Project
PRJNA375882: Comprehensive Mouse Transcriptomic BodyMap
Navigation
Downloads
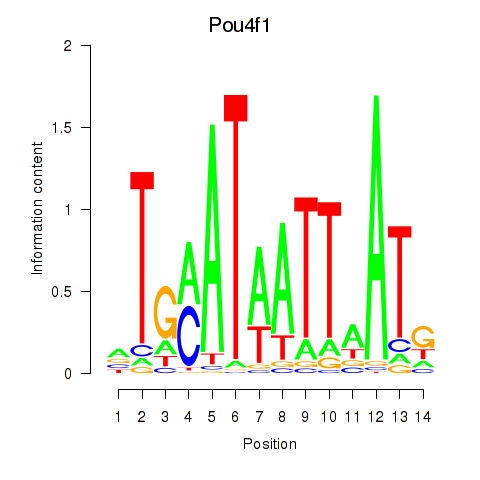
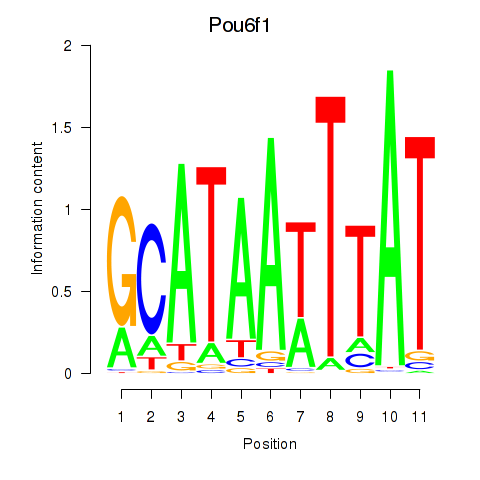
Results for Pou4f1_Pou6f1
Z-value: 0.98
Transcription factors associated with Pou4f1_Pou6f1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Pou4f1
|
ENSMUSG00000048349.10 | Pou4f1 |
|
Pou6f1
|
ENSMUSG00000009739.18 | Pou6f1 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Pou6f1 | mm39_v1_chr15_-_100497863_100497920 | 0.19 | 1.1e-01 | Click! |
| Pou4f1 | mm39_v1_chr14_-_104705420_104705479 | 0.05 | 7.0e-01 | Click! |
Activity profile of Pou4f1_Pou6f1 motif
Sorted Z-values of Pou4f1_Pou6f1 motif
Network of associatons between targets according to the STRING database.
First level regulatory network of Pou4f1_Pou6f1
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr3_-_33136153 | 6.38 |
ENSMUST00000108225.10
|
Pex5l
|
peroxisomal biogenesis factor 5-like |
| chr14_+_27598021 | 5.16 |
ENSMUST00000211684.2
ENSMUST00000210924.2 |
Erc2
|
ELKS/RAB6-interacting/CAST family member 2 |
| chr15_+_92059224 | 5.09 |
ENSMUST00000068378.6
|
Cntn1
|
contactin 1 |
| chr1_-_132318039 | 4.96 |
ENSMUST00000132435.2
|
Tmcc2
|
transmembrane and coiled-coil domains 2 |
| chr9_-_58108988 | 4.44 |
ENSMUST00000163200.3
ENSMUST00000165276.2 ENSMUST00000214647.2 |
Islr2
|
immunoglobulin superfamily containing leucine-rich repeat 2 |
| chr3_-_116047148 | 4.25 |
ENSMUST00000090473.7
|
Gpr88
|
G-protein coupled receptor 88 |
| chr9_-_58109564 | 4.18 |
ENSMUST00000163897.8
ENSMUST00000215950.2 |
Islr2
|
immunoglobulin superfamily containing leucine-rich repeat 2 |
| chrX_-_58211440 | 4.03 |
ENSMUST00000119306.2
|
Fgf13
|
fibroblast growth factor 13 |
| chr2_+_67004178 | 4.00 |
ENSMUST00000239009.2
ENSMUST00000238912.2 |
Xirp2
|
xin actin-binding repeat containing 2 |
| chr2_+_102488985 | 3.87 |
ENSMUST00000080210.10
|
Slc1a2
|
solute carrier family 1 (glial high affinity glutamate transporter), member 2 |
| chr15_+_30457772 | 3.74 |
ENSMUST00000228282.2
|
Ctnnd2
|
catenin (cadherin associated protein), delta 2 |
| chr13_+_42834989 | 3.62 |
ENSMUST00000149235.8
|
Phactr1
|
phosphatase and actin regulator 1 |
| chr5_-_70999547 | 3.43 |
ENSMUST00000199705.2
|
Gabrg1
|
gamma-aminobutyric acid (GABA) A receptor, subunit gamma 1 |
| chr5_-_30619246 | 3.21 |
ENSMUST00000114747.9
ENSMUST00000074171.10 |
Otof
|
otoferlin |
| chr8_+_85065268 | 3.14 |
ENSMUST00000238701.2
|
Cacna1a
|
calcium channel, voltage-dependent, P/Q type, alpha 1A subunit |
| chr11_-_99134885 | 3.13 |
ENSMUST00000103132.10
ENSMUST00000038214.7 |
Krt222
|
keratin 222 |
| chr15_+_18819033 | 3.10 |
ENSMUST00000166873.9
|
Cdh10
|
cadherin 10 |
| chr2_+_83642910 | 3.09 |
ENSMUST00000051454.4
|
Fam171b
|
family with sequence similarity 171, member B |
| chr11_+_92989229 | 2.99 |
ENSMUST00000107859.8
ENSMUST00000107861.8 ENSMUST00000042943.13 ENSMUST00000107858.9 |
Car10
|
carbonic anhydrase 10 |
| chr11_-_42073737 | 2.98 |
ENSMUST00000206085.2
ENSMUST00000020707.12 ENSMUST00000132971.3 |
Gabra1
|
gamma-aminobutyric acid (GABA) A receptor, subunit alpha 1 |
| chr8_+_13034245 | 2.83 |
ENSMUST00000110873.10
ENSMUST00000173006.8 ENSMUST00000145067.8 |
Mcf2l
|
mcf.2 transforming sequence-like |
| chr4_-_41569500 | 2.78 |
ENSMUST00000108049.9
ENSMUST00000108052.10 ENSMUST00000108050.2 |
Fam219a
|
family with sequence similarity 219, member A |
| chr6_-_102441628 | 2.78 |
ENSMUST00000032159.7
|
Cntn3
|
contactin 3 |
| chr4_+_102112189 | 2.65 |
ENSMUST00000106908.9
|
Pde4b
|
phosphodiesterase 4B, cAMP specific |
| chr17_-_50401305 | 2.55 |
ENSMUST00000113195.8
|
Rftn1
|
raftlin lipid raft linker 1 |
| chr6_-_70318437 | 2.53 |
ENSMUST00000196599.2
|
Igkv8-19
|
immunoglobulin kappa variable 8-19 |
| chr11_+_92990110 | 2.52 |
ENSMUST00000107863.4
|
Car10
|
carbonic anhydrase 10 |
| chr2_+_97298002 | 2.43 |
ENSMUST00000059049.8
|
Lrrc4c
|
leucine rich repeat containing 4C |
| chr6_+_96092230 | 2.19 |
ENSMUST00000075080.6
|
Tafa1
|
TAFA chemokine like family member 1 |
| chr3_-_63391300 | 2.19 |
ENSMUST00000192926.2
|
Strit1
|
small transmembrane regulator of ion transport 1 |
| chr2_+_163500290 | 2.10 |
ENSMUST00000164399.8
ENSMUST00000064703.13 ENSMUST00000099105.9 ENSMUST00000152418.8 ENSMUST00000126182.8 ENSMUST00000131228.8 |
Pkig
|
protein kinase inhibitor, gamma |
| chr9_-_58109627 | 2.06 |
ENSMUST00000216231.2
|
Islr2
|
immunoglobulin superfamily containing leucine-rich repeat 2 |
| chr4_+_101365144 | 2.05 |
ENSMUST00000149047.8
ENSMUST00000106929.10 |
Dnajc6
|
DnaJ heat shock protein family (Hsp40) member C6 |
| chr10_+_90412114 | 2.04 |
ENSMUST00000182427.8
ENSMUST00000182053.8 ENSMUST00000182113.8 |
Anks1b
|
ankyrin repeat and sterile alpha motif domain containing 1B |
| chr3_-_19749489 | 2.01 |
ENSMUST00000061294.5
|
Crh
|
corticotropin releasing hormone |
| chr6_+_21215472 | 1.99 |
ENSMUST00000081542.6
|
Kcnd2
|
potassium voltage-gated channel, Shal-related family, member 2 |
| chr12_+_58258558 | 1.94 |
ENSMUST00000110671.3
ENSMUST00000044299.3 |
Sstr1
|
somatostatin receptor 1 |
| chr3_+_68598757 | 1.92 |
ENSMUST00000107816.4
|
Il12a
|
interleukin 12a |
| chr2_+_67935015 | 1.91 |
ENSMUST00000042456.4
|
B3galt1
|
UDP-Gal:betaGlcNAc beta 1,3-galactosyltransferase, polypeptide 1 |
| chr19_-_41065544 | 1.89 |
ENSMUST00000087176.8
|
Opalin
|
oligodendrocytic myelin paranodal and inner loop protein |
| chr9_-_58109526 | 1.83 |
ENSMUST00000216297.2
|
Islr2
|
immunoglobulin superfamily containing leucine-rich repeat 2 |
| chr15_+_99590098 | 1.79 |
ENSMUST00000228185.2
|
Asic1
|
acid-sensing (proton-gated) ion channel 1 |
| chr10_+_29019645 | 1.78 |
ENSMUST00000092629.4
|
Soga3
|
SOGA family member 3 |
| chr5_-_108943211 | 1.74 |
ENSMUST00000004943.2
|
Tmed11
|
transmembrane p24 trafficking protein 11 |
| chr4_-_110144676 | 1.73 |
ENSMUST00000106598.8
ENSMUST00000102723.11 ENSMUST00000153906.2 |
Elavl4
|
ELAV like RNA binding protein 4 |
| chr5_+_27109679 | 1.68 |
ENSMUST00000120555.8
|
Dpp6
|
dipeptidylpeptidase 6 |
| chr1_-_38875757 | 1.66 |
ENSMUST00000147695.9
|
Lonrf2
|
LON peptidase N-terminal domain and ring finger 2 |
| chr3_+_67799510 | 1.64 |
ENSMUST00000063263.5
ENSMUST00000182006.4 |
Iqcj
Iqschfp
|
IQ motif containing J Iqcj and Schip1 fusion protein |
| chr3_-_152687877 | 1.64 |
ENSMUST00000044278.6
|
St6galnac5
|
ST6 (alpha-N-acetyl-neuraminyl-2,3-beta-galactosyl-1,3)-N-acetylgalactosaminide alpha-2,6-sialyltransferase 5 |
| chr5_-_52723700 | 1.61 |
ENSMUST00000039750.7
|
Lgi2
|
leucine-rich repeat LGI family, member 2 |
| chr4_+_152423075 | 1.61 |
ENSMUST00000030775.12
ENSMUST00000164662.8 |
Chd5
|
chromodomain helicase DNA binding protein 5 |
| chr6_+_79995860 | 1.59 |
ENSMUST00000147663.8
ENSMUST00000128718.8 ENSMUST00000126005.8 ENSMUST00000133918.8 |
Lrrtm4
|
leucine rich repeat transmembrane neuronal 4 |
| chr19_+_53781721 | 1.52 |
ENSMUST00000162910.2
|
Rbm20
|
RNA binding motif protein 20 |
| chrX_-_87159237 | 1.49 |
ENSMUST00000113966.8
ENSMUST00000113964.2 |
Il1rapl1
|
interleukin 1 receptor accessory protein-like 1 |
| chr11_-_79421397 | 1.49 |
ENSMUST00000103236.4
ENSMUST00000170799.8 ENSMUST00000170422.4 |
Evi2a
Evi2
|
ecotropic viral integration site 2a ecotropic viral integration site 2 |
| chr4_+_102111936 | 1.49 |
ENSMUST00000106907.9
|
Pde4b
|
phosphodiesterase 4B, cAMP specific |
| chr2_-_148574353 | 1.48 |
ENSMUST00000028926.13
|
Napb
|
N-ethylmaleimide sensitive fusion protein attachment protein beta |
| chr6_+_58808438 | 1.48 |
ENSMUST00000141600.8
ENSMUST00000122981.2 |
Herc3
|
hect domain and RLD 3 |
| chr6_+_79995994 | 1.46 |
ENSMUST00000126399.2
ENSMUST00000136421.2 |
Lrrtm4
|
leucine rich repeat transmembrane neuronal 4 |
| chrX_+_149330371 | 1.43 |
ENSMUST00000066337.13
ENSMUST00000112715.2 |
Alas2
|
aminolevulinic acid synthase 2, erythroid |
| chr8_+_81220410 | 1.42 |
ENSMUST00000063359.8
|
Gypa
|
glycophorin A |
| chr10_+_39296005 | 1.38 |
ENSMUST00000157009.8
|
Fyn
|
Fyn proto-oncogene |
| chr3_+_84859453 | 1.36 |
ENSMUST00000029727.8
|
Fbxw7
|
F-box and WD-40 domain protein 7 |
| chr5_+_57876401 | 1.33 |
ENSMUST00000094783.7
|
Pcdh7
|
protocadherin 7 |
| chr6_+_70640233 | 1.33 |
ENSMUST00000103400.3
|
Igkv3-5
|
immunoglobulin kappa chain variable 3-5 |
| chr17_-_33136277 | 1.32 |
ENSMUST00000234538.2
ENSMUST00000235058.2 ENSMUST00000234759.2 ENSMUST00000179434.8 ENSMUST00000234797.2 |
Cyp4f14
|
cytochrome P450, family 4, subfamily f, polypeptide 14 |
| chr5_-_52723607 | 1.32 |
ENSMUST00000199942.5
|
Lgi2
|
leucine-rich repeat LGI family, member 2 |
| chr13_+_110063364 | 1.31 |
ENSMUST00000117420.8
|
Pde4d
|
phosphodiesterase 4D, cAMP specific |
| chr10_-_63926044 | 1.30 |
ENSMUST00000105439.2
|
Lrrtm3
|
leucine rich repeat transmembrane neuronal 3 |
| chr6_+_58808498 | 1.30 |
ENSMUST00000203714.4
|
Herc3
|
hect domain and RLD 3 |
| chr1_-_60082246 | 1.28 |
ENSMUST00000027172.13
ENSMUST00000191251.7 |
Ica1l
|
islet cell autoantigen 1-like |
| chr13_+_43276323 | 1.28 |
ENSMUST00000136576.8
|
Phactr1
|
phosphatase and actin regulator 1 |
| chr13_+_93441307 | 1.28 |
ENSMUST00000080127.12
|
Homer1
|
homer scaffolding protein 1 |
| chr10_+_79650496 | 1.27 |
ENSMUST00000218857.2
ENSMUST00000220365.2 |
Palm
|
paralemmin |
| chr17_-_52133594 | 1.27 |
ENSMUST00000129667.8
ENSMUST00000169480.8 ENSMUST00000148559.2 |
Satb1
|
special AT-rich sequence binding protein 1 |
| chr9_-_70842090 | 1.25 |
ENSMUST00000034731.10
|
Lipc
|
lipase, hepatic |
| chr6_-_68681962 | 1.23 |
ENSMUST00000103330.2
|
Igkv10-94
|
immunoglobulin kappa variable 10-94 |
| chr6_-_90758954 | 1.23 |
ENSMUST00000238821.2
|
Iqsec1
|
IQ motif and Sec7 domain 1 |
| chr13_+_93440572 | 1.23 |
ENSMUST00000109493.9
|
Homer1
|
homer scaffolding protein 1 |
| chr6_+_68279392 | 1.22 |
ENSMUST00000103322.3
|
Igkv2-109
|
immunoglobulin kappa variable 2-109 |
| chr17_-_33136021 | 1.20 |
ENSMUST00000054174.9
|
Cyp4f14
|
cytochrome P450, family 4, subfamily f, polypeptide 14 |
| chr4_+_102446883 | 1.19 |
ENSMUST00000097949.11
ENSMUST00000106901.2 |
Pde4b
|
phosphodiesterase 4B, cAMP specific |
| chr12_-_113733922 | 1.13 |
ENSMUST00000180013.3
|
Ighv2-9-1
|
immunoglobulin heavy variable 2-9-1 |
| chr8_-_86112938 | 1.13 |
ENSMUST00000137290.2
|
Mylk3
|
myosin light chain kinase 3 |
| chr12_-_72964646 | 1.13 |
ENSMUST00000044000.12
|
4930447C04Rik
|
RIKEN cDNA 4930447C04 gene |
| chr6_-_68609426 | 1.12 |
ENSMUST00000103328.3
|
Igkv10-96
|
immunoglobulin kappa variable 10-96 |
| chr1_-_179572765 | 1.11 |
ENSMUST00000211943.3
ENSMUST00000131716.4 ENSMUST00000221136.2 |
Kif28
|
kinesin family member 28 |
| chr11_-_43792013 | 1.11 |
ENSMUST00000067258.9
ENSMUST00000139906.2 |
Adra1b
|
adrenergic receptor, alpha 1b |
| chr13_+_83652280 | 1.10 |
ENSMUST00000199450.5
|
Mef2c
|
myocyte enhancer factor 2C |
| chr10_+_29020055 | 1.09 |
ENSMUST00000216757.2
|
Soga3
|
SOGA family member 3 |
| chr15_+_100768776 | 1.03 |
ENSMUST00000108909.9
|
Scn8a
|
sodium channel, voltage-gated, type VIII, alpha |
| chr9_+_109865810 | 1.02 |
ENSMUST00000163190.8
|
Map4
|
microtubule-associated protein 4 |
| chr2_+_81883566 | 1.02 |
ENSMUST00000047527.8
|
Zfp804a
|
zinc finger protein 804A |
| chr9_-_103099262 | 1.02 |
ENSMUST00000170904.2
|
Trf
|
transferrin |
| chr3_+_68479578 | 1.01 |
ENSMUST00000170788.9
|
Schip1
|
schwannomin interacting protein 1 |
| chr4_-_138053603 | 0.98 |
ENSMUST00000030536.13
|
Pink1
|
PTEN induced putative kinase 1 |
| chr10_+_7465555 | 0.97 |
ENSMUST00000134346.8
ENSMUST00000019931.12 ENSMUST00000130590.8 |
Lrp11
|
low density lipoprotein receptor-related protein 11 |
| chr11_+_29668563 | 0.97 |
ENSMUST00000060992.6
|
Rtn4
|
reticulon 4 |
| chr6_-_68840015 | 0.95 |
ENSMUST00000103336.2
|
Igkv1-88
|
immunoglobulin kappa chain variable 1-88 |
| chr9_-_70841881 | 0.94 |
ENSMUST00000214995.2
|
Lipc
|
lipase, hepatic |
| chr13_+_83652150 | 0.94 |
ENSMUST00000198199.5
|
Mef2c
|
myocyte enhancer factor 2C |
| chr6_-_70313491 | 0.93 |
ENSMUST00000103388.4
|
Igkv6-20
|
immunoglobulin kappa variable 6-20 |
| chr10_+_127226180 | 0.93 |
ENSMUST00000077046.12
ENSMUST00000105250.9 |
R3hdm2
|
R3H domain containing 2 |
| chr1_-_22386016 | 0.92 |
ENSMUST00000164877.8
|
Rims1
|
regulating synaptic membrane exocytosis 1 |
| chr11_-_54751738 | 0.92 |
ENSMUST00000144164.9
|
Lyrm7
|
LYR motif containing 7 |
| chr10_-_75946790 | 0.91 |
ENSMUST00000120757.2
|
Slc5a4b
|
solute carrier family 5 (neutral amino acid transporters, system A), member 4b |
| chr4_-_119349760 | 0.89 |
ENSMUST00000049994.8
|
Rimkla
|
ribosomal modification protein rimK-like family member A |
| chr4_-_119047202 | 0.88 |
ENSMUST00000239029.2
ENSMUST00000138395.9 ENSMUST00000156746.3 |
Ermap
|
erythroblast membrane-associated protein |
| chr12_-_115172211 | 0.87 |
ENSMUST00000103526.3
|
Ighv1-55
|
immunoglobulin heavy variable 1-55 |
| chr11_+_70410445 | 0.87 |
ENSMUST00000179000.2
|
Gltpd2
|
glycolipid transfer protein domain containing 2 |
| chr17_+_37180437 | 0.87 |
ENSMUST00000060524.11
|
Trim10
|
tripartite motif-containing 10 |
| chr1_-_130589321 | 0.86 |
ENSMUST00000137276.3
|
C4bp
|
complement component 4 binding protein |
| chr8_+_15107646 | 0.85 |
ENSMUST00000033842.4
|
Myom2
|
myomesin 2 |
| chr8_-_68427217 | 0.85 |
ENSMUST00000098696.10
ENSMUST00000038959.16 ENSMUST00000093469.11 |
Psd3
|
pleckstrin and Sec7 domain containing 3 |
| chr6_-_69626340 | 0.84 |
ENSMUST00000198328.2
|
Igkv4-53
|
immunoglobulin kappa variable 4-53 |
| chr13_-_19491721 | 0.84 |
ENSMUST00000103561.3
|
Trgc2
|
T cell receptor gamma, constant 2 |
| chr19_+_24853039 | 0.82 |
ENSMUST00000073080.7
|
Gm10053
|
predicted gene 10053 |
| chr4_-_138053545 | 0.82 |
ENSMUST00000105817.4
|
Pink1
|
PTEN induced putative kinase 1 |
| chr9_-_54381409 | 0.81 |
ENSMUST00000127880.8
|
Dmxl2
|
Dmx-like 2 |
| chr2_+_84564394 | 0.81 |
ENSMUST00000238573.2
ENSMUST00000090729.9 |
Ypel4
|
yippee like 4 |
| chr9_-_71075939 | 0.81 |
ENSMUST00000113570.8
|
Aqp9
|
aquaporin 9 |
| chr4_-_119047167 | 0.80 |
ENSMUST00000030396.15
|
Ermap
|
erythroblast membrane-associated protein |
| chr2_-_168576155 | 0.80 |
ENSMUST00000109175.9
|
Atp9a
|
ATPase, class II, type 9A |
| chr13_+_108996606 | 0.80 |
ENSMUST00000177907.8
|
Pde4d
|
phosphodiesterase 4D, cAMP specific |
| chr17_-_32491339 | 0.80 |
ENSMUST00000237008.2
|
Brd4
|
bromodomain containing 4 |
| chr2_+_67276338 | 0.79 |
ENSMUST00000239060.2
ENSMUST00000028410.4 ENSMUST00000112347.8 ENSMUST00000238878.2 |
Xirp2
|
xin actin-binding repeat containing 2 |
| chr19_-_40175709 | 0.78 |
ENSMUST00000051846.13
|
Cyp2c70
|
cytochrome P450, family 2, subfamily c, polypeptide 70 |
| chr11_-_89732091 | 0.77 |
ENSMUST00000238273.3
|
Ankfn1
|
ankyrin-repeat and fibronectin type III domain containing 1 |
| chr5_+_114427227 | 0.77 |
ENSMUST00000169347.6
|
Myo1h
|
myosin 1H |
| chr7_-_101765915 | 0.77 |
ENSMUST00000084830.2
|
Chrna10
|
cholinergic receptor, nicotinic, alpha polypeptide 10 |
| chr16_-_22258320 | 0.76 |
ENSMUST00000170393.2
|
Etv5
|
ets variant 5 |
| chr4_-_119047180 | 0.75 |
ENSMUST00000150864.3
ENSMUST00000141227.9 |
Ermap
|
erythroblast membrane-associated protein |
| chr4_-_21767116 | 0.75 |
ENSMUST00000029915.6
|
Tstd3
|
thiosulfate sulfurtransferase (rhodanese)-like domain containing 3 |
| chr13_-_32960379 | 0.73 |
ENSMUST00000230119.2
|
Mylk4
|
myosin light chain kinase family, member 4 |
| chr6_+_70549568 | 0.73 |
ENSMUST00000196940.2
ENSMUST00000103397.3 |
Igkv3-10
|
immunoglobulin kappa variable 3-10 |
| chr7_-_85895409 | 0.72 |
ENSMUST00000165771.2
ENSMUST00000233075.2 ENSMUST00000233317.2 ENSMUST00000233312.2 ENSMUST00000233928.2 ENSMUST00000232799.2 ENSMUST00000233744.2 |
Vmn2r76
|
vomeronasal 2, receptor 76 |
| chr18_+_59195354 | 0.71 |
ENSMUST00000165666.9
|
Minar2
|
membrane integral NOTCH2 associated receptor 2 |
| chr13_+_18901459 | 0.71 |
ENSMUST00000072961.6
|
Vps41
|
VPS41 HOPS complex subunit |
| chr14_-_96756503 | 0.71 |
ENSMUST00000022666.9
|
Klhl1
|
kelch-like 1 |
| chr5_-_113311185 | 0.71 |
ENSMUST00000050125.9
|
2900026A02Rik
|
RIKEN cDNA 2900026A02 gene |
| chr3_-_96615838 | 0.70 |
ENSMUST00000029742.9
ENSMUST00000200387.2 ENSMUST00000171249.6 |
Nudt17
|
nudix (nucleoside diphosphate linked moiety X)-type motif 17 |
| chr17_+_85335775 | 0.69 |
ENSMUST00000024944.9
|
Slc3a1
|
solute carrier family 3, member 1 |
| chr2_-_79959178 | 0.69 |
ENSMUST00000102654.11
ENSMUST00000102655.10 |
Pde1a
|
phosphodiesterase 1A, calmodulin-dependent |
| chr5_-_143963413 | 0.69 |
ENSMUST00000031622.13
|
Ocm
|
oncomodulin |
| chr18_+_65022035 | 0.68 |
ENSMUST00000224385.3
ENSMUST00000163516.9 |
Nedd4l
|
neural precursor cell expressed, developmentally down-regulated gene 4-like |
| chr19_-_39729431 | 0.68 |
ENSMUST00000099472.4
|
Cyp2c68
|
cytochrome P450, family 2, subfamily c, polypeptide 68 |
| chr6_-_70121150 | 0.68 |
ENSMUST00000197525.2
|
Igkv8-28
|
immunoglobulin kappa variable 8-28 |
| chr2_-_34990689 | 0.67 |
ENSMUST00000226631.2
ENSMUST00000045776.5 ENSMUST00000226972.2 |
AI182371
|
expressed sequence AI182371 |
| chr16_-_19079594 | 0.66 |
ENSMUST00000103752.3
ENSMUST00000197518.2 |
Iglv2
|
immunoglobulin lambda variable 2 |
| chr1_+_65321215 | 0.66 |
ENSMUST00000140190.8
|
Pth2r
|
parathyroid hormone 2 receptor |
| chr14_+_53941464 | 0.65 |
ENSMUST00000103664.6
|
Trav5-4
|
T cell receptor alpha variable 5-4 |
| chr8_-_25085654 | 0.65 |
ENSMUST00000110667.8
|
Ido1
|
indoleamine 2,3-dioxygenase 1 |
| chr18_+_69726654 | 0.64 |
ENSMUST00000200921.4
|
Tcf4
|
transcription factor 4 |
| chr19_-_39451509 | 0.64 |
ENSMUST00000035488.3
|
Cyp2c38
|
cytochrome P450, family 2, subfamily c, polypeptide 38 |
| chr4_-_156340713 | 0.63 |
ENSMUST00000219393.2
|
Samd11
|
sterile alpha motif domain containing 11 |
| chr13_+_75237939 | 0.63 |
ENSMUST00000022075.6
|
Pcsk1
|
proprotein convertase subtilisin/kexin type 1 |
| chr3_-_58729732 | 0.63 |
ENSMUST00000191233.4
|
Mindy4b-ps
|
MINDY lysine 48 deubiquitinase 4B, pseudogene |
| chr1_+_179928709 | 0.62 |
ENSMUST00000133890.8
|
Cdc42bpa
|
CDC42 binding protein kinase alpha |
| chr2_+_181408833 | 0.62 |
ENSMUST00000108756.8
|
Myt1
|
myelin transcription factor 1 |
| chrX_-_138683102 | 0.61 |
ENSMUST00000101217.4
|
Ripply1
|
ripply transcriptional repressor 1 |
| chr2_-_79959802 | 0.61 |
ENSMUST00000102653.8
|
Pde1a
|
phosphodiesterase 1A, calmodulin-dependent |
| chrX_-_72080709 | 0.61 |
ENSMUST00000114540.4
ENSMUST00000051569.7 |
Pnma5
|
paraneoplastic antigen family 5 |
| chr4_-_119047146 | 0.61 |
ENSMUST00000124626.9
|
Ermap
|
erythroblast membrane-associated protein |
| chr7_-_104018989 | 0.60 |
ENSMUST00000164410.2
|
Trim30b
|
tripartite motif-containing 30B |
| chr11_+_71640739 | 0.60 |
ENSMUST00000150531.2
|
Wscd1
|
WSC domain containing 1 |
| chr6_-_69753317 | 0.60 |
ENSMUST00000103366.3
|
Igkv5-45
|
immunoglobulin kappa chain variable 5-45 |
| chr5_-_89605622 | 0.60 |
ENSMUST00000049209.13
|
Gc
|
vitamin D binding protein |
| chr1_+_128069677 | 0.59 |
ENSMUST00000187023.7
|
R3hdm1
|
R3H domain containing 1 |
| chr5_-_104225458 | 0.59 |
ENSMUST00000198485.5
ENSMUST00000164471.8 ENSMUST00000178967.2 |
Gm17660
|
predicted gene, 17660 |
| chr1_+_128069716 | 0.59 |
ENSMUST00000187557.2
|
R3hdm1
|
R3H domain containing 1 |
| chr2_-_77349909 | 0.59 |
ENSMUST00000111830.9
|
Zfp385b
|
zinc finger protein 385B |
| chr9_-_78016302 | 0.59 |
ENSMUST00000001402.14
|
Fbxo9
|
f-box protein 9 |
| chr6_-_39397334 | 0.57 |
ENSMUST00000031985.13
|
Mkrn1
|
makorin, ring finger protein, 1 |
| chr9_-_99302205 | 0.56 |
ENSMUST00000123771.2
|
Mras
|
muscle and microspikes RAS |
| chr7_+_46496506 | 0.56 |
ENSMUST00000209984.2
|
Ldha
|
lactate dehydrogenase A |
| chr7_-_140461769 | 0.56 |
ENSMUST00000106048.10
ENSMUST00000147331.9 ENSMUST00000137710.2 |
Sirt3
|
sirtuin 3 |
| chr17_+_18269686 | 0.56 |
ENSMUST00000176802.3
|
Vmn2r124
|
vomeronasal 2, receptor 124 |
| chr6_+_41369290 | 0.55 |
ENSMUST00000049079.9
|
Gm5771
|
predicted gene 5771 |
| chr18_+_56565188 | 0.55 |
ENSMUST00000070166.6
|
Gramd3
|
GRAM domain containing 3 |
| chr6_-_69037208 | 0.55 |
ENSMUST00000103343.4
|
Igkv4-78
|
immunoglobulin kappa variable 4-78 |
| chr5_-_123804745 | 0.54 |
ENSMUST00000149410.2
|
Clip1
|
CAP-GLY domain containing linker protein 1 |
| chr10_-_45346297 | 0.54 |
ENSMUST00000079390.7
|
Lin28b
|
lin-28 homolog B (C. elegans) |
| chr2_+_175971970 | 0.54 |
ENSMUST00000135430.2
|
2210418O10Rik
|
RIKEN cDNA 2210418O10 gene |
| chr6_-_130003917 | 0.53 |
ENSMUST00000074056.3
|
Klra6
|
killer cell lectin-like receptor, subfamily A, member 6 |
| chr6_-_69704122 | 0.53 |
ENSMUST00000103364.3
|
Igkv5-48
|
immunoglobulin kappa variable 5-48 |
| chr2_+_22785534 | 0.53 |
ENSMUST00000053729.14
|
Pdss1
|
prenyl (solanesyl) diphosphate synthase, subunit 1 |
| chr1_-_130589349 | 0.53 |
ENSMUST00000027657.14
|
C4bp
|
complement component 4 binding protein |
| chrX_-_149242125 | 0.53 |
ENSMUST00000173996.8
|
Tmem29
|
transmembrane protein 29 |
| chr1_-_87322443 | 0.52 |
ENSMUST00000113212.4
|
Kcnj13
|
potassium inwardly-rectifying channel, subfamily J, member 13 |
| chr1_-_171885140 | 0.52 |
ENSMUST00000059794.4
|
Nhlh1
|
nescient helix loop helix 1 |
| chr7_+_28455563 | 0.52 |
ENSMUST00000178767.3
|
Ccer2
|
coiled-coil glutamate-rich protein 2 |
| chr13_-_56696310 | 0.52 |
ENSMUST00000062806.6
|
Lect2
|
leukocyte cell-derived chemotaxin 2 |
| chr6_-_66537080 | 0.51 |
ENSMUST00000079584.3
ENSMUST00000227014.2 |
Vmn1r32
|
vomeronasal 1 receptor 32 |
| chr15_-_100497863 | 0.50 |
ENSMUST00000073837.13
|
Pou6f1
|
POU domain, class 6, transcription factor 1 |
| chr15_+_79575046 | 0.50 |
ENSMUST00000046463.10
|
Gtpbp1
|
GTP binding protein 1 |
| chr7_+_46496552 | 0.49 |
ENSMUST00000005051.6
|
Ldha
|
lactate dehydrogenase A |
| chr6_-_70364222 | 0.48 |
ENSMUST00000103392.3
ENSMUST00000195945.2 |
Igkv8-16
|
immunoglobulin kappa variable 8-16 |
| chr14_+_52962756 | 0.48 |
ENSMUST00000181483.3
|
Trav6d-3
|
T cell receptor alpha variable 6D-3 |
| chr8_-_84184978 | 0.48 |
ENSMUST00000081506.11
|
Scoc
|
short coiled-coil protein |
| chr16_+_49620883 | 0.47 |
ENSMUST00000229640.2
|
Cd47
|
CD47 antigen (Rh-related antigen, integrin-associated signal transducer) |
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.8 | 6.4 | GO:0016560 | protein import into peroxisome matrix, docking(GO:0016560) |
| 0.8 | 3.1 | GO:0021750 | cerebellar molecular layer development(GO:0021679) vestibular nucleus development(GO:0021750) |
| 0.7 | 2.0 | GO:0008628 | hormone-mediated apoptotic signaling pathway(GO:0008628) |
| 0.6 | 4.5 | GO:0061743 | motor learning(GO:0061743) |
| 0.6 | 4.0 | GO:0045196 | establishment or maintenance of neuroblast polarity(GO:0045196) establishment of neuroblast polarity(GO:0045200) |
| 0.6 | 7.4 | GO:1901898 | negative regulation of relaxation of cardiac muscle(GO:1901898) |
| 0.6 | 3.9 | GO:0070777 | D-aspartate transport(GO:0070777) D-aspartate import(GO:0070779) |
| 0.4 | 2.2 | GO:0034371 | chylomicron remodeling(GO:0034371) |
| 0.4 | 1.1 | GO:0010705 | meiotic DNA double-strand break processing involved in reciprocal meiotic recombination(GO:0010705) |
| 0.4 | 1.1 | GO:0001982 | baroreceptor response to decreased systemic arterial blood pressure(GO:0001982) |
| 0.3 | 3.1 | GO:0032596 | protein transport into membrane raft(GO:0032596) |
| 0.3 | 2.1 | GO:0072318 | clathrin coat disassembly(GO:0072318) |
| 0.3 | 1.8 | GO:1904925 | neuron intrinsic apoptotic signaling pathway in response to hydrogen peroxide(GO:0036482) positive regulation of macromitophagy(GO:1901526) positive regulation of mitochondrial electron transport, NADH to ubiquinone(GO:1902958) regulation of hydrogen peroxide-induced neuron intrinsic apoptotic signaling pathway(GO:1903383) negative regulation of hydrogen peroxide-induced neuron intrinsic apoptotic signaling pathway(GO:1903384) positive regulation of mitophagy in response to mitochondrial depolarization(GO:1904925) |
| 0.3 | 1.8 | GO:0050915 | sensory perception of sour taste(GO:0050915) |
| 0.3 | 1.4 | GO:1903378 | positive regulation of oxidative stress-induced neuron intrinsic apoptotic signaling pathway(GO:1903378) |
| 0.3 | 2.3 | GO:0006682 | galactosylceramide biosynthetic process(GO:0006682) galactolipid biosynthetic process(GO:0019375) |
| 0.2 | 5.6 | GO:0016082 | synaptic vesicle priming(GO:0016082) |
| 0.2 | 0.6 | GO:0036269 | swimming behavior(GO:0036269) |
| 0.2 | 1.9 | GO:2000510 | positive regulation of dendritic cell chemotaxis(GO:2000510) |
| 0.2 | 0.6 | GO:1903045 | neural crest cell migration involved in sympathetic nervous system development(GO:1903045) |
| 0.2 | 1.3 | GO:0060160 | negative regulation of dopamine receptor signaling pathway(GO:0060160) |
| 0.2 | 1.0 | GO:0048687 | positive regulation of sprouting of injured axon(GO:0048687) positive regulation of axon extension involved in regeneration(GO:0048691) positive regulation of retinal ganglion cell axon guidance(GO:1902336) |
| 0.2 | 0.8 | GO:0015851 | nucleobase transport(GO:0015851) pyrimidine nucleobase transport(GO:0015855) |
| 0.2 | 1.6 | GO:0098532 | histone H3-K27 trimethylation(GO:0098532) |
| 0.2 | 1.1 | GO:0002528 | regulation of vascular permeability involved in acute inflammatory response(GO:0002528) |
| 0.2 | 2.0 | GO:1900383 | regulation of synaptic plasticity by receptor localization to synapse(GO:1900383) |
| 0.2 | 1.4 | GO:1902951 | negative regulation of dendritic spine maintenance(GO:1902951) |
| 0.2 | 0.9 | GO:1902728 | positive regulation of skeletal muscle satellite cell proliferation(GO:1902724) positive regulation of growth factor dependent skeletal muscle satellite cell proliferation(GO:1902728) |
| 0.2 | 2.0 | GO:0003172 | primary heart field specification(GO:0003138) sinoatrial valve development(GO:0003172) sinoatrial valve morphogenesis(GO:0003185) |
| 0.2 | 3.1 | GO:0097113 | AMPA glutamate receptor clustering(GO:0097113) |
| 0.2 | 6.4 | GO:0007214 | gamma-aminobutyric acid signaling pathway(GO:0007214) |
| 0.1 | 12.9 | GO:0045773 | positive regulation of axon extension(GO:0045773) |
| 0.1 | 1.0 | GO:0051012 | microtubule sliding(GO:0051012) |
| 0.1 | 0.7 | GO:1902774 | late endosome to lysosome transport(GO:1902774) |
| 0.1 | 1.4 | GO:0019659 | glucose catabolic process to lactate(GO:0019659) glycolytic fermentation(GO:0019660) glucose catabolic process to lactate via pyruvate(GO:0019661) |
| 0.1 | 1.2 | GO:0098728 | germ-line stem cell division(GO:0042078) male germ-line stem cell asymmetric division(GO:0048133) germline stem cell asymmetric division(GO:0098728) |
| 0.1 | 0.8 | GO:0010587 | miRNA catabolic process(GO:0010587) |
| 0.1 | 1.0 | GO:0070447 | positive regulation of oligodendrocyte progenitor proliferation(GO:0070447) |
| 0.1 | 0.4 | GO:0002631 | regulation of granuloma formation(GO:0002631) negative regulation of granuloma formation(GO:0002632) regulation of toll-like receptor 5 signaling pathway(GO:0034147) negative regulation of toll-like receptor 5 signaling pathway(GO:0034148) negative regulation of nucleotide-binding oligomerization domain containing 1 signaling pathway(GO:0070429) negative regulation of CD40 signaling pathway(GO:2000349) |
| 0.1 | 2.5 | GO:2001256 | regulation of store-operated calcium entry(GO:2001256) |
| 0.1 | 0.4 | GO:0010133 | proline catabolic process to glutamate(GO:0010133) |
| 0.1 | 1.0 | GO:0009414 | response to water deprivation(GO:0009414) |
| 0.1 | 1.0 | GO:0071787 | endoplasmic reticulum tubular network assembly(GO:0071787) |
| 0.1 | 1.4 | GO:0047484 | regulation of response to osmotic stress(GO:0047484) |
| 0.1 | 1.5 | GO:0097105 | presynaptic membrane assembly(GO:0097105) |
| 0.1 | 0.3 | GO:0051970 | negative regulation of transmission of nerve impulse(GO:0051970) |
| 0.1 | 1.6 | GO:0009312 | oligosaccharide biosynthetic process(GO:0009312) |
| 0.1 | 2.1 | GO:2000480 | negative regulation of cAMP-dependent protein kinase activity(GO:2000480) |
| 0.1 | 1.4 | GO:0042541 | hemoglobin biosynthetic process(GO:0042541) |
| 0.1 | 1.2 | GO:1904659 | glucose transmembrane transport(GO:1904659) |
| 0.1 | 0.2 | GO:1905150 | regulation of voltage-gated sodium channel activity(GO:1905150) |
| 0.1 | 5.7 | GO:0042982 | amyloid precursor protein metabolic process(GO:0042982) |
| 0.1 | 0.9 | GO:0034551 | respiratory chain complex III assembly(GO:0017062) mitochondrial respiratory chain complex III assembly(GO:0034551) |
| 0.1 | 0.4 | GO:0002729 | positive regulation of natural killer cell cytokine production(GO:0002729) |
| 0.1 | 4.7 | GO:0019228 | neuronal action potential(GO:0019228) |
| 0.1 | 0.3 | GO:0000973 | posttranscriptional tethering of RNA polymerase II gene DNA at nuclear periphery(GO:0000973) |
| 0.1 | 0.4 | GO:0007621 | negative regulation of female receptivity(GO:0007621) |
| 0.1 | 0.9 | GO:0050957 | equilibrioception(GO:0050957) |
| 0.1 | 1.3 | GO:0043374 | CD8-positive, alpha-beta T cell differentiation(GO:0043374) |
| 0.1 | 4.5 | GO:0019369 | arachidonic acid metabolic process(GO:0019369) |
| 0.1 | 1.1 | GO:0032525 | somite rostral/caudal axis specification(GO:0032525) |
| 0.1 | 1.3 | GO:0046069 | cGMP catabolic process(GO:0046069) |
| 0.1 | 0.8 | GO:0070072 | vacuolar proton-transporting V-type ATPase complex assembly(GO:0070072) |
| 0.1 | 5.8 | GO:0010765 | positive regulation of sodium ion transport(GO:0010765) |
| 0.1 | 2.8 | GO:0035025 | positive regulation of Rho protein signal transduction(GO:0035025) |
| 0.1 | 2.1 | GO:0032011 | ARF protein signal transduction(GO:0032011) regulation of ARF protein signal transduction(GO:0032012) |
| 0.1 | 0.4 | GO:0031296 | B cell costimulation(GO:0031296) |
| 0.1 | 0.9 | GO:0071688 | striated muscle myosin thick filament assembly(GO:0071688) |
| 0.1 | 0.8 | GO:0044154 | histone H3-K14 acetylation(GO:0044154) |
| 0.0 | 0.3 | GO:0046826 | negative regulation of protein export from nucleus(GO:0046826) |
| 0.0 | 0.6 | GO:0034983 | peptidyl-lysine deacetylation(GO:0034983) |
| 0.0 | 1.1 | GO:0071392 | cellular response to estradiol stimulus(GO:0071392) |
| 0.0 | 0.1 | GO:0035874 | amiloride transport(GO:0015898) cellular response to copper ion starvation(GO:0035874) response to azide(GO:0097184) cellular response to azide(GO:0097185) |
| 0.0 | 0.3 | GO:0006083 | acetate metabolic process(GO:0006083) |
| 0.0 | 0.5 | GO:0008228 | opsonization(GO:0008228) |
| 0.0 | 3.7 | GO:0060997 | dendritic spine morphogenesis(GO:0060997) |
| 0.0 | 1.0 | GO:0001553 | luteinization(GO:0001553) |
| 0.0 | 4.4 | GO:0055008 | cardiac muscle tissue morphogenesis(GO:0055008) |
| 0.0 | 0.4 | GO:0045919 | positive regulation of cytolysis(GO:0045919) |
| 0.0 | 0.5 | GO:0061635 | regulation of protein complex stability(GO:0061635) |
| 0.0 | 0.3 | GO:2001181 | positive regulation of interleukin-10 secretion(GO:2001181) |
| 0.0 | 9.9 | GO:0002377 | immunoglobulin production(GO:0002377) |
| 0.0 | 1.0 | GO:0042118 | endothelial cell activation(GO:0042118) |
| 0.0 | 0.3 | GO:2000911 | positive regulation of cholesterol import(GO:1904109) positive regulation of sterol import(GO:2000911) |
| 0.0 | 0.3 | GO:0001514 | selenocysteine incorporation(GO:0001514) translational readthrough(GO:0006451) |
| 0.0 | 5.1 | GO:0043149 | contractile actin filament bundle assembly(GO:0030038) stress fiber assembly(GO:0043149) |
| 0.0 | 0.1 | GO:0034552 | respiratory chain complex II assembly(GO:0034552) mitochondrial respiratory chain complex II assembly(GO:0034553) mitochondrial respiratory chain complex II biogenesis(GO:0097032) |
| 0.0 | 0.4 | GO:0001867 | complement activation, lectin pathway(GO:0001867) |
| 0.0 | 0.2 | GO:0019614 | catechol-containing compound catabolic process(GO:0019614) catecholamine catabolic process(GO:0042424) |
| 0.0 | 4.4 | GO:0006958 | complement activation, classical pathway(GO:0006958) |
| 0.0 | 0.6 | GO:0046597 | negative regulation of viral entry into host cell(GO:0046597) |
| 0.0 | 1.7 | GO:0033120 | positive regulation of RNA splicing(GO:0033120) |
| 0.0 | 0.1 | GO:0023016 | signal transduction by trans-phosphorylation(GO:0023016) |
| 0.0 | 0.2 | GO:0006032 | chitin metabolic process(GO:0006030) chitin catabolic process(GO:0006032) |
| 0.0 | 0.3 | GO:0006122 | mitochondrial electron transport, ubiquinol to cytochrome c(GO:0006122) |
| 0.0 | 0.1 | GO:1901187 | regulation of ephrin receptor signaling pathway(GO:1901187) |
| 0.0 | 0.6 | GO:0060539 | diaphragm development(GO:0060539) |
| 0.0 | 0.6 | GO:0050910 | detection of mechanical stimulus involved in sensory perception of sound(GO:0050910) |
| 0.0 | 0.6 | GO:0016486 | peptide hormone processing(GO:0016486) |
| 0.0 | 0.4 | GO:0001778 | plasma membrane repair(GO:0001778) |
| 0.0 | 0.2 | GO:0014827 | intestine smooth muscle contraction(GO:0014827) |
| 0.0 | 0.5 | GO:0006744 | ubiquinone biosynthetic process(GO:0006744) |
| 0.0 | 1.7 | GO:1900006 | positive regulation of dendrite development(GO:1900006) |
| 0.0 | 0.1 | GO:0045659 | negative regulation of neutrophil differentiation(GO:0045659) |
| 0.0 | 2.8 | GO:0019236 | response to pheromone(GO:0019236) |
| 0.0 | 0.1 | GO:0002331 | pre-B cell allelic exclusion(GO:0002331) |
| 0.0 | 0.3 | GO:1902455 | negative regulation of stem cell population maintenance(GO:1902455) |
| 0.0 | 0.2 | GO:0009082 | branched-chain amino acid biosynthetic process(GO:0009082) leucine biosynthetic process(GO:0009098) valine biosynthetic process(GO:0009099) |
| 0.0 | 0.3 | GO:0033327 | Leydig cell differentiation(GO:0033327) |
| 0.0 | 0.1 | GO:1900110 | establishment of protein localization to chromatin(GO:0071169) negative regulation of histone H3-K9 dimethylation(GO:1900110) |
| 0.0 | 0.1 | GO:0033313 | meiotic cell cycle checkpoint(GO:0033313) |
| 0.0 | 0.8 | GO:0006890 | retrograde vesicle-mediated transport, Golgi to ER(GO:0006890) |
| 0.0 | 0.2 | GO:0021796 | cerebral cortex regionalization(GO:0021796) |
| 0.0 | 0.3 | GO:0000712 | resolution of meiotic recombination intermediates(GO:0000712) |
| 0.0 | 0.0 | GO:0070175 | positive regulation of enamel mineralization(GO:0070175) |
| 0.0 | 2.4 | GO:0050770 | regulation of axonogenesis(GO:0050770) |
| 0.0 | 0.7 | GO:0007628 | adult walking behavior(GO:0007628) |
| 0.0 | 1.6 | GO:0032436 | positive regulation of proteasomal ubiquitin-dependent protein catabolic process(GO:0032436) |
| 0.0 | 2.0 | GO:0007156 | homophilic cell adhesion via plasma membrane adhesion molecules(GO:0007156) |
| 0.0 | 0.5 | GO:0061014 | positive regulation of mRNA catabolic process(GO:0061014) |
| 0.0 | 2.9 | GO:0010506 | regulation of autophagy(GO:0010506) |
| 0.0 | 0.6 | GO:0051180 | vitamin transport(GO:0051180) |
| 0.0 | 0.3 | GO:0007095 | mitotic G2 DNA damage checkpoint(GO:0007095) |
| 0.0 | 0.4 | GO:0045746 | negative regulation of Notch signaling pathway(GO:0045746) |
| 0.0 | 0.8 | GO:0072384 | organelle transport along microtubule(GO:0072384) |
| 0.0 | 0.1 | GO:0032277 | negative regulation of gonadotropin secretion(GO:0032277) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.8 | 6.4 | GO:0017071 | intracellular cyclic nucleotide activated cation channel complex(GO:0017071) |
| 0.3 | 1.4 | GO:1990452 | Parkin-FBXW7-Cul1 ubiquitin ligase complex(GO:1990452) |
| 0.3 | 0.8 | GO:0043291 | RAVE complex(GO:0043291) |
| 0.2 | 1.5 | GO:0070044 | synaptobrevin 2-SNAP-25-syntaxin-1a complex(GO:0070044) |
| 0.2 | 3.2 | GO:0048787 | presynaptic active zone membrane(GO:0048787) |
| 0.2 | 1.0 | GO:0005892 | acetylcholine-gated channel complex(GO:0005892) |
| 0.2 | 5.3 | GO:0000930 | gamma-tubulin complex(GO:0000930) |
| 0.2 | 0.9 | GO:0098831 | presynaptic active zone cytoplasmic component(GO:0098831) |
| 0.1 | 1.8 | GO:0005742 | mitochondrial outer membrane translocase complex(GO:0005742) |
| 0.1 | 0.3 | GO:0071007 | U2-type catalytic step 2 spliceosome(GO:0071007) |
| 0.1 | 5.2 | GO:0048786 | presynaptic active zone(GO:0048786) |
| 0.1 | 1.7 | GO:0042788 | polysomal ribosome(GO:0042788) |
| 0.1 | 3.0 | GO:1902711 | GABA-A receptor complex(GO:1902711) |
| 0.1 | 1.3 | GO:0032591 | dendritic spine membrane(GO:0032591) |
| 0.1 | 1.0 | GO:0097433 | dense body(GO:0097433) |
| 0.1 | 0.3 | GO:1904511 | cortical microtubule plus-end(GO:1903754) cytoplasmic microtubule plus-end(GO:1904511) |
| 0.1 | 0.7 | GO:0033263 | CORVET complex(GO:0033263) |
| 0.1 | 5.3 | GO:0005891 | voltage-gated calcium channel complex(GO:0005891) |
| 0.1 | 1.0 | GO:0000801 | central element(GO:0000801) |
| 0.1 | 2.0 | GO:0043196 | varicosity(GO:0043196) |
| 0.1 | 0.6 | GO:0030485 | smooth muscle contractile fiber(GO:0030485) |
| 0.1 | 3.9 | GO:0030673 | axolemma(GO:0030673) |
| 0.1 | 2.2 | GO:0043034 | costamere(GO:0043034) |
| 0.1 | 3.1 | GO:0032281 | AMPA glutamate receptor complex(GO:0032281) |
| 0.1 | 1.8 | GO:0005859 | muscle myosin complex(GO:0005859) |
| 0.1 | 3.6 | GO:0034707 | chloride channel complex(GO:0034707) |
| 0.1 | 0.3 | GO:0044614 | nuclear pore cytoplasmic filaments(GO:0044614) |
| 0.1 | 0.8 | GO:0008024 | cyclin/CDK positive transcription elongation factor complex(GO:0008024) |
| 0.1 | 2.0 | GO:0097038 | perinuclear endoplasmic reticulum(GO:0097038) |
| 0.1 | 1.6 | GO:0090545 | NuRD complex(GO:0016581) CHD-type complex(GO:0090545) |
| 0.1 | 1.0 | GO:0001518 | voltage-gated sodium channel complex(GO:0001518) |
| 0.1 | 0.5 | GO:0044352 | pinosome(GO:0044352) macropinosome(GO:0044354) |
| 0.1 | 2.2 | GO:0034364 | high-density lipoprotein particle(GO:0034364) |
| 0.0 | 0.2 | GO:0032437 | cuticular plate(GO:0032437) |
| 0.0 | 3.9 | GO:0031941 | filamentous actin(GO:0031941) |
| 0.0 | 1.4 | GO:0035686 | sperm fibrous sheath(GO:0035686) |
| 0.0 | 1.3 | GO:0044291 | cell-cell contact zone(GO:0044291) |
| 0.0 | 5.1 | GO:0014704 | intercalated disc(GO:0014704) |
| 0.0 | 1.8 | GO:0043198 | dendritic shaft(GO:0043198) |
| 0.0 | 0.5 | GO:0000177 | cytoplasmic exosome (RNase complex)(GO:0000177) |
| 0.0 | 0.4 | GO:0000235 | astral microtubule(GO:0000235) |
| 0.0 | 0.3 | GO:0031080 | nuclear pore outer ring(GO:0031080) |
| 0.0 | 1.0 | GO:0071782 | endoplasmic reticulum tubular network(GO:0071782) |
| 0.0 | 6.6 | GO:0031225 | anchored component of membrane(GO:0031225) |
| 0.0 | 3.2 | GO:0005882 | intermediate filament(GO:0005882) |
| 0.0 | 7.3 | GO:0099572 | postsynaptic density(GO:0014069) postsynaptic specialization(GO:0099572) |
| 0.0 | 3.0 | GO:0042571 | immunoglobulin complex, circulating(GO:0042571) |
| 0.0 | 7.9 | GO:0045211 | postsynaptic membrane(GO:0045211) |
| 0.0 | 0.3 | GO:0005750 | mitochondrial respiratory chain complex III(GO:0005750) |
| 0.0 | 0.2 | GO:0031466 | Cul5-RING ubiquitin ligase complex(GO:0031466) |
| 0.0 | 0.1 | GO:0030896 | checkpoint clamp complex(GO:0030896) |
| 0.0 | 0.3 | GO:0071203 | WASH complex(GO:0071203) |
| 0.0 | 1.7 | GO:0008076 | voltage-gated potassium channel complex(GO:0008076) |
| 0.0 | 1.3 | GO:0005720 | nuclear heterochromatin(GO:0005720) |
| 0.0 | 3.1 | GO:0030027 | lamellipodium(GO:0030027) |
| 0.0 | 1.1 | GO:0005871 | kinesin complex(GO:0005871) |
| 0.0 | 0.2 | GO:0005662 | DNA replication factor A complex(GO:0005662) |
| 0.0 | 0.3 | GO:0030008 | TRAPP complex(GO:0030008) |
| 0.0 | 0.5 | GO:0016235 | aggresome(GO:0016235) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.1 | 6.4 | GO:0005052 | peroxisome matrix targeting signal-1 binding(GO:0005052) |
| 0.7 | 2.2 | GO:0052870 | tocopherol omega-hydroxylase activity(GO:0052870) alpha-tocopherol omega-hydroxylase activity(GO:0052871) |
| 0.6 | 3.9 | GO:0015501 | glutamate:sodium symporter activity(GO:0015501) |
| 0.5 | 2.0 | GO:0051431 | corticotropin-releasing hormone receptor 2 binding(GO:0051431) |
| 0.5 | 1.9 | GO:0047275 | glucosaminylgalactosylglucosylceramide beta-galactosyltransferase activity(GO:0047275) |
| 0.5 | 1.4 | GO:0003870 | 5-aminolevulinate synthase activity(GO:0003870) N-succinyltransferase activity(GO:0016749) |
| 0.4 | 2.2 | GO:0035478 | chylomicron binding(GO:0035478) |
| 0.4 | 1.6 | GO:0061628 | H3K27me3 modified histone binding(GO:0061628) |
| 0.4 | 2.0 | GO:0005250 | A-type (transient outward) potassium channel activity(GO:0005250) |
| 0.4 | 1.9 | GO:0005143 | interleukin-12 receptor binding(GO:0005143) |
| 0.4 | 3.0 | GO:1904315 | transmitter-gated ion channel activity involved in regulation of postsynaptic membrane potential(GO:1904315) |
| 0.3 | 1.9 | GO:0004994 | somatostatin receptor activity(GO:0004994) |
| 0.3 | 3.9 | GO:0031802 | type 5 metabotropic glutamate receptor binding(GO:0031802) |
| 0.3 | 1.5 | GO:0005483 | soluble NSF attachment protein activity(GO:0005483) |
| 0.3 | 1.1 | GO:0004937 | alpha1-adrenergic receptor activity(GO:0004937) |
| 0.2 | 1.7 | GO:0031750 | D3 dopamine receptor binding(GO:0031750) |
| 0.2 | 1.4 | GO:0050816 | phosphothreonine binding(GO:0050816) |
| 0.2 | 0.6 | GO:0033754 | indoleamine 2,3-dioxygenase activity(GO:0033754) |
| 0.2 | 1.9 | GO:0004687 | myosin light chain kinase activity(GO:0004687) |
| 0.2 | 8.7 | GO:0004115 | 3',5'-cyclic-AMP phosphodiesterase activity(GO:0004115) |
| 0.2 | 1.8 | GO:0055131 | C3HC4-type RING finger domain binding(GO:0055131) |
| 0.2 | 0.9 | GO:0072590 | N-acetyl-L-aspartate-L-glutamate ligase activity(GO:0072590) |
| 0.2 | 1.6 | GO:0001665 | alpha-N-acetylgalactosaminide alpha-2,6-sialyltransferase activity(GO:0001665) |
| 0.2 | 0.8 | GO:0015254 | purine nucleobase transmembrane transporter activity(GO:0005345) pyrimidine nucleobase transmembrane transporter activity(GO:0005350) nucleobase transmembrane transporter activity(GO:0015205) glycerol channel activity(GO:0015254) |
| 0.2 | 0.8 | GO:0034211 | GTP-dependent protein kinase activity(GO:0034211) |
| 0.2 | 0.6 | GO:0032093 | SAM domain binding(GO:0032093) |
| 0.2 | 3.2 | GO:0035612 | AP-2 adaptor complex binding(GO:0035612) |
| 0.1 | 1.8 | GO:0015280 | ligand-gated sodium channel activity(GO:0015280) |
| 0.1 | 0.6 | GO:1902271 | D3 vitamins binding(GO:1902271) |
| 0.1 | 1.0 | GO:0015091 | ferric iron transmembrane transporter activity(GO:0015091) trivalent inorganic cation transmembrane transporter activity(GO:0072510) |
| 0.1 | 3.4 | GO:0004890 | GABA-A receptor activity(GO:0004890) GABA receptor binding(GO:0050811) |
| 0.1 | 0.5 | GO:0050347 | trans-hexaprenyltranstransferase activity(GO:0000010) trans-octaprenyltranstransferase activity(GO:0050347) |
| 0.1 | 1.4 | GO:0004459 | L-lactate dehydrogenase activity(GO:0004459) |
| 0.1 | 0.7 | GO:0004792 | thiosulfate sulfurtransferase activity(GO:0004792) |
| 0.1 | 3.1 | GO:0008331 | high voltage-gated calcium channel activity(GO:0008331) |
| 0.1 | 4.8 | GO:0008157 | protein phosphatase 1 binding(GO:0008157) |
| 0.1 | 0.6 | GO:0034875 | oxidoreductase activity, acting on CH or CH2 groups, quinone or similar compound as acceptor(GO:0033695) caffeine oxidase activity(GO:0034875) |
| 0.1 | 0.3 | GO:0019807 | aspartoacylase activity(GO:0019807) |
| 0.1 | 1.2 | GO:0005412 | glucose:sodium symporter activity(GO:0005412) |
| 0.1 | 0.4 | GO:0001733 | galactosylceramide sulfotransferase activity(GO:0001733) |
| 0.1 | 0.7 | GO:0004991 | parathyroid hormone receptor activity(GO:0004991) |
| 0.1 | 5.7 | GO:0051393 | alpha-actinin binding(GO:0051393) |
| 0.1 | 0.4 | GO:0030144 | alpha-1,6-mannosylglycoprotein 6-beta-N-acetylglucosaminyltransferase activity(GO:0030144) |
| 0.1 | 2.1 | GO:0004862 | cAMP-dependent protein kinase inhibitor activity(GO:0004862) |
| 0.1 | 2.8 | GO:0005545 | 1-phosphatidylinositol binding(GO:0005545) |
| 0.1 | 0.2 | GO:0016206 | catechol O-methyltransferase activity(GO:0016206) |
| 0.1 | 2.0 | GO:0003680 | AT DNA binding(GO:0003680) |
| 0.1 | 0.3 | GO:0003831 | beta-N-acetylglucosaminylglycopeptide beta-1,4-galactosyltransferase activity(GO:0003831) |
| 0.1 | 0.9 | GO:0031730 | CCR5 chemokine receptor binding(GO:0031730) |
| 0.1 | 0.4 | GO:0005134 | interleukin-2 receptor binding(GO:0005134) |
| 0.1 | 0.4 | GO:0045505 | dynein intermediate chain binding(GO:0045505) |
| 0.1 | 4.0 | GO:0048487 | beta-tubulin binding(GO:0048487) |
| 0.1 | 1.0 | GO:0001087 | transcription factor activity, sequence-specific DNA binding, RNA polymerase recruiting(GO:0001011) transcription factor activity, TFIIB-class binding(GO:0001087) |
| 0.1 | 0.2 | GO:0032422 | purine-rich negative regulatory element binding(GO:0032422) |
| 0.1 | 0.2 | GO:0071614 | linoleic acid epoxygenase activity(GO:0071614) |
| 0.0 | 2.1 | GO:0005086 | ARF guanyl-nucleotide exchange factor activity(GO:0005086) |
| 0.0 | 0.4 | GO:0061578 | Lys63-specific deubiquitinase activity(GO:0061578) |
| 0.0 | 0.1 | GO:0052600 | diamine oxidase activity(GO:0052597) histamine oxidase activity(GO:0052598) methylputrescine oxidase activity(GO:0052599) propane-1,3-diamine oxidase activity(GO:0052600) |
| 0.0 | 0.3 | GO:0004556 | alpha-amylase activity(GO:0004556) |
| 0.0 | 1.7 | GO:0017091 | AU-rich element binding(GO:0017091) |
| 0.0 | 0.1 | GO:0005148 | prolactin receptor binding(GO:0005148) |
| 0.0 | 4.3 | GO:0004860 | protein kinase inhibitor activity(GO:0004860) |
| 0.0 | 0.1 | GO:0016314 | phosphatidylinositol-3,4,5-trisphosphate 3-phosphatase activity(GO:0016314) |
| 0.0 | 1.4 | GO:0005245 | voltage-gated calcium channel activity(GO:0005245) |
| 0.0 | 1.0 | GO:1905030 | voltage-gated ion channel activity involved in regulation of postsynaptic membrane potential(GO:1905030) |
| 0.0 | 0.7 | GO:0019871 | sodium channel inhibitor activity(GO:0019871) |
| 0.0 | 0.8 | GO:0051010 | microtubule plus-end binding(GO:0051010) |
| 0.0 | 2.1 | GO:0046875 | ephrin receptor binding(GO:0046875) |
| 0.0 | 0.6 | GO:0045499 | chemorepellent activity(GO:0045499) |
| 0.0 | 0.6 | GO:0070403 | NAD+ binding(GO:0070403) |
| 0.0 | 5.9 | GO:0003774 | motor activity(GO:0003774) |
| 0.0 | 0.3 | GO:0016308 | 1-phosphatidylinositol-4-phosphate 5-kinase activity(GO:0016308) |
| 0.0 | 0.8 | GO:0070330 | aromatase activity(GO:0070330) |
| 0.0 | 5.2 | GO:0030165 | PDZ domain binding(GO:0030165) |
| 0.0 | 1.6 | GO:0031624 | ubiquitin conjugating enzyme binding(GO:0031624) |
| 0.0 | 0.2 | GO:0004568 | chitinase activity(GO:0004568) |
| 0.0 | 0.8 | GO:0035259 | glucocorticoid receptor binding(GO:0035259) |
| 0.0 | 0.3 | GO:0035368 | selenocysteine insertion sequence binding(GO:0035368) |
| 0.0 | 0.8 | GO:0004012 | phospholipid-translocating ATPase activity(GO:0004012) |
| 0.0 | 3.0 | GO:0034987 | immunoglobulin receptor binding(GO:0034987) |
| 0.0 | 1.7 | GO:0015459 | potassium channel regulator activity(GO:0015459) |
| 0.0 | 2.8 | GO:0016503 | pheromone receptor activity(GO:0016503) |
| 0.0 | 2.7 | GO:0008013 | beta-catenin binding(GO:0008013) |
| 0.0 | 0.2 | GO:0052656 | branched-chain-amino-acid transaminase activity(GO:0004084) L-leucine transaminase activity(GO:0052654) L-valine transaminase activity(GO:0052655) L-isoleucine transaminase activity(GO:0052656) |
| 0.0 | 0.1 | GO:0004594 | pantothenate kinase activity(GO:0004594) |
| 0.0 | 0.3 | GO:0017070 | U6 snRNA binding(GO:0017070) |
| 0.0 | 0.2 | GO:1990825 | sequence-specific mRNA binding(GO:1990825) |
| 0.0 | 6.2 | GO:0030246 | carbohydrate binding(GO:0030246) |
| 0.0 | 0.4 | GO:0017075 | syntaxin-1 binding(GO:0017075) |
| 0.0 | 0.0 | GO:0002113 | interleukin-33 binding(GO:0002113) |
| 0.0 | 0.6 | GO:0030742 | GTP-dependent protein binding(GO:0030742) |
| 0.0 | 0.2 | GO:0051378 | serotonin binding(GO:0051378) |
| 0.0 | 0.2 | GO:0044323 | retinoic acid-responsive element binding(GO:0044323) |
| 0.0 | 0.1 | GO:0031821 | G-protein coupled serotonin receptor binding(GO:0031821) |
| 0.0 | 0.4 | GO:0005242 | inward rectifier potassium channel activity(GO:0005242) |
| 0.0 | 0.1 | GO:0034594 | phosphatidylinositol trisphosphate phosphatase activity(GO:0034594) |
| 0.0 | 0.2 | GO:0045125 | bioactive lipid receptor activity(GO:0045125) |
| 0.0 | 0.4 | GO:0071949 | FAD binding(GO:0071949) |
| 0.0 | 0.3 | GO:0017056 | structural constituent of nuclear pore(GO:0017056) |
| 0.0 | 0.3 | GO:0005184 | neuropeptide hormone activity(GO:0005184) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 2.8 | PID IL27 PATHWAY | IL27-mediated signaling events |
| 0.1 | 1.4 | PID SYNDECAN 3 PATHWAY | Syndecan-3-mediated signaling events |
| 0.0 | 2.9 | PID CDC42 REG PATHWAY | Regulation of CDC42 activity |
| 0.0 | 4.0 | PID NOTCH PATHWAY | Notch signaling pathway |
| 0.0 | 1.2 | PID IL12 STAT4 PATHWAY | IL12 signaling mediated by STAT4 |
| 0.0 | 2.0 | PID MAPK TRK PATHWAY | Trk receptor signaling mediated by the MAPK pathway |
| 0.0 | 1.2 | PID ARF6 PATHWAY | Arf6 signaling events |
| 0.0 | 0.9 | PID LIS1 PATHWAY | Lissencephaly gene (LIS1) in neuronal migration and development |
| 0.0 | 0.6 | PID HDAC CLASSIII PATHWAY | Signaling events mediated by HDAC Class III |
| 0.0 | 0.3 | PID ARF6 DOWNSTREAM PATHWAY | Arf6 downstream pathway |
| 0.0 | 1.0 | PID P75 NTR PATHWAY | p75(NTR)-mediated signaling |
| 0.0 | 0.7 | PID TRKR PATHWAY | Neurotrophic factor-mediated Trk receptor signaling |
| 0.0 | 0.4 | PID INTEGRIN A4B1 PATHWAY | Alpha4 beta1 integrin signaling events |
| 0.0 | 0.4 | PID CD40 PATHWAY | CD40/CD40L signaling |
| 0.0 | 0.8 | SIG INSULIN RECEPTOR PATHWAY IN CARDIAC MYOCYTES | Genes related to the insulin receptor pathway |
| 0.0 | 2.7 | NABA ECM GLYCOPROTEINS | Genes encoding structural ECM glycoproteins |
| 0.0 | 0.5 | PID INTEGRIN3 PATHWAY | Beta3 integrin cell surface interactions |
| 0.0 | 1.0 | PID LYSOPHOSPHOLIPID PATHWAY | LPA receptor mediated events |
| 0.0 | 4.1 | NABA SECRETED FACTORS | Genes encoding secreted soluble factors |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 7.8 | REACTOME DARPP 32 EVENTS | Genes involved in DARPP-32 events |
| 0.1 | 3.0 | REACTOME GABA A RECEPTOR ACTIVATION | Genes involved in GABA A receptor activation |
| 0.1 | 5.1 | REACTOME ACTIVATED NOTCH1 TRANSMITS SIGNAL TO THE NUCLEUS | Genes involved in Activated NOTCH1 Transmits Signal to the Nucleus |
| 0.1 | 2.0 | REACTOME SEMA3A PLEXIN REPULSION SIGNALING BY INHIBITING INTEGRIN ADHESION | Genes involved in SEMA3A-Plexin repulsion signaling by inhibiting Integrin adhesion |
| 0.1 | 3.1 | REACTOME ADHERENS JUNCTIONS INTERACTIONS | Genes involved in Adherens junctions interactions |
| 0.1 | 0.9 | REACTOME ACETYLCHOLINE NEUROTRANSMITTER RELEASE CYCLE | Genes involved in Acetylcholine Neurotransmitter Release Cycle |
| 0.1 | 1.4 | REACTOME METABOLISM OF PORPHYRINS | Genes involved in Metabolism of porphyrins |
| 0.1 | 0.8 | REACTOME PASSIVE TRANSPORT BY AQUAPORINS | Genes involved in Passive Transport by Aquaporins |
| 0.1 | 1.4 | REACTOME PYRUVATE METABOLISM | Genes involved in Pyruvate metabolism |
| 0.1 | 1.7 | REACTOME LYSOSOME VESICLE BIOGENESIS | Genes involved in Lysosome Vesicle Biogenesis |
| 0.1 | 4.6 | REACTOME AMINO ACID AND OLIGOPEPTIDE SLC TRANSPORTERS | Genes involved in Amino acid and oligopeptide SLC transporters |
| 0.1 | 0.9 | REACTOME IL 6 SIGNALING | Genes involved in Interleukin-6 signaling |
| 0.1 | 3.0 | REACTOME MYOGENESIS | Genes involved in Myogenesis |
| 0.0 | 0.4 | REACTOME CREATION OF C4 AND C2 ACTIVATORS | Genes involved in Creation of C4 and C2 activators |
| 0.0 | 0.6 | REACTOME PEPTIDE HORMONE BIOSYNTHESIS | Genes involved in Peptide hormone biosynthesis |
| 0.0 | 0.6 | REACTOME TRYPTOPHAN CATABOLISM | Genes involved in Tryptophan catabolism |
| 0.0 | 1.3 | REACTOME CGMP EFFECTS | Genes involved in cGMP effects |
| 0.0 | 1.3 | REACTOME AMINE LIGAND BINDING RECEPTORS | Genes involved in Amine ligand-binding receptors |
| 0.0 | 2.0 | REACTOME VOLTAGE GATED POTASSIUM CHANNELS | Genes involved in Voltage gated Potassium channels |
| 0.0 | 2.7 | REACTOME CLASS B 2 SECRETIN FAMILY RECEPTORS | Genes involved in Class B/2 (Secretin family receptors) |
| 0.0 | 1.4 | REACTOME ASSOCIATION OF TRIC CCT WITH TARGET PROTEINS DURING BIOSYNTHESIS | Genes involved in Association of TriC/CCT with target proteins during biosynthesis |
| 0.0 | 0.7 | REACTOME CHYLOMICRON MEDIATED LIPID TRANSPORT | Genes involved in Chylomicron-mediated lipid transport |
| 0.0 | 3.0 | REACTOME REGULATION OF INSULIN SECRETION | Genes involved in Regulation of Insulin Secretion |
| 0.0 | 1.0 | REACTOME INTERACTION BETWEEN L1 AND ANKYRINS | Genes involved in Interaction between L1 and Ankyrins |
| 0.0 | 0.3 | REACTOME PD1 SIGNALING | Genes involved in PD-1 signaling |
| 0.0 | 0.4 | REACTOME REGULATION OF INSULIN LIKE GROWTH FACTOR IGF ACTIVITY BY INSULIN LIKE GROWTH FACTOR BINDING PROTEINS IGFBPS | Genes involved in Regulation of Insulin-like Growth Factor (IGF) Activity by Insulin-like Growth Factor Binding Proteins (IGFBPs) |
| 0.0 | 0.5 | REACTOME SIGNAL REGULATORY PROTEIN SIRP FAMILY INTERACTIONS | Genes involved in Signal regulatory protein (SIRP) family interactions |
| 0.0 | 0.4 | REACTOME N GLYCAN ANTENNAE ELONGATION | Genes involved in N-Glycan antennae elongation |
| 0.0 | 0.3 | REACTOME DEGRADATION OF THE EXTRACELLULAR MATRIX | Genes involved in Degradation of the extracellular matrix |
| 0.0 | 2.3 | REACTOME PEPTIDE LIGAND BINDING RECEPTORS | Genes involved in Peptide ligand-binding receptors |
| 0.0 | 0.1 | REACTOME GLUCURONIDATION | Genes involved in Glucuronidation |
| 0.0 | 0.4 | REACTOME NOD1 2 SIGNALING PATHWAY | Genes involved in NOD1/2 Signaling Pathway |