Project
PRJNA375882: Comprehensive Mouse Transcriptomic BodyMap
Navigation
Downloads
Results for Pitx1
Z-value: 1.27
Transcription factors associated with Pitx1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Pitx1
|
ENSMUSG00000021506.8 | Pitx1 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Pitx1 | mm39_v1_chr13_-_55983946_55984005 | 0.31 | 8.7e-03 | Click! |
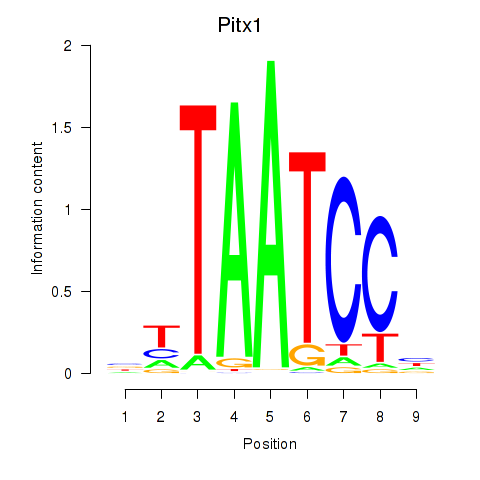
Activity profile of Pitx1 motif
Sorted Z-values of Pitx1 motif
Network of associatons between targets according to the STRING database.
First level regulatory network of Pitx1
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr7_+_43361930 | 11.93 |
ENSMUST00000066834.8
|
Klk13
|
kallikrein related-peptidase 13 |
| chr11_-_99383938 | 6.34 |
ENSMUST00000006969.8
|
Krt23
|
keratin 23 |
| chr2_+_121188195 | 6.17 |
ENSMUST00000125812.8
ENSMUST00000078222.9 ENSMUST00000125221.3 ENSMUST00000150271.8 |
Ckmt1
|
creatine kinase, mitochondrial 1, ubiquitous |
| chr11_-_100012384 | 5.82 |
ENSMUST00000007275.3
|
Krt13
|
keratin 13 |
| chr4_-_41697040 | 5.73 |
ENSMUST00000102962.10
|
Cntfr
|
ciliary neurotrophic factor receptor |
| chr18_-_32271224 | 5.11 |
ENSMUST00000234657.2
ENSMUST00000234386.2 ENSMUST00000234651.2 |
Proc
|
protein C |
| chr19_+_8595369 | 5.07 |
ENSMUST00000010250.4
|
Slc22a6
|
solute carrier family 22 (organic anion transporter), member 6 |
| chr18_-_43925932 | 4.96 |
ENSMUST00000237926.2
ENSMUST00000096570.4 |
Gm94
|
predicted gene 94 |
| chr3_+_106020545 | 4.68 |
ENSMUST00000079132.12
ENSMUST00000139086.2 |
Chia1
|
chitinase, acidic 1 |
| chr11_-_100026754 | 4.36 |
ENSMUST00000107411.3
|
Krt15
|
keratin 15 |
| chr11_+_115225557 | 4.06 |
ENSMUST00000106543.8
ENSMUST00000019006.5 |
Otop3
|
otopetrin 3 |
| chr15_-_79048674 | 4.04 |
ENSMUST00000230261.2
ENSMUST00000040019.5 |
Sox10
|
SRY (sex determining region Y)-box 10 |
| chr7_-_80053063 | 3.85 |
ENSMUST00000147150.2
|
Furin
|
furin (paired basic amino acid cleaving enzyme) |
| chr11_+_60956624 | 3.82 |
ENSMUST00000041944.9
ENSMUST00000108717.3 |
Kcnj12
|
potassium inwardly-rectifying channel, subfamily J, member 12 |
| chr10_+_69761784 | 3.79 |
ENSMUST00000181974.8
ENSMUST00000182795.8 ENSMUST00000182437.8 |
Ank3
|
ankyrin 3, epithelial |
| chr2_+_14393127 | 3.62 |
ENSMUST00000114731.8
ENSMUST00000082290.8 |
Slc39a12
|
solute carrier family 39 (zinc transporter), member 12 |
| chr2_+_102488985 | 3.44 |
ENSMUST00000080210.10
|
Slc1a2
|
solute carrier family 1 (glial high affinity glutamate transporter), member 2 |
| chr18_+_36498826 | 3.38 |
ENSMUST00000144158.2
|
Cystm1
|
cysteine-rich transmembrane module containing 1 |
| chr10_+_69761597 | 3.30 |
ENSMUST00000182269.8
ENSMUST00000183261.8 ENSMUST00000183074.8 |
Ank3
|
ankyrin 3, epithelial |
| chr7_+_43418321 | 3.25 |
ENSMUST00000107970.8
|
Klk12
|
kallikrein related-peptidase 12 |
| chr3_-_33137165 | 3.23 |
ENSMUST00000078226.10
ENSMUST00000108224.8 |
Pex5l
|
peroxisomal biogenesis factor 5-like |
| chr10_-_125164399 | 3.22 |
ENSMUST00000063318.10
|
Slc16a7
|
solute carrier family 16 (monocarboxylic acid transporters), member 7 |
| chr1_+_74582044 | 3.22 |
ENSMUST00000113749.8
ENSMUST00000067916.13 ENSMUST00000113747.8 ENSMUST00000113750.8 |
Plcd4
|
phospholipase C, delta 4 |
| chr1_-_119576347 | 3.18 |
ENSMUST00000027632.14
ENSMUST00000187194.2 |
Epb41l5
|
erythrocyte membrane protein band 4.1 like 5 |
| chr7_+_57069417 | 2.99 |
ENSMUST00000085240.11
|
Gabrb3
|
gamma-aminobutyric acid (GABA) A receptor, subunit beta 3 |
| chr9_+_32135540 | 2.84 |
ENSMUST00000168954.9
|
Arhgap32
|
Rho GTPase activating protein 32 |
| chr2_+_91090167 | 2.82 |
ENSMUST00000138470.2
|
Pacsin3
|
protein kinase C and casein kinase substrate in neurons 3 |
| chr4_-_139974062 | 2.81 |
ENSMUST00000039331.9
|
Igsf21
|
immunoglobulin superfamily, member 21 |
| chr19_+_29923182 | 2.79 |
ENSMUST00000025724.9
|
Il33
|
interleukin 33 |
| chr4_-_49593875 | 2.79 |
ENSMUST00000151542.2
|
Pgap4
|
post-GPI attachment to proteins GalNAc transferase 4 |
| chr5_-_86780277 | 2.78 |
ENSMUST00000116553.9
|
Tmprss11f
|
transmembrane protease, serine 11f |
| chr19_-_47680528 | 2.75 |
ENSMUST00000026045.14
ENSMUST00000086923.6 |
Col17a1
|
collagen, type XVII, alpha 1 |
| chr2_+_14393245 | 2.71 |
ENSMUST00000133258.2
|
Slc39a12
|
solute carrier family 39 (zinc transporter), member 12 |
| chr1_-_160405464 | 2.71 |
ENSMUST00000238289.2
|
Gpr52
|
G protein-coupled receptor 52 |
| chrX_-_42256694 | 2.65 |
ENSMUST00000115058.8
ENSMUST00000115059.8 |
Tenm1
|
teneurin transmembrane protein 1 |
| chr11_+_70350725 | 2.61 |
ENSMUST00000147289.2
|
Zmynd15
|
zinc finger, MYND-type containing 15 |
| chr7_+_43418404 | 2.60 |
ENSMUST00000014063.6
|
Klk12
|
kallikrein related-peptidase 12 |
| chr6_-_138398376 | 2.57 |
ENSMUST00000163065.8
|
Lmo3
|
LIM domain only 3 |
| chr3_-_10366229 | 2.54 |
ENSMUST00000119761.2
ENSMUST00000029043.13 |
Fabp12
|
fatty acid binding protein 12 |
| chr4_-_155095441 | 2.51 |
ENSMUST00000105631.9
ENSMUST00000139976.9 ENSMUST00000145662.9 |
Plch2
|
phospholipase C, eta 2 |
| chr10_+_69761314 | 2.49 |
ENSMUST00000182692.8
ENSMUST00000092433.12 |
Ank3
|
ankyrin 3, epithelial |
| chr6_+_115111872 | 2.36 |
ENSMUST00000009538.12
ENSMUST00000203450.2 |
Syn2
|
synapsin II |
| chr3_-_33039225 | 2.23 |
ENSMUST00000108221.2
|
Pex5l
|
peroxisomal biogenesis factor 5-like |
| chr14_+_68321302 | 2.23 |
ENSMUST00000022639.8
|
Nefl
|
neurofilament, light polypeptide |
| chr10_+_69761630 | 2.21 |
ENSMUST00000182029.8
|
Ank3
|
ankyrin 3, epithelial |
| chr1_-_119576632 | 2.18 |
ENSMUST00000163147.8
ENSMUST00000052404.13 ENSMUST00000191046.7 |
Epb41l5
|
erythrocyte membrane protein band 4.1 like 5 |
| chr19_+_46120327 | 2.18 |
ENSMUST00000043739.6
ENSMUST00000237098.2 |
Elovl3
|
elongation of very long chain fatty acids (FEN1/Elo2, SUR4/Elo3, yeast)-like 3 |
| chr3_-_123484499 | 2.14 |
ENSMUST00000154668.8
|
Ndst3
|
N-deacetylase/N-sulfotransferase (heparan glucosaminyl) 3 |
| chr15_-_101801351 | 2.10 |
ENSMUST00000100179.2
|
Krt76
|
keratin 76 |
| chr10_+_59239466 | 2.09 |
ENSMUST00000009790.14
|
Pla2g12b
|
phospholipase A2, group XIIB |
| chr19_+_38252984 | 2.07 |
ENSMUST00000198518.5
ENSMUST00000199812.5 |
Lgi1
|
leucine-rich repeat LGI family, member 1 |
| chr9_+_32135781 | 2.07 |
ENSMUST00000183121.2
|
Arhgap32
|
Rho GTPase activating protein 32 |
| chr2_-_9888804 | 2.05 |
ENSMUST00000114915.3
|
9230102O04Rik
|
RIKEN cDNA 9230102O04 gene |
| chr10_+_128061699 | 2.04 |
ENSMUST00000026455.8
|
Mip
|
major intrinsic protein of lens fiber |
| chr16_-_17732923 | 2.04 |
ENSMUST00000012279.6
ENSMUST00000232493.2 |
Gsc2
|
goosecoid homebox 2 |
| chr8_-_96615138 | 2.04 |
ENSMUST00000034097.8
|
Got2
|
glutamatic-oxaloacetic transaminase 2, mitochondrial |
| chr17_-_57366795 | 1.99 |
ENSMUST00000040280.14
|
Slc25a23
|
solute carrier family 25 (mitochondrial carrier; phosphate carrier), member 23 |
| chr15_+_18819033 | 1.97 |
ENSMUST00000166873.9
|
Cdh10
|
cadherin 10 |
| chr2_-_23939401 | 1.97 |
ENSMUST00000051416.12
|
Hnmt
|
histamine N-methyltransferase |
| chr5_+_87148697 | 1.96 |
ENSMUST00000031186.9
|
Ugt2b35
|
UDP glucuronosyltransferase 2 family, polypeptide B35 |
| chr7_+_24206431 | 1.95 |
ENSMUST00000176880.2
|
Zfp428
|
zinc finger protein 428 |
| chr18_+_61058684 | 1.94 |
ENSMUST00000102888.10
ENSMUST00000025519.11 |
Camk2a
|
calcium/calmodulin-dependent protein kinase II alpha |
| chr3_+_136376440 | 1.91 |
ENSMUST00000056758.9
|
Ppp3ca
|
protein phosphatase 3, catalytic subunit, alpha isoform |
| chr3_+_94217396 | 1.89 |
ENSMUST00000049822.10
|
Them4
|
thioesterase superfamily member 4 |
| chr12_+_36042899 | 1.88 |
ENSMUST00000020898.12
|
Agr2
|
anterior gradient 2 |
| chr4_-_116024788 | 1.85 |
ENSMUST00000030465.10
ENSMUST00000143426.2 |
Tspan1
|
tetraspanin 1 |
| chr7_+_30252687 | 1.83 |
ENSMUST00000044048.8
|
Hspb6
|
heat shock protein, alpha-crystallin-related, B6 |
| chr9_+_107217786 | 1.82 |
ENSMUST00000042581.4
|
6430571L13Rik
|
RIKEN cDNA 6430571L13 gene |
| chr7_-_109092834 | 1.81 |
ENSMUST00000106739.8
|
Trim66
|
tripartite motif-containing 66 |
| chr18_-_60724855 | 1.81 |
ENSMUST00000056533.9
|
Myoz3
|
myozenin 3 |
| chr10_+_80165787 | 1.80 |
ENSMUST00000105358.8
ENSMUST00000105357.2 ENSMUST00000105354.8 ENSMUST00000105355.8 |
Reep6
|
receptor accessory protein 6 |
| chr7_+_30399208 | 1.79 |
ENSMUST00000013227.8
|
2200002J24Rik
|
RIKEN cDNA 2200002J24 gene |
| chr6_-_136150491 | 1.77 |
ENSMUST00000111905.8
ENSMUST00000152012.8 ENSMUST00000143943.8 ENSMUST00000125905.2 |
Grin2b
|
glutamate receptor, ionotropic, NMDA2B (epsilon 2) |
| chr10_-_107330580 | 1.76 |
ENSMUST00000044210.5
|
Myf6
|
myogenic factor 6 |
| chr11_+_70350963 | 1.76 |
ENSMUST00000126105.2
|
Zmynd15
|
zinc finger, MYND-type containing 15 |
| chr1_+_167426019 | 1.75 |
ENSMUST00000111386.8
ENSMUST00000111384.8 |
Rxrg
|
retinoid X receptor gamma |
| chr13_-_116446166 | 1.74 |
ENSMUST00000036060.13
|
Isl1
|
ISL1 transcription factor, LIM/homeodomain |
| chr6_+_115111860 | 1.73 |
ENSMUST00000169345.4
|
Syn2
|
synapsin II |
| chr7_-_4527228 | 1.72 |
ENSMUST00000154913.2
|
Tnni3
|
troponin I, cardiac 3 |
| chr18_+_73996743 | 1.72 |
ENSMUST00000134847.2
|
Mro
|
maestro |
| chr10_+_85763545 | 1.71 |
ENSMUST00000170396.3
|
Ascl4
|
achaete-scute family bHLH transcription factor 4 |
| chr6_-_35285045 | 1.70 |
ENSMUST00000031868.5
|
Slc13a4
|
solute carrier family 13 (sodium/sulfate symporters), member 4 |
| chr1_-_110905082 | 1.69 |
ENSMUST00000094626.5
|
Cdh19
|
cadherin 19, type 2 |
| chr1_+_66360865 | 1.68 |
ENSMUST00000114013.8
|
Map2
|
microtubule-associated protein 2 |
| chr15_+_4404965 | 1.67 |
ENSMUST00000061925.5
|
Plcxd3
|
phosphatidylinositol-specific phospholipase C, X domain containing 3 |
| chr6_-_142268692 | 1.66 |
ENSMUST00000081380.10
|
Slco1a5
|
solute carrier organic anion transporter family, member 1a5 |
| chr6_-_122317156 | 1.65 |
ENSMUST00000159384.8
|
Phc1
|
polyhomeotic 1 |
| chr1_+_99700490 | 1.64 |
ENSMUST00000086738.10
|
Cntnap5b
|
contactin associated protein-like 5B |
| chr6_+_104469751 | 1.62 |
ENSMUST00000161446.2
ENSMUST00000161070.8 ENSMUST00000089215.12 |
Cntn6
|
contactin 6 |
| chr1_-_173195236 | 1.62 |
ENSMUST00000005470.5
ENSMUST00000111220.8 |
Cadm3
|
cell adhesion molecule 3 |
| chrX_+_55825033 | 1.61 |
ENSMUST00000114772.9
ENSMUST00000114768.10 ENSMUST00000155882.8 |
Fhl1
|
four and a half LIM domains 1 |
| chr14_-_28691423 | 1.58 |
ENSMUST00000225985.2
|
Cacna2d3
|
calcium channel, voltage-dependent, alpha2/delta subunit 3 |
| chr11_+_67586140 | 1.55 |
ENSMUST00000021290.2
|
Rcvrn
|
recoverin |
| chr1_-_136158027 | 1.54 |
ENSMUST00000150163.8
ENSMUST00000144464.7 |
Inava
|
innate immunity activator |
| chr17_-_85397627 | 1.54 |
ENSMUST00000072406.5
ENSMUST00000171795.9 ENSMUST00000234676.2 |
Prepl
|
prolyl endopeptidase-like |
| chr7_-_4517559 | 1.54 |
ENSMUST00000163538.8
|
Tnnt1
|
troponin T1, skeletal, slow |
| chr3_+_136375839 | 1.52 |
ENSMUST00000070198.14
|
Ppp3ca
|
protein phosphatase 3, catalytic subunit, alpha isoform |
| chr17_+_58076565 | 1.50 |
ENSMUST00000076038.7
|
Cntnap5c
|
contactin associated protein-like 5C |
| chr6_-_29380423 | 1.48 |
ENSMUST00000147483.3
|
Opn1sw
|
opsin 1 (cone pigments), short-wave-sensitive (color blindness, tritan) |
| chr12_-_109019507 | 1.47 |
ENSMUST00000185745.2
ENSMUST00000239108.2 |
Begain
|
brain-enriched guanylate kinase-associated |
| chr10_+_80165961 | 1.47 |
ENSMUST00000186864.7
ENSMUST00000040081.7 |
Reep6
|
receptor accessory protein 6 |
| chr8_-_106863423 | 1.46 |
ENSMUST00000146940.2
|
Esrp2
|
epithelial splicing regulatory protein 2 |
| chr17_-_35351026 | 1.46 |
ENSMUST00000025249.7
|
Apom
|
apolipoprotein M |
| chr2_+_22958956 | 1.45 |
ENSMUST00000226571.2
ENSMUST00000114529.10 ENSMUST00000114526.9 ENSMUST00000228050.2 |
Acbd5
|
acyl-Coenzyme A binding domain containing 5 |
| chr2_+_22958179 | 1.44 |
ENSMUST00000227663.2
ENSMUST00000028121.15 ENSMUST00000227809.2 ENSMUST00000144088.2 |
Acbd5
|
acyl-Coenzyme A binding domain containing 5 |
| chr6_-_83504471 | 1.44 |
ENSMUST00000141904.8
|
Actg2
|
actin, gamma 2, smooth muscle, enteric |
| chrX_+_55824797 | 1.44 |
ENSMUST00000114773.10
|
Fhl1
|
four and a half LIM domains 1 |
| chr4_-_137493785 | 1.42 |
ENSMUST00000139951.8
|
Alpl
|
alkaline phosphatase, liver/bone/kidney |
| chr14_-_31362835 | 1.39 |
ENSMUST00000167066.8
ENSMUST00000127204.9 |
Hacl1
|
2-hydroxyacyl-CoA lyase 1 |
| chr14_-_70666513 | 1.39 |
ENSMUST00000226426.2
ENSMUST00000048129.6 |
Piwil2
|
piwi-like RNA-mediated gene silencing 2 |
| chr14_-_64654397 | 1.38 |
ENSMUST00000210428.2
|
Msra
|
methionine sulfoxide reductase A |
| chr8_-_106863521 | 1.37 |
ENSMUST00000115979.9
|
Esrp2
|
epithelial splicing regulatory protein 2 |
| chr2_-_101459274 | 1.36 |
ENSMUST00000099682.9
|
Iftap
|
intraflagellar transport associated protein |
| chr1_+_167425953 | 1.35 |
ENSMUST00000015987.10
|
Rxrg
|
retinoid X receptor gamma |
| chr7_-_101517874 | 1.35 |
ENSMUST00000150184.2
|
Folr1
|
folate receptor 1 (adult) |
| chr10_+_18283405 | 1.35 |
ENSMUST00000037341.14
|
Nhsl1
|
NHS-like 1 |
| chr17_-_23805187 | 1.32 |
ENSMUST00000227952.2
ENSMUST00000115516.11 |
Zfp13
|
zinc finger protein 13 |
| chr19_-_8196196 | 1.31 |
ENSMUST00000113298.9
|
Slc22a29
|
solute carrier family 22. member 29 |
| chr6_-_29380467 | 1.30 |
ENSMUST00000080428.13
|
Opn1sw
|
opsin 1 (cone pigments), short-wave-sensitive (color blindness, tritan) |
| chr6_+_58810674 | 1.30 |
ENSMUST00000041401.11
|
Herc3
|
hect domain and RLD 3 |
| chr11_-_69127848 | 1.29 |
ENSMUST00000021259.9
ENSMUST00000108665.8 ENSMUST00000108664.2 |
Gucy2e
|
guanylate cyclase 2e |
| chr7_-_106423873 | 1.26 |
ENSMUST00000208895.3
|
Olfr702
|
olfactory receptor 702 |
| chr7_+_143792455 | 1.26 |
ENSMUST00000239495.2
|
Shank2
|
SH3 and multiple ankyrin repeat domains 2 |
| chr1_-_13730732 | 1.26 |
ENSMUST00000027071.7
|
Lactb2
|
lactamase, beta 2 |
| chr2_-_101459303 | 1.25 |
ENSMUST00000111231.10
|
Iftap
|
intraflagellar transport associated protein |
| chr1_+_66361252 | 1.24 |
ENSMUST00000123647.8
|
Map2
|
microtubule-associated protein 2 |
| chr7_-_101518217 | 1.23 |
ENSMUST00000123321.8
|
Folr1
|
folate receptor 1 (adult) |
| chr6_+_121277186 | 1.23 |
ENSMUST00000064580.14
|
Slc6a13
|
solute carrier family 6 (neurotransmitter transporter, GABA), member 13 |
| chr7_+_107702486 | 1.20 |
ENSMUST00000104917.4
|
Olfr483
|
olfactory receptor 483 |
| chr7_-_4517608 | 1.18 |
ENSMUST00000166959.8
|
Tnnt1
|
troponin T1, skeletal, slow |
| chr17_+_12803019 | 1.17 |
ENSMUST00000046959.9
ENSMUST00000233066.2 |
Slc22a2
|
solute carrier family 22 (organic cation transporter), member 2 |
| chr6_-_93890659 | 1.16 |
ENSMUST00000093769.8
|
Magi1
|
membrane associated guanylate kinase, WW and PDZ domain containing 1 |
| chr3_-_26207456 | 1.16 |
ENSMUST00000191835.2
|
Nlgn1
|
neuroligin 1 |
| chr12_+_119356318 | 1.16 |
ENSMUST00000221866.2
|
Macc1
|
metastasis associated in colon cancer 1 |
| chr5_-_66161621 | 1.16 |
ENSMUST00000201351.4
|
9130230L23Rik
|
RIKEN cDNA 9130230L23 gene |
| chr6_-_93890520 | 1.15 |
ENSMUST00000203688.3
|
Magi1
|
membrane associated guanylate kinase, WW and PDZ domain containing 1 |
| chr1_+_74830675 | 1.15 |
ENSMUST00000006718.15
|
Wnt10a
|
wingless-type MMTV integration site family, member 10A |
| chr18_-_3299452 | 1.12 |
ENSMUST00000126578.8
|
Crem
|
cAMP responsive element modulator |
| chr16_+_8288627 | 1.11 |
ENSMUST00000046470.16
ENSMUST00000150790.2 ENSMUST00000142899.2 |
Mettl22
|
methyltransferase like 22 |
| chr18_+_61058716 | 1.09 |
ENSMUST00000115297.8
|
Camk2a
|
calcium/calmodulin-dependent protein kinase II alpha |
| chr12_+_78243846 | 1.08 |
ENSMUST00000188791.2
|
Gm6657
|
predicted gene 6657 |
| chr10_+_90412827 | 1.08 |
ENSMUST00000182550.8
ENSMUST00000099364.12 |
Anks1b
|
ankyrin repeat and sterile alpha motif domain containing 1B |
| chr10_+_21869776 | 1.07 |
ENSMUST00000092673.11
|
Sgk1
|
serum/glucocorticoid regulated kinase 1 |
| chr3_-_84178089 | 1.07 |
ENSMUST00000054990.11
|
Trim2
|
tripartite motif-containing 2 |
| chr3_+_135053762 | 1.07 |
ENSMUST00000159658.8
ENSMUST00000078568.12 |
Slc9b1
|
solute carrier family 9, subfamily B (NHA1, cation proton antiporter 1), member 1 |
| chr2_-_121287174 | 1.05 |
ENSMUST00000110613.9
ENSMUST00000056312.10 |
Serinc4
|
serine incorporator 4 |
| chr19_-_12897671 | 1.05 |
ENSMUST00000054737.3
|
Olfr1448
|
olfactory receptor 1448 |
| chr6_+_142359099 | 1.04 |
ENSMUST00000126521.9
ENSMUST00000211094.2 |
Spx
|
spexin hormone |
| chr2_+_136733421 | 1.03 |
ENSMUST00000141463.8
|
Slx4ip
|
SLX4 interacting protein |
| chr3_-_33136153 | 1.03 |
ENSMUST00000108225.10
|
Pex5l
|
peroxisomal biogenesis factor 5-like |
| chr7_+_88079534 | 1.02 |
ENSMUST00000208478.2
|
Rab38
|
RAB38, member RAS oncogene family |
| chr14_+_28740162 | 1.02 |
ENSMUST00000055662.4
|
Lrtm1
|
leucine-rich repeats and transmembrane domains 1 |
| chr11_-_51647290 | 1.02 |
ENSMUST00000109097.9
|
Sec24a
|
Sec24 related gene family, member A (S. cerevisiae) |
| chr8_-_22422724 | 1.01 |
ENSMUST00000062586.3
|
Defb15
|
defensin beta 15 |
| chr11_-_59078565 | 0.98 |
ENSMUST00000170895.3
|
Guk1
|
guanylate kinase 1 |
| chr2_+_38231080 | 0.97 |
ENSMUST00000133661.8
|
Lhx2
|
LIM homeobox protein 2 |
| chr4_+_136349924 | 0.97 |
ENSMUST00000178843.3
|
Lactbl1
|
lactamase, beta-like 1 |
| chr18_-_20192535 | 0.97 |
ENSMUST00000075214.9
ENSMUST00000039247.11 |
Dsc2
|
desmocollin 2 |
| chr4_+_84802592 | 0.96 |
ENSMUST00000102819.10
|
Cntln
|
centlein, centrosomal protein |
| chrX_-_135642025 | 0.96 |
ENSMUST00000155207.8
ENSMUST00000080411.13 ENSMUST00000169418.8 |
Morf4l2
|
mortality factor 4 like 2 |
| chr13_+_96884780 | 0.94 |
ENSMUST00000208758.2
|
Ankrd31
|
ankyrin repeat domain 31 |
| chr2_+_69500444 | 0.94 |
ENSMUST00000100050.4
|
Klhl41
|
kelch-like 41 |
| chr11_-_70350783 | 0.92 |
ENSMUST00000019064.9
|
Cxcl16
|
chemokine (C-X-C motif) ligand 16 |
| chrX_-_166907286 | 0.92 |
ENSMUST00000239138.2
|
Frmpd4
|
FERM and PDZ domain containing 4 |
| chr9_-_100368841 | 0.91 |
ENSMUST00000098458.4
|
Il20rb
|
interleukin 20 receptor beta |
| chr1_-_171050004 | 0.91 |
ENSMUST00000147246.2
ENSMUST00000111326.8 ENSMUST00000138184.8 |
Tomm40l
|
translocase of outer mitochondrial membrane 40-like |
| chr15_-_79138929 | 0.91 |
ENSMUST00000039752.4
|
Slc16a8
|
solute carrier family 16 (monocarboxylic acid transporters), member 8 |
| chr4_-_139560831 | 0.91 |
ENSMUST00000030508.14
|
Pax7
|
paired box 7 |
| chr2_-_113047313 | 0.90 |
ENSMUST00000208135.2
ENSMUST00000134358.9 |
Ryr3
|
ryanodine receptor 3 |
| chr11_+_4587733 | 0.90 |
ENSMUST00000070257.14
ENSMUST00000109930.3 |
Ascc2
|
activating signal cointegrator 1 complex subunit 2 |
| chr1_+_133249613 | 0.90 |
ENSMUST00000007433.5
|
Kiss1
|
KiSS-1 metastasis-suppressor |
| chr4_-_43823866 | 0.89 |
ENSMUST00000215406.2
ENSMUST00000079234.6 ENSMUST00000214843.2 |
Olfr156
|
olfactory receptor 156 |
| chr18_-_3299536 | 0.88 |
ENSMUST00000129435.8
ENSMUST00000122958.8 |
Crem
|
cAMP responsive element modulator |
| chr10_-_78143384 | 0.87 |
ENSMUST00000150828.2
ENSMUST00000239481.2 |
Agpat3
ENSMUSG00000118646.2
|
1-acylglycerol-3-phosphate O-acyltransferase 3 |
| chr11_-_43792013 | 0.87 |
ENSMUST00000067258.9
ENSMUST00000139906.2 |
Adra1b
|
adrenergic receptor, alpha 1b |
| chr6_-_23650205 | 0.86 |
ENSMUST00000115354.2
|
Rnf133
|
ring finger protein 133 |
| chr6_+_42873387 | 0.85 |
ENSMUST00000058668.4
|
Olfr448
|
olfactory receptor 448 |
| chr4_+_101407608 | 0.83 |
ENSMUST00000094953.11
ENSMUST00000106933.2 |
Dnajc6
|
DnaJ heat shock protein family (Hsp40) member C6 |
| chr6_+_136931519 | 0.82 |
ENSMUST00000137768.2
|
Pde6h
|
phosphodiesterase 6H, cGMP-specific, cone, gamma |
| chr9_-_54408780 | 0.81 |
ENSMUST00000118600.8
ENSMUST00000118163.8 |
Dmxl2
|
Dmx-like 2 |
| chr14_+_65504151 | 0.81 |
ENSMUST00000169656.3
ENSMUST00000226005.2 |
Fbxo16
|
F-box protein 16 |
| chr17_-_29768531 | 0.80 |
ENSMUST00000168339.3
ENSMUST00000114683.10 ENSMUST00000234620.2 |
Tmem217
|
transmembrane protein 217 |
| chr1_-_171050077 | 0.80 |
ENSMUST00000005817.9
|
Tomm40l
|
translocase of outer mitochondrial membrane 40-like |
| chrX_+_92718695 | 0.79 |
ENSMUST00000045898.4
|
Pcyt1b
|
phosphate cytidylyltransferase 1, choline, beta isoform |
| chr11_+_29668563 | 0.78 |
ENSMUST00000060992.6
|
Rtn4
|
reticulon 4 |
| chr1_-_86317066 | 0.77 |
ENSMUST00000212058.2
|
Nmur1
|
neuromedin U receptor 1 |
| chr17_-_29768586 | 0.76 |
ENSMUST00000234305.2
ENSMUST00000234648.2 ENSMUST00000234979.2 |
Gm17657
Tmem217
|
predicted gene, 17657 transmembrane protein 217 |
| chr17_+_35188888 | 0.76 |
ENSMUST00000173680.2
|
Gm20481
|
predicted gene 20481 |
| chr2_+_80447389 | 0.76 |
ENSMUST00000028384.5
|
Dusp19
|
dual specificity phosphatase 19 |
| chr2_-_113047397 | 0.76 |
ENSMUST00000080673.13
ENSMUST00000208151.2 ENSMUST00000208290.2 |
Ryr3
|
ryanodine receptor 3 |
| chr2_+_124994425 | 0.75 |
ENSMUST00000110494.9
ENSMUST00000110495.3 ENSMUST00000028630.9 |
Slc12a1
|
solute carrier family 12, member 1 |
| chr16_-_56748424 | 0.73 |
ENSMUST00000210579.2
|
Lnp1
|
leukemia NUP98 fusion partner 1 |
| chr5_-_87240405 | 0.73 |
ENSMUST00000132667.2
ENSMUST00000145617.8 ENSMUST00000094649.11 |
Ugt2b36
|
UDP glucuronosyltransferase 2 family, polypeptide B36 |
| chr3_+_89427458 | 0.72 |
ENSMUST00000000811.8
|
Kcnn3
|
potassium intermediate/small conductance calcium-activated channel, subfamily N, member 3 |
| chr4_-_35845204 | 0.72 |
ENSMUST00000164772.8
ENSMUST00000065173.9 |
Lingo2
|
leucine rich repeat and Ig domain containing 2 |
| chr17_-_28584117 | 0.70 |
ENSMUST00000233420.2
|
Tulp1
|
tubby like protein 1 |
| chr4_-_109333866 | 0.70 |
ENSMUST00000030284.10
|
Rnf11
|
ring finger protein 11 |
| chr2_-_38816229 | 0.68 |
ENSMUST00000076275.11
ENSMUST00000142130.2 |
Nr6a1
|
nuclear receptor subfamily 6, group A, member 1 |
| chr6_-_89822597 | 0.68 |
ENSMUST00000089419.3
|
Vmn1r42
|
vomeronasal 1 receptor 42 |
| chr17_-_59298338 | 0.67 |
ENSMUST00000025064.14
|
Pdzph1
|
PDZ and pleckstrin homology domains 1 |
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.8 | 5.4 | GO:0007509 | mesoderm migration involved in gastrulation(GO:0007509) |
| 1.4 | 5.7 | GO:0003360 | brainstem development(GO:0003360) |
| 1.1 | 3.4 | GO:0010615 | positive regulation of cardiac muscle adaptation(GO:0010615) positive regulation of cardiac muscle hypertrophy in response to stress(GO:1903244) |
| 1.0 | 3.8 | GO:0090472 | dibasic protein processing(GO:0090472) |
| 0.9 | 11.8 | GO:0010650 | positive regulation of cell communication by electrical coupling(GO:0010650) positive regulation of membrane depolarization during cardiac muscle cell action potential(GO:1900827) |
| 0.9 | 1.7 | GO:0048936 | peripheral nervous system neuron axonogenesis(GO:0048936) |
| 0.8 | 6.5 | GO:0016560 | protein import into peroxisome matrix, docking(GO:0016560) |
| 0.7 | 2.2 | GO:0033693 | neurofilament bundle assembly(GO:0033693) |
| 0.7 | 2.0 | GO:0006533 | fumarate metabolic process(GO:0006106) aspartate catabolic process(GO:0006533) |
| 0.6 | 1.9 | GO:1903896 | positive regulation of IRE1-mediated unfolded protein response(GO:1903896) |
| 0.6 | 5.1 | GO:1903142 | positive regulation of endothelial cell development(GO:1901552) positive regulation of establishment of endothelial barrier(GO:1903142) |
| 0.5 | 4.0 | GO:0010626 | negative regulation of Schwann cell proliferation(GO:0010626) |
| 0.5 | 2.0 | GO:0097274 | urea homeostasis(GO:0097274) |
| 0.5 | 3.4 | GO:0070777 | D-aspartate transport(GO:0070777) D-aspartate import(GO:0070779) |
| 0.5 | 1.5 | GO:0034443 | negative regulation of lipoprotein oxidation(GO:0034443) |
| 0.5 | 3.2 | GO:1901475 | pyruvate transmembrane transport(GO:1901475) |
| 0.5 | 2.7 | GO:0031444 | slow-twitch skeletal muscle fiber contraction(GO:0031444) |
| 0.4 | 6.4 | GO:0015747 | urate transport(GO:0015747) |
| 0.4 | 2.8 | GO:0002282 | microglial cell activation involved in immune response(GO:0002282) |
| 0.4 | 2.0 | GO:0001692 | histamine metabolic process(GO:0001692) |
| 0.4 | 1.2 | GO:0048789 | cytoskeletal matrix organization at active zone(GO:0048789) neurexin clustering involved in presynaptic membrane assembly(GO:0097115) retrograde trans-synaptic signaling by trans-synaptic protein complex(GO:0098942) |
| 0.4 | 1.1 | GO:0021627 | olfactory nerve morphogenesis(GO:0021627) olfactory nerve structural organization(GO:0021629) |
| 0.4 | 1.8 | GO:0099566 | regulation of postsynaptic cytosolic calcium ion concentration(GO:0099566) |
| 0.3 | 1.4 | GO:0000239 | pachytene(GO:0000239) |
| 0.3 | 6.3 | GO:0071578 | zinc II ion transmembrane import(GO:0071578) |
| 0.3 | 1.3 | GO:0010726 | positive regulation of hydrogen peroxide metabolic process(GO:0010726) |
| 0.3 | 1.0 | GO:0046710 | GDP metabolic process(GO:0046710) |
| 0.3 | 2.8 | GO:0006030 | chitin metabolic process(GO:0006030) chitin catabolic process(GO:0006032) |
| 0.3 | 0.9 | GO:0001807 | regulation of type IV hypersensitivity(GO:0001807) |
| 0.3 | 0.9 | GO:0033686 | positive regulation of luteinizing hormone secretion(GO:0033686) |
| 0.3 | 0.9 | GO:0001982 | baroreceptor response to decreased systemic arterial blood pressure(GO:0001982) |
| 0.3 | 1.7 | GO:0001980 | regulation of systemic arterial blood pressure by ischemic conditions(GO:0001980) |
| 0.3 | 2.6 | GO:0061713 | neural crest cell migration involved in heart formation(GO:0003147) anterior neural tube closure(GO:0061713) cellular response to folic acid(GO:0071231) |
| 0.3 | 2.9 | GO:0030242 | pexophagy(GO:0030242) |
| 0.2 | 2.1 | GO:0015014 | heparan sulfate proteoglycan biosynthetic process, polysaccharide chain biosynthetic process(GO:0015014) |
| 0.2 | 3.0 | GO:0098989 | NMDA selective glutamate receptor signaling pathway(GO:0098989) |
| 0.2 | 1.4 | GO:0001561 | fatty acid alpha-oxidation(GO:0001561) |
| 0.2 | 1.8 | GO:0048743 | positive regulation of skeletal muscle fiber development(GO:0048743) |
| 0.2 | 3.3 | GO:0018298 | protein-chromophore linkage(GO:0018298) |
| 0.2 | 0.7 | GO:2000595 | optic nerve formation(GO:0021634) optic chiasma development(GO:0061360) regulation of optic nerve formation(GO:2000595) positive regulation of optic nerve formation(GO:2000597) |
| 0.2 | 0.6 | GO:0002074 | extraocular skeletal muscle development(GO:0002074) |
| 0.2 | 2.2 | GO:0034626 | fatty acid elongation, saturated fatty acid(GO:0019367) fatty acid elongation, unsaturated fatty acid(GO:0019368) fatty acid elongation, monounsaturated fatty acid(GO:0034625) fatty acid elongation, polyunsaturated fatty acid(GO:0034626) |
| 0.2 | 0.5 | GO:1903722 | regulation of centriole elongation(GO:1903722) |
| 0.2 | 0.5 | GO:2001178 | mediator complex assembly(GO:0036034) regulation of mediator complex assembly(GO:2001176) positive regulation of mediator complex assembly(GO:2001178) |
| 0.2 | 0.6 | GO:1903225 | negative regulation of endodermal cell differentiation(GO:1903225) |
| 0.1 | 0.7 | GO:0071475 | cellular hyperosmotic salinity response(GO:0071475) |
| 0.1 | 1.2 | GO:0015722 | canalicular bile acid transport(GO:0015722) |
| 0.1 | 3.3 | GO:0048733 | sebaceous gland development(GO:0048733) |
| 0.1 | 1.0 | GO:0060155 | platelet dense granule organization(GO:0060155) phagosome acidification(GO:0090383) melanosome assembly(GO:1903232) |
| 0.1 | 0.8 | GO:0072318 | clathrin coat disassembly(GO:0072318) |
| 0.1 | 1.3 | GO:0097475 | motor neuron migration(GO:0097475) |
| 0.1 | 0.4 | GO:0006212 | uracil catabolic process(GO:0006212) uracil metabolic process(GO:0019860) |
| 0.1 | 3.0 | GO:0071420 | cellular response to histamine(GO:0071420) |
| 0.1 | 0.6 | GO:0061643 | chemoattraction of axon(GO:0061642) chemorepulsion of axon(GO:0061643) |
| 0.1 | 1.8 | GO:1903546 | protein localization to photoreceptor outer segment(GO:1903546) |
| 0.1 | 0.6 | GO:0097498 | endothelial tube lumen extension(GO:0097498) |
| 0.1 | 1.0 | GO:0086073 | bundle of His cell-Purkinje myocyte adhesion involved in cell communication(GO:0086073) |
| 0.1 | 2.8 | GO:0051044 | positive regulation of membrane protein ectodomain proteolysis(GO:0051044) |
| 0.1 | 0.9 | GO:0051835 | positive regulation of synapse structural plasticity(GO:0051835) |
| 0.1 | 1.1 | GO:1900383 | regulation of synaptic plasticity by receptor localization to synapse(GO:1900383) |
| 0.1 | 1.7 | GO:0030150 | protein import into mitochondrial matrix(GO:0030150) |
| 0.1 | 0.6 | GO:0071286 | cellular response to magnesium ion(GO:0071286) |
| 0.1 | 0.5 | GO:1903566 | ciliary basal body organization(GO:0032053) positive regulation of protein localization to cilium(GO:1903566) |
| 0.1 | 0.8 | GO:0071787 | endoplasmic reticulum tubular network assembly(GO:0071787) |
| 0.1 | 1.0 | GO:2000189 | positive regulation of cholesterol homeostasis(GO:2000189) |
| 0.1 | 0.2 | GO:0032416 | negative regulation of sodium:proton antiporter activity(GO:0032416) |
| 0.1 | 0.3 | GO:0044208 | 'de novo' AMP biosynthetic process(GO:0044208) |
| 0.1 | 3.1 | GO:1901522 | positive regulation of transcription from RNA polymerase II promoter involved in cellular response to chemical stimulus(GO:1901522) |
| 0.1 | 1.8 | GO:2000291 | regulation of myoblast proliferation(GO:2000291) |
| 0.1 | 3.8 | GO:0010107 | potassium ion import(GO:0010107) |
| 0.1 | 3.7 | GO:0010972 | negative regulation of G2/M transition of mitotic cell cycle(GO:0010972) |
| 0.1 | 1.7 | GO:0015721 | bile acid and bile salt transport(GO:0015721) |
| 0.1 | 1.1 | GO:0006883 | cellular sodium ion homeostasis(GO:0006883) glucocorticoid mediated signaling pathway(GO:0043402) |
| 0.1 | 1.7 | GO:0051481 | negative regulation of cytosolic calcium ion concentration(GO:0051481) |
| 0.1 | 3.6 | GO:0007340 | acrosome reaction(GO:0007340) |
| 0.1 | 1.0 | GO:0010457 | centriole-centriole cohesion(GO:0010457) |
| 0.1 | 0.5 | GO:1904306 | positive regulation of gastro-intestinal system smooth muscle contraction(GO:1904306) |
| 0.1 | 1.2 | GO:0015697 | quaternary ammonium group transport(GO:0015697) histamine transport(GO:0051608) |
| 0.1 | 0.6 | GO:0070836 | caveola assembly(GO:0070836) |
| 0.1 | 2.8 | GO:0060445 | branching involved in salivary gland morphogenesis(GO:0060445) |
| 0.1 | 0.8 | GO:0070072 | vacuolar proton-transporting V-type ATPase complex assembly(GO:0070072) |
| 0.1 | 0.9 | GO:0090091 | positive regulation of extracellular matrix disassembly(GO:0090091) |
| 0.1 | 2.1 | GO:0050482 | arachidonic acid secretion(GO:0050482) arachidonate transport(GO:1903963) |
| 0.1 | 0.9 | GO:1901386 | negative regulation of voltage-gated calcium channel activity(GO:1901386) |
| 0.1 | 1.4 | GO:0043968 | histone H2A acetylation(GO:0043968) |
| 0.0 | 4.3 | GO:0007157 | heterophilic cell-cell adhesion via plasma membrane cell adhesion molecules(GO:0007157) |
| 0.0 | 1.9 | GO:1902108 | regulation of mitochondrial membrane permeability involved in apoptotic process(GO:1902108) |
| 0.0 | 0.3 | GO:0043569 | negative regulation of insulin-like growth factor receptor signaling pathway(GO:0043569) |
| 0.0 | 1.8 | GO:0010667 | negative regulation of cardiac muscle cell apoptotic process(GO:0010667) |
| 0.0 | 6.2 | GO:0032091 | negative regulation of protein binding(GO:0032091) |
| 0.0 | 0.9 | GO:0008272 | sulfate transport(GO:0008272) |
| 0.0 | 0.8 | GO:0043508 | negative regulation of JUN kinase activity(GO:0043508) |
| 0.0 | 0.9 | GO:0010818 | T cell chemotaxis(GO:0010818) |
| 0.0 | 1.5 | GO:2000300 | regulation of synaptic vesicle exocytosis(GO:2000300) |
| 0.0 | 1.3 | GO:0071625 | vocalization behavior(GO:0071625) |
| 0.0 | 0.9 | GO:0042572 | retinol metabolic process(GO:0042572) |
| 0.0 | 0.1 | GO:0060368 | regulation of Fc receptor mediated stimulatory signaling pathway(GO:0060368) |
| 0.0 | 0.6 | GO:0016998 | cell wall macromolecule catabolic process(GO:0016998) |
| 0.0 | 0.4 | GO:0002155 | regulation of thyroid hormone mediated signaling pathway(GO:0002155) |
| 0.0 | 1.0 | GO:2000678 | negative regulation of transcription regulatory region DNA binding(GO:2000678) |
| 0.0 | 2.6 | GO:0045600 | positive regulation of fat cell differentiation(GO:0045600) |
| 0.0 | 0.4 | GO:1902224 | ketone body metabolic process(GO:1902224) |
| 0.0 | 0.8 | GO:0007202 | activation of phospholipase C activity(GO:0007202) |
| 0.0 | 2.0 | GO:0006687 | glycosphingolipid metabolic process(GO:0006687) |
| 0.0 | 1.4 | GO:0046677 | response to antibiotic(GO:0046677) |
| 0.0 | 0.6 | GO:0090502 | RNA phosphodiester bond hydrolysis, endonucleolytic(GO:0090502) |
| 0.0 | 0.5 | GO:0050908 | detection of light stimulus involved in visual perception(GO:0050908) detection of light stimulus involved in sensory perception(GO:0050962) |
| 0.0 | 0.4 | GO:0097154 | GABAergic neuron differentiation(GO:0097154) |
| 0.0 | 0.8 | GO:1901186 | positive regulation of epidermal growth factor receptor signaling pathway(GO:0045742) positive regulation of G-protein coupled receptor protein signaling pathway(GO:0045745) positive regulation of ERBB signaling pathway(GO:1901186) |
| 0.0 | 2.9 | GO:0021954 | central nervous system neuron development(GO:0021954) |
| 0.0 | 0.2 | GO:0097577 | intracellular sequestering of iron ion(GO:0006880) sequestering of iron ion(GO:0097577) |
| 0.0 | 1.6 | GO:1904893 | negative regulation of JAK-STAT cascade(GO:0046426) negative regulation of STAT cascade(GO:1904893) |
| 0.0 | 2.0 | GO:0007156 | homophilic cell adhesion via plasma membrane adhesion molecules(GO:0007156) |
| 0.0 | 3.8 | GO:0006836 | neurotransmitter transport(GO:0006836) |
| 0.0 | 1.2 | GO:0071300 | cellular response to retinoic acid(GO:0071300) |
| 0.0 | 6.6 | GO:0016485 | protein processing(GO:0016485) |
| 0.0 | 0.2 | GO:0044336 | canonical Wnt signaling pathway involved in negative regulation of apoptotic process(GO:0044336) |
| 0.0 | 1.6 | GO:0051865 | protein autoubiquitination(GO:0051865) |
| 0.0 | 1.1 | GO:0051453 | regulation of intracellular pH(GO:0051453) |
| 0.0 | 0.1 | GO:0051967 | negative regulation of synaptic transmission, glutamatergic(GO:0051967) |
| 0.0 | 0.3 | GO:0007216 | G-protein coupled glutamate receptor signaling pathway(GO:0007216) |
| 0.0 | 0.6 | GO:0032012 | ARF protein signal transduction(GO:0032011) regulation of ARF protein signal transduction(GO:0032012) |
| 0.0 | 2.1 | GO:0050806 | positive regulation of synaptic transmission(GO:0050806) |
| 0.0 | 2.8 | GO:0007286 | spermatid development(GO:0007286) |
| 0.0 | 0.2 | GO:0090331 | negative regulation of platelet aggregation(GO:0090331) |
| 0.0 | 0.2 | GO:0042574 | retinal metabolic process(GO:0042574) |
| 0.0 | 0.1 | GO:0043585 | nose morphogenesis(GO:0043585) alveolar primary septum development(GO:0061143) |
| 0.0 | 0.7 | GO:0051965 | positive regulation of synapse assembly(GO:0051965) |
| 0.0 | 0.4 | GO:0031424 | keratinization(GO:0031424) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.1 | 5.7 | GO:0097059 | CNTFR-CLCF1 complex(GO:0097059) |
| 1.1 | 3.3 | GO:0044317 | rod spherule(GO:0044317) |
| 0.8 | 6.5 | GO:0017071 | intracellular cyclic nucleotide activated cation channel complex(GO:0017071) |
| 0.8 | 3.0 | GO:0099573 | glutamatergic postsynaptic density(GO:0099573) |
| 0.6 | 2.2 | GO:0033596 | TSC1-TSC2 complex(GO:0033596) |
| 0.5 | 2.0 | GO:0046691 | intracellular canaliculus(GO:0046691) |
| 0.4 | 3.1 | GO:0005955 | calcineurin complex(GO:0005955) |
| 0.4 | 11.8 | GO:0014731 | spectrin-associated cytoskeleton(GO:0014731) |
| 0.3 | 2.9 | GO:0097442 | CA3 pyramidal cell dendrite(GO:0097442) |
| 0.3 | 4.0 | GO:0031315 | extrinsic component of mitochondrial outer membrane(GO:0031315) |
| 0.3 | 1.5 | GO:0034365 | discoidal high-density lipoprotein particle(GO:0034365) |
| 0.3 | 1.4 | GO:0010370 | perinucleolar chromocenter(GO:0010370) |
| 0.3 | 0.8 | GO:0043291 | RAVE complex(GO:0043291) |
| 0.2 | 1.7 | GO:1990584 | cardiac Troponin complex(GO:1990584) |
| 0.2 | 2.7 | GO:0005861 | troponin complex(GO:0005861) |
| 0.2 | 3.8 | GO:0012510 | trans-Golgi network transport vesicle membrane(GO:0012510) |
| 0.2 | 7.9 | GO:0045095 | keratin filament(GO:0045095) |
| 0.2 | 1.4 | GO:0033162 | melanosome membrane(GO:0033162) chitosome(GO:0045009) |
| 0.2 | 1.7 | GO:0030314 | junctional membrane complex(GO:0030314) |
| 0.1 | 1.7 | GO:0005742 | mitochondrial outer membrane translocase complex(GO:0005742) |
| 0.1 | 11.3 | GO:0005882 | intermediate filament(GO:0005882) |
| 0.1 | 2.9 | GO:0017146 | NMDA selective glutamate receptor complex(GO:0017146) |
| 0.1 | 2.8 | GO:0030056 | hemidesmosome(GO:0030056) |
| 0.1 | 1.2 | GO:0001739 | sex chromatin(GO:0001739) |
| 0.1 | 2.8 | GO:1902711 | GABA-A receptor complex(GO:1902711) |
| 0.1 | 2.6 | GO:0031362 | anchored component of external side of plasma membrane(GO:0031362) |
| 0.1 | 1.4 | GO:0065010 | extracellular membrane-bounded organelle(GO:0065010) |
| 0.1 | 3.4 | GO:0030673 | axolemma(GO:0030673) |
| 0.1 | 1.8 | GO:0010369 | chromocenter(GO:0010369) |
| 0.1 | 4.1 | GO:0031201 | SNARE complex(GO:0031201) |
| 0.1 | 0.3 | GO:0035841 | new growing cell tip(GO:0035841) |
| 0.1 | 1.0 | GO:0030127 | COPII vesicle coat(GO:0030127) |
| 0.1 | 1.0 | GO:0031045 | dense core granule(GO:0031045) |
| 0.0 | 0.5 | GO:0000815 | ESCRT III complex(GO:0000815) |
| 0.0 | 0.9 | GO:0031143 | pseudopodium(GO:0031143) |
| 0.0 | 5.2 | GO:0030315 | T-tubule(GO:0030315) |
| 0.0 | 1.4 | GO:0035267 | NuA4 histone acetyltransferase complex(GO:0035267) H4/H2A histone acetyltransferase complex(GO:0043189) H4 histone acetyltransferase complex(GO:1902562) |
| 0.0 | 2.3 | GO:0001917 | photoreceptor inner segment(GO:0001917) |
| 0.0 | 1.1 | GO:0043034 | costamere(GO:0043034) |
| 0.0 | 6.7 | GO:0043204 | perikaryon(GO:0043204) |
| 0.0 | 1.0 | GO:0030057 | desmosome(GO:0030057) |
| 0.0 | 0.1 | GO:0098830 | presynaptic endosome(GO:0098830) |
| 0.0 | 2.8 | GO:0043195 | terminal bouton(GO:0043195) |
| 0.0 | 0.1 | GO:0005892 | acetylcholine-gated channel complex(GO:0005892) |
| 0.0 | 1.1 | GO:0097228 | sperm principal piece(GO:0097228) |
| 0.0 | 0.8 | GO:0071782 | endoplasmic reticulum tubular network(GO:0071782) endoplasmic reticulum subcompartment(GO:0098827) |
| 0.0 | 3.0 | GO:0032587 | ruffle membrane(GO:0032587) |
| 0.0 | 6.6 | GO:0045211 | postsynaptic membrane(GO:0045211) |
| 0.0 | 0.7 | GO:0034451 | centriolar satellite(GO:0034451) |
| 0.0 | 8.8 | GO:0030141 | secretory granule(GO:0030141) |
| 0.0 | 2.7 | GO:0005923 | bicellular tight junction(GO:0005923) |
| 0.0 | 0.2 | GO:0044754 | autolysosome(GO:0044754) |
| 0.0 | 0.6 | GO:0034707 | chloride channel complex(GO:0034707) |
| 0.0 | 4.0 | GO:0016323 | basolateral plasma membrane(GO:0016323) |
| 0.0 | 3.2 | GO:0016324 | apical plasma membrane(GO:0016324) |
| 0.0 | 1.6 | GO:0030018 | Z disc(GO:0030018) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.1 | 6.5 | GO:0005052 | peroxisome matrix targeting signal-1 binding(GO:0005052) |
| 1.0 | 5.1 | GO:0070012 | oligopeptidase activity(GO:0070012) |
| 0.9 | 6.2 | GO:0004111 | creatine kinase activity(GO:0004111) |
| 0.7 | 2.0 | GO:0080130 | L-phenylalanine:2-oxoglutarate aminotransferase activity(GO:0080130) |
| 0.7 | 2.7 | GO:0001588 | dopamine neurotransmitter receptor activity, coupled via Gs(GO:0001588) |
| 0.6 | 5.7 | GO:0004897 | ciliary neurotrophic factor receptor activity(GO:0004897) |
| 0.6 | 3.4 | GO:0033192 | calmodulin-dependent protein phosphatase activity(GO:0033192) |
| 0.6 | 3.4 | GO:0015501 | glutamate:sodium symporter activity(GO:0015501) |
| 0.5 | 3.2 | GO:0050833 | pyruvate transmembrane transporter activity(GO:0050833) |
| 0.4 | 4.5 | GO:0031014 | troponin T binding(GO:0031014) |
| 0.4 | 1.7 | GO:0005219 | ryanodine-sensitive calcium-release channel activity(GO:0005219) |
| 0.4 | 1.2 | GO:1901375 | acetylcholine transmembrane transporter activity(GO:0005277) norepinephrine transmembrane transporter activity(GO:0005333) acetate ester transmembrane transporter activity(GO:1901375) |
| 0.4 | 6.4 | GO:1901702 | urate transmembrane transporter activity(GO:0015143) salt transmembrane transporter activity(GO:1901702) |
| 0.4 | 1.8 | GO:0051373 | FATZ binding(GO:0051373) |
| 0.3 | 1.4 | GO:0008113 | peptide-methionine (S)-S-oxide reductase activity(GO:0008113) |
| 0.3 | 1.0 | GO:0036461 | BLOC-2 complex binding(GO:0036461) |
| 0.3 | 2.9 | GO:0005519 | cytoskeletal regulatory protein binding(GO:0005519) |
| 0.3 | 2.8 | GO:0004568 | chitinase activity(GO:0004568) |
| 0.3 | 3.1 | GO:0004886 | 9-cis retinoic acid receptor activity(GO:0004886) |
| 0.3 | 2.1 | GO:0042328 | heparan sulfate N-acetylglucosaminyltransferase activity(GO:0042328) |
| 0.3 | 0.9 | GO:0042015 | interleukin-20 binding(GO:0042015) |
| 0.3 | 3.3 | GO:0008020 | G-protein coupled photoreceptor activity(GO:0008020) |
| 0.3 | 3.0 | GO:0022851 | GABA-gated chloride ion channel activity(GO:0022851) |
| 0.3 | 1.4 | GO:0034584 | piRNA binding(GO:0034584) |
| 0.3 | 0.8 | GO:0001607 | neuromedin U receptor activity(GO:0001607) |
| 0.3 | 0.8 | GO:0008330 | protein tyrosine/threonine phosphatase activity(GO:0008330) |
| 0.3 | 1.8 | GO:0099529 | neurotransmitter receptor activity involved in regulation of postsynaptic membrane potential(GO:0099529) |
| 0.2 | 2.6 | GO:0051870 | methotrexate binding(GO:0051870) |
| 0.2 | 5.7 | GO:0004435 | phosphatidylinositol phospholipase C activity(GO:0004435) |
| 0.2 | 0.9 | GO:0004937 | alpha1-adrenergic receptor activity(GO:0004937) |
| 0.2 | 3.8 | GO:0048406 | nerve growth factor binding(GO:0048406) |
| 0.2 | 1.7 | GO:0015349 | thyroid hormone transmembrane transporter activity(GO:0015349) |
| 0.2 | 1.4 | GO:0004035 | alkaline phosphatase activity(GO:0004035) |
| 0.2 | 0.8 | GO:0004105 | choline-phosphate cytidylyltransferase activity(GO:0004105) |
| 0.2 | 2.5 | GO:0019855 | calcium channel inhibitor activity(GO:0019855) |
| 0.2 | 3.0 | GO:0009931 | calcium-dependent protein serine/threonine kinase activity(GO:0009931) |
| 0.2 | 0.7 | GO:0035827 | rubidium ion transmembrane transporter activity(GO:0035827) |
| 0.2 | 0.9 | GO:0015129 | lactate transmembrane transporter activity(GO:0015129) |
| 0.2 | 2.2 | GO:0102338 | fatty acid elongase activity(GO:0009922) 3-oxo-arachidoyl-CoA synthase activity(GO:0102336) 3-oxo-cerotoyl-CoA synthase activity(GO:0102337) 3-oxo-lignoceronyl-CoA synthase activity(GO:0102338) |
| 0.2 | 2.0 | GO:0005347 | ATP transmembrane transporter activity(GO:0005347) ADP transmembrane transporter activity(GO:0015217) |
| 0.2 | 1.2 | GO:0005332 | gamma-aminobutyric acid:sodium symporter activity(GO:0005332) |
| 0.2 | 3.0 | GO:0002162 | dystroglycan binding(GO:0002162) |
| 0.2 | 1.7 | GO:0015288 | porin activity(GO:0015288) |
| 0.2 | 6.3 | GO:0005385 | zinc ion transmembrane transporter activity(GO:0005385) |
| 0.1 | 0.4 | GO:0002061 | uracil binding(GO:0002058) pyrimidine nucleobase binding(GO:0002061) |
| 0.1 | 1.1 | GO:0043423 | 3-phosphoinositide-dependent protein kinase binding(GO:0043423) |
| 0.1 | 4.1 | GO:0005212 | structural constituent of eye lens(GO:0005212) |
| 0.1 | 0.4 | GO:0031531 | thyrotropin-releasing hormone receptor binding(GO:0031531) |
| 0.1 | 0.6 | GO:0001010 | transcription factor activity, sequence-specific DNA binding transcription factor recruiting(GO:0001010) |
| 0.1 | 5.8 | GO:1990841 | promoter-specific chromatin binding(GO:1990841) |
| 0.1 | 1.4 | GO:0030976 | thiamine pyrophosphate binding(GO:0030976) |
| 0.1 | 17.4 | GO:0005200 | structural constituent of cytoskeleton(GO:0005200) |
| 0.1 | 4.4 | GO:0005242 | inward rectifier potassium channel activity(GO:0005242) |
| 0.1 | 1.9 | GO:0016290 | palmitoyl-CoA hydrolase activity(GO:0016290) |
| 0.1 | 2.9 | GO:0000062 | fatty-acyl-CoA binding(GO:0000062) |
| 0.1 | 1.5 | GO:0070008 | serine-type exopeptidase activity(GO:0070008) |
| 0.1 | 0.9 | GO:0038062 | protein tyrosine kinase collagen receptor activity(GO:0038062) |
| 0.1 | 0.3 | GO:0016842 | amidine-lyase activity(GO:0016842) |
| 0.1 | 0.7 | GO:0016286 | small conductance calcium-activated potassium channel activity(GO:0016286) |
| 0.1 | 4.0 | GO:0051393 | alpha-actinin binding(GO:0051393) |
| 0.1 | 2.7 | GO:0015020 | glucuronosyltransferase activity(GO:0015020) |
| 0.1 | 17.3 | GO:0008236 | serine-type peptidase activity(GO:0008236) |
| 0.1 | 1.1 | GO:0015299 | solute:proton antiporter activity(GO:0015299) |
| 0.1 | 0.4 | GO:0016716 | oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, another compound as one donor, and incorporation of one atom of oxygen(GO:0016716) |
| 0.1 | 2.1 | GO:0004623 | phospholipase A2 activity(GO:0004623) |
| 0.1 | 1.2 | GO:0042043 | neurexin family protein binding(GO:0042043) |
| 0.1 | 1.0 | GO:0004385 | guanylate kinase activity(GO:0004385) |
| 0.1 | 1.1 | GO:0008276 | protein methyltransferase activity(GO:0008276) |
| 0.1 | 0.6 | GO:0004499 | N,N-dimethylaniline monooxygenase activity(GO:0004499) |
| 0.0 | 4.4 | GO:0097110 | scaffold protein binding(GO:0097110) |
| 0.0 | 0.7 | GO:0016175 | superoxide-generating NADPH oxidase activity(GO:0016175) |
| 0.0 | 0.9 | GO:0008331 | high voltage-gated calcium channel activity(GO:0008331) |
| 0.0 | 0.2 | GO:0004952 | dopamine neurotransmitter receptor activity(GO:0004952) |
| 0.0 | 0.9 | GO:0004745 | retinol dehydrogenase activity(GO:0004745) |
| 0.0 | 4.1 | GO:0048306 | calcium-dependent protein binding(GO:0048306) |
| 0.0 | 0.9 | GO:0003841 | 1-acylglycerol-3-phosphate O-acyltransferase activity(GO:0003841) |
| 0.0 | 0.6 | GO:0003796 | lysozyme activity(GO:0003796) |
| 0.0 | 0.2 | GO:0002046 | opsin binding(GO:0002046) |
| 0.0 | 1.0 | GO:0005184 | neuropeptide hormone activity(GO:0005184) |
| 0.0 | 0.7 | GO:0016503 | pheromone receptor activity(GO:0016503) |
| 0.0 | 0.6 | GO:0032794 | GTPase activating protein binding(GO:0032794) |
| 0.0 | 0.9 | GO:0071855 | neuropeptide receptor binding(GO:0071855) |
| 0.0 | 0.7 | GO:0000983 | transcription factor activity, RNA polymerase II core promoter sequence-specific(GO:0000983) |
| 0.0 | 1.1 | GO:0005109 | frizzled binding(GO:0005109) |
| 0.0 | 0.6 | GO:0017128 | phospholipid scramblase activity(GO:0017128) |
| 0.0 | 4.4 | GO:0042826 | histone deacetylase binding(GO:0042826) |
| 0.0 | 0.9 | GO:0005044 | scavenger receptor activity(GO:0005044) |
| 0.0 | 0.2 | GO:0005432 | calcium:sodium antiporter activity(GO:0005432) |
| 0.0 | 11.5 | GO:0005198 | structural molecule activity(GO:0005198) |
| 0.0 | 0.2 | GO:0016724 | ferroxidase activity(GO:0004322) oxidoreductase activity, oxidizing metal ions, oxygen as acceptor(GO:0016724) |
| 0.0 | 2.0 | GO:0008170 | N-methyltransferase activity(GO:0008170) |
| 0.0 | 0.5 | GO:0047555 | 3',5'-cyclic-GMP phosphodiesterase activity(GO:0047555) |
| 0.0 | 0.6 | GO:0005086 | ARF guanyl-nucleotide exchange factor activity(GO:0005086) |
| 0.0 | 0.2 | GO:0043522 | leucine zipper domain binding(GO:0043522) |
| 0.0 | 1.1 | GO:0017022 | myosin binding(GO:0017022) |
| 0.0 | 0.4 | GO:0046965 | retinoid X receptor binding(GO:0046965) |
| 0.0 | 0.7 | GO:0004521 | endoribonuclease activity(GO:0004521) |
| 0.0 | 1.5 | GO:0016209 | antioxidant activity(GO:0016209) |
| 0.0 | 5.2 | GO:0005549 | odorant binding(GO:0005549) |
| 0.0 | 1.6 | GO:0003705 | transcription factor activity, RNA polymerase II distal enhancer sequence-specific binding(GO:0003705) |
| 0.0 | 0.7 | GO:0004879 | RNA polymerase II transcription factor activity, ligand-activated sequence-specific DNA binding(GO:0004879) transcription factor activity, direct ligand regulated sequence-specific DNA binding(GO:0098531) |
| 0.0 | 0.7 | GO:0046875 | ephrin receptor binding(GO:0046875) |
| 0.0 | 0.8 | GO:0008081 | phosphoric diester hydrolase activity(GO:0008081) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 5.1 | PID INTEGRIN2 PATHWAY | Beta2 integrin cell surface interactions |
| 0.1 | 4.4 | ST WNT CA2 CYCLIC GMP PATHWAY | Wnt/Ca2+/cyclic GMP signaling. |
| 0.1 | 5.2 | PID TCR CALCIUM PATHWAY | Calcium signaling in the CD4+ TCR pathway |
| 0.1 | 5.9 | PID A6B1 A6B4 INTEGRIN PATHWAY | a6b1 and a6b4 Integrin signaling |
| 0.0 | 2.9 | PID LKB1 PATHWAY | LKB1 signaling events |
| 0.0 | 3.3 | PID P75 NTR PATHWAY | p75(NTR)-mediated signaling |
| 0.0 | 4.1 | PID TRKR PATHWAY | Neurotrophic factor-mediated Trk receptor signaling |
| 0.0 | 1.8 | PID ERBB4 PATHWAY | ErbB4 signaling events |
| 0.0 | 1.5 | SIG REGULATION OF THE ACTIN CYTOSKELETON BY RHO GTPASES | Genes related to regulation of the actin cytoskeleton |
| 0.0 | 1.0 | PID DELTA NP63 PATHWAY | Validated transcriptional targets of deltaNp63 isoforms |
| 0.0 | 1.1 | PID PI3KCI PATHWAY | Class I PI3K signaling events |
| 0.0 | 0.2 | PID TXA2PATHWAY | Thromboxane A2 receptor signaling |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.6 | 9.0 | REACTOME GAMMA CARBOXYLATION TRANSPORT AND AMINO TERMINAL CLEAVAGE OF PROTEINS | Genes involved in Gamma-carboxylation, transport, and amino-terminal cleavage of proteins |
| 0.4 | 6.2 | REACTOME ORGANIC CATION ANION ZWITTERION TRANSPORT | Genes involved in Organic cation/anion/zwitterion transport |
| 0.3 | 3.3 | REACTOME OPSINS | Genes involved in Opsins |
| 0.2 | 4.1 | REACTOME DOPAMINE NEUROTRANSMITTER RELEASE CYCLE | Genes involved in Dopamine Neurotransmitter Release Cycle |
| 0.2 | 11.8 | REACTOME INTERACTION BETWEEN L1 AND ANKYRINS | Genes involved in Interaction between L1 and Ankyrins |
| 0.2 | 7.0 | REACTOME CREB PHOSPHORYLATION THROUGH THE ACTIVATION OF CAMKII | Genes involved in CREB phosphorylation through the activation of CaMKII |
| 0.2 | 3.4 | REACTOME BILE SALT AND ORGANIC ANION SLC TRANSPORTERS | Genes involved in Bile salt and organic anion SLC transporters |
| 0.1 | 1.9 | REACTOME NEGATIVE REGULATION OF THE PI3K AKT NETWORK | Genes involved in Negative regulation of the PI3K/AKT network |
| 0.1 | 3.0 | REACTOME GABA A RECEPTOR ACTIVATION | Genes involved in GABA A receptor activation |
| 0.1 | 5.7 | REACTOME STRIATED MUSCLE CONTRACTION | Genes involved in Striated Muscle Contraction |
| 0.1 | 1.2 | REACTOME PASSIVE TRANSPORT BY AQUAPORINS | Genes involved in Passive Transport by Aquaporins |
| 0.1 | 1.7 | REACTOME SYNTHESIS SECRETION AND INACTIVATION OF GIP | Genes involved in Synthesis, Secretion, and Inactivation of Glucose-dependent Insulinotropic Polypeptide (GIP) |
| 0.1 | 1.0 | REACTOME ABACAVIR TRANSPORT AND METABOLISM | Genes involved in Abacavir transport and metabolism |
| 0.1 | 2.0 | REACTOME AMINO ACID SYNTHESIS AND INTERCONVERSION TRANSAMINATION | Genes involved in Amino acid synthesis and interconversion (transamination) |
| 0.1 | 2.2 | REACTOME SYNTHESIS OF VERY LONG CHAIN FATTY ACYL COAS | Genes involved in Synthesis of very long-chain fatty acyl-CoAs |
| 0.1 | 2.0 | REACTOME ADHERENS JUNCTIONS INTERACTIONS | Genes involved in Adherens junctions interactions |
| 0.1 | 2.1 | REACTOME HS GAG BIOSYNTHESIS | Genes involved in HS-GAG biosynthesis |
| 0.0 | 0.6 | REACTOME INWARDLY RECTIFYING K CHANNELS | Genes involved in Inwardly rectifying K+ channels |
| 0.0 | 1.4 | REACTOME MUSCLE CONTRACTION | Genes involved in Muscle contraction |
| 0.0 | 2.8 | REACTOME COLLAGEN FORMATION | Genes involved in Collagen formation |
| 0.0 | 1.4 | REACTOME PEROXISOMAL LIPID METABOLISM | Genes involved in Peroxisomal lipid metabolism |
| 0.0 | 3.8 | REACTOME NUCLEAR RECEPTOR TRANSCRIPTION PATHWAY | Genes involved in Nuclear Receptor transcription pathway |
| 0.0 | 1.1 | REACTOME AMINE LIGAND BINDING RECEPTORS | Genes involved in Amine ligand-binding receptors |
| 0.0 | 0.8 | REACTOME SYNTHESIS OF PC | Genes involved in Synthesis of PC |
| 0.0 | 0.8 | REACTOME LYSOSOME VESICLE BIOGENESIS | Genes involved in Lysosome Vesicle Biogenesis |
| 0.0 | 0.9 | REACTOME TRIGLYCERIDE BIOSYNTHESIS | Genes involved in Triglyceride Biosynthesis |
| 0.0 | 0.4 | REACTOME PYRIMIDINE CATABOLISM | Genes involved in Pyrimidine catabolism |
| 0.0 | 0.5 | REACTOME NCAM1 INTERACTIONS | Genes involved in NCAM1 interactions |
| 0.0 | 0.3 | REACTOME PURINE RIBONUCLEOSIDE MONOPHOSPHATE BIOSYNTHESIS | Genes involved in Purine ribonucleoside monophosphate biosynthesis |
| 0.0 | 1.6 | REACTOME L1CAM INTERACTIONS | Genes involved in L1CAM interactions |
| 0.0 | 0.5 | REACTOME ENDOSOMAL SORTING COMPLEX REQUIRED FOR TRANSPORT ESCRT | Genes involved in Endosomal Sorting Complex Required For Transport (ESCRT) |
| 0.0 | 3.3 | REACTOME TRANSMISSION ACROSS CHEMICAL SYNAPSES | Genes involved in Transmission across Chemical Synapses |
| 0.0 | 0.7 | REACTOME DARPP 32 EVENTS | Genes involved in DARPP-32 events |
| 0.0 | 0.6 | REACTOME SIGNALING BY ROBO RECEPTOR | Genes involved in Signaling by Robo receptor |
| 0.0 | 1.1 | REACTOME CLASS B 2 SECRETIN FAMILY RECEPTORS | Genes involved in Class B/2 (Secretin family receptors) |