Project
PRJNA375882: Comprehensive Mouse Transcriptomic BodyMap
Navigation
Downloads
Results for Pou4f3
Z-value: 0.50
Transcription factors associated with Pou4f3
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Pou4f3
|
ENSMUSG00000024497.5 | Pou4f3 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Pou4f3 | mm39_v1_chr18_+_42527604_42527640 | -0.19 | 1.0e-01 | Click! |
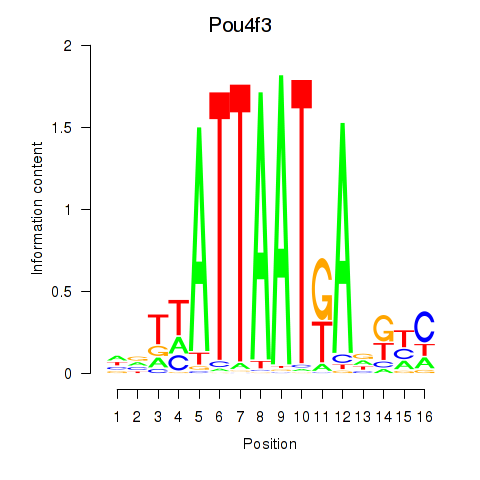
Activity profile of Pou4f3 motif
Sorted Z-values of Pou4f3 motif
Network of associatons between targets according to the STRING database.
First level regulatory network of Pou4f3
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr2_+_172994841 | 4.11 |
ENSMUST00000029017.6
|
Pck1
|
phosphoenolpyruvate carboxykinase 1, cytosolic |
| chr13_+_24023428 | 3.60 |
ENSMUST00000091698.12
ENSMUST00000110422.3 ENSMUST00000166467.9 |
Slc17a3
|
solute carrier family 17 (sodium phosphate), member 3 |
| chr18_-_68562385 | 2.69 |
ENSMUST00000052347.8
|
Mc2r
|
melanocortin 2 receptor |
| chr13_+_24023386 | 2.57 |
ENSMUST00000039721.14
|
Slc17a3
|
solute carrier family 17 (sodium phosphate), member 3 |
| chr9_-_99302205 | 2.36 |
ENSMUST00000123771.2
|
Mras
|
muscle and microspikes RAS |
| chr13_-_56444118 | 2.27 |
ENSMUST00000224801.2
|
Cxcl14
|
chemokine (C-X-C motif) ligand 14 |
| chr12_-_25147139 | 2.17 |
ENSMUST00000221761.2
|
Id2
|
inhibitor of DNA binding 2 |
| chr2_-_140513320 | 2.10 |
ENSMUST00000056760.4
|
Flrt3
|
fibronectin leucine rich transmembrane protein 3 |
| chr9_-_15212849 | 1.96 |
ENSMUST00000034414.10
|
4931406C07Rik
|
RIKEN cDNA 4931406C07 gene |
| chr19_+_29902506 | 1.83 |
ENSMUST00000120388.9
ENSMUST00000144528.8 ENSMUST00000177518.8 |
Il33
|
interleukin 33 |
| chr1_+_139429430 | 1.70 |
ENSMUST00000027615.7
|
F13b
|
coagulation factor XIII, beta subunit |
| chr9_-_15212745 | 1.59 |
ENSMUST00000217042.2
|
4931406C07Rik
|
RIKEN cDNA 4931406C07 gene |
| chr9_+_21634779 | 1.44 |
ENSMUST00000034713.9
|
Ldlr
|
low density lipoprotein receptor |
| chr18_+_23886765 | 1.34 |
ENSMUST00000115830.8
|
Mapre2
|
microtubule-associated protein, RP/EB family, member 2 |
| chrX_+_70860378 | 1.33 |
ENSMUST00000114575.4
|
Vma21
|
VMA21 vacuolar H+-ATPase homolog (S. cerevisiae) |
| chr14_-_73613385 | 1.32 |
ENSMUST00000227454.2
|
Itm2b
|
integral membrane protein 2B |
| chr12_-_103660916 | 1.31 |
ENSMUST00000117053.8
|
Serpina1f
|
serine (or cysteine) peptidase inhibitor, clade A, member 1F |
| chr17_+_7246365 | 1.30 |
ENSMUST00000232245.2
|
Rnaset2b
|
ribonuclease T2B |
| chrX_+_70860357 | 1.27 |
ENSMUST00000114576.9
|
Vma21
|
VMA21 vacuolar H+-ATPase homolog (S. cerevisiae) |
| chr10_+_101994841 | 1.26 |
ENSMUST00000020039.13
|
Mgat4c
|
MGAT4 family, member C |
| chr17_+_7246289 | 1.24 |
ENSMUST00000179728.2
|
Rnaset2b
|
ribonuclease T2B |
| chr17_-_8366536 | 1.20 |
ENSMUST00000231927.2
|
Rnaset2a
|
ribonuclease T2A |
| chr2_+_36120438 | 1.13 |
ENSMUST00000062069.6
|
Ptgs1
|
prostaglandin-endoperoxide synthase 1 |
| chr8_-_84184978 | 1.10 |
ENSMUST00000081506.11
|
Scoc
|
short coiled-coil protein |
| chr7_-_138511221 | 1.05 |
ENSMUST00000130500.8
ENSMUST00000106112.2 |
Bnip3
|
BCL2/adenovirus E1B interacting protein 3 |
| chr10_-_76562002 | 1.03 |
ENSMUST00000001147.5
|
Col6a1
|
collagen, type VI, alpha 1 |
| chrX_+_9751861 | 1.01 |
ENSMUST00000067529.9
ENSMUST00000086165.4 |
Sytl5
|
synaptotagmin-like 5 |
| chr11_+_109434519 | 0.96 |
ENSMUST00000106696.2
|
Arsg
|
arylsulfatase G |
| chr1_-_126758369 | 0.89 |
ENSMUST00000112583.8
ENSMUST00000094609.10 |
Nckap5
|
NCK-associated protein 5 |
| chr4_-_108158242 | 0.85 |
ENSMUST00000043616.7
|
Zyg11b
|
zyg-ll family member B, cell cycle regulator |
| chr11_+_114566257 | 0.83 |
ENSMUST00000045779.6
|
Ttyh2
|
tweety family member 2 |
| chr5_-_44139099 | 0.81 |
ENSMUST00000061299.9
|
Fgfbp1
|
fibroblast growth factor binding protein 1 |
| chr18_+_56695515 | 0.78 |
ENSMUST00000130163.8
ENSMUST00000132628.8 |
Phax
|
phosphorylated adaptor for RNA export |
| chr17_-_33252341 | 0.78 |
ENSMUST00000087654.5
|
Zfp763
|
zinc finger protein 763 |
| chr6_+_41498716 | 0.75 |
ENSMUST00000070380.5
|
Prss2
|
protease, serine 2 |
| chr5_-_44139121 | 0.74 |
ENSMUST00000199894.2
|
Fgfbp1
|
fibroblast growth factor binding protein 1 |
| chrX_-_74423647 | 0.74 |
ENSMUST00000114085.9
|
F8
|
coagulation factor VIII |
| chr8_+_22329942 | 0.72 |
ENSMUST00000006745.4
|
Defb2
|
defensin beta 2 |
| chr16_-_26345493 | 0.66 |
ENSMUST00000165687.3
|
Tmem207
|
transmembrane protein 207 |
| chr14_+_69585036 | 0.65 |
ENSMUST00000064831.6
|
Entpd4
|
ectonucleoside triphosphate diphosphohydrolase 4 |
| chr16_+_11224481 | 0.61 |
ENSMUST00000122168.8
|
Snx29
|
sorting nexin 29 |
| chr10_-_95158827 | 0.59 |
ENSMUST00000220279.2
|
Cradd
|
CASP2 and RIPK1 domain containing adaptor with death domain |
| chr7_-_101486983 | 0.57 |
ENSMUST00000185929.2
ENSMUST00000165052.8 |
Inppl1
|
inositol polyphosphate phosphatase-like 1 |
| chr5_-_38649291 | 0.54 |
ENSMUST00000129099.8
|
Slc2a9
|
solute carrier family 2 (facilitated glucose transporter), member 9 |
| chr11_-_74243447 | 0.48 |
ENSMUST00000141134.2
ENSMUST00000214769.2 |
Olfr411
|
olfactory receptor 411 |
| chr1_+_179788675 | 0.47 |
ENSMUST00000076687.12
ENSMUST00000097450.10 ENSMUST00000212756.2 |
Cdc42bpa
|
CDC42 binding protein kinase alpha |
| chr7_-_99629637 | 0.46 |
ENSMUST00000080817.6
|
Rnf169
|
ring finger protein 169 |
| chr2_+_132689640 | 0.42 |
ENSMUST00000124836.8
ENSMUST00000154160.2 |
Crls1
|
cardiolipin synthase 1 |
| chr12_-_113802603 | 0.41 |
ENSMUST00000103458.3
ENSMUST00000193652.2 |
Ighv5-16
|
immunoglobulin heavy variable 5-16 |
| chr4_+_101843823 | 0.39 |
ENSMUST00000106914.8
|
Gm12789
|
predicted gene 12789 |
| chr7_-_46321952 | 0.37 |
ENSMUST00000049298.15
|
Tph1
|
tryptophan hydroxylase 1 |
| chr11_-_101066266 | 0.35 |
ENSMUST00000062759.4
|
Ccr10
|
chemokine (C-C motif) receptor 10 |
| chr9_-_19275301 | 0.34 |
ENSMUST00000214810.2
|
Olfr846
|
olfactory receptor 846 |
| chrX_+_94942639 | 0.34 |
ENSMUST00000082183.8
|
Zc3h12b
|
zinc finger CCCH-type containing 12B |
| chr4_+_63477018 | 0.32 |
ENSMUST00000077709.11
|
Tmem268
|
transmembrane protein 268 |
| chr17_+_34866160 | 0.31 |
ENSMUST00000173984.2
|
Atf6b
|
activating transcription factor 6 beta |
| chr5_-_120750623 | 0.29 |
ENSMUST00000140554.2
ENSMUST00000031599.9 ENSMUST00000177800.8 |
Rita1
|
RBPJ interacting and tubulin associated 1 |
| chr10_+_101994719 | 0.28 |
ENSMUST00000138522.8
ENSMUST00000163753.8 ENSMUST00000138016.8 |
Mgat4c
|
MGAT4 family, member C |
| chr17_+_34866090 | 0.27 |
ENSMUST00000015605.15
|
Atf6b
|
activating transcription factor 6 beta |
| chr10_+_97318223 | 0.26 |
ENSMUST00000163448.4
|
Dcn
|
decorin |
| chr2_-_91854844 | 0.25 |
ENSMUST00000028663.5
|
Creb3l1
|
cAMP responsive element binding protein 3-like 1 |
| chr1_-_36982747 | 0.24 |
ENSMUST00000185964.3
|
Tmem131
|
transmembrane protein 131 |
| chr3_-_86827664 | 0.23 |
ENSMUST00000194452.2
ENSMUST00000191752.6 |
Dclk2
|
doublecortin-like kinase 2 |
| chr17_+_37977879 | 0.22 |
ENSMUST00000215811.2
|
Olfr118
|
olfactory receptor 118 |
| chr9_-_19799300 | 0.21 |
ENSMUST00000079660.5
|
Olfr862
|
olfactory receptor 862 |
| chr11_+_73851643 | 0.20 |
ENSMUST00000213134.2
ENSMUST00000216291.2 |
Olfr397
|
olfactory receptor 397 |
| chr7_-_103394150 | 0.19 |
ENSMUST00000213906.2
|
Olfr629
|
olfactory receptor 629 |
| chr2_-_86257093 | 0.17 |
ENSMUST00000217481.2
|
Olfr1062
|
olfactory receptor 1062 |
| chr13_+_28200187 | 0.17 |
ENSMUST00000021779.8
|
Prl4a1
|
prolactin family 4, subfamily a, member 1 |
| chr7_+_43077088 | 0.17 |
ENSMUST00000239023.2
|
Gm38999
|
predicted gene, 38999 |
| chr15_+_98350469 | 0.17 |
ENSMUST00000217517.2
|
Olfr281
|
olfactory receptor 281 |
| chr1_+_45834645 | 0.16 |
ENSMUST00000147308.2
|
Wdr75
|
WD repeat domain 75 |
| chr2_+_85835884 | 0.14 |
ENSMUST00000111589.3
|
Olfr1032
|
olfactory receptor 1032 |
| chr10_+_129493563 | 0.14 |
ENSMUST00000217094.2
|
Olfr800
|
olfactory receptor 800 |
| chr13_+_19526322 | 0.14 |
ENSMUST00000184430.2
|
Trgj4
|
T cell receptor gamma joining 4 |
| chr16_-_59169609 | 0.13 |
ENSMUST00000207927.3
|
Olfr206
|
olfactory receptor 206 |
| chr11_+_117700479 | 0.12 |
ENSMUST00000026649.14
ENSMUST00000177131.8 ENSMUST00000120928.2 ENSMUST00000175737.2 ENSMUST00000132298.2 |
Syngr2
Gm20708
|
synaptogyrin 2 predicted gene 20708 |
| chr19_-_12313274 | 0.12 |
ENSMUST00000208398.3
|
Olfr1438-ps1
|
olfactory receptor 1438, pseudogene 1 |
| chr7_+_106630381 | 0.10 |
ENSMUST00000213623.2
|
Olfr713
|
olfactory receptor 713 |
| chr5_+_13448833 | 0.09 |
ENSMUST00000137798.10
|
Sema3a
|
sema domain, immunoglobulin domain (Ig), short basic domain, secreted, (semaphorin) 3A |
| chr2_-_86589298 | 0.09 |
ENSMUST00000215991.2
ENSMUST00000217043.3 |
Olfr1090
|
olfactory receptor 1090 |
| chr5_+_13448647 | 0.09 |
ENSMUST00000125629.8
|
Sema3a
|
sema domain, immunoglobulin domain (Ig), short basic domain, secreted, (semaphorin) 3A |
| chr10_-_129107354 | 0.08 |
ENSMUST00000204573.3
|
Olfr777
|
olfactory receptor 777 |
| chr2_+_36958319 | 0.08 |
ENSMUST00000120704.2
|
Olfr360
|
olfactory receptor 360 |
| chr11_-_4045343 | 0.07 |
ENSMUST00000004868.6
|
Mtfp1
|
mitochondrial fission process 1 |
| chr1_+_109911467 | 0.06 |
ENSMUST00000172005.8
|
Cdh7
|
cadherin 7, type 2 |
| chr5_+_87148697 | 0.06 |
ENSMUST00000031186.9
|
Ugt2b35
|
UDP glucuronosyltransferase 2 family, polypeptide B35 |
| chr3_-_86827640 | 0.05 |
ENSMUST00000195561.6
|
Dclk2
|
doublecortin-like kinase 2 |
| chr17_-_50401305 | 0.05 |
ENSMUST00000113195.8
|
Rftn1
|
raftlin lipid raft linker 1 |
| chr3_-_72875187 | 0.04 |
ENSMUST00000167334.8
|
Sis
|
sucrase isomaltase (alpha-glucosidase) |
| chr1_-_126758520 | 0.03 |
ENSMUST00000162646.8
|
Nckap5
|
NCK-associated protein 5 |
| chr2_+_85600147 | 0.03 |
ENSMUST00000065626.3
|
Olfr1013
|
olfactory receptor 1013 |
| chr11_-_73290321 | 0.02 |
ENSMUST00000131253.2
ENSMUST00000120303.9 |
Olfr1
|
olfactory receptor 1 |
| chr17_-_37472385 | 0.01 |
ENSMUST00000219235.3
|
Olfr93
|
olfactory receptor 93 |
| chr17_-_37404764 | 0.01 |
ENSMUST00000087144.5
|
Olfr91
|
olfactory receptor 91 |
| chr4_-_119047180 | 0.00 |
ENSMUST00000150864.3
ENSMUST00000141227.9 |
Ermap
|
erythroblast membrane-associated protein |
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.0 | 4.1 | GO:0061402 | glycerol biosynthetic process(GO:0006114) positive regulation of transcription from RNA polymerase II promoter in response to acidic pH(GO:0061402) |
| 0.7 | 2.2 | GO:0001966 | thigmotaxis(GO:0001966) |
| 0.6 | 2.3 | GO:2000503 | positive regulation of natural killer cell chemotaxis(GO:2000503) |
| 0.4 | 6.2 | GO:0015747 | urate transport(GO:0015747) |
| 0.4 | 1.4 | GO:0090118 | regulation of phosphatidylcholine catabolic process(GO:0010899) receptor-mediated endocytosis of low-density lipoprotein particle involved in cholesterol transport(GO:0090118) positive regulation of lysosomal protein catabolic process(GO:1905167) |
| 0.3 | 1.8 | GO:0002282 | microglial cell activation involved in immune response(GO:0002282) |
| 0.3 | 1.0 | GO:1902109 | response to cobalt ion(GO:0032025) negative regulation of mitochondrial membrane permeability involved in apoptotic process(GO:1902109) |
| 0.3 | 0.8 | GO:0051030 | snRNA transport(GO:0051030) |
| 0.2 | 2.1 | GO:0003344 | pericardium morphogenesis(GO:0003344) |
| 0.2 | 2.6 | GO:0070072 | vacuolar proton-transporting V-type ATPase complex assembly(GO:0070072) |
| 0.2 | 1.3 | GO:0042985 | negative regulation of amyloid precursor protein biosynthetic process(GO:0042985) |
| 0.2 | 1.1 | GO:0035633 | cyclooxygenase pathway(GO:0019371) maintenance of blood-brain barrier(GO:0035633) |
| 0.2 | 1.6 | GO:0045743 | positive regulation of fibroblast growth factor receptor signaling pathway(GO:0045743) |
| 0.1 | 0.7 | GO:0007597 | blood coagulation, intrinsic pathway(GO:0007597) |
| 0.1 | 1.1 | GO:0061635 | regulation of protein complex stability(GO:0061635) |
| 0.1 | 0.6 | GO:0043569 | negative regulation of insulin-like growth factor receptor signaling pathway(GO:0043569) |
| 0.1 | 0.2 | GO:1903045 | neural crest cell migration involved in sympathetic nervous system development(GO:1903045) |
| 0.1 | 0.8 | GO:1990440 | positive regulation of transcription from RNA polymerase II promoter in response to endoplasmic reticulum stress(GO:1990440) |
| 0.0 | 0.4 | GO:1901162 | serotonin biosynthetic process(GO:0042427) primary amino compound biosynthetic process(GO:1901162) |
| 0.0 | 0.4 | GO:0032049 | cardiolipin biosynthetic process(GO:0032049) |
| 0.0 | 0.6 | GO:0006977 | DNA damage response, signal transduction by p53 class mediator resulting in cell cycle arrest(GO:0006977) |
| 0.0 | 0.7 | GO:0031000 | response to caffeine(GO:0031000) |
| 0.0 | 1.0 | GO:0070208 | protein heterotrimerization(GO:0070208) |
| 0.0 | 0.3 | GO:1900747 | negative regulation of vascular endothelial growth factor signaling pathway(GO:1900747) |
| 0.0 | 0.5 | GO:0046415 | urate metabolic process(GO:0046415) |
| 0.0 | 2.7 | GO:0030819 | positive regulation of cAMP biosynthetic process(GO:0030819) |
| 0.0 | 0.5 | GO:2000780 | negative regulation of double-strand break repair(GO:2000780) |
| 0.0 | 1.0 | GO:0048791 | calcium ion-regulated exocytosis of neurotransmitter(GO:0048791) |
| 0.0 | 0.2 | GO:2000234 | positive regulation of ribosome biogenesis(GO:0090070) positive regulation of rRNA processing(GO:2000234) |
| 0.0 | 0.5 | GO:0050911 | detection of chemical stimulus involved in sensory perception of smell(GO:0050911) |
| 0.0 | 0.4 | GO:0007097 | nuclear migration(GO:0007097) establishment of nucleus localization(GO:0040023) |
| 0.0 | 0.8 | GO:0051438 | regulation of ubiquitin-protein transferase activity(GO:0051438) |
| 0.0 | 0.3 | GO:0021860 | pyramidal neuron development(GO:0021860) |
| 0.0 | 2.4 | GO:1990823 | response to leukemia inhibitory factor(GO:1990823) cellular response to leukemia inhibitory factor(GO:1990830) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 1.4 | GO:1990666 | PCSK9-LDLR complex(GO:1990666) |
| 0.2 | 0.6 | GO:0097637 | intrinsic component of autophagosome membrane(GO:0097636) integral component of autophagosome membrane(GO:0097637) |
| 0.1 | 3.7 | GO:0005788 | endoplasmic reticulum lumen(GO:0005788) |
| 0.1 | 0.8 | GO:0031462 | Cul2-RING ubiquitin ligase complex(GO:0031462) |
| 0.0 | 2.1 | GO:0044295 | axonal growth cone(GO:0044295) |
| 0.0 | 1.0 | GO:0031307 | integral component of mitochondrial outer membrane(GO:0031307) |
| 0.0 | 1.3 | GO:0035371 | microtubule plus-end(GO:0035371) |
| 0.0 | 1.3 | GO:0030660 | Golgi-associated vesicle membrane(GO:0030660) |
| 0.0 | 0.8 | GO:0034707 | chloride channel complex(GO:0034707) |
| 0.0 | 0.3 | GO:0098644 | complex of collagen trimers(GO:0098644) |
| 0.0 | 1.0 | GO:0005581 | collagen trimer(GO:0005581) |
| 0.0 | 5.7 | GO:0016324 | apical plasma membrane(GO:0016324) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.4 | 4.1 | GO:0004613 | phosphoenolpyruvate carboxykinase activity(GO:0004611) phosphoenolpyruvate carboxykinase (GTP) activity(GO:0004613) |
| 0.6 | 6.2 | GO:0015562 | efflux transmembrane transporter activity(GO:0015562) |
| 0.4 | 2.7 | GO:0004977 | melanocortin receptor activity(GO:0004977) |
| 0.2 | 1.4 | GO:0030229 | very-low-density lipoprotein particle receptor activity(GO:0030229) |
| 0.1 | 1.5 | GO:0008454 | alpha-1,3-mannosylglycoprotein 4-beta-N-acetylglucosaminyltransferase activity(GO:0008454) |
| 0.1 | 0.4 | GO:0004510 | tryptophan 5-monooxygenase activity(GO:0004510) |
| 0.1 | 2.3 | GO:0045499 | chemorepellent activity(GO:0045499) |
| 0.1 | 0.6 | GO:0045134 | uridine-diphosphatase activity(GO:0045134) |
| 0.1 | 1.0 | GO:0048407 | platelet-derived growth factor binding(GO:0048407) |
| 0.1 | 2.4 | GO:0030742 | GTP-dependent protein binding(GO:0030742) |
| 0.1 | 0.2 | GO:0005148 | prolactin receptor binding(GO:0005148) |
| 0.1 | 2.3 | GO:0008009 | chemokine activity(GO:0008009) |
| 0.1 | 1.0 | GO:0008484 | sulfuric ester hydrolase activity(GO:0008484) |
| 0.0 | 0.8 | GO:0035497 | cAMP response element binding(GO:0035497) |
| 0.0 | 0.8 | GO:0015643 | toxic substance binding(GO:0015643) |
| 0.0 | 1.6 | GO:0017134 | fibroblast growth factor binding(GO:0017134) |
| 0.0 | 0.5 | GO:0015143 | urate transmembrane transporter activity(GO:0015143) salt transmembrane transporter activity(GO:1901702) |
| 0.0 | 0.6 | GO:0046030 | inositol-polyphosphate 5-phosphatase activity(GO:0004445) inositol trisphosphate phosphatase activity(GO:0046030) |
| 0.0 | 0.6 | GO:0070513 | death domain binding(GO:0070513) |
| 0.0 | 3.7 | GO:0004540 | ribonuclease activity(GO:0004540) |
| 0.0 | 0.4 | GO:0016493 | C-C chemokine receptor activity(GO:0016493) |
| 0.0 | 1.3 | GO:0001540 | beta-amyloid binding(GO:0001540) |
| 0.0 | 0.5 | GO:0070530 | K63-linked polyubiquitin binding(GO:0070530) |
| 0.0 | 1.1 | GO:0004601 | peroxidase activity(GO:0004601) |
| 0.0 | 0.4 | GO:0016780 | phosphotransferase activity, for other substituted phosphate groups(GO:0016780) |
| 0.0 | 1.0 | GO:0005544 | calcium-dependent phospholipid binding(GO:0005544) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 4.1 | PID HNF3B PATHWAY | FOXA2 and FOXA3 transcription factor networks |
| 0.0 | 3.2 | PID HIF1 TFPATHWAY | HIF-1-alpha transcription factor network |
| 0.0 | 0.6 | PID EPHA2 FWD PATHWAY | EPHA2 forward signaling |
| 0.0 | 1.0 | NABA BASEMENT MEMBRANES | Genes encoding structural components of basement membranes |
| 0.0 | 2.4 | ST INTEGRIN SIGNALING PATHWAY | Integrin Signaling Pathway |
| 0.0 | 0.6 | PID HIV NEF PATHWAY | HIV-1 Nef: Negative effector of Fas and TNF-alpha |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 4.1 | REACTOME ABACAVIR TRANSPORT AND METABOLISM | Genes involved in Abacavir transport and metabolism |
| 0.1 | 1.0 | REACTOME THE ACTIVATION OF ARYLSULFATASES | Genes involved in The activation of arylsulfatases |
| 0.1 | 1.7 | REACTOME COMMON PATHWAY | Genes involved in Common Pathway |
| 0.1 | 1.5 | REACTOME N GLYCAN ANTENNAE ELONGATION | Genes involved in N-Glycan antennae elongation |
| 0.1 | 0.7 | REACTOME DEFENSINS | Genes involved in Defensins |
| 0.0 | 1.4 | REACTOME CHYLOMICRON MEDIATED LIPID TRANSPORT | Genes involved in Chylomicron-mediated lipid transport |
| 0.0 | 1.3 | REACTOME AMYLOIDS | Genes involved in Amyloids |
| 0.0 | 0.7 | REACTOME INTRINSIC PATHWAY | Genes involved in Intrinsic Pathway |
| 0.0 | 0.5 | REACTOME FACILITATIVE NA INDEPENDENT GLUCOSE TRANSPORTERS | Genes involved in Facilitative Na+-independent glucose transporters |
| 0.0 | 0.3 | REACTOME CS DS DEGRADATION | Genes involved in CS/DS degradation |
| 0.0 | 0.4 | REACTOME ACYL CHAIN REMODELLING OF PG | Genes involved in Acyl chain remodelling of PG |
| 0.0 | 1.1 | REACTOME PHASE1 FUNCTIONALIZATION OF COMPOUNDS | Genes involved in Phase 1 - Functionalization of compounds |
| 0.0 | 0.4 | REACTOME AMINE DERIVED HORMONES | Genes involved in Amine-derived hormones |
| 0.0 | 1.0 | REACTOME NCAM1 INTERACTIONS | Genes involved in NCAM1 interactions |
| 0.0 | 2.7 | REACTOME PEPTIDE LIGAND BINDING RECEPTORS | Genes involved in Peptide ligand-binding receptors |
| 0.0 | 0.8 | REACTOME METABOLISM OF NON CODING RNA | Genes involved in Metabolism of non-coding RNA |
| 0.0 | 0.6 | REACTOME SYNTHESIS OF PIPS AT THE PLASMA MEMBRANE | Genes involved in Synthesis of PIPs at the plasma membrane |