Project
PRJNA375882: Comprehensive Mouse Transcriptomic BodyMap
Navigation
Downloads
Results for Pou5f1
Z-value: 2.94
Transcription factors associated with Pou5f1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Pou5f1
|
ENSMUSG00000024406.17 | Pou5f1 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Pou5f1 | mm39_v1_chr17_+_35816915_35816968 | -0.28 | 1.6e-02 | Click! |
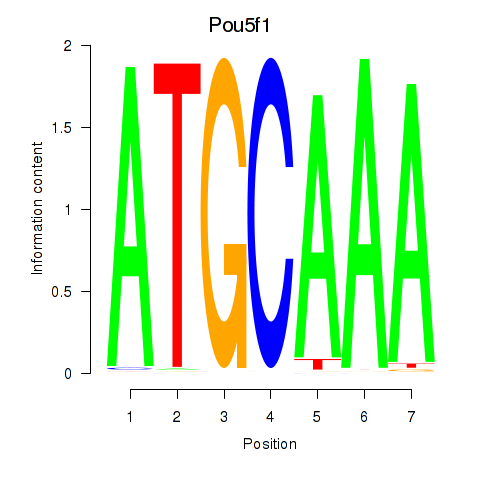
Activity profile of Pou5f1 motif
Sorted Z-values of Pou5f1 motif
Network of associatons between targets according to the STRING database.
First level regulatory network of Pou5f1
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr7_+_121888520 | 27.00 |
ENSMUST00000064989.12
ENSMUST00000064921.5 |
Prkcb
|
protein kinase C, beta |
| chr6_+_68247469 | 23.72 |
ENSMUST00000103321.3
|
Igkv1-110
|
immunoglobulin kappa variable 1-110 |
| chr6_-_70237939 | 22.69 |
ENSMUST00000103386.3
|
Igkv6-23
|
immunoglobulin kappa variable 6-23 |
| chr12_-_114252202 | 22.20 |
ENSMUST00000195124.6
ENSMUST00000103481.3 |
Ighv3-6
|
immunoglobulin heavy variable 3-6 |
| chr6_-_68713748 | 21.73 |
ENSMUST00000183936.2
ENSMUST00000196863.2 |
Igkv19-93
|
immunoglobulin kappa chain variable 19-93 |
| chr6_-_70292451 | 21.32 |
ENSMUST00000103387.3
|
Igkv8-21
|
immunoglobulin kappa variable 8-21 |
| chr6_+_70348416 | 21.15 |
ENSMUST00000103391.4
|
Igkv6-17
|
immunoglobulin kappa variable 6-17 |
| chr6_-_70383976 | 20.98 |
ENSMUST00000103393.2
|
Igkv6-15
|
immunoglobulin kappa variable 6-15 |
| chr6_+_67586695 | 20.87 |
ENSMUST00000103303.3
|
Igkv1-135
|
immunoglobulin kappa variable 1-135 |
| chr6_-_70194405 | 20.78 |
ENSMUST00000103384.2
|
Igkv8-24
|
immunoglobulin kappa chain variable 8-24 |
| chr6_+_68414401 | 19.68 |
ENSMUST00000103324.3
|
Igkv15-103
|
immunoglobulin kappa chain variable 15-103 |
| chr6_-_69792108 | 19.65 |
ENSMUST00000103367.3
|
Igkv12-44
|
immunoglobulin kappa variable 12-44 |
| chr6_+_70495224 | 19.52 |
ENSMUST00000103396.2
|
Igkv3-12
|
immunoglobulin kappa variable 3-12 |
| chr12_-_113552322 | 18.80 |
ENSMUST00000103443.3
|
Ighv2-2
|
immunoglobulin heavy variable 2-2 |
| chr6_+_68026941 | 18.77 |
ENSMUST00000103316.2
|
Igkv9-120
|
immunoglobulin kappa chain variable 9-120 |
| chr6_-_70094604 | 18.62 |
ENSMUST00000103378.3
|
Igkv8-30
|
immunoglobulin kappa chain variable 8-30 |
| chr6_+_68098030 | 18.29 |
ENSMUST00000103317.3
|
Igkv1-117
|
immunoglobulin kappa variable 1-117 |
| chr6_+_68233361 | 17.93 |
ENSMUST00000103320.3
|
Igkv14-111
|
immunoglobulin kappa variable 14-111 |
| chr3_-_50398027 | 17.63 |
ENSMUST00000029297.6
ENSMUST00000194462.6 |
Slc7a11
|
solute carrier family 7 (cationic amino acid transporter, y+ system), member 11 |
| chr6_-_67919524 | 17.57 |
ENSMUST00000196768.2
|
Igkv9-124
|
immunoglobulin kappa chain variable 9-124 |
| chr6_+_67532481 | 17.57 |
ENSMUST00000103302.3
|
Igkv2-137
|
immunoglobulin kappa chain variable 2-137 |
| chr12_-_113823290 | 17.42 |
ENSMUST00000103459.5
|
Ighv5-17
|
immunoglobulin heavy variable 5-17 |
| chr12_-_115876396 | 16.72 |
ENSMUST00000103547.2
|
Ighv1-80
|
immunoglobulin heavy variable 1-80 |
| chr12_-_114355789 | 16.63 |
ENSMUST00000103486.2
|
Ighv6-3
|
immunoglobulin heavy variable 6-3 |
| chr12_-_113561594 | 16.54 |
ENSMUST00000103444.3
|
Ighv5-4
|
immunoglobulin heavy variable 5-4 |
| chr12_-_114398864 | 16.49 |
ENSMUST00000103489.2
|
Ighv6-6
|
immunoglobulin heavy variable 6-6 |
| chr6_-_69741999 | 16.39 |
ENSMUST00000103365.3
|
Igkv12-46
|
immunoglobulin kappa variable 12-46 |
| chr6_-_70313491 | 16.20 |
ENSMUST00000103388.4
|
Igkv6-20
|
immunoglobulin kappa variable 6-20 |
| chr12_-_114752425 | 16.04 |
ENSMUST00000103510.2
|
Ighv1-26
|
immunoglobulin heavy variable 1-26 |
| chr6_-_68907718 | 15.88 |
ENSMUST00000114212.4
|
Igkv13-85
|
immunoglobulin kappa chain variable 13-85 |
| chr6_+_68402550 | 15.78 |
ENSMUST00000103323.3
|
Igkv16-104
|
immunoglobulin kappa variable 16-104 |
| chr12_-_113790741 | 15.77 |
ENSMUST00000103457.3
ENSMUST00000192877.2 |
Ighv5-15
|
immunoglobulin heavy variable 5-15 |
| chr19_-_40982576 | 15.44 |
ENSMUST00000117695.8
|
Blnk
|
B cell linker |
| chr11_-_106205320 | 15.43 |
ENSMUST00000167143.2
|
Cd79b
|
CD79B antigen |
| chr6_-_69282389 | 15.39 |
ENSMUST00000103350.3
|
Igkv4-68
|
immunoglobulin kappa variable 4-68 |
| chr12_-_113589576 | 15.37 |
ENSMUST00000103446.2
|
Ighv5-6
|
immunoglobulin heavy variable 5-6 |
| chr12_-_115258124 | 15.34 |
ENSMUST00000192591.2
|
Ighv8-8
|
immunoglobulin heavy variable 8-8 |
| chr12_-_113912416 | 14.98 |
ENSMUST00000103464.3
|
Ighv4-1
|
immunoglobulin heavy variable 4-1 |
| chr6_+_70640233 | 14.93 |
ENSMUST00000103400.3
|
Igkv3-5
|
immunoglobulin kappa chain variable 3-5 |
| chr6_+_70675416 | 14.93 |
ENSMUST00000103403.3
|
Igkv3-2
|
immunoglobulin kappa variable 3-2 |
| chr6_-_70435020 | 14.77 |
ENSMUST00000198184.2
|
Igkv6-13
|
immunoglobulin kappa variable 6-13 |
| chr6_-_69835868 | 14.68 |
ENSMUST00000103369.2
|
Igkv12-41
|
immunoglobulin kappa chain variable 12-41 |
| chr6_+_70648743 | 14.63 |
ENSMUST00000103401.3
|
Igkv3-4
|
immunoglobulin kappa variable 3-4 |
| chr12_-_114487525 | 14.45 |
ENSMUST00000103495.3
|
Ighv10-3
|
immunoglobulin heavy variable V10-3 |
| chr6_-_70318437 | 14.34 |
ENSMUST00000196599.2
|
Igkv8-19
|
immunoglobulin kappa variable 8-19 |
| chr12_-_113860566 | 14.29 |
ENSMUST00000103474.5
|
Ighv7-1
|
immunoglobulin heavy variable 7-1 |
| chr12_-_113928438 | 13.89 |
ENSMUST00000103478.4
|
Ighv3-1
|
immunoglobulin heavy variable 3-1 |
| chr6_+_68279392 | 13.79 |
ENSMUST00000103322.3
|
Igkv2-109
|
immunoglobulin kappa variable 2-109 |
| chr9_+_50466127 | 13.75 |
ENSMUST00000213916.2
|
Il18
|
interleukin 18 |
| chr6_-_70116066 | 13.67 |
ENSMUST00000103379.3
ENSMUST00000197371.2 |
Igkv6-29
|
immunoglobulin kappa chain variable 6-29 |
| chr5_+_76988444 | 13.55 |
ENSMUST00000120639.9
ENSMUST00000163347.8 ENSMUST00000121851.2 |
Cracd
|
capping protein inhibiting regulator of actin |
| chr12_-_114443071 | 13.51 |
ENSMUST00000103492.2
|
Ighv10-1
|
immunoglobulin heavy variable 10-1 |
| chr12_-_114117264 | 13.28 |
ENSMUST00000103461.5
|
Ighv7-3
|
immunoglobulin heavy variable 7-3 |
| chr6_+_68495964 | 13.19 |
ENSMUST00000199510.5
ENSMUST00000103325.3 |
Igkv14-100
|
immunoglobulin kappa chain variable 14-100 |
| chr12_-_115172211 | 12.86 |
ENSMUST00000103526.3
|
Ighv1-55
|
immunoglobulin heavy variable 1-55 |
| chr5_+_129661233 | 12.84 |
ENSMUST00000031390.10
|
Mmp17
|
matrix metallopeptidase 17 |
| chr6_-_69415741 | 12.82 |
ENSMUST00000103354.3
|
Igkv4-59
|
immunoglobulin kappa variable 4-59 |
| chr4_-_152561896 | 12.59 |
ENSMUST00000238738.2
ENSMUST00000162017.3 ENSMUST00000030768.10 |
Kcnab2
|
potassium voltage-gated channel, shaker-related subfamily, beta member 2 |
| chr6_+_70584375 | 12.57 |
ENSMUST00000197560.2
|
Igkv3-7
|
immunoglobulin kappa variable 3-7 |
| chr12_-_36092475 | 12.46 |
ENSMUST00000020896.17
|
Tspan13
|
tetraspanin 13 |
| chr12_-_114012399 | 12.31 |
ENSMUST00000103468.3
|
Ighv11-2
|
immunoglobulin heavy variable V11-2 |
| chr6_-_68840015 | 12.30 |
ENSMUST00000103336.2
|
Igkv1-88
|
immunoglobulin kappa chain variable 1-88 |
| chr5_+_13448833 | 12.29 |
ENSMUST00000137798.10
|
Sema3a
|
sema domain, immunoglobulin domain (Ig), short basic domain, secreted, (semaphorin) 3A |
| chr12_-_113649535 | 12.28 |
ENSMUST00000103449.4
ENSMUST00000195707.3 |
Ighv2-5
|
immunoglobulin heavy variable 2-5 |
| chr6_+_68547717 | 12.21 |
ENSMUST00000196839.5
ENSMUST00000103327.3 |
Igkv12-98
|
immunoglobulin kappa variable 12-98 |
| chr12_-_115471634 | 12.04 |
ENSMUST00000103535.3
|
Ighv1-64
|
immunoglobulin heavy variable 1-64 |
| chr6_+_67873135 | 12.00 |
ENSMUST00000103310.3
|
Igkv14-126
|
immunoglobulin kappa variable 14-126 |
| chr6_-_70121150 | 11.93 |
ENSMUST00000197525.2
|
Igkv8-28
|
immunoglobulin kappa variable 8-28 |
| chr6_-_70318164 | 11.84 |
ENSMUST00000103389.3
|
Igkv8-19
|
immunoglobulin kappa variable 8-19 |
| chr12_-_114321838 | 11.84 |
ENSMUST00000125484.3
|
Ighv13-2
|
immunoglobulin heavy variable 13-2 |
| chr6_-_55658242 | 11.82 |
ENSMUST00000044767.10
|
Neurod6
|
neurogenic differentiation 6 |
| chr12_-_114672701 | 11.62 |
ENSMUST00000103505.3
ENSMUST00000193855.2 |
Ighv1-19
|
immunoglobulin heavy variable V1-19 |
| chr6_-_70120881 | 11.54 |
ENSMUST00000103380.3
|
Igkv8-28
|
immunoglobulin kappa variable 8-28 |
| chr6_-_70412460 | 11.49 |
ENSMUST00000103394.2
|
Igkv6-14
|
immunoglobulin kappa variable 6-14 |
| chr6_+_70549568 | 11.48 |
ENSMUST00000196940.2
ENSMUST00000103397.3 |
Igkv3-10
|
immunoglobulin kappa variable 3-10 |
| chr6_-_68887957 | 11.36 |
ENSMUST00000200454.2
|
Igkv4-86
|
immunoglobulin kappa variable 4-86 |
| chr12_-_113625906 | 11.29 |
ENSMUST00000103448.3
|
Ighv5-9
|
immunoglobulin heavy variable 5-9 |
| chr12_-_113575237 | 11.21 |
ENSMUST00000178229.3
|
Ighv2-3
|
immunoglobulin heavy variable 2-3 |
| chr12_-_113802603 | 11.14 |
ENSMUST00000103458.3
ENSMUST00000193652.2 |
Ighv5-16
|
immunoglobulin heavy variable 5-16 |
| chr12_-_114878652 | 11.12 |
ENSMUST00000103515.2
|
Ighv1-39
|
immunoglobulin heavy variable 1-39 |
| chr15_-_66703471 | 11.08 |
ENSMUST00000164163.8
|
Sla
|
src-like adaptor |
| chr5_-_140986312 | 11.03 |
ENSMUST00000085786.7
|
Card11
|
caspase recruitment domain family, member 11 |
| chr11_-_3454766 | 10.90 |
ENSMUST00000044507.12
|
Inpp5j
|
inositol polyphosphate 5-phosphatase J |
| chr12_-_113733922 | 10.81 |
ENSMUST00000180013.3
|
Ighv2-9-1
|
immunoglobulin heavy variable 2-9-1 |
| chr12_-_114710326 | 10.78 |
ENSMUST00000103507.2
|
Ighv1-22
|
immunoglobulin heavy variable 1-22 |
| chr16_+_96295011 | 10.74 |
ENSMUST00000233816.2
|
Pcp4
|
Purkinje cell protein 4 |
| chr8_+_66838927 | 10.62 |
ENSMUST00000039540.12
ENSMUST00000110253.3 |
Marchf1
|
membrane associated ring-CH-type finger 1 |
| chr12_-_114621406 | 10.58 |
ENSMUST00000192077.2
|
Ighv1-15
|
immunoglobulin heavy variable 1-15 |
| chr5_+_13449276 | 10.58 |
ENSMUST00000030714.9
ENSMUST00000141968.2 |
Sema3a
|
sema domain, immunoglobulin domain (Ig), short basic domain, secreted, (semaphorin) 3A |
| chr6_-_69678271 | 10.39 |
ENSMUST00000103363.2
|
Igkv4-50
|
immunoglobulin kappa variable 4-50 |
| chr3_-_144555062 | 10.25 |
ENSMUST00000159989.2
|
Clca3b
|
chloride channel accessory 3B |
| chr3_+_14011445 | 10.23 |
ENSMUST00000192209.6
ENSMUST00000171075.8 ENSMUST00000108372.4 |
Ralyl
|
RALY RNA binding protein-like |
| chr6_-_70149254 | 10.17 |
ENSMUST00000197272.2
|
Igkv8-27
|
immunoglobulin kappa chain variable 8-27 |
| chr12_-_115109539 | 10.08 |
ENSMUST00000192554.6
ENSMUST00000103522.3 |
Ighv1-52
|
immunoglobulin heavy variable 1-52 |
| chr13_+_5911481 | 10.02 |
ENSMUST00000000080.8
|
Klf6
|
Kruppel-like factor 6 |
| chr12_-_115766700 | 9.90 |
ENSMUST00000196587.5
ENSMUST00000103543.3 |
Ighv1-74
|
immunoglobulin heavy variable V1-74 |
| chr13_-_95661726 | 9.83 |
ENSMUST00000022185.10
|
F2rl1
|
coagulation factor II (thrombin) receptor-like 1 |
| chr1_-_144052997 | 9.81 |
ENSMUST00000111941.2
ENSMUST00000052375.8 |
Rgs13
|
regulator of G-protein signaling 13 |
| chr6_+_67993691 | 9.74 |
ENSMUST00000103314.3
|
Igkv1-122
|
immunoglobulin kappa chain variable 1-122 |
| chr6_-_68887922 | 9.71 |
ENSMUST00000103337.3
|
Igkv4-86
|
immunoglobulin kappa variable 4-86 |
| chr2_-_66271097 | 9.66 |
ENSMUST00000112371.9
ENSMUST00000138910.3 |
Scn1a
|
sodium channel, voltage-gated, type I, alpha |
| chr6_-_69204417 | 9.65 |
ENSMUST00000103346.3
|
Igkv4-72
|
immunoglobulin kappa chain variable 4-72 |
| chr9_+_75093177 | 9.28 |
ENSMUST00000129281.8
ENSMUST00000148144.8 ENSMUST00000130384.2 |
Myo5a
|
myosin VA |
| chr12_-_115323192 | 8.83 |
ENSMUST00000103531.4
|
Ighv1-61
|
immunoglobulin heavy variable 1-61 |
| chr7_-_44180700 | 8.79 |
ENSMUST00000205506.2
|
Spib
|
Spi-B transcription factor (Spi-1/PU.1 related) |
| chr12_-_115276219 | 8.75 |
ENSMUST00000103529.4
|
Ighv1-58
|
immunoglobulin heavy variable 1-58 |
| chr6_+_68196928 | 8.68 |
ENSMUST00000103318.6
ENSMUST00000103319.3 |
Igkv2-112
|
immunoglobulin kappa variable 2-112 |
| chr11_-_115968745 | 8.62 |
ENSMUST00000156545.2
ENSMUST00000075036.9 ENSMUST00000106451.8 |
Unc13d
|
unc-13 homolog D |
| chr12_-_114815280 | 8.61 |
ENSMUST00000103512.3
|
Ighv1-34
|
immunoglobulin heavy variable 1-34 |
| chr12_-_114226570 | 8.58 |
ENSMUST00000103479.4
ENSMUST00000195619.2 |
Ighv3-5
|
immunoglobulin heavy variable 3-5 |
| chr12_-_34578842 | 8.57 |
ENSMUST00000110819.4
|
Hdac9
|
histone deacetylase 9 |
| chr12_+_71095112 | 8.56 |
ENSMUST00000135709.2
|
Arid4a
|
AT rich interactive domain 4A (RBP1-like) |
| chr12_-_115083839 | 8.48 |
ENSMUST00000103521.3
|
Ighv1-50
|
immunoglobulin heavy variable 1-50 |
| chr3_-_144525255 | 8.42 |
ENSMUST00000029929.12
|
Clca3a2
|
chloride channel accessory 3A2 |
| chr1_-_143879738 | 8.39 |
ENSMUST00000153527.3
|
Rgs2
|
regulator of G-protein signaling 2 |
| chr11_+_110289941 | 8.38 |
ENSMUST00000020949.12
ENSMUST00000100260.2 |
Map2k6
|
mitogen-activated protein kinase kinase 6 |
| chr7_+_28682253 | 8.36 |
ENSMUST00000085835.8
|
Map4k1
|
mitogen-activated protein kinase kinase kinase kinase 1 |
| chr6_-_138398376 | 8.28 |
ENSMUST00000163065.8
|
Lmo3
|
LIM domain only 3 |
| chr17_+_28426752 | 8.10 |
ENSMUST00000002327.6
ENSMUST00000233560.2 ENSMUST00000233958.2 ENSMUST00000233170.2 |
Def6
|
differentially expressed in FDCP 6 |
| chr6_-_69877642 | 8.05 |
ENSMUST00000103370.3
|
Igkv5-39
|
immunoglobulin kappa variable 5-39 |
| chr9_+_89081262 | 7.96 |
ENSMUST00000068569.5
|
Bcl2a1b
|
B cell leukemia/lymphoma 2 related protein A1b |
| chr11_+_56902658 | 7.82 |
ENSMUST00000094179.11
|
Gria1
|
glutamate receptor, ionotropic, AMPA1 (alpha 1) |
| chr3_-_84128160 | 7.74 |
ENSMUST00000154152.2
ENSMUST00000107693.9 ENSMUST00000107695.9 |
Trim2
|
tripartite motif-containing 2 |
| chr12_+_100165694 | 7.72 |
ENSMUST00000110082.11
|
Calm1
|
calmodulin 1 |
| chr1_-_143879877 | 7.69 |
ENSMUST00000127206.8
|
Rgs2
|
regulator of G-protein signaling 2 |
| chr9_-_95697441 | 7.68 |
ENSMUST00000119760.2
|
Pls1
|
plastin 1 (I-isoform) |
| chr11_+_66847446 | 7.68 |
ENSMUST00000211300.2
ENSMUST00000150220.2 |
Tmem238l
|
transmembrane protein 238 like |
| chr6_-_70364222 | 7.60 |
ENSMUST00000103392.3
ENSMUST00000195945.2 |
Igkv8-16
|
immunoglobulin kappa variable 8-16 |
| chr6_+_145067457 | 7.55 |
ENSMUST00000032396.13
|
Lrmp
|
lymphoid-restricted membrane protein |
| chr1_-_64160557 | 7.52 |
ENSMUST00000055001.10
ENSMUST00000114086.8 |
Klf7
|
Kruppel-like factor 7 (ubiquitous) |
| chr13_+_97377604 | 7.50 |
ENSMUST00000041623.9
|
Enc1
|
ectodermal-neural cortex 1 |
| chr6_+_70680515 | 7.48 |
ENSMUST00000103404.2
|
Igkv3-1
|
immunoglobulin kappa variable 3-1 |
| chr5_+_64969679 | 7.36 |
ENSMUST00000166409.6
ENSMUST00000197879.2 |
Klf3
|
Kruppel-like factor 3 (basic) |
| chr10_+_75784126 | 7.19 |
ENSMUST00000000926.3
|
Vpreb3
|
pre-B lymphocyte gene 3 |
| chr11_-_97944239 | 7.17 |
ENSMUST00000017544.9
|
Stac2
|
SH3 and cysteine rich domain 2 |
| chr14_+_79753055 | 7.16 |
ENSMUST00000110835.3
ENSMUST00000227192.2 |
Elf1
|
E74-like factor 1 |
| chr11_-_5787743 | 7.08 |
ENSMUST00000109837.8
|
Polm
|
polymerase (DNA directed), mu |
| chr13_+_102830029 | 7.07 |
ENSMUST00000022124.10
ENSMUST00000171267.2 ENSMUST00000167144.2 ENSMUST00000170878.2 |
Cd180
|
CD180 antigen |
| chr8_+_55407872 | 7.04 |
ENSMUST00000033915.9
|
Gpm6a
|
glycoprotein m6a |
| chr11_+_56902624 | 7.02 |
ENSMUST00000036315.16
|
Gria1
|
glutamate receptor, ionotropic, AMPA1 (alpha 1) |
| chrX_+_165127688 | 7.01 |
ENSMUST00000112223.8
ENSMUST00000112224.8 ENSMUST00000112229.9 ENSMUST00000112228.8 ENSMUST00000112227.9 ENSMUST00000112226.3 |
Gpm6b
|
glycoprotein m6b |
| chrX_+_7744535 | 6.86 |
ENSMUST00000033495.15
|
Pim2
|
proviral integration site 2 |
| chr11_-_115968576 | 6.85 |
ENSMUST00000106450.8
|
Unc13d
|
unc-13 homolog D |
| chr12_-_113666198 | 6.74 |
ENSMUST00000103450.4
|
Ighv5-12
|
immunoglobulin heavy variable 5-12 |
| chr12_-_115459678 | 6.73 |
ENSMUST00000103534.2
|
Ighv1-63
|
immunoglobulin heavy variable V1-63 |
| chr6_-_92458324 | 6.72 |
ENSMUST00000113445.2
|
Prickle2
|
prickle planar cell polarity protein 2 |
| chr6_-_70036183 | 6.72 |
ENSMUST00000197429.5
ENSMUST00000103376.3 |
Igkv7-33
|
immunoglobulin kappa chain variable 7-33 |
| chr10_+_42736771 | 6.67 |
ENSMUST00000105494.8
|
Scml4
|
Scm polycomb group protein like 4 |
| chr12_-_113617344 | 6.63 |
ENSMUST00000103447.2
|
Ighv2-4
|
immunoglobulin heavy variable V2-4 |
| chr5_-_115791032 | 6.61 |
ENSMUST00000121746.8
ENSMUST00000118576.8 |
Bicdl1
|
BICD family like cargo adaptor 1 |
| chr12_-_114477427 | 6.58 |
ENSMUST00000191803.2
|
Ighv1-5
|
immunoglobulin heavy variable V1-5 |
| chr7_+_90739904 | 6.55 |
ENSMUST00000107196.10
ENSMUST00000074273.10 |
Dlg2
|
discs large MAGUK scaffold protein 2 |
| chr18_-_61169262 | 6.51 |
ENSMUST00000025521.9
|
Cdx1
|
caudal type homeobox 1 |
| chr16_+_10306075 | 6.48 |
ENSMUST00000023147.8
|
Ciita
|
class II transactivator |
| chr12_-_113760187 | 6.44 |
ENSMUST00000192911.2
ENSMUST00000103455.3 |
Ighv2-6-8
|
immunoglobulin heavy variable 2-6-8 |
| chr2_+_90927053 | 6.43 |
ENSMUST00000132741.3
|
Spi1
|
spleen focus forming virus (SFFV) proviral integration oncogene |
| chr4_+_123077515 | 6.42 |
ENSMUST00000152194.2
|
Hpcal4
|
hippocalcin-like 4 |
| chr14_+_71127540 | 6.40 |
ENSMUST00000022699.10
|
Gfra2
|
glial cell line derived neurotrophic factor family receptor alpha 2 |
| chr17_+_41121979 | 6.31 |
ENSMUST00000024721.8
ENSMUST00000233740.2 |
Rhag
|
Rhesus blood group-associated A glycoprotein |
| chr6_+_67768007 | 6.30 |
ENSMUST00000196006.5
ENSMUST00000103307.3 |
Igkv14-130
|
immunoglobulin kappa variable 14-130 |
| chr4_-_152213315 | 6.27 |
ENSMUST00000049305.14
|
Espn
|
espin |
| chr3_+_45332831 | 6.25 |
ENSMUST00000193252.2
ENSMUST00000171554.8 ENSMUST00000166126.7 ENSMUST00000170695.4 |
Pcdh10
|
protocadherin 10 |
| chr4_-_132990362 | 6.24 |
ENSMUST00000105908.10
ENSMUST00000030674.8 |
Sytl1
|
synaptotagmin-like 1 |
| chr13_-_49462694 | 6.22 |
ENSMUST00000110087.9
|
Fgd3
|
FYVE, RhoGEF and PH domain containing 3 |
| chr1_-_132318039 | 6.22 |
ENSMUST00000132435.2
|
Tmcc2
|
transmembrane and coiled-coil domains 2 |
| chr3_-_80710097 | 6.15 |
ENSMUST00000075316.10
ENSMUST00000107745.8 |
Gria2
|
glutamate receptor, ionotropic, AMPA2 (alpha 2) |
| chr6_+_68518603 | 6.15 |
ENSMUST00000168090.3
ENSMUST00000103326.3 |
Igkv1-99
|
immunoglobulin kappa variable 1-99 |
| chr4_+_119671688 | 6.15 |
ENSMUST00000106307.9
|
Hivep3
|
human immunodeficiency virus type I enhancer binding protein 3 |
| chr3_-_61273228 | 6.07 |
ENSMUST00000066298.3
|
B430305J03Rik
|
RIKEN cDNA B430305J03 gene |
| chr4_+_3678108 | 6.06 |
ENSMUST00000041377.13
ENSMUST00000103010.4 |
Lyn
|
LYN proto-oncogene, Src family tyrosine kinase |
| chr2_+_72306503 | 6.04 |
ENSMUST00000102691.11
ENSMUST00000157019.2 |
Cdca7
|
cell division cycle associated 7 |
| chr10_+_7708178 | 6.04 |
ENSMUST00000039484.6
|
Zc3h12d
|
zinc finger CCCH type containing 12D |
| chr10_+_115653152 | 6.03 |
ENSMUST00000080630.11
ENSMUST00000179196.3 ENSMUST00000035563.15 |
Tspan8
|
tetraspanin 8 |
| chr18_+_69633741 | 6.00 |
ENSMUST00000207214.2
ENSMUST00000201094.4 ENSMUST00000200703.4 ENSMUST00000202765.4 |
Tcf4
|
transcription factor 4 |
| chr10_+_67373691 | 5.98 |
ENSMUST00000048289.14
ENSMUST00000130933.2 ENSMUST00000105438.9 ENSMUST00000146986.2 |
Egr2
|
early growth response 2 |
| chr12_-_114793177 | 5.97 |
ENSMUST00000103511.2
ENSMUST00000195735.2 |
Ighv1-31
|
immunoglobulin heavy variable 1-31 |
| chr10_+_28544356 | 5.96 |
ENSMUST00000060409.13
ENSMUST00000056097.11 ENSMUST00000105516.9 |
Themis
|
thymocyte selection associated |
| chr2_+_28082943 | 5.89 |
ENSMUST00000113920.8
|
Olfm1
|
olfactomedin 1 |
| chr10_+_85222677 | 5.87 |
ENSMUST00000105307.8
ENSMUST00000020231.10 |
Btbd11
|
BTB (POZ) domain containing 11 |
| chr15_+_82031382 | 5.77 |
ENSMUST00000023100.8
ENSMUST00000229336.2 |
Srebf2
|
sterol regulatory element binding factor 2 |
| chr11_+_97697328 | 5.74 |
ENSMUST00000153520.3
|
Lasp1
|
LIM and SH3 protein 1 |
| chr1_+_93301596 | 5.66 |
ENSMUST00000058682.11
ENSMUST00000186641.7 |
Ano7
|
anoctamin 7 |
| chr17_+_27276262 | 5.65 |
ENSMUST00000049308.9
|
Itpr3
|
inositol 1,4,5-triphosphate receptor 3 |
| chr11_-_115968373 | 5.64 |
ENSMUST00000174822.8
|
Unc13d
|
unc-13 homolog D |
| chr1_-_138103021 | 5.61 |
ENSMUST00000182755.8
ENSMUST00000193650.2 ENSMUST00000182283.8 |
Ptprc
|
protein tyrosine phosphatase, receptor type, C |
| chr11_+_51180747 | 5.60 |
ENSMUST00000102765.9
|
Col23a1
|
collagen, type XXIII, alpha 1 |
| chr11_-_99313078 | 5.59 |
ENSMUST00000017741.4
|
Krt12
|
keratin 12 |
| chr12_-_114843941 | 5.59 |
ENSMUST00000191862.6
ENSMUST00000103513.3 |
Ighv1-36
|
immunoglobulin heavy variable 1-36 |
| chr2_+_143388062 | 5.58 |
ENSMUST00000028905.10
|
Pcsk2
|
proprotein convertase subtilisin/kexin type 2 |
| chr13_+_42834039 | 5.56 |
ENSMUST00000128646.8
|
Phactr1
|
phosphatase and actin regulator 1 |
| chr6_-_69920632 | 5.44 |
ENSMUST00000198880.5
ENSMUST00000103371.3 |
Igkv12-38
|
immunoglobulin kappa chain variable 12-38 |
| chr9_-_51240201 | 5.43 |
ENSMUST00000039959.11
ENSMUST00000238450.3 |
1810046K07Rik
|
RIKEN cDNA 1810046K07 gene |
| chr2_+_122129379 | 5.42 |
ENSMUST00000028656.2
|
Duoxa2
|
dual oxidase maturation factor 2 |
| chr11_-_58504307 | 5.41 |
ENSMUST00000048801.8
|
Lypd8l
|
LY6/PLAUR domain containing 8 like |
| chr2_+_28083105 | 5.41 |
ENSMUST00000100244.10
|
Olfm1
|
olfactomedin 1 |
| chrX_+_92718695 | 5.41 |
ENSMUST00000045898.4
|
Pcyt1b
|
phosphate cytidylyltransferase 1, choline, beta isoform |
| chr3_+_75982890 | 5.37 |
ENSMUST00000160261.8
|
Fstl5
|
follistatin-like 5 |
| chr6_-_65121892 | 5.37 |
ENSMUST00000031982.5
|
Hpgds
|
hematopoietic prostaglandin D synthase |
| chr17_-_15596230 | 5.34 |
ENSMUST00000014917.8
|
Dll1
|
delta like canonical Notch ligand 1 |
| chr10_-_88520877 | 5.31 |
ENSMUST00000138734.2
|
Spic
|
Spi-C transcription factor (Spi-1/PU.1 related) |
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 7.6 | 22.9 | GO:1903045 | neural crest cell migration involved in sympathetic nervous system development(GO:1903045) |
| 3.4 | 17.2 | GO:0043309 | regulation of eosinophil degranulation(GO:0043309) |
| 3.4 | 13.7 | GO:0042231 | interleukin-13 biosynthetic process(GO:0042231) |
| 3.4 | 509.6 | GO:0006910 | phagocytosis, recognition(GO:0006910) |
| 3.0 | 33.1 | GO:0050861 | positive regulation of B cell receptor signaling pathway(GO:0050861) |
| 2.8 | 5.6 | GO:2000473 | regulation of hematopoietic stem cell migration(GO:2000471) positive regulation of hematopoietic stem cell migration(GO:2000473) |
| 2.6 | 21.1 | GO:0002432 | granuloma formation(GO:0002432) |
| 2.3 | 16.1 | GO:0010519 | negative regulation of phospholipase activity(GO:0010519) |
| 2.3 | 6.9 | GO:0002322 | B cell proliferation involved in immune response(GO:0002322) |
| 2.2 | 6.7 | GO:0021660 | rhombomere formation(GO:0021594) rhombomere 3 formation(GO:0021660) rhombomere 5 morphogenesis(GO:0021664) rhombomere 5 formation(GO:0021666) |
| 1.9 | 512.4 | GO:0002377 | immunoglobulin production(GO:0002377) |
| 1.9 | 3.9 | GO:0032741 | positive regulation of interleukin-18 production(GO:0032741) |
| 1.9 | 7.7 | GO:1902896 | terminal web assembly(GO:1902896) |
| 1.9 | 5.6 | GO:0030070 | insulin processing(GO:0030070) |
| 1.9 | 9.3 | GO:0051643 | endoplasmic reticulum localization(GO:0051643) |
| 1.8 | 5.3 | GO:0021698 | cerebellar cortex structural organization(GO:0021698) |
| 1.6 | 6.3 | GO:0072488 | ammonium transmembrane transport(GO:0072488) |
| 1.5 | 5.9 | GO:0017055 | negative regulation of RNA polymerase II transcriptional preinitiation complex assembly(GO:0017055) |
| 1.4 | 15.8 | GO:0089711 | L-glutamate transmembrane transport(GO:0089711) |
| 1.3 | 6.7 | GO:0001830 | trophectodermal cell fate commitment(GO:0001830) |
| 1.3 | 6.7 | GO:0099558 | maintenance of synapse structure(GO:0099558) |
| 1.3 | 6.4 | GO:0045347 | negative regulation of MHC class II biosynthetic process(GO:0045347) |
| 1.3 | 7.5 | GO:0010499 | proteasomal ubiquitin-independent protein catabolic process(GO:0010499) |
| 1.2 | 5.8 | GO:0072368 | regulation of lipid transport by negative regulation of transcription from RNA polymerase II promoter(GO:0072368) |
| 1.1 | 13.4 | GO:0014807 | regulation of somitogenesis(GO:0014807) |
| 1.1 | 6.5 | GO:0045346 | regulation of MHC class II biosynthetic process(GO:0045346) positive regulation of MHC class II biosynthetic process(GO:0045348) |
| 1.1 | 3.2 | GO:0018146 | keratan sulfate biosynthetic process(GO:0018146) |
| 1.0 | 3.9 | GO:0045659 | negative regulation of neutrophil differentiation(GO:0045659) |
| 0.9 | 5.7 | GO:0061589 | calcium activated phosphatidylserine scrambling(GO:0061589) |
| 0.9 | 5.7 | GO:0050916 | sensory perception of sweet taste(GO:0050916) |
| 0.9 | 11.3 | GO:0023041 | neuronal signal transduction(GO:0023041) |
| 0.9 | 4.6 | GO:1905150 | regulation of voltage-gated sodium channel activity(GO:1905150) |
| 0.9 | 3.6 | GO:0072275 | metanephric glomerulus morphogenesis(GO:0072275) metanephric glomerulus vasculature morphogenesis(GO:0072276) metanephric glomerular capillary formation(GO:0072277) |
| 0.9 | 3.6 | GO:0051673 | membrane disruption in other organism(GO:0051673) |
| 0.9 | 4.5 | GO:0048850 | hypophysis morphogenesis(GO:0048850) |
| 0.9 | 9.0 | GO:0097368 | establishment of Sertoli cell barrier(GO:0097368) |
| 0.9 | 3.6 | GO:2000872 | positive regulation of progesterone secretion(GO:2000872) |
| 0.9 | 4.5 | GO:1903378 | positive regulation of oxidative stress-induced neuron intrinsic apoptotic signaling pathway(GO:1903378) |
| 0.9 | 5.4 | GO:2000255 | negative regulation of male germ cell proliferation(GO:2000255) |
| 0.9 | 8.9 | GO:0032264 | IMP salvage(GO:0032264) |
| 0.9 | 7.0 | GO:0051581 | negative regulation of neurotransmitter uptake(GO:0051581) negative regulation of serotonin uptake(GO:0051612) |
| 0.8 | 5.5 | GO:1902748 | positive regulation of lens fiber cell differentiation(GO:1902748) |
| 0.8 | 6.3 | GO:0030046 | parallel actin filament bundle assembly(GO:0030046) |
| 0.8 | 3.8 | GO:0035905 | ascending aorta development(GO:0035905) ascending aorta morphogenesis(GO:0035910) |
| 0.8 | 2.3 | GO:0071929 | alpha-tubulin acetylation(GO:0071929) |
| 0.8 | 3.0 | GO:0055071 | cellular manganese ion homeostasis(GO:0030026) Golgi calcium ion homeostasis(GO:0032468) manganese ion homeostasis(GO:0055071) |
| 0.7 | 3.0 | GO:0006244 | pyrimidine nucleotide catabolic process(GO:0006244) pyrimidine deoxyribonucleotide catabolic process(GO:0009223) |
| 0.7 | 4.4 | GO:0007412 | axon target recognition(GO:0007412) |
| 0.7 | 8.3 | GO:0034983 | peptidyl-lysine deacetylation(GO:0034983) |
| 0.7 | 3.4 | GO:0044727 | DNA demethylation of male pronucleus(GO:0044727) |
| 0.7 | 18.5 | GO:0042118 | endothelial cell activation(GO:0042118) |
| 0.7 | 5.4 | GO:2000609 | regulation of thyroid hormone generation(GO:2000609) |
| 0.7 | 2.7 | GO:0071348 | cellular response to interleukin-11(GO:0071348) |
| 0.7 | 8.6 | GO:0000185 | activation of MAPKKK activity(GO:0000185) |
| 0.7 | 12.6 | GO:0070995 | NADPH oxidation(GO:0070995) |
| 0.7 | 4.6 | GO:0070163 | adiponectin secretion(GO:0070162) regulation of adiponectin secretion(GO:0070163) |
| 0.7 | 3.3 | GO:0002121 | inter-male aggressive behavior(GO:0002121) |
| 0.6 | 9.1 | GO:0098870 | neuronal action potential propagation(GO:0019227) action potential propagation(GO:0098870) |
| 0.6 | 2.6 | GO:0060447 | bud outgrowth involved in lung branching(GO:0060447) |
| 0.6 | 10.7 | GO:0033603 | positive regulation of dopamine secretion(GO:0033603) |
| 0.6 | 4.3 | GO:0060023 | soft palate development(GO:0060023) |
| 0.6 | 3.0 | GO:1903445 | intermembrane transport(GO:0046909) protein transport from ciliary membrane to plasma membrane(GO:1903445) |
| 0.6 | 8.4 | GO:0032308 | positive regulation of prostaglandin secretion(GO:0032308) |
| 0.6 | 8.6 | GO:0051152 | positive regulation of smooth muscle cell differentiation(GO:0051152) |
| 0.6 | 4.5 | GO:2000491 | positive regulation of hepatic stellate cell activation(GO:2000491) |
| 0.6 | 3.4 | GO:1903756 | regulation of transcription from RNA polymerase II promoter by histone modification(GO:1903756) negative regulation of transcription from RNA polymerase II promoter by histone modification(GO:1903758) |
| 0.5 | 2.2 | GO:0044339 | canonical Wnt signaling pathway involved in osteoblast differentiation(GO:0044339) |
| 0.5 | 1.6 | GO:1904501 | regulation of chromatin-mediated maintenance of transcription(GO:1904499) positive regulation of chromatin-mediated maintenance of transcription(GO:1904501) regulation of euchromatin binding(GO:1904793) |
| 0.5 | 6.5 | GO:0003172 | primary heart field specification(GO:0003138) sinoatrial valve development(GO:0003172) sinoatrial valve morphogenesis(GO:0003185) |
| 0.5 | 10.0 | GO:0042756 | drinking behavior(GO:0042756) |
| 0.5 | 2.6 | GO:0048687 | positive regulation of sprouting of injured axon(GO:0048687) positive regulation of axon extension involved in regeneration(GO:0048691) |
| 0.5 | 2.0 | GO:2000395 | regulation of ubiquitin-dependent endocytosis(GO:2000395) positive regulation of ubiquitin-dependent endocytosis(GO:2000397) |
| 0.5 | 10.9 | GO:0031115 | negative regulation of microtubule polymerization(GO:0031115) |
| 0.5 | 11.3 | GO:0031953 | negative regulation of protein autophosphorylation(GO:0031953) |
| 0.5 | 6.4 | GO:0032482 | Rab protein signal transduction(GO:0032482) |
| 0.5 | 3.4 | GO:0021797 | forebrain anterior/posterior pattern specification(GO:0021797) |
| 0.5 | 8.1 | GO:0043383 | negative T cell selection(GO:0043383) |
| 0.5 | 1.4 | GO:1900222 | positive regulation of transcription from RNA polymerase II promoter involved in myocardial precursor cell differentiation(GO:0003257) negative regulation of beta-amyloid clearance(GO:1900222) |
| 0.5 | 2.8 | GO:0090126 | protein complex assembly involved in synapse maturation(GO:0090126) |
| 0.5 | 4.1 | GO:0007196 | adenylate cyclase-inhibiting G-protein coupled glutamate receptor signaling pathway(GO:0007196) |
| 0.4 | 2.2 | GO:0031284 | positive regulation of guanylate cyclase activity(GO:0031284) |
| 0.4 | 0.8 | GO:0034140 | negative regulation of toll-like receptor 3 signaling pathway(GO:0034140) |
| 0.4 | 7.2 | GO:0021527 | spinal cord association neuron differentiation(GO:0021527) |
| 0.4 | 10.2 | GO:0045898 | regulation of RNA polymerase II transcriptional preinitiation complex assembly(GO:0045898) |
| 0.4 | 3.0 | GO:0030422 | production of siRNA involved in RNA interference(GO:0030422) |
| 0.4 | 25.2 | GO:1902476 | chloride transmembrane transport(GO:1902476) |
| 0.4 | 21.0 | GO:0001919 | regulation of receptor recycling(GO:0001919) |
| 0.4 | 2.9 | GO:0042045 | epithelial fluid transport(GO:0042045) |
| 0.3 | 6.6 | GO:0045161 | neuronal ion channel clustering(GO:0045161) |
| 0.3 | 1.9 | GO:0014816 | skeletal muscle satellite cell differentiation(GO:0014816) |
| 0.3 | 3.8 | GO:0010606 | positive regulation of cytoplasmic mRNA processing body assembly(GO:0010606) |
| 0.3 | 0.9 | GO:0001966 | thigmotaxis(GO:0001966) |
| 0.3 | 0.9 | GO:0099566 | regulation of postsynaptic cytosolic calcium ion concentration(GO:0099566) |
| 0.3 | 1.2 | GO:0031959 | mineralocorticoid receptor signaling pathway(GO:0031959) |
| 0.3 | 6.1 | GO:0090129 | positive regulation of synapse maturation(GO:0090129) |
| 0.3 | 2.0 | GO:0006177 | GMP biosynthetic process(GO:0006177) |
| 0.3 | 5.1 | GO:0060056 | mammary gland involution(GO:0060056) |
| 0.3 | 2.0 | GO:0007527 | adult somatic muscle development(GO:0007527) |
| 0.3 | 3.9 | GO:1902004 | positive regulation of beta-amyloid formation(GO:1902004) |
| 0.3 | 2.2 | GO:0031914 | negative regulation of synaptic plasticity(GO:0031914) |
| 0.3 | 2.8 | GO:1903715 | regulation of aerobic respiration(GO:1903715) |
| 0.3 | 10.5 | GO:0035335 | peptidyl-tyrosine dephosphorylation(GO:0035335) |
| 0.3 | 0.8 | GO:0014908 | myotube differentiation involved in skeletal muscle regeneration(GO:0014908) |
| 0.3 | 12.7 | GO:0050853 | B cell receptor signaling pathway(GO:0050853) |
| 0.3 | 1.3 | GO:1990743 | protein sialylation(GO:1990743) |
| 0.3 | 0.8 | GO:1903225 | negative regulation of endodermal cell differentiation(GO:1903225) |
| 0.3 | 9.2 | GO:0008053 | mitochondrial fusion(GO:0008053) |
| 0.3 | 3.9 | GO:0070166 | enamel mineralization(GO:0070166) |
| 0.3 | 7.2 | GO:0050858 | negative regulation of antigen receptor-mediated signaling pathway(GO:0050858) negative regulation of T cell receptor signaling pathway(GO:0050860) |
| 0.2 | 1.5 | GO:0035860 | esophagus smooth muscle contraction(GO:0014846) glial cell-derived neurotrophic factor receptor signaling pathway(GO:0035860) negative regulation of prostatic bud formation(GO:0060686) |
| 0.2 | 6.3 | GO:0043486 | histone exchange(GO:0043486) |
| 0.2 | 3.4 | GO:0007213 | G-protein coupled acetylcholine receptor signaling pathway(GO:0007213) |
| 0.2 | 3.9 | GO:0045199 | maintenance of epithelial cell apical/basal polarity(GO:0045199) |
| 0.2 | 0.9 | GO:2000481 | positive regulation of cAMP-dependent protein kinase activity(GO:2000481) |
| 0.2 | 1.1 | GO:0042271 | susceptibility to natural killer cell mediated cytotoxicity(GO:0042271) |
| 0.2 | 1.0 | GO:1902498 | regulation of protein autoubiquitination(GO:1902498) |
| 0.2 | 1.3 | GO:0060762 | regulation of branching involved in mammary gland duct morphogenesis(GO:0060762) |
| 0.2 | 1.4 | GO:0035426 | extracellular matrix-cell signaling(GO:0035426) |
| 0.2 | 3.0 | GO:0061158 | 3'-UTR-mediated mRNA destabilization(GO:0061158) |
| 0.2 | 2.1 | GO:1901898 | negative regulation of relaxation of cardiac muscle(GO:1901898) |
| 0.2 | 1.0 | GO:0071985 | multivesicular body sorting pathway(GO:0071985) |
| 0.2 | 2.1 | GO:0048312 | intracellular distribution of mitochondria(GO:0048312) |
| 0.2 | 0.6 | GO:0016062 | adaptation of rhodopsin mediated signaling(GO:0016062) light adaption(GO:0036367) |
| 0.1 | 12.1 | GO:0071277 | cellular response to calcium ion(GO:0071277) |
| 0.1 | 2.5 | GO:0048172 | regulation of short-term neuronal synaptic plasticity(GO:0048172) |
| 0.1 | 9.0 | GO:0045744 | negative regulation of G-protein coupled receptor protein signaling pathway(GO:0045744) |
| 0.1 | 1.7 | GO:0016191 | synaptic vesicle uncoating(GO:0016191) |
| 0.1 | 2.8 | GO:0046085 | adenosine metabolic process(GO:0046085) |
| 0.1 | 11.1 | GO:0046847 | filopodium assembly(GO:0046847) |
| 0.1 | 6.2 | GO:0042982 | amyloid precursor protein metabolic process(GO:0042982) |
| 0.1 | 3.1 | GO:0014049 | positive regulation of glutamate secretion(GO:0014049) |
| 0.1 | 1.0 | GO:0016560 | protein import into peroxisome matrix, docking(GO:0016560) |
| 0.1 | 1.7 | GO:0035435 | phosphate ion transmembrane transport(GO:0035435) |
| 0.1 | 10.3 | GO:0038083 | peptidyl-tyrosine autophosphorylation(GO:0038083) |
| 0.1 | 5.6 | GO:0070207 | protein homotrimerization(GO:0070207) |
| 0.1 | 0.7 | GO:0060596 | mammary placode formation(GO:0060596) |
| 0.1 | 0.7 | GO:0097475 | motor neuron migration(GO:0097475) |
| 0.1 | 1.9 | GO:0043568 | positive regulation of insulin-like growth factor receptor signaling pathway(GO:0043568) |
| 0.1 | 2.3 | GO:0046069 | cGMP catabolic process(GO:0046069) |
| 0.1 | 0.4 | GO:0061086 | negative regulation of histone H3-K27 methylation(GO:0061086) |
| 0.1 | 6.8 | GO:0045576 | mast cell activation(GO:0045576) |
| 0.1 | 0.3 | GO:0036091 | positive regulation of transcription from RNA polymerase II promoter in response to oxidative stress(GO:0036091) |
| 0.1 | 15.4 | GO:0042113 | B cell activation(GO:0042113) |
| 0.1 | 1.1 | GO:0070102 | interleukin-6-mediated signaling pathway(GO:0070102) |
| 0.1 | 4.3 | GO:0006284 | base-excision repair(GO:0006284) |
| 0.1 | 8.6 | GO:0051965 | positive regulation of synapse assembly(GO:0051965) |
| 0.1 | 0.7 | GO:0006265 | DNA topological change(GO:0006265) |
| 0.1 | 3.1 | GO:0019835 | cytolysis(GO:0019835) |
| 0.1 | 5.1 | GO:0070830 | bicellular tight junction assembly(GO:0070830) |
| 0.1 | 6.9 | GO:0032091 | negative regulation of protein binding(GO:0032091) |
| 0.1 | 5.0 | GO:0001541 | ovarian follicle development(GO:0001541) |
| 0.1 | 4.0 | GO:0043388 | positive regulation of DNA binding(GO:0043388) |
| 0.1 | 3.5 | GO:0010107 | potassium ion import(GO:0010107) |
| 0.1 | 3.3 | GO:0030514 | negative regulation of BMP signaling pathway(GO:0030514) |
| 0.1 | 4.4 | GO:0045600 | positive regulation of fat cell differentiation(GO:0045600) |
| 0.1 | 1.3 | GO:0046473 | phosphatidic acid metabolic process(GO:0046473) |
| 0.1 | 1.7 | GO:0032456 | endocytic recycling(GO:0032456) |
| 0.1 | 0.4 | GO:1903691 | positive regulation of wound healing, spreading of epidermal cells(GO:1903691) |
| 0.1 | 3.3 | GO:0045494 | photoreceptor cell maintenance(GO:0045494) |
| 0.1 | 1.8 | GO:0007274 | neuromuscular synaptic transmission(GO:0007274) |
| 0.1 | 3.0 | GO:0000038 | very long-chain fatty acid metabolic process(GO:0000038) |
| 0.1 | 1.7 | GO:0030225 | macrophage differentiation(GO:0030225) |
| 0.1 | 1.3 | GO:0014912 | negative regulation of smooth muscle cell migration(GO:0014912) |
| 0.1 | 6.1 | GO:0007156 | homophilic cell adhesion via plasma membrane adhesion molecules(GO:0007156) |
| 0.0 | 1.3 | GO:0038066 | p38MAPK cascade(GO:0038066) |
| 0.0 | 3.0 | GO:0021766 | hippocampus development(GO:0021766) |
| 0.0 | 2.7 | GO:0032611 | interleukin-1 beta production(GO:0032611) |
| 0.0 | 6.3 | GO:1903169 | regulation of calcium ion transmembrane transport(GO:1903169) |
| 0.0 | 5.6 | GO:0043149 | contractile actin filament bundle assembly(GO:0030038) stress fiber assembly(GO:0043149) |
| 0.0 | 1.2 | GO:0035411 | catenin import into nucleus(GO:0035411) |
| 0.0 | 3.8 | GO:0050728 | negative regulation of inflammatory response(GO:0050728) |
| 0.0 | 1.8 | GO:0043631 | mRNA polyadenylation(GO:0006378) RNA polyadenylation(GO:0043631) |
| 0.0 | 0.3 | GO:0006682 | galactosylceramide biosynthetic process(GO:0006682) galactolipid biosynthetic process(GO:0019375) |
| 0.0 | 1.4 | GO:0048821 | erythrocyte development(GO:0048821) |
| 0.0 | 1.6 | GO:0006376 | mRNA splice site selection(GO:0006376) |
| 0.0 | 2.6 | GO:0010923 | negative regulation of phosphatase activity(GO:0010923) |
| 0.0 | 1.2 | GO:0006904 | vesicle docking involved in exocytosis(GO:0006904) |
| 0.0 | 4.7 | GO:0048813 | dendrite morphogenesis(GO:0048813) |
| 0.0 | 1.8 | GO:0030032 | lamellipodium assembly(GO:0030032) |
| 0.0 | 0.8 | GO:0035162 | embryonic hemopoiesis(GO:0035162) |
| 0.0 | 8.4 | GO:0018105 | peptidyl-serine phosphorylation(GO:0018105) |
| 0.0 | 2.0 | GO:0007006 | mitochondrial membrane organization(GO:0007006) |
| 0.0 | 1.5 | GO:0000381 | regulation of alternative mRNA splicing, via spliceosome(GO:0000381) |
| 0.0 | 0.4 | GO:0006622 | protein targeting to lysosome(GO:0006622) |
| 0.0 | 3.0 | GO:0008360 | regulation of cell shape(GO:0008360) |
| 0.0 | 0.1 | GO:0038203 | TORC2 signaling(GO:0038203) |
| 0.0 | 0.3 | GO:0046854 | phosphatidylinositol phosphorylation(GO:0046854) |
| 0.0 | 2.8 | GO:0072659 | protein localization to plasma membrane(GO:0072659) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 4.2 | 12.6 | GO:1990031 | pinceau fiber(GO:1990031) |
| 4.1 | 509.6 | GO:0042571 | immunoglobulin complex, circulating(GO:0042571) |
| 3.0 | 14.8 | GO:0044308 | axonal spine(GO:0044308) |
| 2.3 | 9.3 | GO:0042642 | actomyosin, myosin complex part(GO:0042642) |
| 2.2 | 15.4 | GO:0019814 | immunoglobulin complex(GO:0019814) |
| 2.1 | 21.1 | GO:0033093 | Weibel-Palade body(GO:0033093) |
| 1.5 | 6.1 | GO:0034666 | integrin alpha2-beta1 complex(GO:0034666) |
| 1.1 | 4.5 | GO:1990452 | Parkin-FBXW7-Cul1 ubiquitin ligase complex(GO:1990452) |
| 1.0 | 3.0 | GO:1903095 | microprocessor complex(GO:0070877) ribonuclease III complex(GO:1903095) |
| 0.9 | 7.7 | GO:1990357 | terminal web(GO:1990357) |
| 0.8 | 10.7 | GO:0005883 | neurofilament(GO:0005883) |
| 0.8 | 3.8 | GO:0031251 | PAN complex(GO:0031251) |
| 0.7 | 2.9 | GO:0031088 | platelet dense granule membrane(GO:0031088) |
| 0.7 | 5.8 | GO:0032937 | SREBP-SCAP-Insig complex(GO:0032937) |
| 0.6 | 3.9 | GO:0036019 | endolysosome(GO:0036019) |
| 0.6 | 2.9 | GO:0035976 | AP1 complex(GO:0035976) |
| 0.5 | 4.3 | GO:0035985 | senescence-associated heterochromatin focus(GO:0035985) |
| 0.5 | 3.6 | GO:0072559 | NLRP3 inflammasome complex(GO:0072559) |
| 0.5 | 4.5 | GO:0030485 | smooth muscle contractile fiber(GO:0030485) |
| 0.5 | 4.0 | GO:0033269 | internode region of axon(GO:0033269) |
| 0.5 | 24.0 | GO:0032281 | AMPA glutamate receptor complex(GO:0032281) |
| 0.5 | 9.8 | GO:0031143 | pseudopodium(GO:0031143) |
| 0.4 | 11.0 | GO:0042101 | T cell receptor complex(GO:0042101) |
| 0.4 | 2.2 | GO:0044305 | calyx of Held(GO:0044305) |
| 0.3 | 9.1 | GO:0001518 | voltage-gated sodium channel complex(GO:0001518) |
| 0.3 | 1.7 | GO:1990745 | EARP complex(GO:1990745) |
| 0.3 | 1.6 | GO:0034750 | Scrib-APC-beta-catenin complex(GO:0034750) |
| 0.3 | 4.9 | GO:0043083 | synaptic cleft(GO:0043083) |
| 0.3 | 3.4 | GO:0001940 | male pronucleus(GO:0001940) |
| 0.3 | 6.5 | GO:0005640 | nuclear outer membrane(GO:0005640) |
| 0.3 | 0.8 | GO:0005588 | collagen type V trimer(GO:0005588) |
| 0.3 | 6.6 | GO:0044224 | juxtaparanode region of axon(GO:0044224) |
| 0.3 | 16.2 | GO:0044295 | axonal growth cone(GO:0044295) |
| 0.2 | 6.3 | GO:0032426 | stereocilium tip(GO:0032426) |
| 0.2 | 2.8 | GO:0097197 | tetraspanin-enriched microdomain(GO:0097197) |
| 0.2 | 27.0 | GO:0031526 | brush border membrane(GO:0031526) |
| 0.2 | 18.8 | GO:0005791 | rough endoplasmic reticulum(GO:0005791) |
| 0.2 | 6.7 | GO:0016327 | apicolateral plasma membrane(GO:0016327) |
| 0.2 | 5.4 | GO:0030285 | integral component of synaptic vesicle membrane(GO:0030285) intrinsic component of synaptic vesicle membrane(GO:0098563) |
| 0.2 | 2.3 | GO:0097427 | microtubule bundle(GO:0097427) |
| 0.2 | 17.6 | GO:0043198 | dendritic shaft(GO:0043198) |
| 0.2 | 391.1 | GO:0005615 | extracellular space(GO:0005615) |
| 0.2 | 1.8 | GO:0042382 | paraspeckles(GO:0042382) |
| 0.2 | 5.1 | GO:0030057 | desmosome(GO:0030057) |
| 0.1 | 13.6 | GO:0042734 | presynaptic membrane(GO:0042734) |
| 0.1 | 8.1 | GO:0008180 | COP9 signalosome(GO:0008180) |
| 0.1 | 24.2 | GO:0031225 | anchored component of membrane(GO:0031225) |
| 0.1 | 2.0 | GO:0000813 | ESCRT I complex(GO:0000813) |
| 0.1 | 1.0 | GO:0017071 | intracellular cyclic nucleotide activated cation channel complex(GO:0017071) |
| 0.1 | 1.1 | GO:0070852 | cell body fiber(GO:0070852) |
| 0.1 | 3.8 | GO:0048770 | melanosome(GO:0042470) pigment granule(GO:0048770) |
| 0.1 | 0.7 | GO:0031298 | replication fork protection complex(GO:0031298) |
| 0.1 | 1.3 | GO:0000138 | Golgi trans cisterna(GO:0000138) |
| 0.1 | 7.1 | GO:0045171 | intercellular bridge(GO:0045171) |
| 0.1 | 0.4 | GO:0031313 | extrinsic component of endosome membrane(GO:0031313) |
| 0.1 | 11.9 | GO:0030863 | cortical cytoskeleton(GO:0030863) |
| 0.1 | 25.6 | GO:0009898 | cytoplasmic side of plasma membrane(GO:0009898) |
| 0.1 | 0.6 | GO:0042622 | photoreceptor outer segment membrane(GO:0042622) |
| 0.1 | 16.8 | GO:0032993 | protein-DNA complex(GO:0032993) |
| 0.1 | 8.3 | GO:0035097 | histone methyltransferase complex(GO:0035097) |
| 0.1 | 10.4 | GO:0016605 | PML body(GO:0016605) |
| 0.1 | 7.2 | GO:0005882 | intermediate filament(GO:0005882) |
| 0.1 | 2.1 | GO:0000930 | gamma-tubulin complex(GO:0000930) |
| 0.1 | 7.5 | GO:0016363 | nuclear matrix(GO:0016363) |
| 0.1 | 16.7 | GO:0009897 | external side of plasma membrane(GO:0009897) |
| 0.1 | 3.6 | GO:0005604 | basement membrane(GO:0005604) |
| 0.1 | 1.1 | GO:0035102 | PRC1 complex(GO:0035102) |
| 0.0 | 12.8 | GO:0098857 | membrane raft(GO:0045121) membrane microdomain(GO:0098857) |
| 0.0 | 2.6 | GO:0034707 | chloride channel complex(GO:0034707) |
| 0.0 | 9.2 | GO:0005741 | mitochondrial outer membrane(GO:0005741) |
| 0.0 | 8.0 | GO:0005802 | trans-Golgi network(GO:0005802) |
| 0.0 | 4.3 | GO:0005913 | cell-cell adherens junction(GO:0005913) |
| 0.0 | 6.2 | GO:0043209 | myelin sheath(GO:0043209) |
| 0.0 | 1.1 | GO:0005581 | collagen trimer(GO:0005581) |
| 0.0 | 3.8 | GO:0017053 | transcriptional repressor complex(GO:0017053) |
| 0.0 | 12.9 | GO:0016607 | nuclear speck(GO:0016607) |
| 0.0 | 9.1 | GO:0005911 | cell-cell junction(GO:0005911) |
| 0.0 | 0.8 | GO:0016235 | aggresome(GO:0016235) |
| 0.0 | 2.6 | GO:0005884 | actin filament(GO:0005884) |
| 0.0 | 9.0 | GO:0009986 | cell surface(GO:0009986) |
| 0.0 | 1.8 | GO:0055037 | recycling endosome(GO:0055037) |
| 0.0 | 2.8 | GO:0001669 | acrosomal vesicle(GO:0001669) |
| 0.0 | 1.3 | GO:0008076 | voltage-gated potassium channel complex(GO:0008076) |
| 0.0 | 0.6 | GO:0032420 | stereocilium(GO:0032420) |
| 0.0 | 0.8 | GO:0005801 | cis-Golgi network(GO:0005801) |
| 0.0 | 4.0 | GO:0015629 | actin cytoskeleton(GO:0015629) |
| 0.0 | 1.1 | GO:0016459 | myosin complex(GO:0016459) |
| 0.0 | 1.7 | GO:0043235 | receptor complex(GO:0043235) |
| 0.0 | 0.1 | GO:0061700 | GATOR2 complex(GO:0061700) |
| 0.0 | 0.3 | GO:0005665 | DNA-directed RNA polymerase II, core complex(GO:0005665) |
| 0.0 | 1.7 | GO:0005769 | early endosome(GO:0005769) |
| 0.0 | 0.6 | GO:0001750 | photoreceptor outer segment(GO:0001750) |
| 0.0 | 2.1 | GO:0031252 | cell leading edge(GO:0031252) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 3.8 | 509.6 | GO:0034987 | immunoglobulin receptor binding(GO:0034987) |
| 3.6 | 10.9 | GO:0052658 | inositol-1,4,5-trisphosphate 5-phosphatase activity(GO:0052658) |
| 2.6 | 21.0 | GO:0004971 | AMPA glutamate receptor activity(GO:0004971) |
| 2.5 | 17.6 | GO:0000099 | sulfur amino acid transmembrane transporter activity(GO:0000099) |
| 2.5 | 7.4 | GO:0035717 | chemokine (C-C motif) ligand 7 binding(GO:0035717) |
| 2.5 | 9.8 | GO:0015057 | thrombin receptor activity(GO:0015057) |
| 2.5 | 27.0 | GO:0035184 | histone threonine kinase activity(GO:0035184) |
| 2.3 | 11.3 | GO:0016167 | glial cell-derived neurotrophic factor receptor activity(GO:0016167) |
| 1.9 | 7.7 | GO:0031800 | type 3 metabotropic glutamate receptor binding(GO:0031800) |
| 1.7 | 8.6 | GO:0005220 | inositol 1,4,5-trisphosphate-sensitive calcium-release channel activity(GO:0005220) |
| 1.5 | 6.1 | GO:0005128 | erythropoietin receptor binding(GO:0005128) |
| 1.5 | 5.9 | GO:0002153 | steroid receptor RNA activator RNA binding(GO:0002153) |
| 1.4 | 5.4 | GO:0004105 | choline-phosphate cytidylyltransferase activity(GO:0004105) |
| 1.3 | 5.4 | GO:0004667 | prostaglandin-D synthase activity(GO:0004667) |
| 1.3 | 20.6 | GO:0001087 | transcription factor activity, sequence-specific DNA binding, RNA polymerase recruiting(GO:0001011) transcription factor activity, TFIIB-class binding(GO:0001087) |
| 1.2 | 26.2 | GO:0030215 | semaphorin receptor binding(GO:0030215) |
| 1.2 | 3.6 | GO:0005017 | platelet-derived growth factor-activated receptor activity(GO:0005017) |
| 1.1 | 4.4 | GO:0031694 | alpha-2A adrenergic receptor binding(GO:0031694) |
| 0.9 | 11.0 | GO:0050700 | CARD domain binding(GO:0050700) |
| 0.9 | 8.9 | GO:0047623 | AMP deaminase activity(GO:0003876) adenosine-phosphate deaminase activity(GO:0047623) |
| 0.9 | 28.3 | GO:0005229 | intracellular calcium activated chloride channel activity(GO:0005229) |
| 0.8 | 12.5 | GO:0005068 | transmembrane receptor protein tyrosine kinase adaptor activity(GO:0005068) |
| 0.8 | 3.0 | GO:0015410 | manganese-transporting ATPase activity(GO:0015410) |
| 0.7 | 2.2 | GO:0001565 | phorbol ester receptor activity(GO:0001565) non-kinase phorbol ester receptor activity(GO:0001566) |
| 0.7 | 6.6 | GO:0034452 | dynactin binding(GO:0034452) |
| 0.6 | 3.9 | GO:0045322 | unmethylated CpG binding(GO:0045322) |
| 0.6 | 3.0 | GO:0004525 | ribonuclease III activity(GO:0004525) double-stranded RNA-specific ribonuclease activity(GO:0032296) |
| 0.6 | 33.0 | GO:0005246 | calcium channel regulator activity(GO:0005246) |
| 0.6 | 2.3 | GO:0004337 | dimethylallyltranstransferase activity(GO:0004161) geranyltranstransferase activity(GO:0004337) |
| 0.6 | 2.2 | GO:0034617 | tetrahydrobiopterin binding(GO:0034617) |
| 0.6 | 3.9 | GO:0016312 | inositol bisphosphate phosphatase activity(GO:0016312) |
| 0.5 | 8.4 | GO:0008349 | MAP kinase kinase kinase kinase activity(GO:0008349) |
| 0.5 | 9.3 | GO:0043008 | ATP-dependent protein binding(GO:0043008) |
| 0.5 | 8.6 | GO:0017017 | MAP kinase tyrosine/serine/threonine phosphatase activity(GO:0017017) |
| 0.5 | 2.9 | GO:0001642 | group III metabotropic glutamate receptor activity(GO:0001642) |
| 0.5 | 4.3 | GO:0051575 | 5'-deoxyribose-5-phosphate lyase activity(GO:0051575) |
| 0.5 | 3.2 | GO:0001517 | N-acetylglucosamine 6-O-sulfotransferase activity(GO:0001517) |
| 0.4 | 2.7 | GO:0050816 | phosphothreonine binding(GO:0050816) |
| 0.4 | 6.4 | GO:0051525 | NFAT protein binding(GO:0051525) |
| 0.4 | 4.5 | GO:0016174 | NAD(P)H oxidase activity(GO:0016174) |
| 0.4 | 1.2 | GO:0017082 | mineralocorticoid receptor activity(GO:0017082) |
| 0.4 | 2.3 | GO:0004118 | cGMP-stimulated cyclic-nucleotide phosphodiesterase activity(GO:0004118) |
| 0.4 | 1.2 | GO:0019002 | GMP binding(GO:0019002) |
| 0.4 | 3.4 | GO:0070579 | methylcytosine dioxygenase activity(GO:0070579) |
| 0.4 | 2.6 | GO:0016933 | extracellular-glycine-gated ion channel activity(GO:0016933) extracellular-glycine-gated chloride channel activity(GO:0016934) |
| 0.4 | 2.6 | GO:0045545 | syndecan binding(GO:0045545) |
| 0.4 | 3.6 | GO:1901612 | cardiolipin binding(GO:1901612) |
| 0.4 | 6.6 | GO:0004385 | guanylate kinase activity(GO:0004385) |
| 0.4 | 8.6 | GO:0031078 | histone deacetylase activity (H3-K14 specific)(GO:0031078) NAD-dependent histone deacetylase activity (H3-K14 specific)(GO:0032041) |
| 0.3 | 8.4 | GO:0004708 | MAP kinase kinase activity(GO:0004708) |
| 0.3 | 5.9 | GO:0050897 | cobalt ion binding(GO:0050897) |
| 0.3 | 34.8 | GO:0003823 | antigen binding(GO:0003823) |
| 0.3 | 2.0 | GO:0003938 | IMP dehydrogenase activity(GO:0003938) |
| 0.3 | 8.0 | GO:0051400 | BH domain binding(GO:0051400) |
| 0.3 | 9.1 | GO:0031402 | sodium ion binding(GO:0031402) |
| 0.3 | 19.5 | GO:0001965 | G-protein alpha-subunit binding(GO:0001965) |
| 0.3 | 6.2 | GO:0042043 | neurexin family protein binding(GO:0042043) |
| 0.3 | 3.0 | GO:0019103 | pyrimidine nucleotide binding(GO:0019103) |
| 0.3 | 5.3 | GO:0030957 | Tat protein binding(GO:0030957) |
| 0.3 | 18.6 | GO:0001205 | transcriptional activator activity, RNA polymerase II distal enhancer sequence-specific binding(GO:0001205) |
| 0.3 | 2.6 | GO:0005030 | GPI-linked ephrin receptor activity(GO:0005004) neurotrophin receptor activity(GO:0005030) |
| 0.3 | 2.3 | GO:0004468 | lysine N-acetyltransferase activity, acting on acetyl phosphate as donor(GO:0004468) |
| 0.2 | 0.7 | GO:0097100 | supercoiled DNA binding(GO:0097100) |
| 0.2 | 1.7 | GO:0015315 | hexose phosphate transmembrane transporter activity(GO:0015119) organophosphate:inorganic phosphate antiporter activity(GO:0015315) hexose-phosphate:inorganic phosphate antiporter activity(GO:0015526) glucose 6-phosphate:inorganic phosphate antiporter activity(GO:0061513) |
| 0.2 | 5.6 | GO:0022840 | leak channel activity(GO:0022840) narrow pore channel activity(GO:0022842) |
| 0.2 | 1.4 | GO:0070878 | primary miRNA binding(GO:0070878) |
| 0.2 | 7.1 | GO:0003887 | DNA-directed DNA polymerase activity(GO:0003887) |
| 0.2 | 4.5 | GO:0035497 | cAMP response element binding(GO:0035497) |
| 0.2 | 1.3 | GO:0003835 | beta-galactoside alpha-2,6-sialyltransferase activity(GO:0003835) |
| 0.2 | 1.5 | GO:0008449 | N-acetylglucosamine-6-sulfatase activity(GO:0008449) |
| 0.2 | 3.0 | GO:0031957 | very long-chain fatty acid-CoA ligase activity(GO:0031957) |
| 0.2 | 11.1 | GO:0005070 | SH3/SH2 adaptor activity(GO:0005070) |
| 0.2 | 3.9 | GO:0015095 | magnesium ion transmembrane transporter activity(GO:0015095) |
| 0.2 | 5.2 | GO:0070577 | lysine-acetylated histone binding(GO:0070577) |
| 0.2 | 1.4 | GO:0008093 | cytoskeletal adaptor activity(GO:0008093) |
| 0.2 | 5.6 | GO:0043395 | heparan sulfate proteoglycan binding(GO:0043395) |
| 0.2 | 8.6 | GO:0003785 | actin monomer binding(GO:0003785) |
| 0.2 | 4.6 | GO:0016502 | purinergic nucleotide receptor activity(GO:0001614) nucleotide receptor activity(GO:0016502) |
| 0.2 | 1.0 | GO:0005052 | peroxisome matrix targeting signal-1 binding(GO:0005052) |
| 0.2 | 4.3 | GO:0098641 | cadherin binding involved in cell-cell adhesion(GO:0098641) |
| 0.2 | 3.8 | GO:0004535 | poly(A)-specific ribonuclease activity(GO:0004535) |
| 0.2 | 11.1 | GO:0001540 | beta-amyloid binding(GO:0001540) |
| 0.2 | 0.9 | GO:0099583 | neurotransmitter receptor activity involved in regulation of postsynaptic cytosolic calcium ion concentration(GO:0099583) |
| 0.2 | 2.0 | GO:0046790 | virion binding(GO:0046790) |
| 0.1 | 3.8 | GO:0005540 | hyaluronic acid binding(GO:0005540) |
| 0.1 | 5.6 | GO:0008157 | protein phosphatase 1 binding(GO:0008157) |
| 0.1 | 14.8 | GO:0033613 | activating transcription factor binding(GO:0033613) |
| 0.1 | 11.1 | GO:0015459 | potassium channel regulator activity(GO:0015459) |
| 0.1 | 4.2 | GO:0005242 | inward rectifier potassium channel activity(GO:0005242) |
| 0.1 | 2.8 | GO:0008253 | 5'-nucleotidase activity(GO:0008253) |
| 0.1 | 10.5 | GO:0017022 | myosin binding(GO:0017022) |
| 0.1 | 22.1 | GO:0017137 | Rab GTPase binding(GO:0017137) |
| 0.1 | 4.5 | GO:0005104 | fibroblast growth factor receptor binding(GO:0005104) |
| 0.1 | 0.3 | GO:0004946 | bombesin receptor activity(GO:0004946) |
| 0.1 | 24.8 | GO:0051015 | actin filament binding(GO:0051015) |
| 0.1 | 11.6 | GO:0004222 | metalloendopeptidase activity(GO:0004222) |
| 0.1 | 0.8 | GO:0015016 | [heparan sulfate]-glucosamine N-sulfotransferase activity(GO:0015016) |
| 0.1 | 1.3 | GO:0004143 | diacylglycerol kinase activity(GO:0004143) |
| 0.1 | 0.3 | GO:0047275 | glucosaminylgalactosylglucosylceramide beta-galactosyltransferase activity(GO:0047275) |
| 0.1 | 0.3 | GO:0004430 | 1-phosphatidylinositol 4-kinase activity(GO:0004430) |
| 0.1 | 0.9 | GO:0043495 | protein anchor(GO:0043495) |
| 0.1 | 21.1 | GO:0003779 | actin binding(GO:0003779) |
| 0.1 | 4.8 | GO:0005262 | calcium channel activity(GO:0005262) |
| 0.1 | 1.4 | GO:0051393 | alpha-actinin binding(GO:0051393) |
| 0.1 | 2.7 | GO:0043027 | cysteine-type endopeptidase inhibitor activity involved in apoptotic process(GO:0043027) |
| 0.0 | 15.9 | GO:0005096 | GTPase activator activity(GO:0005096) |
| 0.0 | 2.1 | GO:0043015 | gamma-tubulin binding(GO:0043015) |
| 0.0 | 1.1 | GO:0031435 | mitogen-activated protein kinase kinase kinase binding(GO:0031435) |
| 0.0 | 0.2 | GO:0005005 | transmembrane-ephrin receptor activity(GO:0005005) |
| 0.0 | 0.8 | GO:0019215 | intermediate filament binding(GO:0019215) |
| 0.0 | 4.5 | GO:0005089 | Rho guanyl-nucleotide exchange factor activity(GO:0005089) |
| 0.0 | 4.4 | GO:0030165 | PDZ domain binding(GO:0030165) |
| 0.0 | 7.7 | GO:0005516 | calmodulin binding(GO:0005516) |
| 0.0 | 5.5 | GO:0004519 | endonuclease activity(GO:0004519) |
| 0.0 | 5.8 | GO:0005085 | guanyl-nucleotide exchange factor activity(GO:0005085) |
| 0.0 | 1.4 | GO:0005109 | frizzled binding(GO:0005109) |
| 0.0 | 2.4 | GO:0070491 | repressing transcription factor binding(GO:0070491) |
| 0.0 | 4.8 | GO:0004713 | protein tyrosine kinase activity(GO:0004713) |
| 0.0 | 0.5 | GO:0070742 | C2H2 zinc finger domain binding(GO:0070742) |
| 0.0 | 0.8 | GO:0005201 | extracellular matrix structural constituent(GO:0005201) |
| 0.0 | 1.7 | GO:0005057 | receptor signaling protein activity(GO:0005057) |
| 0.0 | 2.6 | GO:0042393 | histone binding(GO:0042393) |
| 0.0 | 1.7 | GO:0004725 | protein tyrosine phosphatase activity(GO:0004725) |
| 0.0 | 2.7 | GO:0008017 | microtubule binding(GO:0008017) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.1 | 35.4 | PID TCR JNK PATHWAY | JNK signaling in the CD4+ TCR pathway |
| 0.8 | 22.1 | PID ANTHRAX PATHWAY | Cellular roles of Anthrax toxin |
| 0.6 | 21.5 | SA B CELL RECEPTOR COMPLEXES | Antigen binding to B cell receptors activates protein tyrosine kinases, such as the Src family, which ultimate activate MAP kinases. |
| 0.4 | 36.1 | PID BCR 5PATHWAY | BCR signaling pathway |
| 0.3 | 9.8 | ST GA12 PATHWAY | G alpha 12 Pathway |
| 0.3 | 8.6 | PID P38 MKK3 6PATHWAY | p38 MAPK signaling pathway |
| 0.3 | 7.1 | PID DNA PK PATHWAY | DNA-PK pathway in nonhomologous end joining |
| 0.3 | 8.6 | ST GAQ PATHWAY | G alpha q Pathway |
| 0.2 | 9.2 | ST ERK1 ERK2 MAPK PATHWAY | ERK1/ERK2 MAPK Pathway |
| 0.2 | 17.4 | PID IL4 2PATHWAY | IL4-mediated signaling events |
| 0.2 | 5.0 | PID PDGFRA PATHWAY | PDGFR-alpha signaling pathway |
| 0.2 | 12.0 | ST T CELL SIGNAL TRANSDUCTION | T Cell Signal Transduction |
| 0.2 | 7.2 | PID IL2 STAT5 PATHWAY | IL2 signaling events mediated by STAT5 |
| 0.2 | 7.8 | PID NCADHERIN PATHWAY | N-cadherin signaling events |
| 0.2 | 7.2 | PID IL6 7 PATHWAY | IL6-mediated signaling events |
| 0.2 | 5.8 | SA CASPASE CASCADE | Apoptosis is mediated by caspases, cysteine proteases arranged in a proteolytic cascade. |
| 0.2 | 9.1 | SIG REGULATION OF THE ACTIN CYTOSKELETON BY RHO GTPASES | Genes related to regulation of the actin cytoskeleton |
| 0.2 | 8.7 | PID P38 ALPHA BETA DOWNSTREAM PATHWAY | Signaling mediated by p38-alpha and p38-beta |
| 0.2 | 14.9 | PID RAC1 REG PATHWAY | Regulation of RAC1 activity |
| 0.2 | 3.0 | PID IL12 STAT4 PATHWAY | IL12 signaling mediated by STAT4 |
| 0.2 | 4.1 | PID EPHB FWD PATHWAY | EPHB forward signaling |
| 0.1 | 4.9 | PID TCR PATHWAY | TCR signaling in naïve CD4+ T cells |
| 0.1 | 1.3 | ST P38 MAPK PATHWAY | p38 MAPK Pathway |
| 0.1 | 3.4 | PID P38 MK2 PATHWAY | p38 signaling mediated by MAPKAP kinases |
| 0.1 | 4.4 | PID ALPHA SYNUCLEIN PATHWAY | Alpha-synuclein signaling |
| 0.1 | 8.1 | PID HDAC CLASSII PATHWAY | Signaling events mediated by HDAC Class II |
| 0.1 | 11.5 | PID AR PATHWAY | Coregulation of Androgen receptor activity |
| 0.1 | 22.8 | NABA ECM AFFILIATED | Genes encoding proteins affiliated structurally or functionally to extracellular matrix proteins |
| 0.1 | 11.4 | PID PDGFRB PATHWAY | PDGFR-beta signaling pathway |
| 0.1 | 2.0 | PID FCER1 PATHWAY | Fc-epsilon receptor I signaling in mast cells |
| 0.1 | 2.6 | PID INTEGRIN A9B1 PATHWAY | Alpha9 beta1 integrin signaling events |
| 0.1 | 3.6 | PID DELTA NP63 PATHWAY | Validated transcriptional targets of deltaNp63 isoforms |
| 0.1 | 3.9 | PID PS1 PATHWAY | Presenilin action in Notch and Wnt signaling |
| 0.1 | 2.6 | PID SHP2 PATHWAY | SHP2 signaling |
| 0.1 | 2.0 | PID ERBB1 INTERNALIZATION PATHWAY | Internalization of ErbB1 |
| 0.1 | 1.7 | PID HEDGEHOG 2PATHWAY | Signaling events mediated by the Hedgehog family |
| 0.1 | 2.5 | PID ERBB4 PATHWAY | ErbB4 signaling events |
| 0.0 | 0.3 | PID AVB3 INTEGRIN PATHWAY | Integrins in angiogenesis |
| 0.0 | 1.4 | NABA COLLAGENS | Genes encoding collagen proteins |
| 0.0 | 3.1 | PID MYC ACTIV PATHWAY | Validated targets of C-MYC transcriptional activation |
| 0.0 | 0.8 | PID CXCR4 PATHWAY | CXCR4-mediated signaling events |
| 0.0 | 2.2 | PID P53 DOWNSTREAM PATHWAY | Direct p53 effectors |
| 0.0 | 0.7 | PID CASPASE PATHWAY | Caspase cascade in apoptosis |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.0 | 48.0 | REACTOME TRAFFICKING OF GLUR2 CONTAINING AMPA RECEPTORS | Genes involved in Trafficking of GluR2-containing AMPA receptors |
| 1.0 | 24.3 | REACTOME CRMPS IN SEMA3A SIGNALING | Genes involved in CRMPs in Sema3A signaling |
| 0.9 | 48.5 | REACTOME ANTIGEN ACTIVATES B CELL RECEPTOR LEADING TO GENERATION OF SECOND MESSENGERS | Genes involved in Antigen Activates B Cell Receptor Leading to Generation of Second Messengers |
| 0.8 | 7.4 | REACTOME BETA DEFENSINS | Genes involved in Beta defensins |
| 0.6 | 4.3 | REACTOME APOBEC3G MEDIATED RESISTANCE TO HIV1 INFECTION | Genes involved in APOBEC3G mediated resistance to HIV-1 infection |
| 0.6 | 14.9 | REACTOME INSULIN SYNTHESIS AND PROCESSING | Genes involved in Insulin Synthesis and Processing |
| 0.5 | 37.0 | REACTOME MYOGENESIS | Genes involved in Myogenesis |
| 0.4 | 12.8 | REACTOME DEGRADATION OF THE EXTRACELLULAR MATRIX | Genes involved in Degradation of the extracellular matrix |
| 0.4 | 11.1 | REACTOME DOWNSTREAM TCR SIGNALING | Genes involved in Downstream TCR signaling |
| 0.4 | 5.3 | REACTOME TRANSCRIPTIONAL REGULATION OF WHITE ADIPOCYTE DIFFERENTIATION | Genes involved in Transcriptional Regulation of White Adipocyte Differentiation |
| 0.4 | 16.9 | REACTOME BASIGIN INTERACTIONS | Genes involved in Basigin interactions |
| 0.4 | 5.6 | REACTOME PHOSPHORYLATION OF CD3 AND TCR ZETA CHAINS | Genes involved in Phosphorylation of CD3 and TCR zeta chains |
| 0.4 | 5.3 | REACTOME RECEPTOR LIGAND BINDING INITIATES THE SECOND PROTEOLYTIC CLEAVAGE OF NOTCH RECEPTOR | Genes involved in Receptor-ligand binding initiates the second proteolytic cleavage of Notch receptor |
| 0.4 | 8.9 | REACTOME PURINE SALVAGE | Genes involved in Purine salvage |
| 0.3 | 3.9 | REACTOME TRAF6 MEDIATED IRF7 ACTIVATION IN TLR7 8 OR 9 SIGNALING | Genes involved in TRAF6 mediated IRF7 activation in TLR7/8 or 9 signaling |
| 0.3 | 5.7 | REACTOME PYRIMIDINE CATABOLISM | Genes involved in Pyrimidine catabolism |
| 0.3 | 10.9 | REACTOME SYNTHESIS OF PIPS AT THE PLASMA MEMBRANE | Genes involved in Synthesis of PIPs at the plasma membrane |
| 0.3 | 2.8 | REACTOME NOREPINEPHRINE NEUROTRANSMITTER RELEASE CYCLE | Genes involved in Norepinephrine Neurotransmitter Release Cycle |
| 0.3 | 6.8 | REACTOME GENERATION OF SECOND MESSENGER MOLECULES | Genes involved in Generation of second messenger molecules |
| 0.3 | 12.1 | REACTOME SMOOTH MUSCLE CONTRACTION | Genes involved in Smooth Muscle Contraction |
| 0.2 | 5.4 | REACTOME SYNTHESIS OF PC | Genes involved in Synthesis of PC |
| 0.2 | 12.6 | REACTOME VOLTAGE GATED POTASSIUM CHANNELS | Genes involved in Voltage gated Potassium channels |
| 0.2 | 12.1 | REACTOME NCAM1 INTERACTIONS | Genes involved in NCAM1 interactions |
| 0.2 | 2.0 | REACTOME MEMBRANE BINDING AND TARGETTING OF GAG PROTEINS | Genes involved in Membrane binding and targetting of GAG proteins |
| 0.2 | 6.3 | REACTOME AMINE COMPOUND SLC TRANSPORTERS | Genes involved in Amine compound SLC transporters |
| 0.2 | 13.0 | REACTOME NOTCH1 INTRACELLULAR DOMAIN REGULATES TRANSCRIPTION | Genes involved in NOTCH1 Intracellular Domain Regulates Transcription |
| 0.1 | 3.3 | REACTOME OTHER SEMAPHORIN INTERACTIONS | Genes involved in Other semaphorin interactions |
| 0.1 | 5.8 | REACTOME RAP1 SIGNALLING | Genes involved in Rap1 signalling |
| 0.1 | 3.3 | REACTOME NEPHRIN INTERACTIONS | Genes involved in Nephrin interactions |
| 0.1 | 2.0 | REACTOME PURINE RIBONUCLEOSIDE MONOPHOSPHATE BIOSYNTHESIS | Genes involved in Purine ribonucleoside monophosphate biosynthesis |
| 0.1 | 13.7 | REACTOME SIGNALING BY ILS | Genes involved in Signaling by Interleukins |
| 0.1 | 3.0 | REACTOME MICRORNA MIRNA BIOGENESIS | Genes involved in MicroRNA (miRNA) Biogenesis |
| 0.1 | 1.8 | REACTOME SIGNALING BY FGFR1 FUSION MUTANTS | Genes involved in Signaling by FGFR1 fusion mutants |
| 0.1 | 5.3 | REACTOME INTERFERON GAMMA SIGNALING | Genes involved in Interferon gamma signaling |
| 0.1 | 3.1 | REACTOME NITRIC OXIDE STIMULATES GUANYLATE CYCLASE | Genes involved in Nitric oxide stimulates guanylate cyclase |
| 0.1 | 2.6 | REACTOME LIGAND GATED ION CHANNEL TRANSPORT | Genes involved in Ligand-gated ion channel transport |
| 0.1 | 8.6 | REACTOME RESPONSE TO ELEVATED PLATELET CYTOSOLIC CA2 | Genes involved in Response to elevated platelet cytosolic Ca2+ |
| 0.1 | 7.0 | REACTOME NRAGE SIGNALS DEATH THROUGH JNK | Genes involved in NRAGE signals death through JNK |
| 0.1 | 2.3 | REACTOME CHOLESTEROL BIOSYNTHESIS | Genes involved in Cholesterol biosynthesis |
| 0.1 | 2.1 | REACTOME DARPP 32 EVENTS | Genes involved in DARPP-32 events |
| 0.1 | 6.9 | REACTOME TOLL RECEPTOR CASCADES | Genes involved in Toll Receptor Cascades |
| 0.1 | 0.9 | REACTOME UNBLOCKING OF NMDA RECEPTOR GLUTAMATE BINDING AND ACTIVATION | Genes involved in Unblocking of NMDA receptor, glutamate binding and activation |
| 0.1 | 3.0 | REACTOME ION TRANSPORT BY P TYPE ATPASES | Genes involved in Ion transport by P-type ATPases |
| 0.1 | 9.6 | REACTOME PEPTIDE LIGAND BINDING RECEPTORS | Genes involved in Peptide ligand-binding receptors |
| 0.1 | 1.3 | REACTOME N GLYCAN ANTENNAE ELONGATION | Genes involved in N-Glycan antennae elongation |
| 0.0 | 2.4 | REACTOME INTEGRIN CELL SURFACE INTERACTIONS | Genes involved in Integrin cell surface interactions |
| 0.0 | 6.9 | REACTOME G ALPHA I SIGNALLING EVENTS | Genes involved in G alpha (i) signalling events |
| 0.0 | 1.4 | REACTOME INTERACTION BETWEEN L1 AND ANKYRINS | Genes involved in Interaction between L1 and Ankyrins |
| 0.0 | 0.6 | REACTOME ENOS ACTIVATION AND REGULATION | Genes involved in eNOS activation and regulation |
| 0.0 | 0.3 | REACTOME SYNTHESIS OF PIPS AT THE GOLGI MEMBRANE | Genes involved in Synthesis of PIPs at the Golgi membrane |
| 0.0 | 0.9 | REACTOME DOWNSTREAM SIGNAL TRANSDUCTION | Genes involved in Downstream signal transduction |
| 0.0 | 0.6 | REACTOME METABOLISM OF STEROID HORMONES AND VITAMINS A AND D | Genes involved in Metabolism of steroid hormones and vitamins A and D |