Project
PRJNA375882: Comprehensive Mouse Transcriptomic BodyMap
Navigation
Downloads
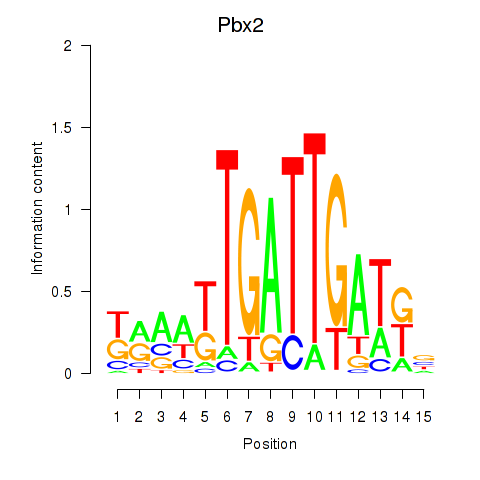
Results for Pbx2
Z-value: 0.80
Transcription factors associated with Pbx2
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Pbx2
|
ENSMUSG00000034673.15 | Pbx2 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Pbx2 | mm39_v1_chr17_+_34812361_34812384 | -0.10 | 4.1e-01 | Click! |
Activity profile of Pbx2 motif
Sorted Z-values of Pbx2 motif
Network of associatons between targets according to the STRING database.
First level regulatory network of Pbx2
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr9_+_95441652 | 4.48 |
ENSMUST00000079597.7
|
Paqr9
|
progestin and adipoQ receptor family member IX |
| chr4_-_61972348 | 4.06 |
ENSMUST00000074018.4
|
Mup20
|
major urinary protein 20 |
| chr8_-_71257623 | 3.54 |
ENSMUST00000212875.2
ENSMUST00000212001.2 ENSMUST00000212757.2 |
Mast3
|
microtubule associated serine/threonine kinase 3 |
| chr17_+_7437500 | 3.21 |
ENSMUST00000024575.8
|
Rps6ka2
|
ribosomal protein S6 kinase, polypeptide 2 |
| chr13_-_113800172 | 3.07 |
ENSMUST00000054650.5
|
Hspb3
|
heat shock protein 3 |
| chr7_-_46392403 | 2.98 |
ENSMUST00000128088.4
|
Saa1
|
serum amyloid A 1 |
| chr8_-_55177510 | 2.92 |
ENSMUST00000175915.8
|
Wdr17
|
WD repeat domain 17 |
| chr13_-_24098981 | 2.65 |
ENSMUST00000110407.4
|
Slc17a4
|
solute carrier family 17 (sodium phosphate), member 4 |
| chr11_-_99134885 | 2.63 |
ENSMUST00000103132.10
ENSMUST00000038214.7 |
Krt222
|
keratin 222 |
| chr10_+_97482946 | 2.59 |
ENSMUST00000105285.4
|
Epyc
|
epiphycan |
| chr7_-_43155351 | 2.52 |
ENSMUST00000191516.7
ENSMUST00000013497.8 ENSMUST00000163619.8 |
4931406B18Rik
|
RIKEN cDNA 4931406B18 gene |
| chr13_-_25454058 | 2.50 |
ENSMUST00000057866.13
|
Nrsn1
|
neurensin 1 |
| chr13_-_24098951 | 2.49 |
ENSMUST00000021769.16
|
Slc17a4
|
solute carrier family 17 (sodium phosphate), member 4 |
| chr3_+_130411097 | 2.47 |
ENSMUST00000166187.8
ENSMUST00000072271.13 |
Etnppl
|
ethanolamine phosphate phospholyase |
| chr2_-_57942844 | 2.45 |
ENSMUST00000090940.6
|
Ermn
|
ermin, ERM-like protein |
| chr10_-_63039709 | 2.39 |
ENSMUST00000095580.3
|
Mypn
|
myopalladin |
| chr11_-_79394904 | 2.35 |
ENSMUST00000164465.3
|
Omg
|
oligodendrocyte myelin glycoprotein |
| chr19_-_5168251 | 2.34 |
ENSMUST00000113728.8
ENSMUST00000113727.8 ENSMUST00000025798.13 |
Klc2
|
kinesin light chain 2 |
| chr15_+_21111428 | 2.29 |
ENSMUST00000075132.8
|
Cdh12
|
cadherin 12 |
| chr2_+_65451100 | 2.28 |
ENSMUST00000144254.6
ENSMUST00000028377.14 |
Scn2a
|
sodium channel, voltage-gated, type II, alpha |
| chr4_+_102843540 | 2.16 |
ENSMUST00000030248.12
ENSMUST00000125417.9 ENSMUST00000169211.3 |
Dynlt5
|
dynein light chain Tctex-type 5 |
| chrX_+_72108393 | 2.13 |
ENSMUST00000060418.8
|
Pnma3
|
paraneoplastic antigen MA3 |
| chr7_+_46401214 | 2.07 |
ENSMUST00000210769.2
ENSMUST00000210272.2 ENSMUST00000075982.4 |
Saa2
|
serum amyloid A 2 |
| chr18_-_23171713 | 2.04 |
ENSMUST00000081423.13
|
Nol4
|
nucleolar protein 4 |
| chr1_+_66426127 | 1.97 |
ENSMUST00000145419.8
|
Map2
|
microtubule-associated protein 2 |
| chr6_+_8259288 | 1.85 |
ENSMUST00000159335.8
|
Umad1
|
UMAP1-MVP12 associated (UMA) domain containing 1 |
| chr4_-_116485118 | 1.73 |
ENSMUST00000030456.14
|
Nasp
|
nuclear autoantigenic sperm protein (histone-binding) |
| chr6_-_129303659 | 1.73 |
ENSMUST00000203159.2
|
Clec2m
|
C-type lectin domain family 2, member m |
| chr3_-_88679881 | 1.67 |
ENSMUST00000090945.5
|
Syt11
|
synaptotagmin XI |
| chr1_+_60948149 | 1.57 |
ENSMUST00000027164.9
|
Ctla4
|
cytotoxic T-lymphocyte-associated protein 4 |
| chr3_+_124114504 | 1.56 |
ENSMUST00000058994.6
|
Tram1l1
|
translocation associated membrane protein 1-like 1 |
| chr7_-_43155118 | 1.55 |
ENSMUST00000168883.2
|
4931406B18Rik
|
RIKEN cDNA 4931406B18 gene |
| chr15_+_16778187 | 1.53 |
ENSMUST00000026432.8
|
Cdh9
|
cadherin 9 |
| chr12_+_108145802 | 1.43 |
ENSMUST00000221167.2
|
Ccnk
|
cyclin K |
| chr1_+_169756217 | 1.43 |
ENSMUST00000094348.4
|
Ccdc190
|
coiled-coil domain containing 190 |
| chr2_-_34951443 | 1.42 |
ENSMUST00000028233.7
|
Hc
|
hemolytic complement |
| chr7_-_3298243 | 1.38 |
ENSMUST00000108653.4
|
Nlrp12
|
NLR family, pyrin domain containing 12 |
| chr9_-_70841881 | 1.36 |
ENSMUST00000214995.2
|
Lipc
|
lipase, hepatic |
| chr11_-_99313078 | 1.34 |
ENSMUST00000017741.4
|
Krt12
|
keratin 12 |
| chr17_-_52140305 | 1.34 |
ENSMUST00000133574.8
|
Satb1
|
special AT-rich sequence binding protein 1 |
| chr17_+_35454049 | 1.34 |
ENSMUST00000130992.2
|
Atp6v1g2
|
ATPase, H+ transporting, lysosomal V1 subunit G2 |
| chr19_+_8897732 | 1.34 |
ENSMUST00000096243.7
|
B3gat3
|
beta-1,3-glucuronyltransferase 3 (glucuronosyltransferase I) |
| chr19_+_39980868 | 1.33 |
ENSMUST00000049178.3
|
Cyp2c37
|
cytochrome P450, family 2. subfamily c, polypeptide 37 |
| chr7_-_66915756 | 1.33 |
ENSMUST00000207715.2
|
Mef2a
|
myocyte enhancer factor 2A |
| chr15_+_92495007 | 1.29 |
ENSMUST00000035399.10
|
Pdzrn4
|
PDZ domain containing RING finger 4 |
| chr1_-_160986880 | 1.28 |
ENSMUST00000135643.8
ENSMUST00000178511.3 |
Tex50
|
testis expressed 50 |
| chr3_-_152687877 | 1.27 |
ENSMUST00000044278.6
|
St6galnac5
|
ST6 (alpha-N-acetyl-neuraminyl-2,3-beta-galactosyl-1,3)-N-acetylgalactosaminide alpha-2,6-sialyltransferase 5 |
| chr16_+_88954838 | 1.22 |
ENSMUST00000062524.6
|
Gm9789
|
predicted gene 9789 |
| chr2_-_110591909 | 1.21 |
ENSMUST00000140777.2
|
Ano3
|
anoctamin 3 |
| chr12_+_103547657 | 1.21 |
ENSMUST00000190664.2
|
Ppp4r4
|
protein phosphatase 4, regulatory subunit 4 |
| chr9_-_70842090 | 1.20 |
ENSMUST00000034731.10
|
Lipc
|
lipase, hepatic |
| chrX_-_43879055 | 1.19 |
ENSMUST00000060481.9
|
Dcaf12l1
|
DDB1 and CUL4 associated factor 12-like 1 |
| chr15_-_100945261 | 1.17 |
ENSMUST00000222611.2
|
Tmdd1
|
transmembrane and death domain 1 |
| chr1_+_85945790 | 1.12 |
ENSMUST00000149469.3
|
Spata3
|
spermatogenesis associated 3 |
| chr6_-_127086480 | 1.11 |
ENSMUST00000039913.9
|
Tigar
|
Trp53 induced glycolysis regulatory phosphatase |
| chr5_-_87288177 | 1.11 |
ENSMUST00000067790.7
|
Ugt2b5
|
UDP glucuronosyltransferase 2 family, polypeptide B5 |
| chr11_+_58311921 | 1.08 |
ENSMUST00000013797.3
|
1810065E05Rik
|
RIKEN cDNA 1810065E05 gene |
| chr16_-_37205277 | 1.07 |
ENSMUST00000114787.8
ENSMUST00000114782.8 ENSMUST00000114775.8 |
Stxbp5l
|
syntaxin binding protein 5-like |
| chr9_+_110948492 | 1.04 |
ENSMUST00000217341.3
|
Lrrfip2
|
leucine rich repeat (in FLII) interacting protein 2 |
| chr17_+_9068805 | 1.03 |
ENSMUST00000115720.8
|
Pde10a
|
phosphodiesterase 10A |
| chr16_-_37205302 | 1.03 |
ENSMUST00000114781.8
ENSMUST00000114780.8 |
Stxbp5l
|
syntaxin binding protein 5-like |
| chr9_+_92191415 | 1.00 |
ENSMUST00000150594.8
ENSMUST00000098477.8 |
1700057G04Rik
|
RIKEN cDNA 1700057G04 gene |
| chr7_-_103778992 | 0.99 |
ENSMUST00000053743.6
|
Ubqln5
|
ubiquilin 5 |
| chr16_+_45355654 | 0.98 |
ENSMUST00000159945.8
|
Slc9c1
|
solute carrier family 9, subfamily C (Na+-transporting carboxylic acid decarboxylase), member 1 |
| chr11_+_5519677 | 0.96 |
ENSMUST00000109856.8
ENSMUST00000109855.8 ENSMUST00000118112.9 |
Ankrd36
|
ankyrin repeat domain 36 |
| chrX_+_115358631 | 0.93 |
ENSMUST00000101269.2
|
Cpxcr1
|
CPX chromosome region, candidate 1 |
| chr1_-_130557349 | 0.93 |
ENSMUST00000142416.2
ENSMUST00000039862.11 ENSMUST00000128128.8 |
Zp3r
|
zona pellucida 3 receptor |
| chrX_+_68988904 | 0.93 |
ENSMUST00000075654.2
|
1700020N15Rik
|
RIKEN cDNA 1700020N15 gene |
| chr2_+_106525938 | 0.91 |
ENSMUST00000016530.14
|
Mpped2
|
metallophosphoesterase domain containing 2 |
| chr12_-_46767619 | 0.90 |
ENSMUST00000219886.2
|
Nova1
|
NOVA alternative splicing regulator 1 |
| chr6_-_3988900 | 0.89 |
ENSMUST00000183682.3
|
Tfpi2
|
tissue factor pathway inhibitor 2 |
| chr4_+_132495636 | 0.85 |
ENSMUST00000102561.11
|
Rpa2
|
replication protein A2 |
| chr14_+_14986011 | 0.85 |
ENSMUST00000164366.8
|
Gm3752
|
predicted gene 3752 |
| chr1_+_60948307 | 0.83 |
ENSMUST00000097720.4
|
Ctla4
|
cytotoxic T-lymphocyte-associated protein 4 |
| chr11_+_88964667 | 0.82 |
ENSMUST00000100619.11
|
Gm525
|
predicted gene 525 |
| chr3_+_122688721 | 0.82 |
ENSMUST00000023820.6
|
Fabp2
|
fatty acid binding protein 2, intestinal |
| chr2_-_151510453 | 0.80 |
ENSMUST00000180195.8
ENSMUST00000096439.4 |
Rad21l
|
RAD21-like (S. pombe) |
| chr11_-_72106418 | 0.79 |
ENSMUST00000021157.9
|
Med31
|
mediator complex subunit 31 |
| chr17_-_53846438 | 0.78 |
ENSMUST00000056198.4
|
Pp2d1
|
protein phosphatase 2C-like domain containing 1 |
| chr15_-_98118858 | 0.77 |
ENSMUST00000142443.8
ENSMUST00000170618.8 |
Gm44579
Olfr287
|
predicted gene 44579 olfactory receptor 287 |
| chr10_+_57521930 | 0.76 |
ENSMUST00000177325.8
|
Pkib
|
protein kinase inhibitor beta, cAMP dependent, testis specific |
| chr3_+_68479578 | 0.75 |
ENSMUST00000170788.9
|
Schip1
|
schwannomin interacting protein 1 |
| chr13_+_21901791 | 0.75 |
ENSMUST00000188775.2
|
H3c10
|
H3 clustered histone 10 |
| chr16_-_38649107 | 0.74 |
ENSMUST00000122078.3
|
Tex55
|
testis expressed 55 |
| chr4_+_147637714 | 0.73 |
ENSMUST00000139784.8
ENSMUST00000143885.8 ENSMUST00000081742.7 |
Zfp985
|
zinc finger protein 985 |
| chr9_-_58277734 | 0.72 |
ENSMUST00000040217.6
|
Tbc1d21
|
TBC1 domain family, member 21 |
| chr13_-_99653045 | 0.70 |
ENSMUST00000064762.6
|
Map1b
|
microtubule-associated protein 1B |
| chr1_+_53100796 | 0.70 |
ENSMUST00000027269.7
ENSMUST00000191197.2 |
Mstn
|
myostatin |
| chrX_-_74621828 | 0.70 |
ENSMUST00000033545.6
|
Rab39b
|
RAB39B, member RAS oncogene family |
| chr16_+_96081998 | 0.69 |
ENSMUST00000099497.4
|
B3galt5
|
UDP-Gal:betaGlcNAc beta 1,3-galactosyltransferase, polypeptide 5 |
| chr15_+_77361241 | 0.69 |
ENSMUST00000060551.9
|
Apol10a
|
apolipoprotein L 10A |
| chrX_-_121307036 | 0.68 |
ENSMUST00000079490.6
|
Nap1l3
|
nucleosome assembly protein 1-like 3 |
| chr6_+_78347636 | 0.68 |
ENSMUST00000204873.3
|
Reg3b
|
regenerating islet-derived 3 beta |
| chr19_-_13126896 | 0.68 |
ENSMUST00000213493.2
|
Olfr1459
|
olfactory receptor 1459 |
| chrX_+_162694397 | 0.65 |
ENSMUST00000140845.2
|
Ap1s2
|
adaptor-related protein complex 1, sigma 2 subunit |
| chr11_-_115599370 | 0.65 |
ENSMUST00000106495.8
|
Grb2
|
growth factor receptor bound protein 2 |
| chrX_-_67936659 | 0.64 |
ENSMUST00000074894.2
|
Gm6812
|
predicted gene 6812 |
| chr11_-_60770098 | 0.64 |
ENSMUST00000062677.12
|
Tmem11
|
transmembrane protein 11 |
| chr2_-_177567397 | 0.63 |
ENSMUST00000108934.9
ENSMUST00000081529.11 |
Zfp972
|
zinc finger protein 972 |
| chr18_+_37858753 | 0.63 |
ENSMUST00000066149.9
|
Pcdhga8
|
protocadherin gamma subfamily A, 8 |
| chr13_-_23929490 | 0.63 |
ENSMUST00000091752.5
|
H3c3
|
H3 clustered histone 3 |
| chr1_+_136552639 | 0.62 |
ENSMUST00000047734.15
ENSMUST00000112046.2 |
Zfp281
|
zinc finger protein 281 |
| chr15_+_77392466 | 0.62 |
ENSMUST00000172191.3
|
Apol11a
|
apolipoprotein L 11a |
| chr17_+_85265420 | 0.61 |
ENSMUST00000080217.14
ENSMUST00000112304.10 |
Ppm1b
|
protein phosphatase 1B, magnesium dependent, beta isoform |
| chr2_-_101451383 | 0.59 |
ENSMUST00000090513.11
|
Iftap
|
intraflagellar transport associated protein |
| chr4_+_146586445 | 0.59 |
ENSMUST00000105735.9
|
Zfp981
|
zinc finger protein 981 |
| chr7_+_18451998 | 0.59 |
ENSMUST00000051973.9
ENSMUST00000208221.2 ENSMUST00000108481.8 |
Psg22
|
pregnancy-specific glycoprotein 22 |
| chr2_+_29014106 | 0.59 |
ENSMUST00000129544.8
|
Setx
|
senataxin |
| chr3_+_20011201 | 0.59 |
ENSMUST00000091309.12
ENSMUST00000108329.8 ENSMUST00000003714.13 |
Cp
|
ceruloplasmin |
| chr9_-_6269846 | 0.59 |
ENSMUST00000051706.6
|
Ddi1
|
DNA-damage inducible 1 |
| chr17_+_24692858 | 0.59 |
ENSMUST00000054946.10
ENSMUST00000238986.2 ENSMUST00000164508.2 |
Bricd5
|
BRICHOS domain containing 5 |
| chr18_+_44960813 | 0.58 |
ENSMUST00000037763.11
|
Ythdc2
|
YTH domain containing 2 |
| chr10_+_57521958 | 0.57 |
ENSMUST00000177473.8
|
Pkib
|
protein kinase inhibitor beta, cAMP dependent, testis specific |
| chr11_+_60244132 | 0.57 |
ENSMUST00000070805.13
ENSMUST00000094140.9 ENSMUST00000108723.9 ENSMUST00000108722.5 |
Drc3
|
dynein regulatory complex subunit 3 |
| chr18_+_3383230 | 0.56 |
ENSMUST00000162301.8
ENSMUST00000161317.2 |
Cul2
|
cullin 2 |
| chr7_+_6048160 | 0.56 |
ENSMUST00000037728.13
ENSMUST00000121583.2 |
Nlrp4c
|
NLR family, pyrin domain containing 4C |
| chr7_-_18165959 | 0.55 |
ENSMUST00000019291.7
|
Psg28
|
pregnancy-specific glycoprotein 28 |
| chr2_-_165242307 | 0.54 |
ENSMUST00000029213.5
|
Ocstamp
|
osteoclast stimulatory transmembrane protein |
| chr8_+_41246310 | 0.54 |
ENSMUST00000056331.8
|
Adam20
|
a disintegrin and metallopeptidase domain 20 |
| chr2_+_128809268 | 0.54 |
ENSMUST00000110320.9
ENSMUST00000110319.3 |
Zc3h6
|
zinc finger CCCH type containing 6 |
| chr7_-_121700958 | 0.54 |
ENSMUST00000139456.2
ENSMUST00000106471.9 ENSMUST00000123296.8 ENSMUST00000033157.10 |
Ndufab1
|
NADH:ubiquinone oxidoreductase subunit AB1 |
| chr6_+_116627635 | 0.52 |
ENSMUST00000204555.2
|
Depp1
|
DEPP1 autophagy regulator |
| chr13_-_86194889 | 0.51 |
ENSMUST00000131011.2
|
Cox7c
|
cytochrome c oxidase subunit 7C |
| chr5_-_122492223 | 0.51 |
ENSMUST00000117263.8
ENSMUST00000049009.7 |
Rad9b
|
RAD9 checkpoint clamp component B |
| chr17_-_35265702 | 0.49 |
ENSMUST00000097338.11
|
Msh5
|
mutS homolog 5 |
| chr7_-_103142086 | 0.49 |
ENSMUST00000055787.8
|
Olfr609
|
olfactory receptor 609 |
| chr1_-_130839178 | 0.46 |
ENSMUST00000027673.11
|
Il20
|
interleukin 20 |
| chr7_+_106737534 | 0.46 |
ENSMUST00000213367.3
ENSMUST00000214819.3 ENSMUST00000216871.3 ENSMUST00000215284.3 ENSMUST00000209942.2 |
Olfr716
|
olfactory receptor 716 |
| chr3_-_107850707 | 0.46 |
ENSMUST00000106681.3
|
Gstm6
|
glutathione S-transferase, mu 6 |
| chr17_+_78815493 | 0.46 |
ENSMUST00000024880.11
ENSMUST00000232859.2 |
Vit
|
vitrin |
| chr2_+_109522781 | 0.46 |
ENSMUST00000111050.10
|
Bdnf
|
brain derived neurotrophic factor |
| chr13_-_43324937 | 0.44 |
ENSMUST00000222160.2
ENSMUST00000179852.9 ENSMUST00000021797.9 |
Tbc1d7
|
TBC1 domain family, member 7 |
| chr6_-_23655130 | 0.44 |
ENSMUST00000104979.2
|
Rnf148
|
ring finger protein 148 |
| chr4_+_146599377 | 0.44 |
ENSMUST00000140089.7
ENSMUST00000179175.3 |
Zfp981
|
zinc finger protein 981 |
| chr11_-_99494134 | 0.44 |
ENSMUST00000072306.4
|
Gm11938
|
predicted gene 11938 |
| chr6_+_8259328 | 0.43 |
ENSMUST00000159378.8
|
Umad1
|
UMAP1-MVP12 associated (UMA) domain containing 1 |
| chr14_+_54429757 | 0.41 |
ENSMUST00000103714.2
|
Traj27
|
T cell receptor alpha joining 27 |
| chr4_+_147106307 | 0.40 |
ENSMUST00000075775.6
|
Rex2
|
reduced expression 2 |
| chr5_-_3697806 | 0.40 |
ENSMUST00000119783.2
ENSMUST00000007559.15 |
Gatad1
|
GATA zinc finger domain containing 1 |
| chrX_+_126301916 | 0.40 |
ENSMUST00000051530.4
|
4921511C20Rik
|
RIKEN cDNA 4921511C20 gene |
| chr8_-_85567256 | 0.40 |
ENSMUST00000003911.13
ENSMUST00000109761.9 ENSMUST00000128035.2 |
Rad23a
|
RAD23 homolog A, nucleotide excision repair protein |
| chr14_+_53194239 | 0.40 |
ENSMUST00000199800.2
|
Trav15d-2-dv6d-2
|
T cell receptor alpha variable 15D-2-DV6D-2 |
| chr14_+_53310461 | 0.40 |
ENSMUST00000103607.3
|
Trav13d-4
|
T cell receptor alpha variable 13D-4 |
| chr16_-_19241884 | 0.40 |
ENSMUST00000206110.4
|
Olfr165
|
olfactory receptor 165 |
| chr18_-_77134939 | 0.40 |
ENSMUST00000137354.8
ENSMUST00000137498.8 |
Katnal2
|
katanin p60 subunit A-like 2 |
| chr18_-_44292952 | 0.39 |
ENSMUST00000181652.9
|
Gm10267
|
predicted gene 10267 |
| chr6_+_41112064 | 0.39 |
ENSMUST00000103272.4
|
Trbv14
|
T cell receptor beta, variable 14 |
| chr7_+_102526329 | 0.39 |
ENSMUST00000098216.2
|
Olfr568
|
olfactory receptor 568 |
| chr5_-_72893941 | 0.38 |
ENSMUST00000169534.6
|
Txk
|
TXK tyrosine kinase |
| chr7_-_12829100 | 0.38 |
ENSMUST00000209822.3
ENSMUST00000235753.2 |
Vmn1r85
|
vomeronasal 1 receptor 85 |
| chr4_+_146093394 | 0.38 |
ENSMUST00000168483.9
|
Zfp600
|
zinc finger protein 600 |
| chr13_+_60749735 | 0.38 |
ENSMUST00000226059.2
ENSMUST00000077453.13 |
Dapk1
|
death associated protein kinase 1 |
| chr7_+_43256593 | 0.37 |
ENSMUST00000120935.2
ENSMUST00000127765.8 ENSMUST00000032661.14 |
Zfp819
|
zinc finger protein 819 |
| chr14_+_53574579 | 0.37 |
ENSMUST00000179580.3
|
Trav13n-3
|
T cell receptor alpha variable 13N-3 |
| chr4_+_145397238 | 0.37 |
ENSMUST00000105738.9
|
Zfp980
|
zinc finger protein 980 |
| chr8_+_22436766 | 0.36 |
ENSMUST00000060587.4
|
Defb13
|
defensin beta 13 |
| chr8_+_66964401 | 0.36 |
ENSMUST00000002025.5
ENSMUST00000183187.2 |
Tktl2
|
transketolase-like 2 |
| chrX_+_18012089 | 0.35 |
ENSMUST00000026018.4
|
Dusp21
|
dual specificity phosphatase 21 |
| chr5_-_6926523 | 0.35 |
ENSMUST00000164784.2
|
Zfp804b
|
zinc finger protein 804B |
| chr2_-_89172742 | 0.35 |
ENSMUST00000216561.2
|
Olfr1233
|
olfactory receptor 1233 |
| chrX_+_8970801 | 0.35 |
ENSMUST00000059967.2
|
4930402K13Rik
|
RIKEN cDNA 4930402K13 gene |
| chr15_+_41573995 | 0.35 |
ENSMUST00000229769.2
|
Oxr1
|
oxidation resistance 1 |
| chr18_+_37813286 | 0.34 |
ENSMUST00000192931.2
|
Pcdhgb1
|
protocadherin gamma subfamily B, 1 |
| chr12_-_103392039 | 0.34 |
ENSMUST00000110001.4
ENSMUST00000223233.2 ENSMUST00000044923.15 ENSMUST00000221211.2 |
Ddx24
|
DEAD box helicase 24 |
| chr17_+_37978659 | 0.34 |
ENSMUST00000216551.2
|
Olfr118
|
olfactory receptor 118 |
| chr8_+_94537910 | 0.33 |
ENSMUST00000138659.9
|
Gnao1
|
guanine nucleotide binding protein, alpha O |
| chr12_+_44268134 | 0.33 |
ENSMUST00000122902.8
|
Pnpla8
|
patatin-like phospholipase domain containing 8 |
| chr14_+_53100756 | 0.33 |
ENSMUST00000103616.5
ENSMUST00000186370.2 |
Trav15d-1-dv6d-1
|
T cell receptor alpha variable 15D-1-DV6D-1 |
| chr11_-_115599418 | 0.32 |
ENSMUST00000021090.14
|
Grb2
|
growth factor receptor bound protein 2 |
| chr7_+_133239414 | 0.31 |
ENSMUST00000128901.9
|
Edrf1
|
erythroid differentiation regulatory factor 1 |
| chr18_-_77134980 | 0.31 |
ENSMUST00000154665.2
ENSMUST00000026486.13 ENSMUST00000123650.2 ENSMUST00000126153.8 |
Katnal2
|
katanin p60 subunit A-like 2 |
| chr11_-_31621727 | 0.29 |
ENSMUST00000109415.2
|
Bod1
|
biorientation of chromosomes in cell division 1 |
| chr3_+_152052102 | 0.29 |
ENSMUST00000117492.9
ENSMUST00000026507.13 ENSMUST00000197748.5 |
Usp33
|
ubiquitin specific peptidase 33 |
| chr1_-_181992051 | 0.29 |
ENSMUST00000227629.2
ENSMUST00000227586.2 |
Vmn1r1
|
vomeronasal 1 receptor 1 |
| chr7_-_26878260 | 0.29 |
ENSMUST00000093040.13
|
Rab4b
|
RAB4B, member RAS oncogene family |
| chr1_-_97689263 | 0.29 |
ENSMUST00000171129.8
|
Ppip5k2
|
diphosphoinositol pentakisphosphate kinase 2 |
| chr11_-_99241924 | 0.29 |
ENSMUST00000017732.3
|
Krt27
|
keratin 27 |
| chr3_-_107851021 | 0.28 |
ENSMUST00000106684.8
ENSMUST00000106685.9 |
Gstm6
|
glutathione S-transferase, mu 6 |
| chr1_-_120198804 | 0.28 |
ENSMUST00000112641.8
|
Steap3
|
STEAP family member 3 |
| chr14_+_78141679 | 0.28 |
ENSMUST00000022591.16
ENSMUST00000169978.2 ENSMUST00000227903.2 |
Epsti1
|
epithelial stromal interaction 1 (breast) |
| chr4_-_147787010 | 0.27 |
ENSMUST00000117638.2
|
Zfp534
|
zinc finger protein 534 |
| chr3_-_106733603 | 0.27 |
ENSMUST00000213616.3
|
Olfr266
|
olfactory receptor 266 |
| chr9_-_20371123 | 0.27 |
ENSMUST00000162438.2
|
Zfp26
|
zinc finger protein 26 |
| chr14_+_54444010 | 0.27 |
ENSMUST00000103729.2
|
Traj12
|
T cell receptor alpha joining 12 |
| chr3_+_20011251 | 0.26 |
ENSMUST00000108328.8
|
Cp
|
ceruloplasmin |
| chr12_-_55349760 | 0.26 |
ENSMUST00000021410.10
|
Ppp2r3c
|
protein phosphatase 2, regulatory subunit B'', gamma |
| chrX_-_77289527 | 0.26 |
ENSMUST00000114026.3
|
Gm14743
|
predicted gene 14743 |
| chr11_+_49138278 | 0.24 |
ENSMUST00000109194.2
|
Mgat1
|
mannoside acetylglucosaminyltransferase 1 |
| chr12_-_15866763 | 0.24 |
ENSMUST00000020922.8
ENSMUST00000221215.2 ENSMUST00000221518.2 |
Trib2
|
tribbles pseudokinase 2 |
| chr4_+_147390131 | 0.24 |
ENSMUST00000148762.4
|
Zfp988
|
zinc finger protein 988 |
| chr17_+_14168635 | 0.24 |
ENSMUST00000088809.6
|
Gm7168
|
predicted gene 7168 |
| chr2_+_111985663 | 0.23 |
ENSMUST00000058176.7
|
Olfr1318
|
olfactory receptor 1318 |
| chr10_-_81335966 | 0.22 |
ENSMUST00000053646.7
|
S1pr4
|
sphingosine-1-phosphate receptor 4 |
| chr11_+_3280771 | 0.22 |
ENSMUST00000136536.8
ENSMUST00000093399.11 |
Pik3ip1
|
phosphoinositide-3-kinase interacting protein 1 |
| chr14_+_123897383 | 0.22 |
ENSMUST00000049681.14
|
Itgbl1
|
integrin, beta-like 1 |
| chrX_+_77230380 | 0.21 |
ENSMUST00000088198.4
|
Obp1b
|
odorant binding protein IB |
| chr19_-_11833365 | 0.21 |
ENSMUST00000079875.4
|
Olfr1418
|
olfactory receptor 1418 |
| chr4_+_147445744 | 0.21 |
ENSMUST00000133078.8
ENSMUST00000154154.2 |
Zfp978
|
zinc finger protein 978 |
| chr18_-_84854841 | 0.21 |
ENSMUST00000236689.2
|
Gm17266
|
predicted gene, 17266 |
| chr7_-_102143980 | 0.21 |
ENSMUST00000058750.4
|
Olfr545
|
olfactory receptor 545 |
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.7 | 4.1 | GO:0008355 | olfactory learning(GO:0008355) |
| 0.5 | 2.6 | GO:0034371 | chylomicron remodeling(GO:0034371) |
| 0.5 | 1.4 | GO:0008588 | release of cytoplasmic sequestered NF-kappaB(GO:0008588) |
| 0.4 | 1.7 | GO:1990927 | negative regulation of membrane invagination(GO:1905154) calcium ion regulated lysosome exocytosis(GO:1990927) |
| 0.4 | 2.4 | GO:0045590 | negative regulation of regulatory T cell differentiation(GO:0045590) |
| 0.4 | 1.1 | GO:1904024 | negative regulation of NAD metabolic process(GO:1902689) negative regulation of glucose catabolic process to lactate via pyruvate(GO:1904024) |
| 0.3 | 1.2 | GO:0080163 | regulation of protein serine/threonine phosphatase activity(GO:0080163) |
| 0.3 | 1.4 | GO:0010760 | negative regulation of macrophage chemotaxis(GO:0010760) |
| 0.3 | 1.3 | GO:0050651 | dermatan sulfate proteoglycan biosynthetic process(GO:0050651) |
| 0.3 | 1.3 | GO:0055005 | ventricular cardiac myofibril assembly(GO:0055005) |
| 0.2 | 3.2 | GO:0002035 | brain renin-angiotensin system(GO:0002035) |
| 0.2 | 1.0 | GO:0035660 | MyD88-dependent toll-like receptor 4 signaling pathway(GO:0035660) |
| 0.2 | 2.3 | GO:0008627 | intrinsic apoptotic signaling pathway in response to osmotic stress(GO:0008627) |
| 0.2 | 0.6 | GO:0031554 | regulation of DNA-templated transcription, termination(GO:0031554) |
| 0.2 | 1.4 | GO:0044828 | negative regulation by host of viral genome replication(GO:0044828) |
| 0.2 | 0.7 | GO:0014732 | skeletal muscle atrophy(GO:0014732) |
| 0.2 | 1.8 | GO:0034723 | DNA replication-dependent nucleosome assembly(GO:0006335) DNA replication-dependent nucleosome organization(GO:0034723) |
| 0.2 | 0.5 | GO:0061193 | taste bud development(GO:0061193) |
| 0.2 | 1.2 | GO:0061588 | calcium activated phospholipid scrambling(GO:0061588) calcium activated phosphatidylcholine scrambling(GO:0061590) calcium activated galactosylceramide scrambling(GO:0061591) |
| 0.1 | 0.6 | GO:2001201 | regulation of transforming growth factor-beta secretion(GO:2001201) |
| 0.1 | 1.0 | GO:0017121 | phospholipid scrambling(GO:0017121) |
| 0.1 | 0.5 | GO:0034241 | positive regulation of macrophage fusion(GO:0034241) |
| 0.1 | 0.3 | GO:0046338 | phosphatidylethanolamine catabolic process(GO:0046338) |
| 0.1 | 2.4 | GO:0048739 | cardiac muscle fiber development(GO:0048739) |
| 0.1 | 2.3 | GO:0022010 | central nervous system myelination(GO:0022010) axon ensheathment in central nervous system(GO:0032291) |
| 0.1 | 1.8 | GO:0009312 | oligosaccharide biosynthetic process(GO:0009312) |
| 0.1 | 1.3 | GO:0061303 | cornea development in camera-type eye(GO:0061303) |
| 0.1 | 0.8 | GO:0097240 | meiotic telomere tethering at nuclear periphery(GO:0044821) meiotic attachment of telomere to nuclear envelope(GO:0070197) chromosome attachment to the nuclear envelope(GO:0097240) |
| 0.1 | 5.4 | GO:0006953 | acute-phase response(GO:0006953) |
| 0.1 | 0.6 | GO:0006499 | N-terminal protein myristoylation(GO:0006499) |
| 0.1 | 0.6 | GO:0044829 | positive regulation by host of viral genome replication(GO:0044829) |
| 0.1 | 1.0 | GO:0098719 | sodium ion import across plasma membrane(GO:0098719) sodium ion import into cell(GO:1990118) |
| 0.1 | 0.2 | GO:0042091 | interleukin-10 biosynthetic process(GO:0042091) regulation of interleukin-10 biosynthetic process(GO:0045074) |
| 0.1 | 1.3 | GO:0043374 | CD8-positive, alpha-beta T cell differentiation(GO:0043374) |
| 0.1 | 1.3 | GO:2000480 | negative regulation of cAMP-dependent protein kinase activity(GO:2000480) |
| 0.1 | 0.2 | GO:0000350 | generation of catalytic spliceosome for second transesterification step(GO:0000350) |
| 0.1 | 0.2 | GO:0006049 | UDP-N-acetylglucosamine catabolic process(GO:0006049) |
| 0.1 | 0.4 | GO:0060332 | positive regulation of response to interferon-gamma(GO:0060332) positive regulation of interferon-gamma-mediated signaling pathway(GO:0060335) |
| 0.1 | 2.7 | GO:0007026 | negative regulation of microtubule depolymerization(GO:0007026) |
| 0.0 | 0.5 | GO:0007262 | STAT protein import into nucleus(GO:0007262) |
| 0.0 | 1.0 | GO:0060670 | branching involved in labyrinthine layer morphogenesis(GO:0060670) |
| 0.0 | 1.3 | GO:0019373 | epoxygenase P450 pathway(GO:0019373) |
| 0.0 | 1.3 | GO:0006298 | mismatch repair(GO:0006298) |
| 0.0 | 0.5 | GO:0000076 | DNA replication checkpoint(GO:0000076) intra-S DNA damage checkpoint(GO:0031573) |
| 0.0 | 5.1 | GO:0035725 | sodium ion transmembrane transport(GO:0035725) |
| 0.0 | 1.0 | GO:0006198 | cAMP catabolic process(GO:0006198) |
| 0.0 | 0.2 | GO:0002188 | translation reinitiation(GO:0002188) |
| 0.0 | 0.3 | GO:0002759 | regulation of antimicrobial humoral response(GO:0002759) |
| 0.0 | 0.5 | GO:0006123 | mitochondrial electron transport, cytochrome c to oxygen(GO:0006123) |
| 0.0 | 0.8 | GO:0001553 | luteinization(GO:0001553) |
| 0.0 | 0.4 | GO:1902018 | negative regulation of cilium assembly(GO:1902018) |
| 0.0 | 0.3 | GO:1902083 | negative regulation of peptidyl-cysteine S-nitrosylation(GO:1902083) |
| 0.0 | 2.1 | GO:0046676 | negative regulation of insulin secretion(GO:0046676) |
| 0.0 | 3.8 | GO:0007156 | homophilic cell adhesion via plasma membrane adhesion molecules(GO:0007156) |
| 0.0 | 0.9 | GO:0046688 | response to copper ion(GO:0046688) |
| 0.0 | 0.3 | GO:0006020 | inositol metabolic process(GO:0006020) |
| 0.0 | 2.4 | GO:0008088 | axo-dendritic transport(GO:0008088) |
| 0.0 | 0.3 | GO:1902572 | regulation of serine-type endopeptidase activity(GO:1900003) negative regulation of serine-type endopeptidase activity(GO:1900004) regulation of serine-type peptidase activity(GO:1902571) negative regulation of serine-type peptidase activity(GO:1902572) |
| 0.0 | 0.2 | GO:0071763 | nuclear membrane organization(GO:0071763) |
| 0.0 | 0.2 | GO:0036444 | calcium ion transmembrane import into mitochondrion(GO:0036444) |
| 0.0 | 0.8 | GO:0048147 | negative regulation of fibroblast proliferation(GO:0048147) |
| 0.0 | 0.1 | GO:0043553 | negative regulation of phosphatidylinositol 3-kinase activity(GO:0043553) |
| 0.0 | 1.3 | GO:0015992 | proton transport(GO:0015992) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 1.4 | GO:0002945 | cyclin K-CDK12 complex(GO:0002944) cyclin K-CDK13 complex(GO:0002945) |
| 0.3 | 2.4 | GO:0033269 | internode region of axon(GO:0033269) |
| 0.2 | 0.9 | GO:0043159 | acrosomal matrix(GO:0043159) |
| 0.2 | 2.0 | GO:0097442 | CA3 pyramidal cell dendrite(GO:0097442) |
| 0.2 | 7.6 | GO:0034364 | high-density lipoprotein particle(GO:0034364) |
| 0.2 | 1.4 | GO:0005579 | membrane attack complex(GO:0005579) |
| 0.1 | 0.8 | GO:0034991 | nuclear meiotic cohesin complex(GO:0034991) |
| 0.1 | 1.7 | GO:0032009 | early phagosome(GO:0032009) |
| 0.1 | 1.0 | GO:0070436 | Grb2-EGFR complex(GO:0070436) |
| 0.1 | 0.5 | GO:0030896 | checkpoint clamp complex(GO:0030896) |
| 0.1 | 0.8 | GO:0070847 | core mediator complex(GO:0070847) |
| 0.1 | 2.3 | GO:0035253 | ciliary rootlet(GO:0035253) |
| 0.1 | 2.3 | GO:0001518 | voltage-gated sodium channel complex(GO:0001518) |
| 0.1 | 0.8 | GO:0045179 | apical cortex(GO:0045179) |
| 0.1 | 2.4 | GO:0045334 | clathrin-coated endocytic vesicle(GO:0045334) |
| 0.1 | 0.6 | GO:0030891 | VCB complex(GO:0030891) |
| 0.0 | 0.3 | GO:0031314 | extrinsic component of mitochondrial inner membrane(GO:0031314) |
| 0.0 | 1.3 | GO:0016471 | vacuolar proton-transporting V-type ATPase complex(GO:0016471) |
| 0.0 | 0.8 | GO:0005662 | DNA replication factor A complex(GO:0005662) |
| 0.0 | 0.5 | GO:0030061 | mitochondrial crista(GO:0030061) |
| 0.0 | 0.7 | GO:0043196 | varicosity(GO:0043196) |
| 0.0 | 0.2 | GO:1990246 | uniplex complex(GO:1990246) |
| 0.0 | 3.5 | GO:0005882 | intermediate filament(GO:0005882) |
| 0.0 | 0.3 | GO:0032593 | insulin-responsive compartment(GO:0032593) |
| 0.0 | 0.6 | GO:0031305 | intrinsic component of mitochondrial inner membrane(GO:0031304) integral component of mitochondrial inner membrane(GO:0031305) |
| 0.0 | 1.2 | GO:1903293 | protein serine/threonine phosphatase complex(GO:0008287) phosphatase complex(GO:1903293) |
| 0.0 | 0.5 | GO:0005614 | interstitial matrix(GO:0005614) |
| 0.0 | 3.1 | GO:0031225 | anchored component of membrane(GO:0031225) |
| 0.0 | 2.4 | GO:0030018 | Z disc(GO:0030018) |
| 0.0 | 3.4 | GO:0030426 | growth cone(GO:0030426) |
| 0.0 | 0.5 | GO:0045271 | mitochondrial respiratory chain complex I(GO:0005747) NADH dehydrogenase complex(GO:0030964) respiratory chain complex I(GO:0045271) |
| 0.0 | 1.3 | GO:0016605 | PML body(GO:0016605) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.8 | 2.5 | GO:0016838 | carbon-oxygen lyase activity, acting on phosphates(GO:0016838) |
| 0.8 | 3.2 | GO:0004711 | ribosomal protein S6 kinase activity(GO:0004711) |
| 0.5 | 2.6 | GO:0035478 | chylomicron binding(GO:0035478) |
| 0.4 | 4.2 | GO:0005186 | pheromone activity(GO:0005186) |
| 0.3 | 5.1 | GO:0005436 | sodium:phosphate symporter activity(GO:0005436) |
| 0.3 | 2.7 | GO:0005519 | cytoskeletal regulatory protein binding(GO:0005519) |
| 0.2 | 1.3 | GO:0015018 | galactosylgalactosylxylosylprotein 3-beta-glucuronosyltransferase activity(GO:0015018) |
| 0.2 | 0.6 | GO:0001160 | transcription termination site sequence-specific DNA binding(GO:0001147) transcription termination site DNA binding(GO:0001160) |
| 0.2 | 0.5 | GO:0031177 | phosphopantetheine binding(GO:0031177) |
| 0.2 | 1.0 | GO:0004118 | cGMP-stimulated cyclic-nucleotide phosphodiesterase activity(GO:0004118) |
| 0.2 | 0.5 | GO:0005169 | neurotrophin TRKB receptor binding(GO:0005169) |
| 0.1 | 2.3 | GO:0043522 | leucine zipper domain binding(GO:0043522) |
| 0.1 | 1.3 | GO:0001665 | alpha-N-acetylgalactosaminide alpha-2,6-sialyltransferase activity(GO:0001665) |
| 0.1 | 5.0 | GO:0042056 | chemoattractant activity(GO:0042056) |
| 0.1 | 0.4 | GO:0004802 | transketolase activity(GO:0004802) |
| 0.1 | 0.7 | GO:0008568 | microtubule-severing ATPase activity(GO:0008568) |
| 0.1 | 1.1 | GO:0004331 | fructose-2,6-bisphosphate 2-phosphatase activity(GO:0004331) |
| 0.1 | 1.0 | GO:0015386 | potassium:proton antiporter activity(GO:0015386) |
| 0.1 | 0.8 | GO:0005324 | long-chain fatty acid transporter activity(GO:0005324) |
| 0.1 | 2.4 | GO:0051371 | muscle alpha-actinin binding(GO:0051371) |
| 0.1 | 2.2 | GO:0017128 | phospholipid scramblase activity(GO:0017128) |
| 0.1 | 0.6 | GO:1990247 | N6-methyladenosine-containing RNA binding(GO:1990247) |
| 0.1 | 0.9 | GO:0016724 | ferroxidase activity(GO:0004322) oxidoreductase activity, oxidizing metal ions, oxygen as acceptor(GO:0016724) |
| 0.1 | 1.0 | GO:0005168 | neurotrophin TRKA receptor binding(GO:0005168) |
| 0.1 | 1.3 | GO:0008553 | hydrogen-exporting ATPase activity, phosphorylative mechanism(GO:0008553) |
| 0.1 | 0.2 | GO:0000386 | second spliceosomal transesterification activity(GO:0000386) |
| 0.1 | 0.9 | GO:0098505 | G-rich strand telomeric DNA binding(GO:0098505) |
| 0.1 | 0.2 | GO:0003827 | alpha-1,3-mannosylglycoprotein 2-beta-N-acetylglucosaminyltransferase activity(GO:0003827) |
| 0.1 | 0.6 | GO:0045545 | syndecan binding(GO:0045545) |
| 0.1 | 1.4 | GO:0008353 | RNA polymerase II carboxy-terminal domain kinase activity(GO:0008353) |
| 0.1 | 1.3 | GO:0004862 | cAMP-dependent protein kinase inhibitor activity(GO:0004862) |
| 0.0 | 0.3 | GO:0000828 | inositol hexakisphosphate kinase activity(GO:0000828) inositol hexakisphosphate 5-kinase activity(GO:0000832) inositol hexakisphosphate 1-kinase activity(GO:0052723) inositol hexakisphosphate 3-kinase activity(GO:0052724) |
| 0.0 | 0.3 | GO:0031821 | G-protein coupled serotonin receptor binding(GO:0031821) |
| 0.0 | 0.5 | GO:0030983 | mismatched DNA binding(GO:0030983) |
| 0.0 | 1.3 | GO:0008392 | arachidonic acid epoxygenase activity(GO:0008392) |
| 0.0 | 0.3 | GO:0047499 | calcium-independent phospholipase A2 activity(GO:0047499) |
| 0.0 | 4.1 | GO:0019905 | syntaxin binding(GO:0019905) |
| 0.0 | 3.5 | GO:0003707 | steroid hormone receptor activity(GO:0003707) |
| 0.0 | 1.7 | GO:0051879 | Hsp90 protein binding(GO:0051879) |
| 0.0 | 2.3 | GO:0019894 | kinesin binding(GO:0019894) |
| 0.0 | 0.1 | GO:0036313 | phosphatidylinositol 3-kinase catalytic subunit binding(GO:0036313) |
| 0.0 | 0.7 | GO:0031489 | myosin V binding(GO:0031489) |
| 0.0 | 1.3 | GO:0035035 | histone acetyltransferase binding(GO:0035035) |
| 0.0 | 0.3 | GO:0017017 | MAP kinase tyrosine/serine/threonine phosphatase activity(GO:0017017) |
| 0.0 | 0.4 | GO:1990381 | ubiquitin-specific protease binding(GO:1990381) |
| 0.0 | 0.6 | GO:0004190 | aspartic-type endopeptidase activity(GO:0004190) |
| 0.0 | 1.4 | GO:0004722 | protein serine/threonine phosphatase activity(GO:0004722) |
| 0.0 | 0.6 | GO:0008378 | galactosyltransferase activity(GO:0008378) |
| 0.0 | 0.2 | GO:0038036 | sphingosine-1-phosphate receptor activity(GO:0038036) |
| 0.0 | 0.7 | GO:0004364 | glutathione transferase activity(GO:0004364) |
| 0.0 | 0.2 | GO:0055106 | ubiquitin-protein transferase regulator activity(GO:0055106) |
| 0.0 | 0.7 | GO:0005160 | transforming growth factor beta receptor binding(GO:0005160) |
| 0.0 | 0.3 | GO:0017160 | Ral GTPase binding(GO:0017160) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 4.2 | ST PHOSPHOINOSITIDE 3 KINASE PATHWAY | PI3K Pathway |
| 0.1 | 1.9 | PID TOLL ENDOGENOUS PATHWAY | Endogenous TLR signaling |
| 0.1 | 2.6 | NABA PROTEOGLYCANS | Genes encoding proteoglycans |
| 0.0 | 0.6 | PID HIF1A PATHWAY | Hypoxic and oxygen homeostasis regulation of HIF-1-alpha |
| 0.0 | 2.0 | PID LKB1 PATHWAY | LKB1 signaling events |
| 0.0 | 2.4 | PID NFAT TFPATHWAY | Calcineurin-regulated NFAT-dependent transcription in lymphocytes |
| 0.0 | 2.3 | PID P75 NTR PATHWAY | p75(NTR)-mediated signaling |
| 0.0 | 0.6 | PID S1P S1P4 PATHWAY | S1P4 pathway |
| 0.0 | 1.1 | PID NETRIN PATHWAY | Netrin-mediated signaling events |
| 0.0 | 1.4 | PID CASPASE PATHWAY | Caspase cascade in apoptosis |
| 0.0 | 1.3 | PID P38 ALPHA BETA DOWNSTREAM PATHWAY | Signaling mediated by p38-alpha and p38-beta |
| 0.0 | 0.8 | PID ATR PATHWAY | ATR signaling pathway |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 3.0 | REACTOME ADVANCED GLYCOSYLATION ENDPRODUCT RECEPTOR SIGNALING | Genes involved in Advanced glycosylation endproduct receptor signaling |
| 0.1 | 3.8 | REACTOME ADHERENS JUNCTIONS INTERACTIONS | Genes involved in Adherens junctions interactions |
| 0.1 | 4.5 | REACTOME ERK MAPK TARGETS | Genes involved in ERK/MAPK targets |
| 0.1 | 2.6 | REACTOME CHYLOMICRON MEDIATED LIPID TRANSPORT | Genes involved in Chylomicron-mediated lipid transport |
| 0.1 | 1.0 | REACTOME CD28 DEPENDENT VAV1 PATHWAY | Genes involved in CD28 dependent Vav1 pathway |
| 0.1 | 2.3 | REACTOME KINESINS | Genes involved in Kinesins |
| 0.1 | 2.4 | REACTOME CTLA4 INHIBITORY SIGNALING | Genes involved in CTLA4 inhibitory signaling |
| 0.1 | 0.8 | REACTOME REMOVAL OF THE FLAP INTERMEDIATE FROM THE C STRAND | Genes involved in Removal of the Flap Intermediate from the C-strand |
| 0.0 | 2.3 | REACTOME INTERACTION BETWEEN L1 AND ANKYRINS | Genes involved in Interaction between L1 and Ankyrins |
| 0.0 | 0.7 | REACTOME NEF MEDIATED DOWNREGULATION OF MHC CLASS I COMPLEX CELL SURFACE EXPRESSION | Genes involved in Nef mediated downregulation of MHC class I complex cell surface expression |
| 0.0 | 1.3 | REACTOME A TETRASACCHARIDE LINKER SEQUENCE IS REQUIRED FOR GAG SYNTHESIS | Genes involved in A tetrasaccharide linker sequence is required for GAG synthesis |
| 0.0 | 1.3 | REACTOME INSULIN RECEPTOR RECYCLING | Genes involved in Insulin receptor recycling |
| 0.0 | 0.7 | REACTOME IRON UPTAKE AND TRANSPORT | Genes involved in Iron uptake and transport |
| 0.0 | 0.4 | REACTOME ROLE OF DCC IN REGULATING APOPTOSIS | Genes involved in Role of DCC in regulating apoptosis |
| 0.0 | 0.6 | REACTOME OXYGEN DEPENDENT PROLINE HYDROXYLATION OF HYPOXIA INDUCIBLE FACTOR ALPHA | Genes involved in Oxygen-dependent Proline Hydroxylation of Hypoxia-inducible Factor Alpha |
| 0.0 | 1.0 | REACTOME RESPIRATORY ELECTRON TRANSPORT | Genes involved in Respiratory electron transport |
| 0.0 | 0.3 | REACTOME ACYL CHAIN REMODELLING OF PE | Genes involved in Acyl chain remodelling of PE |