Project
PRJNA375882: Comprehensive Mouse Transcriptomic BodyMap
Navigation
Downloads
Results for Pou3f2
Z-value: 1.17
Transcription factors associated with Pou3f2
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Pou3f2
|
ENSMUSG00000095139.3 | Pou3f2 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Pou3f2 | mm39_v1_chr4_-_22488296_22488366 | 0.47 | 3.0e-05 | Click! |
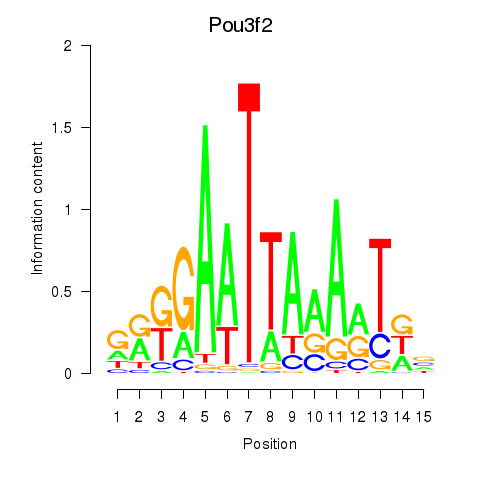
Activity profile of Pou3f2 motif
Sorted Z-values of Pou3f2 motif
Network of associatons between targets according to the STRING database.
First level regulatory network of Pou3f2
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr8_+_21999274 | 12.74 |
ENSMUST00000084042.4
|
Defa20
|
defensin, alpha, 20 |
| chr8_+_21868531 | 12.59 |
ENSMUST00000170275.4
|
Defa2
|
defensin, alpha, 2 |
| chr8_+_21881827 | 12.42 |
ENSMUST00000120874.5
|
Defa33
|
defensin, alpha, 33 |
| chr8_+_22224506 | 12.36 |
ENSMUST00000080533.6
|
Defa24
|
defensin, alpha, 24 |
| chr8_+_22019048 | 12.21 |
ENSMUST00000084041.4
|
Defa32
|
defensin, alpha, 32 |
| chr8_+_22108199 | 10.31 |
ENSMUST00000074343.6
|
Defa26
|
defensin, alpha, 26 |
| chr4_+_11758147 | 10.11 |
ENSMUST00000029871.12
ENSMUST00000108303.2 |
Cdh17
|
cadherin 17 |
| chr2_-_62313981 | 8.48 |
ENSMUST00000136686.2
ENSMUST00000102733.10 |
Gcg
|
glucagon |
| chrX_+_95499968 | 7.98 |
ENSMUST00000079322.12
ENSMUST00000113838.8 |
Heph
|
hephaestin |
| chr8_+_21739729 | 7.57 |
ENSMUST00000098895.3
|
Defa40
|
defensin, alpha, 40 |
| chr11_-_78313043 | 6.69 |
ENSMUST00000001122.6
|
Slc13a2
|
solute carrier family 13 (sodium-dependent dicarboxylate transporter), member 2 |
| chr13_-_110416637 | 6.13 |
ENSMUST00000167824.3
ENSMUST00000224180.2 |
Rab3c
|
RAB3C, member RAS oncogene family |
| chr10_+_115979787 | 6.08 |
ENSMUST00000105271.9
|
Ptprr
|
protein tyrosine phosphatase, receptor type, R |
| chr1_-_162641495 | 6.04 |
ENSMUST00000144916.8
ENSMUST00000140274.2 |
Fmo4
|
flavin containing monooxygenase 4 |
| chr16_-_48232770 | 5.72 |
ENSMUST00000212197.2
|
Gm5485
|
predicted gene 5485 |
| chr2_+_177834868 | 5.68 |
ENSMUST00000103065.2
|
Phactr3
|
phosphatase and actin regulator 3 |
| chr17_-_34694911 | 5.68 |
ENSMUST00000065841.5
|
Btnl4
|
butyrophilin-like 4 |
| chr8_+_21805562 | 5.20 |
ENSMUST00000167683.3
ENSMUST00000168340.2 |
Defa27
|
defensin, alpha, 27 |
| chr11_-_42070517 | 4.94 |
ENSMUST00000206105.2
ENSMUST00000153147.3 |
Gabra1
|
gamma-aminobutyric acid (GABA) A receptor, subunit alpha 1 |
| chr14_-_88708782 | 4.81 |
ENSMUST00000192557.2
ENSMUST00000061628.7 |
Pcdh20
|
protocadherin 20 |
| chr6_+_40605758 | 4.76 |
ENSMUST00000202636.4
ENSMUST00000201148.4 ENSMUST00000071535.10 |
Mgam
|
maltase-glucoamylase |
| chr5_+_14075281 | 4.63 |
ENSMUST00000073957.8
|
Sema3e
|
sema domain, immunoglobulin domain (Ig), short basic domain, secreted, (semaphorin) 3E |
| chr12_+_52746158 | 4.53 |
ENSMUST00000095737.5
|
Akap6
|
A kinase (PRKA) anchor protein 6 |
| chr3_+_55369149 | 4.34 |
ENSMUST00000199585.5
ENSMUST00000070418.9 |
Dclk1
|
doublecortin-like kinase 1 |
| chr4_-_110143777 | 4.31 |
ENSMUST00000138972.8
|
Elavl4
|
ELAV like RNA binding protein 4 |
| chr8_+_22073022 | 4.05 |
ENSMUST00000098887.4
|
Defa28
|
defensin, alpha, 28 |
| chr13_-_51723473 | 3.94 |
ENSMUST00000239056.2
ENSMUST00000223543.3 |
Shc3
|
src homology 2 domain-containing transforming protein C3 |
| chr7_+_51271742 | 3.52 |
ENSMUST00000032710.7
|
Slc17a6
|
solute carrier family 17 (sodium-dependent inorganic phosphate cotransporter), member 6 |
| chr15_+_30457772 | 3.43 |
ENSMUST00000228282.2
|
Ctnnd2
|
catenin (cadherin associated protein), delta 2 |
| chr4_-_149211145 | 3.41 |
ENSMUST00000030815.3
|
Cort
|
cortistatin |
| chr2_+_65760477 | 3.34 |
ENSMUST00000176109.8
|
Csrnp3
|
cysteine-serine-rich nuclear protein 3 |
| chr2_-_79738773 | 3.31 |
ENSMUST00000102652.10
ENSMUST00000102651.10 |
Pde1a
|
phosphodiesterase 1A, calmodulin-dependent |
| chr3_-_144514386 | 3.29 |
ENSMUST00000197013.2
|
Clca3a2
|
chloride channel accessory 3A2 |
| chr13_-_110417421 | 3.19 |
ENSMUST00000223922.2
|
Rab3c
|
RAB3C, member RAS oncogene family |
| chr15_+_39255185 | 3.06 |
ENSMUST00000228839.2
|
Rims2
|
regulating synaptic membrane exocytosis 2 |
| chr2_-_144369261 | 3.05 |
ENSMUST00000163701.2
ENSMUST00000081982.12 |
Dzank1
|
double zinc ribbon and ankyrin repeat domains 1 |
| chr2_-_79738734 | 3.03 |
ENSMUST00000090756.11
|
Pde1a
|
phosphodiesterase 1A, calmodulin-dependent |
| chr8_+_21574411 | 2.96 |
ENSMUST00000095425.2
|
Defa25
|
defensin, alpha, 25 |
| chr7_-_140402037 | 2.95 |
ENSMUST00000106052.2
ENSMUST00000080651.13 |
Zfp941
|
zinc finger protein 941 |
| chr9_-_69945358 | 2.94 |
ENSMUST00000034751.6
|
Gcnt3
|
glucosaminyl (N-acetyl) transferase 3, mucin type |
| chr2_+_65451100 | 2.93 |
ENSMUST00000144254.6
ENSMUST00000028377.14 |
Scn2a
|
sodium channel, voltage-gated, type II, alpha |
| chr14_-_104081119 | 2.93 |
ENSMUST00000227824.2
ENSMUST00000172237.2 |
Ednrb
|
endothelin receptor type B |
| chr2_-_136229849 | 2.93 |
ENSMUST00000035264.9
ENSMUST00000077200.4 |
Pak5
|
p21 (RAC1) activated kinase 5 |
| chr3_+_28038283 | 2.90 |
ENSMUST00000067757.11
ENSMUST00000123539.8 |
Pld1
|
phospholipase D1 |
| chr2_+_20727274 | 2.85 |
ENSMUST00000114607.8
|
Etl4
|
enhancer trap locus 4 |
| chr14_-_54655079 | 2.80 |
ENSMUST00000226753.2
ENSMUST00000197440.5 |
Slc7a7
|
solute carrier family 7 (cationic amino acid transporter, y+ system), member 7 |
| chr11_-_60243695 | 2.73 |
ENSMUST00000095254.12
ENSMUST00000102683.11 ENSMUST00000093048.13 ENSMUST00000093046.13 ENSMUST00000064019.15 ENSMUST00000102682.5 |
Tom1l2
|
target of myb1-like 2 (chicken) |
| chr2_+_134627987 | 2.64 |
ENSMUST00000131552.5
ENSMUST00000110116.8 |
Plcb1
|
phospholipase C, beta 1 |
| chr5_-_99391073 | 2.56 |
ENSMUST00000166484.2
|
Rasgef1b
|
RasGEF domain family, member 1B |
| chr6_-_69394425 | 2.55 |
ENSMUST00000199160.2
|
Igkv4-61
|
immunoglobulin kappa chain variable 4-61 |
| chr6_-_69204417 | 2.41 |
ENSMUST00000103346.3
|
Igkv4-72
|
immunoglobulin kappa chain variable 4-72 |
| chr14_+_60615128 | 2.39 |
ENSMUST00000022561.9
|
Amer2
|
APC membrane recruitment 2 |
| chr6_-_69245427 | 2.39 |
ENSMUST00000103348.3
|
Igkv4-70
|
immunoglobulin kappa chain variable 4-70 |
| chr3_-_157630690 | 2.36 |
ENSMUST00000118539.2
|
Cth
|
cystathionase (cystathionine gamma-lyase) |
| chr14_-_104081827 | 2.27 |
ENSMUST00000022718.11
|
Ednrb
|
endothelin receptor type B |
| chr4_-_138123700 | 2.22 |
ENSMUST00000105032.4
|
Fam43b
|
family with sequence similarity 43, member B |
| chrX_-_144288071 | 2.20 |
ENSMUST00000112835.8
ENSMUST00000143610.3 |
Amot
|
angiomotin |
| chr6_-_138399896 | 2.12 |
ENSMUST00000161450.8
ENSMUST00000163024.8 ENSMUST00000162185.8 |
Lmo3
|
LIM domain only 3 |
| chr5_+_107645626 | 2.07 |
ENSMUST00000152474.8
ENSMUST00000060553.8 |
Btbd8
|
BTB (POZ) domain containing 8 |
| chr10_+_62860291 | 2.06 |
ENSMUST00000020262.5
|
Pbld2
|
phenazine biosynthesis-like protein domain containing 2 |
| chrX_-_42256694 | 2.06 |
ENSMUST00000115058.8
ENSMUST00000115059.8 |
Tenm1
|
teneurin transmembrane protein 1 |
| chr18_+_38429688 | 2.05 |
ENSMUST00000170811.8
ENSMUST00000072376.13 ENSMUST00000237903.2 ENSMUST00000236116.2 ENSMUST00000237824.2 ENSMUST00000236982.2 ENSMUST00000235549.2 ENSMUST00000237416.2 ENSMUST00000237089.2 |
Rnf14
|
ring finger protein 14 |
| chr11_-_99134885 | 2.02 |
ENSMUST00000103132.10
ENSMUST00000038214.7 |
Krt222
|
keratin 222 |
| chrX_-_58211440 | 1.99 |
ENSMUST00000119306.2
|
Fgf13
|
fibroblast growth factor 13 |
| chr19_-_7183626 | 1.93 |
ENSMUST00000025679.11
|
Otub1
|
OTU domain, ubiquitin aldehyde binding 1 |
| chr6_-_69282389 | 1.88 |
ENSMUST00000103350.3
|
Igkv4-68
|
immunoglobulin kappa variable 4-68 |
| chr1_-_134731516 | 1.86 |
ENSMUST00000238280.2
|
Ppp1r12b
|
protein phosphatase 1, regulatory subunit 12B |
| chr6_-_93890520 | 1.84 |
ENSMUST00000203688.3
|
Magi1
|
membrane associated guanylate kinase, WW and PDZ domain containing 1 |
| chr1_+_34160331 | 1.83 |
ENSMUST00000183006.5
|
Dst
|
dystonin |
| chr6_-_69377328 | 1.78 |
ENSMUST00000198345.2
|
Igkv4-62
|
immunoglobulin kappa variable 4-62 |
| chr5_-_66211842 | 1.74 |
ENSMUST00000200852.4
|
Rbm47
|
RNA binding motif protein 47 |
| chr4_-_108637979 | 1.74 |
ENSMUST00000106657.8
|
Zfyve9
|
zinc finger, FYVE domain containing 9 |
| chr7_+_91321500 | 1.73 |
ENSMUST00000238619.2
ENSMUST00000238467.2 |
Dlg2
|
discs large MAGUK scaffold protein 2 |
| chr16_+_44687460 | 1.67 |
ENSMUST00000102805.4
|
Cd200r2
|
Cd200 receptor 2 |
| chr9_+_48073296 | 1.62 |
ENSMUST00000216998.2
ENSMUST00000215780.2 |
Nxpe4
|
neurexophilin and PC-esterase domain family, member 4 |
| chr2_-_84481058 | 1.56 |
ENSMUST00000111670.9
ENSMUST00000111697.9 ENSMUST00000111696.8 ENSMUST00000111678.8 ENSMUST00000111690.8 ENSMUST00000111695.8 ENSMUST00000111677.8 ENSMUST00000111698.8 ENSMUST00000099941.9 ENSMUST00000111676.8 ENSMUST00000111694.8 ENSMUST00000111675.8 ENSMUST00000111689.8 ENSMUST00000111687.8 ENSMUST00000111692.8 ENSMUST00000111685.8 ENSMUST00000111686.8 ENSMUST00000111688.8 ENSMUST00000111693.8 ENSMUST00000111684.8 |
Ctnnd1
|
catenin (cadherin associated protein), delta 1 |
| chr8_-_39128662 | 1.54 |
ENSMUST00000118896.2
|
Sgcz
|
sarcoglycan zeta |
| chr17_-_24382638 | 1.46 |
ENSMUST00000129523.3
ENSMUST00000138685.3 ENSMUST00000040735.12 |
Amdhd2
|
amidohydrolase domain containing 2 |
| chr4_-_108637700 | 1.44 |
ENSMUST00000106658.8
|
Zfyve9
|
zinc finger, FYVE domain containing 9 |
| chr14_+_84680993 | 1.31 |
ENSMUST00000071370.7
|
Pcdh17
|
protocadherin 17 |
| chr6_+_140378688 | 1.27 |
ENSMUST00000203774.3
|
Plekha5
|
pleckstrin homology domain containing, family A member 5 |
| chr6_-_144978557 | 1.24 |
ENSMUST00000136819.3
|
Bcat1
|
branched chain aminotransferase 1, cytosolic |
| chr11_+_95925711 | 1.20 |
ENSMUST00000006217.10
ENSMUST00000107700.4 |
Snf8
|
SNF8, ESCRT-II complex subunit, homolog (S. cerevisiae) |
| chr2_-_84481020 | 1.20 |
ENSMUST00000067232.10
|
Ctnnd1
|
catenin (cadherin associated protein), delta 1 |
| chr9_+_88462944 | 1.15 |
ENSMUST00000164661.4
ENSMUST00000215498.2 ENSMUST00000216686.2 |
Trim43a
|
tripartite motif-containing 43A |
| chr9_-_103569984 | 1.13 |
ENSMUST00000049452.15
|
Tmem108
|
transmembrane protein 108 |
| chr6_+_41582416 | 1.08 |
ENSMUST00000114732.3
|
Ephb6
|
Eph receptor B6 |
| chr3_+_141171347 | 1.04 |
ENSMUST00000130636.8
|
Unc5c
|
unc-5 netrin receptor C |
| chr9_+_74959259 | 1.03 |
ENSMUST00000170310.2
ENSMUST00000166549.2 |
Arpp19
|
cAMP-regulated phosphoprotein 19 |
| chr3_+_141171322 | 1.02 |
ENSMUST00000075282.10
|
Unc5c
|
unc-5 netrin receptor C |
| chr6_-_119940694 | 1.02 |
ENSMUST00000161512.3
|
Wnk1
|
WNK lysine deficient protein kinase 1 |
| chr19_+_31060237 | 1.00 |
ENSMUST00000066039.8
|
Cstf2t
|
cleavage stimulation factor, 3' pre-RNA subunit 2, tau |
| chr8_-_111985824 | 0.98 |
ENSMUST00000169020.8
ENSMUST00000003404.9 |
Glg1
|
golgi apparatus protein 1 |
| chr1_-_150268470 | 0.97 |
ENSMUST00000006167.13
ENSMUST00000097547.10 |
Odr4
|
odr4 GPCR localization factor homolog |
| chr19_-_6910195 | 0.97 |
ENSMUST00000236443.2
|
Kcnk4
|
potassium channel, subfamily K, member 4 |
| chr18_-_36828978 | 0.95 |
ENSMUST00000115682.2
|
E230025N22Rik
|
Riken cDNA E230025N22 gene |
| chr2_-_84481101 | 0.95 |
ENSMUST00000111691.2
|
Ctnnd1
|
catenin (cadherin associated protein), delta 1 |
| chr2_-_84480804 | 0.94 |
ENSMUST00000066177.10
|
Ctnnd1
|
catenin (cadherin associated protein), delta 1 |
| chr19_-_11816583 | 0.86 |
ENSMUST00000214887.2
|
Olfr1417
|
olfactory receptor 1417 |
| chr15_-_100301124 | 0.85 |
ENSMUST00000124324.2
|
Slc11a2
|
solute carrier family 11 (proton-coupled divalent metal ion transporters), member 2 |
| chr11_+_60244132 | 0.84 |
ENSMUST00000070805.13
ENSMUST00000094140.9 ENSMUST00000108723.9 ENSMUST00000108722.5 |
Drc3
|
dynein regulatory complex subunit 3 |
| chr6_-_144993362 | 0.83 |
ENSMUST00000149769.6
|
Bcat1
|
branched chain aminotransferase 1, cytosolic |
| chr10_-_77845571 | 0.82 |
ENSMUST00000020522.9
|
Pfkl
|
phosphofructokinase, liver, B-type |
| chr9_+_54441404 | 0.81 |
ENSMUST00000118413.3
|
Sh2d7
|
SH2 domain containing 7 |
| chr11_+_72580823 | 0.79 |
ENSMUST00000155998.2
|
Ankfy1
|
ankyrin repeat and FYVE domain containing 1 |
| chr7_-_142215595 | 0.73 |
ENSMUST00000145896.3
|
Igf2
|
insulin-like growth factor 2 |
| chr18_-_75094323 | 0.71 |
ENSMUST00000066532.5
|
Lipg
|
lipase, endothelial |
| chr10_+_79978127 | 0.70 |
ENSMUST00000003156.15
|
Atp5d
|
ATP synthase, H+ transporting, mitochondrial F1 complex, delta subunit |
| chr11_-_98620200 | 0.67 |
ENSMUST00000126565.2
ENSMUST00000100500.9 ENSMUST00000017354.13 |
Med24
|
mediator complex subunit 24 |
| chr6_+_67768007 | 0.65 |
ENSMUST00000196006.5
ENSMUST00000103307.3 |
Igkv14-130
|
immunoglobulin kappa variable 14-130 |
| chr6_-_12749192 | 0.64 |
ENSMUST00000172356.8
|
Thsd7a
|
thrombospondin, type I, domain containing 7A |
| chr9_-_88974735 | 0.64 |
ENSMUST00000189557.2
ENSMUST00000167113.8 |
Trim43b
|
tripartite motif-containing 43B |
| chr10_-_130002635 | 0.64 |
ENSMUST00000216530.2
|
Olfr825
|
olfactory receptor 825 |
| chr10_+_79978152 | 0.62 |
ENSMUST00000105366.2
|
Atp5d
|
ATP synthase, H+ transporting, mitochondrial F1 complex, delta subunit |
| chr4_+_129030710 | 0.61 |
ENSMUST00000102600.4
|
Fndc5
|
fibronectin type III domain containing 5 |
| chr9_-_105398346 | 0.61 |
ENSMUST00000176770.8
ENSMUST00000085133.13 |
Atp2c1
|
ATPase, Ca++-sequestering |
| chr6_-_90201420 | 0.60 |
ENSMUST00000076086.3
|
Vmn1r53
|
vomeronasal 1 receptor 53 |
| chr17_+_71511642 | 0.59 |
ENSMUST00000126681.8
|
Lpin2
|
lipin 2 |
| chrX_-_163787825 | 0.58 |
ENSMUST00000114890.3
|
Gm17604
|
predicted gene, 17604 |
| chr10_+_57670431 | 0.56 |
ENSMUST00000151623.8
ENSMUST00000020022.8 |
Smpdl3a
|
sphingomyelin phosphodiesterase, acid-like 3A |
| chr18_+_5593566 | 0.56 |
ENSMUST00000160910.2
|
Zeb1
|
zinc finger E-box binding homeobox 1 |
| chr6_-_12749409 | 0.55 |
ENSMUST00000119581.7
|
Thsd7a
|
thrombospondin, type I, domain containing 7A |
| chr19_+_55169148 | 0.55 |
ENSMUST00000154886.8
ENSMUST00000120936.8 ENSMUST00000025936.12 |
Tectb
|
tectorin beta |
| chr4_+_138926577 | 0.53 |
ENSMUST00000145368.8
|
Capzb
|
capping protein (actin filament) muscle Z-line, beta |
| chr10_-_107747995 | 0.52 |
ENSMUST00000165341.5
|
Otogl
|
otogelin-like |
| chr7_-_132616977 | 0.51 |
ENSMUST00000169570.8
|
Ctbp2
|
C-terminal binding protein 2 |
| chr7_+_27977141 | 0.50 |
ENSMUST00000094651.4
|
Eid2b
|
EP300 interacting inhibitor of differentiation 2B |
| chr10_+_17598961 | 0.50 |
ENSMUST00000038107.9
ENSMUST00000219558.2 ENSMUST00000218370.2 |
Cited2
|
Cbp/p300-interacting transactivator, with Glu/Asp-rich carboxy-terminal domain, 2 |
| chr14_+_50722917 | 0.48 |
ENSMUST00000071932.5
|
Olfr741
|
olfactory receptor 741 |
| chr10_+_56253531 | 0.45 |
ENSMUST00000220194.2
ENSMUST00000218834.2 |
Gja1
|
gap junction protein, alpha 1 |
| chr13_+_96524802 | 0.41 |
ENSMUST00000099295.6
|
Poc5
|
POC5 centriolar protein |
| chr2_-_86067177 | 0.39 |
ENSMUST00000099900.2
|
Olfr1048
|
olfactory receptor 1048 |
| chr8_-_41507808 | 0.39 |
ENSMUST00000093534.11
|
Mtus1
|
mitochondrial tumor suppressor 1 |
| chr19_+_12655487 | 0.37 |
ENSMUST00000215134.2
ENSMUST00000049724.8 |
Olfr1443
|
olfactory receptor 1443 |
| chr2_-_88990751 | 0.36 |
ENSMUST00000216833.2
ENSMUST00000216976.2 |
Olfr1224-ps1
|
olfactory receptor 1224, pseudogene 1 |
| chr8_-_25066313 | 0.36 |
ENSMUST00000121992.2
|
Ido2
|
indoleamine 2,3-dioxygenase 2 |
| chr2_+_144441817 | 0.35 |
ENSMUST00000028917.7
|
Dtd1
|
D-tyrosyl-tRNA deacylase 1 |
| chr16_+_36695479 | 0.34 |
ENSMUST00000023534.7
ENSMUST00000114812.9 ENSMUST00000134616.8 |
Golgb1
|
golgi autoantigen, golgin subfamily b, macrogolgin 1 |
| chr1_+_9978863 | 0.31 |
ENSMUST00000052843.12
ENSMUST00000171802.8 ENSMUST00000125294.9 ENSMUST00000140948.9 |
Mcmdc2
|
minichromosome maintenance domain containing 2 |
| chr12_-_31401432 | 0.26 |
ENSMUST00000110857.5
|
Dld
|
dihydrolipoamide dehydrogenase |
| chr13_-_23041731 | 0.24 |
ENSMUST00000228645.2
|
Vmn1r211
|
vomeronasal 1 receptor 211 |
| chr13_+_25240138 | 0.24 |
ENSMUST00000069614.7
|
Dcdc2a
|
doublecortin domain containing 2a |
| chr17_+_85264134 | 0.23 |
ENSMUST00000112305.10
|
Ppm1b
|
protein phosphatase 1B, magnesium dependent, beta isoform |
| chr3_+_142470806 | 0.21 |
ENSMUST00000029938.10
|
Gtf2b
|
general transcription factor IIB |
| chrX_+_82898974 | 0.21 |
ENSMUST00000239269.2
|
Dmd
|
dystrophin, muscular dystrophy |
| chr11_+_49169131 | 0.19 |
ENSMUST00000213323.2
|
Olfr1393
|
olfactory receptor 1393 |
| chr15_-_83033471 | 0.19 |
ENSMUST00000129372.2
|
Poldip3
|
polymerase (DNA-directed), delta interacting protein 3 |
| chr12_+_51424343 | 0.18 |
ENSMUST00000219434.2
ENSMUST00000021335.7 |
Scfd1
|
Sec1 family domain containing 1 |
| chr6_-_66787481 | 0.17 |
ENSMUST00000228862.2
ENSMUST00000227285.2 ENSMUST00000228008.2 ENSMUST00000228651.2 ENSMUST00000227555.2 |
Vmn1r39
|
vomeronasal 1 receptor 39 |
| chr9_+_38278192 | 0.17 |
ENSMUST00000216168.2
|
Olfr250
|
olfactory receptor 250 |
| chr5_+_122422428 | 0.17 |
ENSMUST00000053426.15
|
Pptc7
|
PTC7 protein phosphatase homolog |
| chr14_+_5894220 | 0.16 |
ENSMUST00000063750.8
|
Rarb
|
retinoic acid receptor, beta |
| chr16_-_36605147 | 0.16 |
ENSMUST00000165531.9
|
Slc15a2
|
solute carrier family 15 (H+/peptide transporter), member 2 |
| chr2_+_80145805 | 0.15 |
ENSMUST00000028392.8
|
Dnajc10
|
DnaJ heat shock protein family (Hsp40) member C10 |
| chrX_+_163202778 | 0.15 |
ENSMUST00000208741.2
ENSMUST00000033754.15 ENSMUST00000208697.2 ENSMUST00000208261.2 |
Piga
|
phosphatidylinositol glycan anchor biosynthesis, class A |
| chr17_+_24026892 | 0.11 |
ENSMUST00000191385.3
|
Srrm2
|
serine/arginine repetitive matrix 2 |
| chr7_-_86046700 | 0.10 |
ENSMUST00000215733.2
|
Olfr303
|
olfactory receptor 303 |
| chr7_+_104226521 | 0.10 |
ENSMUST00000217466.2
ENSMUST00000215585.3 |
Olfr653
|
olfactory receptor 653 |
| chr1_+_90531183 | 0.10 |
ENSMUST00000186750.2
|
Cops8
|
COP9 signalosome subunit 8 |
| chr13_-_105430889 | 0.09 |
ENSMUST00000226044.2
|
Rnf180
|
ring finger protein 180 |
| chr2_-_85917726 | 0.09 |
ENSMUST00000216886.2
ENSMUST00000213333.2 ENSMUST00000216020.2 |
Olfr1037
|
olfactory receptor 1037 |
| chr2_-_87543523 | 0.09 |
ENSMUST00000214209.2
|
Olfr1137
|
olfactory receptor 1137 |
| chr12_+_69288606 | 0.08 |
ENSMUST00000063445.13
|
Klhdc1
|
kelch domain containing 1 |
| chrX_-_95499889 | 0.07 |
ENSMUST00000164693.8
ENSMUST00000119035.9 |
Hsf3
|
heat shock transcription factor 3 |
| chr16_-_58910185 | 0.07 |
ENSMUST00000215647.2
|
Olfr191
|
olfactory receptor 191 |
| chr2_+_86338805 | 0.07 |
ENSMUST00000076263.2
|
Olfr1076
|
olfactory receptor 1076 |
| chr6_-_58418303 | 0.02 |
ENSMUST00000228577.2
ENSMUST00000227466.2 |
Vmn1r30
|
vomeronasal 1 receptor 30 |
| chr2_-_88947627 | 0.01 |
ENSMUST00000217635.2
ENSMUST00000143255.3 ENSMUST00000213404.2 |
Olfr1221
|
olfactory receptor 1221 |
| chr2_-_89084074 | 0.01 |
ENSMUST00000216392.2
|
Olfr1228
|
olfactory receptor 1228 |
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 3.4 | 10.1 | GO:0002314 | germinal center B cell differentiation(GO:0002314) |
| 1.1 | 8.5 | GO:0035948 | positive regulation of gluconeogenesis by positive regulation of transcription from RNA polymerase II promoter(GO:0035948) |
| 1.0 | 6.1 | GO:0038128 | ERBB2 signaling pathway(GO:0038128) |
| 1.0 | 2.9 | GO:0002426 | immunoglobulin production in mucosal tissue(GO:0002426) |
| 0.9 | 2.6 | GO:0035722 | interleukin-12-mediated signaling pathway(GO:0035722) activation of meiosis involved in egg activation(GO:0060466) cellular response to interleukin-12(GO:0071349) response to fluoride(GO:1902617) response to vasopressin(GO:1904116) cellular response to vasopressin(GO:1904117) |
| 0.9 | 5.2 | GO:0014826 | vein smooth muscle contraction(GO:0014826) |
| 0.8 | 4.7 | GO:0060690 | epithelial cell differentiation involved in salivary gland development(GO:0060690) |
| 0.8 | 6.0 | GO:0042737 | drug catabolic process(GO:0042737) |
| 0.8 | 4.5 | GO:1902261 | positive regulation of delayed rectifier potassium channel activity(GO:1902261) |
| 0.7 | 2.2 | GO:0003365 | establishment of cell polarity involved in ameboidal cell migration(GO:0003365) |
| 0.7 | 25.1 | GO:0019731 | antibacterial humoral response(GO:0019731) |
| 0.6 | 2.4 | GO:0019343 | cysteine biosynthetic process via cystathionine(GO:0019343) |
| 0.4 | 3.5 | GO:0098700 | neurotransmitter loading into synaptic vesicle(GO:0098700) |
| 0.3 | 1.2 | GO:0043328 | protein targeting to vacuole involved in ubiquitin-dependent protein catabolic process via the multivesicular body sorting pathway(GO:0043328) |
| 0.3 | 2.4 | GO:0000821 | regulation of arginine metabolic process(GO:0000821) |
| 0.3 | 6.3 | GO:0046069 | cGMP catabolic process(GO:0046069) |
| 0.3 | 2.0 | GO:0045196 | establishment or maintenance of neuroblast polarity(GO:0045196) establishment of neuroblast polarity(GO:0045200) |
| 0.3 | 1.1 | GO:0001806 | type IV hypersensitivity(GO:0001806) |
| 0.3 | 2.9 | GO:0008627 | intrinsic apoptotic signaling pathway in response to osmotic stress(GO:0008627) |
| 0.3 | 2.1 | GO:0009098 | branched-chain amino acid biosynthetic process(GO:0009082) leucine biosynthetic process(GO:0009098) valine biosynthetic process(GO:0009099) |
| 0.2 | 1.0 | GO:0071469 | cellular response to alkaline pH(GO:0071469) |
| 0.2 | 8.8 | GO:0006825 | copper ion transport(GO:0006825) |
| 0.2 | 3.1 | GO:0061669 | spontaneous neurotransmitter secretion(GO:0061669) spontaneous synaptic transmission(GO:0098814) |
| 0.2 | 2.9 | GO:0032534 | regulation of microvillus assembly(GO:0032534) |
| 0.2 | 4.9 | GO:0071420 | cellular response to histamine(GO:0071420) |
| 0.2 | 1.5 | GO:0019262 | N-acetylneuraminate catabolic process(GO:0019262) |
| 0.2 | 1.0 | GO:0023016 | signal transduction by trans-phosphorylation(GO:0023016) |
| 0.2 | 2.1 | GO:0033564 | anterior/posterior axon guidance(GO:0033564) |
| 0.2 | 0.7 | GO:0010982 | regulation of high-density lipoprotein particle clearance(GO:0010982) |
| 0.2 | 12.6 | GO:0050829 | defense response to Gram-negative bacterium(GO:0050829) |
| 0.2 | 3.2 | GO:0007183 | SMAD protein complex assembly(GO:0007183) |
| 0.2 | 0.6 | GO:0055071 | cellular manganese ion homeostasis(GO:0030026) Golgi calcium ion homeostasis(GO:0032468) manganese ion homeostasis(GO:0055071) |
| 0.2 | 0.5 | GO:0060156 | vascular transport(GO:0010232) milk ejection(GO:0060156) |
| 0.1 | 0.8 | GO:0032439 | endosome localization(GO:0032439) |
| 0.1 | 0.5 | GO:0035802 | adrenal cortex development(GO:0035801) adrenal cortex formation(GO:0035802) |
| 0.1 | 3.0 | GO:0008090 | retrograde axonal transport(GO:0008090) |
| 0.1 | 1.4 | GO:0016554 | cytidine to uridine editing(GO:0016554) |
| 0.1 | 0.3 | GO:1990918 | double-strand break repair involved in meiotic recombination(GO:1990918) |
| 0.1 | 4.6 | GO:0071526 | semaphorin-plexin signaling pathway(GO:0071526) |
| 0.1 | 1.0 | GO:0098789 | pre-mRNA cleavage required for polyadenylation(GO:0098789) |
| 0.1 | 1.7 | GO:0045161 | neuronal ion channel clustering(GO:0045161) |
| 0.1 | 4.3 | GO:0021952 | central nervous system projection neuron axonogenesis(GO:0021952) |
| 0.1 | 1.3 | GO:2000807 | regulation of synaptic vesicle clustering(GO:2000807) |
| 0.1 | 0.2 | GO:0021627 | olfactory nerve morphogenesis(GO:0021627) olfactory nerve structural organization(GO:0021629) |
| 0.1 | 0.8 | GO:0006002 | fructose 6-phosphate metabolic process(GO:0006002) |
| 0.1 | 0.3 | GO:0009106 | lipoate metabolic process(GO:0009106) |
| 0.1 | 0.7 | GO:2000467 | positive regulation of glycogen (starch) synthase activity(GO:2000467) |
| 0.1 | 3.3 | GO:0043276 | anoikis(GO:0043276) |
| 0.1 | 7.2 | GO:0019882 | antigen processing and presentation(GO:0019882) |
| 0.1 | 4.9 | GO:0007218 | neuropeptide signaling pathway(GO:0007218) |
| 0.0 | 1.9 | GO:0071108 | protein K48-linked deubiquitination(GO:0071108) |
| 0.0 | 2.1 | GO:0060765 | regulation of androgen receptor signaling pathway(GO:0060765) |
| 0.0 | 1.0 | GO:0045722 | positive regulation of gluconeogenesis(GO:0045722) |
| 0.0 | 0.5 | GO:0051490 | negative regulation of filopodium assembly(GO:0051490) |
| 0.0 | 0.4 | GO:0006450 | regulation of translational fidelity(GO:0006450) |
| 0.0 | 3.7 | GO:0035249 | synaptic transmission, glutamatergic(GO:0035249) |
| 0.0 | 13.3 | GO:0042742 | defense response to bacterium(GO:0042742) |
| 0.0 | 0.6 | GO:0009143 | nucleoside triphosphate catabolic process(GO:0009143) |
| 0.0 | 1.3 | GO:0015986 | energy coupled proton transport, down electrochemical gradient(GO:0015985) ATP synthesis coupled proton transport(GO:0015986) |
| 0.0 | 0.5 | GO:0048386 | positive regulation of retinoic acid receptor signaling pathway(GO:0048386) |
| 0.0 | 0.2 | GO:0006499 | N-terminal protein myristoylation(GO:0006499) |
| 0.0 | 0.6 | GO:0045602 | negative regulation of endothelial cell differentiation(GO:0045602) |
| 0.0 | 9.2 | GO:0002377 | immunoglobulin production(GO:0002377) |
| 0.0 | 0.2 | GO:1904798 | positive regulation of core promoter binding(GO:1904798) |
| 0.0 | 2.5 | GO:0045839 | negative regulation of mitotic nuclear division(GO:0045839) |
| 0.0 | 0.5 | GO:0019321 | pentose metabolic process(GO:0019321) |
| 0.0 | 6.7 | GO:0098656 | anion transmembrane transport(GO:0098656) |
| 0.0 | 0.4 | GO:0046218 | tryptophan catabolic process(GO:0006569) tryptophan catabolic process to kynurenine(GO:0019441) indole-containing compound catabolic process(GO:0042436) indolalkylamine catabolic process(GO:0046218) |
| 0.0 | 4.8 | GO:0007156 | homophilic cell adhesion via plasma membrane adhesion molecules(GO:0007156) |
| 0.0 | 3.3 | GO:0010923 | negative regulation of phosphatase activity(GO:0010923) |
| 0.0 | 0.1 | GO:0010387 | COP9 signalosome assembly(GO:0010387) |
| 0.0 | 7.2 | GO:0007612 | learning(GO:0007612) |
| 0.0 | 0.6 | GO:0014850 | response to muscle activity(GO:0014850) |
| 0.0 | 0.1 | GO:0034975 | protein folding in endoplasmic reticulum(GO:0034975) |
| 0.0 | 0.7 | GO:0030521 | androgen receptor signaling pathway(GO:0030521) |
| 0.0 | 1.0 | GO:1903318 | negative regulation of protein processing(GO:0010955) negative regulation of protein maturation(GO:1903318) |
| 0.0 | 0.2 | GO:0003406 | retinal pigment epithelium development(GO:0003406) |
| 0.0 | 2.4 | GO:0090090 | negative regulation of canonical Wnt signaling pathway(GO:0090090) |
| 0.0 | 0.6 | GO:0019432 | triglyceride biosynthetic process(GO:0019432) |
| 0.0 | 0.2 | GO:1902902 | negative regulation of autophagosome assembly(GO:1902902) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.1 | 4.5 | GO:0014701 | junctional sarcoplasmic reticulum membrane(GO:0014701) |
| 0.4 | 1.2 | GO:0000814 | ESCRT II complex(GO:0000814) |
| 0.3 | 4.7 | GO:0005915 | zonula adherens(GO:0005915) |
| 0.3 | 4.3 | GO:0042788 | polysomal ribosome(GO:0042788) |
| 0.2 | 1.8 | GO:0031673 | H zone(GO:0031673) |
| 0.2 | 4.9 | GO:1902711 | GABA-A receptor complex(GO:1902711) |
| 0.2 | 1.5 | GO:0016012 | sarcoglycan complex(GO:0016012) |
| 0.2 | 0.8 | GO:0005945 | 6-phosphofructokinase complex(GO:0005945) |
| 0.1 | 1.3 | GO:0045261 | mitochondrial proton-transporting ATP synthase complex, catalytic core F(1)(GO:0000275) proton-transporting ATP synthase complex, catalytic core F(1)(GO:0045261) |
| 0.1 | 0.8 | GO:0070826 | paraferritin complex(GO:0070826) |
| 0.1 | 3.5 | GO:0030285 | integral component of synaptic vesicle membrane(GO:0030285) intrinsic component of synaptic vesicle membrane(GO:0098563) |
| 0.1 | 2.9 | GO:0001518 | voltage-gated sodium channel complex(GO:0001518) |
| 0.1 | 0.8 | GO:0044352 | pinosome(GO:0044352) macropinosome(GO:0044354) |
| 0.1 | 3.1 | GO:0060077 | inhibitory synapse(GO:0060077) |
| 0.1 | 0.3 | GO:0043159 | acrosomal matrix(GO:0043159) |
| 0.1 | 3.2 | GO:0031901 | early endosome membrane(GO:0031901) |
| 0.0 | 1.7 | GO:0044224 | juxtaparanode region of axon(GO:0044224) |
| 0.0 | 1.0 | GO:0005847 | mRNA cleavage and polyadenylation specificity factor complex(GO:0005847) |
| 0.0 | 18.1 | GO:0016323 | basolateral plasma membrane(GO:0016323) |
| 0.0 | 2.3 | GO:0008180 | COP9 signalosome(GO:0008180) |
| 0.0 | 0.5 | GO:0097470 | ribbon synapse(GO:0097470) |
| 0.0 | 9.0 | GO:0008021 | synaptic vesicle(GO:0008021) |
| 0.0 | 3.0 | GO:0014704 | intercalated disc(GO:0014704) |
| 0.0 | 1.0 | GO:0005797 | Golgi medial cisterna(GO:0005797) |
| 0.0 | 0.2 | GO:0097550 | transcriptional preinitiation complex(GO:0097550) |
| 0.0 | 1.9 | GO:0031672 | A band(GO:0031672) |
| 0.0 | 0.2 | GO:0060091 | kinocilium(GO:0060091) |
| 0.0 | 8.9 | GO:0099572 | postsynaptic density(GO:0014069) postsynaptic specialization(GO:0099572) |
| 0.0 | 50.3 | GO:0005615 | extracellular space(GO:0005615) |
| 0.0 | 1.6 | GO:0005882 | intermediate filament(GO:0005882) |
| 0.0 | 0.7 | GO:0016592 | mediator complex(GO:0016592) |
| 0.0 | 1.8 | GO:0005923 | bicellular tight junction(GO:0005923) |
| 0.0 | 2.6 | GO:0043209 | myelin sheath(GO:0043209) |
| 0.0 | 0.2 | GO:0090665 | dystrophin-associated glycoprotein complex(GO:0016010) glycoprotein complex(GO:0090665) |
| 0.0 | 0.1 | GO:0034663 | endoplasmic reticulum chaperone complex(GO:0034663) |
| 0.0 | 0.2 | GO:0017119 | Golgi transport complex(GO:0017119) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.5 | 10.1 | GO:0035673 | oligopeptide transmembrane transporter activity(GO:0035673) |
| 1.7 | 5.2 | GO:0004962 | endothelin receptor activity(GO:0004962) |
| 0.8 | 4.8 | GO:0032450 | maltose alpha-glucosidase activity(GO:0032450) |
| 0.7 | 2.9 | GO:0008109 | N-acetyllactosaminide beta-1,6-N-acetylglucosaminyltransferase activity(GO:0008109) |
| 0.7 | 8.0 | GO:0004322 | ferroxidase activity(GO:0004322) oxidoreductase activity, oxidizing metal ions, oxygen as acceptor(GO:0016724) |
| 0.6 | 4.9 | GO:1904315 | transmitter-gated ion channel activity involved in regulation of postsynaptic membrane potential(GO:1904315) |
| 0.6 | 6.3 | GO:0048101 | calmodulin-dependent cyclic-nucleotide phosphodiesterase activity(GO:0004117) calcium- and calmodulin-regulated 3',5'-cyclic-GMP phosphodiesterase activity(GO:0048101) |
| 0.6 | 2.2 | GO:0043532 | angiostatin binding(GO:0043532) |
| 0.5 | 6.0 | GO:0004499 | N,N-dimethylaniline monooxygenase activity(GO:0004499) |
| 0.4 | 2.9 | GO:0070290 | N-acylphosphatidylethanolamine-specific phospholipase D activity(GO:0070290) |
| 0.3 | 9.3 | GO:0031489 | myosin V binding(GO:0031489) |
| 0.3 | 1.0 | GO:0098782 | mechanically-gated potassium channel activity(GO:0098782) |
| 0.3 | 1.9 | GO:0019784 | NEDD8-specific protease activity(GO:0019784) |
| 0.3 | 2.1 | GO:0004084 | branched-chain-amino-acid transaminase activity(GO:0004084) L-leucine transaminase activity(GO:0052654) L-valine transaminase activity(GO:0052655) L-isoleucine transaminase activity(GO:0052656) |
| 0.3 | 2.1 | GO:0005042 | netrin receptor activity(GO:0005042) |
| 0.3 | 4.5 | GO:0043495 | protein anchor(GO:0043495) |
| 0.2 | 4.6 | GO:0030215 | semaphorin receptor binding(GO:0030215) |
| 0.2 | 2.9 | GO:0043522 | leucine zipper domain binding(GO:0043522) |
| 0.2 | 0.8 | GO:0003872 | 6-phosphofructokinase activity(GO:0003872) |
| 0.2 | 0.6 | GO:0015410 | manganese-transporting ATPase activity(GO:0015410) |
| 0.1 | 2.4 | GO:0016846 | carbon-sulfur lyase activity(GO:0016846) |
| 0.1 | 3.5 | GO:0005313 | L-glutamate transmembrane transporter activity(GO:0005313) |
| 0.1 | 5.7 | GO:0008157 | protein phosphatase 1 binding(GO:0008157) |
| 0.1 | 0.8 | GO:0015639 | cadmium ion transmembrane transporter activity(GO:0015086) lead ion transmembrane transporter activity(GO:0015094) vanadium ion transmembrane transporter activity(GO:0015100) ferrous iron uptake transmembrane transporter activity(GO:0015639) |
| 0.1 | 0.4 | GO:0033754 | indoleamine 2,3-dioxygenase activity(GO:0033754) |
| 0.1 | 2.4 | GO:0015174 | basic amino acid transmembrane transporter activity(GO:0015174) |
| 0.1 | 0.7 | GO:0008970 | phosphatidylcholine 1-acylhydrolase activity(GO:0008970) |
| 0.1 | 3.3 | GO:0005229 | intracellular calcium activated chloride channel activity(GO:0005229) |
| 0.1 | 2.6 | GO:0004435 | phosphatidylinositol phospholipase C activity(GO:0004435) |
| 0.1 | 3.4 | GO:0005184 | neuropeptide hormone activity(GO:0005184) |
| 0.1 | 4.0 | GO:0017091 | AU-rich element binding(GO:0017091) |
| 0.1 | 2.9 | GO:0005545 | 1-phosphatidylinositol binding(GO:0005545) |
| 0.1 | 9.5 | GO:0005179 | hormone activity(GO:0005179) |
| 0.1 | 1.3 | GO:0046933 | proton-transporting ATP synthase activity, rotational mechanism(GO:0046933) |
| 0.1 | 1.0 | GO:0019869 | chloride channel inhibitor activity(GO:0019869) |
| 0.1 | 1.7 | GO:0004385 | guanylate kinase activity(GO:0004385) |
| 0.1 | 0.5 | GO:0086075 | gap junction channel activity involved in cardiac conduction electrical coupling(GO:0086075) |
| 0.1 | 1.3 | GO:0010314 | phosphatidylinositol-5-phosphate binding(GO:0010314) |
| 0.1 | 0.5 | GO:0016618 | hydroxypyruvate reductase activity(GO:0016618) glyoxylate reductase (NADP) activity(GO:0030267) |
| 0.0 | 1.1 | GO:0005003 | ephrin receptor activity(GO:0005003) |
| 0.0 | 0.3 | GO:0004738 | pyruvate dehydrogenase activity(GO:0004738) pyruvate dehydrogenase [NAD(P)+] activity(GO:0034603) pyruvate dehydrogenase (NAD+) activity(GO:0034604) |
| 0.0 | 2.1 | GO:0050681 | androgen receptor binding(GO:0050681) |
| 0.0 | 2.5 | GO:0048487 | beta-tubulin binding(GO:0048487) |
| 0.0 | 5.8 | GO:0008013 | beta-catenin binding(GO:0008013) |
| 0.0 | 6.1 | GO:0004725 | protein tyrosine phosphatase activity(GO:0004725) |
| 0.0 | 6.7 | GO:0015293 | symporter activity(GO:0015293) |
| 0.0 | 0.5 | GO:0050693 | LBD domain binding(GO:0050693) |
| 0.0 | 1.0 | GO:0004864 | protein phosphatase inhibitor activity(GO:0004864) |
| 0.0 | 2.7 | GO:0030276 | clathrin binding(GO:0030276) |
| 0.0 | 1.8 | GO:0051393 | alpha-actinin binding(GO:0051393) |
| 0.0 | 1.0 | GO:0017134 | fibroblast growth factor binding(GO:0017134) |
| 0.0 | 1.5 | GO:0019213 | deacetylase activity(GO:0019213) |
| 0.0 | 0.6 | GO:0008195 | phosphatidate phosphatase activity(GO:0008195) |
| 0.0 | 0.4 | GO:0002161 | aminoacyl-tRNA editing activity(GO:0002161) |
| 0.0 | 3.9 | GO:0017137 | Rab GTPase binding(GO:0017137) |
| 0.0 | 1.0 | GO:0001784 | phosphotyrosine binding(GO:0001784) |
| 0.0 | 0.5 | GO:0046966 | thyroid hormone receptor binding(GO:0046966) |
| 0.0 | 0.1 | GO:0017176 | phosphatidylinositol N-acetylglucosaminyltransferase activity(GO:0017176) |
| 0.0 | 0.2 | GO:0002162 | dystroglycan binding(GO:0002162) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 12.7 | PID ARF6 TRAFFICKING PATHWAY | Arf6 trafficking events |
| 0.1 | 8.5 | PID HNF3A PATHWAY | FOXA1 transcription factor network |
| 0.1 | 2.6 | PID IL8 CXCR1 PATHWAY | IL8- and CXCR1-mediated signaling events |
| 0.0 | 2.1 | PID NETRIN PATHWAY | Netrin-mediated signaling events |
| 0.0 | 3.2 | PID TGFBR PATHWAY | TGF-beta receptor signaling |
| 0.0 | 1.3 | PID HIF2PATHWAY | HIF-2-alpha transcription factor network |
| 0.0 | 3.9 | PID TRKR PATHWAY | Neurotrophic factor-mediated Trk receptor signaling |
| 0.0 | 1.8 | PID TAP63 PATHWAY | Validated transcriptional targets of TAp63 isoforms |
| 0.0 | 2.1 | PID MYC ACTIV PATHWAY | Validated targets of C-MYC transcriptional activation |
| 0.0 | 0.5 | PID NCADHERIN PATHWAY | N-cadherin signaling events |
| 0.0 | 0.7 | PID AJDISS 2PATHWAY | Posttranslational regulation of adherens junction stability and dissassembly |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.2 | 4.8 | REACTOME DIGESTION OF DIETARY CARBOHYDRATE | Genes involved in Digestion of dietary carbohydrate |
| 0.4 | 6.7 | REACTOME BILE SALT AND ORGANIC ANION SLC TRANSPORTERS | Genes involved in Bile salt and organic anion SLC transporters |
| 0.4 | 8.5 | REACTOME SYNTHESIS SECRETION AND DEACYLATION OF GHRELIN | Genes involved in Synthesis, Secretion, and Deacylation of Ghrelin |
| 0.2 | 4.9 | REACTOME GABA A RECEPTOR ACTIVATION | Genes involved in GABA A receptor activation |
| 0.2 | 2.6 | REACTOME REGULATION OF INSULIN SECRETION BY ACETYLCHOLINE | Genes involved in Regulation of Insulin Secretion by Acetylcholine |
| 0.2 | 6.3 | REACTOME CGMP EFFECTS | Genes involved in cGMP effects |
| 0.2 | 8.0 | REACTOME METAL ION SLC TRANSPORTERS | Genes involved in Metal ion SLC transporters |
| 0.2 | 4.6 | REACTOME OTHER SEMAPHORIN INTERACTIONS | Genes involved in Other semaphorin interactions |
| 0.2 | 3.9 | REACTOME SIGNAL ATTENUATION | Genes involved in Signal attenuation |
| 0.1 | 4.7 | REACTOME ADHERENS JUNCTIONS INTERACTIONS | Genes involved in Adherens junctions interactions |
| 0.1 | 2.3 | REACTOME BRANCHED CHAIN AMINO ACID CATABOLISM | Genes involved in Branched-chain amino acid catabolism |
| 0.1 | 2.4 | REACTOME SULFUR AMINO ACID METABOLISM | Genes involved in Sulfur amino acid metabolism |
| 0.1 | 5.9 | REACTOME AMINO ACID AND OLIGOPEPTIDE SLC TRANSPORTERS | Genes involved in Amino acid and oligopeptide SLC transporters |
| 0.1 | 2.2 | REACTOME SIGNALING BY HIPPO | Genes involved in Signaling by Hippo |
| 0.1 | 1.3 | REACTOME FORMATION OF ATP BY CHEMIOSMOTIC COUPLING | Genes involved in Formation of ATP by chemiosmotic coupling |
| 0.1 | 2.9 | REACTOME SYNTHESIS OF PA | Genes involved in Synthesis of PA |
| 0.1 | 1.0 | REACTOME TANDEM PORE DOMAIN POTASSIUM CHANNELS | Genes involved in Tandem pore domain potassium channels |
| 0.1 | 2.9 | REACTOME INTERACTION BETWEEN L1 AND ANKYRINS | Genes involved in Interaction between L1 and Ankyrins |
| 0.0 | 1.8 | REACTOME DOWNREGULATION OF TGF BETA RECEPTOR SIGNALING | Genes involved in Downregulation of TGF-beta receptor signaling |
| 0.0 | 1.2 | REACTOME ENDOSOMAL SORTING COMPLEX REQUIRED FOR TRANSPORT ESCRT | Genes involved in Endosomal Sorting Complex Required For Transport (ESCRT) |
| 0.0 | 0.7 | REACTOME REGULATION OF INSULIN LIKE GROWTH FACTOR IGF ACTIVITY BY INSULIN LIKE GROWTH FACTOR BINDING PROTEINS IGFBPS | Genes involved in Regulation of Insulin-like Growth Factor (IGF) Activity by Insulin-like Growth Factor Binding Proteins (IGFBPs) |
| 0.0 | 2.1 | REACTOME NETRIN1 SIGNALING | Genes involved in Netrin-1 signaling |
| 0.0 | 0.6 | REACTOME SYNTHESIS OF PE | Genes involved in Synthesis of PE |
| 0.0 | 2.0 | REACTOME O LINKED GLYCOSYLATION OF MUCINS | Genes involved in O-linked glycosylation of mucins |
| 0.0 | 0.4 | REACTOME TRYPTOPHAN CATABOLISM | Genes involved in Tryptophan catabolism |
| 0.0 | 0.5 | REACTOME GAP JUNCTION DEGRADATION | Genes involved in Gap junction degradation |
| 0.0 | 0.8 | REACTOME GLYCOLYSIS | Genes involved in Glycolysis |
| 0.0 | 0.6 | REACTOME ION TRANSPORT BY P TYPE ATPASES | Genes involved in Ion transport by P-type ATPases |