Project
PRJNA375882: Comprehensive Mouse Transcriptomic BodyMap
Navigation
Downloads
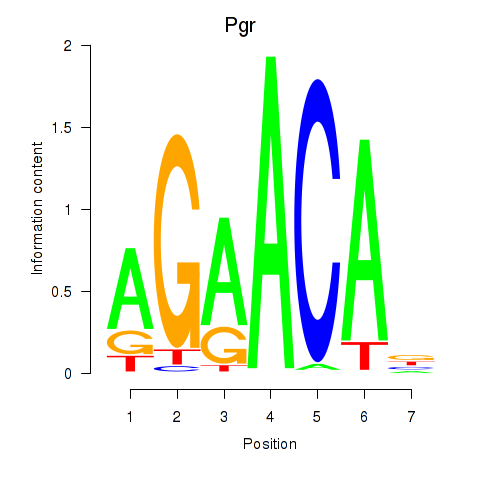
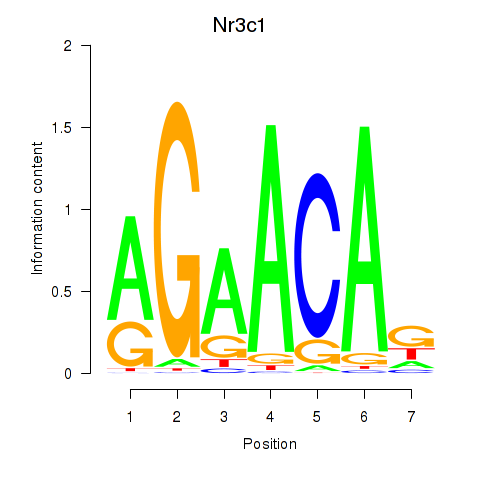
Results for Pgr_Nr3c1
Z-value: 1.20
Transcription factors associated with Pgr_Nr3c1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Pgr
|
ENSMUSG00000031870.17 | Pgr |
|
Nr3c1
|
ENSMUSG00000024431.16 | Nr3c1 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Nr3c1 | mm39_v1_chr18_-_39652468_39652478 | -0.62 | 4.7e-09 | Click! |
| Pgr | mm39_v1_chr9_+_8899829_8899857 | -0.49 | 1.1e-05 | Click! |
Activity profile of Pgr_Nr3c1 motif
Sorted Z-values of Pgr_Nr3c1 motif
Network of associatons between targets according to the STRING database.
First level regulatory network of Pgr_Nr3c1
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr10_+_115653152 | 13.21 |
ENSMUST00000080630.11
ENSMUST00000179196.3 ENSMUST00000035563.15 |
Tspan8
|
tetraspanin 8 |
| chr5_-_103777145 | 12.19 |
ENSMUST00000031263.2
|
Slc10a6
|
solute carrier family 10 (sodium/bile acid cotransporter family), member 6 |
| chr2_-_103133503 | 10.09 |
ENSMUST00000111176.9
|
Ehf
|
ets homologous factor |
| chr2_-_103133524 | 9.73 |
ENSMUST00000090475.10
|
Ehf
|
ets homologous factor |
| chr6_-_124519240 | 9.36 |
ENSMUST00000159463.8
ENSMUST00000162844.2 ENSMUST00000160505.8 ENSMUST00000162443.8 |
C1s1
|
complement component 1, s subcomponent 1 |
| chr1_-_71692320 | 7.08 |
ENSMUST00000186940.7
ENSMUST00000188894.7 ENSMUST00000188674.7 ENSMUST00000189821.7 ENSMUST00000187938.7 ENSMUST00000190780.7 ENSMUST00000186736.2 ENSMUST00000055226.13 ENSMUST00000186129.7 |
Fn1
|
fibronectin 1 |
| chr16_+_34815177 | 6.52 |
ENSMUST00000231589.2
|
Mylk
|
myosin, light polypeptide kinase |
| chr1_+_88066086 | 6.31 |
ENSMUST00000014263.6
|
Ugt1a6a
|
UDP glucuronosyltransferase 1 family, polypeptide A6A |
| chr14_+_99536111 | 6.27 |
ENSMUST00000005279.8
|
Klf5
|
Kruppel-like factor 5 |
| chr11_-_95966407 | 6.01 |
ENSMUST00000107686.8
ENSMUST00000107684.2 |
Atp5g1
|
ATP synthase, H+ transporting, mitochondrial F0 complex, subunit C1 (subunit 9) |
| chr7_-_30623592 | 5.89 |
ENSMUST00000217812.2
ENSMUST00000074671.9 |
Hamp2
|
hepcidin antimicrobial peptide 2 |
| chr6_-_87312743 | 5.65 |
ENSMUST00000042025.12
ENSMUST00000205033.2 |
Antxr1
|
anthrax toxin receptor 1 |
| chr6_+_125298296 | 5.23 |
ENSMUST00000081440.14
|
Scnn1a
|
sodium channel, nonvoltage-gated 1 alpha |
| chr11_+_62711057 | 5.02 |
ENSMUST00000055006.12
ENSMUST00000072639.10 |
Trim16
|
tripartite motif-containing 16 |
| chr6_-_87312681 | 5.02 |
ENSMUST00000204805.3
|
Antxr1
|
anthrax toxin receptor 1 |
| chr6_+_125297596 | 4.98 |
ENSMUST00000176655.8
ENSMUST00000176110.8 |
Scnn1a
|
sodium channel, nonvoltage-gated 1 alpha |
| chr9_+_98372575 | 4.98 |
ENSMUST00000035029.3
|
Rbp2
|
retinol binding protein 2, cellular |
| chr11_+_62711295 | 4.67 |
ENSMUST00000108703.2
|
Trim16
|
tripartite motif-containing 16 |
| chr13_-_55979191 | 4.26 |
ENSMUST00000021968.7
|
Pitx1
|
paired-like homeodomain transcription factor 1 |
| chr4_+_134042423 | 3.97 |
ENSMUST00000105875.8
ENSMUST00000030638.7 |
Trim63
|
tripartite motif-containing 63 |
| chr17_+_43671314 | 3.92 |
ENSMUST00000226087.2
|
Adgrf5
|
adhesion G protein-coupled receptor F5 |
| chr15_+_25843225 | 3.86 |
ENSMUST00000022881.15
|
Retreg1
|
reticulophagy regulator 1 |
| chr11_-_121410152 | 3.76 |
ENSMUST00000092298.6
|
Zfp750
|
zinc finger protein 750 |
| chr8_+_125302843 | 3.72 |
ENSMUST00000093033.6
ENSMUST00000133086.2 |
Capn9
|
calpain 9 |
| chr7_+_30252687 | 3.70 |
ENSMUST00000044048.8
|
Hspb6
|
heat shock protein, alpha-crystallin-related, B6 |
| chrX_+_100427331 | 3.69 |
ENSMUST00000119190.2
|
Gjb1
|
gap junction protein, beta 1 |
| chrX_+_158038778 | 3.58 |
ENSMUST00000126686.8
ENSMUST00000033671.13 |
Rps6ka3
|
ribosomal protein S6 kinase polypeptide 3 |
| chrX_+_156481906 | 3.55 |
ENSMUST00000136141.2
ENSMUST00000190091.7 |
Smpx
|
small muscle protein, X-linked |
| chr5_-_66238313 | 3.53 |
ENSMUST00000202700.4
ENSMUST00000094757.9 ENSMUST00000113724.6 |
Rbm47
|
RNA binding motif protein 47 |
| chr11_-_95966477 | 3.52 |
ENSMUST00000090541.12
|
Atp5g1
|
ATP synthase, H+ transporting, mitochondrial F0 complex, subunit C1 (subunit 9) |
| chr17_-_48739874 | 3.51 |
ENSMUST00000046549.5
|
Apobec2
|
apolipoprotein B mRNA editing enzyme, catalytic polypeptide 2 |
| chrX_+_156482116 | 3.48 |
ENSMUST00000112521.8
|
Smpx
|
small muscle protein, X-linked |
| chr11_+_73244561 | 3.45 |
ENSMUST00000108465.4
|
Olfr20
|
olfactory receptor 20 |
| chr9_+_108356935 | 3.28 |
ENSMUST00000194147.2
ENSMUST00000065014.10 ENSMUST00000195483.6 ENSMUST00000195058.2 |
Lamb2
|
laminin, beta 2 |
| chr7_+_30487322 | 3.27 |
ENSMUST00000189673.7
ENSMUST00000190990.7 ENSMUST00000189962.7 ENSMUST00000187493.7 ENSMUST00000098559.3 |
Krtdap
|
keratinocyte differentiation associated protein |
| chrX_+_158039107 | 3.27 |
ENSMUST00000148570.8
|
Rps6ka3
|
ribosomal protein S6 kinase polypeptide 3 |
| chr2_+_72128239 | 3.26 |
ENSMUST00000144111.2
|
Map3k20
|
mitogen-activated protein kinase kinase kinase 20 |
| chr16_+_48637219 | 3.26 |
ENSMUST00000023328.8
|
Retnlb
|
resistin like beta |
| chrX_+_158038915 | 3.16 |
ENSMUST00000112492.8
|
Rps6ka3
|
ribosomal protein S6 kinase polypeptide 3 |
| chr19_-_45224251 | 3.14 |
ENSMUST00000099401.6
|
Lbx1
|
ladybird homeobox 1 |
| chr15_-_98507913 | 3.08 |
ENSMUST00000226500.2
ENSMUST00000227501.2 |
Adcy6
|
adenylate cyclase 6 |
| chr11_-_3881789 | 3.03 |
ENSMUST00000109992.8
ENSMUST00000109988.2 |
Tcn2
|
transcobalamin 2 |
| chr7_+_44866095 | 3.02 |
ENSMUST00000209437.2
|
Tead2
|
TEA domain family member 2 |
| chr11_-_3881995 | 2.90 |
ENSMUST00000020710.11
ENSMUST00000109989.10 ENSMUST00000109991.8 ENSMUST00000109993.9 |
Tcn2
|
transcobalamin 2 |
| chr6_+_34686373 | 2.86 |
ENSMUST00000115021.8
|
Cald1
|
caldesmon 1 |
| chr2_+_14179324 | 2.82 |
ENSMUST00000077517.9
|
Tmem236
|
transmembrane protein 236 |
| chr6_+_42222841 | 2.77 |
ENSMUST00000031897.8
|
Gstk1
|
glutathione S-transferase kappa 1 |
| chr6_-_3494587 | 2.75 |
ENSMUST00000049985.15
|
Hepacam2
|
HEPACAM family member 2 |
| chr18_-_52662728 | 2.73 |
ENSMUST00000025409.9
|
Lox
|
lysyl oxidase |
| chr10_-_107330580 | 2.67 |
ENSMUST00000044210.5
|
Myf6
|
myogenic factor 6 |
| chr6_+_17491231 | 2.64 |
ENSMUST00000080469.12
|
Met
|
met proto-oncogene |
| chr2_+_91086489 | 2.63 |
ENSMUST00000154959.8
ENSMUST00000059566.11 |
Pacsin3
|
protein kinase C and casein kinase substrate in neurons 3 |
| chr14_+_76082736 | 2.62 |
ENSMUST00000142061.3
|
Tpt1
|
tumor protein, translationally-controlled 1 |
| chr2_+_25179903 | 2.60 |
ENSMUST00000028337.7
|
Lrrc26
|
leucine rich repeat containing 26 |
| chr11_-_3881960 | 2.59 |
ENSMUST00000109990.8
|
Tcn2
|
transcobalamin 2 |
| chr2_-_76812799 | 2.53 |
ENSMUST00000011934.13
ENSMUST00000099981.10 ENSMUST00000099980.10 ENSMUST00000111882.9 ENSMUST00000140091.8 |
Ttn
|
titin |
| chr5_+_31070739 | 2.50 |
ENSMUST00000031055.8
|
Emilin1
|
elastin microfibril interfacer 1 |
| chr2_-_76503382 | 2.46 |
ENSMUST00000002809.14
|
Fkbp7
|
FK506 binding protein 7 |
| chr17_-_46342739 | 2.43 |
ENSMUST00000024747.14
|
Vegfa
|
vascular endothelial growth factor A |
| chr1_+_135727140 | 2.42 |
ENSMUST00000152208.8
ENSMUST00000152075.8 |
Tnni1
|
troponin I, skeletal, slow 1 |
| chr10_+_56253531 | 2.40 |
ENSMUST00000220194.2
ENSMUST00000218834.2 |
Gja1
|
gap junction protein, alpha 1 |
| chr3_-_89294430 | 2.38 |
ENSMUST00000107433.8
|
Zbtb7b
|
zinc finger and BTB domain containing 7B |
| chr6_+_125298168 | 2.38 |
ENSMUST00000176365.2
|
Scnn1a
|
sodium channel, nonvoltage-gated 1 alpha |
| chr13_-_114595122 | 2.32 |
ENSMUST00000231252.2
|
Fst
|
follistatin |
| chr15_-_98796373 | 2.30 |
ENSMUST00000229775.2
ENSMUST00000023737.6 |
Dhh
|
desert hedgehog |
| chr5_+_145063568 | 2.27 |
ENSMUST00000138922.2
|
Arpc1b
|
actin related protein 2/3 complex, subunit 1B |
| chr5_-_31296173 | 2.24 |
ENSMUST00000043475.7
ENSMUST00000201184.2 |
Ucn
|
urocortin |
| chr7_-_109215960 | 2.23 |
ENSMUST00000077909.9
|
Denn2b
|
DENN domain containing 2B |
| chr2_+_91086299 | 2.20 |
ENSMUST00000134699.8
|
Pacsin3
|
protein kinase C and casein kinase substrate in neurons 3 |
| chr5_-_121710768 | 2.16 |
ENSMUST00000200541.5
|
Aldh2
|
aldehyde dehydrogenase 2, mitochondrial |
| chr4_+_99952988 | 2.09 |
ENSMUST00000039630.6
|
Ror1
|
receptor tyrosine kinase-like orphan receptor 1 |
| chr9_-_79626002 | 2.02 |
ENSMUST00000121227.8
|
Col12a1
|
collagen, type XII, alpha 1 |
| chr12_-_73093953 | 1.95 |
ENSMUST00000050029.8
|
Six1
|
sine oculis-related homeobox 1 |
| chr5_-_24806960 | 1.94 |
ENSMUST00000030791.12
|
Smarcd3
|
SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily d, member 3 |
| chr19_-_12098064 | 1.90 |
ENSMUST00000087822.3
|
Olfr76
|
olfactory receptor 76 |
| chr4_+_152410291 | 1.88 |
ENSMUST00000103191.11
ENSMUST00000139685.8 ENSMUST00000188151.2 |
Rpl22
|
ribosomal protein L22 |
| chr10_-_105410280 | 1.82 |
ENSMUST00000061506.9
|
Tmtc2
|
transmembrane and tetratricopeptide repeat containing 2 |
| chr4_-_119279551 | 1.82 |
ENSMUST00000106316.2
ENSMUST00000030385.13 |
Ppcs
|
phosphopantothenoylcysteine synthetase |
| chr11_+_96173475 | 1.78 |
ENSMUST00000168043.2
|
Hoxb8
|
homeobox B8 |
| chr14_+_67953547 | 1.77 |
ENSMUST00000078053.13
|
Kctd9
|
potassium channel tetramerisation domain containing 9 |
| chr9_-_117080869 | 1.77 |
ENSMUST00000172564.3
|
Rbms3
|
RNA binding motif, single stranded interacting protein |
| chr2_-_58050494 | 1.76 |
ENSMUST00000028175.7
|
Cytip
|
cytohesin 1 interacting protein |
| chr6_-_47790272 | 1.70 |
ENSMUST00000077290.9
|
Pdia4
|
protein disulfide isomerase associated 4 |
| chr9_-_79625800 | 1.68 |
ENSMUST00000071750.13
|
Col12a1
|
collagen, type XII, alpha 1 |
| chr6_+_68098030 | 1.64 |
ENSMUST00000103317.3
|
Igkv1-117
|
immunoglobulin kappa variable 1-117 |
| chr5_+_30971915 | 1.61 |
ENSMUST00000031058.15
|
Mapre3
|
microtubule-associated protein, RP/EB family, member 3 |
| chr11_+_100513747 | 1.60 |
ENSMUST00000142993.2
|
Nkiras2
|
NFKB inhibitor interacting Ras-like protein 2 |
| chr3_-_97775557 | 1.58 |
ENSMUST00000107038.6
|
Pde4dip
|
phosphodiesterase 4D interacting protein (myomegalin) |
| chr1_+_135727228 | 1.55 |
ENSMUST00000154463.8
ENSMUST00000139986.8 |
Tnni1
|
troponin I, skeletal, slow 1 |
| chr15_+_102862862 | 1.54 |
ENSMUST00000001701.4
|
Hoxc11
|
homeobox C11 |
| chr2_+_74552322 | 1.53 |
ENSMUST00000047904.4
|
Hoxd4
|
homeobox D4 |
| chr17_-_25300112 | 1.52 |
ENSMUST00000024984.7
|
Tmem204
|
transmembrane protein 204 |
| chr7_+_44866635 | 1.47 |
ENSMUST00000097216.5
ENSMUST00000209343.2 ENSMUST00000209678.2 |
Tead2
|
TEA domain family member 2 |
| chr10_+_99851679 | 1.46 |
ENSMUST00000130190.8
ENSMUST00000218200.2 ENSMUST00000020129.8 |
Kitl
|
kit ligand |
| chr19_-_55304392 | 1.46 |
ENSMUST00000224291.2
ENSMUST00000225495.2 ENSMUST00000076891.7 ENSMUST00000224897.2 |
Zdhhc6
|
zinc finger, DHHC domain containing 6 |
| chr11_+_96173355 | 1.46 |
ENSMUST00000125410.2
|
Hoxb8
|
homeobox B8 |
| chr2_-_126718129 | 1.45 |
ENSMUST00000103224.10
|
Trpm7
|
transient receptor potential cation channel, subfamily M, member 7 |
| chr5_+_24569802 | 1.44 |
ENSMUST00000115090.6
ENSMUST00000030834.7 |
Nos3
|
nitric oxide synthase 3, endothelial cell |
| chr7_-_109215754 | 1.44 |
ENSMUST00000084738.5
|
Denn2b
|
DENN domain containing 2B |
| chr19_+_55882942 | 1.42 |
ENSMUST00000142291.8
|
Tcf7l2
|
transcription factor 7 like 2, T cell specific, HMG box |
| chr16_-_5040125 | 1.41 |
ENSMUST00000050160.6
|
AU021092
|
expressed sequence AU021092 |
| chr15_-_65784103 | 1.38 |
ENSMUST00000079776.14
|
Oc90
|
otoconin 90 |
| chr1_+_135727571 | 1.34 |
ENSMUST00000148201.8
|
Tnni1
|
troponin I, skeletal, slow 1 |
| chr18_+_11766333 | 1.34 |
ENSMUST00000115861.9
|
Rbbp8
|
retinoblastoma binding protein 8, endonuclease |
| chr19_+_4560500 | 1.32 |
ENSMUST00000068004.13
ENSMUST00000224726.3 |
Pcx
|
pyruvate carboxylase |
| chr17_+_35268942 | 1.31 |
ENSMUST00000007257.10
|
Clic1
|
chloride intracellular channel 1 |
| chr1_+_135764092 | 1.29 |
ENSMUST00000188028.7
ENSMUST00000178204.8 ENSMUST00000190451.7 ENSMUST00000189732.7 ENSMUST00000189355.7 |
Tnnt2
|
troponin T2, cardiac |
| chr14_+_67953687 | 1.25 |
ENSMUST00000150768.8
|
Kctd9
|
potassium channel tetramerisation domain containing 9 |
| chr9_+_49014020 | 1.23 |
ENSMUST00000070390.12
ENSMUST00000167095.8 |
Tmprss5
|
transmembrane protease, serine 5 (spinesin) |
| chr9_+_103917821 | 1.19 |
ENSMUST00000216593.2
ENSMUST00000147249.3 |
Nphp3
Gm28305
|
nephronophthisis 3 (adolescent) predicted gene 28305 |
| chr14_+_8293646 | 1.18 |
ENSMUST00000035250.6
|
Olfr720
|
olfactory receptor 720 |
| chr6_-_138056914 | 1.14 |
ENSMUST00000171804.4
|
Slc15a5
|
solute carrier family 15, member 5 |
| chr11_-_117716918 | 1.14 |
ENSMUST00000026661.4
|
Tk1
|
thymidine kinase 1 |
| chr7_+_130179063 | 1.11 |
ENSMUST00000207918.2
ENSMUST00000215492.2 ENSMUST00000084513.12 ENSMUST00000059145.14 |
Tacc2
|
transforming, acidic coiled-coil containing protein 2 |
| chr6_+_68247469 | 1.11 |
ENSMUST00000103321.3
|
Igkv1-110
|
immunoglobulin kappa variable 1-110 |
| chr15_-_65784246 | 1.07 |
ENSMUST00000060522.11
|
Oc90
|
otoconin 90 |
| chr2_+_79538124 | 1.06 |
ENSMUST00000090760.9
ENSMUST00000040863.11 ENSMUST00000111780.3 |
Ppp1r1c
|
protein phosphatase 1, regulatory inhibitor subunit 1C |
| chr14_+_54429757 | 1.03 |
ENSMUST00000103714.2
|
Traj27
|
T cell receptor alpha joining 27 |
| chr5_+_30972067 | 1.01 |
ENSMUST00000200692.4
|
Mapre3
|
microtubule-associated protein, RP/EB family, member 3 |
| chr16_+_90627719 | 0.99 |
ENSMUST00000037539.15
ENSMUST00000231280.2 |
Eva1c
|
eva-1 homolog C (C. elegans) |
| chr3_-_137773149 | 0.99 |
ENSMUST00000053318.4
|
Gm5105
|
predicted gene 5105 |
| chr2_-_153079828 | 0.99 |
ENSMUST00000109795.2
|
Plagl2
|
pleiomorphic adenoma gene-like 2 |
| chr3_+_87704258 | 0.97 |
ENSMUST00000029711.9
ENSMUST00000107582.3 |
Insrr
|
insulin receptor-related receptor |
| chr2_-_126718037 | 0.97 |
ENSMUST00000028843.12
|
Trpm7
|
transient receptor potential cation channel, subfamily M, member 7 |
| chr14_-_50558325 | 0.96 |
ENSMUST00000050928.3
|
Olfr734
|
olfactory receptor 734 |
| chr4_-_52856572 | 0.94 |
ENSMUST00000051520.3
|
Olfr273
|
olfactory receptor 273 |
| chr17_-_34962823 | 0.94 |
ENSMUST00000069507.9
|
C4b
|
complement component 4B (Chido blood group) |
| chr7_+_75259778 | 0.92 |
ENSMUST00000207923.2
|
Akap13
|
A kinase (PRKA) anchor protein 13 |
| chr6_-_54941673 | 0.92 |
ENSMUST00000203837.2
|
Nod1
|
nucleotide-binding oligomerization domain containing 1 |
| chr14_+_67953584 | 0.92 |
ENSMUST00000145542.8
ENSMUST00000125212.2 |
Kctd9
|
potassium channel tetramerisation domain containing 9 |
| chr15_-_81283795 | 0.91 |
ENSMUST00000023039.15
|
St13
|
suppression of tumorigenicity 13 |
| chr14_+_63235512 | 0.91 |
ENSMUST00000100492.5
|
Defb47
|
defensin beta 47 |
| chr4_+_99184137 | 0.91 |
ENSMUST00000094955.3
|
Gm12689
|
predicted gene 12689 |
| chr11_-_110058899 | 0.87 |
ENSMUST00000044850.4
|
Abca9
|
ATP-binding cassette, sub-family A (ABC1), member 9 |
| chr3_-_95811993 | 0.87 |
ENSMUST00000147962.3
ENSMUST00000036181.15 |
Car14
|
carbonic anhydrase 14 |
| chr4_-_43821433 | 0.87 |
ENSMUST00000079465.5
|
Olfr156
|
olfactory receptor 156 |
| chr8_+_47192911 | 0.84 |
ENSMUST00000208433.2
|
Irf2
|
interferon regulatory factor 2 |
| chr14_-_32186721 | 0.82 |
ENSMUST00000191501.2
|
Slc18a3
|
solute carrier family 18 (vesicular monoamine), member 3 |
| chr16_+_38405718 | 0.81 |
ENSMUST00000165631.2
|
Tmem39a
|
transmembrane protein 39a |
| chr9_-_119548098 | 0.80 |
ENSMUST00000084787.6
|
Scn10a
|
sodium channel, voltage-gated, type X, alpha |
| chr16_+_90628001 | 0.79 |
ENSMUST00000099543.10
|
Eva1c
|
eva-1 homolog C (C. elegans) |
| chr15_+_78926085 | 0.78 |
ENSMUST00000058004.4
|
Galr3
|
galanin receptor 3 |
| chr5_+_139338362 | 0.78 |
ENSMUST00000031521.13
|
Cyp2w1
|
cytochrome P450, family 2, subfamily w, polypeptide 1 |
| chr16_+_43067641 | 0.77 |
ENSMUST00000079441.13
ENSMUST00000114691.8 |
Zbtb20
|
zinc finger and BTB domain containing 20 |
| chr17_-_71153283 | 0.76 |
ENSMUST00000156484.2
|
Tgif1
|
TGFB-induced factor homeobox 1 |
| chr4_+_19280850 | 0.76 |
ENSMUST00000102999.2
|
Cngb3
|
cyclic nucleotide gated channel beta 3 |
| chr6_-_146855880 | 0.75 |
ENSMUST00000111622.2
ENSMUST00000036592.15 |
1700034J05Rik
|
RIKEN cDNA 1700034J05 gene |
| chr16_-_50411484 | 0.75 |
ENSMUST00000062439.6
|
Ccdc54
|
coiled-coil domain containing 54 |
| chr12_-_75782406 | 0.74 |
ENSMUST00000220285.2
|
Sgpp1
|
sphingosine-1-phosphate phosphatase 1 |
| chr19_+_53781721 | 0.73 |
ENSMUST00000162910.2
|
Rbm20
|
RNA binding motif protein 20 |
| chr16_+_51852435 | 0.71 |
ENSMUST00000227879.2
|
Cblb
|
Casitas B-lineage lymphoma b |
| chr8_+_47193275 | 0.69 |
ENSMUST00000207571.3
|
Irf2
|
interferon regulatory factor 2 |
| chr10_-_53252210 | 0.68 |
ENSMUST00000095691.7
|
Cep85l
|
centrosomal protein 85-like |
| chr11_-_100830366 | 0.67 |
ENSMUST00000127638.8
|
Stat3
|
signal transducer and activator of transcription 3 |
| chr1_-_75110511 | 0.63 |
ENSMUST00000027405.6
|
Slc23a3
|
solute carrier family 23 (nucleobase transporters), member 3 |
| chr5_+_124065481 | 0.62 |
ENSMUST00000166129.5
|
Gm43518
|
predicted gene 43518 |
| chr8_+_47192767 | 0.62 |
ENSMUST00000034041.9
ENSMUST00000208507.2 ENSMUST00000207105.2 |
Irf2
|
interferon regulatory factor 2 |
| chr19_+_13745671 | 0.60 |
ENSMUST00000061669.3
|
Olfr1495
|
olfactory receptor 1495 |
| chr7_-_28597523 | 0.60 |
ENSMUST00000216863.2
|
Actn4
|
actinin alpha 4 |
| chr13_-_103901010 | 0.59 |
ENSMUST00000210489.2
|
Srek1
|
splicing regulatory glutamine/lysine-rich protein 1 |
| chr16_-_59036978 | 0.56 |
ENSMUST00000099657.5
|
Olfr199
|
olfactory receptor 199 |
| chr7_+_28834391 | 0.56 |
ENSMUST00000160194.8
|
Rasgrp4
|
RAS guanyl releasing protein 4 |
| chr11_+_98277276 | 0.56 |
ENSMUST00000041301.8
|
Pnmt
|
phenylethanolamine-N-methyltransferase |
| chr2_+_153742294 | 0.55 |
ENSMUST00000088955.12
ENSMUST00000135501.3 |
Bpifb6
|
BPI fold containing family B, member 6 |
| chr11_+_117716711 | 0.54 |
ENSMUST00000073388.13
|
Afmid
|
arylformamidase |
| chr7_-_126184935 | 0.53 |
ENSMUST00000084589.11
|
Cln3
|
ceroid lipofuscinosis, neuronal 3, juvenile (Batten, Spielmeyer-Vogt disease) |
| chr5_-_100521343 | 0.53 |
ENSMUST00000182433.8
|
Sec31a
|
Sec31 homolog A (S. cerevisiae) |
| chr3_-_151899470 | 0.53 |
ENSMUST00000050073.13
|
Dnajb4
|
DnaJ heat shock protein family (Hsp40) member B4 |
| chr12_-_118930130 | 0.53 |
ENSMUST00000035515.5
|
Abcb5
|
ATP-binding cassette, sub-family B (MDR/TAP), member 5 |
| chr2_-_93826699 | 0.52 |
ENSMUST00000183110.2
|
Gm27027
|
predicted gene, 27027 |
| chr1_-_126758369 | 0.51 |
ENSMUST00000112583.8
ENSMUST00000094609.10 |
Nckap5
|
NCK-associated protein 5 |
| chr1_-_126758520 | 0.51 |
ENSMUST00000162646.8
|
Nckap5
|
NCK-associated protein 5 |
| chr9_-_119548388 | 0.45 |
ENSMUST00000213392.2
|
Scn10a
|
sodium channel, voltage-gated, type X, alpha |
| chr10_-_63257568 | 0.43 |
ENSMUST00000054760.6
|
Gm7075
|
predicted gene 7075 |
| chr14_-_51433380 | 0.43 |
ENSMUST00000051274.2
|
Ang2
|
angiogenin, ribonuclease A family, member 2 |
| chr6_+_119456629 | 0.42 |
ENSMUST00000032094.7
|
Fbxl14
|
F-box and leucine-rich repeat protein 14 |
| chr11_+_69047815 | 0.41 |
ENSMUST00000036424.3
|
Alox12b
|
arachidonate 12-lipoxygenase, 12R type |
| chr17_+_33418036 | 0.39 |
ENSMUST00000112165.4
|
Olfr239
|
olfactory receptor 239 |
| chr11_+_117716759 | 0.39 |
ENSMUST00000149668.2
|
Afmid
|
arylformamidase |
| chr11_-_120238917 | 0.38 |
ENSMUST00000106215.11
|
Actg1
|
actin, gamma, cytoplasmic 1 |
| chr1_-_65112680 | 0.38 |
ENSMUST00000114064.3
|
Crygc
|
crystallin, gamma C |
| chr10_+_69932930 | 0.36 |
ENSMUST00000147545.8
|
Ccdc6
|
coiled-coil domain containing 6 |
| chr2_-_63014514 | 0.35 |
ENSMUST00000112452.2
|
Kcnh7
|
potassium voltage-gated channel, subfamily H (eag-related), member 7 |
| chr16_-_92118313 | 0.35 |
ENSMUST00000062638.8
|
Fam243
|
family with sequence similarity 243 |
| chr10_+_101994719 | 0.34 |
ENSMUST00000138522.8
ENSMUST00000163753.8 ENSMUST00000138016.8 |
Mgat4c
|
MGAT4 family, member C |
| chr19_+_38121248 | 0.34 |
ENSMUST00000025956.13
|
Pde6c
|
phosphodiesterase 6C, cGMP specific, cone, alpha prime |
| chr10_+_52265049 | 0.34 |
ENSMUST00000219730.2
|
Nepn
|
nephrocan |
| chr10_+_69370038 | 0.32 |
ENSMUST00000182439.8
ENSMUST00000092434.12 ENSMUST00000047061.13 ENSMUST00000092432.12 ENSMUST00000092431.12 ENSMUST00000054167.15 |
Ank3
|
ankyrin 3, epithelial |
| chr17_+_36179273 | 0.31 |
ENSMUST00000190496.2
|
Ppp1r18
|
protein phosphatase 1, regulatory subunit 18 |
| chr9_+_21336300 | 0.29 |
ENSMUST00000172482.8
ENSMUST00000174050.8 |
Dnm2
|
dynamin 2 |
| chr9_+_37903788 | 0.29 |
ENSMUST00000074611.3
|
Olfr881
|
olfactory receptor 881 |
| chr1_-_57011595 | 0.29 |
ENSMUST00000042857.14
|
Satb2
|
special AT-rich sequence binding protein 2 |
| chr11_-_48707763 | 0.29 |
ENSMUST00000140800.2
|
Trim41
|
tripartite motif-containing 41 |
| chr8_+_85957126 | 0.28 |
ENSMUST00000070849.6
|
Olfr371
|
olfactory receptor 371 |
| chr6_+_40548291 | 0.27 |
ENSMUST00000051540.5
|
Olfr460
|
olfactory receptor 460 |
| chr19_-_6134703 | 0.26 |
ENSMUST00000161548.8
|
Zfpl1
|
zinc finger like protein 1 |
| chr10_+_101994841 | 0.25 |
ENSMUST00000020039.13
|
Mgat4c
|
MGAT4 family, member C |
| chr19_+_38121214 | 0.24 |
ENSMUST00000112329.3
|
Pde6c
|
phosphodiesterase 6C, cGMP specific, cone, alpha prime |
| chr17_-_89508103 | 0.23 |
ENSMUST00000035701.6
|
Fshr
|
follicle stimulating hormone receptor |
| chr2_-_77000878 | 0.23 |
ENSMUST00000111833.3
|
Ccdc141
|
coiled-coil domain containing 141 |
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.4 | 7.1 | GO:0071288 | cellular response to mercury ion(GO:0071288) |
| 1.8 | 10.7 | GO:1903054 | negative regulation of extracellular matrix organization(GO:1903054) |
| 1.5 | 5.9 | GO:0034757 | negative regulation of iron ion transport(GO:0034757) negative regulation of iron ion transmembrane transport(GO:0034760) |
| 1.4 | 8.5 | GO:0015889 | cobalamin transport(GO:0015889) |
| 1.3 | 4.0 | GO:0014878 | response to electrical stimulus involved in regulation of muscle adaptation(GO:0014878) |
| 1.3 | 6.5 | GO:0060414 | aorta smooth muscle tissue morphogenesis(GO:0060414) |
| 1.1 | 3.3 | GO:0072313 | metanephric glomerular visceral epithelial cell differentiation(GO:0072248) metanephric glomerular visceral epithelial cell development(GO:0072249) metanephric glomerular epithelial cell differentiation(GO:0072312) metanephric glomerular epithelial cell development(GO:0072313) |
| 1.1 | 4.3 | GO:0014707 | branchiomeric skeletal muscle development(GO:0014707) |
| 1.0 | 3.1 | GO:1904117 | response to vasopressin(GO:1904116) cellular response to vasopressin(GO:1904117) |
| 1.0 | 1.9 | GO:2000729 | positive regulation of mesenchymal cell proliferation involved in ureter development(GO:2000729) |
| 0.9 | 1.7 | GO:1903334 | positive regulation of protein folding(GO:1903334) |
| 0.8 | 2.4 | GO:0038189 | neuropilin signaling pathway(GO:0038189) VEGF-activated neuropilin signaling pathway(GO:0038190) |
| 0.8 | 2.4 | GO:0060156 | vascular transport(GO:0010232) milk ejection(GO:0060156) |
| 0.8 | 2.3 | GO:0030908 | intein-mediated protein splicing(GO:0016539) protein splicing(GO:0030908) |
| 0.7 | 9.7 | GO:0048386 | positive regulation of retinoic acid receptor signaling pathway(GO:0048386) |
| 0.7 | 9.4 | GO:0001867 | complement activation, lectin pathway(GO:0001867) |
| 0.7 | 2.6 | GO:1903031 | regulation of microtubule plus-end binding(GO:1903031) positive regulation of microtubule plus-end binding(GO:1903033) |
| 0.7 | 2.6 | GO:2000383 | regulation of ectoderm development(GO:2000383) negative regulation of ectoderm development(GO:2000384) |
| 0.6 | 2.4 | GO:0043376 | regulation of CD8-positive, alpha-beta T cell differentiation(GO:0043376) |
| 0.6 | 5.0 | GO:0006776 | vitamin A metabolic process(GO:0006776) |
| 0.5 | 7.0 | GO:0016554 | cytidine to uridine editing(GO:0016554) |
| 0.5 | 6.3 | GO:0052697 | flavonoid glucuronidation(GO:0052696) xenobiotic glucuronidation(GO:0052697) |
| 0.5 | 2.5 | GO:0043056 | forward locomotion(GO:0043056) |
| 0.5 | 6.3 | GO:0032534 | regulation of microvillus assembly(GO:0032534) |
| 0.5 | 1.4 | GO:0014805 | smooth muscle adaptation(GO:0014805) |
| 0.4 | 3.9 | GO:0061709 | reticulophagy(GO:0061709) |
| 0.4 | 3.7 | GO:0015868 | purine ribonucleotide transport(GO:0015868) |
| 0.4 | 2.4 | GO:0016340 | calcium-dependent cell-matrix adhesion(GO:0016340) |
| 0.4 | 2.7 | GO:0048743 | positive regulation of skeletal muscle fiber development(GO:0048743) |
| 0.4 | 2.6 | GO:0070495 | regulation of thrombin receptor signaling pathway(GO:0070494) negative regulation of thrombin receptor signaling pathway(GO:0070495) |
| 0.4 | 5.2 | GO:0071073 | positive regulation of phospholipid biosynthetic process(GO:0071073) |
| 0.3 | 1.0 | GO:0034378 | chylomicron assembly(GO:0034378) |
| 0.3 | 1.3 | GO:0032972 | regulation of muscle filament sliding speed(GO:0032972) |
| 0.3 | 2.2 | GO:0035483 | gastric emptying(GO:0035483) negative regulation of gastric acid secretion(GO:0060455) |
| 0.3 | 1.1 | GO:0046104 | thymidine metabolic process(GO:0046104) |
| 0.3 | 9.5 | GO:0015985 | energy coupled proton transport, down electrochemical gradient(GO:0015985) ATP synthesis coupled proton transport(GO:0015986) |
| 0.3 | 1.9 | GO:0003219 | cardiac right ventricle formation(GO:0003219) |
| 0.3 | 2.7 | GO:0048251 | elastic fiber assembly(GO:0048251) |
| 0.3 | 5.3 | GO:0014883 | transition between fast and slow fiber(GO:0014883) |
| 0.3 | 13.0 | GO:0050891 | multicellular organismal water homeostasis(GO:0050891) |
| 0.2 | 1.0 | GO:0071469 | cellular response to alkaline pH(GO:0071469) |
| 0.2 | 1.5 | GO:0070666 | mast cell proliferation(GO:0070662) regulation of mast cell proliferation(GO:0070666) positive regulation of mast cell proliferation(GO:0070668) positive regulation of hematopoietic stem cell proliferation(GO:1902035) |
| 0.2 | 13.2 | GO:1900047 | negative regulation of blood coagulation(GO:0030195) negative regulation of hemostasis(GO:1900047) |
| 0.2 | 6.4 | GO:0021516 | dorsal spinal cord development(GO:0021516) |
| 0.2 | 4.5 | GO:0048368 | lateral mesoderm development(GO:0048368) |
| 0.2 | 0.8 | GO:1901376 | mycotoxin metabolic process(GO:0043385) aflatoxin metabolic process(GO:0046222) organic heteropentacyclic compound metabolic process(GO:1901376) |
| 0.2 | 0.6 | GO:0042414 | epinephrine metabolic process(GO:0042414) |
| 0.2 | 1.4 | GO:0048625 | myoblast fate commitment(GO:0048625) |
| 0.2 | 1.2 | GO:0035469 | determination of pancreatic left/right asymmetry(GO:0035469) maintenance of organ identity(GO:0048496) |
| 0.2 | 1.1 | GO:0061084 | regulation of protein refolding(GO:0061083) negative regulation of protein refolding(GO:0061084) |
| 0.1 | 0.6 | GO:1902396 | protein localization to bicellular tight junction(GO:1902396) |
| 0.1 | 1.3 | GO:0010792 | DNA double-strand break processing involved in repair via single-strand annealing(GO:0010792) |
| 0.1 | 5.1 | GO:0097320 | membrane tubulation(GO:0097320) |
| 0.1 | 2.6 | GO:1903818 | positive regulation of voltage-gated potassium channel activity(GO:1903818) |
| 0.1 | 0.7 | GO:1902724 | positive regulation of skeletal muscle satellite cell proliferation(GO:1902724) positive regulation of growth factor dependent skeletal muscle satellite cell proliferation(GO:1902728) |
| 0.1 | 0.7 | GO:0006668 | sphinganine-1-phosphate metabolic process(GO:0006668) |
| 0.1 | 2.3 | GO:0051798 | positive regulation of hair follicle development(GO:0051798) |
| 0.1 | 0.9 | GO:0071883 | activation of MAPK activity by adrenergic receptor signaling pathway(GO:0071883) |
| 0.1 | 0.9 | GO:0002604 | regulation of dendritic cell antigen processing and presentation(GO:0002604) positive regulation of dendritic cell antigen processing and presentation(GO:0002606) |
| 0.1 | 1.3 | GO:0086028 | bundle of His cell to Purkinje myocyte signaling(GO:0086028) bundle of His cell action potential(GO:0086043) |
| 0.1 | 0.5 | GO:0035752 | lysosomal lumen pH elevation(GO:0035752) |
| 0.1 | 3.7 | GO:0010667 | negative regulation of cardiac muscle cell apoptotic process(GO:0010667) |
| 0.1 | 0.9 | GO:0015670 | carbon dioxide transport(GO:0015670) |
| 0.1 | 1.5 | GO:0060272 | embryonic skeletal joint morphogenesis(GO:0060272) |
| 0.1 | 1.8 | GO:0015937 | coenzyme A biosynthetic process(GO:0015937) |
| 0.1 | 0.9 | GO:0006569 | tryptophan catabolic process(GO:0006569) tryptophan catabolic process to kynurenine(GO:0019441) indole-containing compound catabolic process(GO:0042436) indolalkylamine catabolic process(GO:0046218) |
| 0.1 | 0.2 | GO:0001545 | primary ovarian follicle growth(GO:0001545) |
| 0.1 | 10.0 | GO:0043154 | negative regulation of cysteine-type endopeptidase activity involved in apoptotic process(GO:0043154) |
| 0.1 | 0.7 | GO:0002669 | positive regulation of T cell anergy(GO:0002669) positive regulation of lymphocyte anergy(GO:0002913) |
| 0.1 | 2.1 | GO:0014002 | astrocyte development(GO:0014002) |
| 0.1 | 3.3 | GO:0000186 | activation of MAPKK activity(GO:0000186) |
| 0.1 | 2.5 | GO:0050482 | arachidonic acid secretion(GO:0050482) arachidonate transport(GO:1903963) |
| 0.1 | 1.1 | GO:0030953 | astral microtubule organization(GO:0030953) |
| 0.0 | 1.5 | GO:0018345 | protein palmitoylation(GO:0018345) |
| 0.0 | 0.2 | GO:0071475 | cellular hyperosmotic salinity response(GO:0071475) |
| 0.0 | 2.5 | GO:0070207 | protein homotrimerization(GO:0070207) |
| 0.0 | 2.3 | GO:0034314 | Arp2/3 complex-mediated actin nucleation(GO:0034314) |
| 0.0 | 0.8 | GO:0048387 | negative regulation of retinoic acid receptor signaling pathway(GO:0048387) |
| 0.0 | 1.5 | GO:0001945 | lymph vessel development(GO:0001945) |
| 0.0 | 0.3 | GO:0021902 | commitment of neuronal cell to specific neuron type in forebrain(GO:0021902) |
| 0.0 | 18.9 | GO:0050673 | epithelial cell proliferation(GO:0050673) |
| 0.0 | 0.6 | GO:0046549 | retinal cone cell development(GO:0046549) |
| 0.0 | 2.8 | GO:0006749 | glutathione metabolic process(GO:0006749) |
| 0.0 | 0.5 | GO:0090110 | cargo loading into COPII-coated vesicle(GO:0090110) |
| 0.0 | 1.3 | GO:1900087 | positive regulation of G1/S transition of mitotic cell cycle(GO:1900087) |
| 0.0 | 2.9 | GO:0032092 | positive regulation of protein binding(GO:0032092) |
| 0.0 | 2.5 | GO:0061077 | chaperone-mediated protein folding(GO:0061077) |
| 0.0 | 0.1 | GO:1901252 | regulation of intracellular transport of viral material(GO:1901252) |
| 0.0 | 0.1 | GO:0021691 | cerebellar Purkinje cell layer maturation(GO:0021691) |
| 0.0 | 0.1 | GO:0080154 | regulation of fertilization(GO:0080154) |
| 0.0 | 1.7 | GO:0070527 | platelet aggregation(GO:0070527) |
| 0.0 | 1.7 | GO:0002181 | cytoplasmic translation(GO:0002181) |
| 0.0 | 0.8 | GO:0030818 | negative regulation of cAMP biosynthetic process(GO:0030818) |
| 0.0 | 2.4 | GO:0007586 | digestion(GO:0007586) |
| 0.0 | 0.6 | GO:0032728 | positive regulation of interferon-beta production(GO:0032728) |
| 0.0 | 4.6 | GO:0050907 | detection of chemical stimulus involved in sensory perception(GO:0050907) |
| 0.0 | 1.8 | GO:0045727 | positive regulation of translation(GO:0045727) |
| 0.0 | 6.0 | GO:0015711 | organic anion transport(GO:0015711) |
| 0.0 | 1.9 | GO:0007098 | centrosome cycle(GO:0007098) |
| 0.0 | 3.7 | GO:0008544 | epidermis development(GO:0008544) |
| 0.0 | 0.1 | GO:1902969 | mitotic DNA replication(GO:1902969) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.1 | 4.5 | GO:0071149 | TEAD-2-YAP complex(GO:0071149) |
| 0.9 | 7.0 | GO:0005927 | muscle tendon junction(GO:0005927) |
| 0.8 | 3.3 | GO:0005608 | laminin-3 complex(GO:0005608) |
| 0.8 | 7.1 | GO:0005577 | fibrinogen complex(GO:0005577) |
| 0.6 | 10.7 | GO:0031258 | lamellipodium membrane(GO:0031258) |
| 0.6 | 3.7 | GO:0005593 | FACIT collagen trimer(GO:0005593) |
| 0.6 | 9.5 | GO:0000276 | mitochondrial proton-transporting ATP synthase complex, coupling factor F(o)(GO:0000276) |
| 0.6 | 2.3 | GO:0036284 | tubulobulbar complex(GO:0036284) |
| 0.3 | 6.6 | GO:0005861 | troponin complex(GO:0005861) |
| 0.3 | 1.2 | GO:0097543 | ciliary inversin compartment(GO:0097543) |
| 0.3 | 2.6 | GO:0045298 | tubulin complex(GO:0045298) |
| 0.3 | 9.6 | GO:0034706 | sodium channel complex(GO:0034706) |
| 0.3 | 2.9 | GO:0030478 | actin cap(GO:0030478) |
| 0.2 | 6.1 | GO:0005922 | connexon complex(GO:0005922) |
| 0.2 | 5.2 | GO:0043196 | varicosity(GO:0043196) |
| 0.2 | 0.8 | GO:0030121 | AP-1 adaptor complex(GO:0030121) |
| 0.2 | 3.3 | GO:0042599 | lamellar body(GO:0042599) |
| 0.1 | 1.4 | GO:0070369 | beta-catenin-TCF7L2 complex(GO:0070369) |
| 0.1 | 3.1 | GO:0031528 | microvillus membrane(GO:0031528) |
| 0.1 | 1.7 | GO:0034663 | endoplasmic reticulum chaperone complex(GO:0034663) |
| 0.1 | 2.5 | GO:0005859 | muscle myosin complex(GO:0005859) |
| 0.1 | 1.9 | GO:0071564 | npBAF complex(GO:0071564) |
| 0.1 | 4.9 | GO:0032154 | cleavage furrow(GO:0032154) cell surface furrow(GO:0097610) |
| 0.1 | 9.7 | GO:0016605 | PML body(GO:0016605) |
| 0.1 | 2.6 | GO:0035371 | microtubule plus-end(GO:0035371) |
| 0.1 | 2.5 | GO:0008305 | integrin complex(GO:0008305) |
| 0.0 | 3.5 | GO:0005801 | cis-Golgi network(GO:0005801) |
| 0.0 | 12.0 | GO:0072562 | blood microparticle(GO:0072562) |
| 0.0 | 0.2 | GO:0043564 | Ku70:Ku80 complex(GO:0043564) |
| 0.0 | 0.6 | GO:0031143 | pseudopodium(GO:0031143) |
| 0.0 | 0.5 | GO:0030127 | COPII vesicle coat(GO:0030127) |
| 0.0 | 2.7 | GO:0005581 | collagen trimer(GO:0005581) |
| 0.0 | 2.2 | GO:0009925 | basal plasma membrane(GO:0009925) |
| 0.0 | 0.1 | GO:0000811 | GINS complex(GO:0000811) |
| 0.0 | 1.9 | GO:0022625 | cytosolic large ribosomal subunit(GO:0022625) |
| 0.0 | 2.4 | GO:0005604 | basement membrane(GO:0005604) |
| 0.0 | 5.7 | GO:0005802 | trans-Golgi network(GO:0005802) |
| 0.0 | 0.1 | GO:0002081 | outer acrosomal membrane(GO:0002081) |
| 0.0 | 0.3 | GO:0005614 | interstitial matrix(GO:0005614) |
| 0.0 | 2.8 | GO:0030496 | midbody(GO:0030496) |
| 0.0 | 0.8 | GO:0005788 | endoplasmic reticulum lumen(GO:0005788) |
| 0.0 | 0.2 | GO:0016281 | eukaryotic translation initiation factor 4F complex(GO:0016281) |
| 0.0 | 7.6 | GO:0005743 | mitochondrial inner membrane(GO:0005743) |
| 0.0 | 1.5 | GO:0030175 | filopodium(GO:0030175) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.0 | 12.6 | GO:0015280 | ligand-gated sodium channel activity(GO:0015280) |
| 0.8 | 2.5 | GO:0098640 | integrin binding involved in cell-matrix adhesion(GO:0098640) |
| 0.8 | 2.4 | GO:0043183 | vascular endothelial growth factor receptor 1 binding(GO:0043183) |
| 0.8 | 8.5 | GO:0031419 | cobalamin binding(GO:0031419) |
| 0.7 | 9.7 | GO:0019966 | interleukin-1 binding(GO:0019966) |
| 0.7 | 6.5 | GO:0004687 | myosin light chain kinase activity(GO:0004687) |
| 0.7 | 7.1 | GO:0045340 | mercury ion binding(GO:0045340) |
| 0.7 | 2.6 | GO:0005008 | hepatocyte growth factor-activated receptor activity(GO:0005008) |
| 0.6 | 2.2 | GO:0051431 | corticotropin-releasing hormone receptor 2 binding(GO:0051431) |
| 0.5 | 9.5 | GO:0046933 | proton-transporting ATP synthase activity, rotational mechanism(GO:0046933) |
| 0.5 | 2.7 | GO:0004720 | protein-lysine 6-oxidase activity(GO:0004720) |
| 0.4 | 3.1 | GO:0008294 | calcium- and calmodulin-responsive adenylate cyclase activity(GO:0008294) |
| 0.4 | 3.5 | GO:0004126 | cytidine deaminase activity(GO:0004126) |
| 0.4 | 1.4 | GO:0034617 | tetrahydrobiopterin binding(GO:0034617) |
| 0.3 | 2.4 | GO:0086075 | gap junction channel activity involved in cardiac conduction electrical coupling(GO:0086075) |
| 0.3 | 3.7 | GO:0005243 | gap junction channel activity(GO:0005243) |
| 0.3 | 1.3 | GO:0004736 | pyruvate carboxylase activity(GO:0004736) |
| 0.3 | 1.3 | GO:0030172 | troponin C binding(GO:0030172) |
| 0.3 | 2.2 | GO:0070404 | NADH binding(GO:0070404) |
| 0.3 | 4.8 | GO:0019855 | calcium channel inhibitor activity(GO:0019855) |
| 0.3 | 1.5 | GO:0005173 | stem cell factor receptor binding(GO:0005173) |
| 0.3 | 1.1 | GO:0019136 | deoxynucleoside kinase activity(GO:0019136) |
| 0.3 | 5.0 | GO:0019841 | retinol binding(GO:0019841) |
| 0.3 | 0.8 | GO:1901375 | acetylcholine transmembrane transporter activity(GO:0005277) acetate ester transmembrane transporter activity(GO:1901375) |
| 0.3 | 4.0 | GO:0031432 | titin binding(GO:0031432) |
| 0.2 | 2.5 | GO:0031433 | telethonin binding(GO:0031433) |
| 0.2 | 2.3 | GO:0005113 | patched binding(GO:0005113) |
| 0.2 | 4.5 | GO:0001134 | transcription factor activity, transcription factor recruiting(GO:0001134) |
| 0.2 | 0.7 | GO:0042392 | sphingosine-1-phosphate phosphatase activity(GO:0042392) |
| 0.2 | 10.0 | GO:0043027 | cysteine-type endopeptidase inhibitor activity involved in apoptotic process(GO:0043027) |
| 0.2 | 6.3 | GO:0015020 | glucuronosyltransferase activity(GO:0015020) |
| 0.1 | 2.3 | GO:0048185 | activin binding(GO:0048185) |
| 0.1 | 3.9 | GO:0097602 | cullin family protein binding(GO:0097602) |
| 0.1 | 2.5 | GO:0005527 | macrolide binding(GO:0005527) FK506 binding(GO:0005528) |
| 0.1 | 3.7 | GO:0004198 | calcium-dependent cysteine-type endopeptidase activity(GO:0004198) |
| 0.1 | 1.1 | GO:0004865 | protein serine/threonine phosphatase inhibitor activity(GO:0004865) |
| 0.1 | 4.1 | GO:0005212 | structural constituent of eye lens(GO:0005212) |
| 0.1 | 0.8 | GO:0005223 | intracellular cGMP activated cation channel activity(GO:0005223) |
| 0.1 | 0.9 | GO:0001849 | complement component C1q binding(GO:0001849) |
| 0.1 | 1.1 | GO:0032564 | dATP binding(GO:0032564) |
| 0.1 | 2.8 | GO:0015035 | protein disulfide oxidoreductase activity(GO:0015035) |
| 0.1 | 0.8 | GO:0004966 | galanin receptor activity(GO:0004966) |
| 0.1 | 1.7 | GO:0015037 | peptide disulfide oxidoreductase activity(GO:0015037) |
| 0.1 | 16.5 | GO:0005178 | integrin binding(GO:0005178) |
| 0.1 | 0.4 | GO:0004052 | arachidonate 12-lipoxygenase activity(GO:0004052) |
| 0.1 | 3.3 | GO:0004709 | MAP kinase kinase kinase activity(GO:0004709) |
| 0.1 | 10.1 | GO:0005518 | collagen binding(GO:0005518) |
| 0.1 | 1.3 | GO:0000014 | single-stranded DNA endodeoxyribonuclease activity(GO:0000014) |
| 0.1 | 0.9 | GO:0004691 | cAMP-dependent protein kinase activity(GO:0004691) |
| 0.1 | 3.8 | GO:1990841 | promoter-specific chromatin binding(GO:1990841) |
| 0.1 | 0.9 | GO:0050700 | CARD domain binding(GO:0050700) |
| 0.1 | 1.4 | GO:0070016 | armadillo repeat domain binding(GO:0070016) |
| 0.1 | 0.5 | GO:0015562 | efflux transmembrane transporter activity(GO:0015562) |
| 0.1 | 2.5 | GO:0004623 | phospholipase A2 activity(GO:0004623) |
| 0.1 | 1.9 | GO:0001223 | transcription coactivator binding(GO:0001223) |
| 0.1 | 30.3 | GO:0001077 | transcriptional activator activity, RNA polymerase II core promoter proximal region sequence-specific binding(GO:0001077) |
| 0.1 | 7.9 | GO:0005179 | hormone activity(GO:0005179) |
| 0.1 | 2.1 | GO:0017147 | Wnt-protein binding(GO:0017147) |
| 0.1 | 13.3 | GO:0015293 | symporter activity(GO:0015293) |
| 0.1 | 0.6 | GO:0008454 | alpha-1,3-mannosylglycoprotein 4-beta-N-acetylglucosaminyltransferase activity(GO:0008454) |
| 0.1 | 1.9 | GO:0016922 | ligand-dependent nuclear receptor binding(GO:0016922) |
| 0.1 | 1.8 | GO:0016881 | acid-amino acid ligase activity(GO:0016881) |
| 0.0 | 0.9 | GO:0004089 | carbonate dehydratase activity(GO:0004089) |
| 0.0 | 0.8 | GO:0070410 | co-SMAD binding(GO:0070410) |
| 0.0 | 10.6 | GO:0004252 | serine-type endopeptidase activity(GO:0004252) |
| 0.0 | 1.5 | GO:0019706 | protein-cysteine S-palmitoyltransferase activity(GO:0019706) protein-cysteine S-acyltransferase activity(GO:0019707) |
| 0.0 | 0.2 | GO:0051575 | 5'-deoxyribose-5-phosphate lyase activity(GO:0051575) |
| 0.0 | 0.6 | GO:0043560 | insulin receptor substrate binding(GO:0043560) |
| 0.0 | 0.3 | GO:0031749 | D2 dopamine receptor binding(GO:0031749) |
| 0.0 | 2.6 | GO:0015459 | potassium channel regulator activity(GO:0015459) |
| 0.0 | 5.5 | GO:0001228 | transcriptional activator activity, RNA polymerase II transcription regulatory region sequence-specific binding(GO:0001228) |
| 0.0 | 0.2 | GO:0052629 | phosphatidylinositol-3,5-bisphosphate 3-phosphatase activity(GO:0052629) |
| 0.0 | 0.8 | GO:0008392 | arachidonic acid epoxygenase activity(GO:0008392) |
| 0.0 | 2.2 | GO:0005200 | structural constituent of cytoskeleton(GO:0005200) |
| 0.0 | 0.6 | GO:0042974 | retinoic acid receptor binding(GO:0042974) |
| 0.0 | 0.2 | GO:0008190 | eukaryotic initiation factor 4E binding(GO:0008190) |
| 0.0 | 0.4 | GO:0004993 | G-protein coupled serotonin receptor activity(GO:0004993) |
| 0.0 | 0.2 | GO:0016500 | protein-hormone receptor activity(GO:0016500) |
| 0.0 | 0.5 | GO:0001671 | ATPase activator activity(GO:0001671) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 10.7 | PID ANTHRAX PATHWAY | Cellular roles of Anthrax toxin |
| 0.3 | 7.1 | PID INTEGRIN5 PATHWAY | Beta5 beta6 beta7 and beta8 integrin cell surface interactions |
| 0.3 | 10.0 | SA B CELL RECEPTOR COMPLEXES | Antigen binding to B cell receptors activates protein tyrosine kinases, such as the Src family, which ultimate activate MAP kinases. |
| 0.2 | 2.4 | PID VEGF VEGFR PATHWAY | VEGF and VEGFR signaling network |
| 0.2 | 3.3 | PID INTEGRIN4 PATHWAY | Alpha6 beta4 integrin-ligand interactions |
| 0.1 | 3.1 | PID LPA4 PATHWAY | LPA4-mediated signaling events |
| 0.1 | 6.5 | PID AURORA B PATHWAY | Aurora B signaling |
| 0.1 | 6.3 | PID SYNDECAN 1 PATHWAY | Syndecan-1-mediated signaling events |
| 0.1 | 3.8 | PID FRA PATHWAY | Validated transcriptional targets of AP1 family members Fra1 and Fra2 |
| 0.1 | 2.0 | PID HEDGEHOG 2PATHWAY | Signaling events mediated by the Hedgehog family |
| 0.0 | 2.6 | PID PLK1 PATHWAY | PLK1 signaling events |
| 0.0 | 5.0 | PID RB 1PATHWAY | Regulation of retinoblastoma protein |
| 0.0 | 3.3 | PID IL4 2PATHWAY | IL4-mediated signaling events |
| 0.0 | 0.7 | ST STAT3 PATHWAY | STAT3 Pathway |
| 0.0 | 1.3 | PID CONE PATHWAY | Visual signal transduction: Cones |
| 0.0 | 2.3 | PID BMP PATHWAY | BMP receptor signaling |
| 0.0 | 1.3 | PID ATM PATHWAY | ATM pathway |
| 0.0 | 2.3 | PID RAC1 PATHWAY | RAC1 signaling pathway |
| 0.0 | 1.9 | PID LKB1 PATHWAY | LKB1 signaling events |
| 0.0 | 0.7 | PID CD40 PATHWAY | CD40/CD40L signaling |
| 0.0 | 0.7 | PID S1P META PATHWAY | Sphingosine 1-phosphate (S1P) pathway |
| 0.0 | 1.1 | PID E2F PATHWAY | E2F transcription factor network |
| 0.0 | 0.4 | PID IL23 PATHWAY | IL23-mediated signaling events |
| 0.0 | 0.6 | PID RAS PATHWAY | Regulation of Ras family activation |
| 0.0 | 1.0 | PID SMAD2 3NUCLEAR PATHWAY | Regulation of nuclear SMAD2/3 signaling |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.7 | 12.2 | REACTOME BILE SALT AND ORGANIC ANION SLC TRANSPORTERS | Genes involved in Bile salt and organic anion SLC transporters |
| 0.5 | 9.5 | REACTOME FORMATION OF ATP BY CHEMIOSMOTIC COUPLING | Genes involved in Formation of ATP by chemiosmotic coupling |
| 0.3 | 7.1 | REACTOME P130CAS LINKAGE TO MAPK SIGNALING FOR INTEGRINS | Genes involved in p130Cas linkage to MAPK signaling for integrins |
| 0.3 | 6.1 | REACTOME GAP JUNCTION ASSEMBLY | Genes involved in Gap junction assembly |
| 0.2 | 2.4 | REACTOME VEGF LIGAND RECEPTOR INTERACTIONS | Genes involved in VEGF ligand-receptor interactions |
| 0.2 | 10.3 | REACTOME RECYCLING PATHWAY OF L1 | Genes involved in Recycling pathway of L1 |
| 0.2 | 2.2 | REACTOME ETHANOL OXIDATION | Genes involved in Ethanol oxidation |
| 0.2 | 9.4 | REACTOME SMOOTH MUSCLE CONTRACTION | Genes involved in Smooth Muscle Contraction |
| 0.2 | 3.1 | REACTOME ADENYLATE CYCLASE ACTIVATING PATHWAY | Genes involved in Adenylate cyclase activating pathway |
| 0.2 | 6.4 | REACTOME YAP1 AND WWTR1 TAZ STIMULATED GENE EXPRESSION | Genes involved in YAP1- and WWTR1 (TAZ)-stimulated gene expression |
| 0.1 | 1.8 | REACTOME VITAMIN B5 PANTOTHENATE METABOLISM | Genes involved in Vitamin B5 (pantothenate) metabolism |
| 0.1 | 5.0 | REACTOME METABOLISM OF STEROID HORMONES AND VITAMINS A AND D | Genes involved in Metabolism of steroid hormones and vitamins A and D |
| 0.1 | 1.4 | REACTOME TETRAHYDROBIOPTERIN BH4 SYNTHESIS RECYCLING SALVAGE AND REGULATION | Genes involved in Tetrahydrobiopterin (BH4) synthesis, recycling, salvage and regulation |
| 0.1 | 2.1 | REACTOME TRAF6 MEDIATED IRF7 ACTIVATION | Genes involved in TRAF6 mediated IRF7 activation |
| 0.1 | 0.9 | REACTOME TRYPTOPHAN CATABOLISM | Genes involved in Tryptophan catabolism |
| 0.1 | 2.6 | REACTOME SEMA4D IN SEMAPHORIN SIGNALING | Genes involved in Sema4D in semaphorin signaling |
| 0.1 | 6.3 | REACTOME TRANSCRIPTIONAL REGULATION OF WHITE ADIPOCYTE DIFFERENTIATION | Genes involved in Transcriptional Regulation of White Adipocyte Differentiation |
| 0.1 | 3.7 | REACTOME COLLAGEN FORMATION | Genes involved in Collagen formation |
| 0.0 | 0.7 | REACTOME IL 6 SIGNALING | Genes involved in Interleukin-6 signaling |
| 0.0 | 3.3 | REACTOME INTEGRIN CELL SURFACE INTERACTIONS | Genes involved in Integrin cell surface interactions |
| 0.0 | 0.9 | REACTOME ACTIVATED TAK1 MEDIATES P38 MAPK ACTIVATION | Genes involved in activated TAK1 mediates p38 MAPK activation |
| 0.0 | 1.3 | REACTOME MEIOTIC RECOMBINATION | Genes involved in Meiotic Recombination |
| 0.0 | 0.9 | REACTOME ABCA TRANSPORTERS IN LIPID HOMEOSTASIS | Genes involved in ABCA transporters in lipid homeostasis |
| 0.0 | 2.0 | REACTOME MYOGENESIS | Genes involved in Myogenesis |
| 0.0 | 0.8 | REACTOME CYTOCHROME P450 ARRANGED BY SUBSTRATE TYPE | Genes involved in Cytochrome P450 - arranged by substrate type |
| 0.0 | 0.6 | REACTOME AMINE DERIVED HORMONES | Genes involved in Amine-derived hormones |
| 0.0 | 0.2 | REACTOME HORMONE LIGAND BINDING RECEPTORS | Genes involved in Hormone ligand-binding receptors |
| 0.0 | 0.5 | REACTOME ANTIGEN PRESENTATION FOLDING ASSEMBLY AND PEPTIDE LOADING OF CLASS I MHC | Genes involved in Antigen Presentation: Folding, assembly and peptide loading of class I MHC |
| 0.0 | 0.5 | REACTOME ABC FAMILY PROTEINS MEDIATED TRANSPORT | Genes involved in ABC-family proteins mediated transport |
| 0.0 | 0.6 | REACTOME N GLYCAN ANTENNAE ELONGATION | Genes involved in N-Glycan antennae elongation |
| 0.0 | 0.8 | REACTOME DOWNREGULATION OF SMAD2 3 SMAD4 TRANSCRIPTIONAL ACTIVITY | Genes involved in Downregulation of SMAD2/3:SMAD4 transcriptional activity |
| 0.0 | 2.3 | REACTOME CLASS B 2 SECRETIN FAMILY RECEPTORS | Genes involved in Class B/2 (Secretin family receptors) |
| 0.0 | 0.7 | REACTOME SPHINGOLIPID DE NOVO BIOSYNTHESIS | Genes involved in Sphingolipid de novo biosynthesis |
| 0.0 | 1.9 | REACTOME PEPTIDE CHAIN ELONGATION | Genes involved in Peptide chain elongation |
| 0.0 | 4.7 | REACTOME ANTIGEN PROCESSING UBIQUITINATION PROTEASOME DEGRADATION | Genes involved in Antigen processing: Ubiquitination & Proteasome degradation |
| 0.0 | 0.2 | REACTOME INTEGRATION OF PROVIRUS | Genes involved in Integration of provirus |
| 0.0 | 0.2 | REACTOME DESTABILIZATION OF MRNA BY AUF1 HNRNP D0 | Genes involved in Destabilization of mRNA by AUF1 (hnRNP D0) |
| 0.0 | 0.9 | REACTOME NRAGE SIGNALS DEATH THROUGH JNK | Genes involved in NRAGE signals death through JNK |