Project
PRJNA375882: Comprehensive Mouse Transcriptomic BodyMap
Navigation
Downloads
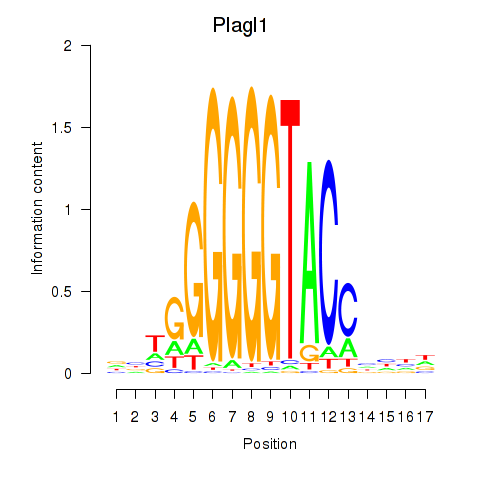
Results for Plagl1
Z-value: 0.64
Transcription factors associated with Plagl1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Plagl1
|
ENSMUSG00000019817.19 | Plagl1 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Plagl1 | mm39_v1_chr10_+_12936248_12936282 | -0.17 | 1.5e-01 | Click! |
Activity profile of Plagl1 motif
Sorted Z-values of Plagl1 motif
Network of associatons between targets according to the STRING database.
First level regulatory network of Plagl1
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr1_+_162466717 | 4.14 |
ENSMUST00000028020.11
|
Myoc
|
myocilin |
| chr15_-_78090591 | 3.53 |
ENSMUST00000120592.2
|
Pvalb
|
parvalbumin |
| chr7_-_118594365 | 2.24 |
ENSMUST00000008878.10
|
Gprc5b
|
G protein-coupled receptor, family C, group 5, member B |
| chr18_+_32510270 | 1.99 |
ENSMUST00000234857.2
ENSMUST00000234496.2 ENSMUST00000091967.13 ENSMUST00000025239.9 |
Bin1
|
bridging integrator 1 |
| chr5_-_136911969 | 1.95 |
ENSMUST00000057497.13
ENSMUST00000111103.2 |
Col26a1
|
collagen, type XXVI, alpha 1 |
| chr6_-_91784405 | 1.86 |
ENSMUST00000162300.8
|
Grip2
|
glutamate receptor interacting protein 2 |
| chr19_-_46327024 | 1.82 |
ENSMUST00000236046.2
ENSMUST00000236980.2 ENSMUST00000235485.2 ENSMUST00000236061.2 ENSMUST00000236236.2 ENSMUST00000236768.2 ENSMUST00000236651.2 |
Cuedc2
|
CUE domain containing 2 |
| chr6_-_112466780 | 1.75 |
ENSMUST00000053306.8
|
Oxtr
|
oxytocin receptor |
| chr19_-_46327071 | 1.74 |
ENSMUST00000235977.2
ENSMUST00000167861.8 ENSMUST00000051234.9 ENSMUST00000236066.2 |
Cuedc2
|
CUE domain containing 2 |
| chr4_+_129030710 | 1.74 |
ENSMUST00000102600.4
|
Fndc5
|
fibronectin type III domain containing 5 |
| chr11_+_69909659 | 1.73 |
ENSMUST00000232002.2
ENSMUST00000134376.10 ENSMUST00000231221.2 |
Dlg4
|
discs large MAGUK scaffold protein 4 |
| chr6_-_52211882 | 1.73 |
ENSMUST00000125581.2
|
Hoxa10
|
homeobox A10 |
| chr4_+_47288057 | 1.72 |
ENSMUST00000140413.8
ENSMUST00000107731.9 |
Col15a1
|
collagen, type XV, alpha 1 |
| chr5_-_139115914 | 1.66 |
ENSMUST00000129851.8
|
Prkar1b
|
protein kinase, cAMP dependent regulatory, type I beta |
| chr7_-_4518958 | 1.65 |
ENSMUST00000163710.8
ENSMUST00000166268.8 ENSMUST00000071798.13 ENSMUST00000178163.8 ENSMUST00000108587.9 |
Tnnt1
|
troponin T1, skeletal, slow |
| chr12_-_15866763 | 1.63 |
ENSMUST00000020922.8
ENSMUST00000221215.2 ENSMUST00000221518.2 |
Trib2
|
tribbles pseudokinase 2 |
| chr16_-_34083200 | 1.55 |
ENSMUST00000114947.2
|
Kalrn
|
kalirin, RhoGEF kinase |
| chr11_-_33793461 | 1.50 |
ENSMUST00000101368.9
ENSMUST00000065970.6 ENSMUST00000109340.9 |
Kcnip1
|
Kv channel-interacting protein 1 |
| chr6_+_42263609 | 1.38 |
ENSMUST00000238845.2
ENSMUST00000031894.13 |
Clcn1
|
chloride channel, voltage-sensitive 1 |
| chr4_+_47288287 | 1.36 |
ENSMUST00000146967.2
|
Col15a1
|
collagen, type XV, alpha 1 |
| chr3_-_89867869 | 1.36 |
ENSMUST00000069805.14
|
Atp8b2
|
ATPase, class I, type 8B, member 2 |
| chrX_+_162694397 | 1.34 |
ENSMUST00000140845.2
|
Ap1s2
|
adaptor-related protein complex 1, sigma 2 subunit |
| chr7_+_45434755 | 1.32 |
ENSMUST00000233503.2
ENSMUST00000120005.10 ENSMUST00000211609.2 |
Lmtk3
|
lemur tyrosine kinase 3 |
| chr6_+_83156550 | 1.29 |
ENSMUST00000113919.10
ENSMUST00000113918.8 ENSMUST00000141680.8 |
Dctn1
|
dynactin 1 |
| chr6_-_91784299 | 1.26 |
ENSMUST00000159684.8
|
Grip2
|
glutamate receptor interacting protein 2 |
| chrX_+_139243012 | 1.23 |
ENSMUST00000208130.2
|
Frmpd3
|
FERM and PDZ domain containing 3 |
| chrX_-_20157966 | 1.21 |
ENSMUST00000115393.3
ENSMUST00000072451.11 |
Slc9a7
|
solute carrier family 9 (sodium/hydrogen exchanger), member 7 |
| chr12_+_16703709 | 1.12 |
ENSMUST00000221049.2
|
Ntsr2
|
neurotensin receptor 2 |
| chr15_-_99149810 | 1.09 |
ENSMUST00000163506.3
ENSMUST00000229671.2 ENSMUST00000229359.2 ENSMUST00000041190.17 |
Mcrs1
|
microspherule protein 1 |
| chr1_-_5089564 | 1.08 |
ENSMUST00000002533.15
|
Rgs20
|
regulator of G-protein signaling 20 |
| chr9_+_113886208 | 1.07 |
ENSMUST00000135338.3
|
Susd5
|
sushi domain containing 5 |
| chr11_+_53410697 | 1.02 |
ENSMUST00000120878.9
ENSMUST00000147912.2 |
Septin8
|
septin 8 |
| chr7_+_49428082 | 1.01 |
ENSMUST00000032715.13
|
Prmt3
|
protein arginine N-methyltransferase 3 |
| chr9_-_52591030 | 0.98 |
ENSMUST00000213937.2
|
AI593442
|
expressed sequence AI593442 |
| chr6_+_42263644 | 0.98 |
ENSMUST00000163936.8
|
Clcn1
|
chloride channel, voltage-sensitive 1 |
| chr10_-_76797622 | 0.96 |
ENSMUST00000001148.11
ENSMUST00000105411.9 |
Pcbp3
|
poly(rC) binding protein 3 |
| chr1_+_135980639 | 0.94 |
ENSMUST00000112064.8
|
Cacna1s
|
calcium channel, voltage-dependent, L type, alpha 1S subunit |
| chrX_+_119199956 | 0.93 |
ENSMUST00000113364.10
ENSMUST00000050239.16 ENSMUST00000113358.10 |
Pcdh11x
|
protocadherin 11 X-linked |
| chr15_-_50753792 | 0.91 |
ENSMUST00000185183.2
|
Trps1
|
transcriptional repressor GATA binding 1 |
| chr5_+_24598633 | 0.91 |
ENSMUST00000138168.3
ENSMUST00000115077.8 |
Abcb8
|
ATP-binding cassette, sub-family B (MDR/TAP), member 8 |
| chr7_+_29003382 | 0.87 |
ENSMUST00000049977.13
|
Dpf1
|
D4, zinc and double PHD fingers family 1 |
| chr2_+_32496990 | 0.87 |
ENSMUST00000095045.9
ENSMUST00000095044.10 ENSMUST00000126636.8 |
St6galnac6
|
ST6 (alpha-N-acetyl-neuraminyl-2,3-beta-galactosyl-1,3)-N-acetylgalactosaminide alpha-2,6-sialyltransferase 6 |
| chr18_-_89787603 | 0.84 |
ENSMUST00000097495.5
|
Dok6
|
docking protein 6 |
| chr19_+_45035942 | 0.83 |
ENSMUST00000237222.2
ENSMUST00000111954.11 |
Sfxn3
|
sideroflexin 3 |
| chr2_-_131987008 | 0.81 |
ENSMUST00000028815.15
|
Slc23a2
|
solute carrier family 23 (nucleobase transporters), member 2 |
| chr11_+_98632953 | 0.80 |
ENSMUST00000153043.8
|
Thra
|
thyroid hormone receptor alpha |
| chr1_+_74627445 | 0.80 |
ENSMUST00000113733.10
ENSMUST00000027358.11 |
Bcs1l
|
BCS1-like (yeast) |
| chr11_-_119910986 | 0.79 |
ENSMUST00000134319.2
|
Aatk
|
apoptosis-associated tyrosine kinase |
| chr14_+_71011744 | 0.78 |
ENSMUST00000022698.8
|
Dok2
|
docking protein 2 |
| chr5_-_143279378 | 0.77 |
ENSMUST00000212715.2
|
Zfp853
|
zinc finger protein 853 |
| chr18_+_35987733 | 0.76 |
ENSMUST00000235337.2
|
Cxxc5
|
CXXC finger 5 |
| chr1_-_74627123 | 0.75 |
ENSMUST00000027315.14
ENSMUST00000113737.8 |
Zfp142
|
zinc finger protein 142 |
| chr11_+_98632696 | 0.72 |
ENSMUST00000103139.11
|
Thra
|
thyroid hormone receptor alpha |
| chr4_+_152423075 | 0.71 |
ENSMUST00000030775.12
ENSMUST00000164662.8 |
Chd5
|
chromodomain helicase DNA binding protein 5 |
| chr9_+_72714156 | 0.71 |
ENSMUST00000055535.9
|
Prtg
|
protogenin |
| chr1_-_65162267 | 0.70 |
ENSMUST00000050047.4
ENSMUST00000148020.8 |
D630023F18Rik
|
RIKEN cDNA D630023F18 gene |
| chr1_+_74627506 | 0.69 |
ENSMUST00000113732.2
|
Bcs1l
|
BCS1-like (yeast) |
| chr11_+_98632631 | 0.69 |
ENSMUST00000064187.12
|
Thra
|
thyroid hormone receptor alpha |
| chr11_-_76289888 | 0.69 |
ENSMUST00000021204.4
|
Nxn
|
nucleoredoxin |
| chr6_+_41520287 | 0.68 |
ENSMUST00000103296.2
|
Trbj2-4
|
T cell receptor beta joining 2-4 |
| chr17_+_46608333 | 0.68 |
ENSMUST00000188223.7
ENSMUST00000061722.13 ENSMUST00000166280.8 |
Dlk2
|
delta like non-canonical Notch ligand 2 |
| chr15_-_99149794 | 0.67 |
ENSMUST00000229926.2
|
Mcrs1
|
microspherule protein 1 |
| chr1_+_134383247 | 0.67 |
ENSMUST00000112232.8
ENSMUST00000027725.11 ENSMUST00000116528.2 |
Klhl12
|
kelch-like 12 |
| chr1_-_74627264 | 0.67 |
ENSMUST00000066986.13
|
Zfp142
|
zinc finger protein 142 |
| chr9_+_108782664 | 0.66 |
ENSMUST00000026740.6
|
Col7a1
|
collagen, type VII, alpha 1 |
| chr6_-_134864731 | 0.66 |
ENSMUST00000203762.4
ENSMUST00000215088.2 ENSMUST00000066107.10 |
Gpr19
|
G protein-coupled receptor 19 |
| chr19_+_45036037 | 0.65 |
ENSMUST00000062213.13
|
Sfxn3
|
sideroflexin 3 |
| chr9_+_75532992 | 0.65 |
ENSMUST00000034702.6
|
Lysmd2
|
LysM, putative peptidoglycan-binding, domain containing 2 |
| chr8_+_94537460 | 0.62 |
ENSMUST00000034198.15
ENSMUST00000125716.8 |
Gnao1
|
guanine nucleotide binding protein, alpha O |
| chr2_-_84500951 | 0.62 |
ENSMUST00000189988.3
ENSMUST00000189636.8 ENSMUST00000102646.4 ENSMUST00000102647.11 ENSMUST00000117299.10 |
Selenoh
|
selenoprotein H |
| chr1_-_128030148 | 0.61 |
ENSMUST00000086614.12
|
Zranb3
|
zinc finger, RAN-binding domain containing 3 |
| chr15_-_43733389 | 0.61 |
ENSMUST00000067469.6
|
Tmem74
|
transmembrane protein 74 |
| chr8_-_14024676 | 0.60 |
ENSMUST00000210414.2
|
Tdrp
|
testis development related protein |
| chr12_+_99850756 | 0.60 |
ENSMUST00000153627.8
|
Tdp1
|
tyrosyl-DNA phosphodiesterase 1 |
| chr5_-_124490296 | 0.59 |
ENSMUST00000111472.6
|
Cdk2ap1
|
CDK2 (cyclin-dependent kinase 2)-associated protein 1 |
| chr5_-_92231517 | 0.58 |
ENSMUST00000202258.4
ENSMUST00000113127.7 |
G3bp2
|
GTPase activating protein (SH3 domain) binding protein 2 |
| chr16_+_31918599 | 0.57 |
ENSMUST00000115168.9
|
Cep19
|
centrosomal protein 19 |
| chr6_+_41519654 | 0.57 |
ENSMUST00000103293.2
|
Trbj2-1
|
T cell receptor beta joining 2-1 |
| chr13_+_74498418 | 0.56 |
ENSMUST00000022063.14
|
Ccdc127
|
coiled-coil domain containing 127 |
| chr4_+_118522716 | 0.55 |
ENSMUST00000102666.5
|
Olfr62
|
olfactory receptor 62 |
| chr18_+_35987791 | 0.55 |
ENSMUST00000235404.2
|
Cxxc5
|
CXXC finger 5 |
| chr7_+_103754080 | 0.53 |
ENSMUST00000214099.2
|
Olfr646
|
olfactory receptor 646 |
| chr9_+_108782646 | 0.52 |
ENSMUST00000112070.8
|
Col7a1
|
collagen, type VII, alpha 1 |
| chr5_-_135518098 | 0.51 |
ENSMUST00000201998.2
|
Hip1
|
huntingtin interacting protein 1 |
| chr6_+_41519884 | 0.49 |
ENSMUST00000103294.2
|
Trbj2-2
|
T cell receptor beta joining 2-2 |
| chrX_+_72818003 | 0.47 |
ENSMUST00000002081.6
|
Srpk3
|
serine/arginine-rich protein specific kinase 3 |
| chr13_+_74498430 | 0.45 |
ENSMUST00000160021.8
ENSMUST00000162672.8 ENSMUST00000162376.2 |
Ccdc127
|
coiled-coil domain containing 127 |
| chr2_+_25318642 | 0.45 |
ENSMUST00000102919.4
|
Abca2
|
ATP-binding cassette, sub-family A (ABC1), member 2 |
| chr7_-_99345016 | 0.44 |
ENSMUST00000107086.9
|
Slco2b1
|
solute carrier organic anion transporter family, member 2b1 |
| chr12_-_13299136 | 0.43 |
ENSMUST00000221623.2
|
Ddx1
|
DEAD box helicase 1 |
| chr7_+_142086749 | 0.42 |
ENSMUST00000038675.7
|
Mrpl23
|
mitochondrial ribosomal protein L23 |
| chr5_-_92231314 | 0.42 |
ENSMUST00000169094.8
ENSMUST00000167918.8 |
G3bp2
|
GTPase activating protein (SH3 domain) binding protein 2 |
| chr1_+_180731843 | 0.42 |
ENSMUST00000027802.9
|
Pycr2
|
pyrroline-5-carboxylate reductase family, member 2 |
| chr10_+_128212953 | 0.40 |
ENSMUST00000014642.10
|
Ankrd52
|
ankyrin repeat domain 52 |
| chr19_-_5538370 | 0.40 |
ENSMUST00000124334.8
|
Mus81
|
MUS81 structure-specific endonuclease subunit |
| chr12_-_13299197 | 0.39 |
ENSMUST00000071103.10
|
Ddx1
|
DEAD box helicase 1 |
| chr17_+_34812361 | 0.38 |
ENSMUST00000174532.2
|
Pbx2
|
pre B cell leukemia homeobox 2 |
| chr2_+_35472841 | 0.37 |
ENSMUST00000135741.8
|
Dab2ip
|
disabled 2 interacting protein |
| chr6_+_41520379 | 0.36 |
ENSMUST00000103297.2
|
Trbj2-5
|
T cell receptor beta joining 2-5 |
| chr2_-_152174565 | 0.36 |
ENSMUST00000028964.14
|
Rbck1
|
RanBP-type and C3HC4-type zinc finger containing 1 |
| chr13_+_104365880 | 0.35 |
ENSMUST00000022227.8
|
Cenpk
|
centromere protein K |
| chr5_-_143255713 | 0.35 |
ENSMUST00000161448.8
|
Zfp316
|
zinc finger protein 316 |
| chr16_+_32096267 | 0.35 |
ENSMUST00000014218.15
ENSMUST00000171474.2 |
Rnf168
|
ring finger protein 168 |
| chr10_-_128383508 | 0.34 |
ENSMUST00000152539.8
ENSMUST00000133458.2 ENSMUST00000040572.10 |
Zc3h10
|
zinc finger CCCH type containing 10 |
| chr11_+_49379915 | 0.33 |
ENSMUST00000214948.2
|
Olfr1385
|
olfactory receptor 1385 |
| chrX_+_73348832 | 0.33 |
ENSMUST00000153141.2
|
Gdi1
|
guanosine diphosphate (GDP) dissociation inhibitor 1 |
| chr3_-_46402355 | 0.32 |
ENSMUST00000195537.2
ENSMUST00000166505.7 ENSMUST00000195436.2 |
Pabpc4l
|
poly(A) binding protein, cytoplasmic 4-like |
| chr17_-_26087696 | 0.31 |
ENSMUST00000236479.2
ENSMUST00000235806.2 ENSMUST00000026828.7 |
Mcrip2
|
MAPK regulated corepressor interacting protein 2 |
| chr15_+_32244947 | 0.29 |
ENSMUST00000067458.7
|
Sema5a
|
sema domain, seven thrombospondin repeats (type 1 and type 1-like), transmembrane domain (TM) and short cytoplasmic domain, (semaphorin) 5A |
| chr19_-_42768374 | 0.28 |
ENSMUST00000069298.13
ENSMUST00000160455.8 ENSMUST00000162004.8 |
Hps1
|
HPS1, biogenesis of lysosomal organelles complex 3 subunit 1 |
| chr6_-_58412879 | 0.27 |
ENSMUST00000078890.5
|
Vmn1r30
|
vomeronasal 1 receptor 30 |
| chr5_-_24782465 | 0.25 |
ENSMUST00000030795.10
|
Abcf2
|
ATP-binding cassette, sub-family F (GCN20), member 2 |
| chr2_-_31031814 | 0.25 |
ENSMUST00000073879.12
ENSMUST00000100208.9 ENSMUST00000100207.9 ENSMUST00000113555.8 ENSMUST00000075326.11 ENSMUST00000113552.9 ENSMUST00000136181.8 ENSMUST00000113564.9 ENSMUST00000113562.9 ENSMUST00000113560.8 |
Fnbp1
|
formin binding protein 1 |
| chr17_+_29879684 | 0.25 |
ENSMUST00000235014.2
ENSMUST00000130423.4 |
Cmtr1
|
cap methyltransferase 1 |
| chr9_+_121232480 | 0.24 |
ENSMUST00000210351.2
|
Trak1
|
trafficking protein, kinesin binding 1 |
| chr4_+_33132502 | 0.24 |
ENSMUST00000029947.6
|
Gabrr1
|
gamma-aminobutyric acid (GABA) C receptor, subunit rho 1 |
| chr14_+_35816874 | 0.24 |
ENSMUST00000226305.2
|
4930474N05Rik
|
RIKEN cDNA 4930474N05 gene |
| chr19_+_4761181 | 0.23 |
ENSMUST00000008991.8
|
Sptbn2
|
spectrin beta, non-erythrocytic 2 |
| chr15_-_102274781 | 0.23 |
ENSMUST00000078508.7
|
Sp7
|
Sp7 transcription factor 7 |
| chr7_-_30062197 | 0.22 |
ENSMUST00000046351.7
|
Lrfn3
|
leucine rich repeat and fibronectin type III domain containing 3 |
| chr12_-_103409912 | 0.22 |
ENSMUST00000055071.9
|
Ifi27l2a
|
interferon, alpha-inducible protein 27 like 2A |
| chr11_-_87788066 | 0.22 |
ENSMUST00000217095.2
ENSMUST00000215150.2 |
Olfr463
|
olfactory receptor 463 |
| chr6_+_67873135 | 0.21 |
ENSMUST00000103310.3
|
Igkv14-126
|
immunoglobulin kappa variable 14-126 |
| chr17_+_29768757 | 0.20 |
ENSMUST00000048677.9
ENSMUST00000150388.3 |
Tbc1d22b
Gm28052
|
TBC1 domain family, member 22B predicted gene, 28052 |
| chr4_-_22488296 | 0.20 |
ENSMUST00000178174.3
|
Pou3f2
|
POU domain, class 3, transcription factor 2 |
| chr6_-_92458324 | 0.17 |
ENSMUST00000113445.2
|
Prickle2
|
prickle planar cell polarity protein 2 |
| chr2_-_9888804 | 0.17 |
ENSMUST00000114915.3
|
9230102O04Rik
|
RIKEN cDNA 9230102O04 gene |
| chr19_+_5524701 | 0.16 |
ENSMUST00000165485.8
ENSMUST00000166253.8 ENSMUST00000167371.8 ENSMUST00000167855.8 ENSMUST00000070118.14 |
Efemp2
|
epidermal growth factor-containing fibulin-like extracellular matrix protein 2 |
| chr7_-_106390206 | 0.16 |
ENSMUST00000065024.2
|
Olfr699
|
olfactory receptor 699 |
| chr2_+_164557219 | 0.16 |
ENSMUST00000103097.10
ENSMUST00000180193.2 |
Spint5
|
serine protease inhibitor, Kunitz type 5 |
| chr4_+_59003121 | 0.16 |
ENSMUST00000095070.4
ENSMUST00000174664.2 |
Dnajc25
Gm20503
|
DnaJ heat shock protein family (Hsp40) member C25 predicted gene 20503 |
| chr14_-_55101505 | 0.16 |
ENSMUST00000142283.4
|
Homez
|
homeodomain leucine zipper-encoding gene |
| chr9_+_46184362 | 0.16 |
ENSMUST00000156440.8
ENSMUST00000114552.4 |
Zpr1
|
ZPR1 zinc finger |
| chr6_-_128414616 | 0.15 |
ENSMUST00000151796.3
|
Fkbp4
|
FK506 binding protein 4 |
| chr7_-_23998735 | 0.15 |
ENSMUST00000145131.8
|
Zfp61
|
zinc finger protein 61 |
| chrX_+_159285149 | 0.14 |
ENSMUST00000112377.8
ENSMUST00000139587.8 |
Phka2
|
phosphorylase kinase alpha 2 |
| chr15_-_37792635 | 0.13 |
ENSMUST00000090150.11
ENSMUST00000150453.2 |
Ncald
|
neurocalcin delta |
| chr13_+_104365432 | 0.13 |
ENSMUST00000070761.10
ENSMUST00000225557.2 |
Cenpk
|
centromere protein K |
| chr10_-_119948890 | 0.13 |
ENSMUST00000020449.12
|
Helb
|
helicase (DNA) B |
| chr8_-_88686188 | 0.12 |
ENSMUST00000109655.9
|
Zfp423
|
zinc finger protein 423 |
| chr4_-_3185358 | 0.11 |
ENSMUST00000105159.5
|
Vmn1r3
|
vomeronasal 1 receptor 3 |
| chr4_-_133930315 | 0.10 |
ENSMUST00000097849.3
|
Zpld2
|
zona pellucida like domain containing 2 |
| chr9_+_38841385 | 0.10 |
ENSMUST00000058789.7
|
Olfr930
|
olfactory receptor 930 |
| chrX_+_159285414 | 0.09 |
ENSMUST00000033652.9
ENSMUST00000112380.9 ENSMUST00000112376.2 |
Phka2
|
phosphorylase kinase alpha 2 |
| chr9_+_39932760 | 0.09 |
ENSMUST00000215956.3
|
Olfr981
|
olfactory receptor 981 |
| chr10_+_43097460 | 0.09 |
ENSMUST00000095725.11
|
Pdss2
|
prenyl (solanesyl) diphosphate synthase, subunit 2 |
| chr11_+_67857268 | 0.08 |
ENSMUST00000021286.11
ENSMUST00000108675.2 |
Stx8
|
syntaxin 8 |
| chr16_-_56537650 | 0.08 |
ENSMUST00000128551.8
|
Tfg
|
Trk-fused gene |
| chr12_-_103641723 | 0.08 |
ENSMUST00000095451.2
|
Serpina16
|
serine (or cysteine) peptidase inhibitor, clade A (alpha-1 antiproteinase, antitrypsin), member 16 |
| chr6_+_67243967 | 0.07 |
ENSMUST00000203436.3
ENSMUST00000205106.3 ENSMUST00000204293.3 ENSMUST00000203077.3 ENSMUST00000204294.3 |
Serbp1
|
serpine1 mRNA binding protein 1 |
| chr16_-_4376471 | 0.07 |
ENSMUST00000230875.2
|
Tfap4
|
transcription factor AP4 |
| chr2_-_36899347 | 0.06 |
ENSMUST00000216437.2
|
Olfr358
|
olfactory receptor 358 |
| chr15_-_102275403 | 0.05 |
ENSMUST00000229464.2
|
Sp7
|
Sp7 transcription factor 7 |
| chr10_+_61431271 | 0.05 |
ENSMUST00000020287.8
|
Npffr1
|
neuropeptide FF receptor 1 |
| chr13_-_99121070 | 0.05 |
ENSMUST00000054425.7
|
H2bl1
|
H2B.L histone variant 1 |
| chr4_+_102835888 | 0.05 |
ENSMUST00000223169.2
|
Dynlt5
|
dynein light chain Tctex-type 5 |
| chr7_-_25315299 | 0.05 |
ENSMUST00000098663.4
ENSMUST00000238895.2 |
Erich4
|
glutamate rich 4 |
| chr11_+_32405367 | 0.04 |
ENSMUST00000051053.5
|
Ubtd2
|
ubiquitin domain containing 2 |
| chr6_-_48422612 | 0.03 |
ENSMUST00000114556.2
|
Zfp467
|
zinc finger protein 467 |
| chr10_+_85707687 | 0.03 |
ENSMUST00000001836.11
|
Pwp1
|
PWP1 homolog, endonuclein |
| chr11_+_59433554 | 0.03 |
ENSMUST00000149126.2
|
Nlrp3
|
NLR family, pyrin domain containing 3 |
| chr17_+_29251602 | 0.02 |
ENSMUST00000130216.3
|
Srsf3
|
serine and arginine-rich splicing factor 3 |
| chr1_+_153750081 | 0.01 |
ENSMUST00000055314.4
|
Teddm1b
|
transmembrane epididymal protein 1B |
| chr6_+_119456629 | 0.01 |
ENSMUST00000032094.7
|
Fbxl14
|
F-box and leucine-rich repeat protein 14 |
| chr15_+_75881712 | 0.00 |
ENSMUST00000187868.3
|
Iqank1
|
IQ motif and ankyrin repeat containing 1 |
| chr8_+_20078070 | 0.00 |
ENSMUST00000084046.6
|
Gm15319
|
predicted gene 15319 |
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.0 | 4.1 | GO:0014734 | skeletal muscle hypertrophy(GO:0014734) |
| 0.6 | 2.2 | GO:0017055 | negative regulation of RNA polymerase II transcriptional preinitiation complex assembly(GO:0017055) |
| 0.5 | 1.6 | GO:0045074 | interleukin-10 biosynthetic process(GO:0042091) regulation of interleukin-10 biosynthetic process(GO:0045074) |
| 0.5 | 2.0 | GO:0060988 | lipid tube assembly(GO:0060988) |
| 0.4 | 3.6 | GO:0010936 | negative regulation of macrophage cytokine production(GO:0010936) |
| 0.3 | 1.3 | GO:0090063 | positive regulation of microtubule nucleation(GO:0090063) |
| 0.3 | 1.8 | GO:1904751 | positive regulation of protein localization to nucleolus(GO:1904751) |
| 0.3 | 1.8 | GO:0001992 | regulation of systemic arterial blood pressure by vasopressin(GO:0001992) |
| 0.3 | 1.6 | GO:0031444 | slow-twitch skeletal muscle fiber contraction(GO:0031444) |
| 0.3 | 0.8 | GO:0015882 | L-ascorbic acid transport(GO:0015882) transepithelial L-ascorbic acid transport(GO:0070904) |
| 0.3 | 3.1 | GO:0014824 | artery smooth muscle contraction(GO:0014824) |
| 0.2 | 1.0 | GO:0019919 | peptidyl-arginine methylation, to asymmetrical-dimethyl arginine(GO:0019919) |
| 0.2 | 1.5 | GO:0061368 | behavioral response to chemical pain(GO:0061366) behavioral response to formalin induced pain(GO:0061368) |
| 0.2 | 2.2 | GO:0060907 | positive regulation of macrophage cytokine production(GO:0060907) |
| 0.2 | 2.4 | GO:0019227 | neuronal action potential propagation(GO:0019227) action potential propagation(GO:0098870) |
| 0.1 | 0.4 | GO:0072429 | response to intra-S DNA damage checkpoint signaling(GO:0072429) |
| 0.1 | 0.5 | GO:2000588 | positive regulation of platelet-derived growth factor receptor-beta signaling pathway(GO:2000588) |
| 0.1 | 1.7 | GO:0098970 | receptor diffusion trapping(GO:0098953) postsynaptic neurotransmitter receptor diffusion trapping(GO:0098970) neurotransmitter receptor diffusion trapping(GO:0099628) protein localization to postsynaptic specialization membrane(GO:0099633) neurotransmitter receptor localization to postsynaptic specialization membrane(GO:0099645) |
| 0.1 | 1.5 | GO:0034551 | respiratory chain complex III assembly(GO:0017062) mitochondrial respiratory chain complex III assembly(GO:0034551) |
| 0.1 | 0.6 | GO:0048478 | replication fork protection(GO:0048478) |
| 0.1 | 1.7 | GO:0090336 | positive regulation of brown fat cell differentiation(GO:0090336) |
| 0.1 | 0.9 | GO:0002074 | extraocular skeletal muscle development(GO:0002074) |
| 0.1 | 1.2 | GO:1990118 | sodium ion import across plasma membrane(GO:0098719) sodium ion import into cell(GO:1990118) |
| 0.1 | 0.7 | GO:0098532 | histone H3-K27 trimethylation(GO:0098532) |
| 0.1 | 0.4 | GO:0071718 | sodium-independent icosanoid transport(GO:0071718) |
| 0.1 | 1.7 | GO:2000480 | negative regulation of cAMP-dependent protein kinase activity(GO:2000480) |
| 0.1 | 0.7 | GO:0048207 | vesicle targeting, rough ER to cis-Golgi(GO:0048207) COPII vesicle coating(GO:0048208) |
| 0.1 | 0.8 | GO:1903608 | protein localization to cytoplasmic stress granule(GO:1903608) |
| 0.1 | 0.4 | GO:0036324 | vascular endothelial growth factor receptor-2 signaling pathway(GO:0036324) |
| 0.1 | 0.9 | GO:0001574 | ganglioside biosynthetic process(GO:0001574) |
| 0.1 | 0.4 | GO:0032383 | regulation of intracellular lipid transport(GO:0032377) regulation of intracellular sterol transport(GO:0032380) regulation of intracellular cholesterol transport(GO:0032383) |
| 0.1 | 0.8 | GO:0032482 | Rab protein signal transduction(GO:0032482) |
| 0.1 | 0.4 | GO:0006561 | proline biosynthetic process(GO:0006561) |
| 0.1 | 1.5 | GO:0045760 | positive regulation of action potential(GO:0045760) |
| 0.0 | 0.2 | GO:0097309 | cap1 mRNA methylation(GO:0097309) |
| 0.0 | 0.2 | GO:0098957 | anterograde axonal transport of mitochondrion(GO:0098957) |
| 0.0 | 0.6 | GO:0000012 | single strand break repair(GO:0000012) |
| 0.0 | 1.0 | GO:0034063 | stress granule assembly(GO:0034063) |
| 0.0 | 1.7 | GO:0060065 | uterus development(GO:0060065) |
| 0.0 | 0.4 | GO:0097039 | protein linear polyubiquitination(GO:0097039) |
| 0.0 | 0.3 | GO:1903232 | melanosome assembly(GO:1903232) |
| 0.0 | 0.3 | GO:0070535 | histone H2A K63-linked ubiquitination(GO:0070535) |
| 0.0 | 0.3 | GO:0048842 | positive regulation of axon extension involved in axon guidance(GO:0048842) |
| 0.0 | 0.2 | GO:0001830 | trophectodermal cell fate commitment(GO:0001830) |
| 0.0 | 0.1 | GO:0006269 | DNA replication, synthesis of RNA primer(GO:0006269) |
| 0.0 | 0.2 | GO:0071931 | Cajal body organization(GO:0030576) positive regulation of transcription involved in G1/S transition of mitotic cell cycle(GO:0071931) |
| 0.0 | 0.9 | GO:1902043 | positive regulation of extrinsic apoptotic signaling pathway via death domain receptors(GO:1902043) |
| 0.0 | 0.3 | GO:0090315 | negative regulation of protein targeting to membrane(GO:0090315) |
| 0.0 | 2.2 | GO:0010923 | negative regulation of phosphatase activity(GO:0010923) |
| 0.0 | 0.2 | GO:0021979 | hypothalamus cell differentiation(GO:0021979) |
| 0.0 | 1.3 | GO:0060612 | adipose tissue development(GO:0060612) |
| 0.0 | 0.7 | GO:0045746 | negative regulation of Notch signaling pathway(GO:0045746) |
| 0.0 | 0.3 | GO:0060218 | hematopoietic stem cell differentiation(GO:0060218) |
| 0.0 | 1.5 | GO:0055072 | iron ion homeostasis(GO:0055072) |
| 0.0 | 0.6 | GO:0060325 | face morphogenesis(GO:0060325) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 4.1 | GO:0035748 | myelin sheath abaxonal region(GO:0035748) |
| 0.2 | 0.8 | GO:0071920 | cleavage body(GO:0071920) |
| 0.2 | 4.1 | GO:0044300 | cerebellar mossy fiber(GO:0044300) |
| 0.1 | 1.7 | GO:0005952 | cAMP-dependent protein kinase complex(GO:0005952) |
| 0.1 | 0.5 | GO:0098890 | extrinsic component of postsynaptic membrane(GO:0098890) |
| 0.1 | 1.6 | GO:0005861 | troponin complex(GO:0005861) |
| 0.1 | 1.3 | GO:0005869 | dynactin complex(GO:0005869) |
| 0.1 | 1.8 | GO:0031011 | Ino80 complex(GO:0031011) |
| 0.1 | 0.4 | GO:0048476 | Holliday junction resolvase complex(GO:0048476) |
| 0.1 | 0.4 | GO:0071797 | LUBAC complex(GO:0071797) |
| 0.1 | 6.0 | GO:0005581 | collagen trimer(GO:0005581) |
| 0.0 | 0.7 | GO:0030127 | COPII vesicle coat(GO:0030127) |
| 0.0 | 4.2 | GO:0043198 | dendritic shaft(GO:0043198) |
| 0.0 | 0.2 | GO:0005964 | phosphorylase kinase complex(GO:0005964) |
| 0.0 | 2.6 | GO:0034707 | chloride channel complex(GO:0034707) |
| 0.0 | 0.2 | GO:0008091 | spectrin(GO:0008091) |
| 0.0 | 0.9 | GO:0071565 | nBAF complex(GO:0071565) |
| 0.0 | 1.2 | GO:0055038 | recycling endosome membrane(GO:0055038) |
| 0.0 | 0.7 | GO:0090545 | NuRD complex(GO:0016581) CHD-type complex(GO:0090545) |
| 0.0 | 3.5 | GO:0043195 | terminal bouton(GO:0043195) |
| 0.0 | 0.2 | GO:0097504 | Gemini of coiled bodies(GO:0097504) |
| 0.0 | 0.2 | GO:1904115 | axon cytoplasm(GO:1904115) |
| 0.0 | 0.1 | GO:0005658 | alpha DNA polymerase:primase complex(GO:0005658) |
| 0.0 | 0.9 | GO:0005891 | voltage-gated calcium channel complex(GO:0005891) |
| 0.0 | 1.3 | GO:0009925 | basal plasma membrane(GO:0009925) |
| 0.0 | 1.8 | GO:0005902 | microvillus(GO:0005902) |
| 0.0 | 0.6 | GO:0043596 | nuclear replication fork(GO:0043596) |
| 0.0 | 1.5 | GO:0008076 | voltage-gated potassium channel complex(GO:0008076) |
| 0.0 | 1.1 | GO:0030117 | membrane coat(GO:0030117) coated membrane(GO:0048475) |
| 0.0 | 0.7 | GO:0044295 | axonal growth cone(GO:0044295) |
| 0.0 | 3.6 | GO:0031965 | nuclear membrane(GO:0031965) |
| 0.0 | 0.3 | GO:0031082 | BLOC complex(GO:0031082) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.6 | 4.1 | GO:0032027 | myosin light chain binding(GO:0032027) |
| 0.6 | 2.2 | GO:0002153 | steroid receptor RNA activator RNA binding(GO:0002153) |
| 0.4 | 1.1 | GO:0016492 | G-protein coupled neurotensin receptor activity(GO:0016492) |
| 0.3 | 0.8 | GO:0033677 | DNA/RNA helicase activity(GO:0033677) |
| 0.3 | 0.8 | GO:0070890 | L-ascorbate:sodium symporter activity(GO:0008520) L-ascorbic acid transporter activity(GO:0015229) sodium-dependent L-ascorbate transmembrane transporter activity(GO:0070890) |
| 0.2 | 1.7 | GO:0031811 | G-protein coupled nucleotide receptor binding(GO:0031811) P2Y1 nucleotide receptor binding(GO:0031812) |
| 0.2 | 1.8 | GO:0010521 | telomerase inhibitor activity(GO:0010521) |
| 0.2 | 0.7 | GO:0061628 | H3K27me3 modified histone binding(GO:0061628) |
| 0.2 | 1.8 | GO:0005000 | vasopressin receptor activity(GO:0005000) |
| 0.2 | 1.6 | GO:0031014 | troponin T binding(GO:0031014) |
| 0.1 | 1.2 | GO:0015386 | potassium:proton antiporter activity(GO:0015386) |
| 0.1 | 1.7 | GO:0008603 | cAMP-dependent protein kinase regulator activity(GO:0008603) |
| 0.1 | 2.4 | GO:0005247 | voltage-gated chloride channel activity(GO:0005247) |
| 0.1 | 1.0 | GO:0035242 | protein-arginine omega-N asymmetric methyltransferase activity(GO:0035242) |
| 0.1 | 0.9 | GO:0001665 | alpha-N-acetylgalactosaminide alpha-2,6-sialyltransferase activity(GO:0001665) |
| 0.1 | 0.4 | GO:0004735 | pyrroline-5-carboxylate reductase activity(GO:0004735) |
| 0.1 | 0.3 | GO:0005093 | Rab GDP-dissociation inhibitor activity(GO:0005093) |
| 0.1 | 1.6 | GO:0055106 | ubiquitin-protein transferase regulator activity(GO:0055106) |
| 0.1 | 0.6 | GO:0031821 | G-protein coupled serotonin receptor binding(GO:0031821) |
| 0.1 | 0.7 | GO:0047134 | protein-disulfide reductase activity(GO:0047134) |
| 0.1 | 0.3 | GO:0035373 | chondroitin sulfate proteoglycan binding(GO:0035373) |
| 0.1 | 1.9 | GO:0048156 | tau protein binding(GO:0048156) |
| 0.1 | 0.4 | GO:0048256 | flap endonuclease activity(GO:0048256) |
| 0.1 | 0.6 | GO:0036310 | annealing helicase activity(GO:0036310) |
| 0.1 | 0.8 | GO:0005068 | transmembrane receptor protein tyrosine kinase adaptor activity(GO:0005068) |
| 0.0 | 0.4 | GO:0035662 | Toll-like receptor 4 binding(GO:0035662) |
| 0.0 | 0.5 | GO:0032051 | clathrin light chain binding(GO:0032051) |
| 0.0 | 0.1 | GO:0017116 | single-stranded DNA-dependent ATP-dependent DNA helicase activity(GO:0017116) |
| 0.0 | 1.5 | GO:0030676 | Rac guanyl-nucleotide exchange factor activity(GO:0030676) |
| 0.0 | 0.2 | GO:0004483 | mRNA (nucleoside-2'-O-)-methyltransferase activity(GO:0004483) |
| 0.0 | 1.3 | GO:0070840 | dynein complex binding(GO:0070840) |
| 0.0 | 1.4 | GO:0004012 | phospholipid-translocating ATPase activity(GO:0004012) |
| 0.0 | 0.9 | GO:0008331 | high voltage-gated calcium channel activity(GO:0008331) |
| 0.0 | 0.2 | GO:0004689 | phosphorylase kinase activity(GO:0004689) |
| 0.0 | 3.1 | GO:0035254 | glutamate receptor binding(GO:0035254) |
| 0.0 | 0.3 | GO:0017151 | DEAD/H-box RNA helicase binding(GO:0017151) |
| 0.0 | 2.2 | GO:0030295 | protein kinase activator activity(GO:0030295) |
| 0.0 | 0.1 | GO:0000010 | trans-hexaprenyltranstransferase activity(GO:0000010) trans-octaprenyltranstransferase activity(GO:0050347) |
| 0.0 | 0.6 | GO:0070182 | DNA polymerase binding(GO:0070182) |
| 0.0 | 0.4 | GO:0015125 | bile acid transmembrane transporter activity(GO:0015125) |
| 0.0 | 1.5 | GO:0015459 | potassium channel regulator activity(GO:0015459) |
| 0.0 | 0.8 | GO:0005158 | insulin receptor binding(GO:0005158) |
| 0.0 | 1.7 | GO:0005179 | hormone activity(GO:0005179) |
| 0.0 | 0.3 | GO:0070530 | K63-linked polyubiquitin binding(GO:0070530) |
| 0.0 | 0.2 | GO:0050811 | GABA receptor binding(GO:0050811) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 6.0 | NABA COLLAGENS | Genes encoding collagen proteins |
| 0.0 | 1.7 | ST GA12 PATHWAY | G alpha 12 Pathway |
| 0.0 | 1.5 | PID ARF6 DOWNSTREAM PATHWAY | Arf6 downstream pathway |
| 0.0 | 1.7 | PID HDAC CLASSIII PATHWAY | Signaling events mediated by HDAC Class III |
| 0.0 | 0.6 | PID S1P S1P4 PATHWAY | S1P4 pathway |
| 0.0 | 2.2 | PID RXR VDR PATHWAY | RXR and RAR heterodimerization with other nuclear receptor |
| 0.0 | 1.7 | PID INTEGRIN A4B1 PATHWAY | Alpha4 beta1 integrin signaling events |
| 0.0 | 1.3 | PID NCADHERIN PATHWAY | N-cadherin signaling events |
| 0.0 | 2.0 | PID ARF6 TRAFFICKING PATHWAY | Arf6 trafficking events |
| 0.0 | 0.7 | PID WNT CANONICAL PATHWAY | Canonical Wnt signaling pathway |
| 0.0 | 1.1 | ST G ALPHA I PATHWAY | G alpha i Pathway |
| 0.0 | 0.8 | PID RET PATHWAY | Signaling events regulated by Ret tyrosine kinase |
| 0.0 | 0.8 | PID ANGIOPOIETIN RECEPTOR PATHWAY | Angiopoietin receptor Tie2-mediated signaling |
| 0.0 | 0.2 | ST GAQ PATHWAY | G alpha q Pathway |
| 0.0 | 1.0 | PID MYC REPRESS PATHWAY | Validated targets of C-MYC transcriptional repression |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 3.1 | REACTOME TRAFFICKING OF GLUR2 CONTAINING AMPA RECEPTORS | Genes involved in Trafficking of GluR2-containing AMPA receptors |
| 0.1 | 1.3 | REACTOME NEF MEDIATED DOWNREGULATION OF MHC CLASS I COMPLEX CELL SURFACE EXPRESSION | Genes involved in Nef mediated downregulation of MHC class I complex cell surface expression |
| 0.1 | 1.1 | REACTOME G ALPHA Z SIGNALLING EVENTS | Genes involved in G alpha (z) signalling events |
| 0.1 | 4.1 | REACTOME COLLAGEN FORMATION | Genes involved in Collagen formation |
| 0.1 | 1.7 | REACTOME PKA MEDIATED PHOSPHORYLATION OF CREB | Genes involved in PKA-mediated phosphorylation of CREB |
| 0.0 | 0.4 | REACTOME TRANSPORT OF ORGANIC ANIONS | Genes involved in Transport of organic anions |
| 0.0 | 0.8 | REACTOME TIE2 SIGNALING | Genes involved in Tie2 Signaling |
| 0.0 | 1.6 | REACTOME STRIATED MUSCLE CONTRACTION | Genes involved in Striated Muscle Contraction |
| 0.0 | 1.4 | REACTOME MITOCHONDRIAL PROTEIN IMPORT | Genes involved in Mitochondrial Protein Import |
| 0.0 | 0.4 | REACTOME ABCA TRANSPORTERS IN LIPID HOMEOSTASIS | Genes involved in ABCA transporters in lipid homeostasis |
| 0.0 | 1.3 | REACTOME ACTIVATION OF CHAPERONE GENES BY XBP1S | Genes involved in Activation of Chaperone Genes by XBP1(S) |
| 0.0 | 0.9 | REACTOME NCAM1 INTERACTIONS | Genes involved in NCAM1 interactions |
| 0.0 | 0.5 | REACTOME DEPOSITION OF NEW CENPA CONTAINING NUCLEOSOMES AT THE CENTROMERE | Genes involved in Deposition of New CENPA-containing Nucleosomes at the Centromere |
| 0.0 | 2.2 | REACTOME NUCLEAR RECEPTOR TRANSCRIPTION PATHWAY | Genes involved in Nuclear Receptor transcription pathway |
| 0.0 | 0.6 | REACTOME DOUBLE STRAND BREAK REPAIR | Genes involved in Double-Strand Break Repair |
| 0.0 | 4.5 | REACTOME G ALPHA Q SIGNALLING EVENTS | Genes involved in G alpha (q) signalling events |
| 0.0 | 0.6 | REACTOME PLC BETA MEDIATED EVENTS | Genes involved in PLC beta mediated events |
| 0.0 | 0.2 | REACTOME GLYCOGEN BREAKDOWN GLYCOGENOLYSIS | Genes involved in Glycogen breakdown (glycogenolysis) |
| 0.0 | 0.3 | REACTOME OTHER SEMAPHORIN INTERACTIONS | Genes involved in Other semaphorin interactions |