO:0005945 | 6-phosphofructokinase complex(GO:0005945) |
| 0.5 | 2.7 | GO:0098831 | presynaptic active zone cytoplasmic component(GO:0098831) |
| 0.4 | 3.1 | GO:0097513 | myosin II filament(GO:0097513) |
| 0.4 | 5.0 | GO:0034993 | microtubule organizing center attachment site(GO:0034992) LINC complex(GO:0034993) |
| 0.4 | 1.2 | GO:0009328 | phenylalanine-tRNA ligase complex(GO:0009328) |
| 0.4 | 2.0 | GO:0000308 | cytoplasmic cyclin-dependent protein kinase holoenzyme complex(GO:0000308) |
| 0.4 | 6.2 | GO:0008290 | F-actin capping protein complex(GO:0008290) |
| 0.4 | 1.1 | GO:0034673 | inhibin-betaglycan-ActRII complex(GO:0034673) inhibin A complex(GO:0043512) |
| 0.3 | 2.2 | GO:0098645 | collagen type IV trimer(GO:0005587) network-forming collagen trimer(GO:0098642) collagen network(GO:0098645) basement membrane collagen trimer(GO:0098651) |
| 0.3 | 1.6 | GO:1902937 | inward rectifier potassium channel complex(GO:1902937) |
| 0.3 | 4.3 | GO:0005952 | cAMP-dependent protein kinase complex(GO:0005952) |
| 0.3 | 2.4 | GO:0000275 | mitochondrial proton-transporting ATP synthase complex, catalytic core F(1)(GO:0000275) proton-transporting ATP synthase complex, catalytic core F(1)(GO:0045261) |
| 0.3 | 1.8 | GO:0005638 | lamin filament(GO:0005638) |
| 0.3 | 2.1 | GO:0033503 | HULC complex(GO:0033503) |
| 0.3 | 1.0 | GO:0070722 | Tle3-Aes complex(GO:0070722) |
| 0.3 | 0.8 | GO:1902636 | kinociliary basal body(GO:1902636) |
| 0.2 | 3.1 | GO:0032584 | growth cone membrane(GO:0032584) |
| 0.2 | 1.9 | GO:0008091 | spectrin(GO:0008091) |
| 0.2 | 1.6 | GO:0031466 | Cul5-RING ubiquitin ligase complex(GO:0031466) |
| 0.2 | 2.2 | GO:1990454 | L-type voltage-gated calcium channel complex(GO:1990454) |
| 0.2 | 1.0 | GO:0061673 | mitotic spindle astral microtubule(GO:0061673) |
| 0.2 | 1.2 | GO:0070876 | SOSS complex(GO:0070876) |
| 0.2 | 11.3 | GO:0043034 | costamere(GO:0043034) |
| 0.2 | 1.1 | GO:0000235 | astral microtubule(GO:0000235) |
| 0.2 | 1.1 | GO:0031310 | integral component of vacuolar membrane(GO:0031166) intrinsic component of vacuolar membrane(GO:0031310) |
| 0.2 | 8.7 | GO:0035861 | site of double-strand break(GO:0035861) |
| 0.2 | 1.9 | GO:0061700 | GATOR2 complex(GO:0061700) |
| 0.2 | 0.5 | GO:1990622 | CHOP-C/EBP complex(GO:0036488) CHOP-ATF3 complex(GO:1990622) |
| 0.1 | 1.3 | GO:0097442 | CA3 pyramidal cell dendrite(GO:0097442) |
| 0.1 | 2.7 | GO:0030673 | axolemma(GO:0030673) |
| 0.1 | 3.8 | GO:0031091 | platelet alpha granule(GO:0031091) |
| 0.1 | 1.5 | GO:0020005 | symbiont-containing vacuole membrane(GO:0020005) |
| 0.1 | 2.3 | GO:0033268 | node of Ranvier(GO:0033268) |
| 0.1 | 13.7 | GO:0016234 | inclusion body(GO:0016234) |
| 0.1 | 2.6 | GO:0071564 | npBAF complex(GO:0071564) |
| 0.1 | 1.0 | GO:0005832 | chaperonin-containing T-complex(GO:0005832) |
| 0.1 | 1.4 | GO:0005869 | dynactin complex(GO:0005869) |
| 0.1 | 2.4 | GO:0033270 | paranode region of axon(GO:0033270) |
| 0.1 | 15.5 | GO:0005901 | caveola(GO:0005901) |
| 0.1 | 0.3 | GO:0070044 | synaptobrevin 2-SNAP-25-syntaxin-1a complex(GO:0070044) |
| 0.1 | 0.8 | GO:0070436 | Grb2-EGFR complex(GO:0070436) |
| 0.1 | 1.0 | GO:0005742 | mitochondrial outer membrane translocase complex(GO:0005742) |
| 0.1 | 2.9 | GO:0000159 | protein phosphatase type 2A complex(GO:0000159) |
| 0.1 | 3.8 | GO:0008180 | COP9 signalosome(GO:0008180) |
| 0.1 | 2.2 | GO:0097225 | sperm midpiece(GO:0097225) |
| 0.1 | 0.8 | GO:0016593 | Cdc73/Paf1 complex(GO:0016593) |
| 0.1 | 0.8 | GO:0030015 | CCR4-NOT core complex(GO:0030015) |
| 0.1 | 19.5 | GO:0043209 | myelin sheath(GO:0043209) |
| 0.1 | 3.3 | GO:0032281 | AMPA glutamate receptor complex(GO:0032281) |
| 0.1 | 4.1 | GO:0000315 | organellar large ribosomal subunit(GO:0000315) mitochondrial large ribosomal subunit(GO:0005762) |
| 0.1 | 0.4 | GO:0005968 | Rab-protein geranylgeranyltransferase complex(GO:0005968) |
| 0.1 | 1.0 | GO:0031618 | nuclear pericentric heterochromatin(GO:0031618) |
| 0.1 | 3.5 | GO:0045171 | intercellular bridge(GO:0045171) |
| 0.1 | 2.7 | GO:0097610 | cleavage furrow(GO:0032154) cell surface furrow(GO:0097610) |
| 0.1 | 3.0 | GO:0005834 | heterotrimeric G-protein complex(GO:0005834) |
| 0.0 | 0.7 | GO:0005736 | DNA-directed RNA polymerase I complex(GO:0005736) |
| 0.0 | 1.4 | GO:0098827 | endoplasmic reticulum subcompartment(GO:0098827) |
| 0.0 | 0.7 | GO:0000940 | condensed chromosome outer kinetochore(GO:0000940) |
| 0.0 | 1.1 | GO:0008074 | guanylate cyclase complex, soluble(GO:0008074) |
| 0.0 | 0.3 | GO:0017071 | intracellular cyclic nucleotide activated cation channel complex(GO:0017071) |
| 0.0 | 2.3 | GO:0014704 | intercalated disc(GO:0014704) |
| 0.0 | 1.7 | GO:0032809 | neuronal cell body membrane(GO:0032809) |
| 0.0 | 0.6 | GO:0036038 | MKS complex(GO:0036038) |
| 0.0 | 1.3 | GO:0005637 | nuclear inner membrane(GO:0005637) |
| 0.0 | 7.0 | GO:0030017 | sarcomere(GO:0030017) |
| 0.0 | 0.2 | GO:0032279 | asymmetric synapse(GO:0032279) |
| 0.0 | 1.6 | GO:0031594 | neuromuscular junction(GO:0031594) |
| 0.0 | 2.6 | GO:0000502 | proteasome complex(GO:0000502) |
| 0.0 | 0.3 | GO:0031464 | Cul4A-RING E3 ubiquitin ligase complex(GO:0031464) |
| 0.0 | 5.8 | GO:0030426 | growth cone(GO:0030426) |
| 0.0 | 2.0 | GO:0001750 | photoreceptor outer segment(GO:0001750) |
| 0.0 | 9.3 | GO:0030141 | secretory granule(GO:0030141) |
| 0.0 | 1.5 | GO:0034707 | chloride channel complex(GO:0034707) |
| 0.0 | 0.4 | GO:0035327 | transcriptionally active chromatin(GO:0035327) |
| 0.0 | 3.2 | GO:0043197 | dendritic spine(GO:0043197) |
| 0.0 | 4.9 | GO:0045121 | membrane raft(GO:0045121) membrane microdomain(GO:0098857) |
| 0.0 | 0.8 | GO:0044665 | MLL1/2 complex(GO:0044665) MLL1 complex(GO:0071339) |
| 0.0 | 2.5 | GO:0001726 | ruffle(GO:0001726) |
| 0.0 | 0.1 | GO:0030670 | phagocytic vesicle membrane(GO:0030670) |
| 0.0 | 2.2 | GO:0071013 | catalytic step 2 spliceosome(GO:0071013) |
| 0.0 | 1.3 | GO:0005811 | lipid particle(GO:0005811) |
| 0.0 | 3.5 | GO:0005938 | cell cortex(GO:0005938) |
| 0.0 | 0.4 | GO:0031307 | integral component of mitochondrial outer membrane(GO:0031307) |
| 0.0 | 1.1 | GO:0030027 | lamellipodium(GO:0030027) |
| 0.0 | 0.2 | GO:1990712 | HFE-transferrin receptor complex(GO:1990712) |
| 0.0 | 1.2 | GO:0031463 | Cul3-RING ubiquitin ligase complex(GO:0031463) |
| 0.0 | 0.6 | GO:0031252 | cell leading edge(GO:0031252) |
| 0.0 | 3.6 | GO:0015629 | actin cytoskeleton(GO:0015629) |
| 0.0 | 1.0 | GO:0031234 | extrinsic component of cytoplasmic side of plasma membrane(GO:0031234) |
| 0.0 | 2.4 | GO:0005759 | mitochondrial matrix(GO:0005759) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.3 | 6.8 | GO:0004082 | bisphosphoglycerate mutase activity(GO:0004082) phosphoglycerate mutase activity(GO:0004619) 2,3-bisphosphoglycerate-dependent phosphoglycerate mutase activity(GO:0046538) |
| 2.2 | 6.5 | GO:0004373 | glycogen (starch) synthase activity(GO:0004373) |
| 2.0 | 10.2 | GO:0030618 | transforming growth factor beta receptor, pathway-specific cytoplasmic mediator activity(GO:0030618) |
| 1.8 | 5.5 | GO:0035939 | microsatellite binding(GO:0035939) |
| 1.7 | 25.1 | GO:0019911 | structural constituent of myelin sheath(GO:0019911) |
| 1.4 | 4.3 | GO:0005026 | transforming growth factor beta receptor activity, type II(GO:0005026) |
| 1.3 | 15.1 | GO:0031433 | telethonin binding(GO:0031433) |
| 1.1 | 19.5 | GO:0017017 | MAP kinase tyrosine/serine/threonine phosphatase activity(GO:0017017) |
| 1.0 | 5.2 | GO:0061649 | ubiquitinated histone binding(GO:0061649) |
| 1.0 | 7.3 | GO:0086056 | voltage-gated calcium channel activity involved in AV node cell action potential(GO:0086056) |
| 1.0 | 3.1 | GO:0036004 | GAF domain binding(GO:0036004) |
| 0.9 | 2.8 | GO:0043183 | vascular endothelial growth factor receptor 1 binding(GO:0043183) |
| 0.8 | 3.8 | GO:0070892 | lipoteichoic acid receptor activity(GO:0070892) |
| 0.7 | 7.3 | GO:0008046 | axon guidance receptor activity(GO:0008046) |
| 0.6 | 6.4 | GO:1903136 | cuprous ion binding(GO:1903136) |
| 0.6 | 5.0 | GO:0004687 | myosin light chain kinase activity(GO:0004687) |
| 0.5 | 8.7 | GO:0009931 | calcium-dependent protein serine/threonine kinase activity(GO:0009931) |
| 0.5 | 1.6 | GO:0033149 | FFAT motif binding(GO:0033149) |
| 0.5 | 2.9 | GO:0000213 | tRNA-intron endonuclease activity(GO:0000213) |
| 0.5 | 1.4 | GO:0005135 | interleukin-3 receptor binding(GO:0005135) |
| 0.5 | 2.8 | GO:0000155 | phosphorelay sensor kinase activity(GO:0000155) |
| 0.5 | 2.3 | GO:0003872 | 6-phosphofructokinase activity(GO:0003872) |
| 0.5 | 4.1 | GO:0004723 | calcium-dependent protein serine/threonine phosphatase activity(GO:0004723) |
| 0.4 | 1.6 | GO:0004921 | interleukin-11 receptor activity(GO:0004921) interleukin-11 binding(GO:0019970) |
| 0.4 | 4.3 | GO:0004691 | cAMP-dependent protein kinase activity(GO:0004691) |
| 0.3 | 1.4 | GO:0004711 | ribosomal protein S6 kinase activity(GO:0004711) |
| 0.3 | 8.4 | GO:0030275 | LRR domain binding(GO:0030275) |
| 0.3 | 1.9 | GO:0070728 | leucine binding(GO:0070728) |
| 0.3 | 4.7 | GO:0005168 | neurotrophin TRKA receptor binding(GO:0005168) |
| 0.3 | 5.1 | GO:0004143 | diacylglycerol kinase activity(GO:0004143) |
| 0.3 | 2.9 | GO:0003910 | DNA ligase activity(GO:0003909) DNA ligase (ATP) activity(GO:0003910) |
| 0.3 | 5.7 | GO:0015095 | magnesium ion transmembrane transporter activity(GO:0015095) |
| 0.3 | 3.7 | GO:0005338 | nucleotide-sugar transmembrane transporter activity(GO:0005338) |
| 0.3 | 7.7 | GO:0003680 | AT DNA binding(GO:0003680) |
| 0.3 | 5.0 | GO:0043495 | protein anchor(GO:0043495) |
| 0.3 | 6.6 | GO:0008253 | 5'-nucleotidase activity(GO:0008253) |
| 0.3 | 1.3 | GO:0004668 | protein-arginine deiminase activity(GO:0004668) |
| 0.2 | 6.2 | GO:0042608 | T cell receptor binding(GO:0042608) |
| 0.2 | 1.5 | GO:0019808 | polyamine binding(GO:0019808) |
| 0.2 | 7.1 | GO:0004683 | calmodulin-dependent protein kinase activity(GO:0004683) |
| 0.2 | 1.2 | GO:0004826 | phenylalanine-tRNA ligase activity(GO:0004826) |
| 0.2 | 1.1 | GO:0004784 | superoxide dismutase activity(GO:0004784) oxidoreductase activity, acting on superoxide radicals as acceptor(GO:0016721) |
| 0.2 | 10.9 | GO:0008307 | structural constituent of muscle(GO:0008307) |
| 0.2 | 1.3 | GO:0043426 | MRF binding(GO:0043426) |
| 0.2 | 1.9 | GO:0019992 | diacylglycerol binding(GO:0019992) |
| 0.2 | 1.1 | GO:0008294 | calcium- and calmodulin-responsive adenylate cyclase activity(GO:0008294) |
| 0.1 | 0.7 | GO:0004045 | aminoacyl-tRNA hydrolase activity(GO:0004045) |
| 0.1 | 1.3 | GO:0005519 | cytoskeletal regulatory protein binding(GO:0005519) |
| 0.1 | 1.9 | GO:0042731 | PH domain binding(GO:0042731) |
| 0.1 | 0.8 | GO:0001226 | RNA polymerase II transcription corepressor binding(GO:0001226) |
| 0.1 | 4.1 | GO:0017128 | phospholipid scramblase activity(GO:0017128) |
| 0.1 | 3.0 | GO:0044548 | S100 protein binding(GO:0044548) |
| 0.1 | 0.4 | GO:0010698 | acetyltransferase activator activity(GO:0010698) |
| 0.1 | 3.1 | GO:0030898 | actin-dependent ATPase activity(GO:0030898) |
| 0.1 | 2.2 | GO:0048407 | platelet-derived growth factor binding(GO:0048407) |
| 0.1 | 1.1 | GO:0034711 | inhibin binding(GO:0034711) |
| 0.1 | 2.4 | GO:0046933 | proton-transporting ATP synthase activity, rotational mechanism(GO:0046933) |
| 0.1 | 3.6 | GO:0070300 | phosphatidic acid binding(GO:0070300) |
| 0.1 | 1.9 | GO:0019855 | calcium channel inhibitor activity(GO:0019855) |
| 0.1 | 1.0 | GO:0047134 | protein-disulfide reductase activity(GO:0047134) |
| 0.1 | 3.9 | GO:0005112 | Notch binding(GO:0005112) |
| 0.1 | 1.0 | GO:0015288 | porin activity(GO:0015288) |
| 0.1 | 3.6 | GO:0008198 | ferrous iron binding(GO:0008198) |
| 0.1 | 2.3 | GO:0042813 | Wnt-activated receptor activity(GO:0042813) |
| 0.1 | 3.0 | GO:0005123 | death receptor binding(GO:0005123) |
| 0.1 | 1.7 | GO:0019841 | retinol binding(GO:0019841) |
| 0.1 | 2.3 | GO:0031402 | sodium ion binding(GO:0031402) |
| 0.1 | 11.5 | GO:0033613 | activating transcription factor binding(GO:0033613) |
| 0.1 | 0.6 | GO:0008568 | microtubule-severing ATPase activity(GO:0008568) |
| 0.1 | 2.9 | GO:0035259 | glucocorticoid receptor binding(GO:0035259) |
| 0.1 | 3.1 | GO:0080025 | phosphatidylinositol-3,5-bisphosphate binding(GO:0080025) |
| 0.1 | 1.6 | GO:0050321 | tau-protein kinase activity(GO:0050321) |
| 0.1 | 2.1 | GO:0003841 | 1-acylglycerol-3-phosphate O-acyltransferase activity(GO:0003841) |
| 0.1 | 2.4 | GO:0005251 | delayed rectifier potassium channel activity(GO:0005251) |
| 0.1 | 0.6 | GO:0001515 | opioid peptide activity(GO:0001515) |
| 0.1 | 3.5 | GO:0003785 | actin monomer binding(GO:0003785) |
| 0.1 | 1.8 | GO:0005521 | lamin binding(GO:0005521) |
| 0.1 | 0.6 | GO:0034452 | dynactin binding(GO:0034452) |
| 0.1 | 0.2 | GO:0015091 | ferric iron transmembrane transporter activity(GO:0015091) trivalent inorganic cation transmembrane transporter activity(GO:0072510) |
| 0.1 | 1.0 | GO:0035497 | cAMP response element binding(GO:0035497) |
| 0.1 | 0.4 | GO:0004663 | Rab geranylgeranyltransferase activity(GO:0004663) |
| 0.1 | 6.8 | GO:0017022 | myosin binding(GO:0017022) |
| 0.1 | 2.0 | GO:0005184 | neuropeptide hormone activity(GO:0005184) |
| 0.1 | 1.9 | GO:0030506 | ankyrin binding(GO:0030506) |
| 0.1 | 2.0 | GO:0008143 | poly(A) binding(GO:0008143) |
| 0.1 | 0.5 | GO:0070290 | N-acylphosphatidylethanolamine-specific phospholipase D activity(GO:0070290) |
| 0.1 | 0.6 | GO:0015467 | G-protein activated inward rectifier potassium channel activity(GO:0015467) |
| 0.1 | 7.8 | GO:0051082 | unfolded protein binding(GO:0051082) |
| 0.1 | 0.3 | GO:0061665 | SUMO ligase activity(GO:0061665) |
| 0.1 | 2.0 | GO:0031435 | mitogen-activated protein kinase kinase kinase binding(GO:0031435) |
| 0.1 | 0.7 | GO:0001055 | RNA polymerase II activity(GO:0001055) |
| 0.1 | 0.4 | GO:0098519 | nucleotide phosphatase activity, acting on free nucleotides(GO:0098519) |
| 0.1 | 0.9 | GO:0038036 | sphingosine-1-phosphate receptor activity(GO:0038036) |
| 0.1 | 4.7 | GO:0002039 | p53 binding(GO:0002039) |
| 0.1 | 2.5 | GO:0030552 | cAMP binding(GO:0030552) |
| 0.0 | 1.0 | GO:0051864 | histone demethylase activity (H3-K36 specific)(GO:0051864) |
| 0.0 | 18.0 | GO:0061659 | ubiquitin protein ligase activity(GO:0061630) ubiquitin-like protein ligase activity(GO:0061659) |
| 0.0 | 0.2 | GO:0003918 | DNA topoisomerase type II (ATP-hydrolyzing) activity(GO:0003918) DNA topoisomerase II activity(GO:0061505) |
| 0.0 | 2.0 | GO:0005086 | ARF guanyl-nucleotide exchange factor activity(GO:0005086) |
| 0.0 | 1.0 | GO:0051011 | microtubule minus-end binding(GO:0051011) |
| 0.0 | 0.7 | GO:0036312 | phosphatidylinositol 3-kinase regulatory subunit binding(GO:0036312) |
| 0.0 | 3.1 | GO:0005546 | phosphatidylinositol-4,5-bisphosphate binding(GO:0005546) |
| 0.0 | 0.2 | GO:0072345 | NAADP-sensitive calcium-release channel activity(GO:0072345) |
| 0.0 | 3.6 | GO:0030276 | clathrin binding(GO:0030276) |
| 0.0 | 0.5 | GO:0035184 | histone threonine kinase activity(GO:0035184) |
| 0.0 | 0.5 | GO:0045294 | alpha-catenin binding(GO:0045294) |
| 0.0 | 2.3 | GO:0048487 | beta-tubulin binding(GO:0048487) |
| 0.0 | 4.3 | GO:0004197 | cysteine-type endopeptidase activity(GO:0004197) |
| 0.0 | 2.0 | GO:0030332 | cyclin binding(GO:0030332) |
| 0.0 | 1.0 | GO:0032266 | phosphatidylinositol-3-phosphate binding(GO:0032266) |
| 0.0 | 0.2 | GO:0046974 | histone methyltransferase activity (H3-K9 specific)(GO:0046974) |
| 0.0 | 2.1 | GO:0050699 | WW domain binding(GO:0050699) |
| 0.0 | 7.9 | GO:0008201 | heparin binding(GO:0008201) |
| 0.0 | 1.6 | GO:0050840 | extracellular matrix binding(GO:0050840) |
| 0.0 | 0.6 | GO:0005527 | macrolide binding(GO:0005527) FK506 binding(GO:0005528) |
| 0.0 | 0.7 | GO:0017075 | syntaxin-1 binding(GO:0017075) |
| 0.0 | 1.6 | GO:0001190 | transcriptional activator activity, RNA polymerase II transcription factor binding(GO:0001190) transcriptional repressor activity, RNA polymerase II activating transcription factor binding(GO:0098811) |
| 0.0 | 1.6 | GO:0070491 | repressing transcription factor binding(GO:0070491) |
| 0.0 | 2.4 | GO:0008146 | sulfotransferase activity(GO:0008146) |
| 0.0 | 0.2 | GO:0016907 | G-protein coupled acetylcholine receptor activity(GO:0016907) |
| 0.0 | 0.5 | GO:0008349 | MAP kinase kinase kinase kinase activity(GO:0008349) |
| 0.0 | 0.2 | GO:0036122 | BMP binding(GO:0036122) |
| 0.0 | 0.7 | GO:0016805 | dipeptidase activity(GO:0016805) |
| 0.0 | 2.2 | GO:0004004 | RNA helicase activity(GO:0003724) ATP-dependent RNA helicase activity(GO:0004004) |
| 0.0 | 2.0 | GO:0004722 | protein serine/threonine phosphatase activity(GO:0004722) |
| 0.0 | 2.0 | GO:0005518 | collagen binding(GO:0005518) |
| 0.0 | 0.2 | GO:0048185 | activin binding(GO:0048185) |
| 0.0 | 2.3 | GO:0003725 | double-stranded RNA binding(GO:0003725) |
| 0.0 | 0.4 | GO:0045028 | G-protein coupled nucleotide receptor activity(GO:0001608) G-protein coupled purinergic nucleotide receptor activity(GO:0045028) |
| 0.0 | 4.1 | GO:0017124 | SH3 domain binding(GO:0017124) |
| 0.0 | 0.1 | GO:0043141 | ATP-dependent 5'-3' DNA helicase activity(GO:0043141) |
| 0.0 | 0.2 | GO:0046527 | glucosyltransferase activity(GO:0046527) |
| 0.0 | 0.6 | GO:0015269 | calcium-activated potassium channel activity(GO:0015269) |
| 0.0 | 2.5 | GO:0030165 | PDZ domain binding(GO:0030165) |
| 0.0 | 0.6 | GO:0043539 | protein serine/threonine kinase activator activity(GO:0043539) |
| 0.0 | 2.9 | GO:0042393 | histone binding(GO:0042393) |
| 0.0 | 3.0 | GO:0004252 | serine-type endopeptidase activity(GO:0004252) |
| 0.0 | 5.1 | GO:0003924 | GTPase activity(GO:0003924) |
| 0.0 | 9.3 | GO:0005509 | calcium ion binding(GO:0005509) |
| 0.0 | 5.9 | GO:0001077 | transcriptional activator activity, RNA polymerase II core promoter proximal region sequence-specific binding(GO:0001077) |
| 0.0 | 1.1 | GO:0017137 | Rab GTPase binding(GO:0017137) |
| 0.0 | 5.0 | GO:0005096 | GTPase activator activity(GO:0005096) |
| 0.0 | 0.8 | GO:0004402 | histone acetyltransferase activity(GO:0004402) |
| 0.0 | 0.4 | GO:0008138 | protein tyrosine/serine/threonine phosphatase activity(GO:0008138) |
| 0.0 | 0.1 | GO:0017034 | Rap guanyl-nucleotide exchange factor activity(GO:0017034) |
| 0.0 | 0.9 | GO:0004725 | protein tyrosine phosphatase activity(GO:0004725) |
| 0.0 | 3.1 | GO:0004842 | ubiquitin-protein transferase activity(GO:0004842) |
| 0.0 | 2.1 | GO:0003735 | structural constituent of ribosome(GO:0003735) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 9.3 | PID LPA4 PATHWAY | LPA4-mediated signaling events |
| 0.3 | 9.5 | PID SMAD2 3PATHWAY | Regulation of cytoplasmic and nuclear SMAD2/3 signaling |
| 0.3 | 2.8 | PID VEGF VEGFR PATHWAY | VEGF and VEGFR signaling network |
| 0.3 | 4.0 | PID S1P S1P4 PATHWAY | S1P4 pathway |
| 0.2 | 8.7 | ST WNT CA2 CYCLIC GMP PATHWAY | Wnt/Ca2+/cyclic GMP signaling. |
| 0.2 | 6.8 | PID INSULIN GLUCOSE PATHWAY | Insulin-mediated glucose transport |
| 0.2 | 7.4 | PID P38 MK2 PATHWAY | p38 signaling mediated by MAPKAP kinases |
| 0.1 | 4.1 | SA B CELL RECEPTOR COMPLEXES | Antigen binding to B cell receptors activates protein tyrosine kinases, such as the Src family, which ultimate activate MAP kinases. |
| 0.1 | 7.0 | PID MAPK TRK PATHWAY | Trk receptor signaling mediated by the MAPK pathway |
| 0.1 | 4.7 | PID ALPHA SYNUCLEIN PATHWAY | Alpha-synuclein signaling |
| 0.1 | 5.2 | PID EPHA FWDPATHWAY | EPHA forward signaling |
| 0.1 | 5.3 | PID IL12 STAT4 PATHWAY | IL12 signaling mediated by STAT4 |
| 0.1 | 4.3 | PID ALK1 PATHWAY | ALK1 signaling events |
| 0.1 | 3.5 | PID P38 ALPHA BETA PATHWAY | Regulation of p38-alpha and p38-beta |
| 0.1 | 0.5 | PID TCR RAS PATHWAY | Ras signaling in the CD4+ TCR pathway |
| 0.1 | 1.1 | PID EPHRINB REV PATHWAY | Ephrin B reverse signaling |
| 0.1 | 2.6 | PID HDAC CLASSIII PATHWAY | Signaling events mediated by HDAC Class III |
| 0.1 | 6.4 | PID P53 REGULATION PATHWAY | p53 pathway |
| 0.1 | 1.3 | PID WNT SIGNALING PATHWAY | Wnt signaling network |
| 0.1 | 1.4 | PID ERBB2 ERBB3 PATHWAY | ErbB2/ErbB3 signaling events |
| 0.1 | 5.5 | PID FGF PATHWAY | FGF signaling pathway |
| 0.1 | 4.4 | NABA COLLAGENS | Genes encoding collagen proteins |
| 0.1 | 6.5 | PID REG GR PATHWAY | Glucocorticoid receptor regulatory network |
| 0.1 | 2.1 | PID INTEGRIN A4B1 PATHWAY | Alpha4 beta1 integrin signaling events |
| 0.1 | 3.3 | PID ARF6 PATHWAY | Arf6 signaling events |
| 0.1 | 3.1 | PID INSULIN PATHWAY | Insulin Pathway |
| 0.1 | 2.1 | PID EPHB FWD PATHWAY | EPHB forward signaling |
| 0.1 | 2.3 | PID HIF2PATHWAY | HIF-2-alpha transcription factor network |
| 0.1 | 3.4 | PID LKB1 PATHWAY | LKB1 signaling events |
| 0.1 | 2.3 | ST WNT BETA CATENIN PATHWAY | Wnt/beta-catenin Pathway |
| 0.0 | 3.8 | PID NOTCH PATHWAY | Notch signaling pathway |
| 0.0 | 0.9 | PID IL6 7 PATHWAY | IL6-mediated signaling events |
| 0.0 | 2.5 | PID HIF1 TFPATHWAY | HIF-1-alpha transcription factor network |
| 0.0 | 0.6 | PID ARF 3PATHWAY | Arf1 pathway |
| 0.0 | 0.3 | ST JAK STAT PATHWAY | Jak-STAT Pathway |
| 0.0 | 0.7 | PID HES HEY PATHWAY | Notch-mediated HES/HEY network |
| 0.0 | 1.0 | ST T CELL SIGNAL TRANSDUCTION | T Cell Signal Transduction |
| 0.0 | 0.4 | PID PRL SIGNALING EVENTS PATHWAY | Signaling events mediated by PRL |
| 0.0 | 0.4 | ST JNK MAPK PATHWAY | JNK MAPK Pathway |
| 0.0 | 1.0 | PID TGFBR PATHWAY | TGF-beta receptor signaling |
| 0.0 | 0.7 | PID RXR VDR PATHWAY | RXR and RAR heterodimerization with other nuclear receptor |
| 0.0 | 1.0 | PID CASPASE PATHWAY | Caspase cascade in apoptosis |
| 0.0 | 0.2 | PID BMP PATHWAY | BMP receptor signaling |
| 0.0 | 0.6 | PID CMYB PATHWAY | C-MYB transcription factor network |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 10.2 | REACTOME SIGNALING BY NODAL | Genes involved in Signaling by NODAL |
| 0.3 | 2.8 | REACTOME VEGF LIGAND RECEPTOR INTERACTIONS | Genes involved in VEGF ligand-receptor interactions |
| 0.3 | 3.8 | REACTOME ROLE OF SECOND MESSENGERS IN NETRIN1 SIGNALING | Genes involved in Role of second messengers in netrin-1 signaling |
| 0.3 | 8.7 | REACTOME CREB PHOSPHORYLATION THROUGH THE ACTIVATION OF CAMKII | Genes involved in CREB phosphorylation through the activation of CaMKII |
| 0.3 | 18.1 | REACTOME MYOGENESIS | Genes involved in Myogenesis |
| 0.3 | 3.9 | REACTOME RECEPTOR LIGAND BINDING INITIATES THE SECOND PROTEOLYTIC CLEAVAGE OF NOTCH RECEPTOR | Genes involved in Receptor-ligand binding initiates the second proteolytic cleavage of Notch receptor |
| 0.2 | 3.2 | REACTOME ROLE OF DCC IN REGULATING APOPTOSIS | Genes involved in Role of DCC in regulating apoptosis |
| 0.2 | 4.3 | REACTOME PLATELET CALCIUM HOMEOSTASIS | Genes involved in Platelet calcium homeostasis |
| 0.2 | 11.1 | REACTOME GLUCONEOGENESIS | Genes involved in Gluconeogenesis |
| 0.2 | 8.9 | REACTOME GLUCOSE METABOLISM | Genes involved in Glucose metabolism |
| 0.2 | 2.8 | REACTOME CYTOSOLIC SULFONATION OF SMALL MOLECULES | Genes involved in Cytosolic sulfonation of small molecules |
| 0.2 | 6.2 | REACTOME CASPASE MEDIATED CLEAVAGE OF CYTOSKELETAL PROTEINS | Genes involved in Caspase-mediated cleavage of cytoskeletal proteins |
| 0.2 | 8.3 | REACTOME SMOOTH MUSCLE CONTRACTION | Genes involved in Smooth Muscle Contraction |
| 0.2 | 5.1 | REACTOME EFFECTS OF PIP2 HYDROLYSIS | Genes involved in Effects of PIP2 hydrolysis |
| 0.2 | 4.0 | REACTOME ERK MAPK TARGETS | Genes involved in ERK/MAPK targets |
| 0.1 | 5.6 | REACTOME EGFR DOWNREGULATION | Genes involved in EGFR downregulation |
| 0.1 | 1.1 | REACTOME REGULATION OF ORNITHINE DECARBOXYLASE ODC | Genes involved in Regulation of ornithine decarboxylase (ODC) |
| 0.1 | 8.6 | REACTOME NCAM1 INTERACTIONS | Genes involved in NCAM1 interactions |
| 0.1 | 2.6 | REACTOME CHONDROITIN SULFATE BIOSYNTHESIS | Genes involved in Chondroitin sulfate biosynthesis |
| 0.1 | 2.4 | REACTOME FORMATION OF ATP BY CHEMIOSMOTIC COUPLING | Genes involved in Formation of ATP by chemiosmotic coupling |
| 0.1 | 1.1 | REACTOME GLYCOPROTEIN HORMONES | Genes involved in Glycoprotein hormones |
| 0.1 | 3.1 | REACTOME SEMA4D INDUCED CELL MIGRATION AND GROWTH CONE COLLAPSE | Genes involved in Sema4D induced cell migration and growth-cone collapse |
| 0.1 | 2.3 | REACTOME TGF BETA RECEPTOR SIGNALING IN EMT EPITHELIAL TO MESENCHYMAL TRANSITION | Genes involved in TGF-beta receptor signaling in EMT (epithelial to mesenchymal transition) |
| 0.1 | 1.5 | REACTOME METABOLISM OF POLYAMINES | Genes involved in Metabolism of polyamines |
| 0.1 | 5.2 | REACTOME VOLTAGE GATED POTASSIUM CHANNELS | Genes involved in Voltage gated Potassium channels |
| 0.1 | 1.1 | REACTOME ACETYLCHOLINE NEUROTRANSMITTER RELEASE CYCLE | Genes involved in Acetylcholine Neurotransmitter Release Cycle |
| 0.1 | 3.9 | REACTOME ACTIVATION OF CHAPERONE GENES BY XBP1S | Genes involved in Activation of Chaperone Genes by XBP1(S) |
| 0.1 | 0.7 | REACTOME COPI MEDIATED TRANSPORT | Genes involved in COPI Mediated Transport |
| 0.1 | 0.6 | REACTOME ACTIVATION OF THE AP1 FAMILY OF TRANSCRIPTION FACTORS | Genes involved in Activation of the AP-1 family of transcription factors |
| 0.1 | 1.1 | REACTOME ADENYLATE CYCLASE ACTIVATING PATHWAY | Genes involved in Adenylate cyclase activating pathway |
| 0.1 | 1.0 | REACTOME RECRUITMENT OF NUMA TO MITOTIC CENTROSOMES | Genes involved in Recruitment of NuMA to mitotic centrosomes |
| 0.1 | 1.9 | REACTOME NCAM SIGNALING FOR NEURITE OUT GROWTH | Genes involved in NCAM signaling for neurite out-growth |
| 0.0 | 4.0 | REACTOME ANTIGEN PROCESSING CROSS PRESENTATION | Genes involved in Antigen processing-Cross presentation |
| 0.0 | 2.1 | REACTOME SYNTHESIS OF PA | Genes involved in Synthesis of PA |
| 0.0 | 2.2 | REACTOME COLLAGEN FORMATION | Genes involved in Collagen formation |
| 0.0 | 3.3 | REACTOME NOTCH1 INTRACELLULAR DOMAIN REGULATES TRANSCRIPTION | Genes involved in NOTCH1 Intracellular Domain Regulates Transcription |
| 0.0 | 0.8 | REACTOME FGFR4 LIGAND BINDING AND ACTIVATION | Genes involved in FGFR4 ligand binding and activation |
| 0.0 | 1.1 | REACTOME LYSOSOME VESICLE BIOGENESIS | Genes involved in Lysosome Vesicle Biogenesis |
| 0.0 | 1.6 | REACTOME SPHINGOLIPID DE NOVO BIOSYNTHESIS | Genes involved in Sphingolipid de novo biosynthesis |
| 0.0 | 0.6 | REACTOME REGULATION OF AMPK ACTIVITY VIA LKB1 | Genes involved in Regulation of AMPK activity via LKB1 |
| 0.0 | 0.7 | REACTOME VIRAL MESSENGER RNA SYNTHESIS | Genes involved in Viral Messenger RNA Synthesis |
| 0.0 | 1.8 | REACTOME MEIOTIC SYNAPSIS | Genes involved in Meiotic Synapsis |
| 0.0 | 8.1 | REACTOME ANTIGEN PROCESSING UBIQUITINATION PROTEASOME DEGRADATION | Genes involved in Antigen processing: Ubiquitination & Proteasome degradation |
| 0.0 | 1.0 | REACTOME GENERATION OF SECOND MESSENGER MOLECULES | Genes involved in Generation of second messenger molecules |
| 0.0 | 0.5 | REACTOME RECYCLING PATHWAY OF L1 | Genes involved in Recycling pathway of L1 |
| 0.0 | 0.5 | REACTOME TRAFFICKING OF GLUR2 CONTAINING AMPA RECEPTORS | Genes involved in Trafficking of GluR2-containing AMPA receptors |
| 0.0 | 0.4 | REACTOME P2Y RECEPTORS | Genes involved in P2Y receptors |
| 0.0 | 0.6 | REACTOME ADHERENS JUNCTIONS INTERACTIONS | Genes involved in Adherens junctions interactions |
| 0.0 | 0.6 | REACTOME CGMP EFFECTS | Genes involved in cGMP effects |
| 0.0 | 0.2 | REACTOME P53 DEPENDENT G1 DNA DAMAGE RESPONSE | Genes involved in p53-Dependent G1 DNA Damage Response |
| 0.0 | 1.7 | REACTOME SIGNALING BY PDGF | Genes involved in Signaling by PDGF |
| 0.0 | 0.3 | REACTOME REGULATION OF IFNG SIGNALING | Genes involved in Regulation of IFNG signaling |
| 0.0 | 0.2 | REACTOME THROMBIN SIGNALLING THROUGH PROTEINASE ACTIVATED RECEPTORS PARS | Genes involved in Thrombin signalling through proteinase activated receptors (PARs) |
| 0.0 | 0.7 | REACTOME BMAL1 CLOCK NPAS2 ACTIVATES CIRCADIAN EXPRESSION | Genes involved in BMAL1:CLOCK/NPAS2 Activates Circadian Expression |
| 0.0 | 3.4 | REACTOME G ALPHA I SIGNALLING EVENTS | Genes involved in G alpha (i) signalling events |
| 0.0 | 0.2 | REACTOME DOWNREGULATION OF TGF BETA RECEPTOR SIGNALING | Genes involved in Downregulation of TGF-beta receptor signaling |
| 0.0 | 2.2 | REACTOME SIGNALING BY RHO GTPASES | Genes involved in Signaling by Rho GTPases |