Project
PRJNA375882: Comprehensive Mouse Transcriptomic BodyMap
Navigation
Downloads
Results for Pou2f1
Z-value: 2.29
Transcription factors associated with Pou2f1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Pou2f1
|
ENSMUSG00000026565.19 | Pou2f1 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Pou2f1 | mm39_v1_chr1_-_165762469_165762605 | 0.04 | 7.4e-01 | Click! |
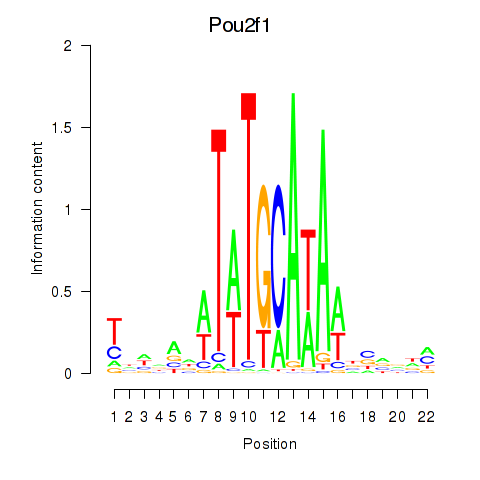
Activity profile of Pou2f1 motif
Sorted Z-values of Pou2f1 motif
Network of associatons between targets according to the STRING database.
First level regulatory network of Pou2f1
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr12_-_114752425 | 22.58 |
ENSMUST00000103510.2
|
Ighv1-26
|
immunoglobulin heavy variable 1-26 |
| chr12_-_115122455 | 20.79 |
ENSMUST00000103523.2
|
Ighv1-53
|
immunoglobulin heavy variable 1-53 |
| chr12_-_115172211 | 19.74 |
ENSMUST00000103526.3
|
Ighv1-55
|
immunoglobulin heavy variable 1-55 |
| chr12_-_113958518 | 19.32 |
ENSMUST00000103467.2
|
Ighv14-2
|
immunoglobulin heavy variable 14-2 |
| chr12_-_114547622 | 19.17 |
ENSMUST00000193893.6
ENSMUST00000103498.3 |
Ighv1-9
|
immunoglobulin heavy variable V1-9 |
| chr6_+_68026941 | 18.13 |
ENSMUST00000103316.2
|
Igkv9-120
|
immunoglobulin kappa chain variable 9-120 |
| chr12_-_114878652 | 17.84 |
ENSMUST00000103515.2
|
Ighv1-39
|
immunoglobulin heavy variable 1-39 |
| chr6_-_69704122 | 17.73 |
ENSMUST00000103364.3
|
Igkv5-48
|
immunoglobulin kappa variable 5-48 |
| chr12_-_114023935 | 17.70 |
ENSMUST00000103469.4
|
Ighv14-3
|
immunoglobulin heavy variable V14-3 |
| chr12_-_115916604 | 17.65 |
ENSMUST00000196991.2
|
Ighv1-82
|
immunoglobulin heavy variable 1-82 |
| chr6_-_69877961 | 17.24 |
ENSMUST00000197290.2
|
Igkv5-39
|
immunoglobulin kappa variable 5-39 |
| chr12_-_115471634 | 16.82 |
ENSMUST00000103535.3
|
Ighv1-64
|
immunoglobulin heavy variable 1-64 |
| chr6_-_69800923 | 16.78 |
ENSMUST00000103368.3
|
Igkv5-43
|
immunoglobulin kappa chain variable 5-43 |
| chr12_-_114710326 | 16.46 |
ENSMUST00000103507.2
|
Ighv1-22
|
immunoglobulin heavy variable 1-22 |
| chr6_-_67919524 | 15.59 |
ENSMUST00000196768.2
|
Igkv9-124
|
immunoglobulin kappa chain variable 9-124 |
| chr6_-_68713748 | 15.08 |
ENSMUST00000183936.2
ENSMUST00000196863.2 |
Igkv19-93
|
immunoglobulin kappa chain variable 19-93 |
| chr12_-_114502585 | 14.55 |
ENSMUST00000103496.4
|
Ighv1-7
|
immunoglobulin heavy variable V1-7 |
| chr14_-_68819544 | 14.20 |
ENSMUST00000022641.9
|
Adamdec1
|
ADAM-like, decysin 1 |
| chr12_-_115876396 | 14.19 |
ENSMUST00000103547.2
|
Ighv1-80
|
immunoglobulin heavy variable 1-80 |
| chr12_-_113649535 | 14.11 |
ENSMUST00000103449.4
ENSMUST00000195707.3 |
Ighv2-5
|
immunoglobulin heavy variable 2-5 |
| chr12_-_114579938 | 14.01 |
ENSMUST00000195469.6
ENSMUST00000109711.4 |
Ighv1-12
|
immunoglobulin heavy variable V1-12 |
| chr12_-_114140482 | 14.00 |
ENSMUST00000103475.2
ENSMUST00000195706.2 |
Ighv14-4
|
immunoglobulin heavy variable 14-4 |
| chr6_+_70648743 | 13.86 |
ENSMUST00000103401.3
|
Igkv3-4
|
immunoglobulin kappa variable 3-4 |
| chr12_-_115109539 | 13.25 |
ENSMUST00000192554.6
ENSMUST00000103522.3 |
Ighv1-52
|
immunoglobulin heavy variable 1-52 |
| chr12_-_114621406 | 12.98 |
ENSMUST00000192077.2
|
Ighv1-15
|
immunoglobulin heavy variable 1-15 |
| chr4_+_11758147 | 12.81 |
ENSMUST00000029871.12
ENSMUST00000108303.2 |
Cdh17
|
cadherin 17 |
| chr3_-_144555062 | 12.77 |
ENSMUST00000159989.2
|
Clca3b
|
chloride channel accessory 3B |
| chr12_-_115798038 | 12.65 |
ENSMUST00000103544.3
|
Ighv1-75
|
immunoglobulin heavy variable 1-75 |
| chr6_-_69415741 | 12.63 |
ENSMUST00000103354.3
|
Igkv4-59
|
immunoglobulin kappa variable 4-59 |
| chr7_-_25176959 | 12.58 |
ENSMUST00000098668.3
ENSMUST00000206687.2 ENSMUST00000206676.2 ENSMUST00000205308.2 ENSMUST00000098669.8 ENSMUST00000206171.2 ENSMUST00000098666.9 |
Ceacam1
|
carcinoembryonic antigen-related cell adhesion molecule 1 |
| chr6_-_69741999 | 12.33 |
ENSMUST00000103365.3
|
Igkv12-46
|
immunoglobulin kappa variable 12-46 |
| chr12_-_115323192 | 12.29 |
ENSMUST00000103531.4
|
Ighv1-61
|
immunoglobulin heavy variable 1-61 |
| chr12_-_114672701 | 11.98 |
ENSMUST00000103505.3
ENSMUST00000193855.2 |
Ighv1-19
|
immunoglobulin heavy variable V1-19 |
| chr12_-_115766700 | 11.65 |
ENSMUST00000196587.5
ENSMUST00000103543.3 |
Ighv1-74
|
immunoglobulin heavy variable V1-74 |
| chr3_+_132335575 | 11.58 |
ENSMUST00000212804.2
ENSMUST00000212852.2 |
Gimd1
|
GIMAP family P-loop NTPase domain containing 1 |
| chr6_-_69877642 | 11.05 |
ENSMUST00000103370.3
|
Igkv5-39
|
immunoglobulin kappa variable 5-39 |
| chr17_-_33136277 | 11.01 |
ENSMUST00000234538.2
ENSMUST00000235058.2 ENSMUST00000234759.2 ENSMUST00000179434.8 ENSMUST00000234797.2 |
Cyp4f14
|
cytochrome P450, family 4, subfamily f, polypeptide 14 |
| chr7_+_43284131 | 10.99 |
ENSMUST00000032663.10
|
Ceacam18
|
carcinoembryonic antigen-related cell adhesion molecule 18 |
| chr6_-_69584812 | 10.98 |
ENSMUST00000103359.3
|
Igkv4-55
|
immunoglobulin kappa variable 4-55 |
| chr12_-_115425105 | 10.67 |
ENSMUST00000103532.3
|
Ighv1-62-3
|
immunoglobulin heavy variable 1-62-3 |
| chr12_-_114579763 | 10.61 |
ENSMUST00000103500.2
|
Ighv1-12
|
immunoglobulin heavy variable V1-12 |
| chr6_-_69553484 | 10.60 |
ENSMUST00000103357.4
|
Igkv4-57
|
immunoglobulin kappa variable 4-57 |
| chr12_-_115811951 | 10.52 |
ENSMUST00000197537.2
|
Ighv1-76
|
immunoglobulin heavy variable 1-76 |
| chr6_+_40619913 | 10.33 |
ENSMUST00000238599.2
|
Mgam
|
maltase-glucoamylase |
| chr12_-_115706126 | 10.29 |
ENSMUST00000166645.2
ENSMUST00000196690.2 |
Ighv1-71
|
immunoglobulin heavy variable 1-71 |
| chr6_-_69835868 | 10.28 |
ENSMUST00000103369.2
|
Igkv12-41
|
immunoglobulin kappa chain variable 12-41 |
| chr6_-_69282389 | 10.23 |
ENSMUST00000103350.3
|
Igkv4-68
|
immunoglobulin kappa variable 4-68 |
| chr12_-_114477427 | 10.13 |
ENSMUST00000191803.2
|
Ighv1-5
|
immunoglobulin heavy variable V1-5 |
| chr6_-_69245427 | 10.00 |
ENSMUST00000103348.3
|
Igkv4-70
|
immunoglobulin kappa chain variable 4-70 |
| chr12_-_115083839 | 9.80 |
ENSMUST00000103521.3
|
Ighv1-50
|
immunoglobulin heavy variable 1-50 |
| chr8_+_21681630 | 9.80 |
ENSMUST00000098896.5
|
Defa31
|
defensin, alpha, 31 |
| chr12_-_115299134 | 9.64 |
ENSMUST00000195359.6
ENSMUST00000103530.3 |
Ighv1-59
|
immunoglobulin heavy variable V1-59 |
| chr7_-_25239229 | 9.53 |
ENSMUST00000044547.10
ENSMUST00000066503.14 ENSMUST00000064862.13 |
Ceacam2
|
carcinoembryonic antigen-related cell adhesion molecule 2 |
| chr12_-_114443071 | 9.31 |
ENSMUST00000103492.2
|
Ighv10-1
|
immunoglobulin heavy variable 10-1 |
| chr1_-_138102972 | 9.28 |
ENSMUST00000195533.6
ENSMUST00000183301.8 |
Ptprc
|
protein tyrosine phosphatase, receptor type, C |
| chr12_-_114815280 | 9.21 |
ENSMUST00000103512.3
|
Ighv1-34
|
immunoglobulin heavy variable 1-34 |
| chr6_+_68279392 | 9.20 |
ENSMUST00000103322.3
|
Igkv2-109
|
immunoglobulin kappa variable 2-109 |
| chr6_-_68907718 | 9.16 |
ENSMUST00000114212.4
|
Igkv13-85
|
immunoglobulin kappa chain variable 13-85 |
| chr1_+_173458458 | 9.01 |
ENSMUST00000056071.13
|
Ifi209
|
interferon activated gene 209 |
| chr17_-_33136021 | 8.98 |
ENSMUST00000054174.9
|
Cyp4f14
|
cytochrome P450, family 4, subfamily f, polypeptide 14 |
| chr12_-_115944754 | 8.97 |
ENSMUST00000103551.2
|
Ighv1-84
|
immunoglobulin heavy variable 1-84 |
| chr6_-_41012435 | 8.83 |
ENSMUST00000031931.6
|
2210010C04Rik
|
RIKEN cDNA 2210010C04 gene |
| chr1_-_138103021 | 8.83 |
ENSMUST00000182755.8
ENSMUST00000193650.2 ENSMUST00000182283.8 |
Ptprc
|
protein tyrosine phosphatase, receptor type, C |
| chr10_-_21036792 | 8.69 |
ENSMUST00000188495.8
|
Myb
|
myeloblastosis oncogene |
| chr12_-_114646685 | 8.63 |
ENSMUST00000194350.6
ENSMUST00000103504.3 |
Ighv1-18
|
immunoglobulin heavy variable V1-18 |
| chr6_+_41515152 | 8.62 |
ENSMUST00000103291.2
ENSMUST00000192856.6 |
Trbc1
|
T cell receptor beta, constant region 1 |
| chr6_-_69678271 | 8.58 |
ENSMUST00000103363.2
|
Igkv4-50
|
immunoglobulin kappa variable 4-50 |
| chr6_-_69753317 | 8.58 |
ENSMUST00000103366.3
|
Igkv5-45
|
immunoglobulin kappa chain variable 5-45 |
| chr6_-_68994064 | 8.54 |
ENSMUST00000103341.4
|
Igkv4-80
|
immunoglobulin kappa variable 4-80 |
| chr12_-_115587215 | 8.36 |
ENSMUST00000199933.5
ENSMUST00000103539.3 |
Ighv1-69
|
immunoglobulin heavy variable 1-69 |
| chr6_-_70036183 | 8.34 |
ENSMUST00000197429.5
ENSMUST00000103376.3 |
Igkv7-33
|
immunoglobulin kappa chain variable 7-33 |
| chr12_-_114451189 | 8.17 |
ENSMUST00000103493.3
|
Ighv1-4
|
immunoglobulin heavy variable 1-4 |
| chr6_-_68840015 | 8.12 |
ENSMUST00000103336.2
|
Igkv1-88
|
immunoglobulin kappa chain variable 1-88 |
| chr3_-_137837117 | 7.84 |
ENSMUST00000029805.13
|
Mttp
|
microsomal triglyceride transfer protein |
| chr10_-_21036824 | 7.82 |
ENSMUST00000020158.9
|
Myb
|
myeloblastosis oncogene |
| chr17_+_56056977 | 7.59 |
ENSMUST00000025004.7
|
Adgre4
|
adhesion G protein-coupled receptor E4 |
| chr6_-_70313491 | 7.50 |
ENSMUST00000103388.4
|
Igkv6-20
|
immunoglobulin kappa variable 6-20 |
| chr5_+_96104775 | 7.34 |
ENSMUST00000023840.7
|
Cxcl13
|
chemokine (C-X-C motif) ligand 13 |
| chr19_+_38995463 | 7.30 |
ENSMUST00000025966.5
|
Cyp2c55
|
cytochrome P450, family 2, subfamily c, polypeptide 55 |
| chr4_+_134238310 | 7.04 |
ENSMUST00000105866.3
|
Aunip
|
aurora kinase A and ninein interacting protein |
| chr12_-_115276219 | 6.88 |
ENSMUST00000103529.4
|
Ighv1-58
|
immunoglobulin heavy variable 1-58 |
| chr12_-_115410489 | 6.50 |
ENSMUST00000194581.2
|
Ighv1-62-2
|
immunoglobulin heavy variable 1-62-2 |
| chr7_+_19741948 | 6.34 |
ENSMUST00000073151.13
|
Nlrp9b
|
NLR family, pyrin domain containing 9B |
| chr7_+_24476597 | 6.29 |
ENSMUST00000038069.9
ENSMUST00000206847.2 |
Ceacam10
|
carcinoembryonic antigen-related cell adhesion molecule 10 |
| chr10_-_102326286 | 6.29 |
ENSMUST00000020040.5
|
Nts
|
neurotensin |
| chr12_-_115459678 | 6.19 |
ENSMUST00000103534.2
|
Ighv1-63
|
immunoglobulin heavy variable V1-63 |
| chr6_-_70116066 | 6.18 |
ENSMUST00000103379.3
ENSMUST00000197371.2 |
Igkv6-29
|
immunoglobulin kappa chain variable 6-29 |
| chr12_-_115964081 | 6.12 |
ENSMUST00000103552.2
|
Ighv1-85
|
immunoglobulin heavy variable 1-85 |
| chr12_-_114843941 | 6.04 |
ENSMUST00000191862.6
ENSMUST00000103513.3 |
Ighv1-36
|
immunoglobulin heavy variable 1-36 |
| chr1_+_67162176 | 5.84 |
ENSMUST00000027144.8
|
Cps1
|
carbamoyl-phosphate synthetase 1 |
| chr12_-_107969853 | 5.74 |
ENSMUST00000066060.11
|
Bcl11b
|
B cell leukemia/lymphoma 11B |
| chr9_+_53678801 | 5.65 |
ENSMUST00000048670.10
|
Slc35f2
|
solute carrier family 35, member F2 |
| chr6_+_70680515 | 5.59 |
ENSMUST00000103404.2
|
Igkv3-1
|
immunoglobulin kappa variable 3-1 |
| chr6_-_68887957 | 5.54 |
ENSMUST00000200454.2
|
Igkv4-86
|
immunoglobulin kappa variable 4-86 |
| chr6_-_69162381 | 5.50 |
ENSMUST00000103344.3
|
Igkv4-74
|
immunoglobulin kappa variable 4-74 |
| chr12_-_107969673 | 5.39 |
ENSMUST00000109887.8
ENSMUST00000109891.3 |
Bcl11b
|
B cell leukemia/lymphoma 11B |
| chr8_+_47070326 | 5.21 |
ENSMUST00000211115.2
ENSMUST00000093517.7 |
Casp3
|
caspase 3 |
| chr11_+_70396070 | 4.99 |
ENSMUST00000019063.3
|
Tm4sf5
|
transmembrane 4 superfamily member 5 |
| chr3_-_88366351 | 4.96 |
ENSMUST00000165898.8
ENSMUST00000127436.8 |
Sema4a
|
sema domain, immunoglobulin domain (Ig), transmembrane domain (TM) and short cytoplasmic domain, (semaphorin) 4A |
| chr12_-_114073050 | 4.93 |
ENSMUST00000103472.4
|
Ighv9-2
|
immunoglobulin heavy variable V9-2 |
| chr6_-_68732577 | 4.93 |
ENSMUST00000103332.2
|
Igkv4-92
|
immunoglobulin kappa variable 4-92 |
| chr14_-_44057096 | 4.92 |
ENSMUST00000100691.4
|
Ear1
|
eosinophil-associated, ribonuclease A family, member 1 |
| chr6_-_129600798 | 4.90 |
ENSMUST00000095412.10
|
Klrk1
|
killer cell lectin-like receptor subfamily K, member 1 |
| chr6_+_41090484 | 4.63 |
ENSMUST00000103267.3
|
Trbv12-1
|
T cell receptor beta, variable 12-1 |
| chr6_-_129600812 | 4.60 |
ENSMUST00000168919.8
|
Klrk1
|
killer cell lectin-like receptor subfamily K, member 1 |
| chr4_-_46413484 | 4.58 |
ENSMUST00000071096.3
|
Hemgn
|
hemogen |
| chr10_-_88520877 | 4.53 |
ENSMUST00000138734.2
|
Spic
|
Spi-C transcription factor (Spi-1/PU.1 related) |
| chr12_-_114793177 | 4.52 |
ENSMUST00000103511.2
ENSMUST00000195735.2 |
Ighv1-31
|
immunoglobulin heavy variable 1-31 |
| chr10_+_26105605 | 4.16 |
ENSMUST00000218301.2
ENSMUST00000164660.8 ENSMUST00000060716.6 |
Samd3
|
sterile alpha motif domain containing 3 |
| chr6_+_129385893 | 4.15 |
ENSMUST00000204860.3
ENSMUST00000164513.8 |
Clec9a
|
C-type lectin domain family 9, member a |
| chr19_-_10952009 | 4.15 |
ENSMUST00000191343.7
|
Ms4a10
|
membrane-spanning 4-domains, subfamily A, member 10 |
| chr3_+_97536120 | 4.14 |
ENSMUST00000107050.8
ENSMUST00000029729.15 ENSMUST00000107049.2 |
Fmo5
|
flavin containing monooxygenase 5 |
| chr1_-_144651157 | 4.09 |
ENSMUST00000027603.4
|
Rgs18
|
regulator of G-protein signaling 18 |
| chr2_-_120867232 | 4.05 |
ENSMUST00000023987.6
|
Epb42
|
erythrocyte membrane protein band 4.2 |
| chr9_+_50476637 | 4.00 |
ENSMUST00000214117.2
|
Il18
|
interleukin 18 |
| chr10_+_101994841 | 3.86 |
ENSMUST00000020039.13
|
Mgat4c
|
MGAT4 family, member C |
| chr6_-_69377328 | 3.86 |
ENSMUST00000198345.2
|
Igkv4-62
|
immunoglobulin kappa variable 4-62 |
| chr6_+_108805594 | 3.84 |
ENSMUST00000089162.5
|
Edem1
|
ER degradation enhancer, mannosidase alpha-like 1 |
| chr6_+_67768007 | 3.78 |
ENSMUST00000196006.5
ENSMUST00000103307.3 |
Igkv14-130
|
immunoglobulin kappa variable 14-130 |
| chr5_+_91222470 | 3.77 |
ENSMUST00000031324.6
|
Ereg
|
epiregulin |
| chrX_+_84913911 | 3.70 |
ENSMUST00000113976.2
|
Tasl
|
TLR adaptor interacting with endolysosomal SLC15A4 |
| chr6_+_67890534 | 3.65 |
ENSMUST00000197406.5
ENSMUST00000103311.3 |
Igkv11-125
|
immunoglobulin kappa variable 11-125 |
| chr18_+_56565188 | 3.63 |
ENSMUST00000070166.6
|
Gramd3
|
GRAM domain containing 3 |
| chr8_-_72178340 | 3.60 |
ENSMUST00000153800.8
ENSMUST00000146100.8 |
Fcho1
|
FCH domain only 1 |
| chr14_+_44340111 | 3.58 |
ENSMUST00000074839.7
|
Ear2
|
eosinophil-associated, ribonuclease A family, member 2 |
| chr12_-_114955196 | 3.56 |
ENSMUST00000194865.2
|
Ighv1-47
|
immunoglobulin heavy variable 1-47 |
| chr14_-_44112974 | 3.56 |
ENSMUST00000179200.2
|
Ear1
|
eosinophil-associated, ribonuclease A family, member 1 |
| chr6_+_129157576 | 3.38 |
ENSMUST00000032260.6
|
Clec2d
|
C-type lectin domain family 2, member d |
| chr10_+_101994719 | 3.37 |
ENSMUST00000138522.8
ENSMUST00000163753.8 ENSMUST00000138016.8 |
Mgat4c
|
MGAT4 family, member C |
| chr8_-_47070201 | 3.33 |
ENSMUST00000210264.2
ENSMUST00000040468.16 ENSMUST00000209787.2 |
Primpol
|
primase and polymerase (DNA-directed) |
| chr10_+_75409282 | 3.30 |
ENSMUST00000006508.10
|
Ggt1
|
gamma-glutamyltransferase 1 |
| chr15_-_57982705 | 3.27 |
ENSMUST00000228783.2
|
Atad2
|
ATPase family, AAA domain containing 2 |
| chr8_-_61407760 | 3.27 |
ENSMUST00000110302.8
|
Clcn3
|
chloride channel, voltage-sensitive 3 |
| chr16_+_36755338 | 3.25 |
ENSMUST00000023531.15
|
Hcls1
|
hematopoietic cell specific Lyn substrate 1 |
| chr4_+_135413593 | 3.23 |
ENSMUST00000074408.7
|
Ifnlr1
|
interferon lambda receptor 1 |
| chr14_-_66071412 | 3.19 |
ENSMUST00000022613.10
|
Esco2
|
establishment of sister chromatid cohesion N-acetyltransferase 2 |
| chr1_-_128520002 | 3.05 |
ENSMUST00000052172.7
ENSMUST00000142893.2 |
Cxcr4
|
chemokine (C-X-C motif) receptor 4 |
| chr6_+_129385816 | 2.98 |
ENSMUST00000058352.15
ENSMUST00000088075.8 |
Clec9a
|
C-type lectin domain family 9, member a |
| chr6_-_69261303 | 2.92 |
ENSMUST00000103349.2
|
Igkv4-69
|
immunoglobulin kappa variable 4-69 |
| chr4_-_149221998 | 2.79 |
ENSMUST00000176124.8
ENSMUST00000177408.2 ENSMUST00000105695.2 |
Cenps
|
centromere protein S |
| chr17_+_56610321 | 2.78 |
ENSMUST00000001258.15
|
Uhrf1
|
ubiquitin-like, containing PHD and RING finger domains, 1 |
| chr6_-_67931692 | 2.72 |
ENSMUST00000103313.3
|
Igkv9-123
|
immunoglobulin kappa variable 9-123 |
| chr4_+_101843823 | 2.71 |
ENSMUST00000106914.8
|
Gm12789
|
predicted gene 12789 |
| chr12_-_114687825 | 2.60 |
ENSMUST00000194968.6
ENSMUST00000103506.3 |
Ighv1-20
|
immunoglobulin heavy variable V1-20 |
| chr12_+_98234884 | 2.60 |
ENSMUST00000075072.6
|
Gpr65
|
G-protein coupled receptor 65 |
| chr7_-_126391657 | 2.60 |
ENSMUST00000032936.8
|
Ppp4c
|
protein phosphatase 4, catalytic subunit |
| chr17_+_56610396 | 2.53 |
ENSMUST00000113038.8
|
Uhrf1
|
ubiquitin-like, containing PHD and RING finger domains, 1 |
| chr4_-_149222057 | 2.51 |
ENSMUST00000030813.10
|
Cenps
|
centromere protein S |
| chr6_-_69377081 | 2.50 |
ENSMUST00000177795.2
|
Igkv4-62
|
immunoglobulin kappa variable 4-62 |
| chr9_+_108820846 | 2.50 |
ENSMUST00000198140.5
ENSMUST00000051873.15 |
Pfkfb4
|
6-phosphofructo-2-kinase/fructose-2,6-biphosphatase 4 |
| chr17_+_23945310 | 2.48 |
ENSMUST00000024701.9
|
Pkmyt1
|
protein kinase, membrane associated tyrosine/threonine 1 |
| chr6_-_130258891 | 2.46 |
ENSMUST00000112020.2
|
Klra10
|
killer cell lectin-like receptor subfamily A, member 10 |
| chr14_-_59632830 | 2.43 |
ENSMUST00000166912.3
|
Phf11c
|
PHD finger protein 11C |
| chr6_-_28397997 | 2.42 |
ENSMUST00000035930.11
|
Zfp800
|
zinc finger protein 800 |
| chr7_-_126391388 | 2.41 |
ENSMUST00000206570.2
|
Ppp4c
|
protein phosphatase 4, catalytic subunit |
| chr12_-_111947487 | 2.37 |
ENSMUST00000190536.2
|
Rd3l
|
retinal degeneration 3-like |
| chr5_+_45650821 | 2.27 |
ENSMUST00000198534.2
|
Lap3
|
leucine aminopeptidase 3 |
| chr5_-_87054796 | 2.23 |
ENSMUST00000031181.16
ENSMUST00000113333.2 |
Ugt2b34
|
UDP glucuronosyltransferase 2 family, polypeptide B34 |
| chr6_+_123099615 | 2.14 |
ENSMUST00000161636.8
ENSMUST00000161365.8 |
Clec4a2
|
C-type lectin domain family 4, member a2 |
| chr5_+_45650716 | 2.14 |
ENSMUST00000046122.11
|
Lap3
|
leucine aminopeptidase 3 |
| chr6_-_69609162 | 2.11 |
ENSMUST00000199437.2
|
Igkv4-54
|
immunoglobulin kappa chain variable 4-54 |
| chr12_-_76756772 | 2.10 |
ENSMUST00000166101.2
|
Sptb
|
spectrin beta, erythrocytic |
| chr3_+_118355811 | 2.09 |
ENSMUST00000149101.3
|
Dpyd
|
dihydropyrimidine dehydrogenase |
| chr3_+_94280101 | 2.08 |
ENSMUST00000029795.10
|
Rorc
|
RAR-related orphan receptor gamma |
| chr8_+_66838927 | 2.06 |
ENSMUST00000039540.12
ENSMUST00000110253.3 |
Marchf1
|
membrane associated ring-CH-type finger 1 |
| chr18_+_67338437 | 2.02 |
ENSMUST00000210564.3
|
Chmp1b
|
charged multivesicular body protein 1B |
| chr13_+_19362068 | 1.97 |
ENSMUST00000103553.3
|
Trgv7
|
T cell receptor gamma, variable 7 |
| chr5_-_113970664 | 1.89 |
ENSMUST00000199109.2
|
Selplg
|
selectin, platelet (p-selectin) ligand |
| chr11_-_82910912 | 1.89 |
ENSMUST00000130822.3
|
Slfn8
|
schlafen 8 |
| chr12_-_115567853 | 1.88 |
ENSMUST00000103538.3
ENSMUST00000198646.2 |
Ighv1-67
|
immunoglobulin heavy variable V1-67 |
| chr14_+_53743184 | 1.86 |
ENSMUST00000103583.5
|
Trav10
|
T cell receptor alpha variable 10 |
| chr6_-_87798613 | 1.86 |
ENSMUST00000204169.2
|
Gm45140
|
predicted gene 45140 |
| chr2_+_120331693 | 1.84 |
ENSMUST00000141181.9
|
Capn3
|
calpain 3 |
| chr5_+_117271632 | 1.81 |
ENSMUST00000179276.8
ENSMUST00000092889.12 ENSMUST00000145640.8 |
Taok3
|
TAO kinase 3 |
| chr17_+_29709723 | 1.77 |
ENSMUST00000024811.9
|
Pim1
|
proviral integration site 1 |
| chr14_+_53048391 | 1.75 |
ENSMUST00000103646.5
|
Trav10d
|
T cell receptor alpha variable 10D |
| chrX_-_50770733 | 1.75 |
ENSMUST00000114871.2
|
Hs6st2
|
heparan sulfate 6-O-sulfotransferase 2 |
| chr17_-_40553176 | 1.70 |
ENSMUST00000026499.6
|
Crisp3
|
cysteine-rich secretory protein 3 |
| chr11_-_82911548 | 1.70 |
ENSMUST00000108152.9
|
Slfn8
|
schlafen 8 |
| chr11_-_82911615 | 1.66 |
ENSMUST00000038141.15
ENSMUST00000092838.11 |
Slfn8
|
schlafen 8 |
| chr16_+_21644692 | 1.65 |
ENSMUST00000232240.2
|
Map3k13
|
mitogen-activated protein kinase kinase kinase 13 |
| chr16_-_19241884 | 1.65 |
ENSMUST00000206110.4
|
Olfr165
|
olfactory receptor 165 |
| chrX_+_59044796 | 1.64 |
ENSMUST00000033477.5
|
F9
|
coagulation factor IX |
| chr16_-_19132814 | 1.60 |
ENSMUST00000216157.2
|
Olfr164
|
olfactory receptor 164 |
| chr6_+_67816777 | 1.57 |
ENSMUST00000200578.5
ENSMUST00000103308.3 |
Igkv9-129
|
immunoglobulin kappa variable 9-129 |
| chr10_+_75868432 | 1.55 |
ENSMUST00000218627.2
ENSMUST00000061617.7 |
Zfp280b
|
zinc finger protein 280B |
| chr10_+_81012465 | 1.51 |
ENSMUST00000047864.11
|
Eef2
|
eukaryotic translation elongation factor 2 |
| chr15_-_89263790 | 1.49 |
ENSMUST00000238996.2
|
Odf3b
|
outer dense fiber of sperm tails 3B |
| chr4_-_131695135 | 1.49 |
ENSMUST00000146443.8
ENSMUST00000135579.8 |
Epb41
|
erythrocyte membrane protein band 4.1 |
| chr9_+_96140750 | 1.48 |
ENSMUST00000186609.7
|
Tfdp2
|
transcription factor Dp 2 |
| chr2_+_121340296 | 1.48 |
ENSMUST00000110603.2
|
Wdr76
|
WD repeat domain 76 |
| chr6_-_69220672 | 1.46 |
ENSMUST00000196201.2
|
Igkv4-71
|
immunoglobulin kappa chain variable 4-71 |
| chr8_+_77628916 | 1.46 |
ENSMUST00000109912.8
ENSMUST00000128862.2 ENSMUST00000109911.8 |
Nr3c2
|
nuclear receptor subfamily 3, group C, member 2 |
| chr6_-_41291634 | 1.46 |
ENSMUST00000064324.12
|
Try5
|
trypsin 5 |
| chr14_-_44161016 | 1.44 |
ENSMUST00000159175.2
|
Ear10
|
eosinophil-associated, ribonuclease A family, member 10 |
| chr10_+_94412116 | 1.40 |
ENSMUST00000117929.2
|
Tmcc3
|
transmembrane and coiled coil domains 3 |
| chr14_+_54453748 | 1.40 |
ENSMUST00000103736.2
|
Traj4
|
T cell receptor alpha joining 4 |
| chr4_-_132260799 | 1.37 |
ENSMUST00000152993.8
ENSMUST00000067496.7 |
Atpif1
|
ATPase inhibitory factor 1 |
| chr6_+_129326927 | 1.36 |
ENSMUST00000065289.6
|
Clec12a
|
C-type lectin domain family 12, member a |
| chr4_-_136626073 | 1.35 |
ENSMUST00000046285.6
|
C1qa
|
complement component 1, q subcomponent, alpha polypeptide |
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 4.5 | 18.1 | GO:2000471 | regulation of hematopoietic stem cell migration(GO:2000471) positive regulation of hematopoietic stem cell migration(GO:2000473) |
| 4.3 | 12.8 | GO:0002314 | germinal center B cell differentiation(GO:0002314) |
| 4.2 | 12.6 | GO:0070237 | positive regulation of activation-induced cell death of T cells(GO:0070237) |
| 3.7 | 11.1 | GO:0097534 | lymphoid lineage cell migration(GO:0097534) lymphoid lineage cell migration into thymus(GO:0097535) |
| 3.3 | 490.2 | GO:0006910 | phagocytosis, recognition(GO:0006910) |
| 3.2 | 9.5 | GO:0030887 | positive regulation of myeloid dendritic cell activation(GO:0030887) |
| 2.0 | 7.8 | GO:0034196 | acylglycerol transport(GO:0034196) triglyceride transport(GO:0034197) |
| 1.8 | 7.3 | GO:2001027 | negative regulation of endothelial cell chemotaxis(GO:2001027) |
| 1.3 | 3.8 | GO:1904154 | positive regulation of retrograde protein transport, ER to cytosol(GO:1904154) |
| 1.1 | 16.5 | GO:0051574 | positive regulation of histone H3-K9 methylation(GO:0051574) |
| 1.1 | 3.3 | GO:0097401 | synaptic vesicle lumen acidification(GO:0097401) |
| 1.1 | 9.5 | GO:0070345 | negative regulation of fat cell proliferation(GO:0070345) |
| 1.0 | 5.2 | GO:0072733 | response to staurosporine(GO:0072733) cellular response to staurosporine(GO:0072734) |
| 1.0 | 3.1 | GO:0035470 | positive regulation of vascular wound healing(GO:0035470) |
| 1.0 | 4.0 | GO:0042231 | interleukin-13 biosynthetic process(GO:0042231) |
| 1.0 | 254.9 | GO:0002377 | immunoglobulin production(GO:0002377) |
| 0.9 | 4.3 | GO:0034421 | post-translational protein acetylation(GO:0034421) |
| 0.8 | 5.8 | GO:0019856 | 'de novo' pyrimidine nucleobase biosynthetic process(GO:0006207) pyrimidine nucleobase biosynthetic process(GO:0019856) |
| 0.8 | 3.8 | GO:0001550 | ovarian cumulus expansion(GO:0001550) fused antrum stage(GO:0048165) |
| 0.7 | 2.1 | GO:0006212 | uracil catabolic process(GO:0006212) uracil metabolic process(GO:0019860) |
| 0.6 | 1.9 | GO:0000494 | box C/D snoRNA 3'-end processing(GO:0000494) box C/D snoRNA metabolic process(GO:0033967) box C/D snoRNA processing(GO:0034963) histone glutamine methylation(GO:1990258) |
| 0.5 | 6.4 | GO:0043651 | linoleic acid metabolic process(GO:0043651) |
| 0.5 | 3.3 | GO:0031936 | negative regulation of chromatin silencing(GO:0031936) |
| 0.5 | 1.8 | GO:1990091 | sodium-dependent self proteolysis(GO:1990091) |
| 0.4 | 3.3 | GO:0031179 | peptide modification(GO:0031179) leukotriene D4 metabolic process(GO:1901748) leukotriene D4 biosynthetic process(GO:1901750) |
| 0.4 | 2.0 | GO:0035660 | MyD88-dependent toll-like receptor 4 signaling pathway(GO:0035660) |
| 0.4 | 1.9 | GO:0050902 | leukocyte adhesive activation(GO:0050902) |
| 0.4 | 1.5 | GO:0031959 | mineralocorticoid receptor signaling pathway(GO:0031959) |
| 0.4 | 0.4 | GO:2000412 | positive regulation of thymocyte migration(GO:2000412) |
| 0.3 | 5.3 | GO:0090308 | regulation of methylation-dependent chromatin silencing(GO:0090308) |
| 0.3 | 4.8 | GO:0000712 | resolution of meiotic recombination intermediates(GO:0000712) |
| 0.3 | 1.7 | GO:0015015 | heparan sulfate proteoglycan biosynthetic process, enzymatic modification(GO:0015015) |
| 0.2 | 15.9 | GO:0019369 | arachidonic acid metabolic process(GO:0019369) |
| 0.2 | 3.3 | GO:0030854 | positive regulation of granulocyte differentiation(GO:0030854) |
| 0.2 | 1.4 | GO:1904925 | positive regulation of macromitophagy(GO:1901526) positive regulation of mitophagy in response to mitochondrial depolarization(GO:1904925) |
| 0.2 | 1.5 | GO:2000767 | positive regulation of cytoplasmic translation(GO:2000767) |
| 0.2 | 4.1 | GO:0043249 | erythrocyte maturation(GO:0043249) |
| 0.2 | 3.4 | GO:0006968 | cellular defense response(GO:0006968) |
| 0.2 | 3.3 | GO:0019985 | translesion synthesis(GO:0019985) |
| 0.2 | 5.0 | GO:0045063 | T-helper 1 cell differentiation(GO:0045063) |
| 0.2 | 1.0 | GO:0015788 | UDP-N-acetylglucosamine transport(GO:0015788) |
| 0.2 | 2.1 | GO:0072615 | interleukin-17 secretion(GO:0072615) |
| 0.2 | 5.0 | GO:0010569 | regulation of double-strand break repair via homologous recombination(GO:0010569) |
| 0.2 | 9.0 | GO:0035458 | cellular response to interferon-beta(GO:0035458) |
| 0.1 | 1.0 | GO:0046116 | queuosine biosynthetic process(GO:0008616) queuosine metabolic process(GO:0046116) negative regulation of protein refolding(GO:0061084) |
| 0.1 | 1.8 | GO:0070561 | vitamin D receptor signaling pathway(GO:0070561) regulation of hematopoietic stem cell proliferation(GO:1902033) |
| 0.1 | 3.1 | GO:0002251 | organ or tissue specific immune response(GO:0002251) mucosal immune response(GO:0002385) |
| 0.1 | 0.5 | GO:0019858 | cytosine metabolic process(GO:0019858) |
| 0.1 | 10.1 | GO:1902476 | chloride transmembrane transport(GO:1902476) |
| 0.1 | 1.4 | GO:0036444 | calcium ion transmembrane import into mitochondrion(GO:0036444) |
| 0.1 | 0.6 | GO:0035617 | stress granule disassembly(GO:0035617) |
| 0.1 | 0.8 | GO:0045200 | establishment or maintenance of neuroblast polarity(GO:0045196) establishment of neuroblast polarity(GO:0045200) |
| 0.1 | 2.0 | GO:1901673 | regulation of mitotic spindle assembly(GO:1901673) |
| 0.1 | 1.1 | GO:0060406 | positive regulation of penile erection(GO:0060406) |
| 0.1 | 0.5 | GO:0071894 | histone H2B conserved C-terminal lysine ubiquitination(GO:0071894) |
| 0.1 | 0.4 | GO:0018343 | protein farnesylation(GO:0018343) |
| 0.1 | 1.7 | GO:2000001 | regulation of DNA damage checkpoint(GO:2000001) |
| 0.1 | 1.8 | GO:0007095 | mitotic G2 DNA damage checkpoint(GO:0007095) |
| 0.1 | 2.6 | GO:0010447 | response to acidic pH(GO:0010447) |
| 0.1 | 0.5 | GO:0051464 | positive regulation of cortisol secretion(GO:0051464) |
| 0.1 | 0.9 | GO:2001288 | positive regulation of caveolin-mediated endocytosis(GO:2001288) |
| 0.1 | 0.8 | GO:0051418 | interphase microtubule nucleation by interphase microtubule organizing center(GO:0051415) microtubule nucleation by microtubule organizing center(GO:0051418) |
| 0.1 | 1.1 | GO:1902231 | positive regulation of intrinsic apoptotic signaling pathway in response to DNA damage(GO:1902231) |
| 0.1 | 0.4 | GO:0034244 | negative regulation of transcription elongation from RNA polymerase II promoter(GO:0034244) |
| 0.1 | 1.2 | GO:0051988 | regulation of attachment of spindle microtubules to kinetochore(GO:0051988) |
| 0.1 | 6.0 | GO:0008542 | visual learning(GO:0008542) |
| 0.0 | 2.1 | GO:0006779 | porphyrin-containing compound biosynthetic process(GO:0006779) |
| 0.0 | 0.2 | GO:2000584 | regulation of platelet-derived growth factor receptor-alpha signaling pathway(GO:2000583) negative regulation of platelet-derived growth factor receptor-alpha signaling pathway(GO:2000584) |
| 0.0 | 0.4 | GO:0046477 | glycosylceramide catabolic process(GO:0046477) |
| 0.0 | 0.3 | GO:2001183 | negative regulation of interleukin-12 secretion(GO:2001183) |
| 0.0 | 0.7 | GO:1904776 | regulation of protein localization to cell cortex(GO:1904776) positive regulation of protein localization to cell cortex(GO:1904778) |
| 0.0 | 0.6 | GO:0043983 | histone H4-K12 acetylation(GO:0043983) |
| 0.0 | 4.9 | GO:0050715 | positive regulation of cytokine secretion(GO:0050715) |
| 0.0 | 4.1 | GO:0008277 | regulation of G-protein coupled receptor protein signaling pathway(GO:0008277) |
| 0.0 | 0.3 | GO:0050957 | equilibrioception(GO:0050957) |
| 0.0 | 0.8 | GO:0032012 | ARF protein signal transduction(GO:0032011) regulation of ARF protein signal transduction(GO:0032012) |
| 0.0 | 1.1 | GO:0033209 | tumor necrosis factor-mediated signaling pathway(GO:0033209) |
| 0.0 | 0.2 | GO:0001682 | tRNA 5'-leader removal(GO:0001682) |
| 0.0 | 4.5 | GO:0042742 | defense response to bacterium(GO:0042742) |
| 0.0 | 0.4 | GO:0031572 | G2 DNA damage checkpoint(GO:0031572) |
| 0.0 | 0.4 | GO:2000269 | regulation of fibroblast apoptotic process(GO:2000269) |
| 0.0 | 0.3 | GO:1904707 | positive regulation of vascular smooth muscle cell proliferation(GO:1904707) |
| 0.0 | 1.2 | GO:0007029 | endoplasmic reticulum organization(GO:0007029) |
| 0.0 | 0.3 | GO:0042640 | anagen(GO:0042640) |
| 0.0 | 0.1 | GO:0042427 | serotonin biosynthetic process(GO:0042427) primary amino compound biosynthetic process(GO:1901162) |
| 0.0 | 0.7 | GO:0018279 | protein N-linked glycosylation via asparagine(GO:0018279) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 4.0 | 493.0 | GO:0042571 | immunoglobulin complex, circulating(GO:0042571) |
| 1.3 | 5.3 | GO:0071821 | FANCM-MHF complex(GO:0071821) |
| 0.7 | 5.0 | GO:0030289 | protein phosphatase 4 complex(GO:0030289) |
| 0.7 | 22.1 | GO:0042101 | T cell receptor complex(GO:0042101) |
| 0.4 | 5.2 | GO:0031264 | death-inducing signaling complex(GO:0031264) |
| 0.3 | 2.1 | GO:0008091 | spectrin(GO:0008091) |
| 0.3 | 7.8 | GO:0031528 | microvillus membrane(GO:0031528) |
| 0.2 | 1.9 | GO:0001652 | granular component(GO:0001652) |
| 0.2 | 4.3 | GO:0031618 | nuclear pericentric heterochromatin(GO:0031618) |
| 0.2 | 1.2 | GO:0097149 | centralspindlin complex(GO:0097149) |
| 0.2 | 1.4 | GO:1990246 | uniplex complex(GO:1990246) |
| 0.2 | 2.0 | GO:0000815 | ESCRT III complex(GO:0000815) |
| 0.1 | 0.4 | GO:0005965 | protein farnesyltransferase complex(GO:0005965) |
| 0.1 | 3.6 | GO:0042581 | specific granule(GO:0042581) |
| 0.1 | 0.8 | GO:0000931 | gamma-tubulin large complex(GO:0000931) gamma-tubulin ring complex(GO:0008274) |
| 0.1 | 1.9 | GO:0031254 | uropod(GO:0001931) cell trailing edge(GO:0031254) |
| 0.1 | 1.7 | GO:0008385 | IkappaB kinase complex(GO:0008385) |
| 0.1 | 31.9 | GO:0009897 | external side of plasma membrane(GO:0009897) |
| 0.1 | 0.4 | GO:0032021 | NELF complex(GO:0032021) |
| 0.1 | 1.5 | GO:0042788 | polysomal ribosome(GO:0042788) |
| 0.1 | 5.3 | GO:0000791 | euchromatin(GO:0000791) |
| 0.1 | 185.1 | GO:0005615 | extracellular space(GO:0005615) |
| 0.1 | 0.9 | GO:0008250 | oligosaccharyltransferase complex(GO:0008250) |
| 0.1 | 4.2 | GO:0042645 | nucleoid(GO:0009295) mitochondrial nucleoid(GO:0042645) |
| 0.1 | 0.4 | GO:0032807 | DNA-dependent protein kinase-DNA ligase 4 complex(GO:0005958) DNA ligase IV complex(GO:0032807) |
| 0.1 | 0.3 | GO:0048476 | Holliday junction resolvase complex(GO:0048476) |
| 0.1 | 0.8 | GO:0000788 | nuclear nucleosome(GO:0000788) |
| 0.0 | 0.5 | GO:0000127 | transcription factor TFIIIC complex(GO:0000127) |
| 0.0 | 0.7 | GO:0043240 | Fanconi anaemia nuclear complex(GO:0043240) |
| 0.0 | 28.0 | GO:0009986 | cell surface(GO:0009986) |
| 0.0 | 6.9 | GO:0000922 | spindle pole(GO:0000922) |
| 0.0 | 0.2 | GO:0000172 | ribonuclease MRP complex(GO:0000172) |
| 0.0 | 0.4 | GO:0070531 | BRCA1-A complex(GO:0070531) |
| 0.0 | 6.3 | GO:0043679 | axon terminus(GO:0043679) |
| 0.0 | 3.6 | GO:0030176 | integral component of endoplasmic reticulum membrane(GO:0030176) |
| 0.0 | 0.4 | GO:0005686 | U2 snRNP(GO:0005686) |
| 0.0 | 0.6 | GO:0000314 | organellar small ribosomal subunit(GO:0000314) mitochondrial small ribosomal subunit(GO:0005763) |
| 0.0 | 0.7 | GO:0099738 | cell cortex region(GO:0099738) |
| 0.0 | 0.4 | GO:0005665 | DNA-directed RNA polymerase II, core complex(GO:0005665) |
| 0.0 | 0.8 | GO:0014704 | intercalated disc(GO:0014704) |
| 0.0 | 1.3 | GO:0030315 | T-tubule(GO:0030315) |
| 0.0 | 0.7 | GO:0022627 | cytosolic small ribosomal subunit(GO:0022627) |
| 0.0 | 0.4 | GO:0030173 | integral component of Golgi membrane(GO:0030173) |
| 0.0 | 2.5 | GO:0005802 | trans-Golgi network(GO:0005802) |
| 0.0 | 0.3 | GO:0042470 | melanosome(GO:0042470) pigment granule(GO:0048770) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 6.7 | 20.0 | GO:0052870 | tocopherol omega-hydroxylase activity(GO:0052870) alpha-tocopherol omega-hydroxylase activity(GO:0052871) |
| 3.7 | 493.0 | GO:0034987 | immunoglobulin receptor binding(GO:0034987) |
| 3.3 | 16.5 | GO:0071987 | WD40-repeat domain binding(GO:0071987) |
| 3.2 | 12.8 | GO:0035673 | oligopeptide transmembrane transporter activity(GO:0035673) |
| 3.2 | 9.5 | GO:0032394 | MHC class Ib receptor activity(GO:0032394) |
| 1.9 | 5.8 | GO:0004087 | carbamoyl-phosphate synthase (ammonia) activity(GO:0004087) carbamoyl-phosphate synthase (glutamine-hydrolyzing) activity(GO:0004088) |
| 1.8 | 7.3 | GO:0031735 | CCR10 chemokine receptor binding(GO:0031735) |
| 1.7 | 5.2 | GO:0016005 | phospholipase A2 activator activity(GO:0016005) |
| 1.7 | 10.3 | GO:0032450 | maltose alpha-glucosidase activity(GO:0032450) |
| 1.6 | 6.4 | GO:0071614 | linoleic acid epoxygenase activity(GO:0071614) |
| 1.1 | 3.3 | GO:0003896 | DNA primase activity(GO:0003896) |
| 1.0 | 12.6 | GO:0046790 | virion binding(GO:0046790) |
| 0.8 | 3.3 | GO:0072320 | volume-sensitive chloride channel activity(GO:0072320) |
| 0.8 | 5.3 | GO:0044729 | hemi-methylated DNA-binding(GO:0044729) |
| 0.7 | 5.0 | GO:0004704 | NF-kappaB-inducing kinase activity(GO:0004704) |
| 0.7 | 2.1 | GO:0002061 | uracil binding(GO:0002058) pyrimidine nucleobase binding(GO:0002061) |
| 0.7 | 7.2 | GO:0008454 | alpha-1,3-mannosylglycoprotein 4-beta-N-acetylglucosaminyltransferase activity(GO:0008454) |
| 0.6 | 1.9 | GO:1990259 | protein-glutamine N-methyltransferase activity(GO:0036009) histone-glutamine methyltransferase activity(GO:1990259) |
| 0.5 | 57.4 | GO:0003823 | antigen binding(GO:0003823) |
| 0.5 | 17.8 | GO:0043395 | heparan sulfate proteoglycan binding(GO:0043395) |
| 0.5 | 1.5 | GO:0017082 | mineralocorticoid receptor activity(GO:0017082) |
| 0.5 | 4.3 | GO:0004468 | lysine N-acetyltransferase activity, acting on acetyl phosphate as donor(GO:0004468) |
| 0.4 | 1.7 | GO:0017095 | heparan sulfate 6-O-sulfotransferase activity(GO:0017095) |
| 0.4 | 3.1 | GO:0032027 | myosin light chain binding(GO:0032027) |
| 0.4 | 9.5 | GO:0015125 | bile acid transmembrane transporter activity(GO:0015125) |
| 0.4 | 4.1 | GO:0003810 | protein-glutamine gamma-glutamyltransferase activity(GO:0003810) |
| 0.3 | 2.1 | GO:0008142 | oxysterol binding(GO:0008142) |
| 0.3 | 4.1 | GO:0004499 | N,N-dimethylaniline monooxygenase activity(GO:0004499) |
| 0.3 | 1.4 | GO:0043532 | angiostatin binding(GO:0043532) |
| 0.3 | 7.8 | GO:0034185 | apolipoprotein binding(GO:0034185) |
| 0.3 | 3.8 | GO:0004571 | mannosyl-oligosaccharide 1,2-alpha-mannosidase activity(GO:0004571) |
| 0.3 | 3.3 | GO:0036374 | glutathione hydrolase activity(GO:0036374) |
| 0.3 | 8.2 | GO:0005229 | intracellular calcium activated chloride channel activity(GO:0005229) |
| 0.2 | 6.3 | GO:0005184 | neuropeptide hormone activity(GO:0005184) |
| 0.2 | 3.4 | GO:0046703 | natural killer cell lectin-like receptor binding(GO:0046703) |
| 0.1 | 0.4 | GO:0008456 | alpha-N-acetylgalactosaminidase activity(GO:0008456) |
| 0.1 | 0.4 | GO:0004660 | protein farnesyltransferase activity(GO:0004660) |
| 0.1 | 1.8 | GO:0043024 | ribosomal small subunit binding(GO:0043024) |
| 0.1 | 1.8 | GO:0031432 | titin binding(GO:0031432) |
| 0.1 | 1.0 | GO:0008479 | queuine tRNA-ribosyltransferase activity(GO:0008479) |
| 0.1 | 14.8 | GO:0004222 | metalloendopeptidase activity(GO:0004222) |
| 0.1 | 3.5 | GO:0004709 | MAP kinase kinase kinase activity(GO:0004709) |
| 0.1 | 1.5 | GO:0008097 | 5S rRNA binding(GO:0008097) |
| 0.1 | 13.5 | GO:0004540 | ribonuclease activity(GO:0004540) |
| 0.1 | 0.8 | GO:0070644 | vitamin D response element binding(GO:0070644) |
| 0.1 | 1.1 | GO:0050700 | CARD domain binding(GO:0050700) |
| 0.1 | 0.9 | GO:0004576 | oligosaccharyl transferase activity(GO:0004576) dolichyl-diphosphooligosaccharide-protein glycotransferase activity(GO:0004579) |
| 0.1 | 0.3 | GO:1904408 | dihydronicotinamide riboside quinone reductase activity(GO:0001512) melatonin binding(GO:1904408) |
| 0.1 | 0.3 | GO:0017108 | 5'-flap endonuclease activity(GO:0017108) |
| 0.1 | 0.4 | GO:0030620 | U2 snRNA binding(GO:0030620) |
| 0.1 | 0.5 | GO:0004966 | galanin receptor activity(GO:0004966) |
| 0.1 | 4.3 | GO:0004177 | aminopeptidase activity(GO:0004177) |
| 0.1 | 0.5 | GO:0070579 | methylcytosine dioxygenase activity(GO:0070579) |
| 0.1 | 0.9 | GO:0019871 | sodium channel inhibitor activity(GO:0019871) |
| 0.0 | 0.8 | GO:0051011 | microtubule minus-end binding(GO:0051011) |
| 0.0 | 0.4 | GO:0003909 | DNA ligase activity(GO:0003909) DNA ligase (ATP) activity(GO:0003910) |
| 0.0 | 10.1 | GO:0001078 | transcriptional repressor activity, RNA polymerase II core promoter proximal region sequence-specific binding(GO:0001078) |
| 0.0 | 1.0 | GO:0005351 | sugar:proton symporter activity(GO:0005351) |
| 0.0 | 0.4 | GO:0001055 | RNA polymerase II activity(GO:0001055) |
| 0.0 | 0.2 | GO:0005223 | intracellular cGMP activated cation channel activity(GO:0005223) |
| 0.0 | 0.1 | GO:0004510 | tryptophan 5-monooxygenase activity(GO:0004510) |
| 0.0 | 0.5 | GO:0035497 | cAMP response element binding(GO:0035497) |
| 0.0 | 0.2 | GO:0005186 | pheromone activity(GO:0005186) |
| 0.0 | 0.7 | GO:0008143 | poly(A) binding(GO:0008143) |
| 0.0 | 3.2 | GO:0004896 | cytokine receptor activity(GO:0004896) |
| 0.0 | 0.5 | GO:0004622 | lysophospholipase activity(GO:0004622) |
| 0.0 | 8.3 | GO:0005525 | GTP binding(GO:0005525) |
| 0.0 | 0.8 | GO:0005086 | ARF guanyl-nucleotide exchange factor activity(GO:0005086) |
| 0.0 | 8.6 | GO:0000978 | RNA polymerase II core promoter proximal region sequence-specific DNA binding(GO:0000978) |
| 0.0 | 3.2 | GO:0017124 | SH3 domain binding(GO:0017124) |
| 0.0 | 0.8 | GO:0004864 | protein phosphatase inhibitor activity(GO:0004864) |
| 0.0 | 5.5 | GO:0005096 | GTPase activator activity(GO:0005096) |
| 0.0 | 0.8 | GO:0017080 | sodium channel regulator activity(GO:0017080) |
| 0.0 | 0.4 | GO:0000993 | RNA polymerase II core binding(GO:0000993) |
| 0.0 | 8.8 | GO:0046982 | protein heterodimerization activity(GO:0046982) |
| 0.0 | 0.2 | GO:0016863 | intramolecular oxidoreductase activity, transposing C=C bonds(GO:0016863) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 5.2 | SA FAS SIGNALING | The TNF-type receptor Fas induces apoptosis on ligand binding. |
| 0.3 | 16.0 | PID IL2 PI3K PATHWAY | IL2 signaling events mediated by PI3K |
| 0.3 | 18.1 | SIG PIP3 SIGNALING IN B LYMPHOCYTES | Genes related to PIP3 signaling in B lymphocytes |
| 0.1 | 4.0 | PID ANTHRAX PATHWAY | Cellular roles of Anthrax toxin |
| 0.1 | 7.3 | PID CXCR3 PATHWAY | CXCR3-mediated signaling events |
| 0.1 | 3.1 | PID S1P S1P3 PATHWAY | S1P3 pathway |
| 0.1 | 1.8 | PID IL5 PATHWAY | IL5-mediated signaling events |
| 0.1 | 6.3 | PID AP1 PATHWAY | AP-1 transcription factor network |
| 0.1 | 1.7 | ST JNK MAPK PATHWAY | JNK MAPK Pathway |
| 0.1 | 1.8 | PID P38 MKK3 6PATHWAY | p38 MAPK signaling pathway |
| 0.1 | 3.3 | PID ERBB4 PATHWAY | ErbB4 signaling events |
| 0.0 | 1.9 | PID AMB2 NEUTROPHILS PATHWAY | amb2 Integrin signaling |
| 0.0 | 1.5 | PID SYNDECAN 2 PATHWAY | Syndecan-2-mediated signaling events |
| 0.0 | 8.7 | NABA ECM AFFILIATED | Genes encoding proteins affiliated structurally or functionally to extracellular matrix proteins |
| 0.0 | 10.6 | NABA ECM REGULATORS | Genes encoding enzymes and their regulators involved in the remodeling of the extracellular matrix |
| 0.0 | 1.2 | PID PLK1 PATHWAY | PLK1 signaling events |
| 0.0 | 1.5 | PID MTOR 4PATHWAY | mTOR signaling pathway |
| 0.0 | 0.4 | PID DNA PK PATHWAY | DNA-PK pathway in nonhomologous end joining |
| 0.0 | 0.2 | PID INTEGRIN2 PATHWAY | Beta2 integrin cell surface interactions |
| 0.0 | 0.8 | PID HES HEY PATHWAY | Notch-mediated HES/HEY network |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.6 | 10.3 | REACTOME DIGESTION OF DIETARY CARBOHYDRATE | Genes involved in Digestion of dietary carbohydrate |
| 1.2 | 18.1 | REACTOME PHOSPHORYLATION OF CD3 AND TCR ZETA CHAINS | Genes involved in Phosphorylation of CD3 and TCR zeta chains |
| 1.0 | 3.1 | REACTOME BINDING AND ENTRY OF HIV VIRION | Genes involved in Binding and entry of HIV virion |
| 0.4 | 5.2 | REACTOME APOPTOSIS INDUCED DNA FRAGMENTATION | Genes involved in Apoptosis induced DNA fragmentation |
| 0.3 | 7.2 | REACTOME N GLYCAN ANTENNAE ELONGATION | Genes involved in N-Glycan antennae elongation |
| 0.3 | 7.5 | REACTOME CHYLOMICRON MEDIATED LIPID TRANSPORT | Genes involved in Chylomicron-mediated lipid transport |
| 0.2 | 3.8 | REACTOME CALNEXIN CALRETICULIN CYCLE | Genes involved in Calnexin/calreticulin cycle |
| 0.2 | 5.0 | REACTOME OTHER SEMAPHORIN INTERACTIONS | Genes involved in Other semaphorin interactions |
| 0.2 | 9.5 | REACTOME IMMUNOREGULATORY INTERACTIONS BETWEEN A LYMPHOID AND A NON LYMPHOID CELL | Genes involved in Immunoregulatory interactions between a Lymphoid and a non-Lymphoid cell |
| 0.1 | 7.3 | REACTOME CHEMOKINE RECEPTORS BIND CHEMOKINES | Genes involved in Chemokine receptors bind chemokines |
| 0.1 | 2.5 | REACTOME CYCLIN A B1 ASSOCIATED EVENTS DURING G2 M TRANSITION | Genes involved in Cyclin A/B1 associated events during G2/M transition |
| 0.1 | 1.6 | REACTOME GAMMA CARBOXYLATION TRANSPORT AND AMINO TERMINAL CLEAVAGE OF PROTEINS | Genes involved in Gamma-carboxylation, transport, and amino-terminal cleavage of proteins |
| 0.1 | 16.0 | REACTOME FACTORS INVOLVED IN MEGAKARYOCYTE DEVELOPMENT AND PLATELET PRODUCTION | Genes involved in Factors involved in megakaryocyte development and platelet production |
| 0.1 | 3.8 | REACTOME SHC1 EVENTS IN ERBB4 SIGNALING | Genes involved in SHC1 events in ERBB4 signaling |
| 0.1 | 3.0 | REACTOME GLUTATHIONE CONJUGATION | Genes involved in Glutathione conjugation |
| 0.1 | 0.8 | REACTOME RECRUITMENT OF NUMA TO MITOTIC CENTROSOMES | Genes involved in Recruitment of NuMA to mitotic centrosomes |
| 0.0 | 12.6 | REACTOME G ALPHA Q SIGNALLING EVENTS | Genes involved in G alpha (q) signalling events |
| 0.0 | 1.7 | REACTOME HS GAG BIOSYNTHESIS | Genes involved in HS-GAG biosynthesis |
| 0.0 | 1.1 | REACTOME GLUCAGON TYPE LIGAND RECEPTORS | Genes involved in Glucagon-type ligand receptors |
| 0.0 | 2.1 | REACTOME INTERACTION BETWEEN L1 AND ANKYRINS | Genes involved in Interaction between L1 and Ankyrins |
| 0.0 | 0.7 | REACTOME PYRIMIDINE CATABOLISM | Genes involved in Pyrimidine catabolism |
| 0.0 | 3.5 | REACTOME NUCLEAR RECEPTOR TRANSCRIPTION PATHWAY | Genes involved in Nuclear Receptor transcription pathway |
| 0.0 | 0.4 | REACTOME INTEGRATION OF PROVIRUS | Genes involved in Integration of provirus |
| 0.0 | 1.1 | REACTOME DOWNREGULATION OF SMAD2 3 SMAD4 TRANSCRIPTIONAL ACTIVITY | Genes involved in Downregulation of SMAD2/3:SMAD4 transcriptional activity |
| 0.0 | 3.8 | REACTOME SIGNALING BY ILS | Genes involved in Signaling by Interleukins |
| 0.0 | 0.2 | REACTOME COMMON PATHWAY | Genes involved in Common Pathway |
| 0.0 | 0.4 | REACTOME VIRAL MESSENGER RNA SYNTHESIS | Genes involved in Viral Messenger RNA Synthesis |
| 0.0 | 1.0 | REACTOME TRANSPORT OF VITAMINS NUCLEOSIDES AND RELATED MOLECULES | Genes involved in Transport of vitamins, nucleosides, and related molecules |
| 0.0 | 3.2 | REACTOME TRANSLATION | Genes involved in Translation |
| 0.0 | 1.1 | REACTOME NRAGE SIGNALS DEATH THROUGH JNK | Genes involved in NRAGE signals death through JNK |