Project
PRJNA375882: Comprehensive Mouse Transcriptomic BodyMap
Navigation
Downloads
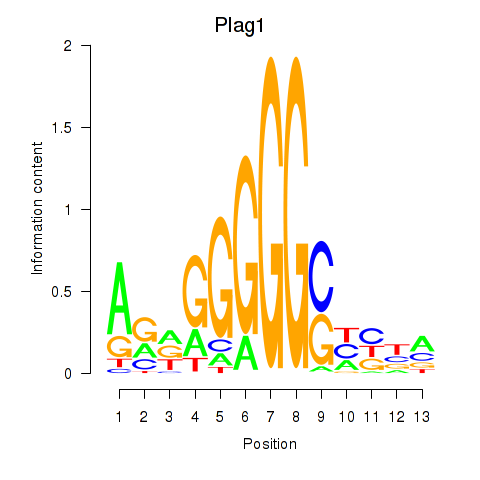
Results for Plag1
Z-value: 1.45
Transcription factors associated with Plag1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Plag1
|
ENSMUSG00000003282.10 | Plag1 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Plag1 | mm39_v1_chr4_-_3938352_3938401 | 0.09 | 4.6e-01 | Click! |
Activity profile of Plag1 motif
Sorted Z-values of Plag1 motif
Network of associatons between targets according to the STRING database.
First level regulatory network of Plag1
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 4.2 | 12.6 | GO:0030824 | negative regulation of cGMP metabolic process(GO:0030824) negative regulation of cGMP biosynthetic process(GO:0030827) negative regulation of guanylate cyclase activity(GO:0031283) |
| 4.1 | 12.3 | GO:0035544 | negative regulation of SNARE complex assembly(GO:0035544) |
| 2.9 | 17.1 | GO:0050758 | thymidylate synthase biosynthetic process(GO:0050757) regulation of thymidylate synthase biosynthetic process(GO:0050758) negative regulation of thymidylate synthase biosynthetic process(GO:0050760) |
| 2.5 | 9.9 | GO:0003050 | regulation of systemic arterial blood pressure by atrial natriuretic peptide(GO:0003050) |
| 2.4 | 14.6 | GO:0016185 | synaptic vesicle budding from presynaptic endocytic zone membrane(GO:0016185) |
| 2.1 | 10.5 | GO:1902163 | negative regulation of DNA damage response, signal transduction by p53 class mediator resulting in transcription of p21 class mediator(GO:1902163) |
| 1.9 | 5.7 | GO:0043181 | vacuolar sequestering(GO:0043181) |
| 1.8 | 11.0 | GO:1902998 | regulation of neuronal signal transduction(GO:1902847) positive regulation of neurofibrillary tangle assembly(GO:1902998) |
| 1.5 | 12.2 | GO:0061368 | behavioral response to chemical pain(GO:0061366) behavioral response to formalin induced pain(GO:0061368) |
| 1.4 | 11.3 | GO:0051612 | negative regulation of neurotransmitter uptake(GO:0051581) negative regulation of serotonin uptake(GO:0051612) |
| 1.2 | 3.7 | GO:1903048 | regulation of acetylcholine-gated cation channel activity(GO:1903048) |
| 1.2 | 4.9 | GO:1904058 | positive regulation of sensory perception of pain(GO:1904058) |
| 1.2 | 3.7 | GO:0002184 | cytoplasmic translational termination(GO:0002184) |
| 1.2 | 4.7 | GO:2001245 | regulation of phosphatidylcholine biosynthetic process(GO:2001245) |
| 1.2 | 3.5 | GO:0006500 | N-terminal protein palmitoylation(GO:0006500) |
| 1.1 | 6.6 | GO:1903566 | positive regulation of protein localization to cilium(GO:1903566) |
| 1.1 | 3.2 | GO:1903412 | response to bile acid(GO:1903412) cellular response to bile acid(GO:1903413) |
| 1.0 | 4.1 | GO:1905051 | regulation of base-excision repair(GO:1905051) positive regulation of base-excision repair(GO:1905053) |
| 1.0 | 2.9 | GO:1900195 | positive regulation of oocyte maturation(GO:1900195) positive regulation of proteasomal ubiquitin-dependent protein catabolic process involved in cellular response to hypoxia(GO:2000777) |
| 1.0 | 2.9 | GO:1902630 | regulation of membrane hyperpolarization(GO:1902630) |
| 0.9 | 9.5 | GO:0045162 | clustering of voltage-gated sodium channels(GO:0045162) |
| 0.9 | 2.7 | GO:1905169 | protein localization to phagocytic vesicle(GO:1905161) regulation of protein localization to phagocytic vesicle(GO:1905169) positive regulation of protein localization to phagocytic vesicle(GO:1905171) |
| 0.9 | 3.5 | GO:0007208 | phospholipase C-activating serotonin receptor signaling pathway(GO:0007208) |
| 0.9 | 5.3 | GO:0071677 | positive regulation of mononuclear cell migration(GO:0071677) |
| 0.8 | 2.5 | GO:0060156 | vascular transport(GO:0010232) milk ejection(GO:0060156) |
| 0.8 | 2.5 | GO:0030070 | insulin processing(GO:0030070) |
| 0.8 | 2.3 | GO:0099578 | regulation of translation at synapse, modulating synaptic transmission(GO:0099547) regulation of translation at postsynapse, modulating synaptic transmission(GO:0099578) positive regulation of intracellular transport of viral material(GO:1901254) |
| 0.8 | 3.1 | GO:0030450 | regulation of complement activation, classical pathway(GO:0030450) |
| 0.8 | 3.8 | GO:1903422 | negative regulation of synaptic vesicle recycling(GO:1903422) regulation of spontaneous neurotransmitter secretion(GO:1904048) |
| 0.7 | 2.2 | GO:0048611 | ectodermal digestive tract development(GO:0007439) embryonic ectodermal digestive tract development(GO:0048611) |
| 0.7 | 2.9 | GO:0021933 | radial glia guided migration of cerebellar granule cell(GO:0021933) negative regulation of cerebellar granule cell precursor proliferation(GO:0021941) |
| 0.7 | 6.6 | GO:0098700 | neurotransmitter loading into synaptic vesicle(GO:0098700) |
| 0.7 | 2.2 | GO:0006546 | glycine catabolic process(GO:0006546) glycine decarboxylation via glycine cleavage system(GO:0019464) |
| 0.7 | 7.2 | GO:2000672 | negative regulation of motor neuron apoptotic process(GO:2000672) |
| 0.7 | 2.8 | GO:0031448 | regulation of fast-twitch skeletal muscle fiber contraction(GO:0031446) positive regulation of fast-twitch skeletal muscle fiber contraction(GO:0031448) |
| 0.7 | 2.8 | GO:1900220 | semaphorin-plexin signaling pathway involved in bone trabecula morphogenesis(GO:1900220) |
| 0.7 | 4.1 | GO:0032423 | regulation of mismatch repair(GO:0032423) |
| 0.7 | 6.8 | GO:1902856 | negative regulation of nonmotile primary cilium assembly(GO:1902856) |
| 0.7 | 2.0 | GO:0090649 | rRNA export from nucleus(GO:0006407) response to oxygen-glucose deprivation(GO:0090649) cellular response to oxygen-glucose deprivation(GO:0090650) |
| 0.7 | 3.4 | GO:0035542 | regulation of SNARE complex assembly(GO:0035542) |
| 0.7 | 2.7 | GO:1902080 | regulation of calcium ion import into sarcoplasmic reticulum(GO:1902080) negative regulation of calcium ion import into sarcoplasmic reticulum(GO:1902081) |
| 0.7 | 3.3 | GO:0010747 | positive regulation of plasma membrane long-chain fatty acid transport(GO:0010747) |
| 0.6 | 3.2 | GO:0071651 | positive regulation of chemokine (C-C motif) ligand 5 production(GO:0071651) |
| 0.6 | 3.2 | GO:0032901 | positive regulation of neurotrophin production(GO:0032901) |
| 0.6 | 2.5 | GO:1900224 | positive regulation of nodal signaling pathway involved in determination of lateral mesoderm left/right asymmetry(GO:1900224) |
| 0.6 | 2.5 | GO:0000239 | pachytene(GO:0000239) |
| 0.6 | 4.4 | GO:0071630 | nucleus-associated proteasomal ubiquitin-dependent protein catabolic process(GO:0071630) |
| 0.6 | 1.8 | GO:0060466 | activation of meiosis involved in egg activation(GO:0060466) |
| 0.6 | 1.8 | GO:0002380 | immunoglobulin secretion involved in immune response(GO:0002380) |
| 0.6 | 18.8 | GO:0035641 | locomotory exploration behavior(GO:0035641) |
| 0.6 | 7.0 | GO:2000467 | positive regulation of glycogen (starch) synthase activity(GO:2000467) |
| 0.6 | 5.8 | GO:0007168 | receptor guanylyl cyclase signaling pathway(GO:0007168) |
| 0.6 | 27.5 | GO:0060074 | synapse maturation(GO:0060074) |
| 0.5 | 3.3 | GO:1903903 | protein localization to plasma membrane raft(GO:0044860) regulation of establishment of T cell polarity(GO:1903903) |
| 0.5 | 6.5 | GO:0098870 | neuronal action potential propagation(GO:0019227) action potential propagation(GO:0098870) |
| 0.5 | 2.2 | GO:0046066 | purine deoxyribonucleoside diphosphate metabolic process(GO:0009182) dGDP metabolic process(GO:0046066) |
| 0.5 | 1.6 | GO:0061193 | taste bud development(GO:0061193) |
| 0.5 | 2.1 | GO:0080163 | regulation of protein serine/threonine phosphatase activity(GO:0080163) |
| 0.5 | 1.6 | GO:1903348 | positive regulation of bicellular tight junction assembly(GO:1903348) |
| 0.5 | 1.6 | GO:0006478 | peptidyl-tyrosine sulfation(GO:0006478) |
| 0.5 | 1.6 | GO:0060748 | tertiary branching involved in mammary gland duct morphogenesis(GO:0060748) |
| 0.5 | 2.1 | GO:0051933 | amino acid neurotransmitter reuptake(GO:0051933) glutamate reuptake(GO:0051935) |
| 0.5 | 2.1 | GO:1903215 | negative regulation of protein targeting to mitochondrion(GO:1903215) |
| 0.5 | 3.6 | GO:0045163 | clustering of voltage-gated potassium channels(GO:0045163) |
| 0.5 | 3.6 | GO:0045113 | regulation of integrin biosynthetic process(GO:0045113) |
| 0.5 | 2.6 | GO:0044268 | multicellular organismal protein metabolic process(GO:0044268) |
| 0.5 | 2.0 | GO:2000983 | regulation of ATP citrate synthase activity(GO:2000983) negative regulation of ATP citrate synthase activity(GO:2000984) |
| 0.5 | 4.9 | GO:2000774 | positive regulation of cellular senescence(GO:2000774) |
| 0.5 | 2.9 | GO:0000160 | phosphorelay signal transduction system(GO:0000160) |
| 0.5 | 1.9 | GO:0071469 | cellular response to alkaline pH(GO:0071469) |
| 0.5 | 1.9 | GO:0019859 | pyrimidine nucleobase catabolic process(GO:0006208) thymine catabolic process(GO:0006210) thymine metabolic process(GO:0019859) |
| 0.5 | 10.8 | GO:0071420 | cellular response to histamine(GO:0071420) |
| 0.5 | 1.9 | GO:2000384 | regulation of ectoderm development(GO:2000383) negative regulation of ectoderm development(GO:2000384) |
| 0.5 | 2.3 | GO:1904694 | negative regulation of vascular smooth muscle contraction(GO:1904694) |
| 0.5 | 1.4 | GO:0016561 | protein import into peroxisome matrix, translocation(GO:0016561) |
| 0.4 | 4.0 | GO:0060373 | regulation of ventricular cardiac muscle cell membrane depolarization(GO:0060373) |
| 0.4 | 1.8 | GO:0010901 | regulation of very-low-density lipoprotein particle remodeling(GO:0010901) |
| 0.4 | 2.6 | GO:0033326 | cerebrospinal fluid secretion(GO:0033326) |
| 0.4 | 4.8 | GO:0051823 | regulation of synapse structural plasticity(GO:0051823) |
| 0.4 | 5.3 | GO:1902474 | positive regulation of protein localization to synapse(GO:1902474) |
| 0.4 | 2.6 | GO:0021633 | optic nerve structural organization(GO:0021633) |
| 0.4 | 3.5 | GO:0005513 | detection of calcium ion(GO:0005513) |
| 0.4 | 3.0 | GO:0014053 | negative regulation of gamma-aminobutyric acid secretion(GO:0014053) |
| 0.4 | 1.3 | GO:1900186 | negative regulation of clathrin-mediated endocytosis(GO:1900186) |
| 0.4 | 3.0 | GO:0045200 | establishment or maintenance of neuroblast polarity(GO:0045196) establishment of neuroblast polarity(GO:0045200) |
| 0.4 | 2.1 | GO:0021965 | spinal cord ventral commissure morphogenesis(GO:0021965) |
| 0.4 | 2.1 | GO:0099558 | maintenance of synapse structure(GO:0099558) |
| 0.4 | 2.0 | GO:2000224 | regulation of testosterone biosynthetic process(GO:2000224) |
| 0.4 | 19.8 | GO:0010107 | potassium ion import(GO:0010107) |
| 0.4 | 1.5 | GO:0097274 | urea homeostasis(GO:0097274) |
| 0.4 | 1.1 | GO:0019405 | alditol catabolic process(GO:0019405) |
| 0.4 | 3.4 | GO:0034316 | negative regulation of Arp2/3 complex-mediated actin nucleation(GO:0034316) |
| 0.4 | 2.6 | GO:0002317 | plasma cell differentiation(GO:0002317) |
| 0.4 | 1.5 | GO:0034436 | glycoprotein transport(GO:0034436) |
| 0.4 | 2.5 | GO:0045629 | negative regulation of T-helper 2 cell differentiation(GO:0045629) |
| 0.4 | 1.1 | GO:1904431 | positive regulation of t-circle formation(GO:1904431) |
| 0.3 | 2.7 | GO:0071313 | cellular response to caffeine(GO:0071313) |
| 0.3 | 1.7 | GO:0014042 | positive regulation of neuron maturation(GO:0014042) |
| 0.3 | 2.4 | GO:0061091 | regulation of phospholipid translocation(GO:0061091) positive regulation of phospholipid translocation(GO:0061092) |
| 0.3 | 2.0 | GO:1903275 | positive regulation of sodium ion export(GO:1903275) positive regulation of sodium ion export from cell(GO:1903278) |
| 0.3 | 1.3 | GO:0060988 | lipid tube assembly(GO:0060988) |
| 0.3 | 1.0 | GO:0031117 | positive regulation of microtubule depolymerization(GO:0031117) |
| 0.3 | 1.0 | GO:0061033 | bronchiole development(GO:0060435) bud outgrowth involved in lung branching(GO:0060447) secretion by lung epithelial cell involved in lung growth(GO:0061033) |
| 0.3 | 2.9 | GO:0006449 | regulation of translational termination(GO:0006449) |
| 0.3 | 1.9 | GO:0031444 | slow-twitch skeletal muscle fiber contraction(GO:0031444) |
| 0.3 | 0.6 | GO:0070358 | actin polymerization-dependent cell motility(GO:0070358) |
| 0.3 | 0.9 | GO:0002946 | tRNA C5-cytosine methylation(GO:0002946) |
| 0.3 | 3.1 | GO:0035021 | negative regulation of Rac protein signal transduction(GO:0035021) |
| 0.3 | 5.2 | GO:0021681 | cerebellar granular layer development(GO:0021681) |
| 0.3 | 2.1 | GO:2001023 | regulation of response to drug(GO:2001023) |
| 0.3 | 1.5 | GO:0050847 | progesterone receptor signaling pathway(GO:0050847) |
| 0.3 | 8.3 | GO:0016339 | calcium-dependent cell-cell adhesion via plasma membrane cell adhesion molecules(GO:0016339) |
| 0.3 | 7.7 | GO:0007614 | short-term memory(GO:0007614) |
| 0.3 | 4.7 | GO:0043562 | cellular response to nitrogen starvation(GO:0006995) cellular response to nitrogen levels(GO:0043562) |
| 0.3 | 0.9 | GO:1990180 | mitochondrial tRNA 3'-end processing(GO:1990180) |
| 0.3 | 17.3 | GO:0061001 | regulation of dendritic spine morphogenesis(GO:0061001) |
| 0.3 | 0.8 | GO:2000184 | glomerular visceral epithelial cell apoptotic process(GO:1903210) regulation of glomerular visceral epithelial cell apoptotic process(GO:1904633) positive regulation of glomerular visceral epithelial cell apoptotic process(GO:1904635) positive regulation of progesterone biosynthetic process(GO:2000184) |
| 0.3 | 6.4 | GO:0070262 | peptidyl-serine dephosphorylation(GO:0070262) |
| 0.3 | 1.9 | GO:1903265 | keratinocyte apoptotic process(GO:0097283) regulation of keratinocyte apoptotic process(GO:1902172) positive regulation of tumor necrosis factor-mediated signaling pathway(GO:1903265) |
| 0.3 | 4.1 | GO:1902715 | positive regulation of interferon-gamma secretion(GO:1902715) |
| 0.3 | 1.6 | GO:0015819 | lysine transport(GO:0015819) |
| 0.3 | 2.4 | GO:0060174 | limb bud formation(GO:0060174) |
| 0.3 | 1.0 | GO:0090063 | positive regulation of microtubule nucleation(GO:0090063) |
| 0.3 | 1.8 | GO:1902035 | positive regulation of hematopoietic stem cell proliferation(GO:1902035) |
| 0.3 | 1.3 | GO:0050651 | dermatan sulfate proteoglycan biosynthetic process(GO:0050651) |
| 0.3 | 1.3 | GO:0042494 | detection of bacterial lipoprotein(GO:0042494) |
| 0.2 | 3.5 | GO:0014051 | gamma-aminobutyric acid secretion(GO:0014051) gamma-aminobutyric acid transport(GO:0015812) |
| 0.2 | 2.7 | GO:0006013 | mannose metabolic process(GO:0006013) |
| 0.2 | 1.0 | GO:0060084 | synaptic transmission involved in micturition(GO:0060084) |
| 0.2 | 1.7 | GO:0030473 | nucleokinesis involved in cell motility in cerebral cortex radial glia guided migration(GO:0021817) nuclear migration along microtubule(GO:0030473) nuclear matrix organization(GO:0043578) nuclear matrix anchoring at nuclear membrane(GO:0090292) |
| 0.2 | 0.7 | GO:0008588 | release of cytoplasmic sequestered NF-kappaB(GO:0008588) |
| 0.2 | 1.9 | GO:0033058 | directional locomotion(GO:0033058) |
| 0.2 | 3.1 | GO:0018095 | protein polyglutamylation(GO:0018095) |
| 0.2 | 1.4 | GO:0035585 | calcium-mediated signaling using extracellular calcium source(GO:0035585) |
| 0.2 | 2.3 | GO:0019243 | methylglyoxal catabolic process to D-lactate via S-lactoyl-glutathione(GO:0019243) methylglyoxal catabolic process(GO:0051596) methylglyoxal catabolic process to lactate(GO:0061727) |
| 0.2 | 1.1 | GO:0071694 | protein localization to extracellular region(GO:0071692) maintenance of protein location in extracellular region(GO:0071694) |
| 0.2 | 0.9 | GO:0036072 | intramembranous ossification(GO:0001957) direct ossification(GO:0036072) |
| 0.2 | 4.0 | GO:0060363 | cranial suture morphogenesis(GO:0060363) |
| 0.2 | 2.4 | GO:0035093 | spermatogenesis, exchange of chromosomal proteins(GO:0035093) |
| 0.2 | 4.1 | GO:0070995 | NADPH oxidation(GO:0070995) |
| 0.2 | 0.4 | GO:0060594 | mammary gland specification(GO:0060594) |
| 0.2 | 0.6 | GO:0019085 | early viral transcription(GO:0019085) |
| 0.2 | 2.6 | GO:0032463 | negative regulation of protein homooligomerization(GO:0032463) |
| 0.2 | 0.4 | GO:1900116 | sequestering of extracellular ligand from receptor(GO:0035581) extracellular regulation of signal transduction(GO:1900115) extracellular negative regulation of signal transduction(GO:1900116) |
| 0.2 | 2.1 | GO:0032264 | IMP salvage(GO:0032264) |
| 0.2 | 3.8 | GO:0031340 | positive regulation of vesicle fusion(GO:0031340) |
| 0.2 | 4.8 | GO:0042178 | xenobiotic catabolic process(GO:0042178) |
| 0.2 | 0.6 | GO:0034970 | histone H3-R2 methylation(GO:0034970) |
| 0.2 | 1.2 | GO:0097210 | response to gonadotropin-releasing hormone(GO:0097210) cellular response to gonadotropin-releasing hormone(GO:0097211) |
| 0.2 | 0.6 | GO:1902277 | negative regulation of pancreatic amylase secretion(GO:1902277) |
| 0.2 | 3.0 | GO:0035721 | intraciliary retrograde transport(GO:0035721) |
| 0.2 | 5.7 | GO:2000311 | regulation of alpha-amino-3-hydroxy-5-methyl-4-isoxazole propionate selective glutamate receptor activity(GO:2000311) |
| 0.2 | 2.2 | GO:0015670 | carbon dioxide transport(GO:0015670) |
| 0.2 | 3.3 | GO:0009437 | carnitine metabolic process(GO:0009437) |
| 0.2 | 4.6 | GO:0033169 | histone H3-K9 demethylation(GO:0033169) |
| 0.2 | 0.4 | GO:0007290 | spermatid nucleus elongation(GO:0007290) |
| 0.2 | 1.7 | GO:0048852 | diencephalon morphogenesis(GO:0048852) |
| 0.2 | 1.7 | GO:1903142 | positive regulation of endothelial cell development(GO:1901552) positive regulation of establishment of endothelial barrier(GO:1903142) |
| 0.2 | 0.7 | GO:0042939 | glutathione transport(GO:0034635) tripeptide transport(GO:0042939) |
| 0.2 | 1.8 | GO:0017185 | peptidyl-lysine hydroxylation(GO:0017185) |
| 0.2 | 1.1 | GO:0002188 | translation reinitiation(GO:0002188) |
| 0.2 | 3.2 | GO:0030388 | fructose 1,6-bisphosphate metabolic process(GO:0030388) |
| 0.2 | 2.3 | GO:0051694 | pointed-end actin filament capping(GO:0051694) |
| 0.2 | 1.1 | GO:1903964 | monounsaturated fatty acid metabolic process(GO:1903964) monounsaturated fatty acid biosynthetic process(GO:1903966) |
| 0.2 | 1.9 | GO:0019244 | lactate biosynthetic process from pyruvate(GO:0019244) |
| 0.2 | 4.7 | GO:0021696 | cerebellar cortex morphogenesis(GO:0021696) |
| 0.2 | 1.5 | GO:0035331 | negative regulation of hippo signaling(GO:0035331) |
| 0.2 | 3.3 | GO:0022027 | interkinetic nuclear migration(GO:0022027) |
| 0.2 | 2.0 | GO:0016139 | glycoside catabolic process(GO:0016139) |
| 0.2 | 0.7 | GO:2000321 | positive regulation of T-helper 17 cell differentiation(GO:2000321) |
| 0.2 | 2.8 | GO:0030049 | muscle filament sliding(GO:0030049) |
| 0.2 | 1.1 | GO:0060332 | heme transport(GO:0015886) positive regulation of response to interferon-gamma(GO:0060332) positive regulation of interferon-gamma-mediated signaling pathway(GO:0060335) |
| 0.2 | 2.4 | GO:0048149 | behavioral response to ethanol(GO:0048149) |
| 0.2 | 1.7 | GO:0071763 | nuclear membrane organization(GO:0071763) |
| 0.2 | 0.8 | GO:0048743 | positive regulation of skeletal muscle fiber development(GO:0048743) |
| 0.2 | 1.2 | GO:0001542 | ovulation from ovarian follicle(GO:0001542) |
| 0.2 | 1.1 | GO:0042769 | DNA damage response, detection of DNA damage(GO:0042769) |
| 0.2 | 0.6 | GO:0072385 | minus-end-directed organelle transport along microtubule(GO:0072385) |
| 0.2 | 1.8 | GO:0030948 | negative regulation of vascular endothelial growth factor receptor signaling pathway(GO:0030948) |
| 0.2 | 0.9 | GO:0034184 | positive regulation of maintenance of mitotic sister chromatid cohesion(GO:0034184) protein localization to site of double-strand break(GO:1990166) |
| 0.2 | 1.1 | GO:0060605 | tube lumen cavitation(GO:0060605) salivary gland cavitation(GO:0060662) |
| 0.2 | 1.7 | GO:1902669 | positive regulation of axon guidance(GO:1902669) |
| 0.1 | 2.2 | GO:2001197 | regulation of basement membrane assembly involved in embryonic body morphogenesis(GO:1904259) positive regulation of basement membrane assembly involved in embryonic body morphogenesis(GO:1904261) basement membrane assembly involved in embryonic body morphogenesis(GO:2001197) |
| 0.1 | 0.9 | GO:0031161 | phosphatidylinositol catabolic process(GO:0031161) |
| 0.1 | 0.7 | GO:0045054 | constitutive secretory pathway(GO:0045054) |
| 0.1 | 2.3 | GO:0043084 | penile erection(GO:0043084) |
| 0.1 | 0.4 | GO:0060112 | generation of ovulation cycle rhythm(GO:0060112) |
| 0.1 | 1.0 | GO:1902748 | positive regulation of lens fiber cell differentiation(GO:1902748) |
| 0.1 | 3.1 | GO:0031290 | retinal ganglion cell axon guidance(GO:0031290) |
| 0.1 | 1.4 | GO:0014807 | regulation of somitogenesis(GO:0014807) |
| 0.1 | 5.2 | GO:0048013 | ephrin receptor signaling pathway(GO:0048013) |
| 0.1 | 0.4 | GO:0042780 | tRNA 3'-end processing(GO:0042780) |
| 0.1 | 1.6 | GO:0032927 | positive regulation of activin receptor signaling pathway(GO:0032927) |
| 0.1 | 5.0 | GO:0048265 | response to pain(GO:0048265) |
| 0.1 | 1.7 | GO:0030828 | positive regulation of cGMP biosynthetic process(GO:0030828) |
| 0.1 | 0.8 | GO:0006742 | NADP catabolic process(GO:0006742) |
| 0.1 | 5.6 | GO:0000266 | mitochondrial fission(GO:0000266) |
| 0.1 | 0.4 | GO:0002355 | detection of tumor cell(GO:0002355) |
| 0.1 | 1.5 | GO:0000301 | retrograde transport, vesicle recycling within Golgi(GO:0000301) |
| 0.1 | 3.4 | GO:2000300 | regulation of synaptic vesicle exocytosis(GO:2000300) |
| 0.1 | 0.4 | GO:0006427 | histidyl-tRNA aminoacylation(GO:0006427) |
| 0.1 | 0.9 | GO:0000733 | DNA strand renaturation(GO:0000733) |
| 0.1 | 0.6 | GO:0051643 | endoplasmic reticulum localization(GO:0051643) |
| 0.1 | 0.4 | GO:0006428 | isoleucyl-tRNA aminoacylation(GO:0006428) |
| 0.1 | 0.4 | GO:0033082 | regulation of extrathymic T cell differentiation(GO:0033082) |
| 0.1 | 0.7 | GO:0015015 | heparan sulfate proteoglycan biosynthetic process, enzymatic modification(GO:0015015) |
| 0.1 | 2.9 | GO:0003417 | growth plate cartilage development(GO:0003417) |
| 0.1 | 0.5 | GO:0055014 | atrial cardiac muscle cell differentiation(GO:0055011) atrial cardiac muscle cell development(GO:0055014) |
| 0.1 | 3.9 | GO:0042551 | neuron maturation(GO:0042551) |
| 0.1 | 2.7 | GO:0048642 | negative regulation of skeletal muscle tissue development(GO:0048642) |
| 0.1 | 5.0 | GO:0060999 | positive regulation of dendritic spine development(GO:0060999) |
| 0.1 | 1.0 | GO:0000492 | box C/D snoRNP assembly(GO:0000492) |
| 0.1 | 1.5 | GO:0045176 | apical protein localization(GO:0045176) |
| 0.1 | 1.0 | GO:0098728 | germ-line stem cell division(GO:0042078) male germ-line stem cell asymmetric division(GO:0048133) germline stem cell asymmetric division(GO:0098728) |
| 0.1 | 1.3 | GO:0097460 | ferrous iron import into cell(GO:0097460) ferrous iron import across plasma membrane(GO:0098707) |
| 0.1 | 1.4 | GO:0002087 | regulation of respiratory gaseous exchange by neurological system process(GO:0002087) |
| 0.1 | 2.0 | GO:0042487 | regulation of odontogenesis of dentin-containing tooth(GO:0042487) |
| 0.1 | 4.2 | GO:0071108 | protein K48-linked deubiquitination(GO:0071108) |
| 0.1 | 2.0 | GO:1903818 | positive regulation of voltage-gated potassium channel activity(GO:1903818) |
| 0.1 | 2.1 | GO:0071392 | cellular response to estradiol stimulus(GO:0071392) |
| 0.1 | 1.8 | GO:0046697 | decidualization(GO:0046697) |
| 0.1 | 0.6 | GO:0001712 | ectodermal cell fate commitment(GO:0001712) |
| 0.1 | 0.9 | GO:0016338 | calcium-independent cell-cell adhesion via plasma membrane cell-adhesion molecules(GO:0016338) |
| 0.1 | 1.5 | GO:0072378 | blood coagulation, fibrin clot formation(GO:0072378) |
| 0.1 | 1.5 | GO:0071391 | cellular response to estrogen stimulus(GO:0071391) |
| 0.1 | 3.1 | GO:0030575 | nuclear body organization(GO:0030575) |
| 0.1 | 1.7 | GO:0046007 | negative regulation of activated T cell proliferation(GO:0046007) |
| 0.1 | 1.2 | GO:0051415 | interphase microtubule nucleation by interphase microtubule organizing center(GO:0051415) microtubule nucleation by microtubule organizing center(GO:0051418) |
| 0.1 | 1.7 | GO:0090435 | protein localization to nuclear envelope(GO:0090435) |
| 0.1 | 0.7 | GO:0051534 | negative regulation of NFAT protein import into nucleus(GO:0051534) |
| 0.1 | 2.1 | GO:0021796 | cerebral cortex regionalization(GO:0021796) |
| 0.1 | 5.0 | GO:0007628 | adult walking behavior(GO:0007628) |
| 0.1 | 1.3 | GO:0042989 | sequestering of actin monomers(GO:0042989) |
| 0.1 | 2.6 | GO:0061318 | renal filtration cell differentiation(GO:0061318) glomerular visceral epithelial cell differentiation(GO:0072112) glomerular epithelial cell differentiation(GO:0072311) |
| 0.1 | 5.2 | GO:0036465 | synaptic vesicle recycling(GO:0036465) |
| 0.1 | 0.3 | GO:0060179 | male mating behavior(GO:0060179) |
| 0.1 | 0.4 | GO:0071376 | response to corticotropin-releasing hormone(GO:0043435) cellular response to corticotropin-releasing hormone stimulus(GO:0071376) |
| 0.1 | 0.3 | GO:0072014 | proximal tubule development(GO:0072014) |
| 0.1 | 1.2 | GO:0036158 | outer dynein arm assembly(GO:0036158) |
| 0.1 | 1.0 | GO:0035754 | B cell chemotaxis(GO:0035754) |
| 0.1 | 0.3 | GO:0019858 | cytosine metabolic process(GO:0019858) |
| 0.1 | 1.5 | GO:0030322 | stabilization of membrane potential(GO:0030322) |
| 0.1 | 0.3 | GO:0035524 | proline transmembrane transport(GO:0035524) glycine import(GO:0036233) |
| 0.1 | 6.6 | GO:0051965 | positive regulation of synapse assembly(GO:0051965) |
| 0.1 | 0.3 | GO:0070650 | actin filament bundle distribution(GO:0070650) |
| 0.1 | 1.0 | GO:0014883 | transition between fast and slow fiber(GO:0014883) |
| 0.1 | 1.5 | GO:0034497 | protein localization to pre-autophagosomal structure(GO:0034497) |
| 0.1 | 4.9 | GO:0010765 | positive regulation of sodium ion transport(GO:0010765) |
| 0.1 | 0.3 | GO:0070346 | positive regulation of fat cell proliferation(GO:0070346) |
| 0.1 | 1.6 | GO:1902187 | negative regulation of viral release from host cell(GO:1902187) |
| 0.1 | 0.4 | GO:0090385 | phagosome-lysosome fusion(GO:0090385) |
| 0.1 | 0.2 | GO:0072429 | response to intra-S DNA damage checkpoint signaling(GO:0072429) |
| 0.1 | 1.4 | GO:0006656 | phosphatidylcholine biosynthetic process(GO:0006656) |
| 0.1 | 0.4 | GO:0060796 | regulation of transcription involved in primary germ layer cell fate commitment(GO:0060796) |
| 0.1 | 5.4 | GO:1901379 | regulation of potassium ion transmembrane transport(GO:1901379) |
| 0.1 | 1.2 | GO:1903546 | protein localization to photoreceptor outer segment(GO:1903546) |
| 0.1 | 0.4 | GO:0050884 | neuromuscular process controlling posture(GO:0050884) |
| 0.1 | 1.4 | GO:0007616 | long-term memory(GO:0007616) |
| 0.1 | 1.5 | GO:0043162 | ubiquitin-dependent protein catabolic process via the multivesicular body sorting pathway(GO:0043162) |
| 0.1 | 1.2 | GO:0015695 | organic cation transport(GO:0015695) |
| 0.1 | 0.8 | GO:0048671 | negative regulation of collateral sprouting(GO:0048671) |
| 0.1 | 3.2 | GO:0060291 | long-term synaptic potentiation(GO:0060291) |
| 0.1 | 0.7 | GO:0014850 | response to muscle activity(GO:0014850) |
| 0.1 | 1.5 | GO:0007530 | sex determination(GO:0007530) |
| 0.1 | 1.1 | GO:0042921 | glucocorticoid receptor signaling pathway(GO:0042921) |
| 0.1 | 3.7 | GO:0045761 | regulation of adenylate cyclase activity(GO:0045761) |
| 0.1 | 0.2 | GO:0032474 | otolith morphogenesis(GO:0032474) |
| 0.1 | 0.5 | GO:1903551 | regulation of extracellular exosome assembly(GO:1903551) |
| 0.1 | 5.7 | GO:0042147 | retrograde transport, endosome to Golgi(GO:0042147) |
| 0.1 | 0.3 | GO:0099563 | modification of synaptic structure(GO:0099563) |
| 0.1 | 1.3 | GO:0006891 | intra-Golgi vesicle-mediated transport(GO:0006891) |
| 0.1 | 0.6 | GO:0006572 | tyrosine catabolic process(GO:0006572) |
| 0.1 | 2.4 | GO:0010573 | vascular endothelial growth factor production(GO:0010573) |
| 0.1 | 1.2 | GO:0048714 | positive regulation of oligodendrocyte differentiation(GO:0048714) |
| 0.1 | 1.5 | GO:0033141 | positive regulation of peptidyl-serine phosphorylation of STAT protein(GO:0033141) |
| 0.1 | 0.7 | GO:0035584 | calcium-mediated signaling using intracellular calcium source(GO:0035584) |
| 0.1 | 12.0 | GO:0050804 | modulation of synaptic transmission(GO:0050804) |
| 0.1 | 1.4 | GO:0042761 | fatty acid elongation(GO:0030497) very long-chain fatty acid biosynthetic process(GO:0042761) |
| 0.1 | 0.8 | GO:0006903 | vesicle targeting(GO:0006903) |
| 0.1 | 0.9 | GO:1990001 | inhibition of cysteine-type endopeptidase activity involved in apoptotic process(GO:1990001) |
| 0.1 | 2.2 | GO:0000184 | nuclear-transcribed mRNA catabolic process, nonsense-mediated decay(GO:0000184) |
| 0.1 | 0.4 | GO:0018344 | protein geranylgeranylation(GO:0018344) |
| 0.1 | 0.2 | GO:0034553 | respiratory chain complex II assembly(GO:0034552) mitochondrial respiratory chain complex II assembly(GO:0034553) mitochondrial respiratory chain complex II biogenesis(GO:0097032) |
| 0.1 | 1.4 | GO:0035767 | endothelial cell chemotaxis(GO:0035767) |
| 0.1 | 0.7 | GO:0045187 | circadian sleep/wake cycle process(GO:0022410) regulation of circadian sleep/wake cycle(GO:0042749) regulation of circadian sleep/wake cycle, sleep(GO:0045187) circadian sleep/wake cycle, sleep(GO:0050802) |
| 0.1 | 1.6 | GO:0015721 | bile acid and bile salt transport(GO:0015721) |
| 0.1 | 0.5 | GO:0007288 | sperm axoneme assembly(GO:0007288) |
| 0.0 | 0.3 | GO:2000507 | positive regulation of energy homeostasis(GO:2000507) |
| 0.0 | 1.2 | GO:2000052 | positive regulation of non-canonical Wnt signaling pathway(GO:2000052) |
| 0.0 | 2.3 | GO:1902017 | regulation of cilium assembly(GO:1902017) |
| 0.0 | 0.6 | GO:0051014 | actin filament severing(GO:0051014) |
| 0.0 | 0.3 | GO:0045218 | zonula adherens maintenance(GO:0045218) |
| 0.0 | 0.7 | GO:0010960 | magnesium ion homeostasis(GO:0010960) |
| 0.0 | 2.0 | GO:0042073 | intraciliary transport(GO:0042073) |
| 0.0 | 3.3 | GO:0065002 | intracellular protein transmembrane transport(GO:0065002) protein transmembrane transport(GO:0071806) |
| 0.0 | 0.9 | GO:0034587 | piRNA metabolic process(GO:0034587) |
| 0.0 | 0.5 | GO:0098706 | ferric iron import(GO:0033216) ferric iron import into cell(GO:0097461) ferric iron import across plasma membrane(GO:0098706) |
| 0.0 | 0.7 | GO:0033617 | mitochondrial respiratory chain complex IV assembly(GO:0033617) |
| 0.0 | 0.2 | GO:0048672 | positive regulation of collateral sprouting(GO:0048672) |
| 0.0 | 1.9 | GO:0046839 | phospholipid dephosphorylation(GO:0046839) |
| 0.0 | 1.9 | GO:0003170 | heart valve development(GO:0003170) |
| 0.0 | 1.3 | GO:0045026 | plasma membrane fusion(GO:0045026) |
| 0.0 | 1.9 | GO:0021766 | hippocampus development(GO:0021766) |
| 0.0 | 0.3 | GO:0031848 | protection from non-homologous end joining at telomere(GO:0031848) |
| 0.0 | 4.7 | GO:0050808 | synapse organization(GO:0050808) |
| 0.0 | 0.3 | GO:0061709 | reticulophagy(GO:0061709) |
| 0.0 | 0.2 | GO:0035694 | mitochondrial protein catabolic process(GO:0035694) |
| 0.0 | 0.2 | GO:0097309 | cap1 mRNA methylation(GO:0097309) |
| 0.0 | 0.4 | GO:0045820 | negative regulation of glycolytic process(GO:0045820) negative regulation of cofactor metabolic process(GO:0051195) negative regulation of coenzyme metabolic process(GO:0051198) |
| 0.0 | 0.2 | GO:0090315 | negative regulation of protein targeting to membrane(GO:0090315) |
| 0.0 | 0.2 | GO:0003413 | chondrocyte differentiation involved in endochondral bone morphogenesis(GO:0003413) |
| 0.0 | 2.6 | GO:0006096 | glycolytic process(GO:0006096) |
| 0.0 | 3.3 | GO:0007224 | smoothened signaling pathway(GO:0007224) |
| 0.0 | 1.2 | GO:0050687 | negative regulation of defense response to virus(GO:0050687) |
| 0.0 | 0.9 | GO:0071539 | protein localization to centrosome(GO:0071539) |
| 0.0 | 1.5 | GO:0071804 | cellular potassium ion transport(GO:0071804) potassium ion transmembrane transport(GO:0071805) |
| 0.0 | 0.9 | GO:0002089 | lens morphogenesis in camera-type eye(GO:0002089) |
| 0.0 | 3.4 | GO:0008366 | ensheathment of neurons(GO:0007272) axon ensheathment(GO:0008366) |
| 0.0 | 2.4 | GO:0022900 | electron transport chain(GO:0022900) |
| 0.0 | 1.8 | GO:0030317 | sperm motility(GO:0030317) |
| 0.0 | 2.3 | GO:0001570 | vasculogenesis(GO:0001570) |
| 0.0 | 0.1 | GO:0043553 | negative regulation of phosphatidylinositol 3-kinase activity(GO:0043553) |
| 0.0 | 0.6 | GO:0043981 | histone H4-K5 acetylation(GO:0043981) histone H4-K8 acetylation(GO:0043982) |
| 0.0 | 1.0 | GO:0003333 | amino acid transmembrane transport(GO:0003333) |
| 0.0 | 0.7 | GO:0071300 | cellular response to retinoic acid(GO:0071300) |
| 0.0 | 0.9 | GO:0050911 | detection of chemical stimulus involved in sensory perception of smell(GO:0050911) |
| 0.0 | 0.2 | GO:0045085 | negative regulation of interleukin-2 biosynthetic process(GO:0045085) |
| 0.0 | 3.0 | GO:0019236 | response to pheromone(GO:0019236) |
| 0.0 | 0.2 | GO:1903298 | regulation of hypoxia-induced intrinsic apoptotic signaling pathway(GO:1903297) negative regulation of hypoxia-induced intrinsic apoptotic signaling pathway(GO:1903298) intrinsic apoptotic signaling pathway in response to hypoxia(GO:1990144) |
| 0.0 | 1.8 | GO:0007156 | homophilic cell adhesion via plasma membrane adhesion molecules(GO:0007156) |
| 0.0 | 0.6 | GO:0042993 | positive regulation of transcription factor import into nucleus(GO:0042993) |
| 0.0 | 0.7 | GO:0015988 | energy coupled proton transmembrane transport, against electrochemical gradient(GO:0015988) ATP hydrolysis coupled proton transport(GO:0015991) ATP hydrolysis coupled transmembrane transport(GO:0090662) |
| 0.0 | 0.0 | GO:0035054 | embryonic heart tube anterior/posterior pattern specification(GO:0035054) |
| 0.0 | 1.1 | GO:0018149 | peptide cross-linking(GO:0018149) |
| 0.0 | 0.1 | GO:0000056 | ribosomal small subunit export from nucleus(GO:0000056) |
| 0.0 | 0.5 | GO:2000649 | regulation of sodium ion transmembrane transporter activity(GO:2000649) |
| 0.0 | 1.2 | GO:0090630 | activation of GTPase activity(GO:0090630) |
| 0.0 | 0.4 | GO:0006379 | mRNA cleavage(GO:0006379) |
| 0.0 | 1.2 | GO:0042273 | ribosomal large subunit biogenesis(GO:0042273) |
| 0.0 | 0.4 | GO:0048814 | regulation of dendrite morphogenesis(GO:0048814) |
| 0.0 | 0.1 | GO:2000381 | negative regulation of mesoderm development(GO:2000381) |
| 0.0 | 0.2 | GO:0006337 | nucleosome disassembly(GO:0006337) |
| 0.0 | 0.3 | GO:0016578 | histone deubiquitination(GO:0016578) |
| 0.0 | 0.2 | GO:0060732 | positive regulation of inositol phosphate biosynthetic process(GO:0060732) |
| 0.0 | 0.1 | GO:0070966 | nuclear-transcribed mRNA catabolic process, no-go decay(GO:0070966) |
| 0.0 | 0.3 | GO:0006890 | retrograde vesicle-mediated transport, Golgi to ER(GO:0006890) |
| 0.0 | 0.2 | GO:0019076 | viral release from host cell(GO:0019076) regulation of viral release from host cell(GO:1902186) |
| 0.0 | 0.2 | GO:0031163 | iron-sulfur cluster assembly(GO:0016226) metallo-sulfur cluster assembly(GO:0031163) |
| 0.0 | 0.1 | GO:0007220 | Notch receptor processing(GO:0007220) |
| 0.0 | 1.0 | GO:0017015 | regulation of transforming growth factor beta receptor signaling pathway(GO:0017015) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 3.6 | 10.9 | GO:0098830 | presynaptic endosome(GO:0098830) |
| 1.8 | 19.5 | GO:1990812 | growth cone filopodium(GO:1990812) |
| 1.6 | 6.3 | GO:0014802 | terminal cisterna(GO:0014802) |
| 1.3 | 6.6 | GO:0044316 | cone cell pedicle(GO:0044316) |
| 1.3 | 11.6 | GO:0008282 | ATP-sensitive potassium channel complex(GO:0008282) |
| 1.3 | 10.1 | GO:0008091 | spectrin(GO:0008091) |
| 1.2 | 3.7 | GO:0018444 | translation release factor complex(GO:0018444) |
| 1.0 | 3.1 | GO:1990031 | pinceau fiber(GO:1990031) |
| 1.0 | 12.0 | GO:0044327 | dendritic spine head(GO:0044327) |
| 1.0 | 2.9 | GO:0098855 | HCN channel complex(GO:0098855) |
| 0.9 | 4.5 | GO:0097058 | CRLF-CLCF1 complex(GO:0097058) |
| 0.7 | 3.5 | GO:0097425 | smooth endoplasmic reticulum membrane(GO:0030868) smooth endoplasmic reticulum part(GO:0097425) |
| 0.7 | 11.0 | GO:0097418 | neurofibrillary tangle(GO:0097418) |
| 0.6 | 1.9 | GO:0005863 | striated muscle myosin thick filament(GO:0005863) |
| 0.6 | 5.7 | GO:0033010 | paranodal junction(GO:0033010) |
| 0.6 | 3.0 | GO:0038039 | G-protein coupled receptor heterodimeric complex(GO:0038039) |
| 0.6 | 1.7 | GO:0098842 | postsynaptic early endosome(GO:0098842) |
| 0.5 | 2.0 | GO:0033596 | TSC1-TSC2 complex(GO:0033596) |
| 0.5 | 2.5 | GO:0010370 | perinucleolar chromocenter(GO:0010370) |
| 0.5 | 4.0 | GO:0032937 | SREBP-SCAP-Insig complex(GO:0032937) |
| 0.5 | 12.9 | GO:1902710 | GABA receptor complex(GO:1902710) GABA-A receptor complex(GO:1902711) |
| 0.4 | 2.2 | GO:0000938 | GARP complex(GO:0000938) EARP complex(GO:1990745) |
| 0.4 | 10.6 | GO:0001518 | voltage-gated sodium channel complex(GO:0001518) |
| 0.4 | 1.1 | GO:0070992 | translation initiation complex(GO:0070992) |
| 0.4 | 17.9 | GO:0032281 | AMPA glutamate receptor complex(GO:0032281) |
| 0.3 | 1.4 | GO:1990415 | Pex17p-Pex14p docking complex(GO:1990415) peroxisomal importomer complex(GO:1990429) |
| 0.3 | 15.9 | GO:0030660 | Golgi-associated vesicle membrane(GO:0030660) |
| 0.3 | 1.2 | GO:0008275 | gamma-tubulin small complex(GO:0008275) |
| 0.3 | 12.6 | GO:0032590 | dendrite membrane(GO:0032590) |
| 0.3 | 4.1 | GO:0000015 | phosphopyruvate hydratase complex(GO:0000015) |
| 0.3 | 3.8 | GO:0030128 | clathrin coat of endocytic vesicle(GO:0030128) |
| 0.3 | 9.1 | GO:0033017 | sarcoplasmic reticulum membrane(GO:0033017) |
| 0.3 | 0.8 | GO:1990843 | subsarcolemmal mitochondrion(GO:1990843) interfibrillar mitochondrion(GO:1990844) |
| 0.3 | 3.5 | GO:0070852 | cell body fiber(GO:0070852) |
| 0.3 | 2.9 | GO:0032584 | growth cone membrane(GO:0032584) |
| 0.2 | 3.7 | GO:0044294 | dendritic growth cone(GO:0044294) |
| 0.2 | 2.1 | GO:0043083 | synaptic cleft(GO:0043083) |
| 0.2 | 2.7 | GO:0097542 | ciliary tip(GO:0097542) |
| 0.2 | 1.8 | GO:0046696 | lipopolysaccharide receptor complex(GO:0046696) |
| 0.2 | 3.8 | GO:0048787 | presynaptic active zone membrane(GO:0048787) |
| 0.2 | 2.4 | GO:0016272 | prefoldin complex(GO:0016272) |
| 0.2 | 3.9 | GO:0031045 | dense core granule(GO:0031045) |
| 0.2 | 4.7 | GO:0005892 | acetylcholine-gated channel complex(GO:0005892) |
| 0.2 | 7.7 | GO:0032809 | neuronal cell body membrane(GO:0032809) |
| 0.2 | 1.2 | GO:0033116 | endoplasmic reticulum-Golgi intermediate compartment membrane(GO:0033116) |
| 0.2 | 5.1 | GO:0043196 | varicosity(GO:0043196) |
| 0.2 | 1.4 | GO:0031095 | platelet dense tubular network membrane(GO:0031095) |
| 0.2 | 3.4 | GO:0048786 | presynaptic active zone(GO:0048786) |
| 0.2 | 43.6 | GO:0031225 | anchored component of membrane(GO:0031225) |
| 0.2 | 1.3 | GO:0071439 | clathrin complex(GO:0071439) |
| 0.2 | 2.4 | GO:0098643 | fibrillar collagen trimer(GO:0005583) banded collagen fibril(GO:0098643) |
| 0.2 | 3.2 | GO:0046581 | intercellular canaliculus(GO:0046581) |
| 0.2 | 1.8 | GO:0042629 | mast cell granule(GO:0042629) |
| 0.2 | 0.7 | GO:0000221 | vacuolar proton-transporting V-type ATPase, V1 domain(GO:0000221) |
| 0.2 | 1.9 | GO:0098839 | postsynaptic density membrane(GO:0098839) |
| 0.2 | 0.7 | GO:0005594 | collagen type IX trimer(GO:0005594) |
| 0.2 | 5.2 | GO:0002080 | acrosomal membrane(GO:0002080) |
| 0.2 | 1.5 | GO:0005577 | fibrinogen complex(GO:0005577) |
| 0.2 | 4.9 | GO:0071565 | nBAF complex(GO:0071565) |
| 0.2 | 4.7 | GO:0005682 | U5 snRNP(GO:0005682) |
| 0.2 | 2.1 | GO:0031315 | extrinsic component of mitochondrial outer membrane(GO:0031315) |
| 0.2 | 3.7 | GO:0035253 | ciliary rootlet(GO:0035253) |
| 0.2 | 5.3 | GO:0000159 | protein phosphatase type 2A complex(GO:0000159) |
| 0.2 | 1.4 | GO:0031428 | box C/D snoRNP complex(GO:0031428) |
| 0.2 | 5.9 | GO:0097440 | apical dendrite(GO:0097440) |
| 0.1 | 10.7 | GO:0042734 | presynaptic membrane(GO:0042734) |
| 0.1 | 2.8 | GO:0005861 | troponin complex(GO:0005861) |
| 0.1 | 0.6 | GO:1902937 | inward rectifier potassium channel complex(GO:1902937) |
| 0.1 | 0.9 | GO:0005672 | transcription factor TFIIA complex(GO:0005672) |
| 0.1 | 1.9 | GO:0036128 | CatSper complex(GO:0036128) |
| 0.1 | 29.5 | GO:0008021 | synaptic vesicle(GO:0008021) |
| 0.1 | 13.0 | GO:0034705 | potassium channel complex(GO:0034705) |
| 0.1 | 2.1 | GO:0097449 | astrocyte projection(GO:0097449) |
| 0.1 | 42.5 | GO:0014069 | postsynaptic density(GO:0014069) |
| 0.1 | 2.2 | GO:0045180 | basal cortex(GO:0045180) |
| 0.1 | 2.0 | GO:0060293 | P granule(GO:0043186) pole plasm(GO:0045495) germ plasm(GO:0060293) |
| 0.1 | 1.3 | GO:0001674 | female germ cell nucleus(GO:0001674) |
| 0.1 | 4.4 | GO:0005581 | collagen trimer(GO:0005581) |
| 0.1 | 3.3 | GO:0035686 | sperm fibrous sheath(GO:0035686) |
| 0.1 | 1.1 | GO:0030991 | intraciliary transport particle A(GO:0030991) |
| 0.1 | 11.6 | GO:0043679 | axon terminus(GO:0043679) |
| 0.1 | 1.5 | GO:0000813 | ESCRT I complex(GO:0000813) |
| 0.1 | 1.7 | GO:0005952 | cAMP-dependent protein kinase complex(GO:0005952) |
| 0.1 | 1.5 | GO:0017119 | Golgi transport complex(GO:0017119) |
| 0.1 | 3.9 | GO:0036379 | myofilament(GO:0036379) |
| 0.1 | 2.5 | GO:0005922 | connexon complex(GO:0005922) |
| 0.1 | 1.3 | GO:1990712 | HFE-transferrin receptor complex(GO:1990712) |
| 0.1 | 3.5 | GO:0030687 | preribosome, large subunit precursor(GO:0030687) |
| 0.1 | 1.5 | GO:0034045 | pre-autophagosomal structure membrane(GO:0034045) |
| 0.1 | 0.7 | GO:0072559 | NLRP3 inflammasome complex(GO:0072559) |
| 0.1 | 2.7 | GO:0034385 | very-low-density lipoprotein particle(GO:0034361) triglyceride-rich lipoprotein particle(GO:0034385) |
| 0.1 | 1.3 | GO:0005869 | dynactin complex(GO:0005869) |
| 0.1 | 0.6 | GO:0070847 | core mediator complex(GO:0070847) |
| 0.1 | 0.5 | GO:0072487 | MSL complex(GO:0072487) |
| 0.1 | 1.5 | GO:0035327 | transcriptionally active chromatin(GO:0035327) |
| 0.1 | 3.3 | GO:0014704 | intercalated disc(GO:0014704) |
| 0.1 | 2.0 | GO:0097228 | sperm principal piece(GO:0097228) |
| 0.1 | 0.9 | GO:0070765 | gamma-secretase complex(GO:0070765) |
| 0.1 | 0.4 | GO:0005968 | Rab-protein geranylgeranyltransferase complex(GO:0005968) |
| 0.1 | 4.7 | GO:0045335 | phagocytic vesicle(GO:0045335) |
| 0.1 | 0.8 | GO:0031209 | SCAR complex(GO:0031209) |
| 0.1 | 3.9 | GO:0016528 | sarcoplasm(GO:0016528) |
| 0.1 | 1.1 | GO:0043205 | microfibril(GO:0001527) fibril(GO:0043205) |
| 0.1 | 2.1 | GO:0060170 | ciliary membrane(GO:0060170) |
| 0.1 | 3.2 | GO:0005905 | clathrin-coated pit(GO:0005905) |
| 0.1 | 1.7 | GO:0071782 | endoplasmic reticulum tubular network(GO:0071782) |
| 0.1 | 2.1 | GO:0005891 | voltage-gated calcium channel complex(GO:0005891) |
| 0.1 | 8.3 | GO:0005913 | cell-cell adherens junction(GO:0005913) |
| 0.1 | 10.7 | GO:0097060 | synaptic membrane(GO:0097060) |
| 0.1 | 1.6 | GO:0097546 | ciliary base(GO:0097546) |
| 0.1 | 4.8 | GO:0031463 | Cul3-RING ubiquitin ligase complex(GO:0031463) |
| 0.1 | 1.9 | GO:0005719 | nuclear euchromatin(GO:0005719) |
| 0.1 | 0.9 | GO:0005662 | DNA replication factor A complex(GO:0005662) |
| 0.0 | 0.8 | GO:0017101 | aminoacyl-tRNA synthetase multienzyme complex(GO:0017101) |
| 0.0 | 0.1 | GO:0030689 | Noc complex(GO:0030689) |
| 0.0 | 5.3 | GO:0005814 | centriole(GO:0005814) |
| 0.0 | 0.9 | GO:0010369 | chromocenter(GO:0010369) |
| 0.0 | 1.5 | GO:0008287 | protein serine/threonine phosphatase complex(GO:0008287) phosphatase complex(GO:1903293) |
| 0.0 | 0.1 | GO:0031372 | UBC13-MMS2 complex(GO:0031372) |
| 0.0 | 1.1 | GO:0005849 | mRNA cleavage factor complex(GO:0005849) |
| 0.0 | 0.3 | GO:0016281 | eukaryotic translation initiation factor 4F complex(GO:0016281) |
| 0.0 | 0.1 | GO:0042567 | insulin-like growth factor ternary complex(GO:0042567) |
| 0.0 | 0.2 | GO:0048476 | Holliday junction resolvase complex(GO:0048476) |
| 0.0 | 0.4 | GO:0008540 | proteasome regulatory particle, base subcomplex(GO:0008540) cytosolic proteasome complex(GO:0031597) |
| 0.0 | 0.5 | GO:0042575 | DNA polymerase complex(GO:0042575) |
| 0.0 | 0.5 | GO:0031430 | M band(GO:0031430) |
| 0.0 | 1.1 | GO:0051233 | spindle midzone(GO:0051233) |
| 0.0 | 0.2 | GO:0089701 | U2AF(GO:0089701) |
| 0.0 | 0.6 | GO:0031201 | SNARE complex(GO:0031201) |
| 0.0 | 2.6 | GO:0005604 | basement membrane(GO:0005604) |
| 0.0 | 0.6 | GO:0005868 | cytoplasmic dynein complex(GO:0005868) |
| 0.0 | 1.1 | GO:0001533 | cornified envelope(GO:0001533) |
| 0.0 | 0.3 | GO:0000124 | SAGA complex(GO:0000124) |
| 0.0 | 0.3 | GO:0044295 | axonal growth cone(GO:0044295) |
| 0.0 | 2.2 | GO:1990204 | oxidoreductase complex(GO:1990204) |
| 0.0 | 0.4 | GO:0043198 | dendritic shaft(GO:0043198) |
| 0.0 | 0.2 | GO:0044322 | endoplasmic reticulum quality control compartment(GO:0044322) |
| 0.0 | 0.2 | GO:0031083 | BLOC-1 complex(GO:0031083) |
| 0.0 | 0.5 | GO:0001741 | XY body(GO:0001741) |
| 0.0 | 0.0 | GO:0005883 | neurofilament(GO:0005883) |
| 0.0 | 3.3 | GO:0005578 | proteinaceous extracellular matrix(GO:0005578) |
| 0.0 | 0.2 | GO:0034663 | endoplasmic reticulum chaperone complex(GO:0034663) |
| 0.0 | 0.5 | GO:0035861 | site of double-strand break(GO:0035861) |
| 0.0 | 0.6 | GO:0005871 | kinesin complex(GO:0005871) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.7 | 13.7 | GO:0015272 | ATP-activated inward rectifier potassium channel activity(GO:0015272) |
| 1.4 | 5.7 | GO:0004574 | oligo-1,6-glucosidase activity(GO:0004574) |
| 1.4 | 7.0 | GO:0005280 | hydrogen:amino acid symporter activity(GO:0005280) |
| 1.4 | 4.1 | GO:0004698 | calcium-dependent protein kinase C activity(GO:0004698) |
| 1.3 | 3.9 | GO:0030348 | syntaxin-3 binding(GO:0030348) |
| 0.9 | 3.5 | GO:0072320 | volume-sensitive chloride channel activity(GO:0072320) |
| 0.8 | 4.8 | GO:0004994 | somatostatin receptor activity(GO:0004994) |
| 0.8 | 3.2 | GO:0031544 | peptidyl-proline 3-dioxygenase activity(GO:0031544) |
| 0.8 | 6.9 | GO:0008597 | calcium-dependent protein serine/threonine phosphatase regulator activity(GO:0008597) |
| 0.7 | 3.7 | GO:0004517 | nitric-oxide synthase activity(GO:0004517) |
| 0.7 | 2.9 | GO:0034739 | histone deacetylase activity (H4-K16 specific)(GO:0034739) |
| 0.7 | 5.1 | GO:0031812 | G-protein coupled nucleotide receptor binding(GO:0031811) P2Y1 nucleotide receptor binding(GO:0031812) |
| 0.7 | 3.6 | GO:0005250 | A-type (transient outward) potassium channel activity(GO:0005250) |
| 0.7 | 4.9 | GO:0004985 | opioid receptor activity(GO:0004985) |
| 0.7 | 3.5 | GO:0034584 | piRNA binding(GO:0034584) |
| 0.7 | 2.7 | GO:0005219 | ryanodine-sensitive calcium-release channel activity(GO:0005219) |
| 0.7 | 2.0 | GO:0004348 | glucosylceramidase activity(GO:0004348) |
| 0.6 | 3.9 | GO:0004706 | JUN kinase kinase kinase activity(GO:0004706) |
| 0.6 | 1.9 | GO:0098782 | mechanically-gated potassium channel activity(GO:0098782) |
| 0.6 | 9.3 | GO:0032050 | clathrin heavy chain binding(GO:0032050) |
| 0.6 | 2.4 | GO:0061628 | H3K27me3 modified histone binding(GO:0061628) |
| 0.6 | 5.2 | GO:0015467 | G-protein activated inward rectifier potassium channel activity(GO:0015467) |
| 0.6 | 18.4 | GO:0048156 | tau protein binding(GO:0048156) |
| 0.6 | 2.2 | GO:0001733 | galactosylceramide sulfotransferase activity(GO:0001733) |
| 0.5 | 3.3 | GO:0004095 | carnitine O-palmitoyltransferase activity(GO:0004095) O-palmitoyltransferase activity(GO:0016416) |
| 0.5 | 3.2 | GO:0090554 | phosphatidylcholine-translocating ATPase activity(GO:0090554) |
| 0.5 | 1.6 | GO:0005169 | neurotrophin TRKB receptor binding(GO:0005169) |
| 0.5 | 1.6 | GO:0008476 | protein-tyrosine sulfotransferase activity(GO:0008476) |
| 0.5 | 2.6 | GO:0003989 | acetyl-CoA carboxylase activity(GO:0003989) |
| 0.5 | 3.0 | GO:0004965 | G-protein coupled GABA receptor activity(GO:0004965) |
| 0.5 | 4.0 | GO:0086006 | voltage-gated sodium channel activity involved in cardiac muscle cell action potential(GO:0086006) |
| 0.5 | 2.9 | GO:0005222 | intracellular cAMP activated cation channel activity(GO:0005222) |
| 0.5 | 11.2 | GO:0051787 | misfolded protein binding(GO:0051787) |
| 0.5 | 2.9 | GO:0000155 | phosphorelay sensor kinase activity(GO:0000155) |
| 0.5 | 4.7 | GO:0070538 | oleic acid binding(GO:0070538) |
| 0.5 | 3.7 | GO:0008079 | translation release factor activity(GO:0003747) translation termination factor activity(GO:0008079) |
| 0.5 | 1.8 | GO:0019970 | interleukin-11 receptor activity(GO:0004921) interleukin-11 binding(GO:0019970) |
| 0.4 | 1.7 | GO:0030297 | transmembrane receptor protein tyrosine kinase activator activity(GO:0030297) |
| 0.4 | 2.6 | GO:0042289 | MHC class II protein binding(GO:0042289) |
| 0.4 | 4.3 | GO:0005127 | ciliary neurotrophic factor receptor binding(GO:0005127) |
| 0.4 | 5.3 | GO:0016151 | nickel cation binding(GO:0016151) |
| 0.4 | 6.4 | GO:0019855 | calcium channel inhibitor activity(GO:0019855) |
| 0.4 | 2.7 | GO:0030160 | GKAP/Homer scaffold activity(GO:0030160) |
| 0.4 | 1.5 | GO:0034437 | glycoprotein transporter activity(GO:0034437) |
| 0.4 | 2.5 | GO:0086075 | gap junction channel activity involved in cardiac conduction electrical coupling(GO:0086075) |
| 0.4 | 11.5 | GO:0016917 | GABA-A receptor activity(GO:0004890) GABA receptor activity(GO:0016917) |
| 0.4 | 2.1 | GO:0043533 | inositol 1,3,4,5 tetrakisphosphate binding(GO:0043533) |
| 0.4 | 1.8 | GO:0060228 | phosphatidylcholine-sterol O-acyltransferase activator activity(GO:0060228) |
| 0.3 | 1.0 | GO:0008396 | oxysterol 7-alpha-hydroxylase activity(GO:0008396) |
| 0.3 | 3.8 | GO:0031749 | D2 dopamine receptor binding(GO:0031749) |
| 0.3 | 1.0 | GO:0044715 | 8-oxo-dGDP phosphatase activity(GO:0044715) |
| 0.3 | 8.2 | GO:0004708 | MAP kinase kinase activity(GO:0004708) |
| 0.3 | 4.1 | GO:0019992 | diacylglycerol binding(GO:0019992) |
| 0.3 | 3.2 | GO:0004332 | fructose-bisphosphate aldolase activity(GO:0004332) |
| 0.3 | 1.6 | GO:0015189 | L-lysine transmembrane transporter activity(GO:0015189) |
| 0.3 | 8.1 | GO:0005540 | hyaluronic acid binding(GO:0005540) |
| 0.3 | 1.8 | GO:0016941 | natriuretic peptide receptor activity(GO:0016941) |
| 0.3 | 4.1 | GO:0004634 | phosphopyruvate hydratase activity(GO:0004634) |
| 0.3 | 2.3 | GO:0033592 | RNA strand annealing activity(GO:0033592) |
| 0.3 | 2.3 | GO:0004416 | hydroxyacylglutathione hydrolase activity(GO:0004416) |
| 0.3 | 1.7 | GO:0010853 | cyclase activator activity(GO:0010853) guanylate cyclase activator activity(GO:0030250) |
| 0.3 | 6.2 | GO:0008510 | sodium:bicarbonate symporter activity(GO:0008510) |
| 0.3 | 0.8 | GO:0047006 | 17-alpha,20-alpha-dihydroxypregn-4-en-3-one dehydrogenase activity(GO:0047006) |
| 0.3 | 2.2 | GO:0031821 | G-protein coupled serotonin receptor binding(GO:0031821) |
| 0.3 | 2.5 | GO:0005519 | cytoskeletal regulatory protein binding(GO:0005519) |
| 0.3 | 10.2 | GO:0030676 | Rac guanyl-nucleotide exchange factor activity(GO:0030676) |
| 0.3 | 7.0 | GO:0005248 | voltage-gated sodium channel activity(GO:0005248) |
| 0.3 | 1.9 | GO:0005111 | type 2 fibroblast growth factor receptor binding(GO:0005111) |
| 0.3 | 1.9 | GO:0031014 | troponin T binding(GO:0031014) |
| 0.3 | 2.8 | GO:0086080 | protein binding involved in heterotypic cell-cell adhesion(GO:0086080) |
| 0.3 | 1.3 | GO:0004998 | transferrin receptor activity(GO:0004998) |
| 0.3 | 3.5 | GO:0051378 | serotonin binding(GO:0051378) |
| 0.2 | 3.2 | GO:0048403 | brain-derived neurotrophic factor binding(GO:0048403) |
| 0.2 | 3.1 | GO:0005001 | transmembrane receptor protein tyrosine phosphatase activity(GO:0005001) transmembrane receptor protein phosphatase activity(GO:0019198) |
| 0.2 | 4.7 | GO:0042166 | acetylcholine binding(GO:0042166) |
| 0.2 | 13.7 | GO:0001540 | beta-amyloid binding(GO:0001540) |
| 0.2 | 6.4 | GO:0005251 | delayed rectifier potassium channel activity(GO:0005251) |
| 0.2 | 2.7 | GO:0004571 | mannosyl-oligosaccharide 1,2-alpha-mannosidase activity(GO:0004571) |
| 0.2 | 4.4 | GO:0005272 | sodium channel activity(GO:0005272) |
| 0.2 | 2.8 | GO:0017154 | semaphorin receptor activity(GO:0017154) |
| 0.2 | 1.1 | GO:0017108 | 5'-flap endonuclease activity(GO:0017108) |
| 0.2 | 2.1 | GO:0047623 | AMP deaminase activity(GO:0003876) adenosine-phosphate deaminase activity(GO:0047623) |
| 0.2 | 1.3 | GO:0015018 | galactosylgalactosylxylosylprotein 3-beta-glucuronosyltransferase activity(GO:0015018) |
| 0.2 | 3.8 | GO:0005242 | inward rectifier potassium channel activity(GO:0005242) |
| 0.2 | 1.6 | GO:0070290 | N-acylphosphatidylethanolamine-specific phospholipase D activity(GO:0070290) |
| 0.2 | 1.0 | GO:0008568 | microtubule-severing ATPase activity(GO:0008568) |
| 0.2 | 1.6 | GO:0015349 | thyroid hormone transmembrane transporter activity(GO:0015349) |
| 0.2 | 1.1 | GO:0035877 | death effector domain binding(GO:0035877) |
| 0.2 | 8.2 | GO:0005246 | calcium channel regulator activity(GO:0005246) |
| 0.2 | 0.6 | GO:0016034 | maleylacetoacetate isomerase activity(GO:0016034) |
| 0.2 | 1.1 | GO:0015232 | heme transporter activity(GO:0015232) |
| 0.2 | 1.1 | GO:0032896 | palmitoyl-CoA 9-desaturase activity(GO:0032896) |
| 0.2 | 0.5 | GO:0030622 | U4atac snRNA binding(GO:0030622) |
| 0.2 | 1.9 | GO:0004972 | NMDA glutamate receptor activity(GO:0004972) |
| 0.2 | 0.9 | GO:0052833 | inositol monophosphate 1-phosphatase activity(GO:0008934) inositol monophosphate 3-phosphatase activity(GO:0052832) inositol monophosphate 4-phosphatase activity(GO:0052833) inositol monophosphate phosphatase activity(GO:0052834) |
| 0.2 | 9.6 | GO:0005245 | voltage-gated calcium channel activity(GO:0005245) |
| 0.2 | 1.9 | GO:0004459 | L-lactate dehydrogenase activity(GO:0004459) |
| 0.2 | 0.7 | GO:0034597 | phosphatidylinositol-3,4-bisphosphate 4-phosphatase activity(GO:0016316) phosphatidylinositol-4,5-bisphosphate 4-phosphatase activity(GO:0034597) |
| 0.2 | 1.7 | GO:0008093 | cytoskeletal adaptor activity(GO:0008093) |
| 0.2 | 3.7 | GO:0008179 | adenylate cyclase binding(GO:0008179) |
| 0.2 | 4.3 | GO:0004435 | phosphatidylinositol phospholipase C activity(GO:0004435) |
| 0.2 | 3.3 | GO:0102391 | decanoate--CoA ligase activity(GO:0102391) |
| 0.2 | 1.1 | GO:0032027 | myosin light chain binding(GO:0032027) |
| 0.2 | 1.6 | GO:0015386 | potassium:proton antiporter activity(GO:0015386) |
| 0.2 | 5.7 | GO:0030506 | ankyrin binding(GO:0030506) |
| 0.2 | 1.8 | GO:0097322 | 7SK snRNA binding(GO:0097322) |
| 0.1 | 0.7 | GO:0016499 | orexin receptor activity(GO:0016499) |
| 0.1 | 1.5 | GO:0005225 | volume-sensitive anion channel activity(GO:0005225) |
| 0.1 | 2.4 | GO:0030957 | Tat protein binding(GO:0030957) |
| 0.1 | 6.4 | GO:0001671 | ATPase activator activity(GO:0001671) |
| 0.1 | 3.4 | GO:0017049 | GTP-Rho binding(GO:0017049) |
| 0.1 | 4.6 | GO:0004683 | calmodulin-dependent protein kinase activity(GO:0004683) |
| 0.1 | 4.7 | GO:0005184 | neuropeptide hormone activity(GO:0005184) |
| 0.1 | 0.3 | GO:0035373 | chondroitin sulfate proteoglycan binding(GO:0035373) |
| 0.1 | 6.6 | GO:0035255 | ionotropic glutamate receptor binding(GO:0035255) |
| 0.1 | 2.0 | GO:0015101 | organic cation transmembrane transporter activity(GO:0015101) |
| 0.1 | 0.9 | GO:0042809 | vitamin D receptor binding(GO:0042809) |
| 0.1 | 1.8 | GO:0022841 | potassium ion leak channel activity(GO:0022841) |
| 0.1 | 2.2 | GO:0044323 | retinoic acid-responsive element binding(GO:0044323) |
| 0.1 | 3.3 | GO:0008195 | phosphatidate phosphatase activity(GO:0008195) |
| 0.1 | 2.9 | GO:0004653 | polypeptide N-acetylgalactosaminyltransferase activity(GO:0004653) |
| 0.1 | 1.7 | GO:0008603 | cAMP-dependent protein kinase regulator activity(GO:0008603) |
| 0.1 | 0.4 | GO:0004821 | histidine-tRNA ligase activity(GO:0004821) |
| 0.1 | 0.4 | GO:0004822 | isoleucine-tRNA ligase activity(GO:0004822) |
| 0.1 | 0.4 | GO:0022865 | transmembrane electron transfer carrier(GO:0022865) |
| 0.1 | 0.7 | GO:0008467 | [heparan sulfate]-glucosamine 3-sulfotransferase 1 activity(GO:0008467) |
| 0.1 | 0.4 | GO:0070740 | tubulin-glutamic acid ligase activity(GO:0070740) |
| 0.1 | 1.0 | GO:0035381 | extracellular ATP-gated cation channel activity(GO:0004931) ATP-gated ion channel activity(GO:0035381) |
| 0.1 | 3.1 | GO:0004709 | MAP kinase kinase kinase activity(GO:0004709) |
| 0.1 | 10.0 | GO:0015459 | potassium channel regulator activity(GO:0015459) |
| 0.1 | 2.0 | GO:0016594 | glycine binding(GO:0016594) |
| 0.1 | 1.7 | GO:0034483 | heparan sulfate sulfotransferase activity(GO:0034483) |
| 0.1 | 1.1 | GO:0016861 | intramolecular oxidoreductase activity, interconverting aldoses and ketoses(GO:0016861) |
| 0.1 | 1.4 | GO:0009931 | calcium-dependent protein serine/threonine kinase activity(GO:0009931) |
| 0.1 | 0.4 | GO:0004663 | Rab geranylgeranyltransferase activity(GO:0004663) |
| 0.1 | 5.3 | GO:0030546 | receptor activator activity(GO:0030546) |
| 0.1 | 0.8 | GO:0005042 | netrin receptor activity(GO:0005042) |
| 0.1 | 1.8 | GO:0032794 | GTPase activating protein binding(GO:0032794) |
| 0.1 | 1.7 | GO:0004089 | carbonate dehydratase activity(GO:0004089) |
| 0.1 | 0.6 | GO:0045322 | unmethylated CpG binding(GO:0045322) |
| 0.1 | 3.6 | GO:0043014 | alpha-tubulin binding(GO:0043014) |
| 0.1 | 5.4 | GO:0017080 | sodium channel regulator activity(GO:0017080) |
| 0.1 | 0.3 | GO:0035939 | microsatellite binding(GO:0035939) |
| 0.1 | 3.0 | GO:0005112 | Notch binding(GO:0005112) |
| 0.1 | 1.1 | GO:0004128 | cytochrome-b5 reductase activity, acting on NAD(P)H(GO:0004128) |
| 0.1 | 4.6 | GO:0051959 | dynein light intermediate chain binding(GO:0051959) |
| 0.1 | 2.7 | GO:0004012 | phospholipid-translocating ATPase activity(GO:0004012) |
| 0.1 | 8.9 | GO:0051082 | unfolded protein binding(GO:0051082) |
| 0.1 | 2.9 | GO:0070330 | aromatase activity(GO:0070330) |
| 0.1 | 1.4 | GO:0008484 | sulfuric ester hydrolase activity(GO:0008484) |
| 0.1 | 0.9 | GO:0036310 | annealing helicase activity(GO:0036310) annealing activity(GO:0097617) |
| 0.1 | 0.2 | GO:0048256 | flap endonuclease activity(GO:0048256) |
| 0.1 | 1.6 | GO:0035259 | glucocorticoid receptor binding(GO:0035259) |
| 0.1 | 4.8 | GO:0046875 | ephrin receptor binding(GO:0046875) |
| 0.1 | 1.8 | GO:0005545 | 1-phosphatidylinositol binding(GO:0005545) |
| 0.1 | 1.6 | GO:0031489 | myosin V binding(GO:0031489) |
| 0.1 | 2.4 | GO:0030742 | GTP-dependent protein binding(GO:0030742) |
| 0.1 | 0.7 | GO:0030346 | protein phosphatase 2B binding(GO:0030346) |
| 0.1 | 1.8 | GO:0001530 | lipopolysaccharide binding(GO:0001530) |
| 0.1 | 0.4 | GO:0046920 | alpha-(1->3)-fucosyltransferase activity(GO:0046920) |
| 0.1 | 0.3 | GO:0008569 | ATP-dependent microtubule motor activity, minus-end-directed(GO:0008569) |
| 0.1 | 2.1 | GO:0019707 | protein-cysteine S-palmitoyltransferase activity(GO:0019706) protein-cysteine S-acyltransferase activity(GO:0019707) |
| 0.1 | 1.5 | GO:0005132 | type I interferon receptor binding(GO:0005132) |
| 0.1 | 10.1 | GO:0000149 | SNARE binding(GO:0000149) |
| 0.1 | 1.5 | GO:0005521 | lamin binding(GO:0005521) |
| 0.1 | 1.2 | GO:0051011 | microtubule minus-end binding(GO:0051011) |
| 0.1 | 0.6 | GO:0035242 | protein-arginine omega-N asymmetric methyltransferase activity(GO:0035242) |
| 0.1 | 6.1 | GO:0019888 | protein phosphatase regulator activity(GO:0019888) |
| 0.1 | 1.1 | GO:0019206 | nucleoside kinase activity(GO:0019206) |
| 0.1 | 0.5 | GO:0001075 | transcription factor activity, RNA polymerase II core promoter sequence-specific binding involved in preinitiation complex assembly(GO:0001075) |
| 0.1 | 0.8 | GO:1990459 | transferrin receptor binding(GO:1990459) |
| 0.1 | 0.2 | GO:0015181 | arginine transmembrane transporter activity(GO:0015181) |
| 0.1 | 0.5 | GO:0008823 | cupric reductase activity(GO:0008823) ferric-chelate reductase (NADPH) activity(GO:0052851) |
| 0.1 | 0.5 | GO:0097642 | calcitonin family receptor activity(GO:0097642) |
| 0.1 | 3.2 | GO:0030507 | spectrin binding(GO:0030507) |
| 0.1 | 0.9 | GO:0004549 | tRNA-specific ribonuclease activity(GO:0004549) |
| 0.1 | 0.5 | GO:0016428 | tRNA (cytosine-5-)-methyltransferase activity(GO:0016428) |
| 0.1 | 0.4 | GO:1990825 | sequence-specific mRNA binding(GO:1990825) |
| 0.0 | 1.1 | GO:0030159 | receptor signaling complex scaffold activity(GO:0030159) |
| 0.0 | 0.2 | GO:0005093 | Rab GDP-dissociation inhibitor activity(GO:0005093) |
| 0.0 | 0.1 | GO:0004980 | melanocyte-stimulating hormone receptor activity(GO:0004980) |
| 0.0 | 0.4 | GO:0036122 | BMP binding(GO:0036122) |
| 0.0 | 0.9 | GO:0019215 | intermediate filament binding(GO:0019215) |
| 0.0 | 0.9 | GO:0030215 | semaphorin receptor binding(GO:0030215) |
| 0.0 | 0.4 | GO:0052629 | phosphatidylinositol-3,5-bisphosphate 3-phosphatase activity(GO:0052629) |
| 0.0 | 0.6 | GO:0043995 | histone acetyltransferase activity (H4-K5 specific)(GO:0043995) histone acetyltransferase activity (H4-K8 specific)(GO:0043996) histone acetyltransferase activity (H4-K16 specific)(GO:0046972) |
| 0.0 | 1.0 | GO:0070402 | NADPH binding(GO:0070402) |
| 0.0 | 3.3 | GO:0003777 | microtubule motor activity(GO:0003777) |
| 0.0 | 1.4 | GO:0005201 | extracellular matrix structural constituent(GO:0005201) |
| 0.0 | 1.3 | GO:0016922 | ligand-dependent nuclear receptor binding(GO:0016922) |
| 0.0 | 1.1 | GO:0034237 | protein kinase A regulatory subunit binding(GO:0034237) |
| 0.0 | 0.3 | GO:0035312 | 5'-3' exodeoxyribonuclease activity(GO:0035312) |
| 0.0 | 0.2 | GO:0005391 | sodium:potassium-exchanging ATPase activity(GO:0005391) |
| 0.0 | 4.5 | GO:0003730 | mRNA 3'-UTR binding(GO:0003730) |
| 0.0 | 4.6 | GO:0004843 | thiol-dependent ubiquitin-specific protease activity(GO:0004843) |
| 0.0 | 1.5 | GO:0080025 | phosphatidylinositol-3,5-bisphosphate binding(GO:0080025) |
| 0.0 | 0.7 | GO:0015095 | magnesium ion transmembrane transporter activity(GO:0015095) |
| 0.0 | 0.3 | GO:0005332 | gamma-aminobutyric acid:sodium symporter activity(GO:0005332) |
| 0.0 | 0.3 | GO:0070579 | methylcytosine dioxygenase activity(GO:0070579) |
| 0.0 | 1.6 | GO:0019894 | kinesin binding(GO:0019894) |
| 0.0 | 2.5 | GO:0015485 | cholesterol binding(GO:0015485) |
| 0.0 | 1.1 | GO:0004198 | calcium-dependent cysteine-type endopeptidase activity(GO:0004198) |
| 0.0 | 1.9 | GO:0016790 | thiolester hydrolase activity(GO:0016790) |
| 0.0 | 0.6 | GO:0005527 | macrolide binding(GO:0005527) FK506 binding(GO:0005528) |
| 0.0 | 0.3 | GO:0004467 | long-chain fatty acid-CoA ligase activity(GO:0004467) |
| 0.0 | 4.3 | GO:0008013 | beta-catenin binding(GO:0008013) |
| 0.0 | 3.5 | GO:0051213 | dioxygenase activity(GO:0051213) |
| 0.0 | 0.3 | GO:0015093 | iron ion transmembrane transporter activity(GO:0005381) ferrous iron transmembrane transporter activity(GO:0015093) |
| 0.0 | 1.4 | GO:0050699 | WW domain binding(GO:0050699) |
| 0.0 | 0.2 | GO:0004483 | mRNA (nucleoside-2'-O-)-methyltransferase activity(GO:0004483) |
| 0.0 | 2.3 | GO:0030971 | receptor tyrosine kinase binding(GO:0030971) |
| 0.0 | 1.3 | GO:0003785 | actin monomer binding(GO:0003785) |
| 0.0 | 0.7 | GO:0016638 | oxidoreductase activity, acting on the CH-NH2 group of donors(GO:0016638) |
| 0.0 | 0.9 | GO:0005104 | fibroblast growth factor receptor binding(GO:0005104) |
| 0.0 | 1.4 | GO:0019003 | GDP binding(GO:0019003) |
| 0.0 | 0.4 | GO:0005523 | tropomyosin binding(GO:0005523) |
| 0.0 | 3.7 | GO:0005178 | integrin binding(GO:0005178) |
| 0.0 | 14.7 | GO:0005509 | calcium ion binding(GO:0005509) |
| 0.0 | 0.1 | GO:0004385 | guanylate kinase activity(GO:0004385) |
| 0.0 | 0.2 | GO:0004991 | parathyroid hormone receptor activity(GO:0004991) |
| 0.0 | 0.3 | GO:0008376 | acetylgalactosaminyltransferase activity(GO:0008376) |
| 0.0 | 0.7 | GO:0070412 | R-SMAD binding(GO:0070412) |
| 0.0 | 0.4 | GO:0017091 | AU-rich element binding(GO:0017091) |
| 0.0 | 0.7 | GO:0046961 | proton-transporting ATPase activity, rotational mechanism(GO:0046961) |
| 0.0 | 3.9 | GO:0008017 | microtubule binding(GO:0008017) |
| 0.0 | 0.7 | GO:0017022 | myosin binding(GO:0017022) |
| 0.0 | 1.5 | GO:0016627 | oxidoreductase activity, acting on the CH-CH group of donors(GO:0016627) |
| 0.0 | 0.2 | GO:0030628 | pre-mRNA 3'-splice site binding(GO:0030628) |
| 0.0 | 1.0 | GO:0031624 | ubiquitin conjugating enzyme binding(GO:0031624) |
| 0.0 | 0.9 | GO:0004693 | cyclin-dependent protein serine/threonine kinase activity(GO:0004693) |
| 0.0 | 0.3 | GO:0005520 | insulin-like growth factor binding(GO:0005520) |
| 0.0 | 0.4 | GO:0008137 | NADH dehydrogenase (ubiquinone) activity(GO:0008137) NADH dehydrogenase (quinone) activity(GO:0050136) |
| 0.0 | 0.9 | GO:0070063 | RNA polymerase binding(GO:0070063) |
| 0.0 | 0.1 | GO:0004565 | beta-galactosidase activity(GO:0004565) |
| 0.0 | 1.5 | GO:0052689 | carboxylic ester hydrolase activity(GO:0052689) |
| 0.0 | 1.4 | GO:0008201 | heparin binding(GO:0008201) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 13.2 | PID ARF6 DOWNSTREAM PATHWAY | Arf6 downstream pathway |
| 0.3 | 3.2 | ST G ALPHA I PATHWAY | G alpha i Pathway |
| 0.2 | 5.1 | PID SYNDECAN 3 PATHWAY | Syndecan-3-mediated signaling events |
| 0.2 | 1.1 | SA G2 AND M PHASES | Cdc25 activates the cdc2/cyclin B complex to induce the G2/M transition. |
| 0.2 | 3.1 | PID S1P S1P4 PATHWAY | S1P4 pathway |
| 0.2 | 15.6 | PID AJDISS 2PATHWAY | Posttranslational regulation of adherens junction stability and dissassembly |
| 0.1 | 2.7 | ST MYOCYTE AD PATHWAY | Myocyte Adrenergic Pathway is a specific case of the generalized Adrenergic Pathway. |
| 0.1 | 6.9 | PID TCR CALCIUM PATHWAY | Calcium signaling in the CD4+ TCR pathway |
| 0.1 | 1.7 | PID S1P S1P2 PATHWAY | S1P2 pathway |
| 0.1 | 5.3 | NABA PROTEOGLYCANS | Genes encoding proteoglycans |
| 0.1 | 13.3 | PID ERA GENOMIC PATHWAY | Validated nuclear estrogen receptor alpha network |
| 0.1 | 3.3 | ST GA12 PATHWAY | G alpha 12 Pathway |
| 0.1 | 5.8 | PID RAC1 REG PATHWAY | Regulation of RAC1 activity |
| 0.1 | 8.2 | PID HNF3B PATHWAY | FOXA2 and FOXA3 transcription factor networks |
| 0.1 | 3.9 | PID P38 MKK3 6PATHWAY | p38 MAPK signaling pathway |
| 0.1 | 2.9 | PID NCADHERIN PATHWAY | N-cadherin signaling events |
| 0.1 | 5.7 | SIG REGULATION OF THE ACTIN CYTOSKELETON BY RHO GTPASES | Genes related to regulation of the actin cytoskeleton |
| 0.1 | 3.8 | PID EPHB FWD PATHWAY | EPHB forward signaling |
| 0.1 | 2.7 | PID NECTIN PATHWAY | Nectin adhesion pathway |
| 0.1 | 0.9 | ST TUMOR NECROSIS FACTOR PATHWAY | Tumor Necrosis Factor Pathway. |
| 0.1 | 2.0 | ST JNK MAPK PATHWAY | JNK MAPK Pathway |
| 0.1 | 0.6 | PID EPO PATHWAY | EPO signaling pathway |
| 0.1 | 3.6 | PID FAS PATHWAY | FAS (CD95) signaling pathway |
| 0.1 | 5.2 | PID CDC42 REG PATHWAY | Regulation of CDC42 activity |
| 0.1 | 16.9 | NABA ECM AFFILIATED | Genes encoding proteins affiliated structurally or functionally to extracellular matrix proteins |
| 0.1 | 7.3 | PID MYC REPRESS PATHWAY | Validated targets of C-MYC transcriptional repression |
| 0.1 | 2.8 | PID REELIN PATHWAY | Reelin signaling pathway |
| 0.1 | 0.7 | SA PTEN PATHWAY | PTEN is a tumor suppressor that dephosphorylates the lipid messenger phosphatidylinositol triphosphate. |
| 0.1 | 1.2 | SA TRKA RECEPTOR | The TrkA receptor binds nerve growth factor to activate MAP kinase pathways and promote cell growth. |
| 0.1 | 1.9 | SIG CD40PATHWAYMAP | Genes related to CD40 signaling |
| 0.1 | 2.1 | PID HDAC CLASSIII PATHWAY | Signaling events mediated by HDAC Class III |
| 0.1 | 1.0 | PID AR NONGENOMIC PATHWAY | Nongenotropic Androgen signaling |
| 0.1 | 1.7 | PID INTEGRIN A4B1 PATHWAY | Alpha4 beta1 integrin signaling events |
| 0.1 | 2.1 | PID RAS PATHWAY | Regulation of Ras family activation |
| 0.1 | 2.0 | SIG BCR SIGNALING PATHWAY | Members of the BCR signaling pathway |
| 0.0 | 1.6 | PID INTEGRIN1 PATHWAY | Beta1 integrin cell surface interactions |
| 0.0 | 3.2 | PID NOTCH PATHWAY | Notch signaling pathway |
| 0.0 | 10.1 | NABA ECM GLYCOPROTEINS | Genes encoding structural ECM glycoproteins |
| 0.0 | 1.9 | PID ECADHERIN STABILIZATION PATHWAY | Stabilization and expansion of the E-cadherin adherens junction |
| 0.0 | 0.2 | PID EPHRINB REV PATHWAY | Ephrin B reverse signaling |
| 0.0 | 1.6 | PID LKB1 PATHWAY | LKB1 signaling events |
| 0.0 | 0.4 | PID NEPHRIN NEPH1 PATHWAY | Nephrin/Neph1 signaling in the kidney podocyte |
| 0.0 | 1.8 | PID FGF PATHWAY | FGF signaling pathway |
| 0.0 | 1.0 | PID IL12 STAT4 PATHWAY | IL12 signaling mediated by STAT4 |
| 0.0 | 0.8 | PID NETRIN PATHWAY | Netrin-mediated signaling events |
| 0.0 | 0.7 | PID VEGFR1 2 PATHWAY | Signaling events mediated by VEGFR1 and VEGFR2 |
| 0.0 | 2.4 | PID HIF1 TFPATHWAY | HIF-1-alpha transcription factor network |
| 0.0 | 0.4 | PID IL8 CXCR1 PATHWAY | IL8- and CXCR1-mediated signaling events |
| 0.0 | 1.4 | PID HDAC CLASSII PATHWAY | Signaling events mediated by HDAC Class II |
| 0.0 | 0.5 | PID ARF 3PATHWAY | Arf1 pathway |
| 0.0 | 0.6 | PID LIS1 PATHWAY | Lissencephaly gene (LIS1) in neuronal migration and development |
| 0.0 | 0.3 | PID RET PATHWAY | Signaling events regulated by Ret tyrosine kinase |
| 0.0 | 1.9 | PID P53 DOWNSTREAM PATHWAY | Direct p53 effectors |
| 0.0 | 0.6 | PID TNF PATHWAY | TNF receptor signaling pathway |
| 0.0 | 5.0 | NABA SECRETED FACTORS | Genes encoding secreted soluble factors |
| 0.0 | 0.3 | PID HNF3A PATHWAY | FOXA1 transcription factor network |
| 0.0 | 3.2 | NABA MATRISOME ASSOCIATED | Ensemble of genes encoding ECM-associated proteins including ECM-affilaited proteins, ECM regulators and secreted factors |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.7 | 10.2 | REACTOME CS DS DEGRADATION | Genes involved in CS/DS degradation |
| 0.7 | 16.1 | REACTOME GABA A RECEPTOR ACTIVATION | Genes involved in GABA A receptor activation |
| 0.5 | 9.5 | REACTOME BOTULINUM NEUROTOXICITY | Genes involved in Botulinum neurotoxicity |
| 0.4 | 19.8 | REACTOME INTERACTION BETWEEN L1 AND ANKYRINS | Genes involved in Interaction between L1 and Ankyrins |
| 0.3 | 6.3 | REACTOME GAP JUNCTION DEGRADATION | Genes involved in Gap junction degradation |
| 0.3 | 4.7 | REACTOME PRESYNAPTIC NICOTINIC ACETYLCHOLINE RECEPTORS | Genes involved in Presynaptic nicotinic acetylcholine receptors |
| 0.3 | 6.9 | REACTOME GABA SYNTHESIS RELEASE REUPTAKE AND DEGRADATION | Genes involved in GABA synthesis, release, reuptake and degradation |
| 0.3 | 3.5 | REACTOME SEROTONIN RECEPTORS | Genes involved in Serotonin receptors |
| 0.3 | 1.7 | REACTOME RETROGRADE NEUROTROPHIN SIGNALLING | Genes involved in Retrograde neurotrophin signalling |
| 0.3 | 6.2 | REACTOME REGULATION OF INSULIN LIKE GROWTH FACTOR IGF ACTIVITY BY INSULIN LIKE GROWTH FACTOR BINDING PROTEINS IGFBPS | Genes involved in Regulation of Insulin-like Growth Factor (IGF) Activity by Insulin-like Growth Factor Binding Proteins (IGFBPs) |
| 0.3 | 17.4 | REACTOME INWARDLY RECTIFYING K CHANNELS | Genes involved in Inwardly rectifying K+ channels |
| 0.3 | 5.9 | REACTOME TRAFFICKING OF GLUR2 CONTAINING AMPA RECEPTORS | Genes involved in Trafficking of GluR2-containing AMPA receptors |
| 0.2 | 14.6 | REACTOME NCAM1 INTERACTIONS | Genes involved in NCAM1 interactions |
| 0.2 | 12.4 | REACTOME VOLTAGE GATED POTASSIUM CHANNELS | Genes involved in Voltage gated Potassium channels |
| 0.2 | 7.3 | REACTOME FATTY ACYL COA BIOSYNTHESIS | Genes involved in Fatty Acyl-CoA Biosynthesis |
| 0.2 | 3.6 | REACTOME INSULIN SYNTHESIS AND PROCESSING | Genes involved in Insulin Synthesis and Processing |
| 0.2 | 3.1 | REACTOME G PROTEIN ACTIVATION | Genes involved in G-protein activation |
| 0.2 | 3.1 | REACTOME NOTCH HLH TRANSCRIPTION PATHWAY | Genes involved in Notch-HLH transcription pathway |
| 0.2 | 8.2 | REACTOME GLUCONEOGENESIS | Genes involved in Gluconeogenesis |
| 0.1 | 1.2 | REACTOME THE ACTIVATION OF ARYLSULFATASES | Genes involved in The activation of arylsulfatases |
| 0.1 | 2.0 | REACTOME REGULATION OF RHEB GTPASE ACTIVITY BY AMPK | Genes involved in Regulation of Rheb GTPase activity by AMPK |
| 0.1 | 2.1 | REACTOME IONOTROPIC ACTIVITY OF KAINATE RECEPTORS | Genes involved in Ionotropic activity of Kainate Receptors |
| 0.1 | 16.1 | REACTOME RESPONSE TO ELEVATED PLATELET CYTOSOLIC CA2 | Genes involved in Response to elevated platelet cytosolic Ca2+ |
| 0.1 | 1.3 | REACTOME COPI MEDIATED TRANSPORT | Genes involved in COPI Mediated Transport |
| 0.1 | 1.9 | REACTOME TANDEM PORE DOMAIN POTASSIUM CHANNELS | Genes involved in Tandem pore domain potassium channels |
| 0.1 | 2.5 | REACTOME FGFR4 LIGAND BINDING AND ACTIVATION | Genes involved in FGFR4 ligand binding and activation |
| 0.1 | 2.1 | REACTOME DSCAM INTERACTIONS | Genes involved in DSCAM interactions |
| 0.1 | 10.4 | REACTOME NRAGE SIGNALS DEATH THROUGH JNK | Genes involved in NRAGE signals death through JNK |
| 0.1 | 3.0 | REACTOME GLUCAGON TYPE LIGAND RECEPTORS | Genes involved in Glucagon-type ligand receptors |
| 0.1 | 1.0 | REACTOME FGFR1 LIGAND BINDING AND ACTIVATION | Genes involved in FGFR1 ligand binding and activation |
| 0.1 | 1.3 | REACTOME NONSENSE MEDIATED DECAY ENHANCED BY THE EXON JUNCTION COMPLEX | Genes involved in Nonsense Mediated Decay Enhanced by the Exon Junction Complex |
| 0.1 | 2.3 | REACTOME UNBLOCKING OF NMDA RECEPTOR GLUTAMATE BINDING AND ACTIVATION | Genes involved in Unblocking of NMDA receptor, glutamate binding and activation |
| 0.1 | 4.4 | REACTOME LATENT INFECTION OF HOMO SAPIENS WITH MYCOBACTERIUM TUBERCULOSIS | Genes involved in Latent infection of Homo sapiens with Mycobacterium tuberculosis |
| 0.1 | 1.7 | REACTOME SYNTHESIS OF PE | Genes involved in Synthesis of PE |
| 0.1 | 2.1 | REACTOME PURINE SALVAGE | Genes involved in Purine salvage |
| 0.1 | 1.1 | REACTOME REGULATION OF THE FANCONI ANEMIA PATHWAY | Genes involved in Regulation of the Fanconi anemia pathway |
| 0.1 | 1.9 | REACTOME PYRUVATE METABOLISM | Genes involved in Pyruvate metabolism |
| 0.1 | 1.9 | REACTOME BRANCHED CHAIN AMINO ACID CATABOLISM | Genes involved in Branched-chain amino acid catabolism |
| 0.1 | 1.2 | REACTOME RECRUITMENT OF NUMA TO MITOTIC CENTROSOMES | Genes involved in Recruitment of NuMA to mitotic centrosomes |
| 0.1 | 2.9 | REACTOME ACTIVATED NOTCH1 TRANSMITS SIGNAL TO THE NUCLEUS | Genes involved in Activated NOTCH1 Transmits Signal to the Nucleus |
| 0.1 | 2.7 | REACTOME SIGNALING BY ROBO RECEPTOR | Genes involved in Signaling by Robo receptor |
| 0.1 | 0.6 | REACTOME CELL DEATH SIGNALLING VIA NRAGE NRIF AND NADE | Genes involved in Cell death signalling via NRAGE, NRIF and NADE |
| 0.1 | 3.7 | REACTOME HEPARAN SULFATE HEPARIN HS GAG METABOLISM | Genes involved in Heparan sulfate/heparin (HS-GAG) metabolism |
| 0.1 | 0.9 | REACTOME P38MAPK EVENTS | Genes involved in p38MAPK events |
| 0.1 | 3.5 | REACTOME PLATELET AGGREGATION PLUG FORMATION | Genes involved in Platelet Aggregation (Plug Formation) |
| 0.1 | 1.0 | REACTOME ENDOGENOUS STEROLS | Genes involved in Endogenous sterols |
| 0.1 | 2.1 | REACTOME PREFOLDIN MEDIATED TRANSFER OF SUBSTRATE TO CCT TRIC | Genes involved in Prefoldin mediated transfer of substrate to CCT/TriC |
| 0.1 | 3.2 | REACTOME ION TRANSPORT BY P TYPE ATPASES | Genes involved in Ion transport by P-type ATPases |
| 0.1 | 2.7 | REACTOME TRANSPORT TO THE GOLGI AND SUBSEQUENT MODIFICATION | Genes involved in Transport to the Golgi and subsequent modification |
| 0.0 | 0.8 | REACTOME SIGNALING BY PDGF | Genes involved in Signaling by PDGF |
| 0.0 | 0.7 | REACTOME THE NLRP3 INFLAMMASOME | Genes involved in The NLRP3 inflammasome |
| 0.0 | 0.7 | REACTOME ORGANIC CATION ANION ZWITTERION TRANSPORT | Genes involved in Organic cation/anion/zwitterion transport |
| 0.0 | 1.9 | REACTOME TIGHT JUNCTION INTERACTIONS | Genes involved in Tight junction interactions |
| 0.0 | 1.7 | REACTOME DARPP 32 EVENTS | Genes involved in DARPP-32 events |
| 0.0 | 1.1 | REACTOME CYTOSOLIC TRNA AMINOACYLATION | Genes involved in Cytosolic tRNA aminoacylation |
| 0.0 | 0.9 | REACTOME CELL SURFACE INTERACTIONS AT THE VASCULAR WALL | Genes involved in Cell surface interactions at the vascular wall |
| 0.0 | 2.4 | REACTOME RECRUITMENT OF MITOTIC CENTROSOME PROTEINS AND COMPLEXES | Genes involved in Recruitment of mitotic centrosome proteins and complexes |
| 0.0 | 1.6 | REACTOME GLUTATHIONE CONJUGATION | Genes involved in Glutathione conjugation |
| 0.0 | 0.7 | REACTOME AMYLOIDS | Genes involved in Amyloids |
| 0.0 | 0.4 | REACTOME VITAMIN B5 PANTOTHENATE METABOLISM | Genes involved in Vitamin B5 (pantothenate) metabolism |
| 0.0 | 1.7 | REACTOME TRANSMISSION ACROSS CHEMICAL SYNAPSES | Genes involved in Transmission across Chemical Synapses |
| 0.0 | 0.5 | REACTOME EGFR DOWNREGULATION | Genes involved in EGFR downregulation |
| 0.0 | 0.8 | REACTOME POST NMDA RECEPTOR ACTIVATION EVENTS | Genes involved in Post NMDA receptor activation events |
| 0.0 | 0.4 | REACTOME TRNA AMINOACYLATION | Genes involved in tRNA Aminoacylation |
| 0.0 | 0.4 | REACTOME SIGNALING BY BMP | Genes involved in Signaling by BMP |
| 0.0 | 0.3 | REACTOME MTORC1 MEDIATED SIGNALLING | Genes involved in mTORC1-mediated signalling |
| 0.0 | 0.7 | REACTOME CELL CELL JUNCTION ORGANIZATION | Genes involved in Cell-cell junction organization |
| 0.0 | 0.5 | REACTOME ACYL CHAIN REMODELLING OF PE | Genes involved in Acyl chain remodelling of PE |
| 0.0 | 0.4 | REACTOME PROCESSING OF INTRONLESS PRE MRNAS | Genes involved in Processing of Intronless Pre-mRNAs |
| 0.0 | 0.5 | REACTOME SIGNALING BY NODAL | Genes involved in Signaling by NODAL |
| 0.0 | 2.4 | REACTOME G ALPHA I SIGNALLING EVENTS | Genes involved in G alpha (i) signalling events |
| 0.0 | 0.6 | REACTOME MRNA SPLICING MINOR PATHWAY | Genes involved in mRNA Splicing - Minor Pathway |
| 0.0 | 0.2 | REACTOME PHOSPHORYLATION OF THE APC C | Genes involved in Phosphorylation of the APC/C |