Project
PRJNA375882: Comprehensive Mouse Transcriptomic BodyMap
Navigation
Downloads
Results for Phox2a
Z-value: 0.59
Transcription factors associated with Phox2a
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Phox2a
|
ENSMUSG00000007946.12 | Phox2a |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Phox2a | mm39_v1_chr7_+_101467512_101467529 | -0.01 | 9.1e-01 | Click! |
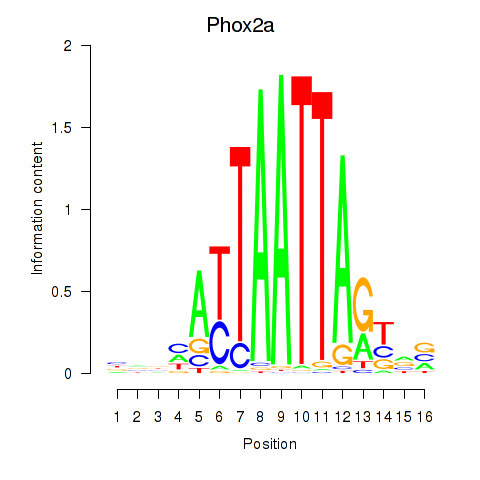
Activity profile of Phox2a motif
Sorted Z-values of Phox2a motif
Network of associatons between targets according to the STRING database.
First level regulatory network of Phox2a
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr3_+_93301003 | 4.60 |
ENSMUST00000045912.3
|
Rptn
|
repetin |
| chr16_-_56688024 | 4.13 |
ENSMUST00000232373.2
|
Tmem45a
|
transmembrane protein 45a |
| chr19_-_47680528 | 2.95 |
ENSMUST00000026045.14
ENSMUST00000086923.6 |
Col17a1
|
collagen, type XVII, alpha 1 |
| chr19_-_45224251 | 2.42 |
ENSMUST00000099401.6
|
Lbx1
|
ladybird homeobox 1 |
| chr4_-_14621805 | 2.38 |
ENSMUST00000042221.14
|
Slc26a7
|
solute carrier family 26, member 7 |
| chr6_+_145879839 | 2.26 |
ENSMUST00000032383.14
|
Sspn
|
sarcospan |
| chr5_+_13448833 | 2.19 |
ENSMUST00000137798.10
|
Sema3a
|
sema domain, immunoglobulin domain (Ig), short basic domain, secreted, (semaphorin) 3A |
| chr1_+_172383499 | 2.17 |
ENSMUST00000061835.10
|
Vsig8
|
V-set and immunoglobulin domain containing 8 |
| chr4_-_14621669 | 2.00 |
ENSMUST00000143105.2
|
Slc26a7
|
solute carrier family 26, member 7 |
| chr1_-_171854818 | 1.89 |
ENSMUST00000138714.2
ENSMUST00000027837.13 ENSMUST00000111264.8 |
Vangl2
|
VANGL planar cell polarity 2 |
| chr19_-_24178000 | 1.82 |
ENSMUST00000233658.3
|
Tjp2
|
tight junction protein 2 |
| chr4_-_14621497 | 1.58 |
ENSMUST00000149633.2
|
Slc26a7
|
solute carrier family 26, member 7 |
| chr6_-_147165623 | 1.56 |
ENSMUST00000052296.9
ENSMUST00000204197.2 |
Pthlh
|
parathyroid hormone-like peptide |
| chr2_+_36120438 | 1.55 |
ENSMUST00000062069.6
|
Ptgs1
|
prostaglandin-endoperoxide synthase 1 |
| chr8_-_85389470 | 1.52 |
ENSMUST00000060427.6
|
Ier2
|
immediate early response 2 |
| chr5_+_13448647 | 1.50 |
ENSMUST00000125629.8
|
Sema3a
|
sema domain, immunoglobulin domain (Ig), short basic domain, secreted, (semaphorin) 3A |
| chr3_+_66127330 | 1.41 |
ENSMUST00000029421.6
|
Ptx3
|
pentraxin related gene |
| chr2_-_164080172 | 1.32 |
ENSMUST00000109374.2
|
Svs2
|
seminal vesicle secretory protein 2 |
| chr9_+_118892497 | 1.30 |
ENSMUST00000141185.8
ENSMUST00000126251.8 ENSMUST00000136561.2 |
Vill
|
villin-like |
| chr13_-_103042554 | 1.24 |
ENSMUST00000171791.8
|
Mast4
|
microtubule associated serine/threonine kinase family member 4 |
| chr1_-_144427302 | 1.18 |
ENSMUST00000184189.3
|
Rgs21
|
regulator of G-protein signalling 21 |
| chr9_-_85631361 | 1.07 |
ENSMUST00000039213.15
|
Ibtk
|
inhibitor of Bruton agammaglobulinemia tyrosine kinase |
| chr2_-_168608949 | 1.07 |
ENSMUST00000075044.10
|
Sall4
|
spalt like transcription factor 4 |
| chrX_+_159551171 | 0.98 |
ENSMUST00000112368.3
|
Rs1
|
retinoschisis (X-linked, juvenile) 1 (human) |
| chr2_-_168609110 | 0.95 |
ENSMUST00000029061.12
ENSMUST00000103074.2 |
Sall4
|
spalt like transcription factor 4 |
| chr4_+_134658209 | 0.92 |
ENSMUST00000030622.3
|
Syf2
|
SYF2 homolog, RNA splicing factor (S. cerevisiae) |
| chr4_+_138606671 | 0.91 |
ENSMUST00000105804.2
|
Pla2g2e
|
phospholipase A2, group IIE |
| chrX_+_159551009 | 0.89 |
ENSMUST00000033650.14
|
Rs1
|
retinoschisis (X-linked, juvenile) 1 (human) |
| chr2_-_89855921 | 0.82 |
ENSMUST00000216616.3
|
Olfr1264
|
olfactory receptor 1264 |
| chr13_-_103042294 | 0.81 |
ENSMUST00000167462.8
|
Mast4
|
microtubule associated serine/threonine kinase family member 4 |
| chr7_-_101486983 | 0.78 |
ENSMUST00000185929.2
ENSMUST00000165052.8 |
Inppl1
|
inositol polyphosphate phosphatase-like 1 |
| chr2_-_111843053 | 0.68 |
ENSMUST00000213559.3
|
Olfr1310
|
olfactory receptor 1310 |
| chrX_+_9751861 | 0.67 |
ENSMUST00000067529.9
ENSMUST00000086165.4 |
Sytl5
|
synaptotagmin-like 5 |
| chr2_-_111820618 | 0.66 |
ENSMUST00000216948.2
ENSMUST00000214935.2 ENSMUST00000217452.2 ENSMUST00000215045.2 |
Olfr1309
|
olfactory receptor 1309 |
| chr7_-_44752508 | 0.40 |
ENSMUST00000209830.2
|
Fcgrt
|
Fc fragment of IgG receptor and transporter |
| chr10_-_129000620 | 0.39 |
ENSMUST00000214271.2
|
Olfr771
|
olfactory receptor 771 |
| chr3_-_72875187 | 0.38 |
ENSMUST00000167334.8
|
Sis
|
sucrase isomaltase (alpha-glucosidase) |
| chr4_-_43710231 | 0.37 |
ENSMUST00000217544.2
ENSMUST00000107862.3 |
Olfr71
|
olfactory receptor 71 |
| chr2_-_168607166 | 0.36 |
ENSMUST00000137536.2
|
Sall4
|
spalt like transcription factor 4 |
| chr13_-_22289994 | 0.35 |
ENSMUST00000227357.2
ENSMUST00000228428.2 |
Vmn1r189
|
vomeronasal 1 receptor 189 |
| chr2_+_89642395 | 0.34 |
ENSMUST00000214508.2
|
Olfr1255
|
olfactory receptor 1255 |
| chr11_+_116734104 | 0.33 |
ENSMUST00000106370.10
|
Mettl23
|
methyltransferase like 23 |
| chr14_+_65612788 | 0.29 |
ENSMUST00000224687.2
|
Zfp395
|
zinc finger protein 395 |
| chr2_-_164427367 | 0.28 |
ENSMUST00000109342.2
|
Wfdc6a
|
WAP four-disulfide core domain 6A |
| chr17_-_37430949 | 0.27 |
ENSMUST00000214994.2
ENSMUST00000216341.2 |
Olfr92
|
olfactory receptor 92 |
| chr2_-_27365633 | 0.23 |
ENSMUST00000138693.8
ENSMUST00000113941.9 ENSMUST00000077737.13 |
Brd3
|
bromodomain containing 3 |
| chr1_+_173924457 | 0.21 |
ENSMUST00000213832.2
|
Olfr427
|
olfactory receptor 427 |
| chr14_+_26722319 | 0.19 |
ENSMUST00000035433.10
|
Hesx1
|
homeobox gene expressed in ES cells |
| chr8_-_3675024 | 0.18 |
ENSMUST00000133459.8
|
Pcp2
|
Purkinje cell protein 2 (L7) |
| chr5_-_34817412 | 0.14 |
ENSMUST00000041364.13
|
Nop14
|
NOP14 nucleolar protein |
| chr7_-_44753168 | 0.14 |
ENSMUST00000211085.2
ENSMUST00000210642.2 ENSMUST00000003512.9 |
Fcgrt
|
Fc fragment of IgG receptor and transporter |
| chr10_-_129965752 | 0.12 |
ENSMUST00000215217.2
ENSMUST00000214192.2 |
Olfr824
|
olfactory receptor 824 |
| chr6_+_37847721 | 0.11 |
ENSMUST00000031859.14
ENSMUST00000120428.8 |
Trim24
|
tripartite motif-containing 24 |
| chr19_-_41921676 | 0.08 |
ENSMUST00000075280.12
ENSMUST00000112123.4 |
Exosc1
|
exosome component 1 |
| chr17_-_37511885 | 0.08 |
ENSMUST00000222190.2
|
Olfr94
|
olfactory receptor 94 |
| chr15_+_98350469 | 0.07 |
ENSMUST00000217517.2
|
Olfr281
|
olfactory receptor 281 |
| chr10_-_53252210 | 0.05 |
ENSMUST00000095691.7
|
Cep85l
|
centrosomal protein 85-like |
| chr19_+_12364643 | 0.05 |
ENSMUST00000217062.3
ENSMUST00000216145.2 ENSMUST00000213657.2 |
Olfr1440
|
olfactory receptor 1440 |
| chr12_+_30934320 | 0.01 |
ENSMUST00000067087.7
|
Alkal2
|
ALK and LTK ligand 2 |
| chr6_-_30936013 | 0.00 |
ENSMUST00000101589.5
|
Klf14
|
Kruppel-like factor 14 |
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.2 | 3.7 | GO:1903045 | neural crest cell migration involved in sympathetic nervous system development(GO:1903045) |
| 0.7 | 6.0 | GO:0019532 | oxalate transport(GO:0019532) |
| 0.6 | 1.9 | GO:0060448 | dichotomous subdivision of terminal units involved in lung branching(GO:0060448) |
| 0.5 | 1.6 | GO:0060618 | nipple development(GO:0060618) |
| 0.5 | 1.4 | GO:0052422 | modulation by symbiont of host molecular function(GO:0052055) modulation of catalytic activity in other organism involved in symbiotic interaction(GO:0052203) modulation by host of symbiont catalytic activity(GO:0052422) |
| 0.5 | 1.9 | GO:0036367 | adaptation of rhodopsin mediated signaling(GO:0016062) light adaption(GO:0036367) |
| 0.3 | 1.8 | GO:2001205 | negative regulation of osteoclast development(GO:2001205) |
| 0.2 | 1.6 | GO:0035633 | cyclooxygenase pathway(GO:0019371) maintenance of blood-brain barrier(GO:0035633) |
| 0.2 | 2.4 | GO:0048664 | neuron fate determination(GO:0048664) |
| 0.1 | 2.4 | GO:0001833 | inner cell mass cell proliferation(GO:0001833) |
| 0.1 | 0.8 | GO:0043569 | negative regulation of insulin-like growth factor receptor signaling pathway(GO:0043569) |
| 0.1 | 0.3 | GO:1901317 | regulation of sperm motility(GO:1901317) |
| 0.0 | 0.9 | GO:0034374 | low-density lipoprotein particle remodeling(GO:0034374) |
| 0.0 | 1.3 | GO:0048240 | sperm capacitation(GO:0048240) |
| 0.0 | 0.9 | GO:0007095 | mitotic G2 DNA damage checkpoint(GO:0007095) |
| 0.0 | 0.2 | GO:0016056 | rhodopsin mediated signaling pathway(GO:0016056) |
| 0.0 | 0.2 | GO:0030916 | otic vesicle formation(GO:0030916) |
| 0.0 | 1.3 | GO:0051693 | actin filament capping(GO:0051693) |
| 0.0 | 0.1 | GO:0070562 | regulation of vitamin D receptor signaling pathway(GO:0070562) |
| 0.0 | 1.5 | GO:0071774 | response to fibroblast growth factor(GO:0071774) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.6 | 1.9 | GO:0060187 | cell pole(GO:0060187) |
| 0.1 | 2.9 | GO:0030056 | hemidesmosome(GO:0030056) |
| 0.1 | 0.9 | GO:0071014 | post-mRNA release spliceosomal complex(GO:0071014) |
| 0.1 | 2.3 | GO:0016010 | dystrophin-associated glycoprotein complex(GO:0016010) glycoprotein complex(GO:0090665) |
| 0.1 | 3.6 | GO:0001533 | cornified envelope(GO:0001533) |
| 0.0 | 1.8 | GO:0005921 | gap junction(GO:0005921) |
| 0.0 | 0.1 | GO:0030689 | Noc complex(GO:0030689) |
| 0.0 | 6.0 | GO:0016323 | basolateral plasma membrane(GO:0016323) |
| 0.0 | 0.1 | GO:0005726 | perichromatin fibrils(GO:0005726) |
| 0.0 | 2.4 | GO:0000792 | heterochromatin(GO:0000792) |
| 0.0 | 1.6 | GO:0001750 | photoreceptor outer segment(GO:0001750) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 6.0 | GO:0019531 | oxalate transmembrane transporter activity(GO:0019531) |
| 0.2 | 3.7 | GO:0038191 | neuropilin binding(GO:0038191) |
| 0.2 | 1.4 | GO:0001849 | complement component C1q binding(GO:0001849) |
| 0.2 | 1.9 | GO:0008429 | phosphatidylethanolamine binding(GO:0008429) |
| 0.1 | 1.1 | GO:0030292 | protein tyrosine kinase inhibitor activity(GO:0030292) |
| 0.1 | 0.4 | GO:0004574 | oligo-1,6-glucosidase activity(GO:0004574) |
| 0.1 | 0.5 | GO:0019770 | IgG receptor activity(GO:0019770) |
| 0.0 | 1.6 | GO:0051428 | peptide hormone receptor binding(GO:0051428) |
| 0.0 | 0.8 | GO:0046030 | inositol-polyphosphate 5-phosphatase activity(GO:0004445) inositol trisphosphate phosphatase activity(GO:0046030) |
| 0.0 | 0.9 | GO:0004623 | phospholipase A2 activity(GO:0004623) |
| 0.0 | 1.6 | GO:0004601 | peroxidase activity(GO:0004601) |
| 0.0 | 0.1 | GO:0034056 | estrogen response element binding(GO:0034056) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 2.9 | NABA COLLAGENS | Genes encoding collagen proteins |
| 0.0 | 1.6 | PID HEDGEHOG 2PATHWAY | Signaling events mediated by the Hedgehog family |
| 0.0 | 1.4 | ST MYOCYTE AD PATHWAY | Myocyte Adrenergic Pathway is a specific case of the generalized Adrenergic Pathway. |
| 0.0 | 0.8 | PID EPHA2 FWD PATHWAY | EPHA2 forward signaling |
| 0.0 | 3.7 | NABA ECM AFFILIATED | Genes encoding proteins affiliated structurally or functionally to extracellular matrix proteins |
| 0.0 | 2.4 | PID BETA CATENIN NUC PATHWAY | Regulation of nuclear beta catenin signaling and target gene transcription |
| 0.0 | 1.6 | PID MYC REPRESS PATHWAY | Validated targets of C-MYC transcriptional repression |
| 0.0 | 1.1 | PID BCR 5PATHWAY | BCR signaling pathway |
| 0.0 | 4.6 | NABA SECRETED FACTORS | Genes encoding secreted soluble factors |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 3.7 | REACTOME SEMA3A PLEXIN REPULSION SIGNALING BY INHIBITING INTEGRIN ADHESION | Genes involved in SEMA3A-Plexin repulsion signaling by inhibiting Integrin adhesion |
| 0.1 | 1.8 | REACTOME APOPTOTIC CLEAVAGE OF CELL ADHESION PROTEINS | Genes involved in Apoptotic cleavage of cell adhesion proteins |
| 0.1 | 0.9 | REACTOME ACYL CHAIN REMODELLING OF PS | Genes involved in Acyl chain remodelling of PS |
| 0.0 | 2.9 | REACTOME COLLAGEN FORMATION | Genes involved in Collagen formation |
| 0.0 | 6.0 | REACTOME TRANSPORT OF INORGANIC CATIONS ANIONS AND AMINO ACIDS OLIGOPEPTIDES | Genes involved in Transport of inorganic cations/anions and amino acids/oligopeptides |
| 0.0 | 1.2 | REACTOME PHASE1 FUNCTIONALIZATION OF COMPOUNDS | Genes involved in Phase 1 - Functionalization of compounds |
| 0.0 | 0.8 | REACTOME SYNTHESIS OF PIPS AT THE PLASMA MEMBRANE | Genes involved in Synthesis of PIPs at the plasma membrane |
| 0.0 | 1.6 | REACTOME CLASS B 2 SECRETIN FAMILY RECEPTORS | Genes involved in Class B/2 (Secretin family receptors) |