Project
PRJNA375882: Comprehensive Mouse Transcriptomic BodyMap
Navigation
Downloads
Results for Pou2f2_Pou3f1
Z-value: 3.46
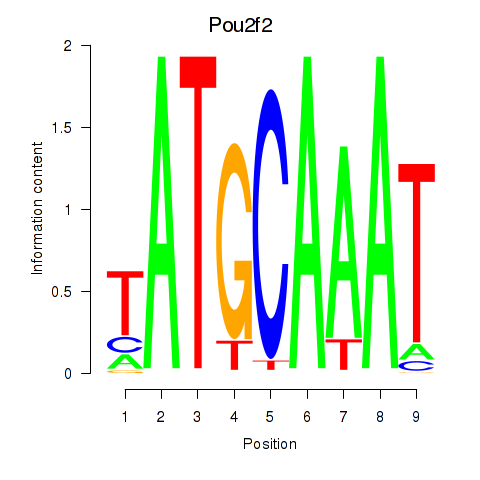
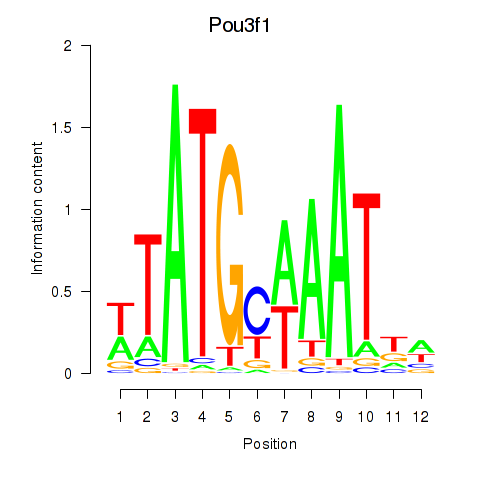
Transcription factors associated with Pou2f2_Pou3f1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Pou2f2
|
ENSMUSG00000008496.20 | Pou2f2 |
|
Pou3f1
|
ENSMUSG00000090125.4 | Pou3f1 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Pou3f1 | mm39_v1_chr4_+_124550600_124550600 | -0.59 | 4.6e-08 | Click! |
| Pou2f2 | mm39_v1_chr7_-_24831892_24831937 | 0.23 | 5.4e-02 | Click! |
Activity profile of Pou2f2_Pou3f1 motif
Sorted Z-values of Pou2f2_Pou3f1 motif
Network of associatons between targets according to the STRING database.
First level regulatory network of Pou2f2_Pou3f1
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr6_-_68713748 | 57.49 |
ENSMUST00000183936.2
ENSMUST00000196863.2 |
Igkv19-93
|
immunoglobulin kappa chain variable 19-93 |
| chr6_+_67586695 | 57.33 |
ENSMUST00000103303.3
|
Igkv1-135
|
immunoglobulin kappa variable 1-135 |
| chr6_-_69877961 | 54.74 |
ENSMUST00000197290.2
|
Igkv5-39
|
immunoglobulin kappa variable 5-39 |
| chr6_+_68247469 | 53.74 |
ENSMUST00000103321.3
|
Igkv1-110
|
immunoglobulin kappa variable 1-110 |
| chr6_-_68609426 | 53.41 |
ENSMUST00000103328.3
|
Igkv10-96
|
immunoglobulin kappa variable 10-96 |
| chr6_+_68098030 | 51.94 |
ENSMUST00000103317.3
|
Igkv1-117
|
immunoglobulin kappa variable 1-117 |
| chr6_-_69704122 | 51.75 |
ENSMUST00000103364.3
|
Igkv5-48
|
immunoglobulin kappa variable 5-48 |
| chr6_+_68026941 | 50.79 |
ENSMUST00000103316.2
|
Igkv9-120
|
immunoglobulin kappa chain variable 9-120 |
| chr6_-_69800923 | 48.72 |
ENSMUST00000103368.3
|
Igkv5-43
|
immunoglobulin kappa chain variable 5-43 |
| chr12_-_114752425 | 45.19 |
ENSMUST00000103510.2
|
Ighv1-26
|
immunoglobulin heavy variable 1-26 |
| chr6_+_70495224 | 44.14 |
ENSMUST00000103396.2
|
Igkv3-12
|
immunoglobulin kappa variable 3-12 |
| chr12_-_113802603 | 43.74 |
ENSMUST00000103458.3
ENSMUST00000193652.2 |
Ighv5-16
|
immunoglobulin heavy variable 5-16 |
| chr6_-_69741999 | 43.08 |
ENSMUST00000103365.3
|
Igkv12-46
|
immunoglobulin kappa variable 12-46 |
| chr6_+_67532481 | 42.70 |
ENSMUST00000103302.3
|
Igkv2-137
|
immunoglobulin kappa chain variable 2-137 |
| chr6_-_69877642 | 42.50 |
ENSMUST00000103370.3
|
Igkv5-39
|
immunoglobulin kappa variable 5-39 |
| chr12_-_115876396 | 41.94 |
ENSMUST00000103547.2
|
Ighv1-80
|
immunoglobulin heavy variable 1-80 |
| chr6_-_68907718 | 41.41 |
ENSMUST00000114212.4
|
Igkv13-85
|
immunoglobulin kappa chain variable 13-85 |
| chr6_-_67919524 | 40.62 |
ENSMUST00000196768.2
|
Igkv9-124
|
immunoglobulin kappa chain variable 9-124 |
| chr12_-_114710326 | 40.58 |
ENSMUST00000103507.2
|
Ighv1-22
|
immunoglobulin heavy variable 1-22 |
| chr12_-_113912416 | 40.47 |
ENSMUST00000103464.3
|
Ighv4-1
|
immunoglobulin heavy variable 4-1 |
| chr12_-_114355789 | 39.23 |
ENSMUST00000103486.2
|
Ighv6-3
|
immunoglobulin heavy variable 6-3 |
| chr12_-_113790741 | 39.19 |
ENSMUST00000103457.3
ENSMUST00000192877.2 |
Ighv5-15
|
immunoglobulin heavy variable 5-15 |
| chr6_+_68402550 | 38.58 |
ENSMUST00000103323.3
|
Igkv16-104
|
immunoglobulin kappa variable 16-104 |
| chr12_-_113823290 | 37.04 |
ENSMUST00000103459.5
|
Ighv5-17
|
immunoglobulin heavy variable 5-17 |
| chr6_+_68916540 | 36.85 |
ENSMUST00000103339.2
|
Igkv13-84
|
immunoglobulin kappa chain variable 13-84 |
| chr12_-_114502585 | 35.74 |
ENSMUST00000103496.4
|
Ighv1-7
|
immunoglobulin heavy variable V1-7 |
| chr12_-_114487525 | 33.81 |
ENSMUST00000103495.3
|
Ighv10-3
|
immunoglobulin heavy variable V10-3 |
| chr6_-_69835868 | 33.34 |
ENSMUST00000103369.2
|
Igkv12-41
|
immunoglobulin kappa chain variable 12-41 |
| chr6_+_70675416 | 33.07 |
ENSMUST00000103403.3
|
Igkv3-2
|
immunoglobulin kappa variable 3-2 |
| chr6_+_68279392 | 32.53 |
ENSMUST00000103322.3
|
Igkv2-109
|
immunoglobulin kappa variable 2-109 |
| chr12_-_113561594 | 32.04 |
ENSMUST00000103444.3
|
Ighv5-4
|
immunoglobulin heavy variable 5-4 |
| chr12_-_113860566 | 28.43 |
ENSMUST00000103474.5
|
Ighv7-1
|
immunoglobulin heavy variable 7-1 |
| chr12_-_113958518 | 28.15 |
ENSMUST00000103467.2
|
Ighv14-2
|
immunoglobulin heavy variable 14-2 |
| chr12_-_113589576 | 27.83 |
ENSMUST00000103446.2
|
Ighv5-6
|
immunoglobulin heavy variable 5-6 |
| chr12_-_114443071 | 27.81 |
ENSMUST00000103492.2
|
Ighv10-1
|
immunoglobulin heavy variable 10-1 |
| chr6_+_67838100 | 27.35 |
ENSMUST00000200586.2
ENSMUST00000103309.3 |
Igkv17-127
|
immunoglobulin kappa variable 17-127 |
| chr6_-_69792108 | 27.24 |
ENSMUST00000103367.3
|
Igkv12-44
|
immunoglobulin kappa variable 12-44 |
| chr3_-_144555062 | 26.19 |
ENSMUST00000159989.2
|
Clca3b
|
chloride channel accessory 3B |
| chr6_+_145091196 | 26.08 |
ENSMUST00000156849.8
ENSMUST00000132948.2 |
Lrmp
|
lymphoid-restricted membrane protein |
| chr12_-_114252202 | 25.93 |
ENSMUST00000195124.6
ENSMUST00000103481.3 |
Ighv3-6
|
immunoglobulin heavy variable 3-6 |
| chr6_+_67873135 | 25.47 |
ENSMUST00000103310.3
|
Igkv14-126
|
immunoglobulin kappa variable 14-126 |
| chr12_-_115766700 | 25.43 |
ENSMUST00000196587.5
ENSMUST00000103543.3 |
Ighv1-74
|
immunoglobulin heavy variable V1-74 |
| chr12_-_115323192 | 24.92 |
ENSMUST00000103531.4
|
Ighv1-61
|
immunoglobulin heavy variable 1-61 |
| chr11_-_106205320 | 24.75 |
ENSMUST00000167143.2
|
Cd79b
|
CD79B antigen |
| chr12_-_115276219 | 23.50 |
ENSMUST00000103529.4
|
Ighv1-58
|
immunoglobulin heavy variable 1-58 |
| chr6_+_68414401 | 23.31 |
ENSMUST00000103324.3
|
Igkv15-103
|
immunoglobulin kappa chain variable 15-103 |
| chr16_-_19079594 | 23.29 |
ENSMUST00000103752.3
ENSMUST00000197518.2 |
Iglv2
|
immunoglobulin lambda variable 2 |
| chr6_+_70584375 | 22.81 |
ENSMUST00000197560.2
|
Igkv3-7
|
immunoglobulin kappa variable 3-7 |
| chr12_-_115916604 | 22.74 |
ENSMUST00000196991.2
|
Ighv1-82
|
immunoglobulin heavy variable 1-82 |
| chr12_-_114878652 | 21.78 |
ENSMUST00000103515.2
|
Ighv1-39
|
immunoglobulin heavy variable 1-39 |
| chr12_-_114023935 | 21.73 |
ENSMUST00000103469.4
|
Ighv14-3
|
immunoglobulin heavy variable V14-3 |
| chr12_-_114012399 | 20.97 |
ENSMUST00000103468.3
|
Ighv11-2
|
immunoglobulin heavy variable V11-2 |
| chr12_-_113625906 | 20.80 |
ENSMUST00000103448.3
|
Ighv5-9
|
immunoglobulin heavy variable 5-9 |
| chr12_-_114321838 | 20.06 |
ENSMUST00000125484.3
|
Ighv13-2
|
immunoglobulin heavy variable 13-2 |
| chrX_-_111608339 | 19.87 |
ENSMUST00000039887.4
|
Pof1b
|
premature ovarian failure 1B |
| chr12_-_114398864 | 19.78 |
ENSMUST00000103489.2
|
Ighv6-6
|
immunoglobulin heavy variable 6-6 |
| chr15_-_66703471 | 18.54 |
ENSMUST00000164163.8
|
Sla
|
src-like adaptor |
| chr12_-_114286421 | 18.53 |
ENSMUST00000103483.3
|
Ighv3-8
|
immunoglobulin heavy variable V3-8 |
| chr12_-_115172211 | 18.11 |
ENSMUST00000103526.3
|
Ighv1-55
|
immunoglobulin heavy variable 1-55 |
| chr3_+_51568588 | 18.10 |
ENSMUST00000099106.10
|
Mgst2
|
microsomal glutathione S-transferase 2 |
| chr6_-_41535322 | 17.98 |
ENSMUST00000193003.2
|
Trbv31
|
T cell receptor beta, variable 31 |
| chr1_-_165762469 | 17.80 |
ENSMUST00000069609.12
ENSMUST00000111427.9 ENSMUST00000111426.11 |
Pou2f1
|
POU domain, class 2, transcription factor 1 |
| chr6_+_68196928 | 17.51 |
ENSMUST00000103318.6
ENSMUST00000103319.3 |
Igkv2-112
|
immunoglobulin kappa variable 2-112 |
| chr6_-_3494587 | 17.34 |
ENSMUST00000049985.15
|
Hepacam2
|
HEPACAM family member 2 |
| chr6_-_69753317 | 17.33 |
ENSMUST00000103366.3
|
Igkv5-45
|
immunoglobulin kappa chain variable 5-45 |
| chr12_-_114226570 | 17.22 |
ENSMUST00000103479.4
ENSMUST00000195619.2 |
Ighv3-5
|
immunoglobulin heavy variable 3-5 |
| chr6_-_67014348 | 17.21 |
ENSMUST00000204369.2
|
Gadd45a
|
growth arrest and DNA-damage-inducible 45 alpha |
| chr6_+_70648743 | 17.12 |
ENSMUST00000103401.3
|
Igkv3-4
|
immunoglobulin kappa variable 3-4 |
| chr12_-_115706126 | 16.74 |
ENSMUST00000166645.2
ENSMUST00000196690.2 |
Ighv1-71
|
immunoglobulin heavy variable 1-71 |
| chr4_+_3678108 | 16.63 |
ENSMUST00000041377.13
ENSMUST00000103010.4 |
Lyn
|
LYN proto-oncogene, Src family tyrosine kinase |
| chr12_-_113666198 | 16.59 |
ENSMUST00000103450.4
|
Ighv5-12
|
immunoglobulin heavy variable 5-12 |
| chr12_-_114621406 | 16.51 |
ENSMUST00000192077.2
|
Ighv1-15
|
immunoglobulin heavy variable 1-15 |
| chr6_+_68495964 | 16.40 |
ENSMUST00000199510.5
ENSMUST00000103325.3 |
Igkv14-100
|
immunoglobulin kappa chain variable 14-100 |
| chr12_-_114117264 | 16.23 |
ENSMUST00000103461.5
|
Ighv7-3
|
immunoglobulin heavy variable 7-3 |
| chr12_-_114843941 | 16.01 |
ENSMUST00000191862.6
ENSMUST00000103513.3 |
Ighv1-36
|
immunoglobulin heavy variable 1-36 |
| chr12_-_115798038 | 15.91 |
ENSMUST00000103544.3
|
Ighv1-75
|
immunoglobulin heavy variable 1-75 |
| chr10_-_88520877 | 15.78 |
ENSMUST00000138734.2
|
Spic
|
Spi-C transcription factor (Spi-1/PU.1 related) |
| chr17_+_29709723 | 15.72 |
ENSMUST00000024811.9
|
Pim1
|
proviral integration site 1 |
| chr12_-_115471634 | 15.54 |
ENSMUST00000103535.3
|
Ighv1-64
|
immunoglobulin heavy variable 1-64 |
| chr12_-_114672701 | 15.22 |
ENSMUST00000103505.3
ENSMUST00000193855.2 |
Ighv1-19
|
immunoglobulin heavy variable V1-19 |
| chr5_-_147244074 | 15.19 |
ENSMUST00000031650.4
|
Cdx2
|
caudal type homeobox 2 |
| chr5_+_137628377 | 15.18 |
ENSMUST00000175968.8
|
Lrch4
|
leucine-rich repeats and calponin homology (CH) domain containing 4 |
| chr6_-_67014383 | 15.15 |
ENSMUST00000043098.9
|
Gadd45a
|
growth arrest and DNA-damage-inducible 45 alpha |
| chr11_-_5787743 | 15.11 |
ENSMUST00000109837.8
|
Polm
|
polymerase (DNA directed), mu |
| chr6_+_67701864 | 14.59 |
ENSMUST00000103304.3
|
Igkv1-133
|
immunoglobulin kappa variable 1-133 |
| chr12_-_113928438 | 14.58 |
ENSMUST00000103478.4
|
Ighv3-1
|
immunoglobulin heavy variable 3-1 |
| chr12_-_113700190 | 14.44 |
ENSMUST00000103452.3
ENSMUST00000192264.2 |
Ighv5-9-1
|
immunoglobulin heavy variable 5-9-1 |
| chr12_-_115964081 | 14.26 |
ENSMUST00000103552.2
|
Ighv1-85
|
immunoglobulin heavy variable 1-85 |
| chr6_+_70640233 | 14.23 |
ENSMUST00000103400.3
|
Igkv3-5
|
immunoglobulin kappa chain variable 3-5 |
| chr12_-_115811951 | 14.17 |
ENSMUST00000197537.2
|
Ighv1-76
|
immunoglobulin heavy variable 1-76 |
| chr7_-_44180700 | 14.13 |
ENSMUST00000205506.2
|
Spib
|
Spi-B transcription factor (Spi-1/PU.1 related) |
| chr6_+_68547717 | 14.11 |
ENSMUST00000196839.5
ENSMUST00000103327.3 |
Igkv12-98
|
immunoglobulin kappa variable 12-98 |
| chr12_-_115832846 | 14.06 |
ENSMUST00000199373.2
|
Ighv1-78
|
immunoglobulin heavy variable 1-78 |
| chr12_-_115109539 | 14.05 |
ENSMUST00000192554.6
ENSMUST00000103522.3 |
Ighv1-52
|
immunoglobulin heavy variable 1-52 |
| chr12_-_115884332 | 14.03 |
ENSMUST00000103548.3
|
Ighv1-81
|
immunoglobulin heavy variable 1-81 |
| chr11_+_95227836 | 13.94 |
ENSMUST00000037502.7
|
Fam117a
|
family with sequence similarity 117, member A |
| chr6_-_67014191 | 13.86 |
ENSMUST00000204282.2
|
Gadd45a
|
growth arrest and DNA-damage-inducible 45 alpha |
| chr9_-_51240201 | 13.65 |
ENSMUST00000039959.11
ENSMUST00000238450.3 |
1810046K07Rik
|
RIKEN cDNA 1810046K07 gene |
| chr6_-_67931692 | 13.42 |
ENSMUST00000103313.3
|
Igkv9-123
|
immunoglobulin kappa variable 9-123 |
| chr6_-_148847854 | 13.26 |
ENSMUST00000139355.8
ENSMUST00000146457.2 ENSMUST00000054080.15 |
Sinhcaf
|
SIN3-HDAC complex associated factor |
| chr14_+_79753055 | 12.89 |
ENSMUST00000110835.3
ENSMUST00000227192.2 |
Elf1
|
E74-like factor 1 |
| chr12_-_113896002 | 12.78 |
ENSMUST00000103463.3
|
Ighv14-1
|
immunoglobulin heavy variable 14-1 |
| chr6_+_70680515 | 12.52 |
ENSMUST00000103404.2
|
Igkv3-1
|
immunoglobulin kappa variable 3-1 |
| chr14_-_67953035 | 12.50 |
ENSMUST00000163100.8
ENSMUST00000132705.8 ENSMUST00000124045.3 |
Cdca2
|
cell division cycle associated 2 |
| chr6_-_41535292 | 12.47 |
ENSMUST00000103300.3
|
Trbv31
|
T cell receptor beta, variable 31 |
| chrX_-_165992145 | 12.41 |
ENSMUST00000112176.8
|
Tmsb4x
|
thymosin, beta 4, X chromosome |
| chr6_-_68840015 | 12.40 |
ENSMUST00000103336.2
|
Igkv1-88
|
immunoglobulin kappa chain variable 1-88 |
| chr1_-_135186176 | 12.19 |
ENSMUST00000185752.2
ENSMUST00000003135.14 |
Elf3
|
E74-like factor 3 |
| chr10_+_75784126 | 12.16 |
ENSMUST00000000926.3
|
Vpreb3
|
pre-B lymphocyte gene 3 |
| chr2_+_118877594 | 12.07 |
ENSMUST00000152380.8
ENSMUST00000099542.9 |
Knl1
|
kinetochore scaffold 1 |
| chr12_-_115944754 | 11.67 |
ENSMUST00000103551.2
|
Ighv1-84
|
immunoglobulin heavy variable 1-84 |
| chr15_-_58261093 | 11.66 |
ENSMUST00000227274.3
|
Anxa13
|
annexin A13 |
| chr10_+_42736644 | 11.59 |
ENSMUST00000105495.8
|
Scml4
|
Scm polycomb group protein like 4 |
| chr12_-_114451189 | 11.58 |
ENSMUST00000103493.3
|
Ighv1-4
|
immunoglobulin heavy variable 1-4 |
| chr5_+_137627431 | 11.41 |
ENSMUST00000176667.8
|
Lrch4
|
leucine-rich repeats and calponin homology (CH) domain containing 4 |
| chr12_-_115611981 | 11.39 |
ENSMUST00000103540.3
ENSMUST00000199266.2 |
Ighv8-12
|
immunoglobulin heavy variable V8-12 |
| chr15_+_75734159 | 11.15 |
ENSMUST00000023238.6
ENSMUST00000230514.2 ENSMUST00000229331.2 |
Gsdmd
|
gasdermin D |
| chr11_+_115714853 | 11.14 |
ENSMUST00000103032.11
ENSMUST00000133250.8 ENSMUST00000177736.8 |
Llgl2
|
LLGL2 scribble cell polarity complex component |
| chr12_-_114477427 | 10.99 |
ENSMUST00000191803.2
|
Ighv1-5
|
immunoglobulin heavy variable V1-5 |
| chr3_-_144525255 | 10.85 |
ENSMUST00000029929.12
|
Clca3a2
|
chloride channel accessory 3A2 |
| chr6_+_67993691 | 10.68 |
ENSMUST00000103314.3
|
Igkv1-122
|
immunoglobulin kappa chain variable 1-122 |
| chr13_-_95661726 | 10.50 |
ENSMUST00000022185.10
|
F2rl1
|
coagulation factor II (thrombin) receptor-like 1 |
| chr12_-_114815280 | 10.38 |
ENSMUST00000103512.3
|
Ighv1-34
|
immunoglobulin heavy variable 1-34 |
| chr17_-_15596230 | 10.35 |
ENSMUST00000014917.8
|
Dll1
|
delta like canonical Notch ligand 1 |
| chr11_+_66847446 | 10.35 |
ENSMUST00000211300.2
ENSMUST00000150220.2 |
Tmem238l
|
transmembrane protein 238 like |
| chr19_-_40260060 | 10.13 |
ENSMUST00000068439.13
|
Pdlim1
|
PDZ and LIM domain 1 (elfin) |
| chr12_-_36092475 | 10.07 |
ENSMUST00000020896.17
|
Tspan13
|
tetraspanin 13 |
| chr6_-_148847633 | 10.04 |
ENSMUST00000132696.8
|
Sinhcaf
|
SIN3-HDAC complex associated factor |
| chr6_-_69920632 | 9.93 |
ENSMUST00000198880.5
ENSMUST00000103371.3 |
Igkv12-38
|
immunoglobulin kappa chain variable 12-38 |
| chr12_-_115083839 | 9.72 |
ENSMUST00000103521.3
|
Ighv1-50
|
immunoglobulin heavy variable 1-50 |
| chr17_+_27276262 | 9.51 |
ENSMUST00000049308.9
|
Itpr3
|
inositol 1,4,5-triphosphate receptor 3 |
| chr6_+_41098273 | 9.50 |
ENSMUST00000103270.4
|
Trbv13-2
|
T cell receptor beta, variable 13-2 |
| chr2_+_118877610 | 9.30 |
ENSMUST00000153300.8
ENSMUST00000028799.12 |
Knl1
|
kinetochore scaffold 1 |
| chr11_-_34724458 | 9.27 |
ENSMUST00000093191.3
|
Spdl1
|
spindle apparatus coiled-coil protein 1 |
| chrX_-_165992311 | 9.26 |
ENSMUST00000112172.4
|
Tmsb4x
|
thymosin, beta 4, X chromosome |
| chr3_+_159201048 | 8.96 |
ENSMUST00000120272.8
|
Depdc1a
|
DEP domain containing 1a |
| chr19_-_40260286 | 8.89 |
ENSMUST00000182432.2
|
Pdlim1
|
PDZ and LIM domain 1 (elfin) |
| chr12_-_114057841 | 8.85 |
ENSMUST00000103471.2
ENSMUST00000195884.2 |
Ighv9-1
|
immunoglobulin heavy variable 9-1 |
| chr9_-_85209340 | 8.62 |
ENSMUST00000187711.2
|
Tent5a
|
terminal nucleotidyltransferase 5A |
| chr12_-_115157739 | 8.49 |
ENSMUST00000103525.3
|
Ighv1-54
|
immunoglobulin heavy variable V1-54 |
| chr9_-_85209162 | 8.47 |
ENSMUST00000034802.15
|
Tent5a
|
terminal nucleotidyltransferase 5A |
| chr6_+_64706101 | 8.25 |
ENSMUST00000101351.6
|
Atoh1
|
atonal bHLH transcription factor 1 |
| chr3_+_159201077 | 7.89 |
ENSMUST00000029825.14
|
Depdc1a
|
DEP domain containing 1a |
| chr12_-_114186874 | 7.82 |
ENSMUST00000103477.4
ENSMUST00000192499.3 |
Ighv7-4
|
immunoglobulin heavy variable 7-4 |
| chr6_+_67890534 | 7.75 |
ENSMUST00000197406.5
ENSMUST00000103311.3 |
Igkv11-125
|
immunoglobulin kappa variable 11-125 |
| chr17_+_27775637 | 7.74 |
ENSMUST00000117254.9
ENSMUST00000231243.2 ENSMUST00000231358.2 ENSMUST00000118570.2 ENSMUST00000231796.2 |
Hmga1
|
high mobility group AT-hook 1 |
| chr8_+_121215155 | 7.73 |
ENSMUST00000034279.16
|
Gse1
|
genetic suppressor element 1, coiled-coil protein |
| chr3_+_159201092 | 7.59 |
ENSMUST00000106041.3
|
Depdc1a
|
DEP domain containing 1a |
| chr1_+_36510670 | 7.52 |
ENSMUST00000153128.2
|
Cnnm4
|
cyclin M4 |
| chr8_+_71069476 | 7.50 |
ENSMUST00000052437.6
|
Lrrc25
|
leucine rich repeat containing 25 |
| chr17_+_27775613 | 7.44 |
ENSMUST00000231780.2
ENSMUST00000232253.2 ENSMUST00000232552.2 ENSMUST00000117600.9 |
Hmga1
|
high mobility group AT-hook 1 |
| chr4_-_3938352 | 7.41 |
ENSMUST00000003369.10
|
Plag1
|
pleiomorphic adenoma gene 1 |
| chr2_+_90927053 | 7.40 |
ENSMUST00000132741.3
|
Spi1
|
spleen focus forming virus (SFFV) proviral integration oncogene |
| chr19_-_46033353 | 7.25 |
ENSMUST00000026252.14
ENSMUST00000156585.9 ENSMUST00000185355.7 ENSMUST00000152946.8 |
Ldb1
|
LIM domain binding 1 |
| chr8_+_91635192 | 7.14 |
ENSMUST00000211403.2
|
Chd9
|
chromodomain helicase DNA binding protein 9 |
| chr16_-_92494203 | 7.14 |
ENSMUST00000113956.10
|
Runx1
|
runt related transcription factor 1 |
| chr6_+_67816777 | 6.77 |
ENSMUST00000200578.5
ENSMUST00000103308.3 |
Igkv9-129
|
immunoglobulin kappa variable 9-129 |
| chr1_+_52047368 | 6.65 |
ENSMUST00000027277.7
|
Stat4
|
signal transducer and activator of transcription 4 |
| chr10_-_79911245 | 6.48 |
ENSMUST00000217972.2
|
Sbno2
|
strawberry notch 2 |
| chr17_+_27775471 | 6.45 |
ENSMUST00000118599.9
ENSMUST00000232265.2 ENSMUST00000232013.2 ENSMUST00000114888.11 ENSMUST00000231874.2 ENSMUST00000119486.9 ENSMUST00000231825.2 ENSMUST00000231866.2 |
Hmga1
|
high mobility group AT-hook 1 |
| chr12_-_115557211 | 6.41 |
ENSMUST00000103537.3
|
Ighv1-66
|
immunoglobulin heavy variable 1-66 |
| chr16_+_23043474 | 6.33 |
ENSMUST00000023601.14
|
St6gal1
|
beta galactoside alpha 2,6 sialyltransferase 1 |
| chr12_-_113542610 | 6.32 |
ENSMUST00000195468.6
ENSMUST00000103442.3 |
Ighv5-2
|
immunoglobulin heavy variable 5-2 |
| chr12_-_115459678 | 6.26 |
ENSMUST00000103534.2
|
Ighv1-63
|
immunoglobulin heavy variable V1-63 |
| chr5_+_93045837 | 6.22 |
ENSMUST00000113051.9
|
Shroom3
|
shroom family member 3 |
| chr14_+_26722319 | 6.08 |
ENSMUST00000035433.10
|
Hesx1
|
homeobox gene expressed in ES cells |
| chr12_-_114073050 | 6.05 |
ENSMUST00000103472.4
|
Ighv9-2
|
immunoglobulin heavy variable V9-2 |
| chr4_-_129590372 | 5.97 |
ENSMUST00000137640.3
|
Tmem39b
|
transmembrane protein 39b |
| chr7_-_4607040 | 5.92 |
ENSMUST00000166650.3
|
Ptprh
|
protein tyrosine phosphatase, receptor type, H |
| chr12_-_101785307 | 5.91 |
ENSMUST00000021603.9
|
Fbln5
|
fibulin 5 |
| chr12_-_101784727 | 5.76 |
ENSMUST00000222587.2
|
Fbln5
|
fibulin 5 |
| chr13_-_23755374 | 5.74 |
ENSMUST00000102969.6
|
H2ac8
|
H2A clustered histone 8 |
| chr6_+_68657317 | 5.69 |
ENSMUST00000198735.2
|
Igkv10-95
|
immunoglobulin kappa variable 10-95 |
| chr13_+_102830029 | 5.67 |
ENSMUST00000022124.10
ENSMUST00000171267.2 ENSMUST00000167144.2 ENSMUST00000170878.2 |
Cd180
|
CD180 antigen |
| chr16_+_44913974 | 5.65 |
ENSMUST00000099498.10
|
Ccdc80
|
coiled-coil domain containing 80 |
| chr13_+_21906214 | 5.59 |
ENSMUST00000224651.2
|
H2bc14
|
H2B clustered histone 14 |
| chrX_+_132751729 | 5.57 |
ENSMUST00000033602.9
|
Tnmd
|
tenomodulin |
| chr6_+_67768007 | 5.56 |
ENSMUST00000196006.5
ENSMUST00000103307.3 |
Igkv14-130
|
immunoglobulin kappa variable 14-130 |
| chr2_-_103133503 | 5.54 |
ENSMUST00000111176.9
|
Ehf
|
ets homologous factor |
| chr6_-_52237765 | 5.48 |
ENSMUST00000147595.7
|
Hoxa13
|
homeobox A13 |
| chr5_-_124003553 | 5.43 |
ENSMUST00000057145.7
|
Hcar2
|
hydroxycarboxylic acid receptor 2 |
| chr12_-_115031622 | 5.36 |
ENSMUST00000194257.2
|
Ighv8-5
|
immunoglobulin heavy variable V8-5 |
| chr12_-_114955196 | 5.31 |
ENSMUST00000194865.2
|
Ighv1-47
|
immunoglobulin heavy variable 1-47 |
| chr2_-_147887810 | 5.24 |
ENSMUST00000109964.8
|
Foxa2
|
forkhead box A2 |
| chrX_+_55500170 | 5.24 |
ENSMUST00000039374.9
ENSMUST00000101553.9 ENSMUST00000186445.7 |
Ints6l
|
integrator complex subunit 6 like |
| chr12_-_34578842 | 5.20 |
ENSMUST00000110819.4
|
Hdac9
|
histone deacetylase 9 |
| chr9_+_50466127 | 5.10 |
ENSMUST00000213916.2
|
Il18
|
interleukin 18 |
| chr18_+_36414122 | 5.09 |
ENSMUST00000051301.6
|
Pura
|
purine rich element binding protein A |
| chr5_+_144127102 | 5.08 |
ENSMUST00000060747.8
|
Bhlha15
|
basic helix-loop-helix family, member a15 |
| chr3_+_96177010 | 4.99 |
ENSMUST00000051089.4
ENSMUST00000177113.2 |
Gm42743
H2bc18
|
predicted gene 42743 H2B clustered histone 18 |
| chr12_-_113945961 | 4.96 |
ENSMUST00000103466.2
|
Ighv11-1
|
immunoglobulin heavy variable 11-1 |
| chr16_+_11131676 | 4.89 |
ENSMUST00000023140.6
|
Tnfrsf17
|
tumor necrosis factor receptor superfamily, member 17 |
| chr6_-_69477770 | 4.85 |
ENSMUST00000197448.2
|
Igkv4-58
|
immunoglobulin kappa variable 4-58 |
| chr12_-_11258973 | 4.83 |
ENSMUST00000049877.3
|
Msgn1
|
mesogenin 1 |
| chr4_+_99184137 | 4.73 |
ENSMUST00000094955.3
|
Gm12689
|
predicted gene 12689 |
| chr7_+_44667377 | 4.62 |
ENSMUST00000044111.10
|
Rras
|
related RAS viral (r-ras) oncogene |
| chr7_-_113716996 | 4.61 |
ENSMUST00000069449.7
|
Rras2
|
related RAS viral (r-ras) oncogene 2 |
| chr11_+_54486925 | 4.59 |
ENSMUST00000218995.2
|
Rapgef6
|
Rap guanine nucleotide exchange factor (GEF) 6 |
| chr7_+_5018453 | 4.56 |
ENSMUST00000086349.5
ENSMUST00000207050.2 |
Zfp524
Gm44973
|
zinc finger protein 524 predicted gene 44973 |
| chr4_-_63965161 | 4.53 |
ENSMUST00000107377.10
|
Tnc
|
tenascin C |
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 7.7 | 1156.0 | GO:0006910 | phagocytosis, recognition(GO:0006910) |
| 5.5 | 16.6 | GO:2000670 | positive regulation of dendritic cell apoptotic process(GO:2000670) |
| 3.5 | 10.5 | GO:0070949 | positive regulation of eosinophil degranulation(GO:0043311) regulation of neutrophil mediated cytotoxicity(GO:0070948) regulation of neutrophil mediated killing of symbiont cell(GO:0070949) positive regulation of renin secretion into blood stream(GO:1900135) positive regulation of eosinophil activation(GO:1902568) |
| 3.5 | 10.4 | GO:0021698 | cerebellar cortex structural organization(GO:0021698) |
| 3.2 | 41.6 | GO:0000185 | activation of MAPKKK activity(GO:0000185) |
| 3.1 | 824.6 | GO:0002377 | immunoglobulin production(GO:0002377) |
| 2.9 | 8.8 | GO:0045013 | carbon catabolite repression of transcription(GO:0045013) negative regulation of transcription by glucose(GO:0045014) |
| 2.8 | 11.1 | GO:0051673 | membrane disruption in other organism(GO:0051673) |
| 2.8 | 11.1 | GO:0016332 | establishment or maintenance of polarity of embryonic epithelium(GO:0016332) |
| 2.8 | 8.3 | GO:0042668 | auditory receptor cell fate determination(GO:0042668) |
| 2.5 | 17.8 | GO:0071698 | olfactory placode formation(GO:0030910) olfactory placode development(GO:0071698) olfactory placode morphogenesis(GO:0071699) |
| 2.4 | 7.3 | GO:0043973 | histone H3-K4 acetylation(GO:0043973) |
| 2.1 | 4.1 | GO:0032741 | positive regulation of interleukin-18 production(GO:0032741) |
| 2.0 | 4.1 | GO:0021502 | neural fold elevation formation(GO:0021502) |
| 2.0 | 12.1 | GO:0060847 | endothelial cell fate specification(GO:0060847) |
| 1.9 | 7.5 | GO:2000872 | positive regulation of progesterone secretion(GO:2000872) |
| 1.8 | 30.6 | GO:0034501 | protein localization to kinetochore(GO:0034501) |
| 1.7 | 15.7 | GO:2000138 | positive regulation of cell proliferation involved in heart morphogenesis(GO:2000138) |
| 1.6 | 6.5 | GO:0071348 | cellular response to interleukin-11(GO:0071348) |
| 1.5 | 18.4 | GO:0014807 | regulation of somitogenesis(GO:0014807) |
| 1.5 | 4.5 | GO:0060447 | bud outgrowth involved in lung branching(GO:0060447) |
| 1.5 | 7.4 | GO:0045347 | negative regulation of MHC class II biosynthetic process(GO:0045347) |
| 1.5 | 8.9 | GO:0050916 | sensory perception of sweet taste(GO:0050916) |
| 1.4 | 4.2 | GO:0060435 | bronchiole development(GO:0060435) trachea cartilage morphogenesis(GO:0060535) intestinal epithelial cell maturation(GO:0060574) |
| 1.4 | 24.4 | GO:0045199 | maintenance of epithelial cell apical/basal polarity(GO:0045199) |
| 1.4 | 21.7 | GO:0051152 | positive regulation of smooth muscle cell differentiation(GO:0051152) |
| 1.3 | 3.9 | GO:0098583 | mastication(GO:0071626) learned vocalization behavior(GO:0098583) |
| 1.3 | 5.1 | GO:0042231 | negative regulation of neutrophil apoptotic process(GO:0033030) interleukin-13 biosynthetic process(GO:0042231) |
| 1.3 | 6.3 | GO:1990743 | protein sialylation(GO:1990743) |
| 1.1 | 5.5 | GO:0002322 | B cell proliferation involved in immune response(GO:0002322) |
| 1.1 | 11.7 | GO:2000121 | regulation of removal of superoxide radicals(GO:2000121) |
| 1.0 | 3.1 | GO:0014908 | myotube differentiation involved in skeletal muscle regeneration(GO:0014908) |
| 0.9 | 4.5 | GO:0003409 | optic cup structural organization(GO:0003409) |
| 0.9 | 4.4 | GO:1902340 | telomeric heterochromatin assembly(GO:0031509) negative regulation of chromosome condensation(GO:1902340) |
| 0.8 | 6.6 | GO:0033484 | nitric oxide homeostasis(GO:0033484) |
| 0.8 | 2.5 | GO:1904172 | positive regulation of bleb assembly(GO:1904172) |
| 0.8 | 2.4 | GO:0021660 | rhombomere formation(GO:0021594) rhombomere 3 formation(GO:0021660) rhombomere 5 morphogenesis(GO:0021664) rhombomere 5 formation(GO:0021666) |
| 0.8 | 3.9 | GO:0014886 | transition between slow and fast fiber(GO:0014886) |
| 0.8 | 18.1 | GO:0006750 | glutathione biosynthetic process(GO:0006750) |
| 0.7 | 3.0 | GO:1902202 | regulation of hepatocyte growth factor receptor signaling pathway(GO:1902202) |
| 0.7 | 3.6 | GO:0046909 | intermembrane transport(GO:0046909) protein transport from ciliary membrane to plasma membrane(GO:1903445) |
| 0.7 | 5.4 | GO:0070162 | adiponectin secretion(GO:0070162) regulation of adiponectin secretion(GO:0070163) |
| 0.7 | 6.1 | GO:0030916 | otic vesicle formation(GO:0030916) |
| 0.7 | 2.0 | GO:0021759 | globus pallidus development(GO:0021759) |
| 0.6 | 12.2 | GO:0060056 | mammary gland involution(GO:0060056) |
| 0.6 | 3.3 | GO:0045084 | positive regulation of interleukin-12 biosynthetic process(GO:0045084) |
| 0.5 | 2.2 | GO:0021993 | initiation of neural tube closure(GO:0021993) |
| 0.5 | 37.0 | GO:1902476 | chloride transmembrane transport(GO:1902476) |
| 0.5 | 5.3 | GO:0030718 | germ-line stem cell population maintenance(GO:0030718) |
| 0.5 | 2.9 | GO:0061589 | calcium activated phosphatidylserine scrambling(GO:0061589) |
| 0.5 | 4.2 | GO:0031666 | positive regulation of lipopolysaccharide-mediated signaling pathway(GO:0031666) |
| 0.5 | 4.2 | GO:0006265 | DNA topological change(GO:0006265) |
| 0.4 | 2.7 | GO:2000053 | regulation of mismatch repair(GO:0032423) regulation of chondrocyte development(GO:0061181) regulation of Wnt signaling pathway involved in dorsal/ventral axis specification(GO:2000053) negative regulation of Wnt signaling pathway involved in dorsal/ventral axis specification(GO:2000054) |
| 0.4 | 7.5 | GO:0015693 | magnesium ion transport(GO:0015693) |
| 0.4 | 5.2 | GO:0034983 | peptidyl-lysine deacetylation(GO:0034983) |
| 0.4 | 2.6 | GO:2000210 | positive regulation of anoikis(GO:2000210) |
| 0.4 | 5.1 | GO:0006268 | DNA unwinding involved in DNA replication(GO:0006268) |
| 0.4 | 4.2 | GO:1902166 | negative regulation of intrinsic apoptotic signaling pathway in response to DNA damage by p53 class mediator(GO:1902166) |
| 0.4 | 7.6 | GO:0033234 | negative regulation of protein sumoylation(GO:0033234) |
| 0.4 | 21.6 | GO:0006284 | base-excision repair(GO:0006284) |
| 0.4 | 2.4 | GO:0071484 | cellular response to light intensity(GO:0071484) cellular response to high light intensity(GO:0071486) retinal rod cell apoptotic process(GO:0097473) |
| 0.4 | 2.0 | GO:0018101 | protein citrullination(GO:0018101) histone citrullination(GO:0036414) |
| 0.4 | 2.3 | GO:0090158 | endoplasmic reticulum membrane organization(GO:0090158) |
| 0.4 | 1.9 | GO:0060686 | negative regulation of prostatic bud formation(GO:0060686) |
| 0.4 | 11.4 | GO:0060252 | positive regulation of glial cell proliferation(GO:0060252) |
| 0.4 | 4.2 | GO:0043568 | positive regulation of insulin-like growth factor receptor signaling pathway(GO:0043568) |
| 0.4 | 2.3 | GO:1900245 | positive regulation of MDA-5 signaling pathway(GO:1900245) |
| 0.4 | 12.9 | GO:0050860 | negative regulation of T cell receptor signaling pathway(GO:0050860) |
| 0.3 | 2.4 | GO:0007527 | adult somatic muscle development(GO:0007527) |
| 0.3 | 1.0 | GO:0021530 | spinal cord oligodendrocyte cell differentiation(GO:0021529) spinal cord oligodendrocyte cell fate specification(GO:0021530) |
| 0.3 | 3.4 | GO:0032000 | positive regulation of fatty acid beta-oxidation(GO:0032000) |
| 0.3 | 1.9 | GO:2000255 | negative regulation of male germ cell proliferation(GO:2000255) |
| 0.3 | 25.4 | GO:0003382 | epithelial cell morphogenesis(GO:0003382) |
| 0.3 | 4.1 | GO:0048023 | positive regulation of melanin biosynthetic process(GO:0048023) positive regulation of secondary metabolite biosynthetic process(GO:1900378) |
| 0.3 | 2.7 | GO:0015712 | hexose phosphate transport(GO:0015712) glucose-6-phosphate transport(GO:0015760) |
| 0.3 | 5.1 | GO:0006851 | mitochondrial calcium ion transport(GO:0006851) |
| 0.3 | 0.5 | GO:1901228 | positive regulation of transcription from RNA polymerase II promoter involved in myocardial precursor cell differentiation(GO:0003257) positive regulation of transcription from RNA polymerase II promoter involved in heart development(GO:1901228) |
| 0.2 | 4.8 | GO:0007379 | segment specification(GO:0007379) |
| 0.2 | 1.0 | GO:2000230 | pancreatic stellate cell proliferation(GO:0072343) regulation of pancreatic stellate cell proliferation(GO:2000229) negative regulation of pancreatic stellate cell proliferation(GO:2000230) |
| 0.2 | 0.9 | GO:0007352 | zygotic specification of dorsal/ventral axis(GO:0007352) positive regulation of nodal signaling pathway involved in determination of lateral mesoderm left/right asymmetry(GO:1900224) |
| 0.2 | 1.4 | GO:0048539 | bone marrow development(GO:0048539) |
| 0.2 | 0.9 | GO:0060800 | regulation of cell differentiation involved in embryonic placenta development(GO:0060800) |
| 0.2 | 1.4 | GO:1903644 | regulation of chaperone-mediated protein folding(GO:1903644) |
| 0.2 | 19.1 | GO:0038083 | peptidyl-tyrosine autophosphorylation(GO:0038083) |
| 0.2 | 24.7 | GO:0030183 | B cell differentiation(GO:0030183) |
| 0.2 | 2.0 | GO:0018230 | peptidyl-L-cysteine S-palmitoylation(GO:0018230) peptidyl-S-diacylglycerol-L-cysteine biosynthetic process from peptidyl-cysteine(GO:0018231) |
| 0.2 | 0.7 | GO:0010982 | regulation of high-density lipoprotein particle clearance(GO:0010982) |
| 0.2 | 4.5 | GO:0002115 | store-operated calcium entry(GO:0002115) |
| 0.1 | 27.3 | GO:0007338 | single fertilization(GO:0007338) |
| 0.1 | 5.3 | GO:0030225 | macrophage differentiation(GO:0030225) |
| 0.1 | 4.6 | GO:0030033 | microvillus assembly(GO:0030033) |
| 0.1 | 3.8 | GO:0002076 | osteoblast development(GO:0002076) |
| 0.1 | 0.8 | GO:0048743 | positive regulation of skeletal muscle fiber development(GO:0048743) |
| 0.1 | 0.5 | GO:0070346 | positive regulation of fat cell proliferation(GO:0070346) |
| 0.1 | 1.5 | GO:0010606 | positive regulation of cytoplasmic mRNA processing body assembly(GO:0010606) |
| 0.1 | 17.3 | GO:0007098 | centrosome cycle(GO:0007098) |
| 0.1 | 7.1 | GO:0060325 | face morphogenesis(GO:0060325) |
| 0.1 | 1.3 | GO:0002002 | regulation of angiotensin levels in blood(GO:0002002) regulation of angiotensin metabolic process(GO:0060177) |
| 0.1 | 0.9 | GO:0006477 | protein sulfation(GO:0006477) |
| 0.1 | 0.8 | GO:0006104 | succinyl-CoA metabolic process(GO:0006104) |
| 0.1 | 0.2 | GO:0031959 | mineralocorticoid receptor signaling pathway(GO:0031959) |
| 0.1 | 6.0 | GO:0035307 | positive regulation of protein dephosphorylation(GO:0035307) |
| 0.1 | 0.2 | GO:0000320 | re-entry into mitotic cell cycle(GO:0000320) |
| 0.1 | 3.0 | GO:0002053 | positive regulation of mesenchymal cell proliferation(GO:0002053) |
| 0.1 | 1.7 | GO:0016180 | snRNA processing(GO:0016180) |
| 0.1 | 1.9 | GO:0097435 | fibril organization(GO:0097435) |
| 0.1 | 4.4 | GO:0010762 | regulation of fibroblast migration(GO:0010762) |
| 0.1 | 1.3 | GO:0015937 | coenzyme A biosynthetic process(GO:0015937) |
| 0.1 | 1.7 | GO:0001967 | suckling behavior(GO:0001967) |
| 0.1 | 0.5 | GO:0097475 | motor neuron migration(GO:0097475) |
| 0.1 | 0.8 | GO:0019321 | pentose metabolic process(GO:0019321) |
| 0.1 | 3.6 | GO:0002260 | lymphocyte homeostasis(GO:0002260) |
| 0.1 | 4.1 | GO:0045576 | mast cell activation(GO:0045576) |
| 0.0 | 0.3 | GO:0061368 | behavioral response to chemical pain(GO:0061366) behavioral response to formalin induced pain(GO:0061368) |
| 0.0 | 1.0 | GO:0007095 | mitotic G2 DNA damage checkpoint(GO:0007095) |
| 0.0 | 0.7 | GO:0048387 | negative regulation of retinoic acid receptor signaling pathway(GO:0048387) |
| 0.0 | 13.6 | GO:0019221 | cytokine-mediated signaling pathway(GO:0019221) |
| 0.0 | 1.8 | GO:0048286 | lung alveolus development(GO:0048286) |
| 0.0 | 0.8 | GO:0075522 | IRES-dependent viral translational initiation(GO:0075522) |
| 0.0 | 0.7 | GO:0010569 | regulation of double-strand break repair via homologous recombination(GO:0010569) |
| 0.0 | 1.8 | GO:0042475 | odontogenesis of dentin-containing tooth(GO:0042475) |
| 0.0 | 3.5 | GO:0006261 | DNA-dependent DNA replication(GO:0006261) |
| 0.0 | 1.0 | GO:0032728 | positive regulation of interferon-beta production(GO:0032728) |
| 0.0 | 1.4 | GO:0045670 | regulation of osteoclast differentiation(GO:0045670) |
| 0.0 | 2.8 | GO:0051028 | mRNA transport(GO:0051028) |
| 0.0 | 4.2 | GO:0030833 | regulation of actin filament polymerization(GO:0030833) |
| 0.0 | 1.0 | GO:0032611 | interleukin-1 beta production(GO:0032611) |
| 0.0 | 1.3 | GO:0007029 | endoplasmic reticulum organization(GO:0007029) |
| 0.0 | 1.0 | GO:0007585 | respiratory gaseous exchange(GO:0007585) |
| 0.0 | 0.9 | GO:0010501 | RNA secondary structure unwinding(GO:0010501) |
| 0.0 | 0.8 | GO:0032088 | negative regulation of NF-kappaB transcription factor activity(GO:0032088) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 9.4 | 1156.0 | GO:0042571 | immunoglobulin complex, circulating(GO:0042571) |
| 4.2 | 16.6 | GO:0034666 | integrin alpha2-beta1 complex(GO:0034666) |
| 3.5 | 24.7 | GO:0019814 | immunoglobulin complex(GO:0019814) |
| 2.7 | 21.6 | GO:0035985 | senescence-associated heterochromatin focus(GO:0035985) |
| 2.3 | 11.7 | GO:0071953 | elastic fiber(GO:0071953) |
| 1.9 | 7.6 | GO:0002111 | BRCA2-BRAF35 complex(GO:0002111) |
| 1.6 | 11.1 | GO:0072559 | NLRP3 inflammasome complex(GO:0072559) |
| 1.2 | 9.5 | GO:0002193 | MAML1-RBP-Jkappa- ICN1 complex(GO:0002193) |
| 0.9 | 3.5 | GO:0008623 | CHRAC(GO:0008623) |
| 0.8 | 2.4 | GO:0042272 | nuclear RNA export factor complex(GO:0042272) |
| 0.8 | 30.6 | GO:0000777 | condensed chromosome kinetochore(GO:0000777) |
| 0.8 | 3.8 | GO:0035976 | AP1 complex(GO:0035976) |
| 0.7 | 4.1 | GO:0036019 | endolysosome(GO:0036019) |
| 0.6 | 8.1 | GO:0000788 | nuclear nucleosome(GO:0000788) |
| 0.6 | 4.2 | GO:0031298 | replication fork protection complex(GO:0031298) |
| 0.6 | 2.3 | GO:0014701 | junctional sarcoplasmic reticulum membrane(GO:0014701) |
| 0.5 | 10.5 | GO:0031143 | pseudopodium(GO:0031143) |
| 0.5 | 4.1 | GO:0032585 | multivesicular body membrane(GO:0032585) |
| 0.5 | 6.3 | GO:0000138 | Golgi trans cisterna(GO:0000138) |
| 0.4 | 14.9 | GO:0030057 | desmosome(GO:0030057) |
| 0.4 | 9.5 | GO:0005640 | nuclear outer membrane(GO:0005640) |
| 0.4 | 11.7 | GO:0005614 | interstitial matrix(GO:0005614) |
| 0.3 | 3.4 | GO:0045179 | apical cortex(GO:0045179) |
| 0.3 | 3.0 | GO:0001674 | female germ cell nucleus(GO:0001674) |
| 0.3 | 688.8 | GO:0005615 | extracellular space(GO:0005615) |
| 0.3 | 41.6 | GO:0017053 | transcriptional repressor complex(GO:0017053) |
| 0.3 | 2.5 | GO:0005826 | actomyosin contractile ring(GO:0005826) |
| 0.3 | 5.1 | GO:0005662 | DNA replication factor A complex(GO:0005662) |
| 0.3 | 1.5 | GO:0031251 | PAN complex(GO:0031251) |
| 0.3 | 3.3 | GO:0033256 | I-kappaB/NF-kappaB complex(GO:0033256) |
| 0.2 | 2.6 | GO:0070822 | Sin3-type complex(GO:0070822) |
| 0.2 | 26.6 | GO:0016605 | PML body(GO:0016605) |
| 0.2 | 0.9 | GO:0032444 | activin responsive factor complex(GO:0032444) SMAD2-SMAD3 protein complex(GO:0071144) |
| 0.2 | 3.4 | GO:0035102 | PRC1 complex(GO:0035102) |
| 0.2 | 28.0 | GO:0030176 | integral component of endoplasmic reticulum membrane(GO:0030176) |
| 0.1 | 2.4 | GO:0070938 | contractile ring(GO:0070938) |
| 0.1 | 2.4 | GO:0032593 | insulin-responsive compartment(GO:0032593) |
| 0.1 | 7.5 | GO:0005604 | basement membrane(GO:0005604) |
| 0.1 | 2.7 | GO:0000178 | exosome (RNase complex)(GO:0000178) |
| 0.1 | 48.0 | GO:0016607 | nuclear speck(GO:0016607) |
| 0.1 | 1.7 | GO:0032039 | integrator complex(GO:0032039) |
| 0.1 | 1.4 | GO:0031227 | intrinsic component of endoplasmic reticulum membrane(GO:0031227) |
| 0.1 | 0.8 | GO:0045252 | oxoglutarate dehydrogenase complex(GO:0045252) |
| 0.1 | 2.2 | GO:0031519 | PcG protein complex(GO:0031519) |
| 0.1 | 12.1 | GO:0005913 | cell-cell adherens junction(GO:0005913) |
| 0.1 | 17.3 | GO:0005819 | spindle(GO:0005819) |
| 0.1 | 5.2 | GO:0035097 | histone methyltransferase complex(GO:0035097) |
| 0.1 | 3.1 | GO:0005801 | cis-Golgi network(GO:0005801) |
| 0.1 | 3.7 | GO:0032420 | stereocilium(GO:0032420) |
| 0.0 | 0.5 | GO:0072669 | tRNA-splicing ligase complex(GO:0072669) |
| 0.0 | 1.5 | GO:0045095 | keratin filament(GO:0045095) |
| 0.0 | 3.8 | GO:0005902 | microvillus(GO:0005902) |
| 0.0 | 2.2 | GO:0005881 | cytoplasmic microtubule(GO:0005881) |
| 0.0 | 4.1 | GO:0000794 | condensed nuclear chromosome(GO:0000794) |
| 0.0 | 7.7 | GO:0098857 | membrane raft(GO:0045121) membrane microdomain(GO:0098857) |
| 0.0 | 1.9 | GO:0005578 | proteinaceous extracellular matrix(GO:0005578) |
| 0.0 | 2.8 | GO:0005681 | spliceosomal complex(GO:0005681) |
| 0.0 | 1.5 | GO:0036064 | ciliary basal body(GO:0036064) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 8.7 | 1156.0 | GO:0034987 | immunoglobulin receptor binding(GO:0034987) |
| 4.5 | 18.1 | GO:0004464 | leukotriene-C4 synthase activity(GO:0004464) |
| 4.2 | 16.6 | GO:0005128 | erythropoietin receptor binding(GO:0005128) |
| 2.6 | 10.5 | GO:0015057 | thrombin receptor activity(GO:0015057) |
| 2.4 | 21.6 | GO:0051575 | 5'-deoxyribose-5-phosphate lyase activity(GO:0051575) |
| 2.4 | 9.5 | GO:0000822 | inositol hexakisphosphate binding(GO:0000822) |
| 2.1 | 22.8 | GO:1901611 | phosphatidylglycerol binding(GO:1901611) |
| 1.4 | 4.2 | GO:0097100 | supercoiled DNA binding(GO:0097100) |
| 1.3 | 9.3 | GO:0043515 | kinetochore binding(GO:0043515) |
| 1.3 | 5.1 | GO:0032422 | purine-rich negative regulatory element binding(GO:0032422) |
| 1.3 | 132.7 | GO:0003823 | antigen binding(GO:0003823) |
| 1.2 | 39.9 | GO:0005229 | intracellular calcium activated chloride channel activity(GO:0005229) |
| 1.2 | 15.7 | GO:0043024 | ribosomal small subunit binding(GO:0043024) |
| 1.1 | 6.3 | GO:0003835 | beta-galactoside alpha-2,6-sialyltransferase activity(GO:0003835) |
| 1.0 | 3.0 | GO:0005171 | hepatocyte growth factor receptor binding(GO:0005171) |
| 0.8 | 2.4 | GO:0016781 | selenide, water dikinase activity(GO:0004756) phosphotransferase activity, paired acceptors(GO:0016781) |
| 0.7 | 19.0 | GO:0098641 | cadherin binding involved in cell-cell adhesion(GO:0098641) |
| 0.6 | 18.3 | GO:0000983 | transcription factor activity, RNA polymerase II core promoter sequence-specific(GO:0000983) |
| 0.6 | 3.6 | GO:0045322 | unmethylated CpG binding(GO:0045322) |
| 0.5 | 10.4 | GO:0030957 | Tat protein binding(GO:0030957) |
| 0.5 | 7.4 | GO:0051525 | NFAT protein binding(GO:0051525) |
| 0.5 | 1.9 | GO:0004667 | prostaglandin-D synthase activity(GO:0004667) |
| 0.5 | 7.3 | GO:0030274 | LIM domain binding(GO:0030274) |
| 0.5 | 15.1 | GO:0003887 | DNA-directed DNA polymerase activity(GO:0003887) |
| 0.5 | 3.7 | GO:0070644 | vitamin D response element binding(GO:0070644) |
| 0.5 | 5.9 | GO:0005001 | transmembrane receptor protein tyrosine phosphatase activity(GO:0005001) transmembrane receptor protein phosphatase activity(GO:0019198) |
| 0.5 | 4.5 | GO:0045545 | syndecan binding(GO:0045545) |
| 0.4 | 4.4 | GO:0031492 | nucleosomal DNA binding(GO:0031492) |
| 0.4 | 8.8 | GO:0001205 | transcriptional activator activity, RNA polymerase II distal enhancer sequence-specific binding(GO:0001205) |
| 0.4 | 2.0 | GO:0004668 | protein-arginine deiminase activity(GO:0004668) |
| 0.4 | 2.7 | GO:0015315 | hexose phosphate transmembrane transporter activity(GO:0015119) organophosphate:inorganic phosphate antiporter activity(GO:0015315) hexose-phosphate:inorganic phosphate antiporter activity(GO:0015526) glucose 6-phosphate:inorganic phosphate antiporter activity(GO:0061513) |
| 0.4 | 7.5 | GO:0015095 | magnesium ion transmembrane transporter activity(GO:0015095) |
| 0.4 | 3.4 | GO:0032052 | long-chain fatty acid transporter activity(GO:0005324) bile acid binding(GO:0032052) |
| 0.4 | 4.8 | GO:0070700 | BMP receptor binding(GO:0070700) |
| 0.3 | 0.9 | GO:0030618 | transforming growth factor beta receptor, pathway-specific cytoplasmic mediator activity(GO:0030618) |
| 0.3 | 7.6 | GO:0000400 | four-way junction DNA binding(GO:0000400) |
| 0.3 | 2.7 | GO:0004126 | cytidine deaminase activity(GO:0004126) |
| 0.3 | 19.1 | GO:0005070 | SH3/SH2 adaptor activity(GO:0005070) |
| 0.3 | 4.5 | GO:0015279 | store-operated calcium channel activity(GO:0015279) |
| 0.3 | 14.4 | GO:0003785 | actin monomer binding(GO:0003785) |
| 0.3 | 2.5 | GO:0015016 | [heparan sulfate]-glucosamine N-sulfotransferase activity(GO:0015016) |
| 0.3 | 39.6 | GO:0000980 | RNA polymerase II distal enhancer sequence-specific DNA binding(GO:0000980) |
| 0.3 | 58.9 | GO:0047485 | protein N-terminus binding(GO:0047485) |
| 0.3 | 1.9 | GO:0008449 | N-acetylglucosamine-6-sulfatase activity(GO:0008449) |
| 0.3 | 2.2 | GO:0051864 | histone demethylase activity (H3-K36 specific)(GO:0051864) |
| 0.2 | 5.2 | GO:0032041 | histone deacetylase activity (H3-K14 specific)(GO:0031078) NAD-dependent histone deacetylase activity (H3-K14 specific)(GO:0032041) |
| 0.2 | 2.4 | GO:0034046 | poly(G) binding(GO:0034046) |
| 0.2 | 1.3 | GO:0004594 | pantothenate kinase activity(GO:0004594) |
| 0.2 | 4.2 | GO:0070577 | lysine-acetylated histone binding(GO:0070577) |
| 0.2 | 4.6 | GO:0070300 | phosphatidic acid binding(GO:0070300) |
| 0.2 | 5.4 | GO:0001614 | purinergic nucleotide receptor activity(GO:0001614) nucleotide receptor activity(GO:0016502) |
| 0.2 | 3.8 | GO:0032454 | histone demethylase activity (H3-K9 specific)(GO:0032454) |
| 0.2 | 0.8 | GO:0034602 | oxoglutarate dehydrogenase (NAD+) activity(GO:0034602) |
| 0.2 | 5.6 | GO:0001968 | fibronectin binding(GO:0001968) |
| 0.1 | 0.9 | GO:0008467 | [heparan sulfate]-glucosamine 3-sulfotransferase 1 activity(GO:0008467) |
| 0.1 | 2.7 | GO:0070411 | I-SMAD binding(GO:0070411) |
| 0.1 | 2.0 | GO:0001161 | intronic transcription regulatory region sequence-specific DNA binding(GO:0001161) |
| 0.1 | 0.7 | GO:0008970 | phosphatidylcholine 1-acylhydrolase activity(GO:0008970) |
| 0.1 | 1.9 | GO:0048407 | platelet-derived growth factor binding(GO:0048407) |
| 0.1 | 2.8 | GO:0005112 | Notch binding(GO:0005112) |
| 0.1 | 15.6 | GO:0005178 | integrin binding(GO:0005178) |
| 0.1 | 5.6 | GO:0048487 | beta-tubulin binding(GO:0048487) |
| 0.1 | 7.5 | GO:0070491 | repressing transcription factor binding(GO:0070491) |
| 0.1 | 1.0 | GO:1901567 | icosanoid binding(GO:0050542) icosatetraenoic acid binding(GO:0050543) arachidonic acid binding(GO:0050544) fatty acid derivative binding(GO:1901567) |
| 0.1 | 2.3 | GO:0019789 | SUMO transferase activity(GO:0019789) |
| 0.1 | 2.6 | GO:0051059 | NF-kappaB binding(GO:0051059) |
| 0.1 | 3.0 | GO:0016814 | hydrolase activity, acting on carbon-nitrogen (but not peptide) bonds, in cyclic amidines(GO:0016814) |
| 0.1 | 2.0 | GO:0019706 | protein-cysteine S-palmitoyltransferase activity(GO:0019706) protein-cysteine S-acyltransferase activity(GO:0019707) |
| 0.1 | 27.0 | GO:0005096 | GTPase activator activity(GO:0005096) |
| 0.1 | 1.5 | GO:0004535 | poly(A)-specific ribonuclease activity(GO:0004535) |
| 0.1 | 1.4 | GO:0070628 | proteasome binding(GO:0070628) |
| 0.1 | 0.8 | GO:1990829 | C-rich single-stranded DNA binding(GO:1990829) |
| 0.1 | 0.2 | GO:0017082 | mineralocorticoid receptor activity(GO:0017082) |
| 0.1 | 4.2 | GO:0002039 | p53 binding(GO:0002039) |
| 0.1 | 3.6 | GO:0019003 | GDP binding(GO:0019003) |
| 0.0 | 2.4 | GO:0001102 | RNA polymerase II activating transcription factor binding(GO:0001102) |
| 0.0 | 0.1 | GO:0051373 | FATZ binding(GO:0051373) |
| 0.0 | 2.8 | GO:0003697 | single-stranded DNA binding(GO:0003697) |
| 0.0 | 1.0 | GO:0004709 | MAP kinase kinase kinase activity(GO:0004709) |
| 0.0 | 5.0 | GO:0004386 | helicase activity(GO:0004386) |
| 0.0 | 1.8 | GO:0005080 | protein kinase C binding(GO:0005080) |
| 0.0 | 0.6 | GO:0043027 | cysteine-type endopeptidase inhibitor activity involved in apoptotic process(GO:0043027) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.1 | 42.6 | PID P38 MKK3 6PATHWAY | p38 MAPK signaling pathway |
| 1.0 | 32.3 | PID IL5 PATHWAY | IL5-mediated signaling events |
| 0.6 | 15.1 | PID DNA PK PATHWAY | DNA-PK pathway in nonhomologous end joining |
| 0.6 | 3.3 | PID NFKAPPAB ATYPICAL PATHWAY | Atypical NF-kappaB pathway |
| 0.4 | 25.0 | PID BCR 5PATHWAY | BCR signaling pathway |
| 0.4 | 33.8 | PID IL4 2PATHWAY | IL4-mediated signaling events |
| 0.4 | 11.7 | PID IL27 PATHWAY | IL27-mediated signaling events |
| 0.4 | 17.2 | PID TCR CALCIUM PATHWAY | Calcium signaling in the CD4+ TCR pathway |
| 0.4 | 11.9 | ST GAQ PATHWAY | G alpha q Pathway |
| 0.3 | 12.9 | PID IL2 STAT5 PATHWAY | IL2 signaling events mediated by STAT5 |
| 0.3 | 10.5 | ST GA12 PATHWAY | G alpha 12 Pathway |
| 0.3 | 17.9 | PID PS1 PATHWAY | Presenilin action in Notch and Wnt signaling |
| 0.2 | 4.6 | PID RAS PATHWAY | Regulation of Ras family activation |
| 0.1 | 2.7 | PID BETA CATENIN DEG PATHWAY | Degradation of beta catenin |
| 0.1 | 4.5 | PID SYNDECAN 4 PATHWAY | Syndecan-4-mediated signaling events |
| 0.1 | 8.2 | PID HEDGEHOG GLI PATHWAY | Hedgehog signaling events mediated by Gli proteins |
| 0.1 | 2.7 | PID FRA PATHWAY | Validated transcriptional targets of AP1 family members Fra1 and Fra2 |
| 0.1 | 7.5 | PID DELTA NP63 PATHWAY | Validated transcriptional targets of deltaNp63 isoforms |
| 0.1 | 10.5 | PID HDAC CLASSI PATHWAY | Signaling events mediated by HDAC Class I |
| 0.1 | 5.4 | PID HES HEY PATHWAY | Notch-mediated HES/HEY network |
| 0.1 | 8.7 | PID CASPASE PATHWAY | Caspase cascade in apoptosis |
| 0.1 | 3.0 | NABA PROTEOGLYCANS | Genes encoding proteoglycans |
| 0.1 | 1.2 | PID HDAC CLASSIII PATHWAY | Signaling events mediated by HDAC Class III |
| 0.1 | 3.8 | PID TCR PATHWAY | TCR signaling in naïve CD4+ T cells |
| 0.0 | 0.9 | PID SMAD2 3PATHWAY | Regulation of cytoplasmic and nuclear SMAD2/3 signaling |
| 0.0 | 11.0 | NABA ECM GLYCOPROTEINS | Genes encoding structural ECM glycoproteins |
| 0.0 | 0.5 | ST GA13 PATHWAY | G alpha 13 Pathway |
| 0.0 | 1.9 | PID SYNDECAN 1 PATHWAY | Syndecan-1-mediated signaling events |
| 0.0 | 1.5 | PID CERAMIDE PATHWAY | Ceramide signaling pathway |
| 0.0 | 1.3 | PID TAP63 PATHWAY | Validated transcriptional targets of TAp63 isoforms |
| 0.0 | 0.5 | PID ANGIOPOIETIN RECEPTOR PATHWAY | Angiopoietin receptor Tie2-mediated signaling |
| 0.0 | 4.3 | NABA ECM AFFILIATED | Genes encoding proteins affiliated structurally or functionally to extracellular matrix proteins |
| 0.0 | 0.8 | PID BMP PATHWAY | BMP receptor signaling |
| 0.0 | 1.7 | PID P53 DOWNSTREAM PATHWAY | Direct p53 effectors |
| 0.0 | 0.9 | WNT SIGNALING | Genes related to Wnt-mediated signal transduction |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 3.1 | 21.6 | REACTOME APOBEC3G MEDIATED RESISTANCE TO HIV1 INFECTION | Genes involved in APOBEC3G mediated resistance to HIV-1 infection |
| 1.4 | 14.0 | REACTOME ELEVATION OF CYTOSOLIC CA2 LEVELS | Genes involved in Elevation of cytosolic Ca2+ levels |
| 1.0 | 16.6 | REACTOME PLATELET ADHESION TO EXPOSED COLLAGEN | Genes involved in Platelet Adhesion to exposed collagen |
| 0.7 | 10.4 | REACTOME RECEPTOR LIGAND BINDING INITIATES THE SECOND PROTEOLYTIC CLEAVAGE OF NOTCH RECEPTOR | Genes involved in Receptor-ligand binding initiates the second proteolytic cleavage of Notch receptor |
| 0.6 | 17.8 | REACTOME RNA POL III TRANSCRIPTION INITIATION FROM TYPE 3 PROMOTER | Genes involved in RNA Polymerase III Transcription Initiation From Type 3 Promoter |
| 0.5 | 25.0 | REACTOME ANTIGEN ACTIVATES B CELL RECEPTOR LEADING TO GENERATION OF SECOND MESSENGERS | Genes involved in Antigen Activates B Cell Receptor Leading to Generation of Second Messengers |
| 0.5 | 9.8 | REACTOME REGULATION OF GENE EXPRESSION IN BETA CELLS | Genes involved in Regulation of gene expression in beta cells |
| 0.4 | 6.6 | REACTOME NOTCH HLH TRANSCRIPTION PATHWAY | Genes involved in Notch-HLH transcription pathway |
| 0.4 | 18.1 | REACTOME GLUTATHIONE CONJUGATION | Genes involved in Glutathione conjugation |
| 0.3 | 4.1 | REACTOME TRAF6 MEDIATED IRF7 ACTIVATION IN TLR7 8 OR 9 SIGNALING | Genes involved in TRAF6 mediated IRF7 activation in TLR7/8 or 9 signaling |
| 0.3 | 4.4 | REACTOME RNA POL I PROMOTER OPENING | Genes involved in RNA Polymerase I Promoter Opening |
| 0.3 | 4.6 | REACTOME SEMA3A PLEXIN REPULSION SIGNALING BY INHIBITING INTEGRIN ADHESION | Genes involved in SEMA3A-Plexin repulsion signaling by inhibiting Integrin adhesion |
| 0.2 | 6.3 | REACTOME TERMINATION OF O GLYCAN BIOSYNTHESIS | Genes involved in Termination of O-glycan biosynthesis |
| 0.2 | 2.4 | REACTOME TRANSCRIPTIONAL REGULATION OF WHITE ADIPOCYTE DIFFERENTIATION | Genes involved in Transcriptional Regulation of White Adipocyte Differentiation |
| 0.2 | 2.4 | REACTOME NEGATIVE REGULATION OF THE PI3K AKT NETWORK | Genes involved in Negative regulation of the PI3K/AKT network |
| 0.1 | 4.1 | REACTOME SIGNAL REGULATORY PROTEIN SIRP FAMILY INTERACTIONS | Genes involved in Signal regulatory protein (SIRP) family interactions |
| 0.1 | 16.9 | REACTOME RESPONSE TO ELEVATED PLATELET CYTOSOLIC CA2 | Genes involved in Response to elevated platelet cytosolic Ca2+ |
| 0.1 | 3.4 | REACTOME HS GAG BIOSYNTHESIS | Genes involved in HS-GAG biosynthesis |
| 0.1 | 5.2 | REACTOME NOTCH1 INTRACELLULAR DOMAIN REGULATES TRANSCRIPTION | Genes involved in NOTCH1 Intracellular Domain Regulates Transcription |
| 0.1 | 1.3 | REACTOME VITAMIN B5 PANTOTHENATE METABOLISM | Genes involved in Vitamin B5 (pantothenate) metabolism |
| 0.1 | 10.5 | REACTOME FACTORS INVOLVED IN MEGAKARYOCYTE DEVELOPMENT AND PLATELET PRODUCTION | Genes involved in Factors involved in megakaryocyte development and platelet production |
| 0.1 | 2.0 | REACTOME ENOS ACTIVATION AND REGULATION | Genes involved in eNOS activation and regulation |
| 0.1 | 10.5 | REACTOME PEPTIDE LIGAND BINDING RECEPTORS | Genes involved in Peptide ligand-binding receptors |
| 0.0 | 1.3 | REACTOME SYNTHESIS SECRETION AND INACTIVATION OF GLP1 | Genes involved in Synthesis, Secretion, and Inactivation of Glucagon-like Peptide-1 (GLP-1) |
| 0.0 | 3.3 | REACTOME ACTIVATION OF NF KAPPAB IN B CELLS | Genes involved in Activation of NF-kappaB in B Cells |
| 0.0 | 5.1 | REACTOME SIGNALING BY ILS | Genes involved in Signaling by Interleukins |
| 0.0 | 4.9 | REACTOME CLASS A1 RHODOPSIN LIKE RECEPTORS | Genes involved in Class A/1 (Rhodopsin-like receptors) |
| 0.0 | 0.9 | REACTOME SIGNALING BY NODAL | Genes involved in Signaling by NODAL |
| 0.0 | 0.8 | REACTOME CITRIC ACID CYCLE TCA CYCLE | Genes involved in Citric acid cycle (TCA cycle) |
| 0.0 | 1.9 | REACTOME NCAM1 INTERACTIONS | Genes involved in NCAM1 interactions |
| 0.0 | 1.6 | REACTOME NEGATIVE REGULATORS OF RIG I MDA5 SIGNALING | Genes involved in Negative regulators of RIG-I/MDA5 signaling |
| 0.0 | 2.3 | REACTOME TOLL RECEPTOR CASCADES | Genes involved in Toll Receptor Cascades |